Abstract
Tumor-reactive CD8 T cells found in cancer patients are frequently dysfunctional, unable to halt tumor growth. Adoptive T cell transfer (ACT), the administration of large numbers of in vitro-generated cytolytic tumor-reactive CD8 T cells, is an important cancer immune therapy being pursued. However, a limitation of ACT is that transferred CD8 T cells often rapidly lose effector function, and despite exciting results in certain malignancies, few ACT clinical trials have shown responses in solid tumors. Here, we developed preclinical cancer mouse models to investigate if and how tumor-specific CD4 T cells can be enlisted to overcome CD8 T cell dysfunction in the setting of ACT. In situ confocal microscopy of color-coded cancer cells, tumor-specific CD8 and CD4 T cells, and antigen presenting cells (APC), combined with functional studies, revealed that the spatial positioning and interactions of CD8 and CD4 T cells, but not their numbers, dictates ACT efficacy and anti-tumor responses. We uncover a new role of antigen-specific CD4 T cells in addition to the known requirement for CD4 T cells during priming/activation of naïve CD8 T cells. CD4 T cells must co-engage with CD8 T cells and APC cross-presenting CD8- and CD4-tumor antigens during the effector phase, forming a three-cell-cluster (triad), to license CD8 T cell cytotoxicity and mediate cancer cell elimination. Triad formation transcriptionally and epigenetically reprogram CD8 T cells, prevent T cell dysfunction/exhaustion, and ultimately lead to the elimination of large established tumors and confer long-term protection from recurrence. When intratumoral triad formation was disrupted, adoptively transferred CD8 T cells could not be reprogrammed, and tumors progressed despite equal numbers of tumor-infiltrating CD8 and CD4 T cells. Strikingly, the formation of CD4 T cell::CD8 T cell::APC triads in tumors of patients with lung cancers treated with immune checkpoint blockade was associated with clinical responses, but not CD4::APC dyads or overall numbers of CD8 or CD4 T cells, demonstrating the importance of triads in non-ACT settings in humans. Our work uncovers intratumoral triads as a key requirement for anti-tumor immunity and a new role for CD4 T cells in CD8 T cell cytotoxicity and cancer cell eradication.
INTRODUCTION
CD8 T cells are powerful components of the adaptive immune system that have the potential to selectively eradicate cancer cells. However, despite the presence of tumor-specific CD8 T cells in tumor-bearing hosts, cancers develop, suggesting that CD8 T cells become dysfunctional and unresponsive to cancer cells over the course of tumorigenesis [1]. Tumor-infiltrating dysfunctional CD8 T cells (also referred to as ‘exhausted’ T cells) commonly express high levels of inhibitory receptors (PD1, LAG3, CTLA4, TIM3) and fail to produce effector cytokines (interferon-γ (IFN-γ), tumor necrosis factor-α (TNF-α)) and cytotoxic molecules (granzymes, perforin). These hallmarks of CD8 T cell dysfunction/exhaustion have been attributed to chronic tumor antigen encounter/TCR signaling and immunosuppressive signals within the tumor microenvironment [1–3].
Adoptive T cell transfer (ACT), the infusion of large numbers (> 109 – 1010 CD8 T cells/patient) of tumor-reactive cytolytic effector CD8 T cells into cancer patients, has emerged as a powerful therapeutic strategy for the treatment of cancers [4]. Tumor-reactive CD8 T cells can either be isolated from patients’ own tumors (tumor-infiltrating lymphocytes (TIL)) or blood, expanded ex vivo and infused back, or engineered in vitro to become tumor-reactive through the introduction of genes encoding T cell receptors (TCR) or chimeric antigen receptors (CAR) specific for tumor antigens [5–11]. Although remarkable successes with ACT have been observed in a subset of cancer patients and cancer types (e.g. leukemia, lymphoma, and melanoma) [12–14], most patients still fail to achieve long-term responses, especially those with (nonmelanoma) solid tumors. Factors which mitigate the efficacy of adoptively transferred CD8 T cells include poor in vivo persistence, poor tumor localization/infiltration, and rapid loss of effector function [13, 15, 16]. Various therapeutic strategies have been identified to improve persistence and tumor infiltration, such as lymphodepletion and/or administration of homeostatic cytokines (IL-2, IL-7, IL-15) [12, 15, 17–21]. However, the loss of effector function of CD8 T cells remains a major roadblock [22, 23]. Thus, the development of immunotherapeutic interventions to prevent or reverse CD8 T cell dysfunction/exhaustion has become the concerted effort of many clinicians and scientists.
While direct cytotoxic activity against cancer cells generally resides within the CD8 T cell compartment, various modes of action have been described for CD4 T cells [24]: (1) productive priming of naïve CD8 T cells in lymphoid tissues through “licensing” and functional maturation of dendritic cells (DC) [25–31], (2) anti-tumor effector functions and elimination of MHC class II-negative cancer cells without CD8 T cells [32–36] through IFN-γ acting on the host stroma, or activation of macrophages and other non-lymphoid tumoricidal effector cells [35, 37–42], and (3) induction of cancer cell senescence rather than cancer cell elimination through the secretion of Th1-cytokines (TNFα, IFNγ) [43, 44]. Moreover, we and others have demonstrated that CD4 T cells might play an important role during CD8 T cell-mediated tumor elimination as well as during autoimmune tissue destruction, however, the mechanisms remained elusive [45–47]. MHC class II-restricted tumor antigens and tumor-specific CD4 T cells have been identified in many cancer patients and cancer types, and their importance in anti-tumor immunity has been recognized [24, 32, 48–52]. If and how tumor-reactive CD4 T cells can be utilized to prevent or reverse CD8 T cell dysfunction/exhaustion leading to tumor eradication is not known. To address this question, we developed a clinically relevant ACT-cancer mouse model. We demonstrate that CD4 T cells mediate tumor-specific CD8 T cell reprogramming within large solid tumors when tumor-reactive CD4 and CD8 T cells form three-cell-type clusters (triads) together with antigen-presenting cells (APC). Triad-formation resulted in the molecular and functional reprogramming of adoptively transferred CD8 T cells, preventing and even reversing T cell exhaustion, leading to tumor destruction. Strikingly, the formation of CD4 T cell-CD8 T cell-APC triads in tumors of patients with mesothelioma treated with immune checkpoint blockade (ICB) was associated with clinical responses, uncovering CD4 T cell-CD8 T cell-APC triads as a key determinant for cancer elimination and ACT therapy efficacy against solid tumors.
RESULTS
Tumor-specific CD4 T cells reverse tumor-specific CD8 T cell dysfunction/exhaustion in solid tumors
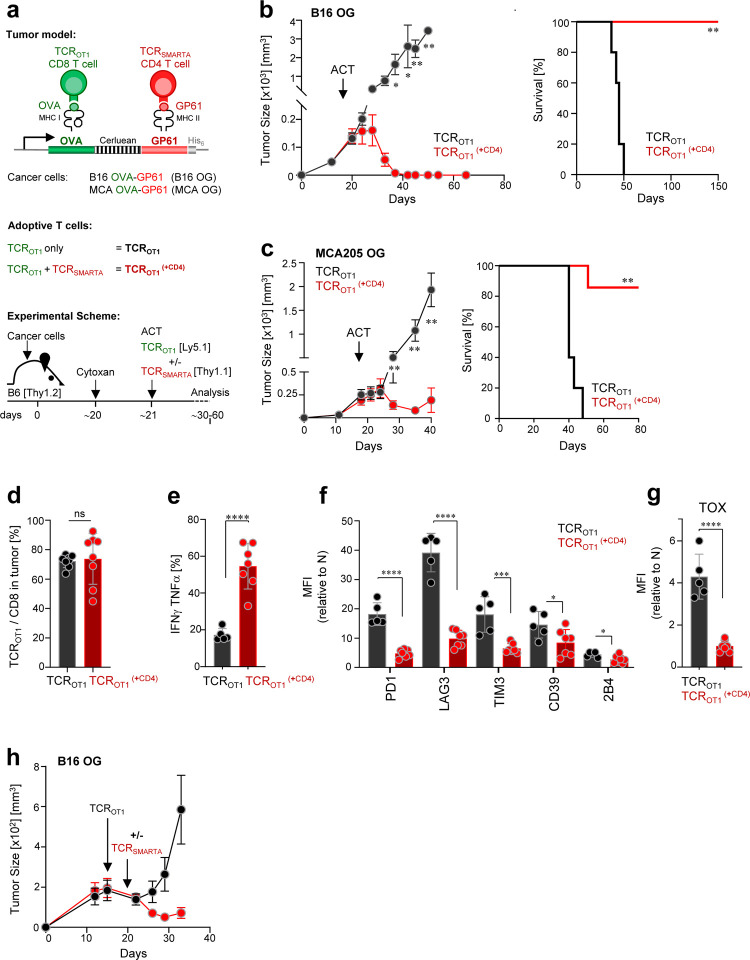
B16 is a highly aggressive murine melanoma cell line; B16 cancer cells injected subcutaneously into immunocompetent C57BL/6 wildtype mice (B6 WT) form large established tumors within 2 weeks, ultimately killing the host, and treatment regiments are generally ineffective. We engineered B16 cancer cells to express the CD8 T cell–recognized epitope from ovalbumin OVA257–264 (SIINFEKL) as well as the CD4 T cell-recognized glycoprotein epitope GP61–80 (GLKGPDIYKGVYQFKSVEFD) from the lymphocytic choriomeningitis virus (LCMV); the vector was constructed to encode the trimeric peptide sequence (SIINFEKL-AAY)3 fused to the fluorescent protein Cerulean, followed by the 19-mer GP61–80 peptide (Fig.1a). The OVA257–264 epitope is presented on the MHC class I molecule H-2Kb and recognized by TCR transgenic OT1 CD8 T cells (TCROT1); the GP61–80 epitope is presented on the MHC class II I-Ab molecule and recognized by TCR transgenic SMARTA CD4 T cells (TCRSMARTA). B16-OVA257–264-GP61–80 cancer cells (B16-OG; 2.5 ×106 cells/host) were injected subcutaneously into B6 WT (CD45.2) mice. Despite the expression of strong CD8- and CD4-T cell tumor antigens, B16-OG tumors grew aggressively, forming large tumors within 2 weeks (Fig. 1b). We then employed an adoptive T cell transfer (ACT) regimen modeled on that used in cancer patients treated with ACT: preconditioning the host and inducing lymphopenia through a nonmyeloablative chemotherapeutic dose of cyclophosphamide followed by the infusion of in vitro generated cytotoxic tumor-specific CD8 T cells (Fig. 1a). Naïve congenic (CD45.1) TCROT1 were activated in vitro for 3–4 days and adoptively transferred into lymphopenic B16-OG tumorbearing mice. Despite the infusion of highly functional effector TCROT1 CD8 T cells, B16-OG tumors progressed, recapitulating the scenario commonly observed in patients with solid tumors receiving ACT (Fig. 1b). Next, we asked whether the simultaneous infusion of in vitro activated effector TCRSMARTA CD4 T cells would mediate anti-tumor responses. Co-transfer of effector TCROT1 together with TCRSMARTA resulted in complete tumor elimination, with 100% long-term tumor-free survival (Fig. 1b). Tumor-bearing mice that received TCRSMARTA alone did not show tumor regression (data not shown), demonstrating that cancer elimination was dependent on both TCROT1 and TCRSMARTA T cells. We confirmed our results in a second tumor model using the fibrosarcoma cell line MCA205 (MCA205-OG) and obtained similar results (Fig. 1c).
Figure 1 |. Tumor-specific CD4 T cells prevent and reverse CD8 T cell dysfunction/exhaustion within solid tumors and mediate tumor elimination.
a. Scheme: tumor models, adoptively transferred effector T cells, and experimental schemes. b. B16 OVA-GP61–80 (B16-OG) tumor growth (right) and Kaplan–Meier survival curve (left) of tumor-bearing B6 WT mice (CD45.2; Thy1.2) receiving effector TCROTI CD8 T cells alone (CD45.1) (black; TCROT1) or together with TCRSMARTA CD4 T cells (Thy1.1) (red; TCROT1(+CD4)) (ACT = adoptive T cell transfer). Data is representative of 5 independent experiments (n=5 mice/group). Values are mean ± SEM. Significance is calculated by multiple t test. Kaplan–Meier curve; **p=0.00021; Mantel–Cox test. c. MCA205 OVA-GP61–80 (MCA-OG) tumor outgrowth and survival in B6 mice treated as described in b; **p=0.0003; Mantel–Cox test. Data is representative of 2 independent experiments (n=5–6 mice/group). d. TCROTI (% of total of CD8+ T cells) within progressing B16-OG tumors 8–9 days post transfer +/− TCRSMARTA CD4 T cells. Data pooled from 2 independent experiments (n=8 mice/group). Each symbol represents an individual mouse. e. IFNγ and TNFα production of TCROTI isolated from B16-OG tumors 8–9 days post transfer +/− TCRSMARTA CD4 T cells. Cytokine production was assessed after 4-hr peptide stimulation ex vivo. Data show 2 pooled independent experiments (n=5–7). f. Inhibitory receptor expression, and g. TOX expression of B16-OG tumor-infiltrating TCROTI isolated 8–9 days post transfer +/− TCRSMARTA. Graphs depict relative MFI normalized to naive TCROTI; two pooled independent experiments (n=5–7mice/group). h. Mice with B16-OG tumors received effector TCROTI CD8 T cells 14 days post tumor transplantation; 9 days later, TCRSMARTA CD4 T cells were adoptively transferred (red); B16-OG tumor growth in mice receiving only TCROT1 are shown in black. Data is representative of 2 independent experiments (n=8 mice/group). Values are mean ± SEM. Significance is calculated by multiple t test.
CD4 T cells are known to enhance CD8 T cell mobilization into peripheral tissues [28]. To understand whether TCRSMARTA enhanced TCROT1 tumor infiltration, we compared the numbers of TCROT1 TIL in mice which received effector TCROT1 alone (TCROT1) or together with TCRSMARTA (TCROT1(+CD4)); we evaluated numbers of TIL 8–9 days post transfer, a time point when tumors are similar in size. Surprisingly, we found equal numbers of TCROT1 TIL in both cohorts (Fig. 1d), suggesting that TCRSMARTA-mediated anti-tumor immunity was not due to an enhancement of TCROT1 tumor infiltration but likely due to functional changes of TCROT1 TIL. Indeed, while TCROT1 TIL were impaired in their ability to produce the effector cytokines IFNγ and TNFα (Fig. 1e), expressed high levels of numerous canonical inhibitory receptors including PD1, LAG3, TIM3, CD39 and 2B4 (Fig. 1f), as well as the transcription factor TOX (Fig. 1g), a critical regulator associated with T cell exhaustion [53–58], TCROT1(+CD4) were able to produce high amounts of IFNγ and TNFα and showed little/no expression of inhibitory receptors and TOX (Fig. 1e–1g). To understand whether these phenotypic and functional differences were already induced in the tumor-draining lymph node (tdLN), we compared phenotype and function of tdLN-TCROT1 and tdLN-TCROT1(+CD4). Interestingly, no differences were observed (Suppl. Fig. 1), thus co-transferred CD4 T cells specifically acted on tumor-specific CD8 T cells within the tumor.
Next, we wanted to understand whether CD4 T cells could not only prevent but also reverse CD8 T cell dysfunction/exhaustion. We adoptively transferred effector TCROT1 into B16-OG tumor-bearing mice, and 10 days later, when TCROT1 TIL were dysfunctional/exhausted, we adoptively transferred effector TCRSMARTA. Remarkably, mice that received TCRSMARTA showed tumor regression while control cohorts did not (Fig. 1h). Thus, tumor-reactive TCRSMARTA CD4 T cells prevent and reverse tumor-induced CD8 T cell dysfunction and mediate tumor regression.
CD4 T cells transcriptionally and epigenetically reprogram tumor-specific CD8 T cells, leading to tumor elimination
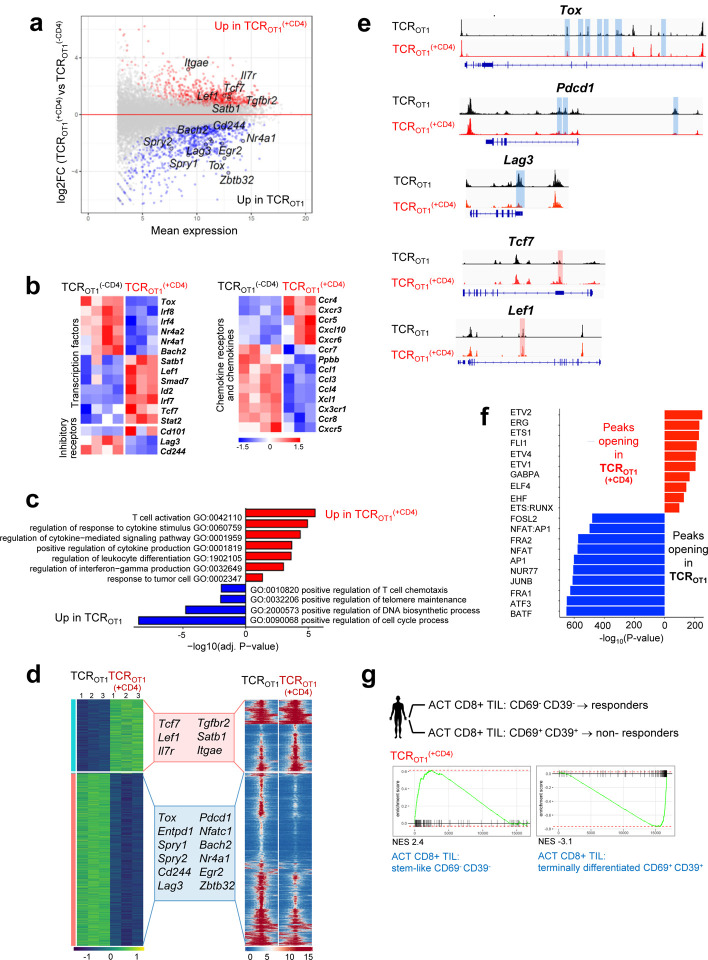
Tumor-specific CD8 T cell dysfunction in mice and humans is associated with global transcriptional and epigenetic dysregulation of genes and pathways important for T cell differentiation and function. To understand how CD4 T cells mediated functional rescue of TCROT1 CD8 T cells, we conducted RNA-seq and ATAC-seq of TCROT1(+CD4) and TCROT1 TIL isolated from size-matched B16-OG tumors 8 days post transfer. 1795 genes were differentially expressed (DEG) including exhaustion/dysfunction-associated TF and inhibitory receptors/activation markers (Tox, Irf4, Pdcd1 (PD1), Havcr2, Lag3, CD160, Cd244 (2B4)) (Fig. 2a and 2b), which were highly expressed in TCROT1. In contrast, TF and molecules associated with stem-like progenitor T cell states were enriched and highly expressed in TCROT1(+CD4) TIL, including genes encoding Tcf7 (TCF1), Il7r, Itgae (CD103), Itga1, and Ifitm3, as well as chemokine receptors such as Ccr5, Ccr4 and Ccr2 [30, 59]. Geneontology (GO) classification revealed that pathways associated with positive cytokine regulation, immune differentiation and responses to tumor cells were enriched in TCROT1(+CD4) but not in TCROT1 (Fig. 2c). ATAC-seq revealed 11,787 differentially accessible regions (DAR), including enhancers in many exhaustion (Tox, Spry1 Spry2, Cd244, Bach2, Egr2) or stem-/progenitor cell state-associated genes (Tcf7, IL7r, Lef1), respectively (Fig. 2d and 2e). Many enhancer peaks with TF motifs associated with terminal differentiation were less accessible in reprogrammed CD8 T cells, which was surprising given that TCROT1(+CD4) and TCROT1 TIL were isolated from equally sized tumors (Fig. 2f). To understand whether reprogrammed TCROT1(+CD4) revealed molecular signatures similar to human CD8 TIL driving clinical responses in the context of ACT, we utilized a data set from a study conducted by the Rosenberg group, using ex vivo-expanded autologous CD8+ TIL from metastatic melanoma lesions for ACT into preconditioned, lymphodepleted patients [60]. The authors identified a CD39-CD69- stem-like TIL subset that was associated with complete cancer regression in ACT-responders but lacking in ACT-non-responders. Gene set enrichment analysis (GSEA) revealed that the same genes were enriched in TCROT1(+CD4) CD8 TIL as in ACT (CD39-CD69-) CD8 TIL responders, and genes in CD8 TIL from ACT (CD39+CD69+) non-responders were enriched in TCROT1 CD8 TIL (Fig. 2g, Suppl. Fig. 2) [60].
Figure 2 |. Tumor-specific CD4 T cells transcriptionally and epigenetically reprogram tumor-specific CD8 T cells and prevent terminal differentiation/exhaustion.
a. MA plot of RNA-seq data showing the relationship between average expression and expression changes of TCROT1 and TCROT1(+CD4) TIL.Statistically significantly DEGs (false discovery rate (FDR) < 0.05) are shown in red and blue, with select genes highlighted for reference. b. Heat map of RNA-seq expression (normalized counts after variance stabilizing transformation, centered and scaled by row for DEGs) (FDR < 0.05) in TCROT1 and TCROT1(+CD4) TIL. c. Selected GO terms enriched for genes up-regulated in TCROT1 (blue) and TCROT1(+CD4) (red) TIL. d. Chromatin accessibility (ATAC-seq); (left) heatmap of log2-transformed normalized read counts transformed with variance stabilization per for regions with differential chromatin accessibility; (right) each row represents one peak (differentially accessible between TCROT1 and TCROT1(+CD4) TIL; FDR < 0.05) displayed over a 2-kb window centered on the peak summit; regions were clustered with k-means clustering. Genes associated with the two major clusters are highlighted. e. ATAC-seq signal profiles across the Tox, Pdcd1, Lag3, Tcf7, and Lef1 loci. Peaks significantly lost or gained are highlighted in red or blue, respectively. f. Top 10 most-significantly enriched transcription factor motifs in peaks with increased accessibility in TCROT1(+CD4) TIL (red) or TCROT1 TIL (blue). g. Enrichment of gene sets in TCROT1 and TCROT1 (+CD4), respectively, described for human tumor infiltrating (TIL) CD8 T cell subsets (CD69- CD39) stem-like CD8 T cells/TIL (responders) or (CD69+ CD39+) terminally differentiated CD8 T cells/TIL (non-responders) from metastatic melanoma patients receiving ex vivo expanded TIL for ACT (S. Krishna et al, Science 2020). TCROT1(+CD4) are enriched in genes observed in CD69- CD39- stem-like T cells/TIL from responders in contrast to TCROT1 which are positively enriched for genes in CD69+ CD39+ terminally differentiated CD8 T cells/TIL from non-responders. NES, normalized enrichment score.
Taken together, tumor-specific TCRSMARTA CD4 T cells transcriptionally and epigenetically reprogram tumor-reactive CD8 TIL within progressing tumors, preventing terminal differentiation and exhaustion, and resulting in tumor elimination.
Spatial positioning of tumor-specific CD8 and CD4 T cells within tumors determine anti-tumor immunity and cancer elimination
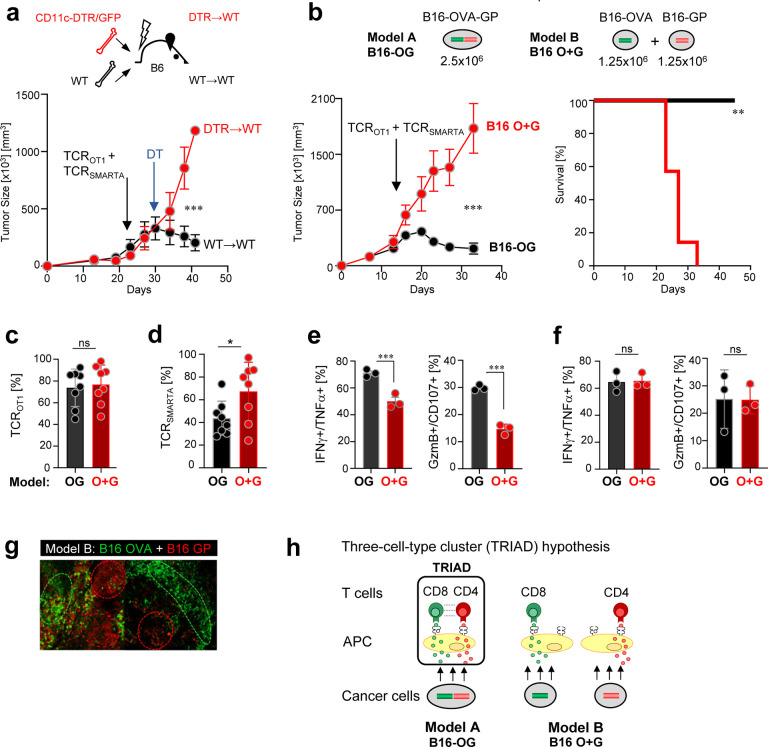
Next, we wanted to understand how TCRSMARTA CD4 T cells prevent CD8 T cell exhaustion within tumors. B16 tumor cells express low level MHC II in vivo (Suppl. Fig. 3a), thus cancer cells could become targets of CD4 T cells. Employing CRISPR/Cas9-mediated gene editing, we generated MHC class II I-Ab-deficient B16-OG cancer cells. Surprisingly, large established B16-OG I-Ab-deficient tumors were eliminated as efficiently as parental MHC class II-expressing B16-OG tumors, demonstrating that cancer elimination does not require CD4 T cell to directly target cancer cells (Suppl. Fig. 3b and 3c). Next, we turned to the tumor stroma, which includes MHC class I- and II-expressing antigen presenting cells (APC) such as CD11c+ dendritic cells (DC) and macrophages. To assess the role of CD11c+ cells, we employed a targeted depletion approach: CD11c+ DC from CD11c-DTR/GFP transgenic mice express the primate diphtheria toxin receptor (DTR) transgene under the CD11c promoter, enabling conditional depletion of CD11c+ cells in vivo upon DT treatment [61]. We generated bone marrow (BM) chimeras by transferring BM cells from CD11c-DTR/GFP (CD11c-DTR) or littermate control (WT) mice into lethally irradiated WT (CD45.1) B6 mice (designated “DTR→WT” and “WT→WT” chimeras). B16-OG tumors were established in DTR→WT and WT→WT BM chimeras, and 2–3 weeks post B16-OG tumor cell transplantation effector TCROTI and TCRSMARTA were adoptively transferred. 5 days post ACT, when TCROTI and TCRSMARTA infiltrated into tumors, mice were treated twice weekly with DT. Depletion of CD11c+ APC prevented tumor elimination in DTR→WT mice but not control WT→WT mice, suggesting that CD11c+ APC within the tumor microenvironment were necessary for TCRSMARTA-mediated TCROTI reprogramming and tumor elimination (Fig. 3a).
Figure 3 |. Tumor elimination requires tumor antigen/epitope linkage and unique spatial orientation of tumor-specific CD8 T cells, CD4 T cells and CD11c+ dendritic cells (DC) within tumors.
a. B16-OG tumor outgrowth in CD11c-DTR/GFP bone marrow (BM) chimeras (scheme, top; DTR→WT or WT→WT) treated with diphtheria toxin (DT). In vitro activated TCROTI and TCRSMARTA were adoptively transferred into lymphodepleted tumor-bearing BM chimeras. 5 days post ACT, mice were treated with DT. Representative of 2 independent experiments (n=3 mice/group). Values are mean ± SEM. Significance is calculated by multiple t test. b. (Top) Experimental scheme of tumor models A and B: 2.5×106 B16-OG cancer cells (B16 OG; model A) or 1.25×106 B16-OVA (B16-O) mixed with 1.25×106 B16-GP61–80 cancer cells (B16 O+G; model B) were transplanted into B6 WT mice. (Bottom), (left) Tumor outgrowth of B16-OG or B16 O+G tumors after TCROTI and TCRSMARTA ACT. Representative of 2 independent experiments (n=7 mice/cohort). Data are shown as mean ± SEM. Significance is calculated by multiple t test. (Right) Kaplan–Meier curve; **p=0.0002; Mantel–Cox test. c. Percentage of TCROT1(+CD4) (out of total CD8+ TIL) 9 days post ACT. d. Percentage of TCRSMARTA (out of total CD4+ TIL) 9 days post ACT. Data represent 2 pooled, independent experiments (n=8 mice/tumor model). Each symbol represents an individual mouse. e. IFNγ, TNFα, CD107, Granzyme B production of TCROT1(+CD4) isolated from B16-OG or B16 O+G tumors, or f. isolated from tumor-draining lymph nodes of B16-OG or B16 O+G tumor-bearing hosts. Cytotoxic molecules and cytokine production assessed after 4-hr peptide stimulation ex vivo. Representative of 2 independent experiments (n=3 mice/tumor). Data are shown as mean ± SEM. *p<0.05, unpaired two-tailed Student’s t test. NS, not significant. g. Mosaic, clonal growth of B16 OVA-EGFP mixed with B16 GP61–80-Cerulean tumor cells (B16 O+G) in B6 WT mice. Shown are confocal microscopy sections of tumors with B16 OVA (green) and B16 GP (red) distinct tumor regions. h. Proposed model: Triad formation (three-cell-type clusters; CD8 T cells::CD4 T cells:: APC) form in B16 OG tumors (Model A) where CD8- and CD4-tumor antigens/epitopes are linked and co-presented on the same APC within tumors; tumor-specific CD8 and CD4 T cells engage on same APC; CD4 T cells reprogram CD8 T cells. Model B: B16 O+G; triads cannot form due to CD8- and CD4-tumor antigens being presented on distinct APC.
Next, we wanted to investigate how TCRSMARTA, TCROT1(+CD4) and stromal cell interactions cause tumor elimination. To answer this question, we modified our tumor model (Fig. 3b): we generated B16 tumor cell lines expressing either the CD8-OVA (B16-O) or CD4-GP (B16-G) tumor antigens. We implanted a mixture of 1.25×106 B16-O and 1.25×106 B16-G cancer cells into WT B6 mice, forming mixed B16 O+G tumors. Control mice received 2.5×106 B16-OG tumor cells as in Figures 1 and 2; thus, both cohorts received the same total number (2.5×106) of cancer cells, expressing similar levels of OVA and GP tumor antigens (data not shown). B16 O+G tumors grew with similar kinetics as B16-OG tumors. 2–3 weeks post tumor transplantation, mice received effector TCROTI and TCRSMARTA. 7 days post ACT, equal numbers of TCROTI and TCRSMARTA TIL were found within progressing B16 O+G and B16-OG tumors (Fig. 3c, 3d).Strikingly, despite the same numbers of tumor cells, equal tumor sizes, and same numbers of TCROT1 and TCRSMARTA TIL, mixed B16 O+G tumors continued to grow, in contrast to B16-OG tumors, which ultimately regressed (Fig. 3b). TCROT1 TIL isolated from B16 O+G tumors revealed a dysfunctional phenotype similar to those described for TCROT1 transferred without CD4 T cells shown in Figure 1 (Fig. 3e). Importantly, these functional differences were only observed within the tumor and not in the tdLN (Fig. 3f).
What are the factors and mechanisms that determine tumor progression or regression if numbers of cancer cells and antigen-specific CD8 and CD4 TIL are equal? We hypothesized that a unique spatial organization of cancer cells, CD4 T cells, CD8 T cells, and DC within tumors likely drove CD8 T cell reprogramming and tumor destruction.
Intratumoral immune triads in mouse and human tumors are required for anti-tumor responses
To define the intratumoral spatial characteristics we conducted confocal microscopic analysis of established B16 O+G tumors. We found regions of either B16-OVA-positive and B16-GP-positive cancer cells, and very few regions that had B16-OVA and B16-GP cancer cells intermingled (Fig. 3g). The mosaic-like appearance of distinct tumor regions is a typical feature of clonally growing cancer cells in transplantation tumor models [45]. Consequently, in B16 O+G tumors CD8 or CD4 antigens are largely presented in distinct regions within the tumor and on distinct DC/APC (Model B), unlike in B16-OG tumors where CD8 and CD4 antigens are co-presented on the same DC/APC through epitope linkage (Model A) (Fig. 3h). Thus, we propose the following model: co-presentation of tumor-specific CD4 and CD8 tumor antigens on the same APC will “force” antigen-specific CD4 and CD8 T cells to form three-cell-type clusters (triads) with APC, and the physical proximity of CD8 T cells with CD4 T cells drives CD4 T cell-mediated CD8 T cell reprogramming and cancer cell destruction (Model A). In Model B, CD8 and CD4 T cells fail to form triads with APC, CD4 T cells are unable to mediate CD8 T cell reprogramming, ultimately allowing tumors to progress. The concept of a ‘three-cell-type cluster’ was first described in 1987: Mitchison and O’Malley suggested that three-cell-type clusters of CD4 T cell-CD8 T cells-APC were required for the cytolytic response of CD8 T cells in an allogeneic transplant setting [62]. However, little is known about their functional relevance in vivo and/or underlying mechanisms.
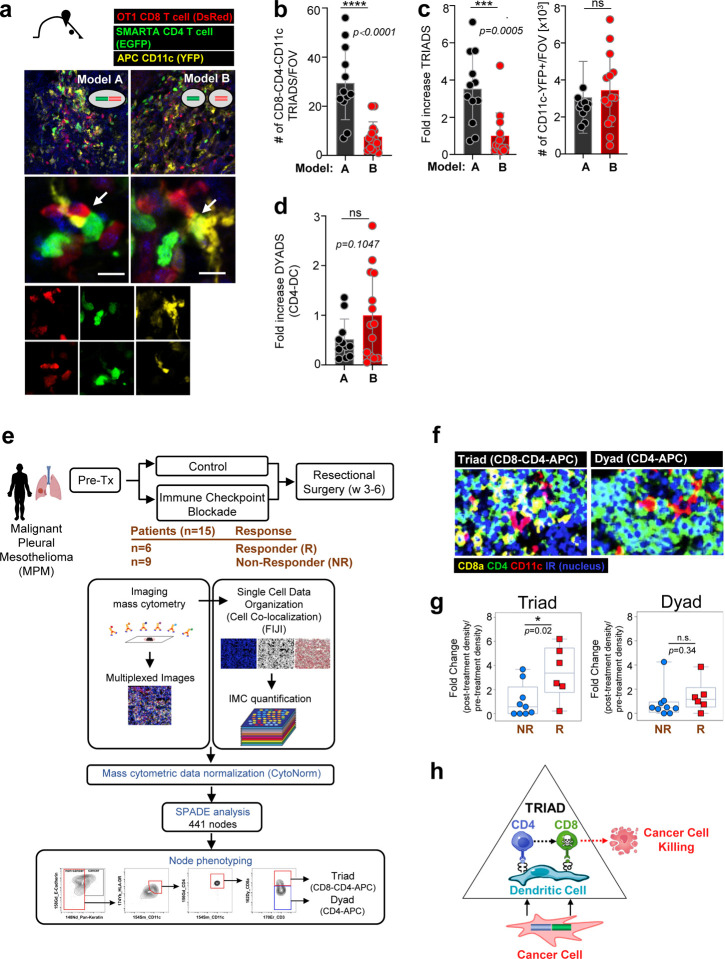
To determine whether triads are indeed a requisite for tumor elimination, we generated color-coded B16 O+G and B16 O-G tumor models: TCRSMARTA transgenic mice were crossed to EGFP transgenic mice, generating EGFP-expressing TCRSMARTA CD4 T cells; TCROT1 were engineered to express the red fluorescent protein (RFP); CD11c-YFP mice were used as hosts (with yellow fluorescent protein (YFP) under the transcriptional control of the CD11c promoter, thereby YFP-labeling CD11c+ host cells). B16-OG, B16-O, and B16-G cancer cells expressed Cerulean. B16-OG or B16 O+G tumors were established in CD11c-YFP mice and effector TCROTI-RFP+ and TCRSMARTA-EGFP+ adoptively transferred (Fig. 4a). Strikingly, 8–9 days post ACT significantly higher numbers of TCROT1::CD11c+YFP+::TCRSMARTA three-cell-clusters/triads (~30 interactions/field (or close apposition)) were present in B16-OG tumors, which eventually regressed, in contrast to B16 O+G tumors (~7 interactions), which eventually progressed (Fig. 4b). When normalized to the total number of infiltrating CD11c+YFP+ cells/field, which remained constant in both tumor models (Fig. 4c, right), we observed a 3.5-fold increase of triads in B16-OG tumors (Fig. 4c, left). Importantly, dyads, two-cell-interactions between TCRSMARTA::CD11c+YFP+ DC, were not significantly different between B16-OG and B16 O+G (Fig. 4d). Thus, CD8 T cell::CD4 T cell::DC triads are associated with tumor-specific CD8 T cell reprogramming and tumor elimination.
Figure 4 |. Intratumoral immune triads (three-cell-types clusters; CD8 T cell::CD4 T cell::APC) are required for CD8 T cell reprogramming and tumor elimination.
a. Color-coded mouse models to determine intratumoral immune triad formation (Models A and B (see Fig. 3)). B16 OG (Model A) or B16 O+G (Model B) tumors were established in CD11c-YFP mice (yellow); effector TCROTI-RFP (red) and TCRSMARTA-EGFP T cells (green) were adoptively transferred into tumor-bearing hosts. Confocal microscopy analysis of frozen tumor tissue sections. Arrows indicate triads. b. Numbers of triads per field of view (FOV), and c. (left) Fold increase of triads normalized to total numbers of CD11c+YFP+ cells/FOV (right). c. Quantification of fold increase of numbers of CD4 T cell-DC dyads normalized to total number of infiltrating CD11c+YFP+ cells/FOV. Each symbol represents an individual frozen tumor section (n=3 mice/group/model). Data are shown as mean ± SEM. *** P <0.001, unpaired two-tailed Student’s t test. (e.-g). Increased triads in patients with Malignant Pleural Mesothelioma (MPM) treated with checkpoint immunotherapy is associated with pathologic responses. e. Treatment regimen and methodology used to determine triads (CD8 T cell::CD4 T cell::APC) and dyads (CD4::APC). Pipeline of co-localization detection by imaging mass cytometry (IMC; see Methods for more details). Briefly, FFPE tumor tissues were stained with 35 target-specific antibodies. Automated cluster detection estimated cluster boundaries by expanding the perimeter of nuclei, identified by Cell ID Intercalator-iridium (191Ir). IMC images were quantified through FIJI, and protein expression data extracted through mean intensity multiparametric measurements performed on individual clusters. Acquired cluster data were normalized with CytoNorm tools, and normalized cytometric data transferred into additional Spanning-tree Progression Analysis of Density-normalized Events (SPADE) to generate automated clustering algorithm and applied cytometric analysis in FlowJo. f. Representative multiplexed mass cytometry images of triads and dyads. g. Fold change of triads and dyads of pre- and post-immune checkpoint therapy (Tx) density (numbers/mm2) in responders (R) and non-responders (NR); *p=0.02; n.s. p=0.34 (not significant). h. Proposed model of TRIAD-associated cancer elimination.
Next, we asked whether CD8 T cell::CD4 T cell::APC triads could be associated with clinical responsiveness in humans. As clinical data assessing spatial characteristics of immune cells within tumors of ACT-treated patients was not available, we turned to patients treated with immune checkpoint blockade (ICB) therapy; ICB therapies have shown efficacy in some cancer patients and cancer types, however most patients remain refractory. The underlying mechanisms determining ICB resistance or responsiveness, as well as predictive biomarkers, remain poorly defined. We assessed the spatial orientation of CD8 T cells, CD4 T cells and APC in patients with malignant pleural mesothelioma (MPM) undergoing ICB therapy [63]. Patients were randomized and treated with Durvalumab (anit-PDL1) mono- or Durvalumab and Tremelimumab (anti-CTLA4) combination therapy. A no ICB group was included as a control cohort. Tumor tissues were obtained both before and after ICB treatment [63]. Evaluable tumors, before and after ICB were available for 15 patients receiving ICB. Out of the 15 patients, 6 patients showed a pathologic response (R; Responders) while 9 patients did not (NR; Non-Responders) (Fig. 4e). Imaging mass cytometry (IMC) and time-of-flight mass cytometry (CyTOF) were performed on all 15 patients’ pre- and post-treatment tumor tissues using 35 markers to determine co-localization of non-TREG CD4 T cells, CD8 T cells, and CD11c+ APC, including the presence of dyads (CD4::APC or CD8::APC) and triads (CD4::CD8::APC) (Fig. 4e and 4f). Strikingly, while neither numbers of tumor-infiltrating CD8 T cells, nor CD4::APC or CD8::APC dyads were associated with a pathologic response and ICB responsiveness, triads were able to demarcate responders from non-responders (Fig. 4g). Our studies reveal triads as critical determinants for anti-tumor immunity and ICB responsiveness in patients with MPM.
DISCUSSION
Here, we demonstrate a new role for CD4 T cells during the effector phase of cytotoxic CD8 T cell- elimination of solid tumors in the setting of ACT. CD4 T cell reprogramming of CD8 T cells and cancer cell elimination is strictly dependent on the formation of immune triads, tumor-specific CD8 T cells and CD4 T cells co-engaged with the same DC, and not on CD4 T cell engagement with cancer cells, important given that most epithelial cancers do not express MHC class II. We demonstrate that the spatial positioning of CD8 and CD4 T cells within tumors, and not the number of intratumoral tumor-specific CD8 and CD4 T cells, is the critical determinant of effective anti-tumor immunity and ACT efficacy. Our data may provide clues as to why ACT clinical trials utilizing predominantly tumor-reactive CD8 T cells have shown only limited responses for the treatment of solid tumors.
It is well established that CD4 T cells are required for CD8 T cell effector differentiation. However, studies have mainly focused on CD4 T cell ‘help’ of naïve CD8 T cells during the priming/activation phase and memory formation in infection and vaccination settings [31, 42, 64–66]. The importance of CD8-CD4 T cell co-operation during the priming/activation phase was elegantly described by the Germain group, demonstrating that nonrandom, chemokine-driven (CCL3, CCL4) recruitment of CCR5+ naïve, antigen-specific CD8 T cells to sites of antigen-specific DC-CD4 T cell interactions within antigen-draining lymph nodes led to optimal CD8 T cell responses during vaccination and early infections [30]. CD4 T cells license DC through CD40L-CD40 interactions, enhancing B7 and CD70 expression on DC; CD28- and CD27- expressing antigen-specific CD8 T cells (ligands for B7 and CD70, respectively) receive optimal costimulatory signals when engaging with DC-CD4 T cells and/or abundant IL2 produced by CD4 T cells. Vaccines relying only on short, single MHC class I-restricted peptides showed reduced clinical benefits compared to synthetic long peptide vaccine platforms containing both MHC class I and class II epitopes, highlighting the importance of guided CD8 and CD4 cooperation [42–46]. Here, we discover that CD4 T cells and triads are critical for cancer cell elimination by cytolytic effector CD8 T cells: antigen-specific CD4 T cells within tumors reprogram antigen-specific effector CD8 T cells, repressing terminal differentiation and preserving stem-like features and effector function. Physical proximity of CD8 T cells with CD4 T cells likely enforces chemokine and/or cytokine signaling, or direct receptor-ligand interactions needed for CD8 T cell reprogramming. Interestingly, chemokine receptors such as Ccr5, Ccr4 and Ccr2 were upregulated on TCROT1(+CD4) encountering DC-CD4 T cells, as well as Il2rg and Ifngr1. Future studies must determine the precise mechanisms by which CD8 T cells resist T cell exhaustion and mediate cancer destruction. Our finding that triads (but not dyads) were associated with a pathogenic anti-tumor response in ICB-treated patients with malignant pleural mesothelioma, suggests that intratumoral immune triads may also be critical for anti-tumor responses in non-ACT settings. Interestingly, and congruent with our findings, a recent study demonstrated that dendritic cell–CD4 T helper cell niches enable CD8 T cell differentiation in patients with hepatocellular carcinoma following PD-1 blockade [67].
Our study reveals a previously unappreciated role of unique cell-cell interactions and spatial positioning within tumors where tumor-specific CD4 T cells empower tumor-specific CD8 T cells to eliminate solid tumors in adoptive T cell therapy. MHC class II-restricted neoantigens or self/tumor antigens and tumor-specific CD4 T cells have been described in human cancers [48–50]. Designing therapeutic interventions that enforce the formation of CD4-CD8-DC triads in tumors might be powerful strategies for the treatment of cancers, including for ICB-, vaccine- and ACT-approaches.
MATERIALS AND METHODS
Mice
B6 mice (C57BL/6J), TCROTI (C57BL/6-Tg(TcraTcrb)1100Mjb/J), TCRSMARTA (B6.Cg-Ptprca Pepcb Tg(TcrLCMV)1Aox/PpmJ), CD11c-YFP (B6.Cg-Tg(Itgax-Venus)1Mnz/J), CD11c-DTR-GFP (B6.FVB-1700016L21RikTg(Itgax-DTR/EGFP)57Lan/J), GFP transgenic (C57BL/6-Tg(CAG-EGFP)1Osb/J), B6 Thy1.1 (B6.PL-Thy1a/CyJ), and B6 CD45.1 (B6.SJL-Ptprca Pepcb/BoyJ) mice were purchased from the Jackson Laboratory. TCRSMARTA mice were crossed to Thy.1.1 mice to generate TCRSMARTA Thy1.1 mice; for Figure 4 imaging studies, TCRSMARTA Thy1.1 mice were crossed to GFP-transgenic mice to generate TCRSMARTA Thy1.1 GFP mice. TCROTI (Thy1.2) mice were crossed to CD45.1 mice to generate TCROTI CD45.1 mice. Both female and male mice were used for experimental studies. Donor and host mice were age- and sex-matched; mice were 7–12 weeks old. All mice were bred and maintained in the animal facility at Memorial Sloan Kettering Cancer Center (MSKCC). Experiments were performed in compliance with the MSKCC Institutional Animal Care and Use Committee (IACUC) regulations.
Antibodies and Reagents
Fluorochrome-conjugated antibodies were purchased from BD Biosciences, eBioscience, and Biolegend. The OVA257–264 and GP61–80 peptides were purchased from GenScript.
Intracellular cytokine staining
Intracellular cytokine staining was performed using the Foxp3 staining kit (BD Biosciences) following the manufacturer’s protocol. Briefly, T cells isolated from lymph nodes or tumors were mixed with 3×106 congenically marked B6 splenocytes and incubated with 1 μg/mL of OVA peptide and/or 2 μg/mL of GP peptide for 4–5h at 37°C in the presence of GolgiPlug (BD Biosciences). After staining for cell surface molecules, cells were fixed, permeabilized and stained with antibodies against IFNγ (XMG1.2) and TNFα (MP6-XT22).
Flow Cytometric Analysis
Flow cytometric analysis was performed using Fortessa X20. Cells were sorted using BD FACS Aria (BD Biosciences) at the MSKCC Flow Core Facility. Flow data were analyzed with FlowJo v.10 software (Tree Star Inc.).
Generation of plasmids and tumor cell lines
Tumor antigen-encoding pMFG-Cerulean vectors
pMFG-OVA257–264-Cerulean, pMFG-GP61–80-Cerulean, and pMFG-OVA257–264-GP61–80-Ceruelan plasmids were constructed by inserting annealed oligonucleotides encoding triple SIINFEKL-AAY repeats, GLKGPDIYKGVYQFKSVEFD, or (SIINFEKL-AAY)3-P2A-GLKGPDIYKGVYQFKSVEFD, respectively, into the NcoI-linearized pMFG-Cerulean vector, as previously described [45]. Restriction enzymes were purchased from New England Biolabs. All constructs were verified by sequence analysis. Phoenix packaging cells (ATCC) were transfected with pMFG constructs; supernatants were used to transduce B16-F10 mouse melanoma tumor cell line to generate B16-F10 OVA257–264-Cerulean, B16-F10GP61–80-Cerulean and B16-F10 OVA257–264-GP61–80-Cerulean, respectively [45]. Transduced bulk cell lines were sorted for similar Cerulean expression levels.
In vitro T cell activation
For the generation of effector TCROT1 CD8 T cells and TCRSMARTA CD4 T cells, single-cell suspensions were prepared from spleens of TCROT1 and TCRSMARTA transgenic mice and cultured in vitro in RPMI 1640 medium supplemented with 10% FBS, 100 IU/ml penicillin, 100 mg/ml streptomycin, nonessential amino acids, 1 mM sodium pyruvate, and 20 mM HEPES, together with 1 μg/mL of OVA257–264 peptide or 2 μg/mL of GP61–80 peptide, respectively, at a concentration of 4–5×106 splenocytes/ml in the presence of 50U/mL IL-2 for 4 days.
Adoptive T cell transfer
For adoptive transfer studies, 2.5×105 in vitro activated TCROT1 (CD45.1) and/or 5×105 in vitro activated TCRSMARTA (Thy1.1) were transferred (i.v.) into tumor-bearing WT B6 mice at indicated time points post tumor transplantation (approximately 2–3 weeks post tumor implantation). Tumor-bearing mice were treated with cyclophosphamide (180mg/kg), and 24h later in vitro activated TCROT1 CD8 T cells and/or TCRSMARTA CD4 T cells were adoptively transferred. At indicated time points, adoptively transferred T cells were isolated from tumor-draining lymph nodes and tumors and prepared for downstream analyses.
B16 and MCA 205 transplantation tumor models
2.5×106 B16 OVA257–264-GP61–80 (B16 OG) tumor cells, or a mixture of 1.25×106 B16 OVA257–264 (B16 O) + 1.25×106 B16 GP61–80 (B16 G) tumor cells (B16 O+G), or MCA OVA257–264-GP61–80 tumor cells were injected subcutaneously into mice. Antigen-specific T cells were adoptively transferred into tumor-bearing mice as described in text and figure legends. For outgrowth experiments, tumors were measured manually with a caliper. Tumor volume was estimated with the formula (L x W x H)/2.
Generation of bone marrow chimeras and depletion of dendritic cells in vivo
B6 WT (CD45.1) mice were irradiated twice with 600 cGy, 6 hours apart. 12–18 hours later, bone marrow (BM) was isolated from femurs and tibias of CD11c-DTR/GFP (CD45.2) mice, and 5–8×106 BM cells were injected i.v. into irradiated CD45.1 mice. BM chimeric were given antibiotics (trimethoprim-sulfamethoxazole) for 2 weeks. BM chimeric were analyzed for successful engraftment and BM reconstitution 6–8 weeks later. For conditional DC depletion, CD11c-DTR/GFP BM chimeric mice were injected (i.p.) with 4–5 ng/g body weight diphtheria toxin (DT, Sigma-Aldrich) every other day for 14 days.
Generation of B16 -I-Ab-deficient tumor cell line
The B16 tumor cells were subjected to CRISPR/Cas9-mediated knockout of I-Ab by transient transfection of a plasmid encoding both Cas9 nuclease and single guide (sg) RNA targeting the I-Ab locus, as well as GFP reporter gene. 2.5×105 B16 cells were plated and transfected with 2μg of Cas9- and sgRNA-encoding plasmid DNA using Lipofectamine 3,000 (Invitrogen) following the manufacturer’s protocol. 3 days post transduction, GFP+ cells were FACS-sorted. Deletion of I-Ab was confirmed by treating GFP+ B16 I-Ab cells with 20 U/ml IFNγ for 48h, followed by flow cytometric analysis of I-Ab expression.
Color-coded tumor model and adoptive transfer of color-coded T cells
CD11c-YFP transgenic mice were injected subcutaneously with 2.5×106 (B16 OG) tumor cells or a mixture of 1.25×106 B16-O + 1.25×106 B16-G tumor cells (B16 O+G). To generate color coded TCROT1 CD8 T cells, TCROT1 splenocytes were transduced to express tRFP using retroviral transduction as previously described [68]. Briefly, Platinum-E cells (ATCC) were transfected with a tRFP-encoding retroviral vector using the Mirus TransIT-LT1 reagent (catalog no. 2305). Viral supernatant was supplemented with polybrene and added to TCROT1 splenocytes, and the cells were transduced via spinfection on two consecutive days. To generate color-coded TCRSMARTA CD4 T cells, splenocytes from TCRSMARTA GFP transgenic mice were used and activated as described above. Tumor-bearing mice were treated with cyclophosphamide (180mg/kg) one day before ACT, and in vitro activated 2.5+105 TCROT1 tRFP+ CD8 T cells and 4X105 cells TCRSMARTA EGFP CD4 T cells were transferred (i.v.) into tumor-bearing mice.
Immunofluorescence staining and confocal imaging
For confocal microscopy analysis, pieces of established tumors were excised and fixed for 18–24 hours in 4% paraformaldehyde solution, followed by dehydration in 20% sucrose, and then embedded in OCT, and stored at −80°C. 30-μm-thick frozen sections were cut on a CM3050S cryostat (Leica) and adhered to Superfrost Plus slides (Thermo Fisher Scientific). Nuclei were labeled using DAPI (Sigma). Slides were mounted with ProLong Diamond Antifade Mountant (Invitrogen) and analyzed on a Leica TCS SP8 confocal microscope. Fiji Is Just ImageJ (FIJI) was utilized for image analysis. 3D reconstitution was performed, and triple contacts/triads were assessed based on color-coded immune subset identification. Analyses was performed as a blinded outcome assessment. To quantify double contacts, after thresholding and binarization of images, the function “analyze particles” has been applied. To precisely estimate only events showing double contact, the mathematical function “AND” was used.
Isolation of adoptively transferred T cells from downstream analyses
Lymph nodes were mechanically disrupted with the back of a 3-mL syringe, filtered through a 100-μm strainer, and red blood cells (RBC) were lysed with ammonium chloride potassium buffer. Cells were washed twice with cold RPMI 1640 media supplemented with 2μM glutamine, 100U/mL penicillin/streptomycin, and 3% fetal bovine serum (FBS). Tumor tissue was mechanically disrupted with a glass pestle and a 150-μm metal mesh in 5mL of cold HBSS with 3% FBS. Cell suspension was filtered through 70-μm strainers. Tumor homogenate was spun down at 400g for 5 minutes at 4°C. Pellet was resuspended in 15 mL HBSS with 3% FBS, 500 μl (500U) heparin, and 8 mL isotonic Percoll (GE), mixed by several inversions, and spun at 500g for 10 min at 4°C. Pellet was lysed with ammonium chloride potassium buffer and cells were further processed for downstream applications.
Sample Preparation for RNA-Seq and ATAC-Seq
TCROT1 CD8 T cells were isolated from tumors (see above); cells were stained for CD8α (clone 53–6.7, eBioscience) and CD45.1+(clone A20, Biolegend). CD8+CD45.1+ cells were sorted by FACS. For RNA-seq, T cells were directly sorted into Trizol LS reagent (Invitrogen, catalog no. 10296010) and stored at −80°7 For ATAC-seq, sorted T cells were resuspended in cold FBS with 10% DMSO and stored at −80°.
RNA-seq
RNA from sorted cells was extracted using the RNeasy Mini Kit (Qiagen; catalog no. 74104) according to instructions provided by the manufacturer. After RiboGreen quantification and quality control by an Agilent BioAnalyzer, total RNA underwent amplification using the SMART-Seq v4 Ultra Low Input RNA Kit (Clontech), and amplified cDNA was used to prepare libraries with the KAPA Hyper Prep Kit (Kapa Biosystems). Samples were barcoded and run on a HiSeq 2500 in a 50-bp/50-bp paired-end run with the HiSeq SBS Kit v4 (Illumina). An average of 50 million paired reads were generated per sample.
ATAC-seq
Profiling of chromatin accessibility was performed by ATAC-seq as previously described (Buenrostro et al., 2013). Briefly, viably frozen, sorted T cells were washed in cold PBS and lysed. The transposition reaction was incubated at 42°C for 45 min. The DNA was cleaned with the MinElute PCR Purification Kit (Qiagen; catalog no. 28004), and material was amplified for five cycles. After evaluation by real-time PCR, 7–13 additional PCR cycles were done. The final product was cleaned by AMPure XP beads (Beckman Coulter, catalog no. A63882) at a 1× ratio, and size selection was performed at a 0.5× ratio. Libraries were sequenced on a HiSeq 2500 or HiSeq 4000 in a 50-bp/50-bp paired-end run using the TruSeq SBS Kit v4, HiSeq Rapid SBS Kit v2, or HiSeq 3000/4000 SBS Kit (Illumina). An average of 100 million paired reads were generated per sample.
Bioinformatics methods
The quality of the sequenced reads was assessed with FastQC and QoRTs (for RNA-seq samples; Hartley and Mullikin, 2015; Andrews, 2010). Unless stated otherwise, plots involving high- throughput sequencing data were created using R version 4.1.0 (R Core Team, 2017) and ggplot2 (Wickham, 2016).
RNA-seq data:
DNA sequencing reads were aligned with default parameters to the mouse reference genome (GRCm38.p6) using STAR v2.6.0c (Dobin et al., 2013). Gene expression estimates were obtained with featureCounts v1.6.2 using composite gene models (union of the exons of all transcript isoforms per gene) from Gencode (version M17; Liao et al., 2014).
DEGs
DEGs were determined using DESeq2 v1.34.0 with Wald tests with a q-value cutoff of 0.05 (Benjamini– Hochberg correction).
Heatmaps
Heatmaps in Fig. 2b were created using DESeq2 normalized read counts after variance stabilizing transformation of genes identified as differentially expressed by DESeq2. Rows were centered and scaled.
Pathway and GO term enrichment analyses
Gene set enrichment analyses (Fig. 2g and Suppl. Fig 1) were done using fgsea v1.20.0 [69] with the fgseaMultilevel function. Genes were ranked based on the DESeq2 Wald statistic. Gene sets with an FDR < 0.05 were considered enriched.
Gene ontology analysis was performed on up- and down-regulated DEGs using the clusterProfiler v4.2.2 R package [70]. Only GO categories enriched using a 0.05 false discovery rate cutoff were considered.
ATAC-seq data:
Alignment and creation of peak atlas
Reads were aligned to the mouse reference genome (version GRCm38) with BWA-backtrack v0.7.17 (Li and Durbin, 2009). Post-alignment filtering was done with samtools v1.8 and Picard tools v2.18.9 (Li et al., 2009) to remove unmapped reads, improperly paired reads, nonunique reads, and duplicates. Peaks were called with MACS2 v2.1.1 (Liu, 2014), and peaks with adjusted P values smaller than 0.01 were excluded. Consensus peak sets were generated for each condition if a peak was found in at least two replicates. Reproducible peaks from each condition were merged with DiffBind v3.4.11 to create an atlas of accessible peaks, which was used for downstream analyses. The peak atlas was annotated using the ChIPseeker v1.30.3 [71] and TxDb.Mmusculus.UCSC.mm10.knownGene [Bioconductor Core Team and Bioconductor Package Maintainer (2019). TxDb.Mmusculus.UCSC.mm10.knownGene: Annotation package for TxDb object(s). R package version 3.10.0.]. Blacklisted regions were excluded (https://sites.google.com/site/anshulkundaje/projects/blacklists).
Differentially accessible regions
Regions where the chromatin accessibility changed between different conditions were identified with DESeq2 v1.34.0, and only Benjamini-Hochberg corrected P values < 0.05 were considered statistically significant.
Coverage files
Genome coverage files were normalized for differences in sequencing depth (RPGC normalization) with bamCoverage from deepTools v3.1.0. Replicates were averaged together using UCSC-tools bigWigMerge. Merged coverage files were used for display in Integrated Genomics Viewer shown in Fig. 2e.
Heatmaps
Heatmaps based on the differentially accessible peaks identified between TCROT1 and TCROT1(+CD4) as shown in Fig. 2d were created using profileplyr v1.10.2 (T. Carroll and D. Barrows (2021). profileplyr: Visualization and annotation of read signal over genomic ranges with profileplyr. R package version 1.10.2.) and ComplexHeatmap v2.15.1 [72], by binning the region +/− 1kb around the peak summits in 20bp bins. To improve visibility, bins with read counts greater than the 75th percentile + 1.5*IQR were capped at that value.
Motif analyses
For identifying motifs enriched in differentially accessible peaks, we utilized HOMER via marge v0.0.4 ([73]; and [Robert A. Amezquita (2021). marge: API for HOMER in R for Genomic Analysis using Tidy Conventions. R package version 0.0.4.9999]). HOMER was run separately on hyper- or hypo-accessible peaks with the flags -size given and -mask. Motifs enriched in hyper- or hypo-accessible peaks were determined by comparing the rank differences (based on P value). The consensus peakset identified by DiffBind was used as the background set.
Human Data (Fig. 4e–4g):
Trial, Patients, Study Design:
For more details on patients, study design, and methodology see Hyun-Sung Lee et al [63]. Briefly, this was a phase II, prospective, randomized window-of-opportunity trial completed at Baylor College of Medicine that enrolled patients with surgically resectable MPM (NCT02592551). Eligible patients underwent a staging procedure that included cervical mediastinoscopy with mediastinal lymph node biopsies and diagnostic laparoscopy with peritoneal lavage and peritoneal biopsies. Thoracoscopy with tumor biopsies was performed for the purpose of the trial. Patients without pathologic nodal or peritoneal disease were randomly assigned in a 2:2:1 ratio to receive (i) one dose of durvalumab (10 mg/kg i.v.), (ii) one dose of durvalumab (1,500 mg) plus one dose of tremelimumab (75 mg i.v.), or (iii) no ICB. ICB was administered 3 days to 3 weeks following the staging procedure and surgical resection was performed 3 to 6 weeks after ICB by extended pleurectomy/decortication (P/D) or extrapleural pneumonectomy (EPP). Tumor and blood were obtained before and after ICB (at thoracoscopy and resection, respectively).
Methods:
Cancer specimens were processed into single-cell suspensions, fresh frozen tissue preparations, samples cryopreserved in optimal cutting temperature (OCT) compound, and formaldehyde-fixed tissues (FFPE).
Imaging mass cytometry (IMC).
FFPE tissue samples were sectioned at a 5-μm thickness for IMC. FFPE tissues on charged slides were stained with 1:100 diluted antibody cocktails (concentration of each antibody=0.5mg/mL) as recommended by the user’s manual. The slides were scanned in the Hyperion Imaging System (Fluidigm). They were scanned at least four regions of interest in >1mm2 at 200 Hz.
IMC analysis.
Fiji was used for cell segmentation and conversion of imaging data into flow cytometric data, with the advantage of fast, robust, unsupervised, automated cell segmentation method. 32-bit TIFF stacked images were loaded in Fiji and novel method of automated cell segmentation that estimates cell boundaries by expanding the perimeter of their nuclei, identified by Cell ID intercalator iridium (191Ir) was used as described in more detail in Hyun-Sung Lee et al [63]. Once images from the IMC methodology were acquired, images were quantified through FIJI’s threshold and watershed tools. Protein expression data were then extracted at the single-cell level through mean intensity multiparametric measurements performed on individual 10 cells and acquired single-cell data were transferred into additional cytometric analysis in FlowJo V10 software (FlowJo, LLC, OR). All protein markers in quantified IMC data are adjusted with 191Ir and 193Ir nucleus intensities and normalized with CytoNorm across IMC regions of interests, a normalization method for cytometry data applicable to large clinical studies that is plugged-in FlowJo. CytoNorm allows reducing mass cytometry signal variability across multiple batches of barcoded samples. Normalized IMC data are combined by using FlowJo. For CyTOF, please see Hyun-Sung Lee et al [63].
Statistical analyses
Statistical analyses on flow cytometric data were performed using unpaired two-tailed Student’s t tests (Prism 7.0, GraphPad Software). A P value of < 0.05 was considered statistically significant. All other statistical testing methods are described in figure legends.
Supplementary Material
Acknowledgements:
We thank the members of the Schietinger lab for helpful discussions, and M. Philip (Vanderbilt University) for critical reading of the manuscript. This work was supported by NIH grants DP2CA225212 and R01CA269733 (A.S.), Lloyd Old STAR Award of the Cancer Research Institute (A.S.), AACR-Bristol Myers Squibb Midcareer Female Investigator Award (A.S.), Pershing Square Sohn Award (A.S.), Josie Robertson Young Investigator Award (A.S.), the Weill Cornell Medicine Core Laboratories Center (P.Z., D.B), Ludwig Cancer Center Postdoctoral Fellowship (G.E.C.); NIH R37 MERIT Award R37CA248478 (B.M.B), Cancer Prevention and Research Institute of Texas Grant CPRIT RP200443 (H.S.L.), Department of Defense Peer Reviewed Cancer Impact Award CA210522 (H.S.L.).We acknowledge the use of the Integrated Genomics Operation Core, funded by the NCI Cancer Center Support Grant (P30 CA08748), Cycle for Survival, and the Marie-Josée and Henry R. Kravis Center for Molecular Oncology.
REFERENCES:
- 1.Philip M. and Schietinger A., CD8(+) T cell differentiation and dysfunction in cancer. Nat Rev Immunol, 2022. 22(4): p. 209–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson K.G., Stromnes I.M., and Greenberg P.D., Obstacles Posed by the Tumor Microenvironment to T cell Activity: A Case for Synergistic Therapies. Cancer Cell, 2017. 31(3): p. 311–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gajewski T.F., Schreiber H., and Fu Y.X., Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol, 2013. 14(10): p. 1014–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang J.C. and Rosenberg S.A., Adoptive T-Cell Therapy for Cancer. Adv Immunol, 2016. 130: p. 279–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leko V. and Rosenberg S.A., Identifying and Targeting Human Tumor Antigens for T Cell-Based Immunotherapy of Solid Tumors. Cancer Cell, 2020. 38(4): p. 454–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmitt T.M., et al. , New Strategies in Engineering T-cell Receptor Gene-Modified T cells to More Effectively Target Malignancies. Clin Cancer Res, 2015. 21(23): p. 5191–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenberg S.A., et al. , Gene transfer into humans--immunotherapy of patients with advanced melanoma, using tumor-infiltrating lymphocytes modified by retroviral gene transduction. N Engl J Med, 1990. 323(9): p. 570–8. [DOI] [PubMed] [Google Scholar]
- 8.Schmitt T.M., Ragnarsson G.B., and Greenberg P.D., T cell receptor gene therapy for cancer. Hum Gene Ther, 2009. 20(11): p. 1240–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morgan R.A., et al. , Cancer regression in patients after transfer of genetically engineered lymphocytes. Science, 2006. 314(5796): p. 126–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morgan R.A., Dudley M.E., and Rosenberg S.A., Adoptive cell therapy: genetic modification to redirect effector cell specificity. Cancer J, 2010. 16(4): p. 336–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosenberg S.A. and Dudley M.E., Cancer regression in patients with metastatic melanoma after the transfer of autologous antitumor lymphocytes. Proc Natl Acad Sci U S A, 2004. 101 Suppl 2(Suppl 2): p. 14639–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenberg S.A., et al. , Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin Cancer Res, 2011. 17(13): p. 4550–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yee C., et al. , Adoptive T cell therapy using antigen-specific CD8+ T cell clones for the treatment of patients with metastatic melanoma: in vivo persistence, migration, and antitumor effect of transferred T cells. Proc Natl Acad Sci U S A, 2002. 99(25): p. 16168–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chapuis A.G., et al. , T cell receptor gene therapy targeting WT1 prevents acute myeloid leukemia relapse post-transplant. Nat Med, 2019. 25(7): p. 1064–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dudley M.E., et al. , Cancer regression and autoimmunity in patients after clonal repopulation with antitumor lymphocytes. Science, 2002. 298(5594): p. 850–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stromnes I.M., et al. , T Cells Engineered against a Native Antigen Can Surmount Immunologic and Physical Barriers to Treat Pancreatic Ductal Adenocarcinoma. Cancer Cell, 2015. 28(5): p. 638–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leen A.M., Rooney C.M., and Foster A.E., Improving T cell therapy for cancer. Annu Rev Immunol, 2007. 25: p. 243–65. [DOI] [PubMed] [Google Scholar]
- 18.Wrzesinski C., et al. , Increased intensity lymphodepletion enhances tumor treatment efficacy of adoptively transferred tumor-specific T cells. J Immunother, 2010. 33(1): p. 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klebanoff C.A., et al. , Sinks, suppressors and antigen presenters: how lymphodepletion enhances T cell-mediated tumor immunotherapy. Trends Immunol, 2005. 26(2): p. 111–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klebanoff C.A., et al. , IL-15 enhances the in vivo antitumor activity of tumor-reactive CD8+ T cells. Proc Natl Acad Sci U S A, 2004. 101(7): p. 1969–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Restifo N.P., Dudley M.E., and Rosenberg S.A., Adoptive immunotherapy for cancer: harnessing the T cell response. Nat Rev Immunol, 2012. 12(4): p. 269–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baulu E., et al. , TCR-engineered T cell therapy in solid tumors: State of the art and perspectives. Sci Adv, 2023. 9(7): p. eadf3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chandran S.S. and Klebanoff C.A., T cell receptor-based cancer immunotherapy: Emerging efficacy and pathways of resistance. Immunol Rev, 2019. 290(1): p. 127–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borst J., et al. , CD4(+) T cell help in cancer immunology and immunotherapy. Nat Rev Immunol, 2018. 18(10): p. 635–647. [DOI] [PubMed] [Google Scholar]
- 25.Bennett S.R., et al. , Help for cytotoxic-T-cell responses is mediated by CD40 signalling. Nature, 1998. 393(6684): p. 478–80. [DOI] [PubMed] [Google Scholar]
- 26.Schoenberger S.P., et al. , T-cell help for cytotoxic T lymphocytes is mediated by CD40-CD40L interactions. Nature, 1998. 393(6684): p. 480–3. [DOI] [PubMed] [Google Scholar]
- 27.Le Saout C., et al. , Memory-like CD8+ and CD4+ T cells cooperate to break peripheral tolerance under lymphopenic conditions. Proc Natl Acad Sci U S A, 2008. 105(49): p. 19414–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakanishi Y., et al. , CD8(+) T lymphocyte mobilization to virus-infected tissue requires CD4(+) T-cell help. Nature, 2009. 462(7272): p. 510–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bos R. and Sherman L.A., CD4+ T-cell help in the tumor milieu is required for recruitment and cytolytic function of CD8+ T lymphocytes. Cancer Res, 2010. 70(21): p. 8368–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castellino F., et al. , Chemokines enhance immunity by guiding naive CD8+ T cells to sites of CD4+ T cell-dendritic cell interaction. Nature, 2006. 440(7086): p. 890–5. [DOI] [PubMed] [Google Scholar]
- 31.Castellino F. and Germain R.N., Cooperation between CD4+ and CD8+ T cells: when, where, and how. Annu Rev Immunol, 2006. 24: p. 519–40. [DOI] [PubMed] [Google Scholar]
- 32.Greenberg P.D., Kern D.E., and Cheever M.A., Therapy of disseminated murine leukemia with cyclophosphamide and immune Lyt-1+,2- T cells. Tumor eradication does not require participation of cytotoxic T cells. J Exp Med, 1985. 161(5): p. 1122–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frey A.B., Rat mammary adenocarcinoma 13762 expressing IFN-gamma elicits antitumor CD4+ MHC class II-restricted T cells that are cytolytic in vitro and tumoricidal in vivo. J Immunol, 1995. 154(9): p. 4613–22. [PubMed] [Google Scholar]
- 34.Mumberg D., et al. , CD4(+) T cells eliminate MHC class II-negative cancer cells in vivo by indirect effects of IFN-gamma. Proc Natl Acad Sci U S A, 1999. 96(15): p. 8633–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monach P.A., et al. , A unique tumor antigen produced by a single amino acid substitution. Immunity, 1995. 2(1): p. 45–59. [DOI] [PubMed] [Google Scholar]
- 36.Perez-Diez A., et al. , CD4 cells can be more efficient at tumor rejection than CD8 cells. Blood, 2007. 109(12): p. 5346–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qin Z. and Blankenstein T., CD4+ T cell--mediated tumor rejection involves inhibition of angiogenesis that is dependent on IFN gamma receptor expression by nonhematopoietic cells. Immunity, 2000. 12(6): p. 677–86. [DOI] [PubMed] [Google Scholar]
- 38.Egilmez N.K., et al. , Human CD4+ effector T cells mediate indirect interleukin-12- and interferongamma-dependent suppression of autologous HLA-negative lung tumor xenografts in severe combined immunodeficient mice. Cancer Res, 2002. 62(9): p. 2611–7. [PubMed] [Google Scholar]
- 39.Broderick L., et al. , Human CD4+ effector memory T cells persisting in the microenvironment of lung cancer xenografts are activated by local delivery of IL-12 to proliferate, produce IFN-gamma, and eradicate tumor cells. J Immunol, 2005. 174(2): p. 898–906. [DOI] [PubMed] [Google Scholar]
- 40.Muranski P., et al. , Tumor-specific Th17-polarized cells eradicate large established melanoma. Blood, 2008. 112(2): p. 362–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Greenberg P.D., Adoptive T cell therapy of tumors: mechanisms operative in the recognition and elimination of tumor cells. Adv Immunol, 1991. 49: p. 281–355. [DOI] [PubMed] [Google Scholar]
- 42.Hung K., et al. , The central role of CD4(+) T cells in the antitumor immune response. J Exp Med, 1998. 188(12): p. 2357–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Braumuller H., et al. , T-helper-1-cell cytokines drive cancer into senescence. Nature, 2013. 494(7437): p. 361–5. [DOI] [PubMed] [Google Scholar]
- 44.Muller-Hermelink N., et al. , TNFR1 signaling and IFN-gamma signaling determine whether T cells induce tumor dormancy or promote multistage carcinogenesis. Cancer Cell, 2008. 13(6): p. 507–18. [DOI] [PubMed] [Google Scholar]
- 45.Schietinger A., et al. , Bystander killing of cancer requires the cooperation of CD4(+) and CD8(+) T cells during the effector phase. J Exp Med, 2010. 207(11): p. 2469–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Espinosa-Carrasco G., et al. , CD4(+) T Helper Cells Play a Key Role in Maintaining Diabetogenic CD8(+) T Cell Function in the Pancreas. Front Immunol, 2017. 8: p. 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alspach E., et al. , MHC-II neoantigens shape tumour immunity and response to immunotherapy. Nature, 2019. 574(7780): p. 696–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sahin U., et al. , Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature, 2017. 547(7662): p. 222–226. [DOI] [PubMed] [Google Scholar]
- 49.Ott P.A., et al. , An immunogenic personal neoantigen vaccine for patients with melanoma. Nature, 2017. 547(7662): p. 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tran E., et al. , Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science, 2014. 344(6184): p. 641–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hunder N.N., et al. , Treatment of metastatic melanoma with autologous CD4+ T cells against NYESO-1. N Engl J Med, 2008. 358(25): p. 2698–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Greenberg P.D., Cheever M.A., and Fefer A., Eradication of disseminated murine leukemia by chemoimmunotherapy with cyclophosphamide and adoptively transferred immune syngeneic Lyt1+2- lymphocytes. J Exp Med, 1981. 154(3): p. 952–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scott A.C., et al. , TOX is a critical regulator of tumour-specific T cell differentiation. Nature, 2019. 571(7764): p. 270–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim K., et al. , Single-cell transcriptome analysis reveals TOX as a promoting factor for T cell exhaustion and a predictor for anti-PD-1 responses in human cancer. Genome Med, 2020. 12(1): p. 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seo H., et al. , TOX and TOX2 transcription factors cooperate with NR4A transcription factors to impose CD8(+) T cell exhaustion. Proc Natl Acad Sci U S A, 2019. 116(25): p. 12410–12415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang X., et al. , TOX promotes the exhaustion of antitumor CD8(+) T cells by preventing PD1 degradation in hepatocellular carcinoma. J Hepatol, 2019. 71(4): p. 731–741. [DOI] [PubMed] [Google Scholar]
- 57.Khan O., et al. , TOX transcriptionally and epigenetically programs CD8(+) T cell exhaustion. Nature, 2019. 571(7764): p. 211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alfei F., et al. , TOX reinforces the phenotype and longevity of exhausted T cells in chronic viral infection. Nature, 2019. 571(7764): p. 265–269. [DOI] [PubMed] [Google Scholar]
- 59.Wu M., Fang H., and Hwang S.T., Cutting edge: CCR4 mediates antigen-primed T cell binding to activated dendritic cells. J Immunol, 2001. 167(9): p. 4791–5. [DOI] [PubMed] [Google Scholar]
- 60.Krishna S., et al. , Stem-like CD8 T cells mediate response of adoptive cell immunotherapy against human cancer. Science, 2020. 370(6522): p. 1328–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jung S., et al. , In vivo depletion of CD11c+ dendritic cells abrogates priming of CD8+ T cells by exogenous cell-associated antigens. Immunity, 2002. 17(2): p. 211–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mitchison N.A. and O’Malley C., Three-cell-type clusters of T cells with antigen-presenting cells best explain the epitope linkage and noncognate requirements of the in vivo cytolytic response. Eur J Immunol, 1987. 17(11): p. 1579–83. [DOI] [PubMed] [Google Scholar]
- 63.Lee H.S., et al. , A Phase II Window of Opportunity Study of Neoadjuvant PD-L1 versus PD-L1 plus CTLA-4 Blockade for Patients with Malignant Pleural Mesothelioma. Clin Cancer Res, 2023. 29(3): p. 548–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Keene J.A. and Forman J., Helper activity is required for the in vivo generation of cytotoxic T lymphocytes. J Exp Med, 1982. 155(3): p. 768–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hu H.M., et al. , Divergent roles for CD4+ T cells in the priming and effector/memory phases of adoptive immunotherapy. J Immunol, 2000. 165(8): p. 4246–53. [DOI] [PubMed] [Google Scholar]
- 66.Gao F.G., et al. , Antigen-specific CD4+ T-cell help is required to activate a memory CD8+ T cell to a fully functional tumor killer cell. Cancer Res, 2002. 62(22): p. 6438–41. [PubMed] [Google Scholar]
- 67.Magen A., et al. , Intratumoral dendritic cell-CD4(+) T helper cell niches enable CD8(+) T cell differentiation following PD-1 blockade in hepatocellular carcinoma. Nat Med, 2023. 29(6): p. 1389–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shakiba M., et al. , TCR signal strength defines distinct mechanisms of T cell dysfunction and cancer evasion. J Exp Med, 2022. 219(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Korotkevich G., et al. , Fast gene set enrichment analysis. bioRxiv, 2021: p. 060012. [Google Scholar]
- 70.Wu T., et al. , clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb), 2021. 2(3): p. 100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu G., Wang L.G., and He Q.Y., ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics, 2015. 31(14): p. 2382–3. [DOI] [PubMed] [Google Scholar]
- 72.Gu Z., Eils R., and Schlesner M., Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics, 2016. 32(18): p. 2847–9. [DOI] [PubMed] [Google Scholar]
- 73.Heinz S., et al. , Simple combinations of lineage-determining transcription factors prime cisregulatory elements required for macrophage and B cell identities. Mol Cell, 2010. 38(4): p. 57689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.