Abstract
The link between bipolar disorder (BP) and immune dysfunction remains controversial. While epidemiological studies have long suggested an association, recent research has found only limited evidence of such a relationship. To clarify this, we investigated the contributions of immune-relevant genetic factors to the response to lithium (Li) treatment and the clinical presentation of BP. First, we assessed the association of a large collection of immune-related genes (4,925) with Li response, defined by the Retrospective Assessment of the Lithium Response Phenotype Scale (Alda scale), and clinical characteristics in patients with BP from the International Consortium on Lithium Genetics (ConLi+Gen, N = 2,374). Second, we calculated here previously published polygenic scores (PGSs) for immune-related traits and evaluated their associations with Li response and clinical features. We found several genes associated with Li response at p < 1×10− 4 values, including HAS3, CNTNAP5 and NFIB. Network and functional enrichment analyses uncovered an overrepresentation of pathways involved in cell adhesion and intercellular communication, which appear to converge on the well-known Li-induced inhibition of GSK-3β. We also found various genes associated with BP’s age-at-onset, number of mood episodes, and presence of psychosis, substance abuse and/or suicidal ideation at the exploratory threshold. These included RTN4, XKR4, NRXN1, NRG1/3 and GRK5. Additionally, PGS analyses suggested serum FAS, ECP, TRANCE and cytokine ligands, amongst others, might represent potential circulating biomarkers of Li response and clinical presentation. Taken together, our results support the notion of a relatively weak association between immunity and clinically relevant features of BP at the genetic level.
Keywords: bipolar disorder, immunity, lithium, polygenic scores, genetics
INTRODUCTION
Bipolar disorder (BP) has been associated with some degree of immune dysfunction. Epidemiological data has linked immune-related medical comorbidities, including autoimmune and metabolic diseases, and chronic low-grade inflammation with BP. In particular, increases in pro-inflammatory cytokines are observed during affective episodes in patients with BP [1]. In addition, genomic studies have revealed weak yet significant genetic correlation between BP and immune-related diseases [2]. Nevertheless, as a number of these observations originated from underpowered studies [3], further investigations are required to elucidate the proposed relationships.
Lithium (Li), mainly used in the treatment of BP, is an effective pharmacological agent in the treatment of an array of psychiatric conditions [4, 5]. In addition to its mood-stabilizing effects, Li shows anti-viral and immune cell regulatory properties [6, 7]. The immune regulatory activity of Li has been partially attributed to the modulation of pro-inflammatory cytokines and GSK-3β. Therefore, it has been suggested that the mechanism through which Li improves symptom progression may be via anti-inflammatory effects [8, 9]. The Retrospective Assessment of the Lithium Response Phenotype Scale (Alda scale) is the most widely used clinical measure of Li response. Most often, it is dichotomized such that individuals with scores ≥ 7 are classified as “responders” and those with scores < 7 as “non-responders” [10, 11]. Using this metric, previous genetic studies have implicated human leukocyte antigen (HLA) and inflammatory cytokine genes in the response to Li treatment in BP [12, 13]. Therefore, we hypothesized that single nucleotide polymorphisms (SNPs) in immune-related genes contribute, to some extent, to Li response and further, may impact specific clinical features within BP. To test our hypothesis, we performed association studies of a comprehensive collection of immune-related genes in 2,374 patients with BP from the International Consortium on Lithium Genetics (ConLi+Gen) [14]. Additionally, we tested associations with published polygenic scores (PGSs) for immune-relevant traits.
METHODS
Since our study follows a candidate approach to selected genes, pathways and networks, a diagram summarizing the methodology employed can be found in the Supplementary Figures: Figure S1.
Study sample
The ConLi+Gen cohort has been previously described in detail [15]. Briefly, peripheral blood samples from individuals with a diagnosis of a bipolar spectrum disorder (in accordance with the criteria established in the Diagnostic and Statistical Manual of Mental Disorders -DSM- versions III or IV) that had taken Li for a minimum of six months (with no additional mood stabilizers), were collected from 2003 to 2013. The isolated DNA was genotyped in two phases. This resulted in two sample batches originally referred to as “GWAS1” and “GWAS2”, comprising 1,162 and 1,401 individuals, respectively. Long-term responses to Li treatment were assessed in both sample batches using the Alda scale. Here, the A subscale rates the degree of response on a 10-point scale, and the B subscale reflects the relationship between improvement and treatment. A total score, ranging from 0–10, is obtained by subtracting the B score from the A score of these subscales. Negative scores are set to 0. Data on age-at-onset (AAO), age (at sample collection and phenotyping), sex and diagnostic subtype were available for both sample batches. Diagnoses included bipolar disorder type I and type II, schizoaffective bipolar disorder and bipolar disorder not otherwise specified. Additionally, information on psychiatric features, namely the number of episodes of depression, mania and hypomania, the presence of psychosis, alcohol and substance abuse, and suicidal ideation, were available for patients in the “GWAS1” batch.
The Ethics Committee at the University of Heidelberg provided central approval for the ConLi+Gen Consortium. Written informed consent from all participants was obtained according to the study protocols of each of the participating sites and their institutions. All procedures were performed in accordance with the guidelines of the Declaration of Helsinki.
Immune gene collection
A comprehensive set of immune-related genes was collated from gene lists available in the online databases MSigDB [16] and InnateDB [17]. From MSigDB (https://www.gsea-msigdb.org/gsea/msigdb/), the following gene sets contained in the C2 “curated gene sets” collection were retrieved: M1036: Reactome-innate immune system, M1058: Reactome-adaptive immune system, M39895: WikiPathways (WP)-neuroinflammation, M39711: WP-cytokines and inflammatory response, and M39641: WP-inflammatory response pathway. From InnateDB (https://www.innatedb.com/index.jsp), the curated gene lists derived from the Immunology Database and Analysis Portal (ImmPort), the Immunogenetic Related Information Source (IRIS) and the Immunome Database, were downloaded. Chromosomal locations were annotated from Ensembl using the hg19 build. Herein, the combined collection is referred to as the ImmuneSet and contained 4,925 autosomal genes to be included in association analyses.
Genotype data
Schubert et al. (2021) [18] have previously described the creation of the genotype dataset used herein. Briefly, DNA samples were originally genotyped using either Affymetrix or Illumina SNP arrays. These genotype data from multiple cohorts were separately imputed using the 1000 Genomes Project reference panel phase 3 v5. Each imputed dataset underwent a basic quality control (QC) step to keep variants with minor allele frequency (MAF) > 0.01, Hardy-Weinberg equilibrium p-value (HWE) ≥ 1×10− 6 and imputation quality score (Rsq) ≥ 0.6. Genotype calls were derived from the imputed dosage scores and all datasets were merged by retaining only common sets of SNPs. To update this dataset and obtain a higher number of good quality variants, we re-imputed the genotype data via the Michigan Imputation Server [19] using the Haplotype Reference Consortium (HRC) panel for European ancestry. The re-imputed genotypes underwent a QC step to keep variants with Rsq ≥ 0.8, MAF ≥ 0.01 and HWE ≥ 1×10− 6. Additionally, individuals were removed if they failed the heterozygosity test and/or showed relatedness, according to the tests performed using the plinkQC R package [20]. In the latter case, one individual from each pair of related individuals (PI-HAT > 0.25) was removed. For analysis of the ImmuneSet, SNPs within each gene’s boundaries (± 0 kb) were retained. The final ImmuneSet genotype datasets contained 701,031 SNPs from 1,024 and 1,350 individuals in “GWAS1” and “GWAS2”, respectively.
Polygenic Scores
A set of 32 published PGSs available at the PGS Catalog [21] were used to approximate markers of inflammation and immune-related phenotypes that were not experimentally measured in the “GWAS1” ConLi+Gen sample. These PGSs, created and evaluated in large samples of predominantly European ancestry, stemmed from three recent publications and corresponded to the following traits: autoimmune disease [22], lymphocyte / monocyte / eosinophil / neutrophil / basophil percentage of white (blood) cells [23], and serum levels of 26 markers of inflammation [24]. After downloading and harmonizing weight files, we performed allelic scoring in ConLi+Gen using the sum method applied in Plink 1.9 [25].
Association analyses
The “GWAS1” (N = 853) and “GWAS2” (N = 1,258) samples were tested separately for associations of 701,031 SNPs in the ImmuneSet with: 1) Li response (responder / non-responder, defined by Alda scores ≥ 7 or < 7, respectively), 2) total Alda score, 3) Alda subscale A, and 4) Alda subscale B (total). Because the most reliable continuous Li response phenotype has been previously shown to be the Alda A score, when excluding individuals with Alda B scores > 4 [10], we tested this as the primary continuous phenotype in our study. All association tests were performed applying an additive model in Plink 1.9, and all models were adjusted for age at recruitment, age-at-onset (AAO), sex, diagnosis and the first principal components (PCs) obtained for each ImmuneSet genotypes dataset. PCA plots were explored to determine the optimal number of PCs to be used as covariates for each sample. Therefore, the first five PCs were used as covariates for “GWAS1” while the first six PCs were used for “GWAS2”. Population stratification due to ancestry was successfully corrected by the selected numbers of PCs (Supplementary Figures: Figure S2). Next, the association results for Li response from “GWAS1” and “GWAS2” samples were meta-analyzed using the weighted-z (METAL) method applied in Plink 1.9. These meta-analysis results were QCed to exclude variants with I2 heterogeneity index (I) > 40 and p-value for Cochran’s Q statistic (Q) < 0.1 (highly heterogeneous). We searched first for associations at the commonly accepted thresholds for GWASs (genome-wide significance, p < 5×10− 8, and suggestive significance, p < 1×10− 5). However, considering this a candidate gene rather than a genome-wide approach, we chose to look further into findings with p < 1×10− 4, a threshold that has been previously used to select association findings for follow-up in GWASs [26] and, therefore, represents an acceptable exploratory threshold.
In “GWAS1”, the ImmuneSet was further tested for associations with other BP clinical phenotypes (i.e. AAO, the number of episodes of depression, mania and hypomania, as well as the presence of psychosis, alcohol and/or substance abuse, and suicidal ideation). These models were adjusted for age at recruitment, AAO (except when AAO was tested as phenotype), sex, diagnosis and the first five PCs. Statistical significance was considered as above.
Associations between PGSs and the various BP clinical phenotypes were tested using linear or binomial regression models, as appropriate, adjusted for age at recruitment, AAO (except when AAO was tested as phenotype), sex, diagnosis and population using the robustbase R package. Significance was set to false discovery rate (FDR) < 0.05. However, we also looked into the nominally significant findings (p < 0.05) for exploratory purposes.
Downstream analyses
All variants under the p < 1×10− 4 threshold were annotated for known regulatory effects on gene expression (i.e. expression quantitative trait loci, eQTLs) in all human brain, blood, spleen and thyroid tissues, as well as in immune cells (e.g. monocytes and macrophages) using Qtlizer [27].
A protein-protein interaction (PPI) network to explore the functional relevance of the identified genes associated with Li response was created using the ReactomeFIViz app [28] for Cytoscape 3.7 [29]. This analysis used as input a list composed of the ImmuneSet genes showing associations at the p < 1×10− 4 threshold with the dichotomous and continuous Li response phenotypes. The network also incorporated “linker” genes (i.e. genes not in the input gene list that create indirect connections between input genes) to increase biological interpretability. Moreover, pathway overrepresentation analysis was performed on the PPI network (including linker genes) using the pathway enrichment network function of the app. Because the linker genes were not drawn from the ImmuneSet collection, we used the standard background genes of the ReactomeFiViz app for this analysis. The resulting overrepresented pathways were filtered to exclude terms that: 1) had FDR > 0.05, 2) corresponded to a specific disease (e.g. bladder cancer, herpes virus infection), 3) had less than two genes overlapping between the pathway set and the network set, and/or 4) the overlap with the pathway set represented less than 3% of genes in the set. Additionally, we repeated the pathway overrepresentation analysis including not only the variant mapped genes, but also the annotated eQTL genes.
For associations with clinical phenotypes in the “GWAS1” sample, functional analyses were performed using the GENE2FUNC tool of the Functional Mapping and Annotation of Genome-Wide Association Studies (FUMA-GWAS) platform [30]. The input gene lists included mapped and eQTL genes annotated for variants below the p < 1×10− 4 threshold for each studied phenotype. Because eQTL genes were not drawn from our ImmuneSet collection, we used all protein-coding genes as background for these analyses. Overrepresented gene sets were those that showed FDR < 0.05, following a hypergeometric test, and a minimum of two overlapping genes. Curated gene sets from pathway databases in the “canonical pathways” category were preferred when available. Otherwise, Gene Ontology biological processes (GO_BPs) or any other available category (including GWAS Catalog trait associations) were taken. For GTEx-based enriched tissues of expression, as our focus is on immune-brain relationships, we kept only those enrichments corresponding to brain expression, as these are the most relevant tissues for the analysis of Li response and clinical features of BP.
In addition, the relative importance for (dichotomous) Li response of the calculated PGSs in “GWAS1” was assessed through a machine learning (ML) screening approach using the Auto Model extension of RapidMiner Studio. This applied various classification algorithms to the raw PGS data. Auto Model provides the following models: Naïve Bayes, Generalized Linear Model, Logistic Regression, Fast Large Margin, Deep Learning, Decision Tree, Random Forest, Gradient Boosted Trees and Support Vector Machines. Because ML algorithms are sensitive to class imbalance, an equal number of responder and non-responder individuals were randomly selected for the analysis (N = 657) using the sample method of the Python’s Pandas library. The resulting file with balanced classes was used as input for the ML screening in Auto Model, where the sample was randomly divided into training (60%) and test (40%) sets. Default parameters for all algorithms were applied. Given that different types of ML algorithms can differ in their feature selection procedure due to their inherent characteristics, here, features were considered important for Li response, with either a positive (i.e. favoring response) or a negative (i.e. favoring non-response) effect, when at least two algorithms selected the same feature with the same effect direction as important for the classification task.
RESULTS
Immune-related genes associated with response to Li treatment in BP
After excluding individuals with missing phenotypic data (age and/or AAO), the effective sample sizes for the association analyses in ConLi+Gen were 853 and 1,258 in “GWAS1” and “GWAS2”, respectively. A basic description of both samples is shown in Table 1. In general, there were more female than male patients in both samples and there were minimal differences in the mean ages at recruitment and disease onset between “GWAS1” and “GWAS2”. Therefore, the total sample size of our meta-analyses of Li response was 2,111, including 606 (28.7%) responders and 1,505 non-responders for the dichotomized variable, which included 1,224 (58%) females and 887 males, with mean age 47 (± 14) years and mean AAO 25 (± 11) years. For the continuous Li response phenotype (i.e. Alda A score, excluding individuals with Alda B score > 4), the effective sample sizes were 828 for “GWAS1” and 1,044 for “GWAS2”. After the post-meta-analysis exclusion of variants with heterogeneous effects between both ConLi+Gen samples, a mean of 556,196 SNPs remained in each set of summary statistics. This was higher for the continuous phenotype, in which 625,818 SNPs remained.
Table 1.
Description of the ConLi+Gen samples.
| Responders (Total Alda ≤ 7) | Non-Responders (Total Alda < 7) | Total | |
|---|---|---|---|
| GWAS1 | |||
| N Effective sample (% from total) | 297 (34.8) | 556 (65.2) | 853 |
| N Females (%) | 178 (59.9) | 337 (60.6) | 515 (60.4) |
| Age (mean ± SD) | 52 ± 14 | 46 ± 14 | 48 ± 14 |
| Age-at-onset (mean ± SD) | 28 ± 11 | 23 ± 11 | 25 ± 11 |
| # Depressive episodes (mean) | 5 | 7 | 6 |
| # Hypomanic episodes (mean) | 2 | 6 | 4 |
| # Manic episodes (mean) | 4 | 6 | 5 |
| N Psychosis cases (%) | 72 (24.2) | 270 (48.6) | 342 (40.1) |
| N Alcohol abuse cases (%) | 18 (6.1) | 122 (21.9) | 140 (16.4) |
| N Substance abuse cases (%) | 30 (10.1) | 105 (18.9) | 135 (15.8) |
| N Suicidal ideation cases (%) | 75 (25.3) | 256 (46.0) | 331 (38.8) |
| GWAS2 | |||
| N Effective sample (% from total) | 309 (24.6) | 949 (75.4) | 1258 |
| Females (%) | 157 (50.8) | 552 (58.2) | 709 (56.4) |
| Age (mean ± SD) | 48 ± 15 | 46 ± 13 | 47 ± 14 |
| Age-at-onset (mean ± SD) | 25 ± 10 | 25 ± 11 | 25 ± 11 |
We found no associations with Li response at the genome-wide GWAS threshold (p < 5×10− 8). At the suggestive threshold for GWAS (p < 1×10− 5), the dichotomous Li response phenotype and Alda B (total) showed associations with FAT3 (best SNP: rs4313539, p = 2.1×10− 6, z = 4.744), and with ADAMTS5 (best SNP: rs162501, p = 9.18×10− 7, z = 4.909) and GRID2 (best SNP: rs62312225, p = 2.71×10− 6, z = 4.692), respectively (Supplementary File 1: Table 1). At the exploratory threshold (p < 1×10− 4), when considering linkage disequilibrium (LD), we identified between 9 and 12 genomic loci in relation with different aspects of Li response (Supplementary File 1: Table 1). The most significant SNPs from the analyses of the dichotomous and continuous phenotypes mapped to FAT3 and BMPR1A, respectively (Table 2). In total, 42 genes were implicated in the response to Li in patients with BP from our exploratory analyses in the ConLi+Gen cohort (Supplementary File 2: Table 1). As expected, there was a number of gene-based overlaps between different aspects of Li response, particularly between the dichotomous variable and total Alda score, and between the continuous variable and the other Alda variables.
Table 2.
Summary findings from the genetic association meta-analyses of Li responses in ConLi+Gen.
| Meta-analysis summary | Response vs No-response | Continuous Li response |
|---|---|---|
| # SNPs after QC | 557,037 | 625,818 |
| # SNPs p < 0.05 | 27,426 | 31,489 |
| # SNPs p < 1×10−4 | 124 | 33 |
| # Lead SNPs | 11 | 11 |
| Top lead SNP | rs4313539 | rs12776537 |
| Effect allele | C | A |
| p-value | 2.1×10−6 | 2.5×10−5 |
| Z | 4.7 | −4.2 |
| Gene | FAT3 | BMPR1A |
| # SNPs p < 1×10−5 | 15 | 0 |
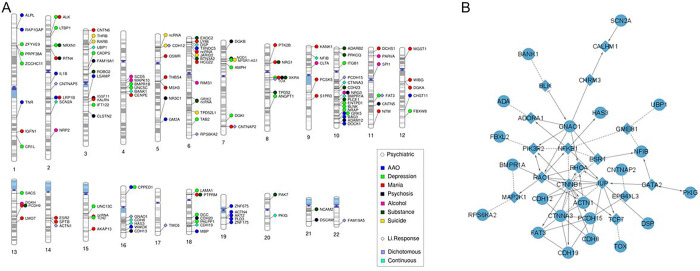
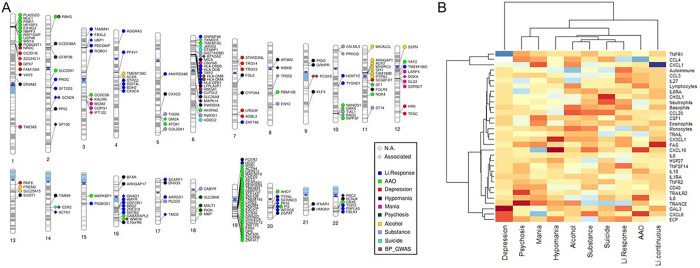
Twenty-four ImmuneSet genes were implicated in the primary Li response phenotypes (i.e. dichotomous and continuous) in our exploratory analysis (Fig. 1A). These were used as input for a network analysis to facilitate biological interpretation of the findings. This network analysis provided known and predicted functional interactions between a subset of 21 input genes from our association results and 16 linkers drawn from the total of protein-coding genes in the background reference of the ReactomeFiViz app (Fig. 1B). Functional analysis of the network uncovered an overrepresentation of crucial developmental pathways and regulatory networks, suggesting the involvement of processes such as assembly and stability of the cell-cell signaling machinery (e.g. adherens junction, E-cadherin signaling, focal adhesion, integrin signaling, L1 cell adhesion molecule signaling), neuronal development and function (e.g. neurotrophic signaling, lysophosphatidic acid receptor mediated events, regulation of pluripotency, Wnt signaling), as well as activation of inflammatory (e.g. sphingolipid signaling, S1P pathways, toll-like receptor signaling) and adaptive immune pathways (e.g. T and B cell receptor signaling) in the biological response to Li. Interestingly, processes such as angiogenesis, long-term potentiation, thyroid hormone signaling pathways, melanogenesis and sensory processing were also overrepresented in our network analysis (Supplementary File 2: Table 2). These observations suggest that variation in immune-related genes and its effects that expand beyond inflammatory and immune responses from early developmental stages contribute to determine the extent of the organism’s response to Li treatment in patients with BP. Moreover, when eQTL genes were incorporated into the analysis, there was a marked overrepresentation of inflammatory and autoimmune disease pathways (e.g. asthma, type 1 diabetes mellitus, autoimmune thyroid disease, inflammatory bowel disease) and of vitamin D metabolism. The Wnt signaling, cell adhesion and adaptive immune pathways remained significantly overrepresented (data not shown). All protein-coding eQTL genes annotated for Li response phenotypes are shown in Fig. 2A.
Figure 1.
Exploratory findings for the ImmuneSet in ConLi+Gen. A) Gene-based phenogram of associations of the ImmuneSet with Li response and clinical features in ConLi+Gen. B) Protein-protein interaction network of Li response phenotype associations in the ImmuneSet. Circles represent input genes and diamonds represent linker genes. Dotted lines denote predicted interactions.
Figure 2.
A) Gene-based phenogram of eQTL annotations for the exploratory-level findings of the ImmuneSet in ConLi+Gen. Those eQTL genes that were different from the mapped gene and those that were the same are presented in circle and diamond shapes, respectively. Only protein-coding genes are shown. In addition, overlaps with BP genetic associations reported in the GWAS Catalog are presented. B) Exploratory findings for the immune-related polygenic scores calculated in ConLi+Gen. Significance of calculated PGSs for BP phenotypes in ConLi+Gen “GWAS1”. The heatmap shows the z-values obtained for each PGS-phenotype pair. Increasing color darkness alludes to increasing effect, with red and blue colors at the negativ and positive extremes, respectively.
Immune-related genes associated with clinical phenotypes in BP
The association analyses of the ImmuneSet with specific clinical features that were available for the “GWAS1” sample showed no associations at the genome-wide GWAS threshold. However, at the suggestive GWAS threshold there were, collectively, 100 associations between the ImmuneSet and AAO (3), number of depressive (54) and manic (14) episodes, and the presence of psychosis (1), substance use disorder (15) and/or suicidal ideation (13). These implicated 17 genes that associated to specific clinical features (i.e. no overlaps were observed at this threshold Table 3). When we moved forward to the exploratory analysis, we identified 786 SNP-phenotype associations in total (Supplementary File 1: Tables 2–9). These implicated 166 immune-related genes (Fig. 1A; Supplementary File 2: Table 1) mostly involved in adaptive immunity and inflammation. Beyond their immune functions, however, these genes play important roles in the development of the nervous system, signal transduction, synaptic processes and cell adhesion (Supplementary File 2: Tables 3–9). In particular, large numbers of associations were found for AAO and mood episodes (Table 3). In addition, 156 eQTL genes were collectively annotated for these phenotypes. Figure 2A shows the protein-coding eQTL genes annotated for each clinical feature. Even when these showed no overlaps among the clinical phenotypes, there were some overlaps with the ImmuneSet genes meaning that, in some instances, the eQTL gene corresponded to the gene mapped to the variant while, in others, the eQTL gene was different from the mapped gene. A summary of exploratory findings for each clinical phenotype studied is shown below.
Table 3.
Summary of findings from the association analyses of the ImmuneSet with clinical characteristics in the ConLi+Gen “GWAS1” sample.
| Phenotype | N | Exploratory threshold (p < 1e−4) | Suggestive GWAS threshold (p < 1e−5) | Top gene | ||||
|---|---|---|---|---|---|---|---|---|
| # SNPs | # Genes | # SNPs | # Genes | Genes | Symbol | Pval | ||
| Age-at-onset | 853 | 54 | 21 | 3 | 2 | GRK5, PLD3 | GRK5 | 3.9×10−6 |
| # Depressive episodes | 692 | 107 | 31 | 54 | 6 | BLNK, PHLPP1, ZCCHC11, SACS, CPPED1, PRPF38A | BLNK | 1.7×10−7 |
| # Manic episodes | 665 | 116 | 32 | 14 | 4 | CNTN6, KALRN, LY86, PTK2B | CNTN6 | 2.4×10−6 |
| Psychosis | 692 | 45 | 13 | 1 | 1 | DSCAM | DSCAM | 7.6×10−6 |
| Alcohol abuse | 835 | 29 | 9 | 0 | 0 | - | SCD5 | 1.8×10−5 |
| Substance abuse | 832 | 78 | 17 | 15 | 3 | TPD52, NOD1, XKR4 | TPD52 | 4.510−7 |
| Suicidal ideation | 660 | 30 | 7 | 13 | 1 | JARID2 | JARID2 | 3.9×10−6 |
Age-at-onset.
Fifty-four associations in 21 ImmuneSet genes were found for AAO in our study (Table 3, Supplementary File 1: Table 2). These genes were enriched for negative regulation of cell death and synaptic transmission, as well as expression in the cerebellum (Supplementary File 2: Table 3). The top variant, rs1248079 (p = 3.9×10− 6, beta = 2.75), mapped to an intronic region in GRK5 (G Protein-Coupled Receptor Kinase 5). Other important genes included PLD3, AKT2 and IL1B. The addition of eQTL genes to the functional enrichment analysis resulted in an additional overrepresentation of processes related to cellular stress responses.
Depression.
With an effective sample of 692 individuals, 107 associations in 31 ImmuneSet genes for the number of depressive episodes were found (Table 3, Supplementary File 1: Table 3). These genes were enriched for synaptic processes, as well as expression in frontal and anterior cingulate cortices (Supplementary File 2: Table 4). While the top variant, rs55975329 (p = 1.7×10− 7, beta = 4.54), localized to an intron in BLNK (B cell linker), other important genes included PHLPP1, ZCCHC11 (TUT4) and CPPED1. The addition of eQTL genes to the functional enrichment analysis resulted in an additional overrepresentation of axonal and synaptic components.
Hypomania.
Although the largest associations were found for the number of hypomanic episodes, these observations were based on only 85 individuals with available data. Therefore, we have excluded this phenotype from the figures and tables shown within this manuscript. All corresponding results are provided in the supplementary material (Supplementary File 1: Table 4, Supplementary File 2: Table 5).
Mania.
With an effective sample of 665 individuals, 116 associations in 32 ImmuneSet genes for the number of manic episodes were found (Table 3, Supplementary File 1: Table 5). These genes were enriched for neuronal development and differentiation, as well as expression in spinal cord and frontal cortex (Supplementary File 2: Table 6). The top variant, rs59134172 (p = 2.4×10− 6, beta = 1.62), localized to an intron in CNTN6 (Contactin 6). Other important genes included KALRN, PCDH9, PTK2B and CNTNAP2. Interestingly, we also found enrichment for response to serotonin re-uptake inhibitors in major depressive disorder (FDR = 0.042) and serum thyroid-stimulating hormone levels (FDR = 0.0028), from the GWAS Catalog trait associations, in mania-associated ImmuneSet genes. The addition of eQTL genes to the functional enrichment analysis had no impact on the overrepresented gene set classes.
Psychosis.
The effective sample for our analysis of the presence of psychosis in BP was 692 individuals. Here, 45 associations in 13 genes were identified (Table 3, Supplementary File 1: Table 6). The implicated genes were enriched for cell adhesion and synapse organization, as well as expression in frontal cortex (Supplementary File 2: Table 7). The top SNP, rs459374 (p = 7.6×10− 6, beta = 1.96), is located in an intronic region of DSCAM (DS Cell Adhesion Molecule). Other interesting genes included CLSTN2, RTN4 and CDH13. Moreover, the GWAS Catalog traits seasonality and depression (FDR = 0.02), and response to amphetamines (FDR = 0.026), as well as obesity-related traits (FDR = 0.015), atrial fibrillation (FDR = 0.027) and diastolic blood pressure (FDR = 0.027) were enriched among the psychosis-associated genes. The addition of eQTL genes to the functional enrichment analysis had no impact on the overrepresented processes. However, this resulted in a considerable increase in overepresented brain tissues of expression, including the hippocampus, amygdala, hipothalamus, anterior cingulate cortex, putamen and substantia nigra.
Alcohol and substance abuse.
The effective sample sizes for alcohol and substance use disorders in ConLi+Gen were 835 and 832, respectively. Twenty-nine SNPs in nine genes were associated with alcohol use, with the rs7698751 SNP in SCD5 (Stearoyl-CoA Desaturase 5) being the top association (p = 1.8×10−5, beta = 2.61). Although no gene set enrichments were found for associations with alcohol abuse, other implicated genes included the BP-associated DGKH, NRG3 and RIMS1 (Supplementary File 1: Table 7). Interestingly, when incorporating the eQTLs genes into this functional analysis, overrepresentation of genes associated with mood swings, loneliness and anxious behaviors in the GWAS Catalog was observed (Supplementary File 2: Table 8). For substance use disorder, 78 associations implicating 17 genes were found (Supplementary File 1: Table 8). Genes were overrepresented in neurogenesis-related processes, with expression in cerebellum as well as frontal and anterior cingulate cortices (Supplementary File 2: Table 8). The top variant, rs7814474 (p = 4.5×10− 7, beta = 6.7), was mapped to an intron in TPD52 (Tumor Protein D52). Other genes included the BP-associated NRG1, the schizophrenia-associated PTPRM, as well as NOD1 and PRKCQ. In addition, the GWAS Catalog traits chronotype (FDR = 0.011) and serum thyroid-stimulating hormone levels (FDR = 0.047) were also overrepresented in substance abuse-associated ImmuneSet genes. The addition of eQTL genes to the functional enrichment analysis resulted in an additional overrepresentation of the phosphatidylinositol signaling system and expression in the amygdala, hippocampus and basal ganglia.
Suicidal ideation.
Information on the presence of suicidal thoughts was available for 660 ConLi+Gen individuals. Based on these, 30 variants in seven genes were associated with suicidal ideation in the “GWAS1” sample (Table 3, Supplementary File 1: Table 9). The top SNP was rs2327882 (p = 3.9×10− 6, beta = 0.55), located in an intron of the JARID2 (Jumonji and AT-Rich Interaction Domain Containing 2) gene. Gene set enrichment analysis found overrepresentation of functions of nuclear receptors (Supplementary File 2: Table 9). Indeed, these terms related to RARB and THRB. The addition of eQTL genes to the functional enrichment analysis had no impact on the overrepresented gene set classes.
Immune-related genes showed pleiotropy for BP phenotypes
Not surprisingly, a number of genes showed shared associations with the different phenotypes included in our exploratory ImmuneSet analyses in ConLi+Gen (Fig. 1A). Therefore, these genes can be prioritized for follow-up studies due to their pleiotropic effects in Li response, clinical features, or both. In this way, we prioritized 21 genes that showed associations with more than one BP phenotype (Table 4). Here, we excluded genes associated with Alda A, total Alda B and/or Total Alda when there was no overlap with the dichotomous and/or continuous Li response phenotypes. However, a complete list of corresponding gene-phenotype associations (197 in total) is shown in Supplementary File 2: Table 1. Five of the prioritized genes (CNTNAP5, DSP, NFIB, BMPR1A, and HAS3) were associated with multiple Li response phenotypes, 10 of them (XKR4, NRXN1, RTN4, NRG1/3, ALK, GRK5, LRP1B, NPSR1-AS1 and CPPED1) were associated with multiple clinical features, and another six genes (BANK1, ROBO2, CNTNAP2, PCDH9, CDH12 and FAT3) were associated with Li response as well as with clinical features.
Table 4.
Prioritized ImmuneSet candidate genes for Li response and clinical characteristics in ConLi+Gen.
| Gene | Chr | Start | End | Priority | Phenotypes |
|---|---|---|---|---|---|
| ALK | 2 | 29415640 | 30144432 | Psychiatric | Depression, Mania |
| NRXN1 | 2 | 50145643 | 51259674 | Psychiatric | Depression, Hypomania, Substance |
| RTN4 | 2 | 55199325 | 55339757 | Psychiatric | Mania, Psychosis |
| CNTNAP5 | 2 | 124025287 | 124915287 | Li response | LiResponse, Alda_B, Alda_Total |
| LRP1B | 2 | 140988992 | 142889270 | Psychiatric | AAO, Mania |
| ROBO2 | 3 | 75906695 | 77649964 | Both | Alda_Total, Hypomania, Substance |
| BANK1 | 4 | 101411286 | 102074812 | Both | Continuous.LiResp, Alda_A, Alda_B, Alda_Total, Hypomania |
| CDH12 | 5 | 21750673 | 22853622 | Both | LiResponse, Suicide |
| DSP | 6 | 7541575 | 7586717 | Li response | Continuous.LiResp, Alda_Total |
| NPSR1-AS1 | 7 | 34386124 | 34911194 | Psychiatric | Depression, Suicide |
| CNTNAP2 | 7 | 146116002 | 148420998 | Both | LiResponse, Alda_Total, Mania |
| NRG1 | 8 | 31496902 | 32622548 | Psychiatric | Mania, Substance |
| XKR4 | 8 | 56014949 | 56454613 | Psychiatric | Depression, Mania, Psychosis, Substance |
| NFIB | 9 | 14081843 | 14398983 | Li response | Continuous.LiResp, Alda_B |
| NRG3 | 10 | 83635070 | 84746935 | Psychiatric | Alcohol, Psychosis |
| BMPR1A | 10 | 86756601 | 86932838 | Li response | Continuous.LiResp, Alda_A |
| GRK5 | 10 | 120967101 | 121215131 | Psychiatric | AAO, Depression |
| FAT3 | 11 | 92352096 | 92896470 | Both | LiResponse, Alda_Total, Depression |
| PCDH9 | 13 | 66302834 | 67230445 | Both | Alda_B, Mania, Substance |
| CPPED1 | 16 | 12756919 | 12897874 | Psychiatric | AAO, Depression |
| HAS3 | 16 | 69105564 | 69118719 | Li response | Continuous.LiResp, Alda_A, Alda_Total |
Polygenic scores for immune-related traits associated with BP phenotypes
In addition to testing associations of the ImmuneSet with BP phenotypes, we calculated a set of 32 previously published immune-related PGSs, namely for: 1) (general) autoimmune disease, 2) the proportions of white blood cell populations and 3) inflammatory marker levels in serum (Supplementary File 2: Table 10). The overlap of variants between the PGS weight files obtained from PGS Catalog and the SNPs available in our ConLi+Gen “GWAS1” sample was, in general, better for the serum levels of inflammatory markers (80.6% in average) than for the other PGSs. The lowest valid SNP overlap was found for the PGS of general autoimmune disease (38.8%), suggesting that this PGS may relatively poorly index autoimmune disease signatures. For the proportion of white blood cell populations, the valid SNP overlap was also not fully satisfactory (49.4% in average). Nevertheless, we were able to obtain some valuable insights by using these scores (Fig. 2B). We found associations (FDR < 0.05) of the PGSs for CXCL1 (C-X-C Motif Chemokine Ligand 1; z = 14.9, p < 2×10− 6), ECP (Eosinophilic Cationic Protein; z = 3.7, p = 2.4×10− 4), FAS (Fas Cell Surface Death Receptor; z= −5.9, p = 5.6×10− 9) and TRANCE/TNFSF11 (TNF-related Activation-induced Cytokine/TNF Superfamily Member 11; z = 3.4, p = 0.0007) with the continuous Li response variable. No associations with the dichotomous variable survived correction for multiple comparisons. However, ML-based PGS ranking for the dichotomized Li response added another level of support to the links with CCL4 and ECP, and suggested relative importance of HSP27 (Heat Shock Protein Family B (Small) Member 1), CHI3L1 (Chitinase 3 Like 1), TNFR1, TRAILR2 and TNFSF14 (TNF Superfamily Member 14) for the prediction of responses to Li treatment in ConLi+Gen (Supplementary Figures: Figure S3). The PGSs for CCL4 (C-C Motif Chemokine Ligand 4; z = 5.4, p = 7.5×10− 8) and TRAIL/TNFSF10 (TNF-related apoptosis-inducing ligand/TNF Superfamily Member 10; z = 3.6, p = 3.7×10− 4) were associated with disease AAO. For the number of episodes of depression, we found associations with the PGSs for CXCL1 (z = 6.5, p = 1.3×10− 10), CXCL6 (C-X-C Motif Chemokine Ligand 6; z= −12.2, p < 2×10− 6), ECP (z= −11.8, p < 2×10− 6), GAL3 (Galectin 3; z= −20.1, p < 2×10− 6), and TNFR1/TNFRSF1A (TNF Receptor Superfamily Member 1A; z = 21.9, p < 2×10− 6). The number of episodes of mania were associated with the PGSs for eosinophils (z= −2.9, p = 0.003), CSF1 (Colony Stimulating Factor 1; z= −3.4, p = 7.7×10− 4), CXCL1 (z= −4.1, p = 4.6×10− 5), CXCL6 (z = 3.9, p = 1.1×10− 4), CXCL16 (z = 3.9, p = 1.2×10− 4), FAS (z= −3.3, p = 0.0012) and TRANCE (z= −3.8, p = 1.5×10− 4). Finally, the PGSs for TRAILR2/TNFRSF10B (TNF Receptor Superfamily Member 10b; z= −4.6, p = 5.1×10− 6) and TRANCE (z= −5.2, p = 2.1×10− 7) were associated with the presence of psychosis in patients with BP. A number of associations between PGSs for immune-relevant blood traits and clinically-relevant BP features remained only suggested, as these did not survive correction for multiple comparisons (Supplementary File 2: Table 10).
DISCUSSION
There is apparent mounting evidence of immune dysregulation in BP and other major psychiatric diseases. Nevertheless, some observations have originated from underpowered studies, resulting in a lack of reproducibility [3]. Therefore, it becomes crucial to gain a better understanding of the relationships between the immune and central nervous systems, and to discern between causes and consequences of disease. In very general terms, it can be assumed that the existence of genetic associations with a given phenotype indicates causal contributions of the associated loci to the studied phenotype. With this in mind, we sought to investigate how genetic factors relevant to immune activity relate to disease phenotypes, such as response to Li treatment, AAO and psychiatric symptoms, in patients with BP. Using an exploratory and extensive candidate gene approach, our study prioritized various genes and inflammatory markers that appeared to represent pleiotropic factors contributing to multiple phenotypes. However, we should note that, here, we refer to pleiotropy as the (suggested) association with multiple traits in the ConLi+Gen cohort and, by no means, have we implied that these features are independent from each other. In fact, it should be expected that, given the important correlation between psychiatric disorders, the features that we have studied in ConLi+Gen are, to some extent, also correlated with each other. Also, because genes can play different roles in different tissues and cell types, we observed widespread enrichments of biological pathways participating in the development and function of the brain. This suggested that the genes implicated in BP phenotypes might affect in parallel both the immune and nervous systems. Nevertheless, because we found no associations at the genome-wide GWAS threshold of significance, and those observed at the suggestive GWAS threshold were limited, our findings are also consistent with a relatively weak effect of immune genetic factors over BP phenotypes.
The results of our exploratory assessment of associations of genetic polymorphisms in immune-related genes with Li response in the ConLi+Gen cohort suggest that variations in inflammatory and adaptive immune processes might contribute to the efficiency of the response to Li treatment in patients with BP. Importantly, our network and gene set enrichment analysis uncovered an involvement of numerous biological pathways that participate in cell adhesion, migration and intercellular communication, helping in the development and maintenance of the central and peripheral nervous systems, as well as of the immune and vascular systems. Interestingly, many of these appear to converge in the participation of GSK-3β (glycogen synthase kinase-3 beta), as assessed through comparative overlap analysis of the enriched KEGG and Reactome gene sets. GSK-3β is involved in multiple major developmental pathways, such as the Wnt, Notch and Hedgehog signaling pathways. Genetic manipulation in mouse models has shown an antidepressant-like behavior upon GSK-3β knockdown in hippocampus, as well as cognitive, behavioral and biochemical changes associated with psychiatric disorders, including Alzheimer’s disease, BP and schizophrenia, upon GSK-3β overexpression [31]. Li possesses a well-known inhibitory effect over GSK-3β [32]. Therefore, our analysis suggests that GSK-3β might be highly relevant for the response to Li treatment in BP. Our findings are also in agreement with other epidemiological and molecular investigations of Li effects. For example, we repeatedly observed overrepresentation of gene sets related to thyroid function, such as thyroid-stimulating hormone signaling and autoimmune thyroid disease. This is in line with the reports of a reversible association of Li treatment with hypothyroidism, particularly in women [33, 34].
Taken together, the encouraging literature supporting our findings sparked our interest in exploring how a genetic measure (PGS) for inflammatory marker levels in serum might associate with Li response in ConLi+Gen. Our results supported a relationship between Li treatment response and the genetic influence on the levels of various inflammatory markers circulating in serum. These included CXCL1, ECP, FAS, TRANCE and CCL4, molecules that regulate the activation and recruitment of immune cells (T and B lymphocytes, monocytes, macrophages, neutrophils and eosinophils).
The results of our exploratory assessment of associations of genetic polymorphisms in the ImmuneSet genes with BP’s AAO, numbers of mood episodes and psychiatric comorbidities in ConLi+Gen identified various candidate genes particularly contributing to mood episodes and substance abuse, including XKR4, NRXN1, GRK5 and NRG1/3. These genes, besides their immune-related functionalities, seem to play important roles in neuronal development and function, according to our gene set enrichment analyses. Indeed, this could be corroborated by the literature in many instances. For example, NRXN1, a cell surface protein involved in cell-cell interactions, exocytosis of secretory granules and regulation of signal transmission, has been associated with autism, schizophrenia and nicotine dependence [35]. GRK5 has a role in the regulation of motility in polymorphonuclear leukocytes and inflammation [36, 37]. It also regulates the activity of various G-protein coupled receptors, including neurotransmitter receptors [38]. XKR4, a phospholipid scramblase strongly expressed in brain tissue and activated by caspases, has been suggested to participate in the remodeling of neural networks by triggering microglial responses to the exposure of phosphatidylserine on axons, dendrites and synapses [39].
It is worth noting that three of our candidate genes for depressive episodes have previously shown associations with either major depression at the gene-level (DCC) [40] or mapped to schizophrenia loci (CR1L and DGKI) [41] in large GWASs. Moreover, one of our candidate genes for manic episodes, ESR2, and one for substance use disorder, BTN3A2, were previously associated at the gene-level with major depression in the large GWAS, while one of our candidates for hypomanic episodes and alcohol dependence, the BP-associated RIMS1 [26], mapped previously to schizophrenia loci. This, together with the overlaps with reported BP-associated genes that we observed, particularly concerning known eQTL genes, provides another level of support to the validity of our findings.
Additionally, the results of our exploratory assessment of PGS associations led to interesting observations. For example, that activation of macrophages and neutrophils, reflected by the associations with the PGSs for various cytokines and chemokines produced by or targeting these cells (e.g. interleukin-8, CX3CL1, CXCL6/16), might contribute to disease AAO in ConLi+Gen. Indeed, these observations are supported by studies that have found increases in neutrophil counts in psychiatric disorders, including BP [42, 43], as well as association of genetic polymorphisms in interleukin-1β, a pro-inflammatory cytokine produced by activated immune cells, including neutrophils and macrophages, with age of onset of depression in geriatric patients [44]. In addition, if we consider that macrophage/neutrophil activation was also a suggested mechanism of Li response in our study, it would be easy to speculate that activation of these cells might be linked with some aspect of the disease onset.
In conclusion, we performed an exploratory study that indicates a relationship between immunity and clinically relevant BP phenotypes at the genetic level, and pinpoints various interesting candidates for follow-up studies. We acknowledge that our study was limited by a relatively small sample size, particularly for the episodes of hypomania, and by incomplete overlap between the variants in the PGSs and our ConLi+Gen dataset. The latter, which likely resulted from a limited overlap among the different SNP arrays initially used to genotype samples in different collection centers, caused an incomplete indexing of the immune phenotypes of interest, and a relatively low rate of survival of correction for multiple comparisons in the PGS-BP phenotype relationships. Nevertheless, despite inherent limitations, we believe that our study provides valuable insight and furthers the understanding of the immune implications in BP. These results complement the evidence coming from epidemiological data and previous findings in ConLi+Gen, and support the hypothesis that, to some extent, immune regulation might represent a feasible strategy to improve the symptomatology and treatment response in patients with BP.
Funding
The project received funding from the programme “Profilbildung 2020”, an initiative of the Ministry of Culture and Science of the State of Northrhine Westphalia. The sole responsibility for the content of this publication lies with the authors.
The primary sources of funding for ConLi+Gen were grants RI 908/7–1, FOR2107 and RI 908/11–1 from the Deutsche Forschungsgemeinschaft (Marcella Rietschel) and grant NO 246/10–1 (Markus M. Nöthen) and grant ZIA-MH00284311 from the Intramural Research Program of the National Institute of Mental Health (ClinicalTrials.gov identifier: NCT00001174). The genotyping was funded in part by the German Federal Ministry of Education and Research through the Integrated Network IntegraMent (Integrated Understanding of Causes and Mechanisms in Mental Disorders), under the auspices of the e:Med Programme (Thomas G. Schulze, Marcella Rietschel and Markus M. Nöthen). The Canadian part of the study was supported by grant #166098 from the Canadian Institutes of Health Research and by a grant from Genome Atlantic/Research Nova Scotia (Martin Alda). Collection and phenotyping of the Australian University of New South Wales sample was funded by Program Grant 1037196 from the Australian National Health and Medical Research Council (Philip B. Mitchell, Peter R. Schofield, Janice M. Fullerton), and acknowledges support from Lansdowne Foundation, Betty Lynch OAM (dec) and the Janette Mary O’Neill Fellowship. Azmeraw T. Amare is supported by the 2019–2021 National Alliance for Research on Schizophrenia and Depression (NARSAD) Young Investigator Grant from the Brain & Behaviour Research Foundation (BBRF) and National Health and Medical Research Council (NHMRC) Emerging Leadership Investigator Grant 2021 – 2008000. The collection of the Barcelona sample was supported by grants PI080247, PI1200906, PI12/00018, 2014SGR1636, 2014SGR398, and MSII14/00030 from the Centro de Investigación en Red de Salud Mental, Institut d’Investigacions Biomèdiques August Pi i Sunyer, the Centres de Recerca de Catalunya Programme/Generalitat de Catalunya, and the Miguel Servet II and Instituto de Salud Carlos III. The Swedish Research Council, the Stockholm County Council, Karolinska Institutet and the Söderström-Königska Foundation supported this research through grants awarded to Lena Backlund, Louise Frisen, Catharina Lavebratt and Martin Schalling. The collection of the Geneva sample was supported by grants Synapsy–The Synaptic Basis of Mental Diseases 51NF40–158776 and 32003B-125469 from the Swiss National Foundation. The work by the French group was supported by INSERM (Institut National de la Santé et de la Recherche Médicale), AP-HP (Assistance Publique des Hôpitaux de Paris), the Fondation FondaMental (RTRS Santé Mentale), and the labex Bio-PSY (Investissements d’Avenir program managed by the ANR under reference ANR-11-IDEX-0004–02). The collection of the Romanian sample was supported by a grant from UEFISCDI, Bucharest, Romania (grants PCCA-89/2012; PCE-203/2021) to Maria Grigoroiu-Serbanescu. The collection of the Czech sample was supported by the project Nr. LO1611 with a financial support from the MEYS under the NPU I program and by the Czech Science Foundation, grant Nr. 17–07070S. Biju Viswanath is funded by the Intermediate (Clinical and PublicHealth) Fellowship (IA/CPHI/20/1/505266) of the DBT/Wellcome Trust India Alliance.
Footnotes
Conflict of interests
Eduard Vieta has received grants and served as consultant, advisor or CME speaker for the following entities: AB-Biotics, Abbvie, Almirall, Allergan, Angelini, AstraZeneca, Bristol-Myers Squibb, Dainippon Sumitomo Pharma, Farmindustria, Ferrer, Forest Research Institute, Gedeon Richter, GH Research, Glaxo-Smith-Kline, Janssen, Lundbeck, Orion, Otsuka, Pfizer, Roche, Rovi, Sanofi-Aventis, Servier, Shire, Sunovion, Takeda, the Brain and Behaviour Foundation, the Spanish Ministry of Science and Innovation (CIBERSAM), the Stanley Medical Research Institute and Viatris. Michael Bauer has received grants from the Deutsche Forschungsgemeinschaft (DFG), and Bundesministeriums für Bildung und Forschung (BMBF), and served as consultant, advisor or CME speaker for the following entities: Allergan, Aristo, Janssen, Lilly, Lundbeck, neuraxpharm, Otsuka, Sandoz, Servier and Sunovion outside the submitted work. Sarah Kittel-Schneider has received grants and served as consultant, advisor or speaker for the following entities: Medice Arzneimittel Pütter GmbH and Takeda. Bernhard Baune has received grants and served as consultant, advisor or CME speaker for the following entities: AstraZeneca, Bristol-Myers Squibb, Janssen, Lundbeck, Otsuka, Servier, the National Health and Medical Research Council, the Fay Fuller Foundation, the James and Diana Ramsay Foundation. Tadafumi Kato received honoraria for lectures, manuscripts, and/or consultancy, from Kyowa Hakko Kirin Co, Ltd, Eli Lilly Japan K.K., Otsuka Pharmaceutical Co, Ltd, GlaxoSmithKline K.K., Taisho Toyama Pharmaceutical Co, Ltd, Dainippon Sumitomo Pharma Co, Ltd, Meiji Seika Pharma Co, Ltd, Pfizer Japan Inc., Mochida Pharmaceutical Co, Ltd, Shionogi & Co, Ltd, Janssen Pharmaceutical K.K., Janssen Asia Pacific, Yoshitomiyakuhin, Astellas Pharma Inc, Wako Pure Chemical Industries, Ltd, Wiley Publishing Japan, Nippon Boehringer Ingelheim Co Ltd, Kanae Foundation for the Promotion of Medical Science, MSD K.K., Kyowa Pharmaceutical Industry Co, Ltd and Takeda Pharmaceutical Co, Ltd. Tadafumi Kato also received a research grant from Takeda Pharmaceutical Co, Ltd. Peter Falkai has received grants and served as consultant, advisor or CME speaker for the following entities Abbott, GlaxoSmithKline, Janssen, Essex, Lundbeck, Otsuka, Gedeon Richter, Servier and Takeda as well as the German Ministry of Science and the German Ministry of Health. Eva Reininghaus has received grants and served as consultant, advisor or CME speaker for the following entities: Janssen and Institut Allergosan. Mikael Landén has received lecture honoraria from Lundbeck. Kazufumi Akiyama has received consulting honoraria from Taisho Toyama Pharmaceutical Co, Ltd. Scott Clark has received grants and served as consultant, advisor or CME speaker for the following entities: Otsuka Austalia, Lundbeck Australia, Janssen-Cilag Australia, Servier Australia. The rest of authors have no conflicts of interest to disclose.
Contributor Information
Marisol Herrera-Rivero, University of Münster.
Anbupalam Thalamuthu, University of New South Wales.
Azmeraw T. Amare, University of Adelaide, AUSTRALIA
Kazufumi Akiyama, Department of Biological Psychiatry and Neuroscience, Dokkyo Medical University.
Nirmala Akula, National Institutes of Health, US Dept of Health & Human Services.
Raffaella Ardau, Hospital University Agency of Cagliari.
Bárbara Arias, Facultat de Biologia and Institut de Biomedicina (IBUB), Universitat de Barcelona, CIBERSAM.
Jean-Michel Aubry, Geneva University Hospitals.
Joanna Biernacka, Mayo Clinic.
Pablo Cervantes, McGill University Health Centre.
Hsi-Chung Chen, National Taiwan University Hospital.
Sven Cichon, Mayo Clinic.
Scott Clark, University of Adelaide.
Cristiana Cruceanu, Max Plank Institute for Psychiatry.
Piotr Czerski, Poznan University of Medical Sciences.
Franziska Degenhardt, University of Bonn.
Maria Del Zompo, University of Cagliari.
J. Raymond DePaulo, Johns Hopkins University.
Peter Falkai, University Hospital LMU.
Andreas J. Forstner, University of Bonn, School of Medicine & University Hospital Bonn
Josef Frank, Central Institute for Mental Health.
Louise Frisen, School of Medicine & University Hospital Bonn.
Mark Frye, Mayo Clinic.
Janice Fullerton, Neuroscience Research Australia.
Maria Grigoroiu-Serbanescu, Alexandru Obregia Clinical Psychiatric Hospital.
Ryota Hashimoto, National Center of Neurology and Psychiatry.
Urs Heilbronner, Institute of Psychiatric Phenomics and Genomics, University Hospital, LMU Munich.
Per Hoffmann, Institute of Human Genetics.
Liping Hou, National Institute of Mental Health Intramural Research Program, National Institutes of Health.
YiHsiang Hsu, Mayo Clinic.
Stéphane Jamain, Univ Paris Est Creteil, INSERM, IMRB.
Layla Kassem, School of Medicine & University Hospital Bonn.
Tadafumi Kato, Laboratory for Molecular Dynamics of Mental Disorders, RIKEN Brain Science Institute, Wako, Saitama 351-0198, Japan.
John Kelsoe, University of California, San Diego.
Po-Hsiu kuo, College of Public Health, National Taiwan University, Taipei, Taiwan.
Ichiro Kusumi, Hokkaido University Graduate School of Medicine.
Mikael Landén, Gothenburg University.
Catharina Lavebratt, Karolinska Institutet.
Mario Maj, University of Naples, Italy.
Mirko Manchia, Dalhousie University.
Cynthia Marie-Claire, INSERM UMR-S 1144.
Lina Martinsson, Karolinska Institutet.
Susan L. McElroy, Lindner Center of Hope / University of Cincinnati
Marina Mitjans, Max Planck Institute of Experimental Medicine, Göttingen, Germany.
Francis Mondimore, Neuroscience Research Australia.
Palmiero Monteleone, University of Salerno, University of Naples SUN.
Caroline Nievergelt, University of California, San Diego.
Tomas Novak, National Institute of Mental Health, Klecany.
Markus Nöthen, School of Medicine & University Hospital Bonn.
Norio Ozaki, Nagoya University.
Sergi Papiol, University Hospital LMU.
Andrea Pfennig, University Hospital Carl Gustav Carus, TU Dresden.
Claudia Pisanu, University of Cagliari.
Andreas Reif, University Hospital Frankfurt, Germany.
Guy Rouleau, McGill University.
Janusz K. Rybakowski, Poznan University of Medical Sciences
Martin Schalling, Karolinska Institutet.
Peter Schofield, Neuroscience Research Australia.
Klaus Oliver Schubert, University of Adelaide.
Eva Schulte, University Hospital, LMU Munich.
Giovanni Severino, University of Cagliari.
Alessio Squassina, Universita degli Studi Di Cagliari.
Thomas Stamm, Charité - Universitätsmedizin Berlin, Campus Charité Mitte.
Fabian Streit, University of Heidelberg.
Alfonso Tortorella, Department of Psychiatry, University of Perugia, Perugia, Italy.
Gustavo Turecki, Douglas Institute, Department of Psychiatry, McGill University.
Eduard Vieta, Hospital Clinic of Barcelona.
Biju Viswanath, National Institute of Mental Health and Neuro Sciences, Bengaluru, Karnataka, India..
Stephanie Witt, University Medical Centre Mannheim.
Peter Zandi, Johns Hopkins University.
Martin Alda, Dalhousie University.
Michael Bauer, University Hospital Carl Gustav Carus.
Francis McMahon, National Institute of Mental Health Intramural Research Program; National Institutes of Health.
Philip Mitchell, University of New South Wales.
Marcella Rietschel, University of Mannheim.
Thomas Schulze, University of Munich.
Bernhard Baune, University of Münster.
References
- 1.Rosenblat JD, McIntyre RS. Bipolar Disorder and Immune Dysfunction: Epidemiological Findings, Proposed Pathophysiology and Clinical Implications. Brain Sci. 2017;7(11):144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tylee DS, Sun J, Hess JL, et al. Genetic correlations among psychiatric and immune-related phenotypes based on genome-wide association data. Am J Med Genet B Neuropsychiatr Genet. 2018;177(7):641–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yuan N, Chen Y, Xia Y, Dai J, Liu C. Inflammation-related biomarkers in major psychiatric disorders: a cross-disorder assessment of reproducibility and specificity in 43 meta-analyses. Transl Psychiatry. 2019;9(1):233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauer M. Lithium: What can we do to overcome the discrepancies between evidence, guideline recommendations and clinical practice? Eur Neuropsychopharmacol. 2022. Jul;60:1–3. [DOI] [PubMed] [Google Scholar]
- 5.Rybakowski JK. Lithium. Eur Neuropsychopharmacol. 2022;57:86–87. [DOI] [PubMed] [Google Scholar]
- 6.Spuch C, López-García M, Rivera-Baltanás T, Rodrígues-Amorím D, Olivares JM. Does Lithium Deserve a Place in the Treatment Against COVID-19? A Preliminary Observational Study in Six Patients, Case Report. Front Pharmacol. 2020;11:557629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Landén M, Larsson H, Lichtenstein P, Westin J, Song J. Respiratory infections during lithium and valproate medication: a within-individual prospective study of 50,000 patients with bipolar disorder. Int J Bipolar Disord. 2021;9(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nassar A, Azab AN. Effects of lithium on inflammation. ACS Chem Neurosci. 2014;5(6):451–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Queissner R, Lenger M, Birner A, et al. The association between anti-inflammatory effects of long-term lithium treatment and illness course in Bipolar Disorder. J Affect Disord. 2021;281:228–234. [DOI] [PubMed] [Google Scholar]
- 10.Manchia M, Adli M, Akula N, et al. Assessment of Response to Lithium Maintenance Treatment in Bipolar Disorder: A Consortium on Lithium Genetics (ConLiGen) Report. PLoS One. 2013;8(6):e65636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nunes A, Trappenberg T, Alda M; international Consortium on Lithium Genetics (ConLiGen). Asymmetrical reliability of the Alda score favours a dichotomous representation of lithium responsiveness. PLoS One. 2020. Jan 27;15(1):e0225353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.International Consortium on Lithium Genetics (ConLi + Gen), Amare AT, Schubert KO, et al. Association of Polygenic Score for Schizophrenia and HLA Antigen and Inflammation Genes With Response to Lithium in Bipolar Affective Disorder: A Genome-Wide Association Study. JAMA Psychiatry. 2018;75(1):65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le Clerc S, Lombardi L, Baune BT, et al. HLA-DRB1 and HLA-DQB1 genetic diversity modulates response to lithium in bipolar affective disorders. Sci Rep. 2021;11(1):17823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schulze TG, Alda M, Adli M, et al. The International Consortium on Lithium Genetics (ConLiGen): an initiative by the NIMH and IGSLI to study the genetic basis of response to lithium treatment. Neuropsychobiology. 2010;62(1):72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hou L, Heilbronner U, Degenhardt F, et al. Genetic variants associated with response to lithium treatment in bipolar disorder: a genome-wide association study. Lancet. 2016;387(10023):1085–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breuer K, Foroushani AK, Laird MR, et al. InnateDB: systems biology of innate immunity and beyond–recent updates and continuing curation. Nucleic Acids Res. 2013;41(Database issue):D1228–D1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schubert KO, Thalamuthu A, Amare AT, et al. Combining schizophrenia and depression polygenic risk scores improves the genetic prediction of lithium response in bipolar disorder patients [published correction appears in Transl Psychiatry. 2022;12(1):278]. Transl Psychiatry. 2021;11(1):606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Das S, Forer L, Schönherr S, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48(10):1284–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyer HV. plinkQC: Genotype quality control in genetic association studies. 2020. DOI 10.5281/zenodo.3934294. [DOI] [Google Scholar]
- 21.Lambert SA, Gil L, Jupp S, et al. The Polygenic Score Catalog as an open database for reproducibility and systematic evaluation. Nat Genet. 2021;53(4):420–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weissbrod O, Kanai M, Shi H, et al. Leveraging fine-mapping and multipopulation training data to improve cross-population polygenic risk scores. Nat Genet. 2022;54(4):450–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Y, Vuckovic D, Ritchie SC, et al. Machine learning optimized polygenic scores for blood cell traits identify sex-specific trajectories and genetic correlations with disease. Cell Genom. 2022;2(1):None. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Folkersen L, Gustafsson S, Wang Q, et al. Genomic and drug target evaluation of 90 cardiovascular proteins in 30,931 individuals. Nat Metab. 2020;2(10):1135–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stahl EA, Breen G, Forstner AJ, et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet. 2019;51(5):793–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Munz M, Wohlers I, Simon E, et al. Qtlizer: comprehensive QTL annotation of GWAS results. Sci Rep. 2020;10(1):20417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu G, Dawson E, Duong A, Haw R, Stein L. ReactomeFIViz: a Cytoscape app for pathway and network-based data analysis. F1000Res. 2014;3:146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8(1):1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaidanovich-Beilin O, Woodgett JR. GSK-3: Functional Insights from Cell Biology and Animal Models. Front Mol Neurosci. 2011;4:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Freland L, Beaulieu JM. Inhibition of GSK3 by lithium, from single molecules to signaling networks. Front Mol Neurosci. 2012;5:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shine B, McKnight RF, Leaver L, Geddes JR. Long-term effects of lithium on renal, thyroid, and parathyroid function: a retrospective analysis of laboratory data. Lancet. 2015;386(9992):461–468. [DOI] [PubMed] [Google Scholar]
- 34.Lieber I, Ott M, Öhlund L, et al. Lithium-associated hypothyroidism and potential for reversibility after lithium discontinuation: Findings from the LiSIE retrospective cohort study. Journal of Psychopharmacology. 2020;34(3):293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ching MS, Shen Y, Tan WH, et al. Deletions of NRXN1 (neurexin-1) predispose to a wide spectrum of developmental disorders. Am J Med Genet B Neuropsychiatr Genet. 2010;153B(4):937–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patial S, Shahi S, Saini Y, et al. G-protein coupled receptor kinase 5 mediates lipopolysaccharide-induced NFκB activation in primary macrophages and modulates inflammation in vivo in mice. J Cell Physiol. 2011;226(5):1323–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Lucia C, Grisanti LA, Borghetti G, et al. G protein-coupled receptor kinase 5 (GRK5) contributes to impaired cardiac function and immune cell recruitment in post-ischemic heart failure. Cardiovasc Res. 2022;118(1):169–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gurevich EV, Gurevich VV. GRKs as Modulators of Neurotransmitter Receptors. Cells. 2020;10(1):52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki J, Imanishi E, Nagata S. Exposure of phosphatidylserine by Xk-related protein family members during apoptosis. J Biol Chem. 2014;289(44):30257–30267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wray NR, Ripke S, Mattheisen M, et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. 2018;50(5):668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Çakır U, Tuman TC, Yıldırım O. Increased neutrophil/lymphoctye ratio in patients with bipolar disorder: a preliminary study. Psychiatr Danub. 2015;27(2):180–184. [PubMed] [Google Scholar]
- 43.Singh D, Guest PC, Dobrowolny H, et al. Changes in leukocytes and CRP in different stages of major depression. J Neuroinflammation. 2022;19(1):74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hwang JP, Tsai SJ, Hong CJ, Yang CH, Hsu CD, Liou YJ. Interleukin-1 beta – 511C/T genetic polymorphism is associated with age of onset of geriatric depression. Neuromolecular Med. 2009;11(4):322–327. [DOI] [PubMed] [Google Scholar]