Abstract
Objective
The indocyanine green retention rate at 15 min (ICG-R15) is a useful tool to evaluate the functional liver reserve before hepatectomy for liver cancer. Taking ICG-R15 as criteria, we investigated the ability of a machine learning (ML)-based radiomics model produced by Gd-EOB-DTPA-enhanced hepatic magnetic resonance imaging (MRI) or contrast-enhanced computed tomography (CT) image in evaluating functional liver reserve of hepatocellular carcinoma (HCC) patients.
Methods
A total of 190 HCC patients with CT, among whom 112 also with MR, were retrospectively enrolled and randomly classified into a training dataset (CT: n = 133, MR: n = 78) and a test dataset (CT: n = 57, MR: n = 34). Then, radiomics features from Gd-EOB-DTPA MRI and CT images were extracted. The features associated with the ICG-R15 classification were selected. Five ML classifiers were used for the ML-model investigation. The accuracy (ACC) and the area under curve (AUC) of receiver operating characteristic (ROC) with 95% confidence intervals (CI) were utilized for ML-model performance evaluation.
Results
A total of 107 different radiomics features were extracted from MRI and CT, respectively. The features related to ICG-R15 which was classified into 10%, 20% and 30% were selected. In MRI groups, classifier XGBoost performed best with its AUC = 0.917 and ACC = 0.882 when the threshold was set as ICG-R15 = 10%. When ICG-R15 = 20%, classifier Random Forest performed best with AUC = 0.979 and ACC = 0.882. When ICG-R15 = 30%, classifier XGBoost performed best with AUC = 0.961 and ACC = 0.941. For CT groups, the classifier XGBoost performed best when ICG-R15 = 10% with AUC = 0.822 and ACC = 0.842. When ICG-R15 = 20%, classifier SVM performed best with AUC = 0.860 and ACC = 0.842. When ICG-R15 = 30%, classifier XGBoost performed best with AUC = 0.938 and ACC = 0.965.
Conclusions
Both the MRI- and CT-based machine learning models are proved to be valuable noninvasive methods for functional liver reserve evaluation.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12880-023-01050-1.
Keywords: Radiomics, Functional liver reserve, Machine learning, Gd-EOB-DTPA-enhanced hepatic MRI, Contrast-enhanced CT
Introduction
Primary liver cancer is the sixth most common cancer and is demonstrated to be the third contributing factor for global cancer death, showing about 906,000 new cases and 830,000 deaths in 2020 [1]. Among primary liver cancer cases, hepatocellular carcinoma (HCC) accounts for the most with about 75-85% [1]. Evidence has shown that Asia and Africa get the highest incidence of HCC in the world [2]. For HCC therapy, partial hepatectomy (PH) is still the optimal choice although most patients have reached an advanced stage because of insidious symptoms [3, 4]. However, it should be noticed that the post-hepatectomy liver failure (PHLF) is one of the important complications, and PHLF is the major cause of postoperative mortality. Normally the incidence of PHLF is 0.7-9.1% and can reach 58.22% when the major hepatectomy is performed [5, 6]. Thus the presurgical evaluation of functional liver reserve seems critical and necessary as the accurate evaluation can help reduce the risk of hepatectomy and avoid PHLF.
In clinical work, liver volumetry and scoring systems based on blood tests, such as ALB, AST, TBIL, and indocyanine green retention at 15 min retention rate (ICG-R15) are classic indexes used for the evaluation of functional liver reserve. Liver volumetry can be obtained by 3D reconstruction technology [7]. The scoring systems contain the MELD score, Child-Turcotte-Pugh (CTP) score, and Albumin-bilirubin (ALBI) grade. By comparing the above indexes, the ICG-R15 has its own advantages. Firstly, it can help doctors detect functional liver reserve abnormality earlier and more accurately. Secondly, it has also been proven to have a positive correlation to liver failure and morbidity after hepatectomy [8]. The ICG-R15 values of the patient in different intervals (threshold: 10%, 20%, 30%) can affect and guide the selection of surgical treatment methods [9]. Thus, the ICG clearance test was considered to be the optimal evaluation of preoperative function liver reserve [10].
Radiomics, which is one emerging methodology in medicine, can depict quantitative computerized algorithm-based features from traditional image materials, like CT or MRI images [11–13]. The medical images were proven related to clinical manifestations and the relation can be identified via machine learning (ML) approaches [14, 15]. In the past studies, the radiomics has been applied to investigating liver diseases, such as diagnosis, staging, liver tumor biological behaviors, and prognosis of primary liver cancer [16, 17]. Radiomics is promising to assist doctors in evaluating hepatic functional reserve, especially in conditions of the lack of ICG equipment in the hospital or other reasons that cause the failure of the ICG clearance test. However, the value of radiomics in evaluating functional liver reserve has been scarcely examined.
Under this direction, our work will evaluate whether the radiomics models derived from Gd-EOB-DTPA-enhanced hepatic MRI or contrast-enhanced CT can assess functional liver reserve and will further compare their performance with ICG-R15 in HCC patients.
Methods
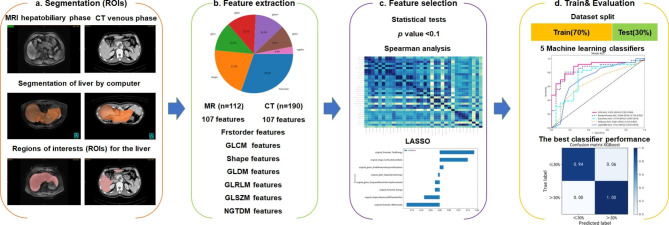
Flowchart
According to the flowchart of this work (Fig. 1), the Gd-EOB-DTPA-enhanced hepatic MRI (MRI) data from 112 patients and contrast-enhanced CT (CT) data from 190 patients were retrospectively collected. The hepatobiliary phase at 15 min of MRI and the portal venous phase of CT were selected. Firstly, the ROI liver region was segmented, and the features were extracted from the MRI and CT images. Next, the features were further screened through the P-value and correlation coefficient. For radiomic model development, the dataset was randomly divided into a training dataset and a test dataset. The training dataset aimed to train the model. Before training, the features associated with the label of ICG-R15 were screened by using the least absolute shrinkage and selection operator (LASSO) algorithm. Multiple ML algorithms were used for training on the training dataset. The training models based on the multiple ML algorithms will be further examined to classify the functional liver reserve on the test dataset. At last, the best model was selected to evaluate functional liver reserve classification.
Fig. 1.
Depiction of the whole procedure for developing and validation of the machine learning models
Patients
By reviewing the patients who were diagnosed with HCC in our hospital from May 2017 to April 2022, the inclusion criteria were set as follows: (1) all patients were diagnosed as HCC; (2) Gd-EOB-DTPA-enhanced hepatic MRI or contrast-enhanced CT in all phases was completed within one week before treatment or surgery; (3) ICG clearance test was completed within one week before treatment or surgery; (4) patients without jaundice during ICG clearance test [18]; (5) all patients had no history of previous liver surgery or radiofrequency ablation(RFA). A total of 190 patients were included in this study. All the 190 patients have CT data while only 112 cases of them have both MRI data. Details are listed in Table 1. The Ethics Committee of the Affiliated Hospital of Qingdao University approved this study with the ethical approval number QYFY-WZLL-27,465.
Table 1.
Demographics and preoperative data of patients
| Characteristics | Categories | Value | Number | ||
|---|---|---|---|---|---|
| MRI | CT | MRI | CT | ||
| dataset | Training dataset (70%) | 78 | 133 | ||
| Test dataset (30%) | 34 | 57 | |||
| Age (mean ± SD) | 58.62 ± 8.62 y | 59.42 ± 8.93 y | |||
| Gender | Female | 25 | 44 | ||
| Male | 87 | 146 | |||
| HBV infection | Yes | 104 | 168 | ||
| No | 8 | 22 | |||
| Liver cirrhosis | Yes | 75 | 122 | ||
| No | 37 | 68 | |||
| BMI (mean ± SE) | 25.02 ± 0.39 kg/m2 | 24.75 ± 0.27 kg/m2 | |||
| TBIL (mean ± SE) | 21.32 ± 1.60 µmol/L | 22.37 ± 1.10 µmol/L | |||
| ALB (mean ± SE) | 36.26 ± 0.66 g/L | 36.99 ± 0.43 g/L | |||
| ALT (mean ± SE) | 48.38 ± 10.21 IU/L | 49.06 ± 5.96 IU/L | |||
| PT (mean ± SE) | 11.34 ± 0.14s | 11.45 ± 0.11s | |||
| AST (mean ± SE) | 52.31 ± 6.81 IU/L | 56.08 ± 5.24 IU/L | |||
| GGT (mean ± SE) | 99.44 ± 16.15 IU/L | 108.12 ± 12.23 IU/L | |||
| ICG-R15 | ICG-R15 ≤ 10% vs. ICG-R15>10% | 62vs128 | 45vs67 | ||
| ICG-R15 ≤ 20% vs. ICG-R15>20% | 126vs64 | 78vs34 | |||
| ICG-R15 ≤ 30% vs. ICG-R15>30% | 160vs30 | 93vs19 | |||
ALT: alanine transaminase, BMI: Body Mass Index, HBV: Hepatitis B Virus; TBIL: total Bilirubin, ALB: albumin, PT: prothrombin time, AST: aspartate aminotransferase, GGT: gamma-glutamyltransferase, ICG-R15: indocyanine green retention rate at 15 min, y: years
ICG Clearance Test
After 6 h of fasting, the patient was in a supine position and injected 0.5 mg/kg ICG (Dandong Yichuang Pharmaceutical Co., Ltd., Liaoning, China) intravenously into a peripheral vein within 10 s. The ICG retention rate was measured with ICG pulse spectrophotometry (DDG 3300 K, Japan) after 15 min of injection. The ICG-R15 value was expressed as the percentage of ICG retention in serum 15 min after injection.
MRI and CT acquisition
The Gd-EOB-DTPA-enhanced hepatic MRI examination and the contrast-enhanced CT examination were conducted by a Siemens Skyra 3.0 T MRI scanner and a Siemens SOMATOM Definition Flash scanner, respectively. Scans were performed from the top of the liver to the pelvis. The MRI scanning parameters were selected as below: the repetitiontime was 3.9 ms, the echotime was 1.4 ms, the matrix was 320 × 256, the field of view was 400 mm × 400 mm and the slice thickness was 3 mm. The Gd-EOB-DTPA (Primovist, Bayer Schering Pharma AG, Berlin, Germany) was used as a contrast agent for enhanced MRI scanning, and the contrast agent (with flow rate of 2.0 ml/s and dose of 0.1ml/kg) was injected through elbow vein using a high-pressure syringe. Afterwards, 20 ml of saline is flushed. The arterial phase, portal venous phase, transitional phase and hepatobiliary phase were acquired at 30 s, 70 s, 180 s and 15 min, respectively. The CT scanning parameters were selected as below: the voltage was 120 kV, the current was 100–400 mA, the layer thickness was 5 mm, the layer spacing was 5 mm, the slice thickness was 1 mm and the matrix was 512 × 512. The iohexol (Yangtze River Pharmaceutical Group, Jiangsu, China) was used as a contrast agent for enhanced CT scanning. The contrast agent (with flow rate of 3.0 ml/s and dose of 1.5 ml/kg) was injected through the peripheral vein using a double-barreled high-pressure syringe. The arterial phase, portal venous phase and equilibrium phase were acquired at 30 s, 60 and 180 s, respectively.
Image segmentation
The computer-assisted surgery system (CAS) (CAS-Lv, Qingdao Hisense Medical Equipment Co., Ltd.) was used to segment the liver contour automatically from the hepatobiliary phase after 15 min of MRI and venous phase of the CT, to obtain the regions of interest (ROI) of the liver. For liver segmentation of MRI and CT, the Dice coefficient was more than 0.95 (this Dice coefficient is the manufacturer reference data of CAS-Lv). Each automatically segmented liver contour was visually inspected and any inaccurate liver contour was manually corrected by a doctor with over a decade of experience. All patients’ images and liver contours were saved as NII format files.
Radiomic feature extraction
Before feature extraction, the images are normalized to reduce the voxel spacing variation effect and are resampled with voxel sizes of 1 mm × 1 mm × 1 mm. Parameters are set as follows: normalizeScale: 1000, interpolations: sitkNearestNeighbor, binWidth: 5. For the type of normalization, we adapt the Min-Max normalization method to scale the pixel values of the image. We extracted two radiomics feature sets from CT and MRI, respectively. Each feature set contains 107 features and was split into seven different groups: (1) first-order statistics of voxel intensity features (n = 18), (2) shape features (n = 14), (3) gray level co-occurrence matrix (GLCM) features (n = 24), (4) gray level dependence matrix (GLDM) features (n = 14), (5) gray level run-length matrix(GLRLM) features (n = 16), (6) gray level size zone matrix (GLSZM) features (n = 16), and (7) neighboring gray tone difference matrix (NGTDM) features (n = 5). The feature extraction process is conducted automatically by using the PyRadiomics package (Python version 3.7). Each feature was named by image type, feature group, feature name and concatenated underlines. For example, original_firstorder_Skewness represents the feature ‘Skewness’ extracted from the original image and firstorder group.
Radiomic feature selection
At first, for each feature, statistical t-test was performed to evaluate differences between different groups. When the two-tailed p-value of the feature was p < 0.1 [19–21], we consider this feature was significantly different between groups and then was retained. Second, to reduce the collinearity of features, spearman correlation analysis was performed. When the correlation coefficient between two features was r>0.9, one feature was randomly retained. At last, the LASSO algorithm was used to reduce the unimportant features and select the features with non-zero coefficient values. The statistical tests, correlation analysis, and LASSO algorithm were implemented by importing the “scipy”, “numpy”, and “sklearn” packages in Python (version 3.7).
Model construction and performance evaluation
Supervised learning was used for training and prediction. More specifically, five ML algorithms were applied to investigate the performance of the model, whereas these classifiers were Support Vector Machines (SVM), Extra-Trees (ET), Random Forest (RF), Light Gradient Boosting Machine (LightGBM) and eXtreme Gradient Boosting (XGBoost). All selected features were used as input to classify the evaluation of functional liver reserve (ICG-R15 ≤ 10% vs. ICG-R15>10%, ICG-R15 ≤ 20% vs. ICG-R15>20%, and ICG-R15 ≤ 30% vs. ICG-R15>30% as 2-class classifier). All patients were randomly split into two cohorts. One was called the training dataset (70%) and the other was called the test dataset (30%). Each model was trained on the training set and then made predictions by using the test set. A total of five models were constructed and compared with each other to find the best performing model. The ROC curves were used to calculate the AUC value which can evaluate the predictive power of these models. The cut-off values of sensitivity and specificity corresponding to the maximum value of the Youden index were calculated. The final prediction results include AUC (95% CI), ACC, sensitivity and specificity. The AUC value was mainly used to evaluate the performance of classification models. We considered the model with the highest AUC as the best model. The detailed model reconstruction and results calculation were achieved with the aid of “pandas” and “sklearn” packages in Python (version 3.7). The detailed hyper-parameters for ML algorithm are shown in Table 2. Additional details of the models are shown in Supplementary S1.
Table 2.
The details of hyper-parameters for ML algorithm
| Num | Algorithms | Hyper-parameters |
|---|---|---|
| 1 | SVM | kernel=’rbf’, degree = 3, C = 1.0, probability = True |
| 2 | Random Forest | n_estimators = 10, max_depth = None, min_samples_split = 2 |
| 3 | Extra Trees | n_estimators = 10, max_depth = None, min_samples_split = 2 |
| 4 | XGBoost | n_estimators = 10, max_depth = 5 |
| 5 | LightGBM | n_estimators = 10, max_depth = 4 |
The definitions of ACC, sensitivity and specificity are as follows:
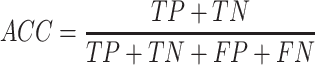 |
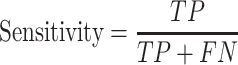 |
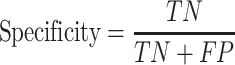 |
Where TP/TN is the true positive/negative value, FP/FN is the false positive/negative value.
Statistical analysis
We used the statistical t-test and Chi-square test to analyze the between-group differences in clinical baseline characteristics (shown in Table 1). Statistical significance was defined as a two-sided p-value < 0.05 (see Supplementary Table S1-S6). Referring to the previous studies [19, 20], we used the statistical t-test to analyze and select the radiomics features with significance to be p-value < 0.1. For features with high repeatability, correlation analysis was performed by Spearman correlation analysis. One of the two features was randomly retained when the correlation coefficient between the two was larger than 0.9. Features were further selected by the LASSO method and were finally used to construct the model. The performance of the classification model was mainly measured by the AUC. Statistical analyses were performed using the “One-key AI” platform (http://www.medai.icu/), which is based on Python (version 3.7).
Results
The features were selected by conducting statistical tests, spearman correlation analysis, and LASSO. And the final selected features and their corresponding LASSO coefficients derived from Gd-EOB-DTPA-enhanced hepatic MRI and contrast-enhanced CT are shown in Table 3.
Table 3.
Feature selection results and LASSO coefficient
| Data | ICG-R15 | Features (Training set) | Coefficient |
|---|---|---|---|
| MRI | ICG-R15 ≤ 10% vs. ICG-R15>10% | original_glszm_ZoneVariance | 0.105246 |
| original_shape_SurfaceVolumeRatio | 0.015642 | ||
| original_firstorder_Minimum | -0.018612 | ||
| original_glrlm_LongRunHighGrayLevelEmphasis | -0.063476 | ||
| original_shape_Sphericity | -0.118149 | ||
| ICG-R15 ≤ 20% vs. ICG-R15>20% | original_firstorder_Kurtosis | 0.111839 | |
| original_shape_SurfaceVolumeRatio | 0.083486 | ||
| original_glszm_ZoneVariance | 0.065786 | ||
| original_glszm_SmallAreaLowGrayLevelEmphasis | 0.063575 | ||
| original_glszm_LargeAreaHighGrayLevelEmphasis | 0.05589 | ||
| original_glcm_InverseVariance | 0.034796 | ||
| original_ngtdm_Contrast | 0.033712 | ||
| original_shape_Sphericity | -0.025289 | ||
| original_firstorder_Minimum | -0.100263 | ||
| ICG-R15 ≤ 30% vs. ICG-R15>30% | original_firstorder_Kurtosis | 0.099461 | |
| original_glszm_ZoneVariance | 0.055393 | ||
| original_glszm_LargeAreaHighGrayLevelEmphasis | 0.029222 | ||
| original_glszm_SmallAreaLowGrayLevelEmphasis | 0.003266 | ||
| original_glszm_SizeZoneNonUniformity | -0.000478 | ||
| original_glszm_SmallAreaEmphasis | -0.008009 | ||
| original_shape_LeastAxisLength | -0.038956 | ||
| CT | ICG-R15 ≤ 10% vs. ICG-R15>10% | original_glcm_Correlation | 0.061806 |
| original_glszm_ZoneEntropy | 0.030831 | ||
| original_firstorder_Energy | -0.096018 | ||
| original_glszm_SmallAreaEmphasis | -0.109115 | ||
| original_firstorder_RootMeanSquared | -0.126984 | ||
| ICG-R15 ≤ 20% vs. ICG-R15>20%* | original_firstorder_TotalEnergy | 0.397737 | |
| original_shape_SurfaceVolumeRatio | 0.316305 | ||
| original_firstorder_Kurtosis | 0.086403 | ||
| original_ngtdm_Strength | 0.074451 | ||
| original_shape_Maximum3DDiameter | -0.074731 | ||
| original_glcm_ClusterShade | -0.088621 | ||
| original_firstorder_Energy | -0.091037 | ||
| original_glszm_SmallAreaEmphasis | -0.112469 | ||
| original_shape_Maximum2DDiameterColumn | -0.12909 | ||
| original_firstorder_RootMeanSquared | -0.354511 | ||
| ICG-R15 ≤ 30% vs. ICG-R15>30% | original_shape_SurfaceVolumeRatio | 0.048541 | |
| original_glszm_SmallAreaLowGrayLevelEmphasis | 0.016216 | ||
| original_shape_Maximum2DDiameterSlice | -0.009696 | ||
| original_firstorder_Energy | -0.010984 | ||
| original_firstorder_90Percentile | -0.021431 |
* Top 10 features are listed in the table for the comparison between group ICG-R15 ≤ 20% and group ICG-R15>20% based on CT images
Under functional liver reserve thresholds (ICG-R15 = 10%, ICG-R15 = 20% and ICG-R15 = 30%), five ML algorithms were used to construct the model and were trained on the training dataset. The trained models were then used to predict the result on the test dataset. The detailed performance of the five models is described in Table 4. For MRI groups, more specifically, the classifier XGBoost achieves the highest performance when ICG-R15 = 10% is used as a threshold, with AUC = 0.917 (95% CI: 0.823–1.000) and ACC = 0.882. And the classifier Random Forest achieves the highest performance with AUC = 0.979 (95% CI: 0.941–1.000) and ACC = 0.882 at threshold ICG-R15 = 20%. For threshold ICG-R15 = 30%, the classifier XGBoost performs the best with AUC = 0.961 (95% CI: 0.890–1.000) and ACC = 0.941. Similar to the results for MRI groups, the classifier XGBoost for CT groups also achieves the best performance when threshold ICG-R15 = 10% (AUC = 0.822, 95% CI: 0.700–0.944, ACC = 0.842), and ICG-R15 = 30% (AUC = 0.938, 95% CI: 0.824–1.000, ACC = 0.965). Under threshold ICG-R15 = 20%, the classifier SVM is observed to perform the best with an AUC value of 0.860 (95% CI: 0.758–0.963) and ACC of 0.842. The detailed information of the best models is listed in Table 5.
Table 4.
Performance comparison among machine learning models
| Data | ICG-R15 | SVM | RF | ExtraTrees | XGBoost | LightGBM | |
|---|---|---|---|---|---|---|---|
| MRI | ICG-R15 ≤ 10% vs. ICG-R15>10% | Test set ACC | 0.824 | 0.765 | 0.824 | 0.853 | 0.794 |
| Test set AUC (95%CI) | 0.802(0.639–0.965) | 0.839(0.703–0.974) | 0.873(0.743–1.000) | 0.899(0.784–1.000) | 0.806(0.650–0.962) | ||
| ICG-R15 ≤ 20% vs. ICG-R15>20% | Test set ACC | 0.824 | 0.882 | 0.735 | 0.824 | 0.824 | |
| Test set AUC (95%CI) | 0.893(0.780–1.000) | 0.979(0.941–1.000) | 0.878(0.739–1.000) | 0.946(0.866–1.000) | 0.833(0.632–1.000) | ||
| ICG-R15 ≤ 30% vs. ICG-R15>30% | Test set ACC | 0.882 | 0.618 | 0.882 | 0.941 | 0.794 | |
| Test set AUC (95%CI) | 0.922(0.802–1.000) | 0.789(0.481–1.000) | 0.945(0.866–1.000) | 0.961(0.890–1.000) | 0.891(0.743–1.000) | ||
| CT | ICG-R15 ≤ 10% vs. ICG-R15>10% | Test set ACC | 0.772 | 0.632 | 0.667 | 0.842 | 0.702 |
| Test set AUC (95%CI) | 0.734(0.590–0.879) | 0.661(0.514–0.807) | 0.723(0.576–0.870) | 0.822(0.700–0.944) | 0.741(0.610–0.872) | ||
| ICG-R15 ≤ 20% vs. ICG-R15>20% | Test set ACC | 0.842 | 0.667 | 0.702 | 0.684 | 0.684 | |
| Test set AUC (95%CI) | 0.860(0.758–0.963) | 0.722(0.591–0.853) | 0.634(0.478–0.789) | 0.709(0.570–0.847) | 0.692(0.552–0.832) | ||
| ICG-R15 ≤ 30% vs. ICG-R15>30% | Test set ACC | 0.982 | 0.912 | 0.807 | 0.965 | 0.982 | |
| Test set AUC (95%CI) | 0.865(0.600–1.000) | 0.871(0.683–1.000) | 0.783(0.471–1.000) | 0.938(0.824–1.000) | 0.925(0.776–1.000) |
The performance of the best model is in boldface
Table 5.
Performance of the best MRI- and CT-based machine learning classification model
| Data | ICG-R15 | Cohort | AUC (95%CI) | Accuracy | Sensitivity | Specificity | model |
|---|---|---|---|---|---|---|---|
| MRI | ICG-R15 ≤ 10% vs. ICG-R15>10% | Training | 0.996(0.989–1.000) | 0.987 | 0.980 | 1.000 | XGBoost |
| Test | 0.899(0.784–1.000) | 0.853 | 0.875 | 0.833 | XGBoost | ||
| ICG-R15 ≤ 20% vs. ICG-R15>20% | Training | 0.995(0.986–1.000) | 0.962 | 0.929 | 0.980 | Random Forest | |
| Test | 0.979(0.941–1.000) | 0.882 | 1.000 | 0.857 | Random Forest | ||
| ICG-R15 ≤ 30% vs. ICG-R15>30% | Training | 0.997(0.991–1.000) | 0.962 | 1.000 | 0.951 | XGBoost | |
| Test | 0.961(0.890–1.000) | 0.941 | 1.000 | 0.968 | XGBoost | ||
| CT | ICG-R15 ≤ 10% vs. ICG-R15>10% | Training | 0.998(0.995–1.000) | 0.970 | 0.957 | 1.000 | XGBoost |
| Test | 0.822(0.700–0.944) | 0.842 | 0.917 | 0.714 | XGBoost | ||
| ICG-R15 ≤ 20% vs. ICG-R15>20% | Training | 0.866(0.781–0.951) | 0.842 | 0.872 | 0.830 | SVM | |
| Test | 0.860(0.758–0.963) | 0.842 | 0.840 | 0.844 | SVM | ||
| ICG-R15 ≤ 30% vs. ICG-R15>30% | Training | 0.997(0.991–1.000) | 0.992 | 1.000 | 0.991 | XGBoost | |
| Test | 0.938(0.824–1.000) | 0.965 | 0.800 | 0.981 | XGBoost |
ICG-R15: indocyanine green retention rate at 15 min, AUC: Area under the ROI curve, ACC: Accuracy
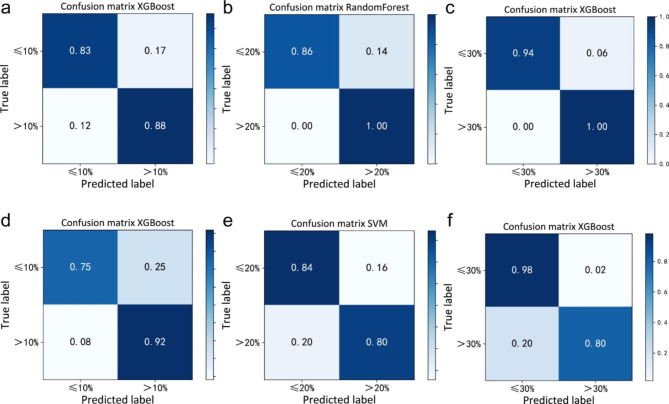
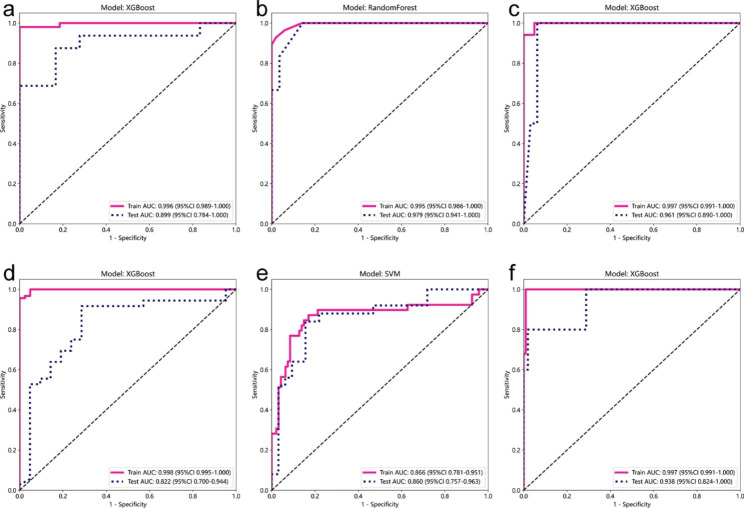
The model confusion matrices and ROC curves are shown in Figs. 2 and 3, respectively. All AUC values are greater than 0.89 for the test dataset from MRI and are greater than 0.82 for the test dataset from CT. The results indicate that both MRI-based and CT-based ML models can achieve the goal of classification in distinguishing the different values of ICG-R15 to some extent, which is promising to become an additional method to predict the functional liver reserve.
Fig. 2.
Based upon Gd-EOB-DTPA-enhanced hepatic MRI, a XGBoost Confusion matrix when ICG-R15 = 10% was selected as a threshold; b Random Forest Confusion matrix when ICG-R15 = 20% was selected as a threshold; c XGBoost Confusion matrix when ICG-R15 = 30% was selected as a threshold. Based upon contrast-enhanced CT, d XGBoost Confusion matrix when ICG-R15 = 10% was selected as a threshold; e SVM Confusion matrix when ICG-R15 = 20% was selected as a threshold; f XGBoost Confusion matrix when ICG-R15 = 30% was selected as a threshold
Fig. 3.
Based upon Gd-EOB-DTPA-enhanced hepatic MRI, a XGBoost ROC curve when ICG-R15 = 10% was selected as a threshold; b Random Forest ROC curve when ICG-R15 = 20% was selected as a threshold; c XGBoost ROC curve when ICG-R15 = 30% was selected as a threshold. Based upon contrast-enhanced CT, d XGBoost ROC curve when ICG-R15 = 10% was selected as a threshold; e SVM ROC curve when ICG-R15 = 20% was selected as a threshold; f XGBoost ROC curve when ICG-R15 = 30% was selected as a threshold
Discussion
Radiomics has shown great value in the diagnosis and therapy of multiple diseases. We consider whether it is possible to use the radiomics method to perform an accurate assessment of functional liver reserve based on Gd-EOB-DTPA-enhanced hepatic MRI and contrast-enhanced CT in HCC patients. Under this perspective, MRI-based and CT-based ML models are developed and validated for distinguishing patients with functional liver reserves of different states. Our results demonstrate that both MRI-based and CT-based models worked satisfactorily in the aspect of the assessment of functional liver reserve.
Recently, there has been an increasing application of imaging techniques in measuring the hepatic function in HCC patients, because it can provide more significant clinical information than an overall assessment [22]. For example, previous studies based on medical image analysis have demonstrated that liver has regional differences in hepatic parenchymal abnormalities [23, 24]. One recent work from Zhaoqi Shi et al. shows that radiomics analysis can be applied in the preoperative assessment of functional liver reserve in HCC patients [25]. However, this research only focused on Gd-EOB-DTPA-enhanced hepatic MRI to predict the ICG classification value to evaluate liver function in HCC patients, and the functional liver reserve thresholds are set to be ICG-R15 = 10%, ICG-R15 = 15%, and ICG-R15 = 20%. The analysis of the CT images is not mentioned. In our work, considering that the contrast-enhanced CT is regarded as the most common modality for patients with HCC, we add CT images to assess functional liver reserve. And we set functional liver reserve thresholds to be ICG-R15 = 10%, ICG-R15 = 20%, and ICG-R15 = 30% according to the requirement of clinical surgical strategies. By referring to the criteria of safe hepatic resection proposed by Makuuchi [9], if ICG-R15<10%, trisegmentectomy and bisegmentectomy of the liver can be performed; If 10%≤ICG-R15<20%, left lobictomy right monosegmentectomy of the liver can be performed; If 20%≤ICG-R15<30%, subsegmentectomy of Couinaud of the liver can be performed; If ICG-R15 ≥ 30%, it is necessary to limit resection or enucleation liver for transplantation [9]. Compared with Zhaoqi’s work, our thresholds are larger in general and are in accordance with the mentioned Makuuchi criteria for safe hepatic resection, which can provide a better reference for clinical surgery. By developing the radiomics model, we could assist doctors to evaluate functional liver reserve in the hospital without ICG equipment.
In our work, Gd-EOB-DTPA-enhanced hepatic MRI and contrast-enhanced CT are selected as data for evaluating presurgical liver function. The hepatobiliary phase after 15 min from Gd-EOB-DTPA-enhanced hepatic MRI is further selected. During hepatobiliary phase, HCCs appear hypointense in Gd-EOB-DTPA-enhanced images because of contrast medium discharges into hepatocytes and bile ducts. Hence, the diagnostic sensitivity and specificity of HCC are dramatically improved [26]. The obtained signals during Gd-EOB-DTPA-enhanced MRI imaging can describe anatomical characteristics of liver and hepatocyte-specific function [26]. And the effectiveness of Gd-EOB-DTPA-enhanced hepatic MRI for evaluating liver function has been evaluated in several works [27]. Besides, for contrast-enhanced CT, we select the portal venous phase to evaluate the presurgical liver function. Intense contrast uptake in the arterial phase was conducted before extracellular contrast washout in portal venous and/or delayed phases [28]. During the portal venous phase, owing to the washout, the CT value of HCCs was lower than liver parenchyma which is beneficial to observe the radiomics features about liver function. Washout here means a relative decrease in the extracellular contrast to the background of the portal venous phase and/or delayed phases [29–32].
In our research, the features in different threshold groups were selected based on shape, voxel intensity, and texture to predict classification. The developed disaggregated model was too complex for clinical practice since it comprises many features. Further research is required to simplify the features and thus we used five classifiers for modeling. Among all classifiers, XGBoost was demonstrated to perform best. The XGBoost is an improved model based on the gradient boosted decision tree (GBDT). It is an ensemble learning method that combines the predictions of multiple weak models to produce a stronger prediction [33]. The XGBoost uses both LASSO and Ridge Regression regularization to penalize the highly complex model and also uses built-in cross-validation to help the algorithm prevent overfitting. The XGBoost has become one of the widely used ML algorithms due to its state-of-the-art performance in many ML tasks, such as classification and regression [34].
However, several limitations should be mentioned in our study. Firstly, the provided research images were obtained from our center, more patients are needed in order to achieve external cohort validation. The baseline characteristics and features from a single center may not conform to the population. Secondly, the analysis of functional liver reserve under specific thresholds is a 2-class classification problem, which cannot cover all of the clinically significant ICG-R15 value intervals mentioned in the Makuuchi criteria for safe hepatic resection [9]. Thirdly, regarding the fact that some patients had CT scans but no MRI scans, we did not make the multimodality evaluation with CT and MRI. In the future, the classification with massive patients and multi-center data will be investigated by different ICG value intervals and multi-class classification methods. The multimodality evaluation with CT, MRI and clinical data will be a focus for our follow-up studies.
Conclusion
Both MRI-based and CT-based ML models are shown to achieve the goal of classification in distinguishing the different values of ICG-R15 to some extent, which are proved to be valuable methods for functional liver reserve evaluation. Among the five classifiers, XGBoost was demonstrated to perform best. Our work provides valuable insights, which can help clinicians to construct an effective prediction model and develop personalized precision treatment strategies.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank all members for constructive advice in manuscript preparation and writing in Shandong Key Laboratory of Digital Medicine and Computer Assisted Surgery, Affiliated Hospital of Qingdao University.
Abbreviations
- ACC
Accuracy
- AST
Aspartate aminotransferase
- ALT
Alanine transaminase
- PT
Prothrombin time
- CT
Computed tomography
- CI
Confidence intervals
- MRI
Magnetic resonance imaging
- HCC
Hepatocellular carcinoma
- RFA
Radiofrequency ablation
- ET
Extra-Trees
- Gd-EOB-DTPA
Gadolinium ethoxybenzyl dimeglumine
- ML
Machine learning
- ICG-R15
Indocyanine green retention rate at 15 min
- ROI
Regions of interest
- RF
Random Forest
- AUC
Area under the ROI curve
- ROC
Receiver operating characteristic
- BMI
Body Mass Index
- HBV
Hepatitis B Virus
- ALB
Albumin
- TBIL
Total Bilirubin
- GGT
Gamma-glutamyltransferase
- SVM
Support Vector Machines
- LASSO
Least absolute shrinkage and selection operator
- XGBoost
eXtreme Gradient Boosting
- LightGBM
Light Gradient Boosting Machine
Author contributions
LZ, QD and CZ devoted themselves to conception and design. LZ, FW, JC and ZL organized the database, LZ wrote the draft of the manuscript, XC, QD, NX and CZ contributed to review and revision. All authors mentioned made a substantial and intellectual contribution to this study directly and approved its publication.
Funding
Our study was funded by the Natural Science Foundation of Shandong Province (grant number ZR2021MH171), Shandong Higher Education Young Science and Technology Support Program (grant number 2020KJL005), and Taishan Scholars Program of Shandong Province (grant number 2019010668).
Data Availability
The data and materials in this study are available through the corresponding authors with permission of the Affiliated Hospital of Qingdao University.
Declarations
Ethics approval and consent to participate
This study was approved by the Ethics Committee of the Affiliated Hospital of Qingdao University. The approval number of the Ethics Review was QYFY-WZLL-27465. The need for informed consent was waived by the Ethics Committee of the Affiliated Hospital of Qingdao University due to its retrospective nature. The methods of this study were carried out in the case of meeting relevant guidelines and regulations.
Consent for publication
Not applicable.
Competing interests
The authors declare that their study was performed without any financial or commercial relation that may be interpreted as a potential conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and Mortality Worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.GLOBOCAN. International Agency for Research on Cancer. Published 2020. Cited 2021. http://gco.iarc.fr/today/home.
- 3.Llovet JM, Zucman-Rossi J, Pikarsky E, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2:16018. doi: 10.1038/nrdp.2016.18. [DOI] [PubMed] [Google Scholar]
- 4.Fu J, Wang H. Precision diagnosis and treatment of liver cancer in China. Cancer Lett. 2018;412:283–88. doi: 10.1016/j.canlet.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 5.van den Broek MA, Olde Damink SW, Dejong CH, et al. Liver failure after partial hepatic resection: definition, pathophysiology, risk factors and treatment. Liver Int. 2008;28(6):767–80. doi: 10.1111/j.1478-3231.2008.01777.x. [DOI] [PubMed] [Google Scholar]
- 6.Li B, Qin Y, Qiu Z, Ji J, Jiang X. A cohort study of hepatectomy-related complications and prediction model for postoperative liver failure after major liver resection in 1,441 patients without obstructive jaundice. Ann Transl Med. 2021;9(4):305. doi: 10.21037/atm-20-5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takamoto T, Ban D, Nara S, et al. Automated Three-Dimensional Liver Reconstruction with Artificial Intelligence for virtual hepatectomy. J Gastrointest Surg. 2022;26(10):2119–27. doi: 10.1007/s11605-022-05415-9. [DOI] [PubMed] [Google Scholar]
- 8.Greco E, Nanji S, Bromberg IL, et al. Predictors of peri-opertative morbidity and liver dysfunction after hepatic resection in patients with chronic liver disease. HPB (Oxford) 2011;13(8):559–65. doi: 10.1111/j.1477-2574.2011.00329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Makuuchi M, Kosuge T, Takayama T, et al. Surgery for small liver cancers. Semin Surg Oncol. 1993;9(4):298–304. doi: 10.1002/ssu.2980090404. [DOI] [PubMed] [Google Scholar]
- 10.De Gasperi A, Mazza E, Prosperi M. Indocyanine green kinetics to assess liver function: ready for a clinical dynamic assessment in major liver surgery? World J Hepatol. 2016;8(7):355–67. doi: 10.4254/wjh.v8.i7.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi JY. Radiomics and Deep Learning in Clinical Imaging: what should we do? Nucl Med Mol Imaging. 2018;52(2):89–90. doi: 10.1007/s13139-018-0514-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peeken JC, Nüsslin F, Combs SE. Radio-oncomics”: the potential of radiomics in radiation oncology. Strahlenther Onkol. 2017;193(10):767–79. doi: 10.1007/s00066-017-1175-0. [DOI] [PubMed] [Google Scholar]
- 13.Vial A, Stirling D, Field M, et al. The role of deep learning and radiomic feature extraction in cancer-specific predictive modelling: a review. Transl Cancer Res. 2018;7(3):803–16. doi: 10.21037/tcr.2018.05.02. [DOI] [Google Scholar]
- 14.Cook TS. The importance of Imaging Informatics and Informaticists in the implementation of AI. Acad Radiol. 2020;27(1):113–16. doi: 10.1016/j.acra.2019.10.002. [DOI] [PubMed] [Google Scholar]
- 15.Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14(12):749–62. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 16.Wei J, Jiang H, Gu D, et al. Radiomics in liver diseases: current progress and future opportunities. Liver Int. 2020;40(9):2050–63. doi: 10.1111/liv.14555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen C, Chen C, Ma M, et al. Classification of multi-differentiated liver cancer pathological images based on deep learning attention mechanism. BMC Med Inform Decis Mak. 2022;22(1):176. doi: 10.1186/s12911-022-01919-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seyama Y, Kokudo N. Assessment of liver function for safe hepatic resection. Hepatol Res. 2009;39(2):107–16. doi: 10.1111/j.1872-034X.2008.00441.x. [DOI] [PubMed] [Google Scholar]
- 19.Aron A, Aron E, Coups E. Statistics for Psychology (6rd ed.). 2023;179–202.
- 20.Zhang Y, Zhang B, Liang F, et al. Radiomics features on non-contrast-enhanced CT scan can precisely classify AVM-related hematomas from other spontaneous intraparenchymal hematoma types. Eur Radiol. 2019;29(4):2157–65. doi: 10.1007/s00330-018-5747-x. [DOI] [PubMed] [Google Scholar]
- 21.Ragin C, Edwards R, Larkins-Pettigrew M, et al. Oral HPV infection and sexuality: a cross-sectional study in women. Int J Mol Sci. 2011;12(6):3928–40. doi: 10.3390/ijms12063928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geisel D, Lüdemann L, Hamm B, et al. Imaging-based liver function Tests–Past, Present and Future. Rofo. 2015;187(10):863–71. doi: 10.1055/s-0035-1553306. [DOI] [PubMed] [Google Scholar]
- 23.Kwon AH, Matsui Y, Ha-Kawa SK, et al. Functional hepatic volume measured by technetium-99m-galactosyl-human serum albumin liver scintigraphy: comparison between hepatocyte volume and liver volume by computed tomography. Am J Gastroenterol. 2001;96(2):541–6. doi: 10.1111/j.1572-0241.2001.03556.x. [DOI] [PubMed] [Google Scholar]
- 24.Ratziu V, Charlotte F, Heurtier A, et al. Sampling variability of liver biopsy in nonalcoholic fatty liver disease. Gastroenterology. 2005;128(7):1898–906. doi: 10.1053/j.gastro.2005.03.084. [DOI] [PubMed] [Google Scholar]
- 25.Shi Z, Cai W, Feng X, et al. Radiomics Analysis of Gd-EOB-DTPA enhanced hepatic MRI for Assessment of Functional Liver Reserve. Acad Radiol. 2022;29(2):213–8. doi: 10.1016/j.acra.2021.04.019. [DOI] [PubMed] [Google Scholar]
- 26.Van Beers BE, Pastor CM, Hussain HK. Primovist, Eovist: what to expect? J Hepatol. 2012;57(2):421–9. doi: 10.1016/j.jhep.2012.01.031. [DOI] [PubMed] [Google Scholar]
- 27.Bae KE, Kim SY, Lee SS, et al. Assessment of hepatic function with Gd-EOB-DTPA-enhanced hepatic MRI. Dig Dis. 2012;30(6):617–22. doi: 10.1159/000343092. [DOI] [PubMed] [Google Scholar]
- 28.Ayuso C, Rimola J, Vilana R, et al. Diagnosis and staging of hepatocellular carcinoma (HCC): current guidelines. Eur J Radiol. 2018;101:72–81. doi: 10.1016/j.ejrad.2018.01.025. [DOI] [PubMed] [Google Scholar]
- 29.Elsayes KM, Hooker JC, Agrons MM, et al. 2017 version of LI-RADS for CT and MRI imaging: an update. Radiographics. 2017;37(7):1994–2017. doi: 10.1148/rg.2017170098. [DOI] [PubMed] [Google Scholar]
- 30.Tang A, Bashir MRI, Corwin MT, et al. Evidence supporting LI-RADS major features for CT- and MRI imaging-based diagnosis of Hepatocellular Carcinoma: a systematic review. Radiology. 2018;286(1):29–48. doi: 10.1148/radiol.2017170554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi JY, Lee JM, Sirlin CB. CT and MRI imaging diagnosis and staging of hepatocellular carcinoma: part II. Extracellular agents, hepatobiliary agents, and ancillary imaging features. Radiology. 2014;273(1):30–50. doi: 10.1148/radiol.14132362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoon JH, Park JW, Lee JM. Noninvasive diagnosis of Hepatocellular Carcinoma: Elaboration on Korean Liver Cancer Study Group-National Cancer Center Korea Practice Guidelines compared with other guidelines and remaining issues. Korean J Radiol. 2016;17(1):7–24. doi: 10.3348/kjr.2016.17.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen T, Guestrin C, XGBoost:. A Scalable Tree Boosting System. ACM, 2016.
- 34.Wang C, Guo J. A data-driven framework for learners’ cognitive load detection using ECG-PPG physiological feature fusion and XGBoost classification. Procedia Comput Sci. 2019;147:338–48. doi: 10.1016/j.procs.2019.01.234. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data and materials in this study are available through the corresponding authors with permission of the Affiliated Hospital of Qingdao University.