Abstract
Background
Central nervous system tumors are the most common pediatric solid tumors and the most frequent cause of cancer-related morbidity in childhood. Significant advances in understanding the molecular features of these tumors have facilitated the development of liquid biopsy assays that may aid in diagnosis and monitoring response to therapy. In this report, we describe our comprehensive liquid biopsy platform for detection of genome-wide copy number aberrations, sequence variants, and gene fusions using cerebrospinal fluid (CSF) from pediatric patients with brain, spinal cord, and peripheral nervous system tumors.
Methods
Cell-free DNA was isolated from the CSF from 55 patients, including 47 patients with tumors and 8 controls.
Results
Abnormalities in cell-free DNA were detected in 24 (51%) patients including 11 with copy number alterations, 9 with sequence variants, and 7 with KIAA1549::BRAF fusions. Positive findings were obtained in patients spanning histologic subtypes, tumor grades, and anatomic locations.
Conclusions
This study demonstrates the feasibility of employing this platform in routine clinical care in upfront diagnostic and monitoring settings. Future studies are required to determine the utility of this approach for assessing response to therapy and long-term surveillance.
Keywords: cerebrospinal fluid , liquid biopsy , pediatric central nervous system tumors
Key Points.
Cerebrospinal fluid liquid biopsy in pediatric central nervous system tumors is feasible for detection of copy number alterations, sequence variants, and fusions.
Importance of the Study.
This study demonstrates the feasibility of employing this cerebrospinal fluid liquid biopsy platform in clinical care for detection of copy number alterations, sequence variants, and fusions in upfront diagnostic and monitoring settings in pediatric central nervous system tumors.
Central nervous system (CNS) tumors are the most common solid tumors affecting children and the most frequent cause of cancer-related death in the pediatric population. The incidence of CNS tumors in children and adolescents (aged 0–19 years) is 6.14 per 100,000 and tumors vary significantly by histology, grade, propensity to invade or metastasize, and response to therapy.1 Over the last several decades there have been significant advances in the understanding of the molecular features of these tumors, leading to refined tumor classification and targeted therapies.
Liquid biopsy assays that utilize profiling of cell-free DNA (cfDNA) to detect circulating-tumor DNA (ctDNA) provide a minimally invasive solution to identify key molecular alterations that may aid in diagnosis, determine prognosis, provide biologic targets for therapy, and detect early relapse.2–9 For example, diffuse midline glioma (DMG) is an inoperable and universally fatal tumor for which a liquid biopsy diagnosis could spare the potential morbidity associated with direct brainstem biopsy, provided acquisition is clinically safe. Liquid biopsy assays may also help to eliminate sampling bias due to intra-tumoral heterogeneity and provide a more accurate assessment of the subclonal composition of tumors. Serial liquid biopsies may also facilitate monitoring of response to therapy and early detection of relapse.10–12 Important insights into tumor evolution may be gained from these studies, which will increase the clinical utility of liquid biopsy-based assays.
Cerebrospinal fluid (CSF) has been shown to yield the highest fraction of ctDNA in patients with brain tumors as compared to plasma.13 Initial liquid biopsy assays in the pediatric neuro-oncology setting employed digital droplet PCR (ddPCR), which demonstrated high sensitivity even with low tumor DNA input and low yield of ctDNA.14–17 Initial studies focused on the histone 3 p.K27M (H3K27M) mutation, which is present in over 70% of pediatric DMG patients and is correlated with extremely poor clinical outcome.18 Panditharatna et al. evaluated a cohort of patients with DMG and successfully detected the H3K27M mutation in ctDNA by ddPCR in 20 of 23 (87%) of CSF samples.15 The variant allele frequency (VAF) of the H3K27M mutation in ctDNA determined by ddPCR from CSF also appeared to be prognostic in patients treated with the novel agent ONC201 for whom a decrease in the VAF correlated with prolonged survival.11
Liquid biopsy studies have utilized hybridization capture or amplicon-based next-generation sequencing (NGS) for the detection of recurrent somatic mutations and structural variants in brain tumors.10,19 Miller et al. evaluated CSF liquid biopsies in pediatric and adolescent and young adult (AYA) patients with primary or recurrent brain tumors using the MSK-IMPACT panel.20 Alterations were detected in 21 of 45 (46%) patients, and those with leptomeningeal disease had higher positivity rates than those without (81.5% vs. 18.5%).20 Feasibility on a larger scale was more recently assessed by Pages et al. who analyzed blood, urine, and CSF from 258 patients with various histologic diagnoses.21 CSF (n = 67) was found to be most enriched for ctDNA compared to other biofluids. The study evaluated copy number alterations (CNAs) by Low-pass whole-genome sequencing (LP-WGS) and point mutations by hybrid gene capture with a 20% and 30% detection rate by each modality, respectively. Notably, CNAs or mutations were only detected in high-grade tumors.21 Liu et al. employed LP-WGS for measurable residual disease (MRD) detection in medulloblastoma and reported positivity rates at diagnosis (postoperative baseline prior to adjuvant therapy) of 54% and 85% in patients with localized and metastatic disease, respectively.12 This study also demonstrated that MRD positivity at the end of therapy was highly correlated with the rate of relapse, underscoring the clinical utility of these assays.12
Taken together, the aforementioned studies demonstrate the feasibility and highlight the potential clinical utility of liquid biopsy assays in the pediatric CNS tumor setting. However, most studies focused on either LP-WGS for detecting copy number aberrations or a targeted panel for mutations. Detection of fusion genes, specifically the KIAA1549::BRAF fusion which has diagnostic and prognostic implications in low-grade gliomas, has been challenging using cfDNA assays. The goal of the present study was to develop an integrated platform for comprehensive detection of genome-wide CNAs, mutations, and gene fusions using CSF liquid biopsy in pediatric nervous system tumors. This included LP-WGS to detect CNAs and a custom capture hybridization-based panel to detect somatic single nucleotide variants (SNVs), indels, and gene fusions.
Materials and Methods
Eligibility, Sample Collection, and DNA Extraction
Patients aged 0–21 years undergoing neurosurgical intervention for tumor resection or treatment of nonmalignant control diagnoses (ex: hydrocephalus, Chiari malformation, etc.) were eligible for this study (Supplemental Table S1). Patients and parents/guardians consented to an Institutional Review Board (IRB) approved study (CHLA-19-00230). For each patient an attempt was made to collect 3–5mL of CSF in sterile specimen tubes. The CSF was centrifuged at 3000 × g for 10 minutes at 4°C to separate the supernatant and CSF pellet (Supplemental Figure S1). Blood was collected in EDTA tubes, centrifuged at 1900 × g for 10 minutes at 4°C, and separated into plasma and buffy coat for further studies. All samples were immediately frozen at −80°C. DNA was extracted from the CSF supernatant using the MagMAX Cell-Free Total Nucleic Acid Isolation Kit (Thermo Fisher, Waltham, MA). CSF findings were compared to available results of chromosomal microarray (CMA; OncoScan or CytoScanHD; Thermo Fisher) or OncoKids® next-generation sequencing panel from the matched primary tumor specimens processed for clinical testing in the Center for Personalized Medicine at Children’s Hospital Los Angeles (CHLA).22
Library Preparation and Sequencing
Libraries were prepared using xGen Prism DNA Library Prep Kit from Integrated DNA Technologies (IDT; Coralville, IA) per the manufacturer’s standard protocol incorporating unique molecular identifiers (UMIs) and using 5 ng of DNA input. Libraries were quantified using the Tape Station High Sensitivity DNA D1000 assay from Agilent (Santa Clara, CA) and split into two aliquots. One library aliquot was used to perform LP-WGS sequenced to an average depth of coverage of 4x. From the second library aliquot, 187 ng from each of eight libraries was combined to create an equimolar pool. To detect mutations and fusions prevalent in pediatric gliomas, the pool of eight samples was hybridized to a custom hybrid gene capture panel manufactured by Twist Bioscience (San Francisco, CA; Supplemental Table 2). Probes were designed to cover specific genomic regions frequently mutated in gliomas, including sequences from introns 8 to 11 in BRAF.23,24 Twist Exome with optional spike-in FastHyb Protocol alpha was used, following the manufacturer’s protocol for hybrid gene capture. Sequencing was performed on the Illumina (San Diego, CA) NextSeq 500 with 2 × 100bp paired-end runs using the Mid Output KT v2 for each pool of eight capture libraries. Coverage depth of the panel averaged 331x.
Bioinformatic Analysis
The sequencing data were processed using Illumina’s Dragen Tool (v3.5.7). Data were first demultiplexed using Dragen’s bcl2fastq module to extract the UMI sequence from the beginning of each sequence and incorporated into the final fastq output. The fastq files were then processed using Dragen’s tumor-only liquid-biopsy variant calling pipeline with default settings and UMI-mode turned on. In the UMI-mode Dragen aligned the data to the human reference genome (build hs37d5) and used the UMIs to generate a consensus read from a set of candidate input reads belonging to the same duplex UMI family. Only read families with two or more supporting reads were used for generating the consensus read. Read families with a lesser number of supporting reads were discarded. In the case of LP-WGS, UMI based read-collapsing was not performed given the low targeted coverage. LP-WGS reads were binned into 500 kb regions across the entire genome and the ichorCNA algorithm was utilized to obtain the segmented calls.25 Tumor fraction was estimated for all samples using the ichorCNA variant calling pipeline as previously described. Overall positivity rates for CNAs were determined based on the ichor-derived tumor fraction higher than 10%. In the case of hybrid gene capture, the resulting vcf file was then uploaded to Alissa Interpret (Agilent) for downstream interpretation of the variants.26 Illumina Manta (version 1.6.0) was used in the “targeted” mode (https://github.com/Illumina/manta) for the detection of structural variants. The presence of mutations and fusions was further verified using Integrative Genomics Viewer (IGV).27
Results
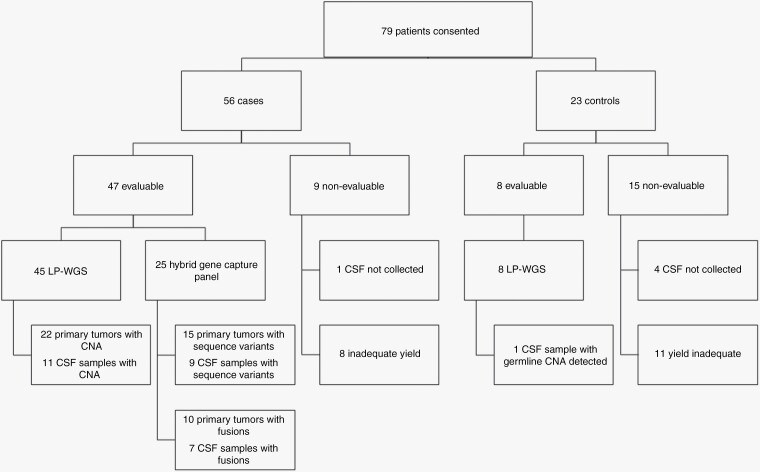
A total of 79 patients including 56 patients with tumors and 23 nontumor controls consented to participate in the study (Figure 1). The nontumor control group was comprised of patients receiving clinically indicated neurosurgical intervention for nonmalignant diagnoses (i.e. hydrocephalus, Chiari malformation). Of the patients enrolled, 9 tumor subjects and 15 controls were nonevaluable and excluded from the study. Specifically, for one case and four controls, CSF could not be collected. For 8 cases and for 11 controls, the cfDNA yield was inadequate to meet the required 5 ng DNA input for library preparation. Tumor cases with inadequate yield included diagnoses of pilocytic astrocytoma (n = 2), tectal glioma (n = 2), ETMR (n = 2), ependymoma (n = 1) and schwannoma (n = 1).
Figure 1.
Summary of patient enrollment, CSF collection, and results.
Patient ages ranged from 2 months to 21 years with a median age of 8 years (Table 1 and Supplemental Table S1). Sex was evenly distributed between male and female patients. The majority of patients with tumor diagnoses (85.1%) had nonmetastatic disease. Tumors were located throughout the brain and spinal cord, with the largest percentage (44.7%) located within the posterior fossa. Diagnoses of evaluable patients included pilocytic astrocytoma (n = 10), ependymoma (n = 6), medulloblastoma (n = 5), craniopharyngioma (n = 5), low-grade glioma (n = 3), DMG (n = 2), choroid plexus carcinoma (n = 2), AT/RT (n = 2), ganglioglioma (n = 2), glioneuronal tumor (n = 2), pilomyxoid astrocytoma (n = 2), ETMR (n = 1), high-grade glioma (n = 1), germ cell tumor (n = 1), neurofibroma (n = 1), hemangioblastoma (n = 1), and metastatic neuroblastoma to the CNS (n = 1).
Table 1.
Evaluable patient demographics and diagnoses.
| Total Sequenced | n = 55 (%) |
|---|---|
| Cases | 47 (85.5) |
| Controls | 8 (14.5) |
| Age | |
| Median age | 8 y |
| Age range | 2 mo to 21 y |
| Sex | |
| Male | 28 (50.9) |
| Female | 27 (49.1) |
| Metastatic | |
| M0 | 40 (85.1) |
| M+ | 7 (14.9) |
| Tumor location | |
| Posterior fossa | 21 (44.7) |
| Sellar or suprasellar | 7 (14.9) |
| Optic pathway | 5 (10.6) |
| Lateral ventricle | 3 (6.4) |
| Brainstem/ midline | 3 (6.4) |
| Frontal lobe | 2 (4.3) |
| Parietal lobe | 2 (4.3) |
| Pineal | 2 (4.3) |
| Temporal lobe | 1 (2.1) |
| Spinal cord | 1 (2.1) |
| Tumor diagnosis | |
| Pilocytic astrocytoma | 10 (21.3) |
| Ependymoma (PFA n = 5) | 6 (12.8) |
| Medulloblastoma | 5 (10.6) |
| Craniopharyngioma | 5 (10.6) |
| Low-grade glioma | 3 (6.4) |
| Diffuse midline glioma | 2 (4.3) |
| Choroid plexus carcinoma | 2 (4.3) |
| AT/RT | 2 (4.3) |
| Ganglioglioma | 2 (4.3) |
| Glioneuronal tumor | 2 (4.3) |
| Pilomyxoid astrocytoma | 2 (4.3) |
| ETMR | 1 (2.1) |
| High-grade glioma | 1 (2.1) |
| Germ cell tumor | 1 (2.1) |
| Neurofibroma | 1 (2.1) |
| Hemangioblastoma | 1 (2.1) |
| Metastatic CNS neuroblastoma | 1 (2.1) |
Of the evaluable cases, 30 liquid biopsies were obtained at diagnosis, 14 at the time of recurrence or progression and three while patients were actively on therapy. In 41 cases, CSF was collected prior to tumor resection and in six cases due to intra-operative factors CSF was collected posttumor resection.
Fifty-five patients including 47 tumor cases and eight nontumor controls had sufficient cfDNA for analysis by LP-WGS (n = 53) and/or the capture panel (n = 25). As expected, tumor cases overall yielded greater quantity of cfDNA compared to controls (mean yield 18.9 and 0.5 ng/mL, respectively). Patients with high grade tumors yielded more cfDNA (mean yield 33.6 ng/mL) compared to those with low grade lesions (mean yield 4.2 ng/mL). The mean CSF volume from tumors with adequate DNA yield and detectable alterations was 5.4mL.
Low-Pass Whole-Genome Sequencing (LP-WGS) Results
CMA analysis of tumor tissue was performed as part of the clinical diagnostic work-up for 45 patients. Twenty-three cases had noninformative results and 22 patients had abnormal CMA profiles. None of the 23 cases with normal copy number profiles in the primary tumor had detectable CNAs by LP-WGS in the cfDNA. In contrast, 11 of the 22 patients with abnormal copy number profiles in the tumor had CNAs detected by LP-WGS, consistent with the presence of ctDNA in the CSF (including AT/RT, DMG, ependymoma, high-grade glioma, medulloblastoma, choroid plexus carcinoma, low-grade glioma, ganglioglioma, glioneuronal tumors, as well as CNS metastatic neuroblastoma). Overall, 24.4% of evaluated tumor cases had CNAs detected in the cfDNA (Figure 1 and Supplemental Table S1). The CNAs included partial and whole chromosome gains and losses in addition to focal amplifications. CSF and primary tumor copy number analyses were highly concordant (Figure 2). Of note, one control patient was found to have a chromosome 1q loss in the cfDNA from CSF. This patient was ultimately diagnosed with a germline 18.9 Mb copy number deletion involving the distal long arm of chromosome 1 (1q42.13q44 region) and complex medical comorbidities including hydrocephalus. Seven additional control patients had negative copy number profiles as expected.
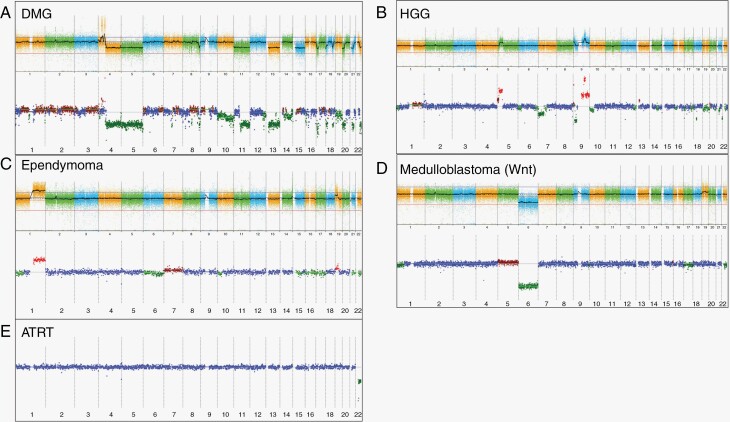
Figure 2.
Representative copy number arrays. Copy number profiling from a variety of histopathological diagnoses. Results of the OncoScan analysis of the primary tumor are displayed above and liquid biopsy iChor copy number plot below for each case. The log2ratio is plotted on the y-axis against chromosome numbers 1-22 on the x-axis. Red indicates a copy number gain, green indicates a copy number loss, and blue indicates a neutral copy number. A Diffuse midline glioma notable for PDGFRA amplification, and a variety of whole and partial chromosome losses. B High-grade glioma with 5p gain and complex chromosome 9 gains and losses as well as additional partial chromosome gains and losses. C Ependymoma with 1q gain, a known poor prognostic marker, also 19p gain. D Medulloblastoma with evidence of monosomy 6, consistent with Wnt subgroup medulloblastoma. E AT/RT with chromosome 22 deletion, harboring SMARCB1 gene.
Sequence Variants Detected by Targeted Gene Capture
Nine cases evaluated with the targeted gene capture panel had detectable sequence variants in cfDNA. Fifteen evaluable patients had primary tumors with known pathogenic or likely pathogenic variants identified using our DNA- and RNA-based targeted next-generation sequencing assay, OncoKids®, in genes that were also captured by the custom glioma panel. Nine of the fifteen (60.0%) samples had identical variants detected in the cfDNA from CSF (Figure 1 and Supplemental Table S1). Notably, four of these six patients had histologically low-grade tumors (ganglioglioma n = 2, low-grade glioma n = 1, pilocytic astrocytoma n = 1). Variants in H3-3A (H3K27M), BRAF, FGFR1, PIK3CA, PIK3R1, ATRX, and TP53 were detected in both low- and high-grade tumors (Supplemental Table S1). The variant allele frequency for the somatic variants was highly variable, ranging from 1.5% to 91.4%. Three patients had germline TP53 mutations which were also identified in cfDNA from the CSF with a median VAF of 50.4%.
Fusion Genes Detected by Targeted Gene Capture
Ten patients with pilocytic or pilomyxoid astrocytoma had primary tumors with KIAA1549::BRAF fusions identified with the RNA component of the OncoKids® assay. Five were in the posterior fossa, four in the optic pathway, and one was suprasellar in location. Only one patient had M+ metastatic disease. Seven of these ten (70.0%) samples also had a detectable fusion in cfDNA from CSF. A representative example of a KIAA1549::BRAF fusion detected in the cfDNA is shown in Figure 3. All seven patients with KIAA1549::BRAF fusion positive cfDNA had nonmetastatic disease (Supplemental Table S1). One of the three patients without a detectable KIAA1549::BRAF fusion in CSF had metastatic disease with tumors that disseminated into the optic chiasm, suprasellar cistern, basal ganglia, mesial temporal lobes, and brainstem. There was no association between tumor location with the ability to detect the fusion in the liquid biopsy specimen.
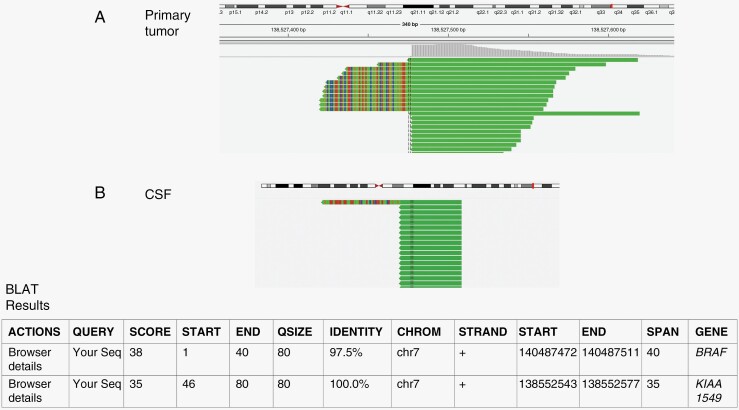
Figure 3.
Representative fusion analysis with detection of split reads. Split read pairs mapping to KIAA1549 (solid green) and BRAF (multicolor) of the primary tumor are displayed above (A) and from the CSF below (B).
Clinical Testing
LP-WGS for CNA detection in CSF, plasma and aqueous humor of the eye as described was recently validated for clinical use at the CLIA-certified Center for Personalized Medicine at CHLA as an aid in diagnosis, prognosis, and detection of residual/recurrent disease. The relevance and utility of this assay is demonstrated by the first brain tumor patient to undergo clinical testing. The patient is an 11-year-old male who initially presented with headache and vomiting and was found to have a posterior fossa mass with disseminated subependymal and leptomeningeal disease, consistent with M2 stage. He underwent resection and histopathologic diagnosis was consistent with medulloblastoma. Clinical CMA analysis showed a pattern of copy number alterations consistent with a group 3/4 tumor including gain of chromosome 7, loss of 8p, loss of 11p and gain of 11q, gain of 14q22.3 and an isodicentric chromosome 17q. DNA methylation profiling was consistent with group 4 medulloblastoma. His treatment included 36Gy craniospinal radiation plus 20Gy boost followed by maintenance chemotherapy which was complicated by myelosuppression and ototoxicity. The chemotherapeutic plan required significant modifications with dose reductions (cyclophosphamide) and eliminations (cisplatin). At the end of therapy, MRI showed a nodular focus of abnormal enhancement at the fourth ventricle compatible with treated disease and CSF cytology was negative, however, LP-WGS of cfDNA demonstrated gain of chromosome 7, loss of chromosome 8, loss of 11p, gain of 11q and isochromosome 17q, consistent with residual disease (Figure 4).
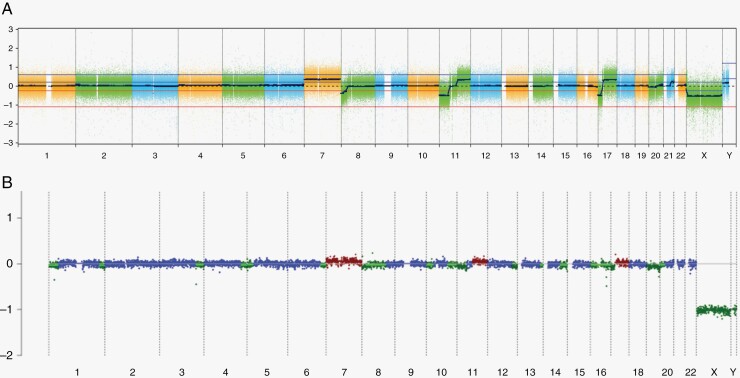
Figure 4.
iChor plot from cfDNA from CSF obtained from a patient with medulloblastoma at the end of therapy. Results of the OncoScan analysis of the primary tumor are displayed above (A) and iChor copy number plot below (B). The log2ratio is plotted on the y-axis against chromosome numbers 1–22 on the x-axis. Red indicates a copy number gain, green indicates a copy number loss, and blue indicates a neutral copy number. This patients CSF demonstrated continued presence of the gain of chromosome 7, loss of the short arm and most of the long arm of chromosome 8, deletion of 11p and gain of 11q, deletion of 16q and an isochromosome 17q.
Given that optimal therapy was limited by toxicity, and given the persistent nodularity on MRI in the setting of positive CSF liquid biopsy results at the end of therapy, and given recently published reports that patients with persistent copy number changes have a significantly higher risk of progression, the clinical decision was made not to stop treatment but rather continue with metronomic therapy and undertake further serial CSF liquid biopsy monitoring.12 The patient is tolerating metronomic therapy well.
Discussion
The molecular characterization of pediatric CNS tumors has led to major advances in understanding and classification, now enabling tumor diagnosis by evaluation of molecular signatures.3 Copy number aberrations, sequence variants, and fusions have clinically important prognostication and treatment implications in pediatric CNS tumors. In our previous studies which utilized OncoKids® or CMA assays, approximately two-thirds of patients had pathogenic or likely pathogenic events, including mutations in TP53, NF1, and BRAF, fusions involving KIAA1549::BRAF, and other significant copy number alterations.22 The primary limitations of those assays include the requirement for at least 20 ng of DNA, and the need to obtain tissue through surgical resection or biopsy.
We have developed a clinical liquid biopsy protocol using CSF from pediatric patients with CNS tumors that only requires 5 ng of DNA and can be used for patients when surgical biopsy is not feasible. The same library is used for both LP-WGS and sequencing with a targeted gene capture panel. This platform allows for comprehensive analysis of CNAs, sequence variants, and detection of diagnostic or prognostic relevant gene fusions.
Diagnostic and prognostic copy number changes detected with LP-WGS from the CSF included monosomy 22 in AT/RT, monosomy 6 in the WNT subgroup of medulloblastoma, PDGFRA amplification in glioma, and 1q gain in ependymoma. A comparison of sequencing results for the ctDNA analysis versus the known pathogenic mutations and fusions identified in the primary tumor using OncoKids® yielded a detection rate of 60% for mutations and 70% for fusions in the CSF, respectively. Overall, pathogenic sequence variants were detected in key drivers including TP53, FGFR1, ATRX, PIK3R1, PIK3CA, H3-3A, and BRAF (V600E) albeit often at a lower VAF than the primary tumors. In several cases, there were slight differences between the CNAs in the primary tumor and CSF, which were most likely due to tumor heterogeneity.
Low-grade gliomas are the most common pediatric CNS tumor and though biologically less aggressive than their high-grade counterparts, continue to pose clinical challenges. Complete surgical resection is generally considered standard of care, however, anatomic location may make some tumors inoperable leading to important morbidity considerations.28 To date, detection of low-grade glioma DNA in liquid biopsy approaches has been technically challenging.
Key drivers in low-grade gliomas include a high frequency of aberrations in the MAP-kinase pathway—predominantly through BRAF (V600E) and KIAA1549::BRAF fusions.29–31 A tandem duplication at 7q34 gives rise to most KIAA1549::BRAF fusions commonly involving BRAF exon 9. Hence, we designed a custom panel tiling BRAF exons and introns from 8—11 and developed bioinformatics methods capable of detecting fusions from a small fraction of tumor-derived cfDNA. Our results demonstrate successful detection of ctDNA in various low-grade lesions, including pilocytic and pilomyxoid astrocytomas. Detection of fusions in ctDNA in CSF has not yet been widely reported in the pediatric setting.
For patients harboring BRAF aberrations, the MAP-kinase pathway or downstream MEK can be inhibited using targeted agents including dabrafenib, vemurafenib, trametinib, and selumetinib.32–35 Selumetinib, a novel kinase inhibitor, is now FDA approved and is in phase III clinical trial evaluating its efficacy as a frontline chemotherapy regimen (NCT04166409, NCT03871257). The ability to monitor ctDNA burden longitudinally may provide further insights into informing therapy response, stratifying risk, and identifying patients who may benefit from adjuvant therapy. Further discernment of the optimal clinical use of these approaches, particularly within each specific disease category would be best answered by incorporating liquid biopsy aims into prospective carefully designed and IRB-approved clinical trial designs.
One limitation of this study is the small cohort size, which did not allow us to stratify results for individual histopathologic diagnostic categories. Secondly, the cfDNA yield from CSF was too low for several patients further limiting the number of patients that could be analyzed. The detection rate may be improved by collecting higher volumes of CSF though this may not be possible in infants, patients small in size, or those who are clinically unstable.
Conclusions
Prospective clinical trials with dedicated liquid biopsy aims will be necessary to continue to evaluate the risk of recurrence and to refine treatment decisions based on findings in the CSF, particularly within disease categories. This pilot study affirms a path forward for future implementation of liquid biopsy evaluation into clinical trial design and clinical care.
Supplementary Material
Acknowledgments
We would like to thank David Ruble and Dolores Estrine from the Center for Personalized Medicine and the Center for Pathology Research Services team at Children’s Hospital Los Angeles for technical assistance.
Contributor Information
Katrina O’Halloran, Cancer and Blood Disease Institute, Children’s Hospital Los Angeles, CA, USA; Keck School of Medicine, University of Southern California, Los Angeles, CA, USA.
Venkata Yellapantula, Division of Pathology and Laboratory Medicine, Children’s Hospital Los Angeles, CA, USA; Keck School of Medicine, University of Southern California, Los Angeles, CA, USA.
Eirini Christodoulou, Division of Pathology and Laboratory Medicine, Children’s Hospital Los Angeles, CA, USA.
Dejerianne Ostrow, Division of Pathology and Laboratory Medicine, Children’s Hospital Los Angeles, CA, USA.
Moiz Bootwalla, Division of Pathology and Laboratory Medicine, Children’s Hospital Los Angeles, CA, USA.
Jianling Ji, Division of Pathology and Laboratory Medicine, Children’s Hospital Los Angeles, CA, USA; Keck School of Medicine, University of Southern California, Los Angeles, CA, USA.
Jennifer Cotter, Division of Pathology and Laboratory Medicine, Children’s Hospital Los Angeles, CA, USA; Keck School of Medicine, University of Southern California, Los Angeles, CA, USA.
Nicholas Chapman, Division of Neurosurgery, Children’s Hospital Los Angeles, CA, USA.
Jason Chu, Keck School of Medicine, University of Southern California, Los Angeles, CA, USA; Division of Neurosurgery, Children’s Hospital Los Angeles, CA, USA.
Ashley Margol, Cancer and Blood Disease Institute, Children’s Hospital Los Angeles, CA, USA; Keck School of Medicine, University of Southern California, Los Angeles, CA, USA.
Mark D Krieger, Keck School of Medicine, University of Southern California, Los Angeles, CA, USA; Division of Neurosurgery, Children’s Hospital Los Angeles, CA, USA.
Peter A Chiarelli, Keck School of Medicine, University of Southern California, Los Angeles, CA, USA; Division of Neurosurgery, Children’s Hospital Los Angeles, CA, USA.
Xiaowu Gai, Division of Pathology and Laboratory Medicine, Children’s Hospital Los Angeles, CA, USA; Keck School of Medicine, University of Southern California, Los Angeles, CA, USA.
Jaclyn A Biegel, Division of Pathology and Laboratory Medicine, Children’s Hospital Los Angeles, CA, USA; Keck School of Medicine, University of Southern California, Los Angeles, CA, USA.
Authorship statement
Experimental design and implementation: KO, VY, EC, DO, MB, JJ, JC, NC, MK, PC XG, JB
Data analysis and interpretation: KO, VY, EC, DO, MB, JJ, JC, NC, MK, PC, XG, JB
Manuscript writing and review and approval: KO, VY, EC, DO, MB, JJ, JC, NC, JC, AM, MK, PC, XG, JB
Previous Presentations
A portion of this work was presented as an oral abstract at the International Symposium of Pediatric Neuro-Oncology (ISPNO) in June 2022.
References
- 1. Ostrom QT, Patil N, Cioffi G, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2013-2017. Neuro Oncol. 2020;22(12 Suppl 2):iv1iv96. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Soffietti R, Bettegowda C, Mellinghoff IK, et al. Liquid biopsy in gliomas: A RANO review and proposals for clinical applications. Neuro Oncol. 2022;24(6):855–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Louis DN, Perry A, Wesseling P, et al. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol. 2021;23(8):1231–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Figarella-Branger D, Appay R, Metais A, et al. The 2021 WHO classification of tumours of the central nervous system. Ann Pathol. 2022;42(5):367–382. [DOI] [PubMed] [Google Scholar]
- 5. Ryall S, Tabori U, Hawkins C.. Pediatric low-grade glioma in the era of molecular diagnostics. Acta Neuropathol Commun. 2020;8(1):30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Selt F, van Tilburg CM, Bison B, et al. Response to trametinib treatment in progressive pediatric low-grade glioma patients. J Neurooncol. 2020;149(3):499–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de Blank P, Fouladi M, Huse JT.. Molecular markers and targeted therapy in pediatric low-grade glioma. J Neurooncol. 2020;150(1):5–15. [DOI] [PubMed] [Google Scholar]
- 8. Liu AP, Northcott PA, Robinson GW, Gajjar A.. Circulating tumor DNA profiling for childhood brain tumors: Technical challenges and evidence for utility. Lab Invest. 2022;102(2):134–142. [DOI] [PubMed] [Google Scholar]
- 9. Jones C, Karajannis MA, Jones DTW, et al. Pediatric high-grade glioma: biologically and clinically in need of new thinking. Neuro Oncol. 2017;19(2):153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Miller AM, Shah RH, Pentsova EI, et al. Tracking tumour evolution in glioma through liquid biopsies of cerebrospinal fluid. Nature. 2019;565(7741):654–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cantor E, Wierzbicki K, Tarapore RS, et al. Serial H3K27M cell-free tumor DNA (cf-tDNA) tracking predicts ONC201 treatment response and progression in diffuse midline glioma. Neuro Oncol. 2022;24(8):1366–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu APY, Smith KS, Kumar R, et al. Serial assessment of measurable residual disease in medulloblastoma liquid biopsies. Cancer Cell. 2021;39(11):1519–1530.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. De Mattos-Arruda L, Mayor R, Ng CKY, et al. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat Commun. 2015;6:8839. doi: 10.1038/ncomms9839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Izquierdo E, Proszek P, Pericoli G, et al. Droplet digital PCR-based detection of circulating tumor DNA from pediatric high grade and diffuse midline glioma patients. Neurooncol Adv. 2021;3(1):vdab013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Panditharatna E, Kilburn LB, Aboian MS, et al. Clinically relevant and minimally invasive tumor surveillance of pediatric diffuse midline gliomas using patient-derived liquid biopsy. Clin Cancer Res. 2018;24(23):5850–5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li D, Bonner ER, Wierzbicki K, et al. Standardization of the liquid biopsy for pediatric diffuse midline glioma using ddPCR. Sci Rep. 2021;11(1):5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mattox AK, Yang B, Douville C, et al. The mutational landscape of spinal chordomas and their sensitive detection using circulating tumor DNA. Neurooncol Adv. 2021;3(1):vdaa173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Castel D, Philippe C, Calmon R, et al. Histone H3F3A and HIST1H3B K27M mutations define two subgroups of diffuse intrinsic pontine gliomas with different prognosis and phenotypes. Acta Neuropathol. 2015;130(6):815–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Piccioni DE, Achrol AS, Kiedrowski LA, et al. Analysis of cell-free circulating tumor DNA in 419 patients with glioblastoma and other primary brain tumors. CNS Oncol. 2019;8(2):Cns34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Miller AM, Szalontay L, Bouvier N, et al. Next-generation sequencing of cerebrospinal fluid for clinical molecular diagnostics in pediatric, Adolescent and Young Adult (AYA) brain tumor patients. Neuro Oncol. 2022;24(10):1763–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pages M, Rotem D, Gydush G, et al. Liquid biopsy detection of genomic alterations in pediatric brain tumors from cell-free DNA in peripheral blood, CSF, and urine. Neuro Oncol. 2022;24(8):1352–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ji J, Kaneva K, Hiemenz MC, et al. Clinical utility of comprehensive genomic profiling in central nervous system tumors of children and young adults. Neurooncol Adv. 2021;3(1):vdab037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jones DT, Kocialkowski S, Liu L, Pearson DM, Bäcklund LM, Ichimura K, Collins VP.. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res. 2008;68(21):8673–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dougherty MJ, Santi M, Brose MS, et al. Activating mutations in BRAF characterize a spectrum of pediatric low-grade gliomas. Neuro Oncol. 2010;12(7):621–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Adalsteinsson VA, Ha G, Freeman SS, et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat Commun. 2017;8(1):1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ryutov A, M B, A G, J B, D M, Ji J. LUBA: a software toolbox for efficiently manipulating and analyzing NGS data. Paper presented at: Am Soc Hum Genet Ann Conf. 2017.
- 27. Robinson JT, Thorvaldsdottir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Qaddoumi I, Sultan I, Gajjar A.. Outcome and prognostic features in pediatric gliomas: a review of 6212 cases from the Surveillance, Epidemiology, and End Results database. Cancer. 2009;115(24):5761–5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pfister S, Janzarik WG, Remke M, et al. BRAF gene duplication constitutes a mechanism of MAPK pathway activation in low-grade astrocytomas. J Clin Invest. 2008;118(5):1739–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schindler G, Capper D, Meyer J, et al. Analysis of BRAF V600E mutation in 1,320 nervous system tumors reveals high mutation frequencies in pleomorphic xanthoastrocytoma, ganglioglioma and extra-cerebellar pilocytic astrocytoma. Acta Neuropathol. 2011;121(3):397–405. [DOI] [PubMed] [Google Scholar]
- 31. Jacob K, Albrecht S, Sollier C, et al. Duplication of 7q34 is specific to juvenile pilocytic astrocytomas and a hallmark of cerebellar and optic pathway tumours. Br J Cancer. 2009;101(4):722–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hargrave DR, Bouffet E, Tabori U, et al. Efficacy and safety of dabrafenib in pediatric patients with braf v600 mutation-positive relapsed or refractory low-grade glioma: results from a phase i/iia study. Clin Cancer Res. 2019;25(24):7303–7311. [DOI] [PubMed] [Google Scholar]
- 33. Nicolaides T, Nazemi KJ, Crawford J, et al. Phase I study of vemurafenib in children with recurrent or progressive BRAF(V600E) mutant brain tumors: Pacific Pediatric Neuro-Oncology Consortium study (PNOC-002). Oncotarget. 2020;11(21):1942–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Perreault S, Larouche V, Tabori U, et al. A phase 2 study of trametinib for patients with pediatric glioma or plexiform neurofibroma with refractory tumor and activation of the MAPK/ERK pathway: TRAM-01. BMC Cancer. 2019;19(1):1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fangusaro J, Onar-Thomas A, Young Poussaint T, et al. Selumetinib in paediatric patients with BRAF-aberrant or neurofibromatosis type 1-associated recurrent, refractory, or progressive low-grade glioma: a multicentre, phase 2 trial. Lancet Oncol. 2019;20(7):1011–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.