ABSTRACT
The emergence of carbapenem-resistant, hypervirulent Klebsiella pneumoniae is a new threat to health care. We studied the molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae isolates in Qatar using whole-genome sequence data. We also characterized the prevalence and genetic basis of hypervirulent phenotypes and established the virulence potential using a Galleria mellonella model. Of 100 Klebsiella isolates studied, NDM and OXA-48 were the most common carbapenemases. Core genome single-nucleotide polymorphism (SNP) analysis indicated the presence of diverse sequence types and clonal lineages; isolates belonging to Klebsiella quasipneumoniae subsp. quasipneumoniae sequence type 196 (ST196) and ST1416 may be disseminated among several health care centers. Ten K. pneumoniae isolates carried rmpA and/or truncated rmpA2, and 2 isolates belonged to KL2, indicating low prevalence of classical hypervirulent isolates. Isolates carrying both carbapenem resistance and hypervirulence genes were confined mainly to ST231 and ST383 isolates. One ST383 isolate was further investigated by MinION sequencing, and the assembled genome indicated that blaNDM was located on an IncHI1B-type plasmid (pFQ61_ST383_NDM-5) which coharbored several virulence factors, including the regulator of the mucoid phenotype (rmpA), the regulator of mucoid phenotype 2 (rmpA2), and aerobactin (iucABCD and iutA), likely resulting from recombination events. Comparative genomics indicated that this hybrid plasmid may be present in two additional Qatari ST383 isolates. Carbapenem-resistant, hypervirulent K. pneumoniae ST383 isolates pose an emerging threat to global health due to their simultaneous hypervirulence and multidrug resistance.
KEYWORDS: carbapenem resistance, genomics, molecular epidemiology, virulence, hybrid plasmid, Klebsiella
INTRODUCTION
Klebsiella pneumoniae is a Gram-negative bacterial pathogen that is widely present in nature and in the human intestine. K. pneumoniae is well known to cause hospital-acquired infections in immunocompromised patients (1, 2), but infections caused by K. pneumoniae can also occur in long-term-care facilities, such as nursing homes, and in the community. Types of infection vary and include hospital-acquired pneumonia, lung abscesses, bloodstream infections, catheter-related infections, wound or surgical site infections, upper and lower urinary tract infections, liver abscesses, and meningitis (1). Based on the genome sequencing data, various related species and subspecies, such as K. aerogenes, K. oxytoca, K. quasipneumoniae, and K. variicola, have been recognized (3–5). In our previous study, in which we had studied 149 carbapenem-resistant Enterobacterales (CRE) isolates in Qatar, K. pneumoniae (54%) and K. quasipneumoniae (16%) isolates were prevalent (6).
CRE infections are a global health priority and are among the most serious antimicrobial resistance (AMR) threats (7). Carbapenem resistance in K. pneumoniae is primarily driven by production of carbapenemases, with extended-spectrum β-lactamases (ESBL) such as CTX-M-2 playing a supplementary role in hydrolyzing cephalosporins in combination with decreased membrane permeability in the cell wall (6, 8–11). In the aforementioned study in Qatar, genes encoding metallo-β-lactamases were detected in 45.8% of the isolates and OXA-48-like enzymes in 40.3% (6).
Hypervirulent K. pneumoniae (hvKp) can cause serious life-threatening infections, such as liver abscesses, and is associated with high mortality and morbidity (12). Several virulence factors contribute to the pathogenicity, including hypermucoviscosity-specific capsular antigens (i.e., K1 and K2 serotypes) and virulence loci. such as mucoid phenotype regulator, encoded by rmpA, and aerobactin (12, 13). Traditionally, multidrug-resistant (MDR) and hypervirulent phenotypes in K. pneumoniae have been associated with distinct lineages. However, MDR lineages acquiring virulence traits or hypervirulent lineages acquiring AMR genes have increasingly been reported in the last decade, especially in South and Southeast Asia (14–18), mostly through dissemination of conjugative and hybrid plasmids harboring both resistance and virulence genes. This may lead to widely disseminated community-acquired infections in healthy people that are difficult to treat.
While carbapenem resistance in Klebsiella is increasingly documented in the Middle East region, there is limited information on hypervirulence and how it intersects with carbapenem resistance. We therefore conducted an in-depth analysis of Klebsiella genomes that were sequenced in the study of CRE in Qatar during 2014 to 2017 (6), with the following three aims: (i) to describe the genetic diversity, AMR genes, and virulence determinants of Klebsiella isolates, (ii) to investigate the molecular epidemiology data of selected sequence types (ST) that had ≥4 isolates, and (iii) to characterize the genetic context and virulence potential of a carbapenem-resistant hvKp strain belonging to ST383.
RESULTS
Epidemiology of sequence types and AMR genes.
Whole-genome sequencing (WGS) had been carried out on 100 carbapenem-resistant Klebsiella isolates, which was part of a larger-scale epidemiology study that included all CRE isolates retrieved from the Hamad Medical Corporation’s microbiology department (Doha, Qatar) from 1 April 2015 to 30 November 2017 (6). As previously described, the species included K. pneumoniae (n = 80), K. quasipneumoniae subsp. quasipneumoniae (n = 14), K. quasipneumoniae subsp. similipneumoniae (n = 2), K. aerogenes (n = 3), and K. oxytoca (n = 1). Among the 40 different STs reported, 23 were represented by a single isolate; 4 isolates were not reported elsewhere and were submitted for assignment of new ST numbers, and 1 isolate (FQ156, ST25-1LV) did not meet the criteria for assignment (see Table S1 in the supplemental material). Common K. pneumoniae STs included ST147 (n = 13), ST231 (n = 7), and ST11 (n = 5). ST147 and ST11 belong to widespread clonal group 147 (CG147) and CG258 (Table 1; Table S1). ST147 isolates were identified from all specimen types (blood, pus, sterile body fluid, urine, and respiratory tract), while ST231 isolates were collected from all specimen types except sterile body fluid. Out of the 14 K. quasipneumoniae subsp. quasipneumoniae isolates (14%) (6), 9 belonged to ST196 and 4 belonged to ST1416. These isolates were identified from blood, pus, and urine specimens (Table 1).
TABLE 1.
Comparison of key features of Klebsiella isolates from different specimens in Qatar
| Group or feature | No. (%) of isolates from: |
Total | ||||
|---|---|---|---|---|---|---|
| Blood (n = 20) | Pus (n = 12) | Fluid and other sources (n = 6) | Urine (n = 37) | RTa (n = 23) | ||
| Species | ||||||
| K. pneumoniae | 13 (65) | 8 (66.7) | 5 (83.3) | 32 (86.4) | 22 (95.7) | 80 |
| K. quasipneumoniae | 6 (30) | 3 (25) | 1 (16.7) | 5 (13.5) | 1 (4.3) | 16 |
| K. aerogenes | 1 (8.3) | 2 (5.4) | 3 | |||
| K. oxytocab | 1 (5) | 1 | ||||
| Major STs (CG) | ||||||
| ST147 (CG147) | 3 (15) | 1 (8.3) | 1 (16.7) | 5 (13.5) | 3 (13) | 13 |
| ST231 (CG2321) | 1 (5) | 1 (8.3) | 1 (2.7) | 4 (17.4) | 7 | |
| ST11 (CG258) | 1 (8.3) | 2 (5.4) | 2 (8.7) | 5 | ||
| ST14/15 (CG15) | 1 (8.3) | 3 (8.1) | 4 | |||
| ST383 | 1 (8.3) | 1 (16.7) | 2 (5.4) | 4 | ||
| ST196 | 3 (15) | 2 (16.7) | 4 (10.8) | 9 | ||
| ST1416 | 2 (10) | 1 (16.7) | 1 (4.3) | 4 | ||
| AMR | ||||||
| NDM | 10 (50) | 5 (43) | 2 (33.3) | 22 (59.5) | 6 (26.1) | 45 |
| OXA-48 | 8 (40) | 3 (25) | 1 (16.7) | 18 (48.6) | 12 (52.2) | 42 |
| KPC | 2 (16.7) | 1 (16.7) | 2 (8.7) | 5 | ||
| CTX-M | 19 (95) | 12 (100) | 5 (83.3) | 34 (91.9) | 21 (91.3) | 91 |
| Virulence | ||||||
| rmpA | 1 (5) | 1 (8.3) | 2 (33.3) | 2 (5.4) | 2 (8.7) | 8 |
| rmpA2 | 1 (8.3) | 2 (33.3) | 4 (10.8) | 2 (8.7) | 9 | |
| iuc | 2 (10) | 3 (25) | 2 (33.3) | 5 (13.5) | 4 (17.4) | 16 |
| iro | 1 (8.3) | 3 (8.1) | 1 (4.3) | 5 | ||
| clb | 1 (8.3) | 1 (16.7) | 1 (4.3) | 3 | ||
| KL2/KL20 | 1 (5) | 4 (10.8) | 2 (8.7) | 7 | ||
RT, respiratory tract.
As K. michiganensis in Kleborate.
As previously reported, carbapenemase genes identified included those encoding NDM-1 (n = 39), OXA-48 (n = 20), OXA-232 (n = 10), and OXA-181 (n = 12), but KPC-2 (n = 3) and KPC-3 (n = 2) were rare. Seven K. pneumoniae isolates carried more than one carbapenemase gene, while 15 isolates did not harbor any carbapenemase gene and instead carried combinations of blaCTX-M genes with mutations in porin genes ompK35 and ompK36 (Table S1), which have previously been linked to carbapenem resistance in Klebsiella (19). In total, 68 out of 100 Klebsiella isolates had ompK35 or ompK36 loss/truncation/mutation, which may contribute to reduced susceptibility to carbapenems (Table S1). blaCTX-M was coharbored by 75 isolates (75/85 [88.2%]), including 66 isolates (77.6%) harboring blaCTX-M-15, 7 isolates (8.2%) harboring blaCTX-M-14b, and 2 isolates (2.3%) harboring blaCTX-M-27. Cocarriage of blaNDM and/or blaOXA-48-type and blaCTX-M was reported for 6 isolates, including 3 ST383 isolates (Table S1).
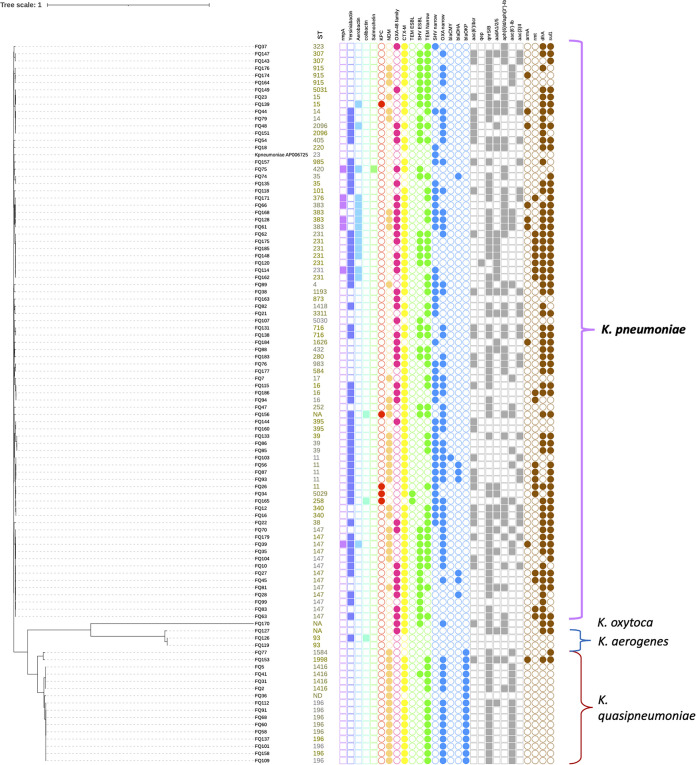
We then studied the genetic relationships among species/isolates using core genome single-nucleotide polymorphism (cgSNP) analysis based on whole-genome alignment. The cgSNP alignment containing 403,061 bases indicated that 93% identity was shared by isolates among K. pneumoniae and K. quasipneumoniae, as well as K. oxytoca and K. aerogenes (Fig. 1; Table S2). cgSNP analysis illustrated that K. pneumoniae and K. quasipneumoniae isolates shared around 98.2% identity (>7,000 cgSNP differences), and they each formed monophyletic clades. There were genetic variations within isolates of K. pneumoniae (0 to 905 cgSNPs), K. quasipneumoniae subsp. quasipneumoniae (0 to 868 cgSNPs), K. quasipneumoniae subsp. similipneumoniae (859 cgSNPs), and K. aerogenes (10 to 1,142 cgSNPs) (Table S2). Genetic variations were also detected within prevalent K. pneumoniae isolates in terms of STs and presence or absence of certain AMR genes. For example, ST147 isolates differed by 2 to 56 cgSNPs, and out of 13 isolates, 6 had blaNDM while 8 had blaOXA-48-like. Similarly, ST231 isolates differed by 2 to 25 cgSNPs, and out of 7 isolates, only 5 had blaOXA-48. ST383 isolates differed by 3 to 12 cgSNPs, and out of 4 isolates, 3 had blaNDM-5. In contrast, K. quasipneumoniae subsp. quasipneumoniae ST196 was highly clonal (0 to 1 cgSNPs), and all isolates carried blaNDM-1 (Fig. 1).
FIG 1.
Genetic relationship of Klebsiella isolates inferred from cgSNPs (Parsnp) overlaid with the presence/absence of AMR and virulence phenotypes.
Although short-read assemblies were fragmented due to repetitive mobile genetic elements like insertion sequences (ISs), we attempted to gain an overview on the spectrum of plasmids associated with carbapenemase genes based on the replicon sequences detected from the same contigs as the carbapenemase genes. blaNDM-1 was associated with the IncFII replicon, while blaOXA-48-like (blaOXA-181 and blaOXA-232) were commonly associated with ColKP3 plasmids (Table 2). Most contigs carrying blaNDM were divergent and had different mobile genetic elements. While blaNDM-1 was linked to IncFIB and IncA/C2 in K. pneumoniae in one isolate each, it was found to be associated with IncFII_1_pKP91 in 7 out of 9 K. quasipneumoniae ST196 isolates. When we aligned the contig carrying blaNDM-1 and blaCTX-M-15 in ST196 to the two homologous contigs in K. quasipneumoniae subsp. quasipneumoniae ST1416 isolates, we found that the overlapping region was highly similar (>90%) and included 6 AMR genes and ISs, suggesting that mobile genetic elements may spread these AMR genes among these STs. The contig bearing blaKPC-3 contained a commonly reported mobile genetic element, Tn4401a, based on annotation (Fig. S1). Overall, among all Klebsiella isolates, FIB was the most common F replicon, found in 89 isolates (89%), followed by FII, in 72 isolates (72%), and FIA, in 21 isolates (21%) (Table S1).
TABLE 2.
Plasmid replicons linked to carbapenemase genes in 24 isolates of K. pneumoniae and K. quasipneumoniae subsp. quasipneumoniae (from Plasmidfinder)
| Isolate | Species | ST | Date of isolation (day/mo/yr) | Carbapenemase | Replicon |
|---|---|---|---|---|---|
| FQ103 | K. pneumoniae | 11 | 16/3/17 | NDM-1 | IncA/C2 |
| FQ7 | K. pneumoniae | 17 | 20/5/15 | NDM-1 | IncFIB (pQil) |
| FQ94 | K. pneumoniae | 16 | 26/2/17 | OXA-181 | ColKP3 |
| FQ115 | K. pneumoniae | 16 | 27/4/17 | OXA-181 | ColKP3 |
| FQ186 | K. pneumoniae | 16 | 24/12/17 | OXA-232 | ColKP3 |
| FQ22 | K. pneumoniae | 38 | 7/7/15 | OXA-232 | ColKP3 |
| FQ27 | K. pneumoniae | 147 | 22/7/15 | OXA-181 | ColKP3 |
| FQ28 | K. pneumoniae | 147 | 25/7/15 | OXA-181 | ColKP3 |
| FQ45 | K. pneumoniae | 147 | 11/11/15 | OXA-181 | ColKP3 |
| FQ70 | K. pneumoniae | 147 | 21/10/16 | OXA-181 | ColKP3 |
| FQ114 | K. pneumoniae | 231 | 25/4/17 | OXA-232 | ColKP3 |
| FQ120 | K. pneumoniae | 231 | 17/5/17 | OXA-232 | ColKP3 |
| FQ148 | K. pneumoniae | 231 | 26/8/17 | OXA-232 | ColKP3 |
| FQ144 | K. pneumoniae | 395 | 20/8/17 | OXA-232 | ColKP3 |
| FQ138 | K. pneumoniae | 716 | 27/7/17 | OXA-48 | ColKP3 |
| FQ151 | K. pneumoniae | 2096 | 5/9/17 | OXA-232 | ColKP3 |
| FQ107 | K. pneumoniae | 5030 | 26/3/17 | OXA-232 | ColKP3 |
| FQ149 | K pneumoniae | 5031 | 28/8/17 | OXA-181 | ColKP3 |
| FQ58 | K. quasipneumoniae subsp. quasipneumoniae | 196 | 3/4/16 | NDM-1 | IncFII_1_pKP91 |
| FQ60 | K. quasipneumoniae subsp. quasipneumoniae | 196 | 22/4/16 | NDM-1 | IncFII_1_pKP91 |
| FQ91 | K. quasipneumoniae subsp. quasipneumoniae | 196 | 12/2/17 | NDM-1 | IncFII_1_pKP91 |
| FQ101 | K. quasipneumoniae subsp. quasipneumoniae | 196 | 14/3/17 | NDM-1 | IncFII_1_pKP91 |
| FQ117 | K. quasipneumoniae subsp. quasipneumoniae | 196 | 28/4/17 | NDM-1 | IncFII_1_pKP91 |
| FQ137 | K. quasipneumoniae subsp. quasipneumoniae | 196 | 26/7/17 | NDM-1 | IncFII_1_pKP91 |
Prevalence of virulence markers.
We used (i) the presence of rmpA or rmpA2 and/or (ii) the presence of aerobactin (iuc) and salmochelin (iro) biomarkers that are associated with hypervirulence to qualify strains that may demonstrate a hypervirulent phenotype (20). According to Kleborate results, 10 isolates (12.5%) had rmpA and/or rmpA2 (rmpA2 was truncated in all isolates), and four of them belonged to ST383. The prevalences of the iuc or iro and rmpA combination in ST147, ST383, ST420, and ST231 were 7.7%, 75%, 100%, and 14.3%, respectively (Table 3). Sixteen (20%) K. pneumoniae isolates carried the aerobactin iuc locus, while only 2 (2.5%) isolates harbored the salmochelin iro locus, and they all belonged to ST420. Two (2.5%) isolates carried the colibactin clb locus, one of which belonged to ST258. The ybt locus, encoding the acquired siderophore yersiniabactin, was detected in 49 (61.3%) K. pneumoniae isolates, representing 20 different STs. Five different ybt locus types and their associated integrative conjugative elements (ICE) were identified, and the most prevalent locus was ybt14 (n = 18 [18.8%]), with ICEKp5 detected in 8 STs, followed by ybt9 with ICEKp3 (n = 12 [12.5%]) and ybt16 with ICEKp12 (n = 11 [11.5%]). All three K. aerogenes isolates carried iro, and one of them (FQ126) also harbored ybt and clb (Table S1). The virulence loci, such as ybt, clb, iro, rmpA, and rmpA2, were not detected in any of the K. oxytoca or K. quasipneumoniae isolates. The capsule biosynthesis (KL) were identified for all isolates, spanning 91 distinct KL types (Table S1). hvKp serotype KL1 was not detected in this study, while KL2, usually associated with invasive liver abscess syndrome, was detected in 5 (5.8%) isolates of different STs. One (FQ44) of them belonged to ST14, and the rest belonged to ST376, ST35, ST39, and ST25-SLV (Table 3). KL20 was detected in two ST420 isolates (Table 3). The most prevalent KL type in K. pneumoniae was KL64 (n = 17 [17%]), followed by KL51 (n = 10 [10%]) and KL46 (n = 9 [9%]). The most prevalent O antigen-type loci were O2 variant 1 (O2v1) (n = 19 [19.8%]), O1v1 (n = 18 [18.8%]), and O1v2 (n = 17 [17.7%]) (Table S1).
TABLE 3.
Notable hypervirulent isolates in this investigation (virulence loci from Kleborate)a
| Isolate | Specimen | Infection | ST | rmpA | rmpA2 | ybt | iuc | iro | clb | K_locus | Carbapenemase(s) | ESBL | Other β-lactamases | Omp mutation(s) and variants | Clinical outcome | Travel (30 days) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FQ44 | Urine | UTI | 14 | ybt14; ICEKp5 | KL2 | NDM-1 | CTX-M-15 | ompK35, 88%; ompK36 GD | Alive | No | ||||||
| FQ139 | Pus | IAI | 15 | iuc5 | KL102 | KPC-2 | CTX-M-14, SHV-28.v1 | ompK35, 21% | Alive | Yes | ||||||
| FQ39 | Other | SSI | 147 | rmp1; KpVP-1 | rmpA2_6*, 47% | ybt9; ICEKp3 | iuc1 | KL64 | NDM-1 | CTX-M-15 | SHV-11.v1 | ompK35, 25% | Alive | Yes | ||
| FQ148 | Pus | SSI | 231 | ybt14; ICEKp5 | iuc unknown | KL51 | OXA-232 | SHV-1 | ompK35, 30% | Alive | No | |||||
| FQ162 | Urine | UTI | 231 | ybt14; ICEKp5 | iuc unknown | KL51 | CTX-M-15 | ompK35, 30%; ompK36 GD | Alive | Yes | ||||||
| FQ62 | Blood | BSI | 231 | ybt14; ICEKp5 | iuc unknown | KL51 | OXA-48 | CTX-M-14; CTX-M-15 | SHV-1 | ompK35, 30%; ompK36, 72% | Dead | No | ||||
| FQ114 | RT | RTI | 231 | rmp1; KpVP-1 | rmpA2_3*, 47% | ybt14; ICEKp5 | iuc unknown | KL51 | OXA-232 | CTX-M-15 | ompK35, 30%; ompK36 GD | Dead | No | |||
| FQ175 | RT | RTI | 231 | ybt14; ICEKp5 | iuc unknown | KL51 | OXA-232 | CTX-M-15 | SHV-1 | ompK35, 30%; ompK36 GD | Alive | Yes | ||||
| FQ185 | RT | RTI | 231 | ybt14; ICEKp5 | iuc unknown | KL51 | CTX-M-15; SHV-106 | ompK35, 30%; ompK36 GD | Alive | Yes | ||||||
| FQ171 | Blood | BSI | 376 | rmp1; KpVP-1 | iuc1 | KL2 | OXA-48 | CTX-M-14 | Dead | Yes | ||||||
| FQ168 | Urine | UTI | 383 | rmpA2_6*, 47% | iuc1 | KL30 | NDM-5; OXA-48 | CTX-M-14; CTX-M-15 | SHV-1 | ompK35, 10% | Alive | No | ||||
| FQ128 | Urine | UTI | 383 | rmp1; KpVP-1 | rmpA2_6, 60% | iuc1 | KL30 | NDM-5; OXA-48 | CTX-M-14; CTX-M-15 | SHV-1 | ompK35, 10% | Alive | No | |||
| FQ66 | Other | SSTI | 383 | rmp1; KpVP-1 | rmpA2_6*, 60% | iuc1 | KL30 | OXA-48 | CTX-M-14 | SHV-1 | ompK35, 10% | Alive | No | |||
| FQ61 | Pus | SSTI | 383 | rmp1; KpVP-1 | rmpA2_6, 60% | iuc1 | KL30 | NDM-5; OXA-48 | CTX-M-14; CTX-M-15 | SHV-1 | ompK35, 10% | Alive | No | |||
| FQ75 | Urine | UTI | 420 | rmp1; KpVP-1 | rmpA2_3, 47% | ybt9; ICEKp3 | iuc1 | iro1 | KL20 | OXA-48 | CTX-M-14 | SHV-75 | Alive | No | ||
| FQ72 | RT | RTI | 420 | rmp1; KpVP-1 | rmpA2_3, 47% | ybt9; ICEKp3 | iuc1 | iro1 | KL20 | OXA-48 | CTX-M-14 | SHV-75 | Alive | No | ||
| FQ48 | Urine | UTI | 2096 | rmpA2_8, 60% | ybt14; ICEKp5 | iuc1 | KL64 | OXA-232 | CTX-M-15, SHV-28.v1 | ompK36 GD | Alive | Yes | ||||
| FQ156 | RT | RTI | ST25-1LV | ybt14; ICEKp5 | clb3 | KL2 | KPC-3; NDM-1 | CTX-M-15 | SHV-11.v1 | ompK36, 50% | Alive | No | ||||
| FQ135 | Urine | UTI | ST35 | ybt16; ICEKp12 | KL2 | OXA-181 | Alive | No | ||||||||
| FQ133 | Urine | UTI | ST39 | ybt16; ICEKp12 | KL2 | NDM-1 | CTX-M-15 | SHV-11.v1 | ompK35, 40%; ompK36, 55% | Alive | No |
UTI, urinary tract infection; IAI, intrabdominal infection; BSI, bloodstream infection; RTI, respiratory tract infection; SSTI, skin and soft tissue infection; ESBL, extended-spectrum β-lactamase. ybt, iuc, iro, and clb encode yersiniabactin, aerobactin, salmochelin, and colibactin, respectively. Percentages in the columns for ompK and rmpA2 represent percent amino acid length from the start codon (truncation).
In-depth investigation of K. pneumoniae ST383.
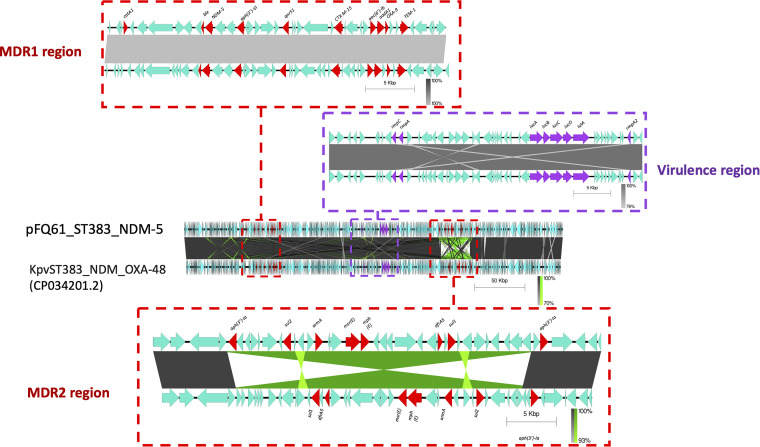
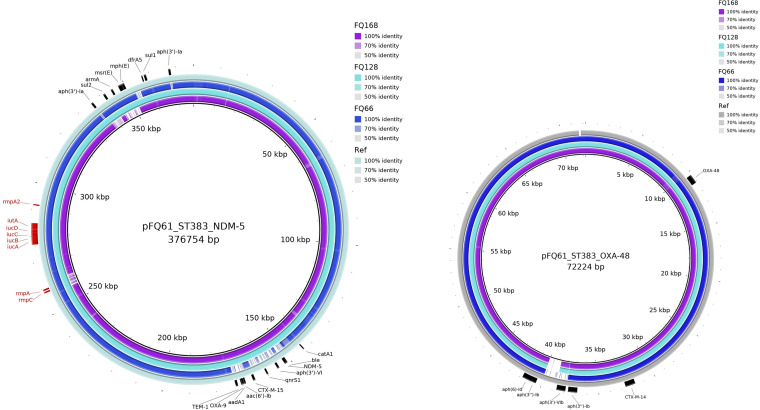
Our collection had four ST383 isolates (FQ61, FQ66, FQ128, and FQ168) collected at different hospitals from April 2016 to October 2017; this ST is not commonly reported. Two isolates (FQ61 and FQ128) carried both rmpA (hypermucoidy locus rmpADC) and truncated rmpA2, as well as blaNDM-5 and blaOXA-48; in contrast, FQ168 had both carbapenemase genes but did not have rmpA, while FQ66 had rmpA and rmpA2 but did not have blaNDM-5. To understand the genetic basis and the plasmids associated with ST383 in Qatar, we sequenced isolate FQ61 using both Illumina and MinION technologies. Hybrid assembly revealed that FQ61 harbored a chromosome and five plasmids, including pFQ61_ST383_NDM-5 (IncHI1B; ~376 kb)-, pFQ61_ST383_OXA-48 (IncL; ~72 kb)-, and Col (phAD28; 5 to 23 kb)-type plasmids. blaNDM-5 was located on pFQ61_ST383_NDM-5 (IncHI1B type), which also carried eight other AMR genes, including blaCTX-M-15, blaOXA-9, blaTEM-1, aac(6′)-Ib, aph(3′)-VI, aph(3′)-Ia, dfrA5, and armA. Based on a BLAST search, plasmid FQ61_ST383_NDM-5 showed high similarity (>99%) with high query coverage (>99%) with pKpvST383L (GenBank accession number CP034201.2), a hybrid virulence/resistance plasmid reported for another ST383 strain in the United Kingdom and carrying multiple ISs as well as AMR and virulence genes (21). Based on the location of AMR and virulence genes, plasmid FQ61_ST383_NDM-5 was divided into three regions: MDR region 1, MDR region 2, and virulence (Fig. 2). Pairwise comparison revealed that MDR region 1 (34,570 bp) was highly similar (99 to 100%) to the homologous region in pKpvST383L (Fig. 2), while evidence of a large-scale inversion and rearrangement event was observed in MDR region 2 adjacent to the repetitive elements (45,327 bp) in comparison to pKpvST383L. The virulence region (42,300 bp) of FQ61_ST383_NDM-5, harboring virulence genes rmpA, rmpA2, iucABCD, and iutA, also exhibited high similarity (>99% identity and 100% query coverage) to pKvpvST383L (UK, 2018) (Fig. 2), as well as pKpvST147B (UK, 2019), pKP-135LU_HIB-FIB (Italy, 2019), pSI0646A-ARMA-Vir-NDM (Italy, 2019), phvKpST395 (Russia, 2019), phvKpST874 (Russia, 2019), and phvKpST147 (Russia, 2017) in various K. pneumoniae isolates (Fig. 2) (22, 23). Based on contig analysis using BANDAGE and BRIG, we identified contigs in FQ128 that were highly homologous to pFQ61_ST383_NDM-5, suggesting that FQ128 may also have a plasmid highly similar to that in FQ61 and carrying both blaNDM-5 and other virulence genes (Fig. 3). FQ168 also carried a similar plasmid that harbored blaNDM-5 and blaCTX-M-15 and the other virulence genes except rmpA; in contrast, the plasmids in FQ66 appeared to be distinct in terms of organization (Fig. 3).
FIG 2.
Genomic comparison of hybrid plasmid harboring blaNDM-5 and virulence phenotypes recovered from FQ61 and pKpvST383L (21).
FIG 3.
Homologous contigs from ST383 isolates FQ66, FQ128, and FQ168 were compared to two major plasmids, pFQ61_ST383_NDM-5 and pFQ61_ST383_OXA-48. BRIG was used to generate a visual representation with pKpvST383_NDM_OXA48 as a reference (CP034201.2 and CP034202.1, respectively). Red and black arcs in the outer ring represent the major well-annotated AMR and virulence genes.
blaOXA-48 was located on pFQ61_ST383_OXA-48 (IncL-type plasmid), together with AMR genes blaCTX-M-14 and aph(3′)-Vib. pFQ61_ST383_OXA-48 was highly similar (>99.95%) to pKpvST383L_2 (CP034202.1) reported from the United Kingdom (21). Contigs homologous to pFQ61_ST383_OXA-48 were also detected in FQ66, FQ128, and FQ168, indicating that these three other ST383 isolates may possess highly similar plasmids that carry blaOXA-48 and blaCTX-M-14b (Fig. 3).
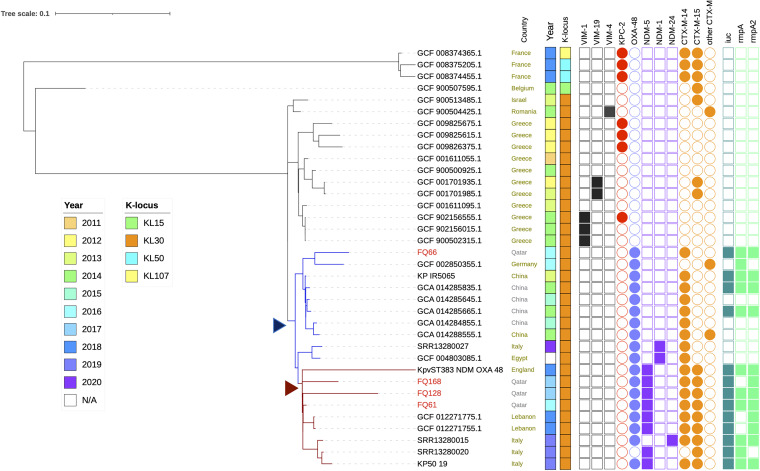
Genomic comparison of our ST383 isolates with global ST383 isolates (Table S3) revealed the genetic compositions as well as different resistomes and virulomes of the isolates which might be linked to mobile genetic elements. Figure 4 illustrates the phylogenetic relatedness of the local ST383 strains together with publicly available ST383 assembled genomes and raw reads (n = 32). The isolates in Qatar (FQ61, FQ128, and FQ168) clustered with those collected in Lebanon, the United Kingdom, and Italy, which also carried blaNDM-5/blaNDM-1, blaOXA-48, blaCTX-M-15, and blaCTX-M-14, as well as several major virulence genes such as iuc, rmpA1, and rmpA2. In contrast, FQ66 nested in a clade which contains isolates from China and Germany mostly carrying blaOXA-48 and blaCTX-M-14. The tree suggested that there could be two independent plasmid acquisition events in the past few years: the first plasmid with blaOXA-48 and blaCTX-M-14, followed by a hybrid plasmid carrying blaNDM-5 and the virulence genes (Fig. 4). However, earlier-reported ST383 isolates from Greece and France carried genes encoding carbapenemases such as KPC, OXA-48, and various VIM types (Fig. 4).
FIG 4.
Phylogenetic tree showing the relationships among K. pneumoniae ST383 isolates from different countries and Qatar using Parsnp (overlaid with presence/absence of key AMR genes and virulence loci). Arrowheads (blue and dark red) indicate the two possible plasmid acquisition events.
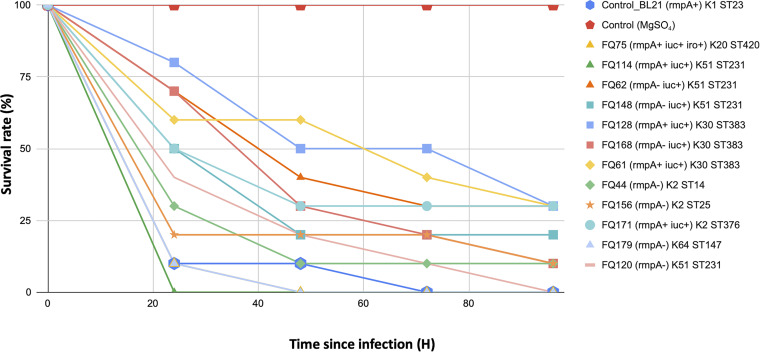
To correlate the presence of virulence genes with virulent phenotype, Galleria mellonella larvae were infected with selected K. pneumoniae isolates. In addition to the ST383 isolates, other isolates were selected based on the presence of KL2/KL20 loci such as rmpA and iuc, which are often associated with virulence. All larvae injected with 10 mM MgSO4 solution only (negative control) survived. With an inoculum of 1.0 × 107 CFU, the survival rates were 0% after 72 h with a classic hypervirulent K1 isolate (BL21, control) and 10% after 96 h with two hypervirulent K2 isolates (FQ44 and FQ156) (Fig. 5). The survival rates were 10 to 30% at 96 h after infection for the ST383 isolates (FQ61, FQ128, and FQ168). Also, the survival rates were 0% after 24 h with FQ114 (ST231; rmpA+ iuc+) and 0% after 48 h with FQ75 (ST420; rmpA+ iuc+ iro+ K20) and FQ179 (ST147; K64) (Fig. 5).
FIG 5.
Virulence potential of selected K. pneumoniae isolates in a Galleria mellonella infection model.
Possible local outbreaks of K. pneumoniae and K. quasipneumoniae.
High-resolution SNP analysis based on read mapping and variant calling together with epidemiological investigation was performed on cases associated with Klebsiella isolates of prevalent STs (n ≥ 4), including ST147, ST231, ST11, ST196, ST383, and ST1416, to identify possible outbreak and transmission events (Table S4).
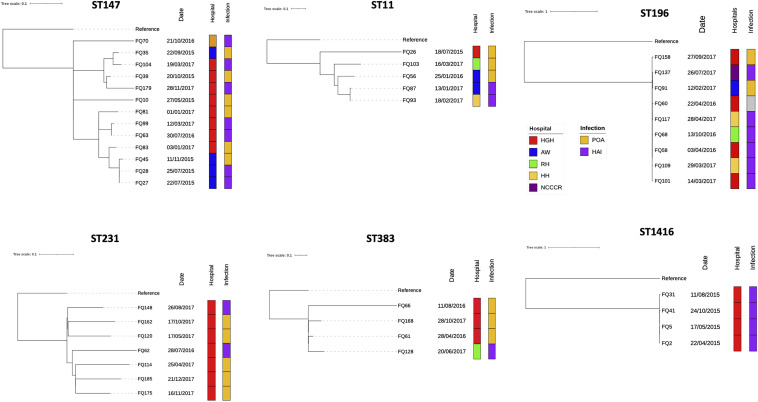
The largest cluster belonged to ST147, with 13 isolates collected from May 2015 and November 2017 (Fig. 6). Over half of the isolates were from hospital-acquired infections (HAI) (n = 7 [54%]), while the rest was present on admission (POA) (n = 6 [46%]). Mean pairwise SNP difference between these 13 isolates was 87.5 SNPs (range, 0 to 139) (Table 4); however, FQ27 and FQ28 differed by 0 SNPs, and the corresponding patients were admitted to the same hospital in different units on different dates, indicating possible intrahospital transmission. Another cluster involved 7 ST231 isolates collected from July 2016 to December 2017. Although most cases were determined to be HAI (n = 5 [71%]) in the same hospital, the SNP differences were large (range, 68 to 105) (Table 4), which was not consistent with intrahospital transmission/dissemination. Similarly, 5 ST11 isolates were collected from July 2015 to March 2017, but the epidemiological information, such as date of admission and hospital systems, and SNP differences (range, 3 to 170) did not suggest intra- or interhospital transmission (Table 4). In contrast, 4 ST383 isolates had relatively large genetic variation (SNP range, 44 to 183), and interestingly, three of them were POA, where the patients had travel and medical histories in Egypt within 6 months of hospital admissions, suggesting that the isolates might have been acquired there (Table S4).
FIG 6.
Phylogenetic tree generated from high-resolution SNPs and linked epidemiological data of prevalent K. pneumoniae and K. quasipneumoniae subsp. quasipneumoniae isolates.
TABLE 4.
Pairwise SNPs differences among K. pneumoniae and K. quasipneumoniae subsp. quasipneumoniae isolates based on high-resolution SNP analysis (using Snippy workflow)
| Organism and sequence type (no. of samples) | No. of characters | Mean pairwise SNPs among samples in Qatar (range, distance to reference genome) | Reference genome used in Snippy pipeline |
|---|---|---|---|
| K. pneumoniae | |||
| ST147 (13) | 485 | 87.5 (0–139) | NZ_CP012745.1.fasta |
| ST231 (7) | 361 | 86.7 (68–105) | GCF_002909775.1_ASM290977v2_genomic.fna |
| ST11 (5) | 348 | 101.6 (3–170) | GCF_003931835.1_ASM393183v1_genomic.fna |
| ST383 (4) | 331 | 112.8 (44–183) | GCF_001611055.1_ASM161105v1_genomic.fna |
| K. quasipneumoniae subsp. quasipneumoniae | |||
| ST196 (9) | 227 | 3.9 (0–10) | GCF_003146655.1_ASM314665v1_genomic.fna |
| ST1416 (4) | 137 | 2 (0–4) | GCF_005503875.1_ASM550387v1_genomic.fna |
Isolates of K. quasipneumoniae subsp. quasipneumoniae ST196 and ST1416 were clustered together with 0 to 1 cgSNPs (among isolates within each ST) in the cgSNP tree (Fig. 1; Table S2), which prompted an investigation to identify possible disease outbreaks. All ST196 and ST1416 isolates had identical capsular and lipopolysaccharide types and similar AMR genes (Fig. 1; Table S1). The high-resolution SNP tree (from mapping and variant calling) indicated that these two subspecies may be clonal, as all ST196 isolates except FQ158 were highly similar, with 0 to 10 high-resolution SNPs (Fig. 6). Most ST196 isolates (66.7%) were associated with HAI. They were collected from various patients in 5 different hospitals in different times and units, but interhospital transmission/dissemination due to transfer may still be possible (Table S4). While four ST1416 isolates were also highly similar, with 0 to 4 high-resolution SNPs (Table 4), and were from the same hospital and were identified as HAI (Fig. 6), there were no epidemiological and clinical associations among the four patients to suspect outbreaks among the isolates, although cryptic transmission events cannot be rule out (Table S4).
The virulence-associated loci such as iuc, clb, iro, and rmpA and rmpA2 were not detected in any of the K. quasipneumoniae subsp. quasipneumoniae isolates. We compared these K. quasipneumoniae subsp. quasipneumoniae isolates (ST1416 and ST196) to the global isolates to determine if they represented local clones. The isolates belonging to ST196 were genetically different from global ST196 isolates (Fig. S2). For instance, previously reported ST196 isolates were mostly KPC producers from Europe and the United States (Table S5), while most K. quasipneumoniae subsp. quasipneumoniae isolates in Qatar were NDM-1 producers. Similarly, K. quasipneumoniae subsp. quasipneumoniae ST1416 were not commonly reported elsewhere; the isolates in Nigeria and China were genetically divergent from the Qatari isolates (Fig. S2), despite the Chinese isolate also carrying blaNDM-1.
DISCUSSION
Klebsiella species are responsible for HAI worldwide and are also increasingly implicated in community-associated infections. The predominant species is K. pneumoniae, but other Klebsiella species, including K. aerogenes, K. michiganensis, K. quasipneumoniae, and K. variicola, also cause human infections. The key clinically relevant attributes of K. pneumoniae are its antimicrobial resistance and virulence, or hypervirulence. However, these aspects are less well studied in Klebsiella species other than K. pneumoniae. Recent reports suggest that these species, like K. pneumoniae, are also sources of antimicrobial resistance and hypervirulence (5, 24, 25). Using a data set of carbapenem-resistant Klebsiella clinical isolates from a hospital in Qatar, we conducted in-depth genomic analysis of carbapenem resistance and its intersection with hypervirulence.
Our data revealed the presence of diverse STs and different lineages across the Klebsiella species. Among K. pneumoniae isolates, ST231, ST147, and ST11 were the most prevalent carbapenem-resistant clones, which were different from those isolated from rectal screening swabs in local pediatric populations (9), among which ST73, ST14, and ST17 were the most common STs. ST147 (CG147) and ST11 (CG258) are international high-risk clones reported mainly from Asia and Europe and have been responsible for nosocomial transmission and various care center outbreaks (26, 27), while ST231 was considered an endemic clone associated with blaOXA-232 in India (28). Based on previous studies, K. pneumoniae CG258 (ST11 and ST258) is the predominant KPC-producing clone reported globally (29); however, only six isolates were reported in our study. In Qatar, substantial proportions of the population are migrant workers from the Indian subcontinent. Pérez-López et al. (9) suggested that CRE in pediatric populations in Qatar were mainly introduced sporadically by asymptomatic carriers who received health care in some nearby countries in which they are endemic. Moreover, consistent with other CRE studies of pediatric and adult patients, genes encoding NDM and OXA-48-type carbapenemases were widely prevalent (9, 27, 30).
Isolates of the next common species, K. quasipneumoniae subsp. quasipneumoniae, mostly belonged to ST196 or ST1416 and carried blaNDM. Outbreaks caused by K. quasipneumoniae have been rare, but ST334 has been reported as a potentially emerging outbreak-associated MDR clone in Pakistan and Cambodia (31, 32, 33). Our K. quasipneumoniae subsp. quasipneumoniae isolates were highly clonal within the STs across hospitals; thus, their potential for regional spread merits monitoring. Two K. quasipneumoniae subsp. similipneumoniae isolates (ST1584 and ST1998) also could raise public health concern on antimicrobial resistance, as they carried blaNDM-7.
Out of three K. aerogenes isolates in Qatar, two belonged to ST93, one of the prevalent STs and the founder genotype in the world (4). The remaining isolate belonged to ST206, which was reported only from Singapore according to the pubMLST database. Though these isolates did not carry known carbapenemase genes, the chromosomal ampC coupled with outer membrane porin alteration may be responsible for the development of carbapenem resistance (34).
In terms of hypervirulent traits, the classical hvKp lineages, including CG23 and CG86, were not among the carbapenem-resistant K. pneumoniae clinical isolates in Qatar. The proportion of hvKp isolates among K. pneumoniae isolates was 10% (rmpA and/or rmpA2) and could be up to 20% when solely aerobactin (iuc)-bearing isolates are also included. The prevalence of rmpA- or rmpA2-positive isolates among K. pneumoniae isolates was lower than in China and Vietnam (>20%) (12) but higher than in the United States or the United Kingdom (35, 36). rmpA- and/or rmpA2-mediated overproduction of capsular polysaccharide has been shown to contribute to hypervirulence (truncation of these loci may reduce virulence) (37), while iuc, iro, ybt, and ent mediate increased siderophore production under iron-limiting conditions (18). In addition, KL1 and KL2 hypermucoviscosity-specific capsular serotypes have been associated with invasive infections and accounted for approximately 70% of hvKp global isolates; however, they were not common in this collection. Convergence of carbapenem resistance and hypervirulence was found in limited STs such as ST231, ST383, and ST410, of which ST231 and ST410 are globally emerging hypervirulent clonal groups associated with virulence plasmids (28, 38, 39).
Our study indicates the emergence and transmission of carbapenem-resistant hvKp ST383 among patients worldwide. Three patients in Qatar infected with ST383 isolates had histories of travel to Egypt, suggesting that they likely acquired the isolates there. This may be in parallel with Kpv_ST383_S1, another ST383 isolate carrying hybrid virulence/resistance plasmids from a patient in Scotland, who had medical treatment in Cairo (21). International travel may play a role in the spread/dissemination of this hybrid plasmid. The wax moth larva virulence study indicated that these ST383 isolates were virulent, even though to a slightly lower degree than the classical hypervirulent KL1 isolate. Russo et al. (40) demonstrated that not all hvKp strains shared the same pathogenic potential in murine model infections. Previously, it was common for K. pneumoniae ST383 isolates to be resistant to carbapenems but not hypervirulent. Due to the acquisition of a hybrid plasmid that contains a fraction of the hvKp virulence plasmid during the evolution of conventional ST383 isolates, these new circulating isolates have become both MDR and hypervirulent and should be regarded as isolates of a superbug that could pose a serious threat to public health. Early K. pneumoniae ST383 isolates carrying blaVIM-4, blaKPC-2, and blaCMY-2 were reported in a Greek hospital during 2009 to 2010 (41). A later study from the Czech Republic demonstrated the presence of blaVIM-19 in isolates of suspected Greek origin (42). More recent studies indicated the presence of blaOXA-48 in clinical isolates in Germany and China, which then went on to acquire another hybrid plasmid that carried both blaNDM and virulence genes (21, 43). Sabivora et al. proposed that the plasticity of the accessory genomes in ST383 isolates may benefit the acquisition of different plasmids (44). Turton et al. (35) studied 31 ST383 isolates in the United Kingdom and identified 5 isolates harboring rmpA or rmpA2 and iutA, which is part of the aerobactin iuc locus. These ST383 isolates have also emerged in Italy recently (43, 45) and in the Middle East, such as in Qatar, Egypt, and Lebanon (Fig. 3). Recently, an ST383 isolate harboring blaKPC and genes encoding various virulence factors was reported in Saudi Arabia (46); however, assembled genome data are not available in the public domain. Tracking the evolution and distribution of ST383 is of major importance due to its ability to acquire carbapenemase genes of different types as well as genes associated with the hv phenotype, but also the ability of mobile genetic elements to spread these genes to different ST types and species. Increasing reports of the presence of hybrid, mosaic plasmid carrying both carbapenem resistance and virulence genes suggest that carbapenem-resistant hvKp isolates are no longer confined to selected clones (16, 22, 23, 47), which will make containment of such isolates challenging.
In conclusion, our study has provided insights into the dynamics and epidemiology of carbapenem-resistant Klebsiella species in Qatar. Analysis of WGS data demonstrated the presence of clonal lineages. Our comparative genomic data also confirmed the emergence of carbapenem-resistant hvKp ST383 in the Middle East and worldwide. Acquisition of virulence-associated loci was reported for at least 10% of the carbapenem-resistant K. pneumoniae isolates, and one of the mechanisms was through the transfer of a carbapenem resistance and hypervirulence hybrid plasmid possibly mediated by mobile genetic elements, as demonstrated for ST383 isolates. Further studies will be required to understand the relationship between the hypervirulent phenotypes, carriage of the hybrid MDR-virulence plasmid, and capsular types.
MATERIALS AND METHODS
Hospital settings, cultures, and antimicrobial susceptibility tests.
Hamad Medical Corporation (HMC) is the major provider of secondary and tertiary health care in Qatar and has 12 hospitals—9 specialist hospitals and 3 community hospitals—as well as the National Ambulance Service and home and residential care services. The isolates were collected, maintained, and identified in the Department of Microbiology in HMC as previously described (6). Also, the antimicrobial susceptibility testing for various antibiotics was performed on BD Phoenix (Becton, Dickinson and Company, USA) using the Clinical and Laboratory Standards Institute breakpoints (6). The study was approved by the institutional review board (MRC-16134/16).
Whole-genome sequencing (WGS) and data analysis.
Briefly, genomic DNA was extracted using a DNeasy blood and tissue kit (Qiagen, Germany), and the DNA libraries were sequenced on the Illumina NextSeq 550 platform using 2 × 150 paired-end reads (PE) at the Microbial Genome Sequencing Center (MiGS; Pittsburgh, PA, USA) as previously described (6). In addition, long reads for isolate FQ61 were generated using an Oxford Nanopore MinION sequencer (SQK-LSK109 and flow cell R9.4.1) at MiGS. MinION reads were generated based on the Guppy software (v4.0.11) available from Oxford Nanopore Technologies (Oxford, UK). The raw reads from Illumina NextSeq were assembled de novo using SPAdes v3.9.0 (48) implemented in shovill (https://github.com/tseemann/shovill) (49). De novo Illumina-Nanopore assemblies were generated with Unicycler v0.4.7 (50).
STs, plasmid replicons, and AMR genes were predicted from the assembled contigs using multilocus sequence typing (MLST) (https://github.com/tseemann/mlst), pubMLST (51), Plasmidfinder v2.1 (52), and ResFinder v3.2 databases implemented in ABRicate v0.9 (https://github.com/tseemann/abricate), based on >70% coverage and 90% sequence identity. Kleborate v2 was used to detect the virulence genes and capsule synthesis (K) and lipopolysaccharide (O) loci, as well as AMR genes and known chromosomal mutations associated with resistance to fluoroquinolones, colistin, and carbapenems (14). Previously unreported STs were submitted to BIGSDB (https://bigsdb.pasteur.fr/klebsiella/) for ST assignment.
Genome assemblies were annotated with Prokka v1.13.3 (53), and the completed plasmid annotations were curated before deposit in GenBank. To clarify and study the plasmid variation among 4 ST383 isolates, assembled genomes of FQ66, FQ128, and FQ168 were manually explored in their assembly graphs using Bandage v0.8.1 (54) and the completed FQ61 plasmids built from Unicycler assembly as a reference. BRIG (55) was used to generate a visual representation of the contigs in FQ66, FQ128, and FQ168 aligned to the major plasmids in FQ61.
Core genome phylogenetic trees were generated using Parsnp (56) and visualized together with associated metadata using iTOL (57). cgSNPs were extracted from the Parsnp alignment to determine the pairwise difference between Klebsiella species and between STs in K. pneumoniae and K. quasipneumoniae (56). Easyfig (58) was used to generate the diagram to compare the plasmid against highly homologous plasmids in the NCBI database. To generate high-resolution SNPs for epidemiological investigation within prevalent STs, the quality-trimmed reads for ST147, ST231, ST11, ST196, and ST1416 were mapped against their respective reference genome and high-quality SNPs were called by using the Snippy pipeline (https://github.com/tseemann/snippy). FastTree was used for phylogenetic analysis (59).
Outbreak analysis.
Cases associated with prevalent Klebsiella STs (≥4 isolates) in this study and forming clusters in the core genome phylogenetic tree were carefully inspected as potential outbreaks. Epidemiological data, antimicrobial susceptibility testing results, and clinical information such as date of patient admission were reviewed to determine whether those isolates represented hospital-acquired infection (HAI) or community-acquired infection based on the National Health Care Safety Network (NHSN) definition published in January 2021 by HMC.
An infection was defined as an HAI if the NHSN site-specific infection occurred on or after the third calendar day of admission to an inpatient location where day of admission was calendar day 1. If the infection was identified within 2 days before admission or the day of admission to an inpatient location (calendar day 1) or calendar day 2 after admission, the infection was considered present on admission (POA).
Virulence study.
The virulence of selected K. pneumoniae isolates, including 3 ST383 isolates, was tested in wax moth (Galleria mellonella) larvae. Briefly, overnight cultures of K. pneumoniae strains were prepared with 10 mM MgSO4 solution and further adjusted to concentrations of 1 × 105 CFU/mL, 1 × 106 CFU/mL, and 1 × 107 CFU/mL. We infected the G. mellonella with the bacteria as described previously (60), and the survival rate of the G. mellonella was recorded every 24 h for 4 days.
Data availability.
Raw sequence reads are available on the NCBI website under BioProject accession number PRJNA656934. The genome sequence of FQ61 was submitted to GenBank under accession numbers CP091813 to CP091818.
ACKNOWLEDGMENTS
This research was funded by the Medical Research Centre (MRC) at Hamad Medical Corporation (HMC) (MRC-16134/16 to F.B.A.) and supported by the National Institutes of Health (R01AI104895, R21AI135522, and R21AI151362 to Y.D.).
We are grateful to clinical laboratory staff at the HMC, Dan Snyder at MiGS (Pittsburgh, PA, USA), and staff at the University of Pittsburgh for technical assistance. We acknowledge the Communicable Disease Center, Hamad Medical Corporation, for performing epidemiological investigations on K. pneumoniae and K. quasipneumoniae isolates. We also thank Prakki Sai Rama Sridatta (NCID, Singapore), Kelly Wyres (Monash University, Australia), and Will Hsiao and Jun Duan (Simon Fraser University, Burnaby, Canada) for their advice and technical support. This research was supported in part through computational resources and services provided by Advanced Research Computing at the University of British Columbia. We thank the Institut Pasteur teams for the curation and maintenance of BIGSdb-Pasteur databases at https://bigsdb.pasteur.fr/.
Footnotes
Supplemental material is available online only.
[This article was published on 13 June 2023 with content missing from the Acknowledgments section. The section was updated in the current version, posted on 17 June 2023.]
Contributor Information
Clement Kin-Ming Tsui, Email: clement_km_tsui@ncid.sg.
Fatma Ben Abid, Email: fabid@hamad.qa.
REFERENCES
- 1.Paczosa MK, Mecsas J. 2016. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev 80:629–661. doi: 10.1128/MMBR.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, Jenney A, Connor TR, Hsu LY, Severin J, Brisse S, Cao H, Wilksch J, Gorrie C, Schultz MB, Edwards DJ, Van Nguyen K, Nguyen TV, Dao TT, Mensink M, Minh VL, Nhu NTK, Schultsz C, Kuntaman K, Newton PN, Moore CE, Strugnell RA, Thomson NR. 2015. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci USA 112:E3574–E3581. doi: 10.1073/pnas.1501049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brisse S, Passet V, Grimont PAD. 2014. Description of Klebsiella quasipneumoniae sp. nov., isolated from human infections, with two subspecies, Klebsiella quasipneumoniae subsp. quasipneumoniae subsp. nov. and Klebsiella quasipneumoniae subsp. similipneumoniae subsp. nov., and demonstration that Klebsiella singaporensis is a junior heterotypic synonym of Klebsiella variicola. Int J Syst Evol Microbiol 64:3146–3152. doi: 10.1099/ijs.0.062737-0. [DOI] [PubMed] [Google Scholar]
- 4.Passarelli-Araujo H, Palmeiro JK, Moharana KC, Pedrosa-Silva F, Dalla-Costa LM, Venancio TM. 2019. Genomic analysis unveils important aspects of population structure, virulence, and antimicrobial resistance in Klebsiella aerogenes. FEBS J 286:3797–3810. doi: 10.1111/febs.15005. [DOI] [PubMed] [Google Scholar]
- 5.Yang J, Long H, Hu Y, Feng Y, McNally A, Zong Z. 2022. Klebsiella oxytoca complex: update on taxonomy, antimicrobial resistance, and virulence. Clin Microbiol Rev 35:e00006-21. doi: 10.1128/CMR.00006-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ben Abid F, Tsui CKM, Doi Y, Deshmukh A, McElheny CL, Bachman WC, Fowler EL, Albishawi A, Mushtaq K, Ibrahim EB, Doiphode SH, Hamed MM, Almaslmani MA, Alkhal A, Butt AA, Omrani AS. 2021. Molecular characterization of clinical carbapenem-resistant Enterobacterales from Qatar. Eur J Clin Microbiol Infect Dis 40:1779–1785. doi: 10.1007/s10096-021-04185-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baker S, Thomson N, Weill F-X, Holt KE. 2018. Genomic insights into the emergence and spread of antimicrobial-resistant bacterial pathogens. Science 360:733–738. doi: 10.1126/science.aar3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Al Mana H, Sundararaju S, Tsui CKM, Perez-Lopez A, Yassine H, Al Thani A, Al-Ansari K, Eltai NO. 2021. Whole-genome sequencing for molecular characterization of carbapenem-resistant Enterobacteriaceae causing lower urinary tract infection among pediatric patients. Antibiotics (Basel) 10:972. doi: 10.3390/antibiotics10080972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pérez-López A, Sundararaju S, Tsui KM, Al-Mana H, Hasan MR, Suleiman M, Al Maslamani E, Imam O, Roscoe D, Tang P. 2021. Fecal carriage and molecular characterization of carbapenemase-producing Enterobacterales in the pediatric population in Qatar. Microbiol Spectr 9:e01122-21. doi: 10.1128/Spectrum.01122-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perez-Lopez A, Sundararaju S, Al-Mana H, Tsui KM, Hasan MR, Suleiman M, Janahi M, Al Maslamani E, Tang P. 2020. Molecular characterization of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae among the pediatric population in Qatar. Front Microbiol 11:581711. doi: 10.3389/fmicb.2020.581711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jamshaid MB, Shahzad A, Zafar A, Kamal I. 2021. Invasive Klebsiella pneumoniae syndrome in Qatar: a case report. Cureus 13:e15015. doi: 10.7759/cureus.15015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Russo TA, Marr CM. 2019. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev 32:e00001-19. doi: 10.1128/CMR.00001-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Russo TA, Olson R, MacDonald U, Beanan J, Davidson BA. 2015. Aerobactin, but not yersiniabactin, salmochelin, or enterobactin, enables the growth/survival of hypervirulent (hypermucoviscous) Klebsiella pneumoniae ex vivo and in vivo. Infect Immun 83:3325–3333. doi: 10.1128/IAI.00430-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wyres KL, Nguyen TNT, Lam MMC, Judd LM, van Vinh Chau N, Dance DAB, Ip M, Karkey A, Ling CL, Miliya T, Newton PN, Lan NPH, Sengduangphachanh A, Turner P, Veeraraghavan B, Vinh PV, Vongsouvath M, Thomson NR, Baker S, Holt KE. 2020. Genomic surveillance for hypervirulence and multi-drug resistance in invasive Klebsiella pneumoniae from South and Southeast Asia. Genome Med 12:11. doi: 10.1186/s13073-019-0706-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hennequin C, Robin F. 2016. Correlation between antimicrobial resistance and virulence in Klebsiella pneumoniae. Eur J Clin Microbiol Infect Dis 35:333–341. doi: 10.1007/s10096-015-2559-7. [DOI] [PubMed] [Google Scholar]
- 16.Yang X, Dong N, Chan EW-C, Zhang R, Chen S. 2021. Carbapenem resistance-encoding and virulence-encoding conjugative plasmids in Klebsiella pneumoniae. Trends Microbiol 29:65–83. doi: 10.1016/j.tim.2020.04.012. [DOI] [PubMed] [Google Scholar]
- 17.Zhang R, Lin D, Chan EW-C, Gu D, Chen G-X, Chen S. 2016. Emergence of carbapenem-resistant serotype K1 hypervirulent Klebsiella pneumoniae strains in China. Antimicrob Agents Chemother 60:709–711. doi: 10.1128/AAC.02173-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wyres KL, Wick RR, Judd LM, Froumine R, Tokolyi A, Gorrie CL, Lam MMC, Duchêne S, Jenney A, Holt KE. 2019. Distinct evolutionary dynamics of horizontal gene transfer in drug resistant and virulent clones of Klebsiella pneumoniae. PLoS Genet 15:e1008114. doi: 10.1371/journal.pgen.1008114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong JLC, Romano M, Kerry LE, Kwong H-S, Low W-W, Brett SJ, Clements A, Beis K, Frankel G. 2019. OmpK36-mediated carbapenem resistance attenuates ST258 Klebsiella pneumoniae in vivo. Nat Commun 10:3957. doi: 10.1038/s41467-019-11756-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lam MMC, Wyres KL, Duchêne S, Wick RR, Judd LM, Gan Y-H, Hoh C-H, Archuleta S, Molton JS, Kalimuddin S, Koh TH, Passet V, Brisse S, Holt KE. 2018. Population genomics of hypervirulent Klebsiella pneumoniae clonal-group 23 reveals early emergence and rapid global dissemination. Nat Commun 9:2703. doi: 10.1038/s41467-018-05114-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turton J, Davies F, Turton J, Perry C, Payne Z, Pike R. 2019. Hybrid resistance and virulence plasmids in “high-risk” clones of Klebsiella pneumoniae, including those carrying blaNDM-5. Microorganisms 7:326. doi: 10.3390/microorganisms7090326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Starkova P, Lazareva I, Avdeeva A, Sulian O, Likholetova D, Ageevets V, Lebedeva M, Gostev V, Sopova J, Sidorenko S. 2021. Emergence of hybrid resistance and virulence plasmids harboring New Delhi metallo-β-lactamase in Klebsiella pneumoniae in Russia. Antibiotics (Basel) 10:691. doi: 10.3390/antibiotics10060691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin MJ, Corey BW, Sannio F, Hall LR, MacDonald U, Jones BT, Mills EG, Harless C, Stam J, Maybank R, Kwak Y, Schaufler K, Becker K, Hübner N-O, Cresti S, Tordini G, Valassina M, Cusi MG, Bennett JW, Russo TA, McGann PT, Lebreton F, Docquier J-D. 2021. Anatomy of an extensively drug-resistant Klebsiella pneumoniae outbreak in Tuscany, Italy. Proc Natl Acad Sci USA 118:e2110227118. doi: 10.1073/pnas.2110227118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harada S, Aoki K, Yamamoto S, Ishii Y, Sekiya N, Kurai H, Furukawa K, Doi A, Tochitani K, Kubo K, Yamaguchi Y, Narita M, Kamiyama S, Suzuki J, Fukuchi T, Gu Y, Okinaka K, Shiiki S, Hayakawa K, Tachikawa N, Kasahara K, Nakamura T, Yokota K, Komatsu M, Takamiya M, Tateda K, Doi Y. 2019. Clinical and molecular characteristics of Klebsiella pneumoniae isolates causing bloodstream infections in Japan: occurrence of hypervirulent infections in health care. J Clin Microbiol 57:e01206-19. doi: 10.1128/JCM.01206-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie M, Chen K, Chan EW-C, Zhang R, Chen S. 2022. Characterisation of clinical carbapenem-resistant K1 Klebsiella quasipneumoniae subsp. similipneumoniae strains harbouring a virulence plasmid. Int J Antimicrob Agents 60:106628. doi: 10.1016/j.ijantimicag.2022.106628. [DOI] [PubMed] [Google Scholar]
- 26.Navon-Venezia S, Kondratyeva K, Carattoli A. 2017. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev 41:252–275. doi: 10.1093/femsre/fux013. [DOI] [PubMed] [Google Scholar]
- 27.Peirano G, Chen L, Kreiswirth BN, Pitout JDD. 2020. Emerging antimicrobial-resistant high-risk Klebsiella pneumoniae clones ST307 and ST147. Antimicrob Agents Chemother 64:e01148-20. doi: 10.1128/AAC.01148-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shankar C, Mathur P, Venkatesan M, Pragasam AK, Anandan S, Khurana S, Veeraraghavan B. 2019. Rapidly disseminating blaOXA-232 carrying Klebsiella pneumoniae belonging to ST231 in India: multiple and varied mobile genetic elements. BMC Microbiol 19:137. doi: 10.1186/s12866-019-1513-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wyres KL, Lam MMC, Holt KE. 2020. Population genomics of Klebsiella pneumoniae. Nat Rev Microbiol 18:344–359. doi: 10.1038/s41579-019-0315-1. [DOI] [PubMed] [Google Scholar]
- 30.Zowawi HM, Sartor AL, Balkhy HH, Walsh TR, Al Johani SM, AlJindan RY, Alfaresi M, Ibrahim E, Al-Jardani A, Al-Abri S, Al Salman J, Dashti AA, Kutbi AH, Schlebusch S, Sidjabat HE, Paterson DL. 2014. Molecular characterization of carbapenemase-producing Escherichia coli and Klebsiella pneumoniae in the countries of the Gulf Cooperation Council: dominance of OXA-48 and NDM producers. Antimicrob Agents Chemother 58:3085–3090. doi: 10.1128/AAC.02050-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wyres KL, Holt KE. 2016. Klebsiella pneumoniae population genomics and antimicrobial-resistant clones. Trends Microbiol 24:944–956. doi: 10.1016/j.tim.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 32.Crellen T, Turner P, Pol S, Baker S, Nguyen Thi Nguyen T, Stoesser N, Day NP, Turner C, Cooper BS. 2019. Transmission dynamics and control of multidrug-resistant Klebsiella pneumoniae in neonates in a developing country. Elife 8:e50468. doi: 10.7554/eLife.50468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heinz E, Ejaz H, Bartholdson Scott J, Wang N, Gujaran S, Pickard D, Wilksch J, Cao H, Haq I-U, Dougan G, Strugnell RA. 2019. Resistance mechanisms and population structure of highly drug resistant Klebsiella in Pakistan during the introduction of the carbapenemase NDM-1. Sci Rep 9:2392. doi: 10.1038/s41598-019-38943-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merhi G, Amayri S, Bitar I, Araj GF, Tokajian S. 2023. Whole genome-based characterization of multidrug resistant Enterobacter and Klebsiella aerogenes isolates from Lebanon. Microbiol Spectr 11:e02917-22. doi: 10.1128/spectrum.02917-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turton JF, Payne Z, Coward A, Hopkins KL, Turton JA, Doumith M, Woodford N. 2018. Virulence genes in isolates of Klebsiella pneumoniae from the UK during 2016, including among carbapenemase gene-positive hypervirulent K1-ST23 and “non-hypervirulent” types ST147, ST15 and ST383. J Med Microbiol 67:118–128. doi: 10.1099/jmm.0.000653. [DOI] [PubMed] [Google Scholar]
- 36.Chou A, Nuila RE, Franco LM, Stager CE, Atmar RL, Zechiedrich L. 2016. Prevalence of hypervirulent Klebsiella pneumoniae-associated genes rmpA and magA in two tertiary hospitals in Houston, TX, USA. J Med Microbiol 65:1047–1048. doi: 10.1099/jmm.0.000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng HY, Chen YS, Wu CY, Chang HY, Lai YC, Peng HL. 2010. RmpA regulation of capsular polysaccharide biosynthesis in Klebsiella pneumoniae CG43. J Bacteriol 192:3144–3158. doi: 10.1128/JB.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shankar C, Veeraraghavan B, Nabarro LEB, Ravi R, Ragupathi NKD, Rupali P. 2018. Whole genome analysis of hypervirulent Klebsiella pneumoniae isolates from community and hospital acquired bloodstream infection. BMC Microbiol 18:6. doi: 10.1186/s12866-017-1148-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eger E, Heiden SE, Becker K, Rau A, Geisenhainer K, Idelevich EA, Schaufler K. 2021. Hypervirulent Klebsiella pneumoniae sequence type 420 with a chromosomally inserted virulence plasmid. Int J Mol Sci 22:9196. doi: 10.3390/ijms22179196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Russo TA, MacDonald U, Hassan S, Camanzo E, LeBreton F, Corey B, McGann P. 2021. An assessment of siderophore production, mucoviscosity, and mouse infection models for defining the virulence spectrum of hypervirulent Klebsiella pneumoniae. mSphere 6:e00045-21. doi: 10.1128/mSphere.00045-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Papagiannitsis CC, Giakkoupi P, Vatopoulos AC, Tryfinopoulou K, Miriagou V, Tzouvelekis LS. 2010. Emergence of Klebsiella pneumoniae of a novel sequence type (ST383) producing VIM-4, KPC-2 and CMY-4 β-lactamases. Int J Antimicrob Agents 36:573–574. doi: 10.1016/j.ijantimicag.2010.07.018. [DOI] [PubMed] [Google Scholar]
- 42.Papagiannitsis CC, Dolejska M, Izdebski R, Giakkoupi P, Skálová A, Chudějová K, Dobiasova H, Vatopoulos AC, Derde LPG, Bonten MJM, Gniadkowski M, Hrabák J. 2016. Characterisation of IncA/C2 plasmids carrying an In416-like integron with the blaVIM-19 gene from Klebsiella pneumoniae ST383 of Greek origin. Int J Antimicrob Agents 47:158–162. doi: 10.1016/j.ijantimicag.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 43.Lorenzin G, Gona F, Battaglia S, Spitaleri A, Saluzzo F, Trovato A, di Marco F, Cichero P, Biancardi A, Nizzero P, Castiglione B, Scarpellini P, Moro M, Cirillo DM. 2022. Detection of NDM-1/5 and OXA-48 co-producing extensively drug-resistant hypervirulent Klebsiella pneumoniae in Northern Italy. J Glob Antimicrob Resist 28:146–150. doi: 10.1016/j.jgar.2022.01.001. [DOI] [PubMed] [Google Scholar]
- 44.Sabirova JS, Xavier BB, Coppens J, Zarkotou O, Lammens C, Janssens L, Burggrave R, Wagner T, Goossens H, Malhotra-Kumar S. 2016. Whole-genome typing and characterization of blaVIM19-harbouring ST383 Klebsiella pneumoniae by PFGE, whole-genome mapping and WGS. J Antimicrob Chemother 71:1501–1509. doi: 10.1093/jac/dkw003. [DOI] [PubMed] [Google Scholar]
- 45.Spaziante M, Venditti C, Butera O, Messina F, Di Caro A, Tonziello G, Lanini S, Cataldo MA, Puro V. 2021. Importance of surveillance of New Delhi metallo-β-lactamase Klebsiella pneumoniae: molecular characterization and clonality of strains isolated in the Lazio region, Italy. Infect Drug Resist 14:3659–3665. doi: 10.2147/IDR.S318717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alghoribi MF, Binkhamis K, Alswaji AA, Alhijji A, Alsharidi A, Balkhy HH, Doumith M, Somily A. 2020. Genomic analysis of the first KPC-producing Klebsiella pneumoniae isolated from a patient in Riyadh: a new public health concern in Saudi Arabia. J Infect Public Health 13:647–650. doi: 10.1016/j.jiph.2020.01.003. [DOI] [PubMed] [Google Scholar]
- 47.Huang Y-H, Chou S-H, Liang S-W, Ni C-E, Lin Y-T, Huang Y-W, Yang T-C. 2018. Emergence of an XDR and carbapenemase-producing hypervirulent Klebsiella pneumoniae strain in Taiwan. J Antimicrob Chemother 73:2039–2046. doi: 10.1093/jac/dky164. [DOI] [PubMed] [Google Scholar]
- 48.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jolley KA, Bray JE, Maiden MCJ. 2018. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res 3:124. doi: 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, Møller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 54.Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31:3350–3352. doi: 10.1093/bioinformatics/btv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alikhan N-F, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Treangen TJ, Ondov BD, Koren S, Phillippy AM. 2014. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol 15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Letunic I, Bork P. 2021. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49:W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Price MN, Dehal PS, Arkin AP. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.O’Hara JA, Ambe LA, Casella LG, Townsend BM, Pelletier MR, Ernst RK, Shanks RMQ, Doi Y. 2013. Activities of vancomycin-containing regimens against colistin-resistant Acinetobacter baumannii clinical strains. Antimicrob Agents Chemother 57:2103–2108. doi: 10.1128/AAC.02501-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download aac.00030-23-s0001.xlsx, XLSX file, 0.09 MB (97.1KB, xlsx)
Supplemental material. Download aac.00030-23-s0002.pdf, PDF file, 0.2 MB (237KB, pdf)
Data Availability Statement
Raw sequence reads are available on the NCBI website under BioProject accession number PRJNA656934. The genome sequence of FQ61 was submitted to GenBank under accession numbers CP091813 to CP091818.