Abstract
A 4.2-kb SphI-BamHI fragment of chromosomal DNA from Streptomyces granaticolor was cloned and shown to encode a protein with significant sequence similarity to the eukaryotic protein serine/threonine kinases. It consists of 701 amino acids and in the N-terminal part contains all conserved catalytic domains of protein kinases. The C-terminal domain of Pkg2 contains seven tandem repeats of 11 or 12 amino acids with similarity to the tryptophan-docking motif known to stabilize a symmetrical three-dimensional structure called a propeller structure. The pkg2 gene was overexpressed in Escherichia coli, and the gene product (Pkg2) has been found to be autophosphorylated at serine and threonine residues. The N- and C-terminal parts of Pkg2 are separated with a hydrophobic stretch of 21 amino acids which translocated a PhoA fusion protein into the periplasm. Thus, Pkg2 is the first transmembrane protein serine/threonine kinase described for streptomycetes. Replacement of the pkg2 gene by the spectinomycin resistance gene resulted in changes in the morphology of aerial hyphae.
Phosphorylation is a key component of a signal transduction network of both eukaryotic and prokaryotic cells. It has been thought for a long time that the main sites of phosphorylation in prokaryotes are histidine and/or asparagine residues, phosphorylated during a signal transduction process mediated by two-component systems (40). Recent results, however, have shown serine/threonine-phosphorylating and tyrosine-phosphorylating enzymes, containing all 11 conserved catalytic domains of eukaryotic protein kinases (15), to be present also in prokaryotes, either in those displaying some kind of differentiation process [Streptomyces coelicolor A3(2) (29, 43), Streptomyces granaticolor (46), Anabaena strain PCC 7120 (47–49), Myxococcus xanthus (16, 30, 42, 50)] or in human pathogens (Yersinia pseudotuberculosis [13] and Mycobacterium tuberculosis [33]).
Streptomycetes are of a great interest due to their ability to produce a vast amount of antibiotics. Their complex life cycle and multicellular differentiation, resembling the growth of filamentous fungi (8), undoubtedly require various levels of regulation and various types of signal transduction mechanisms. In this respect, the identification of protein serine/threonine kinases in Streptomycetes is not surprising. However, as for the kinases of the other organisms mentioned above, little is known about their roles. Five different protein serine/threonine kinases in streptomycetes have been described. One of them, the membrane-associated protein kinase AfsK, was shown to phosphorylate in vitro a global regulatory protein, AfsR, which is involved in secondary metabolism (29). The roles of the other kinases, PkaA and PkaB (43) and Pkg4 and Pkg3 (46), are not known.
In a previous study (23), we showed that in Streptomyces granaticolor cell extracts, the extent and pattern of O-phosphomonoesters were growth stage dependent, suggesting the presence of several protein kinases of eukaryotic type either differentially expressed or activated by unknown stimuli during the cell cycle. This study reports the characterization of the protein serine/threonine kinase gene pkg2, coding for a protein different in structure and other properties from the known homologues. A unique feature of Pkg2 among Streptomyces protein serine/threonine kinases studied so far is its subcellular localization.
MATERIALS AND METHODS
Materials.
[γ-32P]ATP, [α-32P]dCTP, and l-[35S]methionine were purchased from Amersham, and [32P]H3PO4 was obtained from Polatom (Otwock-Swierk, Poland). Restriction enzymes, Klenow polymerase, T4 DNA ligase, T4 DNA polymerase, and T4 polynucleotide kinase were from New England Biolabs or Amersham, Taq polymerase was from Top-Bio (Prague, Czech Republic), and Pfu polymerase was from Stratagene. DNA sequencing was performed with a ThermoSequenase cycle sequencing kit obtained from Amersham. Custom oligonucleotides were purchased from MWG Biotech. Digoxigenin DNA labeling and digoxigenin nucleic acid detection kits were purchased from Boehringer Mannheim.
Bacterial strains and plasmids.
S. granaticolor wild-type strain ETH 7437 (37) was used throughout this study. Media for S. granaticolor were MK medium (1% [wt/vol] Bacto Peptone, 0.2% [wt/vol] yeast extract, 1% [vol/vol] glycerol [pH 7.2]), R2YE medium (20), PPS medium (1% [wt/vol] malt extract, 0.4% [wt/vol] yeast extract, 0.4% [wt/vol] glucose, 2% [wt/vol] agar [pH 7.2]), and SLM3 medium (0.5% [wt/vol] cornsteep, 1% [wt/vol] starch, 0.3% [wt/vol] CaCO3, 0.2% [wt/vol] yeast extract, 2% [wt/vol] agar [pH 7.2]). Escherichia coli XL1-Blue (Stratagene) was used as the recipient strain in most DNA manipulations. E. coli CJ236 was used for the generation of a uracil-containing single-stranded DNA for site-directed mutagenesis. E. coli GM2929 (New England Biolabs) was used for the generation of nonmethylated plasmid DNA for protoplast transformation. E. coli strains were grown in LB medium (1% tryptone, 0.5% yeast extract, 1% NaCl) supplemented with ampicillin (100 μg ml−1), kanamycin (30 μg ml−1), spectinomycin (100 μg ml−1), and hygromycin (50 μg ml−1) when necessary. All antibiotics were purchased from United States Biochemical (USB). E. coli BL21(DE3) (Novagen), used for the T7 RNA polymerase expression system (41), was grown in M9 medium (38) supplemented with 1% Casamino Acids, 1% glucose, and all 20 amino acids except methionine (when cells were labeled with l-[35S]methionine) or in a phosphate-free medium containing 100 mM Tris-HCl (pH 7.4), 92 mM NaCl, 40 mM KCl, 21 mM NH4Cl, 100 mM CaCl2, 160 mM Na2SO4, and 1 mM MgSO4 (when cells were labeled with 32P). E. coli CC118 was the strain used for the alkaline phosphatase assay (28).
Plasmids pTZ18R (USB), pTZ19R (USB), and pMOSBlue (Amersham) were used for cloning, subcloning, and sequencing experiments; plasmid pET24a (Novagen) was used for the expression of pkg2 with T7 RNA polymerase (41); plasmids pHP45Ω (36) and pHP45Ωhyg (5) were used as sources of antibiotic resistance spc and hyg gene cassettes, conferring resistance to spectinomycin and hygromycin, respectively.
DNA manipulations, sequencing, and sequence analysis.
DNA manipulations in E. coli were conducted as described by Sambrook et al. (38), and those in S. granaticolor were performed as described by Hopwood et al. (20). Preparation of S. granaticolor protoplasts was performed as described by Petříček et al. (34). All restriction endonuclease digestions, ligations, and DNA modifications were performed as recommended by the commercial suppliers. When required, DNA restriction fragments or PCR products were separated by agarose gel electrophoresis and purified by using a Gene Clean Spin kit purchased from Bio 101.
The nucleotide sequence was determined in both directions by the dideoxynucleotide chain termination method (39), using double-stranded plasmid DNA and the universal primers or synthetic oligonucleotides as primers designed from newly obtained DNA sequences.
DNASIS software (Hitachi) was used for sequence analysis. The codon usage pattern was determined by FRAME analysis (3). The Fasta3 program at the European Bioinformatics Institute (32) and the BLAST program at the National Center for Biotechnology Information (1) were used to search for local alignments. A prediction of membrane-spanning regions was performed at the EXPASY server with the TMpred program (19).
Construction of the molecular probe.
The molecular probe for detection of protein kinase genes was prepared by PCR. Two degenerate oligonucleotides designed on the basis of the consensus sequence of subdomains VI (DLKP[D/E]N) and VIII (TP[D/E]YM) of eukaryotic protein serine/threonine kinases (15) were used as primers to amplify a fragment of the kinase gene. The sequence of the forward primer was 5′-GACCT(C/G)AAGCC(C/G)GA(C/G)AA-3′; the sequence of reverse primer was 5′-GCCATGTA(C/G)TC(C/G)GG(C/G)GT-3′.
Expression of the pkg2 gene in E. coli.
To place the pkg2 gene under the control of a T7 promoter, a DNA fragment containing the N-terminal part of pkg2 was amplified with Pfu polymerase. The sequences of the two primers used in PCR were 5′-GTGACCACACAGCCCCTCGC-3′ (anneals at positions 844 to 863 near the pkg2 start codon [see Fig. 2]) and 5′-CCACAGAGGCTTCGGGA-3′ (anneals at positions 1946 to 1962). Plasmid p219 was used as the template. Ligation of the PCR product (1,118 bp) in the correct orientation with EcoRV-digested pMOSBlue generated pMOSex with an NdeI site upstream of GTG initiation codon. To shorten the sequenced fragment, pMOSex was digested with BstEII and EcoRI, filled in with Klenow polymerase, and self-circularized; the previously amplified sequence was verified. After sequencing, a 533-bp NdeI-BstEII fragment was ligated with a 2.8-kb BstEII-EcoRI fragment of p219 containing an incomplete pkg2 gene into NdeI/EcoRI-linearized pET24a. The plasmid thus obtained, designated pEX2, was transformed into E. coli BL21(DE3).
FIG. 2.
Nucleotide sequence of the 4.205-bp DNA fragment containing the pkg2 gene and predicted amino acid sequence of the Pkg2 kinase (ORF2). Only the relevant part of the sequence (bases 1 to 3000) is shown. The putative ribosome binding site (bases 833 to 838) is underlined, amino acid residues corresponding to the conserved catalytic domains of eukaryotic-type protein kinases are in boldface, and the TM domain is double underlined.
For l-[35S]methionine labeling, cells were grown in M9 minimal medium (38) supplemented with 1% Casamino Acids and 1% glucose to an optical density at 600 nm of 0.4. Then isopropyl-β-d-thiogalactoside (IPTG) was added to a final concentration of 2 mM, and incubation was continued for 1 h. Cells from 1 ml of culture were harvested by centrifugation, washed with M9 minimal medium, and resuspended in the same medium supplemented with 19 amino acids (all except methionine). Rifampin (400 μg ml−1) was added; after a 30-min incubation, 10 μCi of l-[35S]methionine was added, and incubation continued for 3 h.
Labeling cells with 32P.
To obtain 32P in vivo-labeled Pkg2, E. coli BL21(DE3) harboring pEX2 was grown in phosphate-free medium supplemented with 1% Casamino Acids and 1% glucose to an optical density at 600 nm of 0.4. After the addition of IPTG (2 mM, final concentration) and [32P]H3PO4 (10 mCi ml−1, final concentration), cultivation was continued for 1 h. Then rifampin (400 μg ml−1) was added, and the culture was incubated for a further 3 h. Cells were collected by centrifugation and dissolved in sodium dodecyl sulfate (SDS) sample buffer. Proteins were separated by polyacrylamide gel electrophoresis (PAGE) on a 0.1% SDS–10% polyacrylamide gel; after electrophoresis, gels were soaked in boiling 16% trichloroacetic acid (27), dried under vacuum, and exposed to autoradiography.
Identification of phosphoamino acids.
32P-labeled proteins from a cell extract of E. coli BL21(DE3) harboring pEX2 were separated by PAGE on a 0.1% SDS–10% polyacrylamide gel and transferred onto an Immobilon polyvinylidene difluoride membrane, using a semidry transfer apparatus. Radioactive protein bands were detected by autoradiography, excised, and hydrolyzed in 6 M HCl at 110°C for 90 min (24). The acid-stable phosphoamino acids thus liberated were separated by electrophoresis in the first dimension followed by ascending chromatography in the second dimension as described previously (12). Authentic phosphoamino acids were run simultaneously, and their positions were detected by staining with ninhydrin. Labeled phosphoamino acids were detected by autoradiography.
Site-directed mutagenesis of pkg2.
The target DNA for generation of an amino acid replacement of residue 45 (AAG, Lys) of the Pkg2 protein with Arg (AGG) was constructed as follows. pMOSex was linearized with BstEII, filled in with Klenow polymerase, and redigested with HindIII. A 575-bp fragment thus generated was subcloned into the polylinker of M13mp18 previously digested with XbaI, filled in with Klenow polymerase, and redigested with HindIII. The phage DNA was propagated once in E. coli CJ236 to prepare uracil-containing single-stranded DNA (26). Mutagenic primer 2K45R (5′-CAGCCGTCAGGGTCCTGCG-3′) was annealed with the single-stranded DNA (the mutagenized nucleotide is underlined), and the complementary strand was synthesized by using T4 DNA polymerase and ligase. E. coli XL1-Blue was then transfected with the reaction mixture. The mutation thus generated was confirmed by nucleotide sequencing, and the mutated NdeI-BstEII fragment was replaced with the corresponding wild-type NdeI-BstEII fragment in pEX2. The plasmid thus obtained (pEX2K45R) was transformed into E. coli BL21(DE3).
Construction of the pkg2-phoA fusion gene and alkaline phosphatase assay.
Fusion genes were created as originally described by Udo et al. (42). The DNA fragments with or without the predicted transmembrane (TM) region were amplified by PCR and fused in frame with the lacZ gene of pUC19 and the phoA gene obtained from pCH2 (18). The resulting plasmids expressed the pkg2-phoA fusion gene under the control of the lac promoter. The following oligonucleotides were used as primers: 2TMU (5′-CTAAGCTTGCGCGAGGCGGCCAC-3′), which contains a HindIII site (underlined) and anneals to positions 1701 to 1715; and 2TMD1 (5′-CTCTGCAGACGGCGACGAGGACG-3′) and 2TMD2 (5′-CTCTGCAGGGGTCGCCACGGCGG-3′), which each contain a PstI site (underlined) and anneal to positions 1766 to 1751 and 1901 to 1886, respectively (Fig. 2). The restriction sites ensure that the pkg2 fragments fuse in frame with the lacZ and phoA. For the alkaline phosphatase assay, transformants were induced by 1 mM IPTG for 2 h, and the assay was carried out as described previously (6).
Gene replacement.
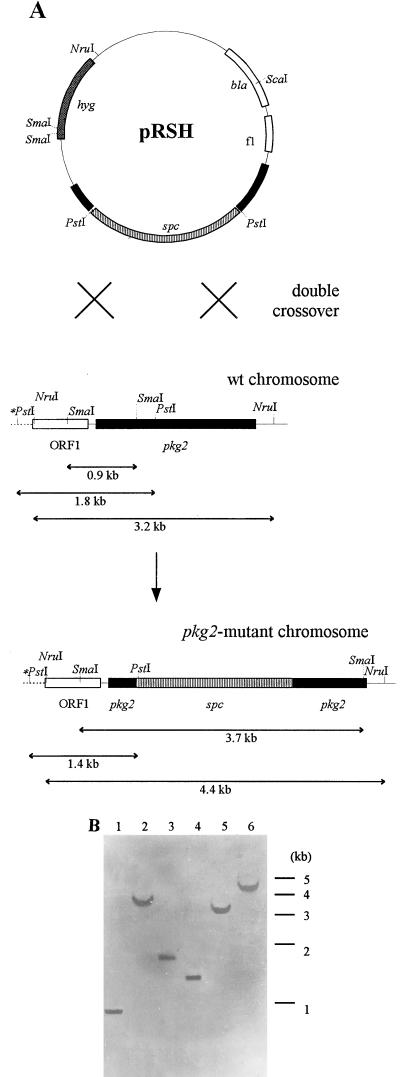
For replacement of the chromosomal pkg2 gene, we used the Ω interposon from pHP45Ω (36), which confers a spectinomycin resistance phenotype in E. coli and Streptomyces and double-stranded circular E. coli plasmid DNA. For construction of this recombinant plasmid, a 443-bp SacII-AluI fragment from plasmid p219, blunt ended with Klenow polymerase, was first cloned in the correct orientation into pTZ18R, previously digested with SmaI to yield pR1. Simultaneously, plasmid p219 was digested with SacI and partially with TaqI, and the 663-bp fragment obtained was cloned between the SacI and AccI sites of pTZ19R to yield pR2. Both plasmids obtained in these parallel steps were cleaved with HindIII and ScaI, and corresponding fragments (i.e., a 2,243-bp fragment from pR1 and a 1,741-bp fragment from pR2) were ligated, yielding pR12. An spc gene was then excised as a 2,014-bp HindIII fragment from pHP45Ω and inserted into a HindIII site of pR12. Similarly, a hyg gene was excised as a 2,274-bp HindIII fragment from pHP45Ωhyg (5), filled in with Klenow polymerase, and inserted into a KpnI site of pR12, previously blunt ended with Klenow polymerase. The resulting plasmid, conferring resistance to both spectinomycin and hygromycin, was named pRSH and transformed into E. coli GM2929 to obtain a nonmethylated plasmid DNA. Prior to transformation of S. granaticolor protoplasts, the plasmid DNA was alkaline denatured by the method of Oh and Chater (31). Spectinomycin-resistant (100 μg ml−1) and hygromycin-sensitive (100 μg ml−1) transformants were selected, and true disruptants were detected by Southern hybridization against the 198-bp SacII-HinPI fragment from p219 as the probe.
Electron microscopy.
Streptomyces aerial mycelium was observed by scanning electron microscopy. For the preparation of specimens, mycelium was fixed in the vapor of aqueous 2% osmium tetroxide in a glass desiccator. After a 2-day exposure to osmium vapor, the samples were transferred onto a stainless steel sieve holder in a beaker containing 200 ml of warm distilled water (40 to 50°C) and incubated for 30 min. The samples were then dehydrated in the vapor of absolute acetone for 2 days. Dehydrated samples were sputter-coated with gold by using a Polaron sputter-coater unit, and the morphology of aerial mycelia was observed in a TESLA BS300 scanning electron microscope operating at 25 kV.
Nucleotide sequence accession number.
The S. granaticolor pkg2 sequence reported in this paper has been submitted to the EMBL database and assigned accession no. AJ000264.
RESULTS
Identification and cloning of the pkg2 gene.
The identification of pkg2 gene was based on our previous work (46), where we carried out PCR using two degenerate primers and chromosomal DNA as a template. PCR yielded three different amplification products containing an open reading frame (ORF) giving an amino acid sequence in which an internal DFG motif, characteristic of domain VII of protein kinases (15), was detected. In this study, a 129-bp fragment designated PKG-2 was chosen for further characterization.
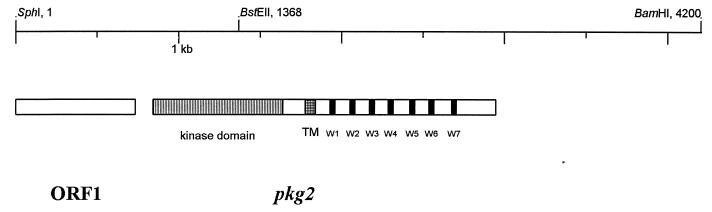
In the next step, we tried to isolate the entire gene corresponding to the PKG-2 sequence. Southern blot analysis using radiolabeled PKG-2 as a probe against the S. granaticolor chromosomal DNA digested with various restriction enzymes revealed a 4.2-kb SphI-BamHI fragment. This fragment was recovered from the gel, purified, ligated to pTZ19R, and transformed into E. coli XL1-Blue. Using the same probe, we screened the transformants by colony hybridization. As a result, a plasmid clone which contained the 4.2-kb SphI-BamHI insert (Fig. 1) was identified and designated p219.
FIG. 1.
Restriction map of the 4.2-kb SphI-BamHI fragment. Only relevant restriction sites are shown. Repeated motifs (W1 to W7) are shown as black boxes in the C-terminal part of the pkg2 gene (see text for details).
Nucleotide sequence analysis of the 4.2-kb SphI-BamHI fragment.
The nucleotide sequence of the 4.2-kb SphI-BamHI fragment (bp 1 to 4205) and predicted amino acid sequence of a complete ORF (ORF2) with a codon usage typical of streptomycetes are shown in Fig. 2. No similarity of the gene product of the truncated ORF1 to any sequence in the databases was discerned.
Sequence analysis of the ORF2 revealed that there are two putative initiation codons that might initiate a gene encoding a protein with sequence similarity to protein serine/threonine kinases, one at positions 757 to 759 and another at positions 844 to 846. However, the latter is most likely the initiation codon, since the percentage of preferred codons between the two putative initiation codons is quite low. An ORF initiating from the second initiation codon GTG (bases 844 to 6) ranges to TAG termination codon (bases 2947 to 9) (Fig. 2). ORF2 contains in the N-terminal part all 11 consensus catalytic subdomains of eukaryotic protein kinases, and the sequence of subdomain VI is characteristic of protein serine/threonine kinases in particular (15). A putative ribosome binding site, GGAGAG, is located 5 bases upstream of the initiation codon. ORF2 should produce a protein, Pkg2, of 701 amino acid residues, with a calculated molecular weight of 74.117 and an estimated pI of 6.80. At approximately 45 amino acid residues downstream of the kinase domain, we detected a hydrophobic stretch of 21 amino acid residues (positions 312 to 332), suggesting the presence of a putative membrane-spanning segment.
A search for homology with proteins registered in the EMBL and GenBank databases using the entire amino acid sequence showed that Pkg2 shared 31% identity in a 729-amino acid (aa) overlap with a S. granaticolor protein serine/threonine kinase Pkg3 (46), 29.6% identity in a 705-aa overlap with a Thermomonospora curvata putative protein kinase PkwA (22), 37.6% identity in a 471-aa overlap with a S. granaticolor protein serine/threonine kinase Pkg4 (46), and 30.4% identity in a 788-aa overlap with a S. coelicolor protein serine/threonine kinase AfsK (29). The C-terminal portion of Pkg2 shows 27.1% identity in a 325-aa overlap with a C-terminal portion of the S. coelicolor protein serine/threonine kinase AfsK (29). Further analysis revealed that in the C-terminal portion of Pkg2 there is a seven-times-repeated motif (Fig. 3 and 4) resembling the sequence of 11 amino acid residues known as a tryptophan-docking motif, previously identified in methanol dehydrogenase (MDH) from Methylobacterium extorquens (14), alcohol dehydrogenase (ADH) from Acetobacter aceti (10), and protein serine/threonine kinases Pkg4 and Pkg3 from S. graniticolor (46). As previously described for the tryptophan-docking motif of MDH (14), the interactions between tryptophan in position 11 and alanine in position 1 or the peptide bond between positions 6 and 7 stabilize a three-dimensional structure known as a propeller or superbarrel structure (2). As shown in Fig. 4, the tryptophan and alanine residues are highly conserved within the repeated motifs of Pkg2.
FIG. 3.
Amino acid sequence comparison between the C-terminal domains of the protein serine/threonine kinases Pkg4 (residues 399 to 761), Pkg3 (residues 399 to 780) (46), AfsK (residues 426 to 790) (29), and Pkg2 (residues 332 to 698). The repeated motifs are in boldface; gaps (indicated by dashes) were introduced to optimize sequence alignment.
FIG. 4.
Amino acid sequence alignment of the repeated motifs of the C-terminal domain of Pkg2. Conserved residues are in boldface. Numbers indicate positions of the tryptophan residues in the amino acid sequence of Pkg2 (for details, see Results).
Expression of Pkg2 in E. coli.
To characterize putative protein kinase Pkg2, the pkg2 gene was cloned under the control of a T7 promoter in pET24a (pEX2) and expressed in E. coli BL21(DE3). To rule out the possibility that the proteins synthesized in E. coli could be phosphorylated by an endogenous protein kinase activity rather than by an autophosphorylation process, the essential lysine residue of catalytic subdomain II was replaced by arginine. This lysine residue is highly conserved among protein kinases and is known to be directly involved in phosphotransfer reaction (7). Therefore, its replacement should abolish any autophosphorylation activity. A pkg2 gene with a Lys-to-Arg change (pkg2K45R) was also cloned into a pET vector (pEX2K45R).
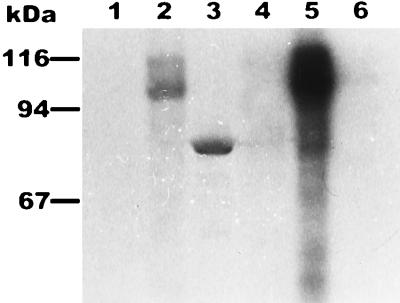
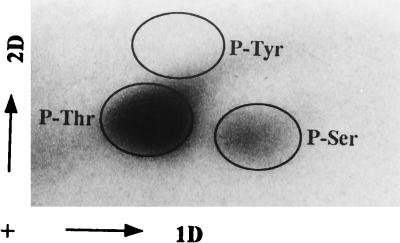
To identify pkg2 and pkg2K45R gene products, the cells were labeled in vivo with l-[35S]methionine in the presence of rifampin after IPTG induction. The cell extracts were analyzed by SDS-PAGE. As shown in Fig. 5 (lane 1), the production of E. coli proteins was almost completely blocked by rifampin. Pkg2 appeared as a major protein band of about 105 kDa (lane 2), which was not in agreement with the calculated molecular mass (74 kDa). When the mutated gene pkg2K45R was expressed in E. coli, Pkg2K45R migrated as a strong and distinct band at a molecular mass of approximately 75 kDa (lane 3), which was in complete agreement with that obtained for Pkg2 from the DNA sequence. Thus, to determine whether the band of approximately 105 kDa obtained after l-[35S]methionine labeling of the wild-type Pkg2 protein does in fact correspond to Pkg2 and whether the slower mobility of this putative Pkg2 protein is due to its phosphorylation status (30), E. coli cells harboring pEX2 were labeled in vivo with 32Pi after IPTG induction as described in Materials and Methods. Total cell proteins were separated by SDS-PAGE, and then a background of nucleic acids was removed by trichloroacetic acid treatment. As shown in lane 5, a fuzzy broad 32P-labeled band migrating at a molecular mass of 100 to 110 kDa appeared; this band corresponded to the band detected by labeling with l-[35S]methionine. Finally, in contrast to the wild-type Pkg2, when Pkg2K45R was labeled in vivo with 32P, no phosphorylated product was detected (lane 6). Phosphoamino acid analysis showed that Pkg2 was phosphorylated by its intrinsic activity at both threonine and serine residues (Fig. 6). These results demonstrated convincingly that Pkg2 is a serine/threonine-specific protein kinase that phosphorylates itself, and a mobility shift on SDS-PAGE arose as a consequence of the presence of a multiphosphorylated form.
FIG. 5.
Expression of the pkg2 gene in E. coli BL21(DE3) cells harboring plasmid pET24a (lanes 1 and 4), pEX2 (lanes 2 and 5), or pEX2K45R (lanes 3 and 6). Lanes 1 to 3, l-[35S]methionine labeling; lanes 4 to 6, 32P labeling.
FIG. 6.
Determination of phosphorylated amino acids of 32P-labeled Pkg2. Positions of the nonradioactive phosphoamino acid standards (P-Ser, phosphoserine; P-Thr, phosphothreonine; P-Tyr, phosphotyrosine) detected by staining with ninhydrin are shown by circles. The first and second dimensions are indicated by the arrows marked 1D and 2D, respectively.
Pkg2 contains a TM domain.
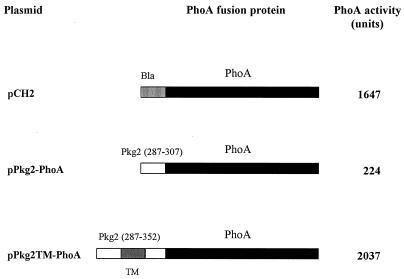
As Pkg2 displayed a putative TM region consisting of a 21-residue apolar stretch flanked by charged residues matching the positive-charge-difference rule (17), we attempted to determine its membrane topology. Fusion proteins were created from fragments either immediately preceding the TM region or encompassing the TM region with flanking sequences on both sides, and E. coli alkaline phosphatase PhoA, as described in Materials and Methods. PCR was used to amplify the corresponding fragments. Each of these two fragments was fused under the control of the lac promoter with a phoA gene. Alkaline phosphatase activities of the cells expressing fusion proteins are shown in Fig. 7. PhoA activity was only slightly increased for the PhoA fusion with the control segment (Pkg2-PhoA) compared to the activity of the PhoA− host strain. However, when the hydrophobic stretch was added to the fusion protein, the PhoA activity of the resulting protein, Pkg2TM-PhoA, increased almost 10-fold, to a level higher than even that of a PhoA+ control strain. Since PhoA is enzymatically active only when it is translocated through the cytoplasmatic membrane into the periplasm (28), these results demonstrated that Pkg2 is a transmembrane protein kinase with its C-terminal region downstream of the TM domain in the periplasmic space and the N-terminal kinase domain upstream of the TM domain in the cytoplasm.
FIG. 7.
Alkaline phosphatase activities of Pkg2-PhoA fusions in E. coli CC118 (PhoA−). Each activity is the average of data from three independent experiments. pCH2, plasmid containing the phoA gene fused with β-lactamase signal sequence (Bla) (18); pPkg2-PhoA, plasmid containing the phoA gene fused with a control segment preceding the putative TM region (residues 287 to 307); pPkg2TM-PhoA, plasmid containing the phoA gene fused with the putative Pkg2 TM region (residues 287 to 352).
Inactivation of the pkg2 gene and mutant strain studies.
To investigate the function of pkg2, we replaced a part of the chromosomal pkg2 gene of S. granaticolor by using the spc gene as a selection marker and a pkg2 sequence for homology to integrate the spc gene by double-crossover events. To select easily for two crossover events, which are necessary for effective gene replacement, another selection marker, the hyg gene, was introduced into a circular E. coli plasmid molecule (i.e., nonreplicating in Streptomyces) as described in Materials and Methods. As denaturation of the donor DNA is known to stimulate the number of transformants obtained by homologous recombination of incoming double-stranded circular DNA with the recipient chromosome, the plasmid DNA was alkaline denaturated prior to protoplast transformation (31). Transformants were collected and screened for their resistance patterns. Spectinomycin-resistant and hygromycin-sensitive colonies were tested for correct replacement of pkg2 on the chromosome of S. granaticolor by Southern hybridization with a part of the pkg2 sequence as a digoxigenin-labeled probe. This procedure yielded a pkg2 disruptant that gave signals with the expected sizes (Fig. 8).
FIG. 8.
Verification of replacement of pkg2 by double-crossover events. (A) Relevant features of genomic organization before and after replacement. Regions of plasmid pRSH corresponding to the wild-type (wt) chromosomal DNA are shown as black boxes. Note that not all restriction sites are shown. The PstI site marked by an asterisk was mapped by Southern analysis of the wild-type chromosomal DNA. (B) Analysis of DNA of a putative double-crossover recombinant (lanes 2, 4, and 6) by Southern blotting. Lanes 1, 3, and 5 contained DNA from the wild-type strain. Samples were digested with SmaI (lanes 1 and 2), PstI (lanes 3 and 4), and NruI (lanes 5 and 6).
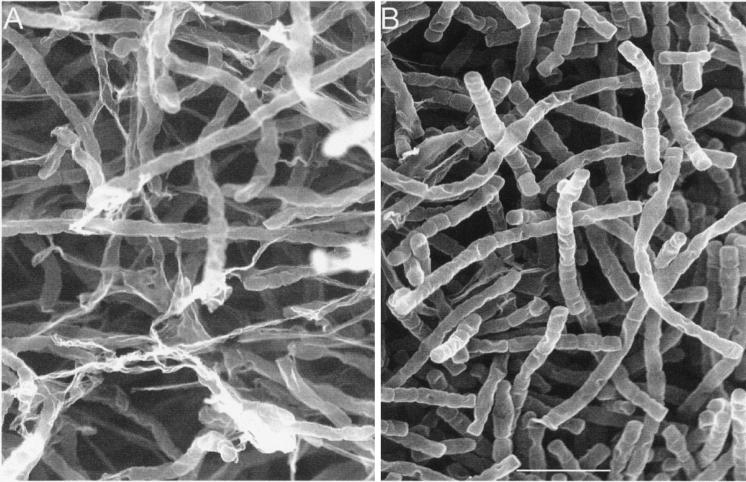
The mutation influenced neither the time course of the growth curve in MK liquid medium nor granaticin production. Wild-type and mutant strains grown on PPS agar plates exhibited no difference in timing of the course of morphological differentiation, as observed by scanning electron microscopy (data not shown). However, a slight difference was revealed in the morphology of aerial hyphae of the wild-type and mutant strains, and it was strongly marked when both strains were grown on SLM3 agar medium (Fig. 9). Photographs were taken so that each showed a representative level of development seen in the visible fields under the electron microscope. The population of hyphae of the wild-type strain was fairly dense and of uniform morphology. The aerial hyphal cells seemed to possess a well-evolved fibrous sheath (i.e., a thin superficial layer which surrounds the aerial hyphae and spores), and the length and diameter of fragmentation elements appeared homogeneous. In contrast, the mycelium of the mutant strain appeared as a heterogeneous network of intact and disintegrating hyphae; the cytological changes characteristic of cellular disintegration (lysis) of the aerial hyphae of the mutant strain are well illustrated in Fig. 9A. The envelopes of disintegrated hyphae tended to remain intact rather than decompose. Moreover, the fragmentation elements were not homogeneous in size, and many hyphae showed no signs of fragmentation, suggesting incomplete septum formation.
FIG. 9.
Electron microscopic observation of the S. granaticolor strains grown on SLM3 agar plates for 6 days at 28°C. The network of intact and disintegrating hyphae of the pkg2 mutant strain (A) is shown in comparison with the wild-type strain as a control (B). Bar, 5 μm. See text for details.
DISCUSSION
In this study, we identified in S. granaticolor and further characterized a new eukaryotic-like protein serine/threonine kinase designated Pkg2. Pkg2 contains a membrane-spanning sequence, as judged by PhoA fusion analysis, and therefore it is the first transmembrane protein serine/threonine kinase found in Streptomyces. In prokaryotes, a large number of transmembrane histidine protein kinases which function as sensors for various external signals have been identified (40). Only recently, several transmembrane protein serine/threonine kinases were found in M. xanthus, a multicellular developmental bacterium, and it was suggested that they could serve as receptors for developmental signals (16, 42, 50).
Using PCR and primers designed from the conserved sequences in the catalytic domains of eukaryotic protein kinases, we identified in S. granaticolor another two protein serine/threonine kinases, Pkg4 and Pkg3 (46), with different enzymatic properties. Both Pkg4 and Pkg3 are likely cytoplasmic proteins. An active form of Pkg4 autophosphorylates at threonine residue(s), whereas Pkg3 does not undergo an autophosphorylation process. If Pkg3 requires phosphorylation to become an active enzyme, then the possibility that Pkg3 can be phosphorylated and so activated by another protein kinase, either cytoplasmatic or membrane localized, cannot be excluded. In vitro phosphorylation experiments performed with partially purified Pkg2 suggested that compared to Pkg4, protein kinase Pkg2 has a broad substrate specificity in S. granaticolor cell extracts (unpublished results). It could mean that Pkg2 targets multiple proteins and so modulates their functions, in which case it might be assumed that Pkg2 occupies a high position in a hierarchy of signaling cascade. It is tempting to speculate that in Streptomyces there are several protein kinases involved in a particular phosphorelay similar to that in eukaryotes constituting a signaling network. Therefore, we concluded that Streptomyces may be a promising model for a study of signal transduction process mediated via protein serine/threonine kinases.
Amino acid residues known to form stacking interactions stabilizing the propeller structure in MDH (14) are highly conserved within the repeated motifs in the C-terminal portion of Pkg2, which is located outside the cell. The propeller structure was identified first in influenza virus neuraminidase (44) and subsequently in sialidase from Salmonella typhimurium LT2 (11), galactose oxidase from Dactylium dendroides (21), methylamine dehydrogenase from Thiobacillus versutus (45), and MDH from Methylobacterium extorquens (14). From amino acid sequences alignment and software modeling, it was also predicted to exist in ADH from A. aceti (10). The propeller structure is made up of topologically identical four-stranded antiparallel β sheets arranged like the blades of a propeller. The number of blades varies from six to eight. There is a little or no homology in parts of the amino acid sequences forming the propeller structures, and the propeller structure is not closely related to the functions of the proteins.
Besides Pkg2, we identified similar repeated motifs in the C-terminal regions of protein serine/threonine kinases Pkg4 and Pkg3 (46) and in the C-terminal part of the protein serine/threonine kinase AfsK from S. coelicolor A3(2) (29) (Fig. 3). Thus, it seems that streptomycetes synthesize at least four protein serine/threonine kinases with homology not only in the protein kinase domains but also in the C-terminal domains, indicating a tighter structural relationship among these signal-transducing enzymes. It is conceivable that the C-terminal regions of these proteins are involved in transmitting signals to or from other proteins through protein-protein interactions.
One can speculate that the C-terminal portion of Pkg2 forms outside the cell a symmetrical structure (probably propeller-like) serving as a bacterial sensor and transducing signals from unknown ligand(s) via its protein kinase domain to an unknown substrate(s), and that the other protein kinases mentioned above somehow participate in this process. Experiments aimed at identifying these ligands and target substrates are in progress.
The pkg2 mutant strain maintained its ability to form aerial mycelium and sporulate when grown on PPS agar plates, and only a few aerial hyphae of the mutant strain showed signs of disintegration. However, the proportion of lysing cells increased strongly when cultivated on SLM3 agar plates. Disintegrating aerial hyphae of the wild-type strain were only occasionally found on both media tested. It seems that the pattern of development of the mutant strain mycelium could be somehow influenced by the composition of the medium. This effect is well known in Streptomyces and illustrates the plasticity of members of this genus and other actinomycetes in response to varied growth conditions. For example, the morphological defect of most of the bld mutants of S. coelicolor, which fail to form aerial hyphae on rich medium, is carbon source dependent (8); recently, a new interpretation for the role of the bld genes in development in Streptomyces suggested that the primary defect in bld mutants is in the regulation of carbon utilization and not specifically in the activation of developmentally regulated genes (35). In this respect, the result of the pkg2 replacement is reminiscent of situation known in bld mutant studies. Nevertheless, the particular steps of the pathway leading from pkg2 gene replacement to scaled-up aerial hyphae disintegration remains to be identified.
It has been shown that apart from a plasmid-encoded secreted protein serine/threonine kinase, YpkA, having an indispensable role in virulence of Y. pseudotuberculosis (13), all prokaryotes for which protein serine/threonine kinases have been described (Streptomyces [29, 43, 46], M. xanthus [16, 30, 42, 50], Anabaena strain PCC 7120 [47–49], and T. curvata [22]) display developmental characteristics comparable to those of multicellular eukaryotes. However, recent results demonstrated that a multigene family of putative protein serine/threonine kinases is commonly present in microorganisms as different as Mycobacterium tuberculosis (9) and Synechocystis sp. (25). On the other hand, E. coli genome sequencing revealed that there are no sequences resembling the eukaryotic-like protein serine/threonine kinases (4). Thus, ongoing sequencing projects followed by sequence analysis of obtained data may divide prokaryotes into two groups: those that need protein serine/threonine kinases for growth and development, and the others. Comparison of these two groups together with the accompanying knowledge of protein serine/threonine kinases’ diverse functions in eukaryotes could be a clue to tracing the pathways of signaling phosphate groups and their significance.
ACKNOWLEDGMENTS
We thank Jean-Luc Pernodet, Miroslav Petříček, and Jana Št’astná for numerous helpful discussions. We thank E. J. Stewart for providing E. coli CC118, J. L. Pernodet for the gift of plasmids pHP45Ω and pHP45Ωhyg, and S. Inouye for the gift of plasmid pCH2. We are grateful to Olga Kofroňová for electron microscopic studies.
This work was supported by the Grant Agency of the Czech Republic (grant 204/96/1236 to P.B.).
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Anthony C, Ghosh M, Blake C C. The structure and function of methanol dehydrogenase and related quinoproteins containing pyrrolo-quinolone quinone. Biochem J. 1994;304:665–674. doi: 10.1042/bj3040665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bibb M J, Findlay P R, Johnson M W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984;30:157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- 4.Blattner F R, Plunkett G R, Bloch C A, Perna N T, Burland V, Riley M, Collado Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 5.Blondelet Rouault M H, Weiser J, Lebrihi A, Branny P, Pernodet J L. Antibiotic resistance gene cassettes derived from the omega interposon for use in E. coli and Streptomyces. Gene. 1997;190:315–317. doi: 10.1016/s0378-1119(97)00014-0. [DOI] [PubMed] [Google Scholar]
- 6.Brickman E, Beckwith J. Analysis of the regulation of Escherichia coli alkaline phosphatase synthesis using deletions and φ80 transducing phages. J Mol Biol. 1975;96:307–316. doi: 10.1016/0022-2836(75)90350-2. [DOI] [PubMed] [Google Scholar]
- 7.Carrera A C, Alexandrov K, Roberts T M. The conserved lysine of the catalytic domain of protein kinases is actively involved in the phosphotransfer reaction and not required for anchoring ATP. Proc Natl Acad Sci USA. 1993;90:442–446. doi: 10.1073/pnas.90.2.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chater K F. Taking a genetic scalpel to the Streptomyces colony. Microbiology. 1998;144:1465–1478. doi: 10.1099/00221287-144-6-1465. [DOI] [PubMed] [Google Scholar]
- 9.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail M A, Rajandream M-A, Rogers J, Rutter S, Seeger K, Skelton J, Squares S, Squares R, Sulston J E, Taylor K, Whitehead S, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 10.Cozier G E, Giles I G, Anthony C. The structure of the quinoprotein alcohol dehydrogenase of Acetobacter aceti modelled on that of methanol dehydrogenase from Methylobacterium extorquens. Biochem J. 1995;308:375–379. doi: 10.1042/bj3080375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crennell S J, Garman E F, Laver W G, Vimr E R, Taylor G L. Crystal structure of a bacterial sialidase (from Salmonella typhimurium LT2) shows the same fold as an influenza virus neuraminidase. Proc Natl Acad Sci USA. 1993;90:9852–9856. doi: 10.1073/pnas.90.21.9852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duclos B, Grangeasse C, Vaganay E, Riberty M, Cozzone A J. Autophosphorylation of a bacterial protein at tyrosine. J Mol Biol. 1996;259:891–895. doi: 10.1006/jmbi.1996.0366. [DOI] [PubMed] [Google Scholar]
- 13.Galyov E E, Hakansson S, Forsberg A, Wolf Watz H. A secreted protein kinase of Yersinia pseudotuberculosis is an indispensable virulence determinant. Nature. 1993;361:730–732. doi: 10.1038/361730a0. [DOI] [PubMed] [Google Scholar]
- 14.Ghosh M, Anthony C, Harlos K, Goodwin M G, Blake C. The refined structure of the quinoprotein methanol dehydrogenase from Methylobacterium extorquens at 1.94 A. Structure. 1995;3:177–187. doi: 10.1016/s0969-2126(01)00148-4. [DOI] [PubMed] [Google Scholar]
- 15.Hanks S K, Quinn A M, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 16.Hanlon W A, Inouye M, Inouye S. Pkn9, a Ser/Thr protein kinase involved in the development of Myxococcus xanthus. Mol Microbiol. 1997;23:459–471. doi: 10.1046/j.1365-2958.1997.d01-1871.x. [DOI] [PubMed] [Google Scholar]
- 17.Hartmann E, Rapoport T A, Lodish H F. Predicting the orientation of eukaryotic membrane-spanning proteins. Proc Natl Acad Sci USA. 1989;86:5786–5790. doi: 10.1073/pnas.86.15.5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffman C S, Wright A. Fusions of secreted proteins to alkaline phosphatase: an approach for studying protein secretion. Proc Natl Acad Sci USA. 1985;82:5107–5111. doi: 10.1073/pnas.82.15.5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hofmann K, Stoffel W. TMbase—a database of membrane spanning protein segments. Biol Chem Hoppe-Seyler. 1993;347:166. [Google Scholar]
- 20.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces. A laboratory manual. Norwich, England: John Innes Foundation; 1985. [Google Scholar]
- 21.Ito N, Phillips S E, Yadav K D, Knowles P F. Crystal structure of a free radical enzyme, galactose oxidase. J Mol Biol. 1994;238:794–814. doi: 10.1006/jmbi.1994.1335. [DOI] [PubMed] [Google Scholar]
- 22.Janda L, Tichý P, Spížek J, Petříček M. A deduced Thermomonospora curvata protein containing serine/threonine protein kinase and WD-repeat domains. J Bacteriol. 1996;178:1487–1489. doi: 10.1128/jb.178.5.1487-1489.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janeček J, Moravec V, Dobrová Z, Janda I, Weiser J. Protein phosphorylation in submerged spores and vegetative mycelium of Streptomyces granaticolor. FEMS Microbiol Lett. 1995;133:91–94. [Google Scholar]
- 24.Kamps M P, Sefton B M. Acid and base hydrolysis of phosphoproteins bound to Immobilon facilitates analysis of phosphoamino acids in gel-fractionated proteins. Anal Biochem. 1989;176:22–27. doi: 10.1016/0003-2697(89)90266-2. [DOI] [PubMed] [Google Scholar]
- 25.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 26.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 27.Manai M, Cozzone A J. Endogenous protein phosphorylation in Escherichia coli extracts. Biochem Biophys Res Commun. 1982;107:981–988. doi: 10.1016/0006-291x(82)90619-2. [DOI] [PubMed] [Google Scholar]
- 28.Manoil C, Beckwith J. TnphoA: a transposon probe for protein export signals. Proc Natl Acad Sci USA. 1985;82:8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsumoto A, Hong S K, Ishizuka H, Horinouchi S, Beppu T. Phosphorylation of the AfsR protein involved in secondary metabolism in Streptomyces species by a eukaryotic-type protein kinase. Gene. 1994;146:47–56. doi: 10.1016/0378-1119(94)90832-x. [DOI] [PubMed] [Google Scholar]
- 30.Munoz-Dorado J, Inouye S, Inouye M. A gene encoding a protein serine/threonine kinase is required for normal development of M. xanthus, a gram-negative bacterium. Cell. 1991;67:995–1006. doi: 10.1016/0092-8674(91)90372-6. [DOI] [PubMed] [Google Scholar]
- 31.Oh S H, Chater K F. Denaturation of circular or linear DNA facilitates targeted integrative transformation of Streptomyces coelicolor A3(2): possible relevance to other organisms. J Bacteriol. 1997;179:122–127. doi: 10.1128/jb.179.1.122-127.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peirs P, DeWit L, Braibant M, Huygen K, Content J. A serine/threonine protein kinase from Mycobacterium tuberculosis. Eur J Biochem. 1997;244:604–612. doi: 10.1111/j.1432-1033.1997.00604.x. [DOI] [PubMed] [Google Scholar]
- 34.Petříček M, Smrčková I, Tichý P. Transformation of Streptomyces granaticolor with natural and recombinant plasmid vectors. Folia Microbiol. 1985;30:474–478. doi: 10.1007/BF02927609. [DOI] [PubMed] [Google Scholar]
- 35.Pope M K, Green B D, Westpheling J. The bld mutants of Streptomyces coelicolor are defective in the regulation of carbon utilization, morphogenesis and cell-cell signalling. Mol Microbiol. 1996;19:747–756. doi: 10.1046/j.1365-2958.1996.414933.x. [DOI] [PubMed] [Google Scholar]
- 36.Prentki P, Krisch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 37.Řičicová A, Řeháček Z. Taxonomic characteristic of the strain ETH 7437 producing granaticin. Folia Microbiol. 1968;13:346–349. doi: 10.1007/BF02909624. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 39.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stock J B, Ninfa A J, Stock A M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 42.Udo H, Munoz-Dorado J, Inouye M, Inouye S. Myxococcus xanthus, a gram-negative bacterium, contains a transmembrane protein serine threonine kinase that blocks the secretion of beta-lactamase by phosphorylation. Genes Dev. 1995;9:972–983. doi: 10.1101/gad.9.8.972. [DOI] [PubMed] [Google Scholar]
- 43.Urabe H, Ogawara H. Cloning, sequencing and expression of serine/threonine kinase-encoding genes from Streptomyces coelicolor A3(2) Gene. 1995;153:99–104. doi: 10.1016/0378-1119(94)00789-u. [DOI] [PubMed] [Google Scholar]
- 44.Varghese J N, Laver W G, Colman P M. Structure of the influenza virus glycoprotein antigen neuraminidase at 2.9 Å resolution. Nature. 1983;303:35–40. doi: 10.1038/303035a0. [DOI] [PubMed] [Google Scholar]
- 45.Vellieux F M, Huitema F, Groendijk H, Kalk K H, Jzn J F, Jongejan J A, Duine J A, Petratos K, Drenth J, Hol W G. Structure of quinoprotein methylamine dehydrogenase at 2.25 Å resolution. EMBO J. 1989;8:2171–2178. doi: 10.1002/j.1460-2075.1989.tb08339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vomastek T, Nádvorník R, Janeček J, Techniková Z, Weiser J, Branny P. Characterisation of two putative protein Ser/Thr kinases from actinomycete Streptomyces granaticolor both endowed with different properties. Eur J Biochem. 1998;257:55–61. doi: 10.1046/j.1432-1327.1998.2570055.x. [DOI] [PubMed] [Google Scholar]
- 47.Zhang C C. A gene encoding a protein related to eukaryotic protein kinases from the filamentous heterocystous cyanobacterium Anabaena PCC 7120. Proc Natl Acad Sci USA. 1993;90:11840–11844. doi: 10.1073/pnas.90.24.11840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang C C, Friry A, Peng L. Molecular and genetic analysis of two closely linked genes that encode, respectively, a protein phosphatase 1/2A/2B homolog and a protein kinase homolog in the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 1998;180:2616–2622. doi: 10.1128/jb.180.10.2616-2622.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang C C, Libs L. Cloning and characterisation of the pknD gene encoding an eukaryotic-type protein kinase in the cyanobacterium Anabaena sp. PCC7120. Mol Gen Genet. 1998;258:26–33. doi: 10.1007/s004380050703. [DOI] [PubMed] [Google Scholar]
- 50.Zhang W D, Inouye M, Inouye S. Reciprocal regulation of the differentiation of Myxococcus xanthus by Pkn5 and Pkn6, eukaryotic-like Ser/Thr protein kinases. Mol Microbiol. 1996;20:435–447. doi: 10.1111/j.1365-2958.1996.tb02630.x. [DOI] [PubMed] [Google Scholar]