Abstract
Purpose
As a member of the C19MC family, miR-526b-5p is mainly expressed in the placental tissue and is a well-known tumor suppressor microRNA. However, its effect on the function of trophoblasts and its role in the development of recurrent spontaneous abortion (RSA) remains unclear.
Methods
Transcriptome sequencing, quantitative real-time polymerase chain reaction (RT-qPCR), Western blot, 5-ethynyl-2′-deoxyuridine (Edu) proliferation analysis, cell counting kit-8 (CCK8) assay, Transwell assays, and wound healing were used to detect the proliferation, migration, and invasion capacity of trophoblasts. Target genes of miR-526b-5p were obtained by the dual luciferase reporter system. The promoter-reporter system and ChIP-qPCR were used to prove that c-Myc positively regulated the expression of Foxp1
Results
The miR-526b-5p levels were significantly higher in patients with RSA than in controls. High expression of miR-526b-5p inhibited the proliferation, migration, and invasion of trophoblast cell line. By contrast, low expression of miR-526b-5p promoted the proliferation and migration of trophoblast cell line. Target genes of miR-526b-5p were c-Myc and Foxp1. c-Myc positively regulated the expression of Foxp1 by binding to the Foxp1 promoter location -146/-135. Finally, miR-526b-5p impeded the proliferation, migration, and invasion of trophoblasts by negatively regulating c-Myc by rescue experiments.
Conclusion
Thus, miR-526b-5p affected the proliferation, migration, and invasion of trophoblasts by targeting c-Myc and Foxp1. Low expression of c-Myc further deactivated the positive transcriptional regulation of c-Myc on Foxp1, which may be the mechanism of RSA. This study provides potential therapeutic targets and clues for the diagnosis and treatment of RSA.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10815-023-02793-0.
Keywords: miR-526b-5p, c-Myc, Foxp1, Trophoblast, RSA
Introduction
Recurrent spontaneous abortion (RSA) refers to three or more spontaneous abortions before 28 weeks of gestation. This phenomenon is one of the common pregnancy complications in obstetrics and gynecology, with an incidence of 3%–5% in reproductive-age couples. RSA seriously impacts the physical and mental health of women of reproductive age [1, 2]. In addition to known factors, such as immunodeficiency, endocrine disorder, and chromosomal abnormalities, the etiology of nearly 50% of patients with RSA remains unknown [3]. The placenta is an essential organ for maintaining the growth and development of the fetus and has the functions of material exchange, defense, synthesis, and immunity. Extravillous trophoblasts are critical cells in the placental tissue and involved in embryo implantation, placenta formation, and immune protection [4]. The proliferation, migration, and invasion of trophoblasts play an essential role in vascular remodeling, oxygen concentration regulation, and endometrial decidualization [5, 6]. Abnormal angiogenesis caused by defects in the proliferation, migration, and invasion of trophoblast cells is one of the most important reasons for RSA. The microRNA (miRNA) is an important factor affecting cell proliferation, migration, and invasion.
miRNAs are noncoding RNAs involved in regulating posttranscriptional gene silencing. These molecules usually inhibit translation or promote mRNA degradation by binding to the 3′ untranslated regions (UTR) or coding sequence (CDS) of target genes [7–9]. During placental development, miRNAs are involved in physiological processes, such as trophoblast proliferation, migration, adhesion, apoptosis, and angiogenesis. The dysregulated expression of miRNAs can cause pregnancy-related complications, such as preeclampsia, inflammation, and RSA [10–12].
The chromosome 19 miRNA cluster (C19MC) is the largest in humans, spanning 100 kb, with 46 highly homologous miRNA genes, and mainly expressed in the placenta tissues [13, 14]. miR-526b-5p is one of the miRNAs encoded by C19MC. Current studies have confirmed that miR-526b-5p is dysregulated in various tumors, plays a tumor suppressor role, and participates in the regulation of tumor cell proliferation, metastasis, apoptosis, and embryonic development [15–17]. The biological behavior of trophoblasts with infiltrating ability has many similarities with tumor cells. However, the effect of miR-526b-5p on the trophoblast cells and its role in the occurrence of RSA is unclear.
Materials and methods
Clinical sample collection
Clinical samples were obtained from the Obstetrics Department of the First Affiliated Hospital of Chongqing Medical University. The control group had voluntary artificial termination of pregnancy. By contrast, the RSA group had three or more unexplained abortions (excluding villous chromosome abnormalities and other maternal-related diseases). The gestational age of the voluntary artificial termination of pregnancy and RSA was between 8 and 12 weeks.
Cell culture and treatment
HTR-8/SVneo was a gift from Professor Charles H. Graham of the Queen’s University in Canada, and JEG3 was provided by Professor Yan-Ling Wang from the Institute of Zoology, Chinese Academy of Sciences in China. HTR-8/SVneo cells were cultured in the cell culture medium supplemented with 10% fetal bovine serum (FBS, Gibco, USA), 90% RPMI 1640 medium (Gibco, USA), and double antibiotics (100 U/mL penicillin and 100 μg/mL streptomycin; Beyotime, China). The JEG3 cells were cultured in the cell culture medium supplemented with 10% FBS, 90% MEM medium (Gibco, USA), and double antibiotics. The human embryonic kidney (HEK) 293T cell line (catalog number: SCSP-502) was obtained from ATCC (Manassas, VA, USA) and cultured in the cell culture medium supplemented with 10% FBS, 90% DMEM (Gibco, USA), and antibiotics. All cells were cultured at 37 °C in 5% CO2.
Transfection
HEK-293T was inoculated in 60 mm dishes. When the cell density reached ~80%, the medium was replaced with a fresh medium. At the ratio of 4:2:1, 8 μg of the plasmid (CD513B-1 [4.57 μg], psPAX2 [2.28 μg], and pVSVG [1.15 μg]) were added into 250 μL of opti-mem. Then, 12 μL of Lipo8000™ was added, and the mixture was added dropwise to the HEK-293T cells. After 8 h, the HEK-293T cells were replaced with fresh medium. After 36 h, the supernatant of HEK-293T cells was collected, filtered through a 0.45 μm filter, and stored at 4 °C for later use.
HTR-8/SVneo cells were seeded in 60 mm dishes. When the cell density reached ~80%, the viral supernatant and HTR-8/SVneo cell culture medium 1:1 were mixed. Then, polybrene was added to a final concentration of 8 μg/mL. After 12 h, the HTR-8/SVneo cells were replaced with fresh medium. After another 12 h, puromycin was added at a final concentration of 6 μg/mL. After 12 h of drug screening, the medium was replaced with fresh medium, and the concentration of puromycin was changed to 1 μg/mL and maintained.
qRT-PCR
RNAiso reagent (TaKaRa, Japan) was used to extract the total cellular RNA, and PrimeScript™ RT kit (TaKaRa, Japan) was used to obtain the cDNA. SYBR Premix Ex Taq II real-time PCR kit (TaKaRa, JAPAN) was used to detect the mRNA expression of the gene. GAPDH was set as the internal control. For the miRNAs, Mir-X™ miRNA first-strand synthesis (TaKaRa, JAPAN) was used for reverse transcription, and TB Green™ qRT-PCR (TaKaRa, JAPAN) was used to amplify the cDNA. U6 was set as the internal control. The relative differential mRNA expression of each gene or miRNA was calculated by 2−ΔΔCt. The primers for qRT-PCR are shown in Table S1.
Western blot
The protein was extracted by BCA (Transgen, China). Western blot was performed according to normal operations [18]. The primary antibodies used for Western blot were c-Myc (E5Q6W, 1:1000 dilution, CST, USA), Foxp1 (A12685, 1:500 dilution, ABclonal, China), and Actin (1:2000 dilution, BOSTER, China).
Dual-luciferase reporter assay
The online website (http://www.mirbase.org) was used to find the mature sequence of miR-526b-5p. The online site (http://asia.ensembl.org/Homo_sapiens/Tools/Blast?db=) was used to find the precursor sequence of miR-526b-5p. A 455-bp fragment containing the seed region of miR-526b-5p was cloned into the pCD513B-1 backbone vector (Promega, USA) by restriction sites EcoRI and BamHI (NEB, USA). Finally, the miR-526b-5p precursor expression vector (miR-526b-5p) was obtained.
NCBI (https://www.ncbi.nlm.nih.gov) was searched to find the 3′-UTR sequence of c-Myc and Foxp1. With human cDNA as a template, a 322-bp fragment of c-Myc and a 365-bp fragment of Foxp1 3′-UTR were amplified as wild-type fragments. Mutant fragments were obtained by overlap extension PCR. These fragments were cloned into the psiCHECK2 backbone vector (Promega, USA) by restriction sites NotI and XhoI. Wild and mutant 3′-UTR expression vectors of c-Myc and Foxp1 were obtained.
HEK-293T cells were seeded in 24-well plates and transfected when the cell density reached 70%. The miR-526b-5p overexpression vector and a gene 3′-UTR expression vector were cotransfected into HEK-293T cells with a ratio of 10:1. After 36 h of cell transfection, the fluorescence activities of firefly and Renilla were measured according to the instructions of the double luciferase reporting system kit. The final result was expressed as dividing Renilla by firefly.
CCK-8 proliferation assay
After transfection, HTR-8/SVneo cells were seeded into 96-well plates with a density of 2.0 × 103 cells/well. The test group was cultured with 90 μL of CCK8 solution (NCM Biotech, China) and 10 μL of DMEM, and the control group was cultured with 100 μL of DMEM. The cells were incubated for 2 h. Finally, absorbance at 450 nm was measured using a microplate reader to assess cell viability. The cell proliferation ability was detected continuously for 72 h with a cycle of 12 h. After 72 h, the cell proliferation curve was plotted by Excel. Each parallel sample had 5 wells, and this experiment was repeated three times.
5-Ethynyl-2′-deoxyuridine (EdU) proliferation assay
HTR-8/SVneo cells were seeded into 96-well plates. According to the manufacturer’s (Beyotime, China) instructions, 50 μM EdU medium and Apollo staining solution were prepared. The cells were incubated with EdU medium for 2 h. After the cells were washed, fixed, and permeabilized, they were stained with Apollo staining solution for 30 min. DAPI staining solution was added for nuclei staining. A fluorescence microscope was used to observe the cells, and ImageJ software was used to quantify the results.
Wound healing assay
Cells were plated in 6-well plates and cultured to 100% confluency. The triangular ruler was wiped with an alcoholic cotton ball and irradiated with UV light. The cells in each well were scratched using a sterile pipette tip and marked with a marker. The marked position was photographed with an inverted microscope. After the scraped cells were replaced with the culture medium, photographs were taken at the 0th, 12th, 24th, and 36th hours. ImageJ software was used to open the picture. Six horizontal lines were drawn randomly, and the average distance between cells was calculated.
Transwell assay
Transwell plates were coated with 60 μL of Matrigel (1:6 dilution of Matrigel with cold basal medium) overnight at 37 °C. Cell culture medium containing 10% FBS was added to the lower chamber. The cells were resuspended at 1 × 105 cells/well density, seeded into the upper chamber, and incubated at 37 °C and 5% CO2 for 24 h. Afterward, cells were fixed in 4% paraformaldehyde for 10 min and stained in 0.5% crystal violet for 10 min. Under the light microscope, five fields of view were randomly selected, and the average number of invading cells in each chamber was calculated. The transwell system has 0.8 μm chambers.
RNA-sequencing analysis
The miR-526b-5p overexpressing HTR8 cell line was used as the experimental group. The pCD513B-1 backbone vector cell line was transfected as the control group and sent to the sequencing company (Novogene, Beijing) for transcriptome sequencing. Thus, the mRNA with polyA structure in the total RNA was enriched by Oligo(dT) magnetic beads. The RNA was broken into fragments of ~300 bp in length by ion interruption. RNA was used as a template with 6-base random primers and reversals. The first strand of cDNA was synthesized by recording enzyme, and the second strand of cDNA was synthesized using the first strand of cDNA as a template. After the library was constructed, the library fragments was enriched by PCR amplification. Then, the library was selected according to the fragment size. The library size was 450 bp. Then, the library was inspected by Agilent 2100 Bioanalyzer. After RNA extraction, purification, and library construction of the samples, the next-generation sequencing (NGS) technology was adopted based on the Illumina sequencing platform. Paired-end (PE) sequencing was performed using these libraries. HTSeq was used to compare the read count value of each gene with the original expression level of the gene. FPKM adopted the standard on express (normalization), and DESeq was used for differences in gene expression analysis. Differentially expressed gene conditions were screened as follows: the expression differences multiples |log2FoldChange|>1 and significant P-value is < 0.05. TopGO was used for GO enrichment analysis. In the analysis, gene list and gene number of each term were calculated using the differentially annotated genes in the GO term. Then, P-value was calculated by the hypergeometric distribution method (P-value<0.05 was the criterion for significant enrichment).
Promoter reporter assay
NCBI (https://www.ncbi.nlm.nih.gov) was used to find the CDS region of c-Myc. The c-Myc overexpression vector was obtained with the pCD513B-1 backbone vector. Through the UCSC website (http://genome.ucsc.edu), the sequence information of the Foxp1 promoter was obtained (~1922 bp before the transcription start site and 171 bp after the transcription start site). Foxp1 promoter reporter vector was constructed with pGL4.10 as the skeleton vector. The mutant Foxp1 promoter reporter vector was constructed by overlapping extension PCR. pGL4.73 was used as the internal reference fluorescence report carrier. The promoter-reporter vector, c-Myc overexpression vector, and pGL4.73 were cotransfected into 293T cells. Cells were collected at 36 h after transfection for promoter activity assay. The luciferase activity was determined according to the instructions of the TransDetect Double Luciferase Report Kit (Transgen, Beijing). The ratio of firefly luciferase to Renilla luciferase is the final result.
Chromatin immunoprecipitation (ChIP) assay
According to the manufacturer’s instructions of the Pierce® Agarose ChIP kit (Thermo, USA), the binding activity of c-Myc and Foxp1 was detected. Thus, long-strand DNA was decomposed into DNA fragments through the unwinding and ultrasound of DNA and protein complexes. DNA fragments were incubated overnight with specific antibody c-Myc (E5Q6W, CST, USA) and control IgG. The next day, antibody-binding magnetic beads were added. Proteinase K was used to elute and digest the immunoprecipitated product. The purified DNA product was used for subsequent PCR detection (Table S2). Mouse IgG was used as the negative control, and total chromatin was used as the input (positive control).
Bioinformatics analysis
The online website (http://www.mirbase.org) was used to find the mature sequence of miR-526b-5p. The online site (http://asia.ensembl.org/Homo_sapiens/Tools/Blast?db=) was used to find the precursor sequence of miR-526b-5p. The online website TargetScan Human 7.2 was used to find the downstream target genes of miR-526b-5p. The NCBI database (https://www.ncbi.nlm.nih.gov) and the UCSC database (http://genome.ucsc.edu) were used to query the DNA sequences. The online website (http://jaspar.genereg.net, http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3) was used to analyze the Foxp1 promoter region transcriptional regulatory elements. Primer Premier 5.0 was used to design the PCR primers.
Statistical analysis
GraphPad Prism 7.0 software was used for data statistics and analysis. ImageJ software was used for cell count and grey scale. Data were presented as the mean ± standard deviation. Student’s t-test or ANOVA was used in this experiment. P < 0.05 (*), P < 0.01 (**), and P < 0.001 (***) were set as statistically significant. All experiments in this study were repeated at least three times.
Results
miR-526b-5p, c-Myc, and Foxp1 were abnormally expressed in the placental villi of patients with RSA
The relative expression of miR-526b-5p was detected by RT-qPCR. The presentation of miR-526b-5p was higher in patients with RSA than those with normal placental villi tissue (P=0.0294) (Fig. 1). The mRNA expression of c-Myc and Foxp1 was also examined. The presentation of c-Myc and Foxp1 was significantly lower in the patients with RSA than in the regular group (P=0.0043; P=0.0001) (Fig. 1). The results suggested that the abnormal expression of miR-526b-5p, c-Myc, and Foxp1 in RSA may affect the function of trophoblast cells and be related to the formation of RSA.
Fig. 1.
miR-526b-5p, c-Myc, and Foxp1 were abnormally expressed in the placental villi of patients with recurrent spontaneous abortion (RSA). The expression of miR-526b-5p, c-Myc, and Foxp1 in normal pregnancy villi and patients with RSA was detected by RT-qPCR (normal group-voluntary artificial termination of pregnancy; patients with RSA-three or more unexplained abortions [excluding villous chromosome abnormalities and other maternal-related diseases]). The results are presented as mean ± standard deviation (SD; n = 18 in the control group; n = 21 in the RSA group). Statistical significance was analyzed by Student’s t-test. *P < 0.05, **P < 0.01, and ***P < 0.001
Overexpression of miR-526b-5p inhibited the proliferation, invasion, and migration of the trophoblasts
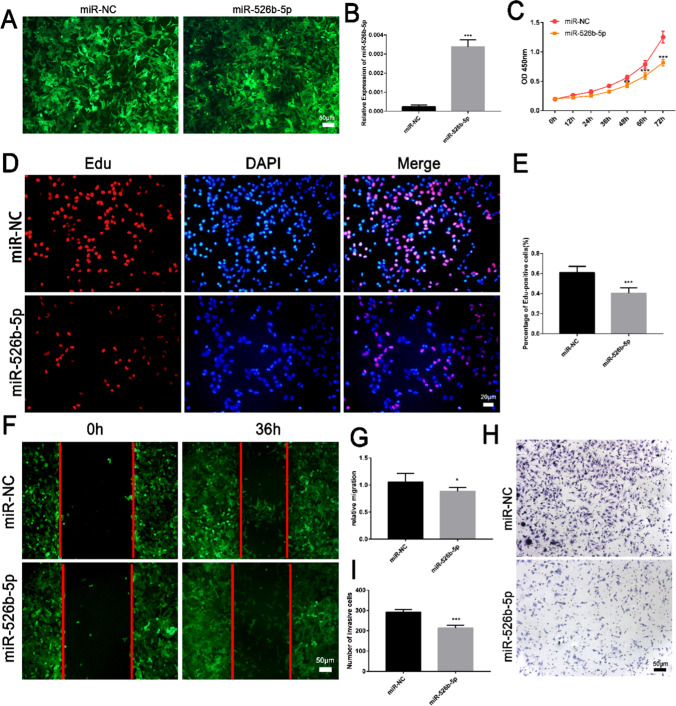
To clarify the function of miR-526b-5p in the development of RSA, the miR-526b-5p overexpression vector was constructed. The vector was transfected into the HTR-8/SVneo trophoblast cells by lentiviral packaging (Fig. 2A). The overexpression effect of miR-526b-5p was as expected (Fig. 2B). The cell proliferation curve drawn by CCK8 assay showed that miR-526b-5p inhibited the proliferation ability of the trophoblast cells (Fig. 2C). miR-526b-5p decreased the number of Edu staining-positive cells (Fig. 2D, E). The migration ability of the trophoblast cells was detected by a wound healing test. The migration ability of the cells after overexpression of miR-526b-5p was lower than the control group (Fig. 2F, G). miR-526b-5p inhibited the invasion of trophoblast cells as shown by the Transwell cell invasion assay (Fig. 2H, I). The results suggested that miR-526b-5p inhibited the proliferation, migration, and invasion of the trophoblasts.
Fig. 2.
Overexpression of miR-526b-5p inhibited trophoblast proliferation, invasion, and migration. A Schematic diagram of the growth state of HTR-8/SVneo trophoblast cells transfected with miR-526b-5p or miR-NC (scale bar = 50 μm). B The expression of miR-525-5p was detected by RT-qPCR. C The proliferation ability of HTR-8/SVneo cells was detected by cell counting kit-8 (CCK8) assay. D The proliferation ability of HTR-8/SVneo cells was detected by 5-ethynyl-2′-deoxyuridine (Edu) staining. EdU-positive cells are shown in red, and cell nuclei are shown in blue (DAPI, 4′,6-diamidino-2-phenylindole; scale bar = 20 μm). E ImageJ software was used to count the number of EdU-positive cells. F, G The migration ability of HTR-8/SVneo cells was detected by wound healing assay (scale bar = 50 μm). Quantitative analysis of the detection results of wound healing assay by ImageJ software. H, I The invasive ability of HTR-8/SVneo cells was detected by Transwell assay (scale bar = 50 μm). Quantitative analysis of the detection results of Transwell assay by ImageJ software. The results are presented as mean ± SD (n = 6). *P < 0.05, **P < 0.01, and ***P < 0.001 (miR-526b-5p, miR-526b-5p precursor expression vector; miR-NC, pCDH513B-1 empty vector)
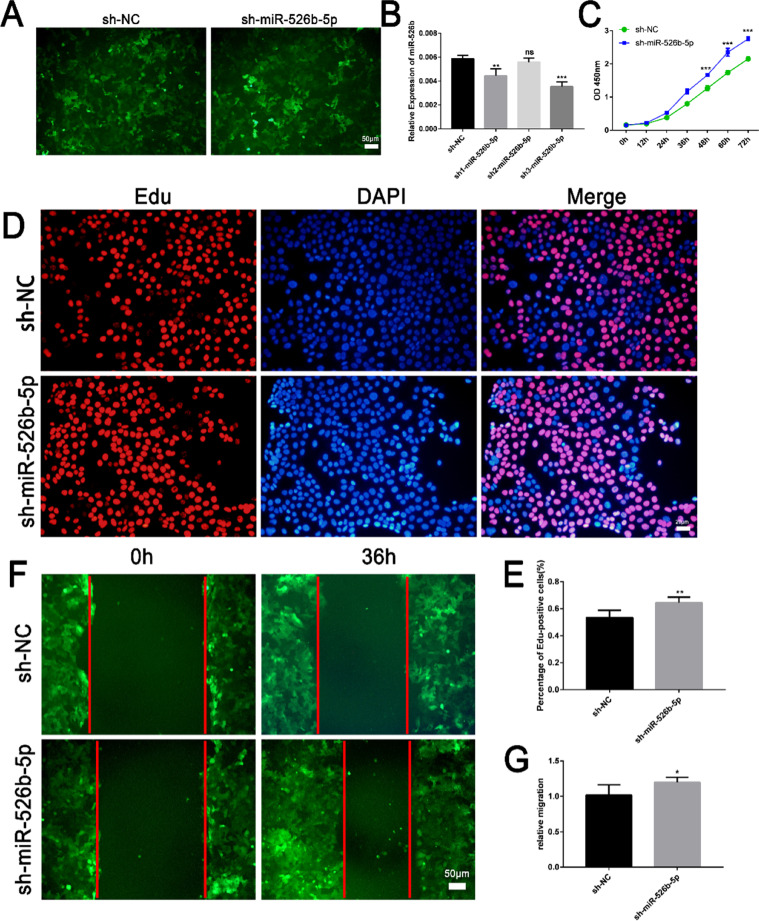
Knockdown of miR-526b-5p promoted the proliferation and migration of trophoblasts
To fully understand the role of miR-526b-5p in the trophoblast, the expression of miR-526b-5p was knocked down in the JEG3 cells (Figs. 3A, and B). The proliferation ability, migration, and invasion ability were detected by CCK8 assay (Fig. 3C), Edu staining (Figs. 3D, and E), and wound healing (Figs. 3F, and G). Given the large cell size of JEG3, the cell chamber was difficult to penetrate. Thus, the invasion ability of JEG3 after miR-526b-5p knockdown was not detected. The results suggested that the knockdown of miR-526b-5p promoted the proliferation and migration ability of the trophoblasts.
Fig. 3.
Knockdown of miR-526b-5p promoted the proliferation and migration of trophoblasts. A Schematic diagram of the growth state of JEG3 trophoblast cells transfected with sh-miR-526b-5p or sh-NC (scale bar = 50 μm). B The expression of miR-525-5p was detected by RT-qPCR. C The proliferation ability of JEG3 cells was detected by CCK8 assay. D The proliferation ability of JEG3 cells was detected by Edu staining. EdU-positive cells are shown in red, and cell nuclei are shown in blue (scale bar = 20 μm). E ImageJ software was used to count the number of EdU-positive cells. F The migration ability of JEG3 cells was detected by wound healing assay (scale bar = 50 μm). G Quantitative analysis of the detection results of wound healing assay by ImageJ software. The results are presented as mean ± SD (n = 6). *P < 0.05, **P < 0.01, and ***P < 0.001 (sh-miR-526b-5p, miR-526b-5p interference vector; sh-NC, pSHI empty vector)
Transcriptome analysis of overexpressed miR-526b-5p in trophoblasts
To verify the function of miR-526b-5p on the trophoblast cells and explore the intrinsic molecular mechanism, transcriptome sequencing was performed on the HTR8 cells overexpressing miR-526b-5p. The miR-526b-5p overexpressing HTR8 cell line was used as the experimental group. The pCD513B-1 empty vector cell line was transfected as the control group. A total of 172 genes were upregulated, and 361 genes were downregulated (Fig. 4A). The differentially expressed genes were enriched in the cell proliferation function as shown by GO analysis (Fig. 4B). RT-qPCR was used to verify the genes related to cell proliferation, migration, and invasion (Fig. 4C), which were consistent with the transcriptome sequencing results.
Fig. 4.
Transcriptome analysis of overexpressed miR-526b-5p in trophoblasts. A Volcano diagram of differentially expressed genes. The abscissa is the log2FoldChange, and the ordinate is −log10 (P-value). The two vertical dashed lines are the 2-fold differential expression threshold. The dotted line indicates the threshold of P-value=0.05. The red dots are the upregulated genes, the blue dots are the downregulated genes, and the grey dots indicate nonsignificant differentially expressed genes. B Bar chart of GO enrichment analysis. The abscissa is the term of Go level2, and the ordinate is the enriched −log10 (P-value) of each term. GO classification was carried out according to molecular function (MF), biological process (BP), and cell component (CC), and the top 10 GO terms with the minimum P-value, namely, the most significant enrichment, were selected for display in each GO classification. C The expression of CDK2, EGFR, P53, and MMP1 was verified by quantitative RT-qPCR. The results are presented as the mean ± SD (n = 6). *P < 0.05, **P < 0.01, and ***P < 0.001 (miR-526b-5p, miR-526b-5p precursor expression vector; miR-NC, pCDH513B-1 empty vector)
miR-526b-5p targeted the 3′-UTR of c-Myc and Foxp1
To obtain the targets of miR-526b-5p, Venn analysis was first performed using the downstream target genes of miR-526b-5p predicted by TargetScan and the downregulated genes in the transcriptome sequencing results (Fig. S1A). The expression of 43 target genes was verified by RT-qPCR. Among these genes, 6 genes (i.e., BMPER, NEDD4L, Foxp1, c-Myc, SASH1, and EMP1) were expressed as expected (Fig. S1B). Therefore, 6 targets of miR-526b-5p were initially determined.
miR-526b-5p, miR-NC, sh-miR-526b-5p, and sh-NC were transfected into the trophoblasts. The results from RT-qPCR (Fig. 5A) and Western blot analysis (Fig. 5B, C) showed that miR-526b-5p negatively regulated the expression of c-Myc and Foxp1. The structure and stability of the binding sites of miR-526b-5p and c-Myc 3′-UTR were predicted by the online software RNAhybrid. The minimum free energy of binding sites was −20.8 kcal/mol (Fig. 5D). The relative luciferase activity of trophoblasts with c-Myc-WT 3′-UTR was significantly reduced by using the dual-luciferase reporter system (Fig. 5E). The TargetScan results showed two binding sites for miR-526b-5p and Foxp1. RNAhybrid predicted the structure and stability of these two binding sites, and their minimum free energies were −17.1 kcal/mol and −20.1 kcal/mol, respectively (Fig. 5F, H). Finally, the dual luciferase reporter system showed that the binding of miR-526b-5p to Foxp1-WT2 decreased the relative fluorescence activity of the trophoblasts (Fig. 5G, I). The results showed that miR-526b-5p was negatively correlated with the expression of c-Myc and Foxp1 and had a targeted binding effect on c-Myc and Foxp1.
Fig. 5.
miR-526b-5p targeted the 3′-UTR of c-Myc and Foxp1. A miR-526b-5p negatively regulated the mRNA expression of c-Myc and Foxp1. Relative mRNA expression of miR-526b-5p, c-Myc, and Foxp1 in HTR-8/SVneo and JEG3 cells measured by RT-qPCR. The results are presented as the mean ± SD (n = 5). *P < 0.05, **P < 0.01, and ***P < 0.001. B, C miR-526b-5p negatively regulated the protein expression of c-Myc and Foxp1. Protein expression of miR-526b-5p, c-Myc, and Foxp1 in HTR-8/SVneo and JEG3 cells measured by Western blot analysis. D Prediction and stability analysis of the target site of miR-526b-5p. Stability analysis of miR-526b-5p and c-Myc binding sites by RNAhybrid. Mfe means the minimum free energy. E c-Myc-WT, c-Myc-Mut with NC (psiCHECK2 empty vector), miR-526b-5p, and miR-NC were separately transfected into HEK293T cells, and the relative luciferase activity was measured. F Prediction and stability analysis of the target site of miR-526b-5p. Stability analysis of miR-526b-5p and Foxp1-WT1 binding sites by RNAhybrid. G Foxp1-WT1, Foxp1-Mut with NC (psiCHECK2 empty vector), miR-526b-5p, and miR-NC were separately transfected into HEK293T cells, and the relative luciferase activity was measured. H Prediction and stability analysis of the target site of miR-526b-5p. Stability analysis of miR-526b-5p and Foxp1-WT2 binding sites by RNAhybrid. I Foxp1-WT2, Foxp1-Mut with NC (psiCHECK2 empty vector), miR-526b-5p, and miR-NC were separately transfected into HEK293T cells, and the relative luciferase activity was measured. The results are presented as the mean ± SD (n = 6). *P < 0.05, **P < 0.01, and ***P < 0.001. NS means no significance (miR-526b-5p, miR-526b-5p precursor expression vector; miR-NC, pCDH513B-1 empty vector; sh-miR-526b-5p, miR-526b-5p interference vector; sh-NC, pSHI empty vector; NC, psiCHECK2 empty vector)
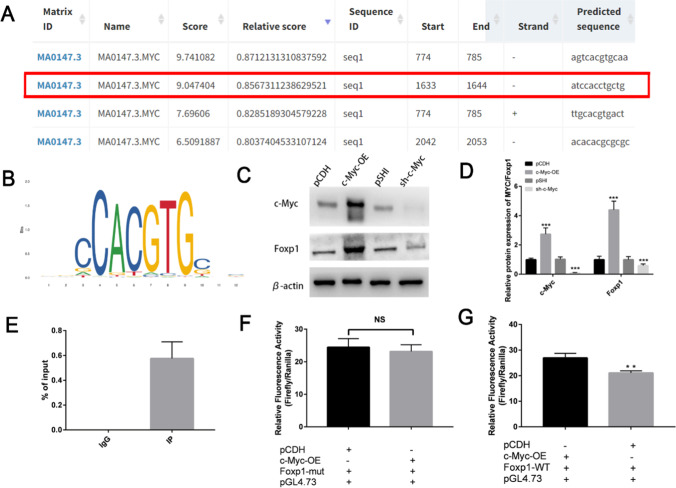
Transcription factor c-Myc positively regulated the expression of Foxp1
The above data indicated that c-Myc and Foxp1 were targets of miR-526b-5p. To explore the relationship between c-Myc and Foxp1, three c-Myc transcription binding sites (the first site was repeated with the third site) were found in the promoter region of Foxp1 through the online software JASPAR (Fig. 6A, B). Then, the correlation between c-Myc and Foxp1 expression was examined. c-Myc could positively regulate the expression of the Foxp1 protein (Fig. 6C, D). For the three predicted binding sites, ChIP-qPCR was performed on the trophoblasts to determine the specific base sites where c-Myc bound to the Foxp1 promoter. The ChIP-qPCR results (Fig. 6E) showed that the Foxp1 promoter region -146/-135 site (the second location, Fig. 6A) was the transcriptional regulatory site of c-Myc. The dual luciferase reporting system further identified the regulatory role of c-Myc on the Foxp1 transcription. Then, the c-Myc transcription binding site -146/-135 was mutated. Upregulation of c-Myc did not significantly alter the activity of the Foxp1 promoter after mutation (Fig. 6F). Upregulation of c-Myc enhanced the activity of the Foxp1 promoter (Fig. 6G). These results indicated that c-Myc positively regulated the expression of Foxp1 by binding to the Foxp1 promoter location -146/-135.
Fig. 6.
Transcription factor c-Myc positively regulated the expression of Foxp1. A, B Binding sequences in Foxp1 promoter and c-Myc motif from JASPAR. C, D c-Myc positively regulated the protein expression of Foxp1. Protein expression of c-Myc and Foxp1 in HTR-8/SVneo measured by Western blot. Quantitative analysis of the detection results of Western blot by ImageJ software. E ChIP-qPCR assay of c-Myc binding to Foxp1 promoter fragment in HTR-8/SVneo. (Negative control, Mouse IgG; an input, total chromatin). F, G The site -146/-135 was mutated from CAGCAGGTGGAT to CAGACTTGATAT. Luciferase reporter assays in c-Myc overexpression by c-Myc-OE plasmids (+). The results showed that the expression of c-Myc had no significant effect on the activity of the mutant Foxp1 promoter. Luciferase reporter assays in c-Myc overexpression by c-Myc-OE plasmids (+). The result indicated that the expression of c-Myc increased Foxp1 promoter activity. The results are presented as the mean ± SD (n = 6). *P < 0.05, **P < 0.01, and ***P < 0.001. NS means no significance
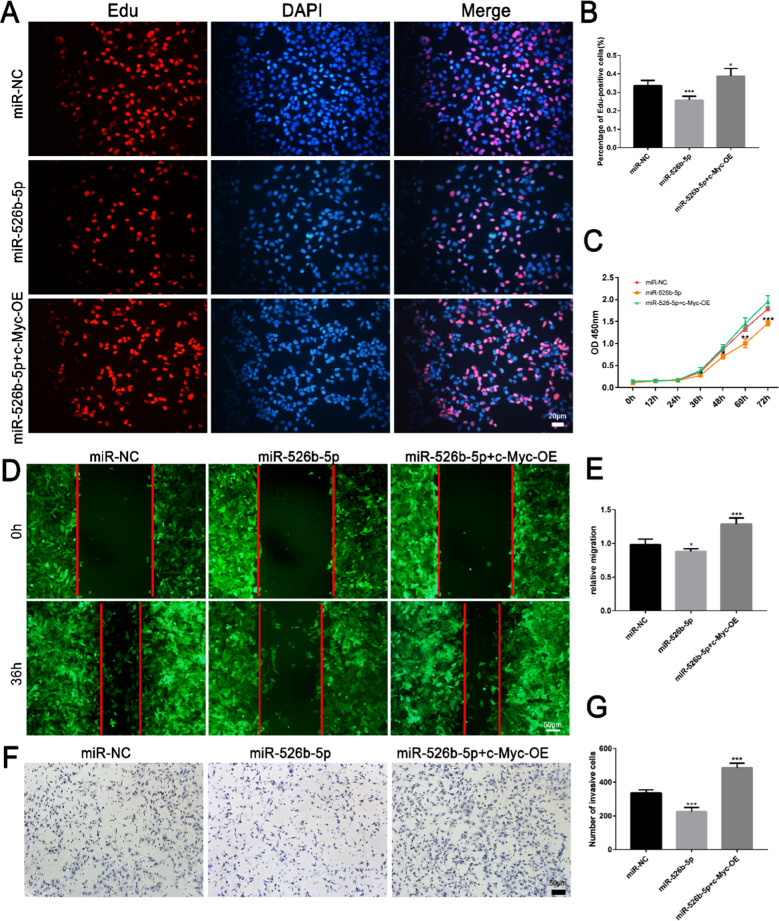
High expression of c-Myc rescued the effect of miR-526b-5p on trophoblast cells
Rescue experiments were performed to clarify whether miR-526b-5p impeded the proliferation, migration, and invasion of HTR-8/SVneo cells via negatively regulating c-Myc. The c-Myc overexpression vector was transfected into miR-526b-5p overexpressing trophoblast cells. Overexpression of c-Myc significantly reduced the proliferation (Fig. 7A–C), migration (Fig. 7D, E), and invasion (Fig. 7F, G) ability of HTR-8/SVneo cells, which decreased due to the upregulation of miR-526b-5p.
Fig. 7.
High expression of c-Myc rescued the effect of miR-526b-5p on the trophoblast cells. A The proliferation ability of HTR-8/SVneo cells was detected by Edu staining. EdU-positive cells are shown in red, and cell nuclei are shown in blue (scale bar = 20 μm). B ImageJ software was used to count the number of EdU-positive cells. C The proliferation ability of JEG3 cells was detected by CCK8 assay. D The migration ability of JEG3 cells was detected by wound healing assay (scale bar = 50 μm). E Quantitative analysis of the detection results of wound healing assay by ImageJ software. F, G The invasive ability of HTR-8/SVneo cells was detected by Transwell assay (scale bar = 50 μm). Quantitative analysis of the detection results of Transwell assay by ImageJ software. The results are presented as the mean ± SD (n = 6). *P < 0.05, **P < 0.01, and ***P < 0.00 (miR-NC, pCDH513B-1 empty vector; miR-526b-5p, miR-526b-5p precursor expression vector; miR-526b-5p+c-Myc-OE, miR-526b-5p precursor expression vector and c-Myc overexpression vector)
Discussion
RSA is a complex pregnancy complication that seriously threatens the health of women health worldwide. Due to the lack of clear and effective diagnostic biomarkers, diagnosing RSA is difficult. Thus, the miRNA may serve as a novel noninvasive biomarker for predicting RSA. Qin et al. [19] collected plasma samples from 27 patients with RSA and 28 healthy pregnant women and found that miRNA-320b, miRNA-146b-5p, and miRNA-221-3p were upregulated, and miRNA-101-3P was downregulated in RSA. Yang et al. [20] found that compared with normal pregnant women, the expression of miRNA-27a-3p, miRNA-29a-3p, miRNA-1005p, and miRNA 23a-3p in the early villi and decidual tissues of patients with RSA was significantly increased. By contrast, the expression of miRNA-127-3p and miRNA-486-5p was significantly decreased. In this study, miR-526b-5p was abnormally highly expressed in RSA. miR-526b-5p can be used as a candidate molecular marker for diagnosing RSA, which can indicate early the possible pregnancy outcome of clinicians and patients.
Nearly 50% of RSA etiologies have yet to be identified. miRNAs can participate in the occurrence and development of RSA by affecting the cellular characteristics of trophoblast cells, angiogenesis, placenta formation, and the immune tolerance microenvironment at the maternal–fetal interface. miRNA-184 upregulates the expression of Fas by targeting WIG1, thereby inhibiting the proliferation of HTR8 cells and promoting the apoptosis of HTR8 cells. These phenomena lead to RSA [21]. miRNA-27a-3p was involved in the occurrence of RSA by negatively regulating the expression of USP25, thereby inhibiting the trophoblast epithelial-mesenchymal transition (EMT), invasion, and migration [22]. In this study, miR-526b-5p was abnormally highly expressed in RSA, indicating that miR-526b-5p may play a role in the pathogenesis of RSA.
As a tumor suppressor miRNA [15], miR-526b-3p can inhibit the proliferation of glioma cells [23], nonsmall-cell lung cancer [24], and rectal cancer cells [25] by targeting genes, such as MAPRE1, WEE1, OSBPL5, and CCND1. At present, the downstream target genes of miR-526b-5p are HGF [26], LYPLA1 [27], HK2 [28], GRK5[29], ADAM15 [16], SERP1 [30], RAB1A [31], and PHLDA1 [32]. miR-526b-3p mainly inhibits cell proliferation and invasion in cervical, liver, melanoma, nonsmall-cell lung cancer, and other tumor cells. However, the function of miR-526b-5p in the placenta and trophoblasts has not been studied. The biological behavior of trophoblasts with infiltrating ability has many similarities with tumor cells. In this study, overexpression of miR-526b-5p inhibited trophoblast proliferation, invasion, and migration by targeting c-Myc and Foxp1.
As a protooncogene, the primary function of c-Myc is to promote the proliferation of tumors and stem cells. For example, c-Myc is abnormally high in patients with Fanconi anemia, thereby promoting the proliferation of hematopoietic stem cells, increasing DNA damage, and leading to bone marrow failure [33]. C-Myc promotes cell proliferation primarily by influencing cycle-related genes, such as cyclinD1, cyclinD2, and cyclinB1, and cyclin-dependent protein 4 (CDK4) [34, 35]. In the functional research of trophoblast, c-Myc binds to the promoter of BMAL1 and induces BMAL1 transcription, further activating matrix metalloproteinase 2/9 (MMP2/9) and facilitating migration and invasion of HTR-8/SVneo cells [36]. A previous study [37] has also shown that c-Myc positively regulated the expression of Prdx2 by targeting the Prdx2 promoter region. After downregulating c-Myc, the expression of Prdx2 was downregulated, and the expression of phosphorylated p53 (p-p53) and p38-MAPK/p21 was upregulated. Finally, cell proliferation was inhibited, apoptosis increased, and c-Myc in RM was significantly lower than that in the control group. These results were consistent with the current results, which showed that the expression of c-Myc in RSA decreased and inhibited the proliferation of trophoblast cells (Fig. 8).
Fig. 8.
Scheme indicating negative effects of miR-526b-5p/c-Myc/Foxp1 axis on proliferation, invasion, and migration of trophoblast cells
As one of the transcription factors of the FOX protein family, Foxp1 is involved in many biological processes, such as embryonic development, cell cycle regulation, glucose and lipid metabolism, immune regulation, and biological aging. The abnormal expression of Foxp1 can inhibit or promote the development of different tumors. Increased Foxp1 expression inactivates the Jak/Stat signaling pathway [38], promotes cell proliferation, and inhibits hippocampal neuronal apoptosis. These processes inhibit hippocampal neuronal damage and epilepsy progression. Foxp1 promotes the proliferation and migration of osteosarcoma cells by regulating the p53-P21/RB signal cascade [39]. In the functional research of trophoblast, a previous study has demonstrated that Foxp1 activated the mTOR signaling pathway by inhibiting the GNG7 expression and upregulated VEGF, thereby promoting trophoblast cell proliferation, invasion, and angiogenesis [40]. In the current study, the overexpression of miR-526b-5p downregulated the expression of c-Myc and Foxp1, which inhibited the proliferation, invasion, and migration of HTR-8/SVneo.
In summary, the results supported that miR-526b-5p inhibited the expression of c-Myc and Foxp1 by targeting the 3′-UTR of c-Myc and Foxp1. In addition, c-Myc positively regulated the expression of Foxp1 by binding to the Foxp1 promoter location -146/-135 and the low expression of c-Myc inhibited Foxp1, which reduced the proliferation, migration, and invasion abilities of trophoblast cells. miR-526b-5p plays a crucial role in placental development by altering the proliferation, migration, and invasion of trophoblasts, ultimately leading to RSA. This study provides a theoretical basis and potential clues for diagnosing and treating RSA. miR-526b-5p has been used as one of the markers for the successful establishment of trophoblast stem cells in multiple studies [41]. Therefore, exploring the effect of miR-526b-5p on the stemness maintenance and differentiation of trophoblast stem cells is the focus of our subsequent work. Nevertheless, the reason for the high expression of miR-526b-5p in trophoblast stem cells is still unclear. Therefore, exploring the upstream regulatory mechanism of miR-526b-5p is also interesting.
Supplementary Information
Below is the link to the electronic supplementary material.
Funding
This study is supported by Chongqing Postdoctoral Science Foundation (cstc2021jcyj-bsh0068), Chongqing Postdoctoral Research Special Funding Project (2021XM3100), and the National Natural Science Foundation of China (82171664).
Data availability
The data that support the findings of this study are available from the corresponding authors upon reasonable request.
Declarations
Ethics approval and consent to participate
The medical ethics committee approved the study of Chongqing Medical University and the First Affiliated Hospital of Chongqing Medical University (Reference Number: 2022138).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Li Luo and Lu Yao contributed equally to this work.
References
- 1.El Hachem H, et al. Recurrent pregnancy loss: current perspectives. Int J Womens Health. 2017;9:331–345. doi: 10.2147/IJWH.S100817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Practice Committee of the American Society for Reproductive, M Evaluation and treatment of recurrent pregnancy loss: a committee opinion. Fertil Steril. 2012;98(5):1103–1111. doi: 10.1016/j.fertnstert.2012.06.048. [DOI] [PubMed] [Google Scholar]
- 3.Li TC, et al. Recurrent miscarriage: aetiology, management and prognosis. Hum Reprod Update. 2002;8(5):463–481. doi: 10.1093/humupd/8.5.463. [DOI] [PubMed] [Google Scholar]
- 4.Lee CQ, et al. What is trophoblast? A combination of criteria define human first-trimester trophoblast. Stem Cell Reports. 2016;6(2):257–272. doi: 10.1016/j.stemcr.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang N, et al. METTL3-Mediated m(6)A RNA methylation of ZBTB4 interferes with trophoblast invasion and maybe involved in RSA. Front Cell Dev Biol. 2022;10:894810. doi: 10.3389/fcell.2022.894810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong X, Yang L, Wang H. miR-520 promotes DNA-damage-induced trophoblast cell apoptosis by targeting PARP1 in recurrent spontaneous abortion (RSA) Gynecol Endocrinol. 2017;33(4):274–278. doi: 10.1080/09513590.2016.1266476. [DOI] [PubMed] [Google Scholar]
- 7.Dragomir MP, Knutsen E, Calin GA. SnapShot: unconventional miRNA functions. Cell. 2018;174(4):1038–1038 e1. doi: 10.1016/j.cell.2018.07.040. [DOI] [PubMed] [Google Scholar]
- 8.Dzobo K, et al. Advances in therapeutic targeting of cancer stem cells within the tumor microenvironment: an updated review. Cells. 2020;9:8. doi: 10.3390/cells9081896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saha S, Ain R. MicroRNA regulation of murine trophoblast stem cell self-renewal and differentiation. Life Sci Alliance. 2020;3:11. doi: 10.26508/lsa.202000674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding J, et al. Extracellular vesicles derived from M1 macrophages deliver miR-146a-5p and miR-146b-5p to suppress trophoblast migration and invasion by targeting TRAF6 in recurrent spontaneous abortion. Theranostics. 2021;11(12):5813–5830. doi: 10.7150/thno.58731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang H, et al. Down-regulation of the Sp1 transcription factor by an increase of microRNA-4497 in human placenta is associated with early recurrent miscarriage. Reprod Biol Endocrinol. 2021;19(1):21. doi: 10.1186/s12958-021-00701-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang H, et al. miR-30-5p-mediated ferroptosis of trophoblasts is implicated in the pathogenesis of preeclampsia. Redox Biol. 2020;29:101402. doi: 10.1016/j.redox.2019.101402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobayashi N, et al. The microRNA cluster C19MC confers differentiation potential into trophoblast lineages upon human pluripotent stem cells. Nat Commun. 2022;13(1):3071. doi: 10.1038/s41467-022-30775-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sadovsky Y, et al. The function of trophomiRs and other microRNAs in the human placenta. Cold Spring Harb Perspect Med. 2015;5(8):a023036. doi: 10.1101/cshperspect.a023036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang R, et al. miR-526b-3p functions as a tumor suppressor in colon cancer by regulating HIF-1alpha. Am J Transl Res. 2016;8(6):2783–2789. [PMC free article] [PubMed] [Google Scholar]
- 16.Lin Q, et al. NCK1-AS1 promotes the progression of melanoma by accelerating cell proliferation and migration via targeting miR-526b-5p/ADAM15 axis. Cancer Cell Int. 2021;21(1):367. doi: 10.1186/s12935-021-02055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang ZY, et al. By downregulating Ku80, hsa-miR-526b suppresses non-small cell lung cancer. Oncotarget. 2015;6(3):1462–1477. doi: 10.18632/oncotarget.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ge L, et al. Mutation in myostatin 3'UTR promotes C2C12 myoblast proliferation and differentiation by blocking the translation of MSTN. Int J Biol Macromol. 2020;154:634–643. doi: 10.1016/j.ijbiomac.2020.03.043. [DOI] [PubMed] [Google Scholar]
- 19.Qin W, et al. Potential role of circulating microRNAs as a biomarker for unexplained recurrent spontaneous abortion. Fertil Steril. 2016;105(5):1247–1254 e3. doi: 10.1016/j.fertnstert.2016.01.028. [DOI] [PubMed] [Google Scholar]
- 20.Yang Q, et al. Association of the peripheral blood levels of circulating microRNAs with both recurrent miscarriage and the outcomes of embryo transfer in an in vitro fertilization process. J Transl Med. 2018;16(1):186. doi: 10.1186/s12967-018-1556-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, et al. MicroRNA-184 promotes apoptosis of trophoblast cells via targeting WIG1 and induces early spontaneous abortion. Cell Death Dis. 2019;10(3):223. doi: 10.1038/s41419-019-1443-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding J, et al. The miR-27a-3p/USP25 axis participates in the pathogenesis of recurrent miscarriage by inhibiting trophoblast migration and invasion. J Cell Physiol. 2019;234(11):19951–19963. doi: 10.1002/jcp.28593. [DOI] [PubMed] [Google Scholar]
- 23.Qiu L, et al. MiR-526b-3p inhibits the resistance of glioma cells to adriamycin by targeting MAPRE1. J Oncol. 2022;2022:2402212. doi: 10.1155/2022/2402212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu R, Yu Y, Wang H. The LMCD1-AS1/miR-526b-3p/OSBPL5 axis promotes cell proliferation, migration and invasion in non-small cell lung cancer. BMC Pulm Med. 2022;22(1):30. doi: 10.1186/s12890-022-01820-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Yan F, et al. Long noncoding RNA HOXD-AS1 promotes the proliferation, migration, and invasion of colorectal cancer via the miR-526b-3p/CCND1 axis. J Surg Res. 2020;255:525–535. doi: 10.1016/j.jss.2020.05.078. [DOI] [PubMed] [Google Scholar]
- 26.Liu Y, et al. Silencing CircEIF3I/miR-526b-5p axis epigenetically targets HGF/c-Met signal to hinder the malignant growth, metastasis and angiogenesis of hepatocellular carcinoma. Biochem Genet. 2023;61(1):48–68. doi: 10.1007/s10528-022-10239-y. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, et al. LncRNA LOXL1-AS1 facilitates the oncogenic character in cervical cancer by the miR-526b-5p /LYPLA1 axis. Biochem Genet. 2022;60(4):1298–1312. doi: 10.1007/s10528-021-10182-4. [DOI] [PubMed] [Google Scholar]
- 28.Zhao N, Hu L, Chen H. Circ_0002762 accelerates glycolysis metabolism to promote cervical cancer progression via the miR-526b-5p/HK2 axis. Gynecol Obstet Invest. 2022;87(6):352–363. doi: 10.1159/000526700. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y, et al. Circ_0001821 knockdown suppresses growth, metastasis, and TAX resistance of non-small-cell lung cancer cells by regulating the miR-526b-5p/GRK5 axis. Pharmacol Res Perspect. 2021;9(4):e00812. doi: 10.1002/prp2.812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu W, et al. hsa_circ_0085539 promotes osteosarcoma progression by regulating miR-526b-5p and SERP1. Mol Ther Oncolytics. 2020;19:163–177. doi: 10.1016/j.omto.2020.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kong Q, et al. CircRNA circUGGT2 contributes to hepatocellular carcinoma development via regulation of the miR-526b-5p/RAB1A axis. Cancer Manag Res. 2020;12:10229–10241. doi: 10.2147/CMAR.S263985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu P, et al. Circ0085539 promotes osteosarcoma progression by suppressing miR-526b-5p and PHLDA1 axis. Front Oncol. 2020;10:1250. doi: 10.3389/fonc.2020.01250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodriguez A, et al. MYC promotes bone marrow stem cell dysfunction in Fanconi anemia. Cell Stem Cell. 2021;28(1):33–47 e8. doi: 10.1016/j.stem.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang C, et al. Long noncoding RNA EMS connects c-Myc to cell cycle control and tumorigenesis. Proc Natl Acad Sci U S A. 2019;116(29):14620–14629. doi: 10.1073/pnas.1903432116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bretones G, Delgado MD, Leon J. Myc and cell cycle control. Biochim Biophys Acta. 2015;1849(5):506–516. doi: 10.1016/j.bbagrm.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 36.Wu L, et al. CRY2 suppresses trophoblast migration and invasion in recurrent spontaneous abortion. J Biochem. 2020;167(1):79–87. doi: 10.1093/jb/mvz076. [DOI] [PubMed] [Google Scholar]
- 37.Wu F, et al. Role of peroxiredoxin2 downregulation in recurrent miscarriage through regulation of trophoblast proliferation and apoptosis. Cell Death Dis. 2017;8(6):e2908. doi: 10.1038/cddis.2017.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng X, et al. Down-regulated microRNA-183 mediates the Jak/Stat signaling pathway to attenuate hippocampal neuron injury in epilepsy rats by targeting Foxp1. Cell Cycle. 2019;18(22):3206–3222. doi: 10.1080/15384101.2019.1671717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H, et al. FOXP1 drives osteosarcoma development by repressing P21 and RB transcription downstream of P53. Oncogene. 2021;40(15):2785–2802. doi: 10.1038/s41388-021-01742-4. [DOI] [PubMed] [Google Scholar]
- 40.Lai W, Yu L. Elevated microRNA 183 impairs trophoblast migration and invasiveness by downregulating FOXP1 expression and elevating GNG7 expression during preeclampsia. Mol Cell Biol. 2020;41:1. doi: 10.1128/MCB.00236-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu X, et al. Reprogramming roadmap reveals route to human induced trophoblast stem cells. Nature. 2020;586(7827):101–107. doi: 10.1038/s41586-020-2734-6. [DOI] [PubMed] [Google Scholar]
- 42.Lv Y, Lu C, Ji X, Miao M, Long W, Ding H, Lv M (2019) Roles of microRNAs in preeclampsia. J Cell Physiol 234(2):1052–61. 10.1002/jcp.27291 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon reasonable request.