Abstract
Background:
Demand for prostate MRI is increasing, but scan times remain long even in abbreviated biparametric MRIs (bpMRI). Deep learning can be leveraged to accelerate T2-weighted imaging (T2WI).
Purpose:
To compare conventional bpMRIs (CL-bpMRI) with bpMRIs including a deep learning-accelerated T2WI (DL-bpMRI) in diagnosing prostate cancer.
Study Type:
Retrospective.
Population:
Eighty consecutive men, mean age 66 years (47–84) with suspected prostate cancer or prostate cancer on active surveillance who had a prostate MRI from December 28, 2020 to April 28, 2021 were included. Follow-up included prostate biopsy or stability of prostate-specific antigen (PSA) for 1 year.
Field Strength and Sequences:
A 3 T MRI. Conventional axial and coronal T2 turbo spin echo (CL-T2), 3-fold deep learning-accelerated axial and coronal T2-weighted sequence (DL-T2), diffusion weighted imaging (DWI) with b = 50 sec/mm2, 1000 sec/mm2, calculated b = 1500 sec/mm2.
Assessment:
CL-bpMRI and DL-bpMRI including the same conventional diffusion-weighted imaging (DWI) were presented to three radiologists (blinded to acquisition method) and to a deep learning computer-assisted detection algorithm (DL-CAD). The readers evaluated image quality using a 4-point Likert scale (1 = nondiagnostic, 4 = excellent) and graded lesions using Prostate Imaging Reporting and Data System (PI-RADS) v2.1. DL-CAD identified and assigned lesions of PI-RADS 3 or greater.
Statistical Tests:
Quality metrics were compared using Wilcoxon signed rank test, and area under the receiver operating characteristic curve (AUC) were compared using Delong’s test. Significance: P = 0.05.
Results:
Eighty men were included (age: 66 ± 9 years; 17/80 clinically significant prostate cancer). Overall image quality results by the three readers (CL-T2, DL-T2) are reader 1: 3.72 ± 0.53, 3.89 ± 0.39 (P = 0.99); reader 2: 3.33 ± 0.82, 3.31 ± 0.74 (P = 0.49); reader 3: 3.67 ± 0.63, 3.51 ± 0.62. In the patient-based analysis, the reader results of AUC are (CL-bpMRI, DL-bpMRI): reader 1: 0.77, 0.78 (P = 0.98), reader 2: 0.65, 0.66 (P = 0.99), reader 3: 0.57, 0.60 (P = 0.52). Diagnostic statistics from DL-CAD (CL-bpMRI, DL-bpMRI) are sensitivity (0.71, 0.71, P = 1.00), specificity (0.59, 0.44, P = 0.05), positive predictive value (0.23, 0.24, P = 0.25), negative predictive value (0.88, 0.88, P = 0.48).
Conclusion:
Deep learning-accelerated T2-weighted imaging may potentially be used to decrease acquisition time for bpMRI.
Evidence Level:
3.
Technical Efficacy:
Stage 2.
Multiparametric MRI (mpMRI) of the prostate has been established as a tool in the diagnosis and management of prostate cancer especially after the development of Prostate Imaging Reporting and Data System (PI-RADS).1–3 It is used not only to detect prostate cancer but also to improve the targeting of prostate lesions on biopsy and to monitor posttreatment patients and patients with low-risk prostate cancer on active surveillance.4
A typical mpMRI protocol includes axial and coronal T2-weighted turbo spin echo (TSE), diffusion-weighted image (DWI), apparent diffusion coefficient (ADC) map, and dynamic contrast-enhanced (DCE) imaging as prescribed by PI-RADS.3 The entire mpMRI may require 30–45 minutes of image acquisition time. The long scan time impinges on patient comfort and also limits patient access to the MRI as fewer patients are able to be imaged.
A few strategies have been employed to decrease scan time. One solution is to remove DCE from the protocol as its use is secondary in PI-RADS.3 This biparametric MRI (bpMRI) protocol has been shown to have similar diagnostic capabilities as the full mpMRI and requires half the scan time.5–7 Another strategy is to accelerate individual sequence acquisitions.
Several MR acceleration techniques such as parallel imaging (PI) have been employed since the advent of MRI to ensure clinically feasible.8 Another example of a more recent novel imaging acceleration technique is compressed sensing (CS).9 Both PI and CS leverage under-sampling of k-space as the main time-saving strategy and rely on reconstruction techniques to recover image quality as under-sampling causes aliasing from not meeting Nyquist criteria.10 A commercial proprietary prototype sequence inspired by PI and CS was created, which under-samples k-space, but uses a deep learning-based reconstruction algorithm instead of traditional PI and CS techniques to create an image of diagnostic quality.
An unrolled variational network was used in the deep learning-based image reconstruction, which performs an iterative optimization inspired by compressed sensing.11,12 The algorithm alternates between data consistency and image regularization steps, with step sizes and image regularizations through convolutional neural networks having trainable parameters. It receives under-sampled k-space data, coilsensitivity maps and a normalization field as input, and provides the reconstructed TSE image as output. Since the reconstruction is designed to keep the original image contrast and to improve signal to noise ratio, the acquisition parameters determining image contrast between CL-T2 and DL-T2 are nearly identical, including echo time, repetition time, and echo train length.
This study aimed to compare the diagnostic ability of a prototype deep learning-accelerated T2-weighted image (DL-T2) against the conventional clinical T2-weighted image (CL-T2) in both a reader study and a study utilizing a commercially developed prototype deep learning-based computer-assisted detection (DL-CAD).
Materials and Methods
This retrospective study was approved by the institutional review board, was HIPAA compliant, and informed consent was waived. Authors associated with industry provided only technical support of maintaining and trouble-shooting DL-T2 acquisition and DL-CAD software and did not have access to data from the study.
Patients
The picture archiving and communication system (PACS) was searched for consecutive patients who had an MRI of the prostate from December 28, 2020 to April 28, 2021 with the indications of suspected prostate cancer or a diagnosed low-risk prostate cancer on active surveillance. Patients were included if their imaging protocol had both the conventional axial T2-weighted image (CL-T2) and the deep learning-accelerated axial T2 (DL-T2). DL-T2 sequence was routinely included in our clinical scan as a backup T2-weighted image in case of artifact. Other exclusion criteria included the presence of a hip arthroplasty, prior treatment of prostate cancer, or no adequate follow-up. Adequate follow up was defined as a prostate biopsy within 1 year of the MRI or stability of PSA of at least 1 year if mpMRI was prospectively determined to be PI-RADS 1 or 2. Patients on active surveillance were included if they demonstrated imaging stability since baseline imaging with associated MR targeted biopsy. Histopathology results were obtained either through MR-targeted transrectal ultrasound fusion biopsy or MR-targeted transperineal ultrasound fusion biopsy. In addition to the targeted biopsies, a 12 core sextant biopsy was also obtained at the same time. Core samples were evaluated by a clinical pathologist utilizing Gleason Grade Group (GG) scores with “clinically significant” prostate cancer (csPCa) defined as GG 2 or greater.13,14
A total of 137 patients were initially included in this retrospective study. The following patients were excluded: 15 for missing DL-T2, 28 for inadequate follow up, 13 for prior prostate cancer treatment, and 1 for presence of hip implant (Fig. 1). Eighty patients were included in the study (mean age 66 years, range 47–84 years), with details in Table 1.
FIGURE 1:
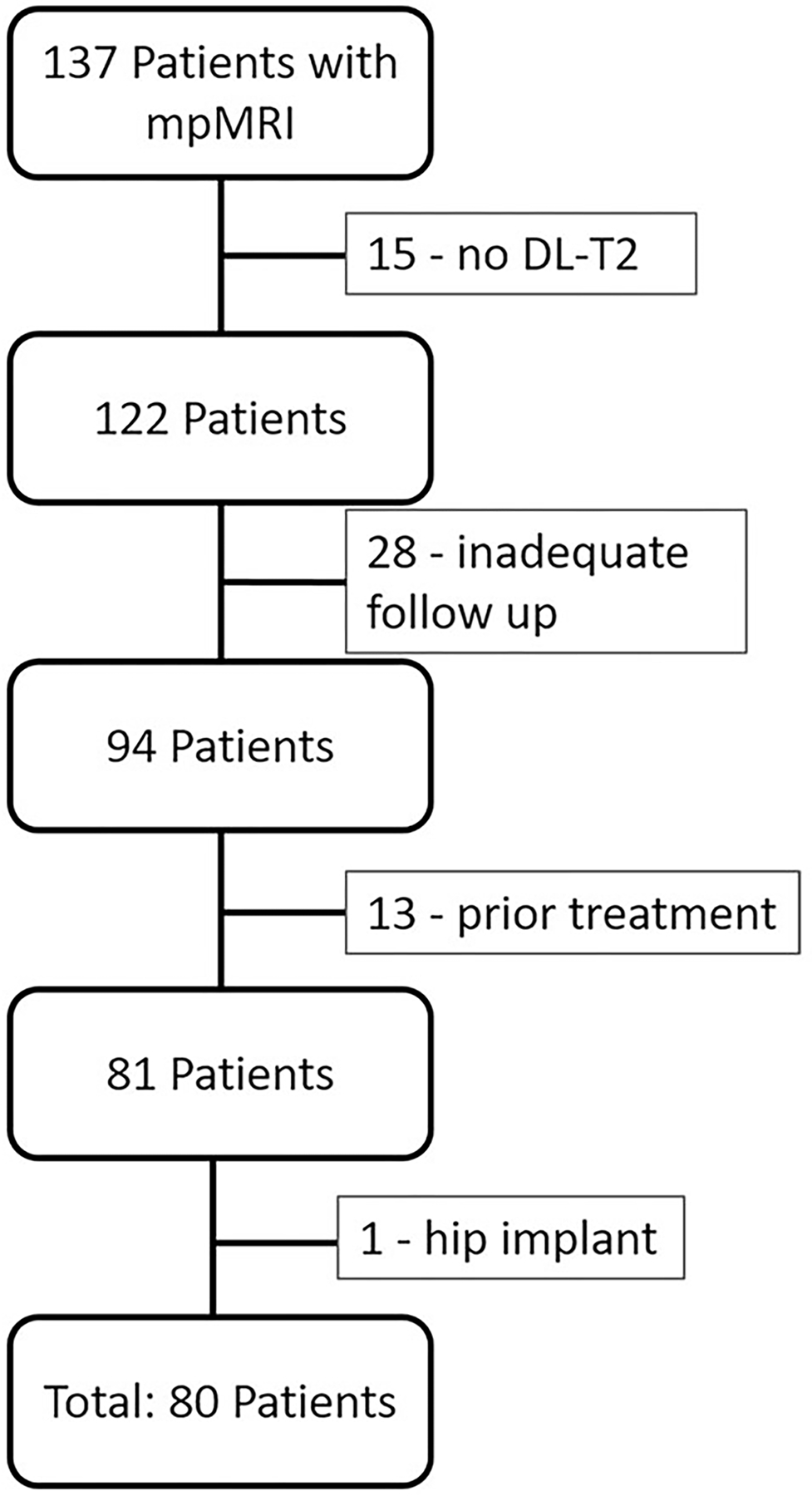
Patient inclusion flowchart. A total of 80 patients were included in the study.
TABLE 1.
Clinical and Demographic Characteristics
| Variable | Total Patients (n = 80) |
|---|---|
| Mean age (years) | 66 ± 9 |
| Number with MRI-targeted biopsies | 51 (64) |
| Number of patients on active surveillance with GG1 prostate cancer | 5 (6) |
| Number of patients classified as PI-RADS 1 with stable or decreasing PSA at 1 year follow-up | 24 (30) |
| Number with csPCa | 17 (21) |
| Number with grade group 2 prostate cancer | 10 (12) |
| Number with grade group 3 prostate cancer | 1 (1) |
| Number with grade group 4 prostate cancer | 5 (6) |
| Number with grade group 5 prostate cancer | 1 (1) |
Except where indicated, data are numbers of participants with percentages in parentheses.
csPCa = clinically significant prostate cancer (Grade Group 2 or greater). GG = grade group. PI-RADS = Prostate Imaging Reporting and Data System v2.1.
Imaging Protocol
All images were acquired on a 3 T MR imaging system, MAG-NETOM Prisma (Siemens Healthcare, Erlangen, Germany). Patients were scanned in supine position with an 18-channel body phased-array coil placed overlying the pelvis. From our institution’s clinical multiparametric prostate MRI protocol, the following sequences were acquired: axial T2 TSE, coronal T2 TSE, axial diffusion-weighted image (DWI) including b = 50 sec/mm2 and 1000 sec/mm2, and calculated DWI b = 1500 sec/mm2. Parameters of the sequences are provided in Table 2.
TABLE 2.
MRI Parameters for Sequences
| Axial | Coronal | DWI (b = 50 mm/2, 1000 mm/2) | |||
|---|---|---|---|---|---|
| CL-T2 | DL-T2 | CL-T2 | DL-T2 | ||
| TR (msec) | 4000 | 5100 | 4000 | 4870 | 6100 |
| TE (msec) | 100 | 100 | 100 | 100 | 80 |
| Averages | 3 | 1 | 2 | 1 | 6 (b50); 14 (b1000) |
| Field of view (mm × mm) | 320 × 224 | 320 × 216 | 320 × 224 | 320 × 116 | 130 × 97 |
| Slice thickness (mm) | 3 | 3 | 3 | 3 | 3 |
| Parallel imaging factor | 2 | 3 | 2 | 3 | 2 |
| Acquisition time (min:sec) | 3:46 | 1:08 | 2:58 | 1:17 | 5:09 |
DWI = diffusion-weighted image; CL-T2 = conventional T2 turbo spin echo; DL-T2 = deep learning; T2 turbo spin echo; TR = repetition time; TE = echo time, b5.
Deep Learning Acceleration Sequence
The deep learning acceleration sequence used in this study is a commercial proprietary prototype sequence trained and developed by Siemens (Erlangen, Germany). The deep learning-based reconstruction algorithm was trained in a supervised manner using more than 10,000 slices of fully sampled T2 TSE acquisitions obtained from volunteers using 1.5 T and 3 T MR scanning systems (MAGNETOM scanners, Siemens Healthcare) of various regions of the body, including head, pelvis, and knee. Different image contrasts were also used to increase the generalization of the obtained training parameters to a broad range of applications. The accelerated training dataset was created retrospectively by under-sampling k-space by a factor of 4 with phase resolution of 75%. The loss function was encompassed by L1-norm and multiscale version of structural similarity (SSIM) content between network prediction and ground truth. The training was implemented in PyTorch and performed using a commercially available GPU cluster with 32 GB of memory. The obtained trained parameters were then converted for prospective use in a scanner integrated inference framework. In actual deployment, there is an average 3 seconds inference time for CPU and 0.5 seconds for GPU. There is an overall ~3-fold acceleration of the axial DL-T2 vs. CL-T2 acquisition (1:08 minutes vs. 3:46 minutes), and a ~2-fold acceleration of the coronal DL-T2 vs. CL-T2 acquisition (1:17 minutes vs. 2:34 minutes).
Deep Learning-Based Computer-Aided Detection
A commercially developed proprietary deep learning-based prototypical computer-aided detection algorithm (DL-CAD) (MR Prostate AI, version 1.3.2, build July 07, 2021, front end build November 06, 2019, Siemens Healthcare) was used as an objective evaluator of both CL-bpMRI and DL-bpMRI. The DL-CAD receives the input of axial T2 TSE and axial DWI with two acquired b values. Preprocessing steps are first performed: 1) constructing a high b = 2000 sec/mm2 and the ADC map from the inputted DWI, 2) segmenting the prostate gland based on a deep learning algorithm, and 3) aligning the T2 and DWI/ADC images.15 Finally, the DL-CAD identifies lesions and produces a heat map of suspicious regions in the prostate. A false positive reduction algorithm is applied. The DL-CAD outputs a final heat map of any worrisome lesions and a PI-RADS-based assessment of 3–5 for each lesion identified. The DL-CAD does not identify PI-RADS 2 lesions as they are presumed to be likely benign.
The DL-CAD is a convolutional neural network based lesion identification and classification tool, which is detailed in the study by Yu et al.16,17 In summary, it was trained and tested on 2170 prostate exams with and without lesions. The training was performed with a hybrid ground truth model either provided through prostate biopsy results if available or PI-RADS grading from radiology reports if prostate biopsy results were not available.17
Image Evaluation
Two studies were created from each patient for a total of 160 studies evaluated by each radiologist reader and by DL-CAD. One of the two studies per patient contained axial and coronal CL-T2 (CL-bpMRI) and the second contained axial and coronal DL-T2 (DL-bpMRI). All exams also included DWI (b = 50 sec/mm2, 1000 sec/mm2), calculated high b = 1500 sec/mm2 image, and an ADC map. All studies were anonymized and randomized.
Radiologist Reader Evaluation
The 160 studies, scrubbed of acquisition method (80 CL-bpMRI and 80 DL-bpMRI), were randomly ordered and presented to three abdominal fellowship trained radiologists (6 [A.V.], 4 [P.S.], and 2 [R.P.] years of experience) via the institutional clinical PACS (Visage Imaging, Berlin, Germany). Readers were asked to identify and grade prostate lesions according to Prostate Imaging Reporting and Data System (PI-RADS) v2.1.3 Readers reported lesion locations based on sextant locations. The lesion locations were then correlated with biopsy results, either with the targeted lesion location or 12 core standard biopsy location.
Readers were also asked to evaluate both axial and coronal CL-T2 and DL-T2 for image quality based on a 4-point Likert scale (1: nondiagnostic, 2: fair, 3: good, 4: excellent) on the following metrics: overall quality, clarity of capsule, clarity transition and peripheral zone boundary, and clarity of periurethral area.18 A 4-point Likert scale was used to force readers to make a decision between favorable and unfavorable quality.
DL-CAD Image Evaluation
Axial images of the 160 studies including 80 with CL-T2 and 80 with DL-T2 were inputted into the DL-CAD for evaluation. The output of the system was lesion location and grade. This was collected for each case.
Statistics
Diagnostic statistics, including sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV), were obtained by evaluating performance of the readers and DL-CAD by study and by lesion. For patient-based analysis, true positive was designated if there was csPCa diagnosed at pathology in a patient where a lesion was identified on imaging as PI-RADS ≥3. For lesion-based evaluation, true positives were designated if an identified PI-RADS ≥3 lesion was in the same sextant as the csPCa on biopsy. In the reader study, the dominant lesion was assessed. All lesions identified by DL-CAD were included in the lesion-based assessment. In addition, the diagnostic statistics (sensitivity, specificity, positive predictive value [PPV], and negative predictive value) were modeled and tested individually by generalized estimating equation (GEE) with logit link and group effect as predictor, and the cluster effect of patient or lesion was adjusted as appropriate. Area under the curve (AUC) of the receiver operating curve (ROC) was obtained for radiology reader study and compared by Delong’s test. The difference and bootstrap confidence interval were calculated for diagnostic statistics with a 5% noninferiority margin. Light’s Kappa was reported for interreader reliability. Quality metrics were compared utilizing two-sided Wilcoxon signed-rank test, and the null hypothesis was that the DL-CAD had equal or higher scores. The significance level of this study was set to be 0.05.
Results
Radiology Reader Results
Image quality results are presented in Table 3. There was no significant difference in overall image quality for readers 1 (axial CL-T2: 3.72 ± 0.53, axial DL-T2: 3.89 ± 0.39, P = 0.99; coronal CL-T2: 3.86 ± 0.35, coronal DL-T2: 3.94 ± 0.25, P = 0.99) and 2 (axial CL-T2: 3.33 ± 0.82, axial DL-T2: 3.31 ± 0.74, P = 0.49; coronal CL-T2: 3.39 ± 0.71, coronal DL-T2: 3.31 ± 0.71, P = 0.20). Reader 3 rated CL-T2 with significantly higher overall image quality, though the difference was small (axial CL-T2: 3.67 ± 0.63, axial DL-T2: 3.51 ± 0.62; coronal CL-T2: 3.73 ± 0.52, coronal DL-T2: 3.48 ± 0.62). Figures 2 and 3 show a peripheral zone (PZ) and a transition zone (TZ) lesion, respectively, on both CL-T2 and DL-T2.
Table 3.
Reader Quality Evaluation Results
| CL-T2 | DL-T2 | P | |
|---|---|---|---|
| Axial | |||
| Reader 1 | |||
| Overall quality | 3.72 ± 0.53 | 3.89 ± 0.39 | 0.99 |
| Clarity of capsule | 3.76 ± 0.51 | 3.91 ± 0.33 | 0.99 |
| Clarity of PZ/TZ boundary | 3.76 ± 0.51 | 3.91 ± 0.33 | 0.99 |
| Clarity of periurethral area | 3.74 ± 0.52 | 3.85 ± 0.39 | 0.96 |
| Reader 2 | |||
| Overall quality | 3.33 ± 0.82 | 3.31 ± 0.74 | 0.49 |
| Clarity of capsule | 3.46 ± 0.75 | 3.52 ± 0.69 | 0.77 |
| Clarity of PZ/TZ boundary | 3.28 ± 0.83 | 3.19 ± 0.81 | 0.20 |
| Clarity of periurethral area | 3.36 ± 0.77 | 3.34 ± 0.75 | 0.41 |
| Reader 3 | |||
| Overall quality | 3.67 ± 0.63 | 3.51 ± 0.62 | 0.03 * |
| Clarity of capsule | 3.70 ± 0.58 | 3.64 ± 0.58 | 0.21 |
| Clarity of PZ/TZ boundary | 3.58 ± 0.63 | 3.34 ± 0.79 | 0.003 * |
| Clarity of periurethral area | 3.96 ± 0.19 | 3.89 ± 0.32 | 0.03 * |
| Coronal | |||
| Reader 1 | |||
| Overall quality | 3.86 ± 0.35 | 3.94 ± 0.25 | 0.99 |
| Clarity of capsule | 3.90 ± 0.30 | 3.95 ± 0.22 | 0.96 |
| Clarity of PZ/TZ boundary | 3.89 ± 0.32 | 3.95 ± 0.22 | 0.98 |
| Clarity of periurethral area | 3.87 ± 0.33 | 3.88 ± 0.32 | 0.73 |
| Reader 2 | |||
| Overall quality | 3.39 ± 0.71 | 3.31 ± 0.71 | 0.20 |
| Clarity of capsule | 3.37 ± 0.72 | 3.41 ± 0.73 | 0.70 |
| Clarity of PZ/TZ boundary | 3.38 ± 0.69 | 3.21 ± 0.76 | 0.06 |
| Clarity of periurethral area | 3.39 ± 0.63 | 3.32 ± 0.73 | 0.26 |
| Reader 3 | |||
| Overall quality | 3.73 ± 0.52 | 3.48 ± 0.62 | 0.003 * |
| Clarity of capsule | 3.81 ± 0.43 | 3.65 ± 0.56 | 0.02 * |
| Clarity of PZ/TZ boundary | 3.08 ± 0.53 | 2.84 ± 0.65 | 0.003 * |
| Clarity of periurethral area | 3.99 ± 0.11 | 3.89 ± 0.32 | 0.01 * |
CL-T2 = conventional T2 turbo spin echo; DL-T2 = deep learning T2 turbo spin echo.
Significant difference.
FIGURE 2:
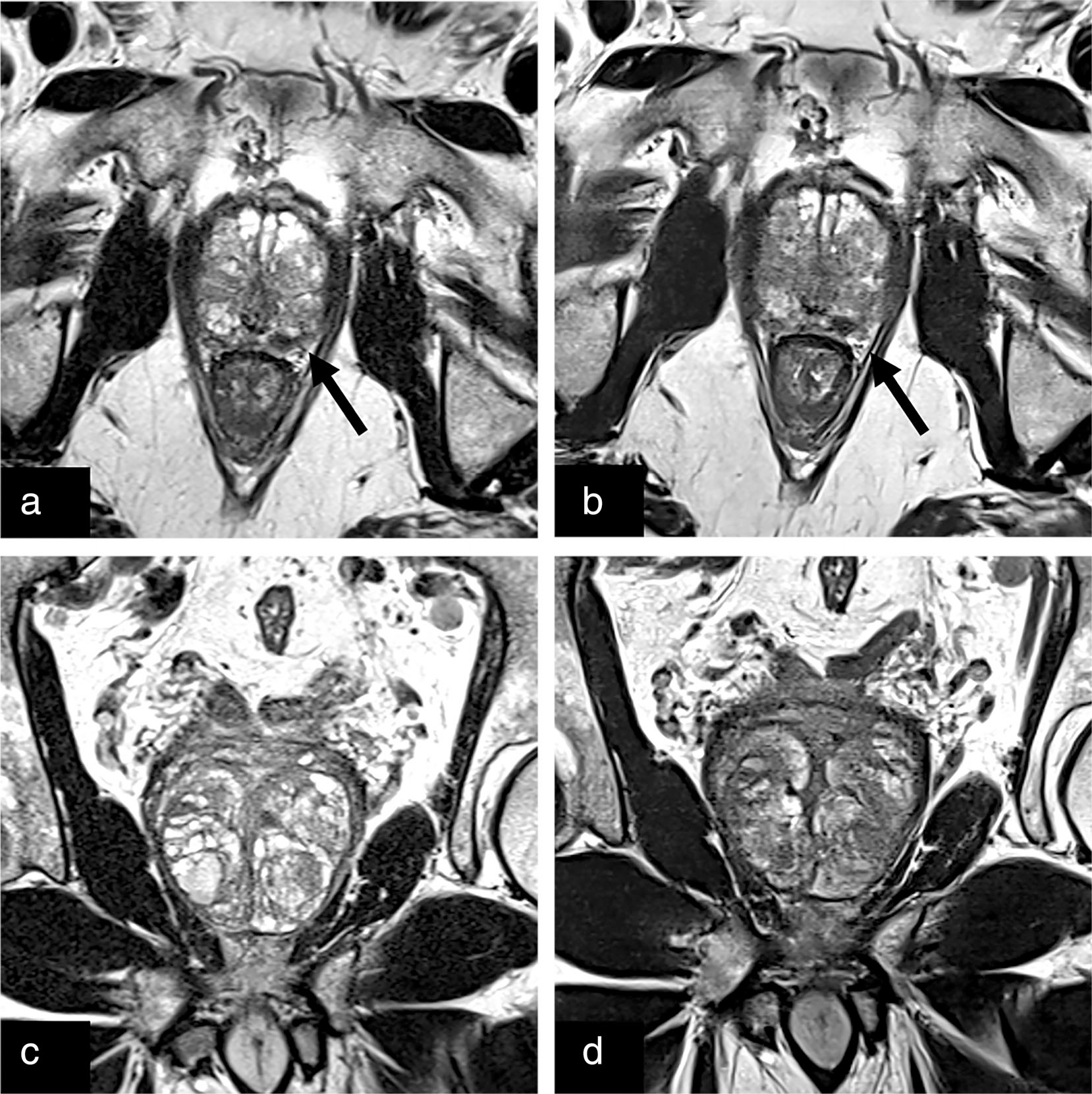
Conventional (a) and deep learning reconstructed (b) axial T2-weighted images of the prostate with an equally well-visualized T2 hypointense lesion in the left posterolateral prostate (black arrow) later biopsied to be a grade group 2 lesion. The adjacent neurovascular bundles can be seen on both clinical and deep learning reconstructed images. Clinical (c) and deep learning reconstructed (d) coronal T2 turbo spin echo with clear prostatic capsule and BPH capsules.
FIGURE 3:
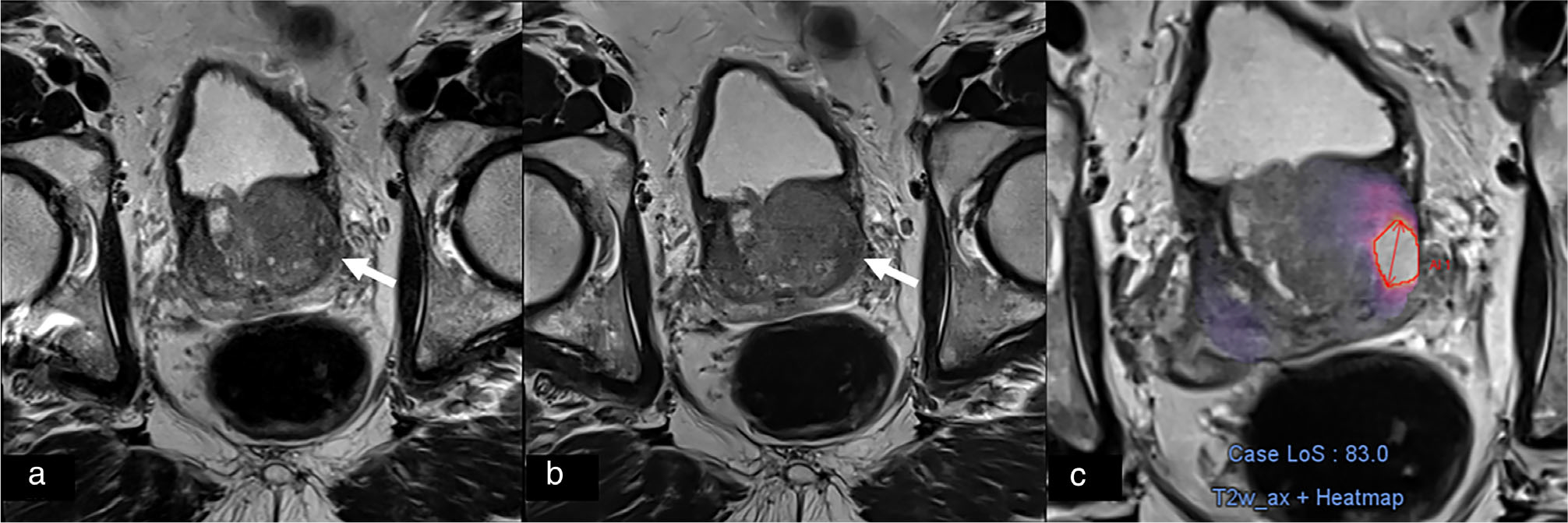
Conventional (a) and deep learning reconstructed (b) T2 turbo spin echo of the prostate with a left lateral transition zone lesion, which was identified with deep learning computer-assisted detection only on deep learning reconstructed T2 as denoted by the heat map (c).
Patient- and lesion-based analysis results are presented in Table 4. There was no significant difference in AUC of the ROC curve between CL-bpMRI and DL-bpMRI in all readers in patient-based analysis: (CL-bpMRI, DL-bpMRI) – reader 1 (0.77, 0.78, P = 0.98); reader 2 (0.65, 0.66, P = 0.95); reader 3 (0.57, 0.60, P = 0.52); lesion-based (CL-bpMRI, DL-bpMRI) – reader 1 (0.71, 0.70, P = 0.92); reader 2 (0.58, 0.62, P = 0.70); reader 3 (0.57, 0.60, P = 0.70). In patient-based analysis, there was no significant difference in AUC of ROC between CL-bpMRI and DL-bpMRI: (CL-bpMRI, DL-bpMRI) – reader 1 (0.71, 0.70, P = 0.92); reader 2 (0.58, 0.62, P = 0.70); reader 3 (0.57, 0.60, P = 0.70). Light’s kappa was fair, measuring 0.35 for interreader variation. In lesion-based analysis results, reader 1 identified a total of 34 lesions on CL-bpMRI (29 PZ, 5 TZ) and 27 lesions on DL-bpMRI (21 PZ, 6 TZ). Reader 2 identified a total of 44 lesions on CL-bpMRI (33 PZ, 11 TZ) and 51 lesions on DL-bpMRI (32 PZ, 19 TZ). Reader 3 identified a total of 17 lesions on CL-bpMRI (16 PZ, 1 TZ) and 15 lesions on DL-bpMRI (12 PZ, 3 TZ).
TABLE 4.
Reader Study Results
| CL-bpMRI | DL-bpMRI | Difference (CI) | P | |
|---|---|---|---|---|
| Patient based | ||||
| Reader 1 | ||||
| Sensitivity | 0.65 | 0.65 | 0.00 (−0.33, 0.29) | 1.00 |
| Specificity | 0.75 | 0.81 | 0.06 (−0.07, 0.19) | 0.25 |
| PPV | 0.41 | 0.41 | 0.00 (−0.05, 0.03) | 0.44 |
| NPV | 0.87 | 0.87 | 0.00 (−0.05, 0.05) | 0.77 |
| AUC | 0.77 | 0.78 | 0.00 (−0.12, 0.13) | 0.98 |
| Reader 2 | ||||
| Sensitivity | 0.59 | 0.65 | 0.06 (−0.18, 0.29) | 0.56 |
| Specificity | 0.62 | 0.51 | −0.11 (−0.26, 0.06) | 0.14 |
| PPV | 0.23 | 0.23 | 0.01 (−0.07, 0.07) | 0.69 |
| NPV | 0.86 | 0.86 | 0.00 (−0.08, 0.08) | 0.93 |
| AUC | 0.65 | 0.66 | 0.00 (−0.13, 0.14) | 0.95 |
| Reader 3 | ||||
| Sensitivity | 0.29 | 0.29 | 0.00 (−0.21, 0.21) | 1.00 |
| Specificity | 0.83 | 0.87 | 0.05 (−0.04, 0.13) | 0.18 |
| PPV | 0.33 | 0.33 | 0.00 (−0.12, 0.11) | 0.42 |
| NPV | 0.81 | 0.81 | 0.00 (−0.03, 0.03) | 0.64 |
| AUC | 0.57 | 0.60 | 0.04 (−0.07, 0.15) | 0.52 |
| Lesion based | ||||
| Reader 1 | ||||
| Sensitivity | 0.53 | 0.53 | 0.00 (−0.26, 0.31) | 1.00 |
| Specificity | 0.72 | 0.79 | 0.07 (−0.06, 0.18) | 0.39 |
| PPV | 0.33 | 0.39 | 0.06 (−0.20, 0.27) | 0.67 |
| NPV | 0.86 | 0.87 | 0.01 (−0.08, 0.11) | 0.85 |
| AUC | 0.71 | 0.70 | −0.01a | 0.92 |
| Reader 2 | ||||
| Sensitivity | 0.47 | 0.59 | 0.12 (−0.14, 0.40) | 0.49 |
| Specificity | 0.60 | 0.50 | −0.1 (−0.24, 0.04) | 0.25 |
| PPV | 0.24 | 0.24 | 0.00 (−0.15, 0.17) | 0.98 |
| NPV | 0.81 | 0.82 | 0.01 (−0.11, 0.15) | 0.92 |
| AUC | 0.58 | 0.62 | 0.04a | 0.73 |
| Reader 3 | ||||
| Sensitivity | 0.29 | 0.29 | 0.00 (−0.28, 0.26) | 1.00 |
| Specificity | 0.83 | 0.87 | 0.05 (−0.04, 0.14) | 0.46 |
| PPV | 0.31 | 0.39 | 0.07 (−0.23, 0.38) | 0.69 |
| NPV | 0.81 | 0.82 | 0.01 (−0.09, 0.12) | 0.90 |
| AUC | 0.57 | 0.60 | 0.04a | 0.70 |
Confidence interval not provided as data for lesion-based AUC is not paired.
CL-bpMRI = conventional biparametric MRI; DL-bpMRI = deep learning based biparametric MRI; CI = confidence interval; PPV = positive predictive value; NPV = negative predictive value; AUC = area under the receiver operating characteristic curve.
DL-CAD Results
DL-CAD did not have significantly different sensitivity in patient-based evaluation or lesion-based evaluation when assessing CL-bpMRI compared to DL-bpMRI but had significantly lower specificity when evaluating DL-bpMRI (Table 5). On lesion-based analysis, DL-CAD identified 22 PZ lesions on CL-bpMRI and 24 PZ lesions on DL-bpMRI and 30 TZ lesions on CL-bpMRI and 41 TZ lesions on DL-bpMRI. Two csPCa were missed on the CL-bpMRI but detected on DL-bpMRI and both were located in the TZ. An example of TZ lesion identified on DL-bpMRI and not on CL-bpMRI can be seen in Fig. 3.
TABLE 5.
Deep Learning Computer-Assisted Detection Algorithm Results
| CL-bpMRI | DL-bpMRI | Difference (CI) | P | |
|---|---|---|---|---|
| Patient based | ||||
| Sensitivity | 0.71 | 0.71 | 0.00 (−0.21, 0.19) | 1.00 |
| Specificity | 0.59 | 0.44 | −0.14 (−0.30, −0.00) | 0.05 * |
| PPV | 0.23 | 0.24 | 0.01 (−0.05, 0.06) | 0.25 |
| NPV | 0.88 | 0.88 | 0.01 (−0.12, 0.09) | 0.48 |
| Lesion based | ||||
| Sensitivity | 0.59 | 0.53 | −0.06 (−0.31, 0.22) | 0.73 |
| Specificity | 0.58 | 0.42 | −0.16 (−0.28, −0.04) | 0.03 * |
| PPV | 0.19 | 0.14 | −0.05 (−0.18, 0.05) | 0.42 |
| NPV | 0.62 | 0.57 | −0.05 (−0.17, 0.05) | 0.05 * |
CL-bpMRI = conventional biparametric MRI; DL-bpMRI = deep learning based biparametric MRI; PPV = positive predictive value; NPV = negative predictive value.
Significant difference.
Discussion
In this study, we assessed a 3-fold accelerated prototype DL-T2 sequence. We compared the image quality and diagnostic ability of CL-bpMRI and DL-bpMRI to identify csPCa in a reader study and with prostate DL-CAD. Two of the three readers rated no significant difference between the overall image quality of CL-T2 and DL-T2. The third reader rated slightly increased overall image quality of CL-T2 over DL-T2. All three readers demonstrated no significant difference in sensitivity, specificity, positive predictive value, negative predictive value, and AUC of the ROC when assessing CL-bpMRI and DL-bpMRI both on a patient-based analysis and lesion-based analysis. DL-CAD demonstrated only slightly increased specificity assessing CL-bpMRI when compared to DL-bpMRI on lesion-based analysis and no significant difference in the remaining diagnostic statistics on patient-based analysis or lesion-based analysis.
Although there was no significant difference in most of the comparisons, when assessing for noninferiority, the difference in diagnostic results between CL-bpMRI and DL-bpMRI were greater than the 5% noninferiority margin set. This indicates that the study may be under-powered. However, these promising preliminary results suggest optimistic results for larger noninferiority studies.
In the last few years, there has been an increase in the number of published studies on deep learning algorithms to decrease MR acquisition time.19 Only a few small studies have focused on the prostate. Gassenmaier et al investigated the image quality and T2 PI-RADS scores of a similar DL-T2 sequence on 30 patients and then on 60 patients with similar results.12,20 The readers in their studies rated the DL-T2 to have significantly higher of overall quality than CL-T2 and no significant difference between the T2 TSE for PI-RADS rating. Our study was more clinically focused and presented readers with full bpMRIs demonstrating no significant difference in the diagnostic abilities of DL-bpMRI and CL-bpMRI for all readers.12,20 Kim et al investigated two versions of a DL-T2 sequence including a high-resolution version (3-fold acceleration) and a low-resolution version (4-fold acceleration). Both demonstrated similar qualitative and quantitative quality and PI-RADS ratings between the two sequences compared to the conventional T2 TSE.21
Johnson et al developed a deep learning-accelerated T2 TSE with 4-fold acceleration; however, they also included a 3-fold accelerated DWI with a fully deep learning-accelerated bpMRI requiring less than 4 minutes of total acquisition time.18 Their pilot study was small including only 20 patients for evaluation image quality and an additional 10 patients evaluating lesion assessment. Three of four readers rated no significant difference in overall quality between DL and CL images, and there was no significant difference in the number of PI-RADS 3 or greater lesions rated in either DL-bpMRI or CL-bpMRI.18
In the current study, the DL-CAD was used as an additional evaluator. Since the DL-CAD was trained only on conventional T2-weighted images and not DL-T2, it is a unique opportunity to assess whether DL-T2 could pass for CL-T2. Only specificity in the lesion-based assessment was significantly higher in CL-bpMRI when compared with DL-bpMRI.
On further investigation of the lesions identified, it was noted that DL-CAD identified more TZ lesions on the DL-bpMRI than the CL-bpMRI, while DL-CAD identified similar numbers of PZ lesions. The discrepancy of the number of TZ lesions identified may potentially be related to the noise profile difference between the two sequences. Since malignant lesions in the TZ are often described as homogeneous moderate T2 hypointense and with an “erased charcoal” appearance, a smoother, decreased noise profile may potentially increase sensitivity to TZ lesions and account for the decreased specificity on evaluating DL-bpMRI.3,22 All readers identified more TZ lesions on DL-bpMRI compared to CL-bpMRI but were more similar in range. Dithering may be a possible solution to achieve similar noise levels, which is under investigation.23
Our DL-T2 can be compared to PI and CS, especially as it draws inspiration and elements from both. DL-T2 undersamples in a similar fashion to parallel imaging and also requires the same coil sensitivity maps.8 Compared to CS, DL-T2 utilizes similar iterative processes but has several advantages. In CS, certain imaging parameters are used, and calibrations are performed on “average” patients or phantoms to be applied in all acquisitions.9 Because of the wide range of patient body habitus and other physical factors specific to each patient, the sequence may not be optimized to produce the best image for every patient.9 DL-T2 is more flexible and is able to adjust in a wider range of inputs correlating with the range of the training dataset. In addition, CS reconstruction requires heavy computing at time of image acquisition, necessitating accessibility to powerful computational hardware at time of scanning.9 Again, the computational heavy portion of DL-T2 is conducted during training, leaving a faster more manageable reconstruction at time of image acquisition.
Replacing conventional axial and coronal CL-T2 with DL-T2 can save 3–4 minutes on a prostate MRI. Although 3–4 minutes may seem small, if compounded over many exams, it can be quite profound. Our institution performs more than 6000 prostate mpMRIs per year and this change would save more than 24,000 minutes equaling 800 full 30 minutes prostate mpMRIs. In societies with long wait times for prostate mpMRIs, this would greatly increase access and impact prostate cancer care.
Limitations
The overall reader results demonstrated lower than expected sensitivity and specificity. This may be attributed to experience level and especially less experience at reading bpMRI studies rather than full mpMRI, as nearly all of the prostate MRIs at our institution are performed with intravenous contrast. The aim of the study, however, was to study the difference between CL-bpMRI and DL-bpMRI, which was demonstrated in our results. In a similar vein, the diagnostic results from DL-CAD were lower than reported in other studies. This may be due to suboptimization of the algorithm to the studies presented as the algorithm was not trained on studies from this institution.
To adequately assess a full range of patients with and without csPCa, we included studies that were prospectively assessed as PI-RADS 1 or 2. Not all of these patients had subsequent associated biopsies; however, those who did not have subsequent biopsies did demonstrate stability in PSA or subsequent MRI. A few studies were of patients in our active surveillance program, and these patients also did not undergo subsequent biopsy but also demonstrated PSA stability This is a single institution study performed on a single scanner type from a single vendor, which limits the evaluation of universal applicability.
Conclusion
This preliminary study demonstrated no significant difference in diagnostic evaluation by readers. DL-T2 may potentially replace CL-T2 in bpMRI, thus decreasing the acquisition time of prostate MRI.
References
- 1.Kasivisvanathan V, Rannikko AS, Borghi M, et al. MRI-targeted or standard biopsy for prostate-cancer diagnosis. N Engl J Med 2018;378(19):1767–1777. 10.1056/NEJMoa1801993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rouviere O, Puech P, Renard-Penna R, et al. Use of prostate systematic and targeted biopsy on the basis of multiparametric MRI in biopsynaive patients (MRI-FIRST): A prospective, multicentre, paired diagnostic study. Lancet Oncol 2019;20(1):100–109. 10.1016/S1470-2045(18)30569-2. [DOI] [PubMed] [Google Scholar]
- 3.Prostate Imaging Reporting Data System version 2.1. American College of Radiology, 2019. [Google Scholar]
- 4.Giganti F, Stabile A, Stavrinides V, et al. Natural history of prostate cancer on active surveillance: Stratification by MRI using the PRECISE recommendations in a UK cohort. Eur Radiol 2021;31(3):1644–1655. 10.1007/s00330-020-07256-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alabousi M, Salameh JP, Gusenbauer K, et al. Biparametric vs multi-parametric prostate magnetic resonance imaging for the detection of prostate cancer in treatment-naive patients: A diagnostic test accuracy systematic review and meta-analysis. BJU Int 2019;124(2):209–220. 10.1111/bju.14759. [DOI] [PubMed] [Google Scholar]
- 6.Cuocolo R, Verde F, Ponsiglione A, et al. Clinically significant prostate cancer detection with biparametric MRI: A systematic review and meta-analysis. AJR Am J Roentgenol 2021;216(3):608–621. 10.2214/AJR.20.23219. [DOI] [PubMed] [Google Scholar]
- 7.Woo S, Suh CH, Kim SY, Cho JY, Kim SH, Moon MH. Head-to-head comparison between biparametric and multiparametric MRI for the diagnosis of prostate cancer: A systematic review and meta-analysis. AJR Am J Roentgenol 2018;211(5):W226–W241. 10.2214/AJR.18.19880. [DOI] [PubMed] [Google Scholar]
- 8.Deshmane A, Gulani V, Griswold MA, Seiberlich N. Parallel MR imaging. J Magn Reson Imaging 2012;36(1):55–72. 10.1002/jmri.23639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng L, Benkert T, Block KT, Sodickson DK, Otazo R, Chandarana H. Compressed sensing for body MRI. J Magn Reson Imaging 2017;45(4):966–987. 10.1002/jmri.25547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maciejewski MW, Mobli M, Schuyler AD, Stern AS, Hoch JC. Data sampling in multidimensional NMR: Fundamentals and strategies. Top Curr Chem 2012;316:49–77. 10.1007/128_2011_185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammernik K, Klatzer T, Kobler E, et al. Learning a variational network for reconstruction of accelerated MRI data. Magn Reson Med 2018;79(6):3055–3071. 10.1002/mrm.26977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gassenmaier S, Afat S, Nickel D, Mostapha M, Herrmann J, Othman AE. Deep learning-accelerated T2-weighted imaging of the prostate: Reduction of acquisition time and improvement of image quality. Eur J Radiol 2021;137:109600. 10.1016/j.ejrad.2021.109600. [DOI] [PubMed] [Google Scholar]
- 13.Humphrey PA. Gleason grading and prognostic factors in carcinoma of the prostate. Mod Pathol 2004;17(3):292–306. 10.1038/modpathol.3800054. [DOI] [PubMed] [Google Scholar]
- 14.Pierorazio PM, Walsh PC, Partin AW, Epstein JI. Prognostic Gleason grade grouping: Data based on the modified Gleason scoring system. BJU Int 2013;111(5):753–760. 10.1111/j.1464-410X.2012.11611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang D, Xu D, Zhou SK, et al. Automatic liver segmentation using an adversarial image-to-image network. In: Maier-Hein KH, Franz A, Jannin P, Duchesne S, Descoteaux M, Collins DL, editors. Medical image computing and computer assisted intervention society - MICCAI 2017 – 20th international conference. Heidelberg, Germany: Springer Verlag; 2017; p 507–515. [Google Scholar]
- 16.Yu X, Lou B, Bibo S, et al. False positive reduction using multiscale contextual features for prostate cancer detection in multi-parametric MRI scans. 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI). Iowa City, IA 2020; p 1355–1359. 10.1109/ISBI45749.2020.9098338. [DOI] [Google Scholar]
- 17.Winkel DJ, Tong A, Lou B, et al. A novel deep learning based computer-aided diagnosis system improves the accuracy and efficiency of radiologists in reading biparametric magnetic resonance images of the prostate: Results of a multireader, multicase study. Invest Radiol 2021;56:605–613. 10.1097/RLI.0000000000000780. [DOI] [PubMed] [Google Scholar]
- 18.Johnson PM, Tong A, Donthireddy A, et al. Deep learning reconstruction enables highly accelerated biparametric MR imaging of the prostate. J Magn Reson Imaging 2021;56:184–195. 10.1002/jmri.28024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y, Schönlieb CB, Liò P, et al. AI-based reconstruction for fast MRI—A systematic review and meta-analysis. Proc IEEE 2022;110(2):224–245. 10.1109/JPROC.2022.3141367. [DOI] [Google Scholar]
- 20.Gassenmaier S, Afat S, Nickel MD, et al. Accelerated T2-weighted TSE imaging of the prostate using deep learning image reconstruction: A prospective comparison with standard T2-weighted TSE imaging. Cancers (Basel) 2021;13(14):3593. 10.3390/cancers13143593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim EH, Choi MH, Lee YJ, Han D, Mostapha M, Nickel D. Deep learning-accelerated T2-weighted imaging of the prostate: Impact of further acceleration with lower spatial resolution on image quality. Eur J Radiol 2021;145:110012. 10.1016/j.ejrad.2021.110012. [DOI] [PubMed] [Google Scholar]
- 22.Purysko AS, Rosenkrantz AB, Barentsz JO, Weinreb JC, Macura KJ. PI-RADS version 2: A pictorial update. Radiographics 2016;36(5):1354–1372. 10.1148/rg.2016150234. [DOI] [PubMed] [Google Scholar]
- 23.Recht MP, Zbontar J, Sodickson DK, et al. Using deep learning to accelerate knee MRI at 3 T: Results of an interchangeability study. AJR Am J Roentgenol 2020;215(6):1421–1429. 10.2214/AJR.20.23313. [DOI] [PMC free article] [PubMed] [Google Scholar]


