Abstract
This article provides recommendations for implementing quantitative susceptibility mapping (QSM) for clinical brain research. It is a consensus of the ISMRM Electro-Magnetic Tissue Properties Study Group. While QSM technical development continues to advance rapidly, the current QSM methods have been demonstrated to be repeatable and reproducible for generating quantitative tissue magnetic susceptibility maps in the brain. However, the many QSM approaches available give rise to the need in the neuroimaging community for guidelines on implementation. This article describes relevant considerations and provides specific implementation recommendations for all steps in QSM data acquisition, processing, analysis, and presentation in scientific publications. We recommend that data be acquired using a monopolar 3D multi-echo GRE sequence, that phase images be saved and exported in DICOM format and unwrapped using an exact unwrapping approach. Multi-echo images should be combined before background removal, and a brain mask created using a brain extraction tool with the incorporation of phase-quality-based masking. Background fields should be removed within the brain mask using a technique based on SHARP or PDF, and the optimization approach to dipole inversion should be employed with a sparsity-based regularization. Susceptibility values should be measured relative to a specified reference, including the common reference region of whole brain as a region of interest in the analysis, and QSM results should be reported with – as a minimum – the acquisition and processing specifications listed in the last section of the article. These recommendations should facilitate clinical QSM research and lead to increased harmonization in data acquisition, analysis, and reporting.
Keywords: Quantitative Susceptibility Mapping, ISMRM Study Group, Clinical Brain Research, Magnetic Resonance Imaging, Data Acquisition, Data Analysis
1. Introduction
Brain quantitative susceptibility mapping (QSM) has been increasingly performed to identify calcifications and study iron, myelin, and oxygen consumption changes associated with normal brain development or aging and with neurological disease. Diseases of interest include hemorrhagic stroke, multiple sclerosis, Alzheimer’s disease, Parkinson’s disease, amyotrophic lateral sclerosis, and tumors1–15. QSM is also increasingly being used in psychiatric disorders such as psychosis16,17 or depression18,19, where it may reflect neurotransmitter or metabolic imbalances. Changes in brain susceptibility have also been associated with alcohol consumption20–22, potentially informing the neural mechanisms through which alcohol affects the brain. Further, QSM has been used for the differentiation between hemorrhages and calcifications23–26, anatomical visualization27–29 and improved segmentation30,31, and for presurgical mapping in deep brain stimulation because it depicts deep gray nuclei targets with exquisite contrast and superior iron source sharpness as compared to other approaches such as T2 and T2*-weighted imaging27,32–36. In conjunction with R2 or R2* mapping, QSM may also be used to separate diamagnetic myelin and calcification from paramagnetic iron within a voxel37–46.
While QSM techniques and their biomedical applications have been extensively reviewed3,6,7,47–56,57(p31),58,59, and the QSM research community has held two challenges attempting to identify the best susceptibility calculation algorithm60–63, there has been no community consensus or white paper on how to best perform brain QSM in the clinical setting. Some vendors have started implementing research-level QSM pipelines for their systems, nevertheless QSM is not yet a product on most magnetic resonance imaging (MRI) systems. On the other hand, demands for a robust “end-to-end” QSM recipe covering acquisition and processing through to presentation in scientific publications have been growing strongly from the neurological imaging community. For example, the need for a QSM consensus method was highlighted at the group discussion of the February 2022 workshop of the North American Imaging in Multiple Sclerosis (NAIMS) Cooperative, as QSM is particularly useful for depicting the paramagnetic rim of chronic active multiple sclerosis lesions64–72. In response to this need, QSM investigators in the Electro-Magnetic Tissue Properties Study Group (EMTP SG) of the International Society for Magnetic Resonance in Medicine (ISMRM) established a QSM Consensus Organization Committee to define the scope of the recommendations and determine the current consensus.
This present article presents the scope of the recommendations, describes the approaches used to obtain consensus, reviews of each step of the QSM reconstruction process, and summarizes its specific consensus recommendations. The technical sections present, first, a general overview of the subject matter in the context of the QSM reconstruction process and then the consensus recommendation. Readers interested only in the consensus recommendations may skip the overview sections and proceed directly to the respective recommendation subsections. Readers interested in the technical details and justification of the recommendations are encouraged to read the overview sections. All consensus recommendations are systematically enumerated to facilitate referencing of the individual recommendations in the literature. Where needed, a sub-section with additional considerations is presented. The paper closes with a summary of the recommendations.
1.1. Scope
This consensus paper was written as a guide on how to implement QSM in clinical research for clinical researchers not familiar with the technical nuances of QSM. The implementation of the recommendations requires the assistance of technical personnel such as medical physicists or vendors with in-depth knowledge of the MRI scanner and with expertise in image processing and analysis. The main purpose of this paper is to increase the use of QSM in clinical trials and in different patient groups. Guidelines for clinical practice are beyond the scope of the current paper and would require further study.
Recommendations represent a consensus among QSM experts and the community of experienced QSM users at the time of publication. For clarity, the committee decided that the emphasis of the paper would be on 3 tesla (T), the field strength most widely used in clinical brain research, and that general guidance would be provided for other field strengths (1.5 T and 7 T) where possible. Due to the focus on clinical applications, including clinical trials, the recommendations prioritize robustness and simplicity over state-of-the-art acquisitions and processing algorithms, and provide a starting point for application-specific improvements. Areas in which the committee could not yet reach a consensus (e.g., due to insufficient evidence) are indicated as such in the paper.
1.2. Organization
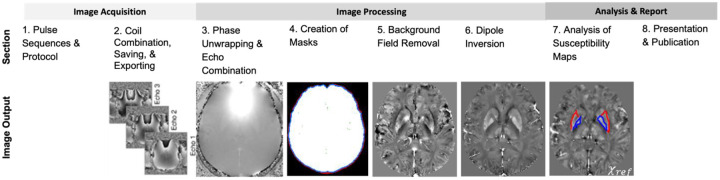
The article is organized into eight sections, covering data acquisition, image processing, image analysis, and presentation of QSM studies in scientific publications (Fig.1). Data acquisition is split into two sections: 1) pulse sequences and protocol, and 2) coil combination, saving, and exporting; image processing is split into four sections corresponding to four processing steps: phase unwrapping and echo combination, creation of masks, background field removal, and dipole inversion; the image analysis section focuses on the analysis of susceptibility maps; and the last section on presentation covers reporting in scientific publications. The contributions of the members of the QSM Consensus Organization Committee are listed in the Supplementary Materials I, Section S1.1 along with an overview of the history and approach of the initiative (Section S1.3). The Acknowledgement section lists all individuals who contributed significantly to the consensus recommendations in verbal or written form.
Figure 1:
This consensus paper comprises eight sections. The first two sections cover image acquisition: 1) pulse sequences and protocol and 2) coil combination, saving, and exporting. The next four sections cover image processing: 3) phase unwrapping & echo combination, 4) creation of masks, 5) background field removal, and 6) dipole inversion. The last two sections cover analysis and presentation in scientific publications: 7) analysis of susceptibility maps, and 8) presentation and publication. The image output from each section is further detailed in the corresponding section.
The paper is accompanied by harmonized pulse sequence protocols for several major platforms (current software versions in 2022) as well as sample code to perform the recommended processing steps (see Supplementary Materials II) using the Sepia toolbox73 [available at https://github.com/kschan0214/sepia]. In addition, curated online resources are provided and will be updated as the field progresses. This information is detailed in the Supplementary Materials.
1.3. Background
Tissue magnetic susceptibility is directly related to tissue chemical composition. A substance gaining a magnetization opposing an externally applied magnetic field, e.g. calcium, is called diamagnetic. Diamagnetism is due to electron orbit perturbation. A substance gaining a magnetization in the same direction as the external field, e.g. iron, is called paramagnetic. Paramagnetism is due to spin alignment of unpaired electrons, which have a magnetic moment 658 times greater than that of the hydrogen nucleus. Most biological tissues, like the human brain, have a susceptibility close to that of water, which is diamagnetic with a value of roughly −9 parts per million (ppm). In the brain, iron deposition makes tissue less diamagnetic than water, and calcium deposition and the presence of myelin make tissue more diamagnetic than water4,11,15,74–76.
The static magnetic field of magnetic resonance imaging (MRI) scanners, B0, affects electrons in tissue and, in linear materials, induces a magnetization proportional to B0 with the proportionality constant defined as magnetic susceptibility χ of the tissue. The induced tissue magnetization generates a field that is nonlocal and extends into the space surrounding the magnetization source, thereby inducing field inhomogeneities. These inhomogeneities can be described mathematically as a convolution of the susceptibility distribution with the unit dipole kernel77–79. During MRI signal generation, water proton spins experience this tissue field. In particular, gradient-recalled echo (GRE) imaging is very sensitive to this tissue field. Consequently, there is complex destructive interfering or dephasing by the tissue field variation within a voxel seen as hypointensities or blooming susceptibility artifacts on the GRE magnitude images, which is known as T2*-weighted or susceptibility-weighted imaging80–82. The GRE phase images, traditionally discarded, represent the tissue field inhomogeneities averaged over the voxel. The process by which tissue magnetic susceptibility is computed from the magnetic field measurement is called quantitative susceptibility mapping (QSM)83,84. Several detailed review papers have been published about the technical foundations and clinical applications of QSM, and we refer the interested reader to these articles for a comprehensive overview of the technique5,13,49,50,52–56,59,85–93.
2. Pulse Sequences and Protocol
This section describes recommendations to robustly acquire data for QSM.
2.1. Overview
Most of the major MRI manufacturers, if not all of them, provide a radiofrequency (RF)-spoiled 3D multi-echo GRE pulse sequence from which one can obtain phase images for QSM in addition to the standard T2*-weighted magnitude images. The GRE sequence is probably the most elementary sequence in the MRI sequence tree, consisting in its simplest form of an RF excitation pulse followed by acquisition of a gradient recalled echo, which for the multi-echo variant is repeated at various echo times (TEs) before the next excitation pulse is applied. This sequence is often used for calibration steps, such as obtaining a field map for B0 shimming.
Optimizing a GRE sequence to maximize QSM contrast shares many of the principles for optimizing T2*-weighted contrast-to-noise ratio or T2* mapping94. We recommend the following principles for designing a QSM acquisition protocol:
Aim to set the last TE equal to at least the T2* value of the tissue of interest. For example, if targeting deep gray matter, the T2* of the putamen can be a good reference (55, 30 and 16 ms at 1.5, 3 and 7 T, respectively95). The first echo time (TE1) should be as short as possible and the spacing between consecutive echoes (ΔTE) should be uniform throughout the echo train.
Use the minimum repetition time (TR) and set the flip angle to the Ernst angle (θErnst = cos−1(e−TR/T1)) for the target region. Most deep gray matter structures have a T1 close to that of white matter, and white matter represents the largest volume fraction in the brain. Hence, the Ernst angle may be calculated for white matter or deep gray matter with T1 = 650, 850 and 1220 ms at 1.5, 3 and 7 T respectively96.
Use three or more monopolar echoes. While QSM can be achieved with one echo and two is the minimum number of echoes needed to separate the intrinsic transmit RF phase from the magnetic field-induced phase (see the Section 3 below), the use of a larger number of echoes (e.g., 5 echoes) will benefit the phase signal-to-noise ratio (SNR) for a range of tissues97. As phase SNR is maximal when TE = T2*, the use of multiple echoes ensures that high SNR field estimates are obtained for both short apparent T2* (venous blood or tissues close to air-tissue interfaces) and other tissues with longer T2* values.
Use the minimum readout bandwidth (BW) that generates acceptable distortions. At 3T, 220 Hz/pixel is often sufficient (two-pixel fat-water shift). Such acquisitions negate the need to use fat suppression for brain applications. Note that as the BW is increased the ratio of the readout duration (1/BW) and echo spacing decreases (as a consequence of the required time to rewind the gradients), which results in a reduction in the SNR of the acquisition. The best trade-off depends on the system’s gradient amplitude and slew rate.
Use isotropic voxels of at most 1 mm to avoid susceptibility underestimation reported to occur at larger voxel sizes98.
Use 3D acquisition instead of 2D acquisition to avoid potential slice-to-slice phase discontinuities in 2D phase maps that mainly occur in interleaved acquisitions and require additional processing or to avoid slice crosstalk in non-interleaved acquisitions99. Note that the 3D spatial coherence of the obtained phase/field maps is of great importance as QSM is ultimately a 3D spatial deconvolution process.
Use a monopolar gradient readout (fly-back) to avoid geometric mismatch and eddy current related phase problems between even and odd echoes in bipolar acquisitions since these require additional correction in the pipeline used100.
Consider using flow compensation when targeting vessels (e.g., oxygenation studies), but note that flow compensation is often only available and guaranteed for the first echo, while flow artifacts increase in later echoes. Flow compensation also reduces the number of echoes achievable as it increases the minimum TE, assuming a fixed BW. In comparison, flow compensation has a much smaller effect on the resulting QSM values than the choice of QSM processing pipeline101.
The following points focus on practical aspects of image orientation, FOV, and acceleration to achieve a whole brain data acquisition in approximately 6 minutes for clinical research on a 3 T system with as few as 8 receive channels using a standard 3D multi-echo RF-spoiled GRE sequence:
Patients should be scanned in the supine position with the head straight to minimize variability in white matter susceptibility related to myelin magnetic susceptibility anisotropy102,103 and microstructural water compartmentation104.
For whole brain acquisition (including cerebellum), it is recommended to use isotropic voxels98 and prescribe the imaging volume with the readout direction along the anterior commissure - posterior commissure (AC-PC) line (oblique axial orientation), which reduces the number of phase encoding steps and, consequently, the total scan time105. Setting the readout in this direction also restricts eye movement artifacts in the left-right direction, outside of the brain. Alternatively, sagittal acquisitions (readout along the head-foot direction) can be beneficial when the region of interest includes the brain stem but will require larger acceleration factors. The use of tilted imaging slab orientations (oblique axial or oblique sagittal) requires that the data header information (see Section 3 below) specifying the exact slab orientation is supplied to the subsequent processing pipeline, otherwise artifacts will occur in the resulting susceptibility map106.
- Parallel imaging and partial k-space coverage can be used to reduce acquisition time, as for any structural MRI acquisition. Below we give general guidelines for GRE brain imaging:
- If accelerating in only one direction, use acceleration factors <3. If available, non-integer acceleration factors allow fine-tuning the acceleration factors to the available equipment.
- If available, elliptical k-space sampling (elliptical k-space shutter) and/or partial k-space acquisition (e.g., 7/8) with zero-filling can be used to reduce scan time but note that this can result in a reduction of the effective spatial resolution.
- If an RF coil with 32 or more channels is available, acceleration in both phase encoding directions by a factor of 2 (2×2) can be used, preferably with CAIPI patterns107.
2.2. Consensus Recommendation
The recommendation in this section is based on protocols currently available on commercial scanners that do not require a special research agreement with the scanner manufacturer:
-
2.a.
3D multi-echo RF-spoiled GRE with monopolar readout. Three or more echoes should be acquired and the TE range should include the T2* times of the target tissues.
-
2.b.
Minimum TR (given selected TEs).
-
2.c.
Flip angle should be set to the Ernst angle for target tissues (e.g., white matter).
-
2.d.
Whole brain coverage.
-
2.e.
Resolution should be isotropic with a voxel edge length of at most 1 mm non-interpolated at 3T.
-
2.f.
Use accelerated imaging methods (e.g., parallel imaging).
-
2.g.
Use coil arrays with a large number of elements covering the whole brain.
The Supplementary Materials I, Section S1.5 contain sample protocols for 1.5T, 3T, and 7T.
2.3. Additional Considerations
Acquisitions for QSM reconstructions can be performed at all clinical field strengths with minimal image distortions using the protocol recommendations provided here, although higher field strengths provide several benefits. Susceptibility contrast and contrast-to-noise ratio (CNR) increase with the static magnetic field once relaxation time changes are considered110, and acquisition can be more time efficient at higher fields due to the shorter T2*. Our recommended protocols implicitly integrate a compensation for lower SNR at lower B0 through lower spatial resolution, while keeping the total acquisition times similar. While useful data can be obtained at all field strengths, finer anatomical details in certain applications, such as brain lesions in multiple sclerosis111, might be difficult to visualize at 1.5T.
Advanced sequences such as segmented 3D-EPI112,113 or Wave-CAIPI114 allow drastic decreases in acquisition time but are not available as commercial sequences on all vendors and may require a research agreement with the scanner manufacturer. Nevertheless, it should be noted that some of these methods may have constraints on the number of echoes that can be acquired and 3D-EPI sequences will have increased spatial distortions that may have to be corrected for and that may result in additional artifacts that subsequent processing steps such as background field removal need to account for.
3. Coil Combination, Saving, and Exporting
This section recommends effective and practical solutions for generating phase images that can be used for QSM and provides guidance on how to save and convert the format of the phase images in preparation for QSM analysis.
3.1. Overview
Modern MRI systems use phased array coils made up typically of 12, 32 or 64 elements, which provide higher SNR than a single birdcage coil and facilitate parallel imaging115–117. However, the images from individual coil elements are sensitive to only a part of the FOV and need to be combined to generate a single image – a process that requires consideration of the differences between the coil signals.
Neglecting phase wraps (see Section 4), the phase measured with a particular RF coil element c in a GRE sequence, φc, is given by: . The first part, φB(TE), is the phase shift caused by the deviation ΔB0 of the magnetic field from the uniform main magnetic field, B0. Neglecting non-linear effects (discussed in Section 9.1.6), φB(TE) evolves linearly over time and is the term that is relevant for QSM. The second part, , is the phase at TE=0, known as the phase offset (or initial phase) of coil element c. This contribution comprises effects that are common to all the RF receive coils in a phased array, such as the phase of the transmit RF field B1+, the effects of tissue conductivity (Liu et al. 2017), gradient delays and eddy current effects, and contributions that are unique to each receive coil, such as the coil sensitivity. Coil-dependent phase offsets must be removed prior to a complex summation of the coil signals, as shown in Figure 2, to avoid destructive interference. Destructive interference leads to reduced SNR and unphysical phase wraps in regions of the image, which cause artifacts in QSM118. Complete destructive interference is often associated with phase wraps referred to as “open-ended fringe lines” or “phase singularities” (see Figure 3, left). These ill-behaved wraps do not represent isophase contours and cannot be fully removed by unwrapping (see Section 3 below).
Figure 2:
Steps in coil combination, saving, and exporting (illustrated for eight example coils from the 64-channel array). Each of the coils generates a phase image (left), which is modified by the coil sensitivity and other terms which make up the initial phase. The initial phase is removed using methods detailed in the text and referenced publications (center left) and phase images are combined in the manufacturer’s reconstruction and saved for export in DICOM format (center right). QSM analysis software may require the DICOM data to be converted, offline, to NIfTI format (right). Images shown were acquired at 3 T (Siemens Healthineers, Erlangen, Germany; Prisma Fit, VE11C) with a head/neck 64 channel coil and the recommended multi-echo GRE sequence (TE1=5.25ms; echo spacing=5.83ms; 5 echoes) using monopolar readout. Additional details are reported in Section 9.1.5. The imaging data and a description of the acquisition protocol may be found in Supplementary Materials II.
Figure 3:
Some scanner manufacturers’ options for processing and saving phase images (like “Sum of Squares”) do not remove coil sensitivities. This may be apparent in the combined phase images having open-ended fringe lines (left). Wraps in phase images combined with the recommended methods are quite symmetric across the brain mid-line (right), and (like contours on a topographic map) either begin and end at the edge of the brain tissues, or form closed loops within the brain.
All major 3T MR system vendors have effective solutions for generating phase images from RF array coils on their current software platforms. Most of these techniques remove individual coil sensitivities, which are estimated by referencing to a coil with a relatively homogenous sensitivity over the object (usually the body coil). The reference data is acquired in a separate, fast, automated measurement, and the coil sensitivity correction is carried out on complex data either in k-space115 or image space119 before extraction of the phase. To combine the signals from each coil element, we recommend using the available ‘on-console’ vendor solutions listed at the end of this section. These methods may not be available on systems running older software versions, in which case phase images can be reconstructed from ‘raw’ (k-space) data offline or, alternatively, phase images can be saved and exported separately for each coil element (e.g., as DICOM-format image data) to allow an appropriate coil combination method to be applied offline. These offline solutions may require additional efforts in 1) dealing with the export and transfer of very large files or a large number of files, which can be problematic in a clinical setting and 2) obtaining research agreements with vendors for proprietary information on the raw data format and on special routines for performing the image reconstruction offline. Several offline possibilities are nonetheless described in the Supplementary Materials III.
A survey of the members of the committee, conducted as part of this study, showed that most respondents use the coil combination methods listed in Section 3.2. A small proportion of users of Siemens systems, particularly with those running older software or those imaging at 7T, use ASPIRE120 or the separate channel coil export with an offline solution. In the broader community, many groups continue to use offline solutions for consistency with 7T (where no body coil-based solution exists) or to provide consistency with older studies.
For all systems, data should be exported in DICOM format. However, most QSM tools require data to be in NIfTI format, and a number of tools are available to perform the necessary conversion. We recommend DCM2NIIX (https://github.com/rordenlab/dcm2niix), which is a well-maintained open-source software with compiled versions for macOS, Linux and Windows. It has been tested on data from a range of scanners121, including the example data provided with this project. The following alternative programs fulfill a similar function: DICM2NII (https://github.com/xiangruili/dicm2nii), MRIConvert (https://lcni.uoregon.edu/downloads/mriconvert/mriconvert-and-mcverter), and Dicomifier (https://github.com/lamyj/dicomifier). In our survey, DCM2NIIX was the most popular conversion tool, but with a significant spread across the others listed. Format conversion with DCM2NIIX and the analysis steps recommended in this paper preserve the correct image orientation. It should be noted, though, that for non-vendor imaging sequences, some conversion tools and non-standard processing steps (or some combination of these) may not save or handle header parameters correctly. If any of these are used, the researcher should check that left-right flips have not been introduced into images (see Section 8.1.1).
If manufacturers’ product susceptibility weighted imaging (SWI) sequences122–124 are used for QSM acquisition, it should be noted that many of these do not allow direct saving or export of unfiltered phase images. Some do allow saving of the processed (e.g., homodyne filtered) phase data, but these are unsuitable for QSM. If raw k-space data are available from SWI scans, it may be possible to generate phase images usable for QSM from these (see Supplementary Materials III). Where available, we recommend that clinical researchers use scanners and sequences which allow saving and exporting of the original unprocessed phase wherever possible, as this greatly facilitates (retrospective) clinical QSM studies and accelerates clinical translation of QSM.
It is important to make sure that phase images are scaled correctly and converted to the correct data type for subsequent analysis. The analysis pipeline supplied with this paper works with a range of data types and arbitrary phase scaling, but some phase unwrapping algorithms and the nonlinear complex fitting algorithm described in Section 4 require phase to be saved as floating-point numbers scaled between −π and π, and this rescaling and data type conversion needs to be performed by the user. Further, note that the sign convention is different for phase values on different vendor systems125 and this needs to be checked and corrected where necessary so that relatively paramagnetic tissues (e.g., iron-rich deep-brain regions) have positive susceptibility values in the resulting susceptibility maps.
3.2. Consensus Recommendations
We provide recommendations for combining phase images from array coils and saving phase data for each of the major manufacturers, listing the software versions for which these solutions have been tested and whether a research agreement is required. Detailed step-by-step descriptions and solutions for older systems are provided in Supplementary Materials III.
-
3.aThe recommended solution for saving phase images are, for
- Canon: SPEEDER, a version of SENSE which is available from MPower version 2.3 onwards and allows phase images to be reconstructed through a vendor-provided service password.
- GE: ASSET, a SENSE-similar solution which reconstructs magnitude, phase, real, and imaginary images without a research key on platforms MR30 onward.
- Philips: SENSE, which provides well-combined phase images116 without the need for a research key from software version 5 onwards.
- Siemens: “Adaptive-combined with prescan normalize”126, which is available from software version VE11 onward in the product GRE sequence.
- United Imaging: an inter-coil referencing and weighted correction approach which is available from software version v9 without the need for a research key.
-
3.b
Exporting data: Data should be exported in (classic) DICOM format.
-
3.c
Format conversion: if the analysis pipeline requires NIfTI data, DICOM data should be converted to NIfTI using DCM2NIIX.
3.3. Additional Considerations
4. Phase Unwrapping and Echo Combination
This section describes the methods used to resolve phase aliasing and calculate a field map from multi-echo GRE data.
4.1. Overview
MRI phase measurements are constrained to an interval of 2π and are, therefore, subject to phase wraps or phase aliasing artifacts, i.e., the measured phase φ = (φB(TE) + φ0) mod 2π. Such phase wraps introduce a phase difference of an integer multiple of 2π between the measured phase, φ, and the true phase φB(TE) + φ0. Phase wraps are usually visible as discontinuous phase jumps in the phase images (Figs. 3 and 4). To a first-order approximation, φB(TE) = 2πγTE ∙ ΔB0, where γ is the proton gyromagnetic ratio. To obtain an accurate estimate of the field shift, ΔB0, for QSM, both phase wraps and the phase offset φ0 need to be removed from the measured phase, φ54. The coil combination methods recommended in the previous section remove the coil-specific contributions to the phase offset, φ0, but leave other non-B0-related contributions in φ0, common to all coils, which should be removed by the QSM processing pipeline. Note that the background field removal step (see Section 6 below) removes only harmonic fields (those which satisfy Laplace’s equation) within the brain region and cannot completely remove φ0 as it contains both harmonic and non-harmonic components55(Schweser et al., 2017). Therefore, φ0 must be explicitly removed for accurate QSM127.
Figure 4.
A) Example wrapped phase images at different echo times after proper coil combination (same images as shown on the right-hand side of Figure 2). More phase wraps can be observed at a later echo (bottom). B) Example unwrapped phase images (using ROMEO template phase unwrapping with MCPC3D-S phase offset correction). C) Total field estimation after echo combination using weighted echo averaging.
Over the years, different phase unwrapping methods have been adapted, refined and applied to MR phase imaging; including time-domain unwrapping methods (with multi-echo acquisition) such as CAMPUS128 and UMPIRE129, and spatial-domain unwrapping methods such as region-based PRELUDE130, SEGUE131, and SPUN132, path-based best-path unwrapping133 and ROMEO134, and Laplacian unwrapping135. The Laplacian unwrapping method is robust and gives wrap-free phase results even with low SNR but can result in high-frequency errors that propagate into susceptibility maps that are hard to detect visually136. It is also noted that Laplacian unwrapping only gives an approximation of the underlying unwrapped phase, especially when using the commonly used Fourier-based implementation, while region-based and path-based unwrapping give quantitatively more accurate estimates of the unwrapped phase54,134. Region-based and path-based methods are termed “exact unwrapping methods” below as they give the exact value of the unwrapped phase134. When comparing exact unwrapping methods to Laplacian unwrapping, unwrapping errors (e.g., in veins and hemorrhages) are observed to be smaller in the former, improving QSM quantification accuracy, e.g., for oxygenation estimation137,138. Therefore, we recommend using exact unwrapping methods.
Multi-echo phase images, e.g., acquired using the recommended protocol (see Section 2), can be combined to achieve a more accurate estimate of the underlying field shift, ΔB0, than can be obtained from single-echo phase images97. This is because combining multi-echo phase images can remove the phase offset contribution and give higher SNR in the estimated tissue field and susceptibility maps (Fig. 4). The optimum approach may depend on the application, but two echo combination methods have been widely used for QSM – nonlinear complex data fitting139 and weighted echo averaging94.
The nonlinear complex data fitting approach takes into account the Gaussian noise in the complex images139 and estimates the field shift, ΔB0, and phase offset, φ0, together as parameters from fitting the complex MR signal over multiple echoes, with the requirement of having acquired three or more echoes139. This approach usually needs spatial phase unwrapping to be performed after the fitting, i.e., on a scaled field-shift estimate, e.g., 2πγ ∙ δTE ∙ ΔB0, with δTE being the echo spacing, wrapped again between −π and +π, as it resolves phase wraps in the temporal dimension. Nonlinear complex data fitting is more robust than linear phase fitting against phase noise at long TEs and around large susceptibility sources, e.g., veins and hemorrhages.
If echoes are acquired over a useful range of TE (depending on T2* values of the tissues of interest, see Section 2), the weighted echo averaging approach gives higher SNR for estimating the field shift, ΔB0, than the complex data fitting approach, which estimates multiple parameters94,140,141. Unlike nonlinear complex data fitting, this approach needs the phase data at each TE to be unwrapped first. It also requires explicit removal of the phase offset through subtraction of the estimated phase offset from the phase measured at each TE (for example as in MCPC-3D-S and ASPIRE, where the phase offset can be estimated by extrapolating the linear phase evolution to zero echo time)120. Unwrapping errors at longer TEs in voxels with large field shifts and more pronounced noise can be reduced by the “template” unwrapping approach used in ROMEO, which performs path-based spatial unwrapping on an early echo and unwraps other echoes on the basis of the expected linear phase evolution134. This, combined with weighted echo averaging, reduces the effect of such errors in the estimated field map.
To improve QSM quality, the spatial noise map generated from non-linear complex fitting139, or the phase “quality map” calculated in path-based unwrapping134 can be used to mask out voxels with unreliable phase (see Section 5).
Most echo combination methods assume linear phase evolution over TE, ignoring non-linear phase evolution due to microstructure-related compartmentalization effects or biased sampling of the sub-voxel field perturbation (see Section 9.1.6), flow, or signal dropout142. Advanced modeling of the phase evolution over time may provide further information about tissue composition and microstructure37,104,143–148, but is beyond the scope of this paper.
4.2. Consensus Recommendations
-
4.a.
Use an exact phase unwrapping method.
-
4.b.
Perform echo combination before background field removal.
-
4.c.
The optimal pipeline for phase unwrapping and echo combination depends on the acquisition and application. We recommend using either nonlinear complex data fitting followed by spatial phase unwrapping, or weighted echo averaging after template phase unwrapping and explicit phase offset removal.
4.3. Additional Considerations
5. Creation of masks
This section provides recommendations on creating masks for background field removal (Section 6), dipole inversion (Section 7), and visualization (Section 9).
5.1. Overview
Masking is often overlooked when describing a QSM pipeline but is a crucial step149, particularly for background field removal (see Section 6). Masking refers to selecting a region of interest (ROI) within the whole field of view and applying a process or function only within this ROI. In QSM, field maps, ΔB0, are masked primarily because most background field removal algorithms require a mask. In general, masks should cover the largest ROI possible to prevent exclusion of brain tissue with a sufficient signal-to-noise ratio to have reliable phase/field values. This is of special concern for studies of the cortex and the brainstem near the brain border or air-tissue interfaces. Unreliable field map data is composed mostly of extremely noisy voxels resulting from phase noise in regions with very low MRI signal or rapid signal decay. The noise distribution in phase images (and hence, field maps) is generally non-Gaussian and depends on the local magnitude of the signal150. In practice, regions with very low MRI signal yield phase noise uniformly distributed throughout the whole −π to π range, obscuring any underlying phase contrast information150.
Masking is a binary operation. Voxels with mask values of 1 (or Boolean “true”) are included in the selected ROI and voxels with mask values of zero (or Boolean “false”) are excluded. Masks may be created by using heuristic thresholding operations on available subject images, including magnitude images, T2* maps, quality maps, or SNR maps. Masks created from differently thresholded images may also be joined or combined to exclude or include regions based on different criteria. In addition, segmentation algorithms may be based on pre-learned shapes or on the optimization of functionals151–157. In particular, the Brain Extraction Tool (BET)158 from the FMRIB Software Library (FSL) is a widely-used method for brain masking (skull stripping), although it may fail when pathologies or injuries are present159,160. BET is a magnitude-image based algorithm that effectively removes non-brain tissues, air, and bone from magnitude images of the head. When acquiring multiple echoes, using the last-echo magnitude image for BET masking is robust to remove regions with rapid signal dropout161, which is undesirable if such regions are of interest. A more balanced approach with a larger ROI selection is achieved by using magnitude images combined across TEs (e.g., using sum of squares or weighted averaging). Alternatively, the magnitude image of the first echo can be used for brain extraction, with the use of a phase-quality map to further exclude voxels with unreliable phase values, as described below. Alternatives to BET include standard template-based brain-extraction162. Deep learning segmentation alternatives may also be considered, as this is a rapidly developing field163–166.
QSM is also vulnerable to errors and artifacts arising from unreliable phase data that may not be directly reflected in the corresponding magnitude data. These may be caused by coil combination errors, flow in vessels, and other factors. For this reason, it has been proposed to use phase-based quality maps in addition to magnitude-based masking to refine masking161. A straightforward method to obtain a phase quality map is to threshold the inverse of the noise map provided by the complex nonlinear multi-echo fitting algorithm (described in Section 4) at its mean value139,161. The thresholding at the mean value maintains an adequate number of voxels, as it is applied to the entire field of view (FOV), and the distribution of values exhibits bimodal characteristics. This approach effectively distinguishes between reliable and unreliable voxels, serving as a suitable initial approximation. However, modifying the threshold factor (e.g., by multiplying it by 1.2) may yield further enhancement in the results. Some exact phase unwrapping algorithms provide an alternative source of phase-based quality maps134,167. Setting a threshold for phase-based quality maps can help to identify voxels within the brain with unreliable phase values, and to provide a better estimation of the brain boundary149,161,168.
Most phase unwrapping and echo combination algorithms do not require masking54 but suppressing extraneous data by masking can speed up some algorithms and improve their robustness. In contrast, almost all background field removal algorithms require masking to define the region of interest, outside which the susceptibility sources are classified as background field sources (see Section 6)55. Notable exceptions (i.e., background field methods that do not require masking) are recent deep learning approaches such as SHARQnet169 and Total Field Inversion (which, however, requires a mask-like preconditioner)170. The performance of background field removal algorithms depends strongly on the mask and poor background field removal can negatively affect the quality of the reconstructed susceptibility maps171,172. Many dipole inversion algorithms use masks to exclude voxels with unreliable field values from the susceptibility computation or use masks for regularization83,139,173. Finally, susceptibility maps should be masked for display purposes to exclude streaks and spurious information outside the brain (see Section 7 on dipole inversion and Section 9 on presentation and publication).
Background field removal methods remove fields induced by all susceptibility sources outside the supplied brain mask. The accuracy of background field removal is lowest at the boundary of the ROI, such as the brain surface, and improves with increasing distance from the brain surface55. Although further erosion of the mask after BFR is not explicitly required, it may be employed in specific cases to eliminate residual artifacts at the boundary. Also, many background removal algorithms are not able to recover a reliable local field over the whole ROI mask55, and further erosion is unavoidable. It should be noted that some background field removal algorithms (e.g., V-SHARP; see Section 6 below) will result in erosion of the brain mask and the resulting eroded mask should then be used in all subsequent operations and for display.
Holes inside the brain mask lead to the elimination of field contributions from the susceptibility sources within the holes during background field removal. Such holes can occur when thresholded (e.g., phase-based quality) maps are used to refine masking. For example, if a pathology (such as a hemorrhage or calcifications) creates unreliable phase data inside the brain, the affected region could be set to zero in the brain mask. Removing the field perturbations from susceptibility sources within the holes during the background elimination step will render these sources undetectable in the final susceptibility maps, which can be a significant problem in the clinical setting. Therefore, it is important that holes within the brain mask are filled before the mask is used for background field removal. Holes can be reintroduced in the mask used for dipole inversion as an effective way to prevent streaking artifacts from regions with unreliable phase values. This procedure has been included in some algorithms139,174. Since dipole inversion is a nonlocal operation, correct susceptibility values may be inferred inside relatively small holes excluded from the dipole inversion mask. To avoid streaking artifacts created by high contrast sources and pathologies, while preserving accurate susceptibility values inside the holes, some recent two-step approaches suggest performing reconstructions with and without holes and then merging both results138,149,175,176. This is useful to improve the accuracy of the reconstructions, and to characterize hemorrhages or calcifications.
Although some recent deep learning-based single-step QSM approaches have shown that explicit masking could be avoided169,172,177–179, these methods require further study and validation to be considered for clinical applications.
5.2. Consensus Recommendations
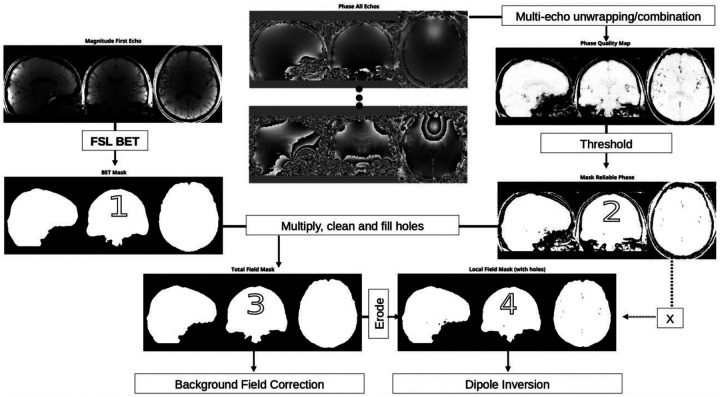
The recommendations below are summarized in Figure 5, and differences between masks (Masks 1–4 in Fig. 5) are highlighted in Figure 6.
Figure 5:
Block diagram of the masking stages. 1) Create an initial mask using FSL BET (Mask 1). 2) Threshold a phase-based quality map to create a reliable phase mask of reliable phase values (Mask 2). 3) Multiply Mask 1 with Mask 2 and fill holes for background field removal. 4) Erode by one or two voxels according to the output of the background field removal algorithm (and optionally reintroduce holes) for dipole inversion. Use Mask 4 without holes filled in for display and reporting susceptibility values. The magnitude and phase images shown are the same as those in Figure 4.
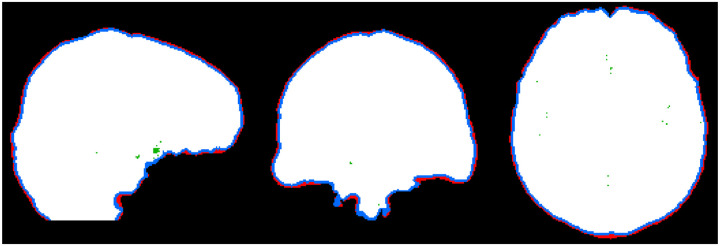
Figure 6:
By using the mask or reliable phase (Mask 2), the initial BET mask (Mask 1) can be further improved by removing unreliable phase data near the boundary (red). Mask 3 is used for background field removal (BFR). After BFR, Mask 3 may need to be further eroded depending on the output of the background field removal algorithms (eroded region shown in blue). This is used for visualization of the results and reporting. Unreliable phase data inside the brain can also be masked out for dipole inversion (holes in green, with the final Mask 4 used for dipole inversion in white).
-
5.a.
Create an initial brain mask (Mask 1) by applying a whole-brain segmentation tool (such as BET) to either the combined (sum of squares) or the first echo magnitude image. The goal of this initial mask is to remove air, skull and other tissues, while preserving cortical areas. Further refinement is performed in the following steps.
-
5.a.
Create a mask of reliable phase values (Mask 2) by thresholding the phase quality map generated by the multi-echo combination method in Section 4. Multiply Mask 1 with Mask 2.
-
5.b.
After multiplication, holes should be filled to obtain the mask to be used as an input to background field removal algorithms (Mask 3).
-
5.c.
Holes from Mask 2 can be reintroduced to avoid streaking artifacts from unreliable phase data within the brain. For increased accuracy of susceptibility values inside pathological regions of low signal, e.g., hemorrhages and calcifications, mixing data from reconstructions with and without the holes can be performed.
-
5.d.
The calculated susceptibility map should be multiplied by the mask used for background field removal (without holes; Mask 3) before display, reporting of susceptibility values or further analysis.
5.3. Additional Considerations
6. Background Field Removal
This section provides recommendations for the background field removal step.
6.1. Overview
In QSM, the background field is defined as the field generated by susceptibility sources outside a chosen ROI55, in our case the brain mask (see previous section). In the brain, the background fields are generated by the tissue and air surrounding the brain. The susceptibility difference between brain tissue and air is approximately 9 ppm76, which is almost two orders of magnitude larger than the naturally occurring susceptibility differences within the brain parenchyma. Therefore, background fields can be significantly larger than the tissue field in the brain, but not always are. Certain pathologies, such as hemorrhages, can create a tissue field that is similar in magnitude locally. The term local field is also often used in the literature for fields generated by tissue within the ROI, but since the field is a nonlocal property, we use the term tissue field here. Removal of the background field from the field map, ΔB0, allows focusing the inversion (see Section 7) on the spatial susceptibility variations located inside the ROI, which generate the so-called tissue field ΔBt (Figure 7). When background fields are not completely removed from ΔB0, most dipole inversion methods will result in shadowing artifacts and/or experience a slow convergence rate.
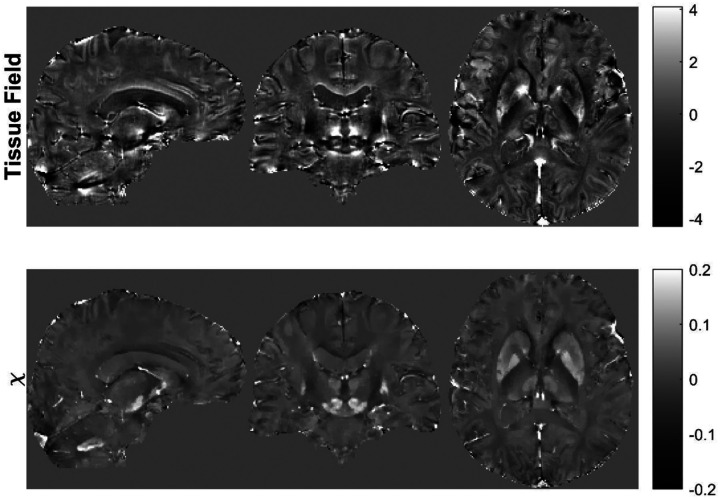
Figure 7.
Process of background field removal estimates the background field component of the total field (first row; same images as shown on the right-hand side of Figure 4; unit is Hz) relative to a chosen region of interest (brain mask, third row) and subtracts it from the total field, resulting in the tissue field (fourth row; unit is Hz). The tissue field encodes the spatially varying susceptibility within the brain but is much smaller than the background field. This is illustrated by showing cross-sections (indicated by the dotted lines in the field images) in the total field (second row) and the tissue field (last row). The background field was calculated using the V-SHARP method.
Because of the spatial smoothness of the background field, spatial high-pass filtering has been a popular method to suppress background fields in the past. However, high-pass filtering also removes the low spatial frequency component, a major signal component, of the tissue field, which is not acceptable to QSM that requires quantitative accuracy of the corrected field maps. Newer methods that directly exploit the harmonic function property of background fields have replaced heuristic filtering methods55. From Maxwell’s equations, it can be derived that the background field is a harmonic field, i.e., it satisfies the Laplace equation within the ROI. A harmonic field is completely determined when it is known on the region boundary. In other words, the solution of the Laplace equation in a region with a given boundary condition is unique55,180.
The SHARP (Sophisticated Harmonic Artifact Reduction for Phase data) method181 and variants thereof use the spherical mean value property of harmonic functions. This property implies that the average of a harmonic function over an arbitrary sphere centered at any location that fits within the region of interest is equal to the value of the harmonic function at that location. In practice, a radius is chosen that is somewhat large compared to the voxel size to overcome discretization effects. This means that the tissue field can only be computed for voxels that are at a distance equal to the chosen radius away from the boundary of the ROI. This limitation leads to an erosion of the region in which susceptibility can be computed (see also previous section). The most common variant of this method is V-SHARP182, which involves multiple partial applications of SHARP with different radii to mitigate the practical implications of the erosion. V-SHARP yields background-corrected field values in the close vicinity of the ROI boundary but the values are not entirely accurate55. E-SHARP183 and other variants of SHARP184,185 overcome the remaining erosion of one voxel required for V-SHARP. Other variants like HARPERELLA186 combine SHARP with phase unwrapping. In general, SHARP-based methods perform less well at the boundary of the region of interest55. In addition, SHARP-based methods include implicit low pass filtering due to the regularized deconvolution inherent in SHARP, which by itself removes slowly varying components187.
The PDF (Projection onto Dipole Fields) method188 finds an effective susceptibility distribution outside the region of interest that mimics the field inside that region. It uses the fact that the field generated by those outside sources are approximately orthogonal to those generated by local sources, allowing background field removal to be formulated as a noise weighted linear least squares problem. Because the orthogonality breaks down at the boundary of the region of interest, like SHARP, this method performs less well at the boundary55,188.
The LBV (Laplacian Boundary Value) method180 assumes that the field at the very boundary of the region of interest is entirely background field and determines a harmonic function that satisfies this boundary condition. An efficient algorithm has been introduced to solve this Laplacian boundary problem180. However, because LBV entirely relies on the field estimate at the mask boundary, it is sensitive to phase SNR at the boundary, rendering accurate masking particularly important. Often a small mask erosion is applied to remove low SNR voxels at the boundary. While LBV can perform better than PDF and SHARP in some situations55, its performance has been observed to be highly dependent on the mask and on the quality of the field estimates at the mask boundary189.
Residual background fields can be suppressed by combining methods, such as applying additional polynomial fitting or V-SHARP after LBV. V-SHARP and PDF typically do not require additional polynomial fitting. B1+ related contributions in the field map, ΔB0 (see Section 4 above) are not removed by background field removal methods, but these are avoided when using multi-echo data combined with field fitting as recommended above.
Susceptibility maps are dimensionless and are conventionally calculated and displayed in parts per million (ppm) (see Section 9.1.4). Assuming that the wrapped input phase was correctly scaled to (−π to π) radians, the corresponding scaling can be done either before (on the tissue field map) or after (on the dipole inversion output) using the scale factor: . When scaling is performed after, care has to be taken to adjust the default regularization parameter when using a total variation based dipole inversion method, as the regularization term scales linearly.
6.2. Consensus Recommendations
-
6.a.
Use V-SHARP to achieve good results in many situations, as it is less sensitive to imperfections in brain masking. This comes at a cost of a one-voxel erosion of the brain mask used for dipole inversion (Mask 4 in Fig. 5) at the brain surface and reduced accuracy at the edge of the brain.
-
6.b.
When whole brain mapping (including the cortex and superficial veins) is desired, use PDF. This method will be slightly more accurate throughout the brain. PDF requires a good brain mask.
-
6.c.
Depending on the application, tissue field quality, i.e., the phase SNR especially near the boundary, must be balanced against mask erosion.
6.3. Additional Considerations
Single step112,170,190–192 or total field inversion methods fit the susceptibility directly to the field map, ΔB0, or even wrapped phase images. These are currently popular for applications of QSM outside the brain but are still under ongoing development to ensure robustness. Residual background field has been tackled for dipole inversion using weak harmonics modeling136. Finally, deep learning52 has found application in background field removal as well and is the subject of ongoing development.
7. Dipole Inversion
This section provides recommendations for the field-to-susceptibility inversion step (Fig. 8), which derives from the tissue field map, ΔBt (with background fields removed; see previous section), a map that is tissue magnetic susceptibility χ(r) (up to a reference value193,194, see Section 8).
Figure 8.
The process of dipole field inversion starts from the tissue field (first row, same images as shown in the bottom row of Figure 7; unit is Hz) and estimates the susceptibility map (second row; unit is ppm).
7.1. Overview
While susceptibility, χ(r), is a local tissue property, the field is a summation of weighted contributions from the distribution of magnetic susceptibility in all space. Mathematically, this summation can be described as a convolution (*) of the susceptibility with the unit dipole kernel 77–79:
| [1] |
Convolution corresponds to multiplication in the spatial frequency domain, which facilitates its fast calculation and is used in most QSM implementations77–79 to accelerate computations. The inversion step performs a deconvolution using the dipole kernel d(r), which reveals the local tissue susceptibility within the region of interest, χt(r), from the background-corrected tissue field, Δ(r): ΔBt(r) *−1 d(r) = χt(r). However, the dipole kernel value is zero at and very small near the cone surface of the magic angle (54. 70) relative to the direction of the main magnetic field, making this deconvolution a poorly conditioned inverse problem75,84,195–197. The measured tissue field, ΔBt, contains deviations from perfect dipole patterns, particularly in regions with small magnitude signal due to lack of water protons (field detectors) or rapid signal decay (largely caused by inhomogeneous fields). While the regions with most extreme deviations are usually eliminated through masking (see Section 5 above), remaining dipole deviations in the estimated field can cause deconvolution errors in the calculated susceptibility map reconstruction, manifesting as streaking and shadowing artifacts53,198. Additional information about the unknown susceptibility map, χ(r), can be incorporated into the solution through regularization to suppress streaking and shadowing artifacts in the solution 25,59,83,84,112,141,173,182,199,200. An optimization approach for incorporating this additional information can be formulated according to Bayesian inference, which is the following minimization problem when approximating the noise in the field as Gaussian:
| [2] |
Here the first term is the data fidelity term with spatially varying noise weighting w and the second term, R(χ), is the regularization term with λ as regularization strength83,141. The minimization problem is iteratively solved with the number of iterations determined by the desired convergence level. The optimal regularization strength (λ) depends on anatomy, susceptibility contrast, and SNR, and should be optimized to balance artifact suppression and image sharpness in each imaging protocol and application by varying λ using, e.g., the L-curve method201,202.
Various regularization strategies have been developed for the inverse problem in QSM25,53,60,83,84,136,141,173,182,192,197,199–201,203–207. Available QSM software packages include FAst Nonlinear Susceptibility Inversion (FANSI)173 (https://gitlab.com/cmilovic/FANSI-toolbox), Morphology Enabled Dipole Inversion (MEDI)201,208 (http://pre.weill.cornell.edu/mri/pages/qsm.html), and STI Suite209 (https://people.eecs.berkeley.edu/~chunlei.liu/software.html).
Total variation regularization has performed favorably in the two QSM reconstruction challenges60,63. Both FANSI and MEDI provide specific implementations of sparsity regularization with openly accessible source code. Specific implementation examples including zero-referencing to the cerebrospinal fluid (CSF) may be included as an extra regularization that provides the CSF-uniformity verification on the QSM output with an additional benefit of further reducing streaking and shadowing artifacts,193,210 but are associated with other limitation as discussed in the next section.
The simple sparse regularizer using L1 norm of the gradient (i.e., the total variation, TV) is a standard approach for brain QSM, as exemplified in MEDI, one of the most popular algorithms. The performance of the TV approach for brain QSM in terms of accuracy and robustness was well established in the 2019 QSM Reconstruction Challenge, particularly in the presence of strong susceptibility sources, such as hemorrhages or calcifications, using nonlinear forward signal modeling60,63. Recent developments in deep learning based QSM reconstruction represent an exciting avenue for improving QSM performance52,179,211,212. However, while there are instances where these methods yield better results than classical methods, the performance of these methods did not reach those of the best classical methods in the QSM challenges potentially due to generalization issues from limited training data60,63.
Some algorithms do not incorporate spatial constraints for suppressing streaking and shadowing artifacts but explicitly modify the dipole kernel instead53,198, for example, the thresholded k-space division192,197,213. Implicit regularizations based on the number of iterations may work, but these methods have limited denoising capabilities and may be less robust than the sparsity regularization optimization approach59,205,214.
7.2. Consensus Recommendation
-
7.a.
Use an optimization approach for dipole inversion with a sparsity type regularization that is commonly used in compressed sensing53. Specific sparsity types include L1-norm, total variation, and generalized total variation, which likely provide similar outcomes. Future algorithm developments and evaluations are needed to provide a more specific consensus on the sparsity type.
-
7.b.
Use the default sparsity type, regularization strength and number of iterations in a QSM software, such as the processing pipelines recommended here (Supplementary Materials II), including FANSI, STI Suite, and MEDI, where these default parameters have been optimized for common brain protocols. If the acquisition protocol recommended here (Supplementary Materials I, Section S1.5) is substantially altered, researchers should perform an L-curve optimization or other method on at least one typical case with the specific study protocol to finetune the regularization strength and iteration number and then fix these parameters for the same protocol.
7.3. Additional Considerations
There may be streaking artifacts coming from strong susceptibility sources near borders and within the brain interior region. Major causes include the breakdown of the Gaussian noise assumption and other errors in the determined field139. These artifacts may be suppressed using methods such as masking out or reducing the weight of less trustworthy voxels in the optimization139. The border streaking can be removed by improving the brain mask149,215,216. The interior streaking can be reduced using techniques to improve convergence such as preconditioning170,217, and using in-painting techniques to compensate for field errors such as MERIT139 and L1 data fidelity174.
There may be shadowing artifacts coming from residual background fields. This shadowing can be reduced by improving background field removal such as harmonic incompatibility removal136,218 and by suppressing slowly varying spatial frequency components through regularization219 or preconditioning53,170.
8. Analysis of Susceptibility Maps
This section provides recommendations for quality control and referencing of susceptibility maps, the quantification of susceptibility values, and the visualization of brain structures on susceptibility maps and derived contrasts in the context of clinical research performing group studies. The physical background for the consensus recommendations is briefly summarized. Possible tools to facilitate susceptibility quantification of brain structures and lesions as well as for group analyses are provided in Supplementary Materials I, Section S1.6. Figure 9 summarizes the recommendations of this section.
Figure 9:
Schematic for susceptibility map analysis in case of a study interested in susceptibility values of the putamen (blue ROI on the susceptibility map, χref) and globus pallidus (red ROI). Data with streaking artifacts that affect the ROIs need to be excluded (or recalculated when applicable to all study data). ROI generation benefits from the inclusion of susceptibility contrast, e.g., by calculation of hybrid images (blue) or use of T1-weighted and susceptibility data (green). Susceptibility maps need to be referenced, then regional average susceptibility values (χROI) can be computed from referenced susceptibility maps (χref). The shown susceptibility map without artifacts is the same as the one in Figure 8.
8.1. Overview
8.1.1. Quality control
Only a few tools for fully automatic QSM calculation and evaluation directly from scanner DICOM data exist to date that perform all steps outlined in Sections 2 to 8149,220. Some of these tools may not be suitable for all possible QSM applications due to assumptions on patient cohorts of the implemented mask generation algorithms (see Section 5 above) or due to the need to adjust reconstruction parameters depending on the data (see Section 7 on dipole inversion). Most QSM applications still require multiple processing steps, which can result in error amplification/propagation or inconsistencies between steps, rendering QSM workflows prone to i) reconstruction artifacts (a list of common reconstruction artifacts is provided in Table 1) and ii) calculation errors of region-specific susceptibility values. Particularly, the use of one or several masks to exclude unreliable phase data and for background field removal during QSM calculation can result in missing areas in computed susceptibility maps especially close to air-tissue interfaces. When voxels in those regions are not properly excluded in subsequent analyses of the susceptibility maps (e.g., in ROI-based analyses), regional mean values may be biased by these erroneously included zero-valued voxels in the susceptibility mean value calculations. The issue can be resolved by incorporating the eroded background-correction mask (Mask 4 without holes in Fig. 5) in the ROI masks. Deviation from the radiological orientation (right-left flip) in the final susceptibility maps (see also Section 3.1) can be a potential issue arising from the combination of different toolboxes when using other tools than those recommended here or as a result of erroneous use. These flips can have detrimental consequences in the clinical setting but can be revealed relatively easily comparing brain features between QSM and the original GRE magnitude images on the scanner console, especially when the subject’s head was tilted to the right or left (about the H-F, A-P or both of these axes) for test purposes (see Section 3.1), a step that should always be done if custom pipelines are used.
Table 1.
Reconstruction artifacts, possible sources, and strategies to identify, mitigate these artifacts and criteria to exclude the data.
| Artifact | Streaking and shadowing artifacts
|
Incorrect susceptibility values
|
(Regional) strong noise
|
|---|---|---|---|
| Typical sources |
|
|
|
| Identification |
|
|
(see streaking and shadowing artifacts) |
| Mitigation |
|
|
|
| Data exclusion |
|
8.1.2. Referencing and choice of reference region
QSM can only assess relative susceptibility differences between tissues as phase data reflect field distortions due to these underlying spatial susceptibility differences. Susceptibility values are therefore given up to a reference56,194. To obtain susceptibility values that are comparable between repeated measurements, subjects, and scanners, consistent referencing of susceptibility maps is required. In QSM, internal reference regions are used. External reference regions are not generally used because it is not currently possible to measure phase differences between disconnected spatial regions separated by noise and perform consistent background field removal for both the brain tissue and the external reference region. The ideal choice of a reference region for brain QSM is still under debate194. Different regions used in the literature come with certain advantages and disadvantages and will lead to different susceptibility values in the resulting susceptibility map. For example, assuming an ROI’s average susceptibility value is 0.010 ppm when computed from a susceptibility map referenced to the whole brain (with assumed mean susceptibility of whole brain −0.001 ppm), this ROI susceptibility value will be 0.008 ppm when computed from a susceptibility map referenced to the CSF (assumed mean susceptibility of CSF 0.001 ppm).
In the case of widespread pathology such as in multiple sclerosis or Alzheimer’s disease, there might not be an ideal choice of reference region. Larger reference regions are generally advantageous over small-sized regions, which are more affected by potential local lesions, reconstruction inhomogeneities and other artifacts (less averaging), which are then propagated to all other regions in the map by the referencing process. This issue reduces statistical power and therefore 3D segmentation of reference regions is advisable to include a greater number of voxels. Consequently, whole-brain referencing is considered stable (largest possible mask) and reproducible (whole-brain mask readily available in all reconstruction pipelines).
The dependence of white matter apparent susceptibility on the fiber orientation with respect to the main magnetic field due to the geometry and complex microstructure of white matter fiber bundles102,104,146,221,222 (see also Section 9.1.6) can be a source of additional variability when using a reference that includes white matter regions. Another challenge of referencing is that pathology or effects of age alter white and gray matter integrity, specifically myelination and brain iron levels, especially in deep gray nuclei223–228. In the case of widespread pathology when no ideal reference region exists, two reference regions could be used to evaluate if the choice of reference region affects the study results. If, for example, whole brain and CSF were used as reference leading to the same significant differences between patients and controls, the results can be assumes with greater confidence to originate from the presence of pathology, instead of being an artifact from susceptibility referencing229.
For local pathology, the use of contralateral or surrounding tissues as reference is an effective strategy to avoid introducing artificial susceptibility differences due to using a reference region affected by pathology.
Table 2 lists advantages and disadvantages of common reference regions. More details on referencing can be found in dedicated literature193,194. We recommend referencing with regions that are commonly used in the literature. In addition, we recommend that studies report the mean and standard deviation of the susceptibility (after referencing) in other regions that are or have been widely used for referencing along with their hypothesis-driven regions of interest. This approach will promote reproducible research as it facilitates the comparison of susceptibility values between studies and enables post hoc re-referencing for meta-analyses. While no normative susceptibility values exist, literature values230 can serve as precedence reference when comparable subjects are studied, e.g. healthy controls of similar age.
Table 2.
Commonly used reference regions in the literature
| Reference region | advantages | disadvantages |
|---|---|---|
| cerebrospinal fluid193,205,230,248–252 |
|
|
| global white matter regions (not restricted to internal capsule)28,91,253 |
|
|
| internal capsule40,254–256 |
|
|
| whole brain229,236,236,257 |
|
|
8.1.3. Effect of segmentation on susceptibility quantification (iron, white matter changes, lesions, vessels, oxygenation)
An accurate segmentation of ROIs is essential to uncover subtle changes in regional susceptibility values that might indicate pathology, or to establish normative values. While manual segmentation of regions by multiple expert readers is the gold standard for quantification of regional susceptibility values, this strategy is very time-consuming and therefore not feasible in larger studies. Many available automated neuroimaging segmentation tools are optimized for use with T1-weighted images or require T1-weighted input data231. However, when using these methods for the analysis of susceptibility maps, the segmentation and registration accuracy in many structures of interest (e.g., basal ganglia) can depend on T1 contrast232, which is also affected by tissue iron233,234, and the generally low visibility of some deep gray matter regions on T1-weighted images235 (depending on sequence parameters). Consequently, ROI-based methods that rely solely on T1-weighted contrast may be biased and suffer from inaccuracies. Previously, it has been shown that the use of a QSM or hybrid QSM-T1-weighted contrasts for template generation improves atlas and voxel-based analyses30,31. Therefore, using multi-contrast segmentation can be considered the best approach to avoid template bias236. A list of recommended tools can be found in the Supplementary Materials I, Section S1.6. Partial volume effects might strongly affect susceptibility quantification both for voxel-based and ROI-based analyses, especially for small structures with relatively high susceptibility values such as veins237. This could be corrected for by eroding of ROIs101, only using high susceptibility voxels (in case of positive susceptibility)238 or using a partial volume map for correction239.
8.2. Consensus Recommendations
-
8.a.
When ROIs are affected by artifacts, exclude data by automated detection of outliers or outlier regions, use of image quality measures or visual inspection.
-
8.b.
Ensure that analysis methods do not include voxels of the susceptibility map with unreliable values, e.g., that lie outside of the eroded background field removal mask (see Section 5 above; Mask 4 without holes in Fig. 5).
-
8.c.
Always reference susceptibility maps to an internal reference region before performing further analyses.
-
8.d.
When choosing a reference region, consider the study design, influence of pathology, how pathology could bias the study findings and discuss accordingly. For widespread pathology, cross-checking results using two different reference regions (e.g., whole brain and CSF) can be considered safe to exclude bias.
-
8.e.
Segment reference regions in 3D.
-
8.f.
Always include commonly used reference regions in your analysis and report mean and standard deviation in these regions along with those in other ROIs.
-
8.g.
Consider incorporating QSM contrast in ROI segmentation or ensure that T1w-based methods are accurate.
8.3. Additional Considerations
9. Presentation and Publication
9.1. Overview
The purpose of the recommendations in this section is to facilitate the interpretation and replicability of future findings with QSM, future meta-analyses, and the comparison among studies. To this end, the general recommendation is to report as much information as possible regarding:
Data acquisition (hardware and scan parameters);
Reconstruction pipeline and analysis procedure; and
Results.
Depending on the study and on the journal in which the study will be published, the degree of information detail that can be reported may vary. The members of the QSM Consensus Organization Committee asked themselves, through a multiple-choice grid form, whether each information entity relevant for QSM should be reported always (a) or only depending (d) on the study and on the journal, or if it is unnecessary (u) to report the entity. Each item in the poll was assigned a score S = (A + 0.5D)/(A + D + U), where A, D and U are the number of a, d and u responses collected for that item, respectively. The reporting of specific items was considered essential if there was unanimous consensus in reporting them among the authors (S = 1). Items that were not considered essential were assigned a “traffic light ranking” (green for 0.75 ≤ S ≤ 1, orange for 0.5 ≤ S ≤ 0.75, and red for S ≤ 0.5). Standardized tables are provided to facilitate the reporting of a broad set of items. For the purpose of availability, unless there is limited space for particular journals, we recommend that items with S>0.5 be reported as other investigators may need this information. It is considered essential to report these items if they vary within the same study (e.g., if different scanners or different software releases are used within the same study). Potential limitations and confounds should always be discussed. The last part of this section reviews some important aspects that should always be considered when interpreting and presenting QSM findings in scientific papers.
9.1.1. Acquisition hardware
Ideally, the acquisition hardware is described in one sentence reporting the scanner field strength, model, vendor, software release version, and type of coil used (including the number of channels). Table 3 provides an overview of consensus recommendations pertaining to acquisition hardware.
Table 3.
Recommendations for reporting of parameters of the acquisition hardware.
| Item | Notes and examples | Recommended |
|---|---|---|
| Field strength | DICOM tag (0018,0087) | essential |
| Vendor | DICOM tag (0008, 0070) | essential |
| Scanner model | DICOM tag (0008, 1090) |
|
| Software release | DICOM tag (0018, 1020) |
|
| Type of coil(s) used, including information on number of channels | e.g. “… a transmitting body-coil and a 64-channel head-and-neck receiving coil” |
|
| Gradient system | e.g. “… a gradient system with maximum amplitude = 50 mT/m and slew rate = 200 mT/m/ms” |
|
9.1.2. Acquisition sequence type and parameters
The QSM Consensus Organization Committee considered it as essential to indicate the acquisition sequence type (e.g. GRE, as recommended in this paper, or EPI; specify if the sequence is 3D or 2D) and several acquisition parameters including number of echoes, TEs, TR, flip angle, bandwidth, resolution and scan duration. Table 4 provides an overview of consensus recommendations pertaining to acquisition sequence type and parameters.
Table 4.
Recommendations for reporting of parameters of the acquisition sequence.
| Item | Notes | Recommended |
|---|---|---|
| Acquisition sequence type | 2D vs 3D; GRE vs EPI etc.; | essential |
| Acquisition sequence commercial name | e.g. “SWAN”, “MERGE”, “SWlp”… |
|
| k-space sampling trajectory scheme | cartesian vs spiral vs radial etc. | essential, if not cartesian |
| Acquisition orientation | pure axial vs sagittal vs oblique |
|
| Number of echoes, TE1:ΔTE:TEmax | e.g. 7 echoes, TE = 5:5:35 ms | essential |
| TR | essential | |
| FA |
|
|
| Pixel Bandwidth or Receiver Bandwidth [Hz] | DICOM tag (0018, 0095) |
|
| Spatial coverage (FOV) and acquisition matrix size | essential | |
| Voxel Size | Attention: it can be different from “FOV divided by matrix size” | essential |
| Monopolar vs bipolar echoes | Indicate if the sequence produces monopolar of bipolar echoes |
|
| Average ± std center frequency [MHz] | In multi-scanner studies, mean ± std center frequency shall be reported for data from each scanner. For example, Siemens “3T” scanners systematically operate at <2.9T. DICOM tag (0018,0084) | essential for multi-scanner studies; unnecessary for single-scanner studies |
| Flow compensation | Yes / no; if yes, please indicate the compensated echo(s): all vs only the first one; and direction (full, phase) |
|
| Acceleration type and factor | Yes / no. If yes: SENSE (or ASSET) vs GRAPPA (or ARC), compressed SENSE, etc.; indicate phase factor and slice factor (if 3D) | essential |
| Partial Fourier factor | Use should be avoided. If used, indicate partial Fourier factors in phase and slice direction | essential, if used |
| Partial echo (GE/Philips) aka Asymmetric echo (Siemens) aka Half echo (Hitachi) | Use should be avoided. | essential, if used |
| Elliptical k-space shutter | Yes / no. |
|
| Phase stabilization | Option available only in particular implementations. If the option is available, indicate Yes / no |
(essential if used) |
| Excitation pulse | Fat-sat vs Water-only |
|
| Scan duration | essential |
9.1.3. Reconstruction pipeline and analysis
It is considered essential to describe the toolbox and reconstruction pipeline and list the algorithms used. The numerical values of parameters used should be listed, even if they were the default parameters. Table 5 provides an overview of consensus recommendations pertaining to the reconstruction and analysis pipelines.
Table 5.
Recommendations for reporting of parameters of the reconstruction and analysis pipelines.
| Item | Notes | Recommended |
|---|---|---|
| Toolbox used | Specify toolbox name and version (or download date), e.g. FANSI, STISuite, MEDI, etc. | mandatory |
| Algorithms used | For each step of the recon pipeline (phase reconstruction, echo combination, masking, phase unwrapping, background field removal, dipole inversion), please specify the algorithm used. Indicate the numerical values of relevant parameters (even if default values were used), e.g. regularization parameters. |
mandatory, at least for non-default algorithms and parameters |
| Further processing | If further processing was necessary to make images compatible with image review environments (such as PACS) used in the study, any data manipulation (including geometrical transformations, interpolation, header data changes, etc.) should be reported |
|
| Referencing | Magnetic susceptibility values should always be reported in either ppm or ppb (parts-per-billion) and the reference region (see the Section 8) should be explicitly stated, even in the case the adopted method did implicit whole brain referencing. When the reference region used in the study is not the whole-brain mask, its [mean ± std] susceptibility value when referenced to the whole-brain mask should be reported, to enable post-hoc re-referencing for meta-analyses. Generally, it should be discussed in the Discussion section how potential pathological changes within the reference region may have biased the study outcome. |
mandatory |
| Data inclusion/exclusion criteria | Details on data inclusion/exclusion criteria should be reported. For example: which artifacts were taken into consideration, and which level of artifact severity was considered as a threshold for inclusion/exclusion. The description of this aspect, which is study-specific, can be supported by images with representative cases in the Supplementary Materials. | Mandatory, in studies where datasets were excluded based on image quality, or when datasets with visible artifacts were deemed acceptable for inclusion |
9.1.4. Displaying figures
When displaying quantitative susceptibility maps, do not use rainbow, jet, or similar types of non-linear colormaps, which introduce the erroneous perception of artificial edges in some parts of the range, hide existing edges in other parts of the range, and lack intuitive perceptual ordering240–242. In the absence of a motivation for doing otherwise in particular studies, the use of a linear, perceptually uniform colormap should be preferred; the use of a linear gray-scale map enables consistency with the vast majority of the published literature. This applies also to phase data, to enable a clear representation of phase wraps and/or possible errors such as open-ended fringe lines. Contrast windowing should be adjusted to avoid saturation of relevant brain areas (i.e., completely black or white appearance). A typical window adapted to healthy brain QSM is [−0.2, +0.2] ppm. The windowing should always be reported, by either using an intensity bar or writing the information in the figure caption. Susceptibility maps should be displayed through the eroded mask used for background field removal (see Section 5), to avoid representation of artifactual data outside the brain.
9.1.5. Sample paragraphs
Representative paragraphs describing data acquisition, processing and QSM calculation in a scientific paper are provided in the following. The description refers to the images shown in Figure 8.
“Data were acquired on a 3 T scanner (Prisma Fit, Siemens Healthcare, Erlangen, Germany; VE11B) using the built-in whole-body RF transmit coil and a 64-channel receive-only head/neck coil. The acquisition sequence was a 3D GRE multi-echo with pure axial orientation with the following scanning parameters: TR = 33 ms, 5 monopolar echoes acquired at TE1 : ΔTE : TE5 = 5.25 : 5.83 : 28.57 ms, flow compensation for the first echo in the readout (AP) and ‘slice’ encoding (HF) direction, FA = 15°, pixel bandwidth = 220 Hz, elliptical k-space shutter, covering a field of view (FOV) of 256(AP)×176(LR)×144(HF) mm3 with matrix size=256×176×144, resulting in isotropic voxels of size 1mm3, with GRAPPA acceleration factor=2 in the phase encoding (LR) direction. Scan duration was 6 minutes 34 seconds. The full QSM reconstruction was performed in Matlab R2021a (MathWorks, Natick, MA, USA) using the SEPIA toolbox73 (v1.2.2.4) for integration of the various processing steps described hereafter. ROMEO total field calculation134 (v3.5.6) was used for echo phase combination. Brain masking was obtained with FSL BET158 on the first-echo magnitude image, using default settings. V-SHARP182 with spherical mean value filtering sizes from12 mm to 1 mm was used for background field removal. Quantitative susceptibility maps were obtained using FANSI173 (v3) with gradient L1 penalty of 0.0005 and gradient consistency weight of 0.05. Susceptibility values are expressed in parts per million (ppm) and have been referenced to the average susceptibility in the brain mask (imposed to zero by the adopted processing pipeline).”
A complete description of the QSM calculation pipeline can be found in the Supplementary Materials II.
9.1.6. Interpretation of results
Potential limitations and confounds related to QSM should always be taken into account.
A potential confound that can affect the extraction of quantitative susceptibility values from MRI phase/frequency data arises from the fact that, even for a uniform voxel-averaged susceptibility distribution, the apparent field measured in a voxel depends on the subvoxel distribution and visibility of water protons (the sensors of the MRI signal) around susceptibility perturbers such as iron and myelin. This can lead to a phase shift resulting from the biased sampling of the fields generated by perturbers when sensor and perturber distributions spatially correlate and such correlation is anisotropic243,244. An example is water in and around myelinated fibers, whose anisotropic distribution leads to a fiber orientation-dependent shift in apparent frequency which can exceed 10 ppb104,144,146,221,243,244. In addition, substantial T1, MT, or T2* weighting may differentially affect water visibility in the various water compartments and render apparent frequency shifts dependent (in a non-linear manner) on TR or TE142,143,145. Especially above 3T, QSM values within and around fibers that run perpendicular to B0 should be interpreted with caution. For example, pathological changes in myelin structure but not myelin content in such fiber bundles may lead to QSM changes without actual changes in tissue susceptibility.
Another point of caution with interpretation of QSM are inaccuracies near the edge of the regions selected for the analysis (see Section 5). A notable example are areas near the surface of the brain, where phase data is unreliable (due to, e.g., the prevalence of paramagnetic blood in pial veins), or unavailable (due to the lack of signal in skull), or the tissue phase was partially removed in the background field removal step. Because of this, QSM values in some of cortical grey matter may be incorrect or have reduced spatial contrast and resolution.
Lastly, it should be realized that, when strong regularizations or prior information are used in QSM dipole inversion, potential smoothing and spatial resolution loss may occur or new features may be added201,245,246 (see Section 7.1). Some anatomical detail, visible in phase or magnitude GRE data, may therefore be lost in the QSM.
9.2. Consensus Recommendations
-
9.a.
Always report at least the essential information regarding the acquisition hardware (Table 3), acquisition sequence type and parameters (Table 4), reconstruction pipeline and analysis (Table 5).
-
9.b.
Representative susceptibility maps and the underlying background-field corrected phase images should be shown in all articles.
To facilitate documenting the reconstruction pipeline, we encourage software developers to enable printing out the values of all relevant parameters in Tables 4 and 5 (including default parameters) and provide suggested descriptions of their toolboxes, which users can re-utilize in their publications.
9.3. Additional Considerations
10. Summary and Conclusion
This consensus paper has been developed by the QSM Consensus Paper Committee with consideration of suggestions from the whole QSM research community (see Acknowledgements and Supplementary Materials I, Section S1.3). The paper provides recommendations for all steps essential in setting up a successful QSM study in a clinical research setting. The recommendations, intended for a robust but not necessarily state-of-the-art QSM, are based on the current understanding (as of 2023) and should be updated as the QSM field progresses.
In summary, we recommend that data be acquired using a monopolar 3D multi-echo GRE sequence, that phase images be saved and exported in DICOM format and unwrapped using a quantitative approach. Echoes should be combined before background removal, and a brain mask created using a brain extraction tool with the incorporation of phase-quality based masking. Background fields within the brain mask should be removed using a SHARP-based or PDF technique and the optimization approach to dipole inversion should be employed with a sparsity type regularization. Susceptibility values should be measured relative to a specified reference, including the common reference region of whole brain as a region of interest in the analysis, and QSM results should be reported with – as a minimum – the acquisition and processing specifications listed in the final section.
The recommended steps for data acquisition, data preparation and post processing are intended to provide a uniform robust reference starting point for a brain-focused QSM study performed with a clinical scanner. Specialty applications such as the depiction of small structures might require spatial resolutions higher than recommended247. In this regard, limitations and further considerations are included in each section, but thorough testing of the processing pipeline is recommended before starting a large patient study.
We hope that the recommendations here will enable many medical research centers to perform comparable QSM studies on scanners from different vendors, and that the standardized acquisition protocols and the processing pipeline provided along with this article will facilitate these studies (see Supplementary Materials I, Section S1.5. and Supplementary Materials II). As more clinical QSM studies are performed, analyzed, and presented in scientific publications, and current and future technical innovations become mature, these QSM recommendations will need to be updated.
Supplementary Material
Acknowledgements
We thank the following individuals for their significant contribution to the development of the initial consensus statements during the study group review phase: Steffen Bollmann (The University of Queensland), Marta Lancione (IRCCS Stella Maris Foundation), Jakob Meineke (Philips Research Hamburg), Xi Peng, Ludovic de Rochefort (Aix-Marseille University), Mathieu Santin (L’Institut du Cerveau et de la Moelle Épinière), Salil Soman (Harvard Medical School).
We thank the following individuals for their significant contribution to the improvement of the manuscript at and following the presentation at the 2022 Joint Workshop on MR phase, magnetic susceptibility and electrical properties mapping held in Lucca, Italy on October 16–19, 2022. Researchers providing general advice on consensus recommendations: Xavier Golay (University College London); MRI physicists supporting clinicians: Wiebke Nordhøy (Oslo University Hospital); MRI physicists working at manufacturers: Brian Burns (GE Healthcare), Kim van de Ven (Philips); Neurologists: Nicolás Crossley K. (The Pontifical Catholic University of Chile).
In addition, we thank the following ISMRM EMPT study group members, industry representatives, and clinicians who provided substantial feedback during the November/December 2022 manuscript review phase: Antje Bishof (University Hospital Münster), Yoshitaka Bito (Industry; Fujifilm), Nicolas Crossley (Pontificia Universidad Catolica de Chile), Andreas Deistung (University Hospital Halle), Cristina Granziera (University of Basel), Mark E. Haacke (Wayne State University), Marta Lancione (IRCCS Stella Maris Foundation), Emelie Lind (Lund University); Anna Lundberg (Lund University); Jie Luo (Shanghai Jiao Tong University), Jakob Meineke (Philips Research Hamburg); Kieran O’Brian (Industry; Siemens Healthineers), Alexander Rauscher (University of British Columbia), Daniel Sodickson (NYU Langone Health), Hongfu Sun (University of Queensland), Anil Man Tuladhar (Radboud University Medical Center), Anja van der Kolk (Donders Institute), Kim van de Ven (Industry; Philips); Alan Wilman (University of Alberta), Ronnie Wirestam (Lund University); Yongquan Ye (Industry; United Imaging); Xiangzhi Zhou (Mayo Clinic).
This publication was supported by the National Institute of Neurological Disorders and Stroke of the National Institutes of Health under Award Number R01 NS114227 (F.S.), R01 NS105144, and R01 NS095562 (Y.W.), the National Center for Advancing Translational Sciences of the National Institutes of Health under Award Number UL1 TR001412 (F.S.), the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health under Award Numbers R01 EB032378, R01 EB028797, R03 EB031175, P41 EB030006, U01 EB026996 (B.B.), U01 EB025162 (B.B., C.L.), and P41 EB031771 (P.V.Z, X.L.), the National Institute on Aging of the National Institutes of Health under Award Numbers R01 AG063842 (X.L.) and R01 AG070826 (C.L.), and the National Institute of Mental Health of the National Institutes of Health under Award Number R01 MH127104 (C.L.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. S.R. was supported by the Austrian Science Fund (FWF): 31452 and the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No. 794298. K.S. is supported by European Research Council consolidator grant DiSCo MRI SFN 770939. M.C. is supported by the Italian Ministry of Health (grant RC and 5×1000 voluntary contributions)
Footnotes
CRediT authorship contribution statement
Berkin Bilgic: Conceptualization, Methodology, Writing – Original Draft Preparation, Writing – Review & Editing. Mauro Costagli: Conceptualization, Methodology, Visualization, Writing – Original Draft Preparation (lead), Writing – Review & Editing. Jeff Duyn: Writing – Review & Editing. Christian Langkammer: Conceptualization, Methodology, Writing – Original Draft Preparation, Writing – Review & Editing. Jongho Lee: Conceptualization, Methodology, Visualization, Writing – Original Draft Preparation (lead), Writing – Review & Editing. Xu Li: Conceptualization, Methodology, Visualization, Writing – Original Draft Preparation (lead), Writing – Review & Editing. Chunlei Liu: Conceptualization, Methodology, Writing – Original Draft Preparation, Writing – Review & Editing. José Marques: Conceptualization, Methodology, Resources, Software, Visualization, Writing – Original Draft Preparation (lead), Writing – Review & Editing. Carlos Milovic: Conceptualization, Methodology, Visualization, Writing – Original Draft Preparation (lead), Writing – Review & Editing. Simon Robinson: Conceptualization, Methodology, Visualization, Writing – Original Draft Preparation (lead), Writing – Review & Editing. Ferdinand Schweser: Conceptualization, Methodology, Project Administration (lead), Supervision (lead), Writing – Original Draft Preparation, Writing – Review & Editing (lead). Kwok-Shing Chan: Conceptualization, Data Curation, Formal Analysis, Methodology, Software (lead). Karin Shmueli: Conceptualization, Methodology, Writing – Original Draft Preparation, Writing – Review & Editing. Pascal Spincemaille: Conceptualization, Methodology, Visualization, Writing – Original Draft Preparation (lead), Writing – Review & Editing. Sina Straub: Conceptualization, Methodology, Visualization, Writing – Original Draft Preparation (lead), Writing – Review & Editing. Peter van Zijl: Conceptualization, Writing – Review & Editing. Yi Wang: Conceptualization, Methodology, Resources, Supervision, Visualization, Writing – Original Draft Preparation (lead), Writing – Review & Editing
References
- 1.Bhattarai A, Egan GF, Talman P, Chua P, Chen Z. Magnetic Resonance Iron Imaging in Amyotrophic Lateral Sclerosis. J Magn Reson Imaging. 2022;55(5):1283–1300. doi: 10.1002/jmri.27530 [DOI] [PubMed] [Google Scholar]
- 2.Duyn JH. Studying brain microstructure with magnetic susceptibility contrast at high-field. NeuroImage. 2018;168(xxxx):152–161. doi: 10.1016/j.neuroimage.2017.02.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eskreis-Winkler S, Zhang Y, Zhang J, et al. The clinical utility of QSM: disease diagnosis, medical management, and surgical planning. NMR Biomed. 2017;30(4):e3668. doi: 10.1002/nbm.3668 [DOI] [PubMed] [Google Scholar]
- 4.Ghassaban K, Liu S, Jiang C, Haacke EM. Quantifying iron content in magnetic resonance imaging. NeuroImage. 2019;187:77–92. doi: 10.1016/j.neuroimage.2018.04.047 [DOI] [PubMed] [Google Scholar]
- 5.Liu C, Wei H, Gong N jie, Cronin MJ, Dibb R, Decker K. Quantitative Susceptibility Mapping: Contrast Mechanisms and Clinical Applications. Tomography. 2015;1(1):3–17. doi: 10.18383/j.tom.2015.00136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nikparast F, Ganji Z, Zare H. Early differentiation of neurodegenerative diseases using the novel QSM technique: what is the biomarker of each disorder? BMC Neurosci. 2022;23(1):48. doi: 10.1186/s12868-022-00725-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nikparast F, Ganji Z, Danesh Doust M, Faraji R, Zare H. Brain pathological changes during neurodegenerative diseases and their identification methods: How does QSM perform in detecting this process? Insights Imaging. 2022;13(1):74. doi: 10.1186/s13244-022-01207-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plantinga BR, Temel Y, Roebroeck A, et al. Ultra-high field magnetic resonance imaging of the basal ganglia and related structures. Front Hum Neurosci. 2014;8(November):876. doi: 10.3389/fnhum.2014.00876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ravanfar P, Loi SM, Syeda WT, et al. Systematic Review: Quantitative Susceptibility Mapping (QSM) of Brain Iron Profile in Neurodegenerative Diseases. Front Neurosci. 2021;15:618435. doi: 10.3389/fnins.2021.618435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ropele S. Applications of susceptibility-weighted imaging and mapping. In: Advances in Magnetic Resonance Technology and Applications. Vol 4. Elsevier; 2021:359–369. doi: 10.1016/B978-0-12-822479-3.00037-3 [DOI] [Google Scholar]
- 11.Ropele S, Langkammer C. Iron quantification with susceptibility. NMR Biomed. 2017;30(4):e3534. doi: 10.1002/nbm.3534 [DOI] [PubMed] [Google Scholar]
- 12.Stüber C, Pitt D, Wang Y. Iron in Multiple Sclerosis and Its Noninvasive Imaging with Quantitative Susceptibility Mapping. Int J Mol Sci. 2016;17(1):100. doi: 10.3390/ijms17010100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vinayagamani S, Sheelakumari R, Sabarish S, et al. Quantitative Susceptibility Mapping: Technical Considerations and Clinical Applications in Neuroimaging. J Magn Reson Imaging. 2021;53(1):23–37. doi: 10.1002/jmri.27058 [DOI] [PubMed] [Google Scholar]
- 14.Wang C, Foxley S, Ansorge O, et al. Methods for quantitative susceptibility and R2* mapping in whole post-mortem brains at 7T applied to amyotrophic lateral sclerosis. NeuroImage. 2020;222:117216. doi: 10.1016/j.neuroimage.2020.117216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Spincemaille P, Liu Z, et al. Clinical quantitative susceptibility mapping (QSM): Biometal imaging and its emerging roles in patient care. J Magn Reson Imaging. 2017;46(4):951–971. doi: 10.1002/jmri.25693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ravanfar P, Syeda WT, Jayaram M, et al. In Vivo 7-Tesla MRI Investigation of Brain Iron and Its Metabolic Correlates in Chronic Schizophrenia. Schizophrenia. 2022;8(1):86. doi: 10.1038/s41537-022-00293-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu M, Guo Y, Cheng J, et al. Brain iron assessment in patients with First-episode schizophrenia using quantitative susceptibility mapping. NeuroImage Clin. 2021;31:102736. doi: 10.1016/j.nicl.2021.102736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang W, Zhou Y, Li Q, et al. Brain Iron Deposits in Thalamus Is an Independent Factor for Depressive Symptoms Based on Quantitative Susceptibility Mapping in an Older Adults Community Population. Front Psychiatry. 2019;10:734. doi: 10.3389/fpsyt.2019.00734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yao S, Zhong Y, Xu Y, et al. Quantitative Susceptibility Mapping Reveals an Association between Brain Iron Load and Depression Severity. Front Hum Neurosci. 2017;11(August):1–8. doi: 10.3389/fnhum.2017.00442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao W, Li W, Han H, et al. Prenatal alcohol exposure reduces magnetic susceptibility contrast and anisotropy in the white matter of mouse brains. NeuroImage. August 2014. doi: 10.1016/j.neuroimage.2014.08.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Juhás M, Sun H, Brown MRG, et al. Deep Grey Matter Iron Accumulation in Alcohol Use Disorder. NeuroImage. 2017;148(January):115–122. doi: 10.1016/j.neuroimage.2017.01.007 [DOI] [PubMed] [Google Scholar]
- 22.Wang C, Martins-Bach AB, Alfaro-Almagro F, et al. Phenotypic and genetic associations of quantitative magnetic susceptibility in UK Biobank brain imaging. Nat Neurosci. 2022;25(6):818–831. doi: 10.1038/s41593-022-01074-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen W, Zhu W, Kovanlikaya I, et al. Intracranial calcifications and hemorrhages: characterization with quantitative susceptibility mapping. Radiology. 2014;270(2):496–505. doi: 10.1148/radiol.13122640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deistung A, Schweser F, Wiestler B, et al. Quantitative Susceptibility Mapping Differentiates between Blood Depositions and Calcifications in Patients with Glioblastoma. Kleinschnitz C, ed. PLoS ONE. 2013;8(3):e57924. doi: 10.1371/journal.pone.0057924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schweser F, Deistung A, Lehr BW, Reichenbach JR. Differentiation Between Diamagnetic and Paramagnetic Cerebral Lesions Based on Magnetic Susceptibility Mapping. Med Phys. 2010;37(10):5165–5178. doi: 10.1118/1.3481505 [DOI] [PubMed] [Google Scholar]
- 26.Straub S, Laun FB, Emmerich J, et al. Potential of quantitative susceptibility mapping for detection of prostatic calcifications. J Magn Reson Imaging. 2017;45(3):889–898. doi: 10.1002/jmri.25385 [DOI] [PubMed] [Google Scholar]
- 27.Cong F, Liu X, Liu CJ, et al. Improved depiction of subthalamic nucleus and globus pallidus internus with optimized high-resolution quantitative susceptibility mapping at 7 T. NMR Biomed. July 2020. doi: 10.1002/nbm.4382 [DOI] [PubMed] [Google Scholar]
- 28.Deistung A, Schäfer A, Schweser F, Biedermann U, Turner R, Reichenbach JR. Toward in vivo histology: a comparison of quantitative susceptibility mapping (QSM) with magnitude-, phase-, and R2*-imaging at ultra-high magnetic field strength. NeuroImage. 2013;65:299–314. doi: 10.1016/j.neuroimage.2012.09.055 [DOI] [PubMed] [Google Scholar]
- 29.Li J, Li Y, Gutierrez L, et al. Imaging the Centromedian Thalamic Nucleus Using Quantitative Susceptibility Mapping. Front Hum Neurosci. 2020;13:447. doi: 10.3389/fnhum.2019.00447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng X, Deistung A, Dwyer MG, et al. An improved FSL-FIRST pipeline for subcortical gray matter segmentation to study abnormal brain anatomy using quantitative susceptibility mapping (QSM). Magn Reson Imaging. 2017;39:110–122. doi: 10.1016/j.mri.2017.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanspach J, Dwyer MG, Bergsland NP, et al. Methods for the computation of templates from quantitative magnetic susceptibility maps (QSM): Toward improved atlas- and voxel-based analyses (VBA). J Magn Reson Imaging. 2017;46(5):1474–1484. doi: 10.1002/jmri.25671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chandran AS, Bynevelt M, Lind CRP. Magnetic resonance imaging of the subthalamic nucleus for deep brain stimulation. J Neurosurg. August 2015:1–10. doi: 10.3171/2015.1.JNS142066 [DOI] [PubMed] [Google Scholar]
- 33.de Hollander G, Keuken MC, Bazin PL, et al. A gradual increase of iron toward the medial-inferior tip of the subthalamic nucleus. Hum Brain Mapp. 2014;00. doi: 10.1002/hbm.22485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu T, Eskreis-Winkler S, Schweitzer AD, et al. Improved Subthalamic Nucleus Depiction with Quantitative Susceptibility Mapping. Radiology. 2013;(7):1–10. doi: 10.1148/radiol.13121991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rasouli J, Ramdhani R, Panov FE, et al. Utilization of Quantitative Susceptibility Mapping for Direct Targeting of the Subthalamic Nucleus During Deep Brain Stimulation Surgery. Oper Neurosurg. 2018;14(4):412–419. doi: 10.1093/ons/opx131 [DOI] [PubMed] [Google Scholar]
- 36.Wei H, Zhang C, Wang T, et al. Precise targeting of the globus pallidus internus with quantitative susceptibility mapping for deep brain stimulation surgery. J Neurosurg. October 2019:1–7. doi: 10.3171/2019.7.JNS191254 [DOI] [PubMed] [Google Scholar]
- 37.Chen J, Gong NJ, Chaim KT, Otaduy MCG, Liu C. Decompose quantitative susceptibility mapping (QSM) to sub-voxel diamagnetic and paramagnetic components based on gradient-echo MRI data. NeuroImage. 2021;242:118477. doi: 10.1016/j.neuroimage.2021.118477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dimov AV, Nguyen TD, Gillen KM, et al. Susceptibility source separation from gradient echo data using magnitude decay modeling. J Neuroimaging. June 2022:jon.13014. doi: 10.1111/jon.13014 [DOI] [PubMed] [Google Scholar]
- 39.Dimov AV, Gillen KM, Nguyen TD, et al. Magnetic Susceptibility Source Separation Solely from Gradient Echo Data: Histological Validation. Tomography. 2022;8(3):1544–1551. doi: 10.3390/tomography8030127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elkady AM, Cobzas D, Sun H, Blevins G, Wilman AH. Discriminative analysis of regional evolution of iron and myelin/calcium in deep gray matter of multiple sclerosis and healthy subjects. J Magn Reson Imaging. 2018;48(3):652–668. doi: 10.1002/jmri.26004 [DOI] [PubMed] [Google Scholar]
- 41.Emmerich J, Bachert P, Ladd ME, Straub S. On the separation of susceptibility sources in quantitative susceptibility mapping: Theory and phantom validation with an in vivo application to multiple sclerosis lesions of different age. J Magn Reson. 2021;330:107033. doi: 10.1016/j.jmr.2021.107033 [DOI] [PubMed] [Google Scholar]
- 42.Li J, Lin H, Liu T, et al. Quantitative susceptibility mapping (QSM) minimizes interference from cellular pathology in R2* estimation of liver iron concentration: QSM Minimizes R2* Cellular Interference. J Magn Reson Imaging. 2018;48(4):1069–1079. doi: 10.1002/jmri.26019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schweser F, Deistung A, Lehr BW, Sommer K, Reichenbach JR. SEMI-TWInS: Simultaneous Extraction of Myelin and Iron using a T2*-Weighted Imaging Sequence. In: Proc Intl Soc Mag Reson Med 19 (2011). Montreal, CA; 2011:120. [Google Scholar]
- 44.Shin HG, Lee J, Yun YH, et al. χ-separation: Magnetic susceptibility source separation toward iron and myelin mapping in the brain. NeuroImage. 2021;240:118371. doi: 10.1016/j.neuroimage.2021.118371 [DOI] [PubMed] [Google Scholar]
- 45.Stüber C, Morawski M, Schäfer A, et al. Myelin and iron concentration in the human brain: A quantitative study of MRI contrast. NeuroImage. 2014;93:95–106. doi: 10.1016/j.neuroimage.2014.02.026 [DOI] [PubMed] [Google Scholar]
- 46.Yablonskiy DA, Wen J, Kothapalli SVVN, Sukstanskii AL. In vivo evaluation of heme and non-heme iron content and neuronal density in human basal ganglia. NeuroImage. 2021;235:118012. doi: 10.1016/j.neuroimage.2021.118012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aimo A, Huang L, Tyler A, et al. Quantitative susceptibility mapping (QSM) of the cardiovascular system: challenges and perspectives. J Cardiovasc Magn Reson. 2022;24(1):48. doi: 10.1186/s12968-022-00883-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bilgic B, Chatnuntawech I, Polak D. Chapter 16 - Quantitative Susceptibility-Mapping Reconstruction. In: Akçakaya M, Doneva M, Prieto C, eds. Advances in Magnetic Resonance Technology and Applications. Vol 7. Magnetic Resonance Image Reconstruction. Academic Press; 2022:441–467. doi: 10.1016/B978-0-12-822726-8.00027-0 [DOI] [Google Scholar]
- 49.Bilgic B, Chatnuntawech I, Langkammer C, Setsompop K. Sparse methods for Quantitative Susceptibility Mapping. SPIE. 2015;9597:959711. doi: 10.1117/12.2188535 [DOI] [Google Scholar]
- 50.Chung J, Ruthotto L. Computational methods for image reconstruction. NMR Biomed. 2017;30(4). doi: 10.1002/nbm.3545 [DOI] [PubMed] [Google Scholar]
- 51.Gozt A, Hellewell S, Ward PGD, Bynevelt M, Fitzgerald M. Emerging Applications for Quantitative Susceptibility Mapping in the Detection of Traumatic Brain Injury Pathology. Neuroscience. 2021;467:218–236. doi: 10.1016/j.neuroscience.2021.05.030 [DOI] [PubMed] [Google Scholar]
- 52.Jung W, Bollmann S, Lee J. Overview of quantitative susceptibility mapping using deep learning: Current status, challenges and opportunities. NMR Biomed. March 2020:e4292. doi: 10.1002/nbm.4292 [DOI] [PubMed] [Google Scholar]
- 53.Kee Y, Liu Z, Zhou L, et al. Quantitative Susceptibility Mapping (QSM) Algorithms: Mathematical Rationale and Computational Implementations. IEEE Trans Biomed Eng. 2017;14(8):1–1. doi: 10.1109/TBME.2017.2749298 [DOI] [PubMed] [Google Scholar]
- 54.Robinson SD, Bredies K, Khabipova D, Dymerska B, Marques JP, Schweser F. An illustrated comparison of processing methods for MR phase imaging and QSM: combining array coil signals and phase unwrapping. NMR Biomed. 2017;30(4):e3601. doi: 10.1002/nbm.3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schweser F, Robinson SD, de Rochefort L, Li W, Bredies K. An illustrated comparison of processing methods for phase MRI and QSM: removal of background field contributions from sources outside the region of interest. NMR Biomed. 2017;30(4):e3604. doi: 10.1002/nbm.3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schweser F, Deistung A, Reichenbach JR. Foundations of MRI phase imaging and processing for Quantitative Susceptibility Mapping (QSM). Z Med Phys. 2016;26(1):6–34. doi: 10.1016/j.zemedi.2015.10.002 [DOI] [PubMed] [Google Scholar]
- 57.Shmueli K. Chapter 31 - Quantitative Susceptibility Mapping. In: Seiberlich N, Gulani V, Calamante F, et al. , eds. Advances in Magnetic Resonance Technology and Applications. Vol 1. Quantitative Magnetic Resonance Imaging. Academic Press; 2020:819–838. doi: 10.1016/B978-0-12-817057-1.00033-0 [DOI] [Google Scholar]
- 58.Uchida Y, Kan H, Sakurai K, Oishi K, Matsukawa N. Quantitative susceptibility mapping as an imaging biomarker for Alzheimer’s disease: The expectations and limitations. Front Neurosci. 2022;16:938092. doi: 10.3389/fnins.2022.938092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Y, Liu T. Quantitative susceptibility mapping (QSM): Decoding MRI data for a tissue magnetic biomarker. Magn Reson Med. 2015;73(1):82–101. doi: 10.1002/mrm.25358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Langkammer C, Schweser F, Shmueli K, et al. Quantitative susceptibility mapping: Report from the 2016 reconstruction challenge. Magn Reson Med. 2018;79(3):1661–1673. doi: 10.1002/mrm.26830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marques JP, Meineke J, Milovic C, et al. QSM reconstruction challenge 2.0: A realistic in silico head phantom for MRI data simulation and evaluation of susceptibility mapping procedures. Magn Reson Med. 2021;86(1):526–542. doi: 10.1002/mrm.28716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Milovic C, Tejos C, Acosta-Cabronero J, et al. The 2016 QSM Challenge: Lessons learned and considerations for a future challenge design. Magn Reson Med. 2020;84(3):1624–1637. doi: 10.1002/mrm.28185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.QSM Challenge 2.0 Organization Committee, Bilgic B, Langkammer C, et al. QSM reconstruction challenge 2.0: Design and report of results. Magn Reson Med. 2021;86(3):1241–1255. doi: 10.1002/mrm.28754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.AlTokhis AI, AlOtaibi AM, Felmban GA, Constantinescu CS, Evangelou N. Iron Rims as an Imaging Biomarker in MS: A Systematic Mapping Review. Diagnostics. 2020;10(11):968. doi: 10.3390/diagnostics10110968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Calvi A, Haider L, Prados F, Tur C, Chard D, Barkhof F. In vivo imaging of chronic active lesions in multiple sclerosis. Mult Scler J. 2022;28(5):683–690. doi: 10.1177/1352458520958589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gillen KM, Mubarak M, Park C, et al. QSM is an imaging biomarker for chronic glial activation in multiple sclerosis lesions. Ann Clin Transl Neurol. March 2021:acn3.51338. doi: 10.1002/acn3.51338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang W, Sweeney EM, Kaunzner UW, Wang Y, Gauthier SA, Nguyen TD. Quantitative susceptibility mapping versus phase imaging to identify multiple sclerosis iron rim lesions with demyelination. J Neuroimaging. 2022;n/a(n/a). doi: 10.1111/jon.12987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jang J, Nam Y, Choi Y, et al. Paramagnetic Rims in Multiple Sclerosis and Neuromyelitis Optica Spectrum Disorder: A Quantitative Susceptibility Mapping Study with 3-T MRI. J Clin Neurol. 2020;16(4):562. doi: 10.3988/jcn.2020.16.4.562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marcille M, Hurtado Rúa S, Tyshkov C, et al. Disease correlates of rim lesions on quantitative susceptibility mapping in multiple sclerosis. Sci Rep. 2022;12(1):4411. doi: 10.1038/s41598-022-08477-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ng Kee Kwong KC, Mollison D, Meijboom R, et al. The prevalence of paramagnetic rim lesions in multiple sclerosis: A systematic review and meta-analysis. Jiang Q, ed. PLOS ONE. 2021;16(9):e0256845. doi: 10.1371/journal.pone.0256845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rahmanzadeh R, Lu PJ, Barakovic M, et al. Myelin and axon pathology in multiple sclerosis assessed by myelin water and multi-shell diffusion imaging. Brain. 2021;144(6):1684–1696. doi: 10.1093/brain/awab088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tolaymat B, W Z, H C, S C, X L, Dm H. Sex-specific differences in rim appearance of multiple sclerosis lesions on quantitative susceptibility mapping. Mult Scler Relat Disord. 2020;45:102317. doi: 10.1016/j.msard.2020.102317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chan KS, Marques JP. SEPIA—Susceptibility mapping pipeline tool for phase images. NeuroImage. 2021;227:117611. doi: 10.1016/j.neuroimage.2020.117611 [DOI] [PubMed] [Google Scholar]
- 74.Duyn JH, van Gelderen P, Li TQ, de Zwart JA, Koretsky AP, Fukunaga M. High-field MRI of brain cortical substructure based on signal phase. Proc Natl Acad Sci U A. 2007;104(28):11796–11801. doi: 10.1073/pnas.0610821104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Haacke EM, Cheng YCN, House MJ, et al. Imaging iron stores in the brain using magnetic resonance imaging. Magn Reson Imaging. 2005;23(1):1–25. doi: 10.1016/j.mri.2004.10.001 [DOI] [PubMed] [Google Scholar]
- 76.Schenck JF. The role of magnetic susceptibility in magnetic resonance imaging: MRI magnetic compatibility of the first and second kinds. Med Phys. 1996;23(6):815–850. [DOI] [PubMed] [Google Scholar]
- 77.Koch KM, Papademetris X, Rothman DL, de Graaf R a. Rapid calculations of susceptibility-induced magnetostatic field perturbations for in vivo magnetic resonance. Phys Med Biol. 2006;51(24):6381–6402. doi: 10.1088/0031-9155/51/24/007 [DOI] [PubMed] [Google Scholar]
- 78.Marques JP, Bowtell RW. Application of a Fourier-based method for rapid calculation of field inhomogeneity due to spatial variation of magnetic susceptibility. Concepts Magn Reson Part B Magn Reson Eng. 2005;25B(1):65–78. doi: 10.1002/cmr.b.20034 [DOI] [Google Scholar]
- 79.Salomir R, de Senneville BD, Moonen CTW. A fast calculation method for magnetic field inhomogeneity due to an arbitrary distribution of bulk susceptibility. Concepts Magn Reson Part B Magn Reson Eng. 2003;19B(1):26–34. doi: 10.1002/cmr.b.10083 [DOI] [Google Scholar]
- 80.Haacke EM, Reichenbach JR, eds. Susceptibility Weighted Imaging in MRI. Wiley-Blackwell; 2011. [Google Scholar]
- 81.Yablonskiy DA, Haacke EM. Theory of NMR Signal Behavior in Magnetically Inhomogeneous Tissues: The Static Dephasing Regime. Magn Reson Med. 1994;32:749–763. [DOI] [PubMed] [Google Scholar]
- 82.McRobbie DW, Moore EA, Graves MJ. MRI from Picture to Proton. Third edition. Cambridge; New York: University Printing House, Cambridge University Press; 2017. [Google Scholar]
- 83.de Rochefort L, Liu T, Kressler B, et al. Quantitative susceptibility map reconstruction from MR phase data using Bayesian regularization: validation and application to brain imaging. Magn Reson Med. 2010;63(1):194–206. doi: 10.1002/mrm.22187 [DOI] [PubMed] [Google Scholar]
- 84.de Rochefort L, Brown R, Prince MR, Wang Y. Quantitative MR susceptibility mapping using piece-wise constant regularized inversion of the magnetic field. Magn Reson Med. 2008;60(4):1003–1009. doi: 10.1002/mrm.21710 [DOI] [PubMed] [Google Scholar]
- 85.Deistung A, Schweser F, Reichenbach JR. Overview of quantitative susceptibility mapping. NMR Biomed. 2017;30(4):e3569. doi: 10.1002/nbm.3569 [DOI] [PubMed] [Google Scholar]
- 86.Duyn JH. MR susceptibility imaging. J Magn Reson. 2013;229:198–207. doi: 10.1016/j.jmr.2012.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Haacke EM, Liu S, Buch S, Zheng W, Wu D, Ye Y. Quantitative susceptibility mapping: current status and future directions. Magn Reson Imaging. 2015;33(1):1–25. doi: 10.1016/j.mri.2014.09.004 [DOI] [PubMed] [Google Scholar]
- 88.Lin F, Prince MR, Spincemaille P, Wang Y. Patents on Quantitative Susceptibility Mapping (QSM) of Tissue Magnetism. Recent Pat Biotechnol. 2019;13(2):90–113. doi: 10.2174/1872208313666181217112745 [DOI] [PubMed] [Google Scholar]
- 89.Liu C, Li W, Tong KA, Yeom KW, Kuzminski S. Susceptibility-weighted imaging and quantitative susceptibility mapping in the brain. J Magn Reson Imaging. 2015;42(1):23–41. doi: 10.1002/jmri.24768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Reichenbach JR. The future of susceptibility contrast for assessment of anatomy and function. NeuroImage. 2012;62(2):1311–1315. doi: 10.1016/j.neuroimage.2012.01.004 [DOI] [PubMed] [Google Scholar]
- 91.Reichenbach JR, Schweser F, Serres B, Deistung A. Quantitative Susceptibility Mapping: Concepts and Applications. Clin Neuroradiol. 2015;25(S2):225–230. doi: 10.1007/s00062-015-0432-9 [DOI] [PubMed] [Google Scholar]
- 92.Ruetten PPR, Gillard JH, Graves MJ. Introduction to Quantitative Susceptibility Mapping and Susceptibility Weighted Imaging. Br J Radiol. 2019;92(1101):20181016. doi: 10.1259/bjr.20181016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shmueli K. Quantitative Susceptibility Mapping. In: Advances in Magnetic Resonance Technology and Applications. Vol 1. Elsevier; 2020:819–838. doi: 10.1016/B978-0-12-817057-1.00033-0 [DOI] [Google Scholar]
- 94.Wu B, Li W, Avram AV, Gho SM, Liu C. Fast and tissue-optimized mapping of magnetic susceptibility and T2* with multi-echo and multi-shot spirals. NeuroImage. 2012;59(1):297–305. doi: 10.1016/j.neuroimage.2011.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Peters AM, Brookes MJ, Hoogenraad FG, et al. T2* measurements in human brain at 1.5, 3 and 7 T. Magn Reson Med. 2007;25(6):748–753. doi: 10.1016/j.mri.2007.02.014 [DOI] [PubMed] [Google Scholar]
- 96.Rooney WD, Johnson GA, Li X, et al. Magnetic field and tissue dependencies of human brain longitudinal 1H2O relaxation in vivo. Magn Reson Med. 2007;57(2):308–318. [DOI] [PubMed] [Google Scholar]
- 97.Biondetti E, Karsa A, Thomas DL, Shmueli K. Investigating the accuracy and precision of TE-dependent versus multi-echo QSM using Laplacian-based methods at 3 T. Magn Reson Med. 2020;84(6):3040–3053. doi: 10.1002/mrm.28331 [DOI] [PubMed] [Google Scholar]
- 98.Karsa A, Punwani S, Shmueli K. The effect of low resolution and coverage on the accuracy of susceptibility mapping. Magn Reson Med. 2019;81(3):1833–1848. doi: 10.1002/mrm.27542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wei H, Zhang Y, Gibbs E, Chen NK, Wang N, Liu C. Joint 2D and 3D phase processing for quantitative susceptibility mapping: Application to 2D echo-planar imaging. NMR Biomed. 2016;(October 2015). doi: 10.1002/nbm.3501 [DOI] [PubMed] [Google Scholar]
- 100.Li J, Chang S, Liu T, et al. Phase-corrected bipolar gradients in multi-echo gradient-echo sequences for quantitative susceptibility mapping. Magn Reson Mater Phy. November 2014. doi: 10.1007/s10334-014-0470-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Berg RC, Preibisch C, Thomas DL, Shmueli K, Biondetti E. Investigating the effect of flow compensation and quantitative susceptibility mapping method on the accuracy of venous susceptibility measurement. NeuroImage. 2021;240:118399. doi: 10.1016/j.neuroimage.2021.118399 [DOI] [PubMed] [Google Scholar]
- 102.Lee J, Shmueli K, Fukunaga M, et al. Sensitivity of MRI resonance frequency to the orientation of brain tissue microstructure. Proc Natl Acad Sci U A. 2010;107(11):5130–5135. doi: 10.1073/pnas.0910222107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu C. Susceptibility tensor imaging. Magn Reson Med. 2010;63(6):1471–1477. doi: 10.1002/mrm.22482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wharton SJ, Bowtell R. Effects of white matter microstructure on phase and susceptibility maps. Magn Reson Med. 2015;73(3):1258–1269. doi: 10.1002/mrm.25189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mennes M, Jenkinson M, Valabregue R, Buitelaar JK, Beckmann C, Smith S. Optimizing full-brain coverage in human brain MRI through population distributions of brain size. NeuroImage. 2014;98:513–520. doi: 10.1016/j.neuroimage.2014.04.030 [DOI] [PubMed] [Google Scholar]
- 106.Kiersnowski OC, Karsa A, Wastling SJ, Thornton JS, Shmueli K. Investigating the Effect of Oblique Image Acquisition on the Accuracy of Quantitative Susceptibility Mapping and a Robust Tilt Correction Method. Magn Reson Med. 2022;epub. doi: 10.1002/mrm.29550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Breuer FA, Blaimer M, Mueller MF, et al. Controlled aliasing in volumetric parallel imaging (2D CAIPIRINHA). Magn Reson Med. 2006;55(3):549–556. doi: 10.1002/mrm.20787 [DOI] [PubMed] [Google Scholar]
- 108.Wang N, Cofer G, Anderson RJ, Qi Y, Liu C, Johnson GA. Accelerating quantitative susceptibility imaging acquisition using compressed sensing. Phys Med Biol. 2018;63(24):245002. doi: 10.1088/1361-6560/aaf15d [DOI] [PubMed] [Google Scholar]
- 109.Lee D, Yoo J, Tak S, Ye JC. Deep Residual Learning for Accelerated MRI Using Magnitude and Phase Networks. IEEE Trans Biomed Eng. 2018;65(9):1985–1995. doi: 10.1109/TBME.2018.2821699 [DOI] [PubMed] [Google Scholar]
- 110.Marques JP, Simonis FFJ, Webb AG. Low-field MRI: An MR physics perspective. J Magn Reson Imaging. 2019;49(6):1528–1542. doi: 10.1002/jmri.26637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cronin MJ, Wharton SJ, Al-Radaideh AM, et al. A comparison of phase imaging and quantitative susceptibility mapping in the imaging of multiple sclerosis lesions at ultrahigh field. Magn Reson Mater Phys Biol Med. 2016:1–15. doi: 10.1007/s10334-016-0560-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Langkammer C, Bredies K, Poser BA, et al. Fast quantitative susceptibility mapping using 3D EPI and total generalized variation. NeuroImage. 2015;111(February 2016):622–630. doi: 10.1016/j.neuroimage.2015.02.041 [DOI] [PubMed] [Google Scholar]
- 113.Stirnberg R, Stöcker T. Segmented K-space blipped-controlled aliasing in parallel imaging for high spatiotemporal resolution EPI. Magn Reson Med. 2021;85(3):1540–1551. doi: 10.1002/mrm.28486 [DOI] [PubMed] [Google Scholar]
- 114.Bilgic B, Ye H, Wald LL, Setsompop K. Simultaneous Time Interleaved MultiSlice (STIMS) for Rapid Susceptibility Weighted acquisition. NeuroImage. 2017;(April):1–10. doi: 10.1016/j.neuroimage.2017.04.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Griswold MA, Jakob PM, Heidemann RM, et al. Generalized autocalibrating partially parallel acquisitions (GRAPPA). Magn Reson Med. 2002;47(6):1202–1210. doi: 10.1002/mrm.10171 [DOI] [PubMed] [Google Scholar]
- 116.Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. SENSE: sensitivity encoding for fast MRI. Magn Reson Med. 1999;42(5):952–962. [PubMed] [Google Scholar]
- 117.Sodickson DK, Manning WJ. Simultaneous acquisition of spatial harmonics (SMASH): Fast imaging with radiofrequency coil arrays. Magn Reson Med. 1997;38(4):591–603. doi: 10.1002/mrm.1910380414 [DOI] [PubMed] [Google Scholar]
- 118.Metere R, Kober T, Möller HE, Schäfer A. Simultaneous Quantitative MRI Mapping of T1, T2* and Magnetic Susceptibility with Multi-Echo MP2RAGE. Grolmusz V, ed. PLOS ONE. 2017;12(1):e0169265. doi: 10.1371/journal.pone.0169265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Roemer PB, Edelstein WA, Hayes CE, Souza SP, Mueller OM. The NMR phased array. Magn Reson Med. 1990;16(2):192–225. [DOI] [PubMed] [Google Scholar]
- 120.Eckstein K, Dymerska B, Bachrata B, et al. Computationally Efficient Combination of Multi-channel Phase Data From Multi-echo Acquisitions (ASPIRE): Combination of Multi-Channel Phase Data from Multi-Echo Acquisitions (ASPIRE). Magn Reson Med. 2018;79(6):2996–3006. doi: 10.1002/mrm.26963 [DOI] [PubMed] [Google Scholar]
- 121.Li X, Morgan PS, Ashburner J, Smith J, Rorden C. The first step for neuroimaging data analysis: DICOM to NIfTI conversion. J Neurosci Methods. 2016;264:47–56. doi: 10.1016/j.jneumeth.2016.03.001 [DOI] [PubMed] [Google Scholar]
- 122.Reichenbach JR, Barth M, Haacke EM, Klarhöfer M, Kaiser WA, Moser E. High-resolution MR venography at 3.0 Tesla. J Comp Assist Tomogr. 2000;24(6):949–957. [DOI] [PubMed] [Google Scholar]
- 123.Reichenbach JR, Venkatesan R, Schillinger D, Kido DK, Haacke EM. Small vessels in the human brain: MR-venography with deoxyhemoglobin as an intrinsic contrast agent. Radiology. 1997;204(1):272–277. [DOI] [PubMed] [Google Scholar]
- 124.Reichenbach JR, Haacke EM. High resolution BOLD venographic imaging: a window into brain function. NMR Biomed. 2001;14(7–8):453–467. doi: 10.1002/nbm.722 [DOI] [PubMed] [Google Scholar]
- 125.Hagberg GE, Welch EB, Greiser A. The sign convention for phase values on different vendor systems: definition and implications for susceptibility-weighted imaging. Magn Reson Imaging. 2010;28(2):297–300. doi: 10.1016/j.mri.2009.06.001 [DOI] [PubMed] [Google Scholar]
- 126.Jell V, Kannengiesser SAR. Adaptive Coil Combination Using a Body Coil Scan as Phase Reference. In: Proc GC Intl Soc Mag Reson Med 17 (2014). 2014:p4406. [Google Scholar]
- 127.Kim DH, Choi N, Gho SM, Shin J, Liu C. Simultaneous imaging of in vivo conductivity and susceptibility. Magn Reson Med. 2014;71(3):1144–1150. doi: 10.1002/mrm.24759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Feng W, Neelavalli J, Haacke EM. Catalytic multiecho phase unwrapping scheme (CAMPUS) in multiecho gradient echo imaging: removing phase wraps on a voxel-by-voxel basis. Magn Reson Med. 2013;70(1):117–126. doi: 10.1002/mrm.24457 [DOI] [PubMed] [Google Scholar]
- 129.Robinson SD, Schödl H, Trattnig S. A method for unwrapping highly wrapped multi-echo phase images at very high field: UMPIRE. Magn Reson Med. 2014;72(1):80–92. doi: 10.1002/mrm.24897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jenkinson M. A Fast, Automated, N-Dimensional Phase-Unwrapping Algorithm. Magn Reson Med. 2003;49:193–197. [DOI] [PubMed] [Google Scholar]
- 131.Karsa A, Shmueli K. SEGUE: a Speedy rEgion-Growing algorithm for Unwrapping Estimated phase. IEEE Trans Med Imaging. 2018:1–1. doi: 10.1109/TMI.2018.2884093 [DOI] [PubMed] [Google Scholar]
- 132.Ye Y, Zhou F, Zong J, et al. Seed prioritized unwrapping (SPUN) for MR phase imaging: SPUN Method. J Magn Reson Imaging. December 2018. doi: 10.1002/jmri.26606 [DOI] [PubMed] [Google Scholar]
- 133.Abdul-Rahman HS, Gdeisat MA, Burton DR, Lalor MJ, Lilley F, Moore CJ. Fast and robust three-dimensional best path phase unwrapping algorithm. Appl Opt. 2007;46(26):6623–6635. [DOI] [PubMed] [Google Scholar]
- 134.Dymerska B, Eckstein K, Bachrata B, et al. Phase unwrapping with a rapid opensource minimum spanning tree algorithm (ROMEO). Magn Reson Med. 2021;85(4):2294–2308. doi: 10.1002/mrm.28563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Schofield MA, Zhu Y. Fast phase unwrapping algorithm for interferometric applications. Opt Lett. 2003;28(14):1194–1196. [DOI] [PubMed] [Google Scholar]
- 136.Milovic C, Bilgic B, Zhao B, Langkammer C, Tejos C, Acosta-Cabronero J. Weak-harmonic regularization for quantitative susceptibility mapping. Magn Reson Med. 2019;81(2):1399–1411. doi: 10.1002/mrm.27483 [DOI] [PubMed] [Google Scholar]
- 137.Hsieh MC, Tsai CY, Liao MC, Yang JL, Su CH, Chen JH. Quantitative susceptibility mapping-based microscopy of magnetic resonance venography (QSM-mMRV) for in vivo morphologically and functionally assessing cerebromicrovasculature in rat stroke model. PLoS ONE. 2016;11(3):1–22. doi: 10.1371/journal.pone.0149602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sun H, Kate M, Gioia LC, Emery DJ, Butcher K, Wilman AH. Quantitative susceptibility mapping using a superposed dipole inversion method: Application to intracranial hemorrhage. Magn Reson Med. 2015;791:781–791. doi: 10.1002/mrm.25919 [DOI] [PubMed] [Google Scholar]
- 139.Liu T, Wisnieff C, Lou M, Chen W, Spincemaille P, Wang Y. Nonlinear formulation of the magnetic field to source relationship for robust quantitative susceptibility mapping. Magn Reson Med. 2013;69(2):467–476. doi: 10.1002/mrm.24272 [DOI] [PubMed] [Google Scholar]
- 140.Chen L, Cai S, van Zijl PCM, Li X. Single-step calculation of susceptibility through multiple orientation sampling. NMR Biomed. 2021;34(7):e4517. doi: 10.1002/nbm.4517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kressler B, de Rochefort L, Liu T, Spincemaille P, Jiang Q, Wang Y. Nonlinear regularization for per voxel estimation of magnetic susceptibility distributions from MRI field maps. IEEE Trans Med Imaging. 2010;29(2):273–281. doi: 10.1109/TMI.2009.2023787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Cronin MJ, Wang N, Decker KS, Wei H, Zhu WZ, Liu C. Exploring the origins of echo-time-dependent quantitative susceptibility mapping (QSM) measurements in healthy tissue and cerebral microbleeds. NeuroImage. 2017;149:98–113. doi: 10.1016/j.neuroimage.2017.01.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lancione M, Donatelli G, Cecchi P, Cosottini M, Tosetti M, Costagli M. Echo-time dependency of quantitative susceptibility mapping reproducibility at different magnetic field strengths. NeuroImage. 2019;197:557–564. doi: 10.1016/j.neuroimage.2019.05.004 [DOI] [PubMed] [Google Scholar]
- 144.Sati P, van Gelderen P, Silva AC, et al. Micro-compartment specific T2* relaxation in the brain. NeuroImage. 2013;77:268–278. doi: 10.1016/j.neuroimage.2013.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sood S, Urriola J, Reutens D, et al. Echo time-dependent quantitative susceptibility mapping contains information on tissue properties: Temporal Quantitative Susceptibility Mapping. Magn Reson Med. 2017;77(5):1946–1958. doi: 10.1002/mrm.26281 [DOI] [PubMed] [Google Scholar]
- 146.Wharton S, Bowtell R. Fiber orientation-dependent white matter contrast in gradient echo MRI. Proc Natl Acad Sci. 2012;109(45):18559–18564. doi: 10.1073/pnas.1211075109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wharton SJ, Bowtell R. Gradient echo based fiber orientation mapping using R2* and frequency difference measurements. NeuroImage. 2013;83:1011–1023. doi: 10.1016/j.neuroimage.2013.07.054 [DOI] [PubMed] [Google Scholar]
- 148.Yablonskiy DA, Sukstanskii AL. Effects of biological tissue structural anisotropy and anisotropy of magnetic susceptibility on the gradient echo MRI signal phase: theoretical background. NMR Biomed. 2017;30(4). doi: 10.1002/nbm.3655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Stewart AW, Robinson SD, O’Brien K, et al. QSMxT: Robust masking and artifact reduction for quantitative susceptibility mapping. Magn Reson Med. 2022;87(3):1289–1300. doi: 10.1002/mrm.29048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gudbjartsson H, Patz S. The rician distribution of noisy mri data. Magn Reson Med. 1995;34(6):910–914. doi: 10.1002/mrm.1910340618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Arrieta C, Sing-Long CA, Mura J, et al. Level set segmentation with shape prior knowledge using intrinsic rotation, translation and scaling alignment. Biomed Signal Process Control. 2021;63:102241. doi: 10.1016/j.bspc.2020.102241 [DOI] [Google Scholar]
- 152.Caselles V, Catté F, Coll T, Dibos F. A geometric model for active contours in image processing. Numer Math. 1993;66(1):1–31. doi: 10.1007/BF01385685 [DOI] [Google Scholar]
- 153.Chan TF, Vese LA. Active contours without edges. IEEE Trans Image Process. 2001;10(2):266–277. doi: 10.1109/83.902291 [DOI] [PubMed] [Google Scholar]
- 154.Cootes TF, Edwards GJ, Taylor CJ. Active appearance models. IEEE Trans Pattern Anal Mach Intell. 2001;23(6):681–685. doi: 10.1109/34.927467 [DOI] [Google Scholar]
- 155.Niessen WJ, Romeny BMTH, Viergever MA. Geodesic deformable models for medical image analysis. IEEE Trans Med Imaging. 1998;17(4):634–641. doi: 10.1109/42.730407 [DOI] [PubMed] [Google Scholar]
- 156.Suri JS, Liu K, Singh S, Laxminarayan SN, Zeng X, Reden L. Shape recovery algorithms using level sets in 2-D/3-D medical imagery: a state-of-the-art review. IEEE Trans Inf Technol Biomed. 2002;6(1):8–28. doi: 10.1109/4233.992158 [DOI] [PubMed] [Google Scholar]
- 157.Tejos C, Irarrazaval P, Cárdenas-Blanco A. Simplex Mesh Diffusion Snakes: Integrating 2D and 3D Deformable Models and Statistical Shape Knowledge in a Variational Framework. Int J Comput Vis. 2009;85(1):19–34. doi: 10.1007/s11263-009-0241-1 [DOI] [Google Scholar]
- 158.Smith SM. Fast robust automated brain extraction. Hum Brain Mapp. 2002;17(3):143–155. doi: 10.1002/hbm.10062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Fennema-Notestine C, Ozyurt IB, Clark CP, et al. Quantitative evaluation of automated skull-stripping methods applied to contemporary and legacy images: Effects of diagnosis, bias correction, and slice location. Hum Brain Mapp. 2006;27(2):99–113. doi: 10.1002/hbm.20161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Speier W, Iglesias JE, El-Kara L, Tu Z, Arnold C. Robust Skull Stripping of Clinical Glioblastoma Multiforme Data. In: Fichtinger G, Martel A, Peters T, eds. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2011. Lecture Notes in Computer Science. Berlin, Heidelberg: Springer; 2011:659–666. doi: 10.1007/978-3-642-23626-6_81 [DOI] [PubMed] [Google Scholar]
- 161.Karsa A, Punwani S, Shmueli K. An optimized and highly repeatable MRI acquisition and processing pipeline for quantitative susceptibility mapping in the head-and-neck region. Magn Reson Med. 2020;84(6):3206–3222. doi: 10.1002/mrm.28377 [DOI] [PubMed] [Google Scholar]
- 162.Lutkenhoff ES, Rosenberg M, Chiang J, et al. Optimized Brain Extraction for Pathological Brains (optiBET). PLOS ONE. 2014;9(12):e115551. doi: 10.1371/journal.pone.0115551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Allioui H, Sadgal M, Elfazziki A. Deep MRI Segmentation: A Convolutional Method Applied to Alzheimer Disease Detection. Int J Adv Comput Sci Appl. 2019;10(11). doi: 10.14569/IJACSA.2019.0101151 [DOI] [Google Scholar]
- 164.Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In: Navab N, Hornegger J, Wells WM, Frangi AF, eds. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. Lecture Notes in Computer Science. Cham: Springer International Publishing; 2015:234–241. doi: 10.1007/978-3-319-24574-4_28 [DOI] [Google Scholar]
- 165.Shrivastava S, Singh N, Mishra U, Chandra A, Verma S. Comparative Study of Deep Learning Models for Segmentation of Corpus Callosum. In: 2020 Fourth International Conference on Computing Methodologies and Communication (ICCMC). 2020:418–423. doi: 10.1109/ICCMC48092.2020.ICCMC-00079 [DOI] [Google Scholar]
- 166.Zhang Y, Wu J, Liu Y, et al. A deep learning framework for pancreas segmentation with multi-atlas registration and 3D level-set. Med Image Anal. 2021;68:101884. doi: 10.1016/j.media.2020.101884 [DOI] [PubMed] [Google Scholar]
- 167.Zhao M, Huang L, Zhang Q, Su X, Asundi A, Kemao Q. Quality-guided phase unwrapping technique: comparison of quality maps and guiding strategies. Appl Opt. 2011;50(33):6214–6224. doi: 10.1364/AO.50.006214 [DOI] [PubMed] [Google Scholar]
- 168.Lindemeyer J, Worthoff WA, Shymanskaya A, Shah NJ. Iterative Restoration of the Fringe Phase (REFRASE) for QSM. Front Neurosci. 2021;15. https://www.frontiersin.org/articles/10.3389/fnins.2021.537666. Accessed October 24, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Bollmann S, Kristensen MH, Larsen MS, et al. SHARQnet – Sophisticated harmonic artifact reduction in quantitative susceptibility mapping using a deep convolutional neural network. Z Für Med Phys. February 2019. doi: 10.1016/j.zemedi.2019.01.001 [DOI] [PubMed] [Google Scholar]
- 170.Liu Z, Kee Y, Zhou D, Wang Y, Spincemaille P. Preconditioned total field inversion (TFI) method for quantitative susceptibility mapping. Magn Reson Med. 2016;00(January). doi: 10.1002/mrm.26331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hagberg GE, Tuzzi E, Loureiro J, et al. Towards robust – and accurate – QSM in cortical and sub-cortical regions of the human brain at 9.4T: the influence of masking. In: Proc Intl Soc Mag Reson Med 27 (2019). 2019:p313. [Google Scholar]
- 172.Zhu X, Gao Y, Liu F, Crozier S, Sun H. BFRnet: A deep learning-based MR background field removal method for QSM of the brain containing significant pathological susceptibility sources. April 2022. doi: 10.48550/arXiv.2204.02760 [DOI] [PubMed] [Google Scholar]
- 173.Milovic C, Bilgic B, Zhao B, Acosta-Cabronero J, Tejos C. Fast nonlinear susceptibility inversion with variational regularization. Magn Reson Med. 2018;80(2):814–821. doi: 10.1002/mrm.27073 [DOI] [PubMed] [Google Scholar]
- 174.Milovic C, Lambert M, Langkammer C, Bredies K, Irarrazaval P, Tejos C. Streaking artifact suppression of quantitative susceptibility mapping reconstructions via L1-norm data fidelity optimization (L1-QSM). Magn Reson Med. 2022;87(1):457–473. doi: 10.1002/mrm.28957 [DOI] [PubMed] [Google Scholar]
- 175.Karsa A, Shmueli K. A New, Simple Two-Pass Masking Approach for Streaking Artifact Removal in Any QSM Pipeline. In: Proc Intl Soc Mag Reson Med 30 (2022). 2022:p2462. [Google Scholar]
- 176.Yaghmaie N, Syeda WT, Wu C, et al. QSMART: Quantitative susceptibility mapping artifact reduction technique. NeuroImage. 2021;231:117701. doi: 10.1016/j.neuroimage.2020.117701 [DOI] [PubMed] [Google Scholar]
- 177.Gao Y, Xiong Z, Fazlollahi A, et al. Instant tissue field and magnetic susceptibility mapping from MRI raw phase using Laplacian enhanced deep neural networks. NeuroImage. 2022;259:119410. doi: 10.1016/j.neuroimage.2022.119410 [DOI] [PubMed] [Google Scholar]
- 178.Hanspach J, Jolla A, Uder M, Hensel B, Bollmann S, Laun FB. Deep learning based quantitative susceptibility mapping (QSM) in the presence of fat by using synthetically generated multi-echo phase data. In: Proc Intl Soc Mag Reson Med 29 (2021). 2021:p3975. [DOI] [PubMed] [Google Scholar]
- 179.Wei H, Cao S, Zhang Y, et al. Learning-based single-step quantitative susceptibility mapping reconstruction without brain extraction. NeuroImage. 2019;202:116064. doi: 10.1016/j.neuroimage.2019.116064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Zhou D, Liu T, Spincemaille P, Wang Y. Background field removal by solving the Laplacian boundary value problem. NMR Biomed. 2014;27(3):312–319. doi: 10.1002/nbm.3064 [DOI] [PubMed] [Google Scholar]
- 181.Schweser F, Deistung A, Lehr BW, Reichenbach JR. Quantitative imaging of intrinsic magnetic tissue properties using MRI signal phase: An approach to in vivo brain iron metabolism? NeuroImage. 2011;54(4):2789–2807. doi: 10.1016/j.neuroimage.2010.10.070 [DOI] [PubMed] [Google Scholar]
- 182.Wu B, Li W, Guidon A, Liu C. Whole brain susceptibility mapping using compressed sensing. Magn Reson Med. 2011;24:1129–1136. doi: 10.1002/mrm.23000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Topfer R, Schweser F, Deistung A, Reichenbach JR, Wilman AH. SHARP edges: Recovering cortical phase contrast through harmonic extension. Magn Reson Med. 2015;73(2):851–856. doi: 10.1002/mrm.25148 [DOI] [PubMed] [Google Scholar]
- 184.Fang J, Bao L, Li X, van Zijl PCM, Chen Z. Background field removal for susceptibility mapping of human brain with large susceptibility variations: Susceptibility Mapping of Human Brain. Magn Reson Med. October 2018. doi: 10.1002/mrm.27492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wen Y, Zhou D, Liu T, Spincemaille P, Wang Y. An iterative spherical mean value method for background field removal in MRI. Magn Reson Med. 2014;72(4):1065–1071. doi: 10.1002/mrm.24998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Li W, Avram AV, Wu B, Xiao X, Liu C. Integrated Laplacian-based phase unwrapping and background phase removal for quantitative susceptibility mapping. NMR Biomed. 2014;27(2):219–227. doi: 10.1002/nbm.3056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Özbay PS, Deistung A, Feng X, Nanz D, Reichenbach JR, Schweser F. A comprehensive numerical analysis of background phase correction with V-SHARP. NMR Biomed. 2017;30(4):e3550. doi: 10.1002/nbm.3550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Liu T, Khalidov I, de Rochefort L, et al. A novel background field removal method for MRI using projection onto dipole fields (PDF). NMR Biomed. 2011;24:1129–1136. doi: 10.1002/nbm.1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Milovic C, Fuchs P, Arsenov O, et al. Exhaustive Comparison of QSM Background Field Removal and Masking using a Realistic Numerical Head Phantom. In: Proc Intl Soc Mag Reson Med 31 (2023). Toronto, ON, Canada. [Google Scholar]
- 190.Chatnuntawech I, Mcdaniel P, Cauley SF, et al. Single-step quantitative susceptibility mapping with variational penalties. 2016;(October 2015). doi: 10.1002/nbm.3570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Sun H, Ma Y, MacDonald ME, Pike GB. Whole head quantitative susceptibility mapping using a least-norm direct dipole inversion method. NeuroImage. 2018;179:166–175. doi: 10.1016/j.neuroimage.2018.06.036 [DOI] [PubMed] [Google Scholar]
- 192.Schweser F, Deistung A, Sommer K, Reichenbach JR. Toward online reconstruction of quantitative susceptibility maps: Superfast dipole inversion. Magn Reson Med. 2013;69(6):1581–1593. doi: 10.1002/mrm.24405 [DOI] [PubMed] [Google Scholar]
- 193.Liu Z, Spincemaille P, Yao Y, Zhang Y, Wang Y. MEDI+0: Morphology enabled dipole inversion with automatic uniform cerebrospinal fluid zero reference for quantitative susceptibility mapping. Magn Reson Med. 2018;79(5):2795–2803. doi: 10.1002/mrm.26946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Straub S, Schneider TM, Emmerich J, et al. Suitable reference tissues for quantitative susceptibility mapping of the brain. Magn Reson Med. 2016;00(9):3–6. doi: 10.1002/mrm.26369 [DOI] [PubMed] [Google Scholar]
- 195.Li LZ, Leigh JS. Quantifying arbitrary magnetic susceptibility distributions with MR. Magn Reson Med. 2004;51(5):1077–1082. doi: 10.1002/mrm.20054 [DOI] [PubMed] [Google Scholar]
- 196.Liu T, Spincemaille P, de Rochefort L, Kressler B, Wang Y. Calculation of susceptibility through multiple orientation sampling (COSMOS): a method for conditioning the inverse problem from measured magnetic field map to susceptibility source image in MRI. Magn Reson Med. 2009;61(1):196–204. doi: 10.1002/mrm.21828 [DOI] [PubMed] [Google Scholar]
- 197.Shmueli K, de Zwart JA, van Gelderen P, Li TQ, Dodd SJ, Duyn JH. Magnetic susceptibility mapping of brain tissue in vivo using MRI phase data. Magn Reson Med. 2009;62(6):1510–1522. doi: 10.1002/mrm.22135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Choi JK, Park HS, Wang S, Wang Y, Seo JK. Inverse Problem in Quantitative Susceptibility Mapping. SIAM J Imag Sci. 2014;7(3):1669–1689. doi: 10.1137/140957433 [DOI] [Google Scholar]
- 199.Bao L, Li X, Cai C, Chen Z, van Zijl PCM. Quantitative Susceptibility Mapping Using Structural Feature Based Collaborative Reconstruction (SFCR) in the Human Brain. IEEE Trans Med Imaging. 2016;35(9):2040–2050. doi: 10.1109/TMI.2016.2544958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Schweser F, Sommer K, Deistung A, Reichenbach JR. Quantitative susceptibility mapping for investigating subtle susceptibility variations in the human brain. NeuroImage. 2012;62(3):2083–2100. doi: 10.1016/j.neuroimage.2012.05.067 [DOI] [PubMed] [Google Scholar]
- 201.Liu J, Liu T, de Rochefort L, et al. Morphology enabled dipole inversion for quantitative susceptibility mapping using structural consistency between the magnitude image and the susceptibility map. NeuroImage. 2012;59(3):2560–2568. doi: 10.1016/j.neuroimage.2011.08.082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Milovic C, Prieto C, Bilgic B, et al. Comparison of parameter optimization methods for quantitative susceptibility mapping. Magn Reson Med. August 2020:mrm.28435. doi: 10.1002/mrm.28435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Bilgic B, Chatnuntawech I, Fan AP, et al. Fast image reconstruction with L2-regularization. J Magn Reson Imaging. 2014;40(1):181–191. doi: 10.1002/jmri.24365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Kressler B, de Rochefort L, Spincemaille P, Liu T, Wang Y. Estimation of Sparse Magnetic Susceptibility Distributions from MRI using Non-linear Regularization. In: Proc Intl Soc Mag Reson Med 16 (2008). Toronto; 2008:1514. [Google Scholar]
- 205.Li W, Wu B, Liu C. Quantitative susceptibility mapping of human brain reflects spatial variation in tissue composition. NeuroImage. 2011;55(4):1645–1656. doi: 10.1016/j.neuroimage.2010.11.088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Wang S, Liu T, Chen W, et al. Noise Effects in Various Quantitative Susceptibility Mapping Methods. IEEE Trans Biomed Eng. 2013;60(12):3441–3448. doi: 10.1109/TBME.2013.2266795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Wei H, Dibb R, Zhou Y, et al. Streaking artifact reduction for quantitative susceptibility mapping of sources with large dynamic range. NMR Biomed. 2015;28(10):1294–1303. doi: 10.1002/nbm.3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Liu T, Liu J, de Rochefort L, et al. Morphology enabled dipole inversion (MEDI) from a single-angle acquisition: Comparison with COSMOS in human brain imaging. Magn Reson Med. 2011;66(3):777–783. doi: 10.1002/mrm.22816 [DOI] [PubMed] [Google Scholar]
- 209.Li W, Wang N, Yu F, et al. A method for estimating and removing streaking artifacts in quantitative susceptibility mapping. NeuroImage. 2014;108:111–122. doi: 10.1016/j.neuroimage.2014.12.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Dimov AV, Nguyen TD, Spincemaille P, et al. Global cerebrospinal fluid as a zero-reference regularization for brain quantitative susceptibility mapping. J Neuroimaging. 2022;32(1):141–147. doi: 10.1111/jon.12923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Bollmann S, Rasmussen KGB, Kristensen M, et al. DeepQSM - using deep learning to solve the dipole inversion for quantitative susceptibility mapping. NeuroImage. 2019;195:373–383. doi: 10.1016/j.neuroimage.2019.03.060 [DOI] [PubMed] [Google Scholar]
- 212.Yoon J, Gong E, Chatnuntawech I, et al. Quantitative susceptibility mapping using deep neural network: QSMnet. NeuroImage. 2018;179:199–206. doi: 10.1016/j.neuroimage.2018.06.030 [DOI] [PubMed] [Google Scholar]
- 213.Wharton SJ, Schäfer A, Bowtell RW. Susceptibility mapping in the human brain using threshold-based k-space division. Magn Reson Med. 2010;63(5):1292–1304. doi: 10.1002/mrm.22334 [DOI] [PubMed] [Google Scholar]
- 214.Polak D, Chatnuntawech I, Yoon J, et al. Nonlinear dipole inversion (NDI) enables robust quantitative susceptibility mapping (QSM). NMR Biomed. 2020;33(12):e4271. doi: 10.1002/nbm.4271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.De A, Sun H, Emery DJ, Butcher KS, Wilman AH. Quantitative susceptibility-weighted imaging in presence of strong susceptibility sources: Application to hemorrhage. Magn Reson Imaging. 2022;92:224–231. doi: 10.1016/j.mri.2022.06.010 [DOI] [PubMed] [Google Scholar]
- 216.Hagberg GE, Eckstein K, Tuzzi E, Zhou J, Robinson S, Scheffler K. Phase-based masking for quantitative susceptibility mapping of the human brain at 9.4T. Magn Reson Med. 2022;88(5):2267–2276. doi: 10.1002/mrm.29368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Wen Y, Spincemaille P, Nguyen T, et al. Multiecho complex total field inversion method (mcTFI) for improved signal modeling in quantitative susceptibility mapping. Magn Reson Med. 2021;86(4):2165–2178. doi: 10.1002/mrm.28814 [DOI] [PubMed] [Google Scholar]
- 218.Bao C, Choi JK, Dong B. Whole Brain Susceptibility Mapping Using Harmonic Incompatibility Removal. ArXiv180512521 Cs Math. May 2018. http://arxiv.org/abs/1805.12521. Accessed January 2, 2019. [Google Scholar]
- 219.Balasubramanian PS, Spincemaille P, Guo L, Huang W, Kovanlikaya I, Wang Y. Spatially Adaptive Regularization in Total Field Inversion for Quantitative Susceptibility Mapping. iScience. 2020;23(10):101553. doi: 10.1016/j.isci.2020.101553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Zachariou V, Bauer CE, Powell DK, Gold BT. Ironsmith: An Automated Pipeline for QSM-based Data Analyses. NeuroImage. December 2021:118835. doi: 10.1016/j.neuroimage.2021.118835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Lancione M, Tosetti M, Donatelli G, Cosottini M, Costagli M. The impact of white matter fiber orientation in single-acquisition quantitative susceptibility mapping. NMR Biomed. 2017;(August):1–8. doi: 10.1002/nbm.3798 [DOI] [PubMed] [Google Scholar]
- 222.Li W, Liu C, Duong TQ, van Zijl PCM, Li X. Susceptibility tensor imaging (STI) of the brain: Review of susceptibility tensor imaging of the brain. NMR Biomed. 2017;30(4):e3540. doi: 10.1002/nbm.3540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Hametner S, Wimmer I, Haider L, Pfeifenbring S, Brück W, Lassmann H. Iron and neurodegeneration in the multiple sclerosis brain. Ann Neurol. 2013;74(6):848–861. doi: 10.1002/ana.23974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Khalil M, Teunissen C, Langkammer C. Iron and Neurodegeneration in Multiple Sclerosis. Mult Scler Int. 2011;2011:1–6. doi: 10.1155/2011/606807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Ropele S, de Graaf WL, Khalil M, et al. MRI assessment of iron deposition in multiple sclerosis. J Magn Reson Imaging. 2011;34(1):13–21. doi: 10.1002/jmri.22590 [DOI] [PubMed] [Google Scholar]
- 226.Staudt M, Krägeloh-Mann I, Grodd W. Die normale Myelinisierung des kindlichen Gehirns in der MRT - eine Metaanalyse. RöFo - Fortschritte Auf Dem Geb Röntgenstrahlen Bildgeb Verfahr. 2000;172(10):802–811. doi: 10.1055/s-2000-7898 [DOI] [PubMed] [Google Scholar]
- 227.Zhang Y, Wei H, Cronin MJ, He N, Yan F, Liu C. Longitudinal atlas for normative human brain development and aging over the lifespan using quantitative susceptibility mapping. NeuroImage. 2018;171(January):176–189. doi: 10.1016/j.neuroimage.2018.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Zhang Y, Wei H, Cronin MJ, He N, Yan F, Liu C. Longitudinal data for magnetic susceptibility of normative human brain development and aging over the lifespan. Data Brief. 2018;20:623–631. doi: 10.1016/j.dib.2018.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Schweser F, Raffaini Duarte Martins AL, Hagemeier J, et al. Mapping of thalamic magnetic susceptibility in multiple sclerosis indicates decreasing iron with disease duration: A proposed mechanistic relationship between inflammation and oligodendrocyte vitality. NeuroImage. 2018;167:438–452. doi: 10.1016/j.neuroimage.2017.10.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Lim IAL, Faria AV, Li X, et al. Human brain atlas for automated region of interest selection in quantitative susceptibility mapping: Application to determine iron content in deep gray matter structures. NeuroImage. 2013;82:449–469. doi: 10.1016/j.neuroimage.2013.05.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Singh MK, Singh KK. A Review of Publicly Available Automatic Brain Segmentation Methodologies, Machine Learning Models, Recent Advancements, and Their Comparison. Ann Neurosci. 2021;28(1–2):82–93. doi: 10.1177/0972753121990175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Derakhshan M, Caramanos Z, Giacomini PS, et al. Evaluation of automated techniques for the quantification of grey matter atrophy in patients with multiple sclerosis. NeuroImage. 2010;52(4):1261–1267. doi: 10.1016/j.neuroimage.2010.05.029 [DOI] [PubMed] [Google Scholar]
- 233.Gelman N, Ewing JR, Gorell JM, Spickler EM, Solomon EG. Interregional variation of longitudinal relaxation rates in human brain at 3.0 T: relation to estimated iron and water contents. Magn Reson Med. 2001;45(1):71–79. [DOI] [PubMed] [Google Scholar]
- 234.Vrenken H, Geurts JJG, Knol DL, et al. Whole-Brain T1 Mapping in Multiple Sclerosis: Global Changes of Normal-appearing Gray and White Matter. Radiology. 2006;240(3):811–820. doi: 10.1148/radiol.2403050569 [DOI] [PubMed] [Google Scholar]
- 235.Ide S, Kakeda S, Ueda I, et al. Internal structures of the globus pallidus in patients with Parkinson’s disease: evaluation with quantitative susceptibility mapping (QSM). Eur Radiol. November 2014. doi: 10.1007/s00330-014-3472-7 [DOI] [PubMed] [Google Scholar]
- 236.Li X, Chen L, Kutten K, et al. Multi-atlas tool for automated segmentation of brain gray matter nuclei and quantification of their magnetic susceptibility. NeuroImage. 2019;191:337–349. doi: 10.1016/j.neuroimage.2019.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Hsieh CY, Cheng YCN, Xie H, Haacke EM, Neelavalli J. Susceptibility and size quantification of small human veins from an MRI method. Magn Reson Imaging. 2015;33(10):1191–1204. doi: 10.1016/j.mri.2015.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Probst J, Rohner M, Zahn M, et al. Quantitative susceptibility mapping in ischemic stroke patients after successful recanalization. Sci Rep. 2021;11(1):16038. doi: 10.1038/s41598-021-95265-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Ward PGD, Fan AP, Raniga P, et al. Improved Quantification of Cerebral Vein Oxygenation Using Partial Volume Correction. Front Neurosci. 2017;11. https://www.frontiersin.org/articles/10.3389/fnins.2017.00089. Accessed February 7, 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Borland D, Taylor II RM. Rainbow Color Map (Still) Considered Harmful. IEEE Comput Graph Appl. 2007;27(2):14–17. doi: 10.1109/MCG.2007.323435 [DOI] [PubMed] [Google Scholar]
- 241.Crameri F, Shephard GE, Heron PJ. The misuse of colour in science communication. Nat Commun. 2020;11(1):5444. doi: 10.1038/s41467-020-19160-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Griswold MA, Sunshine JL, Seiberlich N, Gulani V. Towards Unified Colormaps for Quantitative MRF Data. In: Proc Intl Soc Mag Reson Med 26 (2018). 2018:p2894. [Google Scholar]
- 243.Kiselev VG. Larmor Frequency in Heterogeneous Media. J Magn Reson. 2019;299:168175. [DOI] [PubMed] [Google Scholar]
- 244.Sandgaard AD, Shemesh N, Kiselev VG, Jespersen SN. Larmor frequency shift from magnetized cylinders with arbitrary orientation distribution. NMR Biomed. 2023. doi: 10.1002/nbm.4859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Khabipova D, Wiaux Y, Gruetter R, Marques JP. A Modulated Closed Form solution for Quantitative Susceptibility Mapping - A thorough evaluation and comparison to iterative methods based on edge prior knowledge. NeuroImage. 2014;107:163–174. doi: 10.1016/j.neuroimage.2014.11.038 [DOI] [PubMed] [Google Scholar]
- 246.Wharton SJ, Bowtell RW. Whole-brain susceptibility mapping at high field: a comparison of multiple- and single-orientation methods. NeuroImage. 2010;53(2):515–525. doi: 10.1016/j.neuroimage.2010.06.070 [DOI] [PubMed] [Google Scholar]
- 247.Dimov AV, Gupta A, Kopell BH, Wang Y. High-resolution QSM for functional and structural depiction of subthalamic nuclei in DBS presurgical mapping. J Neurosurg. 2019;131(2):360–367. doi: 10.3171/2018.3.JNS172145 [DOI] [PubMed] [Google Scholar]
- 248.Al-Radaideh AM, Athamneh I, Alabadi H, Hbahbih M. Cortical and Subcortical Morphometric and Iron Changes in Relapsing-Remitting Multiple Sclerosis and Their Association with White Matter T2 Lesion Load. Clin Neuroradiol. 2018. doi: 10.1007/s00062-017-0654-0 [DOI] [PubMed] [Google Scholar]
- 249.Chen W, Zhang Y, Mu K, et al. Quantifying the Susceptibility Variation of NormalAppearing White Matter in Multiple Sclerosis by Quantitative Susceptibility Mapping. Am J Roentgenol. 2017;(October):1–6. doi: 10.2214/AJR.16.16851 [DOI] [PubMed] [Google Scholar]
- 250.Kaunzner UW, Kang Y, Zhang S, et al. Quantitative susceptibility mapping identifies inflammation in a subset of chronic multiple sclerosis lesions. Brain. 2019;142(1):133–145. doi: 10.1093/brain/awy296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Pontillo G, Petracca M, Monti S, et al. Unraveling Deep Gray Matter Atrophy and Iron and Myelin Changes in Multiple Sclerosis. Am J Neuroradiol. April 2021:ajnr;ajnr.A7093v1. doi: 10.3174/ajnr.A7093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Yu FF, Chiang FL, Stephens N, et al. Characterization of normal-appearing white matter in multiple sclerosis using quantitative susceptibility mapping in conjunction with diffusion tensor imaging. Neuroradiology. 2019;61(1):71–79. doi: 10.1007/s00234-018-2137-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Lotfipour AK, Wharton SJ, Schwarz ST, et al. High resolution magnetic susceptibility mapping of the substantia nigra in Parkinson’s disease. J Magn Reson Imaging. 2012;35(1):48–55. doi: 10.1002/jmri.22752 [DOI] [PubMed] [Google Scholar]
- 254.Al-Radaideh AM, Wharton SJ, Lim SY, et al. Increased iron accumulation occurs in the earliest stages of demyelinating disease: an ultra-high field susceptibility mapping study in Clinically Isolated Syndrome. Mult Scler. 2013;19(7):896–903. doi: 10.1177/1352458512465135 [DOI] [PubMed] [Google Scholar]
- 255.Cobzas D, Sun H, Walsh AJ, Lebel RM, Blevins G, Wilman AH. Subcortical gray matter segmentation and voxel-based analysis using transverse relaxation and quantitative susceptibility mapping with application to multiple sclerosis. J Magn Reson Imaging. 2015;42(6):1601–1610. doi: 10.1002/jmri.24951 [DOI] [PubMed] [Google Scholar]
- 256.Feng X, Deistung A, Reichenbach JR. Quantitative susceptibility mapping (QSM) and R2* in the human brain at 3 T. Z Med Phys. 2018;28(1):36–48. doi: 10.1016/j.zemedi.2017.05.003 [DOI] [PubMed] [Google Scholar]
- 257.Zivadinov R, Tavazzi E, Bergsland N, et al. Brain Iron at Quantitative MRI Is Associated with Disability in Multiple Sclerosis. Radiology. 2018;289(2):487–496. doi: 10.1148/radiol.2018180136 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.