Cells respond to a variety of signals, adapting their behavior and pattern of gene expression as a function of the situation. In Escherichia coli, the reactions to environmental stresses—heat shock, starvation, and the like—have been termed global responses. During gradual changes, such as the transition from exponential to stationary phase in complex medium, E. coli gradually adjusts its gene expression and reduces cell size. In addition to signals from the environment, cells also respond to internal signals that allow them to evaluate their physiological state, coordinate their biosynthetic capacities, and determine their readiness to proceed through the cell cycle. Little is known about the nature of these internal signals, although they clearly play a major role in establishing the homeostatic controls and checkpoints that make for balanced growth and a harmonious cell cycle. Cell division, the most visible outward sign of equilibrated growth, is tightly regulated. During balanced growth in given culture conditions, E. coli cells all divide at virtually the same size, but the actual value of this mass is a strong function of the richness of the medium, larger when growth is faster, with up to a 10-fold difference in division mass between very rich and very poor media. In the present review we examine the evidence that the nucleotide ppGpp (guanosine-5′,3′-bis-pyrophosphate), first identified as a signal of amino acid starvation, is also involved in cell division regulation.
When wild-type E. coli cells are starved for an amino acid, they quickly arrest the synthesis of stable RNA. The effector of this stringent response is the nucleotide ppGpp, synthesized on stalled ribosomes by the RelA protein (10). This nucleotide interacts with RNA polymerase to reduce transcription initiation at the promoters of “stringent” operons, including those coding for rRNA and tRNA. The nucleotide ppGpp is also synthesized when the cell is starved for carbon or phosphate. In these circumstances, the active synthetase is the SpoT protein, an interesting bifunctional enzyme which has overlapping synthetase and hydrolase domains (23). However, the role of ppGpp is not limited to stress conditions. It is a positive transcriptional effector of a number of operons, including some coding for amino acid biosynthetic enzymes. In addition, during steady-state growth the ppGpp pool, together with ATP and GTP (20), may contribute to homeostatic regulation of the ribosome concentration, adjusting it to the cell’s ability to produce aminoacyl-tRNA.
It has been suggested that ppGpp might play a role in replication initiation (10). More recently, several observations have hinted at a role for ppGpp in the regulation of cell division as well. First, cells entirely lacking ppGpp tend to filament (63), suggesting a division-promoting role for the nucleotide. A second argument has emerged from studies of penicillin-binding protein 2 (PBP2). E. coli cells, constrained by their rigid peptidoglycan layer, are rod shaped. PBP2, one of the enzymes responsible for the polymerization and insertion of new glycan chains, is required for maintenance of the rod shape (53). When PBP2 activity is inhibited, either genetically or by the specific β-lactam mecillinam, cells become spherical. In rich medium, these spheres stop dividing, increase in diameter, and ultimately die (58). Division in the absence of PBP2 activity can be restored either by increasing the concentration of the cell division proteins FtsQ, FtsA, and FtsZ (42, 58) or, surprisingly, by increasing the ppGpp concentration (33, 57). Since ppGpp is known principally as a transcriptional effector, a simple hypothesis was that it stimulates the transcription of the ftsQAZ operon (58). We examine below the complex regulation of this operon and show that, although ppGpp is clearly involved in division regulation, this simple hypothesis does not explain the extant data.
Of all cell division proteins identified to date in bacteria, the best known is unquestionably FtsZ. This GTP-hydrolyzing tubulin-like protein, found in eubacteria, archæbacteria, and eukaryotic organelles, polymerizes in the presence of GTP (or GDP) to form a ring-like structure at midcell, associated with the cytoplasmic membrane (17, 35). The ring constricts as the septum is synthesized, after which it depolymerizes. Several other division proteins are recruited to the FtsZ ring; these include FtsA (2, 37), ZipA (28), FtsI (PBP3) (59, 62), FtsN (1), FtsW (59), and FtsK (60, 65). The FtsZ/FtsA ratio, approximately 50:1 (36), is important in order for septation to take place: an excess of either protein results in a division block that is relieved by simultaneous amplification of the other (12, 13). In contrast, excess FtsQ, whose function remains to be determined, does not affect division (9).
The FtsZ protein acts early in septation and is required throughout the process. In its absence, no midcell ring is formed; although another protein may be required to trigger FtsZ polymerization and ring formation, it has not been identified. E. coli itself seems to consider the FtsZ step the commitment to division, since FtsZ activity or synthesis is the target of seven endogenous division inhibitors (SulA, SfiC, MinC-MinD, DicB-MinD, Kilrac, DicF, and StfZ).
TRANSCRIPTION OF THE FTSQAZ OPERON
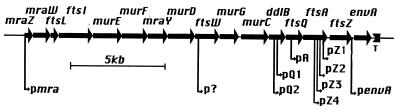
The ftsZ gene lies just downstream of ftsQ and ftsA in a complex operon with a number of promoters (Fig. 1), located at 2 min on the E. coli map. Two promoters upstream of ftsQ (pQ2 and pQ1) transcribe the entire operon, and three within ftsA (pZ4, pZ3, and pZ2) transcribe only ftsZ (4). A promoter within ftsQ, called pA and potentially governing ftsA and ftsZ, has been reported in gene fusion studies (15, 18, 48), although the corresponding mRNA species has not been detected (4). An RNA species whose 5′ end is downstream of pZ2, once attributed to a promoter, “pZ1,” has been shown to result from RNase E processing (8).
FIG. 1.
The 2-min cluster of cell division and cell wall genes. Transcription is from left to right; T represents the only transcriptional terminator acting in this direction. The putative promoter in ftsW is deduced as explained in the text. The envA gene, whose product is involved in lipid A synthesis, is also called lpxC; the ftsI gene, whose product is PBP3, is also called pbpB.
Are there additional promoters required for full ftsQAZ expression? The question is pertinent since there are no transcriptional terminators in this 16-gene cluster of cell division and cell wall genes, all of which are transcribed in the same direction (Fig. 1). Dai and Lutkenhaus (11) reported that the presence at attλ of a λ16-2 prophage, which carries a 10-kb DNA fragment going from the middle of ftsW to beyond envA and thus includes the ftsQAZ region with all neighboring promoters, did not allow them to delete the ftsZ locus at 2 min; they concluded that one or more promoters upstream of the middle of ftsW are required to obtain ftsZ expression sufficient for cell viability. More recently, Hara et al. (30) replaced the pmra promoter, at the very beginning of this cluster, with plac. The resulting strain required isopropyl-β-d-thiogalactopyranoside (IPTG) for growth unless it carried in trans, on a plasmid, the genes from pmra through ftsW. This implies that genes downstream of ftsW have expression sufficient for cell viability without any contribution from promoters upstream of ftsW. Nevertheless, in the plasmid-bearing strain in the absence of IPTG, MurC activity is reduced sixfold and MurG activity is reduced threefold while the amount of FtsZ protein is reduced by 30% (38), strongly suggesting that pmraA does indeed contribute to ftsZ transcription. A similar conclusion was reached very recently by Flärdh et al. (19). These authors show that the activity of an ftsZ::lacZ fusion in situ on the chromosome, with all upstream promoters intact, is reduced threefold when a polar Ω element is inserted within the ddlB gene upstream of pQ2, suggesting that up to two-thirds of ftsQAZ transcription may depend on upstream promoters or activating elements. These upstream transcriptional signals require confirmation and precise mapping; their regulation is for the moment entirely unknown.
The relative contribution of each of the mapped promoters has been measured by a number of groups, using either transcriptional fusions or direct mRNA measurements. Results with lacZ fusions suggest that the upstream promoters, pQ2 plus pQ1, contribute about 40% of the total (18, 61, 64) and that pZ2 is about sixfold stronger than pZ4 plus pZ3 (52). Direct mRNA measurements, in contrast, suggested that only 8% of the total is from pQ2 plus pQ1 (22, 27, 42) and that pZ2 is some 30-fold weaker than pZ4 plus pZ3 (42). Such discrepancies may in part reflect the fact that these promoters are all regulated, and different groups work with different physiological conditions. We therefore turn to the regulation of these promoters.
INVERSE GROWTH RATE REGULATION OF FTSQAZ TRANSCRIPTION
Using a pQ2-pQ1-lacZ transcriptional fusion, Aldea et al. (4) showed that in exponential growth in different media these promoters are more strongly expressed when the growth rate is slower, covering a fivefold range of expression. They further showed that expression of these promoters in complex medium increases five- to sixfold in stationary phase. By S1 mapping they identified the six mRNA species and compared their relative abundance during the transition from exponential to stationary phase; pQ1 showed a strong increase as the growth rate decreased, whereas the other mRNA species did not seem to vary significantly. The increase in expression of pQ1 during the transition from exponential to stationary phase has been confirmed with a pQ1-lacZ fusion lacking pQ2 (21, 51). By calculating at each point the instantaneous growth rate and the theoretical cell volume, which decreases with decreasing growth rate, Aldea et al. (4) concluded that pQ1 activity is adjusted so as to provide a constant number of FtsQAZ molecules per cell at all growth rates. This is intellectually satisfying, since cells, over a nearly 10-fold natural volume range, must make exactly one septum per cell per generation. To indicate this type of regulation, pQ1 was called a “gear box” promoter; similar regulation was found for several other promoters in E. coli, and a possible gear box consensus sequence was proposed (55).
In fact, inverse growth rate regulation has also been observed for the other promoters of the ftsQAZ operon. For pQ2—which has its own regulator, SdiA (see below)—a pQ2-lacZ fusion, cloned on a mini-F plasmid and lacking pQ1, still exhibits increased expression during the transition to stationary phase (21, 51), although less strongly than pQ1 (4). Inverse growth rate regulation was first observed for this operon with pZ4-pZ3-lacZ transcriptional fusions cloned on a λ phage and integrated in the chromosome: β-galactosidase specific activity is higher at lower growth rates, it increases during the transition from exponential to stationary phase (16, 18, 52), and it drops after a nutritional shift-up (16, 47, 52). Larger fusions, including pZ2 and the RNase E processing site “pZ1” (pZ4-pZ3-pZ2-“pZ1”-lacZ), also show increased expression at lower growth rates, during the transition to stationary phase, and after shift-up (3, 21, 52). The β-galactosidase levels with this fusion were about sevenfold higher than those with pZ4-pZ3-lacZ, suggesting that most of the transcription originated at pZ2 and implying that this promoter, like pZ4 and/or pZ3, exhibits inverse growth rate regulation (52). Indeed, the expression of a fusion with just pZ2-“pZ1”-lacZ on a λ phage increases fourfold during the transition from exponential to stationary phase (18).
The above results show clearly that inverse growth rate regulation governs pQ2, pQ1, pZ4 and/or pZ3, and pZ2. Furthermore, it was calculated that the expression from pZ4-pZ3-lacZ and pZ4-pZ3-pZ2-lacZ fusions, like that from pQ1, should provide a constant amount of gene product per cell at all growth rates (15, 52).
POSSIBLE MECHANISMS OF INVERSE GROWTH RATE REGULATION OF THE FTSQAZ OPERON
Several groups have tried to identify the mechanisms responsible for the inverse growth rate regulation and growth phase regulation of the ftsQAZ operon. Two potential transcriptional regulators were the “stationary-phase” sigma factor ςS, product of the rpoS gene and the nucleotide ppGpp. The concentrations of both of these elements increase during the transition to stationary phase, and that of ppGpp also increases with decreasing growth rate in exponential growth (10, 31). These candidates are not independent, since high ppGpp levels stimulate transcription of the rpoS gene (24).
The increase in ςS concentration early during the transition from exponential to stationary phase involves regulation at the levels of transcription, translation, and protein stability (31). Among the 30 or more genes under ςS control, several code for additional transcriptional regulators, making for a cascade of induction. It should be pointed out, however, that some genes whose expression is induced during the transition to stationary phase are not under ςS control, and conversely, ςS may also play a role in gene expression during exponential phase, particularly at low growth rates (31).
The housekeeping sigma factor ς70 is active on all promoters of the ftsQAZ operon. By use of operon fusions, however, ςS has been shown to be responsible for the increase in pQ1 expression during the transition to stationary phase (4a, 51). In contrast, the expression of pQ2-lacZ (51) and pZ4-pZ3-lacZ and pZ4-pZ3-pZ2-lacZ (52) fusions still increases during the transition to stationary phase, even in the absence of ςS. A role for ςS in the response of these promoters to growth rate during exponential growth in different media has not, to our knowledge, been tested.
The nucleotide ppGpp, in addition to stimulating rpoS transcription, could conceivably activate one (or several) of the ftsQAZ promoters directly. This hypothesis was reinforced by the observation by Powell and Court (46) that an increased ppGpp pool can suppress the temperature sensitivity of ftsZ84(Ts) strains, given that overproduction of the mutant FtsZ84 protein can restore division in nonpermissive conditions (45, 61). Indeed, under suppressing conditions (high ppGpp levels), the concentration of mutant FtsZ84 protein was found to increase fourfold (46). However, in the same experiment, expression of a pQ2-pQ1-lacZ operon fusion actually decreased two- to threefold when the ppGpp pool increased while a pZ4-pZ3-pZ2-lacZ fusion showed no change (46). Direct quantification of these five ftsZ mRNA species by Navarro et al. (42) showed no effect of ppGpp, either during the stringent response, when ppGpp levels increase 10-fold, or during steady-state growth of mutants with twofold-higher ppGpp levels. It is unlikely that ppGpp stimulates promoters upstream of pQ2, since no increase in the concentration of FtsZ protein was observed in strains with a permanent twofold-higher ppGpp concentration, although these strains were able to divide in the absence of PBP2 (42). The discrepancy between these two reports may reflect a difference in growth phase in the experiments, exponential for Navarro et al. and early stationary for Powell and Court, or it may be related to the fact that the latter study involved a mutant form of FtsZ.
The results quoted above include an apparent contradiction: increasing the ppGpp level increases the transcription of rpoS, and ςS stimulates pQ1 expression, yet increasing the ppGpp level does not increase pQ1 activity. Assuming that the various reports are all correct, we can only surmise that the answer is quantitative: the increase in ppGpp that was studied, twofold in the case of Navarro et al. (42), although sufficient to permit division in the absence of PBP2, may simply be too little to cause a detectable increase in pQ1 mRNA (and FtsZ concentration) via ςS. This is compatible with the observation that this PBP2-independent division does not require ςS (55a). Other than this, the results are all consistent in finding no effect of ppGpp or RpoS on promoter pQ2, pZ4, pZ3, or pZ2. Furthermore, the fact that the concentration of FtsZ protein did not vary when the ppGpp concentration doubled (42) strongly suggests that ppGpp is not a major transcriptional regulator of ftsZ.
OTHER TRANSCRIPTIONAL REGULATORS OF THE FTSQAZ OPERON
In a selection for multicopy suppression of the division inhibition caused by MinC-MinD, a new gene was picked up, sdiA, amplification of which causes overexpression of FtsZ (61). The target of SdiA action was shown to be pQ2: with 15 copies of an sdiA-bearing plasmid, expression of a pQ2-lacZ plasmid was stimulated 10-fold, resulting in an overall doubling of the FtsZ concentration (61). The sequence of SdiA shows similarity to LuxR-type transcriptional regulators, many of which respond to extracellular signal molecules derived from homoserine lactone; when these substances are secreted into the medium by the cells themselves (autoinducers), they act as quorum-sensing signals, allowing the bacteria to adjust their pattern of gene expression as a function of cell density. To see whether in fact SdiA responds to something excreted into the medium, conditioned medium, in which E. coli had already grown, was tested for an effect on pQ2-lacZ expression. One group reported a four- to fivefold stimulation early in the growth curve (51), whereas another found a slight inhibition (21). The same groups reported, respectively, slight or no stimulation of pQ2-lacZ by addition to the medium of the Vibrio fischeri autoinducer, a homoserine lactone derivative. Thus, although SdiA clearly stimulates pQ2 expression, its precise physiological role remains to be determined. It should be noted that under laboratory conditions, a mutant completely lacking SdiA grows and divides normally (61). This is consistent with the idea that SdiA intervenes primarily at high cell density, possibly regulating the cell division known to occur in stationary phase, even though there is no net increase in cell number (32).
In a selection for genes which, when overexpressed, permit an ftsZ84 mutant to grow in nonpermissive conditions (on Luria-Bertani plates lacking NaCl at 30°C), the rcsB gene was picked up (26). RcsB is the regulator element of a two-component system, RcsB-RcsC, that activates the transcription of a set of genes involved in the synthesis of capsular polysaccharide (colanic acid) (54). Increasing the amount of RcsB in cells causes an increase both in colanic acid production and in the expression of an ftsAZ-lacZ fusion lacking pQ2 and pQ1 but carrying all downstream promoters (26). More recently, RcsB has been shown to stimulate expression from the weak promoter pA located within the ftsQ gene (7a). It is not known at present what stimuli the RcsB-RcsC system responds to. A possible connection between this regulon and cell division regulation remains to be unravelled.
The same selection for multicopy suppressors of ftsZ84 on salt-free Luria-Bertani plates at 30°C picked up several other suppressor genes. These include the relA gene (mentioned in reference 26); assuming that increased RelA increases the ppGpp pool, this is consistent with the observation discussed above that increased ppGpp can suppress the temperature sensitivity of an ftsZ84(Ts) strain (46), although the latter effect was not via ftsZ transcription. Overexpression of the rcsF gene also suppresses ftsZ84, although this seems to be indirect, via RcsB (25). There may also be a multicopy suppressor next to the mutT gene, just beyond the envA end of the cell division cluster (45).
A pA-pZ4-pZ3-lacZ transcriptional fusion on a λ phage was reported to be induced when FtsA protein is depleted in an ftsA(Am) mutant of a strain with a temperature-sensitive amber suppressor, suggesting that FtsA represses one or more of these promoters (16). In other experiments, however, the same fusion was not induced in temperature-sensitive ftsA, ftsZ, or ftsQ mutants at a temperature nonpermissive for cell division (47). Similarly, the FtsZ protein, over a 20-fold concentration range, does not affect its own transcription, as judged from results obtained with an in situ chromosomal transcriptional fusion including all promoters (19).
CELL CYCLE-DEPENDENT REGULATION OF THE FTSQAZ OPERON
Since FtsQ, FtsA, and FtsZ have cell division functions, it seemed possible that they might be synthesized at a specific time during the cell cycle. In a synchronized population obtained by the membrane elution technique, Garrido et al. (22) monitored the ftsZ mRNA level through the cell cycle, using quantitative reverse transcriptase-PCR amplification. Indeed, the total ftsZ mRNA level was found to oscillate over a twofold range. The transcription peak coincided with the calculated time of both initiation and termination of replication. The oscillation was shown to reflect modulation of pZ2 activity (22). Using the same synchronization technique and quantitative S1 mapping, Zhou and Helmstetter (66) obtained similar results. When DNA replication was synchronized by use of a dnaC(Ts) initiation mutant, again oscillation in total ftsZ mRNA levels was observed. The authors conclude that the minimum level of ftsZ expression occurs near the time at which the operon is replicated. This fluctuation did not depend on pZ2 but rather on one or more promoters upstream of pZ3 (66).
Using a completely different synchronization technique, infection with the mutant phage Mugemts2, Ghelardini et al. (27) observed that ftsZ mRNA doubled at the time of division. In a culture synchronized by repeated phosphate starvation, Robin et al. (47) monitored the expression of a pZ4-pZ3-lacZ transcriptional fusion on an integrated λ phage through two cycles. Accumulation of β-galactosidase was found to be linear, with an abrupt doubling in rate shortly after division.
The regulators responsible for cell cycle fluctuation in ftsQAZ transcription have not been identified. It is striking, however, that a harmonious cycle can be obtained in rapid growth when the ftsZ gene is cut off from its natural promoters and placed under control of ptac. With a 40% higher FtsZ concentration than the average in wild-type cells, morphology is normal (44). Cell cycle regulation of ftsZ may provide fine tuning, or it may be more important at slow growth rates.
POSTTRANSCRIPTIONAL REGULATION OF THE FTSQAZ OPERON
The three proteins FtsQ, FtsA, and FtsZ are expressed at very different levels, with about 20, 200, and 10,000 molecules per cell, respectively (36). In addition to different transcription rates for the three genes, there is also a difference in the intrinsic translation efficiency: ftsQ is translated less well than ftsA, which is translated less well than ftsZ (40).
Translation can be modified by antisense RNA. Two such species have been reported, specifically inhibiting the translation of ftsZ mRNA. One is DicF RNA, a 53-nucleotide species complementary to the beginning of the ftsZ mRNA and coded for by a cryptic prophage (6). The second is a sequence spanning the ftsA-ftsZ junction, the stfZ gene, apparently endowed with a promoter and translational stop in the direction opposite to that of ftsQAZ transcription (14). At present, no natural induction conditions have been found for either of these antisense RNAs. When expressed from artificial constructions, however, they are both effective division inhibitors, at least at high temperatures, where they reduce the amount of FtsZ.
A further posttranscriptional level of regulation is mRNA processing, and ftsQAZ mRNA has a double RNase E cleavage site within the ftsA gene, originally mistaken for a proximal promoter (“pZ1”; see Fig. 1). Cleavage at this site would alter the ratio of the protein products and could alter the stability of the resulting mRNA. It is not known whether cleavage is regulated or represents a constant (small) fraction of total ftsZ mRNA. The mRNA studies reported to date have not revealed conditions in which levels of this fraction vary significantly (4, 42), but the question remains open. Growing evidence for control of RNase E activity by medium composition (5) and possibly by growth rate makes controlled cleavage at “pZ1” an attractive possibility for varying the ratio of these division proteins in response to physiological signals.
PROTEIN STABILITY
The FtsZ protein of Caulobacter crescentus is degraded after cell division in the swarmer cell, which does not start its new cycle for some time, but not in the stalk cell, which enters its new cycle immediately after division (34). In E. coli, the FtsZ protein has not been reported to be unstable, although rapid proteolysis at a specific cell age, as in Caulobacter, could escape detection in exponential cultures if special precautions are not taken. A hint of possible instability comes from studies on the interaction of FtsZ with the molecular chaperone DnaK. DnaK was found to copurify with oligomerized FtsZ, suggesting a direct protein-protein interaction (39). Furthermore, the cell division defect of ΔdnaK strains is suppressed by overproduction of FtsZ (7). One interpretation of these observations is that DnaK protects FtsZ from proteolysis in vivo.
PROTEIN MODIFICATIONS
Two division proteins are known to be modified posttranslationally. FtsA is phosphorylated (50), and PBP3 (product of the ftsI gene) loses its 11 C-terminal amino acids (41) through hydrolysis by the periplasmic protease Prc (or Tsp) (29). Recent evidence suggests that there may also be a methylation event required for division, although the target has not been identified (43). It seems likely that FtsZ and other division proteins are modified as well. Surprisingly, there is a dearth of information concerning other modifications of division proteins. Modifications could affect protein localization, as may be the case for FtsA, which is phosphorylated when in the cytoplasm but unphosphorylated in the membrane (50). In the case of FtsZ, a modification at a specific age could permit it to find the nucleation site or trigger polymerization. It is possible that some component of the replication machinery remains associated with the site after cell separation; modification of this protein would then distinguish old (polar) sites from new (midcell) sites, which could be the signal for the Min system to block division preferentially at old sites (49). It will be interesting to establish the complete catalogue of modifications of division proteins: which are modified, in what way, by which enzymes, and whether these modifications take place at a precise cell age.
CELL DIVISION AND PPGPP
Several observations suggested that the nucleotide ppGpp might be a positive regulator of ftsQAZ expression. First of all, ppGpp stimulates the transcription of the rpoS gene, whose product, ςS activates pQ1. As we have seen, this pathway of transcriptional stimulation of the ftsQAZ operon does not seem to be operative in exponential growth. Second, in the absence of PBP2, cell division can be restored either by amplifying FtsQAZ or by increasing the ppGpp concentration. Third, the activities of at least four ftsQAZ promoters increase at low growth rates, when the ppGpp concentration increases. And finally, mutants completely lacking ppGpp tend to form filaments, indicative of a division-promoting role for this nucleotide (63).
Despite these suggestive indices, ppGpp does not activate the five well-characterized promoters of the ftsQAZ operon. It might be thought that ppGpp stimulates another promoter, perhaps upstream of pQ2. However, a doubling of the ppGpp concentration, which is sufficient to restore division in the absence of PBP2, does not increase the concentration of FtsZ protein (42). We are thus led to conclude that ppGpp does not stimulate the synthesis of FtsZ or, presumably, of FtsQ or FtsA.
How then does an increased ppGpp concentration restore division in the absence of PBP2? Several possibilities can be envisaged. The simplest is that ppGpp activates the expression of another gene whose product, at high concentration, allows FtsQAZ to function more efficiently. This putative target could be, for example, another cell division protein or an enzyme catalyzing the formation of a substrate or effector of some division protein.
A second possible mode of action of ppGpp in cell division is not at the transcriptional level but as an effector of one or more division proteins. Vinella et al. (58) tried to test whether ppGpp (or pppGpp) might be an in vivo activator of cell division. They showed that to reestablish division after a period of arrest due to PBP2 inactivation, it is not enough to build up the ppGpp pool; protein synthesis is needed as well. Since the FtsZ concentration was not determined, this leaves open the question as to whether (p)ppGpp activates FtsZ. It should be noted, however, that even if (p)ppGpp activates FtsZ, this is unlikely to be sufficient in itself to restore cell division in the absence of PBP2, since amplification of just FtsZ is insufficient (42).
In fact, additional observations suggest that ppGpp regulates cell division at the transcriptional level. First, the rpoB369 mutant, with an RNA polymerase apparently less sensitive to ppGpp, has a block of cell division at nonpermissive temperatures (Fts− phenotype), and this block is relieved by overproduction of FtsQAZ, by an increase of the ppGpp concentration, or by entry to stationary phase (56). Conversely, there exist mutant RNA polymerases able, in the absence of ppGpp, to transcribe all operons involved in amino acid biosynthesis, including those that normally require ppGpp (10). These are selected as prototrophic derivates of a strain lacking ppGpp (ΔrelA ΔspoT), and certain alleles restore cell division in the absence of PBP2 (55a). Our prediction is therefore that future work will reveal one or more target operons, positively regulated by ppGpp, whose products participate in the cell division process, probably by interacting with FtsZ, FtsA, and FtsQ to make the basal quantity more effective. If this model is correct, it would establish ppGpp as an internal signal involved in cell cycle regulation.
REFERENCES
- 1.Addinall S G, Cao C, Lutkenhaus J. FtsN, a late recruit to the septum in Escherichia coli. Mol Microbiol. 1997;25:303–309. doi: 10.1046/j.1365-2958.1997.4641833.x. [DOI] [PubMed] [Google Scholar]
- 2.Addinall S G, Lutkenhaus J. FtsA is localized to the septum in an FtsZ-dependent manner. J Bacteriol. 1996;178:7167–7172. doi: 10.1128/jb.178.24.7167-7172.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aldea M, Garrido T, Hernández-Chico C, Vicente M, Kushner S R. Induction of a growth-phase-dependent promoter triggers transcription of bolA, an Escherichia coli morphogene. EMBO J. 1989;8:3923–3931. doi: 10.1002/j.1460-2075.1989.tb08573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aldea M, Garrido T, Pla J, Vicente M. Division genes in Escherichia coli are expressed coordinately to cell septum requirements by gearbox promoters. EMBO J. 1990;9:3787–3794. doi: 10.1002/j.1460-2075.1990.tb07592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.Ballesteros M, Kosano S, Ishihama A, Vicente M. The ftsQ1p gearbox promoter of Escherichia coli is a major sigma S-dependent promoter in the ddlB-FtsA region. Mol Microbiol. 1998;30:419–430. doi: 10.1046/j.1365-2958.1998.01077.x. [DOI] [PubMed] [Google Scholar]
- 5.Barlow T, Berkmen M, Georgelis D, Bayr L, Arvidson S, von Gabain A. RNase E, the major player in mRNA degradation, is down-regulated in Escherichia coli during a transient growth retardation (diauxic lag) Biol Chem. 1998;379:33–38. doi: 10.1515/bchm.1998.379.1.33. [DOI] [PubMed] [Google Scholar]
- 6.Bouché F, Bouché J-P. Genetic evidence that DicF, a second division inhibitor encoded by the Escherichia coli dicB operon, is probably RNA. Mol Microbiol. 1989;3:991–994. doi: 10.1111/j.1365-2958.1989.tb00249.x. [DOI] [PubMed] [Google Scholar]
- 7.Bukau B, Walker G C. Cellular defects caused by deletion of the Escherichia coli dnaK gene indicate roles for heat shock protein in normal metabolism. J Bacteriol. 1989;171:2337–2346. doi: 10.1128/jb.171.5.2337-2346.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Cam, K., and J.-P. Bouché. Personal communication.
- 8.Cam K, Rome G, Krisch H M, Bouché J-P. RNase E processing of essential cell division genes mRNA in Escherichia coli. Nucleic Acids Res. 1996;24:3065–3070. doi: 10.1093/nar/24.15.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carson M J, Barondess J, Beckwith J. The FtsQ protein of Escherichia coli: membrane topology, abundance, and cell division phenotypes due to overproduction and insertion mutations. J Bacteriol. 1991;173:2187–2195. doi: 10.1128/jb.173.7.2187-2195.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cashel M, Gentry D R, Hernandez V J, Vinella D. The stringent response. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: ASM Press; 1996. pp. 1458–1496. [Google Scholar]
- 11.Dai K, Lutkenhaus J. ftsZ is an essential cell division gene in Escherichia coli. J Bacteriol. 1991;173:3500–3506. doi: 10.1128/jb.173.11.3500-3506.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dai K, Lutkenhaus J. The proper ratio of FtsZ to FtsA is required for cell division to occur in Escherichia coli. J Bacteriol. 1992;174:6145–6151. doi: 10.1128/jb.174.19.6145-6151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dewar S J, Begg K J, Donachie W D. Inhibition of cell division initiation by an imbalance in the ratio of FtsA to FtsZ. J Bacteriol. 1992;174:6314–6316. doi: 10.1128/jb.174.19.6314-6316.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dewar S J, Donachie W D. Antisense transcription of the ftsZ-ftsA gene junction inhibits cell division in Escherichia coli. J Bacteriol. 1993;175:7097–7101. doi: 10.1128/jb.175.21.7097-7101.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dewar S J, Donachie W D. Regulation of expression of the ftsA cell division gene by sequences in upstream genes. J Bacteriol. 1990;172:6611–6614. doi: 10.1128/jb.172.11.6611-6614.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dewar S J, Kagan-Zur V, Begg K J, Donachie W D. Transcriptional regulation of cell division genes in Escherichia coli. Mol Microbiol. 1989;3:1371–1377. doi: 10.1111/j.1365-2958.1989.tb00118.x. [DOI] [PubMed] [Google Scholar]
- 17.Erickson H P, Taylor D W, Taylor K A, Bramhill D. Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc Natl Acad Sci USA. 1996;93:519–523. doi: 10.1073/pnas.93.1.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flärdh K, Garrido T, Vicente M. Contribution of individual promoters in the ddlB-ftsZ region to the transcription of the essential cell-division gene ftsZ in Escherichia coli. Mol Microbiol. 1997;24:927–936. doi: 10.1046/j.1365-2958.1997.4001762.x. [DOI] [PubMed] [Google Scholar]
- 19.Flärdh K, Palacios P, Vicente M. Cell division genes ftsQAZ in Escherichia coli require distant cis-acting signals upstream of ddlB for full expression. Mol Microbiol. 1998;30:305–315. doi: 10.1046/j.1365-2958.1998.01064.x. [DOI] [PubMed] [Google Scholar]
- 20.Gaal T, Bartlett M S, Ross W, Turnbough C L, Jr, Gourse R L. NTP concentration as a regulator of transcription initiation: control of rRNA synthesis in bacteria. Science. 1997;278:2092–2097. doi: 10.1126/science.278.5346.2092. [DOI] [PubMed] [Google Scholar]
- 21.García-Lara J, Shang L H, Rothfield L I. An extracellular factor regulates expression of sdiA, a transcriptional activator of cell division genes in Escherichia coli. J Bacteriol. 1996;178:2742–2748. doi: 10.1128/jb.178.10.2742-2748.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garrido T, Sánchez M, Palacios P, Aldea M, Vicente M. Transcription of ftsZ oscillates during the cell cycle of Escherichia coli. EMBO J. 1993;12:3957–3965. doi: 10.1002/j.1460-2075.1993.tb06073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gentry D R, Cashel M. Mutational analysis of the Escherichia coli spoT gene identifies distinct but overlapping regions involved in ppGpp synthesis and degradation. Mol Microbiol. 1996;19:1373–1384. doi: 10.1111/j.1365-2958.1996.tb02480.x. [DOI] [PubMed] [Google Scholar]
- 24.Gentry D R, Hernandez V J, Nguyen L H, Jensen D B, Cashel M. Synthesis of the stationary-phase specific sigma factor, ςS, is positively regulated by ppGpp. J Bacteriol. 1993;175:7982–7989. doi: 10.1128/jb.175.24.7982-7989.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gervais F G, Drapeau G R. Identification, cloning, and characterization of rcsF, a new regulator gene for exopolysaccharide synthesis that suppresses the division mutation ftsZ84 in Escherichia coli K-12. J Bacteriol. 1992;174:8016–8022. doi: 10.1128/jb.174.24.8016-8022.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gervais F G, Phoenix P, Drapeau G R. The rcsB gene, a positive regulator of colanic acid biosynthesis in Escherichia coli, is also an activator of ftsZ expression. J Bacteriol. 1992;174:3964–3971. doi: 10.1128/jb.174.12.3964-3971.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghelardini P, Lauri P, Ruberti I, Orlando V, Paolozzi L. Synchronous division induced in Escherichia coli K12 by phage Mu: analysis of DNA topology and gene expression during the cell cycle. Res Microbiol. 1991;142:259–267. doi: 10.1016/0923-2508(91)90039-d. [DOI] [PubMed] [Google Scholar]
- 28.Hale C A, de Boer P A J. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell. 1997;88:175–185. doi: 10.1016/s0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- 29.Hara H, Yamamoto Y, Higashitani A, Suzuki H, Nishimura Y. Cloning, mapping, and characterization of the Escherichia coli prc gene, which is involved in C-terminal processing of penicillin-binding protein 3. J Bacteriol. 1991;173:4799–4813. doi: 10.1128/jb.173.15.4799-4813.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hara H, Yasuda S, Horiuchi K, Park J T. A promoter for the first nine genes of the Escherichia coli mra cluster of cell division and cell envelope biosynthesis genes, including ftsI and ftsW. J Bacteriol. 1997;179:5802–5811. doi: 10.1128/jb.179.18.5802-5811.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hengge-Aronis R. Regulation of gene expression during entry into stationary phase. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: ASM Press; 1996. pp. 1497–1512. [Google Scholar]
- 32.Huisman G W, Siegele D A, Zambrano M M, Kolter R. Morphological and physiological changes during stationary phase. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: ASM Press; 1996. pp. 1672–1682. [Google Scholar]
- 33.Joseleau-Petit D, Thévenet D, D’Ari R. ppGpp concentration, growth without PBP2 activity and growth rate control in Escherichia coli. Mol Microbiol. 1994;13:911–917. doi: 10.1111/j.1365-2958.1994.tb00482.x. [DOI] [PubMed] [Google Scholar]
- 34.Kelly A J, Sackett M J, Din N, Quardokus E, Brun Y V. Cell cycle-dependent transcriptional and proteolytic regulation of FtsZ in Caulobacter. Genes Dev. 1998;12:880–893. doi: 10.1101/gad.12.6.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lutkenhaus J, Addinall S G. Bacterial cell division and the Z ring. Annu Rev Biochem. 1997;66:93–116. doi: 10.1146/annurev.biochem.66.1.93. [DOI] [PubMed] [Google Scholar]
- 36.Lutkenhaus J, Mukherjee A. Cell division. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: ASM Press; 1996. pp. 1615–1626. [Google Scholar]
- 37.Ma X, Ehrhardt D W, Margolin W. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc Natl Acad Sci USA. 1996;93:12998–13003. doi: 10.1073/pnas.93.23.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mengin-Lecreulx D, Ayala J, Bouhss A, van Heijenoort J, Parquet C, Hara H. Contribution of the Pmra promoter to the expression of genes in the Escherichia coli mra cluster of cell envelope biosynthesis and cell division genes. J Bacteriol. 1998;180:4406–4412. doi: 10.1128/jb.180.17.4406-4412.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mukherjee A, Dai K, Lutkenhaus J. Escherichia coli cell division protein FtsZ is a guanine nucleotide binding protein. Proc Natl Acad Sci USA. 1993;90:1053–1057. doi: 10.1073/pnas.90.3.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mukherjee A, Donachie W D. Differential translation of cell division proteins. J Bacteriol. 1990;172:6106–6111. doi: 10.1128/jb.172.10.6106-6111.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nagasawa H, Sakagami Y, Suzuki A, Suzuki H, Hara H, Hirota Y. Determination of the cleavage site involved in C-terminal processing of penicillin-binding protein 3 of Escherichia coli. J Bacteriol. 1989;171:5890–5893. doi: 10.1128/jb.171.11.5890-5893.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Navarro F, Robin A, D’Ari R, Joseleau-Petit D. Analysis of the effect of ppGpp on the ftsQAZ operon in Escherichia coli. Mol Microbiol. 1998;29:815–823. doi: 10.1046/j.1365-2958.1998.00974.x. [DOI] [PubMed] [Google Scholar]
- 43.Newman E B, Budman L I, Chan E C, Greene R C, Lin R T, Woldringh C L, D’Ari R. Lack of S-adenosylmethionine results in a cell division defect in Escherichia coli. J Bacteriol. 1998;180:3614–3619. doi: 10.1128/jb.180.14.3614-3619.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palacios P, Vicente M, Sánchez M. Dependency of Escherichia coli cell-division size, and independency of nucleoid segregation on the mode and level of ftsZ expression. Mol Microbiol. 1996;20:1093–1098. doi: 10.1111/j.1365-2958.1996.tb02549.x. [DOI] [PubMed] [Google Scholar]
- 45.Phoenix P, Drapeau G R. Cell division control in Escherichia coli K-12: some properties of the ftsZ84 mutation and suppression of this mutation by the product of a newly identified gene. J Bacteriol. 1988;170:4338–4342. doi: 10.1128/jb.170.9.4338-4342.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Powell B S, Court D L. Control of ftsZ expression, cell division, and glutamine metabolism in Luria-Bertani medium by the alarmone ppGpp in Escherichia coli. J Bacteriol. 1998;180:1053–1062. doi: 10.1128/jb.180.5.1053-1062.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robin A, Joseleau-Petit D, D’Ari R. Transcription of the ftsZ gene and cell division in Escherichia coli. J Bacteriol. 1990;172:1392–1399. doi: 10.1128/jb.172.3.1392-1399.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson A C, Kenan D J, Hatfull G F, Sullivan N F, Spiegelberg R, Donachie W D. DNA sequence and transcriptional organization of essential cell division genes ftsQ and ftsA of Escherichia coli: evidence for overlapping transcriptional units. J Bacteriol. 1984;160:546–555. doi: 10.1128/jb.160.2.546-555.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rothfield L I, Zhao C R. How do bacteria decide where to divide? Cell. 1996;84:183–186. doi: 10.1016/s0092-8674(00)80971-x. [DOI] [PubMed] [Google Scholar]
- 50.Sánchez M, Valencia A, Ferrándiz M-J, Sander C, Vicente M. Correlation between the structure and biochemical activities of FtsA, an essential cell division protein of the actin family. EMBO J. 1994;13:4919–4925. doi: 10.1002/j.1460-2075.1994.tb06819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sitnikov D M, Schineller J B, Baldwin T O. Control of cell division in Escherichia coli: regulation of transcription of ftsQA involves both rpoS and SdiA-mediated autoinduction. Proc Natl Acad Sci USA. 1996;93:336–341. doi: 10.1073/pnas.93.1.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith R W P, Masters M, Donachie W D. Cell division and transcription of ftsZ. J Bacteriol. 1993;175:2788–2791. doi: 10.1128/jb.175.9.2788-2791.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spratt B G. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K-12. Proc Natl Acad Sci USA. 1975;72:2999–3003. doi: 10.1073/pnas.72.8.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stout V, Gottesman S. RcsB and RcsC: a two-component regulator of capsule synthesis in Escherichia coli. J Bacteriol. 1990;172:659–669. doi: 10.1128/jb.172.2.659-669.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vicente M, Kushner S R, Garrido T, Aldea M. The role of the ’gearbox’ in the transcription of essential genes. Mol Microbiol. 1991;5:2085–2091. doi: 10.1111/j.1365-2958.1991.tb02137.x. [DOI] [PubMed] [Google Scholar]
- 55a.Vinella, D. Unpublished observation.
- 56.Vinella D, D’Ari R. Thermoinducible filamentation in Escherichia coli due to an altered RNA polymerase β subunit is suppressed by high ppGpp. J Bacteriol. 1994;176:966–972. doi: 10.1128/jb.176.4.966-972.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vinella D, D’Ari R, Jaffé A, Bouloc P. Penicillin-binding protein 2 is dispensable in Escherichia coli when ppGpp synthesis is induced. EMBO J. 1992;11:1493–1501. doi: 10.1002/j.1460-2075.1992.tb05194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vinella D, Joseleau-Petit D, Thévenet D, Bouloc P, D’Ari R. Penicillin-binding protein 2 inactivation in Escherichia coli results in cell division inhibition, relieved by FtsZ overexpression. J Bacteriol. 1993;175:6704–6710. doi: 10.1128/jb.175.20.6704-6710.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang L, Khattar M K, Donachie W D, Lutkenhaus J. FtsI and FtsW are localized to the septum in Escherichia coli. J Bacteriol. 1998;180:2810–2816. doi: 10.1128/jb.180.11.2810-2816.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang L, Lutkenhaus J. FtsK is an essential cell division protein that is localized to the septum and induced as part of the SOS response. Mol Microbiol. 1998;29:731–740. doi: 10.1046/j.1365-2958.1998.00958.x. [DOI] [PubMed] [Google Scholar]
- 61.Wang X, de Boer P A J, Rothfield L I. A factor that positively regulates cell division by activating transcription of the major cluster of essential cell division genes of Escherichia coli. EMBO J. 1991;10:3363–3372. doi: 10.1002/j.1460-2075.1991.tb04900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weiss D S, Pogliano K, Carson M, Guzman L-M, Fraipont C, Nguyen-Distèche M, Losick R, Beckwith J. Localization of the Escherichia coli cell division protein FtsI (PBP3) to the division site and cell pole. Mol Microbiol. 1997;25:671–681. doi: 10.1046/j.1365-2958.1997.5041869.x. [DOI] [PubMed] [Google Scholar]
- 63.Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem. 1991;266:5980–5990. [PubMed] [Google Scholar]
- 64.Yi Q-M, Rockenbach S, Ward J E J, Lutkenhaus J. Structure and expression of the cell division genes ftsQ, ftsA and ftsZ. J Mol Biol. 1985;184:399–412. doi: 10.1016/0022-2836(85)90290-6. [DOI] [PubMed] [Google Scholar]
- 65.Yu X-C, Tran A H, Sun Q, Margolin W. Localization of cell division protein FtsK to the Escherichia coli septum and identification of a potential N-terminal targeting domain. J Bacteriol. 1998;180:1296–1304. doi: 10.1128/jb.180.5.1296-1304.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou P, Helmstetter C E. Relationship between ftsZ gene expression and chromosome replication in Escherichia coli. J Bacteriol. 1994;176:6100–6106. doi: 10.1128/jb.176.19.6100-6106.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]