Abstract
Background:
N7-methylguanosine (m7G) modification plays a crucial role in the development and progression of lung cancers. MicroRNAs (miRNAs) are closely involved in programmed cell death and the mechanism of tumor growth. The m7G-associated miRNAs genes in lung adenocarcinoma (LUAD), and their prognosis prediction ability of LUAD, however, had not been investigated.
Methods:
The RNA transcriptomes, clinical indices, and immune scores of LUAD patients were searched and downloaded from The Cancer Genome Atlas (TCGA) and the ESTIMATE database. The miRNAs targeting METTL1 and WDR4 were extracted from the TargetScan database. Differentially expressed m7G-related miRNAs were identified and their prediction power of LUAD prognosis was systematically investigated.
Results:
Among 40 the differentially expressed m7G-related miRNAs in LUAD, five (hsa-miR-31-5p, hsa-miR-5571-3p, hsa-miR-4697-3p, hsa-miR-6858-5p, and hsa-miR-873-3p) demonstrate significant predictive value for prognosis. The risk score constructed by these five miRNAs was an independent prognostic factor (univariate Cox regression results: hazard ratio (HR) = 1.6619, 95% confidential interval (CI) = 1.2103-2.2819, p = 0.0017; multivariate Cox regression results: HR = 1.6004, 95% CI = 1.1633-2.2017, p = 0.0039). The survival curves showed that patients with high-risk scores had a poor prognosis. Calibration curves indicated good predictability in a nomogram constructed combining the miRNA and the clinical indices of age, sex, chemotherapy, radiotherapy, stage, and risk score. GO and KEGG analysis of the overlapping genes showed that the prognostic miRNAs were closely associated with the neuropeptide signaling pathway. Besides, the immune infiltration analysis showed that the expression of the AMPD1 gene was strongly associated with immune cells and immunology functions in LUAD.
Conclusion:
This study identified DE m7G-related miRNAs and demonstrated their prediction ability in the prognosis of LUAD patients. The risk signature based on these miRNAs demonstrates high accuracy in predicting the prognosis of LUAD patients.
Key Words: m7G, miRNA, nomogram, prognosis, immune
Introduction
Lung adenocarcinoma (LUAD), the most common subtype of non-small cell lung cancer (NSCLC) Sung et al., (2021), has the highest incidence and mortality rate in many countries. Therefore, it is an urgent desire to establish a prognostic panel with accurately stratifying risk and high specificity and sensitivity that could assist the selection of treatment strategy, promote the survival rate, and improve the long-term prognosis of LUAD patients.
N7-methylguanosine (m7G) modifies the 5’ caps of tRNAs and rRNAs under the catalyzing of the Methyltransferase complex composed of the Methyltransferase-like 1 (METTL1) and the WD repeat domain 4 (WDR4) in the tRNA variable loop in humans (Alexandrov et al., 2005; Boulias et al., 2019; Lin et al., 2018; Pandolfini et al., 2019). Ma et al., (2021) found in human lung cancer samples that the gene expressions of METTL1 and WDR4 were significantly upregulated and were associated with poor patient prognosis. Chen et al., (2022) study demonstrated the mechanism of the methyltransferase complex of METTL1/WDR4 in promoting the disease progression of HNSCC by impacting the immune microenvironment and regulating the PI3K/AKT/mTOR signaling pathway. Zhao et al., (2021) research shows that METTL1 could promote post-ischemic angiogenesis by increasing the protein production of VEGFA. All these studies indicate that the METTL1/WDR4 complex might also be associated with the mechanism and progression of LUAD.
MicroRNAs (miRNAs) are a category of non-coding RNAs that are universally involved in cell proliferation, differentiation, and apoptosis in tumors (Baltimore et al., 2008; Karrich et al., 2013), their abnormal gene expression has been detected in types of human cancers(El Bezawy et al., 2017; He et al., 2017; Hummel et al., 2014; Wang et al., 2018; Xu et al., 2017). Although miRNAs lack protein-coding capacity, recent studies have revealed their ability in regulating the expression of tumor suppressor genes and oncogenes via binding to tumor-related genomic regions or sensitive areas of the genome (Wu et al., 2021). Previous studies have identified several miRNAs as potential prognostic indicators for LUAD, such as mir-582 and mir-584, the expressions of which were associated with poor prognosis in patients (Siriwardhana et al., 2019). Although miRNAs, as a whole, their associations with the LUAD have been well studied, the impact of m7G-related miRNAs, a subtype of them, had scarcely been studied on LUAD.
The tumor microenvironment (TME) is a blend of tumor cells, immune cells, and other endogenous molecules impacting tumor development such as inflammatory mediators (Hanahan et al., 2012). In the TME, TIICs are critical players in tumor progression and immunotherapeutic response (Binnewies et al., 2018). Close association with the prognosis of lung cancer has been observed in many types of immune cells, such as M1/M2 macrophages, CD8+/CD4+ T cells, mature and immature dendritic cells (DC), and the regulatory T cells (Tregs) (Catacchio et al., 2018). Even some indices of systemic inflammation, such as the high neutrophil to lymphocyte ratio, have also been reported to be associated with the overall survival (OS) and progression free survival (PFS) in cancer patients (Gu et al., 2015). This suggests that the study of the TME, especially immune cells and immunology functions, is critical for the development of therapeutical treatments for cancer patients.
To systematically evaluate the prognostic value of m7G-related miRNAs in LUAD, we designed this study. First, relevant data were extracted from the TCGA, TargetScan, and ESTIMATE databases. Second, through a series of analyses, the DE miRNAs associated with LUAD prognosis were identified and a risk score model was constructed. Third, a prognostic signature for individualized assessment of patient prognosis was established in the combination of the differentially expressed (DE) m7G-related miRNAs and clinical indices, which showed improved prediction accuracy. Finally, DE mRNAs of the LUAD patients were calculated after the grouping of the patients according to their immune scores and risk scores, and their associated biological pathways and immune cell infiltration were further analyzed. The flow chart of the present study is illustrated in Figure 1.
Figure 1.
The Flow Chart of the Overall Study Design
Materials and Methods
Data Collection
The miRNA and mRNA expression data and clinical characteristics of patients with LUAD were downloaded from The Cancer Genome Atlas (TCGA) database (https://www.cancer.gov/tcga). LUAD patients were included only when their clinical information was complete, including survival time, survival status, age, sex, chemotherapy, radiotherapy, and the stages of cancer. The upstream miRNAs of m7G-related genes were extracted from the TargetScan database (http://www.targetscan.org/). Next, the immune scores of the LUAD patients were obtained from the ESTIMATE database (https://bioinformatics.mdanderson.org /estimate/).
Differential expression analysis of m7G associated miRNAs
The differentially expressed (DE) m7G-related miRNAs between LUAD and matched healthy tissues were calculated using the R packages of “limma” and “edgeR”. Heatmaps were drawn using the R package “heatmap”. A statistically significant differential expression was determined when the false discovery rate (FDR) < 0.05 and |log2fold changes (FC)| > 1.
Calculation of risk scores of DE miRNAs and evaluation of their association with prognosis
The relationship between m7G-related DE miRNA and the overall survival (OS) of the patients was investigated using the TCGA-LUAD cohort. Univariate and multivariate Cox regression analyses were performed using the R package “survival” and “forestplot, with a p-value <0.05 as the cutoff criterium. Kaplan-Meier curves were utilized for survival analyses, with log-rank tests and univariate Cox proportional hazard regression models used to generate p-values as well as hazard ratios (HRs) with 95% confidence intervals (CIs). The Lasso regression analysis was conducted using the R package “glmnet”. The DE miRNAs with prognostic relevance resulted in these analyses were selected for further assessment. First, a risk score was defined based on the expression of these prognostic miRNAs for each patient following centralization and standardization with the R “scale” function. The risk scores were calculated using the following formula:
risk score =∑5iXi×Yi
where X and Y are the coefficients and miRNA expression levels, respectively. Second, we then validated the accuracy of the risk scores using principal component analysis (PCA). Patients from the TCGA-LUAD cohort were divided into low- and high-risk groups by the median risk score, and their Oss were compared using the Kaplan-Meier approach. Their 1, 3, and 5-year OS were compared in ROC curves generated by the R packages “survival”, “ROCR”, and “timeROC”. Besides, dot and line plots were used to verify the accuracy of the prognostic model. The prognostic relevance of m7G-associated miRNAs was further assessed by Cox regression analyses. K-M survival analyses were conducted and miRNAs with high prognostic potential were chosen for next stage evaluation.
Construction and validation of a nomogram
Demographics and clinical data, including patients’ age, sex, chemotherapy, radiotherapy, cancer stage, and risk score, were extracted from the TCGA database. Univariate and multivariate Cox regression models were used to analyze the prognostic value of these variables. Then, a prognostic nomogram was created by Cox regression analysis and was subsequently validated by the calibration plots drawn by the R package “rms”.
Calculation of the DE mRNAs associated with immune score and risk score
The immune scores of LUAD patients were obtained from the ESTIMATION database. Based on the risk scores and immune scores of the patients, their mRNA transcriptome data downloaded from the TCGA database were divided into high-risk and low-risk groups. DE genes between the two groups were calculated using the R package “limma” and “edgeR”. A statistically significant differential expression was determined when the false discovery rate (FDR) < 0.05 and |log2fold changes (FC)| > 1.
Functional enrichment analysis of the overlapping genes
The overlapping genes of two groups of DE genes obtained by risk score and immune score were identified and plotted by the R package “VennDiagram”. Then, gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were conducted using the R language.
Functional analysis of the immune correlation
DE genes associated with immune score were analyzed by the ssGESA algorithm, and the correlation coefficients between the immune cells (imc) and the immunology function (imf) scores were calculated. We then analyzed the immune cells and immunology functions in the high- and low- risk groups. Univariate regression analyses were performed for the intersecting genes of statistical significance and the correlations coefficients were calculated among the immune genes, immune cells, and immunology functions.
Results
Differentially expressed m7G-related miRNAs in LUAD
As a result, we identified a total of 443 LUAD patients who meet our criteria in the TCGA database, the clinical characteristics of which were shown in Table 1. Based on the data sets we collected from the TCGA database and the TargetScan databases, we analyzed the expression of m7G-miRNA-associated miRNAs in lung adenocarcinoma. The gene expression differences of miRNAs between LUAD patients and healthy subjects were shown in the volcano plot and heatmap (Figure 2). As a result, there are a total of 40 DE miRNAs, including 34 upregulated and 6 downregulated ones (Figure 2a), and their differential expressions in each sample are shown in the heat map (Figure 2b).
Table 1.
Summary of Basic Clinical Features
| Covariates | Group | Patient number (%) | p-value1 | p-value2 |
|---|---|---|---|---|
| Age | ≤65 | 216 (48.8%) | 0.654 | 0.326 |
| ˃65 | 227 (51.2%) | |||
| Sex | Male | 200 (45.1%) | 0.845 | 0.769 |
| Female | 243 (54.9%) | |||
| Chemotherapy | Yes | 181 (40.9%) | 0.045 | 0.714 |
| No | 262 (59.1%) | |||
| Radiation | Yes | 105 (23.7%) | 1.76E-05 | 0.009 |
| No | 338 (76.3%) | |||
| Stage | Stage I-II | 3539 (79.7%) | 2.14E-07 | 1.01E-05 |
| Stage III-IV | 90 (20.3%) |
1 p-value of univariate Cox regression; 2 p-value of multivariate Cox regression
Figure 2.
Expression of M7G-related DE miRNAs. (a) Volcano plot of the DE miRNAs. Red dots represent upregulated m7G-associated miRNAs, green dots represent downregulated ones, and black dots represent miRNAs with no significant differential expression. (b)The heatmap of top 20 DE miRNAs between the LUAD group and the healthy group
Construction of A multi-miRNA Prediction Model for the Prognostic of LUAD
The prognostic value of these 40 DE miRNAs in LUAD was further evaluated by univariate and multifactorial Cox regression analysis. The univariate Cox regression analysis showed that hsa-miR-31-5p (p=0.0002), hsa-miR-5571-3p (p=0.0298), hsa-miR-4697-3p (p=0.0394), hsa-miR-6858-5p (p=0.0299), and hsa-miR-873-3p (p = 0.0001) have a predictive value for LUAD prognosis (Figure 3a). The multifactorial Cox regression analysis showed that hsa-miR-31-5p (p<0.001), hsa-miR-5571-3p (p=0.0027), hsa-miR-4697-3p (p=0.0417), hsa-miR-6858-5p (p=0.0105), and hsa-miR-873-3p (p=0.0026) might be independent prognostic indicators for LUDA (Figure 3b). Using the lasso regression analysis, five miRNAs with the best prognosis ability, including hsa-miR-31-5p, hsa-miR-5571-3p, hsa-miR-4697-3p, hsa-miR-6858-5p, and hsa-miR-873-3p, were identified as prognostic markers according to the minimum criteria and coefficients (Figure 3c-d). The median risk score in the multifactorial Cox regression analysis was used as the cut-off point to classify LUAD cancer patients into low-risk and high-risk groups for further assessments. To validate this grouping, we first performed a principal components analysis (PAC), which showed good discrimination between the high- and low-risk groups (Figure 3e). Then Kaplan-Meier curves were plotted to compare the survival times between the low-risk patients and high-risk patients. The results showed that the average OS time was significantly longer in low-risk patients than that in high-risk patients (p<0.001, Figure 3g). The area under the ROC curve for time-dependent survival at 1, 3, and 5 years was 0.717, 0642, and 0.582 for the two groups of patients, respectively, indicating the good performance of this risk model in predicting the survival of LUAD patients (Figure 3f). We also plotted the distribution of the risk scores and survival status for each patient (Figure 3h-i). It was observed that along with the increase in patients’ risk scores, the number of patient deaths had a discriminable increase.
Figure 3.
Construction and Validation of the Prognosis Prediction Model. (a-b) Univariate and Multivariate analysis of five miRNAs associated with LUAD prognosis. (c-d) The process of lasso regression analysis to select predictive variables. (e) PCA and (f)ROC curve verified the accuracy of the predictive power of the risk score model. (g)Survival curve compares the survival rate of the high-risk group and low-risk group. (h) Survival status for patients. (i) Distribution of patients’ risk scores
Prognostic Analysis and Clinical Correlation of the Independent Risk Factors
The calculated risk scores of the patients, as well as other clinical features collected from the TCGA database data, including age, sex, chemotherapy, radiotherapy, and cancer stage, were analyzed by the univariate Cox analysis (Table 1). Among them, chemotherapy (p = 0.0452; HR = 1.3827), radiotherapy (p < 0.0001; HR = 2.0463), cancer stage (p < 0.0001; HR = 2.4182), and risk score (p = 0.0017; HR = 1.6619) were used as independent risk factors (Figure 4a). The results of the multivariate Cox analysis showed that radiotherapy (p = 0.0022; HR = 1.7399), stage (p < 0.001; HR = 2.2199), and risk score (p = 0.0039; HR = 1.6004) could be independent risk factors affecting the prognosis of LUAD (Figure 4b). The area under the ROC curve for time-dependent survival of risk score, age, sex, chemotherapy, radiotherapy, and stage were 0.718, 0503, 0.578, 0.524, 0.537, and 0.616, respectively, indicating high prediction accuracy of the risk score and cancer stage (Figure 4c). Next, a prediction model was constructed by the clinical features and the risk scores (*: p < 0.05, **: p < 0.01, *** p < 0.001) (Figure 4e), based on which the total points of each patient were calculated, and their 1-, 3-, and 5-year survival rates were predicted. According to this model, a higher point of a patient predicts a worse prognosis. We also used a calibration curve to test the accuracy of the nomogram. As shown in the calibration curves, the predicted1-, 3-, and 5-years survival curves are very close to the observed survival curves, indicating a high prediction accuracy of the nomogram has (Figure 4d).
Figure 4.
Construction and Validation of a Nomogram. (a-b) Univariate and multivariate Cox regression analysis of the clinical features and the risk scores. (c) The ROC curve of the independent risk factors. (d) Prediction accuracy of the nomogram. The observed and predicted 1-, 3-, and 5-year survival were in good agreement. Y-axis: actual survival; X-axis: nomogram-predicted survival. (e) In the upper part, six lines are drawn for each patient, representing the points of the six predictors in the nomogram, the sum of which is annotated on the ‘Points’ axis. At the bottom, three lines are drawn to display the predicted 1-, 3-, and 5-year overall survival of LUAD
Prognostic significance of the miRNAs and associated DE genes
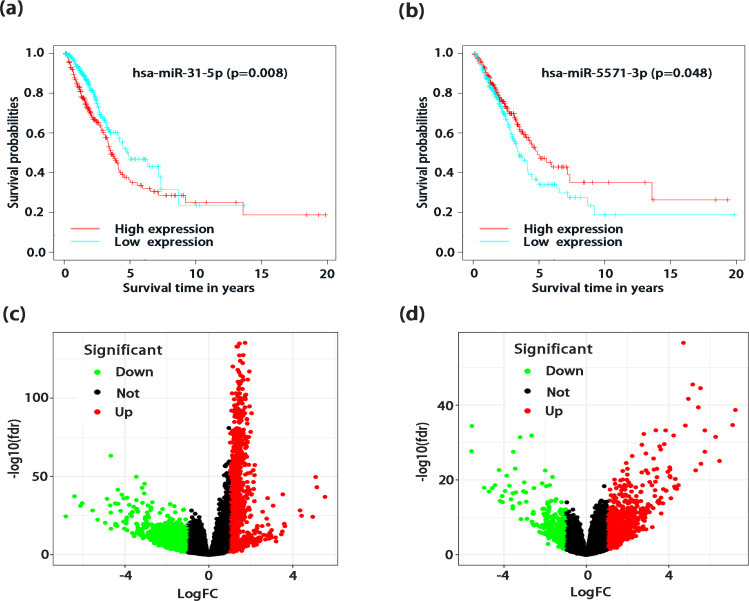
We further screened two miRNAs with survival significance from the five miRNAs used to construct the risk score models and drew their survival curves at p <0.05. As shown in Figure 5, a poor prognosis was observed in the has-miR-315p (p=0.008) (Figure 5a) high expression group and the has-miR-55713p (p=0.048) (Figure 5b) low expression group, suggesting that the high expression of has-miR-315p could promote the occurrence and development of tumor, while the high expression of has-miR-55713p might inhibit the caner exacerbation. Following the grouping of the risk scores, the DE mRNA genes were calculated between the high-risk group and the low-risk group from the TCGA transcriptome data sets. As shown in the volcanic map, there were 1066 DE genes, including 764 up-regulated genes and 302 down-regulated genes (Figure 5c). Similarly, the DE mRNA was also calculated after the patients were grouped by their immune scores obtained from the ESTIMATION database. A total of 1727 DE genes were obtained, of which 864 genes were up-regulated and 863 genes were down-regulated (Figure 5d).
Figure 5.
DE Genes in LUAD Associated with the Grouping by the Risk Score and the Immune Score. (a) Survival curves of differential hsa−miR−31−5p expression. (b)Survival curves of differential hsa-miR-557-3p expression. Volcano plots of the DE genes when patients are grouped by the risk score (c) and the immune score (d). Red dots represent upregulated genes, green dots represent down-regulated genes, and black dots represent genes with nonsignificant differences
Functional Enrichment Analysis of the DE mRNA Genes
To further explore the biological functions associated with these DE mRNA genes, we first obtained 540 DE genes that were associated with the grouping of both the risk score and the immune score, the overlapping of which were displayed in a Venn diagram (Figure 6a). Next, we utilized the GO and KEGG analyses to investigate the related molecular mechanisms of these genes. As a result, in the GO groups of biological processes (BP), cellular components (CC), and molecular function (MF), the “neuropeptide receptor activity”, “antimicrobial humoral response”, and “neuropeptide signaling pathway” were the most significantly terms, respectively (Figure 6b-c). The significantly enriched KEGG pathways were listed in Table 2, which were mainly associated with the pathways of neuroactive ligand-receptor interaction, taste transduction, and protein digestion.
Figure 6.
Enrichment Analysis of the 540 DE Genes. (a), Venn diagram displays the intersection of the two DE gene sets; (b), Bar chart of the GO enrichment analysis; (c), Dot plot of the GO enrichment analysis
Table 2.
KEGG Enrichment Analysis
| miRNA ID | Description | Count | p.adjusted |
|---|---|---|---|
| hsa04080 | Neuroactive ligand-receptor interaction | 32 | 1.57E-10 |
| hsa04742 | Taste transduction | 10 | 0.001002972 |
| hsa04974 | Protein digestion and absorption | 10 | 0.003294531 |
| hsa05033 | Nicotine addiction | 6 | 0.007720087 |
| hsa00140 | Steroid hormone biosynthesis | 7 | 0.009797174 |
| hsa04915 | Estrogen signaling pathway | 10 | 0.017160914 |
| hsa04913 | Ovarian steroidogenesis | 6 | 0.017160914 |
| hsa04950 | Maturity onset diabetes of the young | 4 | 0.046678348 |
| hsa04927 | Cortisol synthesis and secretion | 6 | 0.048272193 |
Immune Infiltration analysis of the DE genes
To further study the immune cells and immune function of associated with these DE genes in LUAD, we drew a heatmap of immune scores calculated using the ssGSEA algorithm (Figure 7a). The heatmap showed that the composition of 16 types of immune cells (imc) and 13 immunology functions (imf) in each sample had significant differential expression. Among the biological cells, T helper cells were highly expressed in each sample, while NK cells were lowly expressed. In immunology functions, HLA, and MHC class I were highly expressed in almost all samples. As shown in Figure 7b, the proportions of differentially infiltrated immune cells were moderately-to-highly-correlated. For instance, the correlation coefficient between the pDCs and the TIL was 0.82 and the correlation coefficient between the T helper cells and the macrophages was 0.8. In the imf correlation matrix, the correlation between T cell co-inhibition and the checkpoint activity reached 0.93 (Figure 7c). Next, we analyzed the differences in immune cells and immunology functions in the high and low risk groups. For the imc, when compared to high-risk groups, low-risk groups had significantly higher aDCs, iDCs, Mast cells (p < 0.001), B cells, Neutrophils (p < 0.01), and TIL (p < 0.05), while had significantly lower infiltration level of NK cells (p < 0.01) (Figure 7d). For the imf, the low-risk group was significantly higher than the high-risk group in HLA (p < 0.001) and type I IFN response (p < 0.01), while significantly lower in MHC class I (p < 0.01) (Figure 7e). Significant DE genes were obtained by univariate Cox regression analysis of all overlapping DE genes (Table 3). Finally, the immune correlation analysis of these genes showed that AMPD1 was positively correlated with most immune cells and immunology functions (Figure 7f), indicating that the role of AMPD1 in LUAD immunity deserves further study. Taken together, these results suggest that differential gene expression plays an important role in the immune regulation of LUAD.
Figure 7.
Analysis of Immune Cells and Immunology Functions Associated with DE Genes. (a) Heatmap of the scores of immune cells and immunology functions. (b-c) Matrix of the correlation coefficients between the immune cells and immunology functions. (d) Immune cell analysis of the high- and low-risk groups. (e) Immunology function analysis of the high- and low-risk groups. (f) Correlation analysis of immune genes with immune cells and immunology functions. The red color represents positive correlations, the blue color represents negative correlations, and the white indicates relationships without a statistical difference
Table 3.
Univariate Cox Regression of Overlapping Genes
| Gene | HR | HR-95Low | HR-95Up | p-value |
|---|---|---|---|---|
| HIST1H2AL | 1.008655118 | 1.004280181 | 1.013049114 | 0.000102011 |
| EBF3 | 1.000362325 | 1.000179036 | 1.000545648 | 0.000106714 |
| HIST1H2AJ | 1.006645254 | 1.003219751 | 1.010082453 | 0.000139917 |
| ANXA13 | 1.000250209 | 1.000121232 | 1.000121232 | 0.000143259 |
| DCAF12L2 | 1.00449623 | 1.002106005 | 1.006892155 | 0.000223595 |
| FAM9A | 1.003356779 | 1.001554714 | 1.005162087 | 0.000258448 |
| HIST1H2BI | 1.005418447 | 1.002484183 | 1.008361301 | 0.000290327 |
| HIST1H4B | 1.004285452 | 1.001956247 | 1.006620071 | 0.000306649 |
| SYT10 | 1.003409143 | 1.00153566 | 1.005286131 | 0.00035801 |
| BARHL2 | 1.003325081 | 1.001479382 | 1.005174181 | 0.000410029 |
| HIST1H2BL | 1.006749345 | 1.002907984 | 1.010605419 | 0.000563306 |
| RBMY1E | 1.116062011 | 1.047945539 | 1.188606054 | 0.000631982 |
| HIST1H3I | 1.008520839 | 1.00361963 | 1.013445983 | 0.000641138 |
| ASIC2 | 1.000816023 | 1.00034647 | 1.001285796 | 0.000657398 |
| HIST1H4C | 1.001254315 | 1.000531476 | 1.001977677 | 0.000669059 |
| HIST1H2AB | 1.006352989 | 1.002676392 | 1.010043067 | 0.000695741 |
| GDPD2 | 1.000434487 | 1.000180611 | 1.000688428 | 0.000794776 |
| LCN15 | 1.000440633 | 1.000179027 | 1.000702307 | 0.000961541 |
| SLC10A2 | 1.000224638 | 1.000089788 | 1.000359506 | 0.001094193 |
| NPY5R | 1.002951746 | 1.001168353 | 1.004738315 | 0.001170847 |
| HIST1H4D | 1.003464179 | 1.001226871 | 1.005706486 | 0.002392559 |
| COL9A1 | 1.002087607 | 1.000707756 | 1.00346936 | 0.003013852 |
| CRABP1 | 1.000073924 | 1.000024559 | 1.000123291 | 0.00333467 |
| AMPD1 | 0.995589173 | 0.992598941 | 0.998588412 | 0.003971938 |
| TGIF2LY | 1.02684375 | 1.008439511 | 1.04558387 | 0.004095319 |
| KRTAP29-1 | 1.005106966 | 1.001617293 | 1.008608796 | 0.004096565 |
| UGT2B28 | 1.002495079 | 1.000766639 | 1.004226505 | 0.0046493 |
| SLC3A1 | 1.000421841 | 1.000127915 | 1.000715853 | 0.00490643 |
| SLC22A10 | 0.906487055 | 0.845723518 | 0.971616329 | 0.005548349 |
| NKAIN1 | 1.00068796 | 1.000198611 | 1.00117755 | 0.005856298 |
| RAB3B | 1.000100968 | 1.000028359 | 1.000173582 | 0.006419937 |
| EPHA5 | 1.000175252 | 1.000044985 | 1.000305536 | 0.008367732 |
| TM4SF20 | 1.000083007 | 1.000020439 | 1.000145579 | 0.009315098 |
| HIST1H4L | 1.020764987 | 1.004927419 | 1.036852153 | 0.009993429 |
| FABP1 | 1.000620756 | 1.000132007 | 1.001109744 | 0.012792349 |
| SCGB1A1 | 0.999975762 | 0.999956361 | 0.999995164 | 0.01434692 |
| CYP17A1 | 0.991524759 | 0.984741181 | 0.998355067 | 0.015099843 |
| PEX5L | 1.000195205 | 1.000036249 | 1.000354186 | 0.01608583 |
| HMX2 | 1.004416694 | 1.000799395 | 1.008047067 | 0.016663167 |
| SCGB3A1 | 0.999992487 | 0.99998633 | 0.999998645 | 0.016785424 |
| CLVS2 | 1.001220833 | 1.000219674 | 1.002222995 | 0.016835501 |
| HIST1H3J | 1.009735816 | 1.001709563 | 1.01782638 | 0.017338146 |
| NPY | 1.000015359 | 1.000002579 | 1.000028139 | 0.018499329 |
| HOXD13 | 1.00082481 | 1.000121615 | 1.0015285 | 0.021500875 |
| TRIM58 | 1.00085058 | 1.000124858 | 1.001576829 | 0.021600885 |
| HIST1H2AH | 1.005753838 | 1.000609254 | 1.010924873 | 0.028325951 |
| CT47B1 | 1.00699033 | 1.000564052 | 1.013457882 | 0.032957884 |
| KRT33A | 1.000628673 | 1.000027531 | 1.001230176 | 0.040388504 |
| GLRA2 | 1.003543926 | 1.000139818 | 1.00695962 | 0.04128971 |
| Gene | HR | HR-95Low | HR-95Up | p-value |
| C8orf86 | 1.023568218 | 1.00086135 | 1.046790243 | 0.041832284 |
| FAM216B | 0.999679134 | 0.999365642 | 0.999992724 | 0.044916679 |
| CYP2B6 | 1.000085411 | 1.000001506 | 1.000169323 | 0.046026185 |
| PRAC1 | 1.002561706 | 1.000025576 | 1.005104268 | 0.047731097 |
| EN1 | 1.000799915 | 1.000006595 | 1.001593865 | 0.048125021 |
Discussion
The 7-methylguanosine (M7G) is one common tRNA methylation modification that occurs at nucleotide position 46 in tRNAs (Guy et al., 2014), it is catalyzed by the methyltransferase complexes formed by METTL1 and WDR4 (Lin et al., 2018). METTL1 is overexpressed in cancers, and studies showed that it was associated with drug resistance to chemotherapies and poor prognosis in the patients (Wang et al., 2021a; Wang et al., 2021b). Ma et al., (2021) found that METTL1 and WDR4 were involved in the development of lung cancers by promoting the m7G tRNA modification and regulating the protein production of cell cycle genes. Shi et al., (2019) found that mir-107 has also been shown to be involved in lung cancer growth and metastasis. However, no study had been reported on the association of m7G-related miRNAs with LUAD and their application in the prognosis prediction of LUAD patients.
A number of studies have proved the prognostic value of miRNA-based signatures for patients with various types of cancers (Lv et al., 2020; Tang et al., 2019). X Xu et al., (2019) identified three miRNAs in patients with hypopharyngeal squamous cell carcinoma, the DE expression of which was significantly correlated with the OS and disease-specific survival (DSS) of the patients. In patients with clear cell renal cell carcinoma, Xie et al., (2018) identified four miRNAs the DE expression of which could be used to predict the survival of patients. Qian et al., (2019) constructed two prediction models based on miRNAs identified in colorectal cancer patients, the prediction from which were in good agreement with the patients’ OS and recurrence-free survival (RFS). Our study identified 40 DE miRNAs in LUAD patients that were associated with m7G, including 34 upregulated and 6 downregulated. Then, we further identified five m7G-related miRNAs (hsa-miR-31-5p, hsa-miR-5571-3p, hsa-miR-4697-3p, hsa-miR-6858-5p, and hsa-miR-873-3p) which show a significant impact on the patients’ prognosis. The prediction performance of this 5-miRNA signature was then evaluated by the K-M survival analysis and ROC analysis in the patients after they were stratified into 2 groups according to the risk score. To establish a more reliable, individualized clinical prediction model, we construct a nomogram by incorporating these five miRNAs with six other independent clinical indices. independent clinical indices. The calibration plots showed that the prediction of the nomogram was in good agreement with the observed patient outcome, and the ROC curve analysis also suggested that this nomogram had higher prognostic efficiency. Furthermore, the K-M survival analysis indicated that has-miR-315p and has-miR-55713p have research significance for the overall survival of LUAD patients. All these results, taken together, strongly suggest that m7G-related miRNAs have a significant prognostic ability in LUAD.
Almost all the five prognostic miRNAs we identified in this study have been previously reported to be associated with cancers. Mi et al., (2020) study showed that upregulated expression of miR-31-5p might promote exacerbation of colon adenocarcinoma by targeting TNS1 thus predicting a poor OS. Lu et al., (2019) study showed that miR-31-5p promoted the progression of oral cancer by enhancing the proliferation of oral epithelial cells. In our study, we found that high expression of miR-31-5p worsen the prognosis of LUAD patients, suggesting miR-31-5p’s involvement in the progression of LUAD. DF Li et al., (2015) found that miR-4697-3p plays an important role in maintaining physiological balance, and its destruction led to the development of gastric cancer. Wang et al., (2021b) study showed that miR-6858-5p plays an essential in melatonin’s inhibiting the malignant biological behaviors of Glioma cells. Some researchers found that mir-873-3p might promote the development of gastric cancer, pancreatic cancer, estrogen-receptor-positive breast cancer, and some other cancers (Deng et al., 2021; Fang et al., 2021; Zhang et al., 2021). The miR-5571-3p we identified was a novel gene that had not been previously studied, our study showed that high expression of miR-5571-3p plays a protective role in LUAD patients and has a good prediction ability of prognosis. It is suggested that miR-5571-3p might inhibit tumor development by regulating m7Gmethylation, which indicates the necessity of future investigations on it as a novel potential therapeutical target of LUAD. These m7G-related miRNAs could be used as predictors of LUAD. Among them, the expression of miR-31-5p and miR-5571-3p affects the prognosis of patients, which is worthy of our further studies on their molecular mechanism.
Furthermore, we analyzed overlapping genes associated with the immune scores and the risk scores using the KEGG and GO enrichment analysis to detect the potential molecular mechanisms. In the KEGG analysis, the neuropeptide receptor activity, antimicrobial humoral response, and neuropeptide signaling pathway were the most significantly enriched pathways. In the groups of biological processes, cellular components, and molecular function, the neuropeptide receptor activity, serine-type endopeptidase inhibitor activity, and neuropeptide signaling pathway were the most significantly enriched gene sets, respectively. The biological functions of neuropeptide receptors have been reported in previous studies. Lee et al., (2020) found that the neuropeptide bombesin receptor stimulates the proliferation of lung cancer cells through the activation of HER3. Some studies have also revealed that immune-related lncRNAs, such as SCHLAP1, could mediate the neuropeptide signaling pathway and the cytokine - cytokine receptor interactions in low-grade gliomas (Li et al., 2019). This indicates that the neuropeptide signaling pathway plays a critical role in the development and progression of cancer, thus suggesting requirement of future studies on the impacting mechanism of the neuropeptide signaling pathway in LUAD.
The number, location, and phenotype of tumor-infiltrating lymphocytes (TILs) play essential roles in the development and progression of tumors (Chen et al., 2017). Tumor immunotherapy is currently the main motive power for personalized precision medicine, on which great efforts are currently being made in treating advanced or metastatic cancers utilizing the functions of the immune system (Ganesh et al., 2019; Szekely et al., 2018; Wang et al., 2017). The tumors’ responses to immunotherapy are closely related to the infiltration of immune cells, including the T helper (Th) cells which are associated with the activation of T lymphocytes (Antony et al., 2005; Hung et al., 1998; Pardoll et al., 1998). Thus, we investigated the role of immune cell infiltration and immunology functions in the immune microenvironment in LUAD. Among the immune cells, T helper cells are highly expressed in each sample, showing that Th may play an important role LUAD. Among the immunology functions, human leukocyte antigen (HLA) and MHC class I were highly expressed in almost every sample. Previous studies have shown that MHC class I is essential for immunology functions in coordinating the killing effect of NK cells and CD4T cells (Shklovskaya et al., 2021). Thereafter, we analyzed the differences in immune cells and immunology functions between the high- and low-risk groups. Our results showed that compared to high-risk groups, low-risk groups had significantly higher infiltration of Dendritic cells, Mast cells, B cells, Neutrophils, and TIL, and significantly lower infiltration of NK cells. The Univariate Cox regression analysis showed that the expression of AMPD1 was positively correlated with most of the immune cells and the immunology functions. Studies on the AMPD1 in LUAD, however, are very limited, thus further studies are required to unveil the impacting mechanism of the AMPD1 gene in LUAD. The above results revealed the possible involvement of immune cell infiltration and immunology functions in LUAD and provide a reference for future studies on the development of novel immunotherapy targeting LUAD.
Besides the novelty and significant findings, we acknowledge certain limitations in the present study. This study integrates the associations between m7G, miRNA, and LUAD prognosis, which has implications for innovation and provides accuracy for clinical prognosis prediction. However, this study only analyzed the data of LUAD patients collected from the TCGA database, the model constructed in this study had not been validated by real world data collected from a purposely designed clinical trial due to a limitation of the research area of our laboratory. Besides, the expression and mechanism of these miRNAs had not been validated using wet-lab experiment, which requires further investigations.
In conclusion, our study is the first in the area to explore the application of m7G-related miRNAs in predicting the prognosis of LUAD. We identified DE m7G-related miRNAs associated with LUAD, five from which with significant prognostic value were extracted to construct an independent prognostic signature for LUAD. Furthermore, we constructed a nomogram by integrating these five m7G-related miRNAs with six other independent clinical indices, which demonstrated a more accurate and reliable predict ability of the prognosis of LUAD patients. In a summary, our study unveiled the prognostic value of m7G-related miRNAs in LUAD, and it should serve as reference for the identification of more m7G-related genes in the future.
Author Contribution Statement
Methodology and Writing – original draft: XiaowenQiu; Data curation:Yongting Chen; Supervision,Writing – review & editing: Yipeng Song and Hongwei Li; Formal analysis:Xingzhuang Zhu,Zheng Gong,Fengyuan Yu and Pengfei Zhang; Final approval of manuscript: All authors.
Acknowledgements
Ethics statement
There were no cell, tissue, or animal studies. No ethical requirements are involved.
Consent for publication
All authors agree to publish the paper.
Availability of data and materials
The datasets in this study are available from TCGA (https://portal.gdc.cancer.gov) TargetScan (http://www.targetscan.org/) and ESTIMATE database (https://bioinformatics.mdanderson.org/estimate/disease.htmlhttps://bioinformatics.mdanderson.org/estimate/disease.html).
Competing interests
The authors have no conflict of interest to declare.
References
- Alexandrov A, Grayhack EJ, Phizicky EM. tRNA m7G methyltransferase Trm8p/Trm82p: evidence linking activity to a growth phenotype and implicating Trm82p in maintaining levels of active Trm8p. Rna. 2005;11:821–30. doi: 10.1261/rna.2030705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antony PA, Piccirillo CA, Akpinarli A, et al. CD8+ T cell immunity against a tumor/self-antigen is augmented by CD4+ T helper cells and hindered by naturally occurring T regulatory cells. J Immunol. 2005;174:2591–601. doi: 10.4049/jimmunol.174.5.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltimore D, Boldin MP, O’Connell RM, et al. MicroRNAs: new regulators of immune cell development and function. Nat Immunol. 2008;9:839–45. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- Binnewies M, Roberts EW, Kersten K, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24:541–50. doi: 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulias K, Greer EL. Put the Pedal to the METTL1: Adding Internal m(7)G Increases mRNA Translation Efficiency and Augments miRNA Processing. Mol Cell. 2019;74:1105–7. doi: 10.1016/j.molcel.2019.06.004. [DOI] [PubMed] [Google Scholar]
- Catacchio I, Scattone A, Silvestris N, et al. Immune Prophets of Lung Cancer: The Prognostic and Predictive Landscape of Cellular and Molecular Immune Markers. Transl Oncol. 2018;11:825–35. doi: 10.1016/j.tranon.2018.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541:321–30. doi: 10.1038/nature21349. [DOI] [PubMed] [Google Scholar]
- Chen J, Li K, Chen J, et al. Aberrant translation regulated by METTL1/WDR4-mediated tRNA N7-methylguanosine modification drives head and neck squamous cell carcinoma progression. Cancer Commun (Lond) 2022;42:223–44. doi: 10.1002/cac2.12273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Xia J, Xu YE. Circular RNA circTP63 enhances estrogen receptor-positive breast cancer progression and malignant behaviors through the miR-873-3p/FOXM1 axis. Colloq Inse. 2021;32:44–52. doi: 10.1097/CAD.0000000000001010. [DOI] [PubMed] [Google Scholar]
- El Bezawy R, Cominetti D, Fenderico N, et al. miR-875-5p counteracts epithelial-to-mesenchymal transition and enhances radiation response in prostate cancer through repression of the EGFR-ZEB1 axis. Cancer Lett. 2017;395:53–62. doi: 10.1016/j.canlet.2017.02.033. [DOI] [PubMed] [Google Scholar]
- Fang L, Wang SH, Cui YG, et al. LINC00941 promotes proliferation and metastasis of pancreatic adenocarcinoma by competitively binding miR-873-3p and thus upregulates ATXN2. Eur Rev Med Pharmacol Sci. 2021;25:1861–8. doi: 10.26355/eurrev_202102_25081. [DOI] [PubMed] [Google Scholar]
- Ganesh K, Stadler ZK, Cercek A, et al. Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat Rev Gastroenterol Hepatol. 2019;16:361–75. doi: 10.1038/s41575-019-0126-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu XB, Tian T, Tian XJ, et al. Prognostic significance of neutrophil-to-lymphocyte ratio in non-small cell lung cancer: a meta-analysis. Sci Rep. 2015;5:12493. doi: 10.1038/srep12493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy MP, Phizicky EM. Two-subunit enzymes involved in eukaryotic post-transcriptional tRNA modification. RNA Biol. 2014;11:1608–18. doi: 10.1080/15476286.2015.1008360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21:309–22. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- He L, Qu L, Wei L, et al. Reduction of miR1323p contributes to gastric cancer proliferation by targeting MUC13. Mol Med Rep. 2017;15:3055–61. doi: 10.3892/mmr.2017.6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel R, Sie C, Watson DI, et al. MicroRNA signatures in chemotherapy resistant esophageal cancer cell lines. World J Gastroenterol. 2014;20:14904–12. doi: 10.3748/wjg.v20.i40.14904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung K, Hayashi R, Lafond-Walker A, et al. The central role of CD4(+) T cells in the antitumor immune response. J Exp Med. 1998;188:2357–68. doi: 10.1084/jem.188.12.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karrich JJ, Jachimowski LC, Libouban M, et al. MicroRNA-146a regulates survival and maturation of human plasmacytoid dendritic cells. Blood. 2013;122:3001–9. doi: 10.1182/blood-2012-12-475087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L, Ramos-Alvarez I, Moody TW, et al. Neuropeptide bombesin receptor activation stimulates growth of lung cancer cells through HER3 with a MAPK-dependent mechanism. Biochim Biophys Acta Mol Cell Res. 2020;1867:118625. doi: 10.1016/j.bbamcr.2019.118625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li DF, Yang MF, Shi SL, et al. TM4SF5-CTD-2354A18 1-miR-4697-3P may play a key role in the pathogenesis of gastric cancer. Bratisl Lek Listy. 2015;116:608–15. doi: 10.4149/bll_2015_118. [DOI] [PubMed] [Google Scholar]
- Li X, Meng Y. Survival analysis of immune-related lncRNA in low-grade glioma. BMC Cancer. 2019;19:813. doi: 10.1186/s12885-019-6032-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Liu Q, Lelyveld VS, et al. Mettl1/Wdr4-Mediated m(7)G tRNA Methylome Is Required for Normal mRNA Translation and Embryonic Stem Cell Self-Renewal and Differentiation. Mol Cell. 2018;71:244–55. doi: 10.1016/j.molcel.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, He Q, Liang J, et al. miR-31-5p Is a Potential Circulating Biomarker and Therapeutic Target for Oral Cancer. Mol Ther Nucleic Acids. 2019;16:471–80. doi: 10.1016/j.omtn.2019.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Y, Duanmu J, Fu X, et al. Identifying a new microRNA signature as a prognostic biomarker in colon cancer. PLoS One. 2020;15:e0228575. doi: 10.1371/journal.pone.0228575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Han H, Huang Y, et al. METTL1/WDR4-mediated m(7)G tRNA modifications and m(7)G codon usage promote mRNA translation and lung cancer progression. Mol Ther. 2021;29:3422–35. doi: 10.1016/j.ymthe.2021.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi B, Li Q, Li T, et al. High miR-31-5p expression promotes colon adenocarcinoma progression by targeting TNS1. Aging (Albany NY) 2020;12:7480–90. doi: 10.18632/aging.103096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandolfini L, Barbieri I, Bannister AJ, et al. METTL1 Promotes let-7 MicroRNA Processing via m7G Methylation. Mol Cell. 2019;74:1278–90. doi: 10.1016/j.molcel.2019.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardoll DM, Topalian SL. The role of CD4+ T cell responses in antitumor immunity. Curr Opin Immunol. 1998;10:588–94. doi: 10.1016/s0952-7915(98)80228-8. [DOI] [PubMed] [Google Scholar]
- Qian J, Zeng L, Jiang X, et al. Novel Multiple miRNA-Based Signatures for Predicting Overall Survival and Recurrence-Free Survival of Colorectal Cancer Patients. Med Sci Monit. 2019;25:7258–71. doi: 10.12659/MSM.916948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Fan S, Wu M, et al. YTHDF1 links hypoxia adaptation and non-small cell lung cancer progression. Nat Commun. 2019;10:4892. doi: 10.1038/s41467-019-12801-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shklovskaya E, Rizos H. MHC Class I Deficiency in Solid Tumors and Therapeutic Strategies to Overcome It. Int J Mol Sci. 2021:22. doi: 10.3390/ijms22136741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- Siriwardhana C, Khadka VS, Chen JJ, et al. Development of a miRNA-seq based prognostic signature in lung adenocarcinoma. BMC Cancer. 2019;19:34. doi: 10.1186/s12885-018-5206-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209–49. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- Szekely B, Bossuyt V, Li X, et al. Immunological differences between primary and metastatic breast cancer. Ann Oncol. 2018;29:2232–39. doi: 10.1093/annonc/mdy399. [DOI] [PubMed] [Google Scholar]
- Tang J, Ma W, Zeng Q, et al. Identification of miRNA-Based Signature as a Novel Potential Prognostic Biomarker in Patients with Breast Cancer. Dis Markers. 2019;2019:3815952. doi: 10.1155/2019/3815952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Wang W, Han X, et al. Methyltransferase-like 1 regulates lung adenocarcinoma A549 cell proliferation and autophagy via the AKT/mTORC1 signaling pathway. Oncol Lett. 2021a;21:330. doi: 10.3892/ol.2021.12591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Zhao Z, Qi Q, et al. miR-6858 plays a key role in the process of melatonin inhibition of the malignant biological behavior of glioma. J Clin Neurosci. 2021b;87:137–46. doi: 10.1016/j.jocn.2021.02.015. [DOI] [PubMed] [Google Scholar]
- Wang R, Wu Y, Huang W, et al. MicroRNA-940 Targets INPP4A or GSK3β and Activates the Wnt/β-Catenin Pathway to Regulate the Malignant Behavior of Bladder Cancer Cells. Oncol Res. 2018;26:145–55. doi: 10.3727/096504017X14902261600566. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang RF, Wang HY. Immune targets and neoantigens for cancer immunotherapy and precision medicine. Cell Res. 2017;27:11–37. doi: 10.1038/cr.2016.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu ZH, Zhong Y, Zhou T, et al. miRNA biomarkers for predicting overall survival outcomes for head and neck squamous cell carcinoma. Genomics. 2021;113:135–41. doi: 10.1016/j.ygeno.2020.12.002. [DOI] [PubMed] [Google Scholar]
- Xie M, Lv Y, Liu Z, et al. Identification and validation of a four-miRNA (miRNA-21-5p, miRNA-9-5p, miR-149-5p, and miRNA-30b-5p) prognosis signature in clear cell renal cell carcinoma. Cancer Manag Res. 2018;10:5759–66. doi: 10.2147/CMAR.S187109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Shao G, Pan Q, et al. MicroRNA-9 regulates non-small cell lung cancer cell invasion and migration by targeting eukaryotic translation initiation factor 5A2. Am J Transl Res. 2017;9:478–88. [PMC free article] [PubMed] [Google Scholar]
- Xu X, Lu Z, Gross N, et al. A 3-miRNA signature predicts survival of patients with hypopharyngeal squamous cell carcinoma after post-operative radiotherapy. J Cell Mol Med. 2019;23:8280–91. doi: 10.1111/jcmm.14702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Hu G, Zhang Z, et al. CircRNA_0043691 sponges miR-873-3p to promote metastasis of gastric cancer. Mamm Genome. 2021;32:476–87. doi: 10.1007/s00335-021-09900-5. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Kong L, Pei Z, et al. m7G Methyltransferase METTL1 Promotes Post-ischemic Angiogenesis via Promoting VEGFA mRNA Translation. Front Cell Dev Biol. 2021;9:642080. doi: 10.3389/fcell.2021.642080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets in this study are available from TCGA (https://portal.gdc.cancer.gov) TargetScan (http://www.targetscan.org/) and ESTIMATE database (https://bioinformatics.mdanderson.org/estimate/disease.htmlhttps://bioinformatics.mdanderson.org/estimate/disease.html).