Abstract
The continued efficacy of glycopeptide antibiotics (GPAs) against Gram-positive bacteria is challenged by the emergence and spread of GPA-resistant pathogens, particularly vancomycin-resistant enterococci (VRE). The growing frequency of GPA resistance propels the need for innovative development of more effective antibiotics. Unlike canonical GPAs like vancomycin, Type V GPAs adopt a distinct mode of action by binding peptidoglycan and blocking the activity of autolysins essential for cell division, rendering them a promising class of antibiotics for further development. In this study, the Type V GPA, rimomycin A, was modified to generate 32 new analogues. Compound 17, derived from rimomycin A through N-terminal acylation and C-terminal amidation, exhibited improved anti-VRE activity and solubility. In a VRE-A neutropenic thigh infection mouse model, compound 17 significantly lowered the bacterial load by 3–4 orders of magnitude. This study sets the stage to develop next-generation GPAs in response to growing VRE infections.
Introduction
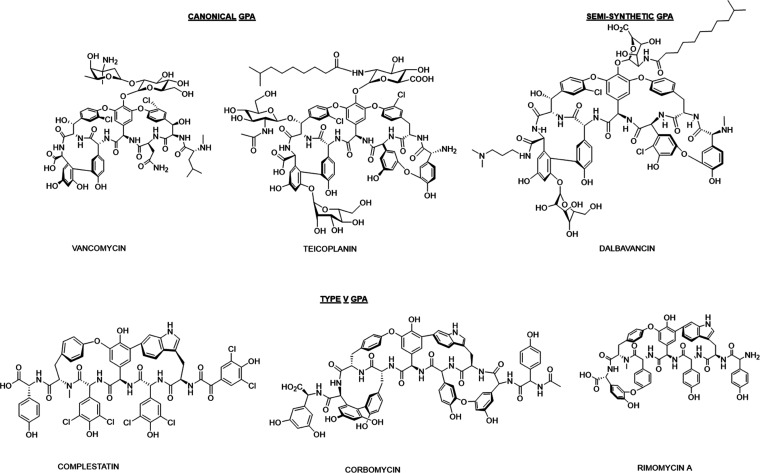
Since the introduction of the natural products vancomycin in the 1950s and teicoplanin 25 years later (Figure 1), glycopeptide antibiotics (GPAs) have been a front-line therapy to treat severe infections caused by Gram-positive bacteria, including methicillin-resistant Staphylococcus aureus (MRSA), Enterococcus sp., Clostridium difficile, and others.1 GPAs block bacterial cell wall biosynthesis by binding the terminal d-Alanine-d-Alanine (d-Ala-d-Ala) dipeptide of the peptidoglycan and lipid II stem peptide, thereby inhibiting essential interstrand crosslinking and extension of the polymer.1−4 The near-ubiquity of the d-Ala-d-Ala dipeptide in bacterial pathogens ensured excellent antibacterial spectrum and the slow emergence of resistance for several decades.5 Nevertheless, some environmental bacteria and clinically dominant GPA-resistant enterococci (VRE) have acquired the vanHAX resistance gene cluster that remodels their peptidoglycan by replacing d-Ala-d-Ala with d-Alanine-d-Lactate (d-Ala-d-Lac).6−8 This isosteric change removes a critical amide bond and introduces an unfavorable electronic clash,9 thereby decreasing the affinity of the antibiotic 1000-fold and elevating the GPA minimum inhibitory concentration (MIC) as much as 100-fold.4 Two major variants of d-Ala-d-Lac-producing VRE differ by the response of the two-component regulatory system, VanR and VanS, that control vanHAX expression.10 VRE-A can be induced by vancomycin and teicoplanin, while VRE-B is responsive only to vancomycin. A second resistance strategy in some rarer enterococcal species, VRE-C, exchanges d-Ala-d-Ala for d-Ala-d-Ser, resulting in up to a 6-fold increase in MIC.11−13 Meanwhile, resistance to vancomycin has also been reported in vancomycin intermediate S. aureus (VISA) through thickening of their cell wall.14
Figure 1.
Glycopeptide antibiotics. Canonical GPAs like vancomycin and teicoplanin and semisynthetic derivatives such as dalbavancin bind lipid II and inhibit peptidoglycan synthesis. In contrast, Type V GPAs such as complestatin, corbomycin, and rimomycin A bind peptidoglycan and inhibit cell division.
The emergence and spread of GPA resistance threaten their clinical efficacy and have fueled the development of next-generation GPAs.15,16 For example, three semisynthetic GPAs, including telavancin,17 dalbavancin,18 and oritavancin,19 with improved antibiotic activity toward some resistant organisms, have been approved for clinical use; however, each has unique pharmacological and toxicity challenges.16 Consequently, there is a growing need for new leads for Gram-positive-directed antibiotics that can overcome GPA resistance.
Using a genome mining approach, we identified several members of a new class of GPA-like antibiotic, previously classified as Type V GPAs (Figure 1).20−23 Unlike canonical GPAs, which are heptapeptides, Type V GPAs range from 7 to 9 amino acids in length, lack sugar modifications, and tend to be more hydrophobic. We established that these antibiotics do not bind d-Ala-d-Ala and consequently retain efficacy against GPA-resistant isolates such as VRE. The Type V GPAs suppress bacterial growth by binding to peptidoglycan and blocking the activity of autolysins, the endogenous cell wall hydrolases essential for cell division.21−23 Furthermore, autolysins play critical roles in biofilm formation and programmed cell death.24 This unique mode of action, which evades known GPA resistance mechanisms, along with their low frequency of resistance (<10–8), position Type V GPAs as promising lead compounds for new antibiotic development. Nevertheless, Type V GPAs suffer from low yield from their natural producers, poor solubility, and limited sites on the molecules suitable for semisynthetic functionalization.
In a recent follow-up genome mining program, we identified a new Type V GPA, rimomycin (Figure 1), active against VRE and mycobacteria.23 The antimycobacterial activity is uniquely observed in rimomycin compared to other characterized Type V GPAs. Rimomycin, like kistamycin,25,26 another Type V GPA, possesses a rare tricyclic heptapeptide scaffold and a free N-terminal primary amine, allowing modification through reductive alkylation or acylation reactions to expand its chemical diversity. Using our glycopeptide antibiotic heterologous expression (GPAHex) synthetic biology platform, rimomycin can be produced on gram scale in the lab, providing an ideal Type V GPA candidate for semisynthesis. Rimomycin’s relatively small size (7 amino acids), the presence of unmodified N- and C-termini that offer nucleophilic sites for chemical derivatization, and the access to gram quantities of the natural product backbone enabled us to embark on a program of semisynthesis to explore the potential of improving rimomycin’s antibiotic and physical properties for future development.
Here, we describe the optimization of rimomycin A production from an engineered rimomycin biosynthetic gene cluster expressed in the GPAHex platform. We report the synthesis of 32 new rimomycin analogues by N- and C-terminal functionalization through reductive alkylation-acylation and carboxylic acid amidation chemistry separately and in combination. We also show that compound 17 offers improved antibacterial activity, solubility, and efficacy in an animal model of VRE infection. This study demonstrates the power of a combined synthetic biology-semisynthesis program in developing next-generation GPAs.
Results and Discussion
Halogenase Deletion Enriches Rimomycin A Production
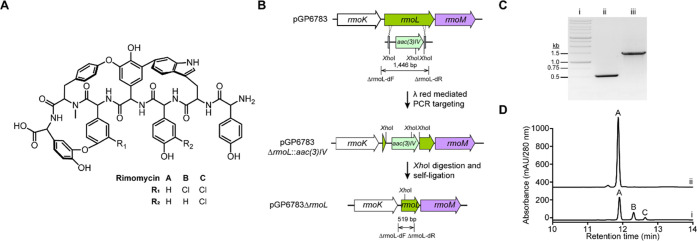
Rimomycins are produced as a mixture of the varyingly chlorinated analogues rimomycins A, B, and C in the heterologous expression strain, Streptomyces coelicolor M1154/pAMX4/pGP6783 (Figure 2A). Rimomycins B and C are derived from rimomycin A through mono-chlorination of 4-hydroxyphenylglycine 5 (Hpg5) and di-chlorination of Hpg3 and Hpg5, respectively. Although halogenation modestly improved the antibacterial activity (2- to 4-fold) of rimomycin,23 the production of a mixture of chlorinated rimomycins makes the isolation of individual halogenated compounds in sufficient quantities for downstream study impractical. In order to drive rimomycin output to a single compound in support of downstream semisynthesis studies and to simplify purification of the parent compound, the halogenase gene-rmoL in the rimomycin BGC was deleted from plasmid pGP6783 (Figure 2B,C) using λ-red-mediated PCR targeting.27 The rmoL deleted version of pGP6783 was mobilized into S. coelicolor M1154/pAMX4 for integration into the chromosome and heterologous expression of rimomycin. As expected, halogenated rimomycins B and C were absent from the high-performance liquid chromatography (HPLC) profile, and only rimomycin A was produced in the engineered strain (Figure 2D).
Figure 2.
Deletion of halogenase-encoding rmoL improves rimomycin A production. (A) Chemical structure of rimomycins A, B, and C. (B) Scheme for in-frame deletion of rmoL gene from pGP6783. (C) DNA gel electrophoresis verifies rmoL deletion. Lane i, 1 kb DNA ladder; lane ii, PCR product amplified from rmoL deletion mutant, pGP6783ΔrmoL; lane iii, PCR product amplified from the wild-type plasmid, pGP6783. (D) HPLC chromatograms of rimomycins. Trace i, S. coelicolor M1154/pAMX4/pGP6783; trace ii, S. coelicolor M1154/pAMX4/pGP6783ΔrmoL.
Semisynthesis and Structure–Activity Relationship (SAR) of Rimomycin Analogues
Compared to most Type V GPAs, which are substituted at the N-terminal amino acid, rimomycin A possesses a free primary amine and a C-terminal carboxylic acid available for chemical modification. Semisynthetic modifications on glycopeptide Type I-IV core to afford compounds with multimodal mechanism of action has been extensively studied and recently reviewed.16,28,29 Our initial studies focused on determining which changes can improve the compounds’ antibacterial activity and solubility. Compounds 1–3 were prepared by alkylation of the free amine and carboxylic acid of rimomycin A with halogenated compounds in the presence of triethylamine (TEA) at 56 °C, for 20 h in dimethyl sulfoxide (DMSO)/N,N-dimethylformamide (DMF) (3:1) (Scheme 1a). These modifications significantly impaired the antibacterial properties compared to the parent rimomycin A. Compounds 5–11 were prepared by reacting either alkyl bromides, substituted alkyl bromides, or various amines with the free carboxylic acid group of N-t-Boc-protected rimomycin A (4) (Scheme 1b,c), followed by Boc deprotection (Scheme 1d). Inspired by the semisynthetic GPA dalbavancin18 (Figure 1), we introduced N-dimethyl propyl diamine at the C-terminus of rimomycin A, which is reported to improve the antimicrobial efficacy of dalbavancin toward Staphylococci.(30) Introducing a positive charge at the C-terminus (6–11) significantly improved activity against Mycobacterium smegmatis and VRE but not MRSA (Table 1).
Scheme 1. Synthetic Route to Rimomycin A Analogues 1–11.
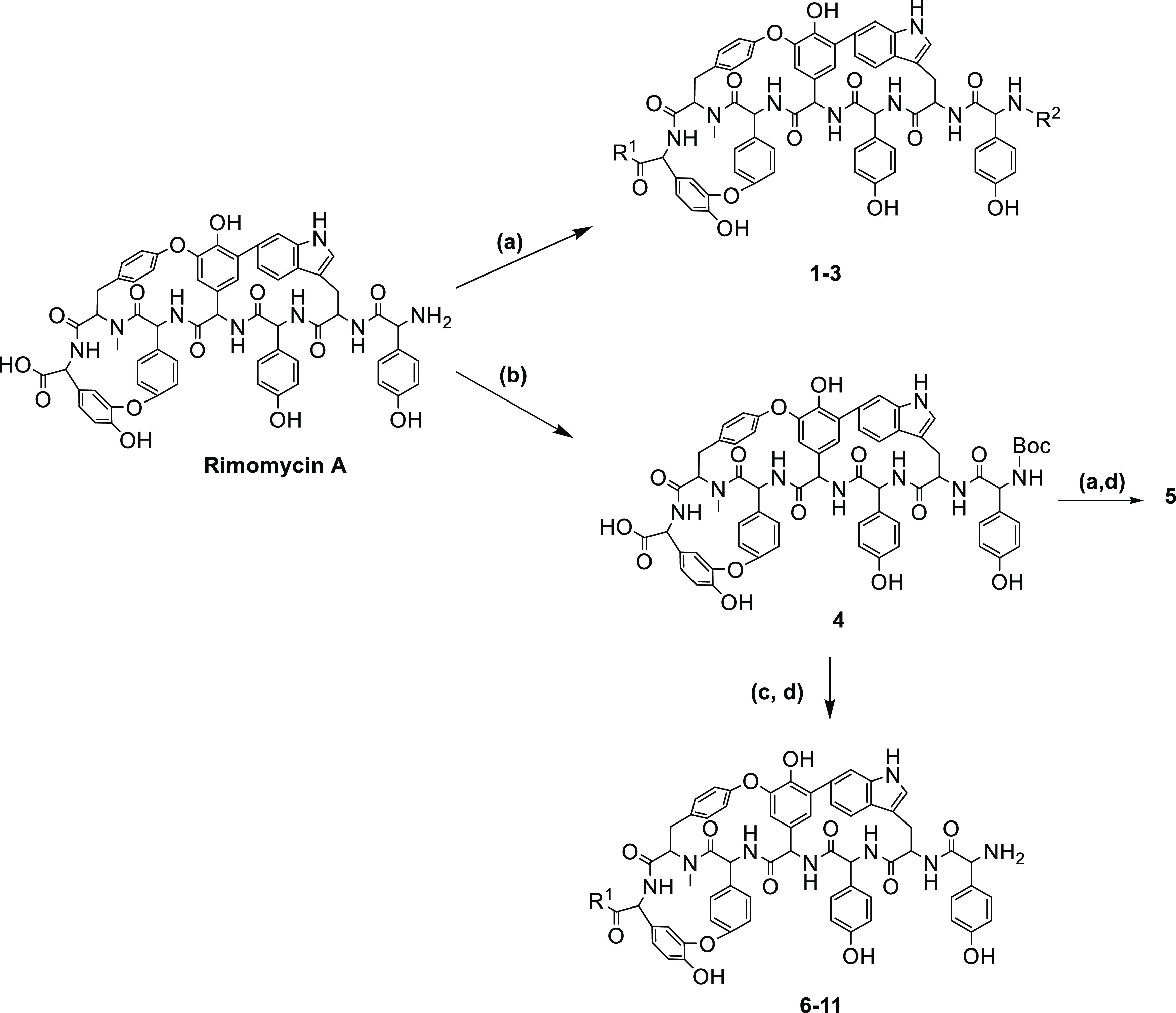
(a) Alkyl halides, TEA, 56 °C, 20 h; (b) (Boc)2O, DMSO/MeOH (1:1), TEA; (c) PyBop, NMM, corresponding amine, 50% DMF/DMSO, 1 h at room temperature (RT); (d) dichloromethane (DCM)/trifluoroacetic acid (TFA) (1:1), 20 min, RT.
Table 1. Antimicrobial Activity of Rimomycin A Analogues 1–17 (μg/mL).
| compound ID | R1 | R2 | S. aureus ATCC 33591 | M. smegmatis mc2155 | VRE-Aa | VRE-Bb | B. subtilis 168 |
|---|---|---|---|---|---|---|---|
| vancomycin | - | - | 2 | 4 | >128 | 32 | 0.5 |
| rimomycin A | OH | H | 8 | 16 | 8–16 | 8–16 | 2 |
| 1 | -O-CH2-CH2-OH | -CH2CH2-OH | 32 | 16 | 32 | 16 | 4 |
| 2 | -O-CH2CH2-SO3H | -CH2CH2-SO3H | >128 | >128 | >128 | >128 | 32 |
| 3 | -O-(CH2)6-B(OH)2 | -CH2-(CH2)5-B(OH)2 | >128 | 16 | 64 | 64 | 32 |
| 4 | OH | Boc | 16 | 128 | 16 | 8 | 4 |
| 5 | -O-(CH2)7CH3 | H | >128 | >128 | >128 | 32 | 64 |
| 6 | -NH-(CH2)3-NH2 | H | 64 | 1 | 1–2 | 4 | 4 |
| 7 | -NH-(CH2)2-NH-CH3 | H | 64 | 0.5–1 | 1–2 | 4 | 4 |
| 8 | -NH-(CH2)3-N(CH3)2 | H | >128 | 0.5 | 2–4 | 16 | 8 |
| 9 | -NH-(CH2)3-N+(CH3)3 | H | >128 | 0.5 | 4 | 16 | 4 |
| 10 | -NH-(CH2)3-NH-(CH2)3-N(CH3)2 | H | >128 | 1 | 2–4 | 16 | 8 |
| 11 | -NH-(CH2)3-N+(CH3)-pyrrolidine | H | >128 | 1 | 8 | 64 | 32 |
| 12 | OH | -(CH2)3-CH3 | 16 | >128 | 32 | 16 | 4 |
| 13 | OH | -(CH2)5-CH3 | 4 | >128 | 4 | 4 | 8 |
| 14 | OH | -(CH2)7-CH3 | 8 | >128 | 16 | 8 | 32 |
| 15 | OH | -(CH2)10-CH3 | 16 | >128 | 16 | 4–8 | 128 |
| 16 | OH | -CO-(CH2)5-NH2 | 8–16 | 16 | 2 | 4 | 1 |
| 17 | -NH-(CH2)3-N(CH3)2 | -CO-(CH2)5-NH2 | 8–16 | 0.5–1 | 0.5 | 1 | 1 |
Enterococcus faecium ATCC 700221.
Enterococcus faecalis ATCC 51299.
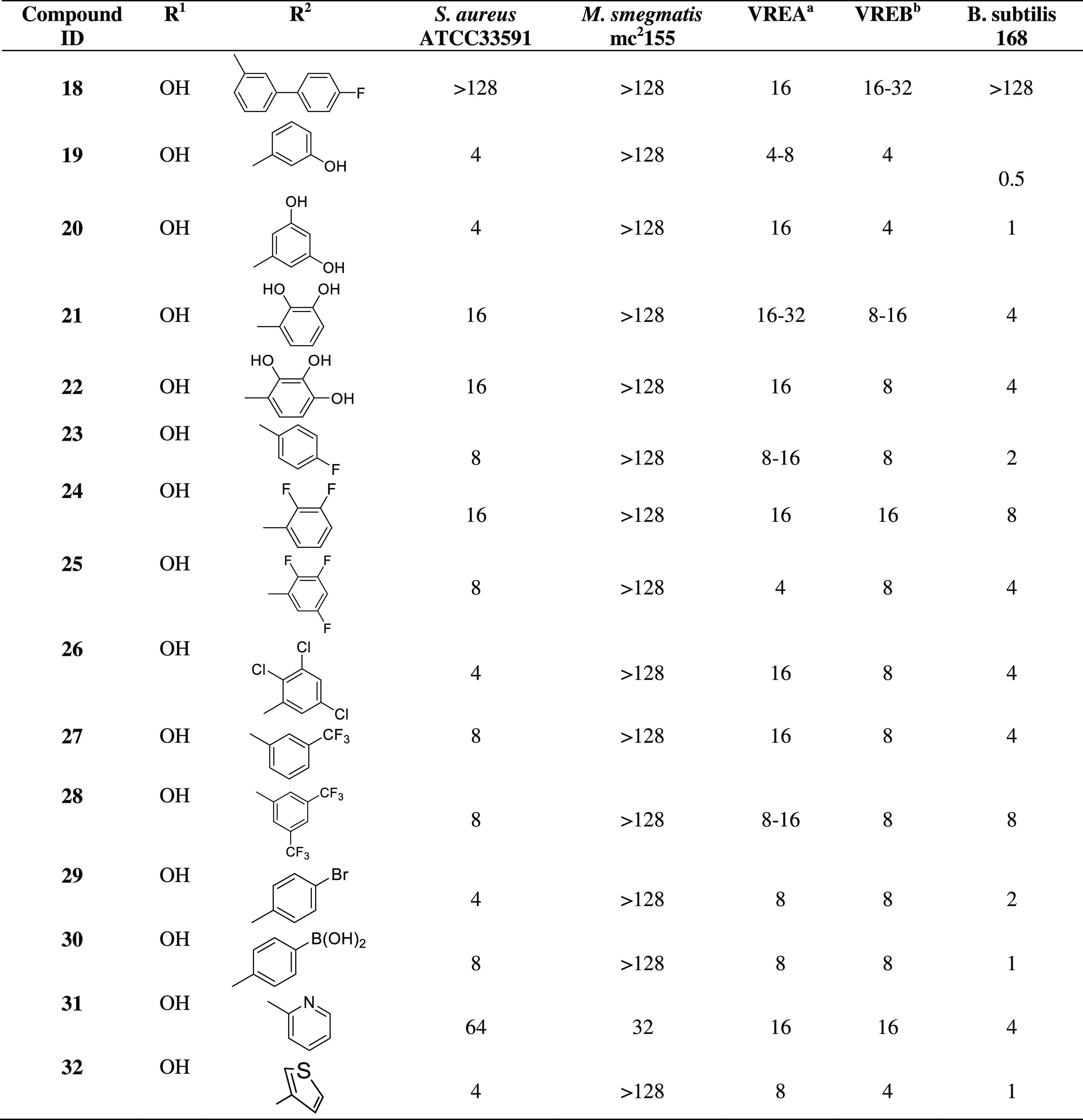
The impact of fatty acids on vancomycin conjugates has been addressed in recent publications.31 Next, we set up to study the effect of different alkyl chain length substituents on the N-terminal by reductive alkylation (Scheme 2, analogues 12–15). Interestingly, compound 13 having hexyl substituent on the N-terminal showed the most promising activity against MRSA and VRE-A but lost the effect of the parent rimomycin A toward the mycobacterial strain used in the study.
Scheme 2. Reductive Alkylation Synthesis of Analogues 12–15 and 18–32.
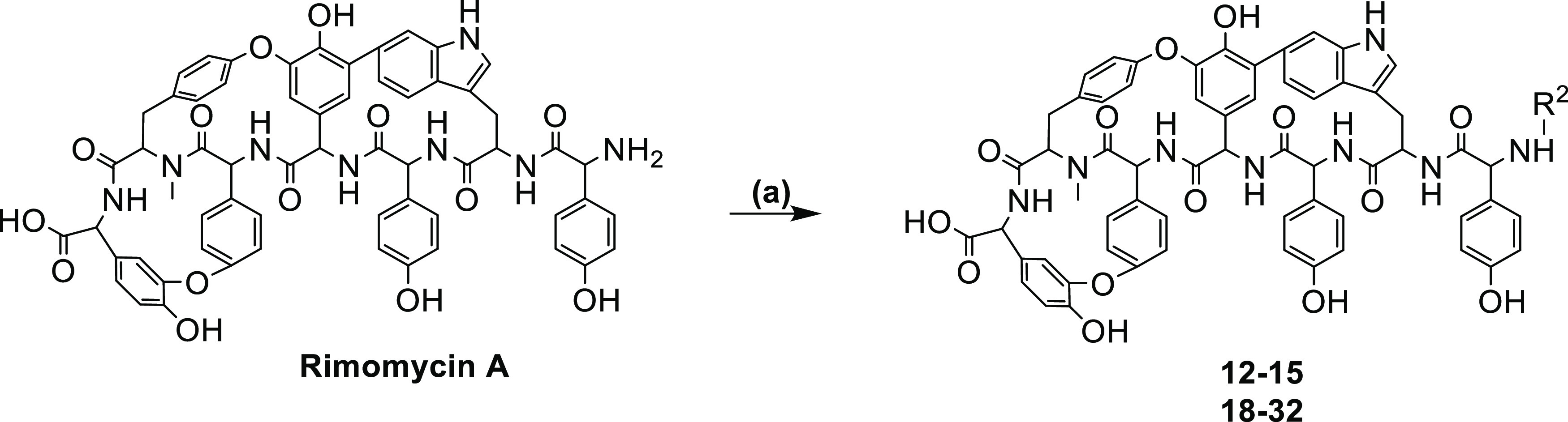
(a) substituted aldehydes, MeOH, NaCNBH3, 4 h 70 °C.
Moreover, the introduction of positive charge-bearing 6-aminohexanoic acid, compound 16, even outperformed the corresponding alkyl analogue 13 against mycobacteria.
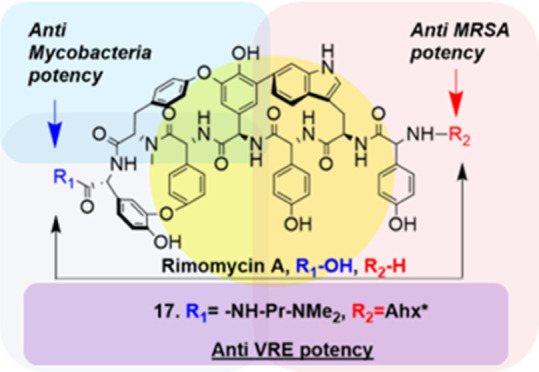
Informed by the superior activities against VRE and mycobacteria exhibited by compounds 8 and 16, compound 17 was prepared by acylation of the primary amino group with Boc-6-aminohexanoic acid-NHS ester (Boc-Ahx-NHS), followed by C-terminal substitution with N,N-dimethylamino propylamine and Boc deprotection (Scheme 3), providing the most promising rimomycin analogue discovered in this study, balancing good anti-VRE and M. smegmatis activity and retaining the inhibition of MRSA.
Scheme 3. Synthesis of Rimomycin A Analogues 16 and 17.
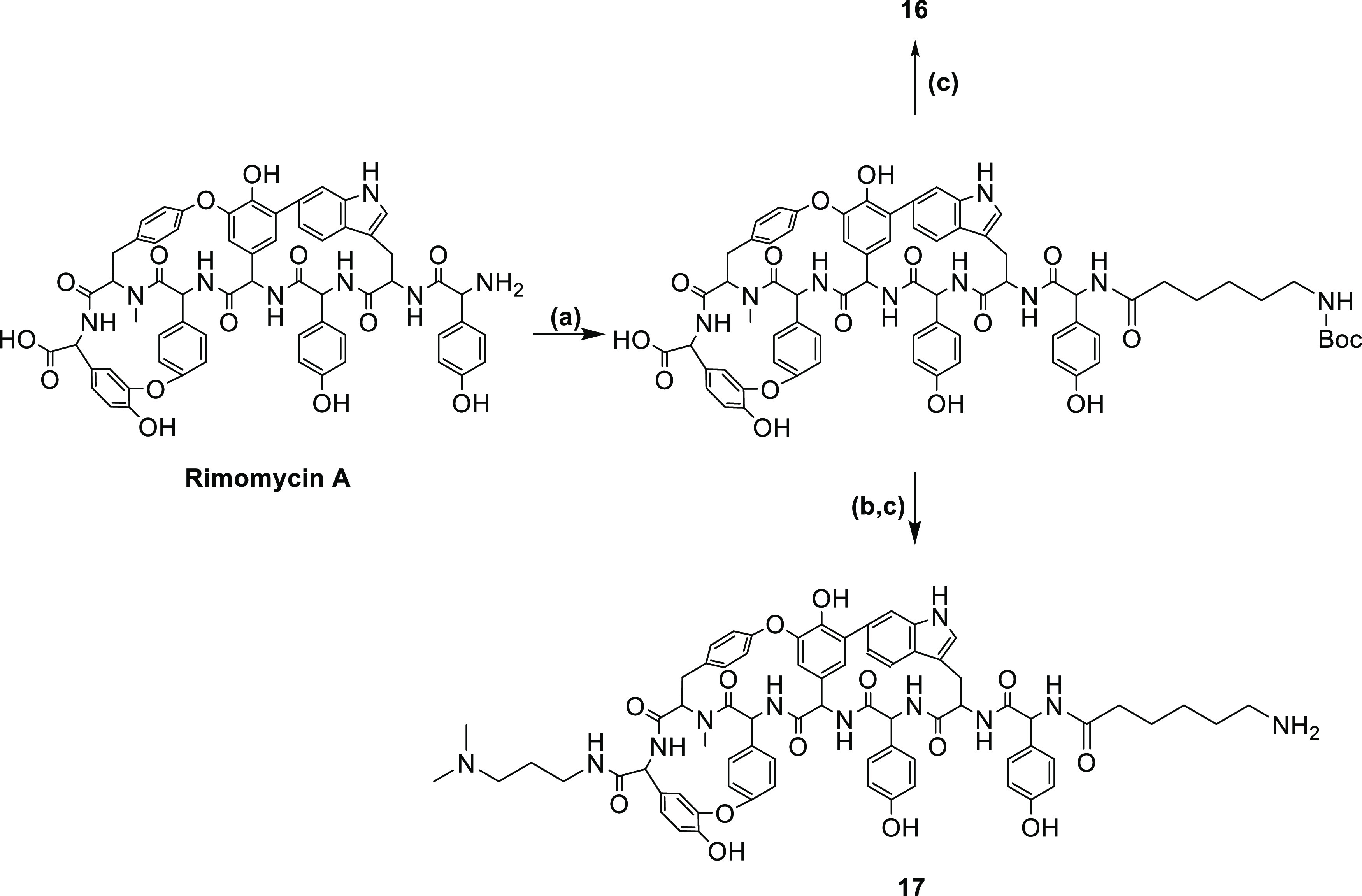
(a) Boc-Ahx-NHS, DMSO/DMF, TEA, 20 h at RT; (b) N,N-dimethylamino propylamine (5 equiv), PyBop, NMM (30 equiv) 2 h at RT; (c) DCM/TFA (1:1), 20 min, RT.
In a next series, reductive alkylation of the N-terminal amine led to the synthesis of compounds 18–32 (Scheme 2),19,32 resulting in varying impacts on antibacterial activity (Table 1). Modifying the N-terminus with alkyl or aryl groups generally abolishes M. smegmatis activity while improving MRSA inhibition with a free carboxy terminus.
Antimicrobial Activity of Rimomycin Analogues
The antimicrobial activity of the rimomycin analogues was tested against S. aureus ATCC 33591 (MRSA), M. smegmatis mc2155, E. faecium ATCC 700221 (VRE-A), E. faecalis ATCC 51299 (VRE-B), and Bacillus subtilis 168. In general, N-terminal mono-substitution with various acyl groups, (halogenated) aromatics, and heterocycles broadly impaired the antibacterial activity of synthetic rimomycin analogues against S. aureus and M. smegmatis mc2155, suggesting that the positive charge on the primary amine is critical for rimomycin activity in these organisms (Table 1). Compounds 13 (hexanyl), 19 (3-hydroxyphenyl), 20 (3,5-dihydroxyphenyl), 26 (2,3,5-trichlorophenyl), 29 (3-bromophenyl), and 32 (thienyl) exhibited a 2-fold improved antibacterial activity against S. aureus ATCC 33591 and compounds 16 (6-amino hexanoyl), 13 (hexanyl), 19 (3-hydroxyphenyl), 25 (2,3,5-trifluorophenyl), and 32 (thienyl) exhibit a similar 2-fold improved antibacterial activity against VRE-A and VRE-B strains (Table 2). The C-terminal propylamine and its N-methylated derivatives substitution (6–11) abolished activity against S. aureus ATCC 33591 but showed a 16- to 32- and 2- to 16-fold improved activity against M. smegmatis mc2155 and VRE-A strains, respectively. Combining the beneficial properties of the 6-amino hexanoyl substitution on the N-termini (16) and N,N-dimethylamino propylamine substitution on the C-termini (8), compound 17 was synthesized to include both substitutions resulting in 16-, 16-, 16-, and 2-fold improved activity against M. smegmatis mc2155, VRE-A, VRE-B, and B. subtilis 168 strains, respectively, representing the best output from our semisynthesis program (Table 1). In contrast to the reported beneficial properties in developing vancomycin analogues (oritavancin),19 3-fluoro-biphenyl (18) and 4-chloro-biphenyl (not shown) substitutions abolished the antibacterial activity of rimomycin (Table 2). This effect likely reflects the distinct MOA of vancomycin and rimomycin and the differences in the scaffold modification sites. Monosubstituted and disubstituted aromatic N-terminal analogues 19–30 showed promiscuous activity against MRSA and VRE strains. The nature and the position of the substituent were also important as meta (3-hydroxyl) (19), 3,5-dihydroxyl (20) and 2,3,5-trichloro (26) phenyl compounds showed the best anti-MRSA activity. From the aromatic heterocycles prepared in this study, the thienyl substituent (32) was better than the pyridyl one (31) specifically against MRSA.
Table 2. Antimicrobial Activity of Rimomycin A Analogues 18–32 (μg/mL).
Enterococcus faecium ATCC 700221.
Enterococcus faecalis ATCC 51299.
Given the superior antibacterial activity of 17 against VRE, we further tested this compound against 21 VRE clinical isolates and VRE-C strains. Compound 17 exhibits an overall 4- to 16-fold improved activity against all tested VRE strains compared to the parent molecule, rimomycin A. The MIC90 values determined for rimomycin A and 17 are 8 and 2, respectively (Table 3).
Table 3. MIC of Rimomycin A and Compound 17 (μg/mL) against Clinical Strains of VRE.
| strains | resistance type | rimomycin A | 17 |
|---|---|---|---|
| E. faecium ATCC 19434 | WT | 8 | 2 |
| E. faecium ATCC 700221 | VRE-A | 8 | 1 |
| E. faecium WCC 0491 | 4 | 1 | |
| E. faecium WCC 0492 | 8 | 2 | |
| E. faecium WCC 0495 | 8 | 1 | |
| E. faecium WCC 0496 | 4–8 | 1–2 | |
| E. faecium WCC 0498 | 8 | 2 | |
| E. faecium WCC 0499 | 8 | 1 | |
| E. faecium WCC 0500 | 8 | 0.5–1 | |
| E. faecium WCC 0502 | 4–8 | 1–2 | |
| E. faecium WCC 0506 | 4–8 | 1–2 | |
| E. faecium WCC 0516 | 8 | 0.5 | |
| E. faecium WCC 0524 | 4–8 | 1 | |
| E. faecium WCC 0528 | 8 | 1 | |
| E. faecium WCC 0533 | 8 | 0.5–1 | |
| E. faecium WCC 0534 | 4–8 | 0.5–1 | |
| E. faecium WCC 0537 | 8 | 2 | |
| E. faecium WCC 0586 | 8 | 1–2 | |
| E. faecium WCC 0541 | VRE-B | 8 | 1 |
| E. faecium WCC 0545 | 8 | 2 | |
| E. faecium WCC 0553 | 8 | 1–2 | |
| E. faecium WCC 0558 | 8 | 1–2 | |
| E. faecium WCC 0587 | 8 | 1 | |
| E. gallinarumATCC49573 | VRE-C | 8 | 2 |
However, compound 17 could not improve the activity of the parent rimomycin A against VISA strains used in the study (Supplementary Table 1).
Antimicrobial Activity against Mycobacteria
The promising antibacterial effect of 17 against M. smegmatis mc2155 encouraged us to test its activity against a panel of mycobacteria (Table 4). Compound 17 exhibits 4-fold improved activity against M. smegmatis mc2155 and Mycobacterium fortuitum ATCC 6891 compared to rimomycin A and significantly outperforms isoniazid in suppressing the growth of M. smegmatis mc2155 and M. fortuitum ATCC 6891. More importantly, rimomycin A and 17 were also modestly active against isoniazid-resistant Mycobacteroides abscessus ATCC 19977, and rimomycin A showed some ability to inhibit Mycobacterium avium subsp. avium ATCC 25291, two notoriously drug-resistant species.
Table 4. Antimycobacterial Activity of Rimomycin A and Compound 17 (μg/mL).
| strains | isoniazid | complestatin | rimomycin A | 17 |
|---|---|---|---|---|
| M. smegmatis mc2155 | 4 | >128 | 8 | 2 |
| M. fortuitum ATCC 6891 | 4 | >128 | 8 | 2 |
| M. abscessus ATCC 19977 | >128 | >128 | 16 | 32–64 |
| M. tuberculosis H37Ra | 0.063 | >64 | 8 | 16 |
| M. bovis BCG Pasteur ATCC 35734 | 0.13 | 64 | 8 | 8 |
| M. avium subsp. avium ATCC 25291 | >128 | >64 | 32 | >128 |
Cytotoxicity and Hemolytic Activity of Rimomycin Analogues
Rimomycin A, compounds 6, 9, 13, 16, 17, and 19 showed no cytotoxicity against HEK293 cells (>256 μg/mL) (Supplementary Table 2). Furthermore, the compounds exhibited no hemolytic activity at similar concentrations (Supplementary Table 3).
Solubility of Selected Rimomycin Analogues
Natural Type V GPAs are hydrophobic compounds largely insoluble in water or water-based buffer systems, which limits formulation and in vivo testing models. Our initial study of corbomycin, for example, focused on a mouse MRSA skin infection model as the compound was not systemically available.21 In addition to expanding antibacterial activity, this study’s orthogonal goal was to improve the physical properties of rimomycin to enable testing in a systemic infection model. The thermodynamic33 solubility of rimomycin analogues was determined in a saline solution by equilibration using an excess amount of solid compound (10 mg/mL) at 25 °C for 24 h with shaking as reported.34 From the tested rimomycin A analogues, compound 17 showed the best solubility in water, approaching 100 μg/mL, compared to rimomycin A, which is limited to 6 μg/mL (Supplementary Table 4). The improved antibacterial activity and solubility of 17 enabled us to assess the in vivo efficacy of Type V GPAs in a mouse systemic infection model for the first time.
In Vivo Efficacy of Compound 17
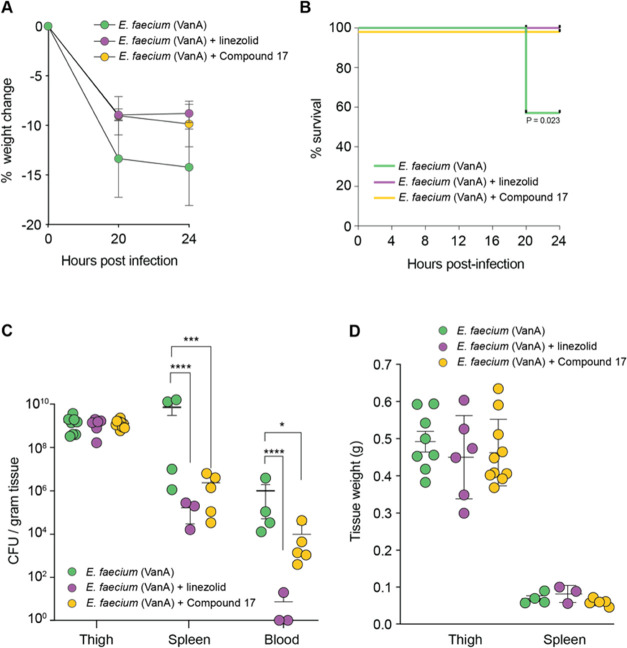
We infected neutropenic mice with the VanA E. faecium ATCC 700221 strain and treated them with compound 17 (4 doses at 100 mg/kg, i.p., n = 5), control (PEG400/DMSO/H2O, i.p., n = 4) or linezolid (4 doses at 50 mg/kg, i.p., n = 3). Linezolid was chosen as a control for its clinical use against VanA bloodstream infections.35 Compared to untreated control mice, linezolid, and compound 17 reduced weight loss and conferred a significant survival benefit against E. faecium (VanA) infection (Figure 3A,B). The bacterial load was lowered by 4 and 3 orders of magnitude in the spleen and blood by compound 17 treatment (Figure 3C). No changes to tissue weights were observed between the treatment groups (Figure 3D).
Figure 3.
Compound 17 reduces E. faecium (Van A) dissemination in a mouse neutropenic thigh infection model. (A) Percent weight change in mice infected with E. faecium ATCC 700221 treated with the indicated compounds. (B) Kaplan–Meier survival curve of infected mice treated with the indicated compounds. Mice were considered to have reached endpoint when 15% of their starting weight had been lost (*P < 0.05, Gehan–Breslow–Wilcoxon test). (C) Tissue E. faecium loads at the experimental endpoint. Each data point represents one tissue sample. Individual data points are shown with means and standard deviation. (D) Tissue weight at the experimental endpoint. Each data point represents one tissue sample. Individual data points are shown with means and standard deviation. For (C) and (D), *P < 0.05, ***P < 0.0005, ****P < 0.0001, two-way ANOVA, Tukey test for multiple comparisons.
Conclusions
Due to the worldwide spread of antimicrobial-resistant pathogens,36 natural product antibiotics with new modes of action that can evade current circulating resistance mechanisms are ideal candidates for antibiotic development. Type V GPAs represent a subclass of GPAs that block the activity of autolysins and are unaffected by the clinically prevalent d-Ala-d-Lac-mediated GPA resistance.21−23 Rimomycin, a recently identified Type V GPA, possesses a tricyclic heptapeptide scaffold and unmodified N- and C-termini that provide an opportunity for chemical derivatization.23 Given the promising antimicrobial activity of rimomycin against MRSA and VRE strains and its unique antimycobacterial activity among known Type V GPAs, rimomycin provides an excellent scaffold to develop new Gram-positive-oriented antibiotics.
While the wild-type rimomycin producer, Streptomyces rimosus WAC06783, yields small quantities of a mixture of rimomycin analogues, application of our GPAHex synthetic biology platform for heterologous production and target deletion of the rmoL halogenase coding gene generated an optimized strain that only produces rimomycin A, greatly facilitating downstream semisynthesis studies.
N- and C- terminal functionalization of rimomycin A through reductive alkylation-acylation and carboxylic acid amidation chemistry separately and, in combination, generated over 30 new rimomycin analogues. SAR studies show that the C-terminal free carboxylic acid group is required for its antibacterial activity against S. aureus; however, the C-terminal amidation with positively charged amines improved its antibacterial activity against mycobacterial and VRE strains. Compound 17, generated through N-terminal acylation by 6-aminohexanoic acid and C-terminal amidation by N,N-dimethylamino propylamine exhibits an improved antibacterial activity against M. smegmatis mc2155, VRE-A, and VRE-B by 16-fold comparing to the parent molecule. Notably, the solubility of 17 was improved 17-fold compared to rimomycin A, enabling us to test the efficacy of Type V GPAs in a systemic infection mouse model for the first time. The effectiveness of 17 in a VRE-A systemic infection mouse model identifies this compound as a promising lead compound to address VRE infections.
Unlike other Type V GPAs such as complestatin, rimomycin showed good activity against various mycobacterial strains, including highly challenging and clinically relevant species such as Mycobacterium tuberculosis, M. abscessus, and M. avium. On the other hand, compound 17 showed poorer activity toward these strains, and these results suggest that a focused campaign to improve antimycobacterial activity may be warranted.
Synthetic biology coupled with semisynthesis offers powerful complementary techniques for discovering new NPs37 and developing new drugs,16,38 respectively. Given the challenge of synthesizing complex NP scaffolds, like GPAs, total synthesis is often impractical for the large-scale preparation of these antibiotics. Given the limited number of scaffold-modifying enzymes in nature, the chemical diversity of NPs is commensurately limited. However, the combination of synthetic biology and semisynthesis takes advantage of the capability of generating complex NP scaffolds through synthetic biology and diversifying through chemical functionalization, generating more diverse NP derivatives for drug discovery and development. Our study demonstrates the usefulness of this approach in developing next-generation antibiotics and sets the stage for developing Type V GPAs as new Gram-positive-targeted antibiotics.
Experimental Section
Synthesis
Material and Methods
All reagents and solvents purchased from commercial vendors were used without further purification. The concentration of reaction mixtures refers to rotary evaporation under reduced pressure at a temperature below 25 °C.
All final products were characterized by 1H NMR, 13C NMR, and high-resolution mass spectrometry (HRMS). The purity of all compounds was >95%, as determined by liquid chromatography–mass spectrometry (LC–MS) and HPLC. High-resolution electrospray ionization mass spectra were acquired using an Agilent 1290 UPLC separation module and a qTOF 6550 mass detector in positive or negative ion modes. For LC separation, an Agilent Eclipse XDB-C18 column (2.1 mm × 150 mm; 3.5 μm) or an XDB C8 column (2.1 mm × 150 mm; 3.5 μm) and the following method were used: from 0 to 1 min 95% A (0.1 v/v formic acid in water), from 1 to 7 min a linear gradient to 100% B (0.1 v/v formic acid in acetonitrile) at a flow rate of 0.4 mL/min. For flash purification, all compounds were purified by loading onto a C18 column (5 g, Teledyne Isco). Products were eluted with a gradient of acetonitrile in water. The dry products appeared as colorless solids. The mass differences are given in Δ, ppm.
All NMR spectra were recorded on an AVIII 700 MHz spectrometer equipped with a cryoprobe. The compounds used in this study were dissolved in the corresponding deuterated solvent (Cambridge Isotope Laboratories Inc) to a concentration of approximately 1.0 to 5.0 mg/mL. Chemical shifts are reported in ppm relative to tetramethylsilane (TMS) using the residual solvent signal at 2.50 ppm and 39.52 ppm for DMSO-d6. Chemical shifts values are expressed in ppm (δ), coupling constants (J, Hz), and peak patterns are reported as broad singlet (bs), singlet (s), doublet (d), triplet (t), quartet (q), pentet (p), and multiplet (m).
Rimomycin A Analogues 1–3 (Scheme 1a)
General Protocol
To a solution of rimomycin A (500 mg, 0.44 mmol, 1 equiv) in DMSO/DMF (50%, 3 mL) was added the corresponding halogenated compound (2 equiv), followed by TEA (4 equiv). The reaction was carried out for 20 h at 56 °C with stirring. Yield 48%.
C-Terminal Modification (5–11) (Scheme 1b–d)39
Boc Protection of N-Terminal Amine (4)
To a solution of rimomycin A in 2 mL of 1:1 DMSO/methanol was added 10 mg of Boc anhydride, followed by 10 μL of TEA. The reaction was carried out for 2 h at RT with stirring and then loaded as it is onto a 15.5 g C18 gold flash column. Fractions 21–40 were pooled and lyophilized. Yield 98%.
N-Boc rimomycin (10 mg, 8 μmol, 1 equiv) in 50% DMSO/DMF (0.3 mL) was treated with the corresponding amine (1 M solution, 5 equiv), NMM (1 M solution, 30 equiv) and PyBop (1 M, 20 equiv). The reaction was carried out for 20 min at RT with stirring and then loaded directly onto a C18 flash column for purification. Yield 80%.
Boc Deprotection Reactions Setup
To the dry material from above (35 mg each) was added 4 mL of 50%TFA in DCM solution, and Boc deprotection was carried out for 40 min at RT with stirring. The solvent was removed under the nitrogen stream, and crude products were lyophilized. C18 flash purification afforded the pure products. Yield: 90%.
Compound 5
To a solution of N-Boc-protected rimomycin A (13) (10 mg, 8 μmol, 1 equiv) in 50% DMSO/DMF (300 μL) was added 1-bromo octane (1.5 mg, 8 μmol, 1 equiv), followed by TEA (3.2 mg, 32 μmol, 4 equiv). The reaction was carried out at 56 °C for 20 h with stirring. Boc deprotection of the crude dry material proceeded as described above, followed by C18 flash purification to afford pure compound 5 (Scheme 1).
Reductive Alkylation (Scheme 2) (12–15 and 18–32)12
General Procedure: The glycopeptide (1 mmol) was dissolved in 150 mL of methanol and treated with aldehyde (1.1 mmol). The resulting solution was heated to 70 °C for 2 h and then treated with NaBH3CN (1.1 mmol). After an additional 2 h at 70 °C, the reaction mixture was cooled, and the solvent was removed in vacuo. The residue was purified by flash chromatography. Yield: 60–80%.
Synthesis of Compounds 16 and 17 (Scheme 3)
To a solution of Rimomycin A (1.1 g, 0.98 mmol) in 10 mL of DMSO and TEA (0.33 mL) was added Boc-Ahx-NHS (620 mg, 1.8 mmol) in 1 mL of DMF. The reaction was carried out for 18 h, and LCMS showed less than 50% conversion. Additional TEA (100 μL) and Boc-Ahx-NHS (200 mg) in DMF (1 mL) were added and reacted for further 6 hours. Lyophilization with C18 resin, followed by C18 flash purification, afforded the product in pure (>95%) form. Yield 46%.
To a solution of Rimomycin-Ahx-Boc (245 mg, 0.18 mmol, 1 equiv) in 3 mL of 50% DMF/DMSO were added N-dimethylamino propylamine (1 M, 1 mL, 5 equiv), NMM (2 M, 1 mL, 10 equiv) and PyBop (1 M, 0.8 mL, 5 equiv). The reaction was completed in 45 min. The final product was purified using flash chromatography. Yield 200 mg, 77%.
The protected compound (200 mg) was stirred in 20 mL of 50% TFA in DCM. The reaction was carried out for 30 min at RT with stirring. The solvent was then removed under reduced pressure, and the residue was lyophilized. The final product was purified on C8 50 g and eluted with 20% ACN at 30 mL/min column flow. Yield 60 mg. Overall yield: 20%.
Analytical Data
Compound 1
1H NMR (700 MHz, DMSO) δ 10.81–10.77 (m, 1H), 9.56 (s, 2H), 9.32 (s, 3H), 9.23 (s, 2H), 9.17 (s, 1H), 8.74 (d, J = 8.6 Hz, 1H), 8.61 (q, J = 6.5 Hz, 1H), 8.32 (s, 2H), 8.12–8.05 (m, 1H), 7.89 (ddq, J = 8.6, 6.2, 2.7 Hz, 2H), 7.82 (dd, J = 8.6, 2.1 Hz, 1H), 7.46–7.33 (m, 2H), 7.07 (s, 1H), 7.22–6.93 (m, 9H), 6.92–6.82 (m, 3H), 6.83 (d, J = 8.4 Hz, 1H), 6.79–6.73 (m, 1H), 6.75–6.67 (m, 3H), 6.64–6.60 (m, 1H), 6.60–6.56 (m, 1H), 6.52–6.42 (m, 2H), 5.56 (dt, J = 16.8, 8.7 Hz, 1H), 5.51–5.43 (m, 1H), 5.41 (dd, J = 9.2, 2.7 Hz, 1H), 5.23 (d, J = 8.5 Hz, 1H), 5.08 (dt, J = 7.6, 3.9 Hz, 1H), 4.92 (s, 2H), 4.80 (dd, J = 7.6, 3.3 Hz, 3H), 4.38 (s, 1H), 4.29 (d, J = 11.8 Hz, 2H), 4.12 (t, J = 4.8 Hz, 3H), 3.98–3.85 (m, 1H), 3.57 (t, J = 4.9 Hz, 3H), 3.48 (s, 1H), 3.17–3.10 (m, 2H), 2.99 (s, 2H), 3.02–2.96 (m, 1H), 2.98 (s, 3H), 2.72–2.61 (m, 1H), 2.47 (s, 8H), 2.23 (ddd, J = 11.8, 7.3, 4.5 Hz, 1H), 2.15–2.08 (m, 1H), 1.17 (s, 2H), 0.76 (s, 2H). 13C NMR (176 MHz, DMSO) δ 170.30, 165.69, 157.06, 150.84, 149.56, 145.26, 135.00, 128.71, 127.91, 124.50, 118.93, 117.04, 115.39, 67.63, 59.21, 54.55, 40.91, 40.50, 40.36, 40.24, 40.12, 40.00, 39.88, 39.77, 39.65, 36.29, 32.83, 1.58. Yield: 48%. HRMS calcd [M + H]+ C65H61N8O16 1209.4200, found 1209.4182 (Δ 1.4 ppm).
Compound 2
1H NMR (700 MHz, DMSO) δ 10.87 (d, J = 2.5 Hz, 1H), 9.73 (s, 1H), 9.55 (s, 1H), 9.39 (s, 1H), 9.21 (s, 2H), 9.18 (s, 1H), 8.95 (s, 1H), 8.77 (dd, J = 15.8, 8.1 Hz, 2H), 8.67 (d, J = 4.9 Hz, 1H), 8.19–8.12 (m, 1H), 7.94 (dd, J = 8.4, 2.0 Hz, 1H), 7.88 (dd, J = 8.6, 2.1 Hz, 1H), 7.53 (d, J = 7.9 Hz, 1H), 7.43 (d, J = 8.4 Hz, 1H), 7.27–7.18 (m, 4H), 7.12–7.03 (m, 3H), 7.00–6.86 (m, 5H), 6.82 (dd, J = 8.4, 1.3 Hz, 1H), 6.80–6.72 (m, 2H), 6.67–6.64 (m, 2H), 6.52 (s, 7H), 6.49 (d, J = 8.5 Hz, 2H), 5.61 (d, J = 9.3 Hz, 1H), 5.50–5.43 (m, 2H), 5.21 (d, J = 8.2 Hz, 1H), 5.12 (d, J = 2.7 Hz, 1H), 4.88 (s, 1H), 4.88–4.83 (m, 2H), 4.39 (dt, J = 11.1, 7.4 Hz, 1H), 4.31 (dt, J = 11.3, 7.3 Hz, 2H), 4.10–4.05 (m, 1H), 3.44 (d, J = 12.4 Hz, 1H), 3.26 (s, 1H), 3.10 (qd, J = 7.2, 4.6 Hz, 1H), 3.06 (s, 2H), 3.04 (s, 3H), 2.94–2.88 (m, 1H), 2.80 (dt, J = 19.6, 7.4 Hz, 4H), 2.54 (s, 2H), 2.07 (s, 1H), 1.23 (s, 1H), 1.17 (t, J = 7.3 Hz, 1H). 13C NMR (176 MHz, DMSO) δ 165.76, 158.41, 150.29, 149.07, 144.67, 139.42, 130.90, 130.16, 127.30, 125.94, 118.04, 115.47, 114.62, 111.89, 110.20, 62.29, 54.29, 49.66, 46.47, 42.24, 40.43, 40.02, 39.88, 39.76, 39.64, 39.52, 39.40, 39.28, 39.16, 32.33, 28.16, 8.63, 1.15. Yield: 48%. HRMS calcd [M – H]− C65H59N8O20S2– 1335.3293, found 667.1634 [M – 2H]2– (Δ 4.1 ppm).
Compound 3
1H NMR (700 MHz, DMSO) δ 10.87–10.78 (m, 1H), 9.58 (s, 1H), 9.44–9.30 (m, 2H), 9.15 (s, 1H), 8.77–8.63 (m, 2H), 8.21–8.12 (m, 2H), 8.09–7.99 (m, 1H), 7.96 (dd, J = 8.7, 1.8 Hz, 1H), 7.85 (dd, J = 8.5, 2.1 Hz, 1H), 7.47 (t, J = 6.2 Hz, 2H), 7.34 (d, J = 17.7 Hz, 7H), 7.27–7.18 (m, 4H), 7.18–6.99 (m, 6H), 6.99–6.84 (m, 3H), 6.84–6.62 (m, 2H), 6.56–6.45 (m, 3H), 5.62 (p, J = 9.9 Hz, 1H), 5.53 (d, J = 7.8 Hz, 1H), 5.48 (dd, J = 5.7, 2.9 Hz, 1H), 5.25 (d, J = 8.3 Hz, 1H), 5.14 (d, J = 2.6 Hz, 1H), 4.91–4.81 (m, 2H), 4.61 (d, J = 2.2 Hz, 0H), 4.40–4.25 (m, 3H), 4.16 (dtd, J = 17.4, 11.1, 6.7 Hz, 2H), 4.11–3.93 (m, 2H), 3.61 (t, J = 6.6 Hz, 1H), 3.52 (td, J = 6.7, 5.4 Hz, 6H), 3.05 (s, 4H), 2.74–2.67 (m, 1H), 2.54 (s, 1H), 2.30–2.22 (m, 1H), 2.22–2.12 (m, 1H), 2.09–2.03 (m, 1H), 1.89 (d, J = 17.8 Hz, 1H), 1.78 (ddt, J = 14.6, 9.4, 6.8 Hz, 6H), 1.69 (dt, J = 14.7, 6.7 Hz, 1H), 1.62 (dq, J = 9.4, 6.8 Hz, 2H), 1.58–1.43 (m, 1H), 1.43–1.18 (m, 24H), 1.10–0.98 (m, 1H), 0.56 (q, J = 6.9 Hz, 8H). 13C NMR (176 MHz, DMSO) δ 170.88, 170.29, 169.71, 169.11, 168.81, 168.30, 163.31, 156.63, 156.42, 154.88, 150.39, 149.08, 144.77, 139.44, 136.29, 134.44, 133.84, 131.13, 130.95, 130.78, 129.58, 128.87, 128.50, 127.48, 127.40, 126.21, 126.13, 124.71, 124.10, 123.22, 121.86, 118.55, 116.57, 114.89, 114.54, 114.18, 113.91, 112.49, 111.99, 110.29, 74.32, 67.32, 65.40, 61.25, 60.75, 60.56, 57.12, 55.76, 54.83, 54.26, 54.15, 47.36, 45.47, 43.05, 42.92, 40.43, 40.01, 39.88, 39.76, 39.64, 39.52, 39.40, 39.28, 39.16, 35.84, 35.28, 35.19, 33.03, 32.48, 32.37, 32.33, 32.31, 32.29, 32.24, 32.18, 32.04, 31.84, 31.76, 31.61, 31.25, 31.13, 28.83, 28.02, 27.96, 27.44, 27.43, 26.51, 26.18, 25.66, 25.50, 25.20, 25.18, 25.08, 24.63, 24.22, 24.16, 24.09, 24.04, 24.03, 23.99, 15.36, 13.40. Yield: 48%. HRMS calcd [M – H]− C73H77B2N8O18– 1375.5547, found 687.2743 [M – 2H]2– (Δ 1.3 ppm).
Compound 4
1H NMR (700 MHz, DMSO) δ 10.84 (d, J = 2.4 Hz, 1H), 9.52 (s, 1H), 9.38 (s, 1H), 9.30 (s, 1H), 9.16 (s, 1H), 8.72–8.50 (m, 2H), 8.15 (t, J = 8.2 Hz, 2H), 7.95 (dd, J = 8.5, 2.0 Hz, 1H), 7.89 (dd, J = 8.6, 2.1 Hz, 1H), 7.46 (d, J = 8.3 Hz, 1H), 7.24–7.14 (m, 4H), 7.14–7.07 (m, 1H), 7.07–6.91 (m, 5H), 6.87 (d, J = 2.3 Hz, 1H), 6.87–6.76 (m, 3H), 6.62–6.47 (m, 14H), 5.61 (d, J = 9.3 Hz, 1H), 5.55–5.44 (m, 2H), 5.13 (dd, J = 5.5, 2.8 Hz, 2H), 5.06 (d, J = 9.0 Hz, 1H), 4.95–4.81 (m, 2H), 4.41–4.32 (m, 1H), 3.95 (d, J = 9.8 Hz, 1H), 3.04 (s, 4H), 2.64 (s, 0H), 2.60 (p, J = 1.9 Hz, 1H), 2.40 (p, J = 1.9 Hz, 1H), 1.34 (s, 9H). 13C NMR (176 MHz, DMSO) δ 171.11, 168.84, 168.18, 157.99, 157.81, 157.64, 157.47, 156.54, 150.29, 149.07, 144.57, 139.44, 129.54, 128.44, 127.42, 126.68, 119.90, 118.19, 116.48, 114.83, 114.64, 61.22, 56.79, 54.48, 40.43, 40.02, 39.88, 39.76, 39.64, 39.52, 39.40, 39.28, 39.16, 32.31, 28.16. Yield 98%. HRMS calcd [M + H]+ C66H61N8O16+ 1221.4200, found 1221.4224 (Δ −1.9 ppm).
Compound 5
1H NMR (700 MHz, DMSO) δ 10.55 (s, 1H), 9.57 (d, J = 14.1 Hz, 2H), 9.45 (s, 1H), 9.20 (s, 1H), 8.75–8.69 (m, 1H), 8.67 (d, J = 5.0 Hz, 1H), 8.56 (s, 1H), 8.06 (dd, J = 9.0, 4.9 Hz, 2H), 7.89 (dt, J = 8.6, 1.3 Hz, 1H), 7.84–7.79 (m, 1H), 7.61 (d, J = 2.6 Hz, 1H), 7.22 (dd, J = 8.2, 2.6 Hz, 2H), 7.21–7.15 (m, 1H), 7.17–7.02 (m, 4H), 7.04–6.94 (m, 2H), 6.94–6.87 (m, 4H), 6.77 (d, J = 8.3 Hz, 1H), 6.76–6.66 (m, 2H), 6.65 (s, 1H), 6.63 (d, J = 3.1 Hz, 1H), 6.54–6.48 (m, 4H), 6.08–6.05 (m, 1H), 5.70 (d, J = 2.4 Hz, 1H), 5.66 (d, J = 8.8 Hz, 1H), 5.36 (dd, J = 18.8, 8.8 Hz, 1H), 5.26 (d, J = 8.4 Hz, 1H), 5.09–5.01 (m, 1H), 4.89 (d, J = 2.2 Hz, 1H), 4.83 (dd, J = 10.1, 3.6 Hz, 1H), 4.71 (s, 1H), 4.31 (dd, J = 11.9, 3.6 Hz, 1H), 4.16 (ddt, J = 32.9, 10.8, 6.5 Hz, 2H), 3.30 (s, 2H), 3.17 (d, J = 12.6 Hz, 1H), 3.12–3.04 (m, 2H), 3.05 (s, 3H), 2.91 (t, J = 12.6 Hz, 1H), 2.54 (s, 1H), 1.72 (d, J = 11.7 Hz, 1H), 1.66–1.56 (m, 1H), 1.61 (s, 2H), 1.53 (s, 1H), 1.35–1.18 (m, 18H), 0.88–0.80 (m, 4H). 13C NMR (176 MHz, DMSO) δ 170.05, 169.63, 169.33, 168.86, 168.57, 168.21, 163.11, 157.71, 157.54, 156.66, 156.39, 155.56, 150.36, 150.07, 144.81, 141.55, 135.41, 134.16, 133.80, 130.79, 129.98, 129.10, 129.00, 128.24, 127.53, 126.26, 126.10, 125.83, 125.52, 124.63, 118.57, 118.26, 116.49, 115.13, 114.60, 107.24, 65.29, 63.69, 61.32, 54.92, 54.41, 54.12, 54.05, 40.43, 40.02, 39.98, 39.90, 39.88, 39.76, 39.64, 39.52, 39.40, 39.28, 39.16, 32.28, 31.22, 31.14, 28.60, 28.53, 27.99, 25.31, 24.64, 22.10, 22.04, 20.72, 13.90. Yield 45%. HRMS calcd [M – H]− C69H67N8O14– 1231.4782, found 1231.4765 (Δ 1.3 ppm).
Compound 6
1H NMR (700 MHz, DMSO) δ 10.52 (s, 1H), 8.63 (d, J = 4.9 Hz, 1H), 8.58 (d, J = 8.3 Hz, 1H), 8.39 (s, 2H), 8.14 (d, J = 8.2 Hz, 1H), 8.10 (d, J = 8.7 Hz, 1H), 8.02 (d, J = 8.8 Hz, 1H), 7.87 (dd, J = 8.6, 2.4 Hz, 2H), 7.58 (d, J = 2.6 Hz, 1H), 7.27 (d, J = 7.9 Hz, 1H), 7.22–7.18 (m, 1H), 7.18–7.10 (m, 5H), 7.09–7.01 (m, 2H), 6.96–6.84 (m, 3H), 6.75 (d, J = 8.3 Hz, 1H), 6.69–6.63 (m, 2H), 6.61 (dd, J = 8.6, 2.1 Hz, 1H), 6.55–6.50 (m, 2H), 6.07 (d, J = 2.5 Hz, 1H), 5.71 (d, J = 2.4 Hz, 1H), 5.63 (d, J = 8.7 Hz, 1H), 5.35 (d, J = 8.6 Hz, 1H), 5.16 (d, J = 8.3 Hz, 1H), 4.87 (d, J = 2.2 Hz, 1H), 4.83 (d, J = 5.3 Hz, 1H), 4.35 (d, J = 11.4 Hz, 1H), 4.26 (s, 1H), 3.24 (dq, J = 13.0, 6.5 Hz, 1H), 3.18–3.09 (m, 2H), 3.03–2.97 (m, 1H), 2.85 (t, J = 12.7 Hz, 1H), 2.73 (t, J = 7.4 Hz, 2H), 2.54 (s, 11H), 2.53 (s, 1H), 1.69 (p, J = 7.0 Hz, 2H). 13C NMR (176 MHz, DMSO) δ 172.29, 170.13, 169.21, 169.08, 169.01, 168.91, 168.28, 165.22, 156.67, 156.55, 156.45, 155.56, 150.26, 150.06, 144.30, 141.54, 135.40, 134.63, 134.31, 133.75, 132.56, 130.97, 130.75, 130.14, 129.06, 128.75, 128.39, 128.18, 128.02, 127.77, 126.30, 125.88, 125.65, 125.47, 124.61, 124.05, 122.66, 121.38, 120.55, 118.52, 118.09, 116.68, 116.26, 114.82, 114.67, 114.21, 111.32, 107.45, 61.28, 57.96, 54.94, 54.36, 54.30, 54.12, 40.43, 40.02, 39.88, 39.76, 39.64, 39.52, 39.40, 39.28, 39.16, 37.13, 35.98, 35.86, 32.35, 28.48, 27.96. Yield: 40% HRMS calcd [M + H]+ C64H61N10O13+ 1177.4414, found 1177.4428 (Δ 1.4 ppm).
Compound 7
1H NMR (700 MHz, DMSO) δ 10.52 (s, 1H), 8.73 (d, J = 6.2 Hz, 1H), 8.63 (d, J = 4.5 Hz, 1H), 8.54 (d, J = 8.3 Hz, 1H), 8.33 (s, 2H), 8.18 (d, J = 8.2 Hz, 1H), 8.10 (d, J = 8.6 Hz, 1H), 8.03 (d, J = 8.9 Hz, 1H), 7.87 (ddd, J = 9.8, 4.0, 1.7 Hz, 2H), 7.58 (d, J = 2.5 Hz, 1H), 7.26 (d, J = 7.9 Hz, 1H), 7.22–7.18 (m, 1H), 7.16 (dd, J = 8.1, 2.6 Hz, 1H), 7.16–7.06 (m, 4H), 7.06–7.01 (m, 2H), 6.99–6.91 (m, 1H), 6.91–6.84 (m, 3H), 6.73 (d, J = 8.3 Hz, 1H), 6.70–6.63 (m, 3H), 6.55–6.50 (m, 2H), 6.07 (d, J = 2.5 Hz, 1H), 5.71 (d, J = 2.5 Hz, 1H), 5.63 (d, J = 8.8 Hz, 1H), 5.35 (d, J = 8.6 Hz, 1H), 5.17 (d, J = 8.3 Hz, 1H), 4.98 (dt, J = 11.9, 8.1 Hz, 1H), 4.88–4.84 (m, 1H), 4.82 (d, J = 5.2 Hz, 1H), 4.37–4.33 (m, 1H), 4.30 (s, 1H), 3.29 (dq, J = 12.4, 6.2 Hz, 1H), 3.24–3.14 (m, 2H), 3.07–3.03 (m, 1H), 3.04 (s, 3H), 3.03–2.96 (m, 1H), 2.86 (t, J = 12.7 Hz, 1H), 2.81–2.76 (m, 0H), 2.71–2.62 (m, 2H), 2.54 (s, 8H), 2.35 (s, 3H). 13C NMR (176 MHz, DMSO) δ 171.86, 170.13, 169.21, 169.05, 168.96, 168.91, 168.21, 164.88, 156.66, 156.56, 156.53, 155.54, 150.19, 150.06, 144.21, 141.54, 135.40, 134.65, 134.34, 133.74, 131.90, 130.98, 130.76, 130.13, 129.05, 128.73, 128.46, 128.19, 128.13, 128.07, 127.75, 126.29, 125.87, 125.66, 125.48, 124.60, 124.07, 122.67, 121.39, 120.56, 118.53, 118.28, 116.65, 116.21, 114.85, 114.66, 114.23, 111.30, 107.44, 61.30, 57.72, 54.92, 54.37, 54.11, 51.35, 49.58, 40.43, 40.01, 39.88, 39.76, 39.64, 39.52, 39.40, 39.28, 39.16, 37.69, 35.84, 34.87, 34.35, 32.36, 27.90. Yield: 45%. HRMS calcd [M + 2H]+ C64H61N10O13+ 1177.4409, found 1177.4387 (Δ 1.8 ppm).
Compound 8
1H NMR (700 MHz, DMSO) δ 10.53–10.50 (m, 1H), 9.45 (s, 4H), 8.63 (d, J = 5.0 Hz, 1H), 8.53 (t, J = 6.1 Hz, 2H), 8.32 (s, 4H), 8.15 (d, J = 8.2 Hz, 1H), 8.09 (d, J = 8.6 Hz, 1H), 8.03 (d, J = 8.8 Hz, 1H), 7.87 (ddd, J = 8.7, 5.1, 1.7 Hz, 2H), 7.58 (d, J = 2.6 Hz, 1H), 7.27 (d, J = 7.9 Hz, 1H), 7.20 (dd, J = 8.4, 2.1 Hz, 1H), 7.18–7.09 (m, 5H), 7.06–7.01 (m, 2H), 6.93 (t, J = 7.5 Hz, 1H), 6.91–6.83 (m, 3H), 6.73 (d, J = 8.3 Hz, 1H), 6.68–6.63 (m, 2H), 6.61 (dd, J = 8.8, 2.1 Hz, 1H), 6.55–6.50 (m, 2H), 6.28 (s, 1H), 6.07 (dd, J = 2.7, 1.1 Hz, 1H), 5.75 (s, 0H), 5.71 (d, J = 2.5 Hz, 1H), 5.63 (d, J = 8.8 Hz, 1H), 5.35 (d, J = 8.6 Hz, 1H), 5.15 (d, J = 8.2 Hz, 1H), 4.97 (dt, J = 12.4, 7.8 Hz, 1H), 4.86 (d, J = 2.1 Hz, 1H), 4.83 (d, J = 5.2 Hz, 1H), 4.37–4.33 (m, 1H), 4.27 (s, 1H), 3.22–3.13 (m, 2H), 3.10–3.03 (m, 1H), 3.03 (s, 3H), 3.00 (t, J = 12.5 Hz, 1H), 2.85 (t, J = 12.7 Hz, 1H), 2.54 (s, 2H), 2.22 (t, J = 7.1 Hz, 2H), 2.11 (s, 6H), 1.57 (p, J = 7.0 Hz, 2H). 13C NMR (176 MHz, DMSO) δ 170.14, 169.20, 168.99, 168.92, 168.79, 168.25, 164.70, 156.67, 156.46, 155.54, 150.23, 150.05, 144.20, 141.54, 135.39, 130.17, 129.06, 128.77, 128.59, 128.18, 128.06, 127.78, 126.29, 125.65, 125.48, 118.16, 116.17, 114.83, 114.66, 114.23, 107.45, 61.28, 57.89, 56.49, 54.95, 54.91, 54.34, 54.12, 45.12, 40.43, 40.02, 39.88, 39.76, 39.64, 39.52, 39.40, 39.28, 39.16, 36.86, 32.34, 26.85. Yield: 45% HRMS calcd [M + H]+ C66H65N10O13+ 1205.4727, found 1205.4712 (Δ 1.2 ppm).
Compound 9
1H NMR (700 MHz, DMSO) δ 10.51 (s, 1H), 8.80 (s, 1H), 8.63 (s, 2H), 8.41 (s, 9H), 8.11 (t, J = 9.6 Hz, 2H), 8.01 (d, J = 8.8 Hz, 1H), 7.87 (d, J = 8.6 Hz, 2H), 7.58 (d, J = 2.6 Hz, 1H), 7.27 (d, J = 7.9 Hz, 1H), 7.21 (d, J = 8.6 Hz, 1H), 7.16 (dd, J = 8.2, 2.6 Hz, 1H), 7.13 (dd, J = 11.2, 7.8 Hz, 4H), 7.04 (d, J = 8.4 Hz, 2H), 6.93 (t, J = 7.6 Hz, 1H), 6.90–6.84 (m, 3H), 6.83 (s, 12H), 6.76 (d, J = 8.2 Hz, 1H), 6.65 (dd, J = 16.3, 8.5 Hz, 3H), 6.53 (d, J = 8.4 Hz, 2H), 6.29 (s, 2H), 6.07 (d, J = 2.5 Hz, 1H), 5.71 (d, J = 2.4 Hz, 1H), 5.63 (d, J = 8.8 Hz, 1H), 5.35 (d, J = 8.6 Hz, 1H), 5.16 (d, J = 8.3 Hz, 1H), 4.97 (s, 1H), 4.87 (s, 1H), 4.83 (d, J = 5.1 Hz, 1H), 4.35 (d, J = 11.6 Hz, 1H), 4.23 (s, 1H), 3.15 (s, 4H), 3.05–3.03 (m, 15H), 2.99 (d, J = 14.3 Hz, 2H), 2.85 (t, J = 12.6 Hz, 1H), 2.54 (s, 2H), 1.88 (s, 3H), 1.24 (s, 2H), 0.85 (t, J = 6.6 Hz, 0H). 13C NMR (176 MHz, DMSO) δ 172.35, 170.23, 169.24, 169.06, 168.98, 168.38, 165.26, 156.75, 156.61, 156.51, 155.62, 150.34, 150.13, 144.43, 141.58, 135.46, 134.64, 134.30, 133.79, 132.53, 131.00, 130.84, 130.15, 129.12, 128.84, 128.31, 128.21, 128.09, 127.82, 126.35, 125.91, 125.72, 125.50, 124.68, 124.11, 122.76, 121.49, 120.62, 118.61, 118.31, 116.76, 116.38, 114.89, 114.74, 114.27, 111.35, 107.53, 63.50, 61.34, 57.98, 55.07, 54.98, 54.39, 54.17, 52.41, 52.39, 52.36, 45.92, 45.89, 40.43, 40.02, 39.88, 39.76, 39.64, 39.52, 39.40, 39.28, 39.16, 35.88, 32.42, 27.97, 25.98, 25.93, 22.66, 19.62, 17.20, 0.04. Yield: 45%. HRMS calcd [M + H]+ C67H67N10O13+ 1219.4884, found 1219.4927 (Δ −3.5 ppm).
Compound 10
1H NMR (700 MHz, DMSO) δ 10.52 (s, 1H), 8.66–8.62 (m, 2H), 8.56 (d, J = 8.3 Hz, 1H), 8.30 (s, 5H), 8.16 (t, J = 7.3 Hz, 1H), 8.10 (d, J = 8.7 Hz, 1H), 8.03 (d, J = 8.9 Hz, 1H), 7.87 (dd, J = 8.5, 1.9 Hz, 2H), 7.58 (d, J = 2.7 Hz, 1H), 7.26 (d, J = 7.9 Hz, 1H), 7.20 (dd, J = 8.4, 2.1 Hz, 1H), 7.16 (dd, J = 8.2, 2.6 Hz, 1H), 7.13 (dd, J = 10.9, 7.8 Hz, 4H), 7.06–7.01 (m, 2H), 6.93 (t, J = 7.5 Hz, 1H), 6.91–6.84 (m, 3H), 6.74 (d, J = 8.3 Hz, 1H), 6.69–6.63 (m, 2H), 6.63 (s, 1H), 6.61 (dd, J = 8.6, 2.1 Hz, 1H), 6.55–6.50 (m, 2H), 6.09–6.06 (m, 1H), 5.71 (d, J = 2.5 Hz, 1H), 5.63 (d, J = 8.8 Hz, 1H), 5.35 (d, J = 8.6 Hz, 1H), 5.16 (d, J = 8.2 Hz, 1H), 4.87 (d, J = 2.1 Hz, 1H), 4.83 (d, J = 5.2 Hz, 1H), 4.35 (d, J = 12.2 Hz, 1H), 4.29 (s, 1H), 3.23 (dt, J = 13.4, 6.4 Hz, 1H), 3.18–3.09 (m, 1H), 3.04 (s, 1H), 3.03–2.95 (m, 1H), 2.91–2.81 (m, 1H), 2.67 (q, J = 7.9 Hz, 5H), 2.54 (s, 0H), 2.25 (t, J = 6.9 Hz, 3H), 2.12 (s, 1H), 2.11 (s, 7H), 1.67 (q, J = 7.2 Hz, 2H), 1.60 (p, J = 7.0 Hz, 2H), 1.46–1.28 (m, 2H), 1.29–1.16 (m, 1H). 13C NMR (176 MHz, DMSO) δ 169.21, 168.99, 168.26, 164.41, 156.52, 150.24, 144.25, 141.54, 135.40, 128.11, 127.77, 126.29, 114.84, 114.66, 107.44, 57.80, 56.71, 54.94, 54.34, 46.54, 45.58, 44.94, 40.02, 39.88, 39.76, 39.64, 39.52, 39.40, 39.28, 39.16, 36.40, 32.34. Yield: 45%. HRMS calcd [M + H]+ C69H72N11O13+ 1262.5306, found 1262.5289 (Δ 1.3 ppm).
Compound 11
1H NMR (700 MHz, DMSO) δ 10.51 (s, 1H), 9.46 (s, 6H), 8.78 (s, 1H), 8.64 (d, J = 4.9 Hz, 2H), 8.41 (s, 3H), 8.11 (t, J = 8.0 Hz, 2H), 8.01 (d, J = 8.9 Hz, 1H), 7.87 (dd, J = 9.3, 4.7 Hz, 2H), 7.58 (d, J = 2.7 Hz, 1H), 7.27 (d, J = 7.9 Hz, 1H), 7.21 (dd, J = 8.4, 2.1 Hz, 1H), 7.17 (dd, J = 8.1, 2.6 Hz, 1H), 7.15–7.09 (m, 4H), 7.07–7.00 (m, 2H), 6.93 (t, J = 7.5 Hz, 1H), 6.91–6.84 (m, 3H), 6.76 (d, J = 8.2 Hz, 1H), 6.69–6.62 (m, 3H), 6.55–6.50 (m, 2H), 6.07 (d, J = 2.5 Hz, 1H), 5.71 (d, J = 2.5 Hz, 1H), 5.63 (d, J = 8.8 Hz, 1H), 5.36 (d, J = 8.6 Hz, 1H), 5.17 (d, J = 8.2 Hz, 1H), 4.88 (d, J = 2.2 Hz, 1H), 4.83 (d, J = 5.2 Hz, 1H), 4.35 (d, J = 11.7 Hz, 1H), 4.23 (s, 1H), 3.49 (dd, J = 10.4, 6.0 Hz, 2H), 3.36–3.29 (m, 18H), 3.17–3.12 (m, 3H), 3.04 (s, 3H), 2.97 (s, 3H), 2.85 (t, J = 12.8 Hz, 1H), 2.54 (s, 1H), 2.07 (dt, J = 16.2, 8.8 Hz, 6H), 1.93–1.87 (m, 2H). 13C NMR (176 MHz, DMSO) δ 172.50, 170.14, 169.19, 168.92, 164.91, 156.37, 150.29, 144.40, 141.52, 129.06, 128.30, 127.96, 127.79, 126.31, 114.80, 114.67, 63.58, 60.98, 58.09, 54.12, 47.54, 40.43, 40.01, 39.88, 39.76, 39.64, 39.52, 39.40, 39.28, 39.16, 35.90, 32.33, 23.35, 21.05. Yield: 45%. HRMS calcd [M + H]+ C69H69N10O13+ 1245.5040, found 1245.5040 (Δ 0 ppm).
Compound 12
1H NMR (700 MHz, DMSO) δ 10.82 (d, J = 2.5 Hz, 1H), 9.37 (s, 2H), 9.21 (s, 2H), 9.07 (s, 1H), 8.59 (d, J = 5.1 Hz, 1H), 8.37 (s, 6H), 8.16 (d, J = 9.3 Hz, 1H), 8.05–8.01 (m, 2H), 7.96–7.91 (m, 2H), 7.48 (t, J = 8.3 Hz, 2H), 7.23 (d, J = 1.3 Hz, 1H), 7.19 (dd, J = 8.3, 2.1 Hz, 1H), 7.14 (dd, J = 6.5, 2.4 Hz, 2H), 7.10–7.06 (m, 3H), 7.07–7.00 (m, 2H), 6.98 (dd, J = 8.1, 1.9 Hz, 1H), 6.96 (s, 4H), 6.91–6.78 (m, 4H), 6.71–6.65 (m, 2H), 6.64 (d, J = 8.3 Hz, 1H), 6.50 (d, J = 2.0 Hz, 1H), 6.49 (s, 1H), 5.59 (d, J = 9.3 Hz, 1H), 5.57–5.51 (m, 2H), 5.15 (d, J = 2.6 Hz, 1H), 4.91–4.85 (m, 2H), 4.55 (d, J = 7.3 Hz, 1H), 4.41–4.36 (m, 1H), 4.06–3.99 (m, 1H), 3.91 (s, 1H), 3.01 (s, 1H), 3.00 (s, 3H), 2.99–2.91 (m, 1H), 2.79–2.67 (m, 1H), 2.54 (s, 1H), 2.48–2.34 (m, 2H), 2.25 (dddd, J = 14.1, 10.5, 7.4, 5.2 Hz, 3H), 2.18–2.09 (m, 1H), 1.92–1.82 (m, 1H), 1.62–1.53 (m, 1H), 1.55–1.39 (m, 3H), 1.41–1.22 (m, 12H), 1.26 (s, 7H), 1.24–1.08 (m, 4H), 1.05–0.98 (m, 1H), 0.99–0.91 (m, 6H), 0.93–0.78 (m, 22H), 0.84 (s, 7H), 0.75 (t, J = 7.3 Hz, 3H). 13C NMR (176 MHz, DMSO) δ 168.86, 164.84, 128.25, 127.39, 114.85, 114.50, 72.60, 72.32, 70.84, 56.87, 55.76, 53.61, 53.50, 47.41, 41.88, 40.01, 39.90, 39.88, 39.76, 39.64, 39.52, 39.40, 39.28, 39.16, 36.20, 35.72, 35.25, 32.44, 31.52, 28.72, 28.61, 21.45, 20.87, 20.07, 20.06, 19.82, 19.72, 19.19, 18.71, 18.48, 17.29, 14.17, 14.12, 13.97, 13.94, 13.78, 13.73, 12.30, 11.56, 10.96. Yield: 80%. HRMS calcd [M – H]− C65H59N8O14– 1175.4156, found 1175.4154 (Δ 0.2 ppm).
Compound 13
1H NMR (700 MHz, DMSO) δ 10.82 (d, J = 2.4 Hz, 1H), 9.38 (s, 1H), 9.35 (s, 1H), 9.17 (s, 1H), 8.61 (dd, J = 12.8, 5.0 Hz, 1H), 8.23–8.16 (m, 2H), 8.12 (s, 1H), 8.02–7.98 (m, 1H), 7.93 (dd, J = 8.4, 2.3 Hz, 1H), 7.47 (dd, J = 9.6, 3.5 Hz, 2H), 7.26–7.20 (m, 1H), 7.19 (dd, J = 8.3, 2.1 Hz, 1H), 7.15 (dt, J = 5.6, 2.7 Hz, 2H), 7.11–6.99 (m, 4H), 6.94 (dd, J = 8.8, 2.4 Hz, 1H), 6.89 (ddd, J = 8.6, 6.0, 2.6 Hz, 1H), 6.87–6.79 (m, 3H), 6.70–6.62 (m, 5H), 6.51 (dd, J = 21.0, 8.6 Hz, 2H), 5.59 (t, J = 9.5 Hz, 1H), 5.56–5.47 (m, 2H), 5.14 (d, J = 2.7 Hz, 1H), 4.90–4.86 (m, 2H), 4.66 (s, 1H), 4.37 (dd, J = 11.8, 3.7 Hz, 1H), 4.03 (ddd, J = 11.0, 8.4, 2.8 Hz, 1H), 3.96 (s, 1H), 3.30–3.19 (m, 2H), 3.00 (s, 3H), 3.03–2.97 (m, 1H), 2.70 (d, J = 12.3 Hz, 1H), 2.54 (s, 1H), 2.26 (dt, J = 13.0, 6.8 Hz, 1H), 2.20–2.13 (m, 1H), 1.40–1.35 (m, 1H), 1.30–1.18 (m, 7H), 1.17–1.07 (m, 3H), 1.03 (ddd, J = 14.1, 9.4, 7.1 Hz, 1H), 0.85 (q, J = 6.7 Hz, 4H). 13C NMR (176 MHz, DMSO) δ 170.82, 169.34, 157.13, 155.26, 149.51, 139.95, 136.77, 131.40, 130.04, 128.84, 127.85, 123.43, 115.35, 115.01, 61.82, 57.49, 56.22, 54.76, 48.01, 40.85, 40.44, 40.30, 40.18, 40.06, 39.94, 39.82, 39.70, 39.58, 32.91, 31.78, 26.71, 22.52, 14.47. Yield: 80% HRMS calcd [M – H]− C67H63N8O14+ 1203.4469, found 1203.4457 (Δ 0.9 ppm).
Compound 14
1H NMR (700 MHz, DMSO) δ 10.82 (d, J = 2.5 Hz, 1H), 9.38 (s, 1H), 9.35 (s, 1H), 9.19 (s, 1H), 9.11 (s, 1H), 8.59 (d, J = 5.0 Hz, 1H), 8.35 (s, 2H), 8.18 (d, J = 9.4 Hz, 1H), 8.06–8.00 (m, 2H), 7.97 (d, J = 8.4 Hz, 1H), 7.93 (dd, J = 8.4, 2.3 Hz, 1H), 7.47 (dd, J = 8.2, 3.9 Hz, 2H), 7.24–7.17 (m, 2H), 7.15 (dq, J = 5.9, 2.9 Hz, 2H), 7.07 (dq, J = 9.5, 2.8 Hz, 3H), 7.05–7.01 (m, 2H), 6.97 (dd, J = 8.5, 2.0 Hz, 1H), 6.89 (dd, J = 8.5, 2.6 Hz, 1H), 6.84 (ddd, J = 15.9, 8.3, 1.8 Hz, 2H), 6.80 (dd, J = 8.4, 2.5 Hz, 1H), 6.76 (s, 8H), 6.68–6.62 (m, 3H), 6.52–6.47 (m, 2H), 5.58 (d, J = 9.3 Hz, 1H), 5.56–5.50 (m, 2H), 5.14 (d, J = 2.6 Hz, 1H), 4.90–4.86 (m, 2H), 4.58 (d, J = 7.4 Hz, 1H), 4.38 (dd, J = 11.8, 3.7 Hz, 1H), 4.02 (ddd, J = 11.1, 8.5, 2.7 Hz, 1H), 3.92 (s, 1H), 3.22 (d, J = 11.8 Hz, 1H), 3.01 (d, J = 4.6 Hz, 0H), 3.00 (s, 3H), 2.97 (d, J = 4.4 Hz, 0H), 2.70 (d, J = 12.4 Hz, 1H), 2.54 (s, 3H), 2.24 (dt, J = 11.2, 6.8 Hz, 1H), 2.12 (dt, J = 11.2, 7.2 Hz, 1H), 1.27 (s, 1H), 1.27 (q, J = 7.0 Hz, 1H), 1.26–1.17 (m, 7H), 1.17–1.12 (m, 2H), 1.14–1.06 (m, 1H), 1.05–0.96 (m, 1H), 0.86 (t, J = 7.2 Hz, 4H). 13C NMR (176 MHz, DMSO) δ 170.43, 168.94, 166.98, 165.05, 156.59, 156.50, 149.68, 149.08, 143.05, 139.54, 136.35, 133.70, 130.98, 128.34, 127.43, 114.90, 114.59, 112.02, 65.63, 61.40, 57.75, 55.80, 47.69, 40.43, 40.02, 39.88, 39.76, 39.64, 39.52, 39.40, 39.28, 39.16, 32.51, 31.39, 29.38, 29.12, 28.72, 26.65, 22.18, 14.05. Yield: 80% HRMS calcd [M – H]− C69H67N8O14– 1231.4782, found 1231.4775 (Δ 0.5 ppm).
Compound 15
1H NMR (700 MHz, DMSO) δ 10.82 (d, J = 2.3 Hz, 1H), 9.38 (s, 1H), 9.35 (s, 1H), 9.18 (s, 1H), 9.10 (s, 1H), 8.59 (d, J = 5.0 Hz, 1H), 8.34 (s, 2H), 8.18 (d, J = 9.4 Hz, 1H), 8.05–8.00 (m, 2H), 7.99–7.91 (m, 2H), 7.81–7.77 (m, 2H), 7.51–7.45 (m, 4H), 7.24–7.17 (m, 2H), 7.16–7.06 (m, 4H), 7.07 (d, J = 2.1 Hz, 1H), 7.06–6.99 (m, 2H), 6.97 (dd, J = 8.6, 2.0 Hz, 1H), 6.91–6.77 (m, 4H), 6.72 (s, 9H), 6.68–6.62 (m, 3H), 6.53–6.47 (m, 2H), 5.58 (d, J = 9.3 Hz, 1H), 5.56–5.51 (m, 2H), 5.14 (d, J = 2.6 Hz, 1H), 4.90–4.86 (m, 2H), 4.58 (d, J = 7.2 Hz, 1H), 4.38 (d, J = 11.7 Hz, 1H), 4.08–3.99 (m, 3H), 3.92 (s, 1H), 3.35 (s, 1H), 3.24–3.20 (m, 1H), 2.99 (s, 3H), 2.70 (d, J = 12.6 Hz, 1H), 2.54 (s, 1H), 2.42 (s, 3H), 2.23 (dt, J = 11.2, 6.7 Hz, 1H), 2.12 (dt, J = 11.3, 7.2 Hz, 1H), 1.81 (p, J = 6.4 Hz, 2H), 1.37 (d, J = 28.5 Hz, 1H), 1.23 (p, J = 7.3 Hz, 25H), 1.17–1.13 (m, 1H), 1.13 (s, 2H), 1.04–0.96 (m, 1H), 0.85 (t, J = 7.0 Hz, 4H). 13C NMR (176 MHz, DMSO) δ 168.93, 164.61, 156.49, 145.10, 143.09, 137.67, 130.27, 128.35, 128.10, 127.66, 127.43, 125.54, 114.90, 114.58, 68.07, 54.34, 47.66, 47.54, 46.89, 40.43, 40.02, 39.88, 39.76, 39.64, 39.52, 39.40, 39.28, 39.16, 32.50, 31.37, 29.34, 29.16, 29.10, 29.05, 28.79, 27.77, 26.64, 22.16, 21.16, 20.83, 14.03, 14.01. Yield: 80%. HRMS calcd [M – H]− C72H73N8O14– 1273.5252, found 1273.5225 (Δ 2.1 ppm).
Compound 16
1H NMR (700 MHz, DMSO) δ 10.52 (s, 1H), 8.60 (s, 1H), 8.34 (s, 1H), 8.29 (s, 1H), 8.22 (s, 2H), 8.17 (dd, J = 22.9, 7.6 Hz, 1H), 8.10 (d, J = 10.8 Hz, 1H), 7.98 (tt, J = 18.3, 9.0 Hz, 2H), 7.86 (s, 2H), 7.58 (s, 1H), 7.48 (dd, J = 21.7, 8.4 Hz, 1H), 7.38 (s, 1H), 7.21 (td, J = 17.6, 8.4 Hz, 3H), 7.19 (s, 3H), 7.18–7.10 (m, 2H), 7.10–7.01 (m, 3H), 7.01–6.97 (m, 2H), 6.97–6.81 (m, 6H), 6.87 (s, 3H), 6.75–6.70 (m, 1H), 6.67 (dd, J = 8.1, 3.6 Hz, 1H), 6.59–6.49 (m, 4H), 6.08 (d, J = 6.2 Hz, 1H), 5.74 (s, 1H), 5.64 (s, 1H), 5.50 (d, J = 14.0 Hz, 1H), 5.44–5.41 (m, 1H), 5.39 (d, J = 8.1 Hz, 1H), 5.33 (d, J = 6.9 Hz, 1H), 5.07 (s, 1H), 4.95 (s, 2H), 4.91 (s, 1H), 4.88 (s, 2H), 4.69–4.63 (m, 2H), 4.39 (s, 2H), 3.93 (s, 1H), 3.04 (dd, J = 6.2, 3.9 Hz, 1H), 3.04–2.98 (m, 6H), 2.96–2.89 (m, 1H), 2.73 (q, J = 8.3 Hz, 6H), 2.61 (d, J = 5.8 Hz, 1H), 2.54 (s, 17H), 2.30–2.26 (m, 2H), 2.18–2.13 (m, 2H), 2.13 (s, 2H), 1.71–1.62 (m, 2H), 1.66 (s, 3H), 1.56–1.49 (m, 1H), 1.51 (s, 8H), 1.51–1.45 (m, 9H), 1.38 (s, 5H), 1.27 (d, J = 7.9 Hz, 1H), 1.25 (s, 4H). 13C NMR (176 MHz, DMSO) δ 176.92, 171.51, 171.39, 171.30, 169.68, 169.15, 168.87, 163.66, 157.81, 157.64, 156.66, 156.57, 156.38, 155.48, 150.00, 149.85, 141.61, 139.49, 135.40, 134.48, 133.69, 131.07, 129.87, 129.05, 128.58, 128.56, 127.50, 127.37, 126.30, 124.08, 119.02, 118.54, 116.53, 115.94, 114.81, 114.75, 114.67, 114.59, 112.33, 107.58, 61.35, 55.10, 54.90, 54.34, 54.05, 41.43, 40.43, 40.02, 39.88, 39.76, 39.64, 39.52, 39.40, 39.28, 39.16, 38.69, 38.62, 36.41, 34.69, 34.64, 32.97, 32.38, 29.98, 29.80, 26.82, 26.80, 26.62, 25.49, 25.11, 24.69, 24.64, 23.93, 22.96. Yield: 45%. HRMS calcd [M + H]+ C67H64N9O15+ 1234.4516, found 1234.4520 (Δ −0.3 ppm).
Compound 17
1H NMR (700 MHz, DMSO) δ 10.52 (s, 1H), 9.56 (s, 4H), 8.64 (d, J = 4.9 Hz, 1H), 8.55 (d, J = 7.4 Hz, 2H), 8.45 (s, 5H), 8.22 (dd, J = 8.1, 3.3 Hz, 2H), 8.09 (d, J = 8.9 Hz, 1H), 7.91–7.85 (m, 3H), 7.63 (s, 1H), 7.58 (d, J = 2.7 Hz, 1H), 7.24–7.18 (m, 2H), 7.16 (dd, J = 8.1, 2.6 Hz, 1H), 7.13 (dd, J = 6.8, 2.3 Hz, 2H), 7.05–7.00 (m, 2H), 7.01 (s, 2H), 7.00–6.95 (m, 2H), 6.94–6.87 (m, 1H), 6.90 (s, 1H), 6.86 (dd, J = 8.6, 2.6 Hz, 1H), 6.74 (d, J = 8.3 Hz, 1H), 6.63–6.54 (m, 3H), 6.54–6.49 (m, 2H), 6.29 (s, 1H), 6.07 (d, J = 2.5 Hz, 1H), 5.71 (d, J = 2.5 Hz, 1H), 5.66 (d, J = 8.8 Hz, 1H), 5.42 (d, J = 8.2 Hz, 1H), 5.33 (d, J = 8.7 Hz, 1H), 5.15 (d, J = 8.2 Hz, 1H), 4.95 (dt, J = 13.3, 7.3 Hz, 1H), 4.85 (dd, J = 18.7, 3.7 Hz, 2H), 4.36 (d, J = 11.4 Hz, 1H), 3.18 (ddd, J = 17.6, 12.3, 8.4 Hz, 2H), 3.09–3.05 (m, 0H), 3.04 (s, 2H), 3.04 (s, 2H), 3.00 (t, J = 12.4 Hz, 1H), 2.92 (t, J = 12.7 Hz, 1H), 2.67 (t, J = 7.5 Hz, 2H), 2.54 (s, 2H), 2.50–2.44 (m, 1H), 2.21 (t, J = 7.2 Hz, 2H), 2.15 (t, J = 7.5 Hz, 2H), 2.10 (s, 6H), 1.56 (p, J = 7.0 Hz, 2H), 1.50–1.43 (m, 4H), 1.25 (dq, J = 15.4, 7.1 Hz, 3H). 13C NMR (176 MHz, DMSO) δ 171.42, 170.11, 169.67, 169.23, 168.89, 168.78, 168.24, 164.86, 156.69, 156.62, 156.45, 155.50, 150.22, 150.03, 144.21, 141.55, 135.39, 134.35, 133.78, 130.98, 130.77, 129.79, 129.05, 128.99, 128.77, 128.54, 128.23, 127.47, 126.31, 125.73, 125.48, 124.07, 122.65, 121.37, 118.53, 116.56, 116.17, 114.76, 114.60, 114.23, 111.31, 107.59, 61.28, 56.50, 55.09, 54.94, 54.34, 54.04, 45.14, 40.43, 40.01, 39.88, 39.76, 39.64, 39.52, 39.40, 39.28, 39.16, 38.95, 36.86, 35.86, 34.73, 32.34, 30.69, 27.55, 27.22, 26.87, 25.54, 24.76. Overall yield: 20%. HRMS calcd [M + H]+ C72H76N11O14+ 1318.5568, found 1318.5546 (Δ 1.6 ppm).
Compound 18
1H NMR (700 MHz, DMSO) δ 10.83 (d, J = 2.5 Hz, 1H), 9.38 (s, 2H), 9.16 (s, 1H), 9.06 (s, 1H), 8.60 (d, J = 5.0 Hz, 1H), 8.34 (s, 3H), 8.21 (d, J = 9.4 Hz, 1H), 8.09 (d, J = 8.5 Hz, 1H), 8.06–8.02 (m, 2H), 7.94 (dd, J = 8.4, 2.3 Hz, 1H), 7.73–7.66 (m, 3H), 7.62 (d, J = 1.9 Hz, 1H), 7.53–7.45 (m, 4H), 7.43–7.31 (m, 2H), 7.33–7.25 (m, 3H), 7.24 (d, J = 1.3 Hz, 1H), 7.19 (dd, J = 8.3, 2.1 Hz, 1H), 7.17–7.12 (m, 2H), 7.11–7.04 (m, 5H), 6.98 (dd, J = 8.6, 2.0 Hz, 1H), 6.89 (dd, J = 8.5, 2.6 Hz, 1H), 6.85 (dt, J = 8.4, 1.8 Hz, 2H), 6.80 (dd, J = 8.4, 2.5 Hz, 1H), 6.72–6.65 (m, 2H), 6.63 (d, J = 8.3 Hz, 1H), 6.49–6.45 (m, 2H), 5.59 (dd, J = 14.3, 8.6 Hz, 2H), 5.53 (d, J = 2.6 Hz, 1H), 5.16 (d, J = 2.7 Hz, 1H), 4.90 (d, J = 5.2 Hz, 1H), 4.87 (d, J = 2.1 Hz, 1H), 4.54 (d, J = 7.1 Hz, 1H), 4.39 (d, J = 12.0 Hz, 1H), 4.21 (s, 3H), 4.07 (s, 1H), 3.99 (s, 1H), 3.78 (s, 1H), 3.47–3.38 (m, 2H), 3.21 (q, J = 7.2 Hz, 8H), 3.12–3.07 (m, 3H), 3.02–2.99 (m, 1H), 3.00 (s, 2H), 2.98 (s, 1H), 2.71 (d, J = 12.7 Hz, 1H), 2.54 (s, 2H), 1.76 (d, J = 11.4 Hz, 1H), 1.71 (d, J = 12.2 Hz, 1H), 1.57 (q, J = 7.8 Hz, 3H), 1.31–1.25 (m, 14H), 1.27–1.23 (m, 1H), 1.23 (s, 3H), 1.16 (tt, J = 7.3, 1.9 Hz, 12H), 1.07 (s, 1H), 0.95 (t, J = 7.1 Hz, 1H), 0.86 (q, J = 7.7 Hz, 4H). 13C NMR (176 MHz, DMSO) δ 170.99, 168.87, 164.32, 162.49, 161.11, 156.62, 149.59, 149.01, 142.91, 141.54, 140.66, 139.51, 138.98, 138.87, 130.19, 128.75, 128.72, 128.67, 128.65, 128.60, 128.34, 127.42, 127.27, 127.08, 126.46, 126.28, 124.87, 115.70, 115.66, 115.58, 115.54, 114.89, 114.56, 64.81, 57.03, 55.95, 54.26, 52.16, 51.93, 51.91, 51.90, 51.08, 45.65, 40.43, 40.02, 39.90, 39.88, 39.76, 39.64, 39.52, 39.40, 39.33, 39.28, 39.16, 32.43, 32.29, 31.16, 28.48, 28.46, 25.81, 24.64, 22.03, 20.90, 13.95, 7.14. 19F NMR (659 MHz, DMSO) δ −115.81. Yield: 60%. HRMS calcd [M – H]− C74H60FN8O14– 1303.4219, found 1303.4234 (Δ −1.1 ppm).
Compound 19
1H NMR (700 MHz, DMSO) δ 10.84 (d, J = 2.5 Hz, 1H), 9.45 (s, 1H), 9.39 (s, 2H), 9.31 (s, 1H), 9.17 (s, 1H), 8.66 (d, J = 5.0 Hz, 1H), 8.52 (s, 1H), 8.18 (d, J = 9.4 Hz, 1H), 8.12 (s, 1H), 7.95 (dd, J = 8.5, 2.2 Hz, 1H), 7.91 (dd, J = 8.6, 2.1 Hz, 1H), 7.47 (dd, J = 12.1, 8.1 Hz, 2H), 7.24 (s, 1H), 7.23–7.14 (m, 3H), 7.10 (dd, J = 8.2, 2.7 Hz, 1H), 7.10–7.01 (m, 5H), 6.93 (dd, J = 8.5, 2.6 Hz, 1H), 6.91–6.80 (m, 4H), 6.75 (d, J = 8.3 Hz, 1H), 6.69 (s, 1H), 6.67–6.60 (m, 3H), 6.58 (s, 2H), 6.54 (dd, J = 12.2, 7.9 Hz, 1H), 6.51–6.44 (m, 2H), 5.61 (d, J = 9.3 Hz, 1H), 5.54 (d, J = 8.0 Hz, 1H), 5.49 (d, J = 2.6 Hz, 1H), 5.14 (d, J = 2.6 Hz, 1H), 5.04–5.01 (m, 1H), 4.91–4.86 (m, 2H), 4.36 (dd, J = 11.7, 3.7 Hz, 1H), 4.03 (ddd, J = 11.0, 8.2, 2.8 Hz, 1H), 3.99 (s, 1H), 3.20–3.15 (m, 1H), 3.06 (d, J = 12.5 Hz, 1H), 3.03 (s, 3H), 2.73 (d, J = 12.7 Hz, 1H), 2.54 (s, 1H), 1.27–1.17 (m, 1H), 0.82 (s, 1H). 13C NMR (176 MHz, DMSO) δ 171.16, 170.94, 169.09, 168.91, 163.18, 157.27, 156.70, 156.47, 154.89, 150.23, 149.13, 139.51, 136.36, 134.50, 133.84, 131.01, 129.84, 129.63, 129.15, 127.50, 126.27, 124.79, 124.15, 123.34, 118.63, 116.43, 114.98, 114.66, 114.26, 113.84, 112.08, 110.37, 61.31, 55.86, 54.91, 54.30, 40.43, 40.02, 39.88, 39.76, 39.64, 39.52, 39.40, 39.28, 39.16, 32.41. Yield: 65%. HRMS calcd [M – H]− C68H57FN8O15– 1225.3949, found 1225.3915 (Δ 2.7 ppm).
Compound 20
1H NMR (700 MHz, DMSO) δ 10.83 (d, J = 2.3 Hz, 1H), 9.38 (s, 2H), 9.25 (s, 1H), 9.10 (s, 3H), 8.60 (d, J = 5.0 Hz, 1H), 8.43 (s, 0H), 8.37 (s, 3H), 8.20 (d, J = 9.4 Hz, 1H), 8.06 (s, 1H), 8.07–8.01 (m, 2H), 7.94 (dd, J = 8.4, 2.3 Hz, 1H), 7.52–7.45 (m, 1H), 7.40 (d, J = 8.0 Hz, 1H), 7.24 (d, J = 1.2 Hz, 1H), 7.21–7.12 (m, 3H), 7.11–7.05 (m, 1H), 7.07–7.01 (m, 3H), 6.97 (dd, J = 8.6, 2.2 Hz, 1H), 6.91–6.78 (m, 4H), 6.65 (dt, J = 8.4, 2.3 Hz, 3H), 6.51 (dd, J = 9.1, 2.7 Hz, 2H), 6.17–6.09 (m, 2H), 6.06 (t, J = 2.2 Hz, 1H), 5.61 (d, J = 9.2 Hz, 1H), 5.57–5.52 (m, 2H), 5.15 (d, J = 2.6 Hz, 1H), 4.90 (d, J = 5.3 Hz, 1H), 4.87 (d, J = 2.0 Hz, 1H), 4.58 (d, J = 7.3 Hz, 1H), 4.39 (d, J = 10.9 Hz, 1H), 4.04–4.00 (m, 1H), 4.00 (s, 1H), 3.38 (s, 0H), 3.33 (d, J = 13.8 Hz, 2H), 3.23–3.15 (m, 2H), 3.01 (s, 1H), 3.00 (s, 2H), 2.98 (s, 1H), 2.74 (d, J = 12.3 Hz, 1H), 2.54 (s, 2H), 1.24 (s, 1H), 0.87 (td, J = 7.2, 2.6 Hz, 1H). 13C NMR (176 MHz, DMSO) δ 171.26, 168.88, 166.91, 165.06, 158.19, 156.49, 154.78, 149.63, 149.02, 142.99, 142.22, 139.51, 136.31, 134.49, 130.23, 128.29, 127.37, 114.83, 114.62, 113.67, 112.16, 105.90, 63.75, 61.32, 57.69, 57.39, 54.89, 50.69, 40.43, 40.02, 39.90, 39.88, 39.76, 39.64, 39.52, 39.40, 39.33, 39.28, 39.16, 32.43. Yield: 60%. HRMS calcd [M – H]− C68H57N8O16– 1241.3898, found 1241.3904 (Δ −0.4 ppm).
Compound 21
1H NMR (700 MHz, DMSO) δ 10.83 (d, J = 13.2 Hz, 1H), 9.38 (s, 3H), 9.16 (s, 1H), 8.66–8.63 (m, 1H), 8.46 (s, 1H), 8.17 (d, J = 10.2 Hz, 1H), 7.98–7.91 (m, 2H), 7.47 (d, J = 8.4 Hz, 1H), 7.23 (s, 1H), 7.22–7.14 (m, 2H), 7.13–7.06 (m, 1H), 7.08–7.00 (m, 1H), 6.93 (ddd, J = 7.7, 5.3, 2.6 Hz, 1H), 6.86 (qd, J = 8.1, 2.7 Hz, 2H), 6.84 (s, 2H), 6.74 (d, J = 8.2 Hz, 1H), 6.64 (s, 1H), 6.63 (t, J = 4.3 Hz, 1H), 6.56–6.49 (m, 2H), 6.44 (s, 2H), 5.61 (d, J = 9.1 Hz, 1H), 5.54 (d, J = 10.9 Hz, 1H), 5.50 (d, J = 2.7 Hz, 1H), 5.16–5.13 (m, 1H), 4.97 (s, 1H), 4.91–4.87 (m, 2H), 4.38 (d, J = 11.2 Hz, 1H), 4.05 (s, 1H), 3.19 (d, J = 11.9 Hz, 1H), 3.06 (s, 0H), 3.03 (s, 3H), 2.77 (t, J = 10.3 Hz, 1H), 2.54 (s, 6H), 1.24 (s, 1H). 13C NMR (176 MHz, DMSO) δ 171.14, 170.86, 169.01, 168.86, 167.83, 163.26, 156.63, 156.43, 154.84, 150.10, 149.06, 144.12, 139.45, 136.27, 134.63, 134.40, 133.79, 131.09, 130.95, 129.82, 129.55, 128.95, 127.39, 126.23, 124.73, 123.98, 123.29, 123.11, 121.69, 118.82, 118.56, 116.27, 115.01, 114.62, 114.20, 113.79, 111.99, 110.32, 61.25, 55.80, 54.85, 54.24, 40.43, 40.02, 39.88, 39.76, 39.64, 39.52, 39.40, 39.28, 39.16, 35.86, 32.35, 22.09, 13.95. Yield: 60%. HRMS calcd [M – H]− C68H57N8O16– 1241.3898, found 1241.3934 (Δ −2.8 ppm).
Compound 22
1H NMR (700 MHz, DMSO) δ 13.31 (s, 2H), 10.86 (d, J = 12.7 Hz, 2H), 9.69–9.63 (m, 2H), 9.51 (s, 3H), 9.40 (d, J = 18.8 Hz, 3H), 9.21–9.15 (m, 3H), 9.06 (s, 1H), 9.01 (s, 1H), 8.95 (s, 1H), 8.79 (s, 1H), 8.74 (s, 1H), 8.75–8.69 (m, 1H), 8.68 (d, J = 5.1 Hz, 2H), 8.63 (d, J = 8.3 Hz, 3H), 8.54 (s, 1H), 8.35 (s, 1H), 8.17 (d, J = 8.9 Hz, 2H), 7.95 (d, J = 8.4 Hz, 3H), 7.89 (dd, J = 8.7, 2.1 Hz, 2H), 7.47–7.41 (m, 2H), 7.35 (d, J = 8.2 Hz, 1H), 7.25 (t, J = 2.3 Hz, 2H), 7.24–7.19 (m, 7H), 7.15 (dd, J = 14.1, 2.6 Hz, 1H), 7.13–7.02 (m, 6H), 7.02–6.96 (m, 4H), 6.93 (dd, J = 8.5, 2.5 Hz, 2H), 6.90 (s, 0H), 6.89–6.82 (m, 6H), 6.83–6.78 (m, 1H), 6.78 (s, 5H), 6.63 (dq, J = 9.3, 6.8 Hz, 4H), 6.57 (d, J = 8.4 Hz, 1H), 6.53 (s, 12H), 6.52–6.47 (m, 4H), 6.35 (d, J = 8.3 Hz, 1H), 6.19 (s, 1H), 5.80 (s, 0H), 5.63–5.59 (m, 2H), 5.47 (dd, J = 15.3, 5.4 Hz, 5H), 5.12 (t, J = 6.2 Hz, 4H), 5.11 (s, 1H), 4.89 (d, J = 12.0 Hz, 5H), 4.74 (s, 1H), 4.38–4.34 (m, 3H), 3.97 (s, 2H), 3.95 (d, J = 2.9 Hz, 0H), 3.89 (s, 1H), 3.78 (s, 3H), 3.78 (d, J = 13.1 Hz, 0H), 3.17 (d, J = 11.6 Hz, 3H), 3.06 (s, 4H), 3.04 (s, 7H), 2.81 (d, J = 12.7 Hz, 2H), 2.54 (s, 10H), 1.24 (s, 1H), 0.83 (s, 1H). 13C NMR (176 MHz, DMSO) δ 171.10, 169.04, 168.83, 168.16, 157.92, 157.75, 157.57, 150.28, 149.06, 144.57, 130.89, 129.40, 127.28, 126.67, 124.75, 123.15, 121.81, 118.17, 116.46, 115.28, 114.63, 61.21, 54.86, 54.48, 40.43, 40.01, 39.88, 39.76, 39.64, 39.52, 39.40, 39.28, 39.16, 32.31. Yield: 60%. HRMS calcd [M – H]− C68H57N8O17– 1257.3847, found 1257.3784 (Δ 5.0 ppm).
Compound 23
1H NMR (700 MHz, DMSO) δ 10.87–10.84 (m, 1H), 9.51 (s, 1H), 9.42 (s, 1H), 9.39 (s, 1H), 9.14 (s, 1H), 8.68 (d, J = 5.0 Hz, 1H), 8.62 (d, J = 8.4 Hz, 1H), 8.18–8.12 (m, 1H), 7.96 (dd, J = 8.6, 1.9 Hz, 1H), 7.90 (dd, J = 8.6, 2.1 Hz, 1H), 7.48 (dd, J = 13.1, 8.1 Hz, 2H), 7.26–7.15 (m, 4H), 7.14–7.02 (m, 6H), 6.94 (dd, J = 8.4, 2.6 Hz, 1H), 6.87 (s, 1H), 6.90–6.81 (m, 2H), 6.80–6.77 (m, 2H), 6.70–6.63 (m, 2H), 6.54 (s, 3H), 6.50–6.44 (m, 2H), 5.62 (d, J = 9.3 Hz, 1H), 5.53 (d, J = 8.1 Hz, 1H), 5.49 (d, J = 2.6 Hz, 1H), 5.16–5.10 (m, 2H), 4.91–4.87 (m, 2H), 4.38–4.34 (m, 1H), 4.07–4.01 (m, 1H), 3.43–3.36 (m, 1H), 3.17 (d, J = 12.6 Hz, 1H), 3.07 (d, J = 12.6 Hz, 1H), 3.04 (s, 3H), 2.73 (d, J = 12.9 Hz, 1H), 2.54 (s, 24H), 1.24 (s, 1H). 13C NMR (176 MHz, DMSO) δ 171.11, 170.89, 169.04, 168.84, 168.14, 157.73, 157.56, 156.43, 154.84, 150.28, 149.07, 144.53, 139.45, 136.29, 133.82, 130.94, 127.46, 126.20, 118.26, 116.55, 115.01, 114.57, 111.93, 55.83, 54.84, 54.22, 40.43, 40.02, 39.88, 39.76, 39.64, 39.52, 39.40, 39.28, 39.16, 32.31. 19F NMR (659 MHz, DMSO) δ −73.43. Yield: 80%. HRMS calcd [M – H]− C68H56N8O14– 1227.3906, found 1227.3915 (Δ −0.7 ppm).
Compound 24
1H NMR (700 MHz, DMSO) δ 10.83 (d, J = 2.5 Hz, 1H), 9.39 (s, 1H), 9.38 (s, 1H), 9.33 (s, 1H), 9.13 (s, 1H), 8.64 (d, J = 5.1 Hz, 1H), 8.44 (s, 1H), 8.20–8.14 (m, 1H), 8.08 (d, J = 8.2 Hz, 1H), 7.96–7.91 (m, 2H), 7.48 (t, J = 8.2 Hz, 2H), 7.28 (dtd, J = 10.1, 7.9, 1.9 Hz, 1H), 7.24 (s, 1H), 7.22–7.14 (m, 3H), 7.09 (dtd, J = 14.9, 8.0, 3.4 Hz, 3H), 7.06–6.99 (m, 4H), 6.92 (dd, J = 8.4, 2.6 Hz, 1H), 6.89–6.81 (m, 4H), 6.73 (d, J = 8.3 Hz, 1H), 6.67–6.62 (m, 2H), 6.61 (s, 1H), 6.48–6.42 (m, 2H), 5.60 (d, J = 9.3 Hz, 1H), 5.54 (d, J = 7.9 Hz, 1H), 5.50 (d, J = 2.6 Hz, 1H), 5.14 (d, J = 2.6 Hz, 1H), 4.98–4.93 (m, 1H), 4.90–4.86 (m, 2H), 4.36 (dd, J = 11.6, 3.7 Hz, 1H), 4.06–3.98 (m, 1H), 4.01 (s, 1H), 3.57–3.49 (m, 1H), 3.48 (s, 1H), 3.22 (s, 0H), 3.21–3.16 (m, 1H), 3.02 (dd, J = 30.0, 12.9 Hz, 1H), 3.02 (s, 3H), 2.72 (d, J = 12.6 Hz, 1H), 2.54 (s, 1H), 1.29–1.20 (m, 1H), 0.81 (s, 1H). 13C NMR (176 MHz, DMSO) δ 170.92, 170.45, 169.07, 168.91, 156.69, 156.62, 156.45, 154.88, 150.13, 149.12, 148.81, 139.51, 136.35, 134.51, 133.81, 131.00, 129.91, 129.62, 129.04, 128.43, 127.46, 126.27, 125.56, 124.78, 124.42, 124.14, 124.03, 123.30, 123.15, 121.75, 118.92, 118.61, 115.76, 115.66, 114.93, 114.59, 114.27, 113.82, 112.07, 110.37, 64.30, 61.32, 57.31, 55.87, 54.90, 54.31, 43.73, 40.43, 40.01, 39.88, 39.76, 39.64, 39.52, 39.40, 39.28, 39.16, 35.89, 32.42, 28.84. Yield: 75%. HRMS calcd [M – H]− C68H55F2N8O14– 1245.3811, found 1245.3822 (Δ −0.8 ppm).
Compound 25
1H NMR (700 MHz, DMSO) δ 10.83 (d, J = 2.4 Hz, 1H), 9.39 (s, 1H), 9.33 (s, 1H), 9.24 (s, 1H), 9.10 (s, 1H), 8.61 (d, J = 5.0 Hz, 1H), 8.22 (s, 1H), 8.17 (t, J = 4.8 Hz, 1H), 8.08 (d, J = 8.3 Hz, 1H), 7.97 (d, J = 8.5 Hz, 1H), 7.93 (dd, J = 8.5, 2.3 Hz, 1H), 7.48 (q, J = 8.1 Hz, 2H), 7.43–7.32 (m, 2H), 7.24 (d, J = 1.2 Hz, 1H), 7.20 (dd, J = 8.4, 2.1 Hz, 1H), 7.16 (dt, J = 5.4, 2.8 Hz, 2H), 7.13–7.08 (m, 1H), 7.10–7.05 (m, 1H), 7.07 (s, 1H), 7.05 (s, 1H), 7.05–6.98 (m, 3H), 6.91 (dd, J = 8.4, 2.7 Hz, 2H), 6.88–6.80 (m, 3H), 6.68 (d, J = 8.3 Hz, 1H), 6.67–6.58 (m, 6H), 6.45–6.39 (m, 2H), 5.61–5.49 (m, 3H), 5.14 (d, J = 2.6 Hz, 1H), 4.88 (d, J = 2.7 Hz, 2H), 4.76 (s, 1H), 4.58 (d, J = 4.9 Hz, 1H), 4.37 (dd, J = 12.2, 3.6 Hz, 1H), 4.03 (s, 1H), 4.06–3.99 (m, 1H), 3.58–3.49 (m, 1H), 3.20 (d, J = 12.1 Hz, 1H), 3.01 (s, 3H), 2.99 (d, J = 6.7 Hz, 1H), 2.71 (d, J = 12.5 Hz, 1H), 2.54 (s, 3H), 1.23 (d, J = 4.5 Hz, 3H), 0.88–0.81 (m, 1H). 13C NMR (176 MHz, DMSO) δ 170.91, 170.47, 168.92, 156.63, 156.45, 154.86, 149.11, 139.54, 136.35, 130.98, 129.84, 128.41, 127.40, 126.26, 114.91, 114.52, 112.09, 110.10, 104.06, 64.26, 57.31, 56.32, 54.92, 43.50, 40.43, 40.02, 39.88, 39.76, 39.64, 39.52, 39.40, 39.28, 39.16, 35.84, 32.46. Yield: 80%. HRMS calcd [M – H]− C68H54F3N8O14– 1263.3717, found 1263.3748 (Δ – 2.4 ppm).
Compound 26
1H NMR (700 MHz, DMSO) δ 10.86 (d, J = 13.0 Hz, 1H), 9.73 (s, 1H), 9.54 (s, 1H), 9.40 (s, 1H), 9.19 (s, 1H), 8.79 (s, 1H), 8.69–8.61 (m, 2H), 8.16 (d, J = 9.4 Hz, 1H), 7.94 (t, J = 9.7 Hz, 1H), 7.89 (d, J = 8.4 Hz, 1H), 7.80–7.72 (m, 1H), 7.64–7.55 (m, 1H), 7.50 (d, J = 8.6 Hz, 1H), 7.44 (dt, J = 16.0, 7.8 Hz, 2H), 7.29–7.14 (m, 4H), 7.11 (dq, J = 11.7, 7.9 Hz, 3H), 6.98 (d, J = 8.4 Hz, 2H), 6.97–6.90 (m, 1H), 6.90–6.75 (m, 5H), 6.66 (d, J = 8.3 Hz, 2H), 6.54 (s, 0H), 6.50–6.41 (m, 2H), 5.61 (d, J = 9.3 Hz, 1H), 5.55–5.44 (m, 2H), 5.37 (s, 0H), 5.14–5.10 (m, 2H), 4.94 (s, 1H), 4.91–4.86 (m, 2H), 4.72 (s, 1H), 4.36 (dd, J = 11.6, 4.1 Hz, 1H), 4.22 (s, 2H), 4.01 (td, J = 8.7, 4.8 Hz, 1H), 3.80 (s, 1H), 3.19–3.14 (m, 1H), 3.06 (t, J = 11.1 Hz, 1H), 3.04 (s, 3H), 2.79–2.71 (m, 1H), 0.81 (s, 1H). 13C NMR (176 MHz, DMSO) δ 171.16, 170.97, 169.10, 168.91, 168.24, 157.98, 157.80, 156.71, 156.46, 154.85, 150.35, 149.14, 144.62, 139.52, 138.49, 136.37, 134.59, 134.46, 133.85, 133.70, 131.21, 131.05, 130.96, 130.90, 130.51, 129.87, 129.52, 129.41, 129.32, 129.04, 128.63, 127.47, 127.39, 126.73, 126.19, 124.80, 124.16, 123.21, 121.87, 118.71, 118.57, 116.59, 115.34, 114.68, 114.29, 113.86, 112.02, 110.33, 62.18, 61.29, 59.31, 57.83, 55.96, 54.93, 54.55, 54.30, 40.43, 40.01, 39.88, 39.76, 39.64, 39.52, 39.40, 39.28, 39.16, 35.94, 32.40, 28.55. Yield: 80%. HRMS calcd [M – H]− C68H54Cl3N8O14– 1311.2831, found 1311.2788 (Δ 3.2 ppm).
Compound 27
1H NMR (700 MHz, DMSO) δ 10.88 (s, 1H), 9.75 (s, 1H), 9.56 (d, J = 56.7 Hz, 3H), 9.39 (s, 1H), 9.18 (s, 1H), 8.82 (d, J = 7.6 Hz, 1H), 8.65 (dd, J = 35.4, 6.7 Hz, 2H), 8.25–8.09 (m, 1H), 7.95 (d, J = 8.4 Hz, 1H), 7.92–7.85 (m, 2H), 7.85–7.77 (m, 3H), 7.77–7.62 (m, 1H), 7.44 (dd, J = 20.4, 8.1 Hz, 2H), 7.31 (d, J = 6.5 Hz, 1H), 7.27–7.24 (m, 1H), 7.24–7.19 (m, 2H), 7.17 (d, J = 2.4 Hz, 1H), 7.10 (dd, J = 8.5, 2.5 Hz, 3H), 6.99–6.90 (m, 3H), 6.87 (d, J = 1.8 Hz, 2H), 6.83 (dd, J = 8.4, 1.3 Hz, 1H), 6.78 (s, 2H), 6.72–6.62 (m, 2H), 6.63–6.43 (m, 14H), 5.93 (s, 1H), 5.61 (d, J = 9.4 Hz, 1H), 5.52–5.43 (m, 2H), 5.22–5.06 (m, 2H), 4.98–4.67 (m, 3H), 4.36 (d, J = 11.6 Hz, 1H), 4.22–3.88 (m, 2H), 3.04 (s, 3H), 2.70–2.57 (m, 1H), 2.50 (p, J = 1.8 Hz, 72H). 13C NMR (176 MHz, DMSO) δ 171.10, 168.84, 168.16, 157.84, 157.67, 157.49, 150.28, 144.57, 138.64, 130.89, 130.49, 130.40, 130.14, 127.23, 126.66, 125.69, 122.79, 120.09, 117.99, 116.29, 115.37, 114.63, 61.12, 55.90, 54.87, 54.47, 54.22, 40.43, 40.01, 39.88, 39.76, 39.64, 39.52, 39.40, 39.28, 39.16, 32.32. Yield: 80%. HRMS calcd [M – H]− C69H56F3N8O14– 1277.3874, found 1277.3922 (Δ – 3.7 ppm).
Compound 28
1H NMR (700 MHz, DMSO) δ 13.36 (s, 1H), 10.89–10.86 (m, 1H), 9.77 (s, 1H), 9.67 (s, 2H), 9.61 (s, 1H), 9.52 (s, 1H), 9.39 (s, 1H), 9.18 (s, 1H), 8.84 (s, 1H), 8.67 (d, J = 5.0 Hz, 1H), 8.63 (d, J = 8.3 Hz, 1H), 8.19 (s, 3H), 8.16 (d, J = 9.5 Hz, 1H), 7.97–7.91 (m, 1H), 7.89 (dd, J = 8.6, 2.1 Hz, 1H), 7.49 (d, J = 7.5 Hz, 1H), 7.43 (d, J = 8.5 Hz, 1H), 7.25 (s, 1H), 7.22–7.17 (m, 2H), 7.15 (d, J = 2.4 Hz, 1H), 7.10 (dd, J = 8.3, 2.7 Hz, 3H), 6.97–6.89 (m, 3H), 6.87 (s, 2H), 6.85–6.81 (m, 1H), 6.78 (s, 1H), 6.68 (d, J = 8.4 Hz, 2H), 6.54 (s, 2H), 6.47 (d, J = 8.1 Hz, 2H), 5.61 (d, J = 9.3 Hz, 1H), 5.49–5.44 (m, 2H), 5.14–5.10 (m, 2H), 4.89 (s, 1H), 4.91–4.86 (m, 2H), 4.38–4.34 (m, 1H), 4.30 (s, 1H), 4.15 (s, 1H), 4.08–4.02 (m, 1H), 3.17 (dt, J = 11.2, 3.7 Hz, 2H), 3.06 (t, J = 10.6 Hz, 1H), 3.04 (s, 3H), 3.01 (tt, J = 5.4, 2.7 Hz, 2H), 2.75 (d, J = 13.0 Hz, 1H), 2.54 (s, 3H), 1.76–1.70 (m, 2H), 0.82 (s, 1H). 13C NMR (176 MHz, DMSO) δ 171.11, 170.90, 169.05, 168.85, 168.17, 157.80, 157.63, 156.66, 156.39, 154.80, 150.30, 149.09, 144.58, 139.47, 136.31, 134.56, 130.91, 129.48, 127.43, 127.27, 126.68, 126.12, 124.76, 118.20, 116.50, 114.63, 113.81, 111.96, 61.22, 55.91, 54.88, 54.49, 54.24, 45.88, 45.85, 40.43, 40.02, 39.88, 39.76, 39.64, 39.52, 39.40, 39.28, 39.16, 32.33, 25.94, 25.90. 19F NMR (659 MHz, DMSO) δ −61.13, −73.70. Yield: 80%. HRMS calcd [M – H]− C70H55F6N8O14– 1345.3747, found 1345.3747 (Δ 0 ppm).
Compound 29
1H NMR (700 MHz, DMSO) δ 10.84 (d, J = 11.2 Hz, 1H), 9.53 (s, 1H), 9.40 (s, 1H), 9.18 (s, 1H), 8.67 (d, J = 5.0 Hz, 1H), 8.63 (d, J = 8.2 Hz, 1H), 8.16 (d, J = 9.4 Hz, 1H), 8.13 (s, 0H), 7.95 (dd, J = 8.5, 2.1 Hz, 1H), 7.89 (dd, J = 8.5, 2.1 Hz, 1H), 7.67–7.60 (m, 1H), 7.56–7.42 (m, 8H), 7.28 (s, 2H), 7.26 (s, 2H), 7.24 (s, 1H), 7.20 (ddd, J = 15.7, 6.9, 2.2 Hz, 3H), 7.18–7.01 (m, 6H), 6.94 (dd, J = 8.5, 2.6 Hz, 1H), 6.90–6.84 (m, 2H), 6.82 (d, J = 8.5 Hz, 1H), 6.78 (s, 2H), 6.68–6.63 (m, 2H), 6.54 (s, 1H), 6.50–6.44 (m, 2H), 5.61 (d, J = 9.3 Hz, 1H), 5.51 (q, J = 7.9 Hz, 1H), 5.48 (d, J = 2.7 Hz, 1H), 5.29 (t, J = 5.7 Hz, 2H), 5.14–5.06 (m, 2H), 4.88 (d, J = 16.9 Hz, 2H), 4.72 (d, J = 5.4 Hz, 1H), 4.46 (d, J = 5.4 Hz, 4H), 4.36 (dd, J = 11.6, 3.8 Hz, 1H), 4.03 (s, 1H), 3.19–3.11 (m, 1H), 3.07 (d, J = 12.5 Hz, 1H), 3.04 (s, 3H), 2.73 (d, J = 13.0 Hz, 1H), 1.36–1.25 (m, 1H), 0.81 (s, 1H). 13C NMR (176 MHz, DMSO) δ 171.16, 170.94, 169.11, 168.91, 168.23, 156.70, 156.48, 154.89, 150.34, 149.14, 144.60, 142.02, 139.51, 138.49, 136.36, 134.46, 133.85, 131.81, 131.20, 131.05, 130.96, 130.51, 129.84, 129.63, 129.32, 128.63, 128.62, 127.50, 126.79, 126.24, 124.80, 124.07, 119.57, 118.71, 118.56, 116.58, 115.12, 114.65, 114.27, 113.86, 111.99, 110.36, 62.18, 61.29, 59.31, 55.89, 54.90, 54.59, 54.29, 40.43, 40.02, 39.88, 39.76, 39.64, 39.52, 39.40, 39.28, 39.16, 35.94, 32.39. Yield: 80%. HRMS calcd [M – H]− C68H56BrN8O14– 1287.3105, found 1287.3083 (Δ 1.7 ppm).
Compound 30
1H NMR (700 MHz, DMSO) δ 10.83 (d, J = 2.4 Hz, 1H), 9.37 (d, J = 27.7 Hz, 2H), 9.25 (s, 1H), 9.15 (s, 1H), 8.66 (s, 1H), 8.62 (d, J = 5.0 Hz, 1H), 8.25 (s, 3H), 8.19 (d, J = 9.9 Hz, 1H), 8.09 (d, J = 8.3 Hz, 1H), 8.00–7.93 (m, 3H), 7.85 (d, J = 7.9 Hz, 1H), 7.68 (d, J = 7.5 Hz, 2H), 7.49 (dd, J = 7.9, 4.2 Hz, 2H), 7.25–7.18 (m, 2H), 7.17 (dd, J = 8.2, 2.5 Hz, 2H), 7.12–7.02 (m, 7H), 6.94–6.80 (m, 5H), 6.71–6.62 (m, 5H), 6.47 (d, J = 8.2 Hz, 2H), 5.60 (d, J = 9.3 Hz, 1H), 5.56 (d, J = 8.0 Hz, 1H), 5.51 (d, J = 2.6 Hz, 1H), 5.15 (d, J = 2.6 Hz, 1H), 4.89 (dd, J = 10.5, 3.6 Hz, 2H), 4.77 (s, 1H), 4.39 (dd, J = 11.7, 4.0 Hz, 1H), 4.09–4.03 (m, 1H), 3.95 (s, 1H), 3.21 (d, J = 11.6 Hz, 1H), 3.02 (s, 3H), 3.00 (d, J = 8.9 Hz, 1H), 2.71 (d, J = 12.5 Hz, 1H), 2.54 (s, 13H). 13C NMR (176 MHz, DMSO) δ 193.59, 170.92, 170.51, 169.00, 168.94, 163.59, 156.71, 156.65, 156.49, 154.84, 149.92, 149.10, 139.53, 137.16, 136.36, 134.58, 134.34, 134.05, 133.78, 131.02, 129.69, 129.08, 128.43, 128.36, 127.55, 127.50, 127.19, 126.28, 124.15, 124.02, 123.32, 123.10, 121.63, 116.02, 114.98, 114.65, 114.25, 113.75, 111.99, 110.41, 64.69, 61.36, 57.09, 55.84, 54.91, 54.30, 51.08, 40.43, 40.02, 39.88, 39.76, 39.64, 39.52, 39.40, 39.28, 39.16, 35.89, 32.47, 28.91. Yield 80%. HRMS calcd [M – H]− C68H58BN8O16– 1253.4069, found 1253.4057 (Δ 0.9 ppm).
Compound 31
1H NMR (700 MHz, DMSO) δ 10.85–10.78 (m, 1H), 9.40–9.33 (m, 2H), 9.21 (d, J = 17.4 Hz, 2H), 8.62 (t, J = 4.1 Hz, 1H), 8.47 (ddd, J = 18.5, 5.0, 1.7 Hz, 1H), 8.21–8.13 (m, 2H), 8.08–8.03 (m, 1H), 8.01–7.89 (m, 2H), 7.67 (td, J = 7.6, 1.9 Hz, 1H), 7.58–7.51 (m, 1H), 7.51–7.39 (m, 1H), 7.27–7.19 (m, 2H), 7.20–7.16 (m, 1H), 7.18–7.03 (m, 7H), 7.05–6.95 (m, 1H), 6.95–6.79 (m, 5H), 6.68 (dd, J = 14.7, 8.0 Hz, 6H), 6.52 (dd, J = 15.5, 8.6 Hz, 2H), 5.60 (d, J = 9.3 Hz, 1H), 5.55 (dd, J = 7.8, 4.0 Hz, 1H), 5.52 (d, J = 2.6 Hz, 1H), 5.16–5.08 (m, 1H), 4.90–4.84 (m, 2H), 4.71 (s, 1H), 4.38 (dd, J = 11.8, 3.7 Hz, 1H), 4.09–4.00 (m, 2H), 3.50 (s, 1H), 3.21 (d, J = 11.3 Hz, 1H), 3.01 (s, 3H), 3.04–2.96 (m, 2H), 2.74 (d, J = 12.2 Hz, 1H), 2.54 (s, 1H), 1.29–1.20 (m, 1H), 0.82 (s, 1H). 13C NMR (176 MHz, DMSO) δ 170.99, 170.90, 168.92, 163.63, 159.02, 156.70, 156.50, 154.84, 149.09, 148.77, 139.54, 136.55, 136.34, 133.74, 131.10, 130.99, 129.73, 128.42, 127.52, 122.04, 121.60, 115.02, 114.68, 113.76, 112.01, 64.90, 61.38, 54.93, 54.33, 52.42, 40.43, 40.02, 39.88, 39.76, 39.64, 39.52, 39.40, 39.28, 39.16, 32.47. Yield 78%: HRMS calcd [M – H]− C67H57N9O14– 1210.3954, found 1210.3957 (Δ 0.2 ppm).
Compound 32
1H NMR (700 MHz, DMSO) δ 10.83 (d, J = 2.5 Hz, 1H), 9.39 (s, 1H), 9.36 (s, 1H), 9.18 (s, 2H), 8.61 (d, J = 5.1 Hz, 1H), 8.21 (s, 1H), 8.16 (dd, J = 18.7, 8.8 Hz, 2H), 8.05 (d, J = 8.4 Hz, 1H), 7.99 (dd, J = 8.6, 2.1 Hz, 1H), 7.94 (dd, J = 8.4, 2.3 Hz, 1H), 7.56–7.44 (m, 2H), 7.41–7.35 (m, 1H), 7.25–7.18 (m, 2H), 7.18–7.14 (m, 2H), 7.14–7.00 (m, 6H), 6.99 (d, J = 2.9 Hz, 1H), 6.95–6.79 (m, 5H), 6.67 (d, J = 8.4 Hz, 3H), 6.51–6.46 (m, 2H), 5.59 (d, J = 9.3 Hz, 1H), 5.56 (d, J = 8.0 Hz, 1H), 5.51 (d, J = 2.6 Hz, 1H), 5.15 (d, J = 2.6 Hz, 1H), 4.90–4.86 (m, 2H), 4.69 (s, 1H), 4.37 (s, 1H), 4.05 (ddd, J = 11.1, 8.5, 2.9 Hz, 1H), 3.93 (s, 1H), 3.50 (s, 1H), 3.23–3.19 (m, 1H), 3.00 (s, 3H), 2.99 (d, J = 3.9 Hz, 1H), 2.72 (d, J = 12.6 Hz, 1H), 2.54 (s, 1H), 0.82 (s, 1H). 13C NMR (176 MHz, DMSO) δ 171.06, 170.92, 170.56, 168.93, 167.19, 163.80, 156.66, 156.52, 154.83, 149.80, 149.10, 142.33, 140.95, 139.95, 139.54, 136.34, 131.14, 131.00, 130.16, 129.72, 128.46, 128.42, 128.31, 128.00, 127.57, 127.47, 126.87, 126.10, 125.76, 123.31, 122.63, 121.84, 114.98, 114.68, 114.25, 111.97, 110.41, 64.72, 61.39, 57.78, 57.07, 55.87, 54.92, 54.33, 51.69, 46.12, 40.43, 40.02, 39.96, 39.88, 39.76, 39.64, 39.52, 39.40, 39.28, 39.16, 35.84, 32.49. Yield 78%: HRMS calcd [M – H]− C66H55N8O14S– 1215.3564, found 1215.3543 (Δ 1.7 ppm).
Bacteria Growth
Escherichia coli strains were grown in LB broth (Bioshop, Canada) at 37 °C, 250 rpm. The Streptomyces strain was grown on soy flour medium (20 g of soy flour, 20 g of d-mannitol, 20 g of agar, tap water 1 L, pH 7.2–7.4) for sporulation and conjugation (supplemented with 20 mM MgCl2). TSBY (30 g tryptic soy broth, BD; 5 g yeast extract; ddH2O 1 L) medium was used to prepare Streptomyces seed culture for fermentation. Streptomyces antibiotic medium (15 g of glucose, 15 g of soytone, 5 g of NaCl, 1 g of yeast extract, 1 g of CaCO3, 2.5 mL of glycerol, ddH2O 1 L, pH 6.8) was used for rimomycin production. Antibiotics were added for selection pressure as required (50 μg/mL kanamycin, 50 μg/mL apramycin, 25 μg/mL thiostrepton, 25 μg/mL nalidixic acid, 50 μg/mL trimethoprim).
Antibiotic Activity Assay
The antibacterial activity of all selected compounds was tested according to standard procedures using Mueller-Hinton Broth (MHB, BD Biosciences) for B. subtilis 168 and S. aureus ATCC 33591, Brain Heart Infusion medium (BHI, BD Biosciences) for Enterococcus, and Middlebrook 7H9 broth (4.7 g Middlebrook 7H9 powder, 2 mL glycerol, 898 mL ddH2O; 100 mL of Middlebrook OADC Enrichment (BD Biosciences) and 0.5 mL of Tween-80 were added before setting up MIC assays) for M. smegmatis mc2155 and other mycobacterial species.
Antimycobacterial Strains MICs
Mycobacterial strains were cultured in standard 7H9 media (Middlebrook 7H9 containing 0.2% glycerol, 10% OADC, and 0.05% Tween-80) and diluted to a final CFU/mL of ∼5 × 105 cells/mL. CFU totals were confirmed to be within the range of 3–8 × 105 cells/mL by plating dilutions of the inoculum on Middlebrook 7H10 (Becton Dickenson) agar containing 0.5% glycerol and 10% OADC. Susceptibility was assessed at 2 days (M. smegmatis, M. fortuitum, M. abscessus) or 7 days (M. tuberculosis H37Ra, M. bovis, M. avium). Cultures were incubated with 30 μg/mL resazurin for one additional day to identify any effect on growth where compound solubility was limiting. Reported data are the averages from at least two experiments.
Target Gene Deletion of rmoL
rmoL deletion on pGP6783 was performed using λ-red-mediated PCR targeting.9 ΔrmoL-F (CGGCAGGGACACCGCGTCCTGATACTGGAAAAGGAATTCTTCCTCGAGtgcagctcacggtaactgat) and ΔrmoL-R (GAGCAGGAACTCGTAGAAGACGCTGTACTCCCGCCGGTAGCGCTCGAGaggaacttatgagctcagcc) primers were used to amplify the aac(3)IV resistance cassette from pCGW. The aac(3)IV amplicon was electroporated into E. coli BW25113/ pKD46/pGP6783 competent cells to replace rmoL through homologous recombination, resulting in pGP6783ΔrmoL::aac(3)IV. The inserted aac(3)IV cassette was removed through XhoI restriction digestion overnight at 37 °C followed by self-ligation at 20 °C using T4 DNA ligase for 4 h, generating the rmoL in-frame deletion mutant plasmid, pGP6783ΔrmoL. pGP6783ΔrmoL mutant plasmid was confirmed by PCR and Sanger sequencing using ΔrmoL-dF (GAATAGCAGGCGGGCAGATA) and ΔrmoL-dR (GACCACCGAGGACTTGAACA) primers. pGP6783ΔrmoL was then transformed into E. coli ET1257 and conjugated into S. coelicolor M1154/pAMX4 through E. coli-Streptomyces interspecies tri-parental mating for pure rimomycin A production.
Preparation of Rimomycin A
S. coelicolor M1154/pAMX4/pGP6783ΔrmoL strain was grown on SFM agar plate (supplemented with 50 μg/mL kanamycin, 25 μg/mL thiostrepton, and 25 μg/mL nalidixic acid) for 5 days at 30 °C. Picked ten single colonies from the agar plate and then inoculated into 100 mL of TSBY in a 250 mL flask (×6) containing 15 5 mm glass beads (Fisher) supplemented with identical antibiotics, growing for 3 days at 30 °C. The seed culture was then inoculated into 650 mL SAM medium (5% inoculum) in a 2.8 L Thomson ULTR YIELD flask (×20), growing for 8 days at 30 °C. Rimomycin A was then purified from the harvested mycelium using an isolation procedure designed for Type V GPAs described in our previous report.40 Yield: 600 mg/L (24-well plate format).
Cytotoxicity Testing
On day 1, HEK293 cells (ATCC CRL-1573; generation 6) were seeded at 7500 cells/well in 384-well tissue culture-treated white plates in 50 μL of Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, 100 units/mL penicillin, and 100 μg/mL streptomycin. Cells were incubated for 18 h at 37 °C under 5% CO2. After 18 h, 500 nL of compound and DMSO were added to the cells using a Labcyte Echo acoustic dispenser (Beckman Coulter) and a combi nL (Thermo Fisher) dispenser. The final DMSO concentration was 1% in all wells. Cells were incubated for an additional 48 h, after which cell viability was assessed using Promega Cell Titer Glo 2.0 reagent (Fisher Scientific). Cell Titer Glo (50 μL) was added directly to the media; the plates were shaken for 2 min and then incubated for 10 min at room temperature. The luminescence was read on a Neo2 plate reader (Biotek) using luminescence fiber. Controls were untreated cells and cells treated with DMSO only. The data were normalized to DMSO-only controls, and the curves were fitted using a four-parameter logistic (4PL) nonlinear regression model constrained to a minimum response of 0 and a maximum response of 1.
Hemolysis Assay
Human blood drawn into K2-EDTA tubes was purchased from BioIVT (New York). The blood was centrifuged at 500g for 5 min, and the plasma was removed. The red blood cells (RBCs) were washed 2 times with 150 mM NaCl at a volume equal to the removed plasma. After the second wash, the RBCs were resuspended in phosphate-buffered saline (PBS) at a pH of 7.4 at a volume equal to the plasma to maintain the hematocrit. A volume of 1 μL of the compound was added to a 96-well V-bottom plate well using a Labcyte Echo acoustic dispenser (Beckman Coulter). DMSO was kept constant at 1% (v/v) final concentration, and a DMSO-only negative control was included for each replicate. The positive control was 10 μL of Triton X-100 (starting at a high concentration of 20% and going down in 2-fold dilutions to 0.02%). The RBCs were diluted 1:50 in pH 7.4 PBS, and 99 μL was added to each well. The plates were incubated at 37 °C for 1 hour and then centrifuged at 500g for 5 min to pellet intact RBCs. A volume of 65 μL of the supernatant was transferred to a clear, flat bottom 96-well plate, and the absorbance was read at 540 nm.
Solubility Assay
Compound solubility was determined in a saline solution by equilibrating an excess of solid compound (10 mg/mL) at 25 °C for 24 h with shaking. The samples were spun down, and the supernatant was analyzed on LC-qTOF (Agilent Technologies) with injection volumes of 1, 5, 10, and 20 μL. Calibration curves were plotted by dissolving the compounds to a final concentration of 1 mg/mL in DMSO. The solutions were further diluted to 100 μg/mL, and different volumes were injected.
Neutropenic Thigh Infection Model
All mouse experiments were performed in the Central Animal Facility at McMaster University under animal use protocol #20-12-41 as approved by the Animal Research Ethics Board according to guidelines set by the Canadian Council on Animal Care. Female mice were used following previously established models.41 Before infection, mice in groups of two or three were randomly relocated from housing cages into treatment and control cages. No animals were excluded from analyses, and blinding was considered unnecessary. All experiments used 6- to 10-week-old female CD-1 mice (Harlan/Envigo #030). The mice were rendered neutropenic with i.p. cyclophosphamide at 150 mg/kg (day 4) and 100 mg/kg (day 1). For bacterial preparation, E. faecium ATCC 700221 was grown overnight in brain heart infusion (BHI) media supplemented with 128 μg/mL vancomycin and subcultured to OD600 ∼0.4. Bacteria were adjusted to ∼1 × 107 CFU/mL with sterile BHI media, and ∼1 × 106 CFU was injected into the right and left thighs of three to five mice per treatment group. Compound 17 was formulated in PEG400/DMSO/H2O (45/5/50, v/v/v). The animals were treated at 1-, 3-, 5-, and 20 h post-infection with either linezolid (50 mg/kg, i.p., n = 3), Compound 17 (100 mg/kg, i.p., n = 5), or control (PEG400/DMSO/H2O, i.p., n = 4). We dosed linezolid (4 doses at 50 mg/kg) 200 mg/kg/day × 0.08 = 16 mg/kg human equivalent dosage. The mice were euthanized 24 h after infection. Thigh tissue was aseptically collected, weighed, and homogenized in 1 mL of PBS for 15 min at 30 rps (Retsch MM400). Homogenates were serially diluted in PBS and plated onto solid BHI supplemented with vancomycin (128 μg/mL). Plates were incubated overnight at 37 °C, and colonies were quantified to determine bacterial load.
Acknowledgments
Special thanks go to Susan McCusker and Tracey Campbell for performing the toxicity and hemolysis assays.
Glossary
Abbreviations Used
- Boc2O
di-tert-butyl decarbonate
- Boc-Ahx-NHS
6-(boc-amino)hexanoic acid N-succinimidyl ester
- CaCO3
calcium carbonate
- CFU
colony-forming unit
- CH3CN
acetonitrile
- DCM
dichloromethane
- DIPEA
diisopropylethyl amine
- DMF
dimethylformamide
- DMSO
dimethyl sulfoxide
- NMM
N-methyl morpholine
- PBS
phosphate-buffered saline
- PEG
poly(ethylene glycol)
- PyBop
benzotriazol-1-yloxytripyrrolidinophosphonium hexafluorophosphate
- RT
room temperature
- TEA
triethylamine
- TFA
trifluoroacetic acid
- THF
tetrahydrofuran
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jmedchem.3c00633.
Author Contributions
∥ K.K. and M.X. contributed equally to this work. K.K., M.X., and G.D.W. conceived and designed the study. K.K. synthesized analogues. M.X. and W.W. purified the starting rimomycin A. M.X. and M.A.C. performed the biological analyses. A.A.F.-C. performed animal experiments. B.K.C. supervised animal studies. All authors discussed the results. M.X., K.K., and G.D.W. wrote the paper.
This research was funded by a Canadian Institutes of Health Research grant (FRN-148463) to G.D.W. and PJT-388472 to B.K.C.
The authors declare no competing financial interest.
Supplementary Material
References
- Zeng D.; Debabov D.; Hartsell T. L.; Cano R. J.; Adams S.; Schuyler J. A.; McMillan R.; Pace J. L. Approved Glycopeptide Antibacterial Drugs: Mechanism of Action and Resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a026989 10.1101/cshperspect.a026989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick G. M.; Jones P. G.; Kennard O.; Williams D. H.; Smith G. A. Structure of vancomycin and its complex with acetyl-D-alanyl-D-alanine. Nature 1978, 271, 223–225. 10.1038/271223a0. [DOI] [PubMed] [Google Scholar]
- Reynolds P. E. Structure, biochemistry and mechanism of action of glycopeptide antibiotics. Eur. J. Clin. Microbiol. Infect. Dis. 1989, 8, 943–950. 10.1007/BF01967563. [DOI] [PubMed] [Google Scholar]
- Pootoolal J.; Neu J.; Wright G. D. Glycopeptide antibiotic resistance. Annu. Rev. Pharmacol. Toxicol. 2002, 42, 381–408. 10.1146/annurev.pharmtox.42.091601.142813. [DOI] [PubMed] [Google Scholar]
- Leclercq R.; Derlot E.; Duval J.; Courvalin P. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N. Engl. J. Med. 1988, 319, 157–161. 10.1056/NEJM198807213190307. [DOI] [PubMed] [Google Scholar]
- Bugg T. D. H.; Dutka-Malen S.; Arthur M.; Courvalin P.; Walsh C. T. Identification of vancomycin resistance protein VanA as a D-alanine:D-alanine ligase of altered substrate specificity. Biochemistry 1991, 30, 2017–2021. 10.1021/bi00222a002. [DOI] [PubMed] [Google Scholar]
- Bugg T. D. H.; Wright G. D.; Dutka-Malen S.; Arthur M.; Courvalin P.; Walsh C. T. Molecular basis for vancomycin resistance in Enterococcus faecium BM4147: biosynthesis of a depsipeptide peptidoglycan precursor by vancomycin resistance proteins VanH and VanA. Biochemistry 1991, 30, 10408–10415. 10.1021/bi00107a007. [DOI] [PubMed] [Google Scholar]
- Wu Z.; Wright G. D.; Walsh C. T. Overexpression, purification, and characterization of VanX, a D-, D-dipeptidase which is essential for vancomycin resistance in Enterococcus faecium BM4147. Biochemistry 1995, 34, 2455–2463. 10.1021/bi00008a008. [DOI] [PubMed] [Google Scholar]
- McComas C. C.; Crowley B. M.; Boger D. L. Partitioning the loss in vancomycin binding affinity for D-Ala-D-Lac into lost H-bond and repulsive lone pair contributions. J. Am. Chem. Soc. 2003, 125, 9314–9315. 10.1021/ja035901x. [DOI] [PubMed] [Google Scholar]
- Arthur M.; Reynolds P.; Courvalin P. Glycopeptide resistance in enterococci. Trends Microbiol. 1996, 4, 401–407. 10.1016/0966-842X(96)10063-9. [DOI] [PubMed] [Google Scholar]
- Leclercq R.; Dutka-Malen S.; Duval J.; Courvalin P. Vancomycin resistance gene vanC is specific to Enterococcus gallinarum. Antimicrob. Agents Chemother. 1992, 36, 2005–2008. 10.1128/AAC.36.9.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billot-Klein D.; Blanot D.; Gutmann L.; van Heijenoort J. Association constants for the binding of vancomycin and teicoplanin to N-acetyl-D-alanyl-D-alanine and N-acetyl-D-alanyl-D-serine. Biochem. J. 1994, 304, 1021–1022. 10.1042/bj3041021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds P. E.; Courvalin P. Vancomycin resistance in enterococci due to synthesis of precursors terminating in D-alanyl-D-serine. Antimicrob. Agents Chemother. 2005, 49, 21–25. 10.1128/AAC.49.1.21-25.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L.; Ma X.; Sato K.; Okuma K.; Tenover F. C.; Mamizuka E. M.; Gemmell C. G.; Kim M. N.; Ploy M. C.; El-Solh N.; et al. Cell wall thickening is a common feature of vancomycin resistance in Staphylococcus aureus. J. Clin. Microbiol. 2003, 41, 5–14. 10.1128/JCM.41.1.5-14.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaskovich M. A. T.; Hansford K. A.; Butler M. S.; Jia Z.; Mark A. E.; Cooper M. A. Developments in Glycopeptide Antibiotics. ACS Infect. Dis. 2018, 4, 715–735. 10.1021/acsinfecdis.7b00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Groesen E.; Innocenti P.; Martin N. I. Recent Advances in the Development of Semisynthetic Glycopeptide Antibiotics: 2014-2022. ACS Infect. Dis. 2022, 8, 1381–1407. 10.1021/acsinfecdis.2c00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leadbetter M. R.; Adams S. M.; Bazzini B.; Fatheree P. R.; Karr D. E.; Krause K. M.; Lam B. M.; Linsell M. S.; Nodwell M. B.; Pace J. L.; et al. Hydrophobic vancomycin derivatives with improved ADME properties: discovery of telavancin (TD-6424). J. Antibiot. 2004, 57, 326–336. 10.7164/antibiotics.57.326. [DOI] [PubMed] [Google Scholar]
- Malabarba A.; Ciabatti R.; Scotti R.; Goldstein B. P.; Ferrari P.; Kurz M.; Andreini B. P.; Denaro M. New semisynthetic glycopeptides MDL 63,246 and MDL 63,042, and other amide derivatives of antibiotic A-40,926 active against highly glycopeptide-resistant VanA enterococci. J. Antibiot. 1995, 48, 869–883. 10.7164/antibiotics.48.869. [DOI] [PubMed] [Google Scholar]
- COOPER R. D. G.; SNYDER N. J.; ZWEIFEL M. J.; STASZAK M. A.; WILKIE S. C.; NICAS T. I.; MULLEN D. L.; BUTLER T. F.; RODRIGUEZ M. J.; HUFF B. E.; THOMPSON R. C. Reductive alkylation of glycopeptide antibiotics: synthesis and antibacterial activity. J. Antibiot. 1996, 49, 575–581. 10.7164/antibiotics.49.575. [DOI] [PubMed] [Google Scholar]
- Nicolaou K. C.; Boddy C. N.; Brase S.; Winssinger N. Chemistry, Biology, and Medicine of the Glycopeptide Antibiotics. Angew. Chem., Int. Ed. 1999, 38, 2096–2152. . [DOI] [PubMed] [Google Scholar]
- Culp E. J.; Waglechner N.; Wang W.; Fiebig-Comyn A. A.; Hsu Y. P.; Koteva K.; Sychantha D.; Coombes B. K.; Van Nieuwenhze M. S.; Brun Y. V.; Wright G. D. Evolution-guided discovery of antibiotics that inhibit peptidoglycan remodelling. Nature 2020, 578, 582–587. 10.1038/s41586-020-1990-9. [DOI] [PubMed] [Google Scholar]
- Xu M.; Wang W.; Waglechner N.; Culp E. J.; Guitor A. K.; Wright G. D. GPAHex-A synthetic biology platform for Type IV-V glycopeptide antibiotic production and discovery. Nat. Commun. 2020, 11, 5232 10.1038/s41467-020-19138-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M.; Wang W.; Waglechner N.; Culp E. J.; Guitor A. K.; Wright G. D. Phylogeny-Informed Synthetic Biology Reveals Unprecedented Structural Novelty in Type V Glycopeptide Antibiotics. ACS Cent. Sci. 2022, 8, 615–626. 10.1021/acscentsci.1c01389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell S. J.; Verma D.; Griswold K. E.; Bailey-Kellogg C. Building blocks and blueprints for bacterial autolysins. PLoS Comput. Biol. 2021, 17, e1008889 10.1371/journal.pcbi.1008889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naruse N.; Oka M.; Konishi M.; Oki T. New antiviral antibiotics, kistamicins A and B. II. Structure determination. J. Antibiot. 1993, 46, 1812–1818. 10.7164/antibiotics.46.1812. [DOI] [PubMed] [Google Scholar]
- Greule A.; Izore T.; Iftime D.; Tailhades J.; Schoppet M.; Zhao Y.; Peschke M.; Ahmed I.; Kulik A.; Adamek M.; et al. Kistamicin biosynthesis reveals the biosynthetic requirements for production of highly crosslinked glycopeptide antibiotics. Nat. Commun. 2019, 10, 2613 10.1038/s41467-019-10384-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko K. A.; Wanner B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 2000, 97, 6640–6645. 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya Y.; Dhanda G.; Sarkar P.; Haldar J. Pursuit of next-generation glycopeptides: a journey with vancomycin. Chem. Commun. 2022, 58, 1881–1897. 10.1039/d1cc06635h. [DOI] [PubMed] [Google Scholar]
- Ottonello A.; Wyllie J. A.; Yahiaoui O.; Sun S.; Koelln R. A.; Homer J. A.; Johnson R. M.; Murray E.; Williams P.; Bolla J. R.; et al. Shapeshifting bullvalene-linked vancomycin dimers as effective antibiotics against multidrug-resistant gram-positive bacteria. Proc. Natl. Acad. Sci. U.S.A. 2023, 120, e2208737120 10.1073/pnas.2208737120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malabarba A.; Goldstein B. P. Origin, structure, and activity in vitro and in vivo of dalbavancin. J. Antimicrob. Chemother. 2005, 55, ii15–ii20. 10.1093/jac/dki005. [DOI] [PubMed] [Google Scholar]
- Mühlberg E.; Umstatter F.; Domhan C.; Hertlein T.; Ohlsen K.; Krause A.; Kleist C.; Beijer B.; Zimmermann S.; Haberkorn U.; et al. Vancomycin-Lipopeptide Conjugates with High Antimicrobial Activity on Vancomycin-Resistant Enterococci. Pharmaceuticals 2020, 13, 110. 10.3390/ph13060110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan R.; Schabel A. A.; Occolowitz J. L.; Counter F. T.; Ott J. L.; Felty-Duckworth A. M. Synthesis and antibacterial evaluation of N-alkyl vancomycins. J. Antibiot. 1989, 42, 63–72. 10.7164/antibiotics.42.63. [DOI] [PubMed] [Google Scholar]
- Veseli A.; Zakelj S.; Kristl A. A review of methods for solubility determination in biopharmaceutical drug characterization. Drug Dev. Ind. Pharm. 2019, 45, 1717–1724. 10.1080/03639045.2019.1665062. [DOI] [PubMed] [Google Scholar]
- Yang X.; Wang Z. P.; Xiang S.; Wang D.; Zhao Y.; Luo D.; Qiu Y.; Huang C.; Guo J.; Dai Y.; et al. Optimization of the Natural Product Calothrixin A to Discover Novel Dual Topoisomerase I and II Inhibitors with Improved Anticancer Activity. J. Med. Chem. 2022, 65, 8040–8061. 10.1021/acs.jmedchem.2c00615. [DOI] [PubMed] [Google Scholar]
- Bloem A.; Bax H. I.; Yusuf E.; Verkaik N. J. New-Generation Antibiotics for Treatment of Gram-Positive Infections: A Review with Focus on Endocarditis and Osteomyelitis. J. Clin. Med. 2021, 10, 1743. 10.3390/jcm10081743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray C. J. L.; Ikuta K. S.; Sharara F.; et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 2022, 399, 629–655. 10.1016/S0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanasov A. G.; Zotchev S. B.; Dirsch V. M.; Supuran C. T.; Natural products in drug discovery: advances and opportunities. Nat. Rev. Drug Discovery 2021, 20, 200–216. 10.1038/s41573-020-00114-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski A. C.; Johnson J. W.; Wright G. D. Evolving medicinal chemistry strategies in antibiotic discovery. Curr. Opin. Biotechnol. 2016, 42, 108–117. 10.1016/j.copbio.2016.04.006. [DOI] [PubMed] [Google Scholar]
- Okano A.; Isley N. A.; Boger D. L. Peripheral modifications of [Psi[CH(2)NH]Tpg(4)]vancomycin with added synergistic mechanisms of action provide durable and potent antibiotics. Proc. Natl. Acad. Sci. U.S.A. 2017, 114, E5052–E5061. 10.1073/pnas.1704125114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M.; Wang W.; Wright G. D.. Glycopeptide antibiotic discovery in the genomic era. In Methods in Enzymology; Elsevier, 2022; Vol. 665, pp 325–346. 10.1016/bs.mie.2021.11.009. [DOI] [PubMed] [Google Scholar]
- MacNair C. R.; Stokes J. M.; Carfrae L. A.; Fiebig-Comyn A. A.; Coombes B. K.; Mulvey M. R.; Brown E. D. Overcoming mcr-1 mediated colistin resistance with colistin in combination with other antibiotics. Nat. Commun. 2018, 9, 458 10.1038/s41467-018-02875-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.