HighLights
-
•
The phylogenetic analyses of the full-length F genes in HRV1 strains collected from various countries were performed.
-
•
HRV1 strains showed three major lineages in time-scaled phylogenetic tree of F genes.
-
•
No positive selection sites were found in F protein, whereas numerous negative selection sites were identified.
-
•
Incompatibilities between predicted epitopes in this study and NT-Ab binding might contribute to HRV1 reinfection.
Keywords: Human respirovirus 1, Molecular evolutionary analyses, Fusion protein gene, B cell conformational epitope
Abstract
Few evolutionary studies of the human respiratory virus (HRV) have been conducted, but most of them have focused on HRV3. In this study, the full-length fusion (F) genes in HRV1 strains collected from various countries were subjected to time-scaled phylogenetic, genome population size, and selective pressure analyses. Antigenicity analysis was performed on the F protein. The time-scaled phylogenetic tree using the Bayesian Markov Chain Monte Carlo method estimated that the common ancestor of the HRV1 F gene diverged in 1957 and eventually formed three lineages. Phylodynamic analyses showed that the genome population size of the F gene has doubled over approximately 80 years. Phylogenetic distances between the strains were short (< 0.02). No positive selection sites were detected for the F protein, whereas many negative selection sites were identified. Almost all conformational epitopes of the F protein, except one in each monomer, did not correspond to the neutralising antibody (NT-Ab) binding sites. These results suggest that the HRV1 F gene has constantly evolved over many years, infecting humans, while the gene may be relatively conserved. Mismatches between computationally predicted epitopes and NT-Ab binding sites may be partially responsible for HRV1 reinfection and other viruses such as HRV3 and respiratory syncytial virus.
1. Introduction
Human respirovirus 1 (formerly called human parainfluenza virus 1, HRV1) is an RNA virus that belongs to the genus Respirovirus of the family Paramyxoviridae. HRV1 is a causative agent of acute respiratory diseases, such as common colds, acute laryngotracheobronchitis (croup), bronchiolitis, and pneumonia, and is distributed world-wide as the most prevalent type of the former human parainfluenza virus as well as HRV3 (Henrickson, 2003; Karron, 2007). Epidemiological studies showed that HRV is a causative agent for croup in children under five years of age, among which approximately 26–74% experience HRV1 infection (Denny et al., 1983). HRV1 reinfection and HRV3 may occur throughout life; however, the reinfection mechanisms are not exactly known (Henrickson, 2003).
The HRV1 genome encodes six genes that are translated into seven proteins (Karron, 2007). Among these, fusion protein (F protein) and haemagglutinin-neuraminidase (HN) proteins are the major viral antigens (Karron, 2007). In particular, the F protein consists of a homotrimer and may be associated with infection of airway epithelial cells in the host (Karron, 2007). Moreover, the existence of two conformations of the F protein, prefusion and postfusion, have been confirmed (Yin et al., 2006). However, detailed F protein structure is not well understood (Shao et al., 2021).
Antibody responses are central to acquired immunity against viral infections. Epitopes are classified into two categories: conformational and linear epitopes (Van Regenmortel, 2001). Linear epitopes are continuous amino acid sequences of the primary antigen structure. Conformational epitopes are composed of discontinuous residues that are in proximity on the protein three-dimensional (3D) structure. Both epitopes are recognised by the immune system, triggering the production of antibodies (Sharon et al., 2014; Collins and Karron, 2013; Van Regenmortel, 2001). A previous report showed that over 90% of B cell epitopes are conformational, and only a few are linear (Van Regenmortel, 2001). In contrast, due to an explicit distinction between antigenicity and immunogenicity, these epitopes in antigenic proteins may not be adequately recognised by a neutralising antibody (NT-Ab) (Sharon et al., 2014; Collins and Karron, 2013). Our previous data suggested that the computationally predicted conformational epitopes in the respiratory syncytial virus (RSV) and HRV F proteins do not correspond to the NT-Ab binding sites of these proteins (Aso et al., 2020; Saito et al., 2021). However, to the best of our knowledge, the relationship between conformational and linear epitopes and the NT-Ab binding sites of the HRV1 F protein is not known. Moreover, the detailed phylogeny of the viral fusion (F) gene is unknown. Therefore, detailed molecular evolutionary analyses of the HRV1 F gene were performed in strains collected globally, using bioinformatic technologies.
2. Materials and methods
2.1. Strains used in this study
To better understand the molecular evolution of the HRV1 F gene, nucleotide sequences, including the full-length coding region of the gene (positions 5060–6727; 1668 nt for the hPIV1/USA/ATCC VR-94/1957 strain; GenBank accession No. JQ901971) was retrieved from GenBank on 11 June 2019. Among these, sequences from strains with confirmed information on the detected/isolated years and regions were selected. In addition, data of strains that displayed ambiguity with undermined sequences (e.g., N, Y, R, and V) were omitted from the dataset, providing data from 71 strains for the analysis. Homologous sequences were identified using Clustal Omega (Sievers and Higgins, 2021). When three or more similar sequences were present, only two among them were randomly retained, which reduced the final sequence set to those from 66 strains.
Temporal signal analysis of the sequences from 66 HRV1 strains was performed to determine whether the dataset was suitable for molecular clock analysis. Maximum likelihood method was used to generate a phylogenetic tree using molecular evolutionary genetics analysis version 7.0 (MEGA 7; for bigger datasets) software. The data were analysed using TempEst software (version 1.5.3) (Rambaut et al., 2016).
All data are presented in Supplementary Table S1. These sequences were aligned using MAFFT version 7 (Katoh and Standley, 2013) and subsequently trimmed to 1668 nt based on the prototype F gene sequences.
2.2. Time-Scaled phylogenetic analysis and phylodynamic analyses using the bayesian markov chain monte carlo method (BMCMC)
To investigate the evolution of HRV1 strains, a time-scaled phylogenetic analysis of full-length sequences of the HRV1 F gene was conducted using the Bayesian Markov chain Monte Carlo (BMCMC) method in BEAST (version 2.4.8) (Bouckaert et al., 2014). To select a suitable substitution model, jModelTest program (version 2.1.10) was used (Darriba et al., 2012). The path-sampling implemented in the BEAST package was used to determine the best of four clock models (strict clock, exponential relaxed clock, log-normal relaxed clock, and random local clock) and three prior tree models (coalescent constant population, coalescent exponential population, and coalescent Bayesian skyline). The TrN + I substitution model, log-normal of the relaxed clock model, and coalescent exponential from the tree prior model were used for BMCMC analysis of all strains. An BMCMC tree was constructed using BEAST software with the obtained strains and selected models. We analysed the MCMC chains for the 100,000,000 steps with sampling performed after every 2000 steps. To confirm convergence, the effective sample sizes (ESS) were evaluated using Tracer (version 1.6), and values above 200 were considered acceptable. After burn-in of the first 10% of the trees, a maximum clade credibility tree was generated using TreeAnnotator (version 2.4.8) in the BEAST package. The BMCMC phylogenetic tree was visualised using FigTree (version 1.4.03), and the 95% highest posterior densities (HPDs) of all internal nodes were computed. Moreover, strain clustering in the constructed phylogenetic tree of the HRV1 F gene followed the illustrated tree topology. Simultaneously, the evolutionary rates of the 66 HRV1 strains and strains of each lineage determined by the BMCMC phylogenetic tree were estimated using the BMCMC method, and the values were confirmed using the Tracer software. The marginal likelihood (ML) values for model selection and the detailed parameters of the BMCMC analyses are shown in Supplemental Tables S2 and S3. The statistics calculated by Tracer for each dataset are listed in Supplementary Tables S2–S6. Statistical analysis for comparing evolutionary rates between the lineages was performed using the Kruskal–Wallis test with the EZR software (Kanda, 2013). The evolutionary rates sampled every 2000 steps from the MCMC chains after discarding the 10% burn-in (45,001 samples) were used for statistical analysis. Statistical significance was defined as p < 0.05. Past genome population dynamics of the HRV1 F gene were examined using Bayesian skyline plots (BSPs) in BEAST. A coalescent Bayesian skyline was selected as the prior tree model.
2.3. Phylogenetic distance calculation
The phylogenetic distances among all HRV1 strains were analysed to estimate F gene diversity. A phylogenetic tree of all HRV1 strains was constructed using the ML method with MEGA7 software (Kumar et al., 2016), and branch reliability was supported by 1000 bootstrap replications. The jModelTest program was used to select the best substitution model for the ML method. Subsequently, the phylogenetic distance of the ML tree was calculated using Patristic (Fourment and Gibbs, 2006).
2.4. Selective pressure analyses
The selective pressure sites for the F protein of HRV1 were analysed by calculating the non-synonymous (dN) and synonymous (dS) substitution rates at each amino acid site using the Datamonkey web server (https://www.datamonkey.org/) (Weaver et al., 2018). Single-likelihood ancestor counting (SLAC), fixed effects likelihood (FEL), internal fixed-effects likelihood (IFEL), fast unconstrained Bayesian approximation (FUBAR) (Murrell et al., 2013), and the mixed-effects model of evolution (MEME) (Murrell et al., 2012) were used to estimate positive selection sites, whereas, SLAC, FEL, IFEL, and FUBAR were used to predict negative selection sites. The positive (dN/dS > 1) and negative (dN/dS < 1) selection was determined based on the p values (p < 0.05) for SLAC, FEL, IFEL, and MEME and on the posterior probability values (> 0.9) for FUBAR.
2.5. Modelling of three-dimensional structure of the HRV1 F protein
Experimentally validated 3D structure of the HRV1 F protein is not available. Hence, we employed a homology modelling method to construct trimeric structural models of the prefusion F protein of HRV1 for representative strains from each group, determined using the BMCMC phylogenetic tree (prototype, ATCC VR-94/USA/1957 strain, JQ901971; lineage 1, HPIV1/WI/629–008/1997 strain, JQ901978; lineage 2, HPIV1/WI/629–007/1997 strain, JQ901979; and lineage 3, HPIV1/USA/629-D02161/2009 strain, KF687308). The cryo-electron microscopy structure of HRV3 F protein (Protein Data Bank accession ID: 6MJZ) was selected as the template based on the results from BLAST web server (Shao et al., 2021). The amino acid sequences of each strain and template were aligned using MAFFT. The percentage sequence identity of each strain to the template was calculated using Clustal Omega. Based on the template sequence, 3D structures were constructed using Modeller software (version 10.2). The generated models were assessed by Ramachandran plot analyses using WinCoot implemented in the CCP4 package, and the models with the best scores were selected. Energy minimisation of the generated structures was performed using GROMOS96, which was implemented in Swiss PDB Viewer (version 4.1.0) (Guex and Peitsch, 1997).
2.6. Analyses of conformational and linear epitopes and amino acid substitution sites
To accurately analyse the pressure of human immune defence against the natural state of the HRV1 F protein, epitopes in the trimeric prefusion state were predicted. The conformational epitopes of the constructed models were analysed using Disco-Tope (version 2.0) (Kringelum et al., 2012), ElliPro (Ponomarenko et al., 2008), SEMA (Shashkova et al., 2022) and SEPPA (version 3.0) (Zhou et al., 2019) with cut-off values of −3.7, 0.5, 0.76, and 0.064, respectively. The accuracy of the analyses was also supported by the consensus sites predicted by more than three of the four methods, and regions with residues close to two of the sites on the trimeric structure models were determined as conformational epitopes. Subsequently, the linear epitopes were analysed using LBtope (Singh et al., 2013) and BECEPS (Ras Carmona et al., 2021), BepiPred (version 2.0) (Galgonek et al., 2017) and ABCpred (Saha and Raghava, 2006). Cut-off values were set as 80% (LBtope), 0.5 (BECEPS), 0.5 (BepiPred 2.0), and 0.51 (ABCpred), respectively.
Regions that had more than 10 continuous amino acids and were estimated in common by at least three of the four methods were regarded as linear epitopes. Finally, the predicted and previously identified epitopes were mapped onto the constructed, pre-fusion F protein models using PyMOL (version 2.3) (WL, 2002.).
3. Results
3.1. Time-Scaled phylogenetic analysis and phylodynamic analysis of the HRV 1 F gene using the BMCMC method
To estimate the time-scaled evolution of the HRV1 F gene, a phylogenetic tree was constructed using the BMCMC method. In this study, we used only the sequences from HRV1 strains (66 strains) that were detected in humans, because sequence data from bovine respiratory virus (BRV) type 1, which may be a common ancestor of both BRV and HRV, were not available. Before constructing the BMCMC tree, the temporal signal of the dataset was estimated using TempEst (version 1.5.3). The plots of root-to-tip genetic distance against sampling time exhibited a positive correlation between genetic divergence and sampling time, and the R square value was calculated as 0.87 (Figure S1). These results suggest that the dataset of the 66 HRV1 strains was adequate for molecular clock analysis. Hence, we used this dataset to carry out the BMCMC method.
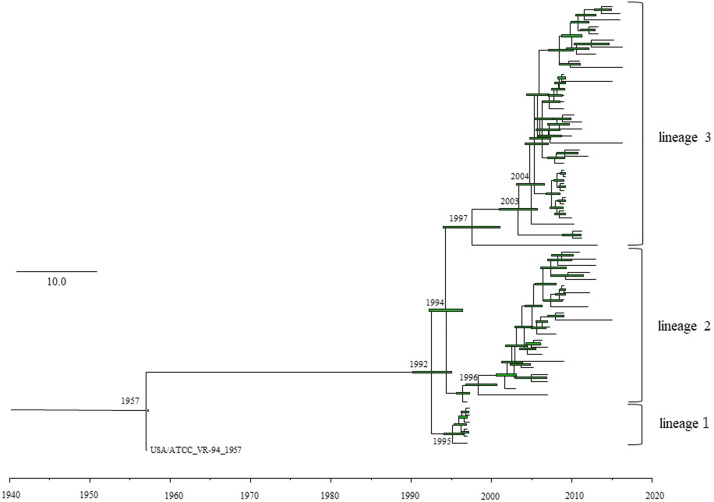
As shown in Fig. 1, a common ancestor of the HRV1 prototype strain (hPIV1/USA/ATCC_VR-94_1957; GenBank accession No. JQ901971) and other existing HRV1 strains diverged in 1957 (95% HPD, 1956–1957), resulting ultimately in the formation of three major lineages 1–3. After the first divergence in 1957, strains belonging to lineage 1 further diverged from a common ancestor of strains belonging to the three lineages in 1992 (95% HPD, 1989–1994), and the opposite side of lineage 1 diverged into lineages 2 and 3 in 1994 (95% HPD, 1991–1996). Currently, strains belonging to lineage 3 are widespread and form several clusters.
Fig. 1.
Time-scaled evolutionary tree of the full length Human respirovirus 1 (HRV1) fusion gene constructed by the Bayesian Markov chain Monte Carlo (BMCMC) method. The scale bar represents time (years). Green bars indicate the 95% highest posterior density (HPD) for each branch year.
Next, the evolutionary rate of HRV1 F gene was estimated (Table 1). The evolutionary rate of all strains was estimated to be 8.504 × 10−4 substitutions/site/year (s/s/y) (95% HPD, 7.003 × 10−4 to 1.0008 × 10−3 s/s/y). The calculations for each of the above lineages showed that the evolutionary rate of strains belonging to lineage 2 was 6.580 × 10−4 s/s/y (95% HPD, 4.784 × 10−4 to 8.4595 × 10−4 s/s/y), and that of strains belonging to lineage 3 was 1.205 × 10−3 s/s/y (95% HPD, 7.159 × 10−4 to 1.6866 × 10−3 s/s/y). The evolutionary rate of the strains in lineage 1 with the same detection year (1997) was not calculated. The evolutionary rate of strains belonging to lineage 3 was significantly higher than that of strains belonging to lineage 2 (p < 2 − 16), which may indicate that strains belonging to lineage 3 are more adapted to humans, although the mechanisms underlying the significance of these values are not known.
Table 1.
Evolutionary rates of all HRV1 strains and each lineage.
| Evolutionary rates (95% HPD) (substitutions/site/year) | Effective sample size | |
|---|---|---|
| All strains (66 strains) | 8.504 × 10−4 (7.003 × 10−4 to 1.0008 × 10−3) | 220 |
| Lineage 1 (6 strains) | ― | ― |
| Lineage 2 (23 strains) | 6.580 × 10−4 (4.784 × 10−4 to 8.4595 × 10−4) | 4053 |
| Lineage 3 (36 strains) | 1.205 × 10−3 (7.159 × 10−4 to 1.6866 × 10−3) | 954 |
3.2. Phylodynamics of the HRV1 F gene using the bayesian skyline plot (BSP) analysis method
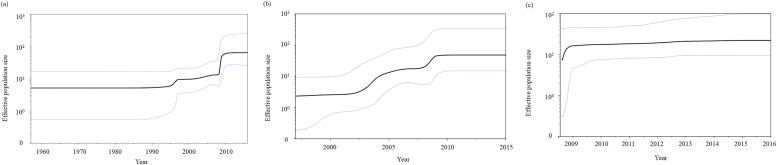
As shown in Fig. 2, the phylodynamics of the F gene in HRV1 strains were analysed using the BSP analysis method to detect fluctuations in effective population size. The genome population size of all the strains doubled between 1995 and 2008 (Fig. 2A). Similarly, strains belonging to lineage 2 showed a two-fold increase in genome population around 2003 and 2008 (Fig. 2B). In contrast, in lineage 3, a steep increase in genome population size was observed, even though it occurred only once around 2008 (Fig. 2C). Because the detection year of the strains belonging to lineage 1 was the same (1997), we could not calculate the genome population size of this lineage. To summarise the results of these BSP analyses, the rapid increase in genome population size around 2008 for all strains was speculated to be mainly due to an increase in the genome population size of lineage 3.
Fig. 2.
Bayesian skyline plot for the Human respirovirus 1 (HRV1) fusion gene. Each panel illustrates the phylodynamics of all 66 strains (a), lineage 2 (b), and lineage 3 (c). Y and x-axes indicate the effective population size and time in years, respectively. Thick black lines show the median values over time; thin blue lines represent the 95% highest posterior density (HPD) intervals.
3.3. Phylogenetic distances calculation of the HRV1 F gene
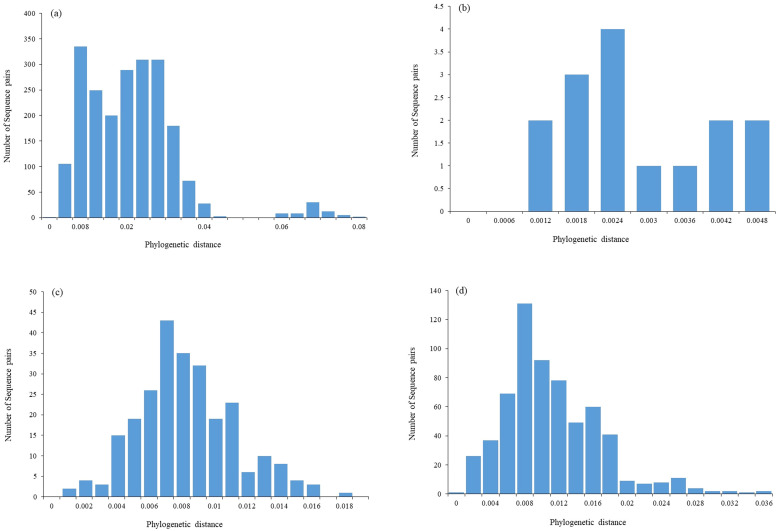
The phylogenetic distance and distribution between strains were evaluated based on their nucleotide sequences. A histogram of the distances between the sequence pairs of all the strains revealed a bimodal distribution (Fig. 3). Furthermore, histograms of lineages 1 and 2 showed bimodal distributions, although the apparent phylogenetic distances of lineage 1 may not represent the phylogenetic distances of the actual lineage, owing to the small sample size (Fig. 3B and 3C). In contrast, the histogram of lineage 3 showed a unimodal pattern (Fig. 3D). The mean distance (± SD) between each pair of F gene sequence in the 66 HRV1 strains examined in this study was 0.018575 ± 0.01227. The results of the study showed that lineages 1, 2 and 3 had phylogenetic distances of 0.0022 ± 0.0012, 0.0073 ± 0.0030, and 0.0092 ± 0.0058, respectively. Thus, the phylogenetic distance for each of the lineage was less than 0.02, suggesting conservation of the F gene sequence.
Fig. 3.
Distribution of phylogenetic distances between the full-length sequences of the fusion gene of all Human respirovirus 1 (HRV1) strains. Each panel illustrates the histogram of all 66 strains (a), lineage 1 (b), lineage 2 (c), and lineage 3 (d). The y-axis and x-axis indicate the number of sequence pairs and phylogenetic distances, respectively.
3.4. Homology modelling
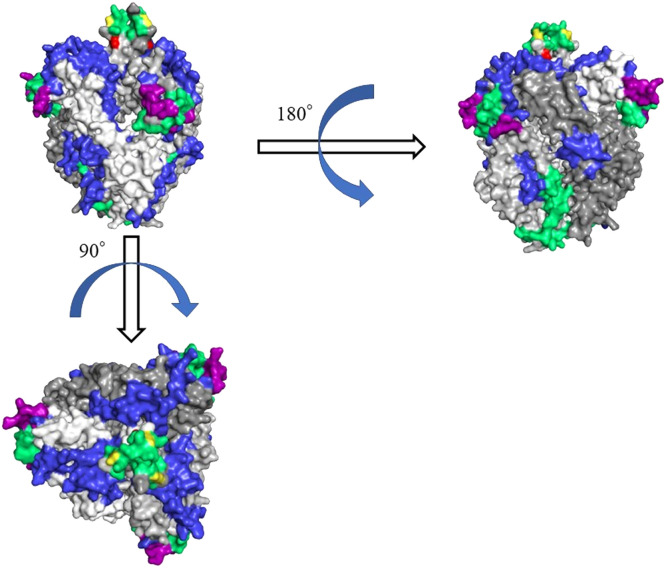
To visualise the relationship between NT-Ab binding sites and predicted B-cell conformational epitopes, we constructed the homotrimer (chains A, B, and C) of the HRV1 pre-fusion protein structure (Fig. 4). The amino acid sequence of the template covered amino acids 24–98 and 126–550 in each strain (Fig. 5). In this range, the amino acid residues on the protein surface were the same between representative strains of lineages 1–3. Moreover, from homologous analysis using Clustal Omega, the percentage sequence identity value of the prototype strain against the representative strains was high (96.8%). Hence, we presented the prototype structural model alone and showed the sites where amino acid substitutions occurred in representative strains from lineages 1 to 3 (Figs. 4 and 5). Both the prototype and lineage 1 representative strains had the same sequence identity (44.3%) as the template.
Fig. 4.
Structural models of the prefusion protein of USA/1957 strain. Chains of the trimeric structures are given in white (chain A), light grey (chain B), and dark grey (chain C). The conformational and linear epitopes have been indicated in green and blue, respectively. Areas where the conformational and linear epitopes overlap are shown in purple. Experimentally identified epitopes (neutralising antibody binding sites) are indicated in red, and those overlapping with the conformational epitope are coloured yellow.
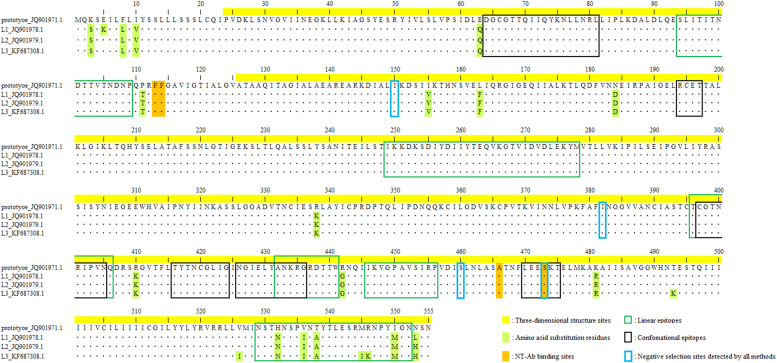
Fig. 5.
The Human respirovirus 1 (HRV1) fusion (F) protein chain A amino acid sequences investigated in this study. The amino acid residue numbers used for constructing HRV1 F protein three-dimensional (3D) structure are highlighted in yellow. The amino acid residues of neutralising antibodies (NT-Ab) binding sites and mutation are highlighted in orange and light green, respectively. The black and green line boxes in the sequences are the conformational epitope and linear epitope sites, respectively. Blue line boxes in the sequence indicate negative selection sites detected by all four methods (single-likelihood ancestor counting, fixed effects likelihood, internal fixed-effects likelihood, and fast unconstrained Bayesian approximation).
3.5. Selective pressure analyses of HRV1 F protein
The rates of dS and dN substitutions were estimated using the DataMonkey web server to identify the positive and negative selection sites of F proteins in all 66 strains (Table S7). Only one method (FUBAR) predicted a positive selection site (amino acid residue 5), and the other four methods identified no positive selection sites. Thus, a positive selection site of the F protein is absent. In contrast, many negative selection sites were identified (Table S7). Among them, four negative selection sites (amino acids 150, 382, 460, and 473) were detected using all methods employed (Fig. 5).
3.6. Analyses of B-Cell epitopes and amino acid substitution sites
The amino acid substitution sites, NT-Ab binding sites, and predicted epitopes are shown in the HRV1 F chain A amino acid sequences and the trimeric structural model of the prototype (Figs. 4 and 5). First, the amino acid substitution sites in the F protein chain A among the prototype strain and the representative strains of lineages 1, 2, and 3 were compared. Seventeen amino acid substitutions were common in lineages 1, 2, and 3 (Fig. 5 and Table S8). An amino acid substitution unique to lineage 1 is present in Glu5Lys. Moreover, four amino acid residues (Thr493Lys, Val526Thr, Met545Ile, and Arg546Lys) showed substitutions that were unique to lineage 3. However, some of the common substitution regions of these amino acid substitution sites are not located on the surface of the 3D structure. Similarly, uncommon substitution residues between lineages 1 and 3 were not present in the 3D structural model. Thus, only seven residues (Glu63Gln, Ile155Val, Leu163Phe, Asn184Asp, Arg338Lys, Arg410Lys, and Arg442Gly) of the common substitution regions in each lineage were located on the surface of the 3D structure model.
Next, conformational and linear B-cell epitopes on HRV1 F protein were analysed. Six conformational epitope sites and seven linear epitope sites were identified for the prototype HRV1 F protein chain A (18 conformational epitopes and 21 linear epitopes in the trimeric structure) (Figs. 4 and 5 and Table S9). No amino acid substitutions were found at these sites of strains in lineages 1, 2, or 3, whereas only one residue substitution (Glu63Gln) was found near one of the predicted conformational epitope sites (Fig. 5). Notably, in the HRV1 F protein chain A, five of the six conformational epitope sites and all the linear epitope sites failed to coincide with the experimentally determined NT-Ab binding sites, whereas only one residue of the conformational epitope (aa 473) coincided (Fig. 5). This mismatch may be a possible mechanism by which HRV1 can reinfect humans.
4. Discussion
Evolutionary studies of HRV have been reported (Bose et al., 2019; Mao et al., 2012; Mizuta et al., 2014), but most of these focus on HRV3 (Mao et al., 2012; Mizuta et al., 2014). A few reports regarding the HRV1 F gene have been published involving domestic or partial F gene analyses (Ambrose et al., 1995; Aso et al., 2020). To study the detailed molecular evolution of the full-length F gene in HRV1 strains from various countries, we performed time-scaled phylogenetic, genome population size, and selective pressure analyses on the gene, as well as antigenicity analysis of the F protein. From the time-scaled phylogenetic tree, constructed using the BMCMC method, it was estimated that the common ancestor of the HRV1 F gene diverged in 1957 and that their progenies continuously evolved and formed three lineages (lineages 1–3, Fig. 1). Strains belonging to lineage 3 predominate in various countries. Second, phylodynamic analyses using the BSP method showed that the genome population size of the F gene doubled over approximately 80 years. Third, phylogenetic distances among the strains were short (< 0.02; Fig. 3). Finally, no positive selection sites were detected in the F protein, whereas many sites were identified as negative selection sites. Moreover, five sites of the six conformational epitopes and all linear epitopes in each chain of the F protein lacked correspondence to the NT-Ab binding sites (Figs. 4 and 5). These results suggest that despite the apparent conservation, the HRV1 F gene has evolved over many years. Yet, the conformational and linear epitopes did not correspond to the NT-Ab binding sites in either the pre or postfusion forms of the protein. This mismatch may be partially responsible for HRV1 reinfection and may extend to related viruses, such as HRV3 and RSV.
A phylogenetic analysis of HRV1 F gene was performed using the BMCMC method. It revealed that this gene continuously evolved and formed three lineages with many clusters (Fig. 1). Our previous report showed full-length F protein genes in HRV1 among patients with acute respiratory infections in Eastern Japan during 2011–2015 (Tsutsui et al., 2017). Phylogenetic analyses using the ML method showed that HRV1 strains formed three lineages and that the lineage 3 strains were dominant during the investigation period (2011–2015). This finding is consistent with the results of the present study. However, the time-scaled phylogeny of the HRV1 F gene was not assessed in the previous study (Tsutsui et al., 2017). Here, time-scaled phylogenetic analyses were done using the BMCMC method. As a result, the divergence year of a common ancestor and each lineage was estimated (Fig. 1). Although the analysed gene was distinct, our previous report suggested that the HRV1 haemagglutinin-neuraminidase (HN) glycoprotein gene (full-length) isolated from Yamagata prefecture, in northern Japan, was classified into two lineages and formed many clusters using different phylogenetic analysis methods, including neighbour-joining and ML methods (Mizuta et al., 2011, 2014). Thus, to the best of our knowledge, the present study may be the first time-scaled phylogenetic analysis of the HRV1 F gene based on full-length sequences from globally collected strains. However, the present study has some limitations, including, the relatively small number of strains used. This is due to the paucity of studies on HRV1 molecular epidemiology. Another limitation is selection bias owing to the limited number of countries studying HRV1 and HRV3 (Aso et al., 2020).
The evolutionary rate of the HRV1 F gene has also been estimated. The mean evolutionary rate was 8.504 × 10−4 s/s/y. This is similar to the rates reported for the F genes of HRV3 and RSV (Aso et al., 2020). Furthermore, the evolutionary rate of HRV1 strains belonging to lineage 3 was found to be faster than that of lineage 2, whereas a previous evolutionary study on the HRV3 F gene did not find this difference (Aso et al., 2020). Moreover, these findings were not found for the RSV F gene, a virus belonging to a different genus and species (Saito et al., 2021). Thus, these findings were only observed for HRV1 F gene, which to the best of our knowledge, is the first report of lineage differences in evolutionary rate. In addition, the rapid evolutionary rate reflects short generation times and/or strong positive selection, which may generate a phenotype that is more adapted to the host (Collins and Karron, 2013). Together, the strains belonging to lineage 3 were more adaptive to humans and could become dominant strains, although our study did not address the mechanisms underlying the difference in the evolutionary rate.
The mean phylogenetic distance of the F gene HRV1 strains was approximately 0.02 (Fig. 3). This agrees with our previous study of Japanese strains, which reported a phylogenetic distance for the HRV1 F gene of 0.026 (Tsutsui et al., 2017). Moreover, the distribution of distances in our study was similar to that in previous reports on the HRV3 F gene (Aso et al., 2020) as well as the HRV3 HN gene (Mao et al., 2012; Takahashi et al., 2018). These data suggest that the diversity of various viruses carrying the F gene may be similar and restricted within each species. With its high conservation and pivotal role in entry, the HRV1 F protein can be an attractive target for prophylaxis therapy, as it does for RSV F protein (Battles and McLellan, 2019).
Phylodynamic analyses of HRV1 F gene (Fig. 2) showed that the genome population of the gene has doubled over approximately 80 years. These fluctuations corresponded to the emergence of strains belonging to lineages 2 and 3 (Figs. 1 and 2). Our previous report showed that the genome population size of HRV3 F gene increased only once between 2000 and 2010 (Aso et al., 2020). Thus, genome fluctuations between HRV1 and 3 were different.
The selective pressure analyses of the HRV1 F gene did not reveal any positive selection sites for the present strains, whereas many negative selection sites were identified. In general, positive selection sites reflect escape from host defence mechanisms, such as cellular or humoral immunity (Barreiro and Quintana-Murci, 2010). Thus, HRV1 F protein may not be affected by such defence mechanisms. Conversely, negative selection sites may act to prevent deterioration of antigenicity (Holmes, 2013; Loewe, 2008). These findings may reflect the essential role the HRV1 F protein plays in host cell infection. Similar findings have been reported for the HRV3 F protein (Henrickson, 2003; Tsutsui et al., 2017). The negative selection sites of the HRV1 F protein were clustered near predicted epitopes, based on the 3D structural modelling. This indicates that these sites play important roles, for example, in the cellular receptor binding domain.
Finally, we analysed the amino acid substitutions and conformational and linear epitopes in the HRV1 F protein and evaluated their relationship with the NT-Ab binding sites (Figs. 4 and 5). Notably, amino acid substitutions were not estimated for computationally predicted epitopes or NT-Ab binding sites. Furthermore, almost all experimentally determined NT-Ab binding sites were incompatible with the computationally estimated conformational epitopes and linear epitopes. Antibody responses play a pivotal role in virus neutralisation or elimination, and antigenicity and immunogenicity differ explicitly (Collins and Karron, 2013). Conformational and linear epitopes may stimulate the production of antiviral antibodies (Lo et al., 2021; Sharon et al., 2014). In contrast, predicted epitope sites on antigenic proteins may not be adequately recognised by NT-Abs, which might mean that these predicted epitopes have weak potential for producing NT-Abs (Collins and Karron, 2013; Lo et al., 2021; Sharon et al., 2014). Thus, incompatibilities between NT-Ab binding sites and predicted epitopes may indicate low immunogenicity of the F protein and may be partially responsible for HRV1 reinfection, as has been reported for reinfections with HRV3 and RSV (Aso et al., 2020; Saito et al., 2021). However, the paucity of antigenic/antibody complex structures may lead to a mismatch between the NT-Ab binding sites and the predicted epitopes. Hence, the interpretation of the computationally conducted epitope analyses in this study may be limited, although conformational and linear epitopes were investigated using multiple computational methods for increased accuracy.
5. Conclusions
In this study, molecular evolutionary analyses of HRV1 F gene were performed based on full-length sequences collected globally. The time-scaled phylogenetic tree generated by the BMCMC method estimated that the common ancestor of the HRV1 F gene diverged in 1957, and that their progenies have continuously evolved and formed three lineages. The phylodynamic analyses using the BSP method showed that the genome population size of the F gene doubled over approximately 80 years. The phylogenetic distances among the strains were short with no positive selection sites. Almost all conformational and linear epitopes in the F protein did not correspond to NT-Ab binding sites. These results showed that the HRV1 F gene has evolved over many years, although the gene may be relatively conserved. Moreover, incompatibility between predicted epitopes and the NT-Ab binding sites in both the pre and postfusion forms of the protein may be responsible for HRV1 virus reinfection, as well as reinfections with HRV3 and RSV.
Funding
This work was supported by a commissioned project for Research on Emerging and Reemerging Infectious Diseases from the Japan Agency for Medical Research and Development (AMED; grant number JP23fk0108661).
Data availability statement
The data sets generated and analysed during the current study are available from the corresponding author upon reasonable request.
CRediT authorship contribution statement
Tomoko Takahashi: Methodology, Data curation, Writing – original draft. Mao Akagawa: Methodology, Visualization. Ryusuke Kimura: Methodology, Visualization. Mitsuru Sada: Methodology, Writing – original draft, Visualization. Tatsuya Shirai: Writing – original draft. Kaori Okayama: Writing – original draft. Yuriko Hayashi: Writing – original draft. Mayumi Kondo: Methodology. Makoto Takeda: Writing – review & editing. Akihide Ryo: Writing – review & editing. Hirokazu Kimura: Conceptualization, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Ms. Miki Kawaji for the skilful support in figure preparation.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.virusres.2023.199142.
Appendix. Supplementary materials
References
- Ambrose M., Hetherington S., Watson A., Scroggs R., Portner A. Molecular evolution of the F glycoprotein of human parainfluenza virus type 1. J. Infect. Diseases. 1995;171(4):851–856. doi: 10.1093/infdis/171.4.851. [DOI] [PubMed] [Google Scholar]
- Aso J., Kimura H., Ishii H., Saraya T., Kurai D., Matsushima Y., Nagasawa K., Ryo A., Takizawa H. Molecular evolution of the fusion protein (F) gene in human respirovirus 3. Front. Microbiol. 2020;10:3054. doi: 10.3389/fmicb.2019.03054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreiro L.B., Quintana-Murci L. From evolutionary genetics to human immunology: how selection shapes host defence genes. Nat. Rev. Genet. 2010;11(1):17–30. doi: 10.1038/nrg2698. [DOI] [PubMed] [Google Scholar]
- Battles M.B., McLellan J.S. Respiratory syncytial virus entry and how to block it. Nat. Rev. Microbiol. 2019;17:233–245. doi: 10.1038/s41579-019-0149-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose M.E., Shrivastava S., He J., Nelson M.I., Bera J., Fedorova N., Halpin R., Town C.D., Lorenzi H.A., Amedeo P., Gupta N., Noyola D.E., Videla C., Kok T., Buys A., Venter M., Vabret A., Cordey S., Henrickson K.J. Sequencing and analysis of globally obtained human parainfluenza viruses 1 and 3 genomes. PLoS. ONE. 2019;14(7) doi: 10.1371/journal.pone.0220057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert R., Heled J., Kühnert D., Vaughan T., Wu C.-H., Xie D., Suchard M.A., Rambaut A., Drummond A.J. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS. Comput. Biol. 2014;10(4) doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins P.L., Karron R.A. In: Fields Virology. 6th ed. Knipe D.M., Howley P., editors. Lippincott Williams & Wilkins; Philadelphia: 2013. Respiratory Syncytial Virus and Metapneumovirus; pp. 1086–1123. [Google Scholar]
- Darriba D., Taboada G.L., Doallo R., Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods. 2012;9(8) doi: 10.1038/nmeth.2109. 772-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny F.W., Murphy T.F., Clyde W.A., Jr, Collier A.M., Henderson F.W., Senior R., Sheaffer C., Conley W., III, Christian R. Croup: an 11-year study in a pediatric practice. Pediatrics. 1983;71(6):871–876. [PubMed] [Google Scholar]
- Fourment M., Gibbs M.J. PATRISTIC: a program for calculating patristic distances and graphically comparing the components of genetic change. BMC. Evol. Biol. 2006;6(1):1–5. doi: 10.1186/1471-2148-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galgonek J., Vymetal J., Jakubec D., Vondrášek J. Amino Acid Interaction (INTAA) web server. Nucl. Acids. Res. 2017;45(W1):W388–W392. doi: 10.1093/nar/gkx352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-Pdb Viewer: an environment for comparative protein modeling. Electrophoresis. 1997;18(15):2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Henrickson K.J. Parainfluenza viruses. Clin. Microbiol. Rev. 2003;16(2):242–264. doi: 10.1128/CMR.16.2.242-264.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E.C. In: Fields Virology. 6th ed. Knipe D.M., Howley P., editors. Lippincott Williams & Wilkins; Philadelphia: 2013. Virus evolution; pp. 286–313. [Google Scholar]
- Kanda Y. Investigation of the freely available easy-to-use software 'EZR' for medical statistics. Bone Marrow Transpl. 2013;48(3):452–458. doi: 10.1038/bmt.2012.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karron R.A. Parainfluenza viruses. Fields Virol. 2007:1497–1526. [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kringelum J.V., Lundegaard C., Lund O., Nielsen M. Reliable B cell epitope predictions: impacts of method development and improved benchmarking. PLoS. Comput. Biol. 2012;8(12) doi: 10.1371/journal.pcbi.1002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Y.-T., Shih T.-C., Pai T.-W., Ho L.-P., Wu J.-L., Chou H.-Y. Conformational epitope matching and prediction based on protein surface spiral features. BMC. Genomics. 2021;22(Suppl_2) doi: 10.1186/s12864-020-07303-5. 116-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewe L. Negative Selection. Nat. Educ. 2008;1(1):59. [Google Scholar]
- Mao N., Ji Y., Xie Z., Wang H., Wang H., An J., Zhang X., Zhang Y., Zhu Z., Cui A., Xu S., Shen K., Liu C., Yang W., Xu W. Human parainfluenza virus-associated respiratory tract infection among children and genetic analysis of HPIV-3 strains in Beijing, China. PLoS. ONE. 2012;7(8):e43893. doi: 10.1371/journal.pone.0043893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuta K., Saitoh M., Kobayashi M., Tsukagoshi H., Aoki Y., Ikeda T., Abiko C., Katsushima N., Itagaki T., Noda M., Kozawa K., Ahiko T., Kimura H. Detailed genetic analysis of hemagglutinin-neuraminidase glycoprotein gene in human parainfluenza virus type 1 isolates from patients with acute respiratory infection between 2002 and 2009 in Yamagata prefecture, Japan. Virol. J. 2011;8 doi: 10.1186/1743-422X-8-533. 533-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuta K., Tsukagoshi H., Ikeda T., Aoki Y., Abiko C., Itagaki T., Nagano M., Noda M., Kimura H. Molecular evolution of the haemagglutinin-neuraminidase gene in human parainfluenza virus type 3 isolates from children with acute respiratory illness in Yamagata prefecture. Japan. J. Med. Microbiol. 2014;63(Pt 4):570–577. doi: 10.1099/jmm.0.068189-0. [DOI] [PubMed] [Google Scholar]
- Murrell B., Moola S., Mabona A., Weighill T., Sheward D., Kosakovsky Pond S.L., Scheffler K. FUBAR: a fast, unconstrained bayesian approximation for inferring selection. Mol. Biol. Evol. 2013;30(5):1196–1205. doi: 10.1093/molbev/mst030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell B., Wertheim J.O., Moola S., Weighill T., Scheffler K., Kosakovsky Pond S.L. Detecting individual sites subject to episodic diversifying selection. PLoS. Genet. 2012;8(7) doi: 10.1371/journal.pgen.1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarenko J., Bui H.-H., Li W., Fusseder N., Bourne P.E., Sette A., Peters B. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC. Bioinformatics. 2008;9(1):1–8. doi: 10.1186/1471-2105-9-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Lam T.T., Max Carvalho L., Pybus O.G. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen) Virus. Evol. 2016;2(1) doi: 10.1093/ve/vew007. vew007-vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ras Carmona A., Pelaez Prestel H.F., Lafuente E.M., Reche P.A. BCEPS: a Web Server to Predict Linear B Cell Epitopes with Enhanced Immunogenicity and Cross-Reactivity. Cells. 2021;10(10) doi: 10.3390/cells10102744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S., Raghava G.P.S. Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins. 2006;65(1):40–48. doi: 10.1002/prot.21078. [DOI] [PubMed] [Google Scholar]
- Saito M., Tsukagoshi H., Sada M., Sunagawa S., Shirai T., Okayama K., Sugai T., Tsugawa T., Hayashi Y., Ryo A. Detailed evolutionary analyses of the F gene in the respiratory syncytial virus subgroup A. Viruses. 2021;13(12):2525. doi: 10.3390/v13122525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao N., Liu B., Xiao Y., Wang X., Ren L., Dong J., Sun L., Zhu Y., Zhang T., Yang F. Genetic characteristics of human parainfluenza virus types 1–4 from patients with clinical respiratory tract infection in China. Front. Microbiol. 2021:12. doi: 10.3389/fmicb.2021.679246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon J., Rynkiewicz M.J., Lu Z., Yang C.Y. Discovery of protective B-cell epitopes for development of antimicrobial vaccines and antibody therapeutics. Immunology. 2014;142(1):1–23. doi: 10.1111/imm.12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shashkova T.I., Umerenkov D., Salnikov M., Strashnov P.V., Konstantinova A.V., Lebed I., Shcherbinin D.N., Asatryan M.N., Kardymon O.L., Ivanisenko N.V. SEMA: antigen B-cell conformational epitope prediction using deep transfer learning. bioRxiv. 2022 doi: 10.3389/fimmu.2022.960985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers, F., Higgins, D.G., 2021. The Clustal Omega Multiple Alignment Package. Vol. 2231. Humana Press, pp. 3–16. [DOI] [PubMed]
- Singh H., Ansari H.R., Raghava G.P.S. Improved method for linear B-cell epitope prediction using antigen's primary sequence. PLoS. ONE. 2013;8(5) doi: 10.1371/journal.pone.0062216. e62216-e62216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M., Nagasawa K., Saito K., Maisawa S.-i., Fujita K., Murakami K., Kuroda M., Ryo A., Kimura H. Detailed genetic analyses of the HN gene in human respirovirus 3 detected in children with acute respiratory illness in the Iwate Prefecture. Japan. Infection,. Genetics. and. Evolution. 2018;59:155–162. doi: 10.1016/j.meegid.2018.01.021. [DOI] [PubMed] [Google Scholar]
- Tsutsui R., Tsukagoshi H., Nagasawa K., Takahashi M., Matsushima Y., Ryo A., Kuroda M., Takami H., Kimura H. Genetic analyses of the fusion protein genes in human parainfluenza virus types 1 and 3 among patients with acute respiratory infections in Eastern Japan from 2011 to 2015. J. Med. Microbiol. 2017;66(2):160–168. doi: 10.1099/jmm.0.000431. [DOI] [PubMed] [Google Scholar]
- Van Regenmortel M.H. Antigenicity and immunogenicity of synthetic peptides. Biologicals. 2001;29:209–213. doi: 10.1006/biol.2001.0308. [DOI] [PubMed] [Google Scholar]
- Weaver S., Shank S.D., Spielman S.J., Li M., Muse S.V., Kosakovsky Pond S.L. Datamonkey 2.0: a modern web application for characterizing selective and other evolutionary processes. Mol. Biol. Evol. 2018;35(3):773–777. doi: 10.1093/molbev/msx335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WL D. DeLano Scientific; San Carlos, CA: 2002. The PyMOL Molecular Graphics System. [Google Scholar]
- Yin H.-S., Wen X., Paterson R.G., Lamb R.A., Jardetzky T.S. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature. 2006;439(7072):38–44. doi: 10.1038/nature04322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Chen Z., Zhang L., Yan D., Mao T., Tang K., Qiu T., Cao Z. SEPPA 3.0—enhanced spatial epitope prediction enabling glycoprotein antigens. Nucleic. Acids. Res. 2019;47(W1):W388–W394. doi: 10.1093/nar/gkz413. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets generated and analysed during the current study are available from the corresponding author upon reasonable request.