Abstract
Ribosomal RNA (rRNA) maturation in archaea is a complex multistep process that requires well-defined endo- and exoribonuclease activities to generate fully mature linear rRNAs. However, technical challenges prevented detailed mapping of rRNA processing steps and a systematic analysis of rRNA maturation pathways across the tree of life. In this study, we used long-read (PCR)-cDNA and direct RNA nanopore-based sequencing to study rRNA maturation in three archaeal model organisms, namely the Euryarchaea Haloferax volcanii and Pyrococcus furiosus and the Crenarchaeon Sulfolobus acidocaldarius. Compared to standard short-read protocols, nanopore sequencing facilitates simultaneous readout of 5′- and 3′-positions, which is required for the classification of rRNA processing intermediates. More specifically, we (i) accurately detect and describe rRNA maturation stages by analysis of terminal read positions of cDNA reads and thereupon (ii) explore the stage-dependent installation of the KsgA-mediated dimethylations in H. volcanii using base-calling and signal characteristics of direct RNA reads. Due to the single-molecule sequencing capacity of nanopore sequencing, we could detect hitherto unknown intermediates with high confidence, revealing details about the maturation of archaea-specific circular rRNA intermediates. Taken together, our study delineates common principles and unique features of rRNA processing in euryarchaeal and crenarchaeal representatives, thereby significantly expanding our understanding of rRNA maturation pathways in archaea.
Keywords: Nanopore sequencing, ribosomal RNA, RNA modifications, RNA processing; archaea
INTRODUCTION
Ribosomes are universally conserved molecular machineries ensuring the efficient and accurate translation of mRNAs into proteins. Accordingly, the assembly of ribosomes is a key regulated housekeeping task enabling the synthesis of functional ribosomal subunits. Ribosome biogenesis, the process by which ribosomal subunits are assembled and matured, is a highly complex process requiring the coordinated action of several ribosome biogenesis factors, the RNA modification machinery, and ribonuclease activities responsible for the processing of the ribosomal RNA precursors (pre-rRNAs).
In contrast to bacteria and eukaryotes, little is known about the ribosomal RNA (rRNA) processing pathway in archaea (Jacob et al. 2013; Yip et al. 2013; Henras et al. 2015; Ferreira-Cerca 2017; Londei and Ferreira-Cerca 2021). Our current knowledge suggests that the primary polycistronic rRNA precursor contains two processing stems formed by the 5′ leader and 3′ trailer sequences surrounding the 16S and 23S rRNAs (Fig. 1A; Tang et al. 2002; Yip et al. 2013, p. 2013; Ferreira-Cerca 2017; Clouet-D'Orval et al. 2018; Londei and Ferreira-Cerca 2021). In Euryarchaeota, the 16S and 23S rRNAs are additionally separated by the presence of an internal tRNA. In most archaea, the 16S and 23S rRNA processing stems contain a bulge-helix-bulge (bhb) motif, which is, in the context of intron-containing tRNAs, recognized by the splicing endonuclease endA (Russell et al. 1999; Tang et al. 2002; Clouet-D'Orval et al. 2018; Schwarz et al. 2020). Similar to intron-containing-tRNA maturation, processing at the bhb motifs is followed by the covalent ligation of the resulting extremities, thereby generating the archaeal-specific circular-pre-16S and circular-pre-23S rRNAs (Tang et al. 2002; Danan et al. 2012; Ferreira-Cerca 2017; Jüttner et al. 2020; Londei and Ferreira-Cerca 2021). The exact molecular mechanisms by which the circular-pre-rRNA intermediates are further processed into linear mature rRNAs remain to be fully characterized (Tang et al. 2002; Ferreira-Cerca 2017; Londei and Ferreira-Cerca 2021).
FIGURE 1.
Exploring rRNA processing and modifications using nanopore-based RNA-seq. (A) Simplified pictogram of rRNA maturation in archaea: The primary transcript containing the 16S (dark gray), internal tRNA (brown; only in Euryarchaeota), and 23S (light gray) is folded at the bulge-helix-bulge (BHB), cleaved at multiple positions (orange triangles), circularized, and bases are modified (pink circles) until the mature rRNA products are formed that ultimately are an integral part of the ribosomal subunits (Tang et al. 2002; Ferreira-Cerca 2017; Londei and Ferreira-Cerca 2021). (B) Nanopore-based library preparation methods for direct sequencing of cDNAs (left, 1) and RNAs (right, 2): For cDNA sequencing, 3′-adapter-ligated RNAs and custom VN-primers were used as input for the DCS109 protocol established by Oxford Nanopore. For RNA sequencing, libraries were prepared from artificially polyadenylated RNAs according to the RNA002 protocol established by Oxford Nanopore. Adapters that carry the motor protein are highlighted with a red pacman symbol. (C) Exemplary direct cDNA coverage profile and (D) single-read plot of the rRNA locus 1 of H. volcanii. Cleavage sites at the bulge-helix-bulge are indicated by orange triangles and dashed vertical lines. (E) RNA base modifications can lead to deviations in the recorded current intensities from the expected signal and/or to systematic base-calling errors, which can be used to detect RNA modifications (Begik et al. 2022).
Deciphering the rRNA processing pathway biochemically can be a time-consuming and tedious task (Deutscher 2015; Henras et al. 2015). Whereas next-generation sequencing has revolutionized sequencing-based approaches in RNA biology, understanding rRNA processing requires the analysis of 5′/3′ ends co-occurrences, a feature that remains challenging to be determined by classical short-read sequencing technologies. Compared to standard short-read protocols, long-read sequencing facilitates simultaneous readout of 5′- and 3′-positions, which is required for pre-rRNA stage classification of individual reads (Garalde et al. 2018; Grünberger et al. 2022). Additionally, single-molecule sequencing is a powerful tool enabling the detection of short-lived intermediates with high confidence. Accordingly, long-read sequencing strategies may facilitate obtaining rapid insights into pre-rRNA processing pathways in poorly studied organisms.
In addition to rRNA processing events, rRNA modifications are known to be timely added at various steps of the rRNA maturation pathway. Similar to processing events, ordering the chemical addition during the maturation is a challenging task and remains mostly unexplored in archaea. Among the known added modifications, the KsgA/Dim1-dependent dimethylation at two universally conserved adenosines at the 3′ end of the SSU are the sole almost universally conserved modifications (type and position) known across the tree of life (Helser et al. 1971; Lafontaine et al. 1994; Connolly et al. 2008). Previous work suggests the existence of different addition timelines across various organisms. In fact, these modifications are added at the late step of rRNA maturation in Escherichia coli and Saccharomyces cerevisiae but at an earlier step of human rRNA maturation (Lafontaine et al. 1998; Mangat and Brown 2008; Xu et al. 2008; Ghalei et al. 2017).
Here, we explored the combination of cDNA and native RNA sequencing to study archaeal rRNA maturation using the long-read sequencing technology offered by Oxford Nanopore Technologies (ONT). We tested the applicability of the ONT-based (PCR)-cDNA and RNA sequencing (i) to analyze pre-ribosomal RNA processing pathways and (ii) to provide information on the timing of base modifications during rRNA maturation. We provide evidence that the long cDNA and RNA reads gathered on the ONT platform allow us to gain new insights into the poorly understood ribosomal RNA (rRNA) maturation pathway in various archaea. Moreover, we provide data that unravel the timely ordering of the KsgA-dependent dimethylations on pre-rRNAs in Haloferax volcanii.
RESULTS
Characterizing archaeal rRNA processing using long-read nanopore sequencing
To reveal characteristics of the rRNA processing pathway(s) in archaea, we applied long-read nanopore sequencing to the total RNA of three archaeal model organisms, namely the Euryarchaea H. volcanii and Pyrococcus furiosus and the Crenarchaeon Sulfolobus acidocaldarius (Fig. 1A). For maturation stage classification, amplification-free direct cDNA sequencing was performed in a poly(A)-independent way using a previously published 3′-adapter protocol to improve the accuracy of 3′ end detection (Fig. 1B–D), while direct RNA sequencing was used for stage-dependent base modification analysis (Fig. 1B,E; Mo et al. 2021; Begik et al. 2022; Grünberger et al. 2022). Note that we previously showed that artificial polyadenylation of prokaryotic RNAs impairs the accuracy of 3′ detection caused by the lower sequencing accuracy of homopolymer regions (Grünberger et al. 2022). Since RNA degradation influences read classification, samples for sequencing were selected based on the presence of rRNA peaks in the Bioanalyzer profiles after RNA pretreatment and adapter ligation (Supplemental Fig. S1A). During library preparation, RNAs were reverse transcribed using a custom primer. Successful reverse transcription (RT) was validated by size estimation of the cDNA product (Supplemental Fig. S1B). Based on the length profiles, we conclude that some RNA species are prone to RT bias rendering absolute quantification impossible. This bias also applies to an in vitro transcribed 16S rRNA, which was used as a control and suggests that problems might occur particularly during RT of 16S rRNAs.
In total, about 90,000–270,000 reads were sequenced per total direct cDNA library with a median q-score of 9.9 and a median read length of 1.2 kb (Supplemental Fig. S2A–C). The length distribution of the sequenced reads corresponds to the expected sizes and enables the analysis of processing events at both extremities of the rRNA intermediates (Supplemental Fig. S2D).
After initial quality control, filtered reads (94.7%, q-score filter: 7) were mapped to the reference genomes. About 99.5% of the reads were successfully aligned with a median read length of about 200 bases shorter than the adapter-containing raw sequence length and a median identity of 92.9%. In total, 71.7% of the reads mapped to rRNAs of the three archaeal species. Coverage profiles of the H. volcanii samples indicated that about twice the number of reads mapped to the 23S rRNA sequence compared to the 16S rRNA although stoichiometric amounts are expected to be incorporated into the ribosome. This unequal ratio was also observed or even more prominent in the other organisms (Supplemental Fig. S3A). After analyzing the distribution of aligned reads and mapped identities, we conclude that many rRNA-originating reads are potentially full-length reads, which are not influenced in their quality by length (Supplemental Fig. S3B,C).
To prevent misidentification of terminal positions by random alignment of adapter sequences, full-length sequenced reads containing SSP and VNP adapters in the correct orientation were strand-oriented, and adapter-trimmed using pychopper. In total, 49% of quality filtered reads contained the adapters in the correct orientation and were selected for subsequent analysis. Comparing the read coverage of full-length sequenced to unfiltered reads, we conclude that the most prominent coverage spikes in the unfiltered data set are not genuine read ends and most likely caused by strand-switching problems (Supplemental Fig. S4). Importantly, trimming allows using the soft-clipping parameter extracted from the CIGAR string as a meaningful parameter during the detection of pre-rRNAs characterized by spliced extremities, which will be described in the following paragraph (Li et al. 2009).
Insights into rRNA maturation in H. volcanii
After stringent filtering and quality control, we aimed to confirm and expand our knowledge on the poorly characterized multistep ribosomal maturation process in archaea by performing enrichment analysis of terminal positions. The following analysis is illustrated using Haloferax as an example before applying the strategy to Pyrococcus and the Crenarchaeon Sulfolobus.
During the analysis, we noticed that some of the observed mature rRNA terminal positions did not match the available annotations at the NCBI databank (https://www.ncbi.nlm.nih.gov/genome/; Supplemental Fig. S5A–C). However, selected examination of the putative mature rRNA extremities obtained by direct cDNA sequencing did match our independent experimental validations by primer extension analysis of the 5′ ends of the 16S and 23S rRNAs of H. volcanii (Supplemental Fig. S5D,E). These results suggest that positions based on our sequencing results most likely represent the genuine mature rRNA extremities. Therefore, the corrected annotation was subsequently used throughout the analysis (Supplemental Table S1; Supplemental Fig. S5).
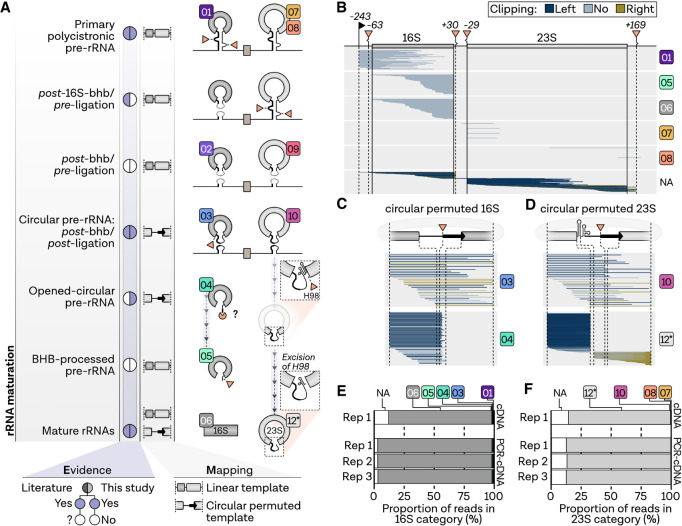
To retrace the multistep rRNA maturation process, we performed a co-occurrence analysis of read start and read end positions. Ribosomal RNA maturation in Haloferax starts with the transcription of a primary transcript from three promoters, which are also visible in a two-dimensional plot of read start and end positions (Fig. 2A,B; Laass et al. 2019). In accordance with the accumulated coverage data (Supplemental Fig. S4), the most significant population of reads mapping to the 23S rRNA represents full-length 23S rRNA reads. In contrast, many of the 16S reads end at the mature 3′ end but have 5′ starts that are shorter than the annotated mature 5′ position. Nevertheless, the 5′ terminus of the mature 16S was significantly enriched and could be detected with high accuracy (Fig. 2C). Processing at the predicted bhb positions, also visible in the scatter plot (Fig. 2B), was analyzed in more detail and is detected with single-nucleotide accuracy (Fig. 2C,D). However, some of the reads starting or ending at the bhb have long soft-clippings originating from bases that are part of the sequenced read but not the alignment. Since the adaptors are trimmed off during pychopper filtering, these reads may correspond to the known circular precursors with covalently connected 5′ and 3′ bhb cleavage sites (Tang et al. 2002; Jüttner et al. 2020). This is also supported by the fact that slightly divergent bhb positions can be explained by sequence similarities with the circularized rRNA. To investigate this in more detail, nanopore reads were remapped to a permuted RNA sequence designed by joining the 3′ bulge with the 5′ bulge of the 16S and 23S rRNA, respectively, to mimic the actual permutated sequence expected for circular-pre-rRNAs (Fig. 2E).
FIGURE 2.
Strategy for the detection of pre-rRNA intermediates shown exemplarily for H. volcanii. (A) 5′ and 3′ terminal positions of the 16S and 23S rRNAs based on previous publications (TSS; Laass et al. 2019), secondary structure prediction, previous analysis (bulge-helix-bulge; Lorenz et al. 2011; Jüttner et al. 2020), and corrected positions of mature rRNAs (compare Supplemental Fig. S5). (B) Co-occurrence analysis of full-length cDNA read start (x-axis) and read end positions (y-axis) mapping to the rRNA locus 1 in H. volcanii (Hvo_3038-3041). Each read is shown as a single point, colored by 2D density. Terminal positions are highlighted by black (transcription start sites) and orange (bulge-helix-bulge sites) triangles, circles (mature 5′ ends) and rectangles (mature 3′ ends). (C) Number of detected 5′ and (D) 3′ ends with respect to the expected terminal positions. Negative distance indicates upstream-mapping of 5′ or 3′ ends. Bars are colored according to the soft-clipping profile in light blue (≤20 nt), dark blue (>20 nt, left) and olive (>20 nt, right). (E) Strategy for the detection of rRNA intermediates: After the detection and co-occurrence analysis of enriched terminal positions (1), reads are analyzed with respect to ends mapping to bulge-helix-bulge ends (2). Potential circular reads are detected by remapping reads to permuted linear rRNA sequences, designed by joining the bulge ends of the 16S and 23S rRNA.
For both, the mapping data based on the genomic sequence and the linear permuted sequence, pre-rRNA-related intermediates were selected based on read coordinates, abundance, biological interpretability, and prior characterization, when available. Finally, the overall findings were used to extract an rRNA maturation pathway model summarized in Figure 3A (Ferreira-Cerca 2017; Jüttner et al. 2020; Londei and Ferreira-Cerca 2021).
FIGURE 3.
rRNA processing in H. volcanii. (A) Proposed rRNA maturation pathway in H. volcanii. (B) Single-track analysis of reads from a representative direct cDNA sample (replicate 1) mapping to genomic linear template, (C) the circular permuted 16S sequence, and (D) the circular permuted 23S sequence grouped by rRNA stage. Each read is shown as a single line sorted according to the read start position, while the relative number is shown on the y-axis. Lines are colored according to the soft-clipping profile in light blue (≤20 nt), dark blue (>20 nt, left), and olive (>20 nt, right). (E) The proportion of reads of the different rRNA processing states in the 16S groups and the (F) 23S read groups are shown as stacked bar charts.
Despite the high sequencing depth, we did not detect a full-length precursor consisting of the 16S leading-16S-tRNA-23S-23S trailing sequences in H. volcanii. Accordingly, the primary pre-rRNA was detected as 3′- (classes 1 and 7) or 5′-processed (class 8) reads (Fig. 3B). Previously, functional cis-acting element analysis suggested that the 16S rRNA bhb-processing occurs prior to internal tRNA and 23S rRNA maturation, leading to a precursor containing the 5′ leader/3′ trailer ligation of the processed 16S pre-rRNA flanking regions linked to the nascent 23S pre-rRNA intermediate (Jüttner et al. 2020). However, we did not observe this particular precursor in the direct cDNA data sets of H. volcanii. Nevertheless, based on the end profiles, we identified four other classes of putative intermediates. In the first class (classes 2 and 9), the pre-rRNA boundaries of these intermediates match the previously described bhb-processing sites located within the 16S and 23S rRNA processing stems, respectively (Jüttner et al. 2020). These intermediates extend from the 5′ to 3′ bulge cleavage sites; however, these extremities are not covalently ligated and may correspond to post-bhb cleavage/pre-ligation pre-rRNA intermediates. An exemplary verification of the 5′ boundary of the post-bhb cleavage/pre-ligation pre-23S rRNA analyzed by primer extension is provided in Supplemental Figure S5E. The second class (classes 3 and 10) correspond to permuted reads covalently connecting the 5′ and 3′ bulge cleavage sites, and are likely observed as the result of random nicking of the circular-pre-rRNA intermediates during sample preparations and are categorized as post-bhb/post-ligation pre-rRNA intermediates (Fig. 3C,D). These intermediates were verified by remapping nanopore reads to a permuted sequence designed by joining the 3′ and 5′ processing site of the bulge. Similarly, we detected a third main class (class 4), which corresponds to a putative pre-16S rRNA intermediate showing an immature 3′ end, which is extended by the typical permuted spacers sequence previously observed in the circular-pre-16S rRNA. This topology possibly results from linearization of the circular-pre-16S rRNA intermediate at the mature 16S rRNA 5′ end (opened-circular-pre-16S rRNA). This putative pre-rRNA intermediate is relatively abundant in H. volcanii (6% of all reads mapping in the 16S rRNA region, Fig. 3E), and strikingly shows a nonrandom 3′ end extremity, matching with the linearization of circ-pre-rRNA at the mature 16S rRNA 5′ end (Fig. 3C). Another previously undescribed intermediate, which is even more abundant in the 16S categories (class 5, 11%), and also present in the 23S rRNA maturation pathway (class 11, 0.7%), is characterized by randomly fragmented 5′ ends and a precise 3′ end at the bhb cleavage site (Fig. 3B). However, a temporal classification of this bhb-processed pre-rRNA intermediate is not possible based on the terminal positions. Both an alternative pathway without circularization (after classes 2 and 9) and processing after opening at the 5′ end (class 4) could account for the formation of this pre-rRNA intermediate (Fig. 3B). Compared to the other stages, all reads classified as mature 16S or 23S rRNA products are expectedly the most abundant (Fig. 3E,F). To confirm the presence of classes 2, 4, and 5 pre-rRNA intermediates in H. volcanii, we performed northern blot analysis after RNase H-dependent cleavage to deconvolute and discriminate between these pre-rRNA intermediates (Supplemental Fig. S6). As shown in Supplemental Figure S6, the presence of the opened-circular-pre-rRNA and classes 2 or 5 are readily observed in H. volcanii.
To test the robustness and reproducibility of the analysis and to validate the presence of low-abundance intermediates, we additionally performed nanopore sequencing of PCR-amplified cDNA libraries from three biological replicates. Although PCR-amplification led to the overamplification of short fragments that did not align to the reference with the selected mapping parameters, the length distribution of the sequenced reads matched our expectations from direct cDNA sequencing (Supplemental Fig. S7A). The coverage profiles of the 16S and 23S rRNA were also in agreement with the results obtained from direct cDNA sequencing, with PCR-amplification leading to a higher coverage of the respective ends, which we already reported in a previous study (Supplemental Fig. S7B; Grünberger et al. 2022). One key advantage is that the PCR-cDNA protocol enriches for full-length reads (92.2% over all samples), which increased the number of primary alignments (Supplemental Table S2). Note that due to this advantage and due to deeper sequencing, about nine times as many reads could be used for rRNA stage analysis in Haloferax (Supplemental Fig. S7C). Amplification of cDNAs did not affect the accuracy of terminal position detection, which was highly reproducible between samples and sequencing methods, also concerning the clipping profiles (Supplemental Fig. S8). Using this approach, the relative ratios from the direct cDNA experiment were confirmed, with significantly more reads being assigned to the individual categories (Fig. 3E,F; absolute number of reads per class shown in Supplemental Fig. S9).
Insights into rRNA maturation in P. furiosus
Similarly, we determined the rRNA maturation stages in P. furiosus and validated the results using deep PCR-cDNA sequencing (Fig. 4; Supplemental Fig. S10). In total, there are three deviations from the processing pathway observed in Haloferax:
FIGURE 4.
rRNA processing in P. furiosus. (A) Proposed rRNA maturation pathway in P. furiosus. (B) Single-track analysis of reads from a representative direct cDNA sample (replicate 1) mapping to genomic linear template, (C) the circular permuted 16S sequence, and (D) the circular permuted 23S sequence grouped by rRNA stage. Each read is shown as a single line sorted according to the read start position, while the relative number is shown on the y-axis. Lines are colored according to the soft-clipping profile in light blue (≤20 nt), dark blue (>20 nt, left), and olive (>20 nt, right). (E) The proportion of reads in the 16S groups and the (F) 23S read groups are shown as stacked bar charts.
First, opened-circular 16S pre-rRNAs (class 4) do not have a distinct 3′ end at the mature start position. This also applies to class 5, characterized by randomly degraded 3′ ends instead of a clear cut at the bhb-processing site. The third variation has been described recently and concerns the mature 23S rRNA (Birkedal et al. 2020). The special feature observed here is that the 23S rRNA is circularly permuted, which is a consequence of the excision of helix 98 after circularization (Fig. 4A). Thus, in this case, not only the pre-rRNAs (classes 3 and 10) but also the matured 23S rRNAs were detected by remapping to permuted sequences (Fig. 4C,D). Similar to the situation in Haloferax, the circular reads correspond to the covalent ligation of the 5′ and 3′ spacers generated by cleavage at the bulge-helix-bulge motifs within the processing stems (Russell et al. 1999; Ferreira-Cerca 2017; Jüttner et al. 2020). To verify this hypothesis, we performed RNA structure prediction of the corresponding double-stranded RNA regions (Supplemental Fig. S11). In agreement with the permuted reads, we could place the corresponding extremities within the bulge-helix-bulge motifs. However, the 23S processing stem does not adopt a canonical bhb motif, but forms an alternative structure similar to the one previously described for the 16S rRNA bhb motif in S. acidocaldarius (Russell et al. 1999; Tang et al. 2002; Qi et al. 2020). We and others could previously demonstrate that this alternative structure is compatible with circular-pre-16S rRNA formation in S. acidocaldarius (Danan et al. 2012; Jüttner et al. 2020). Therefore, these permuted reads likely originate from the random opening of the archaeal-specific circular-pre-rRNA intermediates during sample preparation/sequencing (as observed for H. volcanii) and suggest that circular-pre-rRNA intermediates are also produced in P. furiosus—a prevalent feature in rRNA processing found in many archaea. In fact, circularization is a prerequisite for the subsequent maturation of 23S rRNA by excision of helix 98.
The PCR-cDNA results corresponded to the size distribution and coverage of the direct cDNA results but with a 23-fold higher number of primary mapped reads, which was used to confirm the presence of low-abundant classes 7 and 8 (Supplemental Fig. S10). However, neither PCR nor sequencing depth did affect the accuracy of terminal position detection, where extensive clipping in the 23S positions was caused by the circular permutation (Supplemental Fig. S12). Although our setup does not allow absolute quantification, we find that the relative abundance of 16S and 23S rRNA precursors is reproducible in all samples and lower compared to Haloferax (Fig. 4E,F; Supplemental Fig. S13).
Insights into rRNA maturation in S. acidocaldarius
Next, S. acidocaldarius was analyzed as a representative member of the Crenarchaeota. Regarding rRNA maturation, several features are homologous to the eukaryotic process. One of these features, termed site 0, is located about 70 nt ahead of the 16S mature 5′ end. This feature was suggested to have similarities to the site A0 present in eukaryotic pre-rRNAs (Fig. 5A; Durovic and Dennis 1994). Analyzing the terminal positions in S. acidocaldarius, we did not detect site 0 to be enriched in the cDNA or the PCR-cDNA data sets, where more than 400,000 primary mapped reads could be detected (Fig. 5B; Supplemental Fig. S14). However, this could be because it is most likely one of the earliest cleavage events resulting in a short-lived intermediate that is consequently underrepresented in the samples. Notably, we observed putative deviations in some of the bhb-processing sites (Fig. 5B,C; Supplemental Fig. S15). However, according to secondary structure analysis, this can be explained by sequence similarities after circularization (Fig. 5D,E).
FIGURE 5.
rRNA processing in S. acidocaldarius. (A) 5′ and 3′ ends of mature rRNA in S. acidocaldarius according to Durovic and Dennis (1994). Enriched ends found in this study are highlighted with pink circles. (B) Number of detected 5′ and (C) 3′ ends from a representative direct cDNA sample (replicate 2) with respect to the expected positions. Artificial ends of circular reads are highlighted by dashed circles. Negative distance indicates upstream-mapping of 5′ or 3′ ends. Bars are colored according to the soft-clipping profile in light blue (≤20 nt), dark blue (>20 nt, left), and olive (>20 nt, right). (D) Secondary structure of the bhb sites of 16S and (E) 23S rRNA processing stems. Structures are shown before and after circularization. (F) Proposed rRNA maturation pathway in S. acidocaldarius. (G) Single-track analysis of reads (direct cDNA replicate 2) mapping to genomic linear template, (H) the circular permuted 16S sequence, (I) the circular permuted 23S sequence, and (J) a linear template with excised mature 16S sequence grouped by rRNA stage. Each read is shown as a single line sorted according to the read start position, while the relative number is shown on the y-axis. Lines are colored according to the soft-clipping profile in light blue (≤20 nt), dark blue (>20 nt, left), and olive (>20 nt, right). (K) The proportion of reads in the 16S of the different rRNA processing states and the (L) 23S of the different rRNA processing states are shown as stacked bar charts.
We furthermore considered terminal positions described in Durovic and Dennis (1994) for our rRNA stage classification (Fig. 5F). In addition to the truncated variants of primary RNA (classes 1, 7, and 8), we were also able to detect another intermediate that was already depicted in previous pathways but has not been discussed before (Fig. 5G,J). This intermediate is a result of 16S processing prior to 23S processing, which leads to a post-16S-bhb RNA chimera (class 7*) characterized by a sequence with joined processed 16S leading and trailing sequences linked to the pre-23S rRNA. This state was confirmed by remapping of reads to a 16S excised sequence and is in good agreement with a pre-rRNA intermediate recently described based on cis-acting element analysis in H. volcanii (Jüttner et al. 2020).
As observed for Haloferax and Pyrococcus, circularized forms of the 16S and 23S precursors could also be detected. However, we can find no circumstantial evidence to support the recent suggestion that the mature 23S rRNA is circular in Sulfolobus (Birkedal et al. 2020).
Regarding the other precursors, there are a few additional points worth mentioning:
Similar to Pyrococcus, but different from Haloferax, the 3′ ends of the opened (class 4) and bhb-processed precursors (class 5) do not seem to be the product of a distinct endonucleolytic cleavage event. In contrast, we detected an additional cleavage event in the final steps of 23S rRNA maturation, which prominently accumulates in our data and was also described in other studies (class 11*; Fig. 5A,F,G,K; Durovic and Dennis 1994). In this case, processing occurs at position +10 in the 3′ region of the 23S rRNA, whereas the 5′ ends appear to be exonucleolytically processed as observed for the final rRNAs. In relative quantitative terms, this is also the only precursor found in substantial amounts in any of the samples (Fig. 5K,L; Supplemental Fig. S16).
Taken together, our analysis confirms and expands the number of putative pre-rRNA intermediates in archaea. Moreover, this comprehensive framework provides an additional basis to facilitate further definition of common and specific principles of rRNA maturation in archaea.
Toward stage-dependent analysis of rRNA modifications
In contrast to other sequencing techniques, nanopore-based sequencing offers the possibility to detect base modifications directly as these modifications lead to an electric current signal that differs from the expected theoretical distribution obtained by the unmodified nucleotide sequence (Liu et al. 2019; Smith et al. 2019; Viehweger et al. 2019; Begik et al. 2021, 2022; Jenjaroenpun et al. 2021). In turn, these signal changes might also lead to differences in the base-calling profiles (e.g., systematic errors or a drop in base-calling quality; Begik et al. 2022).
Accordingly, after determining rRNA processing stages via direct cDNA analysis, we aimed to explore the stage-dependent installation of the KsgA-dependent dimethylation of adenosine bases (m62A) at position A1450/A1451 (A1518/A1519 E. coli numbering) in H. volcanii using direct RNA sequencing (Fig. 6). To this end, we analyzed wild-type samples and a deletion mutant of an archaeal KsgA/Dim1 homolog comparing base-calling, as well as signal features in the context of the m62A modification (O'Farrell et al. 2006; Xu et al. 2008; Knüppel et al. 2021). Looking at the entire 16S region, helix 45, which contains the two predicted dimethylated bases, was the region for which the most substantial increase in base-calling errors was detected (Fig. 6A–C). However, single-read analysis showed that the modifications in the wild-type do not result in a uniform change in the profile, suggesting a certain heterogeneity in this rRNA base modification population (Fig. 6B). This is in line with a previous observation: Primer extension data suggested that the KsgA/Dim1-dependent modification is heterogenous in H. volcanii (Knüppel et al. 2021). In addition to describing the percent error of specific bases (ESB), which is used by the algorithm implemented in Eligos2 to detect base modifications based on the sum of alignment errors of direct RNA-seq data, two signal-specific features were also compared, namely the mean dwell values and the mean signal time (Fig. 6D,E; Nookaew et al. 2020; Jenjaroenpun et al. 2021). In our analysis, we observed differences in the signal profiles between the deletion strain and the wild-type, particularly surrounding the m62A modification. Specifically, the mean dwell values showed noticeable changes five bases upstream of the first modified adenosine, while alterations in the ESB were detected three bases downstream and 10 bases upstream of the modification position (Fig. 6F).
FIGURE 6.
Direct RNA-based modification analysis of KsgA-dependent dimethylations in H. volcanii. (A) Heatmap of single-read (y-axis) full-length wild-type (green) and ΔksgA (purple) direct RNA reads combined from two replicates each colored according to their mapping profile in blue (match), green (insertion), yellow (deletion), and pink (mismatch). (B) Hierarchical clustering of combined reads mapping to helix 45 containing the KsgA-dependent dimethylation sites highlighted with pink circles. (C) The ESB is calculated by the sum of all errors (insertion, deletion, and mismatch) per position. The line plot shows the mean ESB values of all reads from the wild-type samples divided by the mean ESB of the ΔksgA samples. (D) Base-specific mean values (two replicates each) of Tombo-calculated dwell time, (E) signal time, and (F) the ESB are shown for the KsgA-dimethylations sequence context. Samples are colored in green (wild-type) and purple (ΔksgA), while the area between the lines is highlighted in gray. (G) Comparison of wild-type and ΔksgA mean ESB values for 16S rRNA (top) and 23S rRNA (bottom) positions. Each point shows a single residue and is color-coded in pink (expected KsgA-dimethylation sequence context) and gray (all other bases). (H) Mean odds ratios calculated by Eligos2 based on the comparison of wild-type and ΔksgA reads grouped according to the categories used during the rRNA processing analysis. Eligos2 uses Fisher's exact test to evaluate the distinction of error for pairwise comparisons. Positions are color-coded in pink (significant: multiple testing corrected [Benjamini–Hochberg] adjusted P-value < 0.01 in all four test/control pairwise comparisons) and gray (not significant). No comparison possible for class 1 caused by the absence of control reads. (I) Mean ESB calculated for wild-type and ΔksgA reads grouped according to the categories used during the rRNA processing state analysis. (J) Proposed model for the stage-dependent installation of the KsgA-dependent dimethylations in H. volcanii.
The robustness of m62A detection through comparative ESB analysis was further supported by the lack of significant ESB deviations in the 23S rRNA (Fig. 6G). Hence, Eligos2 and ESB description were used to explore the potential to detect the introduction of the KsgA-dependent base modifications at different processing stages of rRNA maturation in H. volcanii. Therefore, reads were first sorted according to the main classes of mature and pre-rRNA intermediates described above for H. volcanii. Relative to the wild-type, we detected no significant alterations in the relative quantification of 16S or 23S categories for the KsgA-deletion mutant (Supplemental Fig. S17). Contrary to (PCR)-cDNA sequencing, we found a greater proportion of reads that could not be classified into specific categories and a reduced number of reads in classes 1 and 2, which necessitate the assignment of 5′ positions. This observation can be attributed to the lower sequencing accuracy and the sequencing direction used by the RNA chemistry, which proceeds from the 3′ to 5′ end, causing the loss of reliable 5′ information.
Using Eligos2, we identified the stage-dependent installation of RNA modification with single-base resolution (Fig. 6H; Nookaew et al. 2020; Jenjaroenpun et al. 2021). It is striking that by far the largest value of the odds ratio, which represents the level of error of native sequence over the unmodified RNA, could be assigned to position A1450. In both the mature 16S (class 6) and the opened-circular 16S pre-rRNAs (class 4), one of the highest values is attributed to the second methylation site, position A1451. Although an Eligos2-based comparison could not be performed for the early pre-rRNA (class 1) due to low counts of this precursor in the KsgA-deletion samples, no significant deviations were detected in classes 2 and 3, indicating that the dimethylation modification is not yet installed.
The earliest time point for the KsgA-dependent modifications is thus after opening of the circularized pre-16S (class 4), which was further validated by the ESB analysis (Fig. 6I). Although significant differences were detected for class 5, the lower odds ratio and ESB values could indicate the relative timely assignment of class 5 and suggest an independent pathway from class 2 reads that are processed at the 5′ end without circularization.
Interestingly, deviations in the error profiles were not only observed in positions surrounding the KsgA-dependent modification, but also in a second region, with the most significant difference at position 1350 (Fig. 6G). We performed a stage-dependent comparison for the sequence contexts of the three known additional RNA modifications in the 16S rRNA of H. volcanii that are all KsgA-independent (Grosjean et al. 2008): acp3U (3-amino-3-carboxypropyl) at position 910 (Ec numbering 966), a modified cytidine (C*, unknown modification) at position 1352 (Ec 1404), and m6A at position 1432 (Ec 1500; Supplemental Fig. S18). While no significant deviations between samples were observed for the acp3U context, the percentage of ESB was notably high at the modification site, which is devoid of homopolymer stretches that might otherwise contribute to lower sequence quality. Interestingly, the error rates were considerably lower in the early precursor (class 1) and gradually increased, implying a progressive increase in acp3U modification over time (Supplemental Fig. S18A). As observed in the analysis of total reads, mature reads (class 6) from the wild-type displayed a significantly higher ESB compared to the KsgA-deletion at 17 bases surrounding the unknown C* modification at position 1352, with the highest odds ratio at position 1350 (Supplemental Fig. S18B). In contrast, no significant differences were found in the earlier classes. The m6A modification at position 1432 is in close proximity to the KsgA-dependent site at positions 1450/1451, and the highest odds ratios in this context refer to the upstream region of the dimethylation site at positions 1450/1451, which are significantly different in classes 4 and 6 (Supplemental Fig. S18C).
Taken together, our analysis suggests that despite the current limitations, ONT allows us to discriminate rRNA modifications across rRNA maturation events. Using Eligos2, we effectively detected the stage-dependent installation of KsgA-dependent dimethylation, which we included in our model of 16S rRNA maturation in H. volcanii (Fig. 6J).
DISCUSSION
Insights into rRNA processing in archaea
Ribosomal RNA processing proceeds via the coordinated and defined order of ribonucleases action (exonucleolytic and/or endonucleolytic cleavages), which generate pre-rRNA intermediates with defined premature rRNA sequences (Venema and Tollervey 1995; Deutscher 2015; Henras et al. 2015; Bechhofer and Deutscher 2019). The establishment of faithful rRNA maturation maps in model organisms, like E. coli, S. cerevisiae or human cells, has required numerous analyses over the past decades (Venema and Tollervey 1995; Deutscher 2015; Henras et al. 2015; Bechhofer and Deutscher 2019), and remains a technical challenge. Therefore, methodologies that might accelerate the systematic analysis of rRNA maturation pathways across the tree of life need to be established to ultimately unravel the diversity of rRNA processing strategies in all domains of life. Beyond the identification of processing sites, the order of the processing events, which can be, in part, deduced from co-occurrence analysis of the 5′ and 3′ extremities is of biological relevance.
In this study, we could confirm and expand the presence of pre-rRNA intermediates and processing sites in the different archaeal organisms analyzed, including the archaeal-specific circular-pre-rRNA intermediates (Tang et al. 2002; Ferreira-Cerca 2017; Clouet-D'Orval et al. 2018; Jüttner et al. 2020; Londei and Ferreira-Cerca 2021). Together, our findings are summarized into an updated archaeal rRNA processing model, which is discussed below.
The full-length primary rRNA transcript was not identified in any of the archaeal organisms analyzed. Similarly, this primary rRNA is generally difficult to observe in wild-type E. coli (Nikolaev et al. 1973; Hofmann and Miller 1977). Collectively, these observations suggest that short-lived and/or low-abundant pre-rRNA intermediates escape the detection capacity of the current experimental strategies. Accordingly, it is also difficult to infer differences in rRNA processing features between different (archaeal) organisms by virtue of observed pre-rRNA intermediates absence/presence pattern. In fact, these differences may also be related to organism-specific changes in pre-rRNA intermediates relative levels, which will depend on the sum of the reaction kinetics of the different maturation steps in a given condition. Despite these remaining limitations, it is striking that some general blueprint of the rRNA maturation pathway is conserved across evolutionary distant archaea; however, variations can be readily observed (see P. furiosus; Qi et al. 2020).
Among the identified pre-rRNA intermediates, one precursor (7*), which is observed in S. acidocaldarius and includes ligation at the bhb motif of the upstream region of the 16S leader and downstream region of the 16S trailer sequences and continues to the downstream 23S sequences, is of particular interest. The presence of this ligation event suggests that the 16S rRNA bhb-processing occurs prior to 23S rRNA maturation. Although this ligation event was not identified by nanopore sequencing in H. volcanii and P. furiosus, this observation is in agreement with our recent functional cis-acting element analysis performed in H. volcanii (Tang et al. 2002; Jüttner et al. 2020). In fact, based on this previous analysis, we have proposed a model by which 16S rRNA maturation proceeds and is required for the downstream maturation of the internal tRNA present in Euryarchaea and the 23S rRNA. Moreover, we have hypothesized that ligation of the 16S rRNA leader/trailer resulting from the 16S rRNA bhb maturation process generates a putative new pre-rRNA intermediate for which the corresponding ligation event was observed using direct cDNA sequencing in an evolutionary distant archaeon (Jüttner et al. 2020).
The presence of circular-pre-16S and pre-23S rRNAs and their processing sites could be verified and established in all organisms analyzed in this study. Recently, we determined the functional requirement of the bhb motifs for the formation of circ-pre-rRNAs in H. volcanii (Jüttner et al. 2020). Moreover, in analogy to intron-containing-tRNA splicing, the rRNA bhb motifs are cleaved by the tRNA splicing endonuclease (endA) prior to covalent circularization (Russell et al. 1999; Schwarz et al. 2020). Knockdown of endA has recently been shown to lead to the accumulation of noncleaved pre-rRNAs, presumably corresponding to precursor class 1 and a corresponding decrease in circularized precursors, emphasizing the essential role of this endonuclease in ensuring efficient rRNA maturation in vivo (Schwarz et al. 2020). Although intact circular RNA cannot be directly sequenced by the nanopore approach, we noticed the presence of permuted transcripts by analyzing clipping values, indicative of sequences that do not match to a linear template. Furthermore, most of the permuted reads of the circular rRNA classes 3 and 10 showed heterogenous 5′ and 3′ ends, thereby suggesting that these pre-rRNAs were likely the result of random nicking during sample preparation.
In addition to the circular-pre-rRNAs, we observed pre-rRNA intermediates cleaved at the bhb motifs but not yet ligated to form circular-pre-rRNAs in H. volcanii (post-bhb/pre-ligation pre-rRNAs; classes 2, 9). Whereas the presence of this intermediate processing step is theoretically expected, they were only detectable in larger numbers in the 16S maturation pathway of H. volcanii, suggesting that the maturation kinetics or stability of these pre-rRNA intermediates, prior to covalent ligation, varies among organisms and rRNA species.
Based on our current knowledge, several nonmutually exclusive hypotheses can be drawn for the final processing of circular-pre-rRNA into linear mature rRNAs: (i) opening of the circular-pre-rRNA within the ligated spacer region and subsequent maturation of the 5′ and 3′ end; (ii) opening of the circular-pre-rRNA by first maturation of the 5′ end mature rRNA followed by 3′ end maturation; or (iii) opening of the circular-pre-rRNA by first maturation of the 3′ end mature rRNA followed by 5′ end maturation, as suggested earlier in a Methanogen (Qi et al. 2020).
Putative 16S pre-rRNA intermediates extended in its 3′ end by the presence of the ligated 5′ and 3′ spacers normally observed in the circular-pre-16S rRNAs (class 4), may provide some indications how linearization of the circular-pre-16S rRNA is achieved. This particular configuration is indicative of 5′ end maturation of circular-pre-16S rRNA prior to final 3′ end maturation, thereby generating opened-circular-pre-16S intermediates. Additional properties of this putative intermediate are in agreement with its positioning during rRNA maturation and with the prevalence of 16S rRNA 5′ maturation prior to its 3′ end previously observed in bacteria and eukaryotes (Henras et al. 2015; Bechhofer and Deutscher 2019). After 3′ processing, our data suggest that there may well be organism-specific differences that lead to the final product through the combination of endo- or 3′–5′ exoribonucleolytic pathways. In particular, bhb-processed precursors (class 5) remain difficult to classify, as they could also represent 3′-extended early precursors. Noteworthy, these intermediates were also detected by our northern blot analysis after RNase H-dependent cleavage. Moreover, we could identify a new conserved ribonuclease required for the processing of classes 4/5 pre-rRNA intermediates, suggesting that this step is conserved and might be characteristic of several archaeal rRNA processing pathways. The structure–function analysis of this ribonuclease will be reported elsewhere. Similarly, the archaeal Rio proteins, as well as the endonuclease Nob1, have been implicated in the maturation of the 16S rRNA 3′ end in archaea (Veith et al. 2012; Knüppel et al. 2018; Qi et al. 2020). Based on our previous northern blot analysis it is likely that the pre-rRNA defects observed in vivo after deletion of Rio1 or Rio2 correlates with accumulation of classes 4 and/or 5 pre-rRNAs (Knüppel et al. 2018). Based on the methodological advance reported in this manuscript (nanopore sequencing and RNase H-dependent cleavage), the exact maturation defects of these and other mutants can be now better established and will be the subject of future studies.
Overall, future functional characterization of the cis- and trans-acting elements required for pre-rRNAs maturation will be necessary to further refine our general view on archaeal rRNA processing. In conclusion, despite some intrinsic limitations, we provide evidence that full-length rRNA sequencing can be a useful tool to approach intricate maturation pathways like rRNA maturation and expand our understanding of RNA maturation.
Toward the mapping of stage-dependent installation of rRNA modifications
RNA modifications have been described already in the 1950s–1960s, and have gained significant attention over the last years under the generic term of the epitranscriptome (Littlefield and Dunn 1958; Smith and Dunn 1959; Li and Mason 2014). However, the high-throughput analysis of these post-transcriptional modifications remains challenging and mostly relies on indirect methods, e.g., primer extension stops analysis and/or chemical recoding/derivation strategies (Schwartz and Motorin 2017; Sas-Chen and Schwartz 2019). Moreover, determining the timing of the addition of rRNA modifications during rRNA processing remains challenging. Native RNA sequencing may fill an important gap to systematically analyze RNA modifications on a single-molecule level. However, global strategies enabling the faithful determination of RNA modification identity, position and timing of addition needs to be developed. Several recent analyses have explored different strategies to evaluate the capacity of nanopore sequencing to accurately detect RNA modifications (e.g., m6A; Garalde et al. 2018; Liu et al. 2019; Smith et al. 2019; Lorenz et al. 2020; Begik et al. 2021, 2022; Jenjaroenpun et al. 2021; Leger et al. 2021).
We used a KsgA-deletion strain as a background model, to analyze the installation of the almost universally conserved KsgA-dependent dimethylations (m62A, positions A1450/A1451) at different stages of rRNA maturation in H. volcanii (O'Farrell et al. 2006; Xu et al. 2008; Smith et al. 2019). By analyzing base-calling profiles in wild-type and KsgA-deletion strains, we could unambiguously provide in vivo evidence that the archaeal KsgA-dependent methylations of the 16S rRNA are completed during the later stage of the small ribosomal subunit biogenesis in H. volcanii and may predominantly occur after opening of the circular-pre-16S rRNA (class 4). These results are in good agreement with previous studies in eukaryotes and bacteria (Lafontaine et al. 1998; Connolly et al. 2008; Strunk et al. 2011; Knüppel et al. 2021). In addition, single-molecule sequencing also allowed us to confirm the heterogeneous installation of dimethylation at either one or both expected positions by KsgA (Knüppel et al. 2021).
It was intriguing to observe deviations at other 16S rRNA-modified positions, notably C* at position 1350, which indicated a lower modification in the KsgA-deletion strain. Although it is currently unknown and unlikely that KsgA is responsible for this modification, there may be some dependency related to the recruitment of rRNA-modifying enzymes during ribosome biogenesis or other factors that influence the modification process. Further investigation into these functional dependencies could provide valuable insights into the complex interplay between different RNA modifications and their roles in ribosome assembly and function.
While nanopore sequencing may facilitate RNA modification analysis in general, the exact chemical nature of some modifications cannot be unveiled without prior knowledge and remain a challenging task which greatly benefits from the use of unmodified/hypomodified reference sequences as suggested by a number of comparison-based algorithms (Price et al. 2020; Leger et al. 2021; Pratanwanich et al. 2021). Although the error profiles at the acp3U and C* sites offer some initial insights into the timing, an unmodified reference sequence (in vitro transcript or RNA extracted from mutant strains) would be required to reliably determine the stage-dependent installation of these modifications.
To facilitate high-throughput identification of RNA modifications, future studies will be required to develop and train algorithms improving the de novo identification confidence of diverse RNA/DNA modifications (Hendra et al. 2022).
Limitations of the study and future directions
While nanopore sequencing is a promising tool to speed up the systematic exploration of rRNA maturation in all domains of life, there are several challenges associated with its application, especially in archaea. One of the main features that prevent the absolute quantification of rRNA intermediates is the detection sensitivity toward circular RNAs. In general, it is challenging to detect circular RNAs due to the lack of a clear end point for sequencing. In our case, we could only detect the circular precursors because they appeared to be randomly nicked during purification or library preparation, allowing the ligation of the adapter to the 3′ extremities. While specialized methods are available to capture and sequence circular RNAs effectively, such as RNase R digestion, random nicking and cDNA synthesis for short-read sequencing protocols or rolling circular RT and RNase R treatment for nanopore-based protocols, the joint absolute quantification of linear and circular RNAs remains difficult (Szabo and Salzman 2016; Liu et al. 2021; Zhang et al. 2021).
Quantifying rRNA molecules is also challenged by intrinsic properties, like high GC content, secondary structures and RNA modifications that can cause bias during library preparation and sequencing. In particular, RT is a crucial step that can be optimized by using other reverse transcriptases and reaction conditions. In our experiments, differences in RT efficiency leading to shortened RNA molecules and nonstoichiometric amounts of 16S and 23S rRNA could not be explained by GC content and could be due to more stable RNA structures in the (hyper)-thermophilic model organisms (Gomes-Filho et al. 2019; Sas-Chen et al. 2020). This conclusion is supported by the observation that in Sulfolobus, and to a greater extent in Pyrococcus, there is a significant proportion of reads considered full-length sequenced that are notably short. This inconsistency is not mirrored in the initial RNA lengths, implying that RNA degradation issues may not be the primary cause.
Despite these challenges, our data show that nanopore sequencing can be effective for the high-throughput detection and characterization of rRNA processing intermediates in archaea. Furthermore, we conclude that PCR-based amplification can be used to increase the detection sensitivity and allows the sequencing of samples where biomass might be critical. In the context of rRNA maturation and rRNA modifications, nanopore sequencing is a powerful tool to map the time point for the installation of rRNA modifications in the rRNA maturation pathway, which can illuminate the correlation between processing and modification. Finally, nanopore sequencing of rRNAs from different environmental conditions or strains can help us to gain a deeper understanding of the regulatory principles underlying rRNA maturation.
MATERIALS AND METHODS
Strains and growth conditions
P. furiosus strain DSM 3638 cells were grown anaerobically at one bar excess of nitrogen in 40 mL SME medium (Grünberger et al. 2019) supplemented with 40 mM pyruvate, 0.1% peptone, and 0.1% yeast extract at 95°C to mid-exponential phase and further harvested by centrifugation at 3939g for 45 min at 4°C.
Sulfolobus acidocaldarius strain MW001 was grown in standard Brock medium (Brock et al. 1972; Wagner et al. 2012; Knüppel et al. 2020) at 75°C to mid-exponential phase as described earlier (Knueppel et al. 2017).
Markerless deletion of H. volcanii KsgA (Hvo_2746) was obtained earlier using the pop-in/pop-out procedure as described in Knüppel et al. (2021). Haloferax strains (WT: H26) were grown in Hv-YPC medium at 42°C under agitation to mid-exponential phase as described previously (Knüppel et al. 2020, 2021).
In vitro transcription
First, genomic DNA was isolated from H. volcanii (H26) using standard phenol–chloroform extraction. The DNA was used as template for the PCR-amplification of the 16S region using primers containing sites necessary for in vitro transcription using the T7 RNA polymerase (fwd: TAATACGACTCACTATAGGGATTCCGGTTGATCCTGCCGGA, rev: AGGAGGTGATCCAGCCGCAGA). PCR was conducted using Phusion High-Fidelity DNA polymerase (NEB, M0530, 2000 U/mL) following the manufacturer's guidelines for GC-rich templates (GC buffer, 3% DMSO) using 30-cycles (annealing temperature: 63°C, extension time: 120 sec). PCR products were cleaned up using a PCR Clean-Up System (Promega, A9280). The T7 RiboMax Express Kit (Promega) was used for the in vitro production of 16S rRNA under standard conditions.
RNA isolation
For total RNA isolation, cell pellets were first lysed by the addition of 1 mL TRI Reagent (Zymogen). RNAs from H. volcanii and S. acidocaldarius were further purified using the RNeasy Kit (Qiagen) according to the manufacturer's instructions. Alternatively, RNA from P. furiosus was isolated using the Direct-zol RNA Miniprep Plus protocol (Zymogen) following manufacturer's instructions.
RNA treatment for nanopore sequencing
To prevent secondary structure formation, RNAs were heat incubated at 70°C for 3 min and immediately put on ice before any of the following enzymatic treatments.
RNA treatment for (PCR)-cDNA sequencing
RNAs for direct cDNA and PCR-cDNA sequencing were treated with T4 Polynucleotide Kinase (NEB, M0201) following the manufacturer's nonradioactive phosphorylation protocol to dephosphorylate 3′ termini, which facilitates efficient 3′ ligation. After reaction clean-up using ZYMO RNA Clean & Concentrator-5 Kit (ZYMO, R1013), 5′ triphosphates were removed enzymatically to minimize the influence of different 5′ end properties on the template-switching activity. To that end, 2 µg RNA were incubated for 30 min at 37°C with 2 µL RNA 5′ pyrophosphohydrolase (NEB, M0356, 5000 U/mL). The reactions were stopped by cleaning up the samples following the ZYMO RNA Clean & Concentrator-5 Kit (ZYMO, R1013) protocol. To prevent artificial polyadenylation and improve 3′ end accuracy of nanopore sequencing, we used a custom 3′ cDNA RT primer. Accordingly, a custom 3′ adapter (5′-rAppCTGTAGGCACCATCAAT-NH2-3′, NEB, S1315S) was ligated to all RNAs, following the protocol described in Mo et al. (2021). Briefly, 100 ng pretreated RNA was mixed with 50 pmol 3′ adapter, 2 µL 10× T4 RNA ligase reaction buffer (NEB, M0242), 10 µL 50% PEG 8000 (NEB, M0242), 1 µL 40 U µL−1 RNase Inhibitor (NEB, M0314), and 1 µL T4 RNA ligase 2 (truncated K227Q, NEB, M0242, 200,000 U/mL) and incubated at 16°C for 14 h. Finally, RNAs were cleaned up using the ZYMO RNA Clean & Concentrator Kit (ZYMO, R1013).
RNA treatment and poly(A)-tailing for direct RNA sequencing
RNAs for direct RNA sequencing were treated with T4 Polynucleotide Kinase (NEB, M0201) following the manufacturer's nonradioactive phosphorylation protocol to dephosphorylate 3′ termini, removing blocked ends that would inhibit efficient polyadenylation. After reaction clean-up using the ZYMO RNA Clean & Concentrator-5 Kit (ZYMO, R1013), RNAs were polyadenylated using the E. coli poly(A) polymerase (NEB, M0276) following a recently published protocol (Yan et al. 2018). Briefly, 5 µg RNA, 20 U poly(A) polymerase, 2 µL reaction buffer and 1 mM ATP were incubated for 15 min at 37°C in a total reaction volume of 50 µL. To stop the reaction and to remove the enzyme, the poly(A)-tailed RNA was purified using the ZYMO RNA Clean & Concentrator-5 Kit (ZYMO, R1013).
Nanopore library preparation and sequencing
(PCR)-cDNA sequencing
Libraries for direct cDNA and PCR-cDNA sequencing were prepared following the instructions in the direct cDNA sequencing with native barcoding protocol (SQK-DCS109 with EXP-NBD104) and the PCR-cDNA Barcoding Kit (SQK-PCB109) from Oxford Nanopore Technologies with minor modifications. Briefly, the VN Primer was replaced with a custom 3′ cDNA RT primer (5′-ACTTGCCTGTCGCTCTATCTTCATTGATGGTGCCTACAG-3′, 2 µM). The size of the input RNA and the strand-switched (direct cDNA) or PCR-amplified cDNA (PCR-cDNA) was assessed via a Bioanalyzer (Agilent) run using the RNA 6000 Pico and the High Sensitivity DNA Kit (Agilent), respectively. After RT, samples used for PCR-cDNA sequencing were amplified for 12 cycles at an elongation time of 5 min. During library preparation, the quantity and quality of the samples were tested using standard spectroscopic measurements (Nanodrop One) and using the Qubit 1× dsDNA HS Assay and Qubit RNA HS Assay Kit (Thermo Fisher Scientific). Finally, samples were pooled in equimolar ratios, adapter-ligated or attached following the respective protocols, loaded onto R9.4 flow cells (Oxford Nanopore Technologies) and sequenced on a MK1C device for 48 h.
Direct RNA sequencing
Libraries for direct RNA nanopore sequencing were prepared from poly(A)-tailed RNAs according to the SQK-RNA002 Kit protocol (Oxford Nanopore, Version: DRS_9080_v2_revS_14Aug2019) with minor modifications. Briefly, Agencourt AMPure XP magnetic beads (Beckman Coulter) in combination with 1 µL of RiboGuard RNase Inhibitor (Lucigen) were used instead of the recommended Agencourt RNAclean XP beads to purify samples after enzymatic reactions. Also, the RTA adapter was replaced by custom adapters described in https://github.com/Psy-Fer/deeplexicon to barcode the samples (Smith et al. 2020). RNA was linearized using Maxima RT H Minus enzyme (Thermo Fisher Scientific, #EP0751) and incubated for 30 min at 60°C followed by a heat inactivation step of 5 min at 85°C. After RT reactions, cDNA was quantified using the Qubit DNA HS Assay Kit (Thermo Fisher Scientific), and equimolar amounts of DNA were used for ligation of the RNA Adapter (RMX) in a single tube. Subsequent reactions were performed according to the protocols recommended by Oxford Nanopore Technologies. Finally, libraries were loaded onto R9.4 flow cells (Oxford Nanopore Technologies) and sequenced on a MinION device for 48 h. Direct RNA sequencing was performed for two biological replicates of wild-type and KsgA-deletion samples.
Data analysis
Base-calling, demultiplexing, and trimming of (PCR)-cDNA libraries
All fast5 reads from the (PCR)-cDNA sequencing runs were base-called using guppy (direct cDNA: v. 6.3.2 + bb5453e; PCR-cDNA: v. 6.4.2 + 97a7f06) in high-accuracy mode with a q-score cutoff of 7. Furthermore, base-called files were demultiplexed in a separate step by guppy_barcoder using default parameters (direct cDNA: barcode kit EXP-NBD104; PCR-cDNA: SQK-PCB109) except for disabling barcode trimming. For raw read quality control, relevant information was extracted from the MinKNOW sequencing summary file and the barcode summary file. Demultiplexed (PCR)-cDNA reads were strand-oriented and filtered for full-length sequenced reads containing the custom VN Primer and the strand-switching primer using pychopper with autotuned cutoffs and the recommended edlib backend (v. 2.7.2). Additionally, protocol-specific read rescue was performed for the direct cDNA libraries using the DCS109-specific option in pychopper. All full-length detected reads were merged and used for subsequent steps.
Base-calling, demultiplexing, and trimming of direct RNA libraries
As some bioinformatic tools depend on single-read nanopore files, we converted multiread FAST5 files from the MinKNOW output to single-read FAST5 files using the ont_fast5_api from Oxford Nanopore (https://github.com/nanoporetech/ont_fast5_api). Base-calling was done using guppy (v. 6.4.2 + 97a7f06) in high-accuracy mode (rna_r9.4.1_70bps_hac.cfg). To prevent actual good-quality reads from being discarded (this issue was reported previously; Weirather et al. 2017; Soneson et al. 2019), we disabled q-score filtering during base-calling and used all reads in the following steps of the analysis. Demultiplexing was done by deeplexicon (v. 1.2, https://github.com/Psy-Fer/deeplexicon) with default parameters (Smith et al. 2020). After base-calling and demultiplexing, artificially added poly(A) sequences were removed from the 3′ ends using cutadapt v. 4.2 (−a A{10}, −e 3, −j 0; Martin 2011).
Read alignment
Direct RNA and (PCR)-cDNA reads were mapped using minimap2 (v. 2.24-r1122) with standard parameters suggested for the alignment of noisy direct RNA reads (−ax splice −uf −k14) and nanopore genomic reads (-max map-ont), respectively (Li 2018, 2021). Reads were either aligned to the representative reference genomes downloaded from the NCBI or to circularly permuted sequences (described below).
In each case, –MD tag was used to include the MD tag for calculating mapping identities. Alignment files were converted to BAM files, sorted, and indexed using samtools (v. 1.15.1; Li et al. 2009). Additionally, position-specific read coverages were calculated using samtools depth (−a −J). Read identities were calculated as previously reported (1-NM/aligned_read_length) after calculating the aligned read length by adding the number of M and I characters in the CIGAR string (Li et al. 2009; Soneson et al. 2019; Grünberger et al. 2022).
Detection of rRNA processing sites and classification of rRNA intermediates
Processing site detection was performed by enrichment analysis of start and end positions of reads mapping to the relevant rRNA regions. Sorting was achieved by classifying reads according to enriched and literature-expected terminal positions. For rRNA stage classification, coordinates of 5′ and 3′ ends, clipping information extracted from the CIGAR string, strand information and sequence identity were considered. Briefly, only reads with a mapping identity ≥90% and with soft-clippings ≤20 nt were used for classification based on linear templates (reads mapping to the representative genomes). Finally, stage-sorted reads were plotted in a genome browser-like view and evaluated based on the terminal positions. Enriched and previously undescribed combinations of connected 5′ and 3′ ends were considered in the classification.
Circular RNA detection
Circular reads were initially observed in a subset of reads, which end near/at the 5′-cleavage site of the bhb, but are extensively left-clipped, which happens during mapping if the nucleotides further upstream do not match the 5′-leading, but the 3′-trailing region of the rRNA. The same is true for extensively right-clipped 3′ ends. The accuracy of 5′- and 3′-cleavage site detection using nanopore reads was further evaluated by secondary structure prediction of the potential bhb regions using RNAfold (Lorenz et al. 2011). To investigate circular rRNA reads in more detail, permuted linear sequences were created. These sequences contained 500 nt upstream of the annotated rRNA end to the predicted 3′-cleavage site of the bhb site and were joined with the 5′-cleavage site of the bhb up to 500 nt downstream from the annotated rRNA start. Nanopore reads were remapped to the linear permuted sequences and again categorized by their 5′ ends and 3′ ends as circular (reads cover unconnected mature parts of the rRNA and extend over the bhb) or opened-circular-pre-rRNAs (read extends over bhb, 3′ break at mature rRNA start). Additionally, reads potentially categorized as post-16S-bhb (only present in Sulfolobus, processing step 7*) were detected by mapping to a permuted sequence created by excising the 16S containing the leading and trailing sequences up to the bhb. For final stage classification, circular mapping reads were prioritized over linear templates with each read ID only being assigned to one stage.
Modified base detection
For stage-dependent modified base detection, direct RNA reads were sorted according to the read groups defined using the direct cDNA reads with two minor modifications. The quality threshold was lowered from 90% to 80% and the soft-clipping cutoff was increased from 20 to 30 nt to account for the lower sequencing quality of direct RNA sequencing using nanopore technology (Garalde et al. 2018; Grünberger et al. 2022).
Read IDs were extracted for each stage and direct RNA fastq files sorted using seqtk subseq (v. 1.3-r106). Sorted Fastq files were subsequently mapped and used for calculating the frequency of correct, deleted, inserted and wrong nucleotides at each genomic position using pysamstats (v. v1.1.2). Note that the sum of substitution, deletion and insertion frequencies was defined as the error of specific bases (% ESB). Additionally, modified base detection was performed using Eligos2 in pair_diff_mod (–oddR 0 –esb 0 –pval 1 –adjPval 1) to identify RNA modifications based on the pairwise comparison of each wild-type replicate with each ΔksgA replicate (control sample; Nookaew et al. 2020; Jenjaroenpun et al. 2021). For modified base detection based on signal data, Fast5 files were first sorted to predetermined rRNA stages using fast5_subset (ont_fast5_api). Next, multi-Fast5 files were converted to single-read files using multi_to_single_fast5 (ont_fast5_api). Finally, reads were preprocessed, resquiggled and the raw signals used to extract signal level and dwell value information using Tombo (v. 1.5.1, https://nanoporetech.github.io/tombo). The Tombo pipeline was run for presorted files as well as for unsorted files (Stoiber et al. 2016). Downstream analysis was performed using custom R scripts (R Development Core Team 2011).
Primer extension analysis
Determination of 5′ ends of mature 16S and 23S rRNAs from H. volcanii by primer extension was performed as described previously (Knüppel et al. 2020). In brief, RT was performed with the indicated fluorescently labeled primers (oHv396: 5′-DY682-CCCAATAGCAATGACCTCCG for H. volcanii 16S rRNA 5′ end; oHv622: 5′-DY782-GCTCTCGAGCCGAGCTATCCACC for H. volcanii 23S rRNA 5′ end) and SuperScript III reverse transcriptase using 1 µg of total RNA as template. The resulting cDNAs and reference dideoxy-chain termination sequencing ladder reactions were separated on a denaturing 14% TBE-Urea (6 M)-PAGE. Fluorescence signals (700 nm and 800 nm) were acquired using a Li-COR Odyssey system.
RNase H cleavage assay
Five micrograms of H. volcanii total RNA were incubated with 10 pmol of oHv203 (AGGAGGTGATCCAGCCGC) and oHv254 (GTACTTCCCAGGCGGCTCG) and 1.5 µL 10× RNase H buffer for 5 min at 65°C followed by an oligo annealing step for 10 min at 37°C in a total volume of 14 µL. Two microliters of RNase H (10 U, NEB, M0297) was added followed by an incubation of 20 min at 37°C. Fifteen microliters of 2× formamide buffer (99.95% formamide, 0.05% bromphenol blue) were added and the reaction stopped at 65°C for 15 min. RNase H-cleaved total RNAs were loaded on a denaturing 1.5% agarose gel and further analyzed by northern blotting as described previously (Knueppel et al. 2017) using the following fluorescently labeled probes: Probe 1, targeting the 16S mature rRNA (oHv707: DY682-GAGCTGGTGAGATGTCCGGC); Probe 2, targeting the 16S ligation site (oHv762: DY782-CCGTTCGGATaaggtgtccg); Probe 3, targeting the 16S 3′ trailer (oHv325: DY782-cgtgtgagccaccccgtcgg).
DATA DEPOSITION
All Nanopore sequencing data are available at the European Nucleotide Archive (ENA, https://www.ebi.ac.uk/ena) under project accession number PRJEB57168 (Cummins et al. 2022).
Code availability: Documentation and code of all essential analysis steps (tools and custom Rscripts) are available from https://github.com/felixgrunberger/rRNA_maturation.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank all the members of the Ferreira-Cerca lab and the Grohmann lab, especially Professor Dr. Winfried Hausner and Dr. Robert Reichelt for fruitful discussions. We thank Katharina Vogl for her technical assistance. We furthermore would like to thank Jörg Soppa for the discussion in the early phase of this project. Work in the Grohmann lab was supported by the Deutsche Forschungsgemeinschaft (SFB960-TP7 to D.G.). Research in the SF-C lab is generously supported by the German Research Foundation (DFG): individual research grant (FE1622/2-1) and collaborative research center SFB/CRC 960 grant (SFB960-AP1, SFB960-B13) “RNP biogenesis: assembly of ribosomes and nonribosomal RNPs and control of their function,” as well by the Centre National de la Recherche Scientifique (CNRS) and Ecole Polytechnique de Paris.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.079636.123.
Freely available online through the RNA Open Access option.
REFERENCES
- Bechhofer DH, Deutscher MP. 2019. Bacterial ribonucleases and their roles in RNA metabolism. Crit Rev Biochem Mol Biol 54: 242–300. 10.1080/10409238.2019.1651816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begik O, Lucas MC, Pryszcz LP, Ramirez JM, Medina R, Milenkovic I, Cruciani S, Liu H, Santos Vieira HG, Sas-Chen A, et al. 2021. Quantitative profiling of pseudouridylation dynamics in native RNAs with nanopore sequencing. Nat Biotechnol 39: 1278–1291. 10.1038/s41587-021-00915-6 [DOI] [PubMed] [Google Scholar]
- Begik O, Mattick JS, Novoa EM. 2022. Exploring the epitranscriptome by native RNA sequencing. RNA 28: 1430–1439. 10.1261/rna.079404.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkedal U, Beckert B, Wilson DN, Nielsen H. 2020. The 23S ribosomal RNA from Pyrococcus furiosus is circularly permuted. Front Microbiol 11: 582022. 10.3389/fmicb.2020.582022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock TD, Brock KM, Belly RT, Weiss RL. 1972. Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch Für Mikrobiol 84: 54–68. 10.1007/BF00408082 [DOI] [PubMed] [Google Scholar]
- Clouet-D'Orval B, Batista M, Bouvier M, Quentin Y, Fichant G, Marchfelder A, Maier L-K. 2018. Insights into RNA-processing pathways and associated RNA-degrading enzymes in archaea. FEMS Microbiol Rev 42: 579–613. 10.1093/femsre/fuy016 [DOI] [PubMed] [Google Scholar]
- Connolly K, Rife JP, Culver G. 2008. Mechanistic insight into the ribosome biogenesis functions of the ancient protein KsgA. Mol Microbiol 70: 1062–1075. 10.1111/j.1365-2958.2008.06485.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins C, Ahamed A, Aslam R, Burgin J, Devraj R, Edbali O, Gupta D, Harrison PW, Haseeb M, Holt S, et al. 2022. The European Nucleotide Archive in 2021. Nucleic Acids Res 50: D106–D110. 10.1093/nar/gkab1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danan M, Schwartz S, Edelheit S, Sorek R. 2012. Transcriptome-wide discovery of circular RNAs in archaea. Nucleic Acids Res 40: 3131–3142. 10.1093/nar/gkr1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher MP. 2015. Twenty years of bacterial RNases and RNA processing: how we've matured. RNA 21: 597–600. 10.1261/rna.049692.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durovic P, Dennis PP. 1994. Separate pathways for excision and processing of 16S and 23S rRNA from the primary rRNA operon transcript from the hyperthermophilic archaebacterium Sulfolobus acldocaldarius: similarities to eukaryotic rRNA processing. Mol Microbiol 13: 229–242. 10.1111/j.1365-2958.1994.tb00418.x [DOI] [PubMed] [Google Scholar]
- Ferreira-Cerca S. 2017. Life and death of ribosomes in archaea. In RNA metabolism and gene expression in archaea (ed. Clouet-d'Orval B), pp. 129–158. Springer International Publishing, Cham. [Google Scholar]
- Garalde DR, Snell EA, Jachimowicz D, Sipos B, Lloyd JH, Bruce M, Pantic N, Admassu T, James P, Warland A, et al. 2018. Highly parallel direct RNA sequencing on an array of nanopores. Nat Methods 15: 201–206. 10.1038/nmeth.4577 [DOI] [PubMed] [Google Scholar]
- Ghalei H, Trepreau J, Collins JC, Bhaskaran H, Strunk BS, Karbstein K. 2017. The ATPase Fap7 tests the ability to carry out translocation-like conformational changes and releases Dim1 during 40S ribosome maturation. Mol Cell 67: 990–1000.e3. 10.1016/j.molcel.2017.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes-Filho JV, Randau L, Gomes-Filho JV, Randau L. 2019. RNA stabilization in hyperthermophilic archaea. Ann NY Acad Sci 1447: 88–96. 10.1111/nyas.14060 [DOI] [PubMed] [Google Scholar]
- Grosjean H, Gaspin C, Marck C, Decatur WA, de Crécy-Lagard V. 2008. RNomics and Modomics in the halophilic archaea Haloferax volcanii: identification of RNA modification genes. BMC Genom 9: 470. 10.1186/1471-2164-9-470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünberger F, Reichelt R, Bunk B, Spröer C, Overmann J, Rachel R, Grohmann D, Hausner W. 2019. Next generation DNA-Seq and differential RNA-Seq allow re-annotation of the Pyrococcus furiosus DSM 3638 genome and provide insights into archaeal antisense transcription. Front Microbiol 10: 01603. 10.3389/fmicb.2019.01603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünberger F, Ferreira-Cerca S, Grohmann D. 2022. Nanopore sequencing of RNA and cDNA molecules in Escherichia coli. RNA 28: 400–417. 10.1261/rna.078937.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helser TL, Davies JE, Dahlberg JE. 1971. Change in methylation of 16S ribosomal RNA associated with mutation to kasugamycin resistance in Escherichia coli. Nat New Biol 233: 12–14. 10.1038/newbio233012a0 [DOI] [PubMed] [Google Scholar]
- Hendra C, Pratanwanich PN, Wan YK, Goh WSS, Thiery A, Göke J. 2022. Detection of m6A from direct RNA sequencing using a multiple instance learning framework. Nat Methods 19: 1590–1598. 10.1038/s41592-022-01666-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras AK, Plisson-Chastang C, O'Donohue M-F, Chakraborty A, Gleizes P-E. 2015. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip Rev RNA 6: 225–242. 10.1002/wrna.1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann S, Miller OL. 1977. Visualization of ribosomal ribonucleic acid synthesis in a ribonuclease III-deficient strain of Escherichia coli. J Bacteriol 132: 718–722. 10.1128/jb.132.2.718-722.1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob AI, Köhrer C, Davies BW, RajBhandary UL, Walker GC. 2013. Conserved bacterial RNase YbeY plays key roles in 70S ribosome quality control and 16S rRNA maturation. Mol Cell 49: 427–438. 10.1016/j.molcel.2012.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenjaroenpun P, Wongsurawat T, Wadley TD, Wassenaar TM, Liu J, Dai Q, Wanchai V, Akel NS, Jamshidi-Parsian A, Franco AT, et al. 2021. Decoding the epitranscriptional landscape from native RNA sequences. Nucleic Acids Res 49: e7. 10.1093/nar/gkaa620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jüttner M, Weiß M, Ostheimer N, Reglin C, Kern M, Knüppel R, Ferreira-Cerca S. 2020. A versatile cis-acting element reporter system to study the function, maturation and stability of ribosomal RNA mutants in archaea. Nucleic Acids Res 48: 2073–2090. 10.1093/nar/gkz1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knueppel R, Kuttenberger C, Ferreira-Cerca S. 2017. Toward time-resolved analysis of RNA metabolism in archaea using 4-thiouracil. Front Microbiol 8: 286. 10.3389/fmicb.2017.00286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knüppel R, Christensen RH, Gray FC, Esser D, Strauß D, Medenbach J, Siebers B, MacNeill SA, LaRonde N, Ferreira-Cerca S. 2018. Insights into the evolutionary conserved regulation of Rio ATPase activity. Nucleic Acids Res 46: 1441–1456. 10.1093/nar/gkx1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knüppel R, Fenk M, Jüttner M, Ferreira-Cerca S. 2020. In vivo RNA chemical footprinting analysis in archaea. Methods Mol Biol 2106: 193–208. 10.1007/978-1-0716-0231-7_12 [DOI] [PubMed] [Google Scholar]
- Knüppel R, Trahan C, Kern M, Wagner A, Grünberger F, Hausner W, Quax TEF, Albers S-V, Oeffinger M, Ferreira-Cerca S. 2021. Insights into synthesis and function of KsgA/Dim1-dependent rRNA modifications in archaea. Nucleic Acids Res 49: 1662–1687. 10.1093/nar/gkaa1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laass S, Monzon VA, Kliemt J, Hammelmann M, Pfeiffer F, Förstner KU, Soppa J. 2019. Characterization of the transcriptome of Haloferax volcanii, grown under four different conditions, with mixed RNA-Seq. PLoS One 14: e0215986. 10.1371/journal.pone.0215986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine D, Delcour J, Glasser AL, Desgrès J, Vandenhaute J. 1994. The DIM1 gene responsible for the conserved m62Am62A dimethylation in the 3′-terminal loop of 18 S rRNA is essential in yeast. J Mol Biol 241: 492–497. 10.1006/jmbi.1994.1525 [DOI] [PubMed] [Google Scholar]
- Lafontaine DLJ, Preiss T, Tollervey D. 1998. Yeast 18S rRNA dimethylase Dim1p: a quality control mechanism in ribosome synthesis? Mol Cell Biol 18: 2360–2370. 10.1128/MCB.18.4.2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leger A, Amaral PP, Pandolfini L, Capitanchik C, Capraro F, Miano V, Migliori V, Toolan-Kerr P, Sideri T, Enright AJ, et al. 2021. RNA modifications detection by comparative nanopore direct RNA sequencing. Nat Commun 12: 7198. 10.1038/s41467-021-27393-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34: 3094–3100. 10.1093/bioinformatics/bty191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2021. New strategies to improve minimap2 alignment accuracy. Bioinformatics 37: 4572–4574. 10.1093/bioinformatics/btab705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Mason CE. 2014. The pivotal regulatory landscape of RNA modifications. Annu Rev Genomics Hum Genet 15: 127–150. 10.1146/annurev-genom-090413-025405 [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlefield JW, Dunn DB. 1958. Natural occurrence of thymine and three methylated adenine bases in several ribonucleic acids. Nature 181: 254–255. 10.1038/181254a0 [DOI] [PubMed] [Google Scholar]
- Liu H, Begik O, Lucas MC, Ramirez JM, Mason CE, Wiener D, Schwartz S, Mattick JS, Smith MA, Novoa EM. 2019. Accurate detection of m6A RNA modifications in native RNA sequences. Nat Commun 2019: 1–9. 10.1038/s41467-019-11713-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Tao C, Li S, Du M, Bai Y, Hu X, Li Y, Chen J, Yang E. 2021. circFL-seq reveals full-length circular RNAs with rolling circular reverse transcription and nanopore sequencing. Elife 10: e69457. 10.7554/eLife.69457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londei P, Ferreira-Cerca S. 2021. Ribosome biogenesis in archaea. Front Microbiol 12: 686977. 10.3389/fmicb.2021.686977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R, Bernhart SH, Höner zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. 2011. ViennaRNA package 2.0. Algorithms Mol Biol 6: 26. 10.1186/1748-7188-6-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz DA, Sathe S, Einstein JM, Yeo GW. 2020. Direct RNA sequencing enables m6A detection in endogenous transcript isoforms at base-specific resolution. RNA 26: 19–28. 10.1261/rna.072785.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangat CS, Brown ED. 2008. Ribosome biogenesis; the KsgA protein throws a methyl-mediated switch in ribosome assembly. Mol Microbiol 70: 1051–1053. 10.1111/j.1365-2958.2008.06484.x [DOI] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17: 10. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Mo W, Liu B, Zhang H, Jin X, Lu D, Yu Y, Liu Y, Jia J, Long Y, Deng X, et al. 2021. Landscape of transcription termination in Arabidopsis revealed by single-molecule nascent RNA sequencing. Genome Biol 22: 1–21. 10.1186/s13059-020-02207-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev N, Silengo L, Schlessinger D. 1973. Synthesis of a large precursor to ribosomal RNA in a mutant of Escherichia coli. Proc Natl Acad Sci 70: 3361–3365. 10.1073/pnas.70.12.3361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nookaew I, Jenjaroenpun P, Du H, Wang P, Wu J, Wongsurawat T, Moon SH, Huang E, Wang Y, Boysen G. 2020. Detection and discrimination of DNA adducts differing in size, regiochemistry, and functional group by nanopore sequencing. Chem Res Toxicol 33: 2944–2952. 10.1021/acs.chemrestox.0c00202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Farrell HC, Pulicherla N, Desai PM, Rife JP. 2006. Recognition of a complex substrate by the KsgA/Dim1 family of enzymes has been conserved throughout evolution. RNA 12: 725–733. 10.1261/rna.2310406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratanwanich PN, Yao F, Chen Y, Koh CWQ, Wan YK, Hendra C, Poon P, Goh YT, Yap PML, Chooi JY, et al. 2021. Identification of differential RNA modifications from nanopore direct RNA sequencing with xPore. Nat Biotechnol 39: 1394–1402. 10.1038/s41587-021-00949-w [DOI] [PubMed] [Google Scholar]
- Price AM, Hayer KE, McIntyre ABR, Gokhale NS, Abebe JS, Della Fera AN, Mason CE, Horner SM, Wilson AC, Depledge DP, et al. 2020. Direct RNA sequencing reveals m6A modifications on adenovirus RNA are necessary for efficient splicing. Nat Commun 11: 6016. 10.1038/s41467-020-19787-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L, Li J, Jia J, Yue L, Dong X. 2020. Comprehensive analysis of the pre-ribosomal RNA maturation pathway in a methanoarchaeon exposes the conserved circularization and linearization mode in archaea. RNA Biol 17: 1427–1441. 10.1080/15476286.2020.1771946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. 2011. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org [Google Scholar]
- Russell AG, Ebhardt H, Dennis PP. 1999. Substrate requirements for a novel archaeal endonuclease that cleaves within the 5′ external transcribed spacer of Sulfolobus acidocaldarius precursor rRNA. Genetics 152: 1373–1385. 10.1093/genetics/152.4.1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sas-Chen A, Schwartz S. 2019. Misincorporation signatures for detecting modifications in mRNA: not as simple as it sounds. Methods 156: 53–59. 10.1016/j.ymeth.2018.10.011 [DOI] [PubMed] [Google Scholar]
- Sas-Chen A, Thomas JM, Matzov D, Taoka M, Nance KD, Nir R, Bryson KM, Shachar R, Liman GLS, Burkhart BW, et al. 2020. Dynamic RNA acetylation revealed by quantitative cross-evolutionary mapping. Nature 583: 638–643. 10.1038/s41586-020-2418-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Motorin Y. 2017. Next-generation sequencing technologies for detection of modified nucleotides in RNAs. RNA Biol 14: 1124–1137. 10.1080/15476286.2016.1251543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz TS, Berkemer SJ, Bernhart SH, Weiß M, Ferreira-Cerca S, Stadler PF, Marchfelder A. 2020. Splicing endonuclease is an important player in rRNA and tRNA maturation in archaea. Front Microbiol 11: 594838. 10.3389/fmicb.2020.594838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JD, Dunn DB. 1959. The occurrence of methylated guanines in ribonucleic acids from several sources. Biochem J 72: 294–301. 10.1042/bj0720294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Jain M, Mulroney L, Garalde DR, Akeson M. 2019. Reading canonical and modified nucleobases in 16S ribosomal RNA using nanopore native RNA sequencing. PLoS One 14: e0216709. 10.1371/journal.pone.0216709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MA, Ersavas T, Ferguson JM, Liu H, Lucas MC, Begik O, Bojarski L, Barton K, Novoa EM. 2020. Molecular barcoding of native RNAs using nanopore sequencing and deep learning. Genome Res 30: 1345–1353. 10.1101/gr.260836.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneson C, Yao Y, Bratus-neuenschwander A, Patrignani A, Robinson MD, Hussain S. 2019. A comprehensive examination of nanopore native RNA sequencing for characterization of complex transcriptomes. Nat Commun 10: 1–14. 10.1038/s41467-019-11272-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoiber MH, Quick J, Egan R, Lee JE, Celniker SE, Neely RK, Loman N, Pennacchio LA, Brown J. 2016. De novo identification of DNA modifications enabled by genome-guided nanopore signal processing. bioRxiv 094672. [Google Scholar]
- Strunk BS, Loucks CR, Su M, Vashisth H, Cheng S, Schilling J, Brooks CL, Karbstein K, Skiniotis G. 2011. Ribosome assembly factors prevent premature translation initiation by 40S assembly intermediates. Science 333: 1449–1453. 10.1126/science.1208245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo L, Salzman J. 2016. Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet 17: 679–692. 10.1038/nrg.2016.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang TH, Rozhdestvensky TS, d'Orval BC, Bortolin M-L, Huber H, Charpentier B, Branlant C, Bachellerie J-P, Brosius J, Hüttenhofer A. 2002. RNomics in archaea reveals a further link between splicing of archaeal introns and rRNA processing. Nucleic Acids Res 30: 921–930. 10.1093/nar/30.4.921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veith T, Martin R, Wurm JP, Weis BL, Duchardt-Ferner E, Safferthal C, Hennig R, Mirus O, Bohnsack MT, Wöhnert J, et al. 2012. Structural and functional analysis of the archaeal endonuclease Nob1. Nucleic Acids Res 40: 3259–3274. 10.1093/nar/gkr1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema J, Tollervey D. 1995. Processing of pre-ribosomal RNA in Saccharomyces cerevisiae. Yeast 11: 1629–1650. 10.1002/yea.320111607 [DOI] [PubMed] [Google Scholar]
- Viehweger A, Krautwurst S, Lamkiewicz K, Madhugiri R, Ziebuhr J, Hölzer M, Marz M. 2019. Direct RNA nanopore sequencing of full-length coronavirus genomes provides novel insights into structural variants and enables modification analysis. Genome Res 29: 1545–1554. 10.1101/gr.247064.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M, van Wolferen M, Wagner A, Lassak K, Meyer BH, Reimann J, Albers S-V. 2012. Versatile genetic tool box for the crenarchaeote Sulfolobus acidocaldarius. Front Microbiol 3: 214. 10.3389/fmicb.2012.00214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirather JL, de Cesare M, Wang Y, Piazza P, Sebastiano V, Wang X-J, Buck D, Au KF. 2017. Comprehensive comparison of Pacific Biosciences and Oxford Nanopore Technologies and their applications to transcriptome analysis. F1000Res 6: 100. 10.12688/f1000research.10571.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, O'Farrell HC, Rife JP, Culver GM. 2008. A conserved rRNA methyltransferase regulates ribosome biogenesis. Nat Struct Mol Biol 15: 534–536. 10.1038/nsmb.1408 [DOI] [PubMed] [Google Scholar]
- Yan B, Boitano M, Clark TA, Ettwiller L. 2018. SMRT-Cappable-seq reveals complex operon variants in bacteria. Nat Commun 9: 3676. 10.1038/s41467-018-05997-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip WSV, Vincent NG, Baserga SJ. 2013. Ribonucleoproteins in archaeal pre-rRNA processing and modification. Archaea 2013: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Hou L, Zuo Z, Ji P, Zhang X, Xue Y, Zhao F. 2021. Comprehensive profiling of circular RNAs with nanopore sequencing and CIRI-long. Nat Biotechnol 39: 836–845. 10.1038/s41587-021-00842-6 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.