Abstract
N-Acetylglucosamine (GlcNAc) and N-acetylneuraminic acid (NANA) are good carbon sources for Escherichia coli K-12, whereas N-acetylmannosamine (ManNAc) is metabolized very slowly. The isolation of regulatory mutations which enhanced utilization of ManNAc allowed us to elucidate the pathway of its degradation. ManNAc is transported by the manXYZ-encoded phosphoenolpyruvate-dependent phosphotransferase system (PTS) transporter producing intracellular ManNAc-6-P. This phosphorylated hexosamine is subsequently converted to GlcNAc-6-P, which is further metabolized by the nagBA-encoded deacetylase and deaminase of the GlcNAc-6-P degradation pathway. Two independent mutations are necessary for good growth on ManNAc. One mutation maps to mlc, and mutations in this gene are known to enhance the expression of manXYZ. The second regulatory mutation was mapped to the nanAT operon, which encodes the NANA transporter and NANA lyase. The combined action of the nanAT gene products converts extracellular NANA to intracellular ManNAc. The second regulatory mutation defines an open reading frame (ORF), called yhcK, as the gene for the repressor of the nan operon (nanR). Mutations in the repressor enhance expression of the nanAT genes and, presumably, three distal, previously unidentified genes, yhcJIH. Expression of just one of these downstream ORFs, yhcJ, is necessary for growth on ManNAc in the presence of an mlc mutation. The yhcJ gene appears to encode a ManNAc-6-P-to-GlcNAc-6-P epimerase (nanE). Another putative gene in the nan operon, yhcI, likely encodes ManNAc kinase (nanK), which should phosphorylate the ManNAc liberated from NANA by the NanA protein. Use of NANA as carbon source by E. coli also requires the nagBA gene products. The existence of a ManNAc kinase and epimerase within the nan operon allows us to propose that the pathways for dissimilation of the three amino sugars GlcNAc, ManNAc, and NANA, all converge at the step of GlcNAc-6-P.
Amino sugars are versatile components of the cell surface structures of bacteria. They form the essential backbone of the peptidoglycan in both gram-positive and gram-negative bacteria and are also constituents of the outer membrane lipopolysaccharide (LPS) layer and the polysaccharide capsules of gram-negative bacteria. The glmS-encoded amidotransferase, glucosamine (GlcN) synthase, is responsible for the de novo synthesis of amino sugars in Escherichia coli, producing GlcN-6-P from fructose-6-P and glutamine. The pathway for the conversion of GlcN-6-P to UDP–N-acetylglucosamine (GlcNAc), the first dedicated precursor of the cell wall components, has been recently elucidated (21–23) (Fig. 1). UDP-GlcNAc serves as amino sugar donor in several transferase reactions in the synthesis of peptidoglycan, the core and lipid A moieties of the LPS, enterobacterial common antigen, some O antigens of gram-negative bacteria, and the teichoic acids of gram-positive bacteria (reviewed in references 35, 36, and 44). Some of the UDP-bound amino sugar in enteric bacteria is subsequently converted to the form of N-acetylmannosamine (ManNAc) and N-acetylmannosaminuronic acid for incorporation of the latter into the enterobacterial common antigen.
FIG. 1.
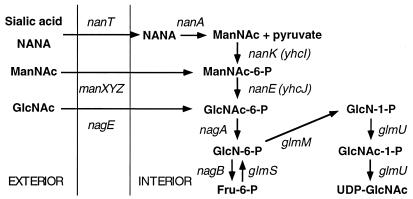
Pathway for the metabolism of GlcNAc and proposed pathway for the degradation of ManNAc and NANA (sialic acid). GlcNAc is transported by both the manXYZ-encoded transporter and its own specific transporter, encoded by nagE, producing intracellular GlcNAc-6-P, which is degraded by the nagA- and nagB-encoded enzymes. The biosynthetic pathway producing UDP-GlcNAc for incorporation into cell wall components involves the glmS, glmM, and glmU gene products. ManNAc is taken up by the manXYZ transporter, producing intracellular ManNAc-6-P. NANA is taken up as the free sugar by a sugar-cation symporter encoded by nanT. Inside the cell, NANA is cleaved by the aldolase encoded by nanA to give ManNAc and pyruvate. The results of this study allow us to propose that intracellular ManNAc is phosphorylated to ManNAc-6-P, which is subsequently converted to GlcNAc-6-P, the substrate of the nagA-encoded deacetylase. Thus, the pathways for degradation of NANA, ManNAc, and GlcNAc converge at the level of GlcNAc-6-P.
In addition to having a structural role, the amino sugars are particularly useful to bacteria as energy sources since they supply both carbon and nitrogen. Both GlcN and GlcNAc are phosphoenolpyruvate-dependent phosphotransferase system (PTS) sugars in E. coli, and the proteins that mediate their uptake (nagE and manXYZ) or degradation (nagBA) have been purified and characterized (4, 9, 26, 40, 50). The genes encoding the GlcNAc-specific PTS transporter (nagE) and the enzymes which convert GlcNAc-6-P to GlcN-6-P and fructose-6-P (nagBA) (Fig. 1) are arranged in divergent operons controlled by the nagC-encoded repressor (33). In addition, the genes for the biosynthesis (glmS) and degradation (nagB) of GlcN-6-P are expressed in a coordinated manner so that in the presence of amino sugars, the catabolic enzymes are induced and the expression of glucosamine synthase is decreased (32).
The amino sugars are ubiquitous and abundant in nature (e.g., chitin is a β1,4-linked homopolymer of GlcNAc) and are present in a range of both simple and complex biopolymers. Most glycoconjugates (glycoproteins and glycolipids) of mammalian cell surfaces contain amino sugars, including sialic acid residues, where the oligosaccharide chains of these conjugates are important ligands for cellular recognition. The sialic acids are a series of N- and O-substituted derivatives of N-acetylneuraminic acid (NANA), a compound which is formed by the condensation of ManNAc and pyruvate (see references 43 and 49 for reviews). Certain bacteria are capable of degrading the complex oligosaccharide chains of glycoconjugates, and many bacteria, including enteric bacteria like E. coli K-12, can use sialic acid as a source of carbon (47) and nitrogen (45). In contrast to this widespread catabolic pathway in bacteria, only relatively few, mostly pathogenic, species are able to synthesize sialic acid for subsequent incorporation into surface structures, e.g., the capsular polysialic acid virulence factors of E. coli K1 and Neisseria meningitidis (46). Although the E. coli genes encoding a sialic acid transporter (nanT) and N-acetylneuraminate lyase (nanA) have been sequenced (19, 29) and characterized both genetically and physiologically (1, 37, 47, 48), the subsequent fate of the ManNAc liberated by the lyase has not been determined for any bacterium. In this communication, we present the results of an investigation of the pathway of ManNAc utilization in E. coli K-12.
MATERIALS AND METHODS
Bacteriological techniques.
The bacterial strains used are listed in Table 1. The ability of bacteria to use different sugars as carbon sources was tested on minimal A plates (24) supplemented with 0.2% GlcNAc, 0.2% ManNAc, or 0.1% sialic acid. Morpholinepropanesulfonic acid (MOPS) minimal medium (27) was used for liquid cultures. The glmS::Tc mutation was constructed by replacing the 700-bp BglII fragment within the glmS gene of pGM7 (8) with a tetracycline resistance cassette and recombining the interrupted gene onto the chromosome as described previously (31). The manXYZ::Tn9 mutation is derived from strain NK6702 (man-6::Tn9) from the E. coli Genetics Stock Center. Although described as a probable mutation in manA (encoding phosphomannose isomerase), the Tn9 is clearly within the manXYZ operon since its introduction into a nagE strain produces a strain that is unable to grow on GlcNAc but still grows (poorly) on mannose. P1 transductions were carried out with P1vir. Lysogenizations with the λ bacteriophage carrying the nagB-lacZ fusion (λRS/EB [Table 2]) and β-galactosidase assays were carried out as described previously (34).
TABLE 1.
Bacterial strains used and growth on ManNAc
| Strain | Growth on ManNAca | Relevant genotypeb | Reference or source |
|---|---|---|---|
| IBPC5321 | +P | thi-1 argG6 argE3 his-4 mtl-1 xyl-5 rpsL ΔlacX74 mlc-1 | 31 |
| IBPC900 | +P | IBPC5321 mlc::Tc | |
| IBPC904 | +++ | IBPC5321 ama-4 | |
| IBPC905 | +++ | IBPC900 ama-5 | |
| IBPC915 | + | IBPC900 mlc+nth1::Km | |
| JM101 | + | F′ traD36 lacIq Δ(lacZ)M15 proA+B+/supE thi Δ(lac-proAB) | |
| IBPC1012 | +P | JM101 mlc::Tc | 30 |
| IBPC1016 | +++ | IBPC1012 ama-6 | |
| Ymc | + | supF lacY | |
| IBPC1013 | +P | Ymc mlc::Tc | |
| IBPC1017 | +++ | IPBC1013 ama-7 | |
| IBPC1047 | + | IBPC1017 mlc+ nth1::Km | |
| IBPC1008 | NT | Ymc nth1::Km | P1(PK2242) × Ymc |
| PK2242 | NT | HfrC Δ1646(dgsA-manA) ptsG tonA22 nth1::Km | P. Kuempel 30 |
| MC4100 | + | araD139 Δ(argF-lac)U169 flbB3501 deoC1 relA1 rbsR rpsL150 ptsF25 | |
| KD622 | +P | MC4100 φ(malK-lacZ)1113 (λplacMu50) mlc::Tn10Km | 6 |
| IBPC1018 | +++ | KD622 ama-8 | |
| IBPC707 | − | IBPC5321 manXYZ::Tn9 | IBPC5321 × P1(NK6702) |
| IBPC567 | − | IBPC5321 ptsM8 | 31 |
| IBPC546 | − | IBPC5321 nagB::Km | 31 |
| IBPC531 | − | IBPC5321 nagA::Cm | 31 |
| IBPC529C | +P | IBPC5321 nagC::Cm | 31 |
| IBPC542 | +P | IBPC5321 nagE::Km | 31 |
| IBPC720 | +P | IBPC5321 ptsG22 zcf229::Tn10 | IBPC5321 × P1(TP2506) |
| IBPC750 | +P | IBPC5321 glmS::Tc | |
| IBPC1000 | +P | IBPC720 mlc::Tn10kan | |
| IBPC1001 | +++ | IBPC1000 ama-1 | |
| IBPC1011 | +P | IBPC1001 Tn10cam11 | IBPC1001 × P1(λNK1324) |
| IBPC1021 | +++ | IBPC1016 Tn10cam11 (ama-6) | IBPC1016 × P1(IBPC1011) |
| IBPC1022 | +P | IBPC1012Tn10cam11 (ama+) | IBPC1016 × P1(IBPC1011) |
| IBPC1023 | +++ | IBPC1017 Tn10cam11 (ama-7) | IBPC1017 × P1(IBPC1011) |
| IBPC1024 | +P | IBPC1013 Tn10cam11 (ama+) | IBPC1017 × P1(IBPC1011) |
| IBPC1027 | +P | JM101 mlc::Tc Tn10kan27 Cms | IBPC1022 × P1(λNK1316) |
| IBPC1028 | +P | Ymc mlc::Tc Tn10kan28c Cms | IBPC1024 × P1(λNK1316) |
| IBPC1029 | +P | Ymc mlc::Tc Tn10kan29 Cms | IBPC1024 × P1(λNK1316) |
| IBPC1033 | +P | JM101 mlc::Tc Tn10kan33 Cms | IBPC1022 × P1(λNK1316) |
| IBPC1059 | +++ | JM101 mlc::Tc ama-7 Tn10cam11 | IBPC1012 × P1(IBPC1023) |
| TP2506 | NT | lacZ ptsG22 glk-7 rha-4 rpsL223 zcf229::Tn10 | 7 |
| NK6702 | NT | man-6::Tn9 IN(rrnD-rrnE)1 | E. coli Genetics Stock Center |
−, no growth after 1 week, +, very slow background growth (tiny colonies after 3 to 4 days); +P, papillae (formation of isolated large colonies during slow background growth); +++, good growth on ManNAc; NT, not tested. Cms, chloramphenicol sensitive.
Sequencing showed that Tn10cam11 is gltD::Tn10cam and that Tn10kan27 and -28 are yhcA::Tn10kan.
TABLE 2.
Phages and plasmids used
| Phage or plasmid | Genotype | Reference |
|---|---|---|
| Phages | ||
| λNK1316 | λ b522 cI857 Pam80 nin5 (mini-Tn10kan/Ptac-ATSa transposase) | 14 |
| λNK1324 | λ b522 cI857 Pam80 nin5 (mini-Tn10cam/Ptac-ATS-transposase) | 14 |
| λRS/EB | λRS45/nagB-lacZ | 34 |
| Plasmids | ||
| pSX600 | nanAT yhcJIH | 19 |
| pTZ(Nan4-5) | plac yhcJ | This work |
| pTZ(Nan4-6) | plac yhcJIH | This work |
| pTZ(Nan1-3) | plac yhcK | This work |
| pBR(Nan4-5) | yhcJ | This work |
| pBR(Nan4-6) | yhcJIH | This work |
| pBR(Nan1-3) | yhcK | This work |
ATS, altered target specificity (14).
Mapping ama mutations affecting use of ManNAc.
Random insertion of mini-Tn10cam or Tn10kan transposons into strain IBPC1001 (IBPC5321 ptsG22 zcf229::Tn10 mlc::Tn10kan ama-1) was performed by transposition from λNK1324 and λNK1316 (14). Transductants were selected on LB-chloramphenicol or LB-kanamycin plates and then gridded onto plates carrying the same antibiotic. The grids were replica plated onto minimal ManNAc medium to screen for those which had lost the capacity for rapid growth on ManNAc. Mutations in nagB, nagA, or manXYZ, besides loss of the mlc or ama mutation, could lead to loss of good growth on ManNAc. Mutations in these genes were eliminated by screening for GlcNAc+ (nagBA+) and Man+ (manXYZ+ in the ptsG background) and mlc::Tn10kan. Of 500 transductants tested, 22 failed to grow on minimal ManNAc plates, but the majority (19) also failed to grow on minimal glucose plates and so probably carried minitransposons producing amino acid auxotrophic mutations. One mini-Tn10cam, Tn10cam11 (IBPC1011), was a candidate for linkage to ama-1. To locate other mini-Tn10 insertions near the ama region and also to avoid the problem of finding ManNAc− mutations due to auxotrophies or mutations in other genes affecting ManNAc metabolism, we performed a second round of mutagenesis, using λNK1316, on strains IBPC1022 and IBPC1024 to search for Tn10kan inserts which had lost the Tn10cam11 insertion. Regions of the chromosome adjacent to the mini-Tn10 insertions were amplified by PCR after digestion with RsaI or TaqI and circularization. The amplified fragments were sequenced by using primers complementary to sequences within the mini-Tn10 (28).
Detection of an mlc mutation in IBPC5321.
The mlc::Tc mutation has been shown to enhance expression of a manX-lacZ fusion threefold (30). However, the introduction of the mlc::Tc mutation into IBPC5321 carrying the same manX-lacZ fusion had no effect on manX expression (approximately 90 U in both strains). Replacing the mlc::Tc mutation with wild-type DNA from IBPC1008, using the adjacent nth1::Km marker, reduced expression of the manX-lacZ fusion to 27 U. This effect on manX expression, together with the observation that IBPC5321 behaves like strains carrying an mlc mutation for growth on ManNAc (formation of papillae), shows that IBPC5321 carries an uncharacterized mlc mutation, mlc-1.
Plasmids.
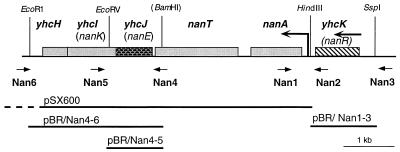
Plasmid pSX600, carrying the nanAT genes and several kilobases of downstream DNA, was described previously (19). Oligonucleotides Nan1, Nan3, Nan4, Nan5, and Nan6 were used to amplify the fragments Nan4-6, Nan4-5, and Nan1-3 (Fig. 2), using Pwo, a thermoresistant polymerase with increased fidelity (Boehringer). The fragments were purified from agarose gels by using Jetsorb (Bioprobe). The Nan4 oligonucleotide includes a BamHI site. The Nan4-5 fragment was digested with BamHI and EcoRV (site near Nan5 [Fig. 2]) and inserted into pTZ18R digested with BamHI and HincII. The Nan4-6 fragment was digested with BamHI and EcoRI (EcoRI site near Nan6) and inserted into pTZ19R digested with the same enzymes. The Nan1-3 fragment was digested with SspI and HindIII and inserted into pTZ18R digested with HincII and HindIII. The cloned fragments in these plasmids were excised as EcoRI-to-HindIII fragments and inserted into pBR322 digested with the same enzymes, to give pBR(Nan4-5) (expressing yhcJ), pBR(Nan4-6) (expressing yhcJIH), and pBR(Nan1-3) (expressing yhcK).
FIG. 2.
Gene organization in the nanAT region of the E. coli chromosome. The relative positions of the ORFs around the nanAT genes as deduced from the E. coli genomic sequence are shown. A promoter, shown by a bent arrow, has been proposed to lie upstream of nanA. The yhcK (nanR) gene is transcribed in the same direction, but no promoter has yet been localized. Locations of the oligonucleotides Nan1 to Nan6 are shown with the arrowhead at the 3′ end. The extent of the cloned DNA in the plasmids is indicated by the horizontal lines. Only restriction enzyme sites used in the clonings are indicated. The BamHI site is not present in the chromosomal sequence but was created from a XhoII site by using the Nan4 oligonucleotide.
Transport assays.
Uptake of sialic acid and ManNAc by different E. coli strains was determined essentially as described previously (47). Briefly, bacteria were grown to an A600 of 0.4 to 0.5 in minimal M63 medium containing 0.4% glycerol as the sole carbon source and then washed twice with an equal volume of M63 before resuspension to a final A600 of approximately 0.5 in the same medium. Transport was initiated by adding 0.1 ml of [9-3H]NANA or [6-3H]ManNAc plus cold carrier sugar to 0.9 ml of bacterial culture at 37°C to give a final concentration of 0.1 mM NANA (7,000 cpm nmol−1) or 0.45 mM ManNAc (9,140 cpm nmol−1). Cultures were incubated for 15 min without shaking, and then the entire contents of each sample were filtered through 0.45-μm-diameter nitrocellulose disks to separate bacteria from free sugar. The filters were washed with a single 4-ml portion of M63 salts prior to drying and liquid scintillation spectrometry to quantify sugar uptake. Data are expressed as nanomoles of sugar taken up in 15 min, normalized to one A600 unit. Radiolabeled ManNAc (15 Ci mmol−1) was purchased from American Radiolabeled Chemicals Inc. (St. Louis, Mo.). CMP-[9-3H]NANA (21.2 Ci mmol−1; New England Nuclear) was a kind gift from Tom Warner. The free, labeled sugar was prepared from its nucleotide derivative by mild acid hydrolysis as previously described (47).
RESULTS
Good growth on ManNAc requires two mutations in E. coli K-12.
Most strains of E. coli K-12 grow very slowly on minimal plates containing the rare amino sugar mannosamine, ManNAc, galactosamine, or N-acetylgalactosamine as the carbon source, with only faint growth seen after 4 to 8 days. However, in the case of strain IBPC5321, and strains derived from it, growing on minimal ManNAc plates, we observed large isolated colonies (papillae) appearing within the smear of slowly growing bacteria. Upon reisolation of papillae on minimal ManNAc medium, the bacteria continued to grow well, producing good-sized colonies after 24 to 36 h. The increased growth rate selected on ManNAc was a stable genetic character; the mutant bacteria retained the ability to grow well on ManNAc medium after several passages on LB or minimal glucose plates (i.e., on nonselective medium).
Strain IBPC5321 was found to carry a spontaneous mutation in gene mlc (see Materials and Methods). Mlc is a DNA binding protein with homology to the NagC regulatory protein (13). It has been shown recently to control the expression of manXYZ, which encodes a PTS transporter with broad sugar specificity including mannose, glucose, fructose, GlcNAc, and GlcN (30). Introduction of an mlc null mutation, mlc::Tc or mlc::Tn10kan, into bacteria from other genetic origins (e.g., JM101, Ymc, or MC4100) allowed the appearance of the fast-growing papillae of ManNAc++ colonies when these bacteria were streaked to minimal ManNAc plates (Table 1). The appearance of fast-growing colonies in the mlc background suggested that a second spontaneous mutation was occurring at high frequency (about 1 in 104); we designated this mutation ama (referring to ManNAc). The doubling times for the different ama strains in minimal ManNAc medium at 37°C were between 90 and 130 min, compared to at least 6 h for JM101. There was no effect of the ama mutations on growth on GlcNAc, with doubling times remaining at 50 min for JM101 and IBPC1016 (mlc ama-6).
ManNAc utilization requires the manXYZ and nagBA gene products.
The observation that an mlc mutation is necessary for good growth on ManNAc suggested that the manXYZ transporter, controlled by Mlc, might be responsible for the entry of ManNAc into the cell. In agreement with this hypothesis, strains derived from IBPC5321 carrying mutations in manXYZ (IBPC567 and IBPC707) failed to produce the fast-growing colonies in the background smear; indeed, there was no background growth (Table 1). Similarly, mutations in nagA and nagB (encoding the two enzymes necessary for degradation of GlcNAc-6-P; strains IBPC546 and IBPC531) also eliminated growth on ManNAc, whereas mutations in nagE, encoding the GlcNAc-specific transporter (strain IBPC542), ptsG, encoding another transporter for glucose and mannose (IBPC720), or nagC, encoding the repressor of the nagE-BA operons (IBPC529C), had no effect on the formation of the papillae (Table 1). These results suggest that after transport by the manXYZ PTS, the intracellular ManNAc-6-P is converted to fructose-6-P by the nagA and nagB gene products.
To confirm our conclusion that the manXYZ and nagBA genes are essential for ManNAc utilization, mutations in nagB, nagA, and manXYZ were transduced into the ama strain IBPC1017. The nagB::Km, nagA::Cm, and manXYZ::Tn9 mutations eliminated all growth on ManNAc. Reintroduction of the mlc+ allele (via cotransduction with the adjacent nth1::Km mutation) eliminated the rapid growth on ManNAc but still allowed slow background growth. The manA gene encodes phosphomannose isomerase, which is necessary for use of mannose as carbon source. The manA gene maps near mlc, and a chromosomal deletion, Δ1646(dgsA-manA) (25, 38), removes both mlc (=dgsA [30]) and manA. Introduction of this deletion into IBPC1017 produced bacteria which were, as expected, unable to grow on mannose but still capable of growth on ManNAc. These data show that (i) ManNAc is taken up by the mannose PTS and (ii) its degradation requires the nagBA gene products and hence proceeds via GlcNAc-6-P and GlcN-6-P, without involvement of mannose-6-P.
Growth on ManNAc induces nag gene expression.
Further evidence for the involvement of GlcNAc-6-P in the utilization of ManNAc comes from the observation that growth of the ManNAc++ strains on ManNAc induces the nag genes. Two ManNAc++ strains (IBPC1016 and IBPC1059) were lysogenized with bacteriophage λRS/EB, carrying a nagB-lacZ fusion. Levels of expression of the fusion were compared during growth on ManNAc and GlcNAc. Growth on ManNAc was clearly slower than growth on GlcNAc, but there was a strong increase in the expression of the nagB-lacZ fusion (Table 3). The inducer for the NagC repressor is GlcNAc-6-P (31), showing that growth on ManNAc produces amounts of GlcNAc-6-P that are sufficient to displace NagC from its operators.
TABLE 3.
Effects of ManNAc and sialic acid on expression of nagB-lacZ fusiona
| Strain | Genotype | β-Galactosidase activity (Miller units)c
|
|||
|---|---|---|---|---|---|
| ManNAcb | Sialic acid | GlcNAc | Glc | ||
| JM101 | Wild type | NT | 1,830 | 3,030 | 68 |
| 1012 | mlc | NT | 1,770 | 3,140 | 69 |
| 1016 | ama-6 mlc | 3,890 | 1,660 | 3,400 | 68 |
| 1059 | ama-7 mlc | 4,300 | 1,330 | 3,400 | 59 |
Bacteria were lysogenized with λRS/EB carrying a nagB-lacZ fusion.
Bacteria were grown in minimal MOPS medium with the carbon sources indicated at 30°C.
Measured at four points during exponential growth. Values are means of at least two independent cultures. NT, not tested.
ManNAc can supply amino sugars for cell wall components.
The glmS gene, encoding GlcN-6-P synthase, is essential for growth of E. coli in the absence of an exogenous supply of amino sugars for peptidoglycan and LPS biosynthesis. IBPC750 (derived from the mlc-1 strain IBPC5321) carries a null mutation in glmS (glmS::Tc) and cannot grow on minimal medium plates in the absence of GlcN or GlcNAc. On minimal ManNAc plates, this strain behaved like IBPC5321, producing papillae in the background growth, characteristic of the ama mutations. On subsequent purification, these colonies continued to grow well on ManNAc but failed to grow on minimal glucose plates. These observations strongly suggest that ManNAc can substitute for GlcN or GlcNAc in a glmS mlc strain and that use of ManNAc proceeds via GlcN-6-P since this is the substrate for the enzymes encoded by glmM and glmU, which convert GlcN-6-P to UDP-GlcNAc, the first committed compound for the cell wall components (22, 23).
Mapping the ama mutations.
To map the ama mutation in IBPC1001 (ama-1), we selected mini-Tn10cam transposons which eliminated the ability of this strain to grow rapidly on minimal ManNAc plates. One mini-Tn10cam isolated by this procedure was a candidate for linkage to ama-1 (strain IBPC1011 [Tn10cam11]). To confirm the linkage between ama and the transposon, P1 phage grown on IBPC1011 was used to transduce the Tn10cam11 marker to five other, independently isolated ManNAc++ (Ama−) strains. All Ama− strains tested (IBPC904, IBPC905, IBPC1016, IBPC1017, and IBPC1018) were transduced to Ama+ with cotransduction frequencies of 30 to 60%.
To locate other mini-Tn10 insertions, a second round of mini-Tn10 mutagenesis was undertaken with P1 grown on pools of mini-Tn10kan to screen for an insert which removed the Tn10cam11. Four such Tn10kan inserts were isolated and designated Tn10kan27, -28, -29, and -33 (Table 1). Tn10kan27, -28, and -29 were found to be about 50% contransducible with ama-7 and 90% cotransducible with Tn10cam11. The locations of three of the mini-Tn10 inserts on the chromosome were found by the method of inverse PCR amplification, after digestion with RsaI and circularization. Regions adjacent to the mini-Tn10 inserts on the amplified fragment were then sequenced with an internal primer (28). This located Tn10cam11 in gltD at 72.4 min and Tn10kan27 and -28 in the nearly adjacent unidentified open reading frame (ORF) yhcA at 72.45 min.
ama mutations map to the nanAT locus.
The nanAT gene encoding NANA lyase (nanA) and the NANA transporter (nanT) are located at 72.7 min, i.e., in the same region of the chromosome to which the ama+ mutations were mapped. It should be recalled that NANA is the condensation product of ManNAc and pyruvate. The sequence of the complete E. coli genome suggested that the nanAT operon also includes three other downstream genes, yhcJ, yhcI, and yhcH (encoding ORFs of 229, 302, and 154 amino acids). Upstream of nanAT there is another unidentified gene, yhcK (263 amino acids) (Fig. 2).
Plasmid pSX600 carries nanAT and several kilobases of downstream DNA (Fig. 2) (19). To determine whether the ama mutations mapped to the nan region, the effect of this plasmid was tested on the growth of a series of wild-type, mlc, and mlc ama strains (JM101, IBPC1012, IBPC1016, and IBPC1013) on ManNAc (Table 4). Interestingly, plasmid pSX600 did not affect the growth of the mlc ama (ManNAc++) strain, IBPC1016, on ManNAc but did allow mlc strains (IBPC1012 and IBPC1013) to grow somewhat better than the wild type on ManNAc. The plasmid seemed to eliminate the papillae and produced uniform slow growth on ManNAc. One possible explanation was that plasmid pSX600 was supplying a function which was also supplied by the ama mutation. Plasmids carrying the equivalent chromosomal region from Salmonella typhimurium (45) were found to have a similar effect on ManNAc growth in E. coli. In particular, a plasmid carrying just the 3′ half of the Salmonella nanT gene and downstream DNA, presumed to carry the three unidentified genes, also conferred good growth on ManNAc to mlc strains IBPC1012 and IBPC1013. It thus seemed possible that the function supplied by the ama mutations was also provided by a plasmid carrying the genes downstream of the nanAT operon. To confirm this result with E. coli DNA, two fragments were cloned from JM101 chromosomal DNA after amplification by PCR. One plasmid, pBR(Nan4-6), carried the three genes yhcJIH, while the second, pBR(Nan4-5), carried just yhcJ (Fig. 2). Both plasmids allowed rapid growth of mlc strains on ManNAc (Table 4).
TABLE 4.
Growth of mlc ama strains on ManNAc in the presence of different nan region plasmidsa
| Plasmid | Growthb
|
||||
|---|---|---|---|---|---|
| JM101 (wild type) | 1012 (mlc) | 1016 (mlc ama-6) | 1013 (mlc) | 1017 (mlc ama-7) | |
| pSX600 | + | ++ | +++ | ++ | NT |
| pBR/Nan4-6 | + | +++ | NT | +++ | NT |
| pBR/Nan4-5 | + | +++ | NT | +++ | NT |
| pBR/Nan1-3 | − | − | − | NT | − |
| pBR322 | + | +P | +++ | +P | +++ |
Bacteria were transformed with the plasmids indicated by selecting for ampicillin resistance on LB plates and then tested for growth on minimal ManNAc plates. Plates used for testing for repression by pBR(Nan1-3) contained 0.1 mg of ampicillin ml−1.
−, no growth; +, slow background growth; ++, medium uniform growth; +P, papillae (formation of isolated large colonies during slow background growth); +++, good growth on ManNAc; NT, not tested.
The ama-7 mutation enhances nanAT expression.
In the presence of an mlc mutation, either an ama mutation or the presence of yhcJ on a multicopy plasmid is sufficient for good growth on ManNAc. One possible explanation of the mode of action of the ama mutations is that they are regulatory mutations activating the expression of genes downstream of the nanAT operon. The high frequency of the ama mutations suggested that they could arise by insertion of a mobile genetic element either knocking out a repressor or supplying a promoter to induce expression of the downstream genes. A promoter with putative −35 and −10 sequences has been postulated to lie immediately upstream of nanA (29), and inspection of the DNA sequence suggests cotranscription of nanAT yhcJIH (Fig. 2). Further upstream is a gene (yhcK) with homology to a series of putative transcriptional regulators including the uxuR gene (the regulator of the uxu operon for the catabolism of uronic acids). On the basis of this homology, it is reasonable to think that YhcK may be involved in regulating the expression of the downstream nan operon. Chromosomal DNA from the ama mutants was screened for insertions of DNA in this region by PCR using oligonucleotides which flanked yhcK and the nanA promoter region (Fig. 2). Chromosomal DNA from IBPC1001 and IBPC1017, carrying the ama-1 and ama-7 mutations, respectively, was found to have an approximately 1.5-kb insertion in the region corresponding to the PCR fragment Nan1-3, whereas the Nan1-2 fragment, covering just the promoter region, was the same size as the wild type. The Nan1-3 fragment from the other four characterized ManNAc++ strains was of wild-type size. This result indicates that in two of the six ama strains, a DNA insertion had occurred in the region defined by oligonucleotides Nan2 and Nan3 and so was likely to inactivate yhcK.
If this insertion was increasing the expression of the nanAT operon, then it should have been measurable as an effect on sialic acid utilization or uptake in cells which had not been induced by growth on NANA. Transport assays were carried out on IBPC1017 (mlc::Tc ama-7) and the related strains Ymc (mlc+), IBPC1013, (mlc::Tc), and IBPC1047 (mlc+ ama-7). The presence of the ama-7 mutation, in either the presence or the absence of the mlc mutation, resulted in at least 20-fold-enhanced NANA uptake, whereas the mlc mutation had no effect on NANA uptake but did give a small increase in uptake of ManNAc, as expected from its role of enhancing manXYZ expression (Table 5). The relatively greater uptake of ManNAc in the mlc strain (IBPC1013) than the mlc ama-7 strain (IBPC1017) could be because there is some loss of label through subsequent metabolism of the ManNAc-6-P in the latter, ManNAc+ strain. The results of the uptake assays confirm our hypothesis that the mlc and ama-7 mutations increase ManNAc uptake and nan operon expression, respectively.
TABLE 5.
Effects of mlc and ama-7 mutations on transport of sialic acid and ManNAc
| Strain | Relevant genotype | Uptakea (nmol/A600)
|
|
|---|---|---|---|
| NANA | ManNAc | ||
| Ymc | mlc+ ama+ | <0.03 | 0.66 |
| IBPC1013 | mlcTc ama+ | <0.03 | 1.39 |
| IBPC1017 | mlc::Tc ama-7 | 0.57 | 1.00 |
| IBPC1047 | mlc+ ama-7 | 0.62 | 0.45 |
Measured on exponentially growing cells as described in Materials and Methods. Values are means of two independent cultures.
The yhcK gene product represses growth on ManNAc.
The yhcK gene, carried on PCR fragment Nan1-3, was cloned from JM101 and tested for an effect on the ama strains. The presence of pBR(Nan1-3) prevented growth of IBPC1016 and IBPC1017 on ManNAc (Table 4), and it, but not pBR322, also eliminated the high-level uptake of NANA in strains IBPC1017 and IBPC1047 (data not shown), showing that it could complement the ama mutation in trans. Furthermore, this plasmid eliminated, or at least drastically reduced, the formation of papillae when IBPC1012 harboring pBR(Nan1-3) was grown on ManNAc. The simplest interpretation of these results is that the ama mutations are predominately mutations in the putative Nan repressor gene, yhcK, which should be renamed nanR.
DISCUSSION
The pathway of ManNAc degradation in E. coli.
Unlike the two common amino sugars GlcN and GlcNAc, ManNAc seems not to be efficiently used as a carbon source by wild-type E. coli K-12. In contrast, experiments described here show that mlc ama double mutants of E. coli are capable of reasonably rapid growth on ManNAc, exhibiting doubling times of about 90 to 130 min at 37°C, depending on the genetic background. This range represents doubling times similar to that on GlcN (90 to 100 min in an mlc background) but greater than that on GlcNAc (50 min). We show here that the efficient utilization of ManNAc depends on the two mutations mlc and ama to activate the otherwise cryptic pathway.
The role of the mlc mutation would appear to be to increase the expression of the manXYZ-encoded transporter (Table 5). Mlc has been shown to be a repressor for this operon, and an mlc mutation enhances manXYZ expression threefold (30). The ManXYZ transporter shows a wide substrate specificity, transporting glucose, mannose, GlcN, and GlcNAc; it is thus not surprising that it can also transport ManNAc. Neither ptsG (PTS transporter of mannose and glucose) nor nagE (GlcNAc-specific transporter) can substitute for manXYZ. It had been established many years ago, by the work of Roseman’s laboratory, that ManNAc is a PTS sugar; it was one of the first sugars used to demonstrate the PTS (15), and mutations in the ptsH and ptsI genes eliminated this phosphorylation (39). The first purified enzyme II preparation was capable of phosphorylating ManNAc, suggesting that it was predominately the manXYZ-encoded complex (16). It is perhaps surprising that a threefold increase in manXYZ expression can produce such a significant increase in growth rate on ManNAc unless the metabolic equipose of this amino sugar is such that a small increase in uptake, coupled with derepression of nanE, is sufficient to stimulate ManNAc catabolism to an extent compatible with the observed growth of mlc ama double mutants. At the moment, we cannot say if the sole role of the mlc mutation in allowing growth on ManNAc is to enhance manXYZ expression or whether it also affects some other genes involved in ManNAc utilization. However, the results of the NANA transport assay in Table 5 seem to exclude a direct role of Mlc on nan operon expression.
The second mutation, ama, required for good growth on ManNAc maps to near the nanAT operon region. The ama mutations occur at high frequency in the mlc background, and all of the six mutations that have been studied have mapped to this same locus near nanAT. The ama mutation can be replaced by a plasmid carrying an ORF from downstream of the nanAT genes (yhcJ). Our interpretation of the data is that the frequently occurring ama mutations are regulatory mutations (either inactivating a repressor or providing a cis-acting promoter mutation) which allow the expression of nanAT and the previously uncharacterized downstream genes of this operon. This interpretation allows us to explain how plasmid-carried genes can replace the ama mutation: the plasmid is an alternative method of enhancing nan operon expression in the absence of either a regulatory mutation or the specific nan operon inducer, presumably sialic acid (47). The ama-7 mutation was shown to increase the expression of the nanT-encoded sialic acid transporter at least 20-fold. This same mutation was shown to be an insertion of 1.5 kb of DNA in the gene for the putative Nan transcriptional repressor, yhcK, which we propose to rename nanR. Plasmids carrying this ORF complemented the Ama− phenotype since they eliminated the good growth on ManNAc. Moreover, they prevented the appearance of all but a few papillae in the mlc strain IBPC1012, implying that the majority of the ama mutations are mutations inactivating the Nan repressor.
What is the function of the uncharacterized gene, yhcJ, whose product is necessary for ManNAc utilization? The experiments described above allow us to deduce the following pathway for ManNAc metabolism (Fig. 1). Transport of ManNAc by the manXYZ PTS transporter produces ManNAc-6-P. The requirement for the nagBA gene products strongly suggests that ManNAc-6-P is converted to GlcNAc-6-P, which is subsequently degraded to GlcN-6-P and then to fructose-6-P and NH3 via the nagA-encoded GlcNAc-6-P deacetylase and nagB-encoded GlcN-6-P deaminase, respectively. The missing step in the pathway for ManNAc degradation, and thus the function potentially supplied by the ama mutation or yhcJ on a plasmid, should be the conversion of GlcNAc-6-P to ManNAc-6-P. The alternative hypothesis that ManNAc-6-P is itself a substrate for the nagA-encoded deacetylase does not seem reasonable. GlcNAc is a C2-N-acetyl-substituted sugar, whereas its 2-epimer, ManNAc, has the other configuration of the N-acetyl-substituted sugar ring, which means that the acetyl group to be removed by the deacetylase is positioned on opposite sides of the molecule in the two sugars. It is unlikely that sugars with these two configurations can be positioned identically in the catalytic site of the deacetylase, and hence ManNAc-6-P should not be a substrate for NagA. Moreover strain IBPC529C, carrying a nagC mutation plus the mlc-1 mutation, grows like IBPC5321, producing papillae on ManNAc. Since the nagC mutation causes a 40-fold induction of the nagA-encoded deacetylase (31), this result indicates that GlcNAc-6-P deacetylase is incapable of degrading ManNAc-6-P. We conclude that the step for which no gene product is assigned for the utilization of ManNAc is the conversion of ManNAc-6-P to GlcNAc-6-P and that the yhcJ gene immediately downstream of the nanT is a candidate to supply this ManNAc-6-P epimerase function. The putative epimerase shows no significant homology with other proteins in the databases except an equivalent ORF (HI0145) in the nan-nag region of the Haemophilus influenzae chromosome and an uncharacterized ORF in the genome of Borrelia burgdorferi. The epimerase could thus be a member of a new family of enzymes.
Over 30 years ago, an enzyme with the function of converting GlcNAc-6-P to ManNAc-6-P was partially purified from Aerobacter cloacae, and the same activity was detected in other bacterial species, including E. coli B and K1 (10). Preliminary experiments show that extracts of strains with YhcJ overproduced from a multicopy plasmid have increased capacity to convert GlcNAc-6-P to ManNAc-6-P (45), and we propose that the yhcJ gene, encoding this epimerase function, be named nanE. It is also relevant to our current findings that the ManNAc epimerase activities which have been detected in mammalian tissues function on the free sugar and not the -6-phosphate derivative (5, 11, 20), whereas E. coli and presumably other prokaryotes lack this activity. In yeast, growth on ManNAc was shown to induce GlcN-6-P deaminase indicating that in this lower eukaryote, ManNAc is also converted to GlcN (2). Biswas et al. (2) also detected a ManNAc epimerase and GlcNAc kinase, suggesting that the pathway involved uptake of the free sugar, followed by its conversion to the GlcNAc epimer prior to its phosphorylation.
Implications for the metabolism of sialic acid.
The nan operon in E. coli potentially contains five genes: nanAT, the putative nanE epimerase gene, yhcJ (described above), plus two other downstream genes (Fig. 1). The first of these other genes, yhcI, encodes a protein homologous to NagC, Mlc, and other proteins of the so-called ROK (repressor, ORF, and kinase) family (42). This family consists of two classes of proteins: the transcriptional regulatory proteins exemplified by NagC and XylR, and the somewhat smaller proteins encoding sugar kinases which are missing the N-terminal helix-turn-helix DNA binding domain present in the transcription factors. The yhcI-encoded protein belongs to this latter class and would be expected to encode a sugar kinase. YhcI is also homologous to the ManNAc kinase domain of the bifunctional UDP-GlcNAc 2-epimerase/ManNAc kinase from mammalian liver (12, 41) and includes (from residues 15 to 30) one of two phosphate binding (ATPase) regions found in a range of hexose kinases. The nanA-encoded aldolase (NANA lyase) generates free intracellular ManNAc and pyruvate from sialic acid. It is tempting to speculate that the substrate for the putative yhcI-encoded kinase is this internally liberated ManNAc, thus generating ManNAc-6-P, the substrate of the epimerase predicted to be encoded by the upstream gene. We propose to name this gene nanK.
The existence of the putative genes for the ManNAc kinase and epimerase function within the nanAT operon allows the metabolic pathway for use of sialic acid to converge with that of ManNAc and GlcNAc at the common intermediate GlcNAc-6-P (Fig. 1). Evidence that the metabolism of sialic acid does pass via the pathway of GlcNAc utilization is that growth on sialic acid, like that of ManNAc, replaces the amino sugar requirement of glmS strains, requires the nagBA genes, and results in strong induction of the nagE and nagB operons as measured by a nagB-lacZ fusion (Table 3). Sialic acid use in viridians streptococci also induces the nag degradative genes (3). Byers et al. (3) proposed a pathway similar to that shown in Fig. 1 but could not distinguish between an epimerization of ManNAc to GlcNAc at the level of the free sugar or on the -6-P form. The experiments described here, plus the early work of Ghosh and Roseman (10), show that in E. coli and probably other bacteria, ManNAc-6-P is the substrate of the epimerase.
Other circumstantial evidence supporting our conclusion that the sialic acid and GlcNAc pathways overlap is that in H. influenzae, the nagBA genes are clustered with the nanA gene (encoding a putative NANA lyase) as well as homologues of the two genes we propose as the ManNAc kinase and epimerase genes, yhcI (HI0144) and yhcJ (HI0145). The gene order in H. influenzae (yhcJ, yhcI, HI0143, nanA, nagB, nagA) is somewhat different from that in E. coli (Fig. 2) but suggests a remarkable grouping of related functions. H. influenzae can grow on sialic acid but is unable to use GlcNAc as a carbon source (17), implying that the nagBA genes may be present solely for the degradation of the GlcNAc-6-P generated intracellularly from sialic acid (18).
The final ORF of the putative nan operon in E. coli (yhcH) shows no strong homology with any proteins in GenBank except an ORF in H. influenzae (HI0227), two ORFs (YigK and YiaL) in E. coli, and an ORF (ORF-1) in Streptococcus pneumoniae. The HI0227 gene is not located in the nan-nag region of H. influenzae. At the moment, we cannot propose any function for YhcH or its homologues.
E. coli seems to be fully equipped to deal with GlcNAc and sialic acid present in the environment, with separate genes for the transport and metabolism of the sugars. GlcNAc is expected to be found in both free-living and animal environments, whereas sialic acid would be present in the host (49). In contrast to the efficient use of these two sugars, ManNAc metabolism seems to rely on part of the sialic acid pathway which itself converges with the GlcNAc pathway. The fact that reasonable growth rates on ManNAc requires mutations in two regulatory loci to increase the expression of the transporter (mlc) and the epimerase (ama) suggests that E. coli is not specially adapted to use ManNAc per se but degrades this sugar only as part of the sialic acid dissimilatory pathway. The proposed pathway for sialic acid utilization requires that ManNAc, generated intracellularly from the action of the nanT and nanA gene products, be a substrate for phosphorylation by the nanK (yhcI) and epimerization by the nanE (yhcJ) gene products. Expression of the operon appears to be subject to negative control by a repressor encoded by the nanR (yhcK) gene located upstream of the operon. Previous results (47, 48) now suggest that sialic acid induces the nan operon by binding to NanR. Since NanR lacks obvious homology to other known sialic acid binding proteins, determining the exact interaction of NANA with the repressor should provide new insight into the structure and function of sialic acid recognition macromolecules. Work is in progress to confirm and extend these predictions through analyses of the nan-encoded proteins.
ACKNOWLEDGMENTS
We thank Valerie Heurgué-Hamard for the gift of strains and for advice on the mini-Tn10 mutagenesis and the inverse PCR procedure.
This work was supported by grants from the CNRS (to UPR9073) and unrestricted funds from the Department of Veterinary Pathobiology (to E.V.) and NIH grant RO1 AI42015 (to E.V.).
REFERENCES
- 1.Aisaka K, Igarashi A, Yamaguchi K, Uwajima T. Purification, crystallization and characterization of N-acetylneuraminate lyase from Escherichia coli. Biochem J. 1991;276:541–546. doi: 10.1042/bj2760541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biswas M, Singh B, Datta A. Induction of N-acetylmannosamine catabolic pathway in yeast. Biochim Biophys Acta. 1979;585:535–542. doi: 10.1016/0304-4165(79)90186-7. [DOI] [PubMed] [Google Scholar]
- 3.Byers H L, Homer K A, Beighton D. Utilization of sialic acid by viridians streptococci. J Dent Res. 1996;75:1564–1571. doi: 10.1177/00220345960750080701. [DOI] [PubMed] [Google Scholar]
- 4.Calcagno M, Campos P J, Mulliert G, Suastegui J. Purification, molecular and kinetic properties of glucosamine-6-phosphate isomerase (deaminase) from E. coli. Biochim Biophys Acta. 1984;787:165–173. doi: 10.1016/0167-4838(84)90076-1. [DOI] [PubMed] [Google Scholar]
- 5.Datta A. Regulatory role of adenosine triphosphate on hog kidney N-acetyl-d-glucosamine 2-epimerase. Biochemistry. 1970;9:3363–3370. doi: 10.1021/bi00819a011. [DOI] [PubMed] [Google Scholar]
- 6.Decker K, Plumbridge J, Boos W. Negative transcriptional regulation of a positive regulator: the expression of malT, encoding the transcriptional activator of the maltose regulon of Escherichia coli, is negatively controlled by Mlc. Mol Microbiol. 1998;27:381–390. doi: 10.1046/j.1365-2958.1998.00694.x. [DOI] [PubMed] [Google Scholar]
- 7.De Reuse H, Danchin A. The ptsH, ptsI, and crr genes of the Escherichia coli phosphoenolpyruvate-dependent phosphotransferase system: a complex operon with several modes of transcription. J Bacteriol. 1988;179:3827–3837. doi: 10.1128/jb.170.9.3827-3837.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dutka-Malen S, Mazodier P, Badet B. Molecular cloning and overexpression of the glucosamine synthetase gene from Escherichia coli. Biochimie. 1988;70:287–290. doi: 10.1016/0300-9084(88)90073-9. [DOI] [PubMed] [Google Scholar]
- 9.Erni B, Zanolari B. The mannose permease of the bacterial phosphotransferase system. Gene cloning and purification of the enzyme IIMan/IIIMan complex of Escherichia coli. J Biol Chem. 1985;260:15495–15503. [PubMed] [Google Scholar]
- 10.Ghosh S, Roseman S. The sialic acids. IV. N-Acyl-d-glucosamine 6-phosphate epimerase. J Biol Chem. 1965;240:1525–1530. [PubMed] [Google Scholar]
- 11.Ghosh S, Roseman S. The sialic acids. V. N-Acyl-d-glucosamine 2-epimerase. J Biol Chem. 1965;240:1531–1536. [PubMed] [Google Scholar]
- 12.Hinderlich S, Stäsche R, Zeitler R, Reutter W. A bifunctional enzyme catalyses the first two steps in N-acetylneuraminic acid biosynthesis of rat liver. Purification and characterization of UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase. J Biol Chem. 1997;272:24313–24318. doi: 10.1074/jbc.272.39.24313. [DOI] [PubMed] [Google Scholar]
- 13.Hosono K, Kakuda H, Ichihara S. Decreasing accumulation of acetate in rich medium by Escherichia coli on introduction of genes on a multicopy plasmid. Biosci Biotechnol Biochem. 1995;59:256–261. doi: 10.1271/bbb.59.256. [DOI] [PubMed] [Google Scholar]
- 14.Kleckner N, Bender J, Gottesman S. Uses of transposons with emphasis on Tn10. Methods Enzymol. 1991;204:139–180. doi: 10.1016/0076-6879(91)04009-d. [DOI] [PubMed] [Google Scholar]
- 15.Kundig W, Ghosh S, Roseman S. Phosphate bound to histidine in a protein as an intermediate in a novel phospho-transferase system. Proc Natl Acad Sci USA. 1964;52:1067–1074. doi: 10.1073/pnas.52.4.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kundig W, Roseman S. Sugar transport. Characterization of constitutive membrane-bound enzymes II of the Escherichia coli phosphotransferase system. J Biol Chem. 1971;246:1407–1418. [PubMed] [Google Scholar]
- 17.Macfadyen L P, Dorocicz I R, Reizer J, Saier M H, Redfield R J. Regulation of competence development and sugar utilization in Haemophilus influenzae Rd by a phosphoenolpyruvate: fructose phosphotransferase system. Mol Microbiol. 1996;21:941–952. doi: 10.1046/j.1365-2958.1996.441420.x. [DOI] [PubMed] [Google Scholar]
- 18.Macfadyen L P, Redfield R J. Life in mucous: sugar metabolism in Haemophilus influenzae. Res Microbiol. 1996;147:541–551. doi: 10.1016/0923-2508(96)84010-1. [DOI] [PubMed] [Google Scholar]
- 19.Martinez J, Steenbergen S, Vimr E. Derived structure of the putative sialic acid transporter from Escherichia coli predicts a novel permease domain. J Bacteriol. 1995;177:6005–6010. doi: 10.1128/jb.177.20.6005-6010.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maru I, Ohta Y, Murata K, Tsukada Y. Molecular cloning and identification of N-acyl-d-glucosamine-2-epimerase from porcine kidney as a renin-binding protein. J Biol Chem. 1996;271:16294–16299. doi: 10.1074/jbc.271.27.16294. [DOI] [PubMed] [Google Scholar]
- 21.Mengin-Lecreulx D, van Heijenoort J. Identification of the glmU gene encoding N-acetylglucosamine-1-phosphate uridyltransferase in Escherichia coli. J Bacteriol. 1993;175:6150–6157. doi: 10.1128/jb.175.19.6150-6157.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mengin-Lecreulx D, van Heijenoort J. Copurification of glucosamine-1-phosphate acetyltransferase and N-acetylglucosamine-1-phosphate uridyltransferase activities of Escherichia coli: characterization of the glmU gene product as a bifunctional enzyme catalyzing two subsequent steps in the pathway for UDP-N-acetylglucosamine synthesis. J Bacteriol. 1994;176:5788–5795. doi: 10.1128/jb.176.18.5788-5795.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mengin-Lecreulx D, van Heijenoort J. Characterization of the essential gene glmM encoding phosphoglucosamine mutase in Escherichia coli. J Biol Chem. 1996;271:32–39. doi: 10.1074/jbc.271.1.32. [DOI] [PubMed] [Google Scholar]
- 24.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 25.Morris P W, Binkley J P, Henson J M, Kuempel P L. Cloning and location of the dgsA gene of Escherichia coli. J Bacteriol. 1985;163:785–786. doi: 10.1128/jb.163.2.785-786.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mukhija S, Erni B. Purification by Ni2+ affinity chromatography and functional reconstitution of the transporter for N-acetylglucosamine of Escherichia coli. J Biol Chem. 1996;271:14819–14824. doi: 10.1074/jbc.271.25.14819. [DOI] [PubMed] [Google Scholar]
- 27.Neidhardt F C, Bloch P L, Smith D F. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ochman H, Medhora M M, Garza D, Hartl D L. Amplification of flanking sequences by inverse PCR. In: Innis M A, et al., editors. PCR protocols: a guide to methods and applications. New York, N.Y: Academic Press; 1990. pp. 219–227. [Google Scholar]
- 29.Ohta Y, Watanabe K, Kimura A. Complete nucleotide sequence of the E. coli N-acetylneuraminate lyase. Nucleic Acids Res. 1985;13:8843–8852. doi: 10.1093/nar/13.24.8843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plumbridge J. Control of the expression of the manXYZ operon in Escherichia coli: Mlc is a negative regulator of the mannose PTS. Mol Microbiol. 1998;27:369–381. doi: 10.1046/j.1365-2958.1998.00685.x. [DOI] [PubMed] [Google Scholar]
- 31.Plumbridge J A. Repression and induction of the nag regulon of Escherichia coli K12: the roles of nagC and nagA in maintenance of the uninduced state. Mol Microbiol. 1991;5:2053–2062. doi: 10.1111/j.1365-2958.1991.tb00828.x. [DOI] [PubMed] [Google Scholar]
- 32.Plumbridge J A. Co-ordinated regulation of aminosugar biosynthesis and degradation: the NagC repressor acts as an activator for the transcription of the glmUS operon and requires two separated NagC binding sites. EMBO J. 1995;14:3958–3965. doi: 10.1002/j.1460-2075.1995.tb00067.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plumbridge J A, Kolb A. DNA loop formation between Nag repressor molecules bound to its two operator sites is necessary for repression of the nag regulon of Escherichia coli in vivo. Mol Microbiol. 1993;10:973–981. doi: 10.1111/j.1365-2958.1993.tb00969.x. [DOI] [PubMed] [Google Scholar]
- 34.Plumbridge J A, Kolb A. Nag repressor-operator interactions: protein-DNA contacts cover more than two turns of the DNA helix. J Mol Biol. 1995;249:809–902. doi: 10.1006/jmbi.1995.0346. [DOI] [PubMed] [Google Scholar]
- 35.Raetz C R H. Bacterial lipopolysaccharides: a remarkable family of bioactive macroamphiphiles. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1035–1063. [Google Scholar]
- 36.Rick P D, Silver R P. Enterobacterial common antigen and capsular polysaccharrides. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 104–122. [Google Scholar]
- 37.Rodriguez-Aparicio L B, Reglero A, Luengo J M. Uptake of N-acetylneuraminic acid by Escherichia coli K-235. Biochemical characterization of the transport system. Biochem J. 1987;246:287–294. doi: 10.1042/bj2460287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roehl R A, Vinopal R T. Genetic locus, distant from ptsM, affecting enzyme IIA/IIB function in Escherichia coli K-12. J Bacteriol. 1980;142:120–130. doi: 10.1128/jb.142.1.120-130.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saier M H, Simoni R D, Roseman S. Sugar transport. Properties of mutant bacteria defective in proteins of the phosphoenolpyruvate phosphotransferase system. J Biol Chem. 1976;251:6584–6597. [PubMed] [Google Scholar]
- 40.Souza J-M, Plumbridge J A, Calcagno M L. N-Acetyl-d-glucosamine-6-phosphate deacetylase from Escherichia coli: purification and molecular and kinetic kinetic characterization. Arch Biochem Biophys. 1997;340:338–346. doi: 10.1006/abbi.1997.9780. [DOI] [PubMed] [Google Scholar]
- 41.Stäsche R, Hinderlich S, Weise C, Effertz K, Lucka L, Moorman P, Reutter W. A bifunctional enzyme catalyzes the first two steps in N-acetylneuraminic acid biosynthesis of rat liver. Molecular cloning and functional expression of UDP-N-acetyl-glucosamine 2-epimerase/N-acetylmannosamine kinase. J Biol Chem. 1997;272:24319–24324. doi: 10.1074/jbc.272.39.24319. [DOI] [PubMed] [Google Scholar]
- 42.Titgemeyer F, Reizer J, Reizer A, Saier M H. Evolutionary relationships between sugar kinases and transcriptional repressors in bacteria. Microbiology. 1994;140:2349–2354. doi: 10.1099/13500872-140-9-2349. [DOI] [PubMed] [Google Scholar]
- 43.Troy F A. Polysialylation from bacteria to brains. Glycobiology. 1992;2:5–23. doi: 10.1093/glycob/2.1.5. [DOI] [PubMed] [Google Scholar]
- 44.van Heijenoort J. Murein synthesis. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: ASM Press; 1996. pp. 1025–1034. [Google Scholar]
- 45.Vimr, E. Unpublished results.
- 46.Vimr E, Steenbergen S, Cieslewicz M. Biosynthesis of the polysialic acid capsule in Escherichia coli K1. J Ind Microbiol. 1995;15:352–360. doi: 10.1007/BF01569991. [DOI] [PubMed] [Google Scholar]
- 47.Vimr E R, Troy F A. Identification of an inducible catabolic system for sialic acids (nan) in Escherichia coli. J Bacteriol. 1985;164:845–853. doi: 10.1128/jb.164.2.845-853.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vimr E R, Troy F A. Regulation of sialic acid metabolism in Escherichia coli: role of N-acylneuraminate pyruvate lyase. J Bacteriol. 1985;164:854–860. doi: 10.1128/jb.164.2.854-860.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Warren L. Bound carbohydrates in nature. Cambridge, England: Cambridge University Press; 1994. [Google Scholar]
- 50.White R J. Control of aminosugar metabolism in Escherichia coli and isolation of mutants unable to degrade amino sugars. Biochem J. 1968;106:847–858. doi: 10.1042/bj1060847. [DOI] [PMC free article] [PubMed] [Google Scholar]