Abstract
Recently, various commercial ark shell products were being sold, and in the case of processed foods, the loss of morphological traits makes species identification visually challenging, which can lead to seafood fraud. Therefore, a multiplex polymerase chain reaction (PCR) assay was developed to simultaneously identify three ark shells. The specific PCR amplicon sizes of the generated species-specific primer pairs were observed to be 99 bp for Anadara kagoshimensis, 148 bp for Anadara broughtonii, and 207 bp for Tegillarca granosa. Specificity was confirmed for 17 fish and shellfish, and only the target was amplified without cross-reactivity. The detection limit for the multiplex PCR assay was 1 pg. Furthermore, 31 commercial products were evaluated to assess the developed assay’s applicability. Therefore, the analytical approach used in this study can rapidly and accurately identify ark shells in commercial food, and may be used as an authentication tool in the seafood industry.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10068-023-01269-2.
Keywords: Anadara kagoshimensis, Tegillarca granosa, Anadara broughtonii, Capillary electrophoresis, Multiplex PCR assay, Foods adulteration
Introduction
Seafood fraud is increasing year by year, attracting substantial attention from society, academics, and governments (Pardo et al., 2016). Seafood fraud ranges from intentional or unintentional species substitution to adulteration and mislabeling. One of the most common fraudulent activities is expensive species substitution with relatively inexpensive ones for economic gain (Ugochukwu et al., 2015). Therefore, seafood species identification is essential for addressing accurate labeling and economic issues (Herrero et al., 2010). Additionally, reliable methods are required for accurate seafood species identification.
Tegillarca granosa, Anadara broughtonii, and Anadara kagoshimensis are consumed and cultivated not only in Korea but also in countries in Southeast Asia, the Indian Ocean, Western Pacific, and others. They are among commercially important fishery resources (Kang et al., 2020; Suh et al., 2017). The aforementioned three Arcidae species are distinguished by the following morphological characteristics: the radial rib number and the size or shape of the shell, and so on. However, ark shells are generally consumed after being processed to ensure safety and extend shelf life. Here, ark shell meats are separated from the shell and marketed as fillets, making species identification difficult (Kim and Yoon, 2018; Kim et al., 2021). Various commercial ark shellfish products are being sold as the market for processed seafood products has expanded in recent years. In the case of processed foods, morphological characteristics are lost and visual identification of species is limited, which might result in seafood fraud. To reliably identify ark shell species and prevent commercial product mislabeling, a sensitive, species-specific, and reliable detection approach is necessary.
Protein- and DNA-based techniques can be used for species authentication. However, protein-based assays are not applicable to processed foods since heat, high pressure, and dry treatments denature proteins (Lee et al., 2022; Soares et al., 2013). In contrast, DNA-based analysis has been widely used to identify species (Kim and Kim, 2019; Naaum et al., 2019; Wilwet et al., 2021) because DNA is resistant to heat and pressure.
One of the DNA-based analysis methods uses polymerase chain reaction (PCR) technology, which can detect trace DNA amounts quickly and with high sensitivity. DNA-based PCR assay techniques, such as multiplex PCR assay (Kang, 2019a; Kim et al., 2021; Lee et al., 2021; Suh et al., 2020; Wilwet et al., 2021), real-time PCR assay (Eischeid et al., 2013; Servusova and Piskata, 2021; Yao et al., 2022), and PCR restriction fragment length polymorphism (Xu et al., 2016; Yao et al., 2020), have been reported for seafood species identification. Among the aforementioned methods, the multiplex PCR assay method has the following advantages: Compared with real-time PCR assay, it does not require expensive machines and trained personnel. This method is also cost-effective because it permits the simultaneous detection of many species in a single reaction, thus saving both time and money (Kim et al., 2016; Salam et al., 2022). Additionally, capillary electrophoresis offers a higher resolution than gel electrophoresis, allowing for precise separation of amplified DNA fragments (Zhang et al., 2018).
In this study, we designed species-specific primer pairs and applied capillary electrophoresis to multiplex PCR analysis, which is the most efficient technique to simultaneously identify three ark shells.
Materials and methods
Samples collection
The National Institute of Biological Resources (Incheon, Korea) provided granular ark (Tegillarca granosa), half-crenate ark (Anadara kagoshimensis), and Broughton’s ribbed ark (Anadara broughtonii) as reference samples. Conversely, venus mactra (Mactra quadrangularis), hard-shelled mussel (Mytilus coruscus), kuruma prawn (Marsupenaeus japonicus), American lobster (Homarus americanus), Japanese abalone (Haliotis discus hannai), Japanese littleneck (Venerupis philippinarum), horned turban (Turbo cornutus), whiteleg shrimp (Litopenaeus vannamei), ocean quahog (Arctica islandica), gazami crab (Portunus trituberculatus), lamellated oyster (Ostrea denselamellosa), Pacific chub mackerel (Scomber japonicus), fleshy prawn (Fenneropenaeus chinensis), and Japanese common squid (Todarodes pacificus) were purchased from Korean markets. The species were identified by morphological features through the National Institute of Fisheries Science’s image database (Busan, Korea). All samples were cut and stored in a freezer at − 20 °C until use.
DNA extraction
A DNeasy tissue kit (Qiagen, Hilden, Germany) was used to extract 17 samples, as reported by Kim et al. (2016). A MaestroNano Micro-Volume Spectrophotometer (Maestro, Las Vegas, NV, USA) was used to determine the DNA purity and concentration.
Primer design
The National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov) provided the sequences of the mitochondrial cytochrome b (cytb) and cytochrome c oxidase subunit I (COI) genes of 17 species. The Clustal Omega alignment system (http://www.ebi.ac.uk/Tools/msa/clustalo/accessed) was used to align the reference sequences. Following that, the Primer Design program version 3.0 (Scientific and Educational Software, Durham, NC, USA) was used to design primer pairs targeting specific regions and synthesized by Bionics (Seoul, Korea). The information on the primer pairs used in this study is listed in Table 1.
Table 1.
Information for primers used in this study
| Target species | Primer name | Sequence | Target gene | Amplicon size (bp) | Concentration (µM) | Reference |
|---|---|---|---|---|---|---|
| Tegillarca granosa | T. granosa-F | GAA GTC CGG CGT TGG ACA TA | COI | 207 | 0.1 | This study |
| T. granosa-R | TTG TCA ACC CCC CAG CCA AT | |||||
| Anadara broughtonii | A. broughtonii-F | CCT TTA GGG GTA TCA ACC TGT | Cyb | 148 | 6 | This study |
| A. broughtonii-R | TGG GAG TTA GAT TAA CGG GA | |||||
| Anadara kagoshimensis | A. kagoshimensis-F | ATA CGT GAC GTT CAA TGT GG | Cyb | 99 | 0.2 | This study |
| A. kagoshimensis-R | TAA ACT CCG ACC AAT GTG AGC |
In silico PCR assay for specificity
Using web-based in silico PCR amplification, the specificity of the developed primer pairs was determined for a total of 42 mitochondrial genomes derived from 30 target and non-target species (Table S1). Newly confirmed specificity for additional Arcidae species and related species.
Single PCR assay for specificity and sensitivity
The specificity of the species-specific primer pairs developed in this study was evaluated with DNA extracted from 17 species of fish and shellfish. Their sensitivity was evaluated by serially diluting a DNA template from 10 ng to 0.01 pg.
A single PCR assay was performed in a 25-μL reaction mixture. The reaction mixture consisted of the following: 10× buffer (Bioneer, Daejeon, Korea), 800-μM dNTP (Bioneer), 0.5-unit Hot Start Taq DNA polymerase (Bioneer), 0.4 μM of each primer, and 10-ng template DNA. PCR conditions were predenaturated at 95 °C for 5 min following 40 denaturation cycles at 95 °C for 30 s, annealing at 95 °C for 30 s, extension at 72 °C for 30 s, and final extension at 72 °C for 5 min using a thermal cycler (Astec, Tokyo, Japan). A DNA 1000 Lab Chip kit (Agilent Technologies, Santa Clara, CA, USA) and an Agilent Bioanalyzer (Agilent Technologies) were used to visualize the amplified products of simple PCR assay through capillary electrophoresis.
Sequencing analysis of PCR amplicon
A QIAquick PCR Purification kit (Qiagen, Valencia, CA, and USA) was used to purify each amplified PCR product. The purified PCR product was then inserted into the pGEM-T easy vector (Promega, Madison, WI, USA). A DNA-spinTM Plasmid DNA Purification kit (Intron Biotechnology, Seongnam, Korea) was used to extract the plasmid DNA into which the PCR product was inserted. An automated DNA sequencer (Applied Biosystems, Foster City, CA, USA) from GenoTech (Daejeon, Korea) was used to perform sequencing. The Basic Local Alignment Search Tool (BLAST) nucleotide sequence analysis in the NCBI database (Ahamad et al., 2017; Suh et al., 2020) was used to validate the nucleotide sequence of the PCR product.
Multiplex PCR assay for specificity and sensitivity
For multiplex PCR assay, specificity and sensitivity tests were conducted after adjusting the primer concentrations to optimize the PCR condition. Specificity was confirmed using DNA extracted from 17 different fish and shellfish species. Sensitivity was determined using a DNA template serially diluted from 10 ng to 0.01 pg in the same manner as in single PCR assay.
The multiplex PCR assay was performed in 25 µL of the reaction mixture comprising the following: 10× buffer (Bioneer), 800 μM of dNTP (Bioneer), 1 unit of Hot Start Taq DNA polymerase (Bioneer), 0.1 μM granular ark primer pair, 6 μM Broughton’s ribbed ark primer pair, 0.2 μM half to crenate ark primer pair, and 10 ng template DNA. Multiplex PCR conditions are the same as single PCR conditions. A DNA 1000 Lab Chip kit (Agilent Technologies) and an Agilent Bioanalyzer (Agilent Technologies) was used to observe the amplified multiplex PCR products through capillary electrophoresis.
Application and intralaboratory validation for commercial ark shell products using the developed multiplex PCR assay
To verify the developed multiplex PCR assay method, 29 commercially processed products were purchased from online markets. Additionally, granular ark and Broughton’s ribbed ark were sterilized (at 121 °C for 20 min) to identify the thermal treatment effects similar to the canned processing conditions of half to crenate ark. In the process of monitoring products, the DNA extraction and multiplex PCR procedures were conducted in the same manner as they were for raw materials. Application experiments were performed using three different PCR machines in the laboratory to confirm the reproducibility of the experiments and validate the developed method.
Results and discussion
In silico PCR assay for specificity
In silico PCR is widely used in PCR applications and can efficiently evaluate the specificity of primer pairs (Yu and Zhang, 2011). A web-based in silico PCR amplification tool (http://insilico.ehu.es/PCR/) was used to confirm the specificity of the designed primer pairs (Ye et al., 2012). 42 mitochondrial genomes were used to evaluate the specificity of the designed primer pairs. Consequently, there was no amplification in 32 mitochondrial genomes of non-target species, indicating no cross-reactivity with other Arcidae species and related species (Table S1).
.
Specificity and sensitivity of primer pairs
The three primer pairs were designed to contrast with the three species of ark shells after analyzing the sequences of the cytb and COI genes in each of the 17 species. Each primer was designed to be at least 40 bp different from the other primer pairs (granular ark, 207 bp; Broughton’s ribbed ark, 148 bp; and half to crenate ark, 99 bp) to enable distinct PCR amplicon detection for each primer by capillary electrophoresis.
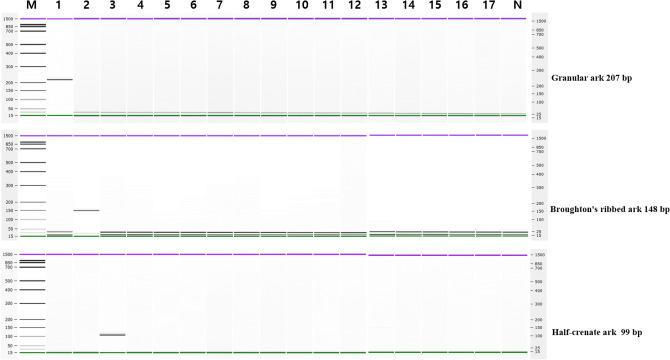
The specificity of each designed primer pairs was evaluated through a single PCR assay. The single PCR assay specificity was validated using DNA extracted from 17 species of fish and shellfish, and species to specific primer pairs amplified only the target species to the expected PCR product size as shown in Fig. 1. This demonstrated that the newly designed primer pair was species-specific by amplifying only the target species and not the other 14 nontarget species. Additionally, the specific target region amplifications of the cytb and COI genes were confirmed by sequencing the amplicons of each PCR product at GenoTech (Daejeon, Korea) (data not shown).
Fig. 1.
Single PCR specificity of each primer set (10 ng of template DNA reacted). Lane M: 100 bp DNA ladder, lane 1: granular ark, lane 2: Broughton’s ribbed ark, lane 3: half-crenate ark, lane 4: venus mactra, lane 5: Japanese littleneck, lane 6: hard-shelled mussel, lane 7: Japanese abalone, lane 8: horned turban, lane 9: ocean quahog, lane 10: lamellated oyster, lane 11: kuruma prawn, lane 12: whiteleg shrimp, lane 13: fleshy prawn, lane 14: gazami crab, lane 15: American lobster, lane 16: Japanese common squid, lane 17: Pacific chub mackerel, lane N: Non-template
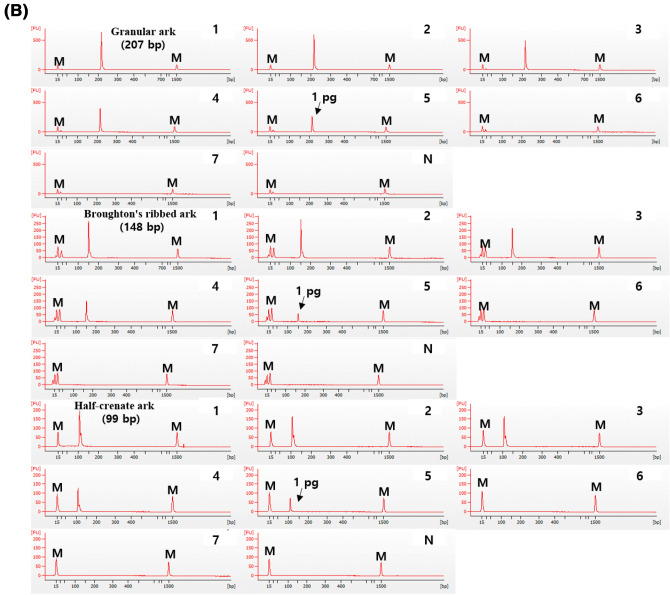
A single PCR assay with DNA diluted from 10 ng to 0.01 pg was used to evaluate the sensitivity of the developed species-specific primer pairs. The sensitivity of all primer pairs for granular ark, Broughton’s ribbed ark, and half-crenate ark was 1 pg (Fig. 2). Wilwet et al. (2021) previously demonstrated a sensitivity of 0.1 ng for the species-specific PCR technique of seven shrimp species in a prior experiment on a specialized seafood PCR-based detection method. Kang (2019a) described a technique for detecting snow crabs with a sensitivity of 0.005 ng. The newly designed primer in this study demonstrated more sensitivity than the primer pairs in a previous comparable study, indicating that it can distinguish three ark shell species even at low concentrations.
Fig. 2.
Single PCR sensitivity of each primer set. (A) Gel, and Lane M: 100 bp DNA ladder, lanes 1–7: 10 ng to 0.01 pg of DNA from target species, and lane N: non-template. (B) Electropherogram, M: alignment marker, lanes 1–7: 10 ng to 0.01 pg of DNA from target species, and lane N: non-template
Specificity and sensitivity of multiplex PCR assay
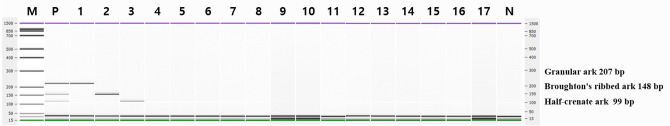
Previous studies on multiplex PCR assays indicated that PCR-related variables, such as Taq DNA polymerase concentration, PCR buffer, dNTPs, and primer pairs, should be adjusted to eliminate nonspecific interactions and maximize sensitivity (Lee et al., 2022; Suh et al., 2020). The optimal concentrations of each primer pair that demonstrated high sensitivity were determined to be 0.1, 6, and 0.2 µM for granular ark, Broughton’s ribbed ark, and half-crenate ark, respectively. The multiplex PCR assay specificity developed for the simultaneous detection of three ark shells was confirmed with 17 types of fish and shellfish (Fig. 3). The capillary electrophoresis principle is to apply a buffer and high voltage to microfluidic channels through an independent electrode and automated pattern for each well, and separation is achieved within 30 min (Ali et al., 2015). Lanes 1–3 in Fig. 3 shows a single band of target PCR products in granular ark, Broughton’s ribbed ark, and half-crenate ark. Cross-reaction to nontarget species was not observed. Consequently, each species-specific primer pair effectively detected just the target species among the ark shell species and demonstrated high specificity for the nontarget species.
Fig. 3.
Multiplex PCR specificity. Lane M: 100 bp DNA ladder, lane P: positive control (10 ng of DNA from target species), lane 1: granular ark, lane 2: Broughton’s ribbed ark, lane 3: half-crenate ark, lane 4: venus mactra, lane 5: Japanese littleneck, lane 6: hard-shelled mussel, lane 7: Japanese abalone, lane 8: horned turban, lane 9: ocean quahog, lane 10: lamellated oyster, lane 11: kuruma prawn, lane 12: whiteleg shrimp, lane 13: fleshy prawn, lane 14: gazami crab, lane 15: American lobster, lane 16: Japanese common squid, lane 17: Pacific chub mackerel, lane N: Non-template
The multiplex PCR assay sensitivity was confirmed by serially diluting 10 times by mixing 10 ng of each DNA extracted from three ark shells. The sensitivity of multiplex PCR assay is defined as the concentration at which all three bands can be detected. Consequently, the multiplex PCR assay sensitivity is 1 pg (Fig. 4), similar to single PCR assay sensitivity. It was found that the developed primer pairs exhibited sensitivity corresponding to single PCR assay without inhibiting with each other in the same reaction solution. Compared with the sensitivity of similar studies on shellfish products (1.6 pg for oyster, mussel, abalone, and clam and 1 ng for four ark shells species), the sensitivity of the developed multiplex PCR assay (1 pg) is exceptional, indicating that it can detect even minute DNA amounts (Kim et al., 2021; Suh et al., 2020). High sensitivity in PCR assay methods is essential for the detection of processed seafood products (Wilwet et al., 2021). As a consequence of these results, the developed method in this study can be applied to the detection of fragmented DNA in processed foods, or on-site detection, or on-site detection using direct DNA extraction.
Fig. 4.
Multiplex PCR sensitivity. (A) Gel, and Lane M: 100 bp DNA ladder, lanes 1–7: 10 ng to 0.01 pg of DNA from target species, and lane N: non-template. (B) Electropherogram, M: alignment marker, lanes 1–7: 10 ng to 0.01 pg of DNA from target species, and lane N: non-template. Numbers 1, 2, and 3 are half-crenate ark, Broughton’s ribbed ark, and granular ark, respectively
Capillary electrophoresis is advantageous in multiplex PCR assays as it can clearly distinguish similarly sized PCR bands. Additionally, it has higher resolution and precision than agarose gel electrophoresis, and even weak bands can be accurately identified through peaks (Butler et al., 1995; Cheng et al., 2016; Lee et al., 2021). Consequently, PCR amplicons were differentiated more clearly, as were bands of different sizes (Fig. 3), using capillary electrophoresis in our multiplex PCR analysis.
Validation of multiplex PCR with processed food
Multiplex PCR analysis applicability was examined on a total of 31 samples; including 29 ark shell processed foods purchased online and 2 granular ark and Broughton’s ribbed ark samples sterilized at 121 °C for 20 min. They were categorized as raw, seasoned, stewed, pickled; autoclaved, canned, fried, or dried (Table 2). The reproducibility and multiplex PCR assays of our experiments were also validated using three different PCR machines in the laboratory.
Table 2.
Application and intra-laboratory validation of the multiplex PCR assay to commercial products
| No. | Labeled species | Product type | Multiplex PCR results | ||
|---|---|---|---|---|---|
| Granular ark | Broughton’s ribbed ark | Half-crenate ark | |||
| 1 | Granular ark | Raw | +++ | ||
| 2 | Granular ark | Raw | +++ | ||
| 3 | Granular ark | Raw | +++ | ||
| 4 | Granular ark | Auto | +++ | ||
| 5 | Granular ark | Dried | +++ | ||
| 6 | Granular ark | Fried | +++ | ||
| 7 | Granular ark | Boiled | +++ | ||
| 8 | Broughton’s ribbed ark | Raw | +++ | ||
| 9 | Broughton’s ribbed ark | Raw | +++ | ||
| 10 | Broughton’s ribbed ark | Auto | +++ | ||
| 11 | Broughton’s ribbed ark | Dried | +++ | ||
| 12 | Broughton’s ribbed ark | Fried | +++ | ||
| 13 | Broughton’s ribbed ark | Boiled | +++ | ||
| 14 | Broughton’s ribbed ark | Boiled | +++ | ||
| 15 | Broughton’s ribbed ark | Seasoned | +++ | ||
| 16 | Broughton’s ribbed ark | Picked | +++ | ||
| 17 | Half-crenate ark | Raw | +++ | ||
| 18 | Half-crenate ark | Raw | +++ | ||
| 19 | Half-crenate ark | Boiled | +++ | ||
| 20 | Half-crenate ark | Boiled | +++ | ||
| 21 | Half-crenate ark | Seasoned | +++ | ||
| 22 | Half-crenate ark | Seasoned | +++ | ||
| 23 | Half-crenate ark | Seasoned | +++ | ||
| 24 | Half-crenate ark | Picked | +++ | ||
| 25 | Half-crenate ark | Picked | +++ | ||
| 26 | Half-crenate ark | Canned | +++ | ||
| 27 | Half-crenate ark | Canned | +++ | ||
| 28 | Non-labeled | Raw | +++ | ||
| 29 | Non-labeled | Seasoned | +++ | ||
| 30 | Non-labeled | Seasoned | +++ | ||
| 31 | Non-labeled | Seasoned | +++ | ||
‘+’ means a positive result. Three different instruments were used to conduct the test independently
In recent years, the relevance of recognizing fish and shellfish species has increased because of the enormous growth of the seafood processed food industry (Pardo and Pérez-Villareal, 2004; So, and Kim, 2013). However, in the case of cockle commercial food, it is offered as seasoned, stewed, pickled, or canned foods and the target species’ visual identification is limited because the original morphological characteristics have been compromised. These limits in visual species identification might result in mislabeling and intentional or unintentional species substitutions.
PCR assay methods based on DNA analysis are mostly used to prevent adulteration. There are reports, however, that if processed foods contain spices, they contain components, such as proteins, carbs, polysaccharides, and lipids, that may negatively affect PCR assay (Focke et al., 2011; Kang, 2019b). Furthermore, it has been reported that the amplification product size must be < 300 bp for it to be applied to the DNA damaged by heat or pressure generated during the manufacturing process (Yao et al., 2022). This study revealed that each of the 27 products included every species listed in Table 2. Only four unidentified species-specific products include the half-crenate ark (Anadara kagoshimensis). The monitoring products used to confirm applicability consisted of raw, seasoned, stewed, pickled, various processing treatments, etc., but all samples showed positive results. Thus, this confirmed that the developed assay was not affected by PCR inhibitors. Moreover, the PCR amplicon size of the primer developed in this study suggests that it is appropriate for the investigation of various processed foods containing DNA that has been degraded to < 300 bp. Multiplex PCR analyses of ark shells reported among DNA-based PCR assay methods were for the following species: Anadara kagoshimensis, Tegillarca granosa, Anadara broughtonii, and Cucullaea labiata (Kim et al., 2021). However, this study’s method demonstrated that primer sensitivity was better than that of the previously reported study and, notably, it is applicable to processed foods.
Therefore, the multiplex PCR assay developed in this study can identify multiple ark shell species quickly and can be used as a low-cost and rapid tool for label validation and commercial food monitoring. Furthermore, this technology is applicable for on-site detection.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This research was supported by the Ministry of Food and Drug Safety in Korea, Grant Number 22193 MFDS471.
Declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ga-Young Lee, Email: gayoung.lee0731@gmail.com.
Seung-Min Yang, Email: ysm9284@gmail.com.
Hae-Yeong Kim, Email: hykim@khu.ac.kr.
References
- Ahamad MNU, Ali ME, Hossain MAM, Asing A, Sultana S, Jahurul MHA. Multiplex PCR assay discriminates rabbit, rat and squirrel meat in food chain. Food Additives & Contaminants: Part a. 2017;34:2043–2057. doi: 10.1080/19440049.2017.1359752. [DOI] [PubMed] [Google Scholar]
- Ali ME, Razzak MA, Hamid SBA, Rahman MM, Amin M Al, Rashid NRA, Asing. Multiplex PCR assay for the detection of five meat species forbidden in Islamic foods. Food Chemistry. 177: 214–224 (2015) [DOI] [PubMed]
- Butler JM, McCord BR, Jung JM, Lee JA, Budowle B, Allen RO. Application of dual internal standards for precise sizing of polymerase chain reaction products using capillary electrophoresis. Electrophoresis. 1995;16:974–980. doi: 10.1002/elps.11501601163. [DOI] [PubMed] [Google Scholar]
- Cheng F, Wu J, Zhang J, Pan A, Quan S, Zhang D, Kim HY, Li X, Zhou S, Yang L. Development and inter-laboratory transfer of a decaplex polymerase chain reaction assay combined with capillary electrophoresis for the simultaneous detection of ten food allergens. Food Chemistry. 2016;199:799–808. doi: 10.1016/j.foodchem.2015.12.058. [DOI] [PubMed] [Google Scholar]
- Eischeid AC, Kim BH, Kasko SM. Two quantitative real-time PCR assays for the detection of penaeid shrimp and blue crab, crustacean shellfish allergens. Journal of Agricultural and Food Chemistry. 2013;61:5669–5674. doi: 10.1021/jf3031524. [DOI] [PubMed] [Google Scholar]
- Focke F, Haase I, Fischer M. DNA-based identification of spices: DNA isolation, whole genome amplification, and polymerase Chain reaction. Journal of Agricultural and Food Chemistry. 2011;59:513–520. doi: 10.1021/jf103702s. [DOI] [PubMed] [Google Scholar]
- Herrero B, Madriñán M, Vieites JM, Espiñeira M. Authentication of atlantic cod (Gadus morhua) Using real time PCR. Journal of Agricultural and Food Chemistry. 2010;58:4794–4799. doi: 10.1021/jf904018h. [DOI] [PubMed] [Google Scholar]
- Kang TS. Rapid and simple identification of two closely-related snow crabs (Chionoecetes opilio and C. japonicus) by direct triplex PCR. Lwt. 99: 562–567 (2019a)
- Kang TS. Basic principles for developing real-time PCR methods used in food analysis: A review. Trends in Food Science and Technology. 2019;91:574–585. doi: 10.1016/j.tifs.2019.07.037. [DOI] [Google Scholar]
- Kang SI, Sohn SK, Choi KS, Kim K-H, Kim YS, Lee JS, Heu MS, Kim J-S. Optimization of the processing of seasoning sauce for seasoned broughton’s ribbed ark Scapharca broughtonii products using response surface methodology. Korean Journal of Fisheries and Aquatic Sciences. 2020;53:334–341. [Google Scholar]
- Kim MJ, Kim HY. A fast multiplex real-time PCR assay for simultaneous detection of pork, chicken, and beef in commercial processed meat products. Lwt. 2019;114:108390. doi: 10.1016/j.lwt.2019.108390. [DOI] [Google Scholar]
- Kim KS, Yoon SJ. Development of Duplex-PCR Method for Rapid Identification of Hard-shelled Mussel, Mytilus Coruscus. Journal of Fishries and Marine Sciences Education. 2018;30:1192–1199. doi: 10.13000/JFMSE.2018.08.30.4.1192. [DOI] [Google Scholar]
- Kim M, Yoo I, Lee SY, Hong Y, Kim HY. Quantitative detection of pork in commercial meat products by TaqMan® real-time PCR assay targeting the mitochondrial D-loop region. Food Chemistry. 2016;210:102–106. doi: 10.1016/j.foodchem.2016.04.084. [DOI] [PubMed] [Google Scholar]
- Kim YH, Han HS, Yun BH, Park JY, Baek IG, Bang IC. Development of rapid identification method for four species of ark shells (Bivalvia: Arcoidae) bivalves using multiplex PCR. The Korean Journal of Malacology. 2021;37:139–148. [Google Scholar]
- Lee YM, Lee S, Kim HY. A multiplex pcr assay combined with capillary electrophoresis for the simultaneous identification of atlantic cod, pacific cod, blue whiting, haddock, and alaska pollock. Foods. 2021;10:2631. doi: 10.3390/foods10112631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YM, Lee GY, Kim HY. Development of a multiplex PCR assay for the simultaneous detection of big blue octopus (Octopus cyanea), giant Pacific octopus (Enteroctopus dofleini), and common octopus (Octopus vulgaris). Food Science and Biotechnology. 31: 497–504 (2022) [DOI] [PMC free article] [PubMed]
- Naaum AM, Hellberg RS, Okuma TA, Hanner RH. Multi-instrument evaluation of a real-time PCR assay for identification of Atlantic salmon: A case study on the use of a pre-packaged Kit for rapid seafood species identification. Food Analytical Methods. 2019;12:2474–2479. doi: 10.1007/s12161-019-01584-7. [DOI] [Google Scholar]
- Pardo MA, Pérez-Villareal B. Identification of commercial canned tuna species by restriction site analysis of mitochondrial DNA products obtained by nested primer PCR. Food Chemistry. 2004;86:143–150. doi: 10.1016/j.foodchem.2003.09.024. [DOI] [Google Scholar]
- Pardo MÁ, Jiménez E, Pérez-Villarreal B. Misdescription incidents in seafood sector. Food Control. 2016;62:277–283. doi: 10.1016/j.foodcont.2015.10.048. [DOI] [Google Scholar]
- Salam MR, Ezaouine A, Zekhnini H, El Mellouli F, Chegdani F, Bennis F. Detection of chicken and turkey in different beef matrix by species-specific multiplex PCR assay. Scientific African. 2022;17:e01338. doi: 10.1016/j.sciaf.2022.e01338. [DOI] [Google Scholar]
- Servusova E, Piskata Z. Identification of selected tuna species in commercial products. Molecules. 2021;26:1137. doi: 10.3390/molecules26041137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So W-H, Kim H-K. Effects of the Enterprise Image in Korean Processed Marine Product Industry on Consumers’ Product Evaluation and Purchase Intention. The Journal of Fisheries Business Administration. 2013;44:1–14. doi: 10.12939/FBA.2013.44.1.001. [DOI] [Google Scholar]
- Soares S, Amaral JS, Oliveira MBPP, Mafra I. A SYBR Green real-time PCR assay to detect and quantify pork meat in processed poultry meat products. Meat Science. 2013;94:115–120. doi: 10.1016/j.meatsci.2012.12.012. [DOI] [PubMed] [Google Scholar]
- Suh J, Kim T, Shin S, Kahng HY, Ahn S, Kim Y, Won N Il. Geochemical Characteristics and Benthos Distribution in the Three Shellfish Farms in Suncheon Bay, Korea. Journal of Environmental Science International. 26: 691–710 (2017)
- Suh SM, Kim MJ, Kim HI, Kim HJ, Kim HY. A multiplex PCR assay combined with capillary electrophoresis for the simultaneous detection of tropomyosin allergens from oyster, mussel, abalone, and clam mollusk species. Food Chemistry. 2020;317:126451. doi: 10.1016/j.foodchem.2020.126451. [DOI] [PubMed] [Google Scholar]
- Ugochukwu AI, Hobbs JE, Phillips PWB, Gray R. An economic analysis of private incentives to adopt DNA barcoding technology for fish species authentication in Canada. Genome. 2015;58:559–567. doi: 10.1139/gen-2015-0033. [DOI] [PubMed] [Google Scholar]
- Wilwet L, Shakila RJ, Sivaraman B, Nayak BB, Kumar HS, Jaiswar AK, Jeyasekaran G. Rapid detection of fraudulence in seven commercial shrimp products by species-specific PCR assays. Food Control. 2021;124:107871. doi: 10.1016/j.foodcont.2021.107871. [DOI] [Google Scholar]
- Xu K, Feng J, Ma X, Wang X, Zhou D, Dai Z. Identification of tuna species (Thunnini tribe) by PCR-RFLP analysis of mitochondrial DNA fragments. Food and Agricultural Immunology. 2016;27:301–313. doi: 10.1080/09540105.2015.1086978. [DOI] [Google Scholar]
- Yao L, Lu J, Qu M, Jiang Y, Li F, Guo Y, Wang L, Zhai Y. Methodology and application of PCR-RFLP for species identification in tuna sashimi. Food Science and Nutrition. 2020;8:3138–3146. doi: 10.1002/fsn3.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao L, Qu M, Jiang Y, Guo Y, Li N, Li F, Tan Z, Wang L. The development of genus-specific and species-specific real-time PCR assays for the authentication of Patagonian toothfish and Antarctic toothfish in commercial seafood products. Journal of the Science of Food and Agriculture. 2022;102:1674–1683. doi: 10.1002/jsfa.11507. [DOI] [PubMed] [Google Scholar]
- Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Zhang C. In Silico PCR Analysis. Vol. 760, pp. 91–107. In: Methods in Molecular Biology. (2011)
- Zhang Y, Zhu L, Zhang Y, He P, Wang Q. Simultaneous detection of three foodborne pathogenic bacteria in food samples by microchip capillary electrophoresis in combination with polymerase chain reaction. Journal of Chromatography a. 2018;1555:100–105. doi: 10.1016/j.chroma.2018.04.058. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.