Abstract
Retinoic acid (RA) is involved in antero-posterior patterning of the chordate body axis and, in jawed vertebrates, has been shown to play a major role at multiple levels of the gene regulatory network (GRN) regulating hindbrain segmentation. Knowing when and how RA became coupled to the core hindbrain GRN is important for understanding how ancient signaling pathways and patterning genes can evolve and generate diversity. Hence, we investigated the link between RA signaling and hindbrain segmentation in the sea lamprey Petromyzon marinus, an important jawless vertebrate model providing clues to decipher ancestral vertebrate features. Combining genomics, gene expression, and functional analyses of major components involved in RA synthesis (Aldh1as) and degradation (Cyp26s), we demonstrate that RA signaling is coupled to hindbrain segmentation in lamprey. Thus, the link between RA signaling and hindbrain segmentation is a pan vertebrate feature of the hindbrain and likely evolved at the base of vertebrates.
Keywords: Hindbrain segmentation, A-P patterning, Hindbrain GRN, Hox genes, Retinoic Acid (RA), RA signaling, Vertebrate Evolution, Lamprey, Jawless vertebrates, Cyp26, Aldh1a2
Introduction
The body plan of most deuterostomes develops using a similar set of transcription factors (TFs) and signaling pathways (e.g., FGF, Wnt) which are expressed in analogous axial domains of different developing embryos1, 2, 3, 4, 5. This suggests the presence of an ancient, conserved core gene regulatory network (GRN) underlying axial patterning, which integrates inputs from developmental TFs and signaling pathways. For example, in the evolution of chordates the coordinated action of FGF and Wnt signaling2, 6, 7 coupled with the Hox family of TFs is essential for establishing and patterning the antero-posterior (A-P) axis5, 8, 9, 10.
In chordate embryos, nested domains of Hox expression play a fundamental role in regulating patterning of the nervous system1, 11, 12, 13, 14. In addition to inputs from FGF and Wnt signaling, evidence from cephalochordate and jawed vertebrate models indicate that retinoic acid (RA) signaling plays a key role in coordinating the regulation of Hox gene expression along the A-P axis5, 7, 15, 16, 17, 18, 19, 20, 21, 22, 23. Regulatory studies have also uncovered the presence of retinoic acid response elements (RAREs) present in conserved positions in an amphioxus and mouse Hox cluster that contribute to nested domains of Hox expression24, 25, 26. This suggests that a direct regulatory link between RA signaling and Hox expression may be an ancient feature of the core GRN coupled to A-P patterning of the body axis in all chordates (Fig. 1a)26.
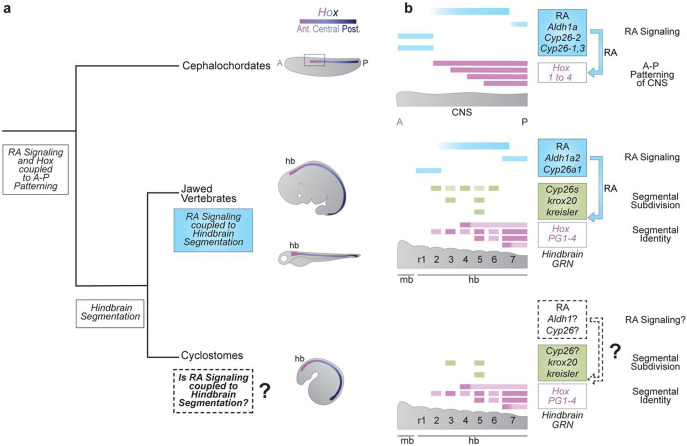
Figure 1: What is the origin of the coupling of RA signaling to vertebrate hindbrain segmentation?
(a) An ancient RA/Hox hierarchy is involved in axial (A-P) patterning in all chordates (e.g., Aldh1as, Cyp26s, Hox 1-4);
(b) Many of these ancestral genes are wired into a complex and dynamic GRN underlying the process of hindbrain segmentation in jawed vertebrates. Some aspects of the GRN have been shown to be highly conserved in sea lamprey (e.g., segmental expression of Hox PG1-4, krox20 and kreisler) suggesting that hindbrain segmentation originated early in vertebrate evolution. However, it remains unknown whether RA signaling plays a role in the GRN for segmentation in sea lamprey. Thus, this project is about using sea lamprey as a jawless vertebrate model, in order to understand the origin of the coupling of RA to the hindbrain GRN for segmentation in the evolution of vertebrates.
In jawed vertebrates this RA/Hox regulatory hierarchy is coupled to hindbrain segmentation, a vertebrate specific feature of the nervous system (Fig. 1a). The hindbrain is a complex coordination center that regulates vital functions and behaviors and is the site of origin of a subset of neural crest cells, whose derivatives form many craniofacial structures14, 27, 28, 29, 30. Hence, processes that form and pattern the hindbrain are believed to be important for the evolution and emergence of a diversified and complex head in vertebrates31, 32, 33.
During early embryogenesis, the hindbrain is transiently organized into segments (rhombomeres), which lays down a ground plan for patterning neural differentiation, circuit formation and head development14, 26, 34. Hindbrain segmentation is regulated by a conserved GRN which can be visualized as a hierarchical series of regulatory modules that govern sequential steps of the cellular and developmental patterning process. The GRN is initiated by signaling cues, including RA, which setup TFs that subdivide the region into segments, followed by a patterning module, which imparts unique properties to each segment (Fig. 1b)14, 19, 35, 36. In jawed vertebrates, RA signaling is essential for the initiation and regulation of multiple modules, and Hox genes subsequently play important functions in specifying the identity of segments (Fig. 1b). The importance of hindbrain segmentation for craniofacial development of jawed vertebrates raises the question of when during vertebrate evolution the ancient RA/Hox hierarchy became coupled to the process of hindbrain segmentation. Exploring the origin of the coupling of RA signaling to hindbrain segmentation is also important for understanding how ancient signaling pathways and patterning genes can evolve their regulatory interactions to generate more complex GRNs that contribute to morphological diversity.
Lampreys and hagfish belong to a group of jawless vertebrates (cyclostomes) that diverged from other vertebrates ~500 million years ago37. Thus, they have a unique position in the vertebrate tree as a sister group to jawed vertebrates and constitute important models for understanding the evolution of vertebrate traits38. It was previously postulated that the lamprey hindbrain employed a rudimentary form of segmentation, which is only partially coupled to Hox expression and does not involve the action of RA signaling, suggesting roles for RA in hindbrain segmentation arose later in vertebrate evolution39, 40. This finding is consistent with the idea that there has been a gain in connectivity between GRNs and signaling pathways in the evolution of jawed vertebrates from their chordate ancestors, which may have contributed to the formation of new cell types and morphological novelties41. However, through gene expression and cross-species regulatory studies we have recently uncovered clear evidence that Hox genes and other TFs of the hindbrain GRN in jawed vertebrates are also coupled to the process of segmentation in the sea lamprey (Petromyzon marinus) (Fig. 1b)42, 43. This conservation implies that hindbrain segmentation is a pan-vertebrate trait. Paradoxically, RA signaling is not thought to be involved in segmental processes in lamprey39, 40, while it plays major roles in regulating hindbrain segmentation in jawed vertebrates. This disparity raises important questions about the origin of the role of RA signaling in hindbrain segmentation and the evolution of the vertebrate hindbrain GRN.
In light of our findings regarding the presence of key regulatory components of the hindbrain GRN in lamprey42, 43, we wanted to re-examine whether or not RA is coupled to this GRN. Thus, we investigated the link between RA signaling and hindbrain segmentation using sea lamprey as a jawless vertebrate model, comparing it to the current knowledge built from other jawed vertebrates. Combining genomic, gene expression, and functional analyses of major components involved in the synthesis (Aldh1a enzymes) and degradation (Cyp26 enzymes) of RA, we demonstrate that RA signaling is coupled to hindbrain segmentation in lamprey. Our findings reveal that the GRN for hindbrain segmentation and the roles for RA signaling in its regulation were already present in the vertebrate ancestor before the split between jawless and jawed vertebrates.
Results
Identification and phylogenetic analysis of sea lamprey Cyp26 and Aldh1a gene families.
To investigate potential roles for RA signaling in hindbrain development of the sea lamprey (Petromyzon marinus, Pm), we first identified members of gene families predicted to encode components of the enzymatic machinery involved in synthesis (Aldh1a) and degradation (Cyp26) of retinoids associated with neural tube development in jawed vertebrates26. We identified three predicted Cyp26-like genes in the sea lamprey germline genome assembly (KPetmar1)44, 45, Cyp26A1, Cyp26B1/C1a, and Cyp26B1/C1b, and two Aldh1a-like genes, Aldh1a1/a2a and Aldh1a1/a2b. The lamprey Aldh1a1/a2a gene corresponds to the previously identified Aldh1a2 gene46. Within the cyclostome group, we also searched the hagfish genome (Eburgeri3.2) and found two Cyp26-like genes and one Aldh1a1 gene. The gene structures and lengths of the predicted proteins of lamprey and hagfish Cyp26 and Aldh1a genes show a high degree of similarity to their putative jawed vertebrate counterpart genes (Supplementary Table 1-Table 4).
To explore properties of the enzymes encoded by the lamprey Cyp26 and Aldh1a genes, we searched for specific catalytic sites essential for the activity of these protein families in other vertebrate species. Protein alignments of the Cyp26 family show conserved amino-acid (AA) sequences for the I/K Helices and the Heme domain in lamprey and hagfish (Supplementary Figure 1)47. Similarly, alignments between Aldh1a proteins reveal that key catalytic domains of this family - the Cysteine (Cys) and Glutamic Acid (Glu) residues - are conserved in the lamprey Aldh1a1/a2 proteins (Supplementary Figure 2)48.
Phylogenetic analyses of predicted protein sequences were performed to examine their evolutionary relationship with jawed vertebrate Aldh1a and Cyp26 genes. All vertebrate Cyp26A1 genes group together as a clade, with high bootstrap support (81) (Fig. 2a). We did not retrieve a Cyp26A1 homologue in the hagfish genome, which may reflect an incomplete assembly or that Cyp26A1 was lost in hagfish. The vertebrate Cyp26B1/C1 genes form a sister clade to the Cyp26A1 clade, with strong bootstrap support (99) (Fig. 2a), indicating a separation between Cyp26A1 and Cyp26B1/C1 genes prior to the divergence of jawed and jawless vertebrates. Within the Cyp26B1/C1 clade, there is support for separate jawed vertebrate Cyp26C1 and Cyp26B1 clades. While lamprey Cyp26B1/C1a and Cyp26B1/C1b appear to group with the two hagfish Cyp26C1-like genes, we are unable to infer any 1:1 orthology between cyclostomes and jawed vertebrates.
Figure 2: Phylogenetic and synteny analyses of the vertebrate Cyp26 and Aldh1a complements.

(a) Phylogenetic analysis of the sea lamprey Cyp26 and Aldh1a complements with Cyp51 and Aldh1a8 used as outgroups respectively. Jawed Vertebrate (J.V.) Cyp26B1/Cyp26C1, J.V. Aldh1a1/Aldh1a2, and Vertebrate (V.) Cyp26A1 clades are indicated with black lines. Trees were generated by Maximum Likelihood using the WAG model with 500 iterations for bootstrap testing, and the resulting supporting value for each node is shown as a percentage. A scale bar for the evolutionary distance is indicated below each tree. Species name abbreviations are indicated on the bottom right of the panel;
(b) Schematic illustrating the synteny analysis of the Aldh1a and Cyp26 gene complements conducted for major jawed vertebrate lineages (J.V.) and the sea lamprey. The corresponding direction of transcription is represented with an arrow. Genes located in proximity to the Cyp26 and Aldh1a genes and used for this analysis as well as their direction of transcription are represented by grey arrows. Synteny groups with gene names are represented at the top of the panel (in bold). Chromosome numbers, scaffold or linkage groups (LG) names are indicated below the loci. Sea lamprey Cyp26 and Aldh1a complements are color-coded in teal and blue respectively.
For the Aldh1a complement, the tree shows clear jawed vertebrate clades for Aldh1a1 and Aldh1a2. However, it does not resolve clear relationships between the cyclostome and jawed vertebrate Aldh1a genes, since the cyclostome Aldh1a genes do not group with either Aldh1a1 or Aldh1a2 clades, nor with each other. In jawed vertebrates, the lineage that led to Aldh1a3 genes appears to have diverged early from the ancestor that led to the Aldh1a1/Aldh1a2 clade49. Hence, we examined whether the Pm Aldh1a1/a2 genes could be related to Aldh1a3. However, including the Aldh1a3 family in this analysis did not help to resolve the relationship of the sea lamprey Aldh1a1/a2a and Aldh1a1/a2b with the vertebrate Aldh1as (Supplementary Figure 3).
Synteny analysis of vertebrate Cyp26 and Aldh1a genomic loci.
To gain further insight into the relationship between the sea lamprey and jawed vertebrate Cyp26 and Aldh1a complements, we looked for evidence of conserved local synteny across vertebrate lineages (Fig. 2b). Despite observing clear syntenic groups in the jawed vertebrate Aldh1a1 and Aldh1a2 genomic loci, we did not find any indication of shared synteny with lamprey Aldh1as (Fig. 2b).
In most jawed vertebrate lineages Cyp26A1 is positioned directly adjacent to a Cyp26C1 gene (Fig. 2b). In sea lamprey, Pm Cyp26A1 and Cyp26B1/C1a are also located adjacent to each other on chromosome 11, and we uncovered evidence of syntenic relationships around the Pm Cyp26A1-Cyp26B1/C1a locus (Myof, Exo6) shared with other vertebrates. These similarities in the genomic organization suggest that these genes arose by tandem duplication before the split between jawed and jawless vertebrates, and that this arrangement was subsequently maintained in most vertebrate lineages. Our analysis of the third Cyp26 gene, Cyp26B1 or lamprey Cyp26B1/C1b, also reveals evidence for shared synteny (Dysf and Exo6b) between lamprey and jawed vertebrates (Fig. 2b).
In summary, the data from phylogenetic and synteny analyses support an early separation of vertebrate Cyp26A1 and Cyp26B1/C1 genes prior to the divergence of jawed and jawless vertebrate lineages. Lamprey Cyp26A1 appears to be a clear ortholog of jawed vertebrate Cyp26A1, but the evolutionary history of the Cyp26B1/C1 group in vertebrates is less clear. This suggests that an ancestral vertebrate had one Cyp26A1 gene and at least one Cyp26B1/C1-like gene. In contrast, the ancestry of the vertebrate Aldh1a genes is not as clear, with cyclostome genes forming basal branches in the phylogeny and not falling within either of the jawed vertebrate Aldh1a1 and Aldh1a2 clades.
Gene expression analysis of sea lamprey Cyp26 and Aldh1a genes.
We next investigated the expression patterns of Cyp26 and Aldh1a genes in sea lamprey embryos. We analyzed existing RNAseq data sets for a series of early developmental stages50, 51 to determine whether any of these genes are expressed during periods relevant to hindbrain segmentation (Fig. 3a). Cyp26A1 and Aldh1a1/a2b are expressed during blastula and gastrulation stages (Tahara stages st7-st17)52 but are downregulated during neurulation and hindbrain segmentation (st17-st24). In contrast, Cyp26B1/C1a and Aldh1a1/a2a are not expressed at the earliest stages but are upregulated during gastrulation, neurulation, and segmentation (Fig. 3a). Cyp26B1/C1b is not expressed at any of these developmental times.
Figure 3: RNA seq profile and gene expression analysis of sea lamprey Cyp26A1, Cyp26B1/C1a and Aldh1a1/a2a.
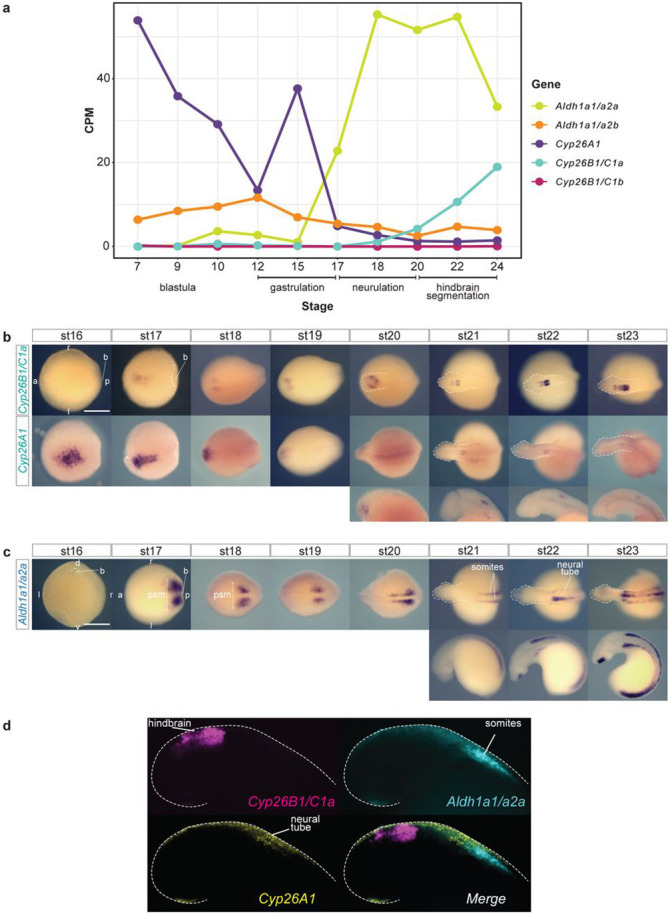
(a) RNAseq profile of sea lamprey Cyp26A1, Cyp26B1/C1a and Aldh1a1/a2a at blastula (~st9), gastrulation (st12-st15), neurulation (st15/16) and hindbrain segmentation (st20-24);
(b) mRNA expression of sea lamprey Cyp26A1, Cyp26B1/C1a revealed by cISH. The orientation of the embryo, anterior (a), posterior (p), left (l) and right (r) is indicated on the image. Cyp26A1 expression from st20 to st23 is shown with both dorsal view of the whole embryo (top) and lateral (bottom) view of the embryo’s head;
(c) mRNA expression of sea lamprey Aldh1a1/a2a revealed by cISH, expression at st21 to st23 is shown with both dorsal (top) and lateral (bottom) view of the whole embryo. Various embryological structures are indicated such as the blastopore (b) and the presomitic mesoderm (psm);
(d) HCR image showing the relative position of Cyp26B1/C1a (magenta), Cyp26A1 (yellow) and Aldh1a1/a2a (cyan) expression domains at stage 22. stage (st).
Based on these data, we focused on Cyp26A1, Cyp26B1/C1a and Aldh1a1/a2a and characterized their spatio-temporal expression patterns in the period between mid-gastrulation and late hindbrain segmentation (st16-st23) by colorimetric in situ hybridization (cISH) (Fig. 3b, c). Cyp26A1 expression is dynamic over these stages. Expression is detected in the anterior neural plate at the beginning of neurulation (st16), persists between st17-st19 and then is progressively down-regulated in the most anterior part of the developing neural tube (Fig. 3b). At st20 and later stages, low levels of Cyp26A1 expression are detected in lateral regions of the head, which appear to correspond to the optic placode, and in the neural tube at the level of the caudal hindbrain.
For Cyp26B1/C1a, between st16-st20, weak expression is observed in small patches lateral to the developing neural tube and at st20 a restricted domain of expression is visible within the hindbrain primordium (Fig. 3b). Between st21-st23, two non-adjacent stripes are observed in the hindbrain and then form a series of stripes of differing intensities that cover most of the hindbrain. This dynamic pattern is reminiscent of the expression of Cyp26B1 and Cyp26C1 in specific rhombomeres of zebrafish and mouse embryos36, 53, 54, suggesting that Cyp26B1/C1a is also coupled to segmentation in lamprey.
A previous study characterized the expression of Aldh1a1/a2a in dorsal interneurons of the lamprey spinal cord (st22-st25) and linked this pattern with an ancient conserved intronic enhancer46. In earlier stages, we find that expression of Aldh1a1/a2a is first visible at st16 around the blastopore. At st17, during the early stages of neurulation, it is highly expressed in two posterior domains corresponding to the presomitic mesoderm (PSM) and this expression is maintained as the embryo undergoes neurulation (Fig. 3c). At st19/20, Aldh1a1/a2a is expressed in the PSM and newly developing somites adjacent to the neural tube, as well as in the lateral plate mesoderm in the posterior part of the trunk. During later stages (st22-st23), Aldh1a1/a2a expression persists in the somitic mesoderm and PSM, but also begins to appear in the dorsal spinal cord and in mouth tissue (Fig. 3c). The expression in somite and PSM domains is highly similar to the expression of Aldh1a2 in zebrafish and mouse embryos55, 56, 57 where it serves as a primary source of RA synthesis required for regulation of patterning in the developing hindbrain14, 36, 53, 58, 59. Aldh1a1/a2a expression in lamprey paraxial mesoderm opens the possibility that it may play a similar role in generating RA important for regulating hindbrain segmentation.
Expression of Cyp26s and Aldh1a genes is linked to lamprey hindbrain segmentation.
Given the dynamic expression of Cyp26A1, Cyp26B1/C1a and Aldh1a1/a2a in mesodermal and neural territories, we next investigated how their spatial and temporal patterns of expression relate to each other and whether any of these are correlated with specific rhombomeres. We optimized a protocol for Hybridization Chain Reaction fluorescent in situ hybridization (HCRv3-FISH)60 (Supplementary Figure 4), to visualize and directly compare the expression of these genes.
During early hindbrain segmentation (st21), Cyp26A1 (yellow) is restricted to the most anterior part of the spinal cord, in a domain abutting the somitic expression of Aldh1a1/a2a (cyan) (Fig. 3d). This spatial relationship is reminiscent of that seen for jawed vertebrate models, where RA is synthesized in the somites by Aldh1a2 and degraded anteriorly in the hindbrain by Cyp26B1 and Cyp26C136, 53, 54, 55, 56, 57. Thus, the expression of lamprey Cyp26A1 in a domain adjacent to Aldh1a1/a2a, positions it for a potential role in modulating RA levels as it spreads more anteriorly in the neural tube.
Cyp26B1/C1a (magenta) is strongly expressed in the hindbrain, anterior to Cyp26A1 (Fig. 3d). In order to determine whether the stripes of Cyp26B1/C1a expression align with developing rhombomeres, we performed HCRv3-FISH assays using probes for Cyp26B1/C1a and krox20, a marker of r3 and r5, which is initially expressed in r3 and then in r3/r5 in lamprey and other vertebrates34, 61, 62 (Fig. 4a). The initial neural domain of Cyp26B1/C1a expression (magenta) (st20) directly overlaps with krox20 (cyan) in r3 and by st21 both display segmentally-restricted expression in r3 and r5. By st22, when segmentation is nearing completion, Cyp26B1/C1a expression covers a region from r1 to r6, with higher levels in r3, r5 and r6 compared to r1-2 and r4 (Fig. 4a, b). This dynamic expression of Cyp26B1/C1a in lamprey shows that it is coupled to segmentation and is reminiscent of zebrafish Cyp26b1 and Cyp26c1 in the r2-r6 region53.
Figure 4: Cyp26B1/C1a is directly coupled to hindbrain segmentation in the sea lamprey.
(a) HCR images showing the individual expression patterns of Cyp26B1/C1a (magenta) and krox20 (cyan) as well as their merged expression during the process of hindbrain segmentation (st20 to st22). Rhombomeres (r) are numbered 2 to 7. st20 is a lateral view of the developing head while st21 and st22 are dorsal views of the developing head.
(b) Cartoon summarizing the segmental expression of Cyp26B1/C1a at stage 22; zoomed HCR image of Cyp26B1/C1a (magenta) and krox20 (cyan) expression at stage 22. stage (st).
Taken together, these patterns of spatio-temporal gene expression in lamprey embryos are consistent with potential roles for the Aldh1a1/a2a, Cyp26A1 and Cyp26B1/C1a genes in regulating levels of RA that impact hindbrain segmentation.
Pharmacological modulation of endogenous RA levels impacts hindbrain segmentation.
To investigate whether RA signaling is functionally involved in regulating hindbrain segmentation in lamprey, we utilized pharmacological inhibitors of the Aldh1a2 and Cyp26 enzymes to perturb endogenous levels of RA. This approach has previously been used to characterize roles for RA signaling during hindbrain segmentation in zebrafish53, 63. Based on the timing of expression of Aldh1a1/a2a and Cyp26 genes (Fig. 3 b-d), we used the following experimental design: batches of early gastrula embryos (st13) were grown until the end of hindbrain segmentation (st23), in media containing: i) 10 μM Talarazole, an inhibitor of Cyp26 enzymes that increases levels of RA; ii) 50 μM DEAB, which inhibits Aldh1a2 enzymes and reduces levels of RA synthesis; or iii) 0.1% DMSO, as a control (Fig. 5a). To assess the effect of these treatments on hindbrain segmentation, we performed cISH on the drug-treated embryos using probes for key genes involved in hindbrain patterning: a) otx and wnt1 mark subdivision of the forebrain and midbrain26; b) krox20 and kreisler are segmental subdivision genes that mark the r3/r5 and r5/r6 territories, respectively34, 42; and c) hoxβ1, hoxα2, hoxα3, hoxζ4 are segmental identity genes each marking different groups of rhombomeres (Fig. 5a)42.
Figure 5: Inhibition of synthesizing and degrading enzymes alters segmental patterning in the sea lamprey.
(a) Cartoons illustrating the different gene markers used to reveal various aspects of hindbrain patterning, as well as the effect of Talarazole in inhibiting the activity of the Cyp26 enzymes leading to increased levels of RA (less degradation) while the use of DEAB inhibits the activity of Aldh1a2, leading to reduced levels of RA synthesis; cISH of key patterning hindbrain markers in embryos treated with DMSO (b), Talarazole (c) and DEAB (d). Otx and wnt define the hindbrain territory within the developing CNS (CNS subdivision), krox20 and kreisler participate in the early process of making segments as part of the segmental subdivision module of the hindbrain GRN and hoxβ1, hoxα2, hoxα3, hoxζ4 contribute to the segmental identity aspect of the hindbrain GRN. For each drug condition, lateral and dorsal views of stage 23 embryos are shown on top and bottom rows respectively (only heads are a shown). For each gene, the most representative ISH phenotype is shown, and numbers of experimental replicate are indicated. Important features are indicated such as the forebrain (fb), midbrain (mb) and rhombomeres (r) of the hindbrain;
In DEAB-treated embryos, the otx and wnt1 expression domains define clear forebrain- and midbrain territories, that appear unaltered compared to control embryos (Fig. 5b, d). In contrast, expression of the segmental subdivision genes, krox20 and kreisler, is no longer detected in the neural tube (Fig. 5d). Furthermore, none of the 4 selected hox genes are expressed in the hindbrain or the spinal cord of DEAB-treated embryos, with the exception of hoxα2, which maintains a faint anterior domain of expression in the hindbrain that may represent an r2-like region (Fig. 5d). These data (summarized in Fig. 6b) imply that proper levels of RA are required for activating and/or maintaining regulatory modules of the hindbrain GRN necessary for subdividing the hindbrain into segments and patterning their distinct identities via the Hox genes, at least posterior to r2.
Figure 6: Consequences of inhibiting RA synthesis and degradation on the hindbrain GRN for segmentation.
(a) HCR-FISH images showing the individual expression patterns of hoxβ1 (magenta), krox20 (cyan) and kreisler (yellow) as well as their merged expression in DMSO and Talarazole stage 23 embryos. Lateral views of the head are shown and rhombomeres (r) are indicated; (b) Cartoon summarising the expression of hindbrain markers based on cISH, HCR-FISH and GFP-reporter analysis in DMSO, Talarazole and DEAB treated embryos. Rectangles with dashed lines are used in DEAB to represent where the expression of the gene would be expected in WT embryos. fb, forebrain; mb, midbrain; rhombomere (r); (c) Image of live Drhoxb3a-GFP sea lamprey reporter embryos in DMSO, Talarazole and DEAB treated embryos. Drhoxb3a-GFP reports activity of the enhancer in rhombomere 5 (r5). Images shown represent the most common phenotype of GFP reporter expression (~50% for DMSO, 50% for Talarazole and 100% for DEAB embryos). Corresponding ratio of positive embryos are indicated (number of embryos showing the phenotype/number of GFP positive embryos).
In Talarazole-treated embryos, otx and wnt1 exhibit major changes in their expression domains (Fig. 5b, c). otx is restricted to a small faint domain of expression in the anterior part of the head, while wnt1 expression is reduced and shifted more anteriorly, such that its anterior boundary now roughly corresponds to the most anterior part of the neural tube. This indicates that elevated levels of RA cause a reduction in size of the forebrain and midbrain territories which are shifted anteriorly to a small region located at the tip of the head.
There are also significant changes in the expression of genes regulating segment formation (Fig. 5c). krox20 is expressed in two small stripes, corresponding to r3- and r5-like territories. The posterior r5-like stripe of expression of both krox20 and kreisler is very faint and difficult to detect, implying a major reduction in the size of the r5-like territory. These data imply that RA may regulate the genetic programs setting up r3 and r5 in different ways, which is in line with evidence in jawed vertebrates indicating that different mechanisms are involved in the regulation of Krox20 in r3 and r562, 64, 65, 66, 67. There are also dramatic perturbations in the normal nested and segmental patterns of hox expression in the hindbrain. In control embryos, hox genes have defined anterior limits of expression in specific rhombomeres (Fig. 5a) but upon exposure to Talazarole, the expression of all 4 hox genes is dramatically upregulated, shifted anteriorly, and lack segmental stripes seen in control embryos (Fig. 5b, c). This is consistent with the malformation of segments observed by loss of expression of segmental subdivision genes. Hence, in lamprey the formation and patterning of the developing fore-, mid- and hind- brain are strongly affected by the inhibition of Cyp26 activity, as summarized in Fig. 6b. Together, these experiments with drug treatments illustrate the need to maintain the proper balance between RA synthesis and degradation for regulating hindbrain segmentation.
Characterization of changes in segmental patterning in drug-treated embryos.
To gain a better understanding of the specific changes in segmental gene expression at the level of r3, r4 and r5 observed in Talarazole-treated embryos we used the adapted lamprey HCRv3 protocol with markers for these rhombomeres: hoxβ1 (r4, magenta), krox20 (r3/r5, cyan) and kreisler (r5, yellow) (Fig. 6a, Supplementary Figure 4). We observe a faint dispersed expression domain of kreisler that overlaps with a weak posterior patch of krox20 expression, indicating that this domain corresponds to a small residual r5-like territory (Fig. 6a). Therefore, the most anterior domain of krox20 expression corresponds to an r3-like territory and is less severely affected by changes in levels of RA than the r5-like domain. hoxβ1 expression in the spinal cord is shifted anteriorly, but we do not detect a clear r4-like stripe adjacent to the r5-like domain of krox20 (Fig. 6a, Supplementary Figure 4), suggesting the putative r4 territory may have adopted an indeterminate fate. These changes are summarized in Fig. 6b and highlight the impact that elevated levels of RA have in altering programs that regulate the formation of segments and segmental identity.
To further investigate the fate of r5, we combined the use of a transgenic GFP-reporter assay and pharmacological treatments. An enhancer from the zebrafish hoxb3a gene (Drhoxb3a) contains binding sites for Kreisler and Krox20 TFs and mediates GFP-reporter activity in r5 of transgenic lamprey embryos (DMSO panel in Fig. 6c)34. In Talarazole-treated transgenic embryos, r5 reporter activity is much fainter and shifted anteriorly (Fig. 6c), consistent with the anterior shifts previously seen for krox20 and kreisler expression (Fig. 5c). Conversely, DEAB-treated transgenic embryos show no reporter activity in r5 (Fig. 6c), consistent with the loss of expression of kreisler and krox20 in DEAB-treated embryos (Fig. 5d). This regulatory assay further supports that a small r5-like domain persists in Talarazole-treated embryos. It also reveals that the impact of RA perturbation on the early steps of segment formation has important consequences on the activation of the molecular and cellular programs that work downstream in the progressive process of segmentation.
Collectively, these pharmacological perturbations highlight the importance of maintaining appropriate levels of RA for regulation of the cellular and molecular processes that govern hindbrain segmentation in lamprey. The experiments suggest that RA plays important roles in multiple aspects of the GRN for hindbrain segmentation in the sea lamprey: 1) in delimiting the future hindbrain territory; 2) in setting up the formation of segments (segmental subdivision via kreisler and krox20 expression); and 3) in defining the molecular identity of each of these segments via hox expression. The changes we observed are analogous to those seen in jawed vertebrate embryos treated with these drugs53, 54, 63, 68, implying that there are similar regulatory inputs from RA into the hindbrain GRN in all vertebrates.
Cyp26A1 and Cyp26B1/C1a are required for hindbrain segmentation in the sea lamprey.
Because pharmacological inhibitors may affect multiple proteins and have off-target or indirect effects, it is important to demonstrate that the changes in hindbrain patterning we observed after treatment with DEAB or Talarazole are directly linked to the Cyp26A1, Cyp26B1/C1a and Aldh1a1/a2a genes encoding these enzymes. Hence, we conducted functional analyses of these genes in sea lamprey by implementing a CRISPR-mediated mutagenesis approach using guide RNAs (gRNAs) designed to target regions that encode key catalytic sites (Fig. 7a)47, 48. We obtained robust and reproducible mutant phenotypes and sequenced the targeted regions of the genes in affected embryos to confirm their genotypes (Supplementary Figure 5 and 6).
Figure 7: Both Cyp26A1 and Cyp26B1/C1a are required for hindbrain segmentation in the sea lamprey.

(a) Schematic representation of the general structure of a Cyp26 protein with its highly conserved enzymatic sites, I helix, K Helix and Heme group; (b) Schematic representation of the sea lamprey Cyp26 locus which includes Cyp26B1/C1a and Cyp26A1 genes, and the sequences targeted by CRISPR/Cas9 gRNA corresponding to the highly conserved enzymatic sites; (c) cISH of key patterning hindbrain markers in CRISPR embryos treated: krox20 and kreisler are used as markers of segmentation (green) while hoxβ1 and hoxζ4 are used as markers of segmental identity (purple). For each gene, the most representative ISH phenotype is shown, and numbers of experimental replicate are indicated. Lateral and dorsal views of stage 23 embryos are shown on the top and bottom rows respectively (only heads are a shown). rhombomeres (r) are indicated; (d) Cartoon summarizing the expression of hindbrain markers in Cyp26 CRISPR (Cr Cyp26s) embryos. Rectangles with dashed lines are used in the Cr Cyp26s panel to represent where the expression of the gene would be expected in WT embryos. fb, forebrain; mb, midbrain; rhombomere (r); Cartoon summarizing the molecular phenotypes of gene markers of hindbrain segmentation obtained by cISH in Talarazole treated embryos (in which the Cyp26s are inactivated) ; fb, forebrain; mb, midbrain.
Following CRISPR-mediated mutagenesis of the Cyp26A1 and Cyp26B1/C1a genes (Fig. 7b; Supplementary Figure 5), we performed cISH analysis on CRISPR (Cr) embryos at st23 using marker genes to investigate the formation of segments (krox20 and kreisler) and segmental identity (hoxβ1 and hoxζ4) (Fig. 7c). When individual genes, CrCyp26A1 or CrCyp26B1/C1a, were targeted all of these markers are shifted anteriorly. The relative levels of krox20 and kreisler appear similar to that observed in control embryos, but there is reduced expression of hoxβ1 and hoxζ4 and loss of a clear r4 stripe of hoxβ1 expression (Fig. 7c). The molecular and morphological phenotypes of the CrCyp26A1 and CrCyp26B1/C1a embryos are very similar, suggesting that these two Cyp26 genes may functionally compensate for each other when one of them is disrupted. Hence, we targeted both Cyp26A1 and Cyp26B1/C1a (CrCyp26s) in individual embryos, which generated a more severe phenotype with a dramatic loss of krox20, kreisler and hoxβ1 expression, and an anterior shift of hoxζ4 (Fig. 7c). This shows that both Cyp26A1 and Cyp26B1/C1a are necessary for regulating endogenous levels of RA in the developing hindbrain, as in the absence of these two genes expression of segmental markers is completely lost (Fig. 7c CrCyp26s). Cyp26 genes also play partially redundant roles in mouse and zebrafish hindbrain patterning36, 53.
The molecular phenotype observed in CrCyp26s embryos is similar to but more severe than the one observed in Talarazole-treated embryos, where the Cyp26 activity is inhibited (Fig. 7d). This difference may be explained by the timing of the drug treatment and/or the degree of impact on enzymatic activity, as compared to the CRISPR embryos, which would be expected to have little or no Cyp26 activity. The extensive molecular phenotypes observed in the CrCyp26s embryos clearly show that Cyp26 genes play key roles in regulating levels of RA that have important inputs into the GRN for hindbrain segmentation in lamprey.
Aldh1a1/a2a contributes to hindbrain segmentation in the sea lamprey.
We targeted Aldh1a1/a2a for functional perturbation by designing gRNAs to delete regions around the Cysteine (Cys) and Glutamic acid (Glu) residues in the conserved catalytic sites responsible for enzymatic activity (Fig. 8a, b; Supplementary Figure 2 and 6)48. CrAldh1a1/a2a embryos exhibit severely truncated heads that morphologically appear very similar to DEAB-treated embryos (Fig. 8c). We detected a posterior shift in the expression domains of krox20 and kreisler and elongation of caudal hindbrain segments. There is also a complete loss of hoxζ4 expression and a posterior shift and elongation of hoxβ1 expression domain in r4. Thus, CRISPR-mediated disruption of Aldh1a1/a2a alters segmental patterning, particularly in the r5-r7 region. This demonstrates roles for RA via Aldh1a1/a2a in regulating hoxζ4 and hoxβ1, as well as providing inputs into the regulation of kreisler and krox20 in r5.
Figure 8: Aldh1a1/a2a influences hindbrain segmentation in the sea lamprey.

(a) Schematic representation of the general structure of Aldh1a2 with its highly conserved catalytic sites, Glutamate (Glu) and Cysteine (Cys); (b) Schematic representation of the sea lamprey Aldh1a1/a2a gene, and the sequences targeted by CRISPR/Cas9 gRNA corresponding to the conserved Glu and Cys sites; (c) cISH of key patterning hindbrain markers in CRISPR embryos treated: krox20 and kreisler are used as markers of segmentation (green) while hoxβ1 and hoxζ4 are used as markers of segmental identity (purple). For each gene, the most representative ISH phenotype is shown, and numbers of experimental replicate are indicated. Lateral and dorsal views of stage 23 embryos are shown on the top and bottom rows respectively (only heads are a shown). rhombomeres (r) are indicated; (d) Cartoon summarizing the expression of hindbrain markers in Aldh1a1/a2a CRISPR (Cr Aldh1a1/a2a) embryos. Rectangles with dashed lines are used in the Cr Aldh1a1/a2a panel to represent where the expression of the gene would be expected in WT embryos. fb, forebrain; mb, midbrain; rhombomere (r).
The genetic disruption of Aldh1a1/a2a induces a relatively milder mis-patterning of the hindbrain compared with DEAB treatment (Fig. 5c, 6b) and the CrAldh1a1/a2a phenotype is very similar to that observed in embryos treated with a lower concentration of DEAB (10 μM) (Supplementary Figure 7). This is analogous to findings in zebrafish, where the nls/raldh2 (Aldh1a2) mutant phenotype is not as severe as those observed in DEAB-treated embryos55, 63. This suggests that zebrafish Aldh1a2 and sea lamprey Aldh1a1/a2a may not be the sole source of RA synthesis, contrasting with mouse where Aldh1a2 appears to be the main source of RA synthesis23. Collectively these results imply the presence of additional Aldh inputs into RA synthesis during hindbrain segmentation that may be inhibited by DEAB acting as a pan-Aldh inhibitor69. In sea lamprey, targeting Aldh1a1/a2b in addition to Aldh1a1/a2a did not result in a more severe perturbation of hindbrain development (data not shown) further suggesting the presence of additional unidentified Aldh inputs into RA synthesis during hindbrain segmentation.
Using gene editing approaches, we have generated independent evidence on functional roles of the Cyp26A1, Cyp26B1/C1a and Aldh1a1/a2a genes that are consistent with conclusions from our analyses using pharmacological treatments. Collectively these data demonstrate that RA signaling is coupled to the GRN for hindbrain segmentation and hox patterning in sea lamprey.
Cyp26A1, Cyp26B1/C1a and Aldh1a1/a2a respond to changes in levels of RA.
In jawed vertebrates, some genes involved in the synthesis and degradation of RA are directly regulated by RA. In zebrafish, mouse and human models, Cyp26A1 is directly regulated by RA via a conserved set of RAREs (R1, R2 and R3) located in its promoter region70, 71, 72, 73. Studies in zebrafish also point to a role for RA in repressing the expression of Aldh1a2, as applying exogenous RA leads to reduced activity of its promoter region and there is an upregulation of Aldh1a2 transcription in the nls/raldh2 mutant 55, 74. These data illustrate that in jawed vertebrates, feedback mechanisms play an important role in regulating levels of RA by controlling its synthesis and degradation. Hence, we investigated whether lamprey Cyp26A1, Cyp26B1/C1a and Aldh1a1/a2a are responsive to changes in levels of RA during hindbrain segmentation. Increasing the levels of RA by exposure to Talarazole leads to reduced expression of Aldh1a1/a2a, while reduced levels of RA synthesis mediated by DEAB lead to increased expression of Aldh1a1/a2a (Fig. 9a). The expression of Cyp26A1 is lost in DEAB-treated embryos and is strongly increased in Talarazole-treated embryos, suggesting that RA has an important and potentially direct role in initiating and maintaining its expression. Cyp26B1/C1a expression is also affected by the drug treatments, as in both cases the domain of expression appears smaller and weaker than in control embryos (Fig. 9b). These changes in expression, summarized in Fig. 9c, are consistent with the predicted differences in segmental organization induced by the drug treatments (Fig. 5 and 6) and hence are likely to be indirect. Our data indicate that the negative regulation of Aldh1a1/a2a and the positive regulation of Cyp26A1 by RA are important aspects of feedback regulation of RA synthesis and degradation during hindbrain segmentation in all vertebrates (Fig. 9d).
Figure 9: Sea lamprey Aldh1a1/a2a, Cyp26A1, Cyp26B1/C1a respond to drug-induced changes in the levels of RA.
cISH of (a) Aldh1a1/a2a and (b) Cyp26A1 and Cyp26B1/C1a in DMSO, Talarazole and DEAB treated embryos. For each drug condition, lateral and dorsal views of stage 23 embryos are shown on the top and bottom rows respectively (only heads are a shown). For each gene, 4 embryos were used per treatment condition, and all showed the phenotype shown in the image; (c) cartoon summarizing the gene expression in each treatment condition. Rectangles with dashed lines are used in the DEAB cartoon to represent where the expression of the gene would be expected in WT embryos. fb, forebrain; mb, midbrain; rhombomere (r); (d) Cartoon illustrating mechanisms of self-regulation of the RA machinery where RA downregulates the expression of Aldh1a1/a2a, while up regulating the expression of Cyp26A1, therefore exerting a control on its own synthesis and degradation.
Discussion
We have investigated the origin and evolution of the coupling of RA to hindbrain segmentation in vertebrates, using sea lamprey as a model. We demonstrated that components of the RA signaling pathway mediating RA synthesis (Aldh1as) and degradation (Cyp26s) are expressed in a spatio-temporal manner consistent with roles in regulating hindbrain segmentation in lamprey. Notably, we found Cyp26B1/C1a expression aligns with specific rhombomeres. Using pharmacological treatments and CRISPR/Cas9 gene editing to perturb the activity of these components we showed that multiple aspects of hindbrain segmentation and patterning in lamprey require RA signaling. Furthermore, lamprey Cyp26A1 and Aldh1a1/a2a genes themselves also appear to be regulated by feedback circuits involving RA signaling. Collectively, these results reveal that an ancestral RA/Hox regulatory circuit for axial patterning became coupled to the process of hindbrain segmentation prior to the split between jawed and jawless vertebrates (Fig. 10). These findings raise interesting questions and avenues for further investigation.
Figure 10: The role of RA signaling in the GRN for vertebrate hindbrain segmentation is conserved to the base of vertebrates.
In this study, using sea lamprey as a jawless vertebrate model, we show that the origin of the coupling of RA to the hindbrain GRN for segmentation occurred before the split between jawed vertebrates and cyclostomes as RA signaling is directly coupled to the hindbrain GRN for segmentation in the sea lamprey.
Our analyses of the Cyp26 genes in lamprey reveal similarities and differences in their coupling to hindbrain segmentation in jawed and jawless vertebrates. In both groups, Cyp26A1 is expressed in the anterior neural plate during gastrulation, while Cyp26B1, Cyp26C1 and Cyp26B1/C1a are expressed later in a dynamic and segmental manner across r2-r6 (Fig. 3 and 4)14, 36, 53, 54. However, rhombomeric patterns vary significantly between vertebrate species. For example, lamprey Cyp26B1/C1a has lower levels of expression in r2/r4 compared to r3/r5/r6 (Fig. 4). In contrast, zebrafish Cyp26b1 and Cyp26c1 expression is higher in r2/r4/r6 than in r3/r5, where they are repressed by Krox2053, 75. This dynamic segmental expression has been linked to important processes in hindbrain segmentation: a) the generation of shifting domains of RA that modulate the anterior limit of expression of genes that prefigure rhombomere boundaries53, 54; b) segmental variation in RA levels across odd versus even rhombomeres that influences formation of sharp segmental boundaries by community signaling75, 76. Thus, variation in Cyp26 rhombomeric domains between vertebrate species may reflect divergence in their precise roles in regulating hindbrain segmentation. It is unknown whether this variation in segmental expression of Cyp26s translates to different patterns of rhombomeric RA concentrations between species. Visualizing or measuring endogenous RA across the developing hindbrain in different models could be enlightening in this regard77.
From an evolutionary perspective, the conserved deployment of A-P patterning genes, such as Hox genes, reflects an ancient and highly conserved GRN for axial patterning that was probably present in the bilaterian ancestor2, 3. FGF and Wnt signaling are important in this axial patterning GRN 2, 6, 7, but the coupling of RA signaling to Hox genes and A-P patterning appears to have arisen later in the deuterostome lineage16, 26. Indeed, invertebrate chordate studies revealed that ancestral chordates employed an RA/Hox regulatory circuit to pattern the neural territory5, 24, 25, 78 and here we show that the role of the ancestral RA/Hox regulatory hierarchy appears to have been elaborated in early vertebrates, becoming coupled to the process of hindbrain segmentation. This could have occurred via evolution of cis-regulatory inputs of RA into gene expression through novel or co-opted RAREs, as has been shown in the Hox clusters17, 25, 26. Supporting this idea, recent comparative genome-wide studies of open chromatin during development revealed more enhancers per gene in zebrafish than in amphioxus, implying an increase in regulatory complexity in vertebrates41, 79. Some of these vertebrate-specific elements respond to multiple signaling pathways (RA, Wnt, and FGF), suggesting that an increase of interactivity between these pathways evolved in early vertebrates and may underlie increased tissue complexity in vertebrates41. Thus, it is likely that novel regulatory inputs from these signaling pathways into target genes played an important role in the evolution of the GRN for vertebrate hindbrain segmentation.
The role of RA signaling in vertebrate hindbrain segmentation could also be linked to the evolution of new expression domains of genes that shape and interpret RA concentrations along the A-P axis, such as Cyp26s, rars, and Crabps26. This idea is supported by the Cyp26 genes, which exhibit shared and derived features between vertebrates and invertebrate chordates, such as amphioxus. For example, there is a conserved early role for Cyp26s acting as anterior sinks for shaping the RA gradient in chordates but their dynamic segmental expression during hindbrain development in vertebrates is not seen in amphioxus80, 81. Since this segmental expression is linked to multiple aspects of rhombomere formation, it is important to understand how this arose in vertebrates as opposed to other chordates.
Cyp26A1 is induced by RA in jawed vertebrates via multiple RAREs70, 72, 82 and we have shown that lamprey Cyp26A1 also requires RA for its expression (Fig. 9). Similar responses were found in amphioxus Cyp26s, and an ancient pan-deuterostome RARE has been implicated in this role81, suggesting that the RA-dependent regulation of Cyp26s may be an ancestral feature of deuterostomes. Functional characterization of RAREs around the tandemly linked Cyp26A1-Cyp26B1/C1a genes in lamprey could help decipher ancestral versus vertebrate-specific RA-dependent regulatory mechanisms.
Feedback-regulation of the RA machinery via Cyp26 genes may constitute an ancestral mechanism that was further elaborated in vertebrates. However, it is also possible that the rhombomere-specific expression of vertebrate Cyp26 genes could be linked to vertebrate-specific inputs from segmentally expressed TFs, such as Krox20. Given the variation in rhombomeric Cyp26 expression between vertebrates, such inputs could vary between models. For example, Cyp26C1 expression in r4 is induced by RA in mouse embryos but not in zebrafish53, 54. In lamprey, we found that Cyp26B1/C1a expression in the hindbrain does not depend upon RA, indicating that it requires other regulatory inputs to generate its expression in rhombomeres. Thus, it will be important to identify such inputs and elucidate how components of the RA machinery became coupled to hindbrain segmentation in lamprey.
It is notable that the vertebrate Cyp26 gene complement has been shaped by both whole genome duplication and tandem gene duplication prior to the split between jawed and jawless vertebrates. This coincides with the evolution of the coupling of RA to hindbrain segmentation. Thus, the duplication of the Cyp26 genes may have been important in the elaboration of their roles in the hindbrain GRN, perhaps by enabling functional regulatory divergence between the resulting paralogs.
In summary, we have shown that the coupling of the RA/Hox regulatory hierarchy to the GRN for hindbrain segmentation is rooted to the base of vertebrates. This raises questions regarding the evolution of regulatory mechanisms linked to the GRN that enabled this coupling. It will also be important to decipher the nature and origin of RA-dependent regulatory mechanisms of components of the RA signaling machinery (e.g., Cyp26 and Aldh1a genes). More generally, this work illustrates how using sea lamprey as a jawless vertebrate model can yield insights into the evolution of GRNs underlying novel vertebrate features.
Methods
Sea lamprey genome and model organism used for comparative studies.
Sea lamprey (Petromyzon marinus) Aldh1a and Cyp26 gene sequences were retrieved from the JBrowse browser and NCBI database, using the most-recent version of the Sea lamprey germline genome (KPetMar1). We found two predicted Aldh1a2-like genes with the following GenBank accession numbers: retinal dehydrogenase 2-like LOC116940317 (‘Aldh1a1/a2a’) and retinal dehydrogenase 2-like-LOC116950758 (‘Aldh1a1/a2b’). We also found three predicted Cyp26 genes with the following GenBank accession numbers: Cyp26A1-LOC116941264, Cyp26B1-like-LOC116941263 (‘Cyp26B1/C1a’) and Cyp26B1-like-LOC116940441 (‘CCyp26B1/C1b’).
Phylogenetic analyses of the sea lamprey Aldh1a and Cyp26 complements were conducted using key representatives of the vertebrate group. In light of the established evolutionary history of the Aldh1a gene family in vertebrates49 and the key position of sea lamprey as part of the early-diverged group of jawless vertebrates, we included both vertebrate Aldh1a1 and Aldh1a2 gene families in the phylogenetic analysis. Within jawed vertebrates, great white shark (Carcharodon carcharias) was used as a representative of the cartilaginous fish group (Chondrichthyes). Spotted gar (Lepisosteus oculatus) rather than zebrafish was used as a representative of the ray-finned fish group (Actinopterygii) because of its slow evolving and non-duplicated genome83. West Indian coelacanth (Latimeria chalumnae) was used as a representative of the early-diverged lobe-finned fish group (Sarcopterygii)84. In tetrapods, platypus (Ornithorhynchus anatinus), grey short-tailed opossum (Monodelphis domestica) and mouse (Mus musculus) were used as representatives of the three mammalian groups (monotreme, non-placental and placental mammals). Inshore hagfish (Eptatretus burgeri) was used as another representative of the cyclostomes (jawless vertebrates) in addition to sea lamprey in the phylogenetic analysis and chicken (Gallus gallus) was included as a bird representative of the tetrapod group for synteny analyses only.
Phylogenetic analysis of sea lamprey Cyp26 and Aldh1a proteins.
Because protein alignments are more sensitive than DNA alignments and to avoid potential artefacts that come with the use of DNA sequences, we opted to perform protein alignments. Protein sequences corresponding to each gene were retrieved using both ENSEMBL and NCBI, focusing on the latest available version of the genome and/or the most complete available genome annotation. Cyp51 and Aldh8a1 gene families were used as outgroups for the analyses of Cyp26 and Aldh1a complements, respectively. Protein sequences were aligned using the CLUSTALW protein alignment algorithm in MEGA1185 and their evolutionary history was inferred using the Maximum Likelihood Tree method and WAG model86 with 500 iterations for bootstrap testing, following Barry G. Hall’s recommended parameters87. Protein accession numbers used to conduct the phylogenetic analyses are indicated in Supplementary Figure 8.
Synteny analysis.
To help resolve the evolutionary history of the sea lamprey Cyp26 and Aldh1a gene families with respect to their jawed vertebrate putative orthologs, we investigated the chromosomal organization of genes (i.e., synteny) located in the vicinity of Cyp26 and Aldh1a genes in different vertebrate models using both ENSEMBL and NCBI Genome Data Viewer. Hagfish was not included in this synteny analysis, as the current version of the genome assembly lacked sufficient detail. The following gene families were selected for the synteny analysis: Cyp26B1 synteny: Znf638, Dysf, Exo6b; Cyp26A1/C1 synteny: Myof, Exo6; Aldh1a2 synteny: Aqp9, Myzap, Cgnl1; Aldh1a1 synteny: Tmc1, Cyp1A/D, Anxa1. mRNA accession numbers of Cyp26 and Aldh1a used to conduct the synteny analyses in different vertebrate models are indicated in Supplementary Figure 9.
RNA-seq profile.
RNAseq profiles corresponding to 1 to 5-day post fertilization (dpf) 50 as well as stages st18 to st24 (Tahara stages)52 51 obtained from P. marinus dorsal neural tube tissues and/or whole embryos and accessible on the KPetMar1 genome browser, were taken into consideration in the design of experiments described below investigating the gene expression of Aldh1a and Cyp26 complements.
Lamprey husbandry and embryo collection.
Sea lamprey embryos were collected following the standard husbandry and culture protocols88. In brief, embryos were cultured in 0.05X Marc's Modified Ringers solution (MMR) embryo media (18 °C), staged according to Tahara stages, fixed in 1X MEMFA (1-part 10X MEMFA Salts, 1-part 37% formaldehyde, 8-parts H2O) at appropriate developmental stages, dehydrated in 100% EtOH and stored at −20 °C for further experimentation.
Colorimetric In Situ Hybridization (cISH).
RNA In Situ probes for P. marinus Aldh1a and Cyp26 genes were designed based on gene sequences predicted by the Refseq model of KPetmar1 (Aldh1a1/a2a: XM_032949973.1; Aldh1a1/a2b: XM_032968791.1; Cyp26A1: XM_032952128.1; Cyp26B1/C1a: XM_032952126.1; Cyp26B1/C1b: XM_032950272.1). Because of the presence of repeat sequences as well as the unique profile of each gene sequence, individual probes were designed to target different gene locations. Aldh1a1/a2a RNA cISH probe was designed to target an internal region spanning from exon 5 to exon 9, based on the previously published mRNA sequence of ‘aldh1a2’ (GenBank: FJ536260.1)46. For Aldh1a1/a2b, Cyp26A1/B1a and Cyp26A1/B1b, probes were designed to target the 3’ UTR sequence, while the RNA probe for Cyp26A1 was designed to target a region overlapping the 5’UTR and the first exon of the gene. The following PCR primers were used to amplify probe templates:
Aldh1a1/a2a (597bp, internal): F 5’- ACTTCACATTCACACGGCAC -3’; R: 5’- GCTTCTCGTCAATCTGTGGC-3’.
Aldh1a1/a2b (490bp, 3’UTR): F 5’- TCCCGAAGAAGAGCTCGTAA-3’; R 5’- CCGCCGTAGAATTTAGAACG-3’.
Cyp26A1 (423bp, 5’UTR): F 5’- AAGAAAACCCCAACACAACG -3’; R 5’- AACTTCTCCCACAGCTTCCA-3’.
Cyp26B1/C1a (487bp, 3’UTR): F 5’- GAGCGTCCTCTACAGCATCC -3’; R 5’- CACCATCTCTCCTCCGTCTC -3’.
Cyp26B1/C1b (402bp, 3’UTR): F 5’- CCTGGGCTCAATAACGATGT -3’; R 5’- GCGCCACACTAAAAATCCAT -3’.
RNA cISH probe sequences for P. marinus hindbrain gene markers otx, wnt1, krox20, kreisler, hoxβ1, hoxα2, hoxα3 and hoxζ4 were described in previous work published from the lab34, 42. Sequences were PCR amplified from P. marinus genomic DNA (gDNA) or st18-st26 embryonic cDNA using KOD Hot Start Master Mix (Millipore Sigma) with primers listed above. PCR products were cloned and sequenced from the PCR4Blunt-Topo Vector (ThermoFisher Scientific). Digoxigenin (DIG)-labelled RNA ISH probes were synthesized from selected clones following standard protocols and purified using the Megaclear Purification of Transcription Kit (Ambion). PCR products were cloned and sequenced from the PCR4Blunt-Topo Vector (ThermoFisher Scientific). DIG-labelled RNA ISH probes were synthesized from selected clones following standard protocols and purified using the Megaclear Purification of Transcription Kit (Ambion). In Situ Hybridization was performed on whole-mount embryos following the standard lamprey cISH protocol88, 89 optimized as recently described from our lab90.
Hybridization-Chain Reaction (HCR) Fluorescent In Situ Hybridization (FISH).
For each gene, an HCR-FISH probe set targeting the coding sequence was custom-designed by Molecular Instruments with the number of probes varying between different sets. For each gene, the accession number, probe set size, amplifier/AlexaFluor dye used, and probe concentration are summarized in Supplementary Table 5. Because the sea lamprey embryo is highly opaque and auto-fluorescent, this HCR-FISH protocol needed to be adapted from the zebrafish HCRv3 FISH protocol60 , with additional steps of bleaching and clearing of the tissue. First, sea lamprey embryos were gradually rehydrated at room temperature from 100% MeOH into 100% PBST (1X PBS + 0.1% Tween) in series of 5-minutes-long washes. Following this, embryos were then bleached using a freshly made solution (ingredients to be mixed in the following order: 5% Formamide, 0.5% SSC, 3% H2O2) then exposed under direct LED light (>10K Lumen) for 2h. This is followed by a few washes in 100% PBST. Embryos were kept at minimal density (about 5 embryos per tube) and were treated in 1ml of proteinase K solution (20μg/ml) for 8’, followed by a few washes in 100% PBST. Embryos were then re-fixed in 4% PFA for 20' (shaking). The detection and amplification stage follows the zebrafish HCRv3 protocol60. The optimal probe input needs to be determined empirically for each probe set and target tissue and is further used to determine the volume of probe to be added to prepare a 2pmol probe/hybridization buffer solution (Supplementary Table 5).
Imaging.
For colorimetric in situ hybridization, embryos were gradually dehydrated in 100% MeOH post in situ hybridization and gradually cleared into 100% glycerol for imaging. Images were taken on a Leica MZ APO microscope using the Lumenera Infinity 3 camera at a magnification of 50X together with the Infinity Analyze software. Images were cropped and post-processed in a consistent way in Adobe Photoshop 2023 for color balance, brightness, and contrast. For HCR-FISH, embryos were cleared for 1h in Optiprep (Stem Cell Technology). For dorsal imaging of the developing head, embryos were mounted dorsally on 0.5%-1% agarose blocks made with a customized inverted mold including a ~250μm lane accommodating the head, and ~400μm accommodating the yolk. Mounted embryos were carefully placed on a 35mm MatTek glass bottom dish. For lateral imaging of the developing head, embryos were mounted laterally on a glass slide in Optiprep and positioned as flat as possible using layers of double-sided tape. Images were taken on a Nikon AT-AT inverted confocal microscope, using a 4X magnification for lateral imaging of the developing head (st20 embryos) and a 10X magnification for dorsal imaging of the developing head (st21-st23.5 embryos). Images were analyzed and post processed for brightness and contrast in Fiji-ImageJ and Adobe Photoshop 2023, then cropped and assembled into montages in Adobe Illustrator 2023.
Drug Treatments.
Sea lamprey embryos were treated with either 0.1 % DiMethyl SulfOxide (DMSO) or with 10/50 μM of the pan-Aldh1 inhibitor DiEthylAminoBenzaldehyde (DEAB) or with 10μM of the Cyp26 inhibitor R115866 (Talarazole). Drug treatment was performed at gastrulation (st13/14) or neurulation (st16/17). Batches of embryos were incubated in a treatment solution made from diluting the drug solution a 1000-fold into 0.05X MMR embryo media, cultured in the dark until fixation at st23. The treatment solution was replaced approximately every 36h with care taken not to agitate embryos. Embryos were then fixed at appropriate developmental stages, dehydrated in 100% EtOH and stored at −20 °C for further experimentation.
GFP-reporter experiments.
A GFP reporter construct for zebrafish hoxb3a enhancer (Drhoxb3a-GFP) was made by PCR amplification of a 928bp enhancer region of zebrafish hoxb3a followed by its cloning into the HLC (Hugo’s Lamprey Construct) as previously described34. The reporter HLC plasmid (20ng μl-1) was digested by 0.5 U I-Sce enzyme in its adequate digestion buffer (NEB) for 30’ at 37°C prior to injections. Sea lamprey embryos were injected at one-cell (st0-st2) with approximately 2nl of the reporter construct and transient transgenesis was mediated by I-SceI meganuclease as previously described34. Approximately 600 embryos were injected with the reporter construct and approximately 200 embryos were treated by each drug. Only embryos displaying GFP fluorescence at st23 were scored. Background GFP activity can be seen in the yolk and can be used as a marker of positively injected embryos. The ratio between the number of embryos displaying GFP fluorescence in the developing neural tube (i.e., positive reporter embryos) and the total number of GFP-positive embryos is listed in Fig. 6c. All representative phenotypes were imaged using Zeiss SteREO Discovery V12, fixed, dehydrated in 100% EtOH and stored at −20 °C for further experimentation. Images were cropped and post-processed in a consistent way in Adobe Photoshop 2023 for color balance, brightness, and contrast.
CRISPR/Cas9 gene editing.
Because of their important expression pattern during neural tube patterning, Aldh1a1/a2a, Cyp26A1 and Cyp26B1/C1a were functionally disrupted using CRISPR/Cas9 gene editing. gRNAs were designed to target highly conserved enzymatic sites shared between sea lamprey and jawed vertebrates. Sea lamprey Aldh1a1/a2a protein sequence was aligned with jawed vertebrate Aldh1a2 protein sequences, jawed vertebrate Cyp26A1 proteins were aligned and sea lamprey Cyp26B1/C1a was aligned with jawed vertebrate Cyp26B1/C1 (Supplementary Figures 1 and 2). Alignments were made using Clustal Omega Multiple Sequence Alignment tool. Conserved amino acid were highlighted in yellow on Snapgene, using a threshold of 95% for conservation. All gRNAs were designed using CRISPOR using KPetMar1 as a target genome and the standard -NGG PAM motif, and carefully selected to avoid off-target cutting. Two different gRNA were designed to target the highly conserved Cysteine and Glutamate residues of Aldh1a1a2a with the following sequences (Nx-PAM):
g1: AAGTTGATCCAGGAGGAGGC-CGG; g2: CCAGGGGGTGTTCTGGAACC-AGG.
Similarly, for each Cyp26 gene, three different gRNA were designed to target the I helix, K helix and Heme Group, three highly conserved enzymatic sites of the Cyp26 family. gRNA sequences are as follows (Nx-PAM):
Cyp26A1 gI: GCCACGGAGCTCCTGTTCGG-GGG;
Cyp26A1 gH1: CGCTTCACCTACATCCCGTT-CGG;
Cyp26A1 gH2: TTCACCTACATCCCGTTCGG-GGG;
Cyp26B1/C1a gK1: CTACCTCGACTGCGTCGTCA-AGG;
Cyp26B1/C1a gK2: GCGTCGTCAAGGAAGTGCTG-CGG;
Cyp26B1/C1a gH: TTCAGCTACCTGCCGTTCGG-CGG.
A gRNA targeting the Xenopus tyrosinase gene was injected in each injected batch of embryos as a negative control (gRNA neg: GGCCCACTGCTCAGAAACCC)91.
Following the IDT protocol ‘Zebrafish embryo microinjection Ribonucleoprotein delivery using the AltR™ CRISPR-Cas9 System’, contributed by Jeffrey Essner, PhD, we created a diluted 3μM guide RNA (gRNA) solution by diluting a 100μM Alt-R CRISPR-Cas9 RNA (crRNA) stock solution (IDT) and 100μM Alt-R Sp. CRISPR-Cas9 tracrRNA stock solution (IDT) in nuclease-free water duplex buffer (IDT). A diluted 0.5 μg/μL Cas9 solution was made by diluting a Cas9 protein stock solution (10μg/μl) (IDT) twenty-fold into Cas9 working buffer (20 mM HEPES; 150 mM KCI, pH 7.5, IDT). For each injection, ribonucleoprotein (RNP) complexes were assembled by combining 1 volume of gRNA and 1 volume of diluted Cas9 solutions into 2 volumes of injection buffer. In addition, 1 μl of 10% Lysinated Rhodamine-Dextran (LRD) tracer was added to each injection solution. The total volume of gRNA and Cas9 solution used varied between 1 to 2 μl according to the quality of the fertilization (i.e., number and quality of fertilized of embryos, death ratio etc.) with a more diluted solution injected in poorer fertilizations. When targeting individual genes (e.g., Cyp26A1 alone), 2 or 3 gRNA were injected, 0.5 μl of each gRNA were used and mixed with 1 μl of Cas9 solution and 2 μl injection buffer. When targeting both Cyp26A1 and Cyp26B1/C1a in the same embryo, we combined the following 4 gRNAs: Cyp26A1 gH1, Cyp26A1 gI, Cyp26B1/C1a gK1 and Cyp26B1/C1a gH and 0.25 μl of each gRNA were used and mixed with 1 μl of Cas9 solution and 2 μl injection buffer. Sea lamprey zygotes (st1-st2) were collected following external fertilization and immediately injected with approximately 3 nanoliters of RNP complex using a standard zebrafish micro-injection setup including injection needles, a microinjection manipulator and a microscope92. Embryo culture dishes were cleared for dead embryos the next day. Embryos showing fluorescence signal were cultured, fixed in MEMFA at appropriate developmental stages, dehydrated in 100% EtOH and stored at −20 °C for further experimentation.
Genotyping of CRISPR embryos.
Following cISH analysis of CRISPR embryos, representative embryos were genotyped to validate their mutant phenotypes. We genotyped CrAldh1a/a2a and CrCyp26s embryos in which both Cyp26A1 and Cyp26B1/C1a were targeted. After rehydration, genomic DNA of CRISPR and control embryos was isolated by placing individual embryos in 45 μL of lysis buffer (10 mM Tris-HCl (pH8.0), 50 mM KCl, 0.3% Tween 20, 0.3% NP40, 4 mM EDTA), and incubated for 10’ at 98°C. Then 5μl of Proteinase K (20 mg/ml) was added and the sample was incubated 55°C for at least 24h followed with a 10’ incubation at 98°C to denature the Proteinase K. The samples of genomic DNA were subsequently diluted in water (1:5). For each gene, primers were designed to amplify a genomic region spanning the targeted sites used in the CRISPR strategy (Supplementary Figures 5 and 6). PCR amplification was performed using KOD Hot Start Master Mix (Millipore Sigma) with primers listed in Supplementary Table 6. PCR products were cloned using the PCR4Blunt-Topo Vector (ThermoFisher Scientific) and sequenced and the resulting sequences were aligned to the reference sequence from control CRISPR embryos. For each series of sequencing, the proportion of mutant loci is listed in Supplementary Figures 5 and 6. We designed multiple sets of primers to amplify the region containing the K helix in the CrCyp26s embryos but were unsuccessful in amplifying this region. Further, sequencing results suggest that Cyp26A1 gI was not very efficient, as the majority of sequenced loci did not show a mutation in the I helix region (Supplementary Figure 5).
Reporting summary.
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Supplementary Material
Acknowledgements.
We thank Brian Slaughter, Jason Morrison for their assistance in generating and visualizing HCR data, Andrew Price for his help in generating the RNAseq graph, Stephen Green, Megan Martik, and Tetsuto Miyashita for assistance with lamprey husbandry, and members of the Krumlauf laboratory for feedback on the experiments and manuscript. This study was conducted in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the NIH and protocols approved by the Institutional Animal Care and Use Committees of the California Institute of Technology (lamprey, MEB Protocol: #IA23-1436. This work was supported by a grant from the Stowers Institute for Medical Research to R.K. (grant #1001) and by a grant to M.E.B. (R35NS111564). This work was conducted to fulfill, in part, the requirements for A.M.H.B.’s thesis research as a Ph.D. student registered with the Open University, and we would like to thank the thesis committee members for their insightful comments and feedback.
Footnotes
Competing Interests. The authors declare no competing interests.
Data availability.
The authors declare that all data supporting the findings of this study are available within the article and its supplementary information files or from the corresponding author upon reasonable request. Original data underlying this manuscript can be accessed from the Stowers Original Data Repository at [http://odr.stowers.org/websimr/].
References
- 1.Lowe CJ, et al. Anteroposterior patterning in hemichordates and the origins of the chordate nervous system. Cell 113, 853–865 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Pani AM, Mullarkey EE, Aronowicz J, Assimacopoulos S, Grove EA, Lowe CJ. Ancient deuterostome origins of vertebrate brain signalling centres. Nature 483, 289–294 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lowe CJ, Clarke DN, Medeiros DM, Rokhsar DS, Gerhart J. The deuterostome context of chordate origins. Nature 520, 456–465 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Yao Y, et al. Cis-regulatory architecture of a brain signaling center predates the origin of chordates. Nat Genet 48, 575–580 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koop D, et al. Retinoic acid signaling targets Hox genes during the amphioxus gastrula stage: insights into early anterior-posterior patterning of the chordate body plan. Dev Biol 338, 98–106 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Darras S, et al. Anteroposterior axis patterning by early canonical Wnt signaling during hemichordate development. PLoS Biol 16, e2003698 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Onai T, et al. Retinoic acid and Wnt/beta-catenin have complementary roles in anterior/posterior patterning embryos of the basal chordate amphioxus. Dev Biol 332, 223–233 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Aronowicz J, Lowe CJ. Hox gene expression in the hemichordate Saccoglossus kowalevskii and the evolution of deuterostome nervous systems. Integr Comp Biol 46, 890–901 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Schubert M, Holland ND, Laudet V, Holland LZ. A retinoic acid-Hox hierarchy controls both anterior/posterior patterning and neuronal specification in the developing central nervous system of the cephalochordate amphioxus. Dev Biol 296, 190–202 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Deschamps J, Duboule D. Embryonic timing, axial stem cells, chromatin dynamics, and the Hox clock. Genes Dev 31, 1406–1416 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wada H, Garcia-Fernandez J, Holland PW. Colinear and segmental expression of amphioxus Hox genes. Dev Biol 213, 131–141 (1999). [DOI] [PubMed] [Google Scholar]
- 12.Nolte C, Krumlauf R. Expression of Hox genes in the nervous system of vertebrates. In: HOX Gene Expression (ed Papageorgiou S). Landes Bioscience and Springer; (2006). [Google Scholar]
- 13.Hunt P, et al. A distinct Hox code for the branchial region of the vertebrate head. Nature 353, 861–864 (1991). [DOI] [PubMed] [Google Scholar]
- 14.Krumlauf R, Wilkinson DG. Segmentation and patterning of the vertebrate hindbrain. Development 148, dev186460 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schubert M, Yu JK, Holland ND, Escriva H, Laudet V, Holland LZ. Retinoic acid signaling acts via Hox1 to establish the posterior limit of the pharynx in the chordate amphioxus. Development 132, 61–73 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Marletaz F, Holland LZ, Laudet V, Schubert M. Retinoic acid signaling and the evolution of chordates. Int J Biol Sci 2, 38–47 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nolte C, De Kumar B, Krumlauf R. Hox genes: Downstream "effectors" of retinoic acid signaling in vertebrate embryogenesis. Genesis 57, e23306 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Bel-Vialar S, Itasaki N, Krumlauf R. Initiating Hox gene expression: in the early chick neural tube differential sensitivity to FGF and RA signaling subdivides the HoxB genes in two distinct groups. Development 129, 5103–5115 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Frank D, Sela-Donenfeld D. Hindbrain induction and patterning during early vertebrate development. Cell Mol Life Sci 76, 941–960 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neijts R, et al. Polarized regulatory landscape and Wnt responsiveness underlie Hox activation in embryos. Genes Dev 30, 1937–1942 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deschamps J, van Nes J. Developmental regulation of the Hox genes during axial morphogenesis in the mouse. Development 132, 2931–2942 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Niederreither K, Dolle P. Retinoic acid in development: towards an integrated view. Nat Rev Genet 9, 541–553 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Niederreither K, Vermot J, Schuhbaur B, Chambon P, Dolle P. Retinoic acid synthesis and hindbrain patterning in the mouse embryo. Development 127, 75–85 (2000). [DOI] [PubMed] [Google Scholar]
- 24.Wada H, Escriva H, Zhang S, Laudet V. Conserved RARE localization in amphioxus Hox clusters and implications for Hox code evolution in the vertebrate neural crest. Dev Dyn 235, 1522–1531 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Manzanares M, Wada H, Itasaki N, Trainor PA, Krumlauf R, Holland PW. Conservation and elaboration of Hox gene regulation during evolution of the vertebrate head. Nature 408, 854–857 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Bedois AMH, Parker HJ, Krumlauf R. Retinoic Acid Signaling in Vertebrate Hindbrain Segmentation: Evolution and Diversification. Diversity 13, 398 (2021). [Google Scholar]
- 27.Lumsden A. Segmentation and compartition in the early avian hindbrain. Mech Dev 121, 1081–1088 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Santagati F, Rijli FM. Cranial neural crest and the building of the vertebrate head. Nat Rev Neurosci 4, 806–818 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Bronner ME, LeDouarin NM. Development and evolution of the neural crest: an overview. Dev Biol 366, 2–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le Douarin N. The Neural Crest. Cambridge University Press; (1983). [Google Scholar]
- 31.Green SA, Simoes-Costa M, Bronner ME. Evolution of vertebrates as viewed from the crest. Nature 520, 474–482 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martik ML, et al. Evolution of the new head by gradual acquisition of neural crest regulatory circuits. Nature 574, 675–678 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gilland E, Baker R. Conservation of neuroepithelial and mesodermal segments in the embryonic vertebrate head. Acta Anat (Basel) 148, 110–123 (1993). [DOI] [PubMed] [Google Scholar]
- 34.Parker HJ, Bronner ME, Krumlauf R. A Hox regulatory network of hindbrain segmentation is conserved to the base of vertebrates. Nature 514, 490–493 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parker HJ, Krumlauf R. A Hox gene regulatory network for hindbrain segmentation. Curr Top Dev Biol 139, 169–203 (2020). [DOI] [PubMed] [Google Scholar]
- 36.White RJ, Schilling TF. How degrading: Cyp26s in hindbrain development. Dev Dyn 237, 2775–2790 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimeld SM, Donoghue PC. Evolutionary crossroads in developmental biology: cyclostomes (lamprey and hagfish). Development 139, 2091–2099 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Shimeld SM, Holland PW. Vertebrate innovations. Proc Natl Acad Sci U S A 97, 4449–4452 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murakami Y, Pasqualetti M, Takio Y, Hirano S, Rijli FM, Kuratani S. Segmental development of reticulospinal and branchiomotor neurons in lamprey: insights into the evolution of the vertebrate hindbrain. Development 131, 983–995 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Murakami Y, Uchida K, Rijli FM, Kuratani S. Evolution of the brain developmental plan: Insights from agnathans. Dev Biol 280, 249–259 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Gil-Galvez A, et al. Gain of gene regulatory network interconnectivity at the origin of vertebrates. Proc Natl Acad Sci U S A 119, e2114802119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parker HJ, Bronner ME, Krumlauf R. An atlas of anterior hox gene expression in the embryonic sea lamprey head: Hox-code evolution in vertebrates. Dev Biol 453, 19–33 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parker HJ, Sauka-Spengler T, Bronner M, Elgar G. A reporter assay in lamprey embryos reveals both functional conservation and elaboration of vertebrate enhancers. PLoS One 9, e85492 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith JJ, et al. The sea lamprey germline genome provides insights into programmed genome rearrangement and vertebrate evolution. Nat Genet 50, 270–277 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Timoshevskaya N, et al. An improved germline genome assembly for the sea lamprey Petromyzon marinus illuminates the evolution of germline-specific chromosomes. Cell Rep 42, 112263 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Castillo HA, et al. Insights into the organization of dorsal spinal cord pathways from an evolutionarily conserved raldh2 intronic enhancer. Development 137, 507–518 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roberts C. Regulating Retinoic Acid Availability during Development and Regeneration: The Role of the CYP26 Enzymes. J Dev Biol 8, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hempel J, Lindahl R. Class III aldehyde dehydrogenase from rat liver: super-family relationship to classes I and II and functional interpretations. Prog Clin Biol Res 290, 3–17 (1989). [PubMed] [Google Scholar]
- 49.Canestro C, Catchen JM, Rodriguez-Mari A, Yokoi H, Postlethwait JH. Consequences of lineage-specific gene loss on functional evolution of surviving paralogs: ALDH1A and retinoic acid signaling in vertebrate genomes. PLoS Genet 5, e1000496 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bryant SA, Herdy JR, Amemiya CT, Smith JJ. Characterization of Somatically-Eliminated Genes During Development of the Sea Lamprey (Petromyzon marinus). Mol Biol Evol 33, 2337–2344 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hockman D, et al. A genome-wide assessment of the ancestral neural crest gene regulatory network. Nat Commun 10, 4689 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tahara Y. Normal stages of development in the lamprey, Lampetra reissneri (Dybowski). Zool Sci 5, 109–118 (1988). [Google Scholar]
- 53.Hernandez RE, Putzke AP, Myers JP, Margaretha L, Moens CB. Cyp26 enzymes generate the retinoic acid response pattern necessary for hindbrain development. Development 134, 177–187 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sirbu IO, Gresh L, Barra J, Duester G. Shifting boundaries of retinoic acid activity control hindbrain segmental gene expression. Development 132, 2611–2622 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Begemann G, Schilling TF, Rauch GJ, Geisler R, Ingham PW. The zebrafish neckless mutation reveals a requirement for raldh2 in mesodermal signals that pattern the hindbrain. Development 128, 3081–3094 (2001). [DOI] [PubMed] [Google Scholar]
- 56.Begemann G, Meyer A. Hindbrain patterning revisited: timing and effects of retinoic acid signalling. BioEssays 23, 981–986 (2001). [DOI] [PubMed] [Google Scholar]
- 57.Niederreither K, McCaffery P, Drager UC, Chambon P, Dolle P. Restricted expression and retinoic acid-induced downregulation of the retinaldehyde dehydrogenase type 2 (RALDH-2) gene during mouse development. Mech Dev 62, 67–78 (1997). [DOI] [PubMed] [Google Scholar]
- 58.Schilling TF, Nie Q, Lander AD. Dynamics and precision in retinoic acid morphogen gradients. Curr Opin Genet Dev 22, 562–569 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.White RJ, Nie Q, Lander AD, Schilling TF. Complex regulation of cyp26a1 creates a robust retinoic acid gradient in the zebrafish embryo. PLoS Biol 5, e304 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choi HMT, et al. Third-generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development 145, dev165753 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilkinson DG, Bhatt S, Chavrier P, Bravo R, Charnay P. Segment-specific expression of a zinc-finger gene in the developing nervous system of the mouse. Nature 337, 461–464 (1989). [DOI] [PubMed] [Google Scholar]
- 62.Schneider-Maunoury S, et al. Disruption of Krox-20 results in alteration of rhombomeres 3 and 5 in the developing hindbrain. Cell 75, 1199–1214 (1993). [DOI] [PubMed] [Google Scholar]
- 63.Begemann G, Marx M, Mebus K, Meyer A, Bastmeyer M. Beyond the neckless phenotype: influence of reduced retinoic acid signaling on motor neuron development in the zebrafish hindbrain. Dev Biol 271, 119–129 (2004). [DOI] [PubMed] [Google Scholar]
- 64.Helmbacher F, et al. Hoxa1 and Krox20 synergize to control the development of rhombomere 3. Development 125, 4739–4748 (1998). [DOI] [PubMed] [Google Scholar]
- 65.Labalette C, et al. Molecular dissection of segment formation in the developing hindbrain. Development 142, 185–195 (2015). [DOI] [PubMed] [Google Scholar]
- 66.Torbey P, et al. Cooperation, cis-interactions, versatility and evolutionary plasticity of multiple cis-acting elements underlie krox20 hindbrain regulation. PLoS Genet 14, e1007581 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thierion E, et al. Krox20 hindbrain regulation incorporates multiple modes of cooperation between cis-acting elements. PLoS Genet 13, e1006903 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maves L, Kimmel CB. Dynamic and sequential patterning of the zebrafish posterior hindbrain by retinoic acid. Dev Biol 285, 593–605 (2005). [DOI] [PubMed] [Google Scholar]
- 69.Morgan CA, Parajuli B, Buchman CD, Dria K, Hurley TD. N,N-diethylaminobenzaldehyde (DEAB) as a substrate and mechanism-based inhibitor for human ALDH isoenzymes. Chem Biol Interact 234, 18–28 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Loudig O, Maclean GA, Dore NL, Luu L, Petkovich M. Transcriptional co-operativity between distant retinoic acid response elements in regulation of Cyp26A1 inducibility. Biochem J 392, 241–248 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hu P, et al. Retinoid regulation of the zebrafish cyp26a1 promoter. Dev Dyn 237, 3798–3808 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Loudig O, Babichuk C, White J, Abu-Abed S, Mueller C, Petkovich M. Cytochrome P450RAI(CYP26) promoter: a distinct composite retinoic acid response element underlies the complex regulation of retinoic acid metabolism. Mol Endocrinol 14, 1483–1497 (2000). [DOI] [PubMed] [Google Scholar]
- 73.Li J, Hu P, Li K, Zhao Q. Identification and characterization of a novel retinoic acid response element in zebrafish cyp26a1 promoter. Anat Rec (Hoboken) 295, 268–277 (2012). [DOI] [PubMed] [Google Scholar]
- 74.Dobbs-McAuliffe B, Zhao Q, Linney E. Feedback mechanisms regulate retinoic acid production and degradation in the zebrafish embryo. Mech Dev 121, 339–350 (2004). [DOI] [PubMed] [Google Scholar]
- 75.Addison M, Xu Q, Cayuso J, Wilkinson DG. Cell Identity Switching Regulated by Retinoic Acid Signaling Maintains Homogeneous Segments in the Hindbrain. Dev Cell 45, 606–620e603 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sosnik J, et al. Noise modulation in retinoic acid signaling sharpens segmental boundaries of gene expression in the embryonic zebrafish hindbrain. Elife 5, e14034 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schilling TF, Sosnik J, Nie Q. Visualizing retinoic acid morphogen gradients. Methods Cell Biol 133, 139–163 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Holland LZ, Holland ND. Expression of AmphiHox-1 and AmphiPax-1 in amphioxus embryos treated with retinoic acid: insights into evolution and patterning of the chordate nerve cord and pharynx. Development 122, 1829–1838 (1996). [DOI] [PubMed] [Google Scholar]
- 79.Marletaz F, et al. Amphioxus functional genomics and the origins of vertebrate gene regulation. Nature 564, 64–70 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Carvalho JE, Lahaye F, Croce JC, Schubert M. CYP26 function is required for the tissue-specific modulation of retinoic acid signaling during amphioxus development. Int J Dev Biol 61, 733–747 (2017). [DOI] [PubMed] [Google Scholar]
- 81.Carvalho JE, et al. Lineage-specific duplication of amphioxus retinoic acid degrading enzymes (CYP26) resulted in sub-functionalization of patterning and homeostatic roles. BMC Evol Biol 17, 24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang Y, Zolfaghari R, Ross AC. Multiple retinoic acid response elements cooperate to enhance the inducibility of CYP26A1 gene expression in liver. Gene 464, 32–43 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Braasch I, et al. The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat Genet 48, 427–437 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nikaido M, et al. Coelacanth genomes reveal signatures for evolutionary transition from water to land. Genome Res 23, 1740–1748 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tamura K, Stecher G, Kumar S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol Biol Evol 38, 3022–3027 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Whelan SaG N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol, (2001). [DOI] [PubMed] [Google Scholar]
- 87.Hall BG. Building phylogenetic trees from molecular data with MEGA. Mol Biol Evol 30, 1229–1235 (2013). [DOI] [PubMed] [Google Scholar]
- 88.Nikitina N, Bronner-Fraser M, Sauka-Spengler T. The sea lamprey Petromyzon marinus: a model for evolutionary and developmental biology. Cold Spring Harb Protoc 2009, pdb emo113 (2009). [DOI] [PubMed] [Google Scholar]
- 89.Sauka-Spengler T, Meulemans D, Jones M, Bronner-Fraser M. Ancient evolutionary origin of the neural crest gene regulatory network. Developmental Cell 13, 405–420 (2007). [DOI] [PubMed] [Google Scholar]
- 90.Parker HJ, De Kumar B, Pushel I, Bronner ME, Krumlauf R. Analysis of lamprey meis genes reveals that conserved inputs from Hox, Meis and Pbx proteins control their expression in the hindbrain and neural tube. Dev Biol 479, 61–76 (2021). [DOI] [PubMed] [Google Scholar]
- 91.Square T, Romasek M, Jandzik D, Cattell MV, Klymkowsky M, Medeiros DM. CRISPR/Cas9-mediated mutagenesis in the sea lamprey Petromyzon marinus: a powerful tool for understanding ancestral gene functions in vertebrates. Development 142, 4180–4187 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pei W, Burgess SM. Microinjection in Zebrafish for Genome Editing and Functional Studies. Methods Mol Biol 1874, 459–474 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the article and its supplementary information files or from the corresponding author upon reasonable request. Original data underlying this manuscript can be accessed from the Stowers Original Data Repository at [http://odr.stowers.org/websimr/].