Abstract
Conventional severity-of-illness scoring systems have shown suboptimal performance for predicting in-intensive care unit (ICU) mortality in patients with severe pneumonia. This study aimed to develop and validate machine learning (ML) models for mortality prediction in patients with severe pneumonia. This retrospective study evaluated patients admitted to the ICU for severe pneumonia between January 2016 and December 2021. The predictive performance was analyzed by comparing the area under the receiver operating characteristic curve (AU-ROC) of ML models to that of conventional severity-of-illness scoring systems. Three ML models were evaluated: (1) logistic regression with L2 regularization, (2) gradient-boosted decision tree (LightGBM), and (3) multilayer perceptron (MLP). Among the 816 pneumonia patients included, 223 (27.3%) patients died. All ML models significantly outperformed the Simplified Acute Physiology Score II (AU-ROC: 0.650 [0.584–0.716] vs 0.820 [0.771–0.869] for logistic regression vs 0.827 [0.777–0.876] for LightGBM 0.838 [0.791–0.884] for MLP; P < 0.001). In the analysis for NRI, the LightGBM and MLP models showed superior reclassification compared with the logistic regression model in predicting in-ICU mortality in all length of stay in the ICU subgroups; all age subgroups; all subgroups with any APACHE II score, PaO2/FiO2 ratio < 200; all subgroups with or without history of respiratory disease; with or without history of CVA or dementia; treatment with mechanical ventilation, and use of inotropic agents. In conclusion, the ML models have excellent performance in predicting in-ICU mortality in patients with severe pneumonia. Moreover, this study highlights the potential advantages of selecting individual ML models for predicting in-ICU mortality in different subgroups.
Subject terms: Diseases, Medical research
Introduction
Pneumonia is among the leading causes of death globally1, and each year, 0.1 million patients with pneumonia require intensive care unit (ICU) admission for mechanical ventilation (MV)2,3. Approximately half of the patients with respiratory failure in the ICU have pneumonia, and a quarter of them are re-admitted to the ICU4. Approximately 20–50% of patients with pneumonia who are admitted to the ICU die4–7. Clinical scoring systems to predict in-ICU mortality in critically ill patients have been introduced, but none of these have been specifically developed for patients admitted to the ICU with severe pneumonia. Therefore, there is an urgent need for severity assessment tools for predicting in-ICU mortality in patients with severe pneumonia8.
There has been recent interest in identifying the factors related to the outcomes of ICU patients. Vital signs9–11; laboratory markers such as lactate, thrombocytopenia, troponin, and bicarbonate levels12–14; and intervention strategies such as the use of vasopressors, MV, and continuous renal replacement therapy have been identified to be closely related to the prognoses of ICU patients15,16. However, previously validated predictive models including CURB-65, CRB-65, Pneumonia Severity Index (PSI), Quick Sequential Organ Failure Assessment (qSOFA), Simplified Acute Physiology Score 3 (SAPS-3), and Acute Physiology and Chronic Health Evaluation II (APACHE II) had suboptimal predictive performance in patients with severe pneumonia at ICU admission or emergency room department visits17–19. Therefore, a new prediction model for in-ICU mortality of patients admitted to the ICU for severe pneumonia needs to be developed.
Machine learning (ML) has demonstrated great potential in various medical fields, including surgery20, cardiology21,22 (early detection of heart failure)23, cancer research24,25, and intensive care medicine (diagnosis or prognosis prediction of critical illness)26–28. Although the integration of ML into the ICU setting is still in its early stages, several studies have already explored its application in managing critically ill patients26. Some studies have used large population datasets to predict length of stay (LOS)29, ICU readmission30, mortality rates31–33, and the risks of sepsis34 or acute respiratory distress syndrome35. Particular, there is increasing evidence that ML models are superior to conventional severity-of-illness scoring systems for predicting mortality in ICU patients36,37 or patients with pneumonia38. In addition, ML models may have good predictive performance for mortality, especially in patients with severe pneumonia39–41. One study demonstrated that the artificial neural network model had strong predictive performance for 14-day hospital readmission in patients with pneumonia42. Another study demonstrated that ML model has good performance for predicting 30-day mortality in sepsis patients28. However, evidence on whether ML models can accurately predict in-ICU mortality in patients with severe pneumonia who require ICU admission is rare.
Thus, the present study aimed to elucidate the performance of ML models in predicting mortality in patients with severe pneumonia who require ICU admission, using the information at the time of ICU admission. Our findings will aid in selecting individual ML models for predicting in-ICU mortality in different subgroups.
Methods
Ethics
This study complied with the Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD) reporting guidelines43. This study was approved by the IRB Committee of Seoul National University Seoul Metropolitan Government (SNU-SMG) Boramae Medical Center (IRB Number: 10-2021-110) on November 16, 2022 and was conducted in accordance with the 1975 ethical guidelines of the Declaration of Helsinki. The requirement for informed consent from the study subjects was waived by the IRB Committee of Seoul National University Seoul Metropolitan Government (SNU-SMG) Boramae Medical Center due to the retrospective study design.
Data collection, study design, and population
This observational study retrospectively evaluated patients admitted to the ICU for pneumonia at the SNU-SMG Boramae Medical Center between January 2016 and December 2021. The inclusion criteria were: (1) age ≥ 18 years; (2) ICU admission; (3) International Classification of Disease 10th edition code for pneumonia as a major diagnosis or detection of pneumonia on chest computed tomography within 1 week of ICU admission; (4) C-reactive protein (CRP) level ≥ 4 mg/dL; and (5) use of antibiotics for pneumonia. The exclusion criteria were: (1) no oxygen requirement; (2) transfer to the general ward within 3 days; and (3) ICU admission due to more serious medical conditions other than pneumonia.
Baseline data including age, sex, body mass index, smoking history, previous underlying disease, and respiratory comorbidities and clinical features were collected. Clinical features included prognostic scores, vital signs, laboratory examination results, and treatment with antibiotics or steroids.
Main outcome measures
The primary outcome measure was the prognostic accuracy of ML models compared with that of conventional severity-of-illness scoring systems for predicting in-ICU mortality in patients who required ICU admission for severe pneumonia. The secondary outcome measures were as follows: (1) the prognostic accuracy of the ML models; (2) the prognostic accuracy of the ML models in different subgroups; (3) the clinical factors contributing to the prediction of in-ICU mortality in patients admitted to the ICU for severe pneumonia. ICU admission due to severe pneumonia was determined as the presence of at least one major criterion or three minor criteria of the Infectious Disease Society of America/American Thoracic Society guidelines3.
The conventional severity-of-illness scoring systems included the SOFA, SAPS II, and APACHE II scores. Among the scoring systems, the best model that showed the strongest performance was used as the baseline comparator. For the statistical and ML models, we tested three popular models: (1) logistic regression with L2 regularization, (2) gradient-boosted decision tree (LightGBM), and (3) multilayer perceptron (MLP).
Data splitting and preprocessing
Variables with more than a 20% missing rate were excluded to generate the available dataset. Approximately 40% of the data were randomly separated with stratification by the outcome and subgrouping variables. The held-out data were used as a test set only for internal validation of the models. The remaining data were used to develop the models as a training set in a tenfold cross-validation scheme.
Missing values were imputed using multivariate imputation by chained equations44. Outliers were detected using an isolation forest45 and subsequently replaced with the closest normal value of the training set. All the variables included in the analysis and their missing rates are listed in Supplementary Table S1.
Variable importance and feature selection
The influence of each variable on the predictive ability of the model was evaluated using the SHapley Additive exPlanations (SHAP) method46. To rank the variables, the mean absolute SHAP values were calculated as the relative importance of the variables. A LightGBM was used for the SHAP evaluation47. The guiding metric for cross-validation performance was the area under the receiver operating characteristic curve (AU-ROC).
Model development
Supplementary Fig. S1 presents the workflow diagram for model development. LightGBM is a gradient-boosted tree-based ensemble model, whereas MLP is a feedforward neural network with a basic architecture comprising fully connected layers. The hyperparameters were tuned using Bayesian optimization to maximize the cross-validation performance. Details of the hyperparameter tuning with package information for all tested models are described in Supplementary Methods and Supplementary Table S2. The models were calibrated using isotonic regression according to the validation data obtained during cross-validation. The methods in the present study were implemented in Python version 3.9.7 (Python Software Foundation, Wilmington, Del, USA), with scikit-learn (version 1.1.2).
Internal validation in different subgroups
The model performance in the different patient subgroups was evaluated using the test set. We prespecified subgroups based on the clinically important phenotypes of pneumonia in the ICU48. Subgroup analyses were performed according to (1) the period from hospital admission to ICU admission, (2) age, (3) APACHE II scores, (4) PaO2/FiO2 ratio, (5) history of chronic respiratory disease, (6) history of cerebrovascular accident (CVA) or dementia, (7) MV, and (8) use of vasopressors.
Statistical analysis
Categorical variables were analyzed using the chi-squared test, while continuous variables were analyzed using the independent t-test or Mann–Whitney U test. We evaluated the AU-ROC as an overall performance measure and compared the models using the Delong method49. Owing to an imbalance in outcome prevalence, the area under the precision-recall curve (AU-PRC) was also evaluated as another overall performance measure50. Furthermore, the performance of the models was evaluated in detail according to sensitivity, positive predictive value, negative predictive value, diagnostic odds ratio, and net reclassification improvement (NRI)51 at three low false-positive rates (FPR) levels of 10%, 20%, and 30%. The recalibration effects were also evaluated using decision curves, which presented a net benefit against different decision thresholds52. The sensitivity at fixed FPR levels was evaluated in the subgroups using the best-performing model for overall performance. Calibration errors were evaluated before and after calibration using the Brier score and calibration curves. The Brier score was calculated as the mean squared error of the predicted probabilities53. P values were adjusted using the Benjamini–Hochberg method for multiple comparisons, and the significance level was set at P < 0.05.
Results
Baseline characteristics and clinical features
In total, 816 patients with pneumonia admitted to the ICU were included in the analysis (Fig. 1). The median patient age was 77 years, and 588 patients (72.1%) were male. Overall, 223 (27.3%) patients died. The median duration of LOS was 6 days (IQR, 2–13 days). The baseline characteristics are summarized in Table 1. There were significant differences in age and smoking history between the survivor and non-survivor groups. The non-survivor group was more likely to be older, involve a higher number of current smokers, and had a higher number of smoking pack-years among ever-smokers (9.9). Regarding comorbidities, the non-survivor group also included more patients with interstitial lung disease, pulmonary tuberculosis, lung cancer, chronic kidney disease, cerebellar vessel disease, cardiovascular disease, chronic heart failure, and metastatic cancer. Further, this group had higher illness severity scores, including the APACHE II, SOFA, and SAPS II scores.
Figure 1.
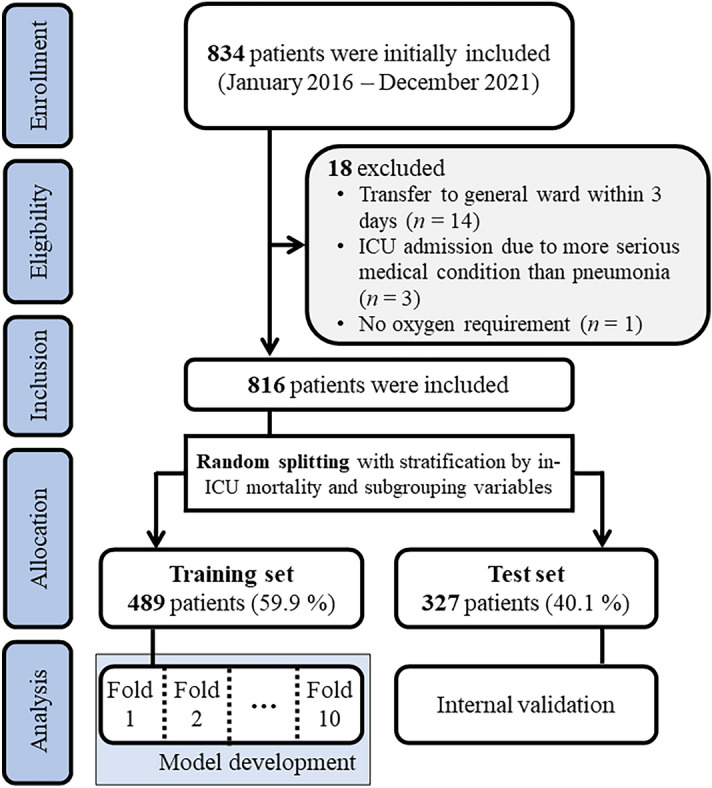
Patient selection flowchart.
Table 1.
Baseline patient characteristics.
| Variable | All | Survivors | Non-survivors | P value |
|---|---|---|---|---|
| (n = 816) | (n = 593, 72.7%) | (n = 223, 27.3%) | ||
| Demographics | ||||
| Age (yrs), median (IQR) | 77 (67–83) | 76 (66–83) | 78 (68–85) | 0.050 |
| Males, n (%) | 588 (72.1%) | 427 (72.0%) | 161 (72.2%) | 1.000 |
| BMI (kg/m2), mean (SD) | 20.7 (4.5) | 20.8 (4.6) | 20.3 (4.1) | 0.255 |
| Smoking | 0.002 | |||
| Ex-smoker, n (%) | 233 (28.6%) | 173 (29.2%) | 60 (26.9%) | |
| Current smoker, n (%) | 43 (5.3%) | 21 (3.5%) | 22 (9.9%) | |
| Pack years in ever-smokers, mean (SD) | 9.9 (19.0) | 9.1 (18.0) | 11.8 (21.1) | 0.083 |
| Underlying comorbidities | ||||
| Hypertension, n (%) | 450 (55.1%) | 334 (56.3%) | 116 (52.0%) | 0.306 |
| Diabetes, n (%) | 319 (39.1%) | 240 (40.5%) | 79 (35.4%) | 0.216 |
| COPD, n (%) | 62 (7.6%) | 42 (7.1%) | 20 (9.0%) | 0.449 |
| Asthma, n (%) | 24 (2.9%) | 16 (2.7%) | 8 (3.6%) | 0.662 |
| Interstitial lung disease, n (%) | 31 (3.8%) | 14 (2.4%) | 17 (7.6%) | < 0.001 |
| Nontuberculous mycobacteria, n (%) | 8 (1.0%) | 4 (0.7%) | 4 (1.8%) | 0.295 |
| Tuberculosis, n (%) | 119 (14.6%) | 82 (13.8%) | 37 (16.6%) | 0.376 |
| Lung cancer, n (%) | 11 (1.3%) | 4 (0.7%) | 7 (3.1%) | 0.017 |
| Chronic kidney disease, n (%) | 121 (14.8%) | 77 (13.0%) | 44 (19.7%) | 0.021 |
| Chronic liver disease, n (%) | 48 (5.9%) | 34 (5.7%) | 14 (6.3%) | 0.898 |
| Cardiovascular disease, n (%) | 14 (1.7%) | 5 (0.8%) | 9 (4.0%) | 0.005 |
| Congestive heart failure, n (%) | 32 (3.9%) | 14 (2.4%) | 18 (8.1%) | < 0.001 |
| Cerebrovascular accident, n (%) | 11 (1.3%) | 5 (0.8%) | 6 (2.7%) | 0.089 |
| Dementia, n (%) | 63 (7.7%) | 44 (7.4%) | 19 (8.5%) | 0.706 |
| HIV, n (%) | 5 (0.6%) | 3 (0.5%) | 2 (0.9%) | 0.893 |
| Metastatic cancer, n (%) | 11 (1.3%) | 3 (0.5%) | 8 (3.6%) | 0.002 |
| Hematologic malignancy, n (%) | 2 (0.2%) | 1 (0.2%) | 1 (0.4%) | 1.000 |
BMI, body mass index; COPD, chronic obstructive pulmonary disease; HIV, human immunodeficiency virus.
The clinical characteristics are presented in Table 2. Regarding vital signs, the survivor group had higher systolic, diastolic, or mean blood pressure, while the non-survivor group had faster heart and respiratory rates and lower urine output. The non-survivor group had lower levels of partial pressure of oxygen (PaO2) or carbon dioxide (HCO3−), lower levels of oxygen saturation (SpO2), and a lower ratio of PaO2/FiO2. For laboratory findings, the levels of urea nitrogen, creatinine, alanine aminotransferase, total bilirubin, and alkaline phosphatase (ALP) and the prothrombin time and international normalized ratio (PT-INR) were higher in the non-survivor group. Meanwhile, the survivor group was more likely to be treated with steroids and vasopressors. The comparison results between the training and test sets for the baseline characteristics and clinical features are presented in Supplementary Tables S3 and S4, respectively.
Table 2.
Clinical patient characteristics.
| Variable | All | Survivors | Non-survivors | P value |
|---|---|---|---|---|
| (n = 816) | (n = 593, 72.7%) | (n = 223, 27.3%) | ||
| Prognostic scores | ||||
| APACHE II, median (IQR) | 21 (16–28) | 20 (15–26) | 25 (19–31) | < 0.001 |
| SOFA, median (IQR) | 10 (7–13) | 9 (7–12) | 12 (8–14) | < 0.001 |
| SAPS II, median (IQR) | 48 (37–62) | 45 (35–58) | 56 (45–71) | < 0.001 |
| Initial vital signs | ||||
| Glasgow Coma Scale, median (IQR) | 11 (6–14) | 11 (6–14) | 10 (6–14) | 0.523 |
| Initial SBP (mmHg), median (IQR) | 122.0 (102.0–146.0) | 126.0 (108.0–151.0) | 110.0 (91.0–132.0) | < 0.001 |
| Initial DBP (mmHg), median (IQR) | 64.0 (54.0–75.2) | 65.0 (54.0–77.0) | 61.0 (50.0–71.5) | < 0.001 |
| Initial MBP, median (IQR) | 84.3 (70.7–98.0) | 86.7 (73.3–100.7) | 78.7 (62.8–89.3) | < 0.001 |
| Initial PR (beats/minute), median (IQR) | 104.0 (86.0–120.0) | 102.0 (84.0–117.0) | 112.0 (94.0–130.0) | < 0.001 |
| Initial RR (breaths/minute), median (IQR) | 22.0 (19.0–27.0) | 22.0 (18.0–26.0) | 24.0 (20.0–28.0) | < 0.001 |
| Body temperature (°C), mean (SD) | 36.9 (1.1) | 36.8 (1.1) | 37.0 (1.2) | 0.100 |
| Urine output (ml/hour), mean (SD) | 66.0 (52.6) | 73.0 (55.1) | 47.3 (39.5) | < 0.001 |
| Laboratory findings | ||||
| pH, median (IQR) | 7.4 (7.3–7.5) | 7.4 (7.3–7.4) | 7.4 (7.2–7.5) | 0.095 |
| pCO2 (mmHg), median (IQR) | 35.3 (29.2–43.7) | 35.9 (29.6–43.3) | 33.4 (27.6–45.3) | 0.122 |
| PaO2 (mmHg), median (IQR) | 68.3 (54.0–85.7) | 70.7 (55.6–88.5) | 63.2 (51.5–76.5) | < 0.001 |
| HCO3- (mEq/L), median (IQR) | 21.0 (17.1–24.7) | 21.7 (17.9–25.3) | 19.6 (15.4–22.9) | < 0.001 |
| Initial SpO2, median (IQR) | 95.0 (90.0–98.0) | 96.0 (92.0–98.0) | 93.0 (87.2–97.0) | < 0.001 |
| PaO2/FiO2 ratio, median (IQR) | 121.6 (78.3–194.0) | 139.7 (88.6–212.4) | 85.7 (63.3–127.0) | < 0.001 |
| White blood cell (103/μL), median (IQR) | 11.2 (7.6–16.3) | 11.1 (7.6–15.5) | 12.0 (7.5–18.7) | 0.148 |
| Neutrophil (103/μL), median (IQR) | 84.0 (75.9–89.2) | 84.0 (76.0–89.4) | 84.1 (75.5–88.8) | 0.665 |
| Lymphocyte (103/μL), median (IQR) | 10.1 (6.6–16.0) | 10.2 (6.9–15.8) | 9.9 (6.0–16.2) | 0.274 |
| Hemoglobin, median (IQR) | 10.9 (9.2–12.5) | 11.1 (9.4–12.6) | 10.6 (8.9–11.9) | 0.003 |
| Platelet (103/μL), median (IQR) | 205.0 (134.0–294.2) | 209.0 (149.0–301.0) | 187.0 (96.0–279.5) | 0.003 |
| Lactate (mmol/L), mean (SD) | 4.3 (5.2) | 3.6 (4.1) | 6.1 (6.8) | < 0.001 |
| C-reactive protein (mg/L), median (IQR) | 13.4 (5.8–22.0) | 11.8 (4.8–20.8) | 17.2 (10.2–24.0) | < 0.001 |
| Procalcitonin (ng/mL), mean (SD) | 2.4 (7.7) | 2.3 (8.4) | 2.7 (5.2) | 0.706 |
| BUN (mg/dL), median (IQR) | 27.0 (15.0–44.0) | 26.0 (15.0–42.0) | 29.0 (17.0–51.5) | 0.031 |
| Creatinine (mg/dL), median (IQR) | 1.2 (0.8–2.1) | 1.1 (0.8–1.9) | 1.4 (0.9–2.3) | 0.006 |
| Na (mEq/L), median (IQR) | 136.0 (132.2–139.8) | 136.2 (132.8–139.9) | 135.1 (131.2–139.1) | 0.027 |
| K (mEq/L), median (IQR) | 4.1 (3.6–4.7) | 4.1 (3.6–4.6) | 4.1 (3.5–4.7) | 0.986 |
| TCO2 (mEq/L), mean (SD) | 20.1 (6.6) | 20.4 (6.5) | 19.3 (6.6) | 0.034 |
| AST (U/L), median (IQR) | 39.0 (26.0–74.0) | 37.0 (25.0–70.0) | 45.0 (28.0–91.0) | 0.005 |
| ALT (U/L), median (IQR) | 20.0 (12.0–38.0) | 20.0 (11.0–37.0) | 19.0 (12.0–38.5) | 0.914 |
| Total bilirubin (mg/dL), median (IQR) | 0.8 (0.6–1.4) | 0.8 (0.6–1.4) | 0.9 (0.6–1.5) | 0.024 |
| ALP (IU/L), median (IQR) | 95.0 (71.8–132.0) | 93.0 (71.0–125.0) | 100.0 (76.0–148.0) | 0.002 |
| PT INR, median (IQR) | 1.2 (1.1–1.4) | 1.2 (1.1–1.4) | 1.3 (1.2–1.5) | < 0.001 |
| Troponin I (µg/L), mean (SD) | 62.3 (570.2) | 56.5 (598.6) | 77.8 (485.9) | 0.649 |
| NT-proBNP, mean (SD) | 7597.3 (10,760.1) | 7527.8 (10,898.5) | 7751.6 (10,445.0) | 0.817 |
| Treatment | ||||
| Steroid, n (%) | 564 (69.1%) | 385 (64.9%) | 179 (80.3%) | < 0.001 |
| Vasopressor, n (%) | 353 (43.3%) | 195 (32.9%) | 158 (70.9%) | < 0.001 |
APACHE, Acute Physiology and Chronic Health Evaluation; ALP, alkaline phosphatase; ALT, alanine aminotransferase; AST, aspartate aminotransferase; BUN, blood urea nitrogen; DBP, diastolic blood pressure; MBP, mean blood pressure; PR, pulse rate; RR, respiratory rate; SBP, systolic blood pressure; NT-proBNP, n-terminal prohormone brain natriuretic peptide; PT INR, international normalized ratio of prothrombin time; SAPS, Simplified Acute Physiology Score; SOFA, Sequential Organ Failure Assessment.
Overall performance of the prediction models
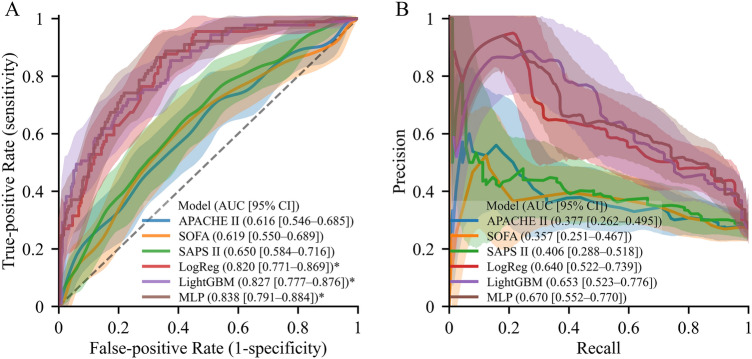
The prognostic performance and accuracy of the prediction models for in-ICU mortality are summarized in Fig. 2. Among the conventional severity-of-illness scoring systems, AU-ROC and AU-PRC were numerically (not statistically) higher in SAPS II than other conventional clinical scoring systems (SAPS II, AU-ROC (0.650 [95% CI, 0.584–0.761]) and AU-PRC (0.406 [0.288–0.518]); SOFA (AU-ROC, 0.619 [0.550–0.689] and AU-PRC, 0.357 [95% CI, 0.251–0.467]), and APACHE II (AU-ROC, 0.616 [95% CI, 0.546–0.685] and AU-PRC, 0.377 [95% CI, 0.262–0.495]) (Supplementary Table S5).
Figure 2.
Overall performance of the models. (A) Receiver operating characteristics curves and (B) precision-recall curves. Solid lines and shades indicate the mean curves and 95% confidence interval areas, respectively. SAPS II is the baseline model, and its confidence intervals are represented with a polka dot pattern. An asterisk (*) indicates a significantly higher area under the receiver operating characteristics curve than the baseline model (P < 0.05, Benjamini–Hochberg corrected).
All the proposed ML models significantly outperformed all scoring systems with respect to AU-ROC (P < 0.001). The MLP model showed better performance than SAPS II, with an AU-ROC of 0.838 [0.791–0.884]), although there was no significant difference compared with the logistic regression (0.820 [0.771–0.869]) and LightGBM (0.827 [0.777–0.876]) models. Furthermore, all tested ML models showed numerically higher AU-PRC values for predicting in-ICU mortality than the SAPS II. The MLP model showed the highest AU-PRC value of 0.670 (95% CI, 0.552–0.770), which was numerically higher than that of the logistic regression (0.640 [0.522–0.739]) and LightGBM (0.653 [0.552–0.770]) models, while SAPS II dropped around 0.406. The cross-validation performance results are presented in Supplementary Table S6. The calibration and decision curves are shown in Supplementary Figs. S2 and S3, respectively. The detailed performance and NRI at the 10%, 20%, and 30% FPR levels are presented in Supplementary Table S7.
Overall performance of ML models according to subgroups
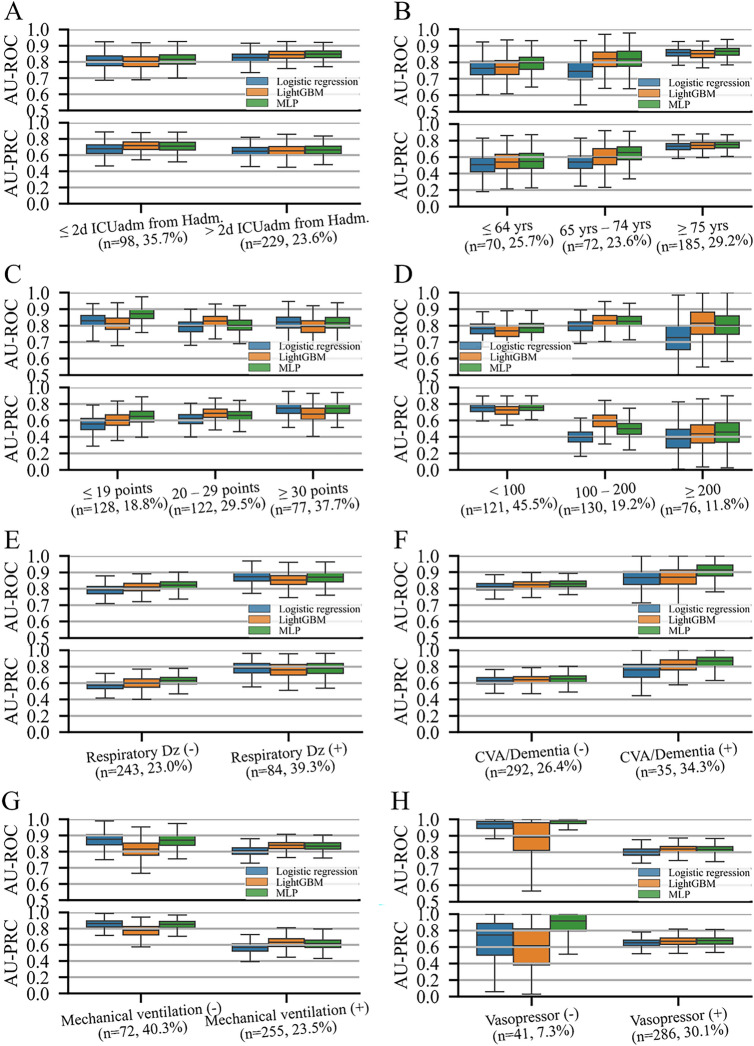
Figure 3 shows the overall performance of the ML models for in-ICU mortality in the different subgroups. The models performed consistently in most of the subgroups. Despite a difference in the in-ICU mortality rate, there was no significant difference in AU-ROC between the models in most of the subgroups except for the age 65–74 years subgroup (logistic regression vs MLP; P = 0.023), APACHE II score ≤ 19 subgroup (logistic regression vs MLP; P = 0.045, LightGBM vs MLP; P = 0.031), and MV treatment subgroup (logistic regression vs MLP; P = 0.044). There was no significant difference in the performance of the ML models in other subgroups.
Figure 3.
Overall performance by subgroup. The area under the receiver operating characteristic curves and precision-recall curves. The notation (n = a, b%) under each subgroup name indicates (A) the number of samples in the test set and (B) the prevalence rate of the outcome of the subgroup. Box plots are plotted with whiskers of 1.5 times the interquartile ranges. The subgrouping variables are (A) the period from hospital admission to ICU admission, (B) age, (C) Acute Physiology and Chronic Health Evaluation (APACH) II scores, (D) PaO2/FiO2 ratio, (E) history of chronic respiratory disease, (F) history of cerebrovascular accident or dementia, (G) mechanical ventilation, and (H) vasopressor use. AU-PRC, area under the precision-recall curve; AU-ROC, area under the receiver operating characteristics curve; CVA, cerebrovascular accident; Dz, disease; Hadm, hospital admission; ICU admission; yrs, years.
In the analysis for NRI, the LightGBM and MLP models showed superior reclassification compared with the logistic regression model in predicting in-ICU mortality in all LOS in the ICU subgroups; all age subgroups; all subgroups with any APACHE II score, PaO2/FiO2 ratio < 200; all subgroups with or without history of respiratory disease; with or without history of CVA or dementia; treatment with MV, and use of inotropic agents (Supplementary Table S8). In most of the above subgroups, the LightGBM model showed higher NRI values than the MLP except for the subgroups aged 65–74 years, with an APACHE II score of ≤ 19, with history of CVA or dementia.
Attributable variables with importance plots and SHAP values
The selected predictors of in-ICU mortality are shown in Fig. 4. From a total of 55 variables, 16 were selected. The selected variables were PaO2/FiO2 ratio, CRP level, lactate level, urine output, initial systolic blood pressure (SBP), white blood cell (WBC) count. Among these variables, the partial SHAP dependence plots for the top six variables with the mean absolute SHAP values are illustrated in Supplementary Fig. S4; those for the other variables in Supplementary Fig. S5. The local interpretability of the LightGBM model is demonstrated in Supplementary Fig. S6, which shows how the model predicts each case of true positive, true negative, false positive, and false negative.
Figure 4.
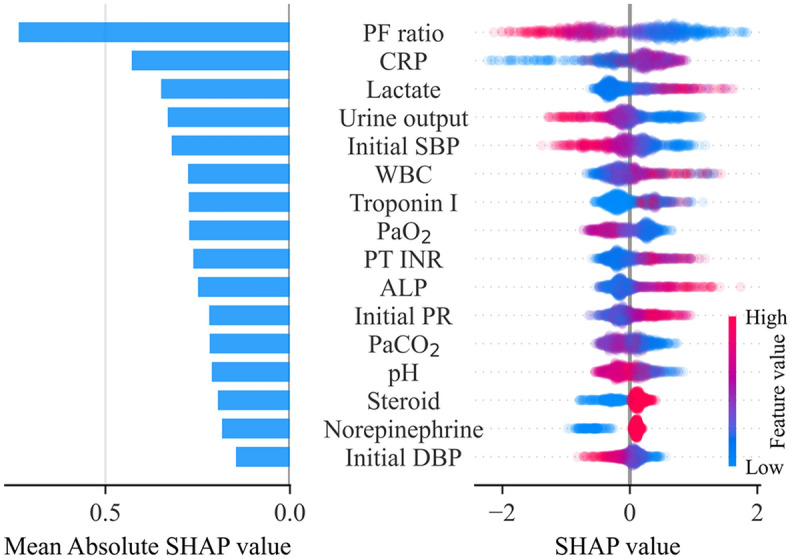
Importance of the selected variables. The individual influences of every value and the overall contributions of each variable to the model prediction are represented as a dot and a bar on the right and left, respectively. In the plot on the right, the red dots indicate high feature values for continuous/ordinal variables or affirmative responses for binary variables. Positive and negative SHAP values indicate that positive contributions result in an increased prediction score and that negative contributions result in a decreased prediction score. ALP, alkaline phosphatase; CRP, C-reactive protein; DBP, diastolic blood pressure; PF ratio, PaO2/FiO2 ratio; PR, pulse rate; PT-INR, international normalized ratio of prothrombin time; SBP, systolic blood pressure; WBC, white blood cell.
Discussion
Our study evaluated the prognostic accuracy of ML models compared with that of conventional severity-of-illness scoring systems for predicting in-ICU mortality in patients with severe pneumonia. All ML models showed excellent performance in predicting in-ICU mortality and were superior to SAPS II. In addition, when the ML models were applied in the different subgroups, the LightGBM and MLP models showed superior reclassification compared with the logistic regression model in predicting in-ICU mortality in all LOS in the ICU subgroups; all age subgroups; all subgroups with any APACHE II score, PaO2/FiO2 ratio < 200; all subgroups with or without history of respiratory disease; with or without history of CVA or dementia; treatment with MV, and use of inotropic agents. Furthermore, the LightGBM model showed higher NRI values than the MLP in most of the above subgroups, except for the subgroups aged 65–74 years, with an APACHE II score of ≤ 19, with history of CVA or dementia. Therefore, ML models have the potential to improve in-ICU mortality prediction in patients with severe pneumonia admitted to the ICU. Moreover, this study shows the potential advantages of individual ML models for predicting in-ICU mortality in different subgroups of patients with severe pneumonia admitted to the ICU.
CURB-65 and PSI are the most commonly used clinical severity-of-illness scoring systems for patients with community-acquired pneumonia. However, CURB-65 and PSI have a limitation in that CURB-65 has low sensitivity, while PSI has low specificity for mortality54. The Clinical Pulmonary Infection Score is a well-validated prediction model for the development of ventilator-associated pneumonia. However, its predictive performance for mortality is inferior to that of the APACHE II score55. Given the limitations of clinical severity-of-illness scoring systems, the usefulness of ML models has recently been studied to predict mortality in patients with pneumonia. The results show that various ML models outperform CURB-6538,39,56 and PSI57 for predicting mortality in patients with severe community- or hospital-acquired pneumonia. However, few previous studies have used ML models to predict the prognosis of patients with severe pneumonia admitted to the ICU. One small study reported that an ML approach had better performance than APACHE II and PSI for predicting mortality in critically ill influenza patients58. In our study, SAPS II showed a numerically higher AU-ROC than APACHE II and SOFA for predicting in-ICU mortality on the first day of ICU admission. Importantly, the ML models outperformed SAPS II, which suggested that the ML model can provide more accurate information for optimal decision-making based on the estimated probability of mortality.
We selected logistic regression, LightGBM, and MLP as the ML predictive models for mortality in patients with severe pneumonia based on several considerations. Logistic regression is a well-established and widely used ML model for binary classification tasks. It offers a simple and interpretable approach to modeling the relationship between predictor variables and the outcome as well as performance of ML59–61. LightGBM is a gradient-boosted decision tree algorithm that has gained popularity for its high performance and efficiency62. Several studies have demonstrated the favorable predictive value of LightGBM in the field of medicine63–65. Another study found that LightGBM had the best predictive ability among other ML models including XGBoost, logistic regression, and naïve Bayes66. MLP-based models are effective in capturing nonlinear relationships, making them ideal candidates for complex and multifactorial disease classification including in stroke67,68 when compared to conventional statistical modeling. Moreover, both LightGBM and MLP have been used in many clinical studies69–71, demonstrating their extensive applicability and promising predictive performance. In the present study, the ML models outperformed conventional severity-of-illness systems. Scoring systems use a limited number of variables, which might restrict their predictive power in individual patients72. ML models are capable of utilizing high-dimensional data, and this could account for their superior performance to conventional scoring systems.
In this study, the LightGBM model showed the highest predictive performance with respect to NRI at 10% FPR. This result supports that decision-tree-based models could be more beneficial than logistic regression models for predicting in-ICU mortality in pneumonia patients at a high cut-off point of 90% specificity. ML models have a strength in capturing the nonlinear relations between the features and the predicted outcomes. We found notable non-linear relationships between in-ICU mortality and several selected variables, including PaO2, WBC count, pH, initial pulse rate, lymphocyte, HCO3−, and PaCO2. This could be the reason for the lower performance of the logistic regression model, a generalized linear model, in predicting the in-ICU mortality of patients with severe pneumonia. Although cross-validation results were not specified to the FPR level, the MLP model had the largest difference in the AU-ROC value between internal validation and cross-validation. This indicates that compared with the other models, the MLP model might be more relatively overfitted to the training set.
Contrary to the performance in different subgroups at the 10% FPR level, our results in the entire test set demonstrated no significant difference in AU-ROC between the ML models and the logistic regression model. In partial dependence plots of the variables that contributed the most to model predictions, linear relationships with in-ICU mortality were observed, especially in those of the PaO2/FiO2 ratio, urine output, and initial SBP. This might be the reason for the lack of statistical significance in these differences. The SHAP model was used to determine the important influencing factors of in-ICU mortality in the ML models, and the results were similar to previous studies: PaO2/FiO273,74, CRP levels75, urine output76, initial SBP73, PaO273,77, and leukopenia78,79 or leukocytosis80,81.
In addition, using the SHAP model, we found that higher ALP levels and prolonged PT-INR were associated with a higher risk of in-ICU mortality. ALP can be elevated as an acute-phase reaction in acute infections82. In community-acquired pneumonia, elevated ALP levels were not associated with mortality83. However, in critically ill patients with septic acute kidney injury (AKI), elevated ALP levels are associated with mortality84. In our study, septic shock was found in > 40% of the critically ill patients with pneumonia, and AKI was a common condition. It appears that ALP is related to the severity of impaired renal function through systemic inflammation caused by pneumonia rather than the severity of pneumonia itself. With respect to prolonged PT-INR, substantial coagulation abnormalities are commonly observed in patients with sepsis or pneumonia85. The excessive production of thrombogenic tissue factors in sepsis pneumonia compared with low levels of tissue factors under normal conditions86 leads to the development of systemic coagulopathy during the period of pneumonia87.
Our study had some limitations. First, as the models were developed using data retrospectively collected in a single center and were not externally validated, the results had limited generalizability. Although our study provides valuable insights into the performance of the ML models, further studies are needed to assess the generalizability and real-time applicability of these ML models in predicting in-ICU mortality in patients with severe pneumonia. Furthermore, these studies should include robust external validation using independent datasets and evaluation of the model performance in prospective clinical practice. Second, owing to the small sample size and the inclusion of patients admitted within a long study period of over 6 years, there is a possibility of heterogeneity with respect to patient characteristics, treatment measures, and potential biases. Thus, it might be challenging to clearly establish the usefulness of ML, which is a non-parametric algorithm. Furthermore, six subgrouping variables were adopted. A large number of stratification variables with a small sample size could lead to optimistic results in the internal validation of the models.
Conclusion
Compared to conventional severity-of-illness scoring systems, the ML models of LightGBM, MLP, and logistic regression have better predictive performance for in-ICU mortality in patients with severe pneumonia. Moreover, this study shows the potential advantages of selecting individual ML models for predicting in-ICU mortality in different subgroups of patients with severe pneumonia.
Supplementary Information
Author contributions
E.T.J. and H.J.L. had full access to all the data in the study and took responsibility for the integrity of the data and the accuracy of the analysis. E.T.J. contributed to study design, data analysis, and manuscript writing. H.J.L. contributed to the data collection, literature review, and manuscript writing. T.Y.P., K.N.J., and B.Y. contributed to data collection and review of the manuscript. H.W.L. and D.H.K. contributed to the study design, data collection, writing of the manuscript, and supervision of the study.
Funding
This research was supported by a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI21C1074).
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Eun-Tae Jeon and Hyo Jin Lee.
Contributor Information
Hyun Woo Lee, Email: athrunzara86@snu.ac.kr, Email: athrunzara86@gmail.com.
Dong Hyun Kim, Email: mi4ri4@gmail.com.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-38765-8.
References
- 1.Mattila JT, Fine MJ, Limper AH, Murray PR, Chen BB, Lin PL. Pneumonia. Treatment and diagnosis. Ann. Am. Thorac. Soc. 2014;11(Suppl 4):S189–S192. doi: 10.1513/AnnalsATS.201401-027PL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spindler C, Ortqvist A. Prognostic score systems and community-acquired bacteraemic pneumococcal pneumonia. Eur. Respir. J. 2006;28(4):816–823. doi: 10.1183/09031936.06.00144605. [DOI] [PubMed] [Google Scholar]
- 3.Mandell LA, Wunderink RG, Anzueto A, Bartlett JG, Campbell GD, Dean NC, et al. Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin. Infect. Dis. 2007;44(Suppl 2):S27–72. doi: 10.1086/511159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee HW, Ji E, Ahn S, Yang HJ, Yoon SY, Park TY, et al. A population-based observational study of patients with pulmonary disorders in intensive care unit. Korean J. Intern. Med. 2020;35(6):1411–1423. doi: 10.3904/kjim.2018.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vallés J, Martin-Loeches I, Torres A, Diaz E, Seijas I, López MJ, et al. Epidemiology, antibiotic therapy and clinical outcomes of healthcare-associated pneumonia in critically ill patients: A Spanish cohort study. Intensive Care Med. 2014;40(4):572–581. doi: 10.1007/s00134-014-3239-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodriguez A, Lisboa T, Blot S, Martin-Loeches I, Solé-Violan J, De Mendoza D, et al. Mortality in ICU patients with bacterial community-acquired pneumonia: When antibiotics are not enough. Intensive Care Med. 2009;35(3):430–438. doi: 10.1007/s00134-008-1363-6. [DOI] [PubMed] [Google Scholar]
- 7.Walden AP, Clarke GM, McKechnie S, Hutton P, Gordon AC, Rello J, et al. Patients with community acquired pneumonia admitted to European intensive care units: An epidemiological survey of the GenOSept cohort. Crit. Care. 2014;18(2):R58. doi: 10.1186/cc13812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Napolitano LM. Use of severity scoring and stratification factors in clinical trials of hospital-acquired and ventilator-associated pneumonia. Clin. Infect. Dis. 2010;51(Suppl 1):S67–80. doi: 10.1086/653052. [DOI] [PubMed] [Google Scholar]
- 9.Vincent JL, Nielsen ND, Shapiro NI, Gerbasi ME, Grossman A, Doroff R, et al. Mean arterial pressure and mortality in patients with distributive shock: A retrospective analysis of the MIMIC-III database. Ann. Intensive Care. 2018;8(1):107. doi: 10.1186/s13613-018-0448-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seo MH, Choa M, You JS, Lee HS, Hong JH, Park YS, et al. Hypoalbuminemia, low base excess values, and tachypnea predict 28-day mortality in severe sepsis and septic shock patients in the emergency department. Yonsei Med. J. 2016;57(6):1361–1369. doi: 10.3349/ymj.2016.57.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen Y, Lou Y, Zhu S. Hyperthermia is a predictor of high mortality in patients with sepsis. Crit. Care. 2020;24(1):543. doi: 10.1186/s13054-020-03263-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim WY, Lee YJ, Yeon Lim S, Ok Koh S, Choi WI, Chan Kim S, et al. Clinical characteristics and prognosis of pneumonia and sepsis: Multicenter study. Minerva Anestesiol. 2013;79(12):1356–1365. [PubMed] [Google Scholar]
- 13.Nichol AD, Egi M, Pettila V, Bellomo R, French C, Hart G, et al. Relative hyperlactatemia and hospital mortality in critically ill patients: A retrospective multi-centre study. Crit. Care. 2010;14(1):R25. doi: 10.1186/cc8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takegawa R, Kabata D, Shimizu K, Hisano S, Ogura H, Shintani A, et al. Serum albumin as a risk factor for death in patients with prolonged sepsis: An observational study. J. Crit. Care. 2019;51:139–144. doi: 10.1016/j.jcrc.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 15.Ren Y, Zhang L, Xu F, Han D, Zheng S, Zhang F, et al. Risk factor analysis and nomogram for predicting in-hospital mortality in ICU patients with sepsis and lung infection. BMC Pulm. Med. 2022;22(1):17. doi: 10.1186/s12890-021-01809-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allegretti AS, Steele DJ, David-Kasdan JA, Bajwa E, Niles JL, Bhan I. Continuous renal replacement therapy outcomes in acute kidney injury and end-stage renal disease: A cohort study. Crit. Care. 2013;17(3):R109. doi: 10.1186/cc12780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carmo TA, Ferreira IB, Menezes RC, Telles GP, Otero ML, Arriaga MB, et al. Derivation and validation of a novel severity scoring system for pneumonia at intensive care unit admission. Clin. Infect. Dis. 2021;72(6):942–949. doi: 10.1093/cid/ciaa183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richards G, Levy H, Laterre PF, Feldman C, Woodward B, Bates BM, et al. CURB-65, PSI, and APACHE II to assess mortality risk in patients with severe sepsis and community acquired pneumonia in PROWESS. J. Intensive Care Med. 2011;26(1):34–40. doi: 10.1177/0885066610383949. [DOI] [PubMed] [Google Scholar]
- 19.Joya-Montosa C, Delgado-Amaya MD, Trujillo-García E, Curiel-Balsera E. Assessment of specific risk scores for patients admitted to the ICU for severe community-acquired pneumonia. Crit. Care. 2015;19(Suppl 1):P9. doi: 10.1186/cc14089. [DOI] [Google Scholar]
- 20.Kassahun Y, Yu B, Tibebu AT, Stoyanov D, Giannarou S, Metzen JH, et al. Surgical robotics beyond enhanced dexterity instrumentation: A survey of machine learning techniques and their role in intelligent and autonomous surgical actions. Int. J. Comput. Assist. Radiol. Surg. 2016;11(4):553–568. doi: 10.1007/s11548-015-1305-z. [DOI] [PubMed] [Google Scholar]
- 21.Lopez-Jimenez F, Attia Z, Arruda-Olson AM, Carter R, Chareonthaitawee P, Jouni H, et al. Artificial intelligence in cardiology: Present and future. Mayo Clin. Proc. 2020;95(5):1015–1039. doi: 10.1016/j.mayocp.2020.01.038. [DOI] [PubMed] [Google Scholar]
- 22.Guo T, Fang Z, Yang G, Zhou Y, Ding N, Peng W, et al. Machine learning models for predicting in-hospital mortality in acute aortic dissection patients. Front. Cardiovasc. Med. 2021;8:727773. doi: 10.3389/fcvm.2021.727773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi E, Schuetz A, Stewart WF, Sun J. Using recurrent neural network models for early detection of heart failure onset. J. Am. Med. Inform. Assoc. 2017;24(2):361–370. doi: 10.1093/jamia/ocw112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang TT, Zawaski JA, Francis KN, Qutub AA, Gaber MW. Image-based classification of tumor type and growth rate using machine learning: A preclinical study. Sci. Rep. 2019;9(1):12529. doi: 10.1038/s41598-019-48738-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tran KA, Kondrashova O, Bradley A, Williams ED, Pearson JV, Waddell N. Deep learning in cancer diagnosis, prognosis and treatment selection. Genome Med. 2021;13(1):152. doi: 10.1186/s13073-021-00968-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gutierrez G. Artificial intelligence in the intensive care unit. Crit. Care. 2020;24(1):101. doi: 10.1186/s13054-020-2785-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo J, Sun Y, Chi J, Liao X, Xu C. A novel deep learning-based method for COVID-19 pneumonia detection from CT images. BMC Med. Inform. Decis. Mak. 2022;22(1):284. doi: 10.1186/s12911-022-02022-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Su Y, Guo C, Zhou S, Li C, Ding N. Early predicting 30-day mortality in sepsis in MIMIC-III by an artificial neural networks model. Eur. J. Med. Res. 2022;27(1):294. doi: 10.1186/s40001-022-00925-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sotoodeh M, Ho JC. Improving length of stay prediction using a hidden Markov model. AMIA Jt. Summits Transl. Sci. Proc. 2019;2019:425–434. [PMC free article] [PubMed] [Google Scholar]
- 30.Lin YW, Zhou Y, Faghri F, Shaw MJ, Campbell RH. Analysis and prediction of unplanned intensive care unit readmission using recurrent neural networks with long short-term memory. PLoS ONE. 2019;14(7):e0218942. doi: 10.1371/journal.pone.0218942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Awad A, Bader-El-Den M, McNicholas J, Briggs J. Early hospital mortality prediction of intensive care unit patients using an ensemble learning approach. Int. J. Med. Inform. 2017;108:185–195. doi: 10.1016/j.ijmedinf.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 32.Holmgren G, Andersson P, Jakobsson A, Frigyesi A. Artificial neural networks improve and simplify intensive care mortality prognostication: A national cohort study of 217,289 first-time intensive care unit admissions. J. Intensive Care. 2019;7:44. doi: 10.1186/s40560-019-0393-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rau CS, Wu SC, Chuang JF, Huang CY, Liu HT, Chien PC, et al. Machine learning models of survival prediction in trauma patients. J. Clin. Med. 2019;8(6):799. doi: 10.3390/jcm8060799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu C, Li L, Huang W, Wu T, Xu Q, Liu J, et al. Interpretable machine learning for early prediction of prognosis in sepsis: A discovery and validation study. Infect. Dis. Ther. 2022;11(3):1117–1132. doi: 10.1007/s40121-022-00628-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maddali MV, Churpek M, Pham T, Rezoagli E, Zhuo H, Zhao W, et al. Validation and utility of ARDS subphenotypes identified by machine-learning models using clinical data: An observational, multicohort, retrospective analysis. Lancet Respir. Med. 2022;10(4):367–377. doi: 10.1016/S2213-2600(21)00461-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shillan D, Sterne JAC, Champneys A, Gibbison B. Use of machine learning to analyse routinely collected intensive care unit data: A systematic review. Crit. Care. 2019;23(1):284. doi: 10.1186/s13054-019-2564-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pirracchio R, Petersen ML, Carone M, Rigon MR, Chevret S, van der Laan MJ. Mortality prediction in intensive care units with the Super ICU Learner Algorithm (SICULA): A population-based study. Lancet Respir. Med. 2015;3(1):42–52. doi: 10.1016/S2213-2600(14)70239-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cilloniz C, Ward L, Mogensen ML, Pericàs JM, Méndez R, Gabarrús A, et al. Machine-Learning Model for Mortality Prediction in Patients With Community-Acquired Pneumonia: Development and Validation Study. Chest. 2022. doi: 10.1016/j.chest.2022.07.005 [DOI] [PubMed]
- 39.Kang SY, Cha WC, Yoo J, Kim T, Park JH, Yoon H, et al. Predicting 30-day mortality of patients with pneumonia in an emergency department setting using machine-learning models. Clin. Exp. Emerg. Med. 2020;7(3):197–205. doi: 10.15441/ceem.19.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang D, He D, Gong L, Yao R, Wang W, Yang L, et al. A prediction model for hospital mortality in patients with severe community-acquired pneumonia and chronic obstructive pulmonary disease. Respir. Res. 2022;23(1):250. doi: 10.1186/s12931-022-02181-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lichtner G, Balzer F, Haufe S, Giesa N, Schiefenhövel F, Schmieding M, et al. Predicting lethal courses in critically ill COVID-19 patients using a machine learning model trained on patients with non-COVID-19 viral pneumonia. Sci. Rep. 2021;11(1):13205. doi: 10.1038/s41598-021-92475-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tey SF, Liu CF, Chien TW, Hsu CW, Chan KC, Chen CJ, et al. Predicting the 14-day hospital readmission of patients with pneumonia using artificial neural networks (ANN) Int. J. Environ. Res. Public Health. 2021;18(10):5110. doi: 10.3390/ijerph18105110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Collins GS, Reitsma JB, Altman DG, Moons KG, The TRIPOD Group Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): The TRIPOD statement. Circulation. 2015;131(2):211–219. doi: 10.1161/CIRCULATIONAHA.114.014508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Buuren S, Groothuis-Oudshoorn K. mice: Multivariate imputation by chained equations in R. J. Stat. Softw. 2011;45(3):1–67. doi: 10.18637/jss.v045.i03. [DOI] [Google Scholar]
- 45.Liu FT, Ting KM, Zhou Z-H. Isolation-based anomaly detection. Acm T. Knowl. Discov. D. 2012;6(1):1–39. doi: 10.1145/2133360.2133363. [DOI] [Google Scholar]
- 46.Lundberg SM, Erion G, Chen H, DeGrave A, Prutkin JM, Nair B, et al. From local explanations to global understanding with explainable AI for trees. Nat. Mach. Intell. 2020;2(1):56–67. doi: 10.1038/s42256-019-0138-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ke GL, Meng Q, Finley T, Wang TF, Chen W, Ma WD, et al. LightGBM: A highly efficient gradient boosting decision tree. Adv. Neural Inf. Process. Syst. 2017;30:3146–3154. [Google Scholar]
- 48.Chalmers JD, Mandal P, Singanayagam A, Akram AR, Choudhury G, Short PM, et al. Severity assessment tools to guide ICU admission in community-acquired pneumonia: Systematic review and meta-analysis. Intensive Care Med. 2011;37(9):1409–1420. doi: 10.1007/s00134-011-2261-x. [DOI] [PubMed] [Google Scholar]
- 49.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics. 1988;44(3):837–845. doi: 10.2307/2531595. [DOI] [PubMed] [Google Scholar]
- 50.Davis, J. & Goadrich, M. (eds.) The relationship between Precision-Recall and ROC curves. In Proceedings of the 23rd International Conference on Machine Learning (2006).
- 51.Leening MJ, Vedder MM, Witteman JC, Pencina MJ, Steyerberg EW. Net reclassification improvement: computation, interpretation, and controversies: A literature review and clinician's guide. Ann. Intern. Med. 2014;160(2):122–131. doi: 10.7326/M13-1522. [DOI] [PubMed] [Google Scholar]
- 52.Vickers AJ, Van Calster B, Steyerberg EW. Net benefit approaches to the evaluation of prediction models, molecular markers, and diagnostic tests. BMJ. 2016;352:i6. doi: 10.1136/bmj.i6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rufibach K. Use of Brier score to assess binary predictions. J. Clin. Epidemiol. 2010;63(8):938–939. doi: 10.1016/j.jclinepi.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 54.Loke YK, Kwok CS, Niruban A, Myint PK. Value of severity scales in predicting mortality from community-acquired pneumonia: Systematic review and meta-analysis. Thorax. 2010;65(10):884–890. doi: 10.1136/thx.2009.134072. [DOI] [PubMed] [Google Scholar]
- 55.Zhou XY, Ben SQ, Chen HL, Ni SS. A comparison of APACHE II and CPIS scores for the prediction of 30-day mortality in patients with ventilator-associated pneumonia. Int. J. Infect. Dis. 2015;30:144–147. doi: 10.1016/j.ijid.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 56.Zhang S, Zhang K, Yu Y, Tian B, Cui W, Zhang G. A new prediction model for assessing the clinical outcomes of ICU patients with community-acquired pneumonia: a decision tree analysis. Ann. Med. 2019;51(1):41–50. doi: 10.1080/07853890.2018.1518580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jones BE, Ying J, Nevers M, Alba PR, He T, Patterson OV, et al. Computerized mortality prediction for community-acquired pneumonia at 117 veterans affairs medical centers. Ann. Am. Thorac. Soc. 2021;18(7):1175–1184. doi: 10.1513/AnnalsATS.202011-1372OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu C-A, Chen C-M, Fang Y-C, Liang S-J, Wang H-C, Fang W-F, et al. Using a machine learning approach to predict mortality in critically ill influenza patients: A cross-sectional retrospective multicentre study in Taiwan. BMJ Open. 2020;10(2):e033898. doi: 10.1136/bmjopen-2019-033898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Christodoulou E, Ma J, Collins GS, Steyerberg EW, Verbakel JY, Van Calster B. A systematic review shows no performance benefit of machine learning over logistic regression for clinical prediction models. J. Clin. Epidemiol. 2019;110:12–22. doi: 10.1016/j.jclinepi.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 60.Lynam AL, Dennis JM, Owen KR, Oram RA, Jones AG, Shields BM, et al. Logistic regression has similar performance to optimised machine learning algorithms in a clinical setting: Application to the discrimination between type 1 and type 2 diabetes in young adults. Diagn. Progn. Res. 2020;4:6. doi: 10.1186/s41512-020-00075-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nusinovici S, Tham YC, Chak Yan MY, Wei Ting DS, Li J, Sabanayagam C, et al. Logistic regression was as good as machine learning for predicting major chronic diseases. J. Clin. Epidemiol. 2020;122:56–69. doi: 10.1016/j.jclinepi.2020.03.002. [DOI] [PubMed] [Google Scholar]
- 62.Ke, G. et al. LightGBM: a highly efficient gradient boosting decision tree. In Proceedings of the 31st International Conference on Neural Information Processing Systems 3149–3157 (Curran Associates Inc., 2017).
- 63.Zeng H, Yang C, Zhang H, Wu Z, Zhang J, Dai G, et al. A LightGBM-based EEG analysis method for driver mental states classification. Comput. Intell. Neurosci. 2019;2019:3761203. doi: 10.1155/2019/3761203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shaker B, Yu MS, Song JS, Ahn S, Ryu JY, Oh KS, et al. LightBBB: computational prediction model of blood–brain-barrier penetration based on LightGBM. Bioinformatics. 2021;37(8):1135–1139. doi: 10.1093/bioinformatics/btaa918. [DOI] [PubMed] [Google Scholar]
- 65.Sayed M, Riaño D, Villar J. Predicting duration of mechanical ventilation in acute respiratory distress syndrome using supervised machine learning. J. Clin. Med. 2021;10(17):3824. doi: 10.3390/jcm10173824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peng X, Li L, Wang X, Zhang H. A machine learning-based prediction model for acute kidney injury in patients with congestive heart failure. Front. Cardiovasc. Med. 2022;9:842873. doi: 10.3389/fcvm.2022.842873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chan KL, Leng X, Zhang W, Dong W, Qiu Q, Yang J, et al. Early identification of high-risk TIA or minor stroke using artificial neural network. Front. Neurol. 2019;10:171. doi: 10.3389/fneur.2019.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Abedi V, Goyal N, Tsivgoulis G, Hosseinichimeh N, Hontecillas R, Bassaganya-Riera J, et al. Novel screening tool for stroke using artificial neural network. Stroke. 2017;48(6):1678–1681. doi: 10.1161/STROKEAHA.117.017033. [DOI] [PubMed] [Google Scholar]
- 69.Lorencin I, Anđelić N, Španjol J, Car Z. Using multi-layer perceptron with Laplacian edge detector for bladder cancer diagnosis. Artif. Intell. Med. 2020;102:101746. doi: 10.1016/j.artmed.2019.101746. [DOI] [PubMed] [Google Scholar]
- 70.Hyland SL, Faltys M, Hüser M, Lyu X, Gumbsch T, Esteban C, et al. Early prediction of circulatory failure in the intensive care unit using machine learning. Nat. Med. 2020;26(3):364–373. doi: 10.1038/s41591-020-0789-4. [DOI] [PubMed] [Google Scholar]
- 71.Barda N, Riesel D, Akriv A, Levy J, Finkel U, Yona G, et al. Developing a COVID-19 mortality risk prediction model when individual-level data are not available. Nat. Commun. 2020;11(1):1–9. doi: 10.1038/s41467-020-18297-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Carter EL, Hutchinson PJ, Kolias AG, Menon DK. Predicting the outcome for individual patients with traumatic brain injury: A case-based review. Br. J. Neurosurg. 2016;30(2):227–232. doi: 10.3109/02688697.2016.1139048. [DOI] [PubMed] [Google Scholar]
- 73.España PP, Capelastegui A, Gorordo I, Esteban C, Oribe M, Ortega M, et al. Development and validation of a clinical prediction rule for severe community-acquired pneumonia. Am. J. Respir. Crit. Care Med. 2006;174(11):1249–1256. doi: 10.1164/rccm.200602-177OC. [DOI] [PubMed] [Google Scholar]
- 74.Luna CM, Famiglietti A, Absi R, Videla AJ, Nogueira FJ, Fuenzalida AD, et al. Community-acquired pneumonia: Etiology, epidemiology, and outcome at a teaching hospital in Argentina. Chest. 2000;118(5):1344–1354. doi: 10.1378/chest.118.5.1344. [DOI] [PubMed] [Google Scholar]
- 75.Huang D, He D, Gong L, Wang W, Yang L, Zhang Z, et al. Clinical characteristics and risk factors associated with mortality in patients with severe community-acquired pneumonia and type 2 diabetes mellitus. Crit. Care. 2021;25(1):419. doi: 10.1186/s13054-021-03841-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.El-Solh AA, Sikka P, Ramadan F, Davies J. Etiology of severe pneumonia in the very elderly. Am. J. Respir. Crit. Care Med. 2001;163(3 Pt 1):645–651. doi: 10.1164/ajrccm.163.3.2005075. [DOI] [PubMed] [Google Scholar]
- 77.Bowton DL, Scuderi PE, Haponik EF. The incidence and effect on outcome of hypoxemia in hospitalized medical patients. Am. J. Med. 1994;97(1):38–46. doi: 10.1016/0002-9343(94)90046-9. [DOI] [PubMed] [Google Scholar]
- 78.Marik PE. The clinical features of severe community-acquired pneumonia presenting as septic shock. Norasept II Study Investigators. J. Crit. Care. 2000;15(3):85–90. doi: 10.1053/jcrc.2000.16460. [DOI] [PubMed] [Google Scholar]
- 79.Paganin F, Lilienthal F, Bourdin A, Lugagne N, Tixier F, Génin R, et al. Severe community-acquired pneumonia: Assessment of microbial aetiology as mortality factor. Eur. Respir. J. 2004;24(5):779–785. doi: 10.1183/09031936.04.00119503. [DOI] [PubMed] [Google Scholar]
- 80.Wang X, Jiao J, Wei R, Feng Y, Ma X, Li Y, et al. A new method to predict hospital mortality in severe community acquired pneumonia. Eur. J. Intern. Med. 2017;40:56–63. doi: 10.1016/j.ejim.2017.02.013. [DOI] [PubMed] [Google Scholar]
- 81.Krüger S, Ewig S, Kunde J, Hartmann O, Marre R, Suttorp N, et al. Assessment of inflammatory markers in patients with community-acquired pneumonia—Influence of antimicrobial pre-treatment: Results from the German competence network CAPNETZ. Clin. Chim. Acta. 2010;411(23):1929–1934. doi: 10.1016/j.cca.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 82.Parker SG. Transient hyperphosphatasaemia in association with acute infection in adults. Postgrad. Med. J. 1991;67(789):638–642. doi: 10.1136/pgmj.67.789.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jinks MF, Kelly CA. The pattern and significance of abnormal liver function tests in community-acquired pneumonia. Eur. J. Intern. Med. 2004;15(7):436–440. doi: 10.1016/j.ejim.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 84.Xia W, Zhao D, Li C, Xu L, Yao X, Hu H. Prognostic significance of albumin to alkaline phosphatase ratio in critically ill patients with acute kidney injury. Clin. Exp. Nephrol. 2022;26(9):917–924. doi: 10.1007/s10157-022-02234-9. [DOI] [PubMed] [Google Scholar]
- 85.Milbrandt EB, Reade MC, Lee M, Shook SL, Angus DC, Kong L, et al. Prevalence and significance of coagulation abnormalities in community-acquired pneumonia. Mol. Med. 2009;15(11–12):438–445. doi: 10.2119/molmed.2009.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rijneveld AW, Weijer S, Bresser P, Florquin S, Vlasuk GP, Rote WE, et al. Local activation of the tissue factor-factor VIIa pathway in patients with pneumonia and the effect of inhibition of this pathway in murine pneumococcal pneumonia. Crit. Care Med. 2006;34(6):1725–1730. doi: 10.1097/01.CCM.0000218807.20570.C2. [DOI] [PubMed] [Google Scholar]
- 87.Abraham E. Coagulation abnormalities in acute lung injury and sepsis. Am. J. Respir. Cell Mol. Biol. 2000;22(4):401–404. doi: 10.1165/ajrcmb.22.4.f184. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.