Abstract
The SORL1 gene has recently emerged as a strong Alzheimer Disease risk gene. Over 500 different variants have been identified in the gene and the contribution of individual variants to AD development and progression is still unknown. Here, we describe a multi-generational family with both early- and late-onset AD in two generations. Exome sequencing identified a coding variant, p.(Arg953Cys), in SORL1 in 4 of 5 individuals affected by AD. Notably, variant carriers had severe AD pathology, including the presence of Aβ plaques in the cerebellum as well as TDP-43 pathology. We further characterized this variant and show that this Arginine substitution occurs at a critical domain position of the SORL1 translation product, SORL1. In vitro studies further show that the SORL1 p.R953C variant has decreased maturation and shedding of the SORL1 protein, and also leads to mis-localization within cells. Together, our analysis suggests that p.R953C is a likely pathogenic variant of SORL1 and sheds light on mechanisms of how missense SORL1 variants may lead to AD.
Keywords: SORL1, Alzheimer’s disease, YWTD-domain, pathogenic variant
Introduction
Alzheimer Disease (AD) is the most common cause of dementia worldwide. The etiology of AD remains elusive, slowing development of disease modifying therapies. Genetic variants in PSEN1, PSEN2 and APP are associated with autosomal dominantly inherited early-onset AD (ADAD), although those families are rare and make up only a very small fraction of all AD. Nevertheless, knowledge gained from studying ADAD has been valuable to our understanding of the clinical, pathological and mechanistic features of AD more broadly. Late onset or “sporadic” AD also has a genetic component and is known to be highly heritable, estimated at 60–80%1. Genome wide association studies (GWAS) as well as genome and exome sequencing studies have revealed the complexity of biological processes contributing to AD risk and progression2. Given that families with AD likely harbor an AD genetic risk factor, they can provide important insight into genetic risk and disease pathogenesis.
The Sortilin-like receptor, SORL1, (protein: SORL1/SORLA) was originally identified as a member of the LDL receptor family, and the SORL1 protein is now classified as one of five mammalian sorting receptors called VPS10 receptors3–7. SORL1 functions as an endosomal receptor to assist cargo sorting out of the endosome to either the cell surface via the recycling pathway or to the trans-Golgi network (TGN) via the retrograde pathway8–12. For sorting of ADrelated cargo, such as APP, SORL1 directly interacts with the multi-sorting complex retromer, itself highly implicated in endo-lysosomal health and neurodegeneration13–15.
Through both candidate gene studies and GWAS, SORL1 was found to be a strong genetic risk factor for AD16–19. Exome-sequencing studies have shown that rare loss-of-function SORL1 alleles, leading to a haploinsufficiency, have been associated with highly penetrant AD20–24, although the full breadth and contribution of SORL1 variants in AD is not well defined. An additional large number (>500) of SORL1 variants have also been identified in patients with AD, but with variable levels of evidence for pathogenicity. Recently, two missense variants have been associated with autosomal dominant AD: p.(Asp1545Val) and p.(Tyr1816Cys) (Bjarnadottir et al., Manuscript in preparation/Jensen et al., Manuscript in preparation). Reported SORL1 variants span the length of the gene and functional domains, but how the pathogenic variants impair the overall functions of SORL1 as an endosomal sorting receptor is not yet clear. Defining the biochemical consequences of SORL1 missense variants can shed light on mechanisms of disease involving SORL1 and other components of the endo-lysosomal network (ELN).
We present here a family with multiple generations of early- and late-onset AD in which affected members were found to carry a novel variant, SORL1 c.2857C>T p.Arg953Cys (R953C; NM_003105.5), which we have classified as likely pathogenic. The family is notable for clinical dementia consistent with AD in two generations, and genetic testing confirmed the SORL1 variant in 4 out of 5 affected individuals tested. Neuropathological studies demonstrated severe AD pathology, including cerebellar amyloid plaques, cortical neurofibrillary tangles, and TDP-43 deposition despite a young age of onset. To further characterize this variant, we turned to in silico studies to predict pathogenicity based on the domain in which the variant occurs and conservation between SORL1 and members of the LDLR family25. We next generated a plasmid containing the p.R953C variant and transfected it into HEK293 and N2a cells. Our in vitro studies show reduced SORL1 maturation and impaired endosomal localization, confirming a functional consequence of the missense variant. This study adds to the growing body of literature supporting a role for SORL1 variants as alleles that may contribute to the missing AD heritability. Mechanisms of pathogenicity may be related to SORL1 cellular localization and its role as an endosomal sorting receptor.
Methods
Study Participants
The family was ascertained by the University of Washington Alzheimer Disease Research Center. The study was approved by the UW Institutional Review Board (IRB) and all participants provided written consents.
Genetic Studies
Genetic analysis was performed by the Northwest Clinical Genomics Laboratory (NCGL), a CLIA certified laboratory at the University of Washington. Samples underwent next-generation exome sequencing and analysis. Libraries were constructed according to the NCGL protocol. The KAPA Hyper Prep DNA library kit (KAPA Biosystems, Wilmington, MA, USA) was used to prepare the libraries, which were subsequently enriched using an in-house, optimized xGen Exome Research Panel v1.0 (Integrated DNA Technologies, Coralville, IA, USA). Paired-end sequencing of the exome-enriched libraries was performed on a HiSeq 4000 instrument (Illumina, San Diego, CA, USA). Greater than 99% of the coding regions and canonical splice sites were sequenced to a read coverage of at least 20X or greater. The average mean depth of coverage was 144 reads. Resulting sequences were aligned to the human genome reference (hg19) using the Burrows-Wheeler Aligner (BWA)26. Variants were identified using the Genome Analysis Toolkit (GATK)27,28 and were annotated using the SnpEff annotation tool29 in combination with various population databases and variant impact scoring tools. 27 genes associated with dementia or risk of dementia were screened, including APP, PSEN1, PSEN2, MAPT, FUS, GRN, and SORL1.
APOE genotyping:
APOE genotyping was performed as previously published30. Briefly, Genomic DNA was amplified in a 9700 Gene Amp PCR System (Applied Biosystems) using primers that amplify APOE gene’s exon 4. This PCR amplicon includes both the codon 112 (ε2/ε3 vs. ε4) and codon 158 (ε2 vs. ε3/ε4) polymorphic sites.
Taqman assay:
SNPs rs429358 (ε2/ε3 vs. ε4) and rs7412 (ε2 vs. ε3/ε4) were genotyped using assay C_3084793_20 and assay C_904973_10 (Thermo Fisher), respectively. All reactions were carried out in a 9700 Gene Amp PCR System with a profile of 50°C for 5 minutes; 95°C for 5 minutes; 50 cycles of 95°C for 15 seconds, and 60°C for 1 minute.
Sanger sequencing:
The PCR reaction/amplicon (1 μl) was used in BigDye sequencing reaction (Thermo Fisher) with a final volume of 10 μl. All reactions were carried out in a 9700 Gene Amp PCR System with a profile of 94°C for 1 minute; 35 cycles of 94°C for 30 seconds, 55°C for 10 seconds, and 60°C for 4 minutes; and a final extension of 60°C for 5 minutes. The PCR generated sequencing products were further purified using EDTA/ethanol precipitation and then subjected to DNA sequencing run using SeqStudio (Thermo Fisher). The sequencing data (electropherograms) were transferred and uploaded onto the Sequencher program (Genecodes) for sequence alignment.
Primer sequences:
APOE_Ex4_F: 5’ TCGGAACTGGAGGAACAACT 3’
APOE_Ex4_R: 5’ GCTCGAACCAGCTCTTGAGG 3’
SORL1 genotyping
Genomic DNA was amplified with Phusion Flash (Thermo Fisher) on a C1000 Touch Thermo cycler (BioRad) using primers that amplify exon 20 in SORL1. Cycle conditions: 98°C for 10s; 98°C for 1s, 65°C for 5s, 72°C for 10s X25 cycles; 72°C for 1 min. Cleaned PCR reactions were sent for Sanger sequencing using GeneWiz (Azenta Life Sciences). Sequences were examined manually using 4 Peaks software.
Primer sequences:
SORL1 F: 5’ GCCTGGGATTTATCGGAGCA 3’
SORL1 R: 5’ TGGCATCCCTCCATAGGCT 3’
Neuropathology
Consent for autopsy was obtained from the donor or from the legal next of kin, according to the protocols approved by the UW Institutional Review Board. At the time of autopsy, the brain was removed in the usual fashion. For patients I-2, II-2, and II-4, the left halves were coronally sectioned and samples were frozen for possible biochemical studies and the right halves were fixed in formalin. For patients II-1 and II-5, the entire brain was fixed in formalin. After fixation, the cerebrum was sectioned coronally, the brainstem was sectioned axially, and the cerebellum was sectioned sagittally.
Representative sections for histology were selected and evaluated according to NIA-AA guidelines31. A microtome was used to cut 4 μm-thick tissue sections from formalin-fixed, paraffin-embedded tissue blocks. Hematoxylin and eosin (H&E), Luxol fast blue (LFB), and Bielschowsky silver-stained slides were prepared. Using previously optimized conditions, immunohistochemistry was performed using a Leica Bond III Fully Automated IHC and ISH Staining System (Leica Biosystems, Wetzlar, Germany). The sections were immunostained with mouse monoclonal antibody against paired helical filament tau (AT8, 1:1,000 dilution) (Pierce Technology, Waltham, MA), mouse monoclonal against β-amyloid (6E10, 1:5,000) (Covance, Princeton, NJ), rat monoclonal against phosphorylated TDP-43 (ser409/ser410, 1:1,000) (Millipore, Burlington, MA), and mouse monoclonal against α-synuclein (LB509, 1:500) (Invitrogen, Carlsbad, CA). Appropriate positive and negative controls were included with each antibody and each run.
Site-directed Mutagenesis
The R953C variant was inserted in SORLA pcDNA3.1 and SORLA-GFP pcDNA3.1 using site directed mutagenesis kit (QuikChange #200521) according to manufacturers’ instruction with the following pair of primers: 5- gga tca cgt tca gtg gcc agc agt gct ctg tca ttc tgg aca acc tcc-3 and 5- gga ggt tgt cca gaa tga cag agc act gct ggc cac tga acg tga tcc-3.
Cell transfection and western blotting
Approximately 5×105 HEK293 and N2a cells were seeded on 6-well plates and transiently transfected with expression constructs for SORLA-WT or SORLA-R953C, using Fugene 6 Transfection Reagent kit (Promega) according to manufacturers’ instructions. 48 hours post transfection, cell medium was changed to serum free conditional medium and after 48 hours, cells and media were harvested. Cells were lysed using lysis buffer (Tris 20mM, EDTA 10mM, TritonX 1%, NP40 1%). Media samples (30ml) and lysate samples(20ug) were mixed with NuPAGE LDS sample buffer (Invitrogen, #2463558) supplemented with β-Mercaptoethanol (Sigma) and separated on SDS-PAGE using 4–12% NuPAGE Bis-Tris gels (Thermo). Proteins were then transferred to nitrocellulose membranes (Thermo) and incubated for 1h at room temperature in Blocking buffer (Tris-Base 0.25M, NaCl 2.5M, skimmed milk powder 2%, tween-20 2%). Next, membranes were incubated overnight at 4°C with LR11 antibody 1:1,000 (BDBiosciences # 612633) to detect SORL1 and Beta actin 1:2,000 (Sigma #A5441) as loading control, followed by three washes for 5 minutes in washing buffer (CaCl2 0.2 mM, MgCl2 0.1 mM, HEPES 1 mM, NaCl 14 mM, skimmed milk powder 0.2%, Tween 20 0.05%) and 1 hour incubation with HRPconjugated secondary antibody (1:1,500, Dako, #P0260) for 1 hour at room temperature. Membranes were washed 5 times for 5 minutes, incubated with FEMTO detection reagent (Thermo #34095) and visualized by iBright1500 scanner. Quantification was performed by densitometric analysis in ImageJ and data were plotted in Graphpad Prism 9.5.0.
Flow cytometry
Cell surface and total receptor level were analyzed by flow cytometry in live, transfected HEK293 and N2a cells. Briefly, HEK293 and N2a cells were transiently transfected with either WT-GFP or R953C-GFP plasmids. Twenty-four hours after transfection, cells were collected by trypsinization, pelleted, and resuspended in phosphate-buffered saline (PBS pH 7.4). After 15min incubation in blocking buffer (PBS pH 7.4, 0.5% BSA), cells were immunostained at 4°C with rabbit anti-soluble-SORL1 primary antibody followed by washing two times with PBS pH 7.4 and 30min incubation with Alexa-flour 647 secondary antibody in the absence of detergent followed by 3 times washing and finally resuspension in FACS buffer (PBS pH 7.4, 2% FBS, 1% Glucose). Cells were analyzed by NovoCyte 3000 flow cytometer equipped with three lasers and 13 fluorescence detectors (Agilent, Santa Clara, CA). GFP and Alexa Flour 647 fluorophores were excited by the 488 and 640 nm lasers, respectively. Results were analyzed using FlowJo™ v10.8.1 Software (BD Life Sciences).
Immunocytochemistry and Confocal Microscopy
Approximately 5×104 HEK293 cells were seeded on poly-L-lysine coated glass coverslips and transfected with expression constructs for SORL1-WT or SORL1-R953C using Fugene 6 Transfection Reagent kit (Promega). 24h post-transfection, cells were fixed with PFA 4% for 10 minutes at room temperature, followed by a wash in PBS pH 7.4. Coverslips were washed twice in PBS with 0.1% Triton-X 100 (for intracellular staining) or only PBS (for membrane staining) and later blocked for 30 minutes at room temperature in blocking buffer (PBS, FBS 10%). Cells were then incubated overnight at 4°C with pAb_5387 (a polyclonal rabbit serum generated for the entire SORLA ectodomain3) antibody alone or with an antibody against markers specific for each intracellular compartment (EEA1 for early endosomes, TFR for recycling endosomes, and Calnexin for ER). Next, cells were washed in PBS with or without Triton-X 0.1 % and incubated in Alexa Flour secondary antibodies (Invitrogen, 1:500) for 1 hour at room temperature. After washing once in PBS, cells were stained with Höechst (Abcam, 1:50,000) for 10 minutes at room temperature. The coverslips were then mounted on glass slides using DAKO fluorescence mounting medium (Agilent) and were imaged using Zeiss LSM800 confocal microscope. Colocalization was quantified using the JACOP plugin in ImageJ software and presented as Mander’s correlation coefficient. Graphing and statistical analysis of the data were performed with GraphPad Prism 9.5.0. Antibodies used were as follows: rabbit polyclonal anti-SORL1 (pAb_5387; Aarhus University) 1:300, mouse monoclonal anti-SORL1 (mAb_AG4; Aarhus University) 1:100, anti EEA1(#610457 BDBiosciences) 1:100, anti TFR 1:100(# A-11130 Invitrogen), anti-Calnexin (1:100) (#610523 BDBiosciences).
Data Availability
The authors confirm that the data supporting the findings of this study are available within the article and/or its supplementary material or available from the corresponding author upon reasonable request.
Results
Clinical Description
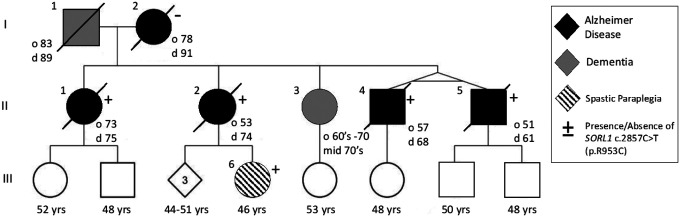
Three generations (Figure 1) of the study family are presented here. Clinical features are reported in Table 1. Both parents (I-1 and I-2) developed late onset dementia and I-1 also demonstrated parkinsonism and aggressive behavior. Of the 5 individuals in the II generation sibship, 4 were clinically diagnosed with AD, with a range of age of onset from 51 years to 74 years. II-4 and II-5, identical twins, developed early onset AD at age 57 years and 51 years, respectively. II-5 developed aphasia and apraxia in addition to memory loss. III-6, daughter of II-2, developed progressive spasticity at age 44. She has also developed evidence of executive dysfunction determined by two neuropsychiatric evaluations. She has not shown any lower motor neuron findings or any other neurological signs. MRI brain did not show atrophy or other abnormality (data not shown).
Figure 1. Pedigree of SORL1 R953C family:
Solid black indicates individuals diagnosed with Alzheimer Disease which was confirmed by neuropathology. Dark grey indicates clinical diagnosis of Dementia. Onset of disease (“o” years) and age at death “d” years is indicated next to the individual when known. Circles indicate female, square indicates male. Diamond is sex unknown to investigators at time of report.
Table 1:
Clinical and Neuropathological Characterization
| Clinical and Neuropathological Characterization | ||||||||
|---|---|---|---|---|---|---|---|---|
| ID | Sex | Age of Onset | Age at Death | Duration (Years) | Clinical Features | Neuropath | SORL1 genotype | APOE genotype |
| I-1 | M | 83 | 89 | 6 | Dx”Severe Dementia” Parkinsonism Aggressive Behavior | N/A | N/A | |
| I-2 | F | 78 | 91 | 13 | Dx Alzheimer Dementia; Age 85 MMSE 23; Age 86 MMSE 15; Age 87 MMSE 11 | CERAD Frequent; Braak VI | WT | 3/3 |
| II-1 | F | 73 | 75 | 2.5 | Dx Alzheimer Dementia; Rapid Progression; Age 74 MOCA 11/30; | A3B3C3; Braak VI; Diffuse LBD; LATE Stage 2; Thal 4; CERAD Freq | p.R953C; | 3/3 |
| II-2 | F | 53 | 74 | 21 | Dx Alzheimer Dementia; Early Memory Loss in 50’s; Age 59 WMS “Profound Impairment”; Age 63 MMSE 26 | A3B3C3; Braak VI; Limbic LBD; LATE Stage 1, Bilateral Hippocampal Sclerosis | p.R953C | 3/3 |
| II-3 | F | Late 60’s | N/A | N/A | Dx Alzheimer Dementia; Current Age Mid 70’s | N/A | N/A | N/A |
| II-4 | M | 57 | 68 | 11 | Dx Alzheimer Dementia; Age 59 MMSE 26; Twin of II-5 | Braak VI; CERAD Freq | p.R953C | 3/3 |
| II-5 | M | 51 | 61 | 10 | Dx Alzheimer Dementia; Aphasia; Apraxia; Age 55 MMSE 9: Twin of II-4 | Braak VI; CERAD Freq | p.R953C | 3/3 |
Neuropathology
Individuals I-2, II-1, II-2, II-4, and II-5 were evaluated at autopsy, and findings are summarized in Table 2. Brain weight in all cases except II-1 was below the 10th percentile for age and sex32. Atherosclerosis was present in all cases, with plaques extending beyond the first branch point of at least one cerebral artery (defined here as moderate); in case II-4, atherosclerotic plaques were also visible on the external surface and thus graded as severe. No other abnormalities were observed grossly in any case.
TABLE 2:
Neuropathologic Findings
| PEDIGREE NUMBER | BRAIN WEIGHT (G) | ATHEROSCLEROSIS | THAL PHASE | BRAAK STAGE | CERAD | ADNC | HIPPOCAMPAL SCELROSIS | LATE STAGE | LEWY BODY DISEASE |
|---|---|---|---|---|---|---|---|---|---|
| I-2 | 900 | Moderate | 4 | VI | Frequent | HIGH | Absent | 0 | Absent |
| II-1 | 1136 | Moderate | 4 | VI | Frequent | HIGH | Absent | 2 | Diffuse |
| II-2 | 867 | Moderate | 5 | VI | Frequent | HIGH | Present | 2 | Limbic |
| II-4 | 950 | Severe | 5 | VI | Frequent | HIGH | Present | 2 | Brainstem |
| II-5 | 1120 | Moderate | 5 | VI | Frequent | HIGH | Absent | 2 | Absent |
Histopathology
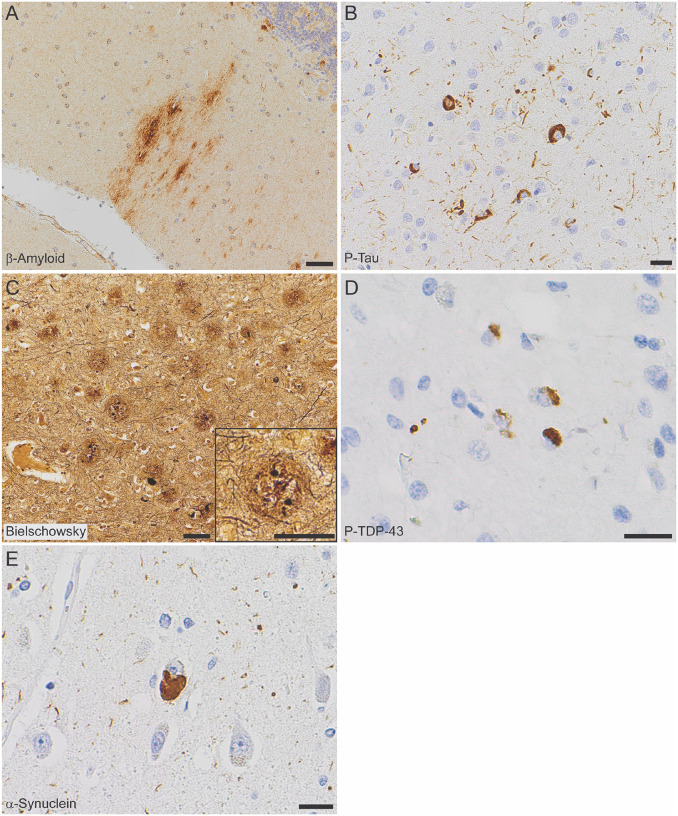
All autopsy cases were evaluated by the standard National Institute of Aging-Alzheimer’s Association (NIA-AA) protocol31,33. β-amyloid plaques progressed to the midbrain in cases I-2 and II-1 (Thal phase 4 of 5), and extended to the cerebellum in cases II-2, II-4, and II-5 (Thal phase 5 of 5, Figure 2A). Tau tangles were present within the calcarine cortex/primary visual cortex in all cases (Braak and Braak stage VI of VI, Figure 2B). Cortical neuritic plaque density in all cases was frequent by Consortium to Establish a Registry for Alzheimer’s Disease (CERAD) criteria (Figure 2C). The features in each case meet criteria for high Alzheimer’s disease neuropathologic change (ADNC) by NIA-AA guidelines31. Additionally, all generation II cases had TDP-43 inclusions in the amygdala and hippocampus, consistent with limbic-predominant age-related TDP-43 encephalopathy neuropathologic change (LATE-NC) stage 234 (Figure 2D); TDP-43 inclusions were not seen in case I-2 (SORL1 variant negative). Hippocampal sclerosis was also seen in cases II-2 and II-4. Varying stages of Lewy body disease (LBD) were also identified, with diffuse (neocortical) LBD diagnosed in II-1, limbic (transitional) LBD in II-2, and brainstempredominant LBD in II-4 (Figure 2E, Supplemental Figure 1).
Figure 2. Neuropathologic evaluation demonstrates high Alzheimer disease pathologic change (ADNC) by NIA-AA criteria in SORL1 R953C cases.
(a) Representative section of cerebellum stained for β-amyloid (6e10), highlighting plaques within the molecular layer and warranting a Thal phase 5. Patient II-5, scale bar = 50 μm. (b) Representative section of calcarine cortex stained for phosphorylated tau (P-Tau; AT8), highlighting neurofibrillary tangles in a background of dystrophic neurites, consistent with Braak and Braak stage VI. Patient II-5, scale bar = 20 μm. (c) Representative section of middle frontal gyrus stained with Bielschowsky silver demonstrating frequent neuritic plaques by CERAD criteria. Insert shows a representative neuritic plaque, composed of brown, targetoid β-amyloid associated with black dystrophic neurites. Patient II-5, scale bars = 50 μm. (d) Representative section of hippocampus stained for phosphorylated TDP-43 (P-TDP43), demonstrating intracytoplasmic inclusions and scattered dystrophic neurites. The pattern is consistent with limbic-predominant age-related TDP-43 encephalopathy (LATE) stage 2, though age < 80 years is atypical for sporadic LATE. Patient II-4, scale bar = 20 μm. (e) Representative section of anterior cingulate gyrus stained for α-synuclein, highlighting the presence of a Lewy body in a background of positive neurites. Though Lewy body disease was present in the majority of SORL1 R953C carriers, the pattern was highly variable. Patient II-2, scale bar = 20 μm.
Genetic Findings
Due to early onset and family history of AD, subject II-5 underwent PSEN1 and APP research genetic testing, which was negative in both genes. Years after the subject’s passing, his sample was included in an early onset AD cohort evaluated by exome sequencing for 27 neurodegeneration genes. II-5 was found to carry a SORL1 missense variant: NM_003105.5 c.2857C>T p.Arg953Cys (R953C). The reported allele frequency of this variant in gnomAD for those of European (non-Finnish) ancestry is (1/113646) or 0.000008799. It has not been reported in anyone of any other ancestry that has been assessed. In silico predictions varied; Polyphen: probably damaging, SIFT: tolerated, REVEL: 0.805, CADD: 25.4, SpliceAI: 0.00 (no_consequence), PrimateAI: 0.633. No other pathogenic or likely pathogenic variants were identified. Next, we screened II-4, II-2 and II-1 by exome sequencing, which revealed that all three also carried the SORL1 R953C variant, and no other pathogenic variants were identified. I-2 did not carry the SORL1 variant, and no DNA samples were available from I-1. III-6 was found to carry the SORL1 variant. C9orf72 gene expansion testing was negative in generation II and III-6. I-2, all individuals in the II generation and III-6 were APOE ε3/ε3 genotype.
Variant characterization
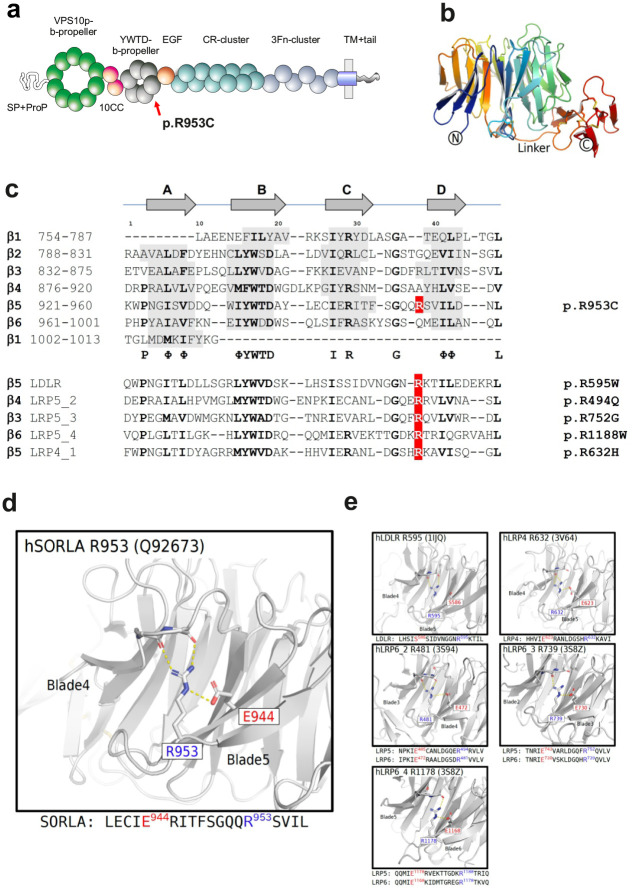
The arginine residue Arg953 is located at domain position 38 of the YWTD-blade repeated sequence, located within the fifth of six repeats that build the 6-bladed β-propeller domain of SORL1 (Figure 3a). We previously undertook a detailed disease-mutation domain-mapping approach to identify the most pathogenic sequence positions for the SORL1 domains and their risk for developing AD25. From this analysis, YWTD-blade sequence position 38 was identified as a high-risk site when arginine substitution occurs, and we identified variant p.Arg953His, (p.R953H) in three early-onset AD patients corresponding to the same SORL1 amino acid. However, the p.R953C variant was not identified in this large exome-sequencing study20.
Figure 3. In silico characterization of SORL1 p. R953C.
(a) Schematic presentation of the mosaic domain structure of the SORL1 protein comprising from the N-terminal end: VPS10p-domain with accompanying 10CCa/b domains, YWTD-repeated β-propeller domain (with p.R953C location indicated) with accompanying EGF-domain, cluster of 11 CR-domains, cluster of 6 3Fn-domains, a transmembrane domain followed by a cytoplasmic tail at the C-terminal end. (b) Three-dimensional model of the SORL1 YWTD-domain folding prepared from coordinates from ModelArchive (Y.Kitago, O.M. Andersen, G.A. Petsko. ModelArchive: https://modelarchiveorg/doi/10.5452/ma-agbg4). (c) Alignment of the ~40 amino acids from each of the six YWTD-repeated sequences corresponding to the blades of the β-propeller with indication of β-strands in grey. The arginine R953 resides at domain position 38 of the sequence located in the loop between strands C and D of the fifth β-blade. Partly conserved domain positions are indicated with bold letters and the consensus residues below the SORL1 alignment. Below 5 sequences of YWTD-repeated sequences from homologous receptor proteins with known pathogenic variants corresponding to arginines at position 38. (d) The side chain of Arg-953 from SORL1 provides structural stabilization of the domain folding by an ionic interaction with the side chain of Glu-943 based on the three-dimensional model of the folded YWTD-domain. (e) Close-up of the Arg-Glu pairs from YWTD-domain crystal structures for residues in LDLR, LRP4 and LRP6 (LRP5 homolog) corresponding to pathogenic variants as listed in panel c
In previous work25, we identified 5 pathogenic variants in homologous proteins corresponding to substitution of an arginine at the YWTD-blade sequence position 38: one in LDLR: p.R595WLDLR in Familial hypercholesterolemia (FHCL1) patients35; three in LRP5: p.R494QLRP5 in patients with Osteoporosis-pseudoglioma (OPPG)36,37; p.R752GLRP5 in a family with Exudative Vitreoretinopathy (EVR4)38; and p.R1188WLRP5, a variant that segregates with disease in large family pedigrees with Polycystic Liver Disease 4 (PCLD4)39; and one in LRP4: p.R632HLRP4 that is causal of sclerosteosis and shows impaired activity documented by functional assays40.
We recently prepared a three-dimensional model of the SORL1 ectodomain including its YWTD-domain using the AlphaFold2 algorithm10. Here, we used this model to investigate the functional role of the arginine side chain (Figure 3b, d). From this model it is observed that the positively charged amino group makes ionic contacts with the side chain of the glutamic acid residue at blade-sequence position 28 (E944 of SORL1) serving to position the long arginine side chain in place to make further hydrogen bonds to two backbone carbonyls in the preceding loop between blades (Figure 3d) thereby strongly contributing to the folding and the stability of the entire β-propeller domain. Interestingly, in four of the five blade-sequences containing the identified disease variants, a glutamic acid is similarly located at blade-sequence position 28 (Figure 3c).
Inspection of a larger alignment of YWTD-repeat sequences revealed that for most blade-sequences, a similar pattern is observed: when an arginine occupies blade-sequence at position 38, then a glutamate resides at blade-sequence position 2825, suggesting this pair of residues may generally be important for the folding of YWTD-domains.
The crystal structures of the YWTD-domains have previously been solved for LDLR41 and LPR442 including R595LDLR and R632LRP4, respectively. The structure of LRP5 has not been determined, but as the crystal structure of the highly homologous LRP6 has been solved43–45, it allowed us to use these YWTD-domain structures to gain insight in the functional role of the arginine side chain for the arginines at blade-sequence position 38 as well as for the LRP5 residues (R494LRP5/R481LRP6; R752LRP5/R739LRP6; R1188LRP5/R1178LRP6) (Figure 3e). Indeed, we found that the arginine side chains in each of the domains are binding backbone carbonyls in the n-1 linker, and for 4 of the 5 structures a salt bridge to a glutamic acid (at domain position 28) assist in keeping the arginine properly positioned to make the main chain interactions to the n-1 linker residue (Figure 3e). This supports a disease mechanism where substitution of the arginine may lead to domain misfolding and destabilization in general, and importantly also for R953 of SORL1.
R953C disrupts SORL1 ectodomain shedding from the cell surface
SORL1 is synthesized in the ER and goes through a complex cellular process of maturation during trafficking in the ER and out of the Golgi into the ELN compartments and to the cell surface. The mature SORL1 isoform has complex-type N-glycosylations, and we previously showed only mature N-glycosylated SORL1 is shed from the cell surface to produce a fragment called soluble SORL1 (sSORL1)46. Mature SORL1 migrates more slowly by SDS-PAGE, thus mature and immature isoforms of SORL1 can be clearly distinguished (Figure 4a). We have recently found that two pathogenic SORL1 missense variants associated with autosomal dominant familial AD are located in either one of the CR-domains (Bjarnadottir et al., Manuscript in preparation; Jensen et al., Manuscript in preparation) or the 3Fn-domains (Jensen et al., Manuscript in preparation) respectively, and both display significantly impaired maturation and shedding. We have also previously observed that sSORL1 is significantly decreased in the CSF from several carriers of other established pathogenic SORL1 variants (Andersen lab, unpublished data). Furthermore, a larger screen of 70 SORL1 coding variants suggested that impaired maturation may be general for dysfunctional proteins although no correlation to AD was established47.
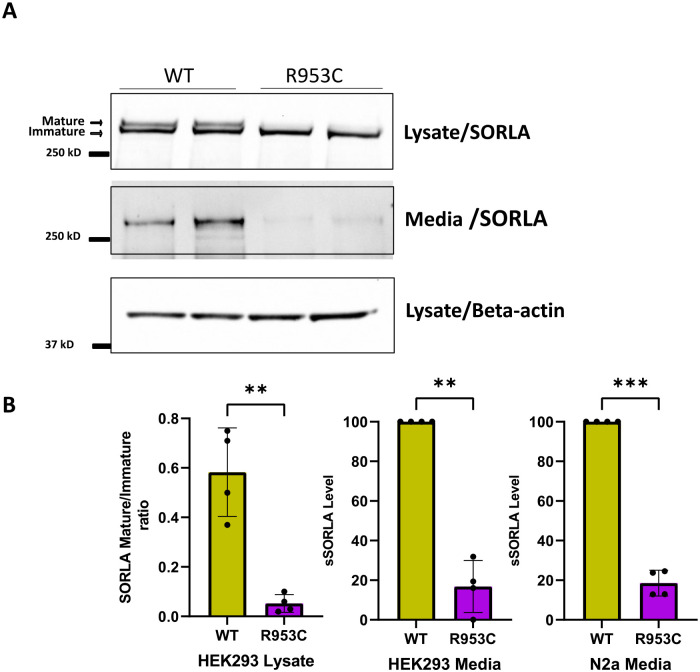
Figure 4. SORL1 R953C cells are defective in maturation and shedding of the SORL1 protein.
(a) Representative western blotting of lysate and media samples from HEK293 cells transiently transfected with SORLA-WT or SORLA-R953C. (b) Densitometric analysis from HEK293 and N2a samples. The signal for the R953C is expressed relative to the WT signal. Results are expressed as Mean± SD and analyzed by parametric two-tailed paired t-test. Significance was defined as a value of **p<0. 01, ***p<0. 001. n= 4 independent experiments
To test whether the p.R953C variant affects SORL1 maturation and shedding, we transfected HEK293 and N2a cells with either the WT SORL1 or a SORL1-R953C variant construct. We performed Western blot analysis to determine the ratio of the mature to immature forms of the protein. We observed significantly decreased levels of mature SORL1 in HEK293 cells transfected with the R953C variant (Figure 4a). We next measured the level of sSORL1 in the culture medium of HEK293 and N2a cells, transiently transfected with expression constructs for SORL1-WT or SORL1-R953C. Compared to cells transfected with WT construct, we observed ~ 80% reduction in the sSORL1 level in the media from both the tested cell types transfected with the R953C construct (Figure 4a, b)
R953C reduces cell surface expression of SORL1
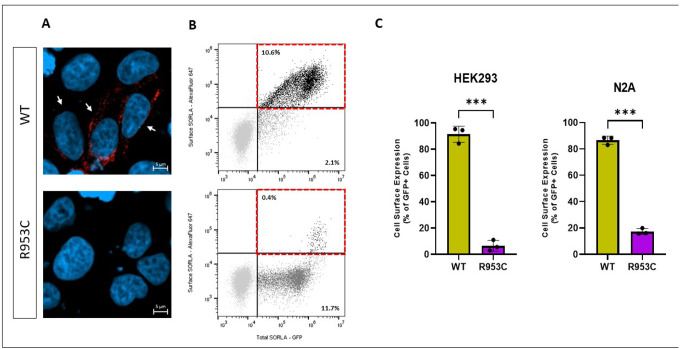
Because we observed a significant decrease in the shedding of SORL1-R953C, we tested whether the cell surface level of SORL1 could also be affected by this variant. We transiently transfected HEK293 cells with either WT SORL1 or SORL1-R953C and first analyzed cell surface levels of SORL1 using immunocytochemistry on unpermeabilized cells, which keeps the membrane intact to allow visualization of SORL1 protein solely located at the cell membrane. Using confocal microscopy, we observed considerably fewer cells expressing SORL1 at the cell surface in cells transfected with SORL1-R953C compared to SORL1-WT (Figure 5a). To quantitatively evaluate cell surface expression of SORL1-R953C relative to the total expression of the receptor in each individual cell, we used flow cytometry. We inserted the R953C variant into a C-terminally GFP tagged SORL1 construct, allowing for the detection of total expression of the receptor in each individual cell. We transfected both HEK293 cells and N2A cells and performed subsequent immunostaining of the transfected cells with anti-sSORL1 primary antibody and an Alexa Flour 647 secondary antibody in the absence of detergent to detect the cell surface expression of the receptor. These experiments demonstrated that more than 80% of the SORL1-R953C cells partially or completely retained SORL1 expression intracellularly compared to ~10–15% of the SORL1-WT cells. Results were consistent in both HEK293 and N2a cells (Figure 5b–c).
Figure 5. SORL1 R953C cells have reduced SORL1 protein localization on the cell surface.
(A) Representative immunocytochemistry from HEK293 cells transiently transfected with SORLA-WT or SORLA-R953C expression construct and stained for SORLA (red) at the cell surface. White arrows show positive cells. (B) Flow cytometry dot plot showing surface (AlexaFluor 647 fluorescence) and total (GFP fluorescence) in live single HEK293 cells expressing WT-GFP and R953C-GFP. Vertical and horizontal lines represent thresholds for GFP and AlexaFluor 647-positive cells, respectively. Represented are GFP-positive cells with AlexaFluor 647 signal above (black, inside red dashed gate) or below threshold (dark grey); and untransfected cells (light grey). Numbers in the plots represent the percentages of the cells inside the gates. (C) Bar plots of AlexaFluor 647 fluorescence in HEK293 and N2A cells expressing WT-GFP or R953C-GFP, generated from population of GFP-positive cells. n=3 independent experiments. Significance was defined as a value of ***p<0. 001.
R953C prevents SORL1 from entering the endosomal sorting pathway
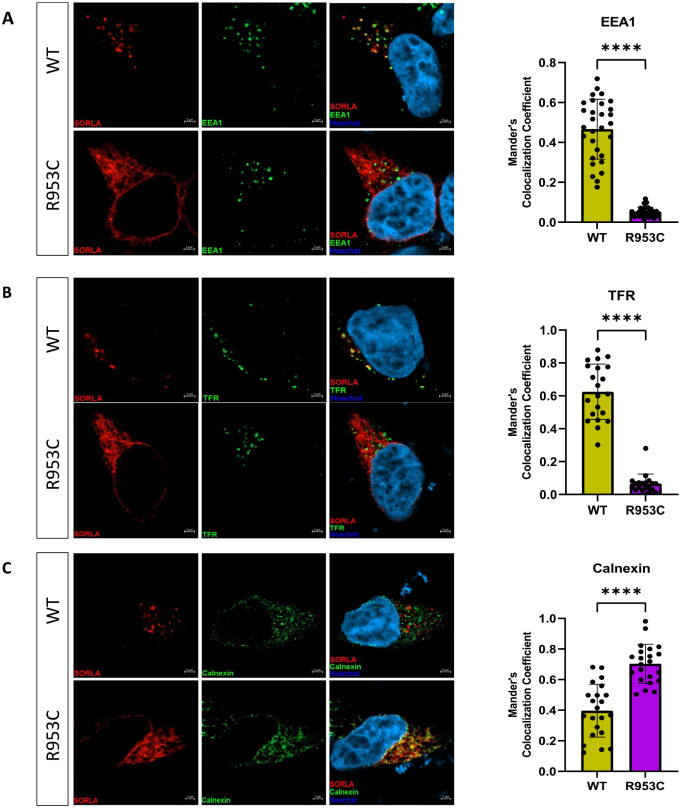
The differential cell surface localization and shedding of the R953C variant compared to WT led us to next investigate for possible changes in the intracellular localization of SORL1. For these experiments we transiently transfected HEK293 cells with either SORL1-WT or SORL1-R953C constructs. We analyzed co-localization of WT and R953C with two well-established endosomal markers, EEA1 (early endosome marker) and TFR (recycling endosome marker) and the ER marker Calnexin 24 hours post-transfection. Using confocal microscopy, we demonstrated that the colocalization of R953C is strongly reduced with both endosomal markers (Figure 6a–b) and significantly increased in the ER (Figure 6c). Taken together, these data suggest that the R953C variant severely disrupts the normal cellular localization trafficking of SORL1 as would be expected if the mutation leads to defective protein folding.
Figure 6. SORL1 R953C cells have reduced localization of the SORL1 protein in early and recycling endosomes.
HEK293 cells transiently expressing WT or R953C (red) are shown for their colocalization with (a) EEA1 (early endosomal marker), (b) TFR (recycling endosomal marker), and (c) Calnexin (ER marker) (green). The nuclei were visualized with Hoechst (blue). Bar graphs on the right panel illustrate quantifications of colocalization between WT and R953C in cells co-stained for (a) EEA1, (b) TFR and (c) Calnexin. In all cases, the quantification of colocalization was represented as Mander’s correlation coefficient. 20–30 images per condition were analyzed. Data are shown as mean±SD and analyzed by parametric two-tailed unpaired t-test. Significance was defined as a value of ****p<0.0001.
Discussion
SORL1 is widely recognized as a strong AD risk gene. Rare frameshift and nonsense variants leading to premature termination codons are associated with highly penetrant AD, although less is known about the AD risk attributable to rare missense variants21,22,48. Here we provide clinicopathological, genetic, and functional data supporting pathogenicity of a novel rare SORL1 missense variant, p.(Arg953Cys) (R953C). This variant was discovered in a family with multi-generational incidence of AD, both early and late onset. Exome sequencing found that the four affected children all carried the SORL1 R953C variant. Tissue from the mother analyzed by Sanger sequencing was found not to harbor R953C and tissue from the father was not available to confirm whether the allele was paternally inherited. Of note one living member of the family has been genotyped and is found to carry the variant (III-6) but is younger than the range of age of onset for the family. It is unknown whether her 3-year course of spasticity is related to the SORL1 variant or is an unrelated case of motor neuron disease.
Neuropathology examination shows the presence of severe AD pathology, including extensive plaque and neurofibrillary tangle distribution. These histologic features typically correlate with advanced clinical disease and are considered sufficient to explain cognitive changes31,33. Interestingly, the presence of LATE-NC correlated exactly with the SORL1 R953C carrier status in this small sample, evidenced by its presence in all generation II individuals (positive of the SORL1 variant) and absence in case I-2 (negative of the SORL1 variant). Although LATE-NC is a common co-pathology identified in AD, the underlying etiology of this TDB-43 pathology is not well understood. Age seems to be the strongest risk factor and it is most frequently noticed in individuals older than 80 years34,49. Similar to other age-related neuropathologic changes, LATE-NC frequently co-occurs with other pathologies such as AD and/or hippocampal sclerosis50, and its presence may accelerate the cognitive decline associated with these disorders51. The cases presented here are particularly interesting in that LATE-NC was seen only in SORL1 R953C carriers, who presented considerably younger than their non-carrier mother without LATE-NC. In a recent meta-analysis of datasets with concurrent neuropathology and genotype data, a number of dementia-associated single nucleotide variants were associated with non-AD pathologies, including a link between variants in SORL1 and LATE-NC. Further analyses suggest that certain SORL1 variants may be specifically linked to the combined AD+LATC-NC phenotype52. Additionally, Lewy body disease (LBD) was frequently observed in SORL1 R953C carriers, though the specific distribution patterns were more varied than that observed with TDP-43 pathology (Supplemental Figure 1). While LBD limited to the amygdala is frequently observed in association with advanced ADNC31,33, other LBD patterns are considerably less common. Together, these co-pathologies suggest that SORL1 R953C may be mechanistically linked to multiple proteinopathies, clinically manifesting as AD but also impacting TDP-43 and α-synuclein histopathology.
The SORL1 protein, SORL1/SORLA, is an endosomal sorting receptor. Decreased SORL1 levels are known to be pathogenic as truncation variants are so far only found in AD cases20,21,48. Furthermore, human neuronal models of SORL1 deficiency show impairments in endosomal trafficking and recycling11,53,54 as does minipigs with only one functional SORL1 allele55. One main function of SORL1 is to sort cargo from the early endosome to either the recycling pathway (cell surface) or the retrograde pathway (TGN) in conjunction with the multi-protein complex retromer14,56. The cellular localization of SORL1 and the cargo it binds depend on the specific isoform: monomer vs. dimer, mature vs. immature. For protein maturation, SORL1 transits through the Golgi and the trans-Golgi network to the cell surface. This process involves glycosylation and may lead to shedding, changes in which can be detected by SDS-PAGE and Western blotting for the full-length protein in cell lysates or cell medium. Here, we demonstrate that the R953C variant of SORLA does not undergo maturation and is not shed from the cell surface. Furthermore, it is likely that this variant cannot function as a normal endosomal receptor, as it fails to enter the endosomal pathway. Instead, it is sequestered in the ER. This in turn may increase the pathogenicity as the variant receptor might still dimerize with the wild-type receptor, thus sequestering more normal SORL1 in the ER as well, potentially acting via a dominant negative mechanism. Structural analysis indicates that this variant occurs at a critical arginine in the YWTD β-propeller domain of SORL1 that appears to be necessary for the proper folding of the domain. When compared against homologous domains in the LDLR receptor family, arginine substitutions at this position strongly suspected to be pathogenic.
Over 500 variants in SORL1 have been identified and recent genetic studies have provided evidence as to which variants may be likely pathogenic or likely benign20. However, with such a large gene (over 2200 amino acids), more variants are likely to be identified. Functional analysis of SORL1 variants will be an important tool to classify these variants based on their cellular pathogenicity and further uncover their contribution to the development of AD.
Acknowledgements
The authors are grateful to the families whose participation made this work possible.
Flow cytometry was performed at the FACS Core Facility, Aarhus University, Denmark. The authors acknowledge AU Health Bioimaging Core Facility for the use of equipment and support of the imaging facility.
The authors thank the members of the University of Washington Medicine Center for Precision Diagnostics for technical support, the Geriatric Research, Education, and Clinical Center at the VA Puget Sound Health Care System, and University of Washington’s Alzheimer Disease Research Center.
We acknowledge Shannon Rose and Harald Frankowski in the Young lab for preparation of gDNA samples for SORL1 variant sequencing and all members of the Young lab for helpful discussions on this work.
Funding
O.M.A is supported by Novo Nordisk Foundation (#NNF20OC0064162), the Alzheimer’s Association (ADSF-21-831378-C), the EU Joint Programme-Neurodegenerative Disease Research (JPND) Working Group SORLA-FIX under the 2019 “Personalized Medicine” call (funded in part by the Danish Innovation Foundation and the Velux Foundation Denmark), and the Danish Alzheimer’s Research Foundation (recipient of the 2022 Basic Research Science Award).
J.E.Y is supported by NIH grants R01 AG062148, K01 AG059841; an Alzheimer’s Association Research Grant 23AARG1022491; a Sponsored Research Agreement from Retromer Therapeutics and a generous gift from the Ellison Foundation (to UW).
D.D.C. is supported by the Alzheimer’s Disease Training Program (ADTP): T32 AG052354-06A1 Clinical and pathological work is supported by the Alzheimer’s Disease Research Center (P30 AG05136)
Funding Statement
O.M.A is supported by Novo Nordisk Foundation (#NNF20OC0064162), the Alzheimer’s Association (ADSF-21-831378-C), the EU Joint Programme-Neurodegenerative Disease Research (JPND) Working Group SORLA-FIX under the 2019 “Personalized Medicine” call (funded in part by the Danish Innovation Foundation and the Velux Foundation Denmark), and the Danish Alzheimer’s Research Foundation (recipient of the 2022 Basic Research Science Award).
J.E.Y is supported by NIH grants R01 AG062148, K01 AG059841; an Alzheimer’s Association Research Grant 23AARG1022491; a Sponsored Research Agreement from Retromer Therapeutics and a generous gift from the Ellison Foundation (to UW).
D.D.C. is supported by the Alzheimer’s Disease Training Program (ADTP): T32 AG052354-06A1 Clinical and pathological work is supported by the Alzheimer’s Disease Research Center (P30 AG05136)
Footnotes
Competing Interests
O.M.A. is a consultant for Retromer Therapeutics and has equity. The other authors report no competing interests.
References
- 1.Jayadev S. Genetics of Alzheimer Disease. Continuum (Minneap Minn). 2022;28(3):852–871. [DOI] [PubMed] [Google Scholar]
- 2.Tosto G, Reitz C. Genome-wide association studies in Alzheimer’s disease: a review. Curr Neurol Neurosci Rep. 2013;13(10):381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jacobsen L, Madsen P, Jacobsen C, Nielsen MS, Gliemann J, Petersen CM. Activation and functional characterization of the mosaic receptor SorLA/LR11. J Biol Chem. 2001;276(25):22788–22796. [DOI] [PubMed] [Google Scholar]
- 4.Jacobsen L, Madsen P, Moestrup SK, et al. Molecular characterization of a novel human hybrid-type receptor that binds the alpha2-macroglobulin receptor-associated protein. J Biol Chem. 1996;271(49):31379–31383. [DOI] [PubMed] [Google Scholar]
- 5.Willnow TE, Petersen CM, Nykjaer A. VPS10P-domain receptors - regulators of neuronal viability and function. Nat Rev Neurosci. 2008;9(12):899–909. [DOI] [PubMed] [Google Scholar]
- 6.Yamazaki H, Bujo H, Saito Y. A novel member of the LDL receptor gene family with eleven binding repeats is structurally related to neural adhesion molecules and a yeast vacuolar protein sorting receptor. J Atheroscler Thromb. 1997;4(1):20–26. [DOI] [PubMed] [Google Scholar]
- 7.Yamazaki H, Bujo H, Kusunoki J, et al. Elements of neural adhesion molecules and a yeast vacuolar protein sorting receptor are present in a novel mammalian low density lipoprotein receptor family member. J Biol Chem. 1996;271(40):24761–24768. [DOI] [PubMed] [Google Scholar]
- 8.Dumanis SB, Burgert T, Caglayan S, et al. Distinct Functions for Anterograde and Retrograde Sorting of SORLA in Amyloidogenic Processes in the Brain. J Neurosci. 2015;35(37):12703–12713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herskowitz JH, Offe K, Deshpande A, Kahn RA, Levey AI, Lah JJ. GGA1-mediated endocytic traffic of LR11/SorLA alters APP intracellular distribution and amyloid-beta production. Mol Biol Cell. 2012;23(14):2645–2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jensen AMG, Kitago Y, Fazeli E, et al. Dimerization of the Alzheimer’s disease pathogenic receptor SORLA regulates its association with retromer. Proc Natl Acad Sci U S A. 2023;120(4):e2212180120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mishra S, Knupp A, Szabo MP, et al. The Alzheimer’s gene SORL1 is a regulator of endosomal traffic and recycling in human neurons. Cell Mol Life Sci. 2022;79(3):162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simoes S, Guo J, Buitrago L, et al. Alzheimer’s vulnerable brain region relies on a distinct retromer core dedicated to endosomal recycling. Cell Rep. 2021;37(13):110182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daly JL, Danson CM, Lewis PA, et al. Multi-omic approach characterises the neuroprotective role of retromer in regulating lysosomal health. Nat Commun. 2023;14(1):3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fjorback AW, Seaman M, Gustafsen C, et al. Retromer binds the FANSHY sorting motif in SorLA to regulate amyloid precursor protein sorting and processing. J Neurosci. 2012;32(4):1467–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reitz C. Retromer Dysfunction and Neurodegenerative Disease. Curr Genomics. 2018;19(4):279–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kunkle BW, Grenier-Boley B, Sims R, et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Abeta, tau, immunity and lipid processing. Nat Genet. 2019;51(3):414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lambert JC, Ibrahim-Verbaas CA, Harold D, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45(12):1452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reitz C, Cheng R, Rogaeva E, et al. Meta-analysis of the association between variants in SORL1 and Alzheimer disease. Arch Neurol. 2011;68(1):99–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rogaeva E, Meng Y, Lee JH, et al. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat Genet. 2007;39(2):168–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holstege H, Hulsman M, Charbonnier C, et al. Exome sequencing identifies rare damaging variants in ATP8B4 and ABCA1 as risk factors for Alzheimer’s disease. Nat Genet. 2022;54(12):1786–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holstege H, van der Lee SJ, Hulsman M, et al. Characterization of pathogenic SORL1 genetic variants for association with Alzheimer’s disease: a clinical interpretation strategy. Eur J Hum Genet. 2017;25(8):973–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pottier C, Hannequin D, Coutant S, et al. High frequency of potentially pathogenic SORL1 mutations in autosomal dominant early-onset Alzheimer disease. Mol Psychiatry. 2012;17(9):875–879. [DOI] [PubMed] [Google Scholar]
- 23.Raghavan NS, Brickman AM, Andrews H, et al. Whole-exome sequencing in 20,197 persons for rare variants in Alzheimer’s disease. Ann Clin Transl Neurol. 2018;5(7):832–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verheijen J, Van den Bossche T, van der Zee J, et al. A comprehensive study of the genetic impact of rare variants in SORL1 in European early-onset Alzheimer’s disease. Acta Neuropathol. 2016;132(2):213–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andersen OM, Monti G, Jensen A.M.G., de Waal M., Hulsman M., Olsen J.G., Holstege H. Relying on the relationship with known disease-causing variants in homologus proteins to predict pathogenicity of SORL1 variants in Alzheimer’s disease. BioRxiv. 2023; 10.1101/2023.02.27.524103. [DOI] [Google Scholar]
- 26.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cingolani P, Platts A, Wang le L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012;6(2):80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee EG, Tulloch J, Chen S, et al. Redefining transcriptional regulation of the APOE gene and its association with Alzheimer’s disease. PLoS One. 2020;15(1):e0227667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montine TJ, Phelps CH, Beach TG, et al. National Institute on Aging-Alzheimer’s Association guidelines for the neuropathologic assessment of Alzheimer’s disease: a practical approach. Acta Neuropathol. 2012;123(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bell MD, Long T, Roden AC, et al. Updating Normal Organ Weights Using a Large Current Sample Database. Arch Pathol Lab Med. 2022;146(12):1486–1495. [DOI] [PubMed] [Google Scholar]
- 33.Hyman BT, Phelps CH, Beach TG, et al. National Institute on Aging-Alzheimer’s Association guidelines for the neuropathologic assessment of Alzheimer’s disease. Alzheimers Dement. 2012;8(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nelson PT, Dickson DW, Trojanowski JQ, et al. Limbic-predominant age-related TDP-43 encephalopathy (LATE): consensus working group report. Brain. 2019;142(6):1503–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santos PC, Morgan AC, Jannes CE, et al. Presence and type of low density lipoprotein receptor (LDLR) mutation influences the lipid profile and response to lipid-lowering therapy in Brazilian patients with heterozygous familial hypercholesterolemia. Atherosclerosis. 2014;233(1):206–210. [DOI] [PubMed] [Google Scholar]
- 36.Gong Y, Slee RB, Fukai N, et al. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. 2001;107(4):513–523. [DOI] [PubMed] [Google Scholar]
- 37.Ai M, Heeger S, Bartels CF, Schelling DK, Osteoporosis-Pseudoglioma Collaborative G. Clinical and molecular findings in osteoporosis-pseudoglioma syndrome. Am J Hum Genet. 2005;77(5):741–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiao X, Ventruto V, Trese MT, Shastry BS, Hejtmancik JF. Autosomal recessive familial exudative vitreoretinopathy is associated with mutations in LRP5. Am J Hum Genet. 2004;75(5):878–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cnossen WR, te Morsche RH, Hoischen A, et al. Whole-exome sequencing reveals LRP5 mutations and canonical Wnt signaling associated with hepatic cystogenesis. Proc Natl Acad Sci U S A. 2014;111(14):5343–5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huybrechts Y, Boudin E, Hendrickx G, et al. Identification of Compound Heterozygous Variants in LRP4 Demonstrates That a Pathogenic Variant outside the Third beta-Propeller Domain Can Cause Sclerosteosis. Genes (Basel). 2021;13(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rudenko G, Henry L, Henderson K, et al. Structure of the LDL receptor extracellular domain at endosomal pH. Science. 2002;298(5602):2353–2358. [DOI] [PubMed] [Google Scholar]
- 42.Zong Y, Zhang B, Gu S, et al. Structural basis of agrin-LRP4-MuSK signaling. Genes Dev. 2012;26(3):247–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahn VE, Chu ML, Choi HJ, Tran D, Abo A, Weis WI. Structural basis of Wnt signaling inhibition by Dickkopf binding to LRP5/6. Dev Cell. 2011;21(5):862–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bourhis E, Wang W, Tam C, et al. Wnt antagonists bind through a short peptide to the first beta-propeller domain of LRP5/6. Structure. 2011;19(10):1433–1442. [DOI] [PubMed] [Google Scholar]
- 45.Cheng Z, Biechele T, Wei Z, et al. Crystal structures of the extracellular domain of LRP6 and its complex with DKK1. Nat Struct Mol Biol. 2011;18(11):1204–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Christensen S, Narimatsu Y., Simoes S., Goth CK., Vaegter CB., Small SA., Clausen H., Andersen OM. Endosomal trafficking is required for glycosylation and normal maturation of the Alzheimer’s-associated protein sorLA. BioRxiv. 2020. [Google Scholar]
- 47.Rovelet-Lecrux A, Feuillette S, Miguel L, et al. Impaired SorLA maturation and trafficking as a new mechanism for SORL1 missense variants in Alzheimer disease. Acta Neuropathol Commun. 2021;9(1):196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scheltens P, De Strooper B, Kivipelto M, et al. Alzheimer’s disease. Lancet. 2021;397(10284):1577–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nelson PT, Lee EB, Cykowski MD, et al. LATE-NC staging in routine neuropathologic diagnosis: an update. Acta Neuropathol. 2023;145(2):159–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Amador-Ortiz C, Lin WL, Ahmed Z, et al. TDP-43 immunoreactivity in hippocampal sclerosis and Alzheimer’s disease. Ann Neurol. 2007;61(5):435–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Josephs KA, Whitwell JL, Tosakulwong N, et al. TAR DNA-binding protein 43 and pathological subtype of Alzheimer’s disease impact clinical features. Ann Neurol. 2015;78(5):697–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Katsumata Y, Shade LM, Hohman TJ, et al. Multiple gene variants linked to Alzheimer’stype clinical dementia via GWAS are also associated with non-Alzheimer’s neuropathologic entities. Neurobiol Dis. 2022;174:105880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hung C, Tuck E, Stubbs V, et al. SORL1 deficiency in human excitatory neurons causes APP dependent defects in the endolysosome-autophagy network. Cell Rep. 2021;35(11):109259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Knupp A, Mishra S, Martinez R, et al. Depletion of the AD Risk Gene SORL1 Selectively Impairs Neuronal Endosomal Traffic Independent of Amyloidogenic APP Processing. Cell Rep. 2020;31(9):107719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Andersen OM, Bogh N, Landau AM, et al. A genetically modified minipig model for Alzheimer’s disease with SORL1 haploinsufficiency. Cell Rep Med. 2022;3(9):100740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Small SA, Kent K, Pierce A, et al. Model-guided microarray implicates the retromer complex in Alzheimer’s disease. Ann Neurol. 2005;58(6):909–919. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and/or its supplementary material or available from the corresponding author upon reasonable request.