Abstract
Multiple distal cis-regulatory elements (CREs) often cooperate to regulate gene expression, and the presence of multiple CREs for a gene has been proposed to provide redundancy and robustness to variation. However, we do not understand how attributes of a gene's distal CRE landscape—the CREs that contribute to its regulation—relate to its expression and function. Here, we integrate three-dimensional chromatin conformation and functional genomics data to quantify the CRE landscape composition genome-wide across ten human tissues and relate their attributes to the function, constraint, and expression patterns of genes. Within each tissue, we find that expressed genes have larger CRE landscapes than nonexpressed genes and that genes with tissue-specific CREs are more likely to have tissue-specific expression. Controlling for the association between expression level and CRE landscape size, we also find that CRE landscapes around genes under strong constraint (e.g., loss-of-function intolerant and housekeeping genes) are not significantly smaller than other expressed genes as previously proposed; however, they do have more evolutionarily conserved sequences than CREs of expressed genes overall. We also show that CRE landscape size does not associate with expression variability across individuals; nonetheless, genes with larger CRE landscapes have a relative depletion for variants that influence expression levels (expression quantitative trait loci). Overall, this work illustrates how differences in gene function, expression, and evolutionary constraint are reflected in features of CRE landscapes. Thus, considering the CRE landscape of a gene is vital for understanding gene expression dynamics across biological contexts and interpreting the effects of noncoding genetic variants.
Keywords: gene regulation, regulatory landscape, cis-regulatory element, tissue-specific gene expression
Significance.
Gene regulation is essential to all cellular and evolutionary processes, from development to speciation. We can now map individual cis-regulatory elements (CREs) genome wide, and many distal CREs often work in combination to regulate gene expression (CRE landscapes). However, we do not yet know how the attributes of CRE landscapes relate to differences in gene expression and function. By integrating diverse genomic data, we define CRE landscapes across ten human tissues and uncover the complex relationships between gene function and CRE landscape size, constraint, and tissue specificity. Understanding regulatory landscapes will allow future work to incorporate these features when interpreting the effects of genetic variation on gene expression and phenotype.
Introduction
Cis-regulatory elements (CREs) regulate gene expression by binding transcription factors and modulating transcription across diverse cell types. Combinations of distal CREs can act additively, synergistically, or redundantly to mediate expression of the same target gene (Dukler et al. 2016; Hay et al. 2016; Shin et al. 2016; Moorthy et al. 2017; Will et al. 2017; Xie et al. 2017; Osterwalder et al. 2018). For example, studies in Drosophila have established that the presence of multiple enhancers for a gene often provides robustness to genetic variation (Hong et al. 2008; Hobert 2010; Cannavò et al. 2016). Redundant “shadow enhancers” can maintain appropriate gene expression when other enhancers regulating the same target gene are inactivated (Hong et al. 2008; Letelier et al. 2018). In other cases, CREs in Drosophila have more complex context- and position-dependent functional relationships (Cannavò et al. 2016; Scholes et al. 2019). Studies of super enhancers, enhancer domains, and CRE redundancy in mice and humans suggest that similar interactions among CREs are widespread in mammalian species (Hay et al. 2016; Shin et al. 2016; Berthelot et al. 2018; Huang et al. 2018; Osterwalder et al. 2018; Wang and Goldstein 2020). The complexity of a gene's regulatory landscape has also recently been proposed to differentially influence power to detect expression quantitative trait loci (eQTL) and associations between genetic variants and traits (Mostafavi et al. 2022). Given the contribution of multiple distal CREs to the expression of many genes, consideration of the full CRE landscape is crucial to interpreting variation in gene expression across tissues, individuals, and even species.
However, previous work on CRE landscapes is largely confined to studies in model organisms and a small number of tissues. As a result, we do not understand how features of the CRE landscape relate to constraints on gene expression or alter the impact of genetic variation in humans. Furthermore, three-dimensional (3D) chromatin looping facilitates CRE activity by modifying the physical proximity of CREs and the genes they regulate. Increasingly high-resolution experimental approaches to map the 3D architecture of the genome are becoming available for a range of human cellular contexts (Kempfer and Pombo 2019). These data provide an additional dimension in which to characterize gene regulatory landscapes by mapping CREs to their putative target genes.
We address these gaps by defining CRE landscapes by integrating 3D chromatin conformation data with functional genomic and evolutionary characterization of human CREs. We then study the relationship between CRE landscapes and variation in gene expression across both tissues and individuals. We consider multiple attributes of CRE landscapes, including the number of active CREs, the physical proximity between CREs and a given target gene, and tissue specificity. We observe that differences in gene function and constraint on gene expression are reflected in features of their CRE landscapes, including the number and tissue specificity of associated CREs. Our results provide a map of CRE landscapes across ten human tissues and demonstrate the importance of considering the CRE landscape when studying gene expression dynamics and interpreting the effects of regulatory genetic variation on expression.
Results
Integration of Histone Modification and Chromatin Conformation Data Reveals Context-Specific Distal CRE Landscapes
We integrated genome-wide 3D contact maps from Hi-C experiments matched with functional genomics data to characterize CRE landscapes across ten cellular contexts. We defined distal CREs by the presence of an H3K27ac histone modification peak not overlapping an H3K4me3 peak from the Roadmap Epigenomics Project. We then combined these with machine-learning–based chromatin loop predictions that integrate data from multiple chromatin conformation assays to account for variable read depth across tissues (Materials and Methods) (Salameh et al. 2020). These loops define 3D domains where CREs and genes are in physical proximity (fig. 1A).
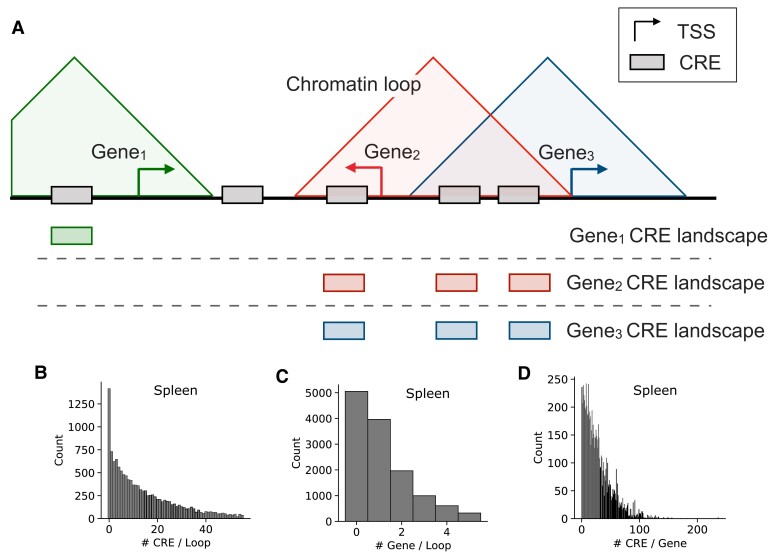
Fig. 1.
Integrated histone modifications and Hi-C loop predictions define CRE landscapes. (A) We define the chromatin loop-based CRE landscape for a gene as the union of all active, distal CREs in a chromatin loop region with the gene's TSS. We define CRE landscapes for each tissue using chromatin conformation and histone modification (H3K27ac without H3K4me3) data. If a gene's TSS overlapped multiple loops, the CREs in all overlapped loops were included. The horizontal line represents the linear genome sequence, and the three triangles represent chromatin loops. CREs are shown using filled rectangles, and the landscapes for each gene are shown underneath the genome line and aligned to match each CRE in the reference schematic. (B) The distribution of the number of CREs per loop in the spleen. (C) The distribution of the number of genes per loop in the spleen. The distributions shown in (B) and (C) have similar shapes for the other nine tissues (supplementary fig. S2, Supplementary Material online). (D) The distribution of the number of CREs per gene in the spleen.
We linked each CRE to a gene if the CRE and the gene's transcription start site (TSS) are within the same chromatin loop. Overlapping loop regions were merged, taking the union of all potential CRE–gene connections, when defining the CRE landscape. Across tissues, this conservative approach links an average of 38% of CREs and 29% of expressed genes; this corresponds to nearly all genes present inside of a loop (96% total, 98% expressed; table 1 and supplementary table S1, Supplementary Material online). Most genes with a chromatin loop–based CRE landscape are linked with multiple CREs (average 18 CREs; fig. 1B and C and supplementary table S2, Supplementary Material online).
Table 1.
Summary Statistics for Loop-Based CRE Landscapes
| Tissue | # Loops | Mean Length (kb) | # CREs | % Linked CREs | # Linked Genes | # Linked Exp Genes | % Linked Exp Genes | # Linked Tissue-specific Genes |
|---|---|---|---|---|---|---|---|---|
| Spleen | 13,753 | 191.2 | 107,591 | 58% | 8,292 | 6,502 | 52% | 455 |
| Liver | 15,059 | 209.9 | 83,547 | 55% | 7,482 | 4,889 | 45% | 443 |
| Heart | 14,177 | 210.3 | 128,298 | 49% | 5,003 | 3,680 | 31% | 270 |
| Hippocampus | 11,425 | 201.4 | 110,817 | 43% | 4,989 | 3,860 | 31% | 431 |
| Lung | 9,184 | 188.8 | 194,643 | 40% | 4,627 | 3,790 | 28% | 319 |
| Pancreas | 11,287 | 172.8 | 90,439 | 34% | 5,011 | 3,288 | 30% | 232 |
| Prefrontal cortex | 7,796 | 189.2 | 175,702 | 30% | 3,517 | 2,806 | 22% | 333 |
| Muscle | 6,811 | 177.4 | 150,967 | 29% | 2,977 | 2,038 | 19% | 156 |
| Small intestine | 10,214 | 173.3 | 172,920 | 27% | 3,128 | 2,462 | 19% | 260 |
| Ovary | 5,646 | 167.6 | 187,690 | 17% | 1,540 | 1,135 | 9% | 76 |
We also considered additional strategies for defining CREs (ChromHMM) and for linking CREs to genes (a contact-based CRE landscape definition and the Activity-By-Contact [ABC] model). Results were similar when using CREs defined by ChromHMM (Supplementary Note S1, Supplementary Material online), so we focus on the larger histone modification–based CREs in the main text. We found that both of the alternative approaches for linking genes and CREs had limitations (Supplementary Note S1 and supplementary figs. S1–S4, Supplementary Material online). The contact-based approach was limited by sequencing depth, which influenced the ability to identify significant chromatin interactions in different tissues. The ABC model was limited by data availability. It could only be applied in six of the available tissues, and it linked a small fraction of CREs to genes. Moreover, the ABC links in our CRE definition were a subset of Peakachu links (supplementary fig. S4, Supplementary Material online). Thus, we focus on the loop-based method in the main text due to its stability across tissues and robustness to differences in sequencing depth. Nonetheless, when possible, we confirmed results using the other linking strategies and report these in the Supplementary material.
Expressed Genes Have a Greater Number of CREs in Their CRE Landscapes
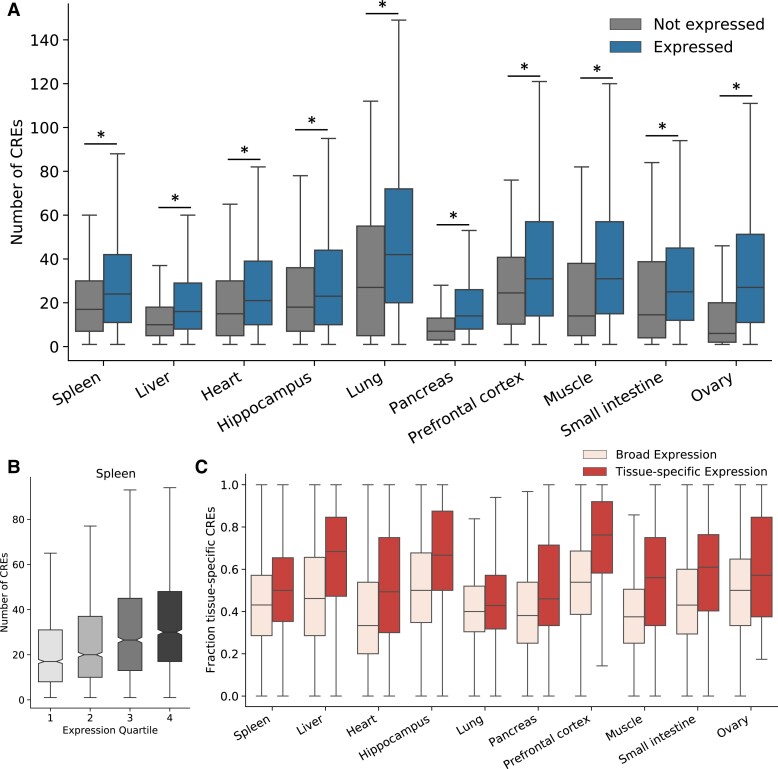
We first evaluated whether properties of a gene's CRE landscape are associated with its expression patterns within and across tissues. Across all ten tissues we considered, genes expressed in a tissue have a larger number of active CREs than genes that are not expressed in that tissue (fig. 2A). In the spleen, for example, the median number of loop-based CREs for expressed genes is 23, compared with 14 for genes not expressed in the spleen (Mann-Whitney U P = 5.0E−51; fig. 2A and supplementary table S3, Supplementary Material online).
Fig. 2.
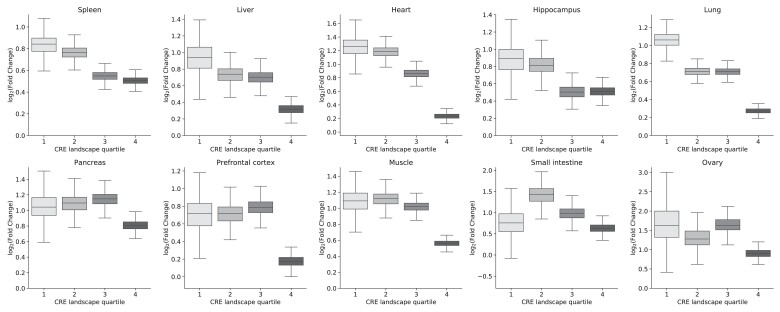
CRE landscapes reflect the expression level and tissue specificity of their associated genes. (A) Genes expressed in a tissue (right) have a larger number of CREs in their landscape active in the same tissue than nonexpressed genes (left). We consider expressed genes as those with a TPM > 1 and highlight significant differences (P < 0.05) with a “*”. This holds across all tissues considered and for contact-based CRE landscape definitions (supplementary fig. S5, Supplementary Material online). (B) Genes with higher expression have larger numbers of CREs in their CRE landscapes. The boxplot shows the number of CREs in the CRE landscape of each gene for genes divided into quartiles based on expression level (in the spleen). This trend is consistent across tissues and CRE landscape definitions (supplementary figs. S6 and S7, Supplementary Material online). (C) Genes with tissue-specific expression patterns have a greater proportion of tissue-specific CREs in their CRE landscapes. Boxplots represent the fraction of tissue-specific CREs in each gene's CRE landscapes for each of the ten tissues. Tissue-specific genes are defined by an expression relative entropy score > 0.3 (Materials and Methods). The numbers of expressed and tissue-specific genes for each tissue are given in table 1. For all boxplots, whiskers extend to 1.5 times the interquartile range. Outliers are not shown.
We also found that expression level among expressed genes is positively correlated with the number of CREs in a gene's landscape (Spearman ρ = 0.17–0.33 across tissues; fig. 2B and supplementary figs. S6 and S7 and table S4, Supplementary Material online). For example, in the spleen (ρ = 0.23, P = 9.6E−81), genes in the lowest expression quartile have a median of 17 CREs in their landscape, while genes in the highest quartile of expression have a median of 30 CREs in their landscapes.
Tissue Specificity of Gene Expression Is Reflected in the Tissue Specificity of the CRE Landscape
We observed variability in the number of CREs associated with genes expressed in each tissue. We hypothesized that the tissue specificity of a gene's expression and the activity patterns of its CREs contribute to this variability. We tested whether genes with more tissue-specific expression patterns had a higher proportion of tissue-specific CREs.
Across all tissues, the proportion of tissue-specific CREs in each landscape is higher for genes with tissue-specific expression than for expressed genes overall (11 more CREs on average; fig. 2C). This trend replicates in the contact-based landscapes for all tissues (supplementary fig. S8Aand table S5, Supplementary Material online). We also tested whether there was a difference in the number of CREs or the level of CRE sequence conservation. In some of the biological contexts, such as the liver, tissue-specific genes also have a greater number of associated CREs (supplementary fig. S8B, Supplementary Material online); genes with tissue-specific expression linked to CREs in the liver have significantly more CREs in their landscapes than genes with broad expression (median 20 CREs v. 15 CREs). However, this trend is not consistent across tissues or landscape definitions (supplementary fig. S8B and C, Supplementary Material online). Similarly, we do not find a consistent difference between the proportion of conserved sequences in CRE landscapes for tissue-specific and broadly expressed genes matched on expression level (supplementary fig. S9, Supplementary Material online). In six of the ten tissues, there is no difference in the level of CRE landscape conservation.
Housekeeping and LoF Intolerant Genes Have Similar CRE Landscape Sizes as Other Genes after Controlling for Expression Level
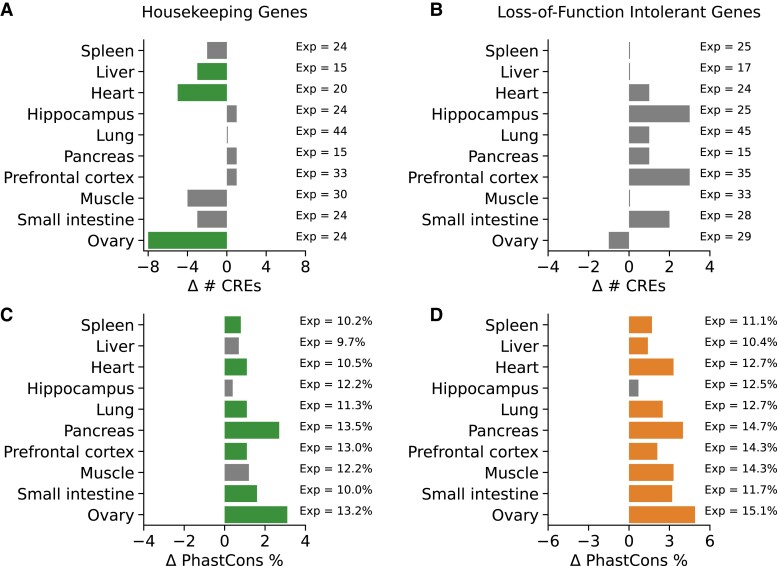
Previous work in model organisms suggests that the number and redundancy of CREs regulating a gene is associated with gene function (Osterwalder et al. 2018). We sought to evaluate the proposed relationship between housekeeping genes and smaller CRE landscapes in human tissues. Given the correlation between CRE landscape size and gene expression levels (fig. 2), we controlled these tests for expression level in each tissue (Materials and Methods). We find that although there may be simpler regulatory control for constitutively active genes in some contexts, it is not a universal pattern across human tissues. After matching on gene expression level and the presence of at least one CRE, we found that housekeeping genes have significantly fewer CREs than expressed genes in three tissues: liver, heart, and ovary (fig. 3A and supplementary table S6, Supplementary Material online). In these tissues, housekeeping genes had an average of 4 fewer CREs than expressed, nonhousekeeping genes. However, the trends were not consistent in the other tissues we considered or when using the contact-based definition (supplementary table S6, Supplementary Material online).
Fig. 3.
CRE landscapes of housekeeping and LoF intolerant genes are more conserved, but not larger, than other expressed genes. (A) In most tissues, housekeeping genes have CRE landscapes of similar size to matched sets of expressed genes (supplementary table S6, Supplementary Material online). However, for three tissues (liver, heart, and ovary), the landscape sizes are significantly smaller. (B) The sizes of CRE landscapes for LoF intolerant genes are not different than other expressed genes (supplementary table S7, Supplementary Material online). For (A) and (B), the x-axis displays the difference in the number of CREs between housekeeping or LoF intolerant genes and a set of matched expressed genes. The number of CREs for the matched expressed genes (Exp) is shown on the right. (C and D) Both housekeeping and LoF intolerant genes have a greater proportion of base pairs in their CRE landscapes in PhastCons conserved elements compared with a matched set of genes expressed in the same tissue. The pattern is present across seven of ten tissues for housekeeping genes and nine of ten tissues for LoF intolerant genes (supplementary tables S6 and S7, Supplementary Material online). The x-axis displays the difference in the proportion of PhastCons elements in the CRE landscapes housekeeping or LoF intolerant genes and a set of matched expressed genes. The proportion of PhastCons elements for the matched genes is shown on the right.
We also hypothesized that genes under strong constraint on their expression, like loss-of-function (LoF) intolerant genes, would have more associated CREs to provide the potential for regulatory buffering or finer control of expression levels (Lek et al. 2016; Karczewski et al. 2020). However, after matching for gene expression level, LoF intolerant genes did not have significantly different numbers of associated CREs than expressed genes in any of the tissues considered (supplementary table S7, Supplementary Material online). For example, in spleen, the median number of CREs in the landscape of both LoF intolerant genes and matched, expressed genes is 25 (fig. 3B and supplementary table S7, Supplementary Material online).
CRE Landscapes of Housekeeping and LoF Intolerant Genes Have Higher Sequence Constraint
Given the lack of consistent trends between constrained gene function and CRE landscape size, we hypothesized that the functional importance of the gene might instead be reflected in the level of DNA sequence constraint in its CRE landscape. Indeed, housekeeping and LoF intolerant genes have a higher proportion of evolutionarily conserved CREs in their landscapes compared with matched sets of expressed genes across most tissues (fig. 3C and D). On average, housekeeping genes have a 14% increase in the proportion of PhastCons elements in their CRE landscapes, while LoF intolerant genes have a 27% increase. The trend also replicates in a smaller number of tissues using the contact-based approach (supplementary tables S6 and S7, Supplementary Material online).
The number of CREs in a landscape is modestly correlated with the proportion of conserved base pairs in the CREs (% PhastCons overlap; Spearman ρ = 0.05–0.17 over all tissues; supplementary table S8, Supplementary Material online). The correlations are similar between the number of CREs and the probability of negative selection measured by the LINSIGHT scores of the CREs (mean LINSIGHT score; Spearman ρ = 0.01–0.15; supplementary table S8, Supplementary Material online). However, these correlations correspond to small changes in the overall conservation score. For example, in the spleen, the median LINSIGHT score for a CRE from a small landscape (first quartile) is 0.16, while the median score for a CRE from a large landscape (fourth quartile) is 0.18 (supplementary fig. S10, Supplementary Material online); the proportion of PhastCons overlap increases from 6% to 9% over the same interval (supplementary fig. S10, Supplementary Material online).
CRE Landscape Size Is Weakly Associated with Variability of Gene Expression across Individuals
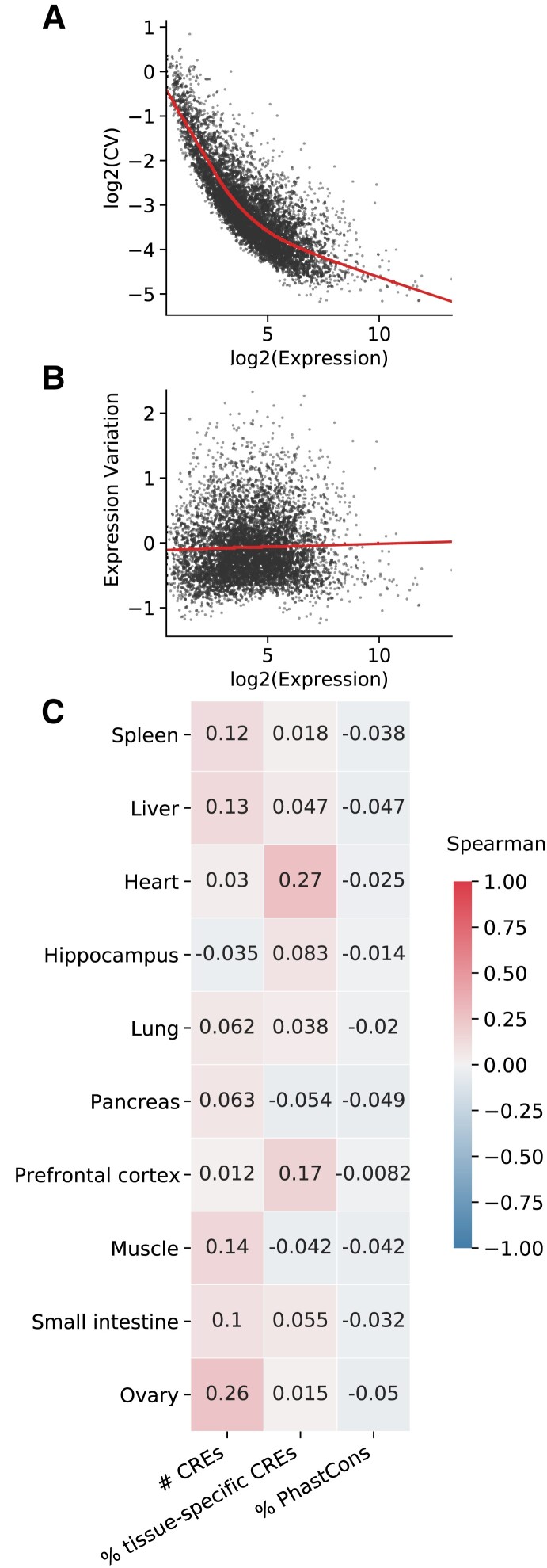
We next explored whether differences in CRE landscapes of expressed genes contribute to the variability in the genes’ expression levels across individuals. We integrated the CRE landscapes with individual-level gene expression data from the Genotype-Tissue Expression (GTEx) project to test whether the number of CREs in a gene's CRE landscape is predictive of the variability of gene expression across individuals. Given that the coefficient of expression variation is highly correlated with expression level (fig. 4A), we first regressed the coefficient of variation on the median expression level, as previously described (fig. 4B and supplementary figs. S11 and S12, Supplementary Material online; Materials and Methods) (Sigalova et al. 2020). We use the residuals from this model as a quantification of “expression variation” for comparison with CRE landscape attributes (fig. 4C).
Fig. 4.
Expression variation across individuals is weakly associated with attributes of the CRE landscape. (A) The median gene expression level is strongly correlated with coefficient of variation (CV) of expression across GTEx individuals. A LOESS regression line is shown over the scatter plot of the log of gene expression (x-axis) versus the log of the CV. (B) To adjust for the correlation between coefficient of variation and overall expression level, we compared the median gene expression level with the “expression variation”—the residuals from the plot in (A). The LOESS regression line is shown over a scatter plot of the log of gene expression (x-axis) versus the expression variation (y-axis). Data in (A) and (B) are shown for spleen, but trends are similar across tissues (supplementary figs. S11 and S12, Supplementary Material online). (C) Heatmap of Spearman correlations between expression variation and attributes of the CRE landscape. Positive correlations are shown in red, and negative correlations are shown in blue. Darker colors indicate a stronger correlation magnitude.
The number of CREs in a gene's landscape is weakly positively correlated with expression variation across individuals in nine of ten tissues (average Spearman ρ = −0.03 to 0.26; fig. 4C and supplementary fig. S13, Supplementary Material online). For example, in the spleen, the correlation between expression variation and the number of CREs in the CRE landscape is 0.12. However, heart and the two brain tissues do not exhibit significant correlations (heart: ρ = 0.03, hippocampus: ρ = −0.03; prefrontal cortex: ρ = 0.01), suggesting that the size of the CRE landscape does not strongly influence gene expression variability across individuals in these tissues. Expression variation weakly negatively correlated with DNA sequence conservation in each tissue and weakly positively correlated with tissue specificity of CREs in eight of ten tissues. However, these correlations are even lower in magnitude than for CRE landscape size. For example, tissue specificity of CREs in the spleen is not significantly correlated with expression variation, while the proportion of PhastCons elements has a weak negative correlation (ρ = −0.04; fig. 4C and supplementary fig. S13 and tables S9 and S10, Supplementary Material online). As expected, tissue specificity of a gene's expression is strongly associated with expression variation across individuals (average Spearman ρ = 0.41–0.61).
eQTL Enrichment Decreases with Increasing CRE Landscape Size
Because CRE landscape size correlated only weakly with evolutionary constraint and expression variation, we tested for a relationship between CRE landscape attributes and expression-associated genetic variants. Due to their role regulating the expression of genes, we expected CREs to be broadly enriched for overlap with eQTL. Indeed, CREs were enriched for overlap with eQTL identified by GTEx in the same cellular context (supplementary fig. S14, Supplementary Material online). However, we also expected that the level of enrichment would be influenced by the size of the CRE landscape of a gene. We hypothesized that genes with large CRE landscapes would be more robust to changes in expression driven by genetic variants, due to the greater potential for redundancy in larger CRE landscapes.
We observed a significant relative depletion for eQTL in landscapes containing more CREs (fig. 5 and supplementary fig. S15 and supplementary table S11, Supplementary Material online). For example, in the spleen, the CREs in the smallest 25% of landscapes are 3.5× enriched for eQTL overlap compared with 1,000 sets of random, length-matched regions. In contrast, CREs in the largest landscape quartile are only 2.7× enriched. The relative depletion for eQTL in the largest CREs held across tissues. This is consistent with previous work that identified a depletion of eQTL targets in genes with large regulatory domains (Wang and Goldstein 2020). In a few tissues (e.g., small intestine), landscapes in the second or third quartiles were slightly more enriched for eQTL than those in the first. This is likely a technical artifact due to the relatively small numbers of CREs in these tissues.
Fig. 5.
Larger CRE landscapes are relatively depleted for overlap with eQTL. Across tissues, CREs are enriched for eQTL from GTEx. However, there is a relative depletion for eQTL in larger landscapes (top 25%) versus CREs in smaller landscapes (bottom 25%). Enrichment is calculated by comparing the observed eQTL overlap to an empirical null distribution generated by random shuffles of length-matched CRE regions (n = 1,000). The fold change is calculated as the observed overlap divided by the expected overlap. This trend is replicated in contact-based CRE landscapes (supplementary fig. S15, Supplementary Material online).
Discussion
Variation in gene regulation plays a large role in both the etiology of complex human disease and the phenotypic differences between closely related species. Here, we leveraged chromatin conformation, functional genomics, and evolutionary data to quantify the CRE landscape composition across ten human tissues and relate landscape attributes to gene expression patterns. For example, genes with a larger number of linked CREs had higher gene expression values, supporting that, in many cases, regulatory elements have an additive effect on transcription levels. We also found that genes with tissue-specific expression patterns had a higher proportion of tissue-specific CREs in their landscapes, and genes under strong functional constraint had greater evolutionary conservation in the sequences of their CRE landscapes, regardless of landscape size. However, other aspects of gene expression and function, for example, variation across individuals and enrichment for expression-associated genetic variation, did not show strong associations with landscape properties. Nonetheless, we found that genes with larger CRE landscapes have relatively lower enrichments for expression-associated genetic variation than those with fewer CREs. This suggests that consideration of the CRE landscape should inform future work on the interpretation of noncoding genetic variants.
Based on previous work (Osterwalder et al. 2018), we hypothesized that tissue-specific genes would require a larger number of CREs to maintain appropriate expression patterns than broadly expressed genes, especially those with housekeeping functions. However, we did not find a clear association between tissue specificity and the total number of CREs in the landscape. Our results suggest that the proportion of the tissue-specific elements is more important to expression specificity than the number of elements overall. Similarly, we did not observe a consistent trend across tissues for the size of the CRE landscape for housekeeping or LoF intolerant genes versus other expressed genes. This contrasts with the consistently lower CRE number for housekeeping genes compared with developmental heart, brain, and limb TFs in mouse (Osterwalder et al. 2018). However, a direct comparison is challenging given that this previous analysis used several different techniques. First, CREs were mapped to genes based on correlated activity patterns. Thus, the power to link CREs to genes was dependent on activity levels, and this introduces a bias when studying tissue-specific genes. Second, the previous analysis did not control for gene expression level. Given the association between overall expression level and number of CREs observed here (fig. 2) and in previous work (Berthelot et al. 2018), controlling for this relationship is essential. Finally, the previous work focused on developmental timepoints, so tissue-specific developmental genes could have different CRE landscapes than adult tissue-specific genes. Overall, our results suggest that there is variability in the size of the CRE landscape required to maintain appropriate levels of gene expression across tissues and that CRE landscape size is not strongly associated with dosage sensitivity.
Our results indicate that the size of the human CRE landscape alone is not a good proxy for functional importance. As suggested by previous work on liver CRE evolution across mammals (Berthelot et al. 2018), we find that considering other landscape attributes, like evolutionary constraint and tissue specificity, is essential. Some elements of a CRE landscape may modulate expression level while others buffer the effects of noncoding genetic variation, thus complicating the associations between CRE landscape attributes, genetic variants, and expression level. Genes that have constraint on their expression levels, such as LoF intolerant genes, do have CRE landscapes with consistently stronger sequence constraint. However, even still, the CRE landscapes of LoF genes vary in CRE size. As previously proposed, redundancy in the CRE landscapes of important genes certainly occurs, and larger CRE landscapes are depleted for eQTL, but not all landscapes with many CREs contain redundant elements. Further work is required to determine the presence of functionally redundant CREs and their attributes.
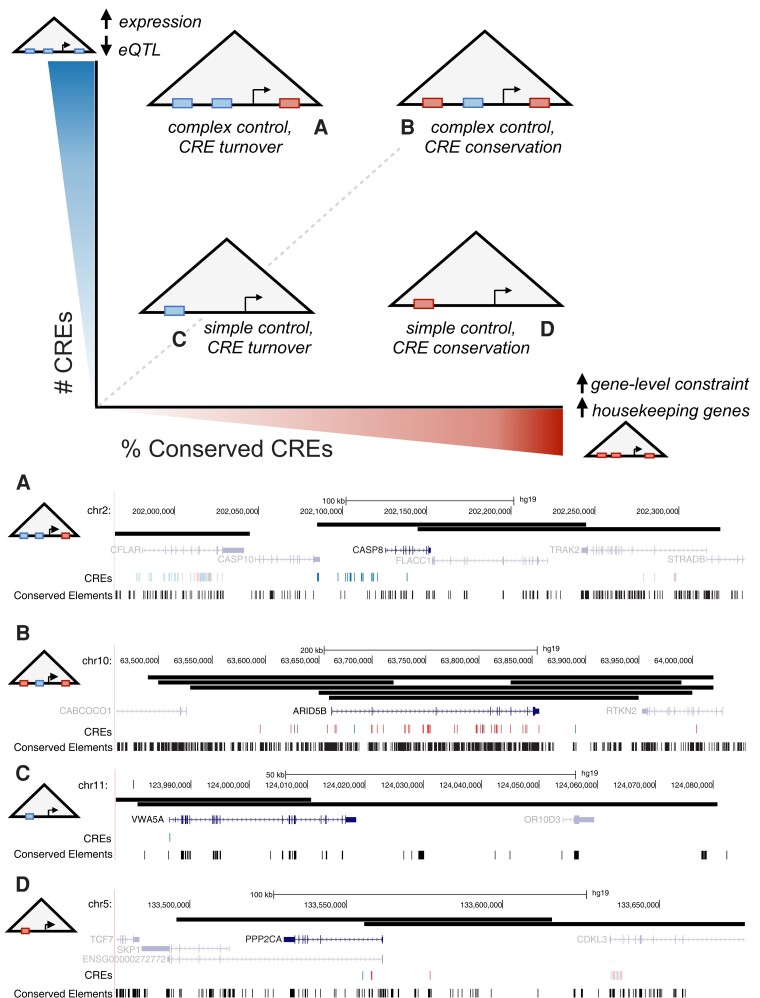
Quantifying the relationships between the composition of the CRE landscape and gene expression patterns will help fill gaps in our understanding of how genetic variation influences gene regulatory architecture and how this mediates relationships with phenotypic outcomes (Krijger and de Laat 2016). We propose a simple model that synthesizes our results on the relationship between CRE landscapes and genomic and functional attributes and provides a foundation for guiding future work. Our model positions CRE landscapes along two main axes of variation: the number of CREs and their evolutionary conservation (fig. 6).
Fig. 6.
CRE landscapes should be characterized in multiple dimensions to understand their function. The x-axis represents an increasing level of evolutionary conservation on the CREs in a landscape (top panel; left to right). The y-axis represents an increasing number of CREs (bottom to top). Genes exist at different locations within this space, and these attributes are related to the genes’ expression patterns, levels, and robustness to variation. Larger CRE landscapes are associated with increased gene expression, but a relative depletion for eQTL, suggesting the potential for gene regulatory buffering. Highly conserved CRE landscapes are associated with stronger gene-level constraint, such as LoF intolerance. Each schematic represents a gene with a CRE landscape along these two continua. Examples of real CRE landscapes in each quadrant are shown in (A)–(D). (A) shows the CRE landscape for CASP8 in the spleen; there are 16 CREs and none overlap conserved elements. (B) shows the CRE landscape for ARID5B in the liver; 34 of the 40 CREs overlap with conserved elements. (C) shows the CRE landscape for VWA5A in the heart; its 1 associated CRE does not overlap a conserved element. (D) shows the CRE landscape for PPP2CA in the liver; two of the three CREs overlap a conserved element. We note that CREs may participate in multiple genes’ landscapes, although our examples highlight one-to-one mappings. The boundaries of predicted chromatin loops defining the CRE landscapes are shown as black bars. CREs overlapping PhastCons conserved elements are shown in red, while those without overlap are shown in blue. Only the canonical transcript of the gene is shown; nearby genes and CRE landscapes are shown at a lower alpha level. These examples are labeled with additional functional annotations in supplementary figure S20, Supplementary Material online.
A large CRE landscape can enable high expression levels and create CRE redundancy that can prevent genetic variation from altering expression levels. This can reduce constraint on some CREs, allowing for evolutionary innovation while still maintaining expression levels (fig. 6A). For example, caspase-8 (CASP8) is a caspase involved in apoptosis and present across a wide range of species (Sakamaki et al. 2014) but has only moderate depletion for LoF variants (gnomAD o/e = 0.51). CASP8 is expressed in nearly all GTEx tissues except the brain, and its strongest expression is in the spleen. The CRE landscape of CASP8 in the spleen is large with 16 linked CREs, nearly half of which are tissue specific. However, none of the CREs overlaps a conserved element. The tissue-specific CREs likely contribute to its strong expression in the spleen compared with other tissues. The large numbers of CREs likely provide expression stability that, paired with the lack of strong constraint on the gene, may result in the absence of sequence conservation. Alternatively, large CRE landscapes can fine-tune expression levels in a way that is not possible with simpler architectures and, when the gene is under strong constraint, result in many conserved CREs (fig. 6B). AT-rich interaction domain 5B (ARID5B) is a DNA-binding protein that encodes a component of the H3K9Me2 demethylase complex in the liver that is involved in chromatin remodeling, adipogenesis, and hematopoiesis. ARID5B is expressed in all GTEx tissues but has substantially different expression levels between tissues. Genetic variation in the gene has been linked to acute lymphoblastic leukemia (ALL) (Baba et al. 2011; Xu et al. 2020; Whitson et al. 2021), and in pediatric patients with ALL, downregulation of ARID5B was associated with relapse and drug resistance, although the exact biological mechanisms remain unknown (Xu et al. 2020). Given its importance, ARID5B is LoF intolerant (gnomAD o/e = 0.02). The CRE landscape of ARID5B in the liver has 40 linked CREs and a high level of overlap with PhastCons conserved elements. We hypothesize that constraint on the regulation of this gene results in its large size and conservation, while the nonconserved but broadly active CREs associated with this gene could contribute to robustness in its overall expression across tissues.
Small CRE landscapes suggest that the genes require relatively simple regulatory control. For example, PPP2CA is a housekeeping gene that codes for part of the protein phosphatase 2A enzyme, which is involved in regulation of cell cycle and division (Janssens and Goris 2001). It is expressed highly in all GTEx tissues and is LoF intolerant (gnomAD o/e = 0.06). Consistent with the trends observed for housekeeping genes, PPP2CA has a small CRE landscape (e.g., 3 CREs in liver), and this landscape is under sequence constraint (fig. 6D). In contrast, the gene von Willebrand factor A domain containing 5A (VWA5A) has no clear annotated function. It has low tissue specificity across GTEx tissues and is lowly expressed in the heart, where it has a CRE landscape composed of 1 CRE (fig. 6C). VWA5A is LoF tolerant (gnomAD o/e = 1.13), and its CRE is not under sequence constraint. Thus, it appears that this gene's landscape is not strongly constrained.
Although these examples illustrate the trends we observe, we emphasize that quantifying either the number of CREs or their conservation alone is not sufficient to understand the CRE landscape of a gene. Indeed, throughout this work, we did not observe a simple relationship between the constraints on gene expression and any single CRE landscape attribute. We also find that the tissue specificity of the CREs is an additional dimension that is often useful to quantify, as well as aspects of the gene itself.
While our analyses provide a framework for studying CREs in the context of the broader CRE landscape of a gene, the results have several limitations that must be considered. First, we have incomplete knowledge of active CREs in complex biological contexts, and there is variation in the quality of data from different contexts. In our previous work, we found that most methods for identifying CREs are similarly enriched for variants identified in genome-wide association studies, eQTL, and sequences validated by massively parallel reporter assays in multiple biological contexts, although we are still likely missing relevant functional CREs in each context (Benton et al. 2019). We also believe that it will be valuable to study variation in the activity of CREs across individuals as more data become available.
We focus on a histone modification–derived approach to identify CREs, which provides greater sensitivity at the expense of specificity. To evaluate the robustness of our main conclusions, we also considered CREs defined by ChromHMM, a machine-learning approach that integrates additional functional genomics markers in its predictions. Our main results on expression levels, tissue specificity, and robustness to variation are similar with ChromHMM CREs, suggesting that our conclusions are robust to different strategies for capturing gene regulatory activity (supplementary figs. S17–S19, supplementary tables S14–S16, Supplementary Material online). Second, we rely on predicted chromatin loops derived from Hi-C to assign putative CREs to their target genes. These loops are less susceptible to bias caused by differences in Hi-C read depth across tissues than contact-based approaches (Salameh et al. 2020) but are also less granular than using individual chromatin interactions. Thus, the resolution of current data precludes us from disentangling the precise gene target of some CREs. Alternatively, the contact-based landscape definition uses individual Hi-C interactions to highlight more precise CRE–gene contacts. However, Hi-C experiments with lower read depth discover fewer robust interactions; thus, those tissues have a small proportion of significant CRE to gene associations. We found that the variation in observed patterns across tissues for contact-based landscapes is frequently correlated with the number of significant Hi-C interactions, suggesting that these differences are likely to reflect technical, rather than biological, differences. Improved data quality could increase our power to detect generalizable trends in the future. Recent approaches, such as the ABC model (Fulco et al. 2019), provide another approach for linking CREs to target genes but require additional experimental data and highlight similar CRE–gene links. As Hi-C and other genomic technologies continue to advance, we anticipate that higher resolution data sets will become available for a wide range of tissues to refine our current results. Our definitions of CRE landscapes are easily extensible to include future high-resolution data sets across biological contexts.
Ultimately, we highlight how differences in gene function, expression, and evolutionary constraint are reflected in the features of CRE landscapes. Our results illustrate that quantifying the CRE landscape of a gene will likely be necessary to understand its expression dynamics across contexts. In the future, we anticipate that quantifying the effects of CRE alteration in the context of our CRE landscape framework will facilitate interpretations of the effects of gene regulatory perturbations to phenotypic variability and disease risk.
Materials and Methods
All analyses were conducted using the GRCh37/hg19 build of the human genome. We used gene and TSS definitions from Ensembl v75 (GRCh37.p13). Analysis scripts were written in Python (v3.6.7) and R (v.4.0.5).
CRE Annotations and Chromatin Interaction Data
We downloaded normalized, 40-kb resolution Hi-C interaction frequency matrices from human samples for ten tissues: brain (prefrontal cortex, hippocampus), heart (left ventricle), liver, lung, ovary, pancreas, psoas muscle, spleen, and small intestine (Schmitt et al. 2016). The matrices were normalized using FitHiC (Ay et al. 2014). The locations of topologically associating domain (TAD) regions were derived from the same Hi-C interaction data by the 3D Genome Browser using the approach described in Dixon et al. (2012). We downloaded predicted chromatin loop anchors for each tissue from Peakachu (Salameh et al. 2020).
For each the ten tissues with Hi-C data, we also downloaded H3K27ac and H3K4me3 ChIP-seq peaks from the Roadmap Epigenomics Consortium (Roadmap Epigenomics Consortium et al. 2015). H3K27ac peaks were called as CREs if the peak was present and overlapped by <50% of its length with an H3K4me3 peak (Villar et al. 2015). We excluded H3K4me3 peaks because these are considered markers of promoters. We filtered putative CREs to remove any overlapping an ENCODE blacklist regions (Amemiya et al. 2019) and those in the top fifth percentile of length (>1.3 kb). The filtering process has little effect on the number of linked CREs and genes in the final analysis (supplementary table S13, Supplementary Material online). To evaluate the robustness of our results to CRE definition, we also downloaded enhancer annotations predicted by the ChromHMM 15-state model (“Enh”: state 7) (Ernst and Kellis 2012; Roadmap Epigenomics Consortium et al. 2015).
Definition of Genome-Wide CRE Landscapes
We considered both loop-based and contact-based approaches to defining a gene's CRE landscape for each tissue. The loop-based CRE landscape considers chromatin loops—the genomic regions between loop anchors predicted by the Peakachu model. Within each loop region, we associated the CREs with the TSSs within the same loop. For each gene, the CRE landscape is the union of all CREs associated with the canonical TSS in a loop region. We excluded any loops that are comprised of >5% ENCODE blacklist regions due to the difficult of mapping putative CREs in those loops.
We also defined contact-based CRE landscapes from evidence of direct chromatin interactions. For each gene, the landscape is based on the combination of CRE, gene, and Hi-C annotations. We considered CREs with evidence of a significant interaction from the Hi-C data (q < 0.05, i.e., the P value of the Hi-C interaction adjusted to a false discovery rate of 5%). The significance of a Hi-C interaction was determined by comparing the frequency of the observed interaction with an empirical null model adjusted for known technical biases. CREs that overlap the anchor of a significant Hi-C interaction (q < 0.05) are assigned to landscapes of genes with a TSS inside the other anchor. Where there are multiple CREs or TSSs within a single anchor, all CREs are linked to all potential gene targets. To account for the known role of TADs in constraining regulatory interactions, we limit the CRE–gene assignment to intra-TAD interactions.
We downloaded CRE to gene links defined using the ABC model for six cell types with matching loop- and contact-based CRE landscapes (liver, heart, muscle, ovary, spleen, and pancreas) (Fulco et al. 2019). We intersected the CREs from these files with our CRE definitions to identify linked genes using the ABC model. We quantified the similarity between the sets of genes linked to the same CRE using a relative Jaccard similarity metric.
Calculating Tissue Specificity of Gene Expression
We downloaded RNA-seq gene expression data from Genotype-Tissue Expression (GTEx, version 8) in transcripts per million (TPM) for ten tissues with matching Hi-C data: prefrontal cortex, hippocampus, heart, liver, lung, ovary, pancreas, skeletal muscle, spleen, and small intestine (Schmitt et al. 2016; GTEx Consortium et al. 2017). We consider expressed genes as those with a TPM > 1 (Uhlén et al. 2015). We calculated the tissue specificity of expression using the relative entropy of each gene's normalized expression profile across tissues compared to the median gene expression distribution across tissues in the sample. We scaled the resulting value between 0 and 1, where genes closer to 0 are broadly expressed and genes closer to 1 are tissue specific. We considered a second tissue specificity metric, τ (Yanai et al. 2005; Ravasi et al. 2010), although tissue-specific genes classified using this score produced similar results to the relative entropy approach. Using the τ metric the distribution was skewed towards tissue specificity (supplementary fig. S16, Supplementary Material online). We defined the final set of “tissue-specific” genes using a threshold on the relative entropy score. We tested multiple cutoffs at varying levels of stringency (supplementary table S12, Supplementary Material online) and selected a cutoff of 0.3 for use in the main text, which classifies approximately 10% of linked genes as tissue specific.
Calculating Tissue Specificity of CREs
We calculated the tissue specificity of CREs using by using the entropy score scaled between 0 and 1. However, because CREs do not have consistent lengths or locations across tissues, we standardized the CRE lengths before calculating the number of tissues where each CRE was active. We tested three possible standard lengths: 1) the median CRE length across tissues with lower quality ChIP-seq data (220 bp), 2) the median CRE length in our data set in the liver (460 bp; no ChIP-seq quality flags), and 3) the median CRE length for the histone-modification-defined liver CREs from Villar et al. (2.5 kb; same CRE definition (Villar et al. 2015). We centered the standardized CRE on the midpoint of the existing annotation and either expanded or truncated each region to the desired length. We selected the most conservative standard length of 220 bp for our final entropy calculations; the scores for the other two were correlated with our chosen threshold (ρ = 0.82 for 460 bp, ρ = 0.53 for 2.5 kb). We then calculated the overlap between standardized CREs across all tissues to determine the number of tissues where each CRE had activity. We calculated the entropy using this number of active tissues and assigned the result back to the original CRE element. The entropy was scaled to create a score between 0 and 1, with low scores indicating broad activity and high scores indicating tissue-specific activity.
Defining Gene Sets With Strong Functional Constraint
We considered two gene sets with higher levels of constraint on their function (and likely their expression) than expressed genes overall: housekeeping genes and LoF intolerant genes. We considered housekeeping genes identified in an earlier study based on consistent gene expression levels in RNA-seq data across sixteen tissues (n = 3,804) (Eisenberg and Levanon 2013). We downloaded a set of likely LoF intolerant genes from gnomAD (v2; n = 2,971) (Karczewski et al. 2020). Following gnomAD guidance, we defined LoF intolerant genes as those with a 90% confidence interval upper bound of the observed/expected (o/e) metric less than 0.35. Lower o/e scores indicate greater intolerance to protein variants.
Quantifying Evolutionary Constraint on CRE Sequences
We used two complementary approaches to define the level of DNA sequence conservation and constraint on CREs. First, we considered conserved elements from vertebrates and primates defined by the two-state hidden Markov model, PhastCons (Siepel et al. 2005). We merged the two sets of PhastCons conserved elements using Bedtools mergeBed (Quinlan and Hall 2010), and then calculated the proportion of each CRE that overlaps one of these elements. Second, we overlapped each CRE region with base-pair-level LINSIGHT scores, which estimate the probability of negative selection on noncoding sequence (Huang et al. 2017). We average the LINSIGHT scores across each CRE to calculate a final score for the element.
Matching Gene Sets on Expression Level
Due to differences in the distribution of gene expression levels across gene functional categories, we used the MatchIt library in R to generate matched sets of control genes for housekeeping, LoF intolerant, and tissue-specific genes (Ho et al. 2011). For each gene category, we matched a set of control genes on gene expression level using the caliper option (0.1). To generate the maximum possible control set, we allowed for up to two matches per gene for housekeeping and LoF intolerant genes, and up to 10 matches for tissue-specific genes.
Quantification of Expression Variation Across Individuals
We downloaded full gene expression matrices from GTEx (v8) for all ten tissues: prefrontal cortex, hippocampus, heart, liver, lung, ovary, pancreas, skeletal muscle, spleen, and small intestine (GTEx Consortium et al. 2017). Using the sample metadata to match subjects with their expression data, we log-transformed the expression values in the individual-by-gene matrix and filtered out samples with low expression (TPM < 1). Genes were required to meet the expression threshold in at least 80% of individuals. We then quantified the coefficient of variation (CV) for the expression of each gene across individuals, which is calculated as the standard deviation divided by the mean. Following previous approaches to account for the strong relationship between expression level and the coefficient of variation, we also calculated a final measure of individual “expression variability” as the residuals from a locally weighted (LOESS) regression of median gene expression on the CV (Sigalova et al. 2020).
Enrichment for GTEx eQTL in Different CRE Landscapes
We downloaded eQTL from GTEx (v8) for all ten tissues (P < 1E-5) (GTEx Consortium et al. 2017). The variant sets were mapped from hg38 to hg19 using LiftOver to match the rest of our annotations (Fujita et al. 2011). We calculated whether CREs were enriched for overlap with eQTL using a permutation-based approach. Briefly, we calculated the amount of overlap between the CREs and eQTL in each tissue using BedTools (Quinlan and Hall 2010), then randomly shuffled the CRE regions throughout the genome and recalculated the amount of overlap with this random set of regions. We performed the random shuffling process 1,000 times, maintaining the original number and length distribution of the CREs, avoiding ENCODE blacklist regions. For the loop-based landscapes, we also required the shuffled regions to fall within a predicted chromatin loop. Finally, we calculated an empirical P value for our observed overlap compared with this null distribution.
In addition, for each tissue and landscape definition, we separated the CRE landscapes into quartiles based on the number of CREs in each landscape. We used the same permutation framework to determine whether the CREs in different CRE landscape quartiles are enriched for overlap with eQTL compared with genomic background. We compared the level of enrichment between the top 25% and bottom 25% using a Mann–Whitney U test.
Supplementary Material
Acknowledgments
We thank E. McArthur, C. Tubbs, L. Colbran, S. Fong, A. Abraham, and other members of the Capra Lab for their helpful discussions and comments on the manuscript. This work was supported by the National Library of Medicine (T32LM012412 to M.L.B); and the National Institute of General Medical Sciences at the National Institutes of Health (R35GM127087 to J.A.C.). This work was conducted in part using the resources from the Advanced Computing Center for Research and Education at Vanderbilt University. The funders did not play any role in the study design, collection, analysis, and interpretation of data or in writing the manuscript.
Contributor Information
Mary Lauren Benton, Department of Computer Science, Baylor University, Waco, Texas, USA.
Douglas M Ruderfer, Departments of Medicine, Psychiatry and Biomedical Informatics, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
John A Capra, Department of Epidemiology and Biostatistics, University of California at San Francisco, USA.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Data Availability
The CRE annotations were derived from ChIP-seq peaks from the Roadmap Epigenomics portal (https://egg2.wustl.edu/roadmap/data/byFileType/peaks/unconsolidated/gappedPeak/). The public 15-state ChromHMM annotations for all tissues were also downloaded from Roadmap Epigenomics (https://egg2.wustl.edu/roadmap/data/byFileType/chromhmmSegmentations/ChmmModels/coreMarks/jointModel/final). The Hi-C interaction matrices for all tissues were obtained from GSE87112. The predicted chromatin loops from Peakachu can be downloaded from the 3D Genome Browser (http://3dgenome.org). The CRE-gene links from the Activity-By-Contact model for six cell types (liver, heart, muscle, ovary, spleen, and pancreas) were downloaded from https://www.engreitzlab.org/abc/. Gene expression matrices for all tissues and tissue-specific eQTL were obtained from the GTEx Portal (version 8). We obtained PhastCons conserved elements from http://hgdownload.soe.ucsc.edu/goldenPath/hg19/phastCons46way/. LINSIGHT scores were downloaded from https://github.com/CshlSiepelLab/LINSIGHT. The analysis scripts and CRE landscape annotations used in this study are available on GitHub: https://github.com/bentonml/cre-landscape.
Literature Cited
- Amemiya HM, Kundaje A, Boyle AP. 2019. The ENCODE blacklist: identification of problematic regions of the genome. Sci Rep. 9:9354. doi: 10.1038/s41598-019-45839-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ay F, Bailey TL, Noble WS. 2014. Statistical confidence estimation for Hi-C data reveals regulatory chromatin contacts. Genome Res. 24:999–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba A, et al. 2011. PKA-dependent regulation of the histone lysine demethylase complex PHF2-ARID5B. Nat Cell Biol. 13:668–675. [DOI] [PubMed] [Google Scholar]
- Benton ML, Talipineni SC, Kostka D, Capra JA. 2019. Genome-wide enhancer annotations differ significantly in genomic distribution, evolution, and function. BMC Genomics 20:511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthelot C, Villar D, Horvath JE, Odom DT, Flicek P. 2018. Complexity and conservation of regulatory landscapes underlie evolutionary resilience of mammalian gene expression. Nat Ecol Evol. 2:152–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannavò E, et al. 2016. Shadow enhancers are pervasive features of developmental regulatory networks. Curr Biol. 26:38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, et al. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485:376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dukler N, Gulko B, Huang Y-F, Siepel A. 2016. Is a super-enhancer greater than the sum of its parts? Nat Genet. 49:2–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg E, Levanon EY. 2013. Human housekeeping genes, revisited. Trends Genet. 29:569–574. [DOI] [PubMed] [Google Scholar]
- Ernst J, Kellis M. 2012. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 9:215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita PA, et al. 2011. The UCSC genome browser database: update 2011. Nucleic Acids Res. 39:D876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulco CP, et al. 2019. Activity-By-Contact model of enhancer–promoter regulation from thousands of CRISPR perturbations. Nat Genet. 51:1664–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium, et al. 2017. Genetic effects on gene expression across human tissues. Nature 550:204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay D, et al. 2016. Genetic dissection of the α-globin super-enhancer in vivo. Nat Genet. 48:895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho DE, Imai K, King G, Stuart EA. 2011. MatchIt: nonparametric preprocessing for parametric causal inference. J Stat Softw. 42:1–28. [Google Scholar]
- Hobert O. 2010. Gene regulation: enhancers stepping out of the shadow. Curr Biol. 20:R697–R699. [DOI] [PubMed] [Google Scholar]
- Hong J-W, Hendrix DA, Levine MS. 2008. Shadow enhancers as a source of evolutionary novelty. Science 321:1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, et al. 2018. Dissecting super-enhancer hierarchy based on chromatin interactions. Nat Commun. 9:943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y-F, Gulko B, Siepel A. 2017. Fast, scalable prediction of deleterious noncoding variants from functional and population genomic data. Nat Genet. 49:618–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens V, Goris J. 2001. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem J. 353:417–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski KJ, et al. 2020. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581:434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempfer R, Pombo A. 2019. Methods for mapping 3D chromosome architecture. Nat Rev Genet. 21:207–226. [DOI] [PubMed] [Google Scholar]
- Krijger PHL, de Laat W. 2016. Regulation of disease-associated gene expression in the 3D genome. Nat Rev Mol Cell Biol. 17:771–782. [DOI] [PubMed] [Google Scholar]
- Kundaje A. 2013. A comprehensive collection of signal artifact blacklist regions in the human genome. … Site/Anshulkundaje/Projects/Blacklists (Last Accessed 30 ….ftp://ftp.ebi.ac.uk/pub/databases/ensembl/encode/supplementary/integration_data_jan2011/byFreeze/jan2011/blacklists/hg19-blacklist-README.pdf%5Cnhttps://sites.google.com/site/anshulkundaje/projects/blacklists.
- Lek M, et al. 2016. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536:285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letelier J, et al. 2018. A conserved Shh cis-regulatory module highlights a common developmental origin of unpaired and paired fins. Nat Genet. 50:504–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorthy SD, et al. 2017. Enhancers and super-enhancers have an equivalent regulatory role in embryonic stem cells through regulation of single or multiple genes. Genome Res. 27:246–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostafavi H, Spence JP, Naqvi S, Pritchard JK. 2022. Limited overlap of eQTLs and GWAS hits due to systematic differences in discovery. BioRxiv. doi: 10.1101/2022.05.07.491045. [DOI] [PubMed]
- Osterwalder M, et al. 2018. Enhancer redundancy provides phenotypic robustness in mammalian development. Nature 554:239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravasi T, et al. 2010. An atlas of combinatorial transcriptional regulation in mouse and man. Cell 140:744–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium, et al. 2015. Integrative analysis of 111 reference human epigenomes. Nature 518:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamaki K, et al. 2014. The apoptotic initiator caspase-8: its functional ubiquity and genetic diversity during animal evolution. Mol Biol Evol. 31:3282–3301. [DOI] [PubMed] [Google Scholar]
- Salameh TJ, et al. 2020. A supervised learning framework for chromatin loop detection in genome-wide contact maps. Nat Commun. 11:3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt AD, et al. 2016. A compendium of chromatin contact maps reveals spatially active regions in the human genome. Cell Rep. 17:2042–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholes C, Biette KM, Harden TT, DePace AH. 2019. Signal integration by shadow enhancers and enhancer duplications varies across the Drosophila embryo. Cell Rep. 26:2407–2418.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin HY, et al. 2016. Hierarchy within the mammary STAT5-driven Wap super-enhancer. Nat Genet. 48:904–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, et al. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15:1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigalova OM, Shaeiri A, Forneris M, Furlong EE, Zaugg JB. 2020. Predictive features of gene expression variation reveal mechanistic link with differential expression. Mol Syst Biol. 16:9539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M, et al. 2015. Tissue-based map of the human proteome. Science 347:1260419. [DOI] [PubMed] [Google Scholar]
- Villar D, et al. 2015. Enhancer evolution across 20 mammalian species. Cell 160:554–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Goldstein DB. 2020. Enhancer domains predict gene pathogenicity and inform gene discovery in complex disease. Am J Hum Genet. 106:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitson RH, Li SL, Zhang G, Larson GP, Itakura K. 2021. Mice with Fabp4-Cre ablation of Arid5b are resistant to diet-induced obesity and hepatic steatosis. Mol Cell Endocrinol. 528:111246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will AJ, et al. 2017. Composition and dosage of a multipartite enhancer cluster control developmental expression of Ihh (Indian hedgehog). Nat Genet. 49:1539–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S, Duan J, Li B, Zhou P, Hon GC. 2017. Multiplexed engineering and analysis of combinatorial enhancer activity in single cells. Mol Cell. 66:285–299.e5. [DOI] [PubMed] [Google Scholar]
- Xu H, et al. 2020. ARID5B influences antimetabolite drug sensitivity and prognosis of acute lymphoblastic leukemia. Clin Cancer Res. 26:256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai I, et al. 2005. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics 21:650–659. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The CRE annotations were derived from ChIP-seq peaks from the Roadmap Epigenomics portal (https://egg2.wustl.edu/roadmap/data/byFileType/peaks/unconsolidated/gappedPeak/). The public 15-state ChromHMM annotations for all tissues were also downloaded from Roadmap Epigenomics (https://egg2.wustl.edu/roadmap/data/byFileType/chromhmmSegmentations/ChmmModels/coreMarks/jointModel/final). The Hi-C interaction matrices for all tissues were obtained from GSE87112. The predicted chromatin loops from Peakachu can be downloaded from the 3D Genome Browser (http://3dgenome.org). The CRE-gene links from the Activity-By-Contact model for six cell types (liver, heart, muscle, ovary, spleen, and pancreas) were downloaded from https://www.engreitzlab.org/abc/. Gene expression matrices for all tissues and tissue-specific eQTL were obtained from the GTEx Portal (version 8). We obtained PhastCons conserved elements from http://hgdownload.soe.ucsc.edu/goldenPath/hg19/phastCons46way/. LINSIGHT scores were downloaded from https://github.com/CshlSiepelLab/LINSIGHT. The analysis scripts and CRE landscape annotations used in this study are available on GitHub: https://github.com/bentonml/cre-landscape.