Abstract
Repeat expansion disorders (REDs) are a devastating group of predominantly neurological diseases. Together they are common, affecting 1 in 3,000 people worldwide with population-specific differences. However, prevalence estimates of REDs are hampered by heterogeneous clinical presentation, variable geographic distributions, and technological limitations leading to under-ascertainment. Here, leveraging whole genome sequencing data from 82,176 individuals from different populations we found an overall carrier frequency of REDs of 1 in 340 individuals. Modelling disease prevalence using genetic data, age at onset and survival, we show that REDs are up to 3-fold more prevalent than currently reported figures. While some REDs are population-specific, e.g. Huntington’s disease type 2, most REDs are represented in all broad genetic ancestries, including Africans and Asians, challenging the notion that some REDs are found only in European populations. These results have worldwide implications for local and global health communities in the diagnosis and management of REDs both at local and global levels.
Repeat expansion disorders (REDs) are a heterogeneous group of conditions which mainly affect the nervous system, and include myotonic dystrophy (DM), Huntington’s disease (HD), and the commonest inherited form of amyotrophic lateral sclerosis and frontotemporal dementia (C9orf72-ALS/FTD)1. REDs are caused by the same underlying mechanism: the expansion of simple repetitive DNA sequences within their respective genes. The mutational process is gradual: normal alleles are usually passed stably from parent to child with rare changes in repeat size; intermediate-size (called “premutation” alleles in some cases) are more likely to expand into the disease range, both in somatic and germline cells. This is relevant for genetic counselling because the offspring of an intermediate allele carrier can be affected by REDs.
Previous studies have estimated that REDs affect 1 in 3,000 people2, with population differences at specific RED loci. Among the most common REDs are myotonic dystrophy type 1 (DM1) and HD. DM1 affects 1 in 8,000 people worldwide, ranging from 1 in 10,000 in Iceland to 1 in 100,000 in Japan3. Similarly, the frequency of HD is 13.7 in 100,000 in the general population, varying between 17.2 in 100,000 in the Caucasian population, and 0.1-2 in 100,000 in Asians and Africans3. A repeat expansion at C9orf72 causes both frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (ALS): in Europeans it is estimated that the prevalence of C9orf72-FTD is 0.04-134 in 100,000, and C9orf72-ALS is 0.5-1.2 in 100,0004. The spinocerebellar ataxias (SCAs) are a group of rare neurodegenerative disorders, with a worldwide prevalence of SCA of 2.7-47 cases per 100,0005 and wide regional variations mainly due to founder effects, with SCA3 being the most common form worldwide, followed by SCA2 and SCA6, SCA1, and SCA6.
Despite their broad distribution in human populations, the few global epidemiological studies that have been performed on these disorders have focused on European populations. In these studies, prevalence estimates are either population-based, in which affected individuals are identified based on clinical presentation, or assessed based on the presence of a relative with a RED who are then genetically tested. Given that one of the most striking features of REDs is that they can present with markedly diverse phenotypes, REDs can remain unrecognised, leading to underestimation of the disease prevalence7.
Large scale analyses of REDs have been limited by repeat expansion profiling techniques, which historically have relied on polymerase chain reaction-based (PCR) assays or Southern blots, which by nature are targeted assays by nature and can be difficult to scale. So far, the largest population study of the genetic frequency REDs involved the analysis of 14,196 individuals of European ancestry8.
In the last few years, bioinformatic tools have been developed to profile DNA repeats from short-read whole genome sequencing data (WGS). We have recently shown that disease-causing repeat expansions can be detected from WGS with high sensitivity and specificity, making large-scale WGS datasets an invaluable resource to analyse the frequency and distribution of disease-causing repeat expansions2.
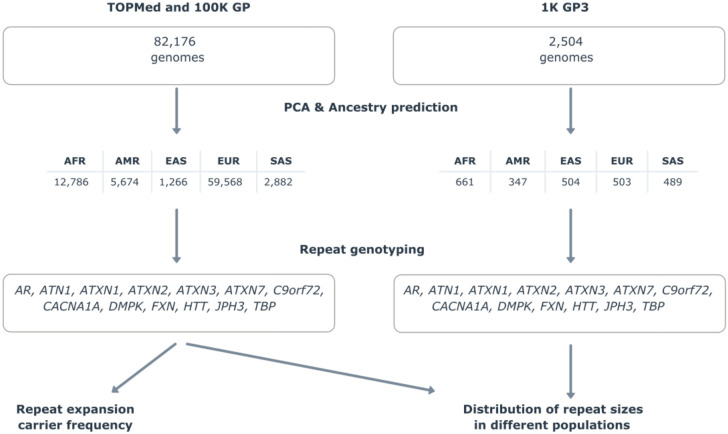
We here analyse disease-causing short tandem repeat (STR) loci in 82,176 individuals from two large-scale medical genomics cohorts with high-coverage WGS and rich phenotypic data: the 100,000 Genomes Project (100K GP) and Trans-Omics for Precision Medicine (TOPMed). The 100K GP is a programme to deliver genome sequencing of people with rare diseases and cancer within the National Health Service (NHS) in the United Kingdom. TOPMed is a clinical and genomic programme focused on elucidating the genetic architecture and risk factors of heart, lung, blood, and sleep disorders from the National Institute of Health (NIH). First, we selected WGS data generated using PCR-free protocols and sequenced with paired-end 150 bp reads (Table S1). To avoid overestimating the carrier frequency of REDs, we excluded individuals with neurological diseases, as their recruitment was driven by the fact that they had a neurological disease potentially caused by an STR expansion. We then performed relatedness and principal component analyses to identify a set of genetically unrelated individuals and predict broad genetic ancestries based on 1,000 Genomes Project super-populations9. The resulting dataset comprised a cross-sectional cohort of 82,176 genomes from unrelated individuals (median age 61, Q1-Q3: 49-70, Table S2, Fig. S1), genetically predicted to be of European (n=59,568), African (n=12,786), American (n=5,674), South Asian (n=2,882), and East Asian (n=1,266) descent (Fig. S2).
We analysed repeats in RED genes for which WGS has been shown to be able to accurately discriminate between normal and pathogenic alleles2, representing a broad spectrum of the most common REDs (Table S3, Table S4). Our analysis workflow (Fig. 1) included profiling each STR locus, followed by quality control of all alleles (visual inspection of pileup plots as previously described2) predicted to be larger than the premutation threshold (Table S5). Clinical and demographic data available on all individuals carrying a pathogenic repeat are listed in Table S6. As our cohort comprises data from different genetically predicted populations, we also compared genotypes generated by WGS and PCR for 1,006 alleles and showed that the accuracy for repeat sizing by WGS was not affected by genetic ancestry (online materials, Table S7, Fig. S3).
Figure 1.
Technical flowchart
In total, there were 242 (0.29%) individuals carrying a fully-expanded repeat, and 798 (0.97%) individuals carrying a repeat in the premutation range (Table S5), meaning that frequency of individuals carrying full-expansion and premutation alleles among this large cohort is 1 in 340 people and 1 in 103 people respectively.
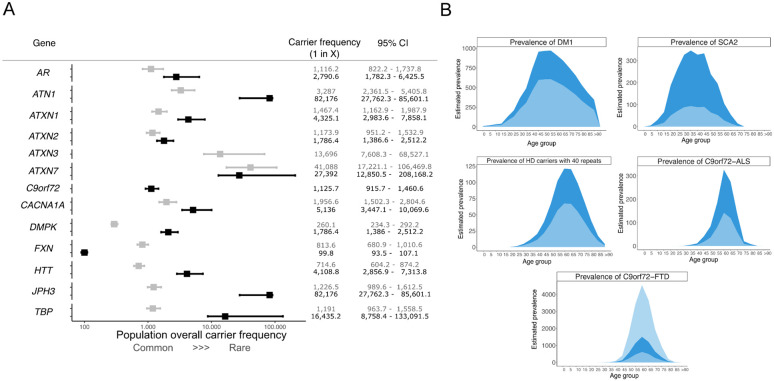
The most common pathogenic expansions in this cohort were those in C9orf72 (1 in 1,126) that cause ALS-FTD, followed by expansions in DMPK causing DM1 (1 in 1,786). Surprisingly, we found expansions in the spinocerebellar ataxia 2 gene ATXN2 to be almost as common as those in DMPK, followed by expansions in AR that cause spinal and bulbar muscular atrophy and in HTT that cause Huntington disease (Fig. 2A). By contrast, expansions in JPH3 that cause Huntington disease-like 2 and in ATN1 that cause dentatorubral-pallidoluysian atrophy were very rare, with only a single individual at each locus identified with a repeat allele in the pathogenic range. No individuals were identified with pathogenic expansions in ATXN3 (SCA3).
Figure 2.
A) Forest plot with combined overall carrier frequency and 95% CI values in the combined 100K GP and TOPMed datasets together. Grey and black boxes show premutation and full-mutation overall carrier frequencies for each locus, respectively. B) Modelling of disease prevalence by age of DM1, SCA2, HD, C9orf72-ALS, and C9orf72-FTD. Age bins are 5-years each. Estimated prevalence (dark blue area) is compared to the reported prevalence from the literature (light blue area). For C9orf72-FTD, given the wide range of the reported disease prevalence12,13,both lower and upper limits are plotted in light blue.
REDs have variable age at onset, disease duration, and penetrance. Therefore, the carrier frequency cannot be directly translated into disease frequency (i.e. prevalence). To estimate the prevalence of REDs using genetic data, we modelled the distribution by age of the most common REDs (C9orf72-ALS/FTD, DM1, HD, and SCA2) in the UK population using data from the Office of National Statistics, taking into account the different age of onset, penetrance, and impact on survival of each RED10. We found an up to three-fold increase in the predicted disease prevalence compared to currently reported figures based on clinical observation, depending on the RED (Fig. 2B). We estimated a prevalence for DM1 of 20 per 100,000, 1.6 times higher than the estimated prevalence from clinical data7. Similarly, prevalence estimates for SCA2 were 3.7 per 100,000, 3.7 times higher than known SCA2 prevalence (1 per 100,000)11. For HD, we estimated an overall prevalence of 6.7 per 100,000. While this figure seems lower than currently reported for HD, the majority of individuals with a pathogenic expansion in HTT in our cohort (12 out of 20) carry alleles with 40 repeats. Given the well-established relationship between HTT repeat length and age at onset, we modelled the HD prevalence taking into account repeat-length and found that 1.3 per 100,000 with 40 repeats are estimated to develop HD (online methods). This is 1.8 times higher than the reported number of affected patients among these carriers (0.72 per 100,000) (personal communication, DHM). Since C9orf72 expansions cause both ALS and FTD, we modelled the prevalence of both diseases separately, providing an estimated disease prevalence of 1.84 per 100,000 for C9orf72-ALS - over two times higher than previous estimates12,13 , and 8.15 per 100,000 for C9orf72-FTD, within the wide reported range of C9orf72-FTD (online methods, Fig. 2B).
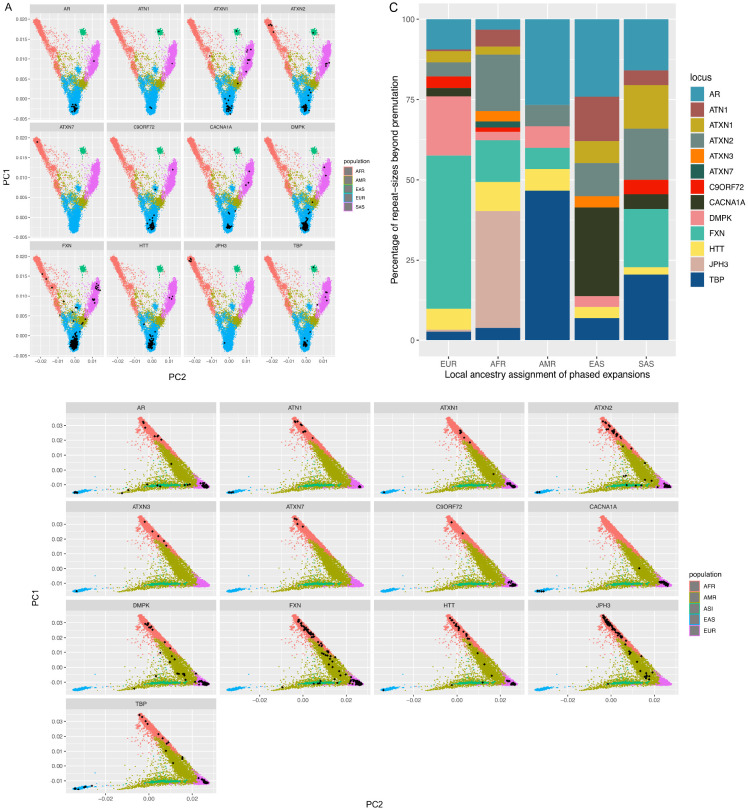
The prevalence of individual REDs varies considerably based on geographic location (Fig. 3A,B). Hence, we then set out to analyse whether these differences are reflected in the broad genetic ancestries of our cohort (Table S8), given the broad representation of different populations in this cohort. For this analysis, we looked at the proportion of abnormal alleles in each population after local ancestry assignment at each RED locus. In agreement with current known epidemiological studies, we observed that the most common abnormal alleles in Europeans are those in DMPK, HTT, and C9orf72, in JPH3 in Africans, TBP, ATN1, and CACNA1A in East Asians, ATXN1 and AR in South Asians. Some expansions like those in ATXN2, ATXN1, and AR are more widely represented across all populations. Surprisingly, we identified pathogenic expansions within C9orf72 and HTT in Africans and South Asians, both of which were previously thought to be found mostly in European populations. Given that the initial ancestry assignments were based on genome-wide data, we performed local ancestry analysis to check for admixture in these individuals, and confirmed that the expanded repeat alleles were carried on haplotypes of African and South Asian ancestry (Fig. 3C, Table S9)
Figure 3.
Principal component (PC) values on all genomes within (A) the 100K GP and B) TOPmed cohorts. Black dots represent genomes having a repeat-size beyond premutation and full-mutation range, split by gene. C) Local ancestry bar plot showing the repeats beyond the premutation threshold by super population within 100K GP and TOPMed cohorts.
We then analysed the complete distribution of repeat sizes in each population in the 100K GP and TOPMed datasets. Here we included WGS data from the 1,000 Genomes Project (1K GP3)9 to reproduce the findings as this cohort includes a broad representation of genetic ancestries. Fig. S4 and S5 show the distribution of repeat sizes across 13 genes among the three genomic datasets. While the overall distributions of repeat sizes are similar when comparing across ancestries for most loci, by contrast, for others such as AR, ATN1, HTT, and TBP, repeat lengths in genomes of African origin are significantly shorter (p<10−16) than other ancestries (Table S10, Fig. S5). This pattern is consistent across all three datasets.
In summary, this study provides the first unbiased, worldwide population-based estimates of carrier frequency and expected disease prevalence of REDs. It shows that: i) the carrier frequency of REDs is approximately ten times higher than the previous estimates based on clinical observations, and that, based on population modelling, we estimate that REDs are predicted to affect up to three times more individuals than are currently recognized clinically; ii) while some REDs are population-specific like JPH3, the majority are observed in all ancestral populations, challenging the notion that some REDs (e.g. C9orf72) are associated with population specific founder effects; iii) an appreciable proportion of the population (1 in 103) carry alleles in the premutation range, and are therefore at risk of having children with REDs.
Different factors are likely to contribute to the increased prevalence estimate in the current study compared to others. First, our study estimates disease prevalence based on carrier frequency in large admixed cohorts, as opposed to studies based on the identification of clinically affected individuals in smaller populations. As REDs have variable clinical presentation and age at onset, it is likely that many individuals with REDs in a given population remain undiagnosed and undetected in clinically based prevalence studies. Second, even in symptomatic patients, there is a significant delay in diagnosis in many individuals with REDs2. The clinical data available on people carrying a pathogenic repeat expansion in this study is not suggestive of the corresponding RED. This might be explained by reduced or age-related penetrance of the mutation, which may go on to present at a later age. This is confirmed by the fact that we observed a large number of individuals carrying repeats in the lower end of the pathogenic range (e.g. HTT and ATXN2, and DMPK Table S5), indicating that they will likely develop milder disease later in life. For example, it is well documented that carriers of small DMPK expansions (50–100 repeats) have milder disease with clinical features that may go unnoticed, especially early in their disease course14. Finally, these individuals may carry genetic modifiers of REDs.
One limitation of this study is that WGS cannot accurately size repeats larger than the sequencing read length (150 bp). For this reason, we did not assess some REDs like Fragile X syndrome, as WGS cannot distinguish between premutation and pathogenic full mutation alleles2. Another limitation of this study is the potential for recruitment bias, especially within the TOPMed cohort: individuals with an overt REDs have a reduced likelihood of being recruited to such studies because of the severity of their disease. For example, we note the absence of expansions in ATXN3, the most commonly reported SCA in patients affected by spinocerebellar ataxia.
Both 100K GP and TOPMed datasets are Euro-centric, comprising over 62% of European samples. TOPMed is more diverse, with 24% and 17% of African and American genomes respectively, which are only present at 3.2% and 2.1% frequency in the 100K GP. East and South Asian backgrounds are underrepresented in both datasets, limiting the ability to detect rarer repeat expansions.
Further analyses on more heterogeneous and diverse large scale WGS datasets are necessary not only to confirm our findings, but also to shed light into additional ancestries. With regards to this, there are multiple ongoing projects with Asian populations15,16. Countries including China, Japan, Qatar, Saudi Arabia, India, Nigeria, and Turkey have all launched their own genomics projects during the last decade17. Analysing genomes from these coming genomic programmes will yield more detail on the prevalence of REDs around the world.
Despite efforts to estimate the frequency globally and locally, there is uncertainty surrounding the true prevalence of REDs, limiting the knowledge of the burden of disease required to secure dedicated resources to support health services, such as the estimation of the numbers of individuals profiting from drug development and novel therapies, or participating in clinical trials.
The finding that REDs are more prevalent than was previously thought has important implications. Clinicians should have a higher index of suspicion when a patient presents with symptoms compatible with a RED, and clinical diagnostic pathways should facilitate genetic testing for REDs. The presence of expansions within HTT and C9orf72 in African and Asian populations supports diagnostic testing for them in people presenting with features of Huntington’s disease and ALS-FTD whatever their ethnicity. There are currently no disease modifying treatments for REDs, however both disease specific treatments, and drugs which target repeat expansions more generally are in development. We have established that the numbers of people who may benefit from such treatments are greater than previously thought.
Supplementary Material
Funding:
Medical Research Council, Department of Health and Social Care, National Health Service England, National Institute for Health Research
REFERENCES
- 1.Paulson H. Repeat expansion diseases. Handb. Clin. Neurol. 147, 105–123 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ibañez K. et al. Whole genome sequencing for the diagnosis of neurological repeat expansion disorders in the UK: a retrospective diagnostic accuracy and prospective clinical validation study. Lancet Neurol. 21, 234–245 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bird T. D. Myotonic dystrophy type 1. GeneReviews®[Internet] (2019). [Google Scholar]
- 4.Gossye H., Engelborghs S., Van Broeckhoven C. & van der Zee J. C9orf72 Frontotemporal Dementia and/or Amyotrophic Lateral Sclerosis. (University of Washington, Seattle, 2020). [Google Scholar]
- 5.Teive H. A. G., Meira A. T., Camargo C. H. F. & Munhoz R. P. The Geographic Diversity of Spinocerebellar Ataxias (SCAs) in the Americas: A Systematic Review. Mov Disord Clin Pract 6, 531–540 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schöls L., Bauer P., Schmidt T., Schulte T. & Riess O. Autosomal dominant cerebellar ataxias: clinical features, genetics, and pathogenesis. Lancet Neurol. 3, 291–304 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Johnson N. E. et al. Population-Based Prevalence of Myotonic Dystrophy Type 1 Using Genetic Analysis of Statewide Blood Screening Program. Neurology 96, e1045–e1053 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gardiner S. L. et al. Prevalence of Carriers of Intermediate and Pathological Polyglutamine Disease-Associated Alleles Among Large Population-Based Cohorts. JAMA Neurol. 76, 650–656 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.1000 Genomes Project Consortium, {fname} et al. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zanovello M. et al. Unexpected frequency of the pathogenic AR CAG repeat expansion in the general population. Brain (2023) doi: 10.1093/brain/awad050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhandari J., Thada P. K. & Samanta D. Spinocerebellar Ataxia. (StatPearls Publishing, 2022). [PubMed] [Google Scholar]
- 12.Van Mossevelde S., Engelborghs S., van der Zee J. & Van Broeckhoven C. Genotype-phenotype links in frontotemporal lobar degeneration. Nat. Rev. Neurol. 14, 363–378 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Hogan D. B. et al. The Prevalence and Incidence of Frontotemporal Dementia: a Systematic Review. Can. J. Neurol. Sci. 43 Suppl 1, S96–S109 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Thornton C. A. Myotonic dystrophy. Neurol. Clin. 32, 705–19, viii (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu D. et al. Large-Scale Whole-Genome Sequencing of Three Diverse Asian Populations in Singapore. Cell 179, 736–749.e15 (2019). [DOI] [PubMed] [Google Scholar]
- 16.GenomeAsia100K Consortium. The GenomeAsia 100K Project enables genetic discoveries across Asia. Nature 576, 106–111 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar R. & Dhanda S. K. Current Status on Population Genome Catalogues in different Countries. Bioinformation 16, 297–300 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.