Abstract
In genetic cardiomyopathies, a frequently described phenomenon is how similar mutations in one protein can lead to discrete clinical phenotypes. One example is illustrated by two mutations in beta myosin heavy chain (β-MHC) that are linked to hypertrophic cardiomyopathy (HCM) (Ile467Val, I467V) and left ventricular non-compaction (LVNC) (Ile467Thr, I467T). To investigate how these missense mutations lead to independent diseases, we studied the molecular effects of each mutation using recombinant human β-MHC Subfragment 1 (S1) in in vitro assays. Both HCM-I467V and LVNC-I467T S1 mutations exhibited similar mechanochemical function, including unchanged ATPase and enhanced actin velocity but had opposing effects on the super-relaxed (SRX) state of myosin. HCM-I467V S1 showed a small reduction in the SRX state, shifting myosin to a more actin-available state that may lead to the “gain-of-function” phenotype commonly described in HCM. In contrast, LVNC-I467T significantly increased the population of myosin in the ultra-slow SRX state. Interestingly, molecular dynamics simulations reveal that I467T allosterically disrupts interactions between ADP and the nucleotide-binding pocket, which may result in an increased ADP release rate. This predicted change in ADP release rate may define the enhanced actin velocity measured in LVNC-I467T, but also describe the uncoupled mechanochemical function for this mutation where the enhanced ADP release rate may be sufficient to offset the increased SRX population of myosin. These contrasting molecular effects may lead to contractile dysregulation that initiates LVNC-associated signaling pathways that progress the phenotype. Together, analysis of these mutations provides evidence that phenotypic complexity originates at the molecular level and is critical to understanding disease progression and developing therapies.
Introduction
The most common genetic cardiomyopathy is Hypertrophic Cardiomyopathy (HCM), typically characterized by thickened ventricular walls and impaired relaxation. This disease affects between 1 in 200 to 1 in 500 people in the United States and is the most common cause of sudden cardiac death in people under 30 years old. A leading cause of HCM are mutations in sarcomeric protein genes, and mutations in MYH7 (also known as cardiac myosin heavy chain; β-MHC) account for ~30% of HCM [1, 2]. Until recently, HCM lacked any pharmaceutical intervention that targeted the cause of disease with existing treatments addressing symptoms of the disease by improving cardiac relaxation [3]. However, FDA approval of Camzyos©, a myosin-inhibitor, provided the first sarcomere-targeted therapeutic for the treatment of obstructive HCM[4].
Another form of cardiomyopathy that can be caused by mutations in MYH7 is left ventricular non-compaction (LVNC). In contrast to HCM, LVNC is a much less understood and studied disease. It is characterized by abnormal growth of cardiac tissue, with excessive deposition but reduced compaction of trabeculae, resulting in a non-compacted, spongiform-like myocardial wall. The introduction of cardiac MRI in the clinic improved the identification of this non-compacted phenotype from thickened, dense muscular wall commonly seen in HCM. While this morphology is characteristic of the disease, the functional phenotype is highly variable. LVNC may present with preserved cardiac function, systolic dysfunction, diastolic dysfunction, or a combination of both[5]. Thus, clinical management of disease is dependent on the clinical manifestation of the LVNC phenotype. Despite these variable presentations, the natural history of disease has been frequently associated with arrhythmias, heart failure, cardiac transplants, and risk of sudden cardiac death. Additionally, due to the spongiform-like myocardium, patients are at risk of thromboembolic events and typically require blood thinners as preventative treatment[6, 7].
Mutations in the MYH7 gene (β-cardiac myosin heavy chain; β-MHC) have also been linked to other genetic cardiomyopathies including LVNC[8–10] and dilated cardiomyopathy, although those mutations are relatively rare[11, 12]. Though the cause of disease has been identified in many cases, there still is no consensus on how mutations lead to specific cardiomyopathies. Some mutations have been shown to alter the mechanochemical function of myosin, typically enhancing ATPase activity of the motor domain [13–15]. Other mutations increase the number of myosin heads that are available to bind to actin and form forcegenerating cross bridges[16–18]. This dysregulation of the relaxed states of myosin may be driven by structural rearrangements of myosin heads[19–23] and/or biochemical function of myosin[13, 24–26]. Specifically, myosin is known to exist in two distinct states known as the actin-available, disordered (DRX) state and the energy-conserving, super-relaxed (SRX) state of myosin [24, 27, 28]. Multiple studies have shown a mutation-specific and disease-dependent shift in the SRX/DRX ratio, providing an additional biophysical mechanism for altered contractility and energetic expenditure in these cardiomyopathies [17, 21, 24]. Many studies have investigated how HCM mutations lead to disease, but the molecular mechanisms that underlie morphogenetic remodeling and altered cardiac function in LVNC are not well understood. Of note, one study investigated the primary insults caused by LVNC-associated β-MHC M531R, describing an increased power output driven by enhanced mechanochemical activity of the motor[29], despite reduced contractile performance reported in patients expressing this mutation[30].
Here, we investigate how two missense mutations at the same position in the β myosin heavy chain lead to two different diseases. Specifically, we investigate the biochemical mechanisms that govern how I467V (Ile467Val) causes HCM while I467T (Ile467Thr) causes LVNC. We performed a variety of biochemical assays that probed how these mutations affect intrinsic enzymatic function, actin sliding, and myosin-based relaxation properties. We also connect our experimental results to molecular dynamics simulations of the β-myosin motor domain carrying the I467T mutation to understand how a small structural perturbation can alter emergent ensemble properties that give rise to biochemical differences.
Results
Actin-Activated ATPase Activity
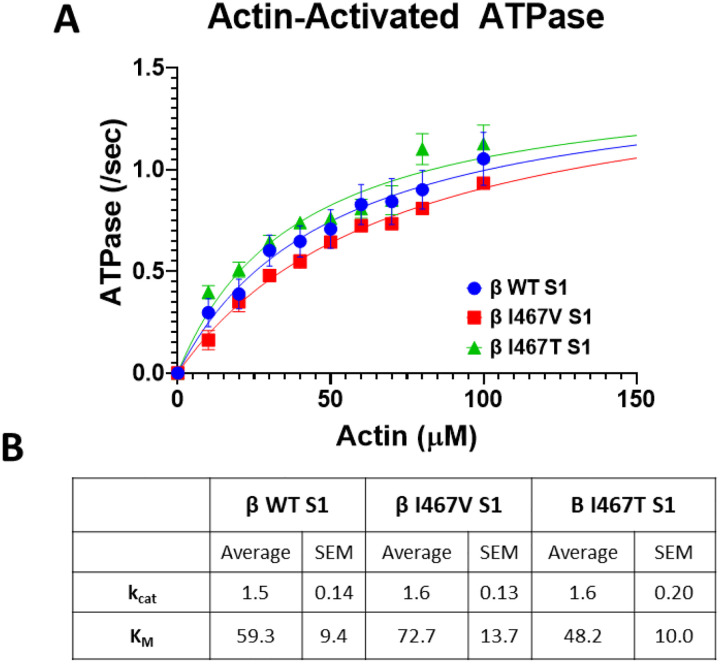
The enzymatic function of cardiac muscle contraction is driven by the mechanochemical activity of actomyosin interactions. To assess whether these similar point mutations alter the enzymatic function of the myosin motor, we performed an NADH-coupled actin-activated ATPase assay to measure maximal ATPase activity and actin affinity (Figure 1A). The maximum ATPase activity of the HCM-I467V and LVNC-I467T S1 constructs were unchanged as compared to both β-MHC WT S1 (1.6 ± 0.13/second, 1.6 ± 0.20/second, and 1.5 ± 0.14/second, respectively) (Figure 1B). The actin affinity for LVNC I467T S1 was increased, albeit not significantly, as compared to both β-MHC WT S1 and HCM-I467V S1 (48.2 ± 10.0 μM, 59.3 ± 9.4 μM, and 72.7 ± 13.7 μM, respectively). These data suggest that though these mutations are in close proximity to the ATP binding pocket, they do not alter the ATPase activity of the myosin motor domain.
Figure 1. Actin-activated ATPase Activity of myosin constructs.
(A) ATPase activity of myosin S1 constructs was measured over time with increasing actin concentrations. Data points were fitted using Michaelis Menten kinetics to obtain maximum ATPase activity (kcat) and actin affinity (KM). (B) Measured values of kcat and KM for HCM-linked β-MHC I467V and LVNC-linked β-MHC I467T S1 mutations. Neither mutation significantly altered maximal ATPase activity nor actin affinity. Data are represented as mean ± SEM. A one-way ANOVA was used to assess statistical significance (p<0.05) compared to β-MHC WT S1. N=6–11 curves per myosin S1 construct.
Single Turnover ATPase Activity
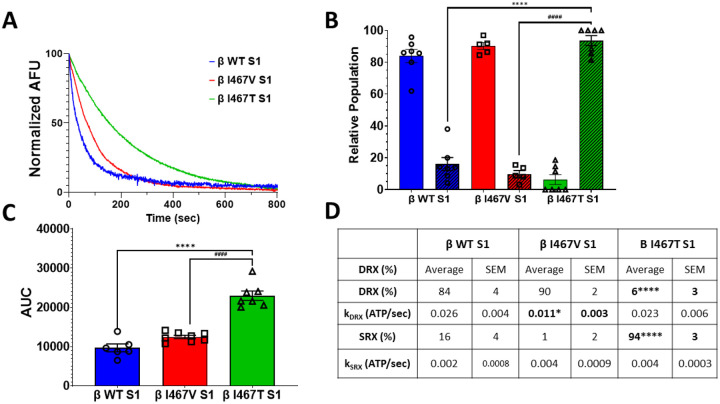
A growing number of studies support the role of myosin in regulating the relaxation of cardiac muscle by adapting two states known as the disordered relaxed (DRX) and super-relaxed (SRX) states. The DRX state is associated with an ATPase rate that is nearly 100-fold slower than the actin-activated ATPase rate while the SRX state is defined by an ATPase activity that is 1000-fold slower. Using a modified single-turnover ATPase assay, we measured the relative proportions of the DRX and SRX states of myosin in the presence of HCM-I467V and LVNC-I467T mutations (Figure 2). Like other HCM-associated mutations, I467V S1 displayed a slight shift of myosin heads to the DRX state of myosin as compared to WT S1 (90±2 %, 84±4%, respectively) (Figure 2B, 2D). Interestingly, the I467T mutation increased the SRX state of myosin (93±4%) (Figure 2B, 2D), a near complete shift of myosin heads to the ultra-slow state. The ATPase rate that is associated with the SRX and DRX states of myosin was also measured. HCM-I467V S1 decreased the DRX rate by nearly 2-fold as compared to β-MHC WT S1 (0.011±0.0003* s−1 and 0.026±0.004 s−1, respectively) (Figure 2D). Alternatively, LVNC-I467T S1 did not affect kfast (0.023±0.006 s−1) (Figure 2D). Both HCM-I467V S1 and LVNC-I467T mutations showed an increase in kslow as compared to β-MHC WT S1(0.0037± 0.0009 s−1, 0. 0.0038± 0.0003 s−1, 0.0021±0.0008 s−1, respectively) (Figure 2D), although not significant. Finally, the area under the curve (AUC) was significantly increased for LVNC-I467T S1 (Figure 2C), suggesting that energetic efficiency was improved in the presence of this mutation. Unlike the intrinsic ATPase activity described above, these two mutations oppositely affected myosin regulation, suggesting that a primary driver for the distinct diseases may be the propensity to adopt the SRX state.
Figure 2. Single turnover ATPase assay of myosin S1.
(A) Representative double-exponential fluorescence decay curves from the single-turnover mant-ATP dependent ATPase assay for myosin S1 constructs (WT: blue, β-I467V: red, β-I467T: green). (B) The disordered (DRX, solid bars) and super-relaxed (SRX, dashed lines) states were calculated from the double-exponential fit of decay curves. (C) Area under the curve (AUC) was calculated for each decay curve. (D) Summary table of the single turnover assay measurements. ATPase rates associated with the DRX and SRX states (kDRX and kSRX, respectively) were calculated from the double exponential decay fits. Values are calculated from 5–7 decay curves per mutation. Data are represented as mean ± SEM. A one-way ANOVA was used to assess statistical significance (* = p<0.05 vs. β-WT S1) and adjusted with Tukey’s multiple comparisons to assess differences between mutations (# = p<0.05 vs. β-I467V S1). N= 5–7 curves per myosin S1 construct.
In vitro Motility
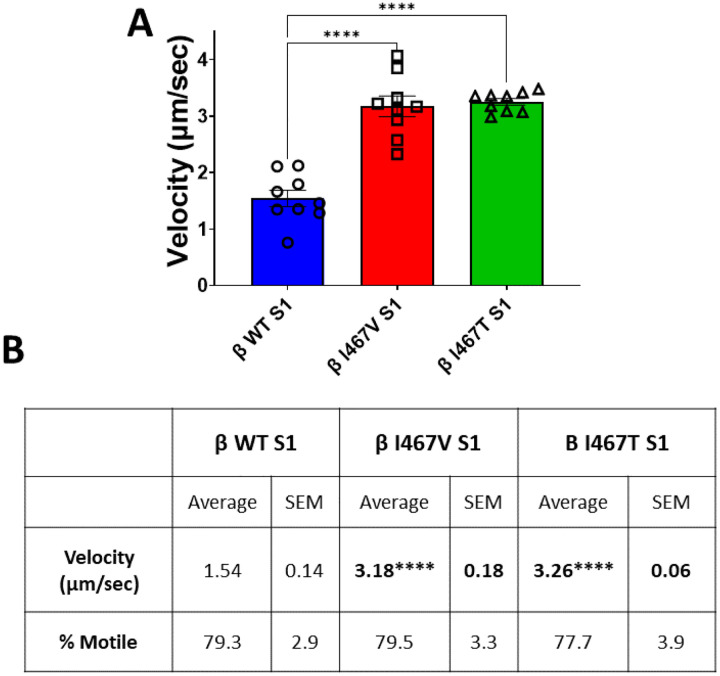
The velocity by which myosin moves actin is a highly characterized parameter to understand the effects of mutations on the mechanical function of myosin. To assess the effects of the HCM-I467V and LVNC-I467T mutations, we utilized an unloaded in vitro motility assay and measured the velocity of actin movement for each S1 mutation. HCM-I467V and LVNC-I467T S1 increased actin velocity approximately >2-fold each, (HCM- I467V S1: 3.18 ± 0.18 μm/sec and β-I467T S1: 3.26 ± 0.06 μm/sec) as compared to WT-S1 (β-MHC WT S1: 1.543 ± 0.14 μm/sec) (Figure 3A and 3B). These data suggest that despite these two mutations leading to distinct cardiomyopathies, they alter the mechanical function of myosin in a similar manner. In combination with the ATPase activity, these data suggest that HCM-I467V and LVNC-I467T do not uniquely alter the intrinsic function of the myosin motor or its ability to translocate actin; rather, these mutations cause different cardiomyopathies in a mechanochemical independent manner, but dependent on myosin regulation.
Figure 3. Unloaded Actin Sliding Velocity.
(A) In vitro motility rates for myosin S1 constructs were measured using TRITC-labeled actin filaments. Average actin sliding velocity was calculated from actin filaments that were ≥1 μm in length and mobile in >15 frames per video. (B) Summary values for In vitro motility actin sliding velocity assay. Values are an average of three separate purifications per S1 construct, and 7–9 videos per construct. % motile was calculated from the ratio of actin filaments moving in >15 frames relative to total number of actin filaments tracked. Values are reported as average ± standard error. A one-way ANOVA was used to assess statistical significance (p<0.05) compared to β-MHC WT S1. N = 7–8 videos per myosin S1 construct.
Molecular Dynamics Simulations of I467T
Differences in motility can result from changes in step size or the ADP release rate. Given that I467T increases the in vitro motility of the β-cardiac myosin motor, we hypothesized that its ADP pocket would be in a more open state that facilitates ADP release. The I467T mutation introduces a polar residue at a cluster of hydrophobic residues near the active site that we hypothesized would allosterically weaken interactions between the active site and ADP, thereby accelerating ADP release. In support of this hypothesis, previous studies have shown that mutations in Switch II can allosterically affect the nucleotide binding pocket in myosin V[31]. Therefore, we sought to test if a similar trend is observed in β-cardiac myosin that may facilitate faster ADP release.
We used molecular dynamics simulations to investigate the effects of I467T on myosin dynamics. We ran long simulations of wild-type human β-cardiac myosin and I467T starting from a homology model of a pre-powerstroke crystal structure (PDB ID: 5N6A) bound to ADP and Pi. We ran 8 trajectories with an average length of 1300 ns (aggregate time = 10.5 μs) for WT and 47 trajectories with an average length of 1000 ns (aggregate time = 47.7 μs) for I467T. To understand the effect of the mutation on ADP and Pi contacts, we calculated contact probabilities for ADP to the nucleotide binding pocket and for Pi to neighboring residues. Additionally, we calculated the distribution of distances between atoms near the mutation and within the nucleotide binding pocket.
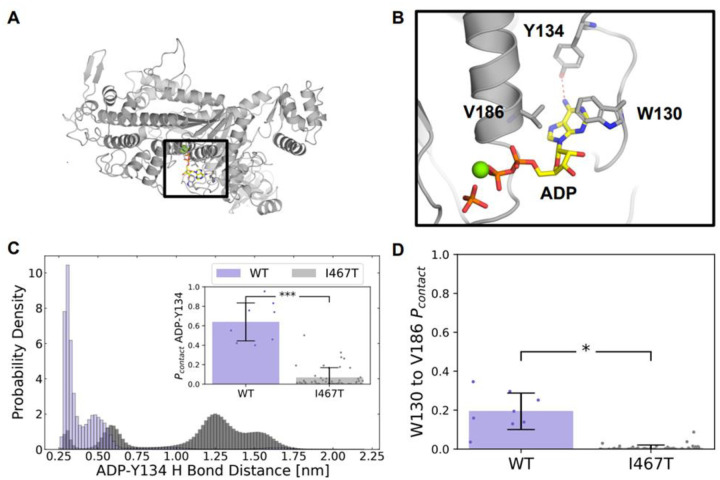
Although I467T is in Switch II, we found that I467T allosterically disrupts interactions between ADP and the nucleotide-binding pocket. In nucleotide-bound crystal structures of β-cardiac myosin, the nucleotide’s base forms a hydrogen bond with Y134 (Figure 4B). In simulations of WT β-cardiac myosin, this hydrogen bond typically remains formed (pcontact= 63.9%, Figure 4C). In contrast, in simulations of I467T, this hydrogen bond is more likely to be broken than formed (pcontact= 6.6%, p < 0.001, Figure 4C). Furthermore, in nucleotide-bound crystal structures of β-cardiac myosin, there are two residues, W130 and V186, that form a lid above the nucleotide’s base (Figure 4D), likely preventing the nucleotide from leaving the active site. Our simulations suggest that the interaction between W130 and V186 is more likely to break in the I467T than in the WT ensemble. The contact probability for W130 and V186 is 19.4% in WT but only 0.7% for I467T (Figure 4B, p < 0.001). Simulations of I467T sample partial dissociation events where the ADP is far from its crystal pose (e.g., ADP-N6 to Y134-OH distances of over 1 nm, Figure 4C). Taken together, these results suggest that ADP is not as tightly bound in the nucleotide binding pocket of β-cardiac myosin harboring the I467T mutation, providing a molecular mechanism for the changes in in vitro motility.
Figure 4. I467T disrupts interactions between ADP and the nucleotide-binding pocket in molecular dynamics simulations.
(A) Homology model structure of human beta-cardiac myosin, (B) focused in on the ADP-Pi binding site, with ADP depicted in yellow and orange, Pi depicted in orange, and Mg2+ in green. The red dotted line indicates a hydrogen bond between adenine and Y134 that is seen in nucleotide-bound crystal structures. (C) A native hydrogen bond the ADP base (ADP-N6) makes with a binding site residue (TYR134-OH) is disrupted in I467T. Additionally, the inset in (C) shows that the probability of a contact between ADP and TYR134 is significantly lower in I467T (p < 0.0001). (D) The contact probability between W130 and V186, two residues that form a lid over the adenine nucleobase, is significantly lower in I467T (p = 0.0011).
Discussion
In this work, we investigated two similar missense mutations in β-myosin heavy chain: HCM-linked Ile467Val (I467V) and LVNC-linked Ile467Thr (I467T). To investigate how these mutations lead to unique pathologies, we studied the molecular effects of each mutation using recombinantly expressed myosin in various in vitro assays. Neither I467V nor I467T altered the maximal actin-activated ATPase activity (Figure 1), while both mutations showed a >2-fold increase in actin sliding velocity (Figure 3), respectively. Finally, I467T showed a significant increase in the propensity to adopt the super-relaxed (SRX) state of myosin compared to WT and I467V (Figure 2). Thus, both I467T and I467V uncouple the canonical mechanochemical function of myosin described by the intrinsic enzymatic (ATPase) activity and the mechanical (actin sliding) properties of the motor, while only LVNC-I467T alters the regulation of the relaxation states of myosin.
While hundreds of sarcomeric mutations have been identified and linked to HCM[32, 33], the molecular mechanism by which these mutations converge on disease has not been identified. Historically, HCM was described as a gain-of-function disease as many mutations in β-MHC lead to enhanced mechanochemical function of myosin. HCM-I467V showed no change in the actin-activated ATPase activity, suggesting that the mutation does not alter the enzymatic function of the motor domain. However, HCM-I467V S1 did significantly increase the velocity by which myosin moves actin filaments. It is important to note that these assays are dependent on distinct steps of the mechanochemical cycle. The ATPase assay is an attachment limited assay in which phosphate release is the rate-limiting step, while the actin sliding assay is detachment limited, rate-limited by the release of ADP[34]. Though this mutation is located in the phosphate leaving tunnel of the myosin motor [35, 36]), the unchanged kcat reported for this mutation suggests that phosphate release in unchanged in this mutation, whereas the enhanced actin velocity in the HCM-I467V S1 mutation is likely due to an increase in ADP release rate. However, direct investigation of the kinetics of ATP product release is necessary to confirm mutation-specific changes.
While the clinical manifestation of HCM is hypercontractile function, one of the primary drivers of disease is an inability to relax properly[37]. Recent studies have shown that many HCM-linked mutations in β-MHC disrupt the off-state of myosin, leading to an imbalance in the number of myosin heads available to bind actin during relaxation[17, 24, 25]. The trending increase in the DRX state in HCM-I467V S1 suggests that this mutation may also increase the number of heads that are biochemically available to interact with actin (Figure 2A and 2B). Importantly, a small molecule therapy selective for cardiac myosin heavy chain, Camzyos ® (formerly mavacamten) has been shown to reduce the number of myosin heads that are available to bind to actin [24, 38–40]. Thus, based on the data herein, Camzyos may be potential therapy for treatment of HCM caused by β-MHC I467V.
LVNC-I467T S1 exhibited similar mechanochemical function to the HCM-I467V S1 mutation, including unchanged ATPase activity and enhanced actin velocity (Figures 1A and 3A). Computational modeling by Charobarkti et al suggested that I467T does not alter the rate of ATP hydrolysis into ADP and phosphate [35]. This is supported by the unchanged ATPase activity of I467T measured above. Similar to HCM-I467V S1, LVNC-I467T uncoupled biochemical and mechanical assays; furthermore, LVNC-I467T is predicted to enhance ADP release rate while having no effect on phosphate release. Given that I467T increases the in vitro motility of the β-cardiac myosin motor, we hypothesized that its ADP pocket would be in a more open state that facilitates ADP release. Previous studies have shown that sequence variation in myosin motors modulates the probabilities of conformations primed for specific functional roles [41, 42]. Though we did not observe complete ADP dissociation in simulations, we found that motors carrying the I467T mutation were more likely to adopt conformations primed for ADP release (i.e., structures with an open ‘lid’ and broken hydrogen bonds between ADP and myosin). This result strongly suggests that although the threonine does not directly interact with ADP, it allosterically disrupts interactions between ADP and the nucleotide-binding pocket.
Interestingly, Charkborati et al. also found that I467T stabilized the post-rigor state of myosin, preventing the structural rearrangements necessary for the recovery stroke and pre-power stroke structure. While it is thought stabilization of the pre-power stroke state of myosin is required for increasing SRX, multiple studies have suggested additional SRX structural states that have not yet been identified[40, 43, 44]. Thus, one can imagine that by slowing the recovery stroke of myosin, I467T induces an unknown structural state that increases the ultra-slow state of myosin. These simulations are supported by the significant increase the in SRX state of myosin that was measured in LVNC-I467T S1 (Figure 2). Additionally, the family affected by this mutation presents clinically with reduced systolic function, suggesting that the LVNC-linked I467T initiates disease, in part, by stabilizing the SRX state of myosin and reducing the availability of myosin heads for force-generation. This complex combination of altered mechanochemical function coupled to the reduced number of heads available to interact with actin may trigger a downstream signaling pathway distinct from both HCM and dilated cardiomyopathy, leading to the development of LVNC.
Development of non-compaction has been linked to unregulated Notch signaling which is sufficient to induce hypertrabeculation. Interestingly, one study showed that single molecule forces of around 4 pico-Newtons are sufficient to induce Notch signaling [45], forces that are within the range produced by a single molecule of cardiac myosin [46]. The strain on the cardiac wall induced by hypercontractile I467T motors may be sufficient to induce Notch signaling, leading to enhanced trabeculation. However, because fewer heads are available to bind actin at any given time, the inconsistent mechanoactivation may disrupt the normal signaling pathway, resulting in improper compaction. However, additional investigation is necessary to understand the cellular signaling disruptions that arise in the presence of I467T that lead to the non-compaction phenotype.
While further studies are required to define the precise mechanisms by which these mutations lead to complex cardiac diseases, our work suggests that phenotypic complexity originates at the molecular level. Thus, a molecular understanding is critical to developing targeted therapies to treat discrete diseases that arise from similar point mutations. By directly modulating the molecular progression of disease, myosin- and/or sarcomere-targeted small molecule therapies offer a path to treat many patients.
Methods
Recombinant Myosin Construct Generation and Protein Purification
Human recombinant β myosin S1 constructs (residues 1–842) were used to assess the functional consequences of mutations located within the motor domain. Site-directed mutagenesis was performed using the Agilent Quikchange II XL kit (Agilent Technologies, Santa Clara, CA) was used to individually introduce the I467V and I467T mutations into the S1 motor domain and confirmed via cDNA sequencing. Constructs were tagged with a C-terminal affinity tag complementary to the PDZ binding domain[47]. Viral construct, protein generation, and protein purification were performed as described by Lee et al[48]. Tissue purified rabbit skeletal actin was also purified as previously described[48].
ATPase Assay
Steady-state actin activated ATPase rates were measured using purified β-MHC S1 fragments expressing either the I467V or I467T mutations using an NADH-coupled regenerative ATPase assay. Purified wild-type (WT) β-MHC S1 was used as a control in all experiments. β-MHC S1 (F.C. = 0.4μM) was mixed with filamentous actin (concentration range: 0–100 μM) in the following solution: 20mM HEPES pH 7.0, 25mM KCl, 5mM MgCl2, 2mM ATP, 5mM DTT, 3mM phosphoenol pyruvate, 1mM NADH, 0.8mM pyruvate kinase/lactate dehydrogenase. Absorbance of NADH was monitored at 340nm over 60 minutes as a surrogate for ATP hydrolysis by the recombinant myosin. Linear absorbance ranges were fitted with Michaelis-Menten kinetics using GraphPad Prism 9 software. Basal ATPase activity (0 μM actin) was subtracted from ATPase rates were reported as kcat (/second) and actin affinity reported as Km (μM).
SRX/DRX ATPase Assay
In order to assess the ATPase activity that is associated with the SRX and DRX myosin states, purified recombinant S1 proteins were run in a modified ATPase assay. To deplete ATP from the myosin S1 proteins, proteins were individually dialyzed exhaustively against a buffer containing: 10mM Tris pH 7.4, 30mM Potassium Acetate, 1mM EDTA, and 4mM MgCl2. Myosin S1 (f.c. = 0.4μM) was rapidly mixed with 0.4μM mant-ATP (2’-(or-3’)-O-(N-Methylanthraniloyl) Adenosine 5’-Triphosphate) and the reaction aged for 60 seconds to allow for mant-ATP + S1 binding and ATP hydrolysis. The reaction was then rapidly mixed with 4mM Na-ATP and decay of mant-ATP signal was immediately monitored using a BMG ClarioSTAR plate reader (BMG Labtech, North Carlina, USA). Specifically, mant-ATP was excited at 385nm and emission was monitored at 450nm at 1 Hz for 1000 seconds. The normalized decay curves were fitted with a two-exponential decay (plateau = 0) using GraphPad Prism 9.0. The DRX populations (%fast) and SRX populations (100-%fast) were calculated for β-MHC I467T and I467V and compared to β-MHC WT and the ATPase rates for the DRX (kfast; ATP/second) and SRX (kslow; ATP/second) populations were calculated. The area under the curve was also calculated for all constructs to assess ATP consumption.
Molecular Dynamics Simulations
A homology model of beta-cardiac myosin in the pre-powerstroke state from Meller et al. was used as the starting structure for simulations of WT and I467T.
All simulations were performed in the GROMACS simulation software with the CHARMM36m force field[49, 50]. Each simulation was prepared by solvating the myosin motor domain with TIP3P water in a dodecahedron box extending 1 nm around the protein. The system was then neutralized and raised to a salt concentration of 0.1 M NaCl. Energy minimization was performed using steepest descent till the maximum force on any atom was below 1000 kJ/(mol × nm). Equilibration was performed using the Bussi-Parrinello thermostat and the Parrinello-Rahman barostat with all heavy atoms restrained at 300 K.
Production runs were performed at 310 K with the leapfrog integrator, Bussi-Parrinello thermostat, and Parrinello-Rahman barostat. A 12 Å cutoff distance was utilized with a force-based switching function starting at 10 Å. We employed periodic boundary conditions to mimic the effects of bulk solution. The PME method with a grid density greater than 1.2 Å3 was used to calculate long-range electrostatics. All hydrogen bonds were constrained with the LINCS algorithm to enable the use of a constant integration timestep of 2 fs. All simulations were performed on our in-house compute cluster consisting of NVIDIA A5000 nodes.
Simulation Analysis
We used MDTraj’s contacts function, which takes a subset of residues and reports any contacts between their atoms if any two atoms are within a cutoff threshold distance of each other; we defined the cutoff distance as 4 Å and used heavy atoms only. For each trajectory, we calculated the amount of time a contact exists between two residues and divided that by the length of the trajectory, yielding a probability of contact between those residues. The scripts for calculating contacts, statistical testing, and graphing the data can be found here:https://github.com/ssolieva/i467t.
In vitro Motility
In vitro motility was used to assess the effect of I467T and I467V mutations on myosin’s ability to translocate actin. Purified recombinant S1 mutants and WT proteins were mixed with filamentous actin in a 1:3 ratio and 1mM ATP then subsequently spun at 90,000rpm for 25 minutes at 4°C to remove inactive myosin molecules. The active myosin S1 molecules were diluted to a final concentration of 0.35μM in a 1X buffer containing: 20mM HEPES pH 7.5, 25mM potassium chloride, 5mM MgCl2, 1mM EGTA, and 10mM DTT. Glass cover slips were dipped in 0.2% nitrocellulose and adhered to a glass microscope slide to create a flow-cell for the experiment. To begin, 3 μM SNAP-PDZ was flowed into the cell and incubated for 2 minutes at room temperature. 1mg/mL BSA was then added and incubated for one minute at room temperature to block all non-specific binding sites. 0.35μM myosin S1 solution was added to the flow chamber and incubated for three minutes to ensure proper binding to the surface mobilized SNAP-PDZ domain. Excess myosin was washed out and phalloidin-labeled filamentous actin (f.c. 2–5nM) was added and incubated for one minute. Finally, activating solution containing 20mM HEPES pH 7.5, 25mM potassium chloride, 5mM MgCl2, 1mM EGTA, and 10mM DTT, 15mM ATP, 4mg/mL glucose, 0.135mg/mL glucose oxidase, 0.0215mg/mL catalase, and 0.5% methylcellulose was added to the flow chamber and incubated for 30 seconds at room temperature. Using an inverted wide-field microscope (Nikon Inc. New York, USA), movement of the phalloidin-labeled actin was monitored over time. Videos were collected between one and three frames per second and a total of 30 frames were collected for each movie. Using MATLAB, actin filament velocity was calculated for individual filaments and averaged across videos. At least 50 filaments were measured for each mutation, across three separately purified samples for β-MHC WT, I467T, and I467V S1.
Statistical Analysis
For actin-activated ATPase assays, single turnover ATPase assays, in vitro motility assays, a one-way ANOVA was used to assess significant differences. Tukey’s multiple comparisons test was applied post-hoc to determine the mutation-specific effects in these assays. For simulation analysis, we conducted a student’s t-Test when the variances were approximately equal and Welch’s t-Test when the variances were unequal. For all comparisons, significance was assigned at p>0.05.
Acknowledgements
We thank Ariana Combs for producing recombinant myosin protein. We thank Dr. Suman Nag for valuable discussions and insight. We thank Dr. Jian Wei Tay for motility analysis script generation and the BioFrontiers Institute Advanced Light Microscopy Core for use of microscopy equipment and for imaging support.
Funding and additional information
This work was supported by the National Institutes of Health (NIH grant R01GM029090; to L. A. Leinwand), NIH training grant (grant no.: T32 HL007822-20; to S. J. Lehman), NIH grant R01HL141086; to M. J. G., and NIH grants R01GM124007 and RF1AG067194; to G. R. B.). A. M. was supported by the National Institutes of Health F30 Fellowship (1F30HL162431-01A1). This work was also supported by the National Science Foundation (NSF grant DGE-2139839 to J. M. L.; and NSF CAREER Award MCB-1552471; to G. R. B.) G. R. B. holds a Packard Fellowship for Science and Engineering from The David & Lucile Packard Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH, the National Science Foundation, or the European Commission.
Funding Statement
This work was supported by the National Institutes of Health (NIH grant R01GM029090; to L. A. Leinwand), NIH training grant (grant no.: T32 HL007822-20; to S. J. Lehman), NIH grant R01HL141086; to M. J. G., and NIH grants R01GM124007 and RF1AG067194; to G. R. B.). A. M. was supported by the National Institutes of Health F30 Fellowship (1F30HL162431-01A1). This work was also supported by the National Science Foundation (NSF grant DGE-2139839 to J. M. L.; and NSF CAREER Award MCB-1552471; to G. R. B.) G. R. B. holds a Packard Fellowship for Science and Engineering from The David & Lucile Packard Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH, the National Science Foundation, or the European Commission.
References
- 1.Buvoli M., et al. , Bioinformatics assessment of beta-myosin mutations reveals myosin’s high sensitivity to mutations. Trends Cardiovasc Med, 2008. 18(4): p. 141–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garfinkel A.C., Seidman J.G., and Seidman C.E., Genetic Pathogenesis of Hypertrophic and Dilated Cardiomyopathy. Heart Failure Clinics, 2018. 14(2): p. 139–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tuohy C.V., et al. , Hypertrophic cardiomyopathy: the future of treatment, in European Journal of Heart Failure. 2020, John Wiley and Sons Ltd. p. 228–240. [DOI] [PubMed] [Google Scholar]
- 4.Green E.M., et al. , A small-molecule inhibitor of sarcomere contractility suppresses hypertrophic cardiomyopathy in mice. Science, 2016. 351(6273): p. 617–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Towbin J.A., Ballweg J., and Johnson J., Chapter 20 - Left Ventricular Noncompaction Cardiomyopathy, in Heart Failure in the Child and Young Adult, Jefferies J.L., et al. , Editors. 2018, Academic Press: Boston. p. 269–290. [Google Scholar]
- 6.Ross S.B., et al. , A systematic review and meta-analysis of the prevalence of left ventricular non-compaction in adults. Eur Heart J, 2020. 41(14): p. 1428–1436. [DOI] [PubMed] [Google Scholar]
- 7.Jenni R., Rojas J., and Oechslin E., Isolated Noncompaction of the Myocardium. New England Journal of Medicine, 1999. 340(12): p. 966–967. [DOI] [PubMed] [Google Scholar]
- 8.Hoedemaekers Y.M., et al. , The importance of genetic counseling, DNA diagnostics, and cardiologic family screening in left ventricular noncompaction cardiomyopathy. Circ Cardiovasc Genet, 2010. 3(3): p. 232–9. [DOI] [PubMed] [Google Scholar]
- 9.Takasaki A., et al. , Sarcomere gene variants act as a genetic trigger underlying the development of left ventricular noncompaction. Pediatric Research, 2018. 84(5): p. 733–742. [DOI] [PubMed] [Google Scholar]
- 10.Klaassen S., et al. , Mutations in sarcomere protein genes in left ventricular noncompaction. Circulation, 2008. 117(22): p. 2893–901. [DOI] [PubMed] [Google Scholar]
- 11.Tanjore R., et al. , Genetic variations of β-MYH7 in hypertrophic cardiomyopathy and dilated cardiomyopathy. Indian J Hum Genet, 2010. 16(2): p. 67–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McNally E.M. and Mestroni L., Dilated Cardiomyopathy. Circulation Research, 2017. 121(7): p. 731–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adhikari A.S., et al. , β-Cardiac myosin hypertrophic cardiomyopathy mutations release sequestered heads and increase enzymatic activity. Nature Communications, 2019. 10(1): p. 2685–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spudich James A., Hypertrophic and Dilated Cardiomyopathy: Four Decades of Basic Research on Muscle Lead to Potential Therapeutic Approaches to These Devastating Genetic Diseases. Biophysical Journal, 2014. 106(6): p. 1236–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palmiter K.A., et al. , R403Q and L908V mutant β-cardiac myosin from patients with familial hypertrophic cardiomyopathy exhibit enhanced mechanical performance at the single molecule level. Journal of Muscle Research & Cell Motility, 2000. 21(7): p. 609–620. [DOI] [PubMed] [Google Scholar]
- 16.Sommese R.F., et al. , Molecular consequences of the R453C hypertrophic cardiomyopathy mutation on human β-cardiac myosin motor function. Proceedings of the National Academy of Sciences, 2013. 110(31): p. 12607–12612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alamo L., et al. , Effects of myosin variants on interacting-heads motif explain distinct hypertrophic and dilated cardiomyopathy phenotypes. eLife, 2017. 6: p. e24634–e24634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nag S., et al. , The myosin mesa and the basis of hypercontractility caused by hypertrophic cardiomyopathy mutations. Nat Struct Mol Biol, 2017. 24(6): p. 525–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alamo L., et al. , Conserved Intramolecular Interactions Maintain Myosin Interacting-Heads Motifs Explaining Tarantula Muscle Super-Relaxed State Structural Basis. Journal of Molecular Biology, 2016. 428(6): p. 1142–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woodhead J.L., et al. , Atomic model of a myosin filament in the relaxed state. Nature, 2005. 436(7054): p. 1195–1199. [DOI] [PubMed] [Google Scholar]
- 21.Rasicci D.V., et al. , Dilated cardiomyopathy mutation E525K in human beta-cardiac myosin stabilizes the interacting-heads motif and super-relaxed state of myosin. Elife, 2022. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma W., et al. , Structural OFF/ON transitions of myosin in relaxed porcine myocardium predict calcium-activated force. Proceedings of the National Academy of Sciences, 2023. 120(5): p. e2207615120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robert-Paganin J., Auguin D., and Houdusse A., Hypertrophic cardiomyopathy disease results from disparate impairments of cardiac myosin function and auto-inhibition. Nature Communications, 2018. 9(1): p. 4019–4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anderson R.L., et al. , Deciphering the super relaxed state of human β-cardiac myosin and the mode of action of mavacamten from myosin molecules to muscle fibers. Proceedings of the National Academy of Sciences, 2018. 115(35): p. E8143–E8143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vander Roest A.S., et al. , Hypertrophic cardiomyopathy β-cardiac myosin mutation (P710R) leads to hypercontractility by disrupting super relaxed state. Proc Natl Acad Sci U S A, 2021. 118(24). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hooijman P., Stewart Melanie A., and Cooke R., A New State of Cardiac Myosin with Very Slow ATP Turnover: A Potential Cardioprotective Mechanism in the Heart. Biophysical Journal, 2011. 100(8): p. 1969–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nag S. and Trivedi D.V., To lie or not to lie: Super-relaxing with myosins. eLife, 2021. 10: p. e63703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmid M. and Toepfer C.N., Cardiac myosin super relaxation (SRX): a perspective on fundamental biology, human disease and therapeutics. Biol Open, 2021. 10(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aksel T., et al. , Ensemble force changes that result from human cardiac myosin mutations and a small-molecule effector. Cell Rep, 2015. 11(6): p. 910–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaneda T., et al. , A novel beta-myosin heavy chain gene mutation, p.Met531Arg, identified in isolated left ventricular non-compaction in humans, results in left ventricular hypertrophy that progresses to dilation in a mouse model. Clin Sci (Lond), 2008. 114(6): p. 431–40. [DOI] [PubMed] [Google Scholar]
- 31.Trivedi D.V., et al. , Switch II mutants reveal coupling between the nucleotide- and actin-binding regions in myosin V. Biophys J, 2012. 102(11): p. 2545–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alejandro E.d.F., Andrew E.K., and Jason R.B., Sarcomere mutation negative hypertrophic cardiomyopathy is associated with ageing and obesity. Open Heart, 2021. 8(1): p. e001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ho C.Y., et al. , Genotype and Lifetime Burden of Disease in Hypertrophic Cardiomyopathy. Circulation, 2018. 138(14): p. 1387–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siemankowski R.F. and White H.D., Kinetics of the interaction between actin, ADP, and cardiac myosin-S1. Journal of Biological Chemistry, 1984. 259(8): p. 5045–5053. [PubMed] [Google Scholar]
- 35.Chakraborti A., et al. , Investigation of the Recovery Stroke and ATP Hydrolysis and Changes Caused Due to the Cardiomyopathic Point Mutations in Human Cardiac β Myosin. J Phys Chem B, 2021. 125(24): p. 6513–6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Llinas P., et al. , How actin initiates the motor activity of Myosin. Dev Cell, 2015. 33(4): p. 401–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ho C.Y., Hypertrophic Cardiomyopathy: Preclinical and Early Phenotype. Journal of Cardiovascular Translational Research, 2009. 2(4): p. 462–470. [DOI] [PubMed] [Google Scholar]
- 38.Gollapudi S.K., et al. , TWO CLASSES OF MYOSIN INHIBITORS, BLEBBISTATIN AND MAVACAMTEN, STABILIZE -CARDIAC MYOSIN IN DIFFERENT STRUCTURAL AND FUNCTIONAL STATES. bioRxiv, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gollapudi S.K., et al. , Synthetic thick filaments: A new avenue for better understanding the myosin super-relaxed state in healthy, diseased, and mavacamten-treated cardiac systems. The Journal of biological chemistry, 2021. 296: p. 100114–100114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rohde J.A., et al. , Mavacamten stabilizes an autoinhibited state of two-headed cardiac myosin. Proceedings of the National Academy of Sciences, 2018. 115(32): p. E7486–E7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Porter J.R., et al. , Conformational distributions of isolated myosin motor domains encode their mechanochemical properties. eLife, 2020. 9: p. e55132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meller A., et al. , Drug specificity and affinity are encoded in the probability of cryptic pocket opening in myosin motor domains. eLife, 2023. 12: p. e83602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Knupp C., Morris E., and Squire J.M., The Interacting Head Motif Structure Does Not Explain the X-Ray Diffraction Patterns in Relaxed Vertebrate (Bony Fish) Skeletal Muscle and Insect (Lethocerus) Flight Muscle. Biology, 2019. 8(3): p. 67–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brunello E., et al. , Myosin filament-based regulation of the dynamics of contraction in heart muscle. Proceedings of the National Academy of Sciences, 2020. 117(14): p. 8177–8177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chowdhury F., et al. , Defining Single Molecular Forces Required for Notch Activation Using Nano Yoyo. Nano Letters, 2016. 16(6): p. 3892–3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Finer J.T., Simmons R.M., and Spudich J.A., Single myosin molecule mechanics: piconewton forces and nanometre steps. Nature, 1994. 368(6467): p. 113–119. [DOI] [PubMed] [Google Scholar]
- 47.Walkup W.G.t. and Kennedy M.B., PDZ affinity chromatography: a general method for affinity purification of proteins based on PDZ domains and their ligands. Protein Expr Purif, 2014. 98: p. 46–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee L.A., et al. , Functional divergence of the sarcomeric myosin, MYH7b, supports species-specific biological roles. Journal of Biological Chemistry, 2023. 299(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abraham M.J., et al. , GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX, 2015. 1: p. 19–25. [Google Scholar]
- 50.Huang J., et al. , CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nature Methods, 2017. 14(1): p. 71–73. [DOI] [PMC free article] [PubMed] [Google Scholar]