Abstract
The SRP9/SRP14 heterodimer is a central component of signal recognition particle (SRP) RNA (7SL) processing and Alu retrotransposition. In this study, we sought to establish the role of nuclear SRP9/SRP14 in the transcriptional regulation of 7SL and BC200 RNA. 7SL and BC200 RNA steady-state levels, rate of decay, and transcriptional activity were evaluated under SRP9/SRP14 knockdown conditions. Immunofluorescent imaging, and subcellular fractionation of MCF-7 cells, revealed a distinct nuclear localization for SRP9/SRP14. The relationship between this localization and transcriptional activity at 7SL and BC200 genes was also examined. These findings demonstrate a novel nuclear function of SRP9/SRP14 establishing that this heterodimer transcriptionally regulates 7SL and BC200 RNA expression. We describe a model in which SRP9/SRP14 cotranscriptionally regulate 7SL and BC200 RNA expression. Our model is also a plausible pathway for regulating Alu RNA transcription and is consistent with the hypothesized roles of SRP9/SRP14 transporting 7SL RNA into the nucleolus for posttranscriptional processing, and trafficking of Alu RNA for retrotransposition.
Keywords: Alu RNA, Alu-like RNA, bromouridine immunoprecipitation chase (BRIC), chromatin immunoprecipitation (ChIP), immunofluorescence
INTRODUCTION
The signal recognition particle (SRP) is a ribonucleoprotein complex responsible for regulating the translation of ∼30% of the human proteome (Akopian et al. 2013). In humans, this complex is assembled in nucleoli and consists of six proteins (SRP9/SRP14 heterodimer, SRP19, SRP54, and SRP68/SRP72 heterodimer) and one 7SL RNA (Politz et al. 2000). The 7SL RNA consists of two main domains, the Alu domain, which is bound by the SRP9/SRP14 heterodimer, and the S domain, which interacts with SRP19, SRP54, and SRP68/SRP72 heterodimers (Fig. 1A; Walter and Johnson 1994). The SRP9/SRP14 heterodimer is vital for arresting translational elongation by directly binding the ribosome (Halic et al. 2004), while SRP19, SRP54, and SRP68/SRP72 are important for the signal peptide and SRP receptor binding (Siegel and Walter 1986). Neither SRP9 nor SRP14 can bind to 7SL RNA individually, as the SRP9/SRP14 RNA-binding surface is formed at the interface of the heterodimer (Strub and Walter 1990). The same is not true for SRP68/SRP72, because each have the capabilities to interact with 7SL RNA in vitro (Lutcke et al. 1993; Iakhiaeva et al. 2005). Together, the SRP complex arrests the translation of secretory and membrane proteins and targets them to the endoplasmic reticulum where translation is resumed (Akopian et al. 2013).
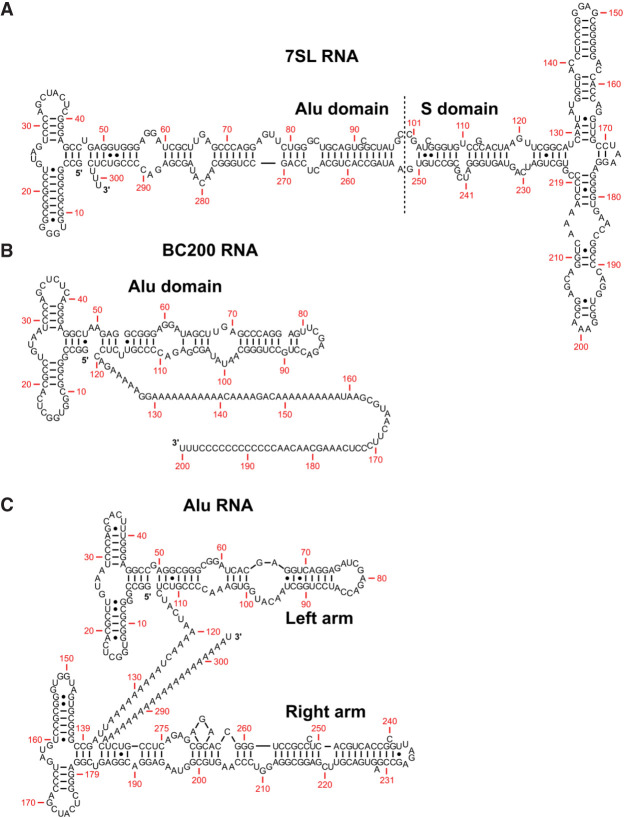
FIGURE 1.
Secondary structure of Alu and Alu-like RNA. (A) Secondary structure of 7SL RNA as determined by enzymatic digestions (Sinnett et al. 1991) and adapted from work by Häsler and Strub (2006). The RNA is segmented into Alu and S domains. SRP9/SRP14 recognize the Alu domain, while SRP19, SRP54, and SRP68/SRP72 recognize the S domain. (B) Predicted secondary structure of BC200 RNA based on conservation of secondary structure motifs compared to 7SL RNA (Labuda and Zitkiewicz 1994). The structure is composed of an Alu domain followed by a polyadenosine tract and a unique sequence. (C) Secondary structure of an AluY RNA found on intron four of the α-fetoprotein gene based on enzymatic digestions of an Alu RNA (Sinnett et al. 1991) and adapted from Häsler and Strub (2006). The structure is composed of two distinct arms with a demarcating internal polyadenosine tract.
SRP9/SRP14 are also protein binding partners of the primate-specific noncoding RNA transcription product of the BCYRN1 gene, referred to as BC200 (Bovia et al. 1997). BC200 is a 200 nt RNA polymerase III transcript with its first 120 nt possessing homology with an AluJ sequence, followed by 40 nt adenosine-rich and 40 nt cytosine-rich regions (Fig. 1B; Martignetti and Brosius 1993; Tiedge et al. 1993). There are 14 validated protein binding partners of BC200 RNA. Each region of BC200 RNA possesses distinct protein binding partners, including SRP9/SRP14 for the Alu region, CSDE1 (cold shock domain-containing E1) for the adenosine-rich region, and HNRNPK (heterogeneous nuclear riboprotein K) for the cytosine-rich region (Booy et al. 2018). BC200 RNA is normally expressed at high levels in brain tissue, but aberrantly expressed in multiple tumor cell types (Chen et al. 1997; Iacoangeli et al. 2004; Hu and Lu 2015; Booy et al. 2017; Samson et al. 2018). Although the functional role and cellular mechanism of BC200 RNA have yet to be determined, it is evidently critical for tumor cell viability and migration, and enhances global translational activity (Booy et al. 2017, 2021).
Alu RNA are a homologous, but diverse family of RNA with 225 reported subfamilies (Price et al. 2004). Alu RNA are transcription products of Alu elements, primate-specific, ∼300 bp DNA elements consisting of two similar but distinct monomers, each originating from processed forms of the 7SL gene (Ullu and Tschudi 1984; Quentin 1992a,b). Alu elements are extremely successful retrotransposons, composing ∼10.5% of the human genome and are large contributors to primate evolution (Lander et al. 2001; Jurka 2004; Daniel et al. 2014). Each Alu element consists of internal RNA polymerase III promoters (A- and B-boxes), an internal polyadenosine tract linking the two monomers, and a terminal polyadenosine tract (Fig. 1C).
The three different classes of RNA polymerase III transcripts are defined by their differing internal RNA polymerase III promoter compositions, where Alu DNA are class II (tRNA-like) RNA polymerase III transcribed genes (Geiduschek and Tocchini-Valentini 1988). These sequences exist as individual genes, as well as embedded in protein-coding genes within introns and exons (Häsler et al. 2007). Although there are over 1 million copies of Alu DNA, an estimated 99% of Alu DNA are epigenetically repressed by methylation (Lander et al. 2001; Stenz 2021). During development, aging, and tumorigenesis, Alu DNA methylation is dynamic with hypomethylation being a precursor to the accumulation of Alu RNA (Yoder et al. 1997; Luo et al. 2014). Alu RNA accumulation and general dysregulation are observed in a variety of human diseases, such as cancer, through many diverse mechanisms (Gussakovsky and McKenna 2021).
The first ∼50 nt of 7SL and BC200 RNA contain SRP9/SRP14 binding sites that are conserved across evolution and all transcribed Alu RNA (Strub et al. 1991; Weichenrieder et al. 1997). The conservation of the primary sequence of the first ∼50 nt is a prerequisite for proper secondary structure formation, where a G24C point mutation in 7SL is sufficient to alter its overall structure and abrogate efficient SRP9/SRP14 binding (Chang et al. 1997). These findings have been characterized with analyses of high-resolution structures of SRP9/SRP14 and the 5′ end of 7SL RNA (Birse et al. 1997; Weichenrieder et al. 2001). Appropriate 7SL RNA secondary structure has been observed to be required for efficient transcription of 7SL RNA (Emde et al. 1997). Previous studies have demonstrated that the knockdown of SRP9/SRP14 in human cells significantly reduced 7SL RNA steady-state levels (Lakkaraju et al. 2007; Mary et al. 2010). Together, this suggests that the efficient transcription and stabilization of Alu-like RNA is dependent on its ability to interact with SRP9/SRP14.
Investigation of the role of SRP9/SRP14 in the nucleus has been limited. SRP9/SRP14 were reported to be involved in the export of nucleolar 7SL RNA in Xenopus oocytes and Saccharomyces cerevisiae to the cytoplasm (He et al. 1994; Grosshans et al. 2001; Sommerville et al. 2005). 7SL RNA found in the nucleolus has its three terminal uridines removed, and a terminal adenine added, but mutants unable to interact with SRP9/SRP14 could not be adenylated in vitro (Chen et al. 1998; Perumal et al. 2001). SRP9/SRP14 are also proposed to play a major role in the propagation of Alu elements by trafficking nascent Alu RNA to a ribosome actively translating the human LINE1 protein (Dewannieux et al. 2003). In vitro, the SRP9/SRP14 heterodimer was determined to be required for successful retrotransposition, but the specific role in the nucleus was not confirmed (Bennett et al. 2008). In this study, we sought to establish the role of nuclear SRP9/SRP14 in the transcriptional regulation of 7SL and BC200 RNA. The effect of SRP9/SRP14 knockdown on 7SL and BC200 RNA steady-state levels was assessed. 7SL and BC200 RNA decay and synthesis was evaluated under SRP9/SRP14 knockdown conditions. Immunofluorescent imaging and subcellular fractionation of MCF-7 cells revealed a distinct nuclear localization for SRP9/SRP14. The relationship between this localization and transcriptional activity at 7SL and BC200 genes was examined. These findings demonstrate a novel nuclear function of SRP9/SRP14, establishing that this heterodimer transcriptionally regulates 7SL and BC200 RNA expression. We describe a model in which SRP9/SRP14 cotranscriptionally regulate 7SL and BC200 RNA expression. Our model is a plausible pathway for regulating Alu RNA transcription and is consistent with the hypothesized roles of SRP9/SRP14 transporting 7SL RNA into the nucleolus for posttranscriptional processing (Massenet 2019), and trafficking of Alu RNA for retrotransposition (Dewannieux et al. 2003).
RESULTS
SRP9 and SRP14 knockdowns reduce 7SL and BC200 RNA steady-state levels
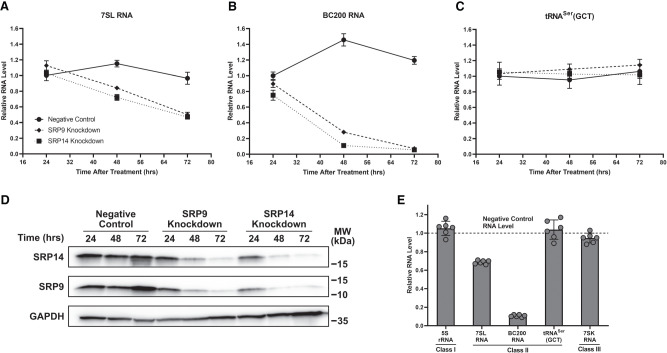
In addition to previous work demonstrating SRP9/SRP14 knockdown reducing 7SL RNA steady-state levels (Lakkaraju et al. 2007; Mary et al. 2010), our preliminary findings indicated a substantial reduction in BC200 RNA expression upon SRP9/SRP14 knockdown. To investigate this further, MCF-7 cells were transfected with a negative control, SRP9, or SRP14 siRNA, and 24, 48, and 72 h later whole-cell lysates were collected for RNA and protein extractions. Experiments were performed in MCF-7 cells due to the relatively high expression of 7SL and BC200 RNA (Booy et al. 2017, 2018). Using extracted RNA as template, RT-qPCR was performed with primers specific to 7SL, BC200, and tRNASer(GCT) (non-Alu-like class II RNA polymerase III transcript) (Fig. 2A–C). For 7SL RNA, significant reductions were observed after 48 h of both SRP9 (10%) and SRP14 (23%) siRNA treatments, with larger reductions detected 72 h after the respective knockdowns (40% for both siRNA) (Fig. 2A). BC200 RNA behaved similarly to 7SL RNA, but with more significant reductions. After 24 h of SRP9 and SRP14 siRNA treatment, the cells exhibited a decline of 13% and 35%, respectively, in BC200 RNA levels (Fig. 2B). After 48 h, BC200 RNA levels decreased by over 80% in both SRP9 and SRP14 siRNA treated cells. Seventy-two hours after, either SRP9 or SRP14 siRNA treatment BC200 RNA was essentially eliminated with a 95% reduction. These declines were not detected for tRNASer(GCT), with its RNA levels remaining stable under all siRNA transfection conditions and time points, suggesting specificity for the Alu-like class II RNA polymerase III transcripts (Fig. 2C).
FIGURE 2.
SRP9 and SRP14 knockdowns reduce 7SL and BC200 RNA steady-state levels. (A) MCF-7 cells were transfected with negative control, SRP9, or SRP14 siRNA and harvested following the labeled time points. Whole-cell lysate RNA extraction was performed, and expression was assessed by RT-qPCR with 25 ng of RNA per reaction. Primers were specific to 7SL, and measurements were presented as the mean of three biological replicates measured in duplicate with error represented as ±SD. (B) Same as (A) but with primers specific to BC200. (C) Same as (A) but with primers specific to tRNASer(GCT). (D) MCF-7 cells for each described transfection condition were harvested and isolated for protein. Whole-cell lysate protein was analyzed by SDS–PAGE and western blot with antibodies specific to SRP14 and SRP9 for monitoring knockdown efficiency, and GAPDH as a loading control. Molecular weight (MW) markers from the protein ladder are presented beside the blot. Representative western blot of three biological replicates was presented. (E) Whole-cell lysate RNA extracted from 48-h negative control and SRP14 siRNA transfected MCF-7 cells were analyzed by RT-qPCR with primers specific to various classes of RNA polymerase III transcripts (I—5S rRNA; II—7SL RNA, BC200 RNA, tRNASer(GCT); and III—7SK RNA). Data were presented as the mean relative RNA level of the SRP14 siRNA samples compared to the negative control across three biological replicates measured in duplicate. Error was shown as ±SD.
Previous efforts to immunodeplete or knockdown SPR9/SRP14 were performed by targeting both proteins simultaneously, or when targeted individually, the protein levels of the heterodimer partners were not considered (Lakkaraju et al. 2007; Mary et al. 2010; Berger et al. 2014; Nabet et al. 2017). Protein extracted from cells collected in Figure 2A–C was subjected to SDS–PAGE followed by a western blot using antibodies specific to SRP14, SRP9, and GAPDH (as a loading control) (Fig. 2D). No significant decreases were observed for either SRP9 or SRP14 protein in the negative control siRNA transfected cells across all time points. Meanwhile, in the independent knockdowns of either SRP9 or SRP14, a precipitous decline in both SRP9 and SRP14 protein was observed over time. Therefore, the knockdown of either SRP9 or SRP14 led to a knockdown of its heterodimer partner.
To determine the specificity of these RNA reductions upon SRP9/SRP14 knockdown, we conducted RT-qPCR on several RNA polymerase III transcripts to compare RNA levels after 48 h of SRP14 siRNA treatment relative to the negative control. These conditions were chosen due to the greater efficiency of the SRP14 knockdown compared to SRP9 knockdown and minimization of secondary effects that may occur at later time points. We assessed the RNA levels of non-Alu RNA polymerase III transcripts including 5S rRNA (class I), tRNASer(GCT) (class II), and 7SK RNA (class III). Only the Alu-like transcripts (7SL and BC200 RNA) were significantly reduced upon SRP9/SRP14 depletion (Fig. 2E). Using the same samples as template, we also investigated whether RNA transcribed by other RNA polymerases were impacted. It was determined only RNA polymerase III transcripts were directly influenced by SRP9/SRP14 protein levels as there was no significant difference detected in the RNA levels of 28S rRNA (RNA polymerase I transcript), hTERC RNA (noncoding RNA polymerase II transcript), TP53 mRNA (exonic Alu RNA possessing RNA polymerase II transcript), or SRP9 mRNA (exonic Alu RNA lacking RNA polymerase II transcript) after SRP9/SRP14 knockdown (Supplemental Fig. S1). To be noted, SRP9 mRNA levels were stable 48 h after SRP14 siRNA treatment, but protein levels were greatly reduced (Fig. 2D, Supplemental Fig. S1).
To ensure these results were not specific to MCF-7 cells, we subjected T-47D, A2780, A549, HEK293T, and HeLa cells to negative control or SRP14 siRNA transfections for 72 h prior to RNA and protein collection and extraction. The 72-h time point was selected due to this time point corresponding to the maximal reductions in 7SL and BC200 RNA in MCF-7 cells. RT-qPCR was performed using these samples as template with primers specific to 7SL RNA, BC200 RNA, and tRNASer(GCT) (Supplemental Fig. S2A). 7SL and BC200 RNA steady-state levels were significantly reduced in all cell lines, while tRNASer(GCT) levels were stable. Also, BC200 RNA was more significantly reduced than 7SL RNA. Western blots were performed with antibodies specific to SRP14, SRP9, and GAPDH (as a loading control) (Supplemental Fig. S2B). In each of the six cell lines, the 72-h SRP9/SRP14 knockdown was sufficient in eliminating the vast majority of SRP9 and SRP14 protein. From the results presented, it was clear the presence of SRP9/SRP14 is required to maintain normal steady-state levels of 7SL and BC200 RNA.
7SL and BC200 RNA half-lives are unaffected by SRP9/SRP14 knockdown
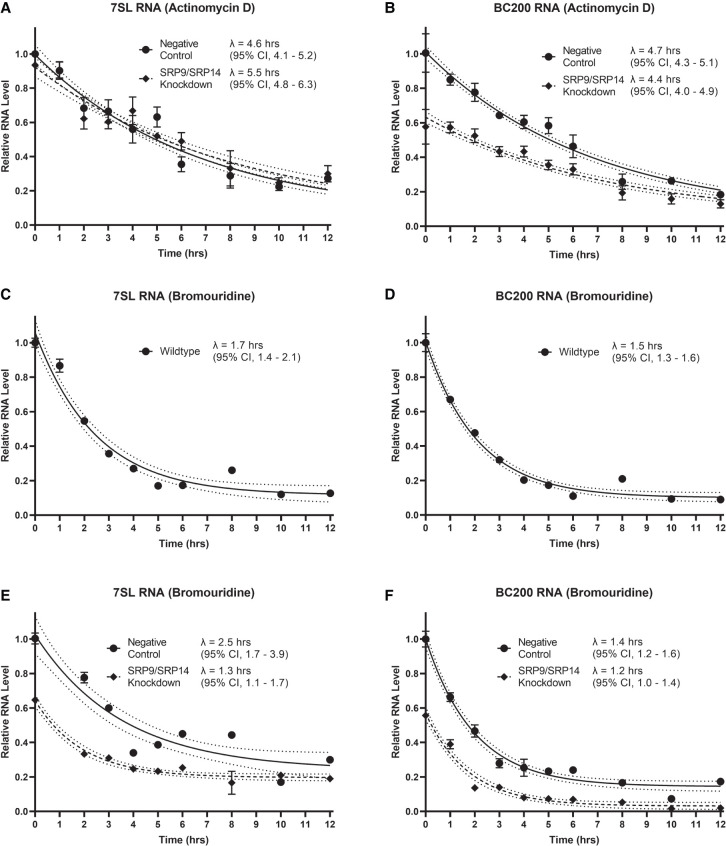
As RNA steady-state levels are dependent upon both the rate of degradation and the rate of transcription, we assessed the impact of SRP9/SRP14 knockdown on 7SL and BC200 RNA half-lives. MCF-7 cells were transfected with either negative control or SRP14 siRNA (as seen in Fig. 1D, SRP14 siRNA provides a knockdown of both SRP9 and SRP14) for 24 h prior to treatment with actinomycin D, an RNA polymerase II/RNA polymerase III transcription inhibitor (Bensaude 2011). Since transcription was blocked, measuring 7SL and BC200 RNA levels from the harvested cells over time allowed for the calculation of their rate of decay. RT-qPCR was performed on RNA collected in both conditions using primers specific to 7SL or BC200. The half-life (λ) of 7SL RNA was not significantly different between negative control and SRP9/SRP14 knockdown samples (Fig. 3A). BC200 RNA half-life was also unaffected by SRP9/SRP14 knockdown compared to the negative control (Fig. 3B). The knockdown was successful as seen by the reduction in BC200 RNA levels (42%) compared to the negative control at t = 0.
FIGURE 3.
7SL and BC200 RNA half-lives are unaffected by SRP9/SRP14 knockdown. (A) MCF-7 cells were transfected with either negative control or SRP14 siRNA (SRP9/SRP14 knockdown) for 24 h prior to treatment with 5 µg/mL actinomycin D. At indicated time points, an equal number of cells were harvested and whole-cell lysate RNA extraction was performed. An equal volume of RNA was measured by RT-qPCR with primers specific to 7SL. Data represent the mean of three biological replicates measured in duplicate relative to t = 0 for the negative control, with error represented as ±SD. Half-lives were calculated by fitting the data to a one-phase decay equation with a constraint of plateau = 0.05 using GraphPad Prism 9 software. Confidence bands demonstrate the likely location of the true curve with a 95% confidence interval. Error was presented as a 95% asymmetrical (profile-likelihood) confidence interval. (B) Same as in (A) but with primers specific to BC200. (C) MCF-7 cells were grown for 24 h with 150 µM 5′-bromouridine. After 24 h, cells were provided fresh media without 5′-bromouridine and collected at the indicated time points for bromouridine immunoprecipitation chase (BRIC). An equal volume of RNA was measured by RT-qPCR with primers specific to 7SL. Data are representative of three biological replicates, presented as the mean of three technical replicates relative to t = 0, with error bars indicating ±SD. Half-lives were calculated by fitting the data to a one-phase decay equation with no constraints using GraphPad Prism 9 software. Confidence bands and error presentation as in (A). (D) Same as in (C) but with primers specific to BC200. (E) Same as in (C) but transfected with either negative control or SRP14 siRNA (SRP9/SRP14 knockdown) for 24 h prior to removal of 5′-bromouridine. Data points are relative to t = 0 for the negative control. (F) Same as in (E) but with primers specific to BC200.
To further confirm this result, we repeated the evaluation of the RNA half-lives using BRIC. MCF-7 cells were treated with 5′-bromouridine for 24 h, prior to being chased with fresh media lacking 5′-bromouridine. Cells were harvested at indicated time points, RNA extracted, and equal amounts of input RNA immunoprecipitated with a 5′-bromouridine binding antibody. Immunoprecipitated RNA was purified, and RT-qPCR was performed with primers specific to 7SL and BC200. This provided us with wild type MCF-7 half-lives for 7SL RNA of 1.7 h (95% confidence interval, 1.4–2.1 h) (Fig. 3C) and BC200 RNA of 1.5 h (95% confidence interval, 1.3–1.6 h) (Fig. 3D). This experiment was repeated with a transfection of a negative control, or SRP14 siRNA 24 h prior collection. For both 7SL and BC200 RNA, their half-lives determined in wild type, negative control or SRP9/SRP14 knockdown conditions were all within error of each other (Fig. 3E,F). The knockdown was deemed to have been successful due to a reduction in both 7SL and BC200 RNA levels among all of the immunoprecipitated SRP9/SRP14 knockdown samples compared to the negative control. Both independent methods determined 7SL and BC200 RNA half-lives were unaffected by SRP9/SRP14 knockdown.
SRP9/SRP14 knockdown reduces transcriptional activity at 7SL and BC200 genetic loci
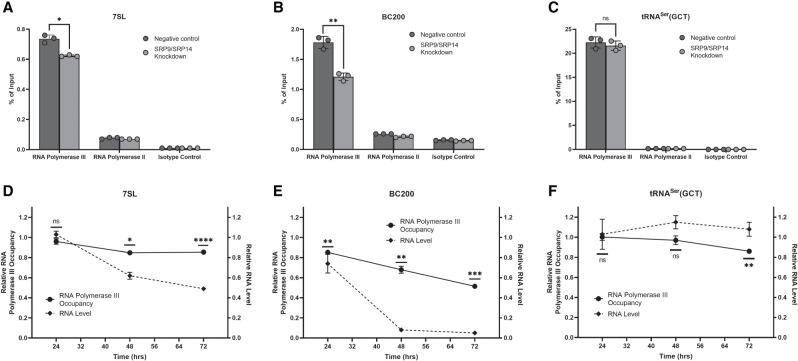
The lack of change in half-lives upon SRP9/SRP14 knockdown led us to investigate its effect on the transcriptional activity at 7SL and BC200 genes. Chromatin immunoprecipitations (ChIPs) were performed with antibodies specific to RNA polymerase III (RNA polymerase for 7SL and BC200), RNA polymerase II (RNA polymerase not specific to 7SL nor BC200), and an isotype control. MCF-7 cells were transfected with either negative control or SRP14 siRNA for 24, 48, or 72 h prior to crosslinking and collection. Chromatin immunoprecipitated DNA was analyzed by qPCR with primers specific to 7SL, BC200, and tRNASer(GCT). As a representative set of data, the 48-h transfection time point is presented (Fig. 4A–C). Unsurprisingly, in both negative control and SRP9/SRP14 knockdown conditions, RNA polymerase III significantly enriched 7SL, BC200, and tRNASer(GCT) DNA, while RNA polymerase II and the isotype control were not enriched. This was repeated for transfections after 24 and 72 h, and consistent results were observed (Supplemental Fig. S3).
FIGURE 4.
SRP9/SRP14 knockdown reduces transcriptional activity at 7SL and BC200 genetic loci. (A) qPCR analysis of ChIP enrichment of RNA polymerase III, RNA polymerase II, and a negative control at the 7SL gene locus in MCF-7 cells after 48-h negative control or SRP14 siRNA (SRP9/SRP14 knockdown) transfections. Data were representative of three independent biological replicates, presented as the mean of three technical replicates and error shown as ±SD. Percent of input was calculated from the measurements of 25 ng input DNA. P-values were calculated using an unpaired two-tailed t-test with statistical significance indicated by (*) P < 0.05, (**) P < 0.01, (***) P < 0.001. (B) Same as for (A) but with primers specific to BC200. (C) Same as for (A) but with primers specific to tRNASer(GCT). (D) Relative RNA polymerase III occupancy (solid line) compared RNA polymerase III ChIP enrichments after SRP9/SRP14 knockdown relative to the negative control at each respective time point. Relative RNA level (dashed line) was assessed from whole-cell lysate RNA extracted from MCF-7 cells transfected with negative control, or SRP14 siRNA following the labeled time points. Expression was assessed by RT-qPCR with 25 ng of RNA per reaction. Primers were specific to 7SL, and measurements were presented as the mean of three biological replicates measured in duplicate with error presented as ±SD. (E) Same as for (D) but with primers specific to BC200. (F) Same as for (D) but with primers specific to tRNASer(GCT).
Evaluating enrichment as an indicator of occupancy at the target genes and consequently transcriptional activity, we compared RNA polymerase III occupancy under SRP9/SRP14 knockdown relative to the negative control at each of the indicated time points. The relative RNA polymerase III occupancy and RNA steady-state levels (as demonstrated in Fig. 2) were plotted for 7SL in Figure 4D. RNA polymerase III occupancy was stable after 24 h (P = 1.0 × 10−1), but was significantly reduced by 15% after 48 h (P = 1.3 × 10−2). This was a plateau as after 72 h the reduction remained at 15% (P = 6.5 × 10−5). At the same time, 7SL RNA steady-state levels were stable up to 24 h after siRNA transfection, but reduced 38% and 51% after 48 and 72 h of siRNA treatment, respectively. In the analysis of BC200 RNA polymerase III occupancy after siRNA treatment, a consistent reduction was observed across the 24- (15%, P = 6.8 × 10−3), 48- (32%, P = 2.0 × 10−3), and 72- (48%, P = 1.9 × 10−4) h time points (Fig. 4E). BC200 RNA steady-state levels correlated with RNA polymerase III occupancy, but with a larger reduction observed at each of the time points (24 h: 26%, 48 h: 92%, 72 h: 95%). Conducting the same analysis on tRNASer(GCT) demonstrates a relatively stable class II RNA polymerase III transcript, with RNA polymerase III occupancy stable over the first 48 h (24 h, P = 9.1 × 10−1; 48 h, P = 4.9 × 10−1) and significantly reduced only after 72 h (14%, P = 2.2 × 10−3) (Fig. 4F). RNA steady-state levels were slightly elevated over time, maximally increased by 15% after 48 h. The data suggest SRP9/SRP14 play a role in transcriptionally regulating 7SL and BC200 genes.
SRP9 and SRP14 are heavily localized in the nucleus
As SRP9/SRP14 knockdown reduced transcriptional activity at 7SL and BC200 genetic loci, we wished to determine if this was occurring through a regulatory function of the heterodimer within the nucleus. To that end, SRP9 and SRP14 localization was determined using immunofluorescent imaging and western blots of protein from subcellular compartments. To ensure the antibodies we used for immunofluorescent imaging were specific, we visualized logarithmically growing MCF-7 cells after 72 h of negative control, or SRP14 siRNA transfections. To allow for quantitative comparisons, we maintained identical imaging settings across all samples. While observing ∼100 cells in each condition with filters specific to DNA (DAPI), SRP9 (CY5), and SRP14 (CY5), the overall signal of SRP9 and SRP14 was significantly reduced after SRP9/SRP14 knockdown (Fig. 5A,B). The images were also evaluated for cellular localization, where both SRP9 and SRP14 had cytoplasmic and nuclear signal but was in general more intense in the nucleus compared to the cytoplasm. Due to the limitation of epitope masking in immunofluorescent analysis, we further validated these results by subjecting protein extracted from different subcellular fractions to SDS–PAGE followed by a western blot. Using antibodies specific to ACTB (cytoplasmic marker), LRP5 (membrane-bound marker), and MYC (nuclear marker) demonstrated the precision of this fractionation. Blotting with antibodies specific to SRP9 and SRP14 revealed the two proteins are in high abundance in cytoplasmic, membrane bound, and nuclear fractions (Fig. 5C). The presence of SRP9/SRP14 in the nucleus is consistent with a functional role involving transcription.
FIGURE 5.
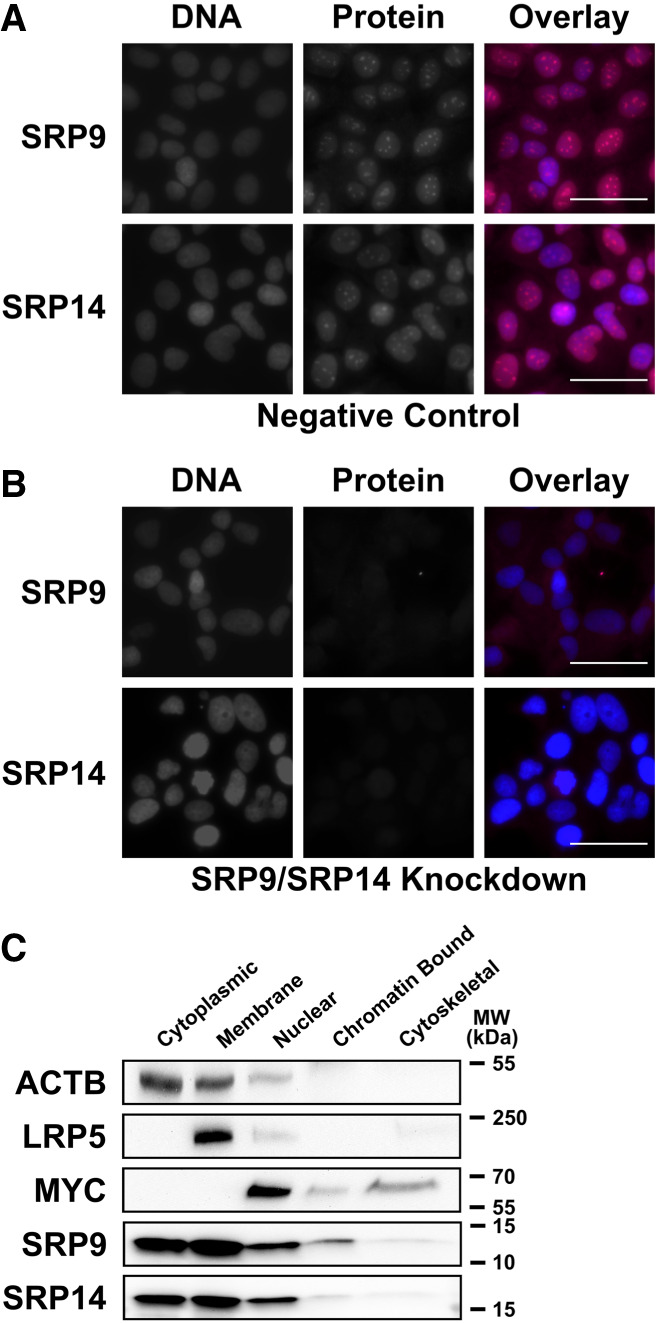
SRP9 and SRP14 are heavily localized in the nucleus. (A) Immunofluorescent analysis of representative logarithmically growing MCF-7 cells transfected with a negative control siRNA for 72 h prior to fixation. Cells were probed with both anti-SRP9 (rabbit) and anti-SRP14 (rabbit) antibodies. Anti-rabbit IgG Alexa Fluor 647 conjugate (CY5 filter) was used for visualization. Cells were counterstained with DAPI. Capturing settings were identical for all images to allow for quantitative analysis. The overlay images are composed of the DNA (DAPI—blue), SRP9 (CY5—red), or SRP14 (CY5—red) channels overlayed. Scale bars indicate 50 µm. (B) Same as for (A) but with SRP14 siRNA (SRP9/SRP14 knockdown). (C) Western blot analysis of the subcellular distribution of SRP9 and SRP14. Subcellular markers were used for loading and fraction specificity with antibodies specific to ACTB (cytoplasmic), LRP5 (membrane bound), and MYC (nuclear).
SRP9/SRP14 associate with chromatin
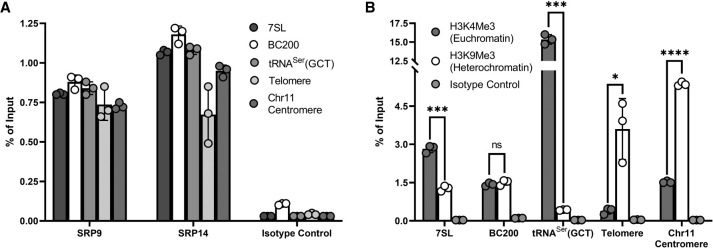
Since we observed substantial nuclear localization of SRP9/SRP14, we set out to determine if SRP9/SRP14 were chromatin bound and were in close proximity to 7SL and BC200 genetic loci. We collected MCF-7 cells for ChIPs performed with antibodies specific to SRP9, SRP14, H3K4Me3 (euchromatin marker, active transcription), H3K9Me3 (heterochromatin marker, repressed transcription), and an isotype control. Chromatin immunoprecipitated DNA was analyzed by qPCR with primers specific to 7SL, BC200, tRNASer(GCT), fixed-length copy of telomere DNA (Cawthon 2009), and a specific chromosome 11 centromere (Contreras-Galindo et al. 2017). These primer sets were specific to a mixture of likely euchromatic [7SL, BC200, tRNASer(GCT)] and heterochromatic (telomere, Chr11 centromere) regions. To assess enrichment comparatively, the measurements were represented as a percent of input. As observed in Figure 6A, regardless of the genomic region assayed, the enrichment of either SRP9 or SRP14 was relatively similar and significantly larger than the isotype control. H3K4Me3 (euchromatin marker) was significantly more enriched than H3K9Me3 (heterochromatin marker) for 7SL (2.8%, 1.3%; P = 1.6 × 10−4) and tRNASer(GCT) (15.4%, 0.43%; P = 6.1 × 10−4) genetic loci (Fig. 6B). As expected, H3K9Me3 (heterochromatin marker) had a greater enrichment for the telomere (3.6%, 0.39%; P = 4.2 × 10−2) and Chr11 centromere (5.4%, 1.5%; P = 7.8 × 10−6) primer sets than H3K4Me3 (euchromatin marker). Both H3K4Me3 and H3K9Me3 are similarly enriched at the BC200 genetic locus (1.4%, 1.5%; P = 3.6 × 10−1). This confirmed SRP9 and SRP14 were enriched in actively transcribing, and repressed chromatin. In all cases, the samples were significantly enriched relative to the isotype control, ranging from eightfold to 510-fold, establishing the specificity of the ChIPs. The association of SRP9/SRP14 with chromatin in close proximity to 7SL and BC200 genetic loci further increases the likelihood of the heterodimer performing a regulatory function at the transcriptional level.
FIGURE 6.
SRP9/SRP14 associated with chromatin. (A) qPCR analysis of ChIP enrichment of SRP9, SRP14, and an isotype control at genomic regions associated with the 7SL, BC200, and tRNASer(GCT) genes, a fixed-length telomere, and chromosome 11 centromere. Data were representative of three independent biological replicates, presented as the mean of three technical replicates and error shown as ±SD. Percent of input was calculated from the measurements of 25 ng input DNA. (B) Same as for (A) but with enrichment of H3K4Me3 (euchromatin marker), H3K9Me3 (heterochromatin marker), and an isotype control. P-values were calculated by an unpaired two-tailed t-test. P-values are reported as (ns) P > 0.05, (*) P ≤ 0.05, (**) P ≤ 0.01, (***) P ≤ 0.001, or (****) P ≤ 0.0001.
Association of SRP9/SRP14 to chromatin is independent of transcription
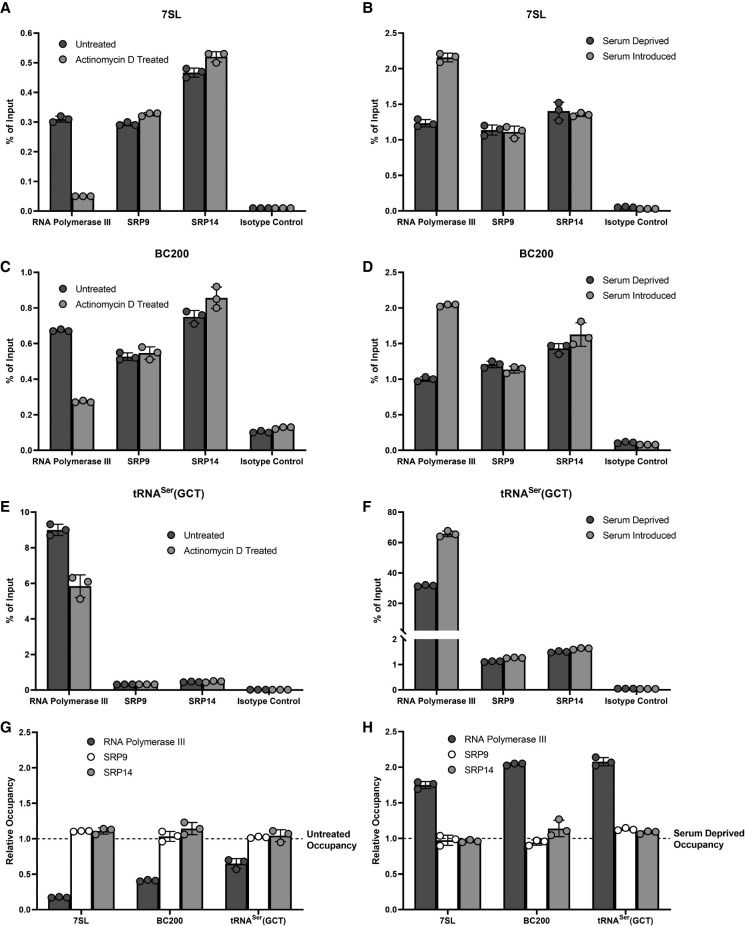
Since SRP9/SRP14 are in close proximity to 7SL and BC200 genetic loci, and SRP9/SRP14 abundance impacts the transcriptional activity of these genes, we sought to establish whether the association of SRP9/SRP14 with chromatin was dependent on transcription. We assessed this by monitoring SRP9/SRP14 chromosomal association after decreasing transcriptional activity with actinomycin D, an RNA polymerase II/RNA polymerase III transcriptional inhibitor (Bensaude 2011), and restoring transcriptional activity under serum-deprived (repressed) and serum-stimulated (activated) conditions. We performed ChIPs in MCF-7 cells with antibodies specific to RNA polymerase III, SRP9, SRP14, and an isotype control. The resultant DNA collected was analyzed by qPCR with primers specific to 7SL, BC200, and tRNASer(GCT). RNA polymerase III occupancy was used as a proxy for the transcriptional activity of the gene probed. At the 7SL genetic locus, RNA polymerase III occupancy was significantly reduced (83%) after actinomycin D treatment, while SRP9 and SRP14 occupancies were relatively unchanged (Fig. 7A). Similarly, after the first serum-depriving cells, RNA polymerase III occupancy was significantly increased (75%) with the reintroduction of 10% serum for 2 h (Fig. 7B). The reintroduction of serum had no impact on the occupancy of SRP9 and SRP14 at the 7SL genetic locus. This pattern was replicated for BC200 and tRNASer(GCT) as observed in Figure 7C–F. In all cases, the enrichment of RNA polymerase III, SRP9, and SRP14 was significantly larger than the isotype control, supporting the specificity of the ChIP. We summarize this in Figure 7G (repression) and Figure 7H (activation). Despite significant alteration of RNA polymerase III activity, the association of SRP9 and SRP14 to chromosomal DNA was unchanged.
FIGURE 7.
Association of SRP9/SRP14 to chromatin is independent of transcription. (A) qPCR analysis of ChIP enrichment of RNA polymerase III, SRP9, SRP14, and an isotype control at the 7SL gene locus in untreated, or 90- min 5 µg/mL actinomycin D treated MCF-7 cells. Data were representative of three independent biological replicates, presented as the mean of three technical replicates and error shown as ±SD. Percent of input was calculated from the measurements of 25 ng input DNA. (B) Same as for (A) but with MCF-7 cells serum deprived for 72 h prior to no treatment (repressed), or addition of 10% serum for 2 h (activated). (C) Same as for (A) but with primers specific to BC200. (D) Same as for (B) but with primers specific to BC200. (E) Same as for (A) but with primers specific to tRNASer(GCT). (F) Same as for (B) but with primers specific to tRNASer(GCT). (G) Relative occupancy comparing the enrichment of each protein after 90 min of actinomycin D treatment relative to untreated. Data were representative of three independent biological replicates, presented as the mean of three technical replicates and error shown as ±SD. (H) Same as for (G) but with relative occupancy comparing enrichment of each protein after addition of 10% serum for 2 h compared to untreated.
DISCUSSION
The first ∼50 nt of Alu and Alu-like RNA are known to be conserved across evolution (Fig. 1; Strub et al. 1991; Weichenrieder et al. 1997). Simultaneously, the proper structural folding of these sequences is required for SRP9/SRP14 binding (Birse et al. 1997; Bovia et al. 1997; Weichenrieder et al. 2001). It was previously suggested that efficient 7SL transcription was reliant on the proper secondary structure of the SRP9/SRP14 binding site (Emde et al. 1997), and that cellular 7SL RNA steady-state levels were reduced upon SRP9/SRP14 knockdown (Lakkaraju et al. 2007; Mary et al. 2010). Our results support these observations by demonstrating RNA steady-state levels of 7SL, and another Alu-like RNA, BC200, being significantly reduced after SRP9/SRP14 knockdown (Fig. 2). The reduction in BC200 RNA steady-state levels after SRP9/SRP14 knockdown was significantly larger than it was for 7SL RNA. One major difference between the two RNAs is their non-Alu structure and their non-SRP9/SRP14 protein binding partners. 7SL nucleotides are almost entirely engaged in the secondary structure, while BC200 may possess a partially accessible 80 nt 3′ end (Fig. 1A,B; Booy et al. 2016). 7SL RNA is bound by additional SRP components (SRP19, SRP54, SRP68/SRP72) on its S domain (Walter and Johnson 1994), while BC200 RNA is bound by up to 11 different protein binding partners on its adenosine and cytosine-rich regions (Booy et al. 2018). SRP9/SRP14 are capable of binding synthetic mRNA, but SRP9/SRP14 knockdown had no impact on the exonic Alu containing p53 mRNA levels (Supplemental Fig. S1; Hsu et al. 1995), instead only affecting Alu-like class II RNA polymerase III transcripts (Fig. 2E). This suggests SRP9/SRP14 play a role in regulating levels of Alu-containing RNA, but differences in RNA steady-state level reductions could be attributable to the unique secondary structure of the RNA and the non-SRP9/SRP14 protein partners these structures are bound by.
The method utilizing actinomycin D to measure RNA decay (Fig. 3A,B) generates larger half-life values for both 7SL and BC200 RNA than the BRIC method (Fig. 3C–F). This is possibly due to incomplete inhibition of transcription, or altered cell physiology in the presence of actinomycin D. Nonetheless, although there are major structural differences between 7SL and BC200, it appears using both methodologies that 7SL and BC200 share similar decay rates. This raises the possibility that the RNA have a common method of degradation due to their shared structure of the Alu domain (Fig. 1A,B), primarily the first ∼50 nt required for SRP9/SRP14 binding (Strub et al. 1991; Birse et al. 1997; Bovia et al. 1997; Weichenrieder et al. 1997, 2001). Considering this information with the observation that SRP9/SRP14 knockdown has no impact on the rate of degradation of either 7SL or BC200 RNA (Fig. 3) but reduces transcriptional activity at 7SL and BC200 genetic loci (Fig. 4), this portends SRP9/SRP14 possess a cotranscriptional regulatory role. In this model, nascent transcripts cotranscriptionally bound by SRP9/SRP14 are stabilized and maintain the original half-life, while those not bound by SRP9/SRP14 are rapidly degraded. Neither described method for measuring the rate of decay would be capable of identifying rapidly degraded nascent transcripts. Consequently, the reduction in SRP9/SRP14 levels upon knockdown would only impact nascent transcript accumulation, progressively reducing the RNA steady-state levels, consistent with our data (Fig. 2). The differing levels of reduction in transcriptional activity after SRP9/SRP14 knockdown between 7SL and BC200 genetic loci could be due to a variety of factors. Some hypotheses include differences in secondary structure and non-SRP9/SRP14 protein binding partners impacting the stabilization of the RNA, abundance of SRP9/SRP14 near the genetic loci, or differences in regulatory feedback loops halting wasteful transcription. While our results do not conclusively suggest the precise mechanism, they suggest an enticing route for future investigation.
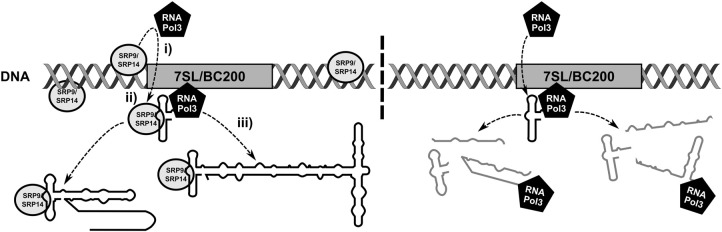
We propose a model in which a substantial number of SRP9/SRP14 molecules associate with chromosomal DNA nonspecifically, to promote RNA polymerase III transcription initiation, and cotranscriptionally regulate 7SL, BC200, and other Alu or Alu-like RNA (Fig. 8). This accounts for the observations that (i) SRP9/SRP14 are heavily localized in the nucleus (Fig. 5); (ii) SRP9/SRP14 are in close proximity to chromatin in actively transcribed and repressed regions (Fig. 6); and (iii) SRP9/SRP14 association with chromatin is independent of transcriptional activity (Fig. 7). In this model, SRP9/SRP14 promote RNA polymerase III transcription initiation, and binds the first ∼50 nt of the nascent RNA as it is being synthesized. Without SRP9/SRP14 stabilizing the nascent transcript, rapid degradation would ensue. This model is consistent with previous reports suggesting the proper secondary structure of the SRP9/SRP14 binding site is required for efficient transcription of the 7SL RNA (Emde et al. 1997). It is also consistent with export (He et al. 1994; Grosshans et al. 2001; Sommerville et al. 2005), posttranscriptional processing of 7SL (Chen et al. 1998; Perumal et al. 2001), and successful retrotransposition of Alu RNA requiring SRP9/SRP14 (Bennett et al. 2008). The SRP9/SRP14 heterodimer exists in a 20-fold molar excess compared to the SRP complex (Bovia et al. 1995), and our model provides a functional role of the heterodimer that is consistent with its abundance.
FIGURE 8.
Model of cotranscriptional regulation of 7SL and BC200 RNA by SRP9/SRP14. Schematic depicting genetic loci of 7SL and BC200 (rectangle), DNA (double helix), RNA polymerase III and associated transcription machinery (pentagon), SRP9/SRP14 (oval), and 7SL and BC200 RNA (outlines of the RNA from those observed in Fig. 1). Proteins, RNA, DNA, and genes are not to scale. SRP9/SRP14 are distributed nonspecifically across the DNA at high abundance. (i) SRP9/SRP14 promote the initiation of RNA polymerase III and associated transcription machinery to the indicated genetic locus; (ii) SRP9/SRP14 associated with DNA at close proximity to the specified genetic locus interact with the evolutionarily conserved structure formed by the first ∼50 nt of 7SL and BC200 RNA while transcription is ongoing (Strub et al. 1991; Emde et al. 1997; Weichenrieder et al. 1997); and (iii) full-length nascent 7SL and BC200 RNA interacting with SRP9/SRP14 are synthesized, while those lacking SRP9/SRP14 are degraded and transcription is aborted. This model is consistent with the proposed functions of SRP9/SRP14 in Alu retrotransposition (Dewannieux et al. 2003; Bennett et al. 2008). Nascent 7SL and BC200 RNA are then capable of continuing their normal life cycle, which could include export (He et al. 1994; Grosshans et al. 2001; Sommerville et al. 2005), posttranscriptional processing (Chen et al. 1998; Perumal et al. 2001; Massenet 2019), among other functions.
The major complications in studying Alu RNA are their high homology with 225 subfamilies (Price et al. 2004), high abundance with Alu elements composing ∼10.5% of the human genome (Lander et al. 2001), and short length (Fig. 1C). Common RNA detection techniques such as northern blots or primer-based amplifications are limited by cross-reactivity (Liu et al. 1994), masking the true identity and quantity of the transcript, impeding study on generalized functions of Alu RNA. Since the SRP9/SRP14 binding site of Alu, 7SL, and BC200 RNA share a ∼50 nt evolutionarily conserved sequence and structure (Strub et al. 1991; Birse et al. 1997; Bovia et al. 1997; Weichenrieder et al. 1997, 2001), the study of the regulatory role of SRP9/SRP14 on 7SL and BC200 RNA likely provides insight to a generalized Alu RNA synthesis mechanism. Overall, our study uncovered a functional role of nuclear SRP9/SRP14 in transcriptionally regulating 7SL and BC200 RNA. From the combination of our results and the available literature, we have generated a model for SRP9/SRP14 cotranscriptional regulation of 7SL and BC200 RNA, that would be consistent with Alu RNA, and proposed mechanisms of these RNA (Dewannieux et al. 2003; Massenet 2019).
MATERIALS AND METHODS
Cell culture and reagents
The MCF-7, T-47D, A549, and HeLa cell lines were a gift from Dr. Spencer Gibson. The HEK293T cell line was a gift from Dr. Thomas Klonisch, and the A2780 cell line was a gift from Dr. Sabine Kuss. Cell culture conditions were performed as previously published (Booy et al. 2016). Serum deprivation of cells was conducted in Gibco Dulbecco's Modified Eagle Medium with high glucose, no glutamine, and no phenol red (Thermo Fisher Scientific) DNA primers were purchased from Integrated DNA Technologies. All standard laboratory chemicals and reagents were purchased from Thermo Fisher Scientific.
siRNA transfection
siRNA were transfected using Lipofectamine RNAiMAX (Thermo Fisher Scientific) according to the manufacturer's protocol. Reverse transfections were performed by combining 50 pmole of siRNA with 7.5 µL Lipofectamine RNAiMAX in 250 µL Opti-MEM media (Thermo Fisher Scientific) per well of a six-well plate. A total of 2 mL of cell suspension was added such that cells were ∼80% confluent after the indicated treatment time. siRNA and nontargeting control (#4390844) were purchased from Thermo Fisher Scientific (Silencer Select). Transfections into other cell culture plates were scaled accordingly by volume. The siRNA targets and corresponding sequences were as follows: SRP9: 5′-GCGCUGCCGAGAAGCUUUATT and SRP14: 5′-CGGUCGAACCAAACCCAUUTT.
RNA purification and RT-qPCR
Whole-cell RNA extraction from cultured cells was performed using the GeneJET RNA Purification kit (Thermo Fisher Scientific) according to the manufacturer's protocol. Cells were lysed by resuspending cell pellets in 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% (v/v) NP-40, 0.1% (v/v) SDS, 0.5% (v/v) sodium deoxycholate (RIPA buffer) supplemented with Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific), and RiboLock RNase Inhibitor (Thermo Fisher Scientific). Resuspended samples were subjected to a brief vortex period, 10-min incubation on ice, and collection of the supernatant after centrifugation at 21,000g at 4°C for 5 min. RNA extraction after lysis was performed using the GeneJET RNA Concentration and Cleanup Micro Kit (Thermo Fisher Scientific) according to the manufacturer's protocol. Isolated RNA was quantified using a NanoDrop 2000c Spectrophotometer (Thermo Fisher Scientific). Reverse transcription quantitative PCR (RT-qPCR) was performed using an Applied Biosystems StepOnePlus instrument (Thermo Fisher Scientific) with the iTaq Universal SYBR Green One-Step kit (Bio-Rad) according to the manufacturer's protocol. A total of 25 ng RNA was used as template for every reaction. Primers used for all qPCR reactions are listed in Supplemental Table S1.
SDS–PAGE, western blotting, and antibodies
Protein isolation from cultured cells was conducted by lysing the cells with RIPA buffer as described above. Protein quantification was performed by a standard Bradford assay. SDS–PAGE and western blotting were performed as previously described (Booy et al. 2012). Subcellular fractionation was performed with the Subcellular Fraction Kit for Cultured Cells (Thermo Fisher Scientific) according to the manufacturer's protocol. The following antibodies were purchased from Thermo Fisher Scientific: Rabbit anti-SRP14 (PA5-88866); Mouse anti-GAPDH (MABC004). The Mouse anti-RNA polymerase II (05-623) and negative control antibody raised against Protein I from Neisseria gonorrhoeae (isotype control) (MABC004) antibodies were purchased from Sigma-Aldrich. The following antibodies were purchased from Proteintech: Rabbit anti-SRP9 (11195-1-AP); Rabbit anti-SRP14 (11528-1-AP). The Rabbit anti-RNA polymerase III (12825) and Mouse anti-H3K9Me3 (5327) antibodies were purchased from Cell Signalling Technologies. The Rabbit anti-H3K4Me3 (ab8580) antibody was purchased from Abcam. The Mouse anti-BrU (MI-11-3, clone 2B1) antibody was purchased from Medical & Biological Laboratories Co., Ltd.
Actinomycin D and bromouridine immunoprecipitation chase rate of decay measurements
To measure the impact of SRP9/SRP14 on the stability of 7SL and BC200 RNA, MCF-7 cells were transfected with either negative control or SRP14 siRNA 24 h prior to treatment with actinomycin D (5 µg/mL) to inhibit RNA polymerase III (Bensaude 2011). Following the treatment, an equal number of cells were collected for all indicated time points. RNA was extracted using the GeneJET RNA Purification kit (Thermo Fisher Scientific) according to the manufacturer's protocol. 7SL and BC200 RNA levels were measured by RT-qPCR as described above as the mean of two technical replicates for three biological replicates per time point. Error bars indicate ±SD. Decay rate constant (K) was calculated by fitting the data with GraphPad Prism 9 to the one-phase decay equation
where the plateau was set to 0.05. Half-life (λ) was calculated as λ = ln(2)/K. Error was presented as a 95% asymmetrical (profile-likelihood) confidence interval.
As a second method to measure the impact of SRP9/SRP14 on the stability of 7SL and BC200, we performed BRIC as described before with slight variations (Tani et al. 2012a,b). To summarize, MCF-7 cells were seeded at a cell density of 7.80 × 106 cells per 100 mm tissue culture dish to achieve ∼70% confluence after 24 h of growth. The cells were incubated at 37°C in the presence of 150 µM 5′-bromouridine (Thermo Fisher Scientific) for 24 h prior to collection. At indicated time points after replacing the treated medium with fresh, non-5′-bromouridine treated medium, cells were harvested. Cells were lysed with RIPA buffer as described above, and RNA purified with GeneJET RNA Concentration and Cleanup Micro Kit. A total of 6000 ng of total RNA was denatured at 80°C for 2 min and cooled on ice. Each sample was then diluted to a final buffer concentration of 20 mM HEPES pH 7.9, 4 mM KCl, 0.4 mM MgCl2, 80 mM NaCl, and 0.2% Igepal. A total of 2 µg of anti-BrU antibody was mixed with each sample end-over-end 4°C overnight. After antibody incubation, 10 µL of pre-equilibrated Pierce protein A/G magnetic beads (Thermo Fisher Scientific) were added to the RNA-antibody sample and mixed end-over-end at 4°C for 2 h. Following incubation, beads were subjected to four washes of 25 mM HEPES pH 7.9, 5 mM KCl, 0.5 mM MgCl2, 100 mM NaCl, and 0.25% Igepal (wash buffer). RNA from each BRIC sample was purified using a GeneJET RNA Concentration and Cleanup Micro Kit according to the manufacturer's protocol. Immunoprecipitated RNA was quantified with RT-qPCR using 2.5% of each immunoprecipitation as template for every reaction. 7SL and BC200 RNA levels were measured by RT-qPCR as described above as the mean of three technical replicates at each time point and are representative of three biological replicates. Error bars indicate ±SD. Half-life was calculated as above, except for no constraints on plateau values. Error was presented as a 95% asymmetrical (profile-likelihood) confidence interval.
Chromatin immunoprecipitation
ChIP analyses were performed in MCF-7 cells that were ∼80% confluent at the time of collection. To achieve this, MCF-7 cells were seeded for 24-, 48-, or 72-h growth periods at cell densities of 2.40 × 107, 1.56 × 107, or 1.14 × 107 cells per 150 mm tissue culture dish, respectively. Two 150 mm tissue culture dishes were used per condition analyzed. For assessment of actinomycin D impact on ChIP results, MCF-7 cells were grown for 72 h and left untreated or treated with 5 µg/mL for 90 min prior to collection. For collection, cells were crosslinked with 1% formaldehyde in phosphate-buffered saline (PBS) with rocking for 14 min. Formaldehyde was quenched with a final concentration of 125 mM glycine with rocking for 5 min. Cells were then scraped with cold PBS with 1% bovine serum albumin (BSA) and consolidated prior to centrifugation at 1000g at 4°C for 5 min. These pellets were washed with PBS three times prior to lysis with 1 mL of RIPA buffer supplemented with Halt protease and phosphatase inhibitor cocktail. Samples were sonicated with 10 cycles of 15 sec on and 15 sec off with 40% output (Branson Ultrasonics Sonifier SFX150 Cell Disruptor, Thermo Fisher Scientific) in order to shear genomic DNA to an average of ∼500 bp. Cell debris was pelleted by centrifugation at 21,000g at 4°C for 10 min. Protein content was measured with a standard Bradford assay and normalized to a concentration of 2.5 mg/mL. Antibodies utilized included anti-RNA polymerase III (12825) (1:40 dilution), isotype control (MABC004) (3 µg), anti-RNA polymerase II (05-623) (3 µg), anti-SRP9 (11195-1-AP) (3 µg), anti-SRP14 (11528-1-AP) (8 µg), anti-H3K4Me3 (ab8580) (3 µg), and anti-H3K9Me3 (5327) (3 µg). Antibodies were mixed with a total of 750 µg of lysate overnight with end-over-end mixing at 4°C. DNA input samples were collected at this stage and also incubated overnight at 4°C. DNA input samples were mixed with 100 mM sodium bicarbonate, 1% (v/v) SDS (elution buffer), made fresh to a final concentration of 55 mM sodium bicarbonate, 0.55% (v/v) SDS, 200 mM NaCl, and 2 µg of RNase A (Thermo Fisher Scientific). DNA input samples were incubated overnight at 65°C with shaking. The following day, DNA input samples were incubated with 100 µg of proteinase K (Thermo Fisher Scientific) for 1 h at 55°C with shaking. Input DNA was purified using a GeneJET PCR Purification kit (Thermo Fisher Scientific) according to the manufacturer's protocol. After antibody incubation, 40 µL of pre-equilibrated Pierce protein A/G magnetic beads were added to the lysate and mixed end-over-end at 4°C for 2 h. Following incubation, beads were subjected to two consecutive washes of each buffer listed in order: RIPA buffer, 20 mM Tris-HCl, pH 8.1, 150 mM NaCl, 1% (v/v) Triton-X-100, 0.1% (v/v) SDS, 2 mM EDTA (low salt wash buffer), 20 mM Tris-HCl, pH 8.1, 500 mM NaCl, 1% (v/v) Triton-X-100, 0.1% (v/v) SDS, 2 mM EDTA (high salt wash buffer), and 10 mM Tris-HCl, pH 8.1, 250 mM LiCl, 1% (v/v) NP-40, 1% (v/v) sodium deoxycholate, 1 mM EDTA (LiCl wash buffer). The samples were then resuspended in RIPA buffer and magnetic beads pelleted prior to elution with 200 µL of elution buffer. Eluted samples were incubated overnight at 65°C with shaking. The following day 2 µg of RNase A was added, and samples were incubated for 30 min at 37°C with shaking prior to addition of 100 µg proteinase K and incubation for 1 h at 55°C with shaking. DNA from each ChIP was collected using a GeneJET PCR Purification kit according to the manufacturer's protocol. Input and chromatin immunoprecipitated DNA samples were quantified with qPCR analyses using an Applied Biosystems StepOnePlus instrument with the SsoAdvanced Universal SYBR Green Supermix kit (Bio-Rad) according to the manufacturer's protocol. A total of 25 ng of input DNA, or 5% of each immunoprecipitation was used as template for every reaction.
Immunofluorescence
Cells used for immunofluorescence experiments were grown on #1.5 thickness glass coverslips (Thermo Fisher Scientific) in a 24-well dish. Coverslips were removed from the cell culture once confluence reached ∼80% and washed once with PBS prior to fixation with 4% formaldehyde in PBS for 10 min at room temperature. The cells were washed three times in PBS prior to permeabilization with 0.1% Triton-X 100 in PBS for 10 min at room temperature. An additional three washes in PBS for 5 min at room temperature were then conducted. Blocking was performed in PBS containing 1% BSA and 22.5 mg/mL glycine for 30 min at room temperature. Coverslips were then incubated in primary antibody in PBS containing 1% BSA for 1 h at 37°C. The anti-SRP9 (11195-1-AP, Proteintech) diluted 1:100 and anti-SRP14 (11528-1-AP, Proteintech) diluted 1:100 antibodies were utilized. Following incubation, coverslips were washed four times in PBS for 10 min at room temperature. Coverslips were then incubated with secondary antibodies in PBS containing 1% BSA for 1 h at 37°C. Goat anti-Rabbit IgG Alexa Fluor 647 conjugate (111-605-003, Jackson ImmunoResearch Laboratories) antibody diluted 1:300 was utilized. Following incubation, the coverslips were washed in PBS four times for 10 min at room temperature. The coverslips were then incubated in PBS with 0.1 µg/mL diamidino-2-phenylindole (DAPI) (62248, Thermo Fisher Scientific) for 5 min at room temperature. Coverslips were mounted onto Vectashield Antifade Mounting Media (Vector Laboratories). Cells were imaged with a 40× objective lens on an EVOS FL Auto Imaging System (Thermo Fisher Scientific).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
This work was supported by a Canadian Institutes of Health Research Project Grant (427781). D.G. was supported by the Natural Sciences and Engineering Research Council of Canada CGS-D.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.079649.123.
Freely available online through the RNA Open Access option.
MEET THE FIRST AUTHOR
Daniel Gussakovsky.

Meet the First Author(s) is an editorial feature within RNA, in which the first author(s) of research-based papers in each issue have the opportunity to introduce themselves and their work to readers of RNA and the RNA research community. Daniel Gussakovsky is the first author of this paper, “Nuclear SRP9/SRP14 heterodimer transcriptionally regulates 7SL and BC200 RNA expression.” Daniel is a PhD candidate from the McKenna laboratory in the chemistry department at the University of Manitoba. The focus of his research involves RNA–protein interactions in the context of human disease states.
What are the major results described in your paper and how do they impact this branch of the field?
The major result in this paper was the determination that the SRP9/SRP14 heterodimer was critical for, and promoted transcription of its RNA-binding partners, the Alu-like 7SL and BC200 RNA. This is a very interesting result as it summarized many of the findings in the literature regarding the importance of the first ∼50 nt of Alu and Alu-like RNA in proper transcription, posttranscriptional processing, and evolutionary conservation.
What led you to study RNA or this aspect of RNA science?
We believe that the field of noncoding RNA is understudied, with many biologically important RNA yet to be elucidated. Our laboratory has studied BC200 RNA extensively, providing a better functional understanding of the RNA, building on top of previous studies on its high RNA levels correlating with poor prognosis of several human cancers. With this study, we learned more about BC200 RNA and what factors may contribute to its initial transcription. We believe that by studying its transcription, we aid in the understanding of its initial overexpression in human cancers.
What are some of the landmark moments that provoked your interest in science or your development as a scientist?
My experiences as a summer undergraduate research student were critical in my development as a scientist. When I first began, I believed that research was simple—a hypothesis led to a result and results culminated in a story. As a summer undergraduate research student, I learned very quickly that to be successful in science, researchers had to be persistent through all the failures that occur and withstand the temptation to chase after all of the “rabbit holes” that appear in the midst of projects.
What are your subsequent near- or long-term career plans?
I hope that after I graduate from my doctoral program, I will work in a biotechnology company that researches and provides treatments for human disease states. My expertise in RNA research and technology would be an asset in any biotechnology company that leverages RNA for the purposes of novel treatments. The recent success of mRNA vaccines against COVID-19 has further supported the idea that the field of RNA therapeutics will be coveted in the future, and I am excited to contribute to it.
REFERENCES
- Akopian D, Shen K, Zhang X, Shan SO. 2013. Signal recognition particle: an essential protein-targeting machine. Annu Rev Biochem 82: 693–721. 10.1146/annurev-biochem-072711-164732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett EA, Keller H, Mills RE, Schmidt S, Moran JV, Weichenrieder O, Devine SE. 2008. Active Alu retrotransposons in the human genome. Genome Res 18: 1875–1883. 10.1101/gr.081737.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensaude O. 2011. Inhibiting eukaryotic transcription: Which compound to choose? How to evaluate its activity? Transcription 2: 103–108. 10.4161/trns.2.3.16172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger A, Ivanova E, Gareau C, Scherrer A, Mazroui R, Strub K. 2014. Direct binding of the Alu binding protein dimer SRP9/14 to 40S ribosomal subunits promotes stress granule formation and is regulated by Alu RNA. Nucleic Acids Res 42: 11203–11217. 10.1093/nar/gku822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birse DEA, Kapp U, Strub K, Cusack S, Aberg A. 1997. The crystal structure of the signal recognition particle Alu RNA binding heterodimer, SRP9/14. EMBO J 16: 3757–3766. 10.1093/emboj/16.13.3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booy EP, Meier M, Okun N, Novakowski SK, Xiong S, Stetefeld J, McKenna SA. 2012. The RNA helicase RHAU (DHX36) unwinds a G4-quadruplex in human telomerase RNA and promotes the formation of the P1 helix template boundary. Nucleic Acids Res 40: 4110–4124. 10.1093/nar/gkr1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booy EP, McRae EKS, Howard R, Deo SR, Ariyo EO, Dzananovic E, Meier M, Stetefeld J, McKenna SA. 2016. RNA helicase associated with AU-rich element (RHAU/DHX36) interacts with the 3′-tail of the long non-coding RNA BC200 (BCYRN1). J Biol Chem 291: 5355–5372. 10.1074/jbc.M115.711499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booy EP, McRae EKS, Koul A, Lin F, McKenna SA. 2017. The long non-coding RNA BC200 (BCYRN1) is critical for cancer cell survival and proliferation. Mol Cancer 16: 109. 10.1186/s12943-017-0679-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booy EP, McRae EKS, Ezzati P, Choi T, Gussakovsky D, McKenna SA. 2018. Comprehensive analysis of the BC200 ribonucleoprotein reveals a reciprocal regulatory function with CSDE1/UNR. Nucleic Acids Res 46: 11575–11591. 10.1093/nar/gky860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booy EP, Gussakovsky D, Choi T, McKenna SA. 2021. The noncoding RNA BC200 associates with polysomes to positively regulate mRNA translation in tumor cells. J Biol Chem 296: 100036. 10.1074/jbc.RA120.015775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovia F, Fornallaz M, Leffers H, Strub K. 1995. The SRP9/14 subunit of the signal recognition particle (SRP) is present in more than 20-fold excess over SRP in primate cells and exists primarily free but also in complex with small cytoplasmic Alu RNAs. Mol Biol Cell 6: 471–484. 10.1091/mbc.6.4.471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovia F, Wolff N, Ryser S, Strub K. 1997. The SRP9/14 subunit of the human signal recognition particle binds to a variety of Alu-like RNAs and with higher affinity than its mouse homolog. Nucleic Acids Res 25: 318–325. 10.1093/nar/25.2.318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawthon RM. 2009. Telomere length measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Res 37: 1–7. 10.1093/nar/gkn1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DY, Newitt JA, Hsu K, Bernstein HD, Maraia RJ. 1997. A highly conserved nucleotide in the Alu domain of SRP RNA mediates translation arrest through high affinity binding to SRP9/14. Nucleic Acids Res 25: 1117–1122. 10.1093/nar/25.6.1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Böcker W, Brosius J, Tiedge H. 1997. Expression of neural BC200 RNA in human tumours. J Pathol 183: 345–351. [DOI] [PubMed] [Google Scholar]
- Chen Y, Sinha K, Perumal K, Gu J, Reddy R. 1998. Accurate 3′ end processing and adenylation of human signal recognition particle RNA and Alu RNA in vitro. J Biol Chem 273: 35023–35031. 10.1074/jbc.273.52.35023 [DOI] [PubMed] [Google Scholar]
- Contreras-Galindo R, Fischer S, Saha AK, Lundy JD, Cervantes PW, Mourad M, Wang C, Qian B, Dai M, Meng F, et al. 2017. Rapid molecular assays to study human centromere genomics. Genome Res 27: 2040–2049. 10.1101/gr.219709.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel C, Silberberg G, Behm M, Öhman M. 2014. Alu elements shape the primate transcriptome by cis-regulation of RNA editing. Genome Biol 15: R28. 10.1186/gb-2014-15-2-r28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewannieux M, Esnault C, Heidmann T. 2003. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet 35: 41–48. 10.1038/ng1223 [DOI] [PubMed] [Google Scholar]
- Emde G, Frontzek A, Benecke B-J. 1997. Secondary structure of the nascent 7SL RNA mediates efficient transcription by RNA polymerase III. RNA 3: 538–549. [PMC free article] [PubMed] [Google Scholar]
- Geiduschek EP, Tocchini-Valentini GP. 1988. Transcription by RNA polymerase III. Annu Rev Biochem 57: 873–914. 10.1146/annurev.bi.57.070188.004301 [DOI] [PubMed] [Google Scholar]
- Grosshans H, Deinert K, Hurt E, Simos G. 2001. Biogenesis of the signal recognition particle (SRP) involves import of SRP proteins into the nucleolus, assembly with the SRP-RNA, and Xpo1p-mediated export. J Cell Biol 153: 745–761. 10.1083/jcb.153.4.745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gussakovsky D, McKenna SA. 2021. Alu RNA and their roles in human disease states. RNA Biol 18: 574–585. 10.1080/15476286.2021.1989201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halic M, Becker T, Pool MR, Spahn CMT, Grassucci RA, Frank J, Beckmann R. 2004. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature 427: 808–814. 10.1038/nature02342 [DOI] [PubMed] [Google Scholar]
- Häsler J, Strub K. 2006. Alu RNP and Alu RNA regulate translation initiation in vitro. Nucleic Acids Res 34: 2374–2385. 10.1093/nar/gkl246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häsler J, Samuelsson T, Strub K. 2007. Useful “junk”: Alu RNAs in the human transcriptome. Cell Mol Life Sci 64: 1793–1800. 10.1007/s00018-007-7084-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XP, Bataillé N, Fried HM. 1994. Nuclear export of signal recognition particle RNA is a facilitated process that involves the Alu sequence domain. J Cell Sci 107: 903–912. 10.1242/jcs.107.4.903 [DOI] [PubMed] [Google Scholar]
- Hsu K, Chang DY, Maraia RJ. 1995. Human signal recognition particle (SRP) Alu-associated protein also binds Alu interspersed repeat sequence RNAs. J Biol Chem 270: 10179–10186. 10.1074/jbc.270.17.10179 [DOI] [PubMed] [Google Scholar]
- Hu T, Lu YR. 2015. BCYRN1, a c-MYC-activated long non-coding RNA, regulates cell metastasis of non-small-cell lung cancer. Cancer Cell Int 15: 1–8. 10.1186/s12935-015-0183-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacoangeli A, Lin Y, Morley EJ, Muslimov IA, Bianchi R, Reilly J, Weedon J, Diallo R, Böcker W, Tiedge H. 2004. BC200 RNA in invasive and preinvasive breast cancer. Carcinogenesis 25: 2125–2133. 10.1093/carcin/bgh228 [DOI] [PubMed] [Google Scholar]
- Iakhiaeva E, Yin J, Zwieb C. 2005. Identification of an RNA-binding domain in human SRP72. J Mol Biol 345: 659–666. 10.1016/j.jmb.2004.10.087 [DOI] [PubMed] [Google Scholar]
- Jurka J. 2004. Evolutionary impact of human Alu repetitive elements. Curr Opin Genet Dev 14: 603–608. 10.1016/j.gde.2004.08.008 [DOI] [PubMed] [Google Scholar]
- Labuda D, Zitkiewicz E. 1994. Evolution of secondary structure in the family of 7SL-like RNAs. J Mol Evol 39: 506–518. 10.1007/BF00173420 [DOI] [PubMed] [Google Scholar]
- Lakkaraju AKK, Luyet PP, Parone P, Falguières T, Strub K. 2007. Inefficient targeting to the endoplasmic reticulum by the signal recognition particle elicits selective defects in post-ER membrane trafficking. Exp Cell Res 313: 834–847. 10.1016/j.yexcr.2006.12.003 [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, Fitzhugh W, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- Liu WM, Maraia RJ, Rubin CM, Schmid CW. 1994. Alu transcripts: cytoplasmic localisation and regulation by DNA methylation. Nucleic Acids Res 22: 1087–1095. 10.1093/nar/22.6.1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Lu X, Xie H. 2014. Dynamic Alu methylation during normal development, aging, and tumorigenesis. Biomed Res Int 2014: 784706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutcke H, Prehn S, Ashford AJ, Remus M, Frank R, Dobberstein B. 1993. Assembly of the 68- and 72-kD proteins of signal recognition particle with 7S RNA. J Cell Biol 121: 977–985. 10.1083/jcb.121.5.977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martignetti JA, Brosius J. 1993. BC200 RNA: a neural RNA polymerase III product encoded by a monomeric Alu element. Proc Natl Acad Sci 90: 11563–11567. 10.1073/pnas.90.24.11563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mary C, Scherrer A, Huck L, Lakkaraju AKK, Thomas Y, Johnson AE, Strub K. 2010. Residues in SRP9/14 essential for elongation arrest activity of the signal recognition particle define a positively charged functional domain on one side of the protein. RNA 16: 969–979. 10.1261/rna.2040410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massenet S. 2019. In vivo assembly of eukaryotic signal recognition particle: a still enigmatic process involving the SMN complex. Biochimie 164: 99–104. 10.1016/j.biochi.2019.04.007 [DOI] [PubMed] [Google Scholar]
- Nabet BY, Qiu Y, Shabason JE, Wu TJ, Yoon T, Kim BC, Benci JL, DeMichele AM, Tchou J, Marcotrigiano J, et al. 2017. Exosome RNA unshielding couples stromal activation to pattern recognition receptor signaling in cancer. Cell 170: 352–366.e13. 10.1016/j.cell.2017.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perumal K, Sinha K, Henning D, Reddy R. 2001. Purification, characterization, and cloning of the cDNA of human signal recognition particle RNA 3′-adenylating enzyme. J Biol Chem 276: 21791–21796. 10.1074/jbc.M101905200 [DOI] [PubMed] [Google Scholar]
- Politz JC, Yarovoi S, Kilroy SM, Gowda K, Zwieb C, Pederson T. 2000. Signal recognition particle components in the nucleolus. Proc Natl Acad Sci 97: 55–60. 10.1073/pnas.97.1.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Eskin E, Pevzner PA. 2004. Whole-genome analysis of Alu repeat elements reveals complex evolutionary history. Genome Res 14: 2245–2252. 10.1101/gr.2693004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quentin Y. 1992a. Fusion of a free left Alu monomer and a free right Alu monomer at the origin of the Alu family in the primate genomes. Nucleic Acids Res 20: 487–493. 10.1093/nar/20.3.487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quentin Y. 1992b. Origin of the Alu family: a family of Alu-like monomers gave birth to the left and the right arms of the Alu elements. Nucleic Acids Res 20: 3397–3401. 10.1093/nar/20.13.3397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson J, Cronin S, Dean K. 2018. BC200 (BCYRN1)—the shortest, long, non-coding RNA associated with cancer. Non-coding RNA Res 3: 131–143. 10.1016/j.ncrna.2018.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel V, Walter P. 1986. Removal of the Alu structural domain from signal recognition particle leaves its protein translocation activity intact. Nature 320: 81–84. 10.1038/320081a0 [DOI] [PubMed] [Google Scholar]
- Sinnett D, Richer C, Deragon JM, Labuda D. 1991. Alu RNA secondary structure consists of two independent 7 SL RNA-like folding units. J Biol Chem 266: 8675–8678. 10.1016/S0021-9258(18)31497-2 [DOI] [PubMed] [Google Scholar]
- Sommerville J, Brumwell CL, Ritland Politz JC, Pederson T. 2005. Signal recognition particle assembly in relation to the function of amplified nucleoli of Xenopus oocytes. J Cell Sci 118: 1299–1307. 10.1242/jcs.01726 [DOI] [PubMed] [Google Scholar]
- Stenz L. 2021. The L1-dependant and Pol III transcribed Alu retrotransposon, from its discovery to innate immunity. Mol Biol Rep 48: 2775–2789. 10.1007/s11033-021-06258-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strub K, Walter P. 1990. Assembly of the Alu domain of the signal recognition particle (SRP): dimerization of the two protein components is required for efficient binding to SRP RNA. Mol Cell Biol 10: 777–784. 10.1128/mcb.10.2.777-784.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strub K, Moss J, Walter P. 1991. Binding sites of the 9- and 14-kilodalton heterodimeric protein subunit of the signal recognition particle (SRP) are contained exclusively in the Alu domain of SRP RNA and contain a sequence motif that is conserved in evolution. Mol Cell Biol 11: 3949–3959. 10.1128/mcb.11.8.3949-3959.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani H, Imamachi N, Salam KA, Mizutani R, Ijiri K, Irie T, Yada T, Suzuki Y, Akimitsu N. 2012a. Identification of hundreds of novel UPF1 target transcripts by direct determination of whole transcriptome stability. RNA Biol 9: 1370–1379. 10.4161/rna.22360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani H, Mizutani R, Salam KA, Tano K, Ijiri K, Wakamatsu A, Isogai T, Suzuki Y, Akimitsu N. 2012b. Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. Genome Res 22: 1382. 10.1101/gr.130559.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedge H, Chen W, Brosius J. 1993. Primary structure, neural-specific expression, and dendritic location of human BC200 RNA. J Neurosci 13: 2382–2390. 10.1523/JNEUROSCI.13-06-02382.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullu E, Tschudi C. 1984. Alu sequences are processed 7SL RNA genes. Nature 312: 171–172. 10.1038/312171a0 [DOI] [PubMed] [Google Scholar]
- Walter P, Johnson AE. 1994. Signal sequence recognition and protein targeting. Annu Rev Cell Biol 10: 87–119. 10.1146/annurev.cb.10.110194.000511 [DOI] [PubMed] [Google Scholar]
- Weichenrieder O, Kapp U, Cusack S, Strub K. 1997. Identification of a minimal Alu RNA folding domain that specifically binds SRP9/14. RNA 3: 1262–1274. [PMC free article] [PubMed] [Google Scholar]
- Weichenrieder O, Stehlin C, Kapp U, Birse DE, Timmins PA, Strub K, Cusack S. 2001. Hierarchical assembly of the Alu domain of the mammalian signal recognition particle. RNA 7: 731–740. 10.1017/S1355838201010160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JA, Walsh CP, Bestor TH. 1997. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet 13: 335–340. 10.1016/S0168-9525(97)01181-5 [DOI] [PubMed] [Google Scholar]