ABSTRACT
Full-genome sequences of seven mycobacteriophages isolated from environmental soil samples are presented. These bacteriophages, with their respective clusters or subclusters are Duplo (A2), Dynamo (P1), Gilberta (A11), MaCh (A11), Nikao (K1), Phloss (N), and Skinny (M1). All had siphovirus-like morphologies, with genome sizes ranging from 43,107 to 82,071 bp.
ANNOUNCEMENT
Bacteriophages are viruses that exclusively infect bacteria, exhibiting obligate intracellular replication and a limited host range (1, 2). Bacteriophages are the most numerous entities in the biosphere, totaling >1031 particles (3). The bacterium-bacteriophage relationship exhibits constant bidirectional selective pressure, with bacteria evolving to resist viral infection and bacteriophages coevolving to maintain their replicative ability within their hosts (4, 5). The prevalence of antibiotic-resistant bacterial infections has propelled a resurgent interest in phage therapy (6–10). Here, we report seven novel lysogenic bacteriophages.
All bacteriophages were isolated from environmental soil samples collected from various locations in east Texas during 2020 to 2021 (Table 1), using standard methods (5). In short, the soil samples were washed in Middlebrook 7H9 medium prior to centrifugation and supernatant filtration (pore size, 0.22 μm). The filtrates were subsequently inoculated with Mycobacterium smegmatis mc2155 cells and incubated at 25°C for 3.5 days with shaking. Filtered samples of each culture were plated with M. smegmatis cells in 7H9 top agar. After purification through three rounds of plating at 37°C for 48 h, the observed plaque morphologies of the various phages ranged from clear to turbid (Table 1). Negative-stain transmission electron microscopy showed all these bacteriophages to exhibit a siphovirus morphotype, with isometric capsids (diameter, ~51.76 to 71.53 nm) and noncontractile tails (length, ~124.23 to 326.19 nm) (Fig. 1 and Table 1), measured using ImageJ (11–13).
TABLE 1.
Properties of seven mycobacteriophages isolated in east Texas, USA, in 2020 and 2021
| Characteristic | Data for phage:a |
||||||
|---|---|---|---|---|---|---|---|
| Duplo | Dynamo | Gilberta | MaCh | Nikao | Phloss | Skinny | |
| Yr found | 2020 | 2021 | 2021 | 2020 | 2020 | 2020 | 2021 |
| Location found | Big Sandy, TX | Longview, TX | Longview, TX | Longview, TX | Longview, TX | Longview, TX | Longview, TX |
| Soil sampling location | 32.588034 N,95.063959 W | 32.46402 N,94.728226 W | 32.468338 N,94.726844 W | 32.465237 N,94.727035 W | 32.54954 N, 94.821309 W | 32.4675 N, 94.725 W | 32.467601 N,94.723749 W |
| Lysate titer (PFU/mL) | 3.0 × 109 | 1.0 × 1010 | 1.0 × 109 | 1.6 × 1010 | 2.17 × 109 | 8.7 × 1011 | 1.18 × 109 |
| Plaque morphology after 48 h at 37°C | Slightly turbid with defined edges | Clear with defined edges | Clear with defined edges | Clear with defined edges | Slightly turbid with defined edges | Clear with defined edges | Clear with defined edges |
| Plaque size (mm) | 3.3 | 0.93 | 1.5 | 2.7 | 1.25 | 2.0 | 1.0 |
| Approx coverage (×) | 1,516 | 1,499 | 5,472 | 705 | 414 | 2,005 | 499 |
| Genome size (bp) | 52,781 | 46,673 | 51,470 | 52,616 | 59,052 | 43,107 | 82,071 |
| GC content (%) | 63.5 | 67.2 | 63.7 | 63.6 | 67.2 | 66.3 | 61.5 |
| Overhang sequence | CGGTCGGTTA | CCCGCCCCCCGA | CGGTCGGTTA | CGGTCGGTTA | CTCGGGGGCAT | CCCGCCGCAATGG | ACCTCCTGCAA |
| Overhang length (bases) | 10 | 12 | 10 | 10 | 11 | 13 | 11 |
| Cluster | A | P | A | A | K | N | M |
| Subcluster | A2 | P1 | A11 | A11 | K1 | M1 | |
| GenBank accession no. | OP297553 | OP434454 | OP297532 | OP297549 | OP297530 | OP297540 | OP297551 |
| SRA accession no. | SRX19690833 | SRX19690834 | SRX19690836 | SRX19690844 | SRX19690847 | SRX19690850 | SRX19690858 |
| Total no. of reads | 560,502 | 485,417 | 1,968,556 | 261,625 | 172,343 | 608,118 | 285,773 |
| No. of predicted genes | 99 | 78 | 99 | 100 | 80 | 70 | 163 |
| No. of predicted tRNAs | 5 | 0 | 1 | 1 | 0 | 0 | 18 |
| tRNA type(s) | Asn, Gln, Glu, Trp, Tyr | Trp | Trp | Trp, Asn, other (Arg/Ala), Tyr, Gln, Pro, Ser, Phe, Met, Arg, His, Leu, Lys, Gly, Val, Thr, Asp, Glu | |||
| No. of genes with predicted functions | 33 | 32 | 36 | 43 | 41 | 28 | 46 |
| % of genes with predicted functions | 33 | 41 | 36 | 43 | 51 | 40 | 28 |
| Key predicted lysogenic life cycle genes | Integrase, excise, immunity repressor | Integrase, excise, immunity repressor | Integrase, immunity repressor | Immunity repressor | Integrase, excise, immunity repressor | Integrase, excise, immunity repressor | Integrase |
| No. of orphams | 2 | 0 | 1 | 0 | 0 | 0 | 2 |
| Capsid size (nm [n value]) | 64.48 (9) | 60.90 (7) | 52.09 (3) | 58.67 (21) | 67.04 (5) | 51.76 (7) | 71.53 (3) |
| Avg tail length (nm [n value]) | 124.23 (9) | 205.57 (6) | 125.54 (3) | 130.93 (21) | 210.16 (5) | 172.04 (7) | 326.19 (3) |
| Isolated by | Skylar M. Weiss, Jimena H. Segovia | Christina A. Holder, Kaitlyn J. Menard, Brady E. Tyler | Hattie R. Mills, Ashlyn B. Collier, Kalista J. Rivera, Claire P. Martinez | Matthew S. Adams, Camryn L. Hill | Kezia K. Happy | Summer L. Apostalo, Gavin J. Meyer | Jenna F. Curran, Kristen N. Rose |
All phages were isolated using the enriched isolation method (5) and purified through three sequential 48-h rounds of plating at 37°C. The genomes were sequenced using the Illumina shotgun sequencing method with 150-base single-end reads using the NEB Ultra II Library sequencing kit. All had 3′ sticky overhang genome ends. Genomic termini were identified through buildups of read start positions and variations in genome-wide coverage and verified manually using Consed v29 (14, 15). All phages had a siphovirus morphotype and were predicted to be temperate based on the presence of predicted lysogeny-related genes.
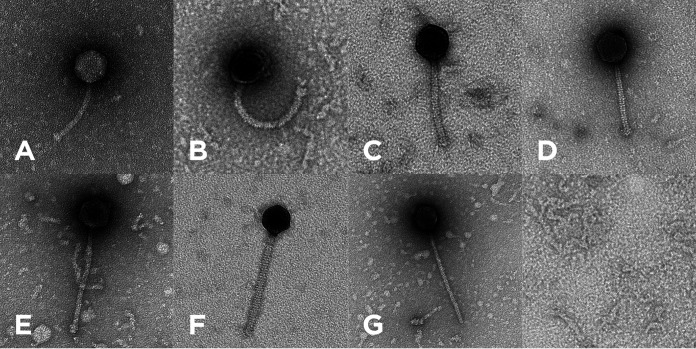
FIG 1.
Transmission electron micrographs of the seven bacteriophages, Duplo (A), Dynamo (B), Gilberta (C), MaCh (D), Nikao (E), Phloss (F), and Skinny (G). Capsid sizes and tail lengths are provided in Table 1. Bacteriophage particles were added to 300-mesh carbon–Formvar-coated copper grids (Ted Pella Inc., Redding, CA), stained with 1% (wt/vol) uranyl acetate, and imaged at the University of Arkansas for Medical Sciences Digital Microscopy Laboratory.
Genomic DNA was extracted from lysates of various titers (Table 1) using the Promega Wizard DNA cleanup kit. Preparation for sequencing using the Illumina MiSeq platform (v3 reagents) was conducted with the NEBNext Ultra II library prep kit. Assembly and verification of the untrimmed reads were performed using Newbler v2.9 (16) and Consed v29 (14, 15). Sequencing revealed genomes ranging in length from 43,107 bp (phage Phloss) to 82,071 bp (phage Skinny) (Table 1). All had 3′ sticky overhangs (10 to 13 bp long) and an average GC content of 64.7% (range, 61.5% to 67.2%), comparable to the 67.4% GC content of their isolation host, Mycobacterium smegmatis mc2155 (17). The seven phages were assigned to subclusters A2, A11, K1, M1, and P1 and cluster N (Table 1) based on ≥35% gene content similarity (GCS) to other phages, using the GCS tool in PhagesDB (18, 19).
Genome annotation was accomplished using DNA Master v5.23.6; Starterator; Phamerator (20); BLASTp with NCBI GenBank and PhagesDB (21, 22); GeneMark v2.5p (23); HHpred, with the PDB_mmCIF70_17_Apr, Pfam-A_v35, UniProt-SwissProt-viral70_3_Nov_2021, and NCBI_Conserved_Domains_v3.19 databases (24, 25); Glimmer v3.02 (26); TMHMM v.2.0 (27); SOSUI v1.11 (28); tRNAscan-SE v2.0 (29, 30); and ARAGORN v1.2.41 (31). Default settings were used for all programs (32). An average of 98.0 putative protein-coding genes (range, 70 to 163) and 3.6 tRNAs (range, 0 to 18) were predicted (Table 1). Functions could only be predicted for 28% to 51% of the putative genes in the phages (Table 1). All phages had at least one of the three key genes associated with a lysogenic life cycle. Duplo, Dynamo, Nikao, and Phloss had the integrase, excise, and immunity repressor genes; Gilberta had both the integrase and immunity repressor genes, while Skinny and MaCh had only the integrase and immunity repressor genes, respectively (Table 1).
Data availability.
The raw reads of all seven reported mycobacteriophages are available in the Sequence Read Archive (SRA), and their complete genome sequences are available at GenBank. The SRA and GenBank accession numbers are provided in Table 1. High-titer lysates of the phages are archived at the University of Pittsburgh Bacteriophage Institute.
ACKNOWLEDGMENTS
We thank Graham Hatfull for his enduring commitment to and leadership of the Science Education Alliance—Phage Hunters Advancing Genomics and Evolutionary Science (SEA-PHAGES) program, Debbie Jacobs-Sera and Viknesh Sivanathan for their expertise and support, and Daniel Russell and Rebecca Garlena of the University of Pittsburgh Bacteriophage Institute for sequencing and assembling the phage genomes. We acknowledge Brianna Mack and Jonathan Matthews for preparing the laboratory materials used. We thank Matthew Adams, Summer Apostalo, Camryn Hill, Ashlyn Collier, Jenna Curran, Gavin Meyer, Christina Holder, Hattie Mills, Jimena Segovia, Brady Tyler, Kalista Rivera, and Kristen Rose, who helped isolate the phages. We also thank Jeff Kamykowski of the University of Arkansas for Medical Sciences Digital Microscopy Laboratory for capturing the transmission electron micrographs for these phages.
We acknowledge the Howard Hughes Medical Institute for their support of the SEA-PHAGES program and LeTourneau University School of Arts and Sciences for supporting and facilitating this undergraduate bacteriophage research.
Contributor Information
Frederick N. Baliraine, Email: FredBaliraine@letu.edu.
Kenneth M. Stedman, Portland State University
REFERENCES
- 1.Jacobs-Sera D, Marinelli LJ, Bowman C, Broussard GW, Guerrero Bustamante C, Boyle MM, Petrova ZO, Dedrick RM, Pope WH, Modlin RL, Hendrix RW, Hatfull GF, Science Education Alliance Phage Hunters Advancing Genomics and Evolutionary Science (SEA-PHAGES) Program . 2012. On the nature of mycobacteriophage diversity and host preference. Virology 434:187–201. doi: 10.1016/j.virol.2012.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boeckaerts D, Stock M, Criel B, Gerstmans H, De Baets B, Briers Y. 2021. Predicting bacteriophage hosts based on sequences of annotated receptor-binding proteins. Sci Rep 11:1467. doi: 10.1038/s41598-021-81063-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Comeau AM, Hatfull GF, Krisch HM, Lindell D, Mann NH, Prangishvili D. 2008. Exploring the prokaryotic virosphere. Res Microbiol 159:306–313. doi: 10.1016/j.resmic.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 4.Naureen Z, Dautaj A, Anpilogov K, Camilleri G, Dhuli K, Tanzi B, Maltese PE, Cristofoli F, De Antoni L, Beccari T, Dundar M, Bertelli M. 2020. Bacteriophages presence in nature and their role in the natural selection of bacterial populations. Acta Biomed 91:e2020024. doi: 10.23750/abm.v91i13-S.10819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poxleitner M, Pope W, Jacobs-Sera D, Sivanathan V, Graham H. 2018. Phage discovery guide. Howard Hughes Medical Institute, Chevy Chase, MD. https://seaphagesphagediscoveryguide.helpdocsonline.com/home. Accessed 12 May 2023. [Google Scholar]
- 6.Azam AH, Tan XE, Veeranarayanan S, Kiga K, Cui L. 2021. Bacteriophage technology and modern medicine. Antibiotics 10:999. doi: 10.3390/antibiotics10080999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y, Batra H, Dong J, Chen C, Rao VB, Tao P. 2019. Genetic engineering of bacteriophages against infectious diseases. Front Microbiol 10:954. doi: 10.3389/fmicb.2019.00954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hatfull GF, Dedrick RM, Schooley RT. 2022. Phage therapy for antibiotic-resistant bacterial infections. Annu Rev Med 73:197–211. doi: 10.1146/annurev-med-080219-122208. [DOI] [PubMed] [Google Scholar]
- 9.Dedrick RM, Guerrero-Bustamante CA, Garlena RA, Russell DA, Ford K, Harris K, Gilmour KC, Soothill J, Jacobs-Sera D, Schooley RT, Hatfull GF, Spencer H. 2019. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat Med 25:730–733. doi: 10.1038/s41591-019-0437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dedrick RM, Smith BE, Cristinziano M, Freeman KG, Jacobs-Sera D, Belessis Y, Whitney Brown A, Cohen KA, Davidson RM, van Duin D, Gainey A, Garcia CB, Robert George CR, Haidar G, Ip W, Iredell J, Khatami A, Little JS, Malmivaara K, McMullan BJ, Michalik DE, Moscatelli A, Nick JA, Tupayachi Ortiz MG, Polenakovik HM, Robinson PD, Skurnik M, Solomon DA, Soothill J, Spencer H, Wark P, Worth A, Schooley RT, Benson CA, Hatfull GF. 2023. Phage therapy of Mycobacterium infections: compassionate use of phages in 20 patients with drug-resistant mycobacterial disease. Clin Infect Dis 76:103–112. doi: 10.1093/cid/ciac453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, Arena ET, Eliceiri KW. 2017. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18:529. doi: 10.1186/s12859-017-1934-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gordon D, Green P. 2013. Consed: a graphical editor for next-generation sequencing. Bioinformatics 29:2936–2937. doi: 10.1093/bioinformatics/btt515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Russell DA. 2018. Sequencing, assembling, and finishing complete bacteriophage genomes. Methods Mol Biol 1681:109–125. doi: 10.1007/978-1-4939-7343-9_9. [DOI] [PubMed] [Google Scholar]
- 16.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen Y-J, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer MLI, Jarvie TP, Jirage KB, Kim J-B, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, et al. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohan A, Padiadpu J, Baloni P, Chandra N. 2015. Complete genome sequences of a Mycobacterium smegmatis laboratory strain (MC2 155) and isoniazid-resistant (4XR1/R2) mutant strains. Genome Announc 3:e01520-14. doi: 10.1128/genomeA.01520-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Russell DA, Hatfull GF. 2017. PhagesDB: the actinobacteriophage database. Bioinformatics 33:784–786. doi: 10.1093/bioinformatics/btw711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pope WH, Mavrich TN, Garlena RA, Guerrero-Bustamante CA, Jacobs-Sera D, Montgomery MT, Russell DA, Warner MH, Hatfull GF, Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science (SEA-PHAGES) . 2017. Bacteriophages of Gordonia spp. display a spectrum of diversity and genetic relationships. mBio 8:e01069-17. doi: 10.1128/mBio.01069-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cresawn SG, Bogel M, Day N, Jacobs-Sera D, Hendrix RW, Hatfull GF. 2011. Phamerator: a bioinformatic tool for comparative bacteriophage genomics. BMC Bioinformatics 12:395. doi: 10.1186/1471-2105-12-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 22.Ismail HD. 2022. Bioinformatics: a practical guide to NCBI databases and sequence alignments. Chapman and Hall, New York, NY. [Google Scholar]
- 23.Borodovsky M, McIninch J. 1993. Recognition of genes in DNA sequence with ambiguities. Biosystems 30:161–171. doi: 10.1016/0303-2647(93)90068-n. [DOI] [PubMed] [Google Scholar]
- 24.Zimmermann L, Stephens A, Nam SZ, Rau D, Kübler J, Lozajic M, Gabler F, Söding J, Lupas AN, Alva V. 2018. A completely reimplemented MPI Bioinformatics Toolkit with a new HHpred server at its core. J Mol Biol 430:2237–2243. doi: 10.1016/j.jmb.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 25.Gabler F, Nam SZ, Till S, Mirdita M, Steinegger M, Söding J, Lupas AN, Alva V. 2020. Protein sequence analysis using the MPI Bioinformatics Toolkit. Curr Protoc Bioinformatics 72:e108. doi: 10.1002/cpbi.108. [DOI] [PubMed] [Google Scholar]
- 26.Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res 27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krogh A, Larsson B, Von Heijne G, Sonnhammer ELL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 28.Hirokawa T, Boon-Chieng S, Mitaku S. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- 29.Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res 44:W54–W57. doi: 10.1093/nar/gkw413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chan PP, Lin BY, Mak AJ, Lowe TM. 2021. TRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucleic Acids Res 49:9077–9096. doi: 10.1093/nar/gkab688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laslett D, Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res 32:11–16. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pope W, Jacobs-Sera D, Russell D, Cresawn S, Hatfull G. 2017. SEA-PHAGES bioinformatics guide. https://seaphagesbioinformatics.helpdocsonline.com/home. Accessed 12 May 2023.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw reads of all seven reported mycobacteriophages are available in the Sequence Read Archive (SRA), and their complete genome sequences are available at GenBank. The SRA and GenBank accession numbers are provided in Table 1. High-titer lysates of the phages are archived at the University of Pittsburgh Bacteriophage Institute.