ABSTRACT
The precursors to mammalian autophagosomes originate from preexisting membranes contributed by a number of sources, and subsequently enlarge through intermembrane lipid transfer, then close to sequester the cargo, and merge with lysosomes to degrade the cargo. Using cellular and in vitro membrane fusion analyses coupled with proteomic and biochemical studies we show that autophagosomes are formed from a hybrid membrane compartment referred to as a prophagophore or HyPAS (hybrid preautophagosomal structure). HyPAS is initially LC3-negative and subsequently becomes an LC3-positive phagophore. The prophagophore emerges through fusion of RB1CC1/FIP200-containing vesicles, derived from the cis-Golgi, with endosomally derived ATG16L1 membranes. A specialized Ca2+-responsive apparatus controls prophagophore biogenesis and can be modulated by pharmacological agents such as SIGMAR1 agonists and antagonists including chloroquine. Autophagic prophagophore formation is inhibited during SARS-CoV-2 infection and is recapitulated by expression of SARS-CoV-2 nsp6. These findings show that mammalian autophagosomal prophagophores emerge via the convergence of secretory and endosomal pathways in a process that is targeted by microbial factors including coronaviral membrane proteins.
Abbreviations: CLEM, correlative light and electron microscopy; CQ, chloroquine; HyPAS, hybrid preautophagosomal; strcuture/prophagophore; LC3, microtubule associated protein 1 light chain 3; RUPEX, a combination of RUSH and APEX2 systems; SARS-CoV-2, SARS-CoV-2 virus, causative agent of COVID19.
KEYWORDS: Autophagy, calcium, COVID19, endosome, endoplasmic reticulum, golgi, HyPAS prophagophore, LC3, syntaxin
Canonical macroautophagy/autophagy is a cytoplasmic quality control and metabolic process with physiological roles in health and disease. The mammalian canonical autophagy pathway is controlled by several protein modules, the RB1CC1/FIP200 complex acting as the conduit for regulation by MTOR and AMPK, and a protein lipidation system which includes ATG16L1 and results in membrane ATG8ylation by mammalian Atg8 proteins (mATG8s) including MAP1LC3B/LC3B. Autophagosomes originate from preexisting membranes coming from a number of putative sources. Autophagosomes enlarge through lipid transfer and fuse with lysosomes to degrade the sequestered cargo.
We hypothesized that RB1CC1 and ATG16L1 compartments fuse during autophagy induction [1]. In full medium, RB1CC1 colocalizes with the cis-Golgi but partially disperses upon starvation and colocalizes with ATG16L1 endosomal profiles that derive from the plasma membrane and can be labeled with cholera toxin B/CtxB. By CLEM ultrastructural analysis, the RB1CC1+ ATG16L1+ compartment appears as a combination of vesicular and cisternal profiles. STX17 is a Qa-SNARE participating in autophagosome-lysosome fusion but additionally acts much earlier in the pathway during initiation stages and influences RB1CC1-containing initiation complexes. STX17 affects fusion of RB1CC1 cis-Golgi (MAN2A1/ManII+) and ATG16L1 endosomal (TFRC+) membranes, demonstrated in an in vitro fusion high-content assay/IvitHC and using a novel RUPEX technique for content mixing that combines RUSH methodology with proximity biotinylation by APEX2. The resulting hybrid preautophagosomal structure (HyPAS) has been shown via a series of genetic knockout tests to represent a precursor, termed the prophagophore, to canonical autophagosomes and is not engaged in noncanonical autophagy-related processes. HyPAS is responsive to ATG9A inactivation under basal conditions. Key autophagy initiation markers, CDIPT/phosphatidylinositol synthase, ZFYVE1/DFCP1 and WIPI2B, colocalize early with the HyPAS prophagophore. The prophagophore formation is independent of the six mATG8s inactivated in HexaKO cells (MAP1LC3s: LC3A, LC3B, LC3C, and GABARAPs: GABARAP, GABARAPL1 and GABARAPL2) because HyPAS forms in the mATG8s’ absence. Nevertheless, the HyPAS prophagophore eventually converts into an LC3B+ phagophore.
Unbiased proteomic analyses with STX17 uncovered key interactors of STX17 necessary for HyPAS prophagophore formation (Figure 1). This includes the longin R-SNARE VAMP7, and possibly the Qb,c-SNARE SNAP47. Interactions of STX17 with VAMP7 (but not with VAMP8, which is implicated in lysosomal fusion) are promoted via TBK1-dependent phosphorylation of STX17 at S202. Another STX17 binding partner is the ER calcium pump ATP2A2/SERCA2 and increased cytoplasmic Ca2+ is needed for optimal HyPAS formation. A further key partner of STX17 is ESYT2, of the ESYT family of ER proteins related to synaptotagmins with a role in Ca2+-dependent membrane tethering. ESYT2-KO prevents HyPAS formation. Synaptotagmins in principle control SNARE complexes, and a mutational analysis, based on prototypical SYT-STX relationships, shows that ESYT2 interacts with STX17 to regulate HyPAS formation. Another ER-localized interactor of STX17 is SIGMAR1 and HyPAS formation is reduced in SIGMAR1 KO cells. The apparatus consisting of the above STX17 interactors and functional partners are necessary for canonical autophagy of diverse conventional autophagic cargos.
Figure 1.
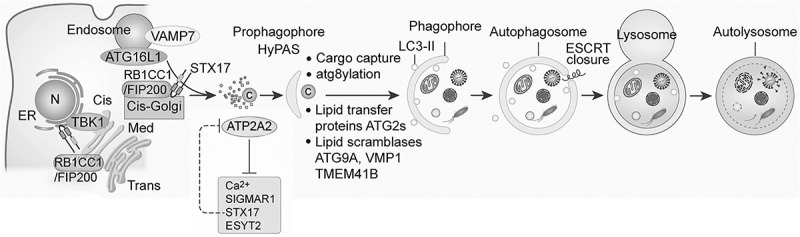
Formation of mammalian autophagosomes via a hybrid preautophagosomal structure (HyPAS) or prophagophore. Details of different components and stages are given in the main text. Modified after figures in Kumar et al. 2021 (Ref 1) and Deretic and Lazarou, J Cell Biol (2022) 221 (7): e202203083.
SIGMAR1 is a target for a series of pharmacological agonists and antagonists including chloroquine (CQ), which is a ligand for SIGMAR1 in addition to its role in neutralizing acidified compartments. CQ inhibits HyPAS formation in vitro whereas bafilomycin A1 does not. Additional agonists (cutamesine) and antagoniosts (BD1047) modulate HyPAS formation. CQ targeting of SIGMAR1 specifically inhibits STX17 and ESYT2 interactions but not STX17 and ATP2A2 interactions. Thus, CQ interferes with autophagy at the point of HyPAS formation.
Autophagy intersects morphologically with coronavirus biogenesis, but recent studies indicate that it does not inhibit SARS-CoV-2. Partially explaining these observations, HyPAS is inhibited during SARS-CoV-2 infection. The SARS-CoV-2 ORF1 polyprotein encodes nsp6, an integral membrane protein that affects the size of LC3B puncta. Unbiased proximity proteomic analysis using APEX2-SARS-CoV-2-nsp6 have revealed that nsp6 interacts with VAMP7, ESYT2, ATP2A2, and TBK1. Expression of SARS-CoV-2 nsp6, but not other tested SARS-CoV-2 proteins, reduces HyPAS yields during starvation-induced autophagy both in cells and in vitro fusion high-content assays. SARS-CoV-2 nsp6 interferes with early formation of autophagosomes at the HyPAS stage.
In conclusion, the HyPAS prophagophore represents a critical step in the biogenesis of canonical autophagosomes in mammalian cells. Mammalian cells commit to autophagy via intermixing of two membrane trafficking pathways, one vectorially acting in secretion and the other flowing in the opposite direction via the endocytic pathway. This is compatible with the majority of other studies and the well-established steps of the canonical autophagy pathway.
Funding Statement
This work was supported by the National Institute of Allergy and Infectious Diseases [R37AI042999, R01AI111935]; National Institute of General Medical Sciences [P20GM121176].
Disclosure statement
No potential conflict of interest was reported by the authors.
Reference
- [1].Kumar S, Javed R, Mudd M, et al. Mammalian hybrid pre-autophagosomal structure HyPAS generates autophagosomes. Cell. 2021;184(24):5950–69 e22. [DOI] [PMC free article] [PubMed] [Google Scholar]


