Summary
Organization of protein sequences into domain families is a foundation for cataloging and investigating protein functions. However, long-standing strategies based on primary amino acid sequences are blind to the possibility that proteins with dissimilar sequences could have comparable tertiary structures. Building on our recent findings that in-silico structural predictions of BEN family DNA binding domains closely resemble their experimentally determined crystal structures, we exploited the AlphaFold2 database for comprehensive identification of BEN domains. Indeed, we identified numerous novel BEN domains, including members of new subfamilies. For example, while no BEN domain factors had previously been annotated in C. elegans, this species actually encodes multiple BEN proteins. These include key developmental timing genes of orphan domain status, sel-7 and lin-14, the latter being the central target of the founding miRNA lin-4. We also reveal that the domain of unknown function 4806 (DUF4806), which is widely distributed across metazoans, is structurally similar to BEN and comprises a new subtype. Surprisingly, we find that BEN domains resemble both metazoan and non-metazoan homeodomains in 3D conformation and preserve characteristic residues, indicating that despite their inability to be aligned by conventional methods, these DNA binding modules are probably evolutionarily related. Finally, we broaden the application of structural homology searches, by revealing novel human members of DUF3504, which exists on diverse proteins with presumed or known nuclear functions. Overall, our work strongly expands this recently-identified family of transcription factors, and illustrates the value of 3D structural predictions to annotate protein domains and interpret their functions.
Keywords: structure prediction, AlphaFold, domain annotation, BEN domain, transcription factor, lin-14, homeodomain
eTOC Blurb
Pan et al. annotate novel BEN DNA-binding domains through in silico structural predictions and comparisons
Introduction
Assignment of amino acid strings into different domain families is a basic step to investigate protein functions. Accordingly, the prediction of protein domains from primary structures has been intensely studied. Traditional clustering methods using multiple sequence alignments are mainly based on similarity of linear protein sequences. However, as protein three-dimensional (3D) structures are more conserved than primary sequences1, 2, proteins with limited sequence similarity can nonetheless harbor similar functional domains. Recent advances in accurate prediction of protein structures provide an unprecedented expansion of available protein 3D structures, and open many potential avenues to classify protein domains and infer their functions3
Sequence-specific transcription factors orchestrate all aspects of biological functions, the annotation of which has long been a central problem in molecular biology. While there remain numerous orphan DNA motifs that lack cognate binding proteins, the identification of novel DNA binding domains has become increasingly rare in past decades. We and others recently recognized the BEN domain as a novel sequence-specific DNA binding module family in both Drosophila and mammals4–8. Collectively, BEN domain factors target cis-elements throughout genomes and function as transcriptional repressors, insulators or activators6, 8–10. In particular, BEN domain proteins play essential roles in diverse Drosophila and mammalian developmental processes, including embryonic stem cell differentiation, neurogenesis, spermiogenesis, and developmental patterning.
BEN domains are recognized across metazoan species, and can even be found in certain animal viruses, likely reflecting their co-option during evolution4. Curiously, BEN domain factors are sporadically distributed across metazoan species. BEN domain factors have been annotated in sponges, insects and mammals. However, C. elegans, which shares most molecular mechanisms and cellular pathways with humans, is not known to encode BEN domain factors. Sequencing of the C. elegans genome 25 years ago revealed that it is indeed missing essential genes preserved in other metazoans11, 12, which may reflect its highly derived state. On the other hand, it remained formally possible that certain conserved genes lay within unsequenced portions, or remained hidden in plain sight due to limitations of current protein annotation strategies.
The BEN domain is initially defined by the presence of four α-helices and a few preserved amino acids at helix-loop boundaries. However, BEN domains from different proteins do not share strong evolutionary conservation. For example, the Drosophila Insensitive BEN (Insv-BEN) domain contains five α-helices separated by a long middle loop between α3 and α4. Structural and biochemical analysis indicates the Insv-BEN interacts with DNA through residues in this long loop and α5 (termed the type I BEN domain). In contrast, the fourth BEN domain (BEN4) of mammalian BEND3 protein contains six α-helices. Although it also includes the loop connecting α3 and α4, it interacts with DNA bases mainly through residues on α5 and α6 (termed the type II BEN domain). Notably, the helix α6 was not considered a part of the BEN domain based on the current BEN model. To date, studies on the BEN domain have been focusing on limited BEN members from Drosophila and mammals. A more comprehensive characterization of BEN domains across different organisms is thus necessary to further understand the function of BEN domains and the mechanism of gene regulation.
The last two years have witnessed monumental advances in protein structure prediction from primary sequences, which has reconfigured the role of in-silico protein modeling in biochemistry13, 14. Accordingly, we were motivated to utilize AlphaFold2 and RoseTTAFold, in combination with solved BEN-DNA complexes, to reveal distinct strategies for DNA binding by different types of BEN domains6.
The AlphaFold database now includes the predicted structures for most proteins in the UniProt 2021 release15, allowing us to use structure comparisons to further investigate domain homology and interpret protein functions. In this study, we compared experimentally solved BEN domains with AlphaFold2-determined protein models in different organisms and demonstrated that BEN domain proteins are more numerous and more phylogenetically broad than previously supposed. We reveal that certain orphan proteins that were long known as critical developmental regulators, such as LIN-14 and SEL7 in C. elegans, contain typical BEN domains. Moreover, we are able to recognize additional subclasses of probable DNA binding BEN domains, such as DUF4806, thus functionalizing an extensive set of previously unstudied proteins. Finally, we reveal structural homology between BEN and homeodomains, uniting this branch of transcription factors that likely initially originated from the helix-turn-helix motif. Overall, our work validates a strategy to cluster protein domains and interpret protein function through structural homology.
Results
Uneven distribution of BEN domain proteins across metazoan species
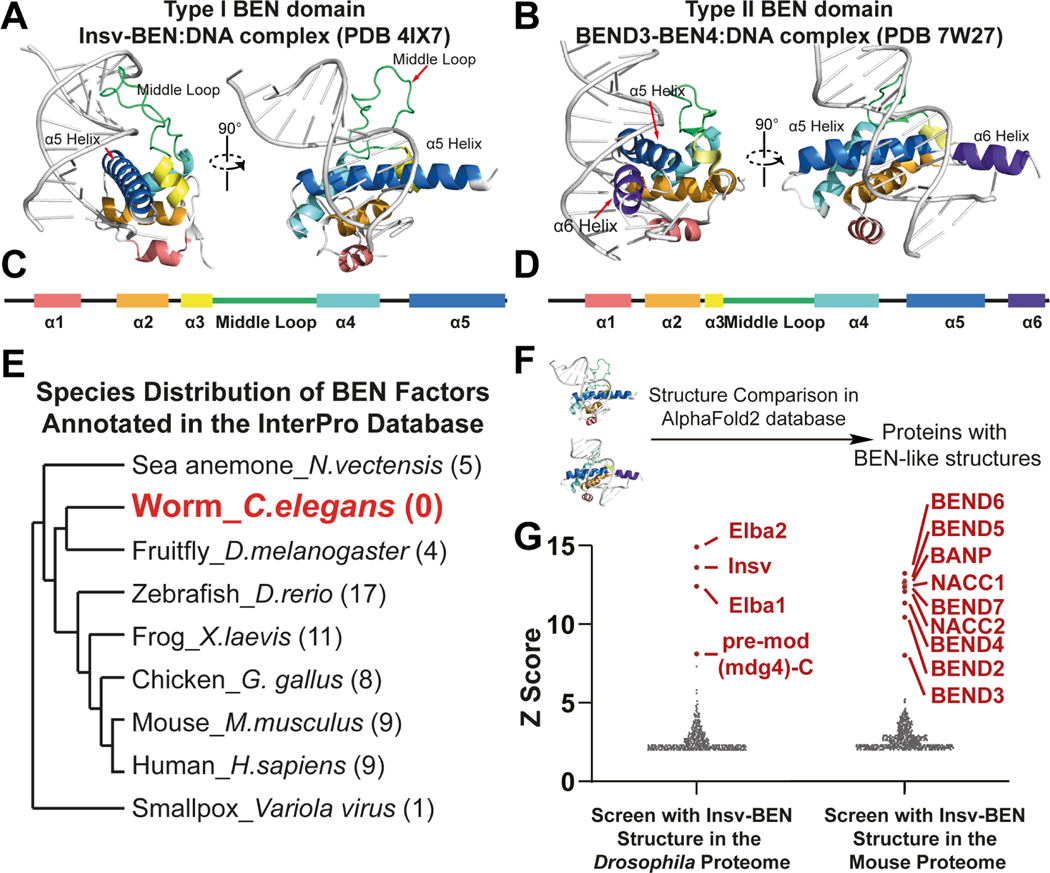
To develop a strategy to utilize AI-predicted protein structures for domain annotation, we chose the BEN domain as a model. Bioinformatics and crystallographic analyses indicate that the BEN domain contains five or six α-helices with a long middle loop, ranging from 13aa in BEND3-BEN4 to 28aa in PG067-BEN1 of Vaccinia virus, between the α3 and α4 helix (Figure 1A–D) 6,16 . In addition, the helix-loop boundaries prefer aliphatic group residues, which are considered to be important for stabilizing protein structures.
Figure 1. BEN domain factors are unevenly distributed across species.
(A) The overall structure of the Drosophila Insv-BEN domain (monomer) in the DNA-bound state (PDB:4IX7). The BEN domain interacts with specific DNA bases through its middle loop (green) and α5 helix.
(B) The overall structure of the human BEND3-BEN4 domain in the DNA-bound state (PDB: 7W27). The BEN structure interacts with DNA bases with its α5 and α6 helices.
(C) The overall secondary structure of Insv-BEN, a representative type I BEN domain.
(D) The overall secondary structure of BEND3-BEN4, a representative type II BEN domain.
(E) BEN factors are unevenly distributed in different species.
(F) Schematic diagram of BEN-like structure screening in the AlphaFold2 database.
(G) Structure screening with BEND3-BEN4 in the Drosophila proteome and with Insv-BEN in the mouse proteome. All known BEN domain-containing proteins were revealed and highlighted with red. Note that linear searches (blastp or PSI-BLAST) do not identify statistically significant hits when using these baits to search the respective non-cognate species.
We first retrieved BEN-containing proteins annotated by multiple alignments from the PROSITE database of different organisms17. BEN domain factors are present from the scarlet sea anemone (Nematostella vectensis) to human, demonstrating an ancient metazoan origin of this protein domain (Figure 1E). However, as previously reported4, the phylogenetic distribution of BEN domains is sporadic as none were identified in C. elegans (Figure 1E). In addition, BEN-containing factors are also absent in many other species of phylum Nematoda, such as ascarids (Ascaris lumbricoides, 29203 proteins in the UniprotKB), hookworms (Ancylostoma duodenale, 27480 proteins in the UniprotKB), and pinworms (Enterobius vermicularis, 13493 proteins in the UniprotKB). By conventional thinking, one might therefore assume that Nematodes generally lost BEN domain factors. However, it remains formally possible that BEN-encoding loci exist in these species, but cannot be recognized by linear matching.
Systematic surveys of AlphaFold structural predictions reveal BEN domain proteins
BEN domains from different proteins usually have low alignment rates by primary sequences. Moreover, the BEN domain annotation matrix is generated from only 46 BEN domains of 36 BEN-containing proteins, many of which are homologous copies in different species. We hypothesized that the current BEN domain matrix does not capture all proteins that bear structural hallmarks of BEN domains. We thus performed structural comparisons using experimentally resolved BEN structures and computer-determined models in the AlphaFold2 database (Figure 1F). From multiple established bioinformatic tools18–20, we chose the DALI server due to its sensitivity21.
The power of structural similarity searches over linear matching was obvious from the outset. For example, we recently characterized the X-ray crystal structures of the solo-BEN domain of Drosophila Insv and the 4th BEN domain of human BEND3 (BEND3-BEN4). The mouse BEND3-BEN4 domain has also been determined in complex with DNA8. These works showed that they represent archetypes of structurally distinct BEN domains, namely type I (Insv- BEN) and type II (BEND3-BEN4). Mammals have members of both BEN subclasses, but Drosophila only bears type I members. Accordingly, when searching the Drosophila proteome with the type II BEN domain BEND3-BEN4 using BLASTP and PSI-BLAST, no hits are returned. However, when searching 3D predictions using the BEND3-BEN4 structure (PDB: 7W27), all four known Drosophila BEN domain factors (Insv, Elba1, Elba2 and mod(mdg4)-C isoform) were returned as the top hits (defined as top 10% of hits with Z-score ≥3. Figure 1G, S1 and Data S1A). Conversely, BLAST-type searches of the mouse proteome using the Drosophila Insv-BEN domain sequence yields no significant hits, while structural searches using the solved Insv-BEN domain (PDB: 4IX7) retrieved all known mouse BEN domain proteins (i.e., BANP, BEND2, BEND3, BEND4, BEND5, BEND6, BEND7, NACC1 and NACC2) (Figure 1G, S1 and Data S1B). Importantly, the top mouse hit to Insv was BEND6, which we previously showed are functional analogs that are expressed in neurons and antagonize Notch signaling 7,22 . Indeed, transgenic expression of mouse BEND6 in flies can compensate for insv mutation. Thus, their structural similarity can rationalize their functional overlap. To be noticed, Insv-BEN and BEND3-BEN4 queries yielded different results. The Insv-BEN search revealed more hits than BEND3-BEN4 in both the Drosophila and mouse proteomes. Notably, in the type I Insv-BEN screening results, the type II BEND3 factor was ranked lower than all other type I candidate BEN factors (Figure S1 and Data S1B). Taken together, structural comparisons accurately capture highly diverged protein modules with conformational similarity.
C. elegans harbors numerous unannotated BEN domain factors
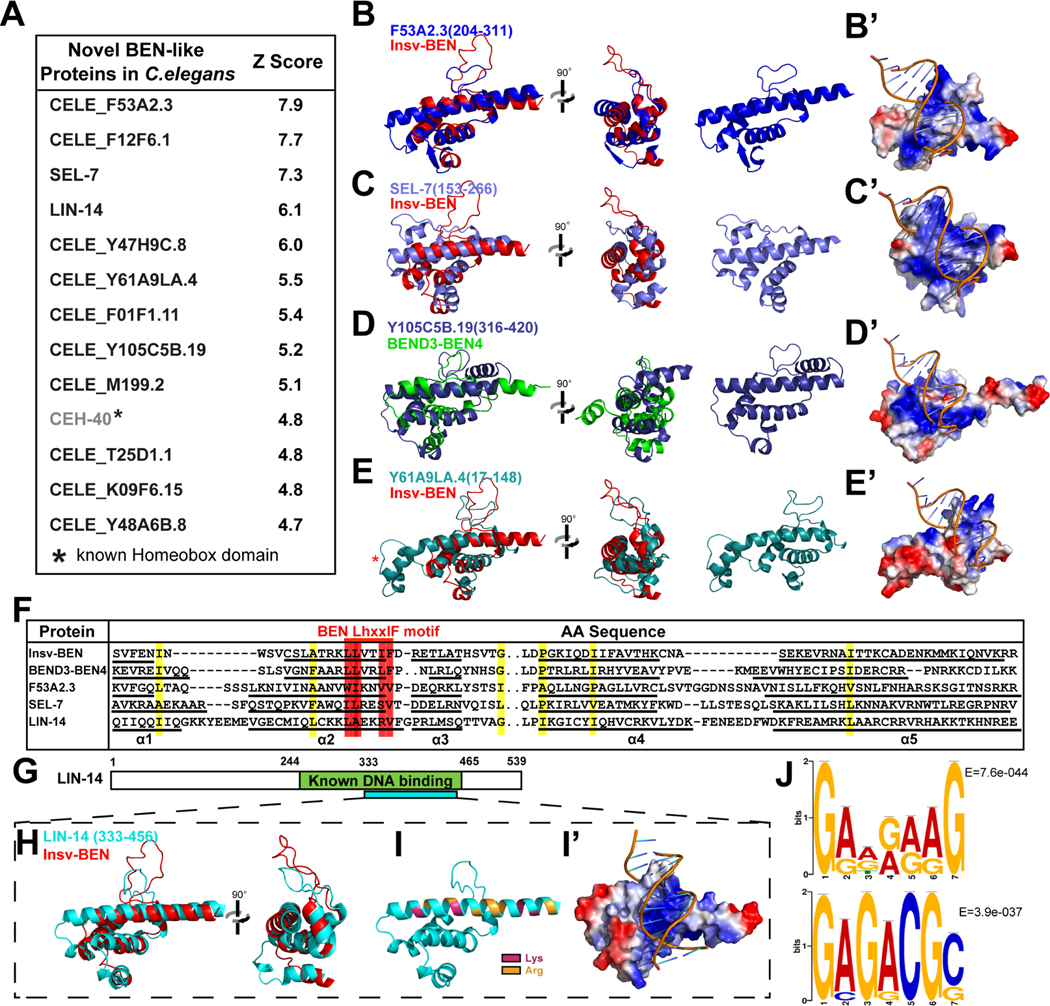
We next investigated whether animals lacking known BEN factors, such as most Nematode species, might still harbor proteins with characteristic BEN domain structures. We first compared Insv-BEN structure (PDB: 4IX7) with predicted protein models of C. elegans in the AlphaFold2 database with the DALI algorithm and retrieved 161 structural neighbors with a similarity score (Z-score) higher than 3.0 (Data S1C). The DALI algorithm Z-score assesses the similarity of substructures mainly through the match of equivalence residues21, which will not determine whether the hit possesses the defining features of a BEN domain, namely, the presence of five α helices and a middle loop connecting helix 3 and 4. We then manually inspected the degree of similarity of each hit to the BEN domain via structure superimposition using PyMOL. Except for CEH-40, a known PBX group homeodomain protein sharing similarities with BEN domain in 3D structure (Figure S2A and S2B), the remaining 12 out of the top 13 proteins (Z-score≥4.7, Figure 2A) exhibit typical globular BEN domain structures, which consists of five core helices with a 13~21aa middle loop separating α3 and α4. By comparison, the middle loop of known human and fly BEN domains ranges from 15 (BANP) to 23 (Insv) residues. Furthermore, BEN-like proteins can also be identified in other Nematoda with this method. For example, the TTRE_0000654801 protein of Trichuris trichiura (whipworm), a medically relevant human parasite, contains a typical BEN-like structure at amino acids 144~266 (Figure S2C and S2D).
Figure 2. Structure comparison identified novel BEN domain proteins in C. elegans.
(A) Structure comparison reveals novel BEN-like modules in C.elegans.
(B-E) Superposition of solved BEN structures and AlphaFold-predicted C.elegans BEN-like modules. F53A2.3 and SEL-7 resemble type I BEN (Insv-BEN) conformation, while Y105C5B contains an α6 helix and is closer to the type II BEN (BEND3-BEN4). Y61A9LA also resembles a Type I BEN domain but has extra residues at the N-terminus part of the α5 helix (star).
(B’-E’) Electrostatic surface representation of AlphaFold-predicted BEN-like modules in the C. elegans proteome. Color density represents the positive (blue) and negative (red) charges.
(F) Alignment of the C.elegans BEN-like modules with BEN domains from Drosophila Insv and human BEND3-BEN4 reveals the conservation of critical residues. The color highlights the conserved hydrophobic position. The red highlights the conserved LhxxIF motif.
(G) The LIN-14 BEN-like structure (333~456) is within a known DNA binding region (244~465).
(H) Superposition of predicted LIN-14 (333~456) and solved Insv-BEN (red) structures.
(I-I’) Electrostatic surface visualization of LIN-14 BEN-like structure (333~456), with blue representing positive charges and red representing negative charges. The Lys and Arg residues in α5 helix are highlighted in (I).
(J) De novo LIN-14 targeting consensus DNA sequences revealed with LIN-14 ChIP-seq data from the modENCODE database.
We reported that BEN domains recognize DNA through two different strategies: type I BEN domains (typified by the Insv-BEN) contain five α-helices and interact with DNA base through residues on α5 helix and the middle loop (Figure 1A and 1C)16. In contrast, type II BEN domains (typified by the BEND3-BEN4) harbor an extra C-terminal α6 helix, which collaborates with α5 to target DNA bases (Figure 1B and 1D)6. Based on the features of the 3D structure, we organize the twelve C.elegans BEN-like proteins into three groups. The first group (CELE_F53A2.3, SEL-7, LIN-14 and CELE_Y47H9C.8) are most similar to Insv-BEN and other type I BEN domains (Figure 2B, 2C and Data S3A). Group II proteins (CELE_F12F6.1,CELE_Y105C5B.19, CELE_M199.2 and CELE_T25D1.1) are more similar to type II BEN as they contain an α6 helix in the C-terminus, which might be involved in DNA binding (Figure 2D). Finally, the third group of proteins (CELE_Y61A9LA.4, CELE_F01F1.11, CELE_K09F6.15 and CELE_Y48A6B.8) resemble type I BEN in putative DNA binding regions (i.e. the presence of a long middle loop and the lack of an α6 helix), but harbor additional residues in the N-terminal portion of α5 helix (Figure 2E). BEN domain folds generate a positively charged binding surface along DNA grooves, providing a structural basis for BEN-DNA interaction. Similarly, the electrostatic surface visualization showed that all three groups of C.elegans BEN-like domains also have basic positive surfaces that are consistent with DNA loading (Figure 2B’–2E’).
In addition, the C. elegans BEN-like modules share similar conservation patterns as the BEN, particularly those hydrophobic residues at the helix-loop boundaries (Figure 2F). Previous work has identified the LhxxlF motif (l, aliphatic; x, any residue) in the α2-helix as the most characteristic feature of the BEN domain4. Research has also suggested that residues within this motif play a role in protein-protein interactions23 . However, the precise function of the LhxxlF motif remains largely unknown. This motif is present, albeit in a modified form, in C. elegans BEN-like proteins, with Leu and Phe often being replaced by other hydrophobic residues (Figure 2F). Together, these newly identified BEN proteins closely resemble known BEN domains by primary sequence, tertiary structure, and electrostatic charge, revealing a novel family of likely DNA binding proteins in C. elegans.
The novel C. elegans BEN domain proteins recognize specific DNA sequences
Among these identified proteins, LIN-14 and SEL-7 have long been known to play essential roles in the development of C. elegans. Based on their nuclear localization and demonstrated in vitro DNA binding activities, both were putatively designated as transcription factors24–27. In particular, lin-14 is perhaps most famous as the critical target of the founding miRNA lin-4, and LIN-14 drives early temporal identity (L1 larval stage) that must be downregulated by lin-4 to permit transition to the L2 stage28. SEL-7 was isolated as a genetic modifier of the Notch signaling pathway26. Nevertheless, despite their fundamental activities, both remained as orphan proteins lacking apparent homologs in most other metazoan species.
Importantly, a previous study found that LIN-14 interacts with DNA through its C-terminal residues 244–46525, while our comparison analysis revealed that the 333–456 region has a similar spatial arrangement to the Insv-BEN domain (Figure 2G and 2H). Similar to other DNA binding BEN domains, the α5 helix of LIN-14 is enriched with Arg and Lys (Figure 2I and S3A). In addition, the electrostatic surface visualization shows that LIN-14 333–456 region contains a positive surface allowing DNA loading (Figure 2I, 2I’ and S3A). In contrast, LIN-14 244–332 is predicted to be a negatively charged disordered region, making it unlikely to bind DNA (Figure S3B). These observations indicate that LIN-14 333–456 comprises a novel sequence-specific DNA binding BEN domain. To investigate whether LIN-14 binds specific DNA sequences, we performed de novo motif discovery on LIN-14 ChIP-seq data from the modENCODE database29. We analyzed all 11329 peaks and identified 31 significant motifs, among which the top-ranked ones were GADRAAG (7399/11329 sites, E = 7.6e-044) and GAGACGS(1401/11329 sites, E = 3.9e-037 (Figure 2J). In addition, these motifs can be found in previously characterized LIN-14 targeting regions at nlp-45 and dma-1 gene loci 30,31. More importantly, the motif sites are also conserved in other Nematodes, such as C.brenneri, C.japonica and C. tropocalis (Figure S3C). Thus, although different from previous LIN-14 binding consensus DNA sequences revealed by SELEX (GAACRY) and ChIP-seq (TGGAR) 25, 30, the collected results suggest that LIN-14 prefers GA-enriched motifs. These findings highlight the potential for our structural analysis approach to uncover novel functional domains in proteins and help interpret their functions.
Structural comparisons identify novel BEN domain proteins in Drosophila and zebrafish
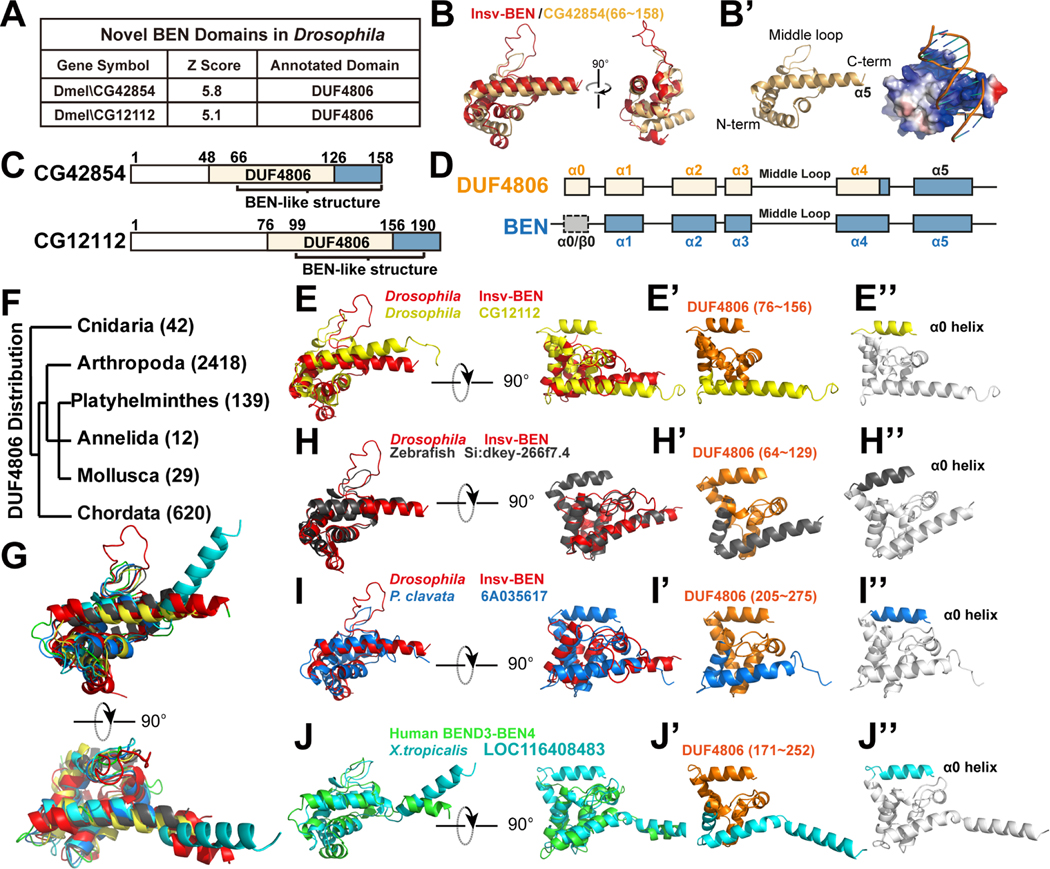
Next, we expand our analysis to other model organisms. In the Drosophila proteome, we identified the four known BEN-containing proteins, including Insv, Elba1, Elba2, and pre-mod(mdg4)-C (Figure 1G, S1 and Data S1C). Furthermore, our structural comparison uncovered two additional novel BEN candidates, CG42854 and CG12112 (Figure 3A). Upon overlaying these two proteins with Insv-BEN, we found them to align well. In addition, the positively charged surfaces of both CG42854 and CG12112 indicate that they are likely to bind DNA (Figure 3B, 3B’, S4A and S4A’).
Figure 3. The DUF4806 motif represents a subgroup of BEN domain with an upstream α0 helix.
(A) Structure comparison shows Drosophila CG42854 and CG12112 to be similar to BEN domains, with BEN-like regions overlapping with a presumed DUF4806 motif.
(B-B') Superposition of the Insv-BEN structure (PDB: 4IX7, red) and the predicted model of CG42854. (B') shows the electrostatic surface potential of the CG42854 BEN-like domain with Insv-BEN targeting DNA.
(C) The domain organization of Drosophila CG42854 and CG12112 proteins, with BEN-like structures overlapping with uncharacterized DUF4806 motifs.
(d) A schematic view of DUF4806 and BEN domains. DUF4806 contains five α-helices (α0-α4) preceding a downstream unrecognized helix α5, and the α1-α4 helices in BEN domains correspond well with DUF4806. In contrast, there could be either α-helices (BEND3-BEN3, PDB:7V9H) or β-sheets (Insv-BEN, PDB: 4IX7) at the upstream of BEN domains.
(E-E”) Superposition of Insv-BEN (PDB: 4IX7, red), and the predicted structure of CG12112 BEN-like region, showing that DUF4806 closely resemble Insv-BEN domains. (E’) shows the cartoon view of predicted structures of CG12112 regions containing α0-α5 helices, with residues corresponding to DUF4806 colored in orange. Note that DUF4806 ends in the middle of helix α4. (E”) shows the α0-α5 helices of CG12112, with core BEN domain structure (α1-α5 helices) colored in white and helix α0 highlighted with yellow.
(F) Phylum distribution of annotated DUF4806-containing proteins in the InterPro database.
(G-J) Superposition of Insv-BEN (PDB: 4IX7, red), BEND3-BEN4 (PDB: 7W27, green), and predicted DUF4806 models, showing that DUF4806 modules closely resemble solved BEN domains.
(H'-J') Cartoon view of predicted structures of regions containing α0-α5 helices, with residues corresponding to annotated DUF4806 colored in orange. Note that DUF4806 ends in the middle of helix α4.
(H''-J'') Cartoon view of predicted structures of regions containing α0-α5 helices, with core BEN domain structure (α1-α5/6 helix) colored in white and helix α0 highlighted with corresponding colors.
Our comparative analysis of the zebrafish proteome revealed homologs of most human BEND proteins, including zebrafish BANP, BEND2 (LOC569178), BEND3, BEND4, BEND5, BEND7, NACC1, and NACC2 (Data S1D). Furthermore, we identified nine novel BEN domain proteins (Figure S4B and Data S1D). Most of these novel BEN-like modules superpose well with Insv-BEN, and some have an additional helix downstream of the core five-helices region, mimicking type II BEND3-BEN4 (Figure S4C, S4C’, S4D, and S4D’).
DUF4806 comprises a previously unrecognized subtype of BEN domain
We observed that ten out of the eleven novel BEN domains in the Drosophila and zebrafish overlap with a predicted DUF4806 motif (Pfam ID: 16064) (Figure 3A and S4B). To further validate the similarity between DUF4806 and known BEN domains, we performed comparisons for proteins similar to CG12112 and CG42854 in the mouse proteome. In both cases, the search returned most BEN-containing proteins as top hits (Figure S4E, S4F, Data S3B and S3C). DUF4806 is an uncharacterized protein motif which has not been experimentally validated. It has a length of approximately 80 amino acids and consists of five predicted α-helices (designated α0~α4 thereafter). Our analysis showed that the last four helices of DUF4806 align with predicted BEN-like regions (Figure 3B–D). Significantly, the unannotated regions downstream of DUF4806 in CG42854 and CG12112 proteins also consist of α-helices (Figure 3C and 3D), and exhibit close structural similarity to the α5-helix in the Insv-BEN domain (Figure 3E and S4A).
According to the InterPro database32, there are 3260 proteins containing DUF4806, which are found across diverse metazoans ranging from Hydra to X. tropicalis (Figure 3F). This distribution pattern is similar to that of BEN domains. Moreover, DUF4806 proteins from different species all closely resemble the structure of either Insv-BEN (e.g. zebrafish si:dkey-266f7.4, Drosophila CG12112 and red coral (P. clavata) 6A035617, Figure 3E and G-I) or BEND3-BEN4 (e.g. X. tropocalis LOC116408483, Figure 3G and J). All the DUF4806 motifs in these proteins terminate before the α4 and missed α5 helix of the BEN domain, while the unannotated regions downstream of DUF4806 correspond to the remaining α5 and α6 helices of the BEN domain. (Figure 3E’ and 3H'–3J'). Previous studies have shown that the basic residues in the α5 helix directly interact with DNA bases6, 7. Thus, to achieve sequence-specific DNA binding, different BEN domains choose distinct numbers and types of amino acids in the C-terminus, explaining why DUF4806 does not cover this region. In the N-terminus, the α0 helix is not identified in Insv-BEN (Figure 3E'' and 3H”–3J''). Structural analysis shows that the Insv and Elba1 (PDB: 4X0G) BEN domains are preceded with two β-sheets, while the BEND3-BEN3 (PDB:7V9H) has an upstream α-helix7, 8, 16. Together, these observations consolidate that DUF4806 comprises a subtype of BEN domain, although the definition DUF4806 should to be amended to include the present findings.
Multiple sequences alignment reveals the conservation pattern of BEN domains.
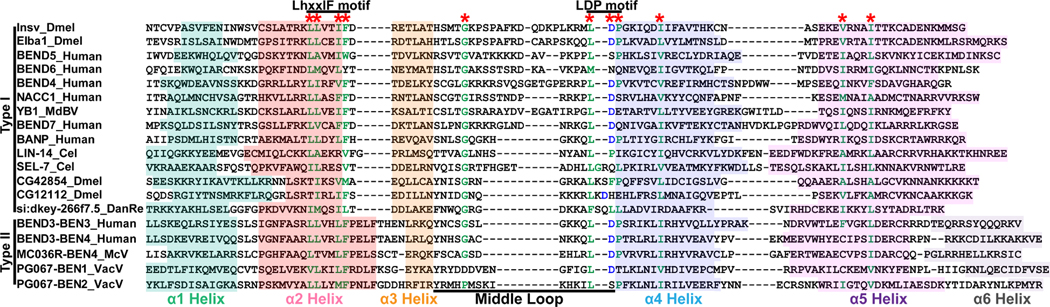
In order to identify characteristic residues of BEN domains, we performed multiple sequence alignment for both known and novel BEN domains, including those from human and Drosophila proteins, novel C.elegans BEN factors, DUF4806, and BEN domains from viruses (i.e. PG067/E5R from Variola virus, MC036R from Molluscum contagiosum virus, and YB1 from Microplitis demolitor bracovirus) (Figure 4).
Figure 4. Multiple sequences alignment revealed the preserved residues of BEN domains.
BEN domains from Drosophila (i.e. BEN domains from Drosophila Insv and Elba1), human (BEN domains from human NACC1, BANP, BEND3, BEND4, BEND5, BEND6 and BEND7), viruses (i.e. BEN domains from YB1 from Microplitis demolitor bracovirus, MC036R from Molluscum contagiosum virus, and PG067/E5R from Variola virus), C.elegans (i.e. BEN domains from LIN- 14 and SEL-7), DUF4806 factors (i.e. BEN domains from Drosophila CG12112 and CG42854 and zebrafish si:dkey-266f7.5) are used for multiple sequences alignment. The alignment reveals the conserved residues (marked with stars) and motifs (LhxxlF and LDP). The conserved hydrophobic resides are labeled with green, and the acidic residues with blue.
We observed that helix 2, helix 3, and the N-terminus part of helix 4 are more conserved compared to other regions. Helix 2 contains a core sequence of 12 amino acids, with the C-terminal boundary having the LhxxlF motif, which is conserved in most BEN members. The helix 3 contains six residues in type I BEN domains and four in type II BENs. The N-terminal boundary of helix 4 has an LDP-rich motif, which interestingly, is less conserved in DUF4806 and SEL-7. The middle loops have varying lengths and compositions, but the fifth residue of this loop is one of the most conserved sites and strongly tends towards glycine.
In addition, we observed differences between type I and type II BEN domains beyond the presence of an additional helix in type II. Specifically, type II BEN domains have a longer helix 2 and a shorter helix 3 compared to type I domains. Type II BENs are rare in metazoans, and the only type II BEN factor in Drosophila and human is BEND3. To be noticed, all the mammalian viral BEN domains show features of type II BEN domains, and share extraordinary sequence similarity with BEND3-BEN3 or BEN4 domains. The mechanism of this preference requires further investigation.
Structure comparison reveals similarities between the BEN domain and homeodomain
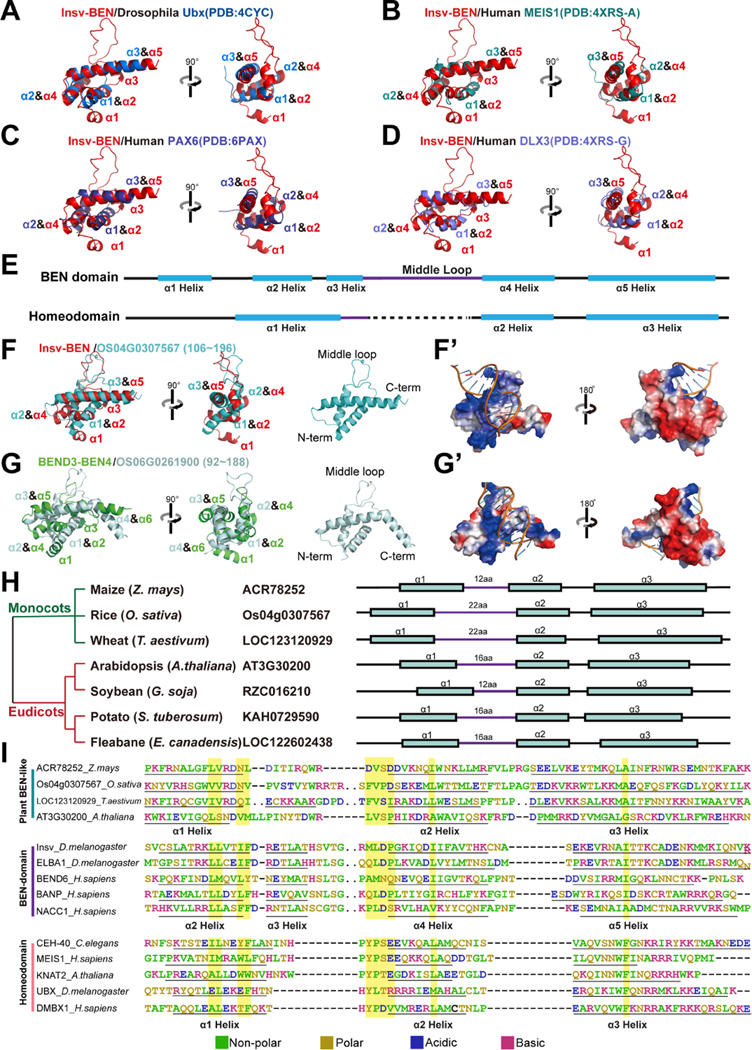
During our structural screening for BEN-like domains, we noticed that helix-turn-helix (HTH) motifs were detected amongst the hits with high to intermediate scores. These hits included homeodomains, FF domains (e.g., mouse Tcerg1 and Tcerg1l, Drosophila CG31367 and Fip1), PH domain (e.g., Drosophila PNPase) and regions lacking annotated domains but exhibiting HTH conformation (e.g., Drosophila CG17341). Among these HTH motifs, homeodomains were the most frequently identified candidates in the mouse, Drosophila and C. elegans proteomes (Figure 1G, S1, S2A, S2B, S5, Data S1A, S1B and S1C). To further demonstrate the similarity between the BEN domain and homeodomain, we screened the proteome for proteins with 3D similarity to homeodomains and found that BEN domains are frequently observed as intermediate-scoring hit. Importantly, this similarity is not only observed in predicted models (Data S2A), but also confirmed in solved structures (Data S2B). The homeodomain is an ancient DNA binding domain present in many eukaryotes, including diverse species that lack annotated BEN domains (e.g., plants and fungi)33. They are essential for a variety of biological processes, including development, growth and metabolism.
The homeodomain is composed of ~60 residues that fold into three α helices connected by short loops. When superimposing the BEN domain onto homeodomains, we found that the BEN helix α2, α4 and α5 resemble the assignment of homeodomain α1~3 helices (Figure 5A–5E). The homeodomain binds DNA through its HTH motif with its α3-helix entering the DNA major groove and serving as the recognition helix, which is the same as the α5-helix in the BEN domain (Figure 5A–5D). Notably, BEN domains exhibit comparable similarities to TALE group homeodomains (i.e., Meis1, Figure 5B) and non-TALE homeodomains. TALE factors comprise an ancient subgroup of homeodomain proteins that are shared across animals, plants, and fungi, whereas other groups in these searches were born in metazoans. Since structural similarity analysis was capable of distinguishing functional orthologs from amongst highly dissimilar BEN proteins (Figure 1G), the general similarity of BEN domains to both TALE and non-TALE homeodomains suggest that metazoan BEN domains likely derived from homeodomain-containing transcriptional regulators at an early evolutionary stage. Overall, homeodomains and BEN domains share a conserved DNA binding strategy, in which a recognition helix steps into the major groove and plays a pivotal role in DNA base-specific targeting.
Figure 5. Structure comparisons reveal similarities between BEN domain, homeodomain and plant BEN-like modules.
(A-D) The superposition of the solved structure of Insv-BEN (red) and the predicted model of homeodomains highlights the similarities in their 3D conformation.
(E) The overall secondary structure of both the BEN domain and Homeodomain. The BEN domain α5 helix and homeodomain α3 helix serve as the central DNA binding elements. Compared with the homeodomain, the BEN domain has more helices and longer middle loops (purple).
(F-G’) Structure screening identifies BEN-like protein domains in plants. (F) shows the superposition of the solved structure of Insv-BEN (PDB:4IX7) and the predicted model of the rice OS04G0307567 protein BEN-like region. (g) shows the superposition of the solved structure of BEND3-BEN4 (PDB:7W27) and the predicted model of the rice OS06G0261900 BEN-like region. (F’ and G’) shows the electrostatic surface representation of AlphaFold-predicted BEN-like modules of rice BEN-like proteins. Color density represents the positive (blue) and negative (red) charges.
(H) The plant BEN-like structures are conserved in both monocots and eudicots. The structures are comprised of three α-helices, with some members having extended middle loops.
(I) Comparisons of the conservation pattern of plant BEN-like domains, homeodomains and BEN domains. The analysis reveals the conserved residues are present in all three different groups of domains (highlighted).
See also Figure S5, S6, Data S1E, S2A–S2C, S4A, S4L and S4M.
Identification of BEN-like structures in plants
The BEN and DUF4806 were predicted to be present exclusively in metazoans. In line with this, we did not discern any proteins with BEN-like structural conformation in yeast (S. cerevisiae and S. pombe), bacteria (E. coli and S. pneumonia), or protozoan (L. infantum and P. Falciparum) species. Surprisingly, when we subjected plant proteomes to the same analyses, we identified a large group of proteins with BEN-like structures. The rice OS04G0307567 protein, for example, has a globular structure resembling Insv-BEN conformation (Figure 5F). As observed in Insv-BEN, OS04G0307567 has a long positively charged middle loop, suggesting its involvement in DNA base interactions (Figure 5F’). The rice OS06G0261900 protein, on the other hand, has an additional positively charged C-terminal α-helix, which is more likely to interact with DNA through type II strategy (Figure 5G and 5G’). Both OS04G0307567 and OS06G0261900 have basic positive surfaces that suggests a preference for nucleic acid binding, and negatively charged surfaces facing away from the predicted DNA (Figure 5F’ and 5G’). OS04G0307567 and OS06G0261900 BEN-like regions are highly similar to each other by primary sequence. In addition, their homologous genes are not only identified in other species of the monocot grass family (e.g., wheat and maize), but also found in eudicots (e.g., Arabidopsis, soybean, potato, and fleabane) (Figure 5H). While none of these BEN-like-region-containing plant genes have been characterized, some have been predicted to play a role in transcription and chromatin regulation. For example, soybean RZC016210 and Artemisia PWA79539 proteins are predicted to have Bromo-like regions, indicating key roles in chromatin regulation.
Interestingly, we did not identify any members of this protein family in basal angiosperms or gymnosperms, suggesting they originated in the common ancestor of monocots and eudicots. This supports a scenario in which different eukaryotic kingdoms independently acquired BEN- like domain factors connected to transcriptional regulation.
The relationship between BEN domains, homeodomains and plant BEN-like regions
We next attempted to establish the evolutionary relationship between protein modules with BEN-like conformations. Our multiple sequence alignments revealed preserved residues in these domains (Figure 5I). For example, the positively charged Arg and Lys (Figure 5I) are highly enriched in the C-terminal helix, which is crucial for direct DNA binding. Previous studies have established that the homeodomain uses conserved residues to maintain its hydrophobic core, including the LxxLxxxF sequence (Figure 5I) in α1 helix and the hXXWF motif (Figure 5I) in α3 helix33. These signatures are also conserved in both BEN domain and plant BEN-like regions (Figure 5I), with substitution occurring with amino acids of the same properties.
Both phylogenetic analysis and structural features indicate that the plant BEN-like regions are more similar to the homeodomains compared with metazoan BEN domains. For example, homeodomains and plant BEN-like modules both have three helices, while BEN domains have five or six helices, implying the independent origin of the plant BEN-like models from a homeodomain or another HTH-containing protein in the ancestor of current monocot and eudicot species (Figure 5H). However, some plant BEN-like domains have a middle loop that is ~20 amino acids long (e.g., rice Os04g0307567; wheat LOC123120929, and fleabane LOC122602438. Figure 5H), which is a characteristic feature for BEN domains. These observations suggest differences between the metazoan BENs and plant BEN-like models. As such, additional research will be necessary to establish the origin of these differences and shed further light on the evolutionary relationship between these domains.
Contextual analysis of novel BEN-containing proteins indicates their co-occurrence with transcription/chromatin regulating modules
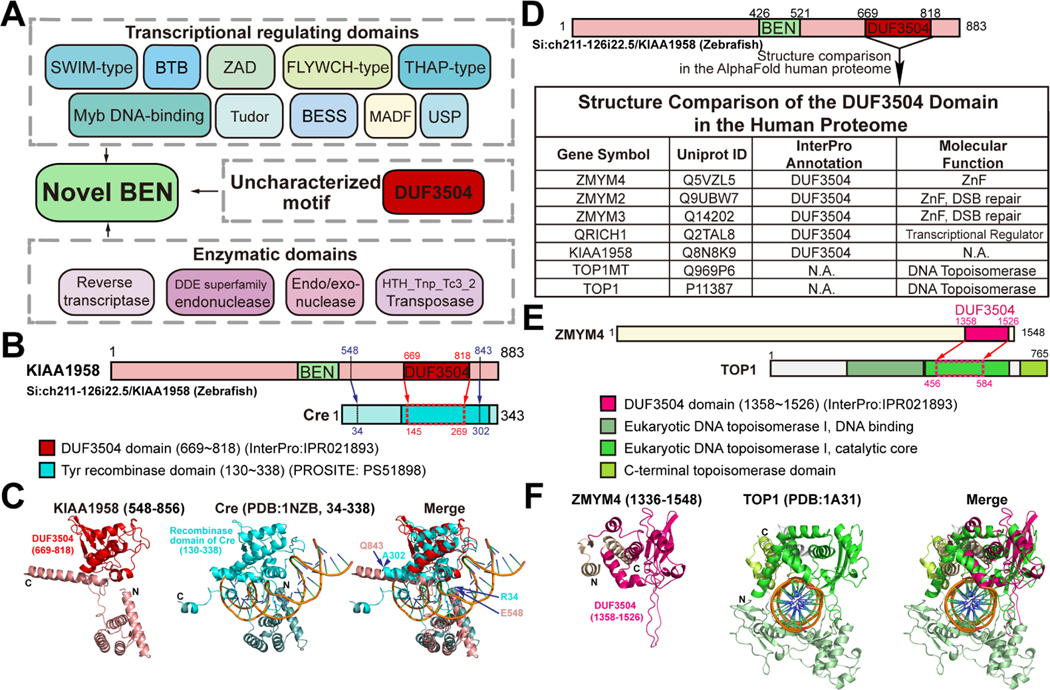
It was previously reported that BEN domains, then of unknown function, sometimes co-occur with other motifs involved in gene regulation4. With our highly expanded set of BEN superfamily factors, we re-evaluated the domain context of BEN-containing proteins. We found novel BEN modules are linked with chromatin and transcriptional regulation domains, such as BTB/POZ (PROSITE: PS50097), Myb-DNA binding (Pfam: PF13837), ZAD (PROSITE: PS51915) and a variety of Zinc Finger domains including SWIM (PROSITE: PS50966), FLYWCH (Pfam: PF04500) and THAP (PROSITE: PS50950) (Figure 6A). These observations are consistent with the known DNA binding activity of BEN domains. Interestingly, some BEN factors also contain nuclease domains, such as reverse transcriptase (PROSITE: PS50878), DDE endonuclease (Pfam: PF13358), Endo/exonuclease (Pfam: PF13359), and HTH_Tnp_Tc3_2_Transposase (Pfam: PF01498), suggesting as yet unknown roles of BEN domain in nucleotide processing and gene translocation (Figure 6A).
Figure 6. Contextual analysis indicates transcriptional roles of novel BEN-containing factors.
(A) The contextual graph illustrates domains identified in novel BEN-containing factors, including those related to transcriptional regulation and nucleotides processing activity, as well as an understudied DUF3504 motif.
(B-C) The structural similarity between the Cre recombinase (PDB: 1NZB) and the predicted model of DUF3504 motif in the zebrafish Si:ch211–126i22/KIAA1958 protein is demonstrated through a superposition. DUF3504 only resembles the central part of Cre recombinase domain (145~269). But the flanking region (548~843) of DUF3504 overlays with the majority of Cre recombinase (34~302).
(D) Structure comparison of AlphaFold determined DUF3504 and human proteome models identified human proteins with structures similar to DUF3504.
(E) The domain architecture graph of human TOP1 protein displays regions with 3D structure similar to DUF3504.
(F) Superposition of the predicted models of human ZMYM4-DUF3504 motif and solved structure of the TOP1 topoisomerase region. The ZMYM4 DUF3504 (1358~1526) resembles amino acids 456~584 of TOP1.
3D comparisons identify novel DUF3504-containing proteins
We noticed that some novel BEN proteins harbor the uncharacterized DUF3504 motif. In order to expand the potential applications of our 3D-comparison approach beyond the study of BEN domains, we conducted a structural similarity analysis of DUF3504 and inferred its putative functions. DUF3504 is a highly conserved predicted protein motif that shares sequence homology with Crypton family tyrosine-recombinase-encoding DNA transposons but lacks recombinase activity34 . To determine the presence of proteins structurally similar to DUF3504, we conducted a screening in the PDB database. Our results showed that DUF3504 in BEN-containing proteins (zebrafish KIAA1958) closely resembles the core DNA binding and processing region of the Cre recombinase (PDB: 1NZB, Figure 6B and 6C). Interestingly, the regions flanking DUF3504 are also structured and overlay well with the Core-binding domain of the Cre protein (Figure 6B and 6C).
In the human proteome of the AlphaFold database, our search successfully identified five DUF3504-containing proteins (ZMYM4, ZMYM2, ZMYM3, QRICH1 and KIAA1958) and two additional hits (TOP1MT and TOP1) (Figure 6D). ZMYM is an MYM-type zinc-finger protein family with essential biological functions35–38. At the molecular level, ZMYM proteins are known transcriptional regulators. In addition, ZMYM2 and ZMYM3 have been implicated in DNA double-strand breaks (DSBs) repair39, 40. Interestingly, structural comparisons revealed the presence of previously unrecognized DUF3504-like structures in TOP1 and TOP1MT, two type I DNA topoisomerases (Figure 6D). Superimposition of the predicted DUF3504 and crystallography-determined TOP1 structures (PDB: 1A31) demonstrates that DUF3504 closely resembles the 3D conformation of the catalytic core of TOP1 enzyme (Figure 6E and 6F). Notably, unlike the above mentioned KIAA1958, the structured regions flanking DUF3504 in ZMYM4 do not share similarity with other regions of TOP1 enzyme. Together, DUF3504 is a potential transcriptional regulator domain.
Discussion
Broadly existing BEN-containing factors in metazoans
The annotation of protein domains is of critical importance for determining protein function. In this article, we establish a method to annotate protein domains by exploiting AlphaFold-predicted protein modules. Through systematic structure comparisons, we identify novel BEN domains and potential BEN-like DNA binding structures in diverse proteomes. We find that C. elegans, previously considered to lack BEN factors, actually contains multiple BEN-containing proteins. We also demonstrate that the previously uncharacterized DUF4806 motif, which has thousands of family members in various phyla, is a type of BEN domain. Further contextual analysis suggests these novel BEN-containing factors as transcriptional regulators. Thus, these results substantially enhance the breadth of BEN factors and provide hypothesis-based directions to study the functions of additional BEN factors (Figure 7). As a whole, this family has only recently begun to be studied in earnest, but studies to date already show that they fulfill several "hidden" features in cis-regulatory gene regulation4–10. Undoubtedly, the functionalization of additional members of this family, which could not be recognized by linear pattern matching, opens new doors into their study.
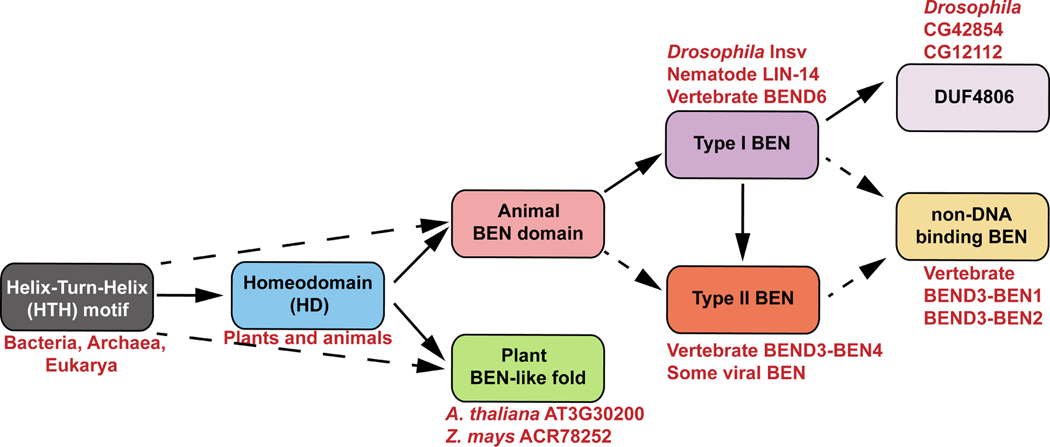
Figure 7. The evolutionary relationship of protein domains/motifs revealed by structural comparisons.
Our structural comparison implies that animal BEN and plant BEN-like domains may have originated from the homeodomain (solid) or HTH motifs (dash). The BEN domain can be categorized into two types based on structural features and DNA binding strategies. Type I BEN domain (e.g. Drosophila Insv, nematode LIN-14 and vertebrate BEND6) is defined by the presence of five α helices. Type II BEN domain (e.g. vertebrate BEND3-BEN4 and some viral BEN domains) has a downstream helix α6 involved in direct DNA binding. In contrast, DUF4806 (e.g. Drosophila CG12112 and CG42854) is more similar to the type I BEN domain but has an upstream α helix. These structural features suggest that both type II BEN and DUF4806 might have originated from ancestral type I BEN. Previous biochemical analyses have shown that vertebrate BEND3-BEN1 and BEND3-BEN2 lack DNA binding activities, despite both domains possessing features characteristic of type I BEN domains6. However, it remains possible that there are unrecognized type II BEN domains that do not bind DNA.
lin-14 and sel-7 have long been characterized as essential transcriptional regulators for C.elegans development24, 26. In particular, lin-14 gained fame as the central target of the founding miRNA lin-4, and it is an executive regulator of early temporal fate. However, neither of them was considered to have homologous genes in other model organisms, and this in particular has stymied insights into lin-14 function despite its genetic identification from profound mutant phenotypes41 nearly 40 years ago, and its molecular cloning 35 years ago24. Our study clarifies that LIN-14 and SEL-7 are in fact typical BEN domain factors in C. elegans, which actually harbors twice as many BEN proteins as D. melanogaster.
Recent studies, including our own and others, have demonstrated that BEN-containing proteins have important biological functions in regulating cell fate specification. For example, BEND6 is exclusively expressed in the postmitotic neurons and is important for neuron specification22. Mammalian BEND3 protein functions to prevent premature gene expression8. These observations, in conjunction with our discovery that LIN-14 and SEL-7 are BEN-containing proteins, highlight the notion that BEN domain proteins play widespread roles in transcriptional control of temporal development and cell fate decisions across metazoans.
Shared 3D conformation between BEN domains, homeodomains, and plant BEN-like structures
Our structure comparison reveals striking similarities between the BEN domain and the homeodomain. They both utilize a central α-helix enriched with basic residues to recognize DNA bases within the DNA major grooves. Homeodomains are widely considered to have derived from the more ancient HTH DNA binding domain, which is found even in bacteria. Interestingly, the BEN domain α3~5 helices also mimicking the conformation of an HTH module except for a much longer middle loop, indicating they may be evolutionarily related. The two domains are also similar in biological functions. Homeodomain proteins are known to play essential roles in regulating spatiotemporal development, similar to the known biological function of BEN factors. The conservation pattern in primary sequences between the two domains suggests that they are ancestrally related. Interestingly, previous studies have shown that many members of the extended homeodomain family exhibit a preference for CpG-methylated DNA sequences 42. This interaction is thought to occur through hydrogen bonds between hydrophobic amino acids, such as Ala, Ile and Val, and the methyl groups present on the CpG dinucleotide. In contrast, recent findings have shown that the BEN domains of BANP and BEND3-BEN4 are DNA binding domains that are actually repelled by mCpG sites 8,10. Notably, the DNA binding helix (α5) of BEN domains is more variable compared to homeodomains. Further research is needed to determine whether other BEN factors exhibit mCpG sensitivity.
Furthermore, we identified a protein model structurally resembling both BEN domain and homeodomain in various plants. These observations, combined with the results of phylogenetic analysis using primary amino acid sequences, suggest that the metazoan BEN domain and plant BEN-like models independently descended from the homeodomain or other HTH motif proteins, but exhibit similar DNA binding strategies. To date, none of these plant proteins with BEN-like regions have been functionally characterized. Our structure comparison and contextual analysis suggest they likely function as DNA binding proteins (Figure 7).
AlphaFold as an emerging tool for domain annotation and biochemical research
The past two years have seen spectacular progress in structure modeling. In our previous study, we found that in-silico prediction algorithms, including AlphaFold and Rosetta, precisely determined the structure of the human BEND3 protein BEN domain. We also used a combination of X-ray solved structures and predicted models to classify BEN domains into two classes: a type I Insv-like class and a type II BEND3-BEN4-like subset6. Another recent study used structure comparisons to reveal remote homologous relationship in the AlphaFold Database version 143. In this article, we utilized the high-accuracy protein models in the massively expanding AlphaFold2 database and revealed novel BEN domains more broadly. We demonstrated that this method can be applied to other protein modules. Previous sequence alignment analysis suggests DUF3504 is a homolog of Crypton family recombinase34. We confirm that the predicted DUF3504 structure closely resembles solved Cre recombinase. In addition, through structure comparison, we identified unrecognized DUF3504 in human TOP1MT and TOP1 proteins.
Overall, our method provides a revised foundation to elucidate how BEN domains and other understudied protein modules regulate biological processes at the molecular level.
STAR★Methods
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Yang Yu (yuy@ibms.pumc.edu.cn).
Materials availability
This study did not generate new unique reagents.
Data and code availability
This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available form the lead contact upon request.
Key Resource Table
| REAGENT or RESOURCE RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| LIN-14 ChIP-seq in L1 stage C.elegans | modENCODE Luo et al. 29 |
Experiment: ENCSR714ALL; Bed file: ENCFF941FVK |
| Software and algorithms | ||
| PyMOL | https://pymol.org/2/ | RRID:SCR_000305 |
| AlphaFold2 | https://alphafold.ebi.ac.uk/ | Varadi et al. 15 |
| ColabFold | https://colab.research.google.com/ | Mirdita et al. 45 |
| UCSF ChimeraX | https://www.cgl.ucsf.edu/chimerax/ | RRID:SCR_015872 Pettersen et al. 46 |
| MEME-ChIP | https://meme-suite.org/meme/ | Machanick and Bailey 49 |
METHOD DETAILS
Acquisition and generation of structural models
All experimentally determined 3D structures were obtained from the RCSB Protein Data Bank (https://rcsb.org/). Water and DNA molecules were removed with PyMOL (https://pymol.org/2/)44 to generate clean BEN domain monomer structure files. In-silico predicted protein models were obtained from the EMBL AlphaFold2 database (https://alphafold.ebi.ac.uk/)15. Other predicted protein structures were generated by AlphaFold2 algorithm in ColabFold (colab.research.google.com)45. The confidence of the predicted structures was assessed using the predicted local distance difference test (pLDDT) scores. To visualized the pLDDT scores with structures, the UCSF ChimeraX 46 was used with “palette alphafold” settings.
Proteome-wide homologous structure searching and structural analysis
The DALI server (http://ekhidna2.biocenter.helsinki.fi/dali/)21 was used to compare the PDB experimentally solved protein structures with AlphaFold2 models. We defined the “top hits” as the top 10% of the hits with a Z-score≥3. The annotated domains were retrieved from the InterPro database.
To determine potential novel BEN domains, we manually inspected the candidate structures obtained from the DALI screening using PyMOL. Specifically, we checked whether each candidate structure possessed the characteristic features of a BEN domain, including five core α helices and a middle loop spanning at least 13 residues.
Multiple sequences alignment
Given BEN domains are variable, we aligned the domains mainly with equivalent amino acids based the 3D conformation rather than directly aligning the sequences. Because the α2~3 helices and the N-term boundary of helix4 are highly conserved, these regions were first aligned separately with PRALINE47 and validated with PyMOL. In contrast, the helix α1, α5 and the middle loop are variable in both length and composition. These regions were first aligned using primary sequences, and then adjusted based on structurally equivalent amino acids. To compare BEN, homeodomains and plant BEN-like domains, we first aligned the three domains separately. We then align the three group of domains based on equivalent residues at 3D conformation.
De novo motif discovery
The LIN-14 ChIP-seq data was obtained from the modENCODE project (Experiment: ENCSR714ALL; Bed file: ENCFF941FVK)29, 48. DNA sequences of binding peaks were fetched through bedtools (v2.30.0) using the C.elegans reference genome 11 from UCSC (https://hgdownload.soe.ucsc.edu/goldenPath/ce11/bigZips/ce11.fa.gz). De novo motif finding was performed using MEME-ChIP (v 5.5.0)49 .
Quantification and Statistical Analysis
To obtain the pLDDT score per residue for all predicted models used in the main figures, we retrieved the data from the AlphaFold database. We calculated the average pLDDT for middle loops by first determining the middle loop residues using PyMOL and then calculating the mean pLDDT in Excel.
Supplementary Material
Structure comparisons are performed between X-ray determined BEN domain structures and AlphaFold predicted models of different species. (A) Screening with Insv-BEN structure (PDB: 4IX7) in the Drosophila proteome. (B) Screening with the Insv-BEN structure in the mouse proteome (C) Screening with the Insv-BEN structure in the C.elegans proteome. (D) Screening with the Insv-BEN structure in the zebrafish proteome (E) Screening with the Insv-BEN structure in the rice proteome. (F) Screening with the BEND3-BEN4 structure (PDB: 7W27) in the Drosophila proteome. (G) Screening with the BEND3-BEN4 structure in the mouse proteome.
Structure comparisons are performed between the X-ray determined homeodomains and AlphaFold predicted models of different species. Both homeodomain proteins and BEN factors are revealed. (A) Screening with the Drosophila Ftz homeodomain structure (PDB: 1ftz) in the mouse proteome. (B) Screening with the Drosophila UBX homeodomain structure (PDB: 4cyc) in the Drosophila proteome. (C) Screening with the human PAX6 homeodomain structure (PDB: 6pax) in the C.elegans proteome.
(A) Screening with the predicted structure of LIN-14 BEN domain in the mouse proteome. (B) Screening with the predicted structure of Drosophila CG12112 DUF4806 motif. (C) Screening with the predicted structure of Drosophila CG42854 DUF4806 motif in the mouse proteome.
(A) The r.m.s.d. value for each structure alignment presented in the figures. (B-W) The predicted local distance difference test (pLDDT) score for each predicted structure is retrieved from the AlphaFold2 database. This test measures the per residue confidence of the predicted model. (B) The pLDDT score for F53A2.3. (C) The pLDDT score for SEL-7. (D) The pLDDT score for Y105C5B.19. (E) The pLDDT score for Y61A9LA.4. (F) The pLDDT score for LIN-14 (G) The pLDDT score for CG42854. (H) The pLDDT score for CG12112. (I) The pLDDT score for SI:DKEY266f7.4. (J) The pLDDT score for 6A035617. (K) The pLDDT score for LOC116408483. (L) The pLDDT score for OS04G0307567. (M) The pLDDT score for OS06G0261900. (N) The pLDDT score for KIAA1958. (O) The pLDDT score for ZMYM4. (P) The pDDT score for CEH-40. (Q) The pLDDT score for TTRE_0000654801. (R) The pLDDT score for SI:CH211–126I22.5. (S) The pLDDT score for SI:CH211–262H13.3. (T) The pLDDT score for CG17341. (U) The pLDDT score for PNPase. (V) The pLDDT score for NOL4. (W) The pLDDT score for TCERG1.
Highlights.
3D comparisons using predicted protein models reveal numerous novel BEN factors
3D homology searches uncover BEN domains in C. elegans LIN-14 and SEL-7
DUF4806 represents a BEN domain subgroup
BEN domains, homeodomains and plant BEN-like folds show 3D resemblances
Acknowledgments
Work in Y.Y.’s group was supported by the National Natural Science Foundation of China (32170550), State Key Laboratory Special Fund (2060204), the Special Research Fund for Central Universities, Peking Union Medical College (2021-RC310–001 and 2022-RC310–07), and CAMS (Chinese Academy of Medical Sciences) Innovation Fund for Medical Sciences (CIFMS) 2021-I2M-1–019. Work in E.C.L.’s group was supported by NIH grants R01GM083300 and R01NS083833 and by the MSK Core Grant P30CA008748.
The authors would like to acknowledge Dr. Thomas Bürglin from the University of Basel, Dr. Bing Liu from the Institute of Botany of the Chinese Academy of Sciences and Dr. Pu Gao from the Institute of Biophysics of the Chinese Academy of Sciences for their valuable advices. Additionally, the authors extend their thanks to the ENCODE and modENCODE consortium, as well as the Kevin White laboratory, for generating the LIN-14 ChIP-seq data.
Footnotes
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Illergard K, Ardell DH, and Elofsson A (2009). Structure is three to ten times more conserved than sequence--a study of structural response in protein cores. Proteins 77, 499–508. 10.1002/prot.22458. [DOI] [PubMed] [Google Scholar]
- 2.Carpentier M, and Chomilier J (2019). Protein multiple alignments: sequence-based versus structure-based programs. Bioinformatics 35, 3970–3980. 10.1093/bioinformatics/btz236. [DOI] [PubMed] [Google Scholar]
- 3.Bayly-Jones C, and Whisstock JC (2022). Mining folded proteomes in the era of accurate structure prediction. PLoS Comput Biol 18, e1009930. 10.1371/journal.pcbi.1009930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abhiman S, Iyer LM, and Aravind L (2008). BEN: a novel domain in chromatin factors and DNA viral proteins. Bioinformatics 24, 458–461. 10.1093/bioinformatics/btn007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aoki T, Sarkeshik A, Yates J, and Schedl P (2012). Elba, a novel developmentally regulated chromatin boundary factor is a hetero-tripartite DNA binding complex. Elife 1, e00171. 10.7554/eLife.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng L, Liu J, Niu L, Kamran M, Yang AWH, Jolma A, Dai Q, Hughes TR, Patel DJ, Zhang L, et al. (2022). Distinct structural bases for sequence-specific DNA binding by mammalian BEN domain proteins. Genes Dev 36, 225–240. 10.1101/gad.348993.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai Q, Ren A, Westholm JO, Duan H, Patel DJ, and Lai EC (2015). Common and distinct DNA-binding and regulatory activities of the BEN-solo transcription factor family. Genes Dev 29, 48–62. 10.1101/gad.252122.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J, Zhang Y, You Q, Huang C, Zhang T, Wang M, Zhang T, Yang X, Xiong J, Li Y, et al. (2022). Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation. Science 375, 1053–1058. 10.1126/science.abm0730. [DOI] [PubMed] [Google Scholar]
- 9.Ueberschar M, Wang H, Zhang C, Kondo S, Aoki T, Schedl P, Lai EC, Wen J, and Dai Q (2019). BEN-solo factors partition active chromatin to ensure proper gene activation in Drosophila. Nat Commun 10, 5700. 10.1038/s41467-019-13558-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grand RS, Burger L, Grawe C, Michael AK, Isbel L, Hess D, Hoerner L, Iesmantavicius V, Durdu S, Pregnolato M, et al. (2021). BANP opens chromatin and activates CpG-island-regulated genes. Nature 596, 133–137. 10.1038/s41586-021-03689-8. [DOI] [PubMed] [Google Scholar]
- 11.Consortium, C.e.S. (1998). Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282, 2012–2018. 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 12.Baker EA, and Woollard A (2019). How Weird is The Worm? Evolution of the Developmental Gene Toolkit in Caenorhabditis elegans. J Dev Biol 7. 10.3390/jdb7040019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baek M, DiMaio F, Anishchenko I, Dauparas J, Ovchinnikov S, Lee GR, Wang J, Cong Q, Kinch LN, Schaeffer RD, et al. (2021). Accurate prediction of protein structures and interactions using a three-track neural network. Science 373, 871–876. 10.1126/science.abj8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tunyasuvunakool K, Adler J, Wu Z, Green T, Zielinski M, Zidek A, Bridgland A, Cowie A, Meyer C, Laydon A, et al. (2021). Highly accurate protein structure prediction for the human proteome. Nature 596, 590–596. 10.1038/s41586-021-03828-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, Yuan D, Stroe O, Wood G, Laydon A, et al. (2022). AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res 50, D439–D444. 10.1093/nar/gkab1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dai Q, Ren A, Westholm JO, Serganov AA, Patel DJ, and Lai EC (2013). The BEN domain is a novel sequence-specific DNA-binding domain conserved in neural transcriptional repressors. Genes Dev 27, 602–614. 10.1101/gad.213314.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sigrist CJ, de Castro E, Cerutti L, Cuche BA, Hulo N, Bridge A, Bougueleret L, and Xenarios I (2013). New and continuing developments at PROSITE. Nucleic Acids Res 41, D344–347. 10.1093/nar/gks1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guzenko D, Burley SK, and Duarte JM (2020). Real time structural search of the Protein Data Bank. PLoS Comput Biol 16, e1007970. 10.1371/journal.pcbi.1007970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okuno T, Ito M, Nakano S, Hattori H, Fujii T, Go T, and Mikawa H (1989). Carbamazepine therapy and long-term prognosis in epilepsy of childhood. Epilepsia 30, 57–61. 10.1111/j.1528-1157.1989.tb05281.x. [DOI] [PubMed] [Google Scholar]
- 20.van Kempen M, Kim SS, Tumescheit C, Mirdita M, Gilchrist CLM, Söding J, and Steinegger M (2022). 10.1101/2022.02.07.479398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holm L (2022). Dali server: structural unification of protein families. Nucleic Acids Res 50, W210–215. 10.1093/nar/gkac387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dai Q, Andreu-Agullo C, Insolera R, Wong LC, Shi SH, and Lai EC (2013). BEND6 is a nuclear antagonist of Notch signaling during self-renewal of neural stem cells. Development 140, 1892–1902. 10.1242/dev.087502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pitchai GP, Hickson ID, Streicher W, Montoya G, and Mesa P (2016). Characterization of the NTPR and BD1 interacting domains of the human PICH-BEND3 complex. Acta Crystallogr F Struct Biol Commun 72, 646–651. 10.1107/S2053230X16010724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruvkun G, and Giusto J (1989). The Caenorhabditis elegans heterochronic gene lin- 14 encodes a nuclear protein that forms a temporal developmental switch. Nature 338, 313–319. 10.1038/338313a0. [DOI] [PubMed] [Google Scholar]
- 25.Hristova M, Birse D, Hong Y, and Ambros V (2005). The Caenorhabditis elegans heterochronic regulator LIN-14 is a novel transcription factor that controls the developmental timing of transcription from the insulin/insulin-like growth factor gene ins- 33 by direct DNA binding. Mol Cell Biol 25, 11059–11072. 10.1128/MCB.25.24.11059-11072.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen J, Li X, and Greenwald I (2004). sel-7, a positive regulator of lin-12 activity, encodes a novel nuclear protein in Caenorhabditis elegans. Genetics 166, 151–160. 10.1534/genetics.166.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia D, Huang X, and Zhang H (2009). The temporally regulated transcription factor sel-7 controls developmental timing in C. elegans. Dev Biol 332, 246–257. 10.1016/j.ydbio.2009.05.574. [DOI] [PubMed] [Google Scholar]
- 28.Wightman B, Ha I, and Ruvkun G (1993). Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75, 855–862. 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 29.Luo Y, Hitz BC, Gabdank I, Hilton JA, Kagda MS, Lam B, Myers Z, Sud P, Jou J, Lin K, et al. (2020). New developments on the Encyclopedia of DNA Elements (ENCODE) data portal. Nucleic Acids Res 48, D882–D889. 10.1093/nar/gkz1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun H, and Hobert O (2021). Temporal transitions in the post-mitotic nervous system of Caenorhabditis elegans. Nature 600, 93–99. 10.1038/s41586-021-04071-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki N, Zou Y, Sun H, Eichel K, Shao M, Shih M, Shen K, and Chang C (2022). Two intrinsic timing mechanisms set start and end times for dendritic arborization of a nociceptive neuron. Proc Natl Acad Sci U S A 119, e2210053119. 10.1073/pnas.2210053119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blum M, Chang HY, Chuguransky S, Grego T, Kandasaamy S, Mitchell A, Nuka G, Paysan-Lafosse T, Qureshi M, Raj S, et al. (2021). The InterPro protein families and domains database: 20 years on. Nucleic Acids Res 49, D344–D354. 10.1093/nar/gkaa977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burglin TR, and Affolter M (2016). Homeodomain proteins: an update. Chromosoma 125, 497–521. 10.1007/s00412-015-0543-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kojima KK, and Jurka J (2011). Crypton transposons: identification of new diverse families and ancient domestication events. Mob DNA 2, 12. 10.1186/1759-8753-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smedley D, Hamoudi R, Lu YJ, Cooper C, and Shipley J (1999). Cloning and mapping of members of the MYM family. Genomics 60, 244–247. 10.1006/geno.1999.5918. [DOI] [PubMed] [Google Scholar]
- 36.Groza T, Gomez FL, Mashhadi HH, Munoz-Fuentes V, Gunes O, Wilson R, Cacheiro P, Frost A, Keskivali-Bond P, Vardal B, et al. (2023). The International Mouse Phenotyping Consortium: comprehensive knockout phenotyping underpinning the study of human disease. Nucleic Acids Res 51, D1038–D1045. 10.1093/nar/gkac972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marenne G, Hendricks AE, Perdikari A, Bounds R, Payne F, Keogh JM, Lelliott CJ, Henning E, Pathan S, Ashford S, et al. (2020). Exome Sequencing Identifies Genes and Gene Sets Contributing to Severe Childhood Obesity, Linking PHIP Variants to Repressed POMC Transcription. Cell Metab 31, 1107–1119 e1112. 10.1016/j.cmet.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu X, Shen B, Liao S, Ning Y, Ma L, Chen J, Lin X, Zhang D, Li Z, Zheng C, et al. (2017). Gene knockout of Zmym3 in mice arrests spermatogenesis at meiotic metaphase with defects in spindle assembly checkpoint. Cell Death Dis 8, e2910. 10.1038/cddis.2017.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee D, Apelt K, Lee SO, Chan HR, Luijsterburg MS, Leung JWC, and Miller KM (2022). ZMYM2 restricts 53BP1 at DNA double-strand breaks to favor BRCA1 loading and homologous recombination. Nucleic Acids Res 50, 3922–3943. 10.1093/nar/gkac160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leung JW, Makharashvili N, Agarwal P, Chiu LY, Pourpre R, Cammarata MB, Cannon JR, Sherker A, Durocher D, Brodbelt JS, et al. (2017). ZMYM3 regulates BRCA1 localization at damaged chromatin to promote DNA repair. Genes Dev 31, 260–274. 10.1101/gad.292516.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ambros V, and Horvitz HR (1984). Heterochronic mutants of the nematode Caenorhabditis elegans. Science 226, 409–416. 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- 42.Yin Y, Morgunova E, Jolma A, Kaasinen E, Sahu B, Khund-Sayeed S, Das PK, Kivioja T, Dave K, Zhong F, et al. (2017). Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 356. 10.1126/science.aaj2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holm L, Laiho A, Toronen P, and Salgado M (2023). DALI shines a light on remote homologs: One hundred discoveries. Protein Sci 32, e4519. 10.1002/pro.4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guindon S, Lethiec F, Duroux P, and Gascuel O (2005). PHYML Online--a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res 33, W557–559. 10.1093/nar/gki352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mirdita M, Schutze K, Moriwaki Y, Heo L, Ovchinnikov S, and Steinegger M (2022). ColabFold: making protein folding accessible to all. Nat Methods 19, 679–682. 10.1038/s41592-022-01488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, and Ferrin TE (2021). UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci 30, 70–82. 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bawono P, and Heringa J (2014). PRALINE: a versatile multiple sequence alignment toolkit. Methods Mol Biol 1079, 245–262. 10.1007/978-1-62703-646-7_16. [DOI] [PubMed] [Google Scholar]
- 48.Consortium EP (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Machanick P, and Bailey TL (2011). MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27, 1696–1697. 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Structure comparisons are performed between X-ray determined BEN domain structures and AlphaFold predicted models of different species. (A) Screening with Insv-BEN structure (PDB: 4IX7) in the Drosophila proteome. (B) Screening with the Insv-BEN structure in the mouse proteome (C) Screening with the Insv-BEN structure in the C.elegans proteome. (D) Screening with the Insv-BEN structure in the zebrafish proteome (E) Screening with the Insv-BEN structure in the rice proteome. (F) Screening with the BEND3-BEN4 structure (PDB: 7W27) in the Drosophila proteome. (G) Screening with the BEND3-BEN4 structure in the mouse proteome.
Structure comparisons are performed between the X-ray determined homeodomains and AlphaFold predicted models of different species. Both homeodomain proteins and BEN factors are revealed. (A) Screening with the Drosophila Ftz homeodomain structure (PDB: 1ftz) in the mouse proteome. (B) Screening with the Drosophila UBX homeodomain structure (PDB: 4cyc) in the Drosophila proteome. (C) Screening with the human PAX6 homeodomain structure (PDB: 6pax) in the C.elegans proteome.
(A) Screening with the predicted structure of LIN-14 BEN domain in the mouse proteome. (B) Screening with the predicted structure of Drosophila CG12112 DUF4806 motif. (C) Screening with the predicted structure of Drosophila CG42854 DUF4806 motif in the mouse proteome.
(A) The r.m.s.d. value for each structure alignment presented in the figures. (B-W) The predicted local distance difference test (pLDDT) score for each predicted structure is retrieved from the AlphaFold2 database. This test measures the per residue confidence of the predicted model. (B) The pLDDT score for F53A2.3. (C) The pLDDT score for SEL-7. (D) The pLDDT score for Y105C5B.19. (E) The pLDDT score for Y61A9LA.4. (F) The pLDDT score for LIN-14 (G) The pLDDT score for CG42854. (H) The pLDDT score for CG12112. (I) The pLDDT score for SI:DKEY266f7.4. (J) The pLDDT score for 6A035617. (K) The pLDDT score for LOC116408483. (L) The pLDDT score for OS04G0307567. (M) The pLDDT score for OS06G0261900. (N) The pLDDT score for KIAA1958. (O) The pLDDT score for ZMYM4. (P) The pDDT score for CEH-40. (Q) The pLDDT score for TTRE_0000654801. (R) The pLDDT score for SI:CH211–126I22.5. (S) The pLDDT score for SI:CH211–262H13.3. (T) The pLDDT score for CG17341. (U) The pLDDT score for PNPase. (V) The pLDDT score for NOL4. (W) The pLDDT score for TCERG1.
Data Availability Statement
This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available form the lead contact upon request.
Key Resource Table
| REAGENT or RESOURCE RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| LIN-14 ChIP-seq in L1 stage C.elegans | modENCODE Luo et al. 29 |
Experiment: ENCSR714ALL; Bed file: ENCFF941FVK |
| Software and algorithms | ||
| PyMOL | https://pymol.org/2/ | RRID:SCR_000305 |
| AlphaFold2 | https://alphafold.ebi.ac.uk/ | Varadi et al. 15 |
| ColabFold | https://colab.research.google.com/ | Mirdita et al. 45 |
| UCSF ChimeraX | https://www.cgl.ucsf.edu/chimerax/ | RRID:SCR_015872 Pettersen et al. 46 |
| MEME-ChIP | https://meme-suite.org/meme/ | Machanick and Bailey 49 |