Abstract
Background:
Chronic lung injury and dysregulated cellular homeostasis in response to particulate matter (PM) exposure are closely associated with adverse health effects. However, an effective intervention for preventing the adverse health effects has not been developed.
Objectives:
This study aimed to evaluate the protective effects of nicotinamide mononucleotide (NMN) supplementation on lung injury and elucidate the mechanism by which NMN improved immune function following subchronic PM exposure.
Methods:
Six-week-old male C57BL/6J mice were placed in a real-ambient PM exposure system or filtered air-equipped chambers (control) for 16 wk with or without NMN supplementation in drinking water (regarded as , , Con-NMN and Exp-NMN groups, respectively) in Shijiazhuang City, China (/group). The effects of NMN supplementation () on PM-induced chronic pulmonary inflammation were assessed, and its mechanism was characterized using single-cell transcriptomic sequencing (scRNA-seq) analysis of whole lung cells.
Results:
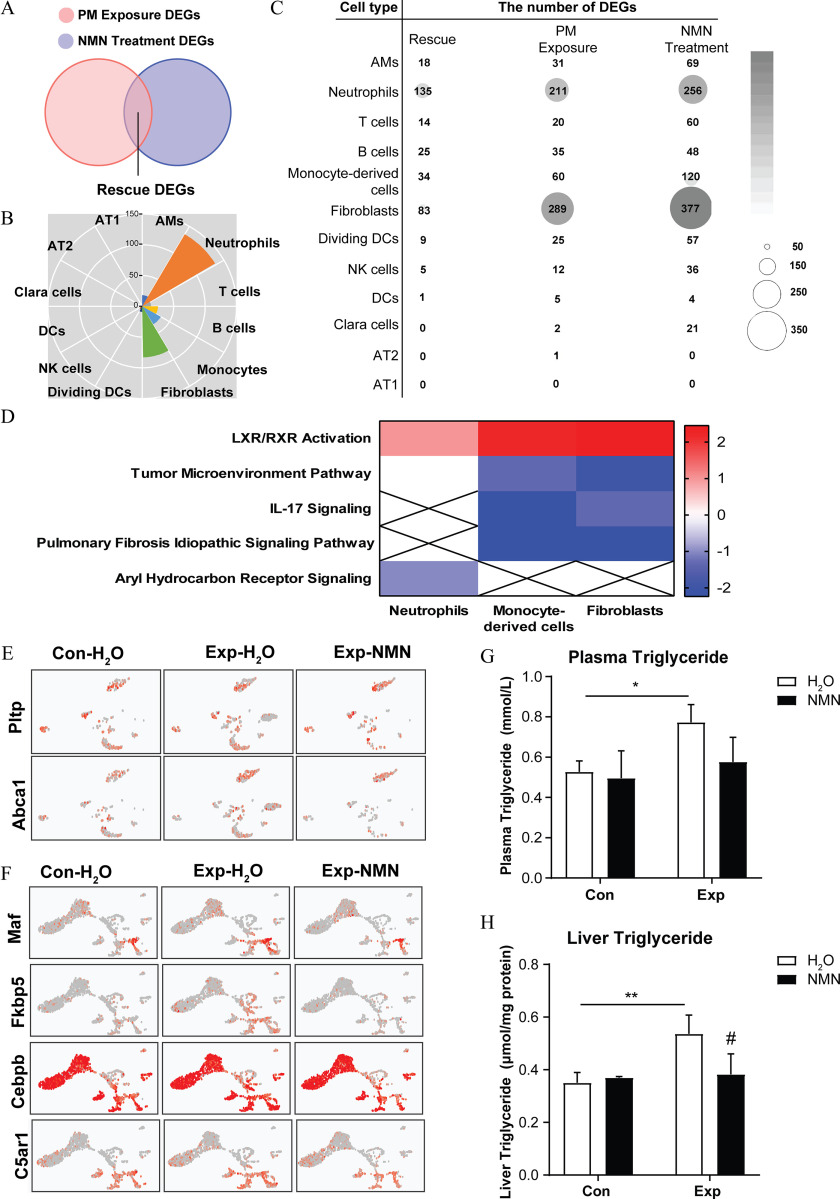
The NMN-treated mice exhibited higher levels in multiple tissues. Following 16-wk PM exposure, slightly less pulmonary inflammation and less collagen deposition were noted in mice with NMN supplementation in response to real-ambient PM exposure (Exp-NMN group) compared with the group (all ). Mouse lung tissue isolated from the Exp-NMN group was characterized by fewer neutrophils, monocyte-derived cells, fibroblasts, and myeloid-derived suppressor cells induced by subchronic PM exposure as detected by scRNA-seq transcriptomic analysis. The improved immune functions were further characterized by interleukin-17 signaling pathway inhibition and lower secretion of profibrotic cytokines in the Exp-NMN group compared with the group. In addition, reduced proportions of differentiated myofibroblasts and profibrotic interstitial macrophages were identified in the NMN-supplemented mice in response to PM exposure. Furthermore, less immune function suppression and altered differentiation of pathological cell phenotypes NMN was related to intracellular lipid metabolism activation.
Discussion:
Our novel findings suggest that NMN supplementation mitigated PM-induced lung injury by regulating immune functions and improving lipid metabolism in male mice, providing a putative intervention method for prevention of human health effects associated with PM exposure. https://doi.org/10.1289/EHP12259
Introduction
Long-term exposure to fine particulate matter (PM) with an aerodynamic diameter of () has been associated with increased mortality and morbidity with diverse human diseases, particularly respiratory diseases.1,2 Great effort has been made in seeking an effective means for interventions to prevent respiratory diseases. Supplementation with nutrients, such as probiotics and vitamins E and C, intake of antioxidants,3 and caloric restriction (CR),4 have been reported to reduce PM-induced acute lung injury (ALI) in mice. However, the effects of continuous vitamin intake on human health remain controversial,5 and the side effects, such as constant hunger and low body temperature, have limited CR diets from promotion in large populations.6 Thus, efficient therapeutics to reverse PM-induced initiation and the progression of chronic lung injury are still needed.
Several years ago, we installed a real-ambient PM exposure system in Shijiazhuang City, China,7 located in the area with the highest ambient concentrations. The mean daily concentration of reached (range: 9.00–) in the winter season in 2019–2020.8 With the advantages of round-the-clock PM exposure and simulating the natural state without further processing of PM, this system enables us to observe the health effects of subchronic PM exposure with sustained high PM exposure levels. Recently, we demonstrated that the PM-induced immunosuppressive microenvironment contributes to the initiation of chronic lung inflammation in male C57/BL6J mice (6-wk-old).8 In the course of PM-induced lung injury, we found that PM exposure led to a depletion of nicotinamide adenine dinucleotide (). is the oxidized form of the NAD(H) coenzyme, which plays a critical role in maintaining cellular homeostasis,9 improving stress resistance by regulating energy metabolism10 and immune functions, including M1/M2 polarization,11 natural killer (NK) cell activation,12 and T-cell antitumor activity.13
A significant decrease in level was observed in peripheral blood from elderly men ( y of age), compared with younger counterparts ( y of age) in a cross-sectional study conducted on a population in Tangshan, China.14 Increased consumption and decreased content (concentration normalized to weights or protein content of tissue) were reported in several mouse models that included aging-related diseases,15 endotoxin-induced ALI, and bleomycin-induced pulmonary fibrosis.11 Previous studies have also shown that the restoration of could lead to a protective effect against xenobiotic-induced lung injury in the mouse pulmonary fibrosis model induced by treatment with bleomycin.16 In addition, a reduction in consumption improved immune functions in aged mice.17 Taken together, we speculated that the maintenance of homeostasis might play an important role in maintaining immune balance and attenuating chronic lung injury upon PM exposure.
Nicotinamide mononucleotide (NMN), naturally found in several kinds of foods, such as broccoli and avocado, is one of the most studied precursors.18 NMN is involved in the salvage biosynthesis pathway, which is responsible for restoration in most tissues, including the lung, brain, and spleen.19 Apart from boosting levels, long-term NMN intervention has been shown to improve cellular glucose and lipid metabolism, enhance mitochondrial functions, and afford resistance against stress.18,20 Owing to the heterogeneity of pulmonary cells and the ubiquitously expressed NMN/nicotinic acid mononucleotide (NaMN) adenylyltransferase (NMNAT) family, which mediates the cellular uptake of NMN,21 research on the protective effects of NMN intervention has been limited to altered cytokine secretion or rejuvenated pulmonary epithelial cells.22 Thus, the critical cell types responsive to continuous NMN supplementation and the underlying potential mechanism that modifies the immune microenvironment have yet to be characterized in the context of subchronic PM exposure (multiple or continuous exposures occurring for of a species’ lifetime, which, in rodents, is usually over a 3-month period).
Chronic lung injury induced in a mouse model was reported to be regulated by the interplay between immune dysfunction and metabolic homeostasis impairment.23 Both immune and metabolic functions could be affected by intracellular content, which could be effectively boosted by NMN supplementation in multiple tissue. We hypothesized that oral NMN supplementation might exert potentially protective effects on PM-induced lung injury by improving immune function and metabolic homeostasis. To seek an effective intervention and delineate the potential molecular mechanism, we carried out an experiment of 16-wk PM exposure in mice with NMN supplementation in a real-ambient PM exposure system using single-cell transcriptomic sequencing (scRNA-seq) analysis.
Methods
NMN Administration in Male C57BL/6J Mice
All animal procedures were conducted in accordance with the guidelines of the Animal Care and Protection Committee of Sun Yat-sen University and Hebei Medical University. Six-week-old male C57BL/6J mice were purchased from Beijing Vital River Laboratories. The mice were randomly divided into control [water ()] and NMN-treated (NMN) groups ( for each group, 5 mice/cage, 100 mice in total). The mice were maintained at 20–25°C and 40%–60% humidity under 12/12-h light/dark cycles with ad libitum food and water. The experimental design of both NMN treatment and PM exposure is illustrated in Figure 1. Of the 100 mice used in this experiment, 10 mice and 10 NMN-treated mice were assigned to investigate the baseline (BS) biological and metabolic profile following 18-wk NMN supplementation (regarded as and NMN-BS groups, respectively). They were housed in the barrier system of the animal facility of the School of Public Health, Sun Yat-sen University in Guangzhou, China, without exposure to PM. The remaining 80 mice were used to investigate the effects of NMN supplementation on PM-induced injury in mice housed in Shijiazhuang, Hebei Province, China (detailed information for animal grouping is provided in the section “Real-Ambient PM Exposure”). The powder with purity of 99.9% was purchased from Hygieia Biotechnology Co., Ltd. (XJY01220618). Based on the range of NMN concentration in healthy human subjects24 and the levels of measured in mice with NMN administration, we chose a dose of NMN of . The NMN was administered orally in drinking water ad libitum and was freshly prepared every 3 d. Average water consumption and food intake were monitored every 3 d, and body weights were measured weekly.
Figure 1.
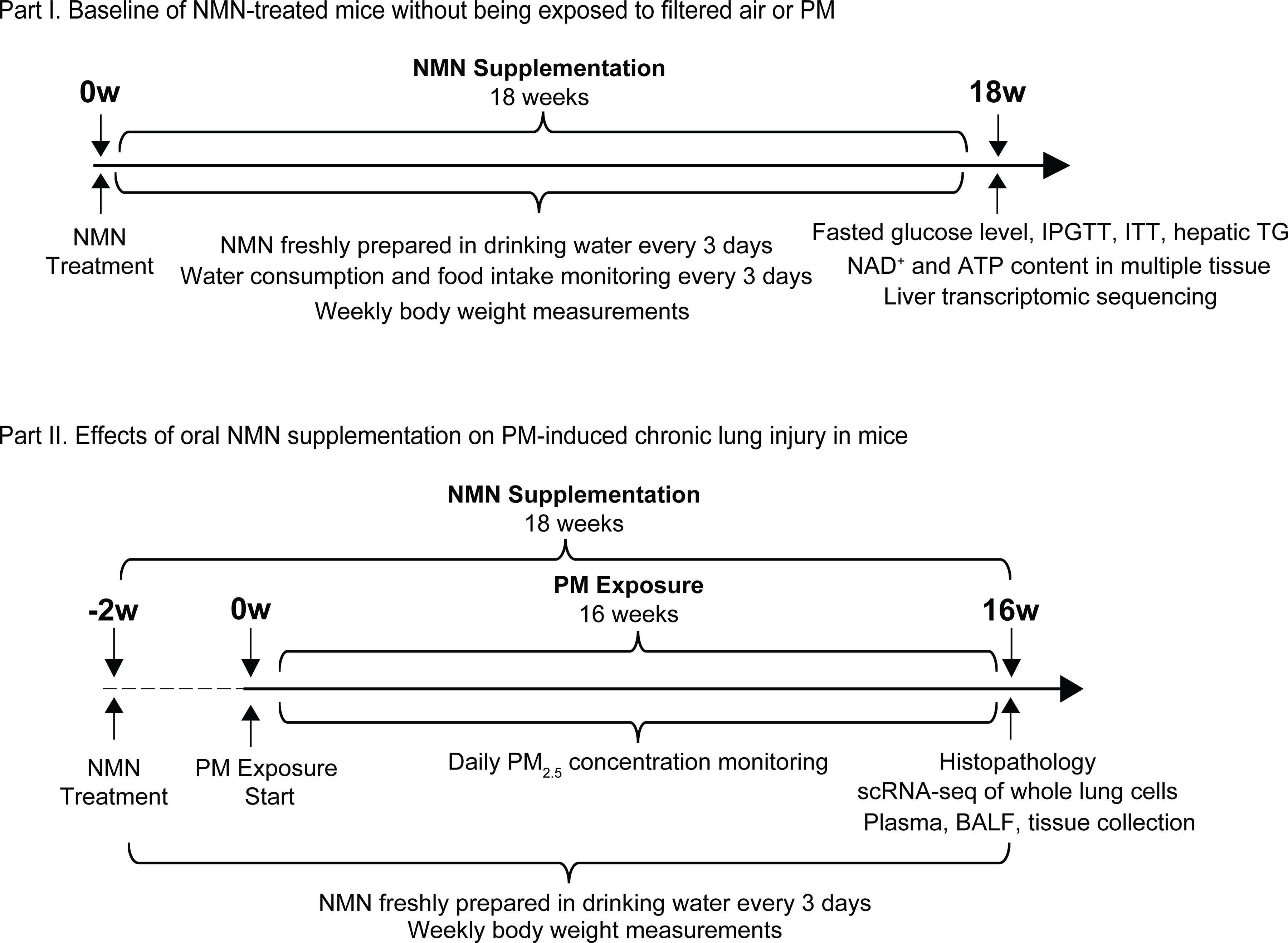
A schematic diagram illustrates the experimental design in this study. The experiment was conducted in two parts: Part I, Characterization of metabolic alterations in mice receiving 18-wk NMN supplementation; and Part II, Effects of NMN supplementation on PM exposure-induced chronic lung injury (NMN supplementation started 2 wk prior to 16-wk PM exposure). Note: ATP, adenosine triphosphate; BALF, bronchoalveolar lavage fluid; IPGTT, intraperitoneal glucose tolerance testing; ITT, insulin tolerance testing; , nicotinamide adenine dinucleotide; NMN, nicotinamide mononucleotide; PM, particulate matter; scRNA-seq, single-cell RNA transcriptomic sequencing; TG, triglyceride.
Real-Ambient PM Exposure
Real-ambient PM exposure was conducted in a system equipped with individual ventilated cage (IVC) chambers in Shijiazhuang7 from 27 November 2019 to 26 March 2020. In brief, the real-ambient PM exposure system was remodeled from a conventional IVC system and consists of a stainless-steel metal pipe, an auto temperature control system, and IVC chambers. Three layers of high-efficiency particulate air (HEPA) filters were installed in the chambers for the air-filtered control group, whereas no HEPA filters were introduced in the PM exposure chambers. concentration in the air-filtered control and the PM exposure chambers was monitored using a TSI Aerosol Detector DUSTTRAKTM II (Shoreview). The detailed methods of collection, extraction, and component analysis have been reported previously.7 In brief, High-Volume Air Samples (Thermo Fischer Scientific) equipped with Teflon or quartz filters were used to collect for 24 h/d for 4 consecutive months at a flow rate of .
Four major categories of extracted organic components, including polycyclic aromatic hydrocarbons (PAHs), nitro-derivatives of PAHs, polychlorinated biphenyls (PCBs), and polychlorinated dibenzo-dioxins (PCDDs) were measured. PAHs, nitrated-PAHs, PCBs, and PCDDs were analyzed using a gas chromatograph (GC) (Agilent 5890) coupled with an Agilent 5973 mass spectrometer (MS) [Agilent 7890B GC equipped with Xevo TQ-Striple-quadrupole MS (Waters Corporation), high-resolution GC/MS (MAT95XL; Thermo Finnigan), respectively]. The water-soluble fraction was determined by inductively coupled plasma MS (ICP-MS) (ELEMENT2; Thermo Finnigan). Meteorological conditions, including temperature, airflow rate, pressure, ventilation rate, humidity, noise, and potential harmful microorganisms were monitored to ensure the system was maintained at a relatively constant condition. Eighty mice in total were randomly divided into four groups ( for each group, 5 mice/cage) in the PM exposure experiment as mentioned above. NMN administration was started 2 wk prior to PM exposure and maintained throughout the 16-wk PM exposure experiment. Mice exposed to filtered air and PM without NMN treatment are referred to as the and the groups, respectively, whereas the mice exposed to filtered air or PM with NMN supplementation are referred to as the Con-NMN or the Exp-NMN group, respectively.
At the end of the 16-wk PM exposure, the mice were anesthetized with 1% sodium pentobarbital, and blood samples were drawn from the inferior vena cava and collected in tubes spray-coated with dipotassium ethylenediaminetetraacetic acid (). Mouse lung, liver, spleen, bone marrow tissue, bronchoalveolar lavage fluid (BALF), and plasma were harvested for subsequent experiment and analysis. Three mice per group in the , , and Exp-NMN groups were selected for scRNA-seq analysis. The plasma was carefully isolated from blood by centrifugation for 10 min at room temperature (RT). The plasma and BALF derived from each group were collected for further analysis. The plasma samples were collected for examination of the content of , triglycerides (TGs), and cytokines [interleukin-1 beta (), tumor necrosis factor-alpha (), and IL-17A]. BALF samples were collected for analysis of total protein (TP), lactate dehydrogenase (LDH) release, albumin content, and total cell numbers. The tissues isolated from the lung, liver, spleen, and bone marrow were frozen in liquid nitrogen or prepared as suspensions for further experiments. A summary of the number of mice, the sample collection, and the corresponding examination is listed in Excel Table S1.
and Adenosine Triphosphate Detection
Plasma or fresh tissue, including lung, liver, spleen, and bone marrow, were isolated from mice in the , NMN-BS, , and groups (/group). In brief, of freshly isolated lung, liver, and spleen tissue were lysed in extraction buffer. Bone marrow tissue was flushed out from both sides of a femur with ice-cold phosphate-buffered saline (PBS) and collected by centrifugation at for 7 min at 4°C and lysed in extraction buffer for further examination. The extraction buffer was included in the Assay Kit with WST-8 (S0175; Beyotime Biotechnology). levels in lung, liver, spleen, and bone marrow tissue and plasma were determined using the Assay Kit with WST-8 according to the manufacturer’s instructions. For adenosine triphosphate (ATP) detection, of freshly isolated lung, liver, and spleen tissue and the collected bone marrow tissue (/group) was lysed in of ATP extraction buffer. ATP levels in multiple tissue were examined using the luciferase-based Enhanced ATP Assay Kit (ATP extraction buffer included; S0027; Beyotime Biotechnology). The protein concentrations in tissue lysates in or ATP extraction buffer were measured using the Bicinchoninic Acid (BCA) Protein Assay Kit (P0012S; Beyotime Biotechnology). Both the content of and ATP levels were normalized to the protein concentration and expressed as picograms per milligram protein.
Fasted Glucose Level Detection, Intraperitoneal Glucose Tolerance Testing, and Insulin Tolerance Testing
Analyses of fasted glucose, intraperitoneal glucose tolerance testing (IPGTT), and insulin tolerance testing (ITT) levels were conducted only in and NMN-BS groups at the end of 18-wk NMN supplementation. Fasted glucose levels were detected in tail-vein blood collected from mice in the control () and NMN-treated groups (/group) using a JPS-7 glucometer (Yicheng). For IPGTT, the mice were injected intraperitoneally (IP) with 20% glucose solution (wt/vol; dissolved in PBS) at a dose of (/group) after being fasted for 15 h. Peripheral blood samples were collected for glucose detections at 15, 30, 60, 90, and 120 min, respectively, post injection. ITT measurement was performed in the two groups of mice following 4 h fasting and IP injection of insulin (P3376; Beyotime Biotechnology). The blood glucose levels were measured subsequently at 15, 30, 60, and 90 min, respectively, post injection (/group). The test values of IPGTT and ITT at each time point were normalized to that at the initial time point (percentage blood glucose relative to initial) and areas under the curve were calculated.
Examination of Hepatic and Plasma TG Levels
Liver tissue isolated from mice in the , NMN-BS, , , Con-NMN, and Exp-NMN groups were precisely weighed () and subjected to TG content determination (/group). TG levels in the liver tissue and plasma from each group were examined using the Triglyceride Assay kit (A110-1-1; Nanjing Jiancheng Bioengineering Institute). In brief, frozen liver tissue was homogenized in ice-cold 0.9% sodium chloride (NaCl) buffer at a ratio of 1:9 (wt/vol; liver tissue/0.9% NaCl buffer) and centrifuged at at 4°C for 10 min to harvest the supernatant for further analysis. The concentration of the TG standard was , and double-distilled, deionized water was used as a blank control. An aliquot of from supernatants, plasma samples, the TG standard, and the blank control were added into a 96-well plate. Next, of working solution was added into the plate and incubated at 37°C for 10 min. The absorbance was examined at using a microplate reader (800TS; Bio-tek). The TG concentrations (in millimoles per liter) were calculated as below: TG concentrations . Hepatic TG levels were normalized to the wet weight of liver tissue and expressed as TG (in micromoles per milligram protein).
Liver Transcriptomic Sequencing
Three mice from the and NMN-BS groups (/group) were randomly selected and subjected to transcriptomic RNA sequencing in the mouse liver tissue. Briefly, of frozen liver tissue was subjected to RNA isolation and complementary DNA (cDNA) library construction. One milliliter of TRIzol was added into the frozen liver tissue and homogenized with an electric homogenizer (IKA). After mixing with of chloroform, the lysates were shaken vigorously for 30 s, rested at RT for 5 min, and centrifuged at at 4°C for 15 min. Six hundred microliters of supernatant was collected and mixed with of isopropanol. After incubation at RT for 10 min, the mixtures were centrifuged at at 4°C for 10 min, and the supernatant was discarded. The pellet was washed with of 75% ethanol and centrifuged at at 4°C for 5 min. The supernatant was discarded, and the pellet was dried at RT and dissolved in of diethyl pyrocarbonate (DEPC) water. RNA purity and concentration was determined using a NanoPhotometer spectrophotometer (IMPLEN) and Qubit 2.0 Flurometer (Life Technologies), respectively. The integrity of the RNA samples was assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies).
After harvesting the qualified RNA samples, a RNA sample was used as the input to generate cDNA sequencing libraries using the NEBNext Ultra RNA Library Prep Kit for Illumina (E7530; NEB) following the manufacturer’s recommendations. Polymerase chain reaction (PCR) was performed as follows: 98°C for 1 min, 13 cycles of 98°C for 10 s, 60°C for 30 s, 72°C for 30 s, and 72°C for 5 min. The libraries were purified (AMPure XP system; Beckman Coulter) and quality was assessed on the Agilent Bioanalyzer 2100 system. The RNA sequencing was performed using an Illumina NovaSeq6000, and 150 bp paired-end reads were generated. After obtaining high-quality clean data, an index of the reference genome was built using Bowtie (version 2.2.3), and paired-end clean reads were aligned to the reference genome using TopHat (version 2.0.12). The expected number of fragments per kilobase of transcript sequence per million base pairs sequenced (FPKM) was used to quantify the expression levels of genes. Differential expression analysis was performed using the DESeq R package (version 1.18.0) and the -values were adjusted using the Benjamini and Hochberg method.25 Differentially expressed genes (DEGs) were identified and restricted with the log to the base 2 absolute value of fold change (), . The enriched pathways were annotated based on these DEGs using Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment programs.
BALF Analysis
At the end of the PM exposure experiment, the mice in the four groups (, , Con-NMN, and Exp-NMN) were anesthetized with an IP injection of sodium pentobarbital, and the BALF was collected by injecting of ice-cold PBS through the cannulated trachea for three times (/group). The cell pellets and supernatant were separated by centrifugation at at 4°C for 10 min. The TP, LDH release, and albumin content in the BALF supernatant were examined. Twenty-microliter aliquots of supernatant of BALF were used for measuring TP using the BCA Protein Assay Kit (P0012S; Beyotime Biotechnology). Analysis of LDH activity was performed using the LDH Cytotoxicity Assay Kit (C0016; Beyotime Biotechnology) according to the manufacturer’s instructions. In brief, of the LDH working solution was mixed with of the BALF supernatant in a 96-well plate. The mixture was incubated in the dark at RT for 30 min. LDH activity was calculated based on the absorbance at and the plotted LDH standard curve. Albumin content was determined in aliquots of the supernatant using the Albumin Assay Kit (A028-2-1; Nanjing Jiancheng Bioengineering Institute) according to the manufacturer’s instructions. Supernatants were mixed with reagent 1 and incubated for 10 min at RT, and the absorbance was measured at . Albumin content was calculated as follows: albumin content . Total cell number in pellets of BALF were resuspended in of ice-cold PBS, and the total cell number in pellets of BAL was determined in a blood cell counting chamber. The remainder of the BALF supernatant was stored at for subsequent cytokine analysis.
Histopathological Analysis
The mouse tissues were isolated and fixed in 4% paraformaldehyde (PFA) overnight at RT. For histopathological analysis of lung tissues, sections of paraffin-embedded tissues were obtained and stained with hematoxylin and eosin (H&E) and Masson‘s trichrome (/group, 3 sections/mouse were stained and analyzed). Ten-micrometer-thick sections of liver tissues were cryosectioned on the slides after the fixed liver tissue was embedded in Tissue-Tek O.C.T. Compound (4583; Sakura Finetek) and stained with Oil Red O staining to visualize hepatic lipid accumulation. TissueFAXS (Tissuegnostics) was used to scan the entire area of the sections. The extent of ALI was scored according to the published American Thoracic Society report.26 In particular, five histological observations that included neutrophils in the alveolar space, neutrophils in the interstitial space, hyaline membranes, proteinaceous debris filling in the airspaces, and alveolar septal thickening in at least 20 random fields for each section were adopted to evaluate the extent of ALI. Using these quantitative data, we calculated the ALI scores. Pulmonary collagen content was indicated by the ratio of the blue-labeled area to the total area of the lung section (percentage per micrometer squared total area) automatically calculated upon Masson’s trichrome staining using HistoQuest software (Tissuegnostics). The modified Ashcroft score system,27 which evaluates the severity of lesions appearing in alveolar septa and lung structure, was simultaneously leveraged in assessing the fibrotic changes in the lung sections. A combination of collagen content (percentage) and the modified Ashcroft score system was applied to indicate the extent of pulmonary fibrosis. Red-labeled areas in Oil Red O–stained sections were identified as positive regions, and the hepatic lipid content (percentage) was quantified by automatically calculating the ratio of positive regions to total area of liver section (percentage per micrometer squared total area) using HistoQuest software (Tissuegnostics).
Hydroxyproline Content Determination
Twenty to of lung tissue derived from the , , Con-NMN, and Exp-NMN groups (/group) was used to examine the hydroxyproline (Hyp) content by a Hydroxyproline Assay Kit (A030-2-1; Nanjing Jiancheng Bioengineering Institute), which indicated the deposited collagen content. The frozen tissue was soaked in of alkaline hydrolysis buffer and heated at 95°C for 20 min. The pH of the reaction mixtures was adjusted to 6.0–6.8 (indicated by yellow color) and the total volume was adjusted to . After removing the color by active carbon, the mixtures were centrifuged at at RT for 10 min, and the supernatant was collected for further analysis. Reagents 1, 2, and 3 were added to the mixtures sequentially, and the mixtures were incubated at 60°C for 15 min. After centrifugation at at RT for 10 min, the absorbance of the supernatant was examined at , and the Hyp content were calculated by the following formula. Hyp content . The protein concentrations of the lung tissue lysates were measured using the BCA Protein Assay Kit (P0012S; Beyotime Biotechnology). Hyp content in mouse lung tissues were normalized to the protein concentration and expressed as Hyp (in micrograms per milligram).
Cytokine Analysis
Cytokines, including and , in plasma (/group) were examined using the Pro-inflammatory Panel 1 Kit (mouse V-PLEX) purchased from MSD (K15049D-1) according to the manufacturer’s instructions. Mouse lung tissue (/group) was homogenized, and the total cellular protein of the lung tissue was extracted by adding of radio immunoprecipitation assay (RIPA) lysis buffer (P0013B; Beyotime Biotechnology). The lysates were centrifuged at at 4°C for 30 min to remove the debris. Then the supernatant was collected for tissue cytokine analysis and quantification of protein concentrations as described above. The plasma level of IL-17A, as well as IL-17A, , and levels in mouse lung tissue (/group) were examined using the QuantiCyto enzyme-linked immunosorbent assay (ELISA) kits (EMC008.96, EMC001b.96, and EMC107b.96; Neobioscience Technology Co., Ltd.). For measurement, of reagent A (activation reagent) was added into of diluted samples (homogenized lung tissue was diluted 1:20) and incubated at 4°C for 60 min, and the reaction was stopped by adding of reagent B. The steps for examination followed the same protocols as for IL-17A or examination.
Briefly, of diluted samples (for IL-17A or measurements, homogenized lung tissue was diluted at 1:25 and plasma was diluted at 1:2 by sample dilution buffer) or the activated diluted samples for measurement, were added into a 96-well plate that was precoated with mouse primary antibodies against IL-17A, , or . IL-17A standards (), standards (), and standards () were prepared according to the manufacturer’s instructions. After incubation at 37°C for 90 min in the dark, the 96-well plate was washed five times with washing buffer. Next, of biotinylated antibodies was added into the plate and incubated at 37°C for 60 min in the dark. After repeating the washing step, of enzyme-conjugated antibodies was added and the plate was incubated at 37°C for 30 min in the dark. After the last washing step, of chromogenic substrate, 3,3′,5,5′-tetramethylbenzidine (TMB), was added and the plate was incubated at 37°C for 15 min in the dark. At the end of the incubation, of stop reaction buffer was added into the plate, and the absorbance was detected at using a microplate reader (800TS; Bio-tek). The absolute concentrations were calculated based on the respective standard curves. The protein concentrations of lung tissue lysates were measured using the BCA Protein Assay Kit (P0012S; Beyotime Biotechnology). Cytokines secreted in mouse lung tissues were normalized to the protein concentration based on the equation below: Cytokines in tissue .
Malondialdehyde Content Determination
Mouse lung tissue and plasma samples from , , Con-NMN, and Exp-NMN groups (/group) were subjected to malondialdehyde (MDA) detection using the Lipid Peroxidation MDA Assay Kit (S0131S; Beyotime Biotechnology). Mouse lung tissue was lysed and total cellular protein of lung tissue was extracted as we described in the section “Cytokine Analysis.” Then the supernatant was harvested for tissue MDA analysis. The MDA standards () were diluted with distilled water. In brief, of MDA standards, diluted samples (homogenized lung tissue was diluted at 1:5 by RIPA lysis buffer) or plasma samples, were mixed and incubated with of MDA detection working solution at 100°C for 15 min. After centrifugation at at RT for 10 min, of supernatant was added into a 96-well plate and the absorbance was detected at using a microplate reader (800TS; Bio-tek). The quantification of the protein concentrations was performed as described above using the BCA Protein Assay Kit (P0012S; Beyotime Biotechnology). MDA content in mouse lung tissues were normalized to the protein concentration and expressed as MDA (in micromoles per gram protein).
Comet Assay
Mouse whole blood samples derived from , , Con-NMN, and Exp-NMN groups (/group) were subjected to a comet assay to evaluate DNA damage in the whole blood cells. The comet assay was performed according to the detailed method described previously.28 Briefly, low-melting-point agarose (LMA) was dissolved in distilled water to generate 1% LMA at 65°C. LMA-precoated slides were prepared by dipping the 1% LMA onto the slides and allowing the slides to air dry. Five microliters of mouse whole blood was added to of 1% LMA at 37.5°C and mixed well. Thirty microliters of the mixtures were pipetted onto the LMA-precoated slides, followed by placing the slides at 4°C for 20 min. Then, the slides were submerged overnight in the alkaline lysis solution ( NaCl, disodium ethylenediaminetetraacetic acid (), and Tris, ) at 4°C.
After overnight lysis, the slides were rinsed in the electrophoresis solution ( sodium hydroxide and , ) in the dark at RT for 20 min before starting the electrophoresis. The slides were placed in an electrophoresis chamber filled with fresh electrophoresis solution ( above the top of the slides) and the process of electrophoresis was conducted at 4°C for 25 min at a voltage of (25 V, 300 mA). After rinsing the slides with distilled water, the slides were submerged in the neutralizing solution ( Tris-HCl, ) for 10 min twice, dehydrated in ice-cold absolute ethanol for 10 min and allowed to air dry. The slides were stained with propidium iodide in the dark for 25 min. After being rinsed with distilled water, 15 images/slide were captured by a fluorescence microscope (TE2000U; Nikon) with a objective and filter set for green-light excitation ( excitation and emission). Tail moment was analyzed and calculated by CASP software66 based on the images.
Lung Dissociation and Cell Isolation
At the end of the 16-wk PM exposure, nine mice in total were subjected to scRNA-seq analysis. Fresh whole lung tissue from , , and Exp-NMN groups (/group) was immediately perfused with ice-cold PBS. The detailed protocol of the lung dissociation has been described previously.8 Briefly, the lungs were finely minced into pieces and incubated in cold digestion medium [Dulbecco’s Modified Eagle Medium (DMEM; 11965092; Gibco) supplemented with collagenase/dispase (COLLDISP-RO; Roche) and DNase I (10104159001; Sigma Aldrich)] at 37°C for 40 min. After red blood cell lysis at 4°C for 7 min and filtration through 70- and cell strainers (352350 and 352340; BD Falcon), we resuspended the single-cell suspensions in PBS supplemented with 0.5% bovine serum albumin at a density of for further examination. Viable cell numbers were counted using 0.4% trypan blue staining using a Cellometer AUTO T4 Bright Field Cell Counter (Dakewe). Cell viabilities should be between 80% and 90%.
10x Genomics scRNA-Seq
Each qualified sample ( viable cells) that consisted of cells was used to build cDNA libraries for the scRNA-seq data based on the manufacturer’s protocol with Chromium Single Cell 3′ v3 Reagent Kit (PN-1000075; 10x Genomics). The workflow of building cDNA libraries was as follows. After preparing the Master Mix on ice, qualified cell suspensions were added to the Master Mix and of the mixtures were loaded into each of the wells in row 1 of the 10x Chip Wells. Forty microliters of Single Cell 3′ v3 Gel Beads were dispensed into each of the wells in row 2. Partitioning Oil was added in row 3 in two aliquots, followed by running the Chromium Single Cell B program in the Chromium Controller. Next, reverse transcription incubation was performed according to the following protocol: 53°C for 45 min, 85°C for 5 min, and 4°C hold. Nine libraries were sequenced on NovaSeq6000 (Illumina) separately at the sequencing depth of 500 million reads per cell. Detailed information about scRNA-seq cDNA libraries is provided in Table S1.
Data Processing
The protocol for scRNA-seq data processing has been described in a previous study.8 Briefly, Cellranger count pipelines (version 3.0; 10x Genomics) were applied for analysis of alignment, filtering, barcode counting, unique molecular identifier counting. Cellranger aggr pipelines were applied to combine three samples from the same group after elimination of batch effects. Further analysis of obtained count matrixes was performed by Seurat R package (version 3.1). After quality control (exclusion of cells with or detected genes and mitochondrial genes, and elimination of genes detected in only cells), data normalization and standardization, identification of highly variable genes with default parameters, we integrated three aggregated expression matrixes using canonical correlation analysis affiliated to the Seurat R package to compare the difference between , , and Exp-NMN groups.
Visualization was performed using Uniform Manifold Approximation and Projection (UMAP) following dimensionality reduction that was accomplished by principal components analysis (PCA) [ with a resolution ]. Cell types were manually assigned to each cluster based on the conserved markers identified by FindConservedMarkers function of the Seurat R package. The specificity and discriminative power of the conserved markers were visualized in the form of a dot plot using the DotPlot function. Feature plots for markers indicating chronic pulmonary inflammation and excessive collagen deposition were accomplished using the FeaturePlot function.
DEGs Identification and Ingenuity Pathway Analysis
DEGs (, ) of each cell type between different group were identified using the FindMarkers function. Precisely, “PM Exposure DEGs” were defined as DEGs between the and the groups, whereas “NMN Treatment DEGs” were defined as DEGs between and Exp-NMN groups. To investigate the rescue effects of NMN treatment on PM-induced lung injury, we applied the concept of “rescue DEGs” by identifying the overlapped DEGs with opposite changes in “PM Exposure DEGs” and “NMN Treatment DEGs.”29 Identification of overlapped DEGs and Venn plots were performed using Venny (version 2.1; BioinfoBP). Then, canonical pathway analysis and disease and function analysis were conducted based on “PM Exposure DEGs,” “NMN Treatment DEGs,” and “rescue DEGs” using Ingenuity Pathway Analysis (IPA) software (Qiagen).
Cell–Cell Communication Analysis
Cell–cell communication indicated by alterations in ligand–receptor pairs was analyzed using the iTALK R package. “PM Exposure DEGs” and “NMN Treatment DEGs” identified using the default Wilcoxon rank sum test in iTALK R package were assigned to calculate the significant shifted ligand–receptor pairs. The top 30 significant ligand–receptor pairs in “cytokines” were visualized by using the LRPlot function of iTALK (detailed information of cell–cell communication analysis is shown in Excel Table S2).
Cell Reclustering and Pseudotime Trajectory Analysis
To illustrate the protective effects of NMN on specific cell types, fibroblasts and monocyte-derived cells were explored and reclustered using the Seurat R package () with resolution values of 0.2 and 0.15, respectively. Reclustering visualization, cell subclusters annotation, and feature plots were conducted as described above. Pseudotime trajectory analysis was carried out using the monocle R package (version 2.9) and displayed in plots colored by cell states, Seurat clusters, and pseudotime scores that were calculated using the plot cell trajectory function of monocle R package.
Flow Cytometry Analysis
The freshly isolated blood, bone marrow, lung, and spleen cells were prepared and stained as previously reported.8 In brief, whole blood cells (/group) were harvested after two times of red blood cell lysis for 7 min at 4°C and passing the lysate through a cell strainer (352350; BD Falcon). The spleen tissue (/group) was thoroughly ground using the end of a syringe in ice-cold PBS supplemented with 2% fetal bovine serum (FBS) and passed through a cell strainer (352350; BD Falcon). The lung cells (/group) were isolated in cold digestion buffer (DMEM supplemented with collagenase/dispase and DNase I) at 37°C for 40 min and collected after being passed through a cell strainer. Bone marrow content (/group) was flushed out from both sides of a femur with ice-cold PBS supplemented with 2% FBS. The isolated cells were passed through a cell strainer. The isolated cells from the lung, spleen, and bone marrow tissue were treated with red blood cell lysis buffer and collected by centrifugation at for 7 min at 4°C after removing the supernatant. Cells derived from blood, bone marrow, lungs, and spleens were resuspended in PBS supplemented with 2% FBS and incubated with the antibody cocktail for 30 min in the dark to quantify the number and the proportion of myeloid-derived suppressor cells (MDSCs).
Cells for MDSC identification were stained with the following antibodies: Fixable Viability Dye (FVD) eFluor 780 (1:1,000; 65-0865-14; FVD eFluor 780; eBioscience), anti-CD45 [1:200; 103110; clone 30-F11; phycoerythrin (PE)/cyanine5; Biolegend], anti-CD11c [1:100; 117306; clone N418; fluorescein isothiocyanate (FITC); Biolegend], anti-CD11b [1:200; 101212; clone M1/70; allophycocyanin (APC); Biolegend], anti-Gr-1 (1:200; 108408; clone RB6-8C5; PE; Biolegend), anti-Ly6C (1:200; 128008; clone HK1.4; PE; Biolegend), and anti-Ly6G (1:200; 127618; clone 1A8; PE/cyanine7; Biolegend). Flow cytometry analysis was performed using a Cytoflex (Beckman Coulter), and the data were analyzed using CytExpert software (version 2.3.1; Beckman Coulter).
Functional Assays for MDSCs
Bone marrow-derived MDSCs (/group) were isolated and sorted based on the staining strategy as described above. Antibodies used for T cells sorting was used as follows: FVD eFluor 780 (1:1,000), anti-CD3 (1:100; 100236; clone 17A2; APC; Biolegend), and anti-CD25 (1:200; 101904; clone 3C7; PE; Biolegend). Sorted T cells were stained using CellTrace carboxyfluorescein succinimidyl ester (CFSE; C34554; Invitrogen) at 37°C for 20 min and washed with PBS supplemented with 2% FBS. Next, T cells were seeded in a round-bottomed 96-well plate coated with anti-CD3e monoclonal antibody (16-0031-82; clone 145-2C11, functional grade; eBioscience) and cultivated in complete culture medium [DMEM supplemented with 10% FBS, 1% penicillin/streptomycin, antibiotic–antimycotic (15240062; Gibco), 2-mercaptoethanol (21985023; Gibco), and anti-CD28 soluble antibody (16-0281-82; clone 37.51, functional grade; eBioscience)]. The sorting of MDSCs and T cells was performed using a BD FACS Aria IIII cell sorter. To determine the effects of NMN on regulating the immunosuppressive activity of MDSCs, T cells were cocultured with MDSCs at the ratio of 1:1 (MDSCs: T cells) for 72 h at 37°C, 5% carbon dioxide. At the end of the coculture, T cells proliferation was indicated by the discrete peaks of CFSE in the gate of cells shown in the histograms created using CytExpert (version 2.3.1) (Beckman), which represented the immunosuppressive function of MDSCs. The in vitro experiment was carried out in three replicates.
Quantitative Real-Time PCR
Total RNA was extracted from the homogenized lung tissue from each group (/group) or liver tissue from and NMN-BS groups using TRIzol (15596026; Thermo Fisher) as described in the section “Liver Transcriptomic Sequencing.” The purity and concentration of extracted RNA were assessed by NanoDrop ND-1000 Spectrophotometer (Thermo Fisher Scientific). The cDNA products were obtained by reverse transcription by using the PrimeScript RT-for-PCR Kit (RR014A; Takara). Reagent mixture 1 consisting of deoxynucleoside triphosphate (dNTP) mixture and Oligo dT primer was added to the RNA samples, and the volume of the reaction mixture was adjusted to by DEPC water. The processes of denaturation and annealing were performed at 65°C for 5 min. Reagent mixture 2 consisting of PrimeScript buffer, RNase inhibitor, and PrimeScript RTase was added to the reaction mixture. The reverse transcription was performed at 42°C for 30 min and inactivation at 70°C for 15 min. The cDNA products were quantified by NanoDrop ND-1000 Spectrophotometer and used for the quantitative real-time PCR (qPCR) using SYBR Green PCR Master Mix (QPK-201; Toyobo). The PCR program was set up as follows: 95°C 10 min, followed by 40 cycles of 95°C for 5 s, 60°C for 20 s, and 70°C for 10 s. The value from each sample was normalized to the internal control , and the results were calculated using the method. The primers (Generay Biotechnology) used for qPCR are listed in Table S2.
Statistical Analysis
ScRNA-seq data and RNA-seq of liver tissues were analyzed by R (version 3.6.3; R Development Core Team). Rose diagrams showing the numbers of “PM Exposure DEGs,” “NMN Treatment DEGs,” and “rescue DEGs” were generated with Excel (Microsoft), and the bubble matrix diagram comparing the three groups of DEGs was produced using Origin 2022 (OriginLab). The other types of figures were generated with GraphPad Prism (version 7). Post hoc two-sided power analysis for sample sizes was performed using G*Power program (release 3.1.9.7).30,31 All statistical analyses were performed with GraphPad Prism (version 7) and confirmed by SPSS 22.0 statistical software (SPSS Inc.). The results were expressed in the form of deviation (SD). Normality tests were performed using the Shapiro-Wilk W test for the data in all analyses.
The data in Part I ( content, ATP content, quantification of Oil Red O–stained sections, and validation of DEGs in liver tissue by qPCR) was analyzed using Student’s -test or the Wilcoxon rank sum test where appropriate. The differences of the data presented in Part II, including histopathological scoring, BALF analysis, Hyp content, cytokine analysis, cell phenotyping, functional assay, plasma and hepatic TG content, plasma and pulmonary MDA content, and comet assay, and so on, were compared using one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test or the Kruskal–Wallis test, followed by Dunn’s multiple comparison where appropriate. Significant differences were considered at using two-tailed tests.
Results
Overview of the Experimental Design
As shown in Figure 1, the experimental design consisted of two parts. The first part was conducted in 20 male mice (strain C57BL/6J, 10 mice/group) to characterize the metabolic alterations in mice treated with NMN in drinking water for 18 wk ( and NMN-BS groups). The second part aimed to investigate the effects of NMN supplementation on PM exposure-induced lung injury. Eighty male C57BL/6J mice were subjected to PM exposure in a real-ambient exposure system installed in Shijiazhuang. NMN supplemented in drinking water was given 2-wk prior to exposure and throughout the experiment. The 80 mice were randomly divided into four groups (, , Con-NMN, and Exp-NMN, respectively) and placed in air-filtered control and PM exposure chambers. The exposure duration was from 27 November 2019 to 26 March 2020 with a mean concentration of in the ambient air and in the PM exposure chamber, respectively (detailed information of chemical components has been described previously8 and the representative components are displayed in Table S3).
and ATP Content in Multiple Tissues following PM Exposure
Previously, we observed a significantly lower content in PM-exposed mice, accompanied by chronic inflammatory injury in multiple organs in response to PM exposure. The content was significantly lower by 39.86% (lung), 37.28% (liver), and 41.20% (spleen) following the 16-wk PM exposure (Figure 2A). Consistent with the lower levels, significantly lower ATP content (suggestive of higher ATP consumption) was evident in mouse lung, liver, and bone marrow upon PM exposure (Figure 2B). In addition, hepatic TG deposition in the PM exposure group was 4.50-fold higher than in the control group (Figure S1A,B).
Figure 2.
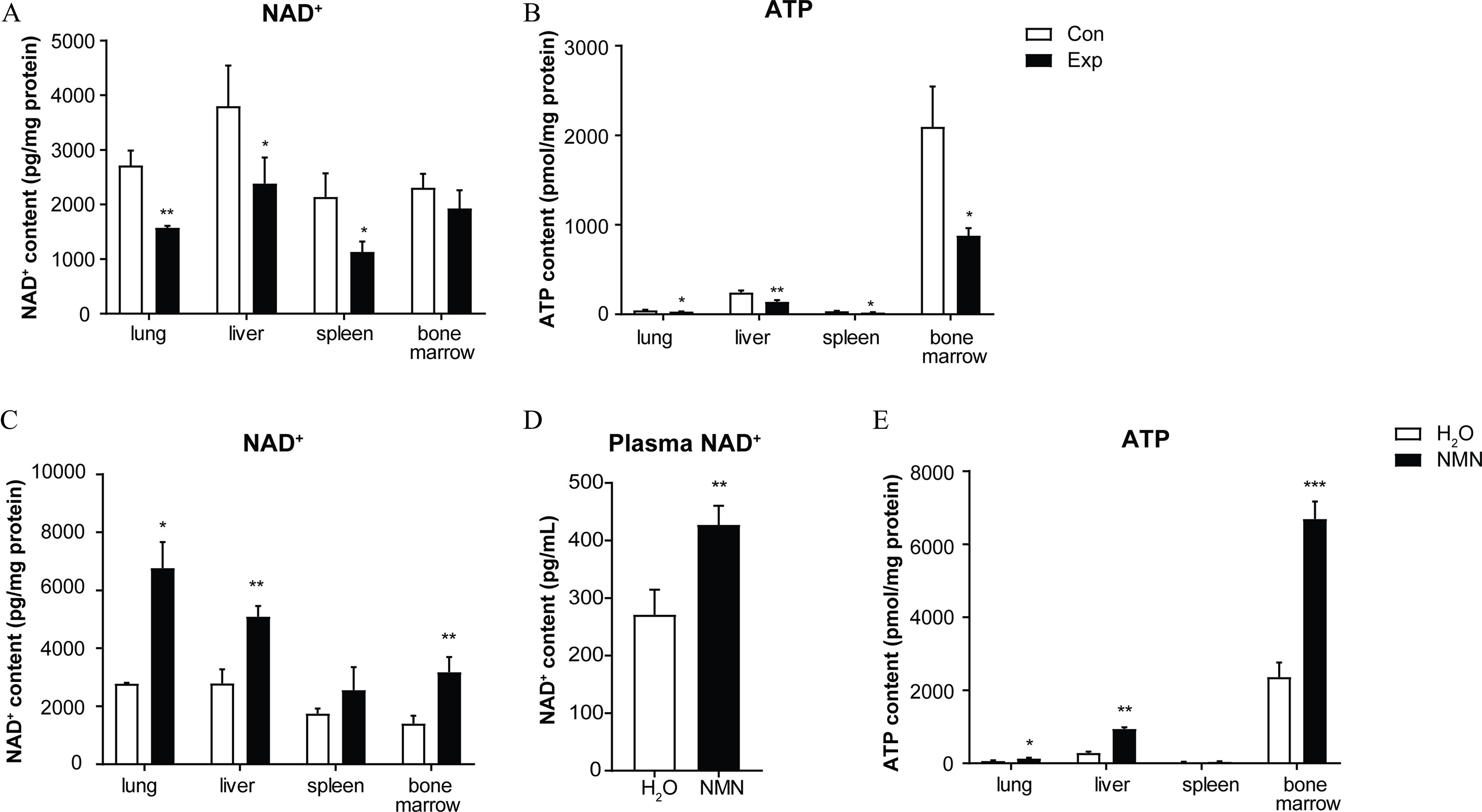
NMN supplementation and and ATP content in mice exposed to PM. (A) levels measured in lung, liver, spleen, and bone marrow tissue in mice with (PM-exposed group, Exp) or without (air-filtered control group, Con) 16-wk PM exposure (/group). (B) ATP content was analyzed in lung, liver, spleen, and bone marrow tissue of Con and Exp groups at the end of PM exposure (/group). levels were analyzed in (C) lung, liver, spleen, and bone marrow tissue (/group) and (D) plasma (/group) of and NMN-BS groups. (E) ATP levels were examined in lung, liver, spleen, and bone marrow tissue (/group) of and NMN-BS groups. Data were analyzed using Student’s -test or the Wilcoxon rank sum test. The data are expressed as . The mean, SD, and SEM values for data are shown in Table S5. *, **, and *** compared with the corresponding control mice. -Values for all tests are reported in Table S6. Note: ATP, adenosine triphosphate; BS, baseline; Con, air-filtered control group; Exp, PM exposure group; , water; , nicotinamide adenine dinucleotide; NMN, nicotinamide mononucleotide; PM, particulate matter; SD, standard deviation; SEM, standard error of mean.
Characterization of Metabolic Alterations in Mice following 18-Wk Oral NMN Supplementation
To test whether replenishment of could effectively protect mice against PM-induced lung injury, we administrated NMN to 60-wk-old C57BL/6J mice at a dose of in drinking water for 18 wk to improve levels without PM exposure. No significant differences in body weight, daily food intake, or water consumption were observed between the NMN administered (NMN-BS) and control () groups (Figure S2A–C). At the end of the 18-wk experiment, 2.43-, 1.83-, 2.26-, and 1.58-fold higher content were detected in mouse lung, liver, bone marrow, and plasma (Figure 2C,D), respectively, compared with mice without NMN supplementation. Correspondingly, markedly higher ATP content appeared in mouse lung, liver, and bone marrow (Figure 2E). Notably, transcriptomic analysis revealed differences in functional regulation annotated to systemic metabolism, particularly lipid metabolism, and cellular response to stimulus in mice with NMN supplementation for 18 wk (Figures S3 and S4A, detailed information is available in Excel Table S3). Although no significant difference was found in the fasted glucose level between NMN-treated and control mice, NMN treatment led to a greater insulin sensitivity and lower content of hepatic TG (Figure S4B–G).
NMN Supplementation and Outcomes Related to Pulmonary Injury in PM-Exposed Mice
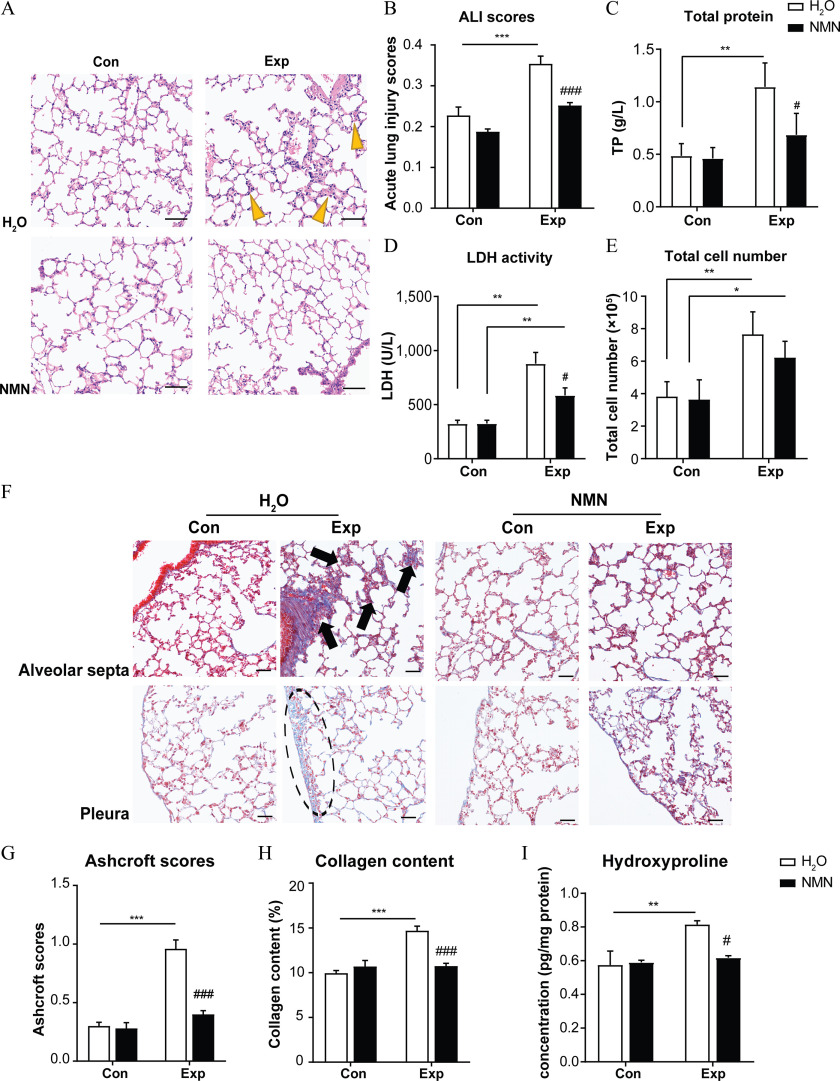
Lung characteristics following the 16-wk PM exposure included excessive collagen deposition, interstitial neutrophil infiltration, and thickened alveolar septa, suggestive of lung injury. An ALI scoring system was applied to quantify the extent of lung injury. Although lung injury was evident in both and Exp-NMN groups, we found a slighter extent of lung injury in PM-exposed mice supplemented with NMN (Exp-NMN group) than in those mice without NMN treatment ( group) (Figure 3A). In particular, the ALI scores in the and groups were and , respectively. In contrast, even when the ALI score in the Exp-NMN group was higher compared with its corresponding control (Con-NMN) group, no statistical significance was observed (Figure 3B). Corroborating the pathological changes, levels of inflammation-associated indicators in BALF—including TP, release of LDH, and total cell numbers—were higher following PM exposure without NMN supplementation compared with the group. However, these inflammatory indicators in BALF derived from PM-exposed mice with NMN treatment were lower compared with those of the group (Figure 3C–E; Figure S5A). Moreover, we observed that the secretion of was significantly lower (Figure S5B) and the mRNA expression of pro-inflammatory cytokines, such as Il1b, S100a9, and p65, was lower in the lung tissue of mice with NMN supplementation following PM exposure, although only the difference in S100a9 reached statistical significance (Figure S5C–E). Along with the milder inflammatory responses compared with the group, pulmonary MDA content and plasma MDA concentration (indicative of lung and systemic oxidative stress) were lower in the NMN-treated mice upon PM exposure. In contrast, higher levels of MDA were observed in the mice exposed to PM absent NMN administration (Figure S5F,G). In addition, the tail moment of the comet assay (a DNA damage index) conducted on peripheral blood was lower in the Exp-NMN group compared with that of the group (Figure S5H).
Figure 3.
NMN supplementation and markers of lung injury in mice exposed to PM. (A) Representative images of H&E-stained lung sections in , , Con-NMN, and Exp-NMN groups, respectively, following 16-wk PM or filtered air exposure. Yellow bold arrows indicate an interstitial neutrophilic infiltration (/group, slides/mouse). Magnifications: . Scale bar: . (B) The acute lung injury (ALI) scores were calculated in different groups (/group). Five histological findings, including neutrophils in the alveolar space, neutrophils in the interstitial space, hyaline membranes, proteinaceous debris filling in the airspaces, and alveolar septal thickening were evaluated in this scoring system. The (C) total protein (TP) content, (D), levels of lactate dehydrogenase (LDH) activity, and (E) and total cell number in the bronchoalveolar lavage fluid (BALF) (/group). (F) Representative images of Masson’s trichrome-stained lung sections indicate collagen deposition in four groups of mice, as described above. Black bold arrows indicate interstitial fibrosis with knot-like formation, and a circle with a dashed line represents pleural plaque (/group, slides/mouse). Magnifications: . Scale bar: . (G) Ashcroft scores were calculated in four groups of mice (/group) by assessing the histopathological changes mainly in alveolar septa and lung structure. (H) Pulmonary collagen content (percentage) was calculated and expressed as the ratio of the blue-labeled areas to the total area of the lung section (percentage per micrometer squared) based on Masson’s trichrome staining (/group). (I) Hydroxyproline content measured in lung tissue from mice in four groups following 16-wk PM exposure with or without NMN supplementation (/group). The significance of differences among data in this figure were analyzed using one-way ANOVA followed by Tukey’s multiple comparison post hoc test. The results in the bar graphs are presented as . The summary data (mean, SD, and SEM values) of bar graphs in this figure are shown in Table S5. *, **, and *** compared with the corresponding control mice. #, ##, and ### compared with the mice (Exp-NMN vs. ). -Values for all tests are reported in Table S6. Note: ALI, acute lung injury; ANOVA, analysis of variance; Con, air-filtered control group; Exp, PM exposure group; , water; H&E, hematoxylin and eosin; NMN, nicotinamide mononucleotide; PM, particulate matter; SD, standard deviation; SEM, standard error of mean.
Moreover, the pathological changes associated with excessive collagen deposition, such as gentle fibrotic changes, isolated knot-like formations, moderate peribronchiolar fibrosis, and pleural plaques, were milder in the mice administered NMN supplementation and PM exposure compared with the PM-exposed mice, whereas no significant difference was found between the and Con-NMN groups (Figure 3F). In line with these observations, Ashcroft scores were markedly lower in the Exp-NMN group () compared with the group (). In addition, the content of both collagen and Hyp were significantly lower in the lung tissue of the mice in the Exp-NMN group compared with that of the mice in the group (Figure 3G–I). In particular, the mean collagen deposition quantified by the ratio of the blue-labeled area to the total area of the lung section (percentage per micrometer squared total area) was in the Exp-NMN group and in the group. Moreover, the mean Hyp content in lung tissue was pg/mg protein in the Exp-NMN group and pg/mg protein in the group. In agreement with these findings, we found significantly lower mRNA expression of profibrotic factors, including Acta2, Tgfb1, and Il17a (Figure S5I–K), and less secretion of IL-17A in the Exp-NMN group compared with the group (Figure S5L).
scRNA Analysis of Whole Lung Cells with NMN Supplementation upon PM Exposure
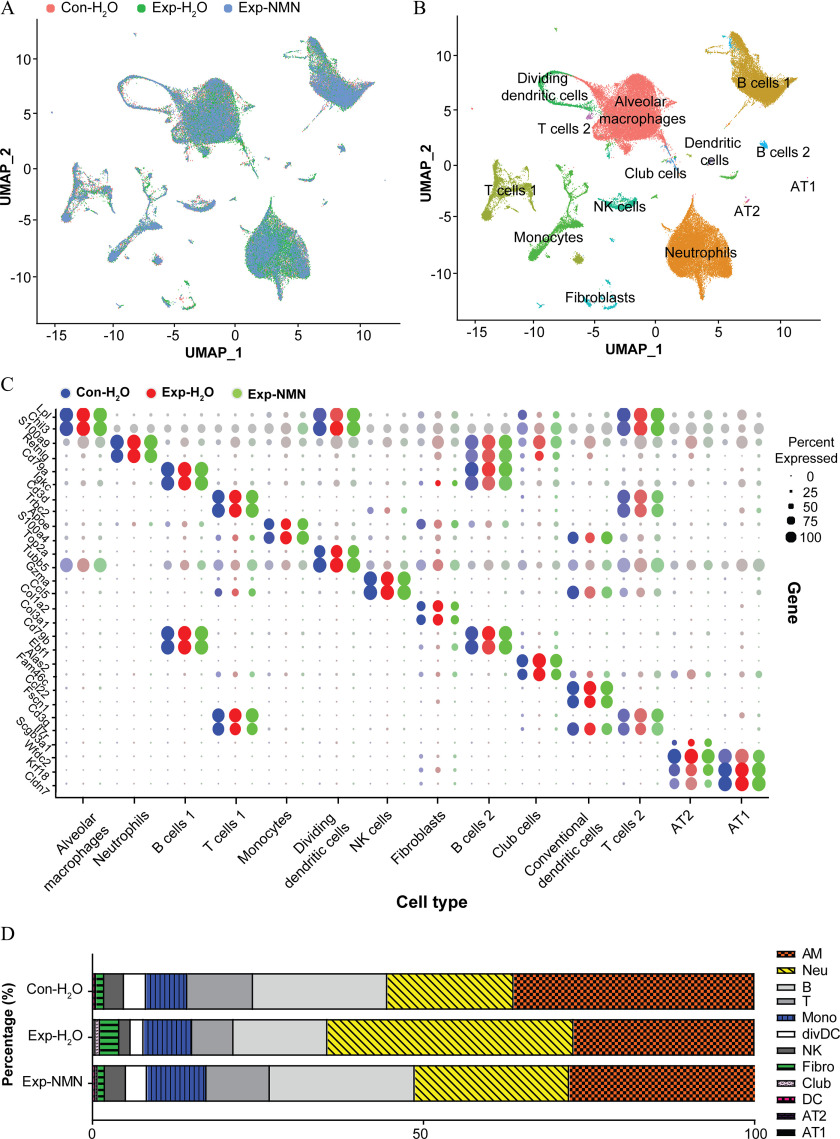
To delineate alterations in cell type composition in response to PM exposure and NMN supplementation, we generated scRNA-seq profiles conducted on whole lung samples from the , , and Exp-NMN groups (/group) following 16-wk PM exposure for comprehensive analysis. We obtained the integrated data set containing 26,745, 26,532, and 21,495 cells in the , , and Exp-NMN groups, respectively, after going through steps of quality control, preprocessing, and aggregation. Based on the annotations of diverse cell types in the mouse lung, we were able to depict a comprehensive transcriptomic landscape of whole lung cells in response to PM exposure and NMN supplementation. The whole lung cell atlas of the aggregated data set was depicted by unsupervised clustering and UMAP embedding visualization (Figure 4A).
Figure 4.
NMN supplementation and cellular composition in lungs of mice exposed to PM. (A) scRNA-seq analysis was conducted in whole lung cells of mice in , , and Exp-NMN groups (/group). UMAP plots of whole lung cells in , , and Exp-NMN groups. Cells from whole lungs of , , and Exp-NMN groups are indicated by red, green, and turquoise color, respectively. The corresponding data are summarized in Table S1. (B) Fourteen clusters with 12 major cell types identified in the integrated data set are displayed on the UMAP plot. (C) The dot plot represents the expression level of the specific marker genes (dot color shades) and the percentage of cells expressing marker genes (dot size). The expression levels of marker genes in the , , and Exp-NMN groups are discriminated by blue, red, and green color, respectively. The expression levels of marker genes in each group are shown in Excel Table S8. (D) The stacked bar plot illustrates the percentage of each type of cells in mouse lung tissue derived from the , , and Exp-NMN groups, respectively. The percentages of 12 major cell clusters in each group are shown in Table S7. Note: AM, alveolar macrophages; AT1, type 1 alveolar epithelial cells; AT2, type 2 alveolar epithelial cells; B, B cells; Club, club cells; Con, air-filtered control group; DC, dendritic cells; divDC, dividing dendritic cells; Exp, PM exposure group; Fibro, fibroblasts; , water; Mono, monocyte-derived cells; Neu, neutrophils; NK, NK cells; NMN, nicotinamide mononucleotide; PM, particulate matter; scRNA-seq, single-cell RNA transcriptomic sequencing; T, T cells; UMAP, Uniform Manifold Approximation and Projection.
Fourteen cell clusters were identified as 12 major cell types in whole lung cells by cell type annotations based on the online databases PanglaodB and the single-cell Mouse Cell Atlas (scMCA) (Table S4). The cell types comprised alveolar macrophages (AMs; cluster 0), neutrophils (cluster 1), B cells (clusters 2 and 8), T cells (clusters 3 and 11), monocyte-derived cells (monocytes; cluster 4), dividing dendritic cells (divDCs; cluster 5), NK cells (cluster 6), fibroblasts (cluster 7), Club cells (cluster 9), dendritic cells (DCs; cluster 10), type 2 alveolar epithelial cells (AT2; cluster 12), and type 1 alveolar epithelial cells (AT1; cluster 13) (Figure 4B), which were confirmed by two specific cell marker genes (Figure 4C).
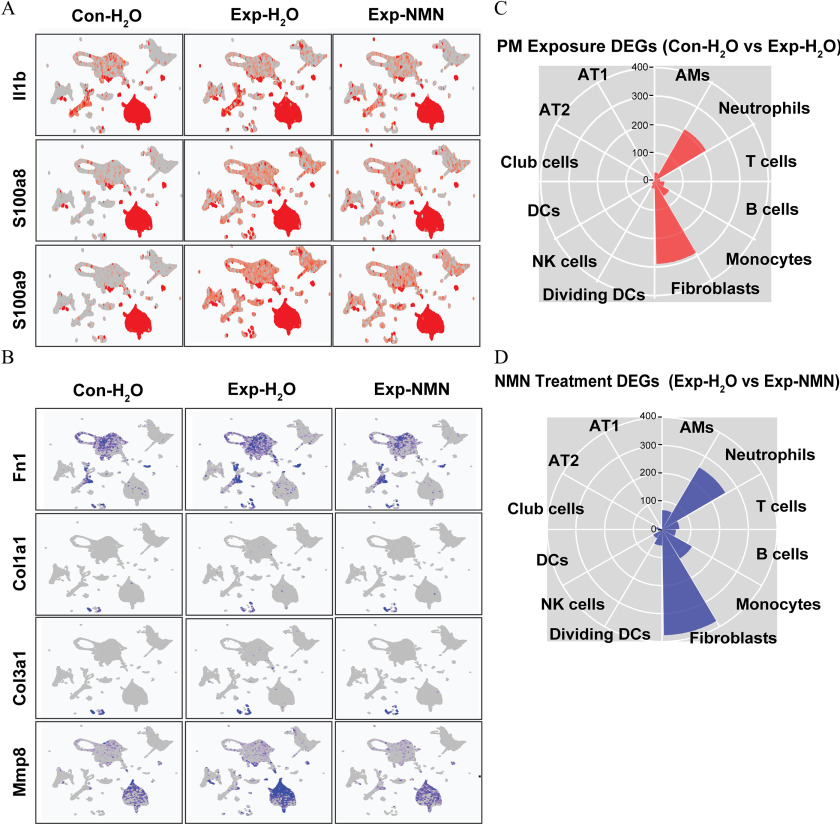
The states of chronic inflammatory and profibrotic phenotypes following subchronic PM exposure were characterized by significantly smaller numbers or proportions of the majority of immune cells—AMs, B cells, T cells, divDCs, NK cells, and DCs—and epithelial cells. However, PM-exposed mice supplemented with NMN displayed higher cell numbers or larger proportions of the immune cells noted above compared with the group (Figure 4D). Notably, the numbers of neutrophils, monocyte-derived cells, and fibroblasts were 1.93-, 1.17-, and 2.33-fold higher in the group compared with the group, whereas the numbers of neutrophils and fibroblasts were markedly lower in the Exp-NMN group compared with the group. In accordance with the pathological alterations, lower expression levels of chronic inflammatory markers—Il1b, S100a8, and S100a9— in all cell clusters from PM-exposed mouse lungs were observed in the Exp-NMN group compared with the group (Figure 5A). The expression of profibrotic genes—including Fn1, Col1a1, Col3a1 and Mmp8—which was up-regulated in PM-exposed mice was down-regulated in the whole lung cells in the Exp-NMN group (Figure 5B).
Figure 5.
Expression of pro-inflammatory and pro-fibrotic marker genes and illustration of DEGs (A) Expression levels of pro-inflammatory genes including Il1b, S100a8, and S100a9 on UMAP plots split by , , and Exp-NMN groups. (B) Expression levels of profibrotic genes including Fn1, Col1a1, Col3a1, and Mmp8 on UMAP plots split by , , and Exp-NMN groups. The numbers of (C) “PM Exposure DEGs” ( vs. ) and (D) “NMN Treatment DEGs” (Exp-NMN vs. ) identified in each cell cluster are displayed in rose diagrams. The corresponding data of (A) and (B) are listed in Excel Table S9. The detailed information of the genes summarized in (C) and (D) are provided in Excel Tables S4 and S5. Note: AM, alveolar macrophages; AT1, type 1 alveolar epithelial cells; AT2, type 2 alveolar epithelial cells; Con, air-filtered control group; DC, dendritic cells; DEGs, differentially expressed genes; dividing DC, dividing dendritic cells; Exp, PM exposure group; , water; Monocytes, monocyte-derived cells; NK, NK cells; NMN, nicotinamide mononucleotide; PM, particulate matter; UMAP, Uniform Manifold Approximation and Projection.
To further assess the effects of PM exposure and NMN supplementation on different types of pulmonary cells, we defined DEGs between the and groups as “PM Exposure DEGs,” and DEGs between the and Exp-NMN groups as “NMN Treatment DEGs” by FindMarkers function analysis (Figure 5C,D). Among these cell clusters, neutrophils, fibroblasts, and monocyte-derived cells possessed the most substantial number of identified DEGs detected under both conditions of PM exposure and NMN treatment (detailed information of the DEGs is listed in Excel Tables S4 and S5). Next, we performed DEG analysis using IPA software. Notably, an up-regulated IL-17 signaling pathway was identified in these three cell clusters following subchronic PM exposure and which was accompanied by the activation of pulmonary fibrosis idiopathic signaling pathway in monocyte-derived cells and fibroblasts. Notably, these alterations were altered by NMN supplementation as indicated in the results of pathway enrichment analysis conducted on “NMN Treatment DEGs” (Figures S6 and S7).
NMN Supplementation and Pulmonary Immune Functions in PM-Exposed Mice
Based on the alterations in cell numbers and pathway enrichment, we identified that neutrophils, fibroblasts, and monocyte-derived cells were the most affected cell types. The pathways related to immune functions were suppressed, whereas chronic inflammation, fibrosis, and tumor-related pathways were predicted to be activated in these three cell clusters in response to PM exposure (Figure S6). In addition, we showed that the up-regulated liver X receptor/retinoid X receptor (LXR/RXR) activation pathway was concurrent with an inhibition of acute phase response signaling, as well as the IL-17 pathway, Pulmonary Fibrosis Idiopathic Signaling Pathway, and Regulation of the Epithelial Mesenchymal Transition by Growth Factors Pathway in monocyte-derived cells and fibroblasts (Figure S7B,C), suggesting the relevance to milder inflammation and fewer immune function alterations.
To assess the effects of NMN supplementation on potential interactions between these cells, we carried out cell–cell interaction analysis on the three critical cells described above using the iTALK R package. The results showed the enhanced interactions of ligand–receptor pairs, particularly Ccl2–Ccr2 and Il1b–Il1r2 between fibroblasts and monocyte-derived cells, Ppbp–Cxcr2, Il10–Il10rb, and Il6–F3 between monocyte-derived cells and neutrophils, as well as Il1b–Il1rap and Il6–F3 between fibroblasts and neutrophils in the group, indicated by the ligand–receptor prediction based on higher expression levels of “PM Exposure DEGs” (Figure 6A). Importantly, these interactions, directly or indirectly, were associated with the induction, accumulation, and activation of MDSCs, contributing to the immune dysfunction in the lung. Notably, these interactions were all predicted to be weakened based on relatively lower expression of both the ligands and the receptors using “NMN Treatment DEGs” (Figure 6B).
Figure 6.
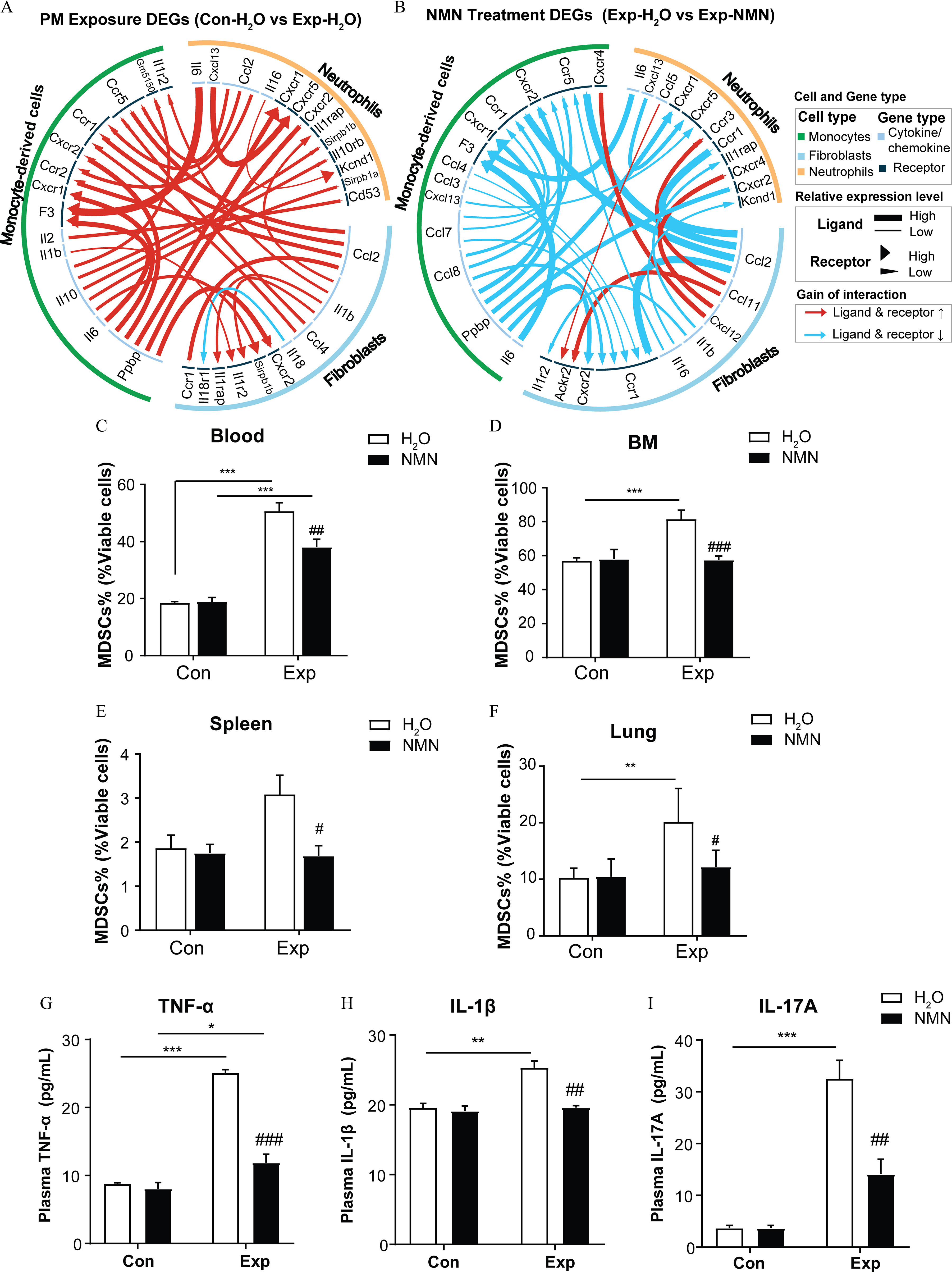
NMN supplementation and markers of immunosuppression in mice exposed to PM. Circos plots are visualized for significant alterations in cellular cytokine interactions among neutrophils, monocyte-derived cells, and fibroblasts based on (A) “PM Exposure DEGs” ( vs. ) and (B) “NMN Treatment DEGs” (Exp-NMN vs. ). The summary data of the top 30 ligand–receptors displayed in (A) and (B) are shown in Excel Table S2. The proportions of MDSCs measured in the viable cells of (C) peripheral blood (/group), (D) bone marrow (/group), (E) spleens (/group), and (F) lungs (/group) of mice in control and PM-exposed groups with or without NMN supplementation. (G) (/group), (H) (/group), and (I) IL-17A (/group) (I) in mouse plasma from , , Con-NMN, and Exp-NMN groups as determined by ELISA assays. Data presented in (C–I) were analyzed using one-way ANOVA followed by Tukey’s multiple comparison post hoc test or the Kruskal–Wallis test followed by Dunn’s multiple comparisons test where appropriate. The results in the bar graphs are presented as . The summary data (mean, SD, and SEM values) of the bar graphs in this figure are shown in Table S5. *, **, and ***. #, ##, and ### compared with the mice (Exp-NMN vs. ). -Values for all tests are reported in Table S6. Note: ANOVA, analysis of variance; BM, bone marrow; Con, air-filtered control group; DEGs, differentially expressed genes; ELISA, enzyme-linked immunosorbent assay; Exp, PM exposure group; , water; MDSCs, myeloid-derived suppressor cells; NMN, nicotinamide mononucleotide; PM, particulate matter; SD, standard deviation; SEM, standard error of mean; TNF, tumor necrosis factor.
Consistent with these results, MDSC proportions in blood, bone marrow, spleen, and lung, were all significantly lower in PM-exposed mice supplemented by NMN compared with the mice. The mean MDSC proportions in blood, bone marrow, spleen, and lung in the Exp-NMN group were , , , and , respectively. Comparatively, the mean MDSC proportions were , , , and in the blood, bone marrow, lung, and spleen of mice in the group (Figure 6C–F). The functional assays showed that the immunosuppressive activity of MDSCs in bone marrow was markedly lower in mice with NMN supplementation upon PM exposure than in those mice exposed to PM without NMN supplementation (Figure S8A). Moreover, the secretion levels of MDSCs recruitment and activation-related cytokines , , and IL-17A in mouse plasma upon PM exposure were 2.86-, 1.29-, and 8.88-fold higher than the levels in plasma isolated from mice exposed to filtered air compared with plasma from mice in the group, whereas the secretion levels of these three cytokines in plasma isolated from the Exp-NMN group were significantly lower compared with levels from the group (Figure 6G–I).
Phenotypes of PM-Induced Fibroblast and Monocyte-Derived Cell in Mice with NMN Supplementation
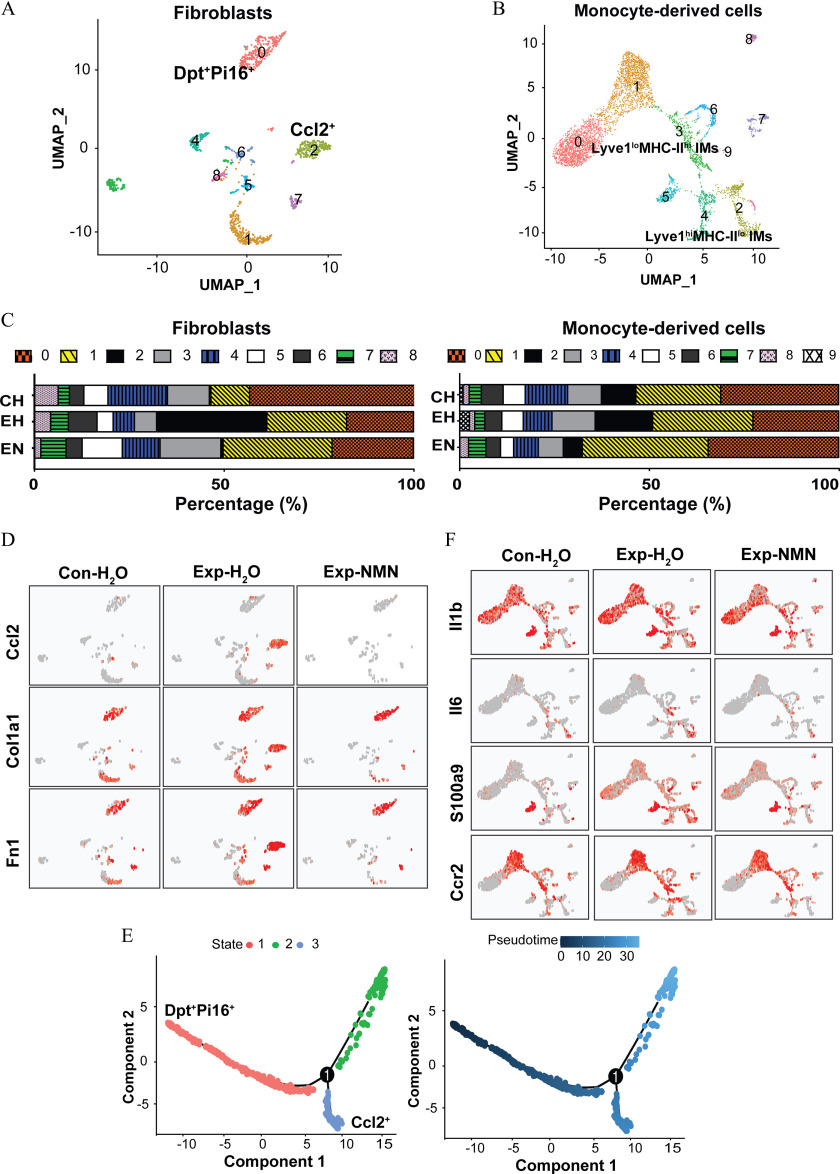
As described above, we have identified specific signaling pathways that were involved in suppressing pulmonary fibrosis in monocyte-derived cells and fibroblasts. To further address the effects of NMN intervention on the interplay among critical cell types, we reclustered them to respectively clarify the altered cellular functions and potential intercellular interactions in response to PM exposure with or without NMN supplementation (Figure 7A,B). As shown in Figure 7A, annotated by the specific markers, cluster 0 in fibroblasts subset (fibroblasts 0) expressed the markers of universal fibroblasts in steady-state including dermatopontin (Dpt) and peptidase inhibitor 16 (Pi16), whereas cluster 2 (fibroblasts 2) expressed higher levels of proliferating fibrocytes, such as Ccl2, Top2a, and Mki67. fibroblasts (fibroblasts 2) that were uniquely induced and recruited in the group upon 16-wk PM exposure were rarely observed in the PM-exposed mice with NMN supplementation (Figure 7C, the left panel). Moreover, the steady-state universal fibroblasts (fibroblasts 0) continued to occupy in the lungs from mice in the Exp-NMN and groups.
Figure 7.
NMN supplementation and cell phenotypes in differentiated fibroblast and monocyte-derived cells in mice exposed to PM. (A) Subclusters of fibroblasts, including universal fibroblasts (fibroblasts 0) and activated fibroblasts (fibroblasts 2), were identified on the UMAP plot of the fibroblast subset. (B) Subclusters of monocyte-derived cells, including IMs (monocytes 2) and IMs (monocytes 3), were visualized on the UMAP plot. (C) The stacked bar plots illustrate the percentage of fibroblast subclusters (in the left panel) and monocyte-derived cell subclusters (in the right panel) in , , and Exp-NMN groups, respectively. The corresponding data of the percentages of fibroblast and monocyte-derived cell subclusters are summarized in Table S8. (D) Expression levels of profibrotic genes, including Ccl2, Col1a1, and Fn1, on UMAP plots split by , , and Exp-NMN groups. (E) Pseudotime trajectory analysis illustrates the differentiation from universal fibroblasts (shown in red color) into activated fibroblasts (shown in blue color, in the left panel) with pseudotime scores from the lowest to the highest in fibroblasts (colored from dark blue to light blue in the right panel). The numbers and pseudotime scores of fibroblasts in the three branches are reported in Excel Table S10. (F) The expressions of Il1b, Il6, S100a9, and Ccr2 in monocyte-derived cell subsets of mouse lungs from , , and Exp-NMN groups are shown in UMAP plots and are displayed by the colors from gray to red. The expression levels of genes in (D) and (F) are listed in Excel Table S11. Note: CH, ; Con, air-filtered control group; Dpt, dermatopontin; EH, ; EN, Exp-NMN; Exp, PM exposure group; , water; Hi, high; IMs, interstitial macrophages; lo, low; Lyve, lymphatic vessel endothelial hyaluronan receptor; MHC, major histocompatibility complex; NMN, nicotinamide mononucleotide; Pi16, peptidase inhibitor 16; PM, particulate matter; UMAP, Uniform Manifold Approximation and Projection.
Notably, the activated fibroblasts that highly expressed profibrotic genes, including Col1a1 and Fn1, were exclusively found in the lungs from the group, whereas they were barely detected in the lungs from the and Exp-NMN groups (Figure 7D). Based on pseudotime trajectory analysis, fibroblasts (fibroblasts 2), were proposed to be one branch of terminal states that were differentiated from steady-state universal fibroblasts (fibroblasts 0). In parallel, the differentiation toward this emerging subcluster of fibroblasts was markedly lower in the Exp-NMN group compared with the group (Figure 7E).
With regard to subclustering of monocyte-derived cells, we identified that PM-induced elevated proportions of lymphatic vessel endothelial hyaluronan receptor-1 (high)/major histocompatibility complex-II (low) () interstitial macrophages (IMs; monocytes 2) and IMs (monocytes 3) were remarkably lower in the PM-exposed mice upon NMN treatment (Figure 7C, the right panel). As shown in Figure 7F, lower expression levels of pro-inflammatory genes, including such as Il1b, Il6, S100a9, and Ccr2, were induced in both IMs and IMs in the mice of the Exp-NMN group compared with those in the group.
Evaluation of Mechanistic Pathways Involved in the Action of NMN in Mice Chronically Exposed to PM
To investigate the potential mechanism of the NMN-mediated attenuation of lung injury, we identified “rescue” DEGs in each cell type through generating the overlapped DEGs from both “PM Exposure DEGs” and “NMN Treatment DEGs” (Figure 8A). The results showed that 135, 34, and 83 DEGs were discriminated in neutrophils, monocyte-derived cells, and fibroblasts, respectively (Figure 8B,C; a detailed summary of “rescue” DEGs is listed in Excel Table S6). Notably, the LXR/RXR activation pathway, which was closely associated with regulating lipid homeostasis and inflammation and maintaining neutrophil homeostasis, was annotated and predicted to be up-regulated in three critical cell clusters, based on the fold changes of DEGs expression between the Exp-NMN and groups (Figure 8D; Figure S8B–D). In addition, the activation of peroxisome proliferator-activated receptor (PPAR) and signaling was also annotated based on a canonical pathway analysis of “NMN Treatment DEGs,” particularly in fibroblasts and monocyte-derived cells (Figure S7A–C). In agreement with these results, “Disease and Function” analysis conducted based on “rescue” DEGs using IPA software revealed the inflammation-related lipid metabolism process, including synthesis of lipid and synthesis of eicosanoid in the Exp-NMN group. In addition, as indicators of pro-inflammatory progress, the synthesis of nitric oxide and the production of reactive oxygen species were predicted to be inhibited in monocyte-derived cells upon NMN supplementation (detailed information is listed in Excel Table S7).
Figure 8.
NMN supplementation and markers of lipid metabolism in critical cell clusters in mice exposed to PM. (A) Schematic Venn diagram of overlapping “rescue” DEGs identified by integration analysis between “PM Exposure DEGs” ( vs. ) and “NMN Treatment DEGs” (Exp-NMN vs. ). The corresponding data are shown in Excel Table S6. (B) Identified “rescue” DEGs of each cell cluster are visualized in a rose diagram. (C) The bubble matrix displays the numbers of “rescue” DEGs (Rescue), “PM Exposure DEGs” (PM Exposure) and “NMN Treatment DEGs” (NMN Treatment). (D) The heatmap illustrates the overlapping enriched canonical pathways analyzed using Ingenuity Pathway Analysis (IPA) software (Qiagen) among neutrophils, monocyte-derived cells, and fibroblasts based on “rescue” DEGs. The -scores, from the lowest to the highest, are colored from blue to red, whereas a cross indicates when -scores were unable to be calculated owing to a lack of sufficient evidence. The corresponding -scores for each enriched pathway of the respective cell clusters are displayed in Table S9. (E) The expression of Pltp and Abca1 in fibroblast subsets of mouse lungs from , , and Exp-NMN groups are shown in UMAP plots. (F) The expression of Maf, Fkbp5, Cebpb, and C5ar1 in monocyte-derived cell subsets of mouse lungs from , , and Exp-NMN groups are shown in UMAP plots. The triglyceride (TG) levels examined in (G) mouse plasma (/group) and (H) mouse liver tissues (/group) from the , , Con-NMN, and Exp-NMN groups, respectively, at the end of the subchronic PM exposure. The expression levels of genes in (E) and (F) are listed in Excel Table S11. Data in (G) and (H) were analyzed using one-way ANOVA followed by Tukey’s multiple comparison post hoc test. The results are presented as . The mean, SD, and SEM values of the bar graphs in (G) and (H) are summarized in Table S5. **, compared with mice. # compared with the mice (Exp-NMN vs. ). -Values for all tests are shown in Table S6. Note: AM, alveolar macrophages; ANOVA, analysis of variance; AT1, type 1 alveolar epithelial cells; AT2, type 2 alveolar epithelial cells; Con, air-filtered control group; DC, dendritic cells; DEGs, differentially expressed genes; dividing DC, dividing dendritic cells; Exp, PM exposure group; , water; IL, interleukin; LXR, liver X receptor; NMN, nicotinamide mononucleotide; PM, particulate matter; RXR, retinoid X receptor; SD, standard deviation; SEM, standard error of the mean; UMAP, Uniform Manifold Approximation and Projection.
Furthermore, the universal fibroblasts (fibroblast 0) in both and Exp-NMN groups expressed higher levels of Pltp and Abca1 compared with those in the group, indicating more activated lipoprotein remodeling and less cellular cholesterol overload in universal fibroblasts in and Exp-NMN groups (Figure 8E). Based on the “rescue” DEGs (Excel Table S6), the expression levels of genes related to cellular lipid concentration and lipid metabolism, including Maf, Fkbp5, Cebpb, and C5ar1 were markedly lower in the profibrotic IMs (monocytes 3) identified in the Exp-NMN group, compared with the corresponding subclusters in the group (Figure 8F). Interestingly, lower lipid concentration and less neutrophils recruitment were also predicted in AMs with 18 “rescue” DEGs (Excel Table S7). Moreover, the significantly higher TG levels in plasma and liver in response to PM exposure were lower in the NMN-treated mice (Figure 8G,H).
Discussion
Both epidemiological32,33 and experimental34 evidence has revealed that long-term exposure is positively associated with the increased incidence and mortality of multiple diseases, even at a low exposure level. As a nutritional intervention, NMN supplementation has been shown to potentially attenuate the diverse detrimental health effects related to aging18 and xenobiotics-induced injury20,35 in mice. However, the protective effects of NMN on PM-induced chronic lung injury and the underlying mechanisms have yet to be defined. In this study, we established, to our knowledge for the first time, that NMN effectively improved immune functions and lipid metabolism to mitigate markers of lung injury following ambient PM exposure in a mouse model. These novel findings uncover the potential mechanisms of the protective effects of NMN intervention against PM-induced chronic lung injury.
NMN supplementation has been previously employed to enhance content in both human and mice as an anti-aging supplement.36 NMN has also shown efficacy in aging-related or metabolic dysfunctions, including diabetes20 and cognitive impairments35 in mice. Most studies on the long-term beneficial effects of NMN treatment were performed by repeated oral gavage24 or IP injection,37 treatment modalities associated with stress (including hormone disruption and increased blood pressure) and chronic inflammation.38 Moreover, the absorbance of NMN and its activated forms upon IP injection could be distinctly different from its oral intake.15 Hence, NMN supplementation in drinking water ad libitum mimics the oral modality of NMN intake in humans absent extra stress. Notably, ( for a subject weighing ) of orally NMN administration has been used in most clinical trials in healthy human subjects without any observed adverse health effects.39–41 After conversion from the human equivalent dose to the mouse dose according to the U.S. Food and Drug Administration (FDA) guidelines, the dose we chose for NMN supplementation in mice was concordant with the actual dose range of NMN treatment in humans ().42,43 In this study, NMN supplementation on healthy mice by ad libitum for 18 wk significantly boosted content in multiple tissues without any observed effects on body weight and food and water intake, which was consistent with the previous studies on safety of NMN administration on rodents.24
According to the previous studies conducted on mice and mouse cell lines, NMN supplementation has been reported to effectively mitigate ALI induced by LPS,11 as well as bleomycin-induced pulmonary fibrosis22 by enhancing cellular storage, and in turn, reducing oxidative stress and genotoxicity.44 lung and systemic injury was reported to be closely associated with oxidative stress and pro-inflammatory cytokines in numerous studies conducted in rodents,45 but an effective intervention strategy and its mechanisms have rarely been investigated. In this study, with higher content in multiple tissues, 18-wk NMN supplementation led to anti-inflammatory and anti-fibrotic effects on PM-exposed mouse lungs by reducing oxidative stress, chronic inflammation, and excessive collagen deposition. Our previous study illustrated that the activated IL-17 signaling pathway was likely induced by chronic inflammation and participated in MDSC recruitment, contributing to the profibrotic immunosuppressive microenvironment in PM-exposed lungs.8 Here, we also observed that overlapped annotations of inhibited IL-17 signaling pathway in critical cell clusters upon PM exposure and NMN treatment. This consistent result suggested that NMN supplementation can regulate immune functions following subchronic PM exposure to attenuate chronic inflammation mediated by immunosuppression.
The immune functions of diverse immune cells as well as their interactions were at least in part regulated by altered metabolism in the progression of chronic inflammation.9 However, the therapeutic efficacy of NMN administration and the effect of altered metabolism on the modification of the immune microenvironment and rectifying immune dysfunctions represented by increased MDSCs has remained controversial. Direct administration has been shown to afford protective effects in autoimmune diseases by recruiting MDSCs,46 whereas improved restoration efficiently activated T cells and lowered MDSC levels to prevent cellular senescence.17,47 Collectively, the reversed immunosuppressive microenvironment mediated by decreased MDSCs and reduced secretion of profibrotic cytokines regulated by inhibited IL-17 signaling pathway might be attributable to the NMN-induced anti-inflammatory effects noted in our study. In addition, it has been recognized that the immunosuppressive microenvironment with higher levels of several cytokines, such as and , reshape the phenotypes of critical cells and their interactions, which could be rescued by NMN supplementation.48
Specific interactions between tissue-specific fibroblasts and macrophages in diverse tissues were reported to participate in diverse chronic disorders, including fibrosis49 and cancer development.50 The interactions between fibroblasts and macrophages have yet to be clearly defined in the spectrum of inflammatory to fibrotic diseases in response to PM exposure. Here, scRNA-seq analysis enabled us to examine changes in cell numbers of monocyte-derived cells (particularly IMs) and fibroblasts and to characterize the phenotypes and functions of activated profibrotic fibroblasts, pro-inflammatory and profibrotic IMs phenotypes, as previously reported.51–53 The weakened interactions between ligand–receptor pairs (including Ccl2-Ccr2 and Il1b-Il1r2) in single-cell resolution and the altered cell phenotypes were in line with previous studies, suggesting that NMN regulated the interactions between cancer-associated fibroblasts and tumor-associated macrophages to promote recruitment of MDSCs.50,54 These findings suggest that NMN supplementation reversed pro-inflammatory and profibrotic phenotype differentiation through regulation of the interactions between interstitial cells and fibroblasts.
Both in humans55 and in rodents,11 higher cellular content upon NMN administration has been shown to afford enhanced resistance and to stabilize systemic and cellular metabolism in response to stress. However, the interactions between metabolic regulation and the protective effects of NMN intervention have yet to be characterized. Here, we described a comprehensive metabolic profile in mice following NMN replenishment. The enhanced systemic and hepatic lipid metabolism prior to xenobiotics stimulation upon NMN treatment might confer mice with resistance to lipid accumulation caused by acute oxidative stress. Although it has been reported that prolonged oxidative stress and pro-inflammatory cytokines secreted from the lung triggers dyslipidemia, hepatic lipid accumulation, and subsequent extra-pulmonary injury,56,57 it is unclear whether NMN intervention leads to attenuation in PM-induced inflammation through the modulation of lipid metabolism. Our results revealed that NMN supplementation effectively lowered the proportions of recruited and activated MDSCs and prevented profibrotic phenotype differentiation. These observations are consistent with previous studies illustrating that systemic and hepatic lipid metabolism disturbance augmented pro-inflammatory monocytes recruitment and impaired neutrophils homeostasis in bone marrow compartment, which are involved in promotion of the inflammation.58,59 Consistent with improved lipid metabolism induced by NMN treatment, studies on cancer immunotherapy have demonstrated that increased exogenous lipid uptake and intracellular lipid accumulation enhanced the immunosuppressive function of MDSCs.46,60
At the beginning of the inflammatory phase, intracellular lipids function as pro-inflammatory signaling molecules to initiate anti-inflammatory gene transcription.59 With reduced interactions between pro-inflammatory and profibrotic effectors, we identified the up-regulated pathways related to lipid metabolism in fibroblasts and IMs in Exp-NMN mice including LXR/RXR activation, as well as the PPAR pathway and the adenosine monophosphate kinase (AMPK) pathway, to name a few. These pathways are closely related to cellular lipid biosynthesis, cholesterol accumulation, and cholesterol efflux, which play important roles in maintaining cellular lipid homeostasis and regulating the progressive inflammation in ALI and pulmonary fibrosis.61–63 Down-regulation of these pathways has been reported upon persistent inflammation and to simultaneously play a key role in aggravating inflammation.64,65 However, additional studies are needed to validate the casual linkage between improved lipid homeostasis, less myofibroblast differentiation, and reduced phagocytic dysfunction of macrophages following NMN supplementation. Here, we showed a correlation between NMN supplementation-attributable lipid homeostasis and the immune functions, and pathological phenotypes, as well as the interactions between critical cell clusters with an advantage of scRNA-seq analysis. Taken together, these findings suggest that increased levels provide a metabolic microenvironment that facilitates the intercellular communication for anti-inflammation, and, in turn, confers cells with resistance to PM-induced adverse health effects.
In conclusion, we demonstrated that NMN supplementation exerted therapeutic efficacy against PM-induced lung injury in mice by regulating immune functions and lipid metabolism. These novel findings provide new insights into effective intervention strategies for dietary nutritional supplements and protection of human health, specifically lung injury, against adverse environmental stimuli.
Supplementary Material
Acknowledgments
Rui Zhang, S.C., and Z.W. contributed equally to this work. D.L. and W.C. conceived the study. Rong Zhang and Y.N. provided technical assistance. Rong Zhang optimized scRNA-seq protocols, generated and analyzed the scRNA-seq data set. Rui Zhang, S.C., Z.W., D.L., and W.C. prepared the manuscript. D.L. and W.C. supervised the study. We thank Z. Xie for the support in computational analysis and L. Ye, Y. Jiang, M. Li, X. Jiang, H. Peng, Z. Guo, and L. Chen for their assistance. This work was supported by the Major Research Plan of National Natural Science Foundation of China (91943301, 91543208, to W.C.), Guangdong Provincial Natural Science Foundation Team Project (2018B030312005, to W.C.), National Institute of Environmental Health Sciences (NIEHS) (R01 ES10563 and R01 ES07331, to M.A.), Guangdong Basic and Applied Basic Research Foundation (2020A1515111114, to S.C.) and Beijing Key Laboratory of Environmental Toxicology (2022BKLET02, to S.C.).
References
- 1.Xing YF, Xu YH, Shi MH, Lian YX. 2016. The impact of PM2.5 on the human respiratory system. J Thorac Dis 8(1):E69–E74, PMID: , 10.3978/j.issn.2072-1439.2016.01.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liang F, Liu F, Huang K, Yang X, Li J, Xiao Q, et al. 2020. Long-term exposure to fine particulate matter and cardiovascular disease in China. J Am Coll Cardiol 75(7):707–717, PMID: , 10.1016/j.jacc.2019.12.031. [DOI] [PubMed] [Google Scholar]
- 3.Panebianco C, Eddine FBN, Forlani G, Palmieri G, Tatangelo L, Villani A, et al. 2019. Probiotic Bifidobacterium lactis, anti-oxidant vitamin E/C and anti-inflammatory DHA attenuate lung inflammation due to PM2.5 exposure in mice. Benef Microbes 10(1):69–75, PMID: , 10.3920/BM2018.0060. [DOI] [PubMed] [Google Scholar]
- 4.Li D, Chen S, Li Q, Chen L, Zhang H, Li H, et al. 2020. Caloric restriction attenuates C57BL/6 J mouse lung injury and extra-pulmonary toxicity induced by real ambient particulate matter exposure. Part Fibre Toxicol 17(1):22, PMID: , 10.1186/s12989-020-00354-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Péter S, Holguin F, Wood LG, Clougherty JE, Raederstorff D, Antal M, et al. 2015. Nutritional solutions to reduce risks of negative health impacts of air pollution. Nutrients 7(12):10398–10416, PMID: , 10.3390/nu7125539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Speakman JR, Mitchell SE. 2011. Caloric restriction. Mol Aspects Med 32(3):159–221, PMID: , 10.1016/j.mam.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 7.Li D, Zhang R, Cui L, Chu C, Zhang H, Sun H, et al. 2019. Multiple organ injury in male C57BL/6J mice exposed to ambient particulate matter in a real-ambient PM exposure system in Shijiazhuang, China. Environ Pollut 248:874–887, PMID: , 10.1016/j.envpol.2019.02.097. [DOI] [PubMed] [Google Scholar]
- 8.Zhang R, Chen S, Chen L, Ye L, Jiang Y, Peng H, et al. 2022. Single-cell transcriptomics reveals immune dysregulation mediated by IL-17A in initiation of chronic lung injuries upon real-ambient particulate matter exposure. Part Fibre Toxicol 19(1):42, PMID: , 10.1186/s12989-022-00483-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Navas LE, Carnero A. 2021. NAD+ metabolism, stemness, the immune response, and cancer. Signal Transduct Target Ther 6(1):2, PMID: , 10.1038/s41392-020-00354-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garten A, Schuster S, Penke M, Gorski T, de Giorgis T, Kiess W. 2015. Physiological and pathophysiological roles of NAMPT and NAD metabolism. Nat Rev Endocrinol 11(9):535–546, PMID: , 10.1038/nrendo.2015.117. [DOI] [PubMed] [Google Scholar]
- 11.Liu J, Zong Z, Zhang W, Chen Y, Wang X, Shen J, et al. 2021. Nicotinamide mononucleotide alleviates LPS-induced inflammation and oxidative stress via decreasing COX-2 expression in macrophages. Front Mol Biosci 8:702107, PMID: , 10.3389/fmolb.2021.702107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takeda K, Okumura K. 2021. Nicotinamide mononucleotide augments the cytotoxic activity of natural killer cells in young and elderly mice. Biomed Res 42(5):173–179, PMID: , 10.2220/biomedres.42.173. [DOI] [PubMed] [Google Scholar]
- 13.Kar A, Mehrotra S, Chatterjee S. 2020. CD38: T cell immuno-metabolic modulator. Cells 9(7):1716, PMID: , 10.3390/cells9071716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang F, Deng X, Yu Y, Luo L, Chen X, Zheng J, et al. 2022. Association of human whole blood NAD+ contents with aging. Front Endocrinol (Lausanne) 13:829658, PMID: , 10.3389/fendo.2022.829658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katsyuba E, Romani M, Hofer D, Auwerx J. 2020. NAD+ homeostasis in health and disease. Nat Metab 2(1):9–31, PMID: , 10.1038/s42255-019-0161-5. [DOI] [PubMed] [Google Scholar]
- 16.Oh GS, Lee SB, Karna A, Kim HJ, Shen A, Pandit A, et al. 2016. Increased cellular NAD+ level through NQO1 enzymatic action has protective effects on bleomycin-induced lung fibrosis in mice. Tuberc Respir Dis (Seoul) 79(4):257–266, PMID: , 10.4046/trd.2016.79.4.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Camacho-Pereira J, Tarragó MG, Chini CCS, Nin V, Escande C, Warner GM, et al. 2016. CD38 dictates age-related NAD decline and mitochondrial dysfunction through an SIRT3-dependent mechanism. Cell Metab 23(6):1127–1139, PMID: , 10.1016/j.cmet.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mills KF, Yoshida S, Stein LR, Grozio A, Kubota S, Sasaki Y, et al. 2016. Long-term administration of nicotinamide mononucleotide mitigates age-associated physiological decline in mice. Cell Metab 24(6):795–806, PMID: , 10.1016/j.cmet.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshino J, Baur JA, Imai SI. 2018. NAD+ intermediates: the biology and therapeutic potential of NMN and NR. Cell Metab 27(3):513–528, PMID: , 10.1016/j.cmet.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshino J, Mills KF, Yoon MJ, Imai S. 2011. Nicotinamide mononucleotide, a key NAD+ intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metab 14(4):528–536, PMID: , 10.1016/j.cmet.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amjad S, Nisar S, Bhat AA, Shah AR, Frenneaux MP, Fakhro K, et al. 2021. Role of NAD+ in regulating cellular and metabolic signaling pathways. Mol Metab 49:101195, PMID: , 10.1016/j.molmet.2021.101195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang T, Yang J, Liu L, Xiao H, Wei X. 2021. Nicotinamide mononucleotide ameliorates senescence in alveolar epithelial cells. MedComm (2020) 2(2):279–287, PMID: , 10.1002/mco2.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gu XY, Chu X, Zeng XL, Bao HR, Liu XJ. 2017. Effects of PM2.5 exposure on the notch signaling pathway and immune imbalance in chronic obstructive pulmonary disease. Environ Pollut 226:163–173, PMID: , 10.1016/j.envpol.2017.03.070. [DOI] [PubMed] [Google Scholar]
- 24.Turner J, Licollari A, Mihalcea E, Tan A. 2021. Safety evaluation for Restorin® NMN, a NAD+ precursor. Front Pharmacol 12:749727, PMID: , 10.3389/fphar.2021.749727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57(1):289–300, 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 26.Matute-Bello G, Downey G, Moore BB, Groshong SD, Matthay MA, Slutsky AS, et al. 2011. An official American Thoracic Society workshop report: features and measurements of experimental acute lung injury in animals. Am J Respir Cell Mol Biol 44(5):725–738, PMID: , 10.1165/rcmb.2009-0210ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hübner RH, Gitter W, El Mokhtari NE, Mathiak M, Both M, Bolte H, et al. 2008. Standardized quantification of pulmonary fibrosis in histological samples. Biotechniques 44(4):507–511, PMID: , 10.2144/000112729. [DOI] [PubMed] [Google Scholar]
- 28.Olive PL, Banáth JP. 2006. The comet assay: a method to measure DNA damage in individual cells. Nat Protoc 1(1):23–29, PMID: , 10.1038/nprot.2006.5. [DOI] [PubMed] [Google Scholar]
- 29.Ma S, Sun S, Geng L, Song M, Wang W, Ye Y, et al. 2020. Caloric restriction reprograms the single-cell transcriptional landscape of Rattus norvegicus aging. Cell 180(5):984–1001.e22, PMID: , 10.1016/j.cell.2020.02.008. [DOI] [PubMed] [Google Scholar]
- 30.Faul F, Erdfelder E, Lang AG, Buchner A. 2007. G*Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav Res Methods 39(2):175–191, PMID: , 10.3758/bf03193146. [DOI] [PubMed] [Google Scholar]
- 31.Faul F, Erdfelder E, Buchner A, Lang AG. 2009. Statistical power analyses using G*Power 3.1: tests for correlation and regression analyses. Behav Res Methods 41(4):1149–1160, PMID: , 10.3758/BRM.41.4.1149. [DOI] [PubMed] [Google Scholar]
- 32.Shi L, Zanobetti A, Kloog I, Coull BA, Koutrakis P, Melly SJ, et al. 2016. Low-concentration PM2.5 and mortality: estimating acute and chronic effects in a population-based study. Environ Health Perspect 124(1):46–52, PMID: , 10.1289/ehp.1409111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li T, Hu R, Chen Z, Li Q, Huang S, Zhu Z, et al. 2018. Fine particulate matter (PM2.5): the culprit for chronic lung diseases in China. Chronic Dis Transl Med 4(3):176–186, PMID: , 10.1016/j.cdtm.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Riva DR, Magalhães CB, Lopes AA, Lanças T, Mauad T, Malm O, et al. 2011. Low dose of fine particulate matter (PM2.5) can induce acute oxidative stress, inflammation and pulmonary impairment in healthy mice. Inhal Toxicol 23(5):257–267, PMID: , 10.3109/08958378.2011.566290. [DOI] [PubMed] [Google Scholar]
- 35.Yoo KH, Tang JJ, Rashid MA, Cho CH, Corujo-Ramirez A, Choi J, et al. 2021. Nicotinamide mononucleotide prevents cisplatin-induced cognitive impairments. Cancer Res 81(13):3727–3737, PMID: , 10.1158/0008-5472.CAN-20-3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nadeeshani H, Li J, Ying T, Zhang B, Lu J. 2022. Nicotinamide mononucleotide (NMN) as an anti-aging health product—promises and safety concerns. J Adv Res 37:267–278, PMID: , 10.1016/j.jare.2021.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Margier M, Kuehnemann C, Hulo N, Morales J, Ashok Kumaar PV, Cros C, et al. 2022. Nicotinamide mononucleotide administration prevents doxorubicin-induced cardiotoxicity and loss in physical activity in mice. Cells 12(1):108, PMID: , 10.3390/cells12010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arantes-Rodrigues R, Henriques A, Pinto-Leite R, Faustino-Rocha A, Pinho-Oliveira J, Teixeira-Guedes C, et al. 2012. The effects of repeated oral gavage on the health of male CD-1 mice. Lab Anim (NY) 41(5):129–134, PMID: , 10.1038/laban0512-129. [DOI] [PubMed] [Google Scholar]
- 39.Okabe K, Yaku K, Uchida Y, Fukamizu Y, Sato T, Sakurai T, et al. 2022. Oral administration of nicotinamide mononucleotide is safe and efficiently increases blood nicotinamide adenine dinucleotide levels in healthy subjects. Front Nutr 9:868640, PMID: , 10.3389/fnut.2022.868640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yi L, Maier AB, Tao R, Lin Z, Vaidya A, Pendse S, et al. 2023. The efficacy and safety of β-nicotinamide mononucleotide (NMN) supplementation in healthy middle-aged adults: a randomized, multicenter, double-blind, placebo-controlled, parallel-group, dose-dependent clinical trial. Geroscience 45(1):29–43, PMID: , 10.1007/s11357-022-00705-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fukamizu Y, Uchida Y, Shigekawa A, Sato T, Kosaka H, Sakurai T. 2022. Safety evaluation of β-nicotinamide mononucleotide oral administration in healthy adult men and women. Sci Rep 12(1):14442, PMID: , 10.1038/s41598-022-18272-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nair A, Morsy MA, Jacob S. 2018. Dose translation between laboratory animals and human in preclinical and clinical phases of drug development. Drug Dev Res 79(8):373–382, PMID: , 10.1002/ddr.21461. [DOI] [PubMed] [Google Scholar]
- 43.FDA (U.S. Food and Drug Administration). 2005. Guidance for Industry: Estimating the Maximum Safe Starting Dose in Initial Clinical Trials for Therapeutics in Adult Healthy Volunteers. https://www.fda.gov/regulatory-information/search-fda-guidance-documents/estimating-maximum-safe-starting-dose-initial-clinical-trials-therapeutics-adult-healthy-volunteers [accessed 6 July 2005].
- 44.Soma M, Lalam SK. 2022. The role of nicotinamide mononucleotide (NMN) in anti-aging, longevity, and its potential for treating chronic conditions. Mol Biol Rep 49(10):9737–9748, PMID: , 10.1007/s11033-022-07459-1. [DOI] [PubMed] [Google Scholar]
- 45.Feng S, Gao D, Liao F, Zhou F, Wang X. 2016. The health effects of ambient PM2.5 and potential mechanisms. Ecotoxicol Environ Saf 128:67–74, PMID: , 10.1016/j.ecoenv.2016.01.030. [DOI] [PubMed] [Google Scholar]
- 46.Wang JL, Li B, Tan GJ, Gai XL, Xing JN, Wang JQ, et al. 2020. NAD+ attenuates experimental autoimmune encephalomyelitis through induction of CD11b+ gr-1+ myeloid-derived suppressor cells. Biosci Rep 40(4):BSR20200353, PMID: , 10.1042/BSR20200353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chatterjee S, Daenthanasanmak A, Chakraborty P, Wyatt MW, Dhar P, Selvam SP, et al. 2018. CD38-NAD+ axis regulates immunotherapeutic anti-tumor T cell response. Cell Metab 27(1):85–100 e8, PMID: , 10.1016/j.cmet.2017.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buechler MB, Fu W, Turley SJ. 2021. Fibroblast-macrophage reciprocal interactions in health, fibrosis, and cancer. Immunity 54(5):903–915, PMID: , 10.1016/j.immuni.2021.04.021. [DOI] [PubMed] [Google Scholar]
- 49.Zhang W, Ohno S, Steer B, Klee S, Staab-Weijnitz CA, Wagner D, et al. 2018. S100a4 is secreted by alternatively activated alveolar macrophages and promotes activation of lung fibroblasts in pulmonary fibrosis. Front Immunol 9:1216, PMID: , 10.3389/fimmu.2018.01216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang X, Lin Y, Shi Y, Li B, Liu W, Yin W, et al. 2016. FAP promotes immunosuppression by cancer-associated fibroblasts in the tumor microenvironment via STAT3-CCL2 signaling. Cancer Res 76(14):4124–4135, PMID: , 10.1158/0008-5472.CAN-15-2973. [DOI] [PubMed] [Google Scholar]
- 51.Tsukui T, Sun KH, Wetter JB, Wilson-Kanamori JR, Hazelwood LA, Henderson NC, et al. 2020. Collagen-producing lung cell atlas identifies multiple subsets with distinct localization and relevance to fibrosis. Nat Commun 11(1):1920, PMID: , 10.1038/s41467-020-15647-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gibbings SL, Thomas SM, Atif SM, McCubbrey AL, Desch AN, Danhorn T, et al. 2017. Three unique interstitial macrophages in the murine lung at steady state. Am J Respir Cell Mol Biol 57(1):66–76, PMID: , 10.1165/rcmb.2016-0361OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ogawa T, Shichino S, Ueha S, Bando K, Matsushima K. 2022. Profibrotic properties of C1q+ interstitial macrophages in silica-induced pulmonary fibrosis in mice. Biochem Biophys Res Commun 599:113–119, PMID: , 10.1016/j.bbrc.2022.02.037. [DOI] [PubMed] [Google Scholar]
- 54.Biffi G, Tuveson DA. 2021. Diversity and biology of cancer-associated fibroblasts. Physiol Rev 101(1):147–176, PMID: , 10.1152/physrev.00048.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhong O, Wang J, Tan Y, Lei X, Tang Z. 2022. Effects of NAD+ precursor supplementation on glucose and lipid metabolism in humans: a meta-analysis. Nutr Metab (Lond) 19(1):20, PMID: , 10.1186/s12986-022-00653-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu MX, Ge CX, Qin YT, Gu TT, Lou DS, Li Q, et al. 2019. Prolonged PM2.5 exposure elevates risk of oxidative stress-driven nonalcoholic fatty liver disease by triggering increase of dyslipidemia. Free Radic Biol Med 130:542–556, PMID: , 10.1016/j.freeradbiomed.2018.11.016. [DOI] [PubMed] [Google Scholar]
- 57.Zeng YQ, Chang LY, Guo C, Lin C, Bo Y, Wong MCS, et al. 2022. Chronic fine particulate matter exposure, habitual exercise, and dyslipidemia: a longitudinal cohort study. Environ Epidemiol 6(1):e190, PMID: , 10.1097/EE9.0000000000000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stiekema LCA, Willemsen L, Kaiser Y, Prange KHM, Wareham NJ, Boekholdt SM, et al. 2021. Impact of cholesterol on proinflammatory monocyte production by the bone marrow. Eur Heart J 42(42):4309–4320, PMID: , 10.1093/eurheartj/ehab465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tall AR, Yvan-Charvet L. 2015. Cholesterol, inflammation and innate immunity. Nat Rev Immunol 15(2):104–116, PMID: , 10.1038/nri3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yan D, Adeshakin AO, Xu M, Afolabi LO, Zhang G, Chen YH, et al. 2019. Lipid metabolic pathways confer the immunosuppressive function of myeloid-derived suppressor cells in tumor. Front Immunol 10:1399, PMID: , 10.3389/fimmu.2019.01399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kotlyarov S, Kotlyarova A. 2021. Anti-inflammatory function of fatty acids and involvement of their metabolites in the resolution of inflammation in chronic obstructive pulmonary disease. Int J Mol Sci 22(23):12803, PMID: , 10.3390/ijms222312803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Suryadevara V, Ramchandran R, Kamp DW, Natarajan V. 2020. Lipid mediators regulate pulmonary fibrosis: potential mechanisms and signaling pathways. Int J Mol Sci 21(12):4257, PMID: , 10.3390/ijms21124257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mamazhakypov A, Schermuly RT, Schaefer L, Wygrecka M. 2019. Lipids—two sides of the same coin in lung fibrosis. Cell Signal 60:65–80, PMID: , 10.1016/j.cellsig.2019.04.007. [DOI] [PubMed] [Google Scholar]
- 64.Hong C, Tontonoz P. 2008. Coordination of inflammation and metabolism by PPAR and LXR nuclear receptors. Curr Opin Genet Dev 18(5):461–467, PMID: , 10.1016/j.gde.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shi Y, Chen Q, Yan H, Gu W. 2016. The effect of a liver-X-receptor ligand on bleomycin induced pulmonary fibrosis in mice. Int Immunopharmacol 41:116–121, PMID: , 10.1016/j.intimp.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 66.Końca K, Lankoff A, Banasik A, Lisowska H, Kuszewski T, Góźdź S, et al. 2003. A cross-platform public domain PC image-analysis program for the comet assay. Mutat Res - Genet Toxicol Environ Mutagen 534(1–2):15–20, 10.1016/S1383-5718(02)00251-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.