ABSTRACT
It has been widely appreciated that numerous bacterial species express chitinases for the purpose of degrading environmental chitin. However, chitinases and chitin-binding proteins are also expressed by pathogenic bacterial species during infection even though mammals do not produce chitin. Alternative molecular targets are therefore likely present within the host. Here, we will describe our current understanding of chitinase/chitin-binding proteins as virulence factors that promote bacterial colonization and infection. The targets of these chitinases in the host have been shown to include immune system components, mucins, and surface glycans. Bacterial chitinases have also been shown to interact with other microorganisms, targeting the peptidoglycan or chitin in the bacterial and fungal cell wall, respectively. This review highlights that even though the name “chitinase” implies activity toward chitin, chitinases can have a wide diversity of targets, including ones relevant to host infection. Chitinases may therefore be useful as a target of future anti-infective therapeutics.
KEYWORDS: bacterial chitinase, chitinases, chitin-binding protein, lytic polysaccharide monooxygenase, host-pathogen interaction, glycans
INTRODUCTION
Chitin is the second-most-abundant biopolymer in nature. It is virtually omnipresent, as it represents a major component of the cuticle of insects and crustaceans and can also be found in the fungal cell wall (1). Given the abundance of chitin, it is not surprising that enzymes degrading this biopolymer can be found throughout the tree of life, from bacteria to mammals (2). Chitin-degrading enzymes are called chitinases. They are glycosyl hydrolases that function by liberating N-acetylglucosamine (GlcNAc) subunits in chitin polymers (3). Many bacterial species produce chitinases to degrade this environmental chitin as a source of nutrients (4). Some bacterial chitinases have activity toward the chitin present in fungi and insects and have been proposed as a method to protect crops from pests (5). Chitinases are therefore clearly an important area of study in environmental microbiology and biotechnology. However, bacteria that infect mammalian hosts also express chitinases even though mammals do not produce chitin. These bacterial chitinases may interact with chitin present in the bacterium’s environmental niche outside the human host. However, chitinase activity inside the human host suggests that the mammalian host provides alternative molecular targets for these chitinase domains. Indeed, an increasing number of studies have revealed that chitinases can target other GlcNAc-containing molecules, such as peptidoglycan and mammalian glycans (6–9). In this review, we will summarize the current knowledge regarding the roles of bacterial chitinases during mammalian infection of different body sites, including skin, gut, airway, and systemic infection (Fig. 1). The biotechnological applications of bacterial chitinases have been recently discussed in other reviews (5, 10).
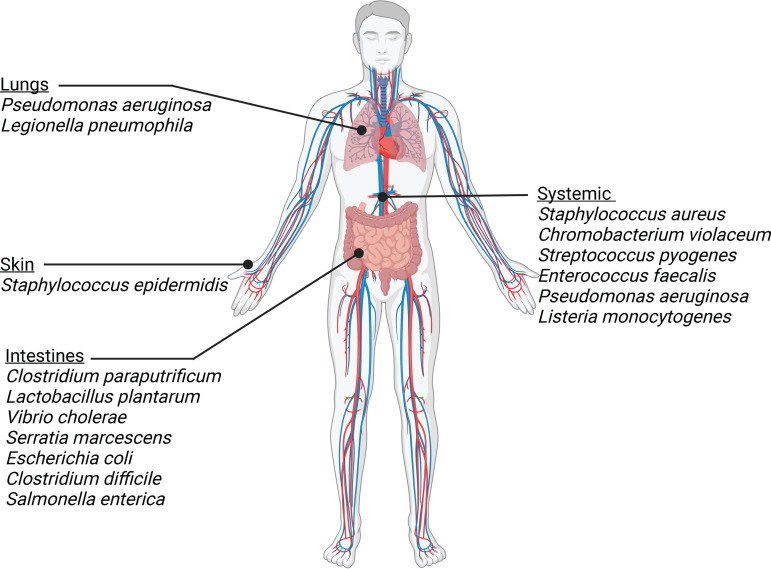
FIG 1.
Bacterial species that express chitinases/CBP during colonization or infection of various locations of the human body.
CHITINASE CLASSIFICATION AND FUNCTION
Chitinases are generally classified in the glycosyl hydrolase (GH) family 18 or 19 (3, 11). This classification is usually not based on experimental evidence. Rather, GH families classify enzymes that hydrolyze carbohydrate polymers based on their amino acid sequence similarities (12). Even though certain glycosyl hydrolases share homology and are classified in the same GH family, that does not necessarily mean that they share the same substrate specificities. Members of the same GH families can demonstrate divergent substrate specificity, while there are also examples of members of separate GH families sharing similar substrate specificity (12). For example, the GH48 family mostly contains cellulases; however, one GH48 enzyme produced by the beetle Gastrophysa atrocyanea demonstrates chitinase activity (13). For this reason, glycosyl hydrolases are further classified after experimental analysis based on their target substrate, catalytic mechanism, or the location on the glycosidic chain that is targeted (12). GH18 chitinases cleave chitin polymers and retain the configuration of the anomeric carbon (retaining mechanism), while GH19 chitinases cleave the polymers with an inversion of the anomeric configuration (inverting mechanism) (14, 15). Chitinases can have exo-activity, cleaving glycosidic bonds on the ends of chitin polymers and oligomers, or endo-activity, cleaving internal glycosidic bonds (3). Chitin-binding proteins (CBP) maintain chitin-binding ability but lack catalytic activity. Most chitin-binding proteins contain either the carbohydrate-binding module (CBM) 33 or CBM5/12. While CBM33 was originally thought to contain no chitinase activity, some CBPs with this domain have been shown to break chitin chains by oxidative cleavage. These CBPs have been reclassified as lytic polysaccharide monooxygenases (LMPOs) (4, 16).
BACTERIAL CHITIN UTILIZATION
A variety of bacterial species produce chitinases for the purpose of degrading chitin. Some of these bacterial species can also colonize human tissue as pathogenic invaders (4). The ability to utilize environmental chitin as a carbon source via chitinase expression is likely important for the persistence and transmittance of these pathogens outside hosts. The bacterial pathogens Listeria monocytogenes (17–19), Vibrio cholerae (20), Enterococcus faecalis (21–23), Escherichia coli (24), and Serratia marcescens (25, 26) all express chitinolytic machinery for the purpose of using chitin as a carbon source. The opportunistic pathogen S. marcescens expresses a complex set of machinery for the degradation of chitin polymers, involving three GH18 chitinases (ChiA, ChiB, and ChiC) and a lytic polysaccharide monooxygenase (CBP21) (25, 26). Expression is controlled by ChiR, a member of the LysR-type transcriptional regulator family (27). In order to degrade extracellular chitin, these chitinases have to be secreted from the bacterial cell. In S. marcescens, secretion of the chitinase machinery was shown to require proteins that partially destabilize the bacterial cell membrane. Without the holin-like protein ChiW and the l-alanyl-d-glutamate endopeptidase ChiX, chitinases cannot be released into the supernatant (28). ChiW likely facilitates the translocation of ChiX to the periplasm, where it can access peptidoglycan (28). ChiX demonstrates catalytic activity toward bacterial peptidoglycan and could therefore facilitate chitinase secretion (29). L. monocytogenes also expresses chitinases, ChiA and ChiB, and the lytic polysaccharide monooxygenase LPMO10. All three proteins are involved with chitin degradation in vitro, but LPMO10 may not be important for in vivo chitin degradation since it is not expressed in chitin-containing media (17–19). V. cholerae expresses chitinolytic machinery that allows growth on a chitin-containing medium (30). Chitin utilization in Vibrio species is thought to involve the coordination of secreted chitinases, porins, transporters, and carbohydrate-specific enzymes (20). The expression of a number of these factors was shown to be regulated by the two-component sensor/kinase ChiS, which senses environmental chitin (30, 31). The zoonotic pathogen Francisella tularensis subsp. novicida forms biofilms on chitinous surfaces and uses chitin as a carbon source via the production of chitinases (32). F. tularensis type B expresses an additional chitinase (ChiD) that promotes infection of ticks, one of the main vectors of transmission, potentially by utilizing chitin present in their exoskeleton as a carbon source (33). E. faecalis expresses two chitinolytic enzymes, EfChiA18A and EndoE, and a chitin-binding protein, EfCBM33A, that work in tandem to degrade chitin and utilize it as a carbon source (21–23). Lastly, E. coli can also utilize chitin as a carbon source through the expression of chitinase machinery (24). The ability of these bacterial pathogens to utilize environmental chitin likely does not impact pathogenesis in the host environment. However, chitinases have also been shown to be directly important for mammalian infection.
CHITINASE ROLES DURING INFECTION
Over recent years, bacterial chitinases have been shown to exhibit alternative roles not involving the direct cleavage of chitin polymers (16, 34) (Fig. 2). A meta-genomic analysis of pathogenic species revealed that the chitin-binding domain 3, a common domain in chitinases, is evolutionarily conserved in virulence factors and co-occurs with peptidase, glycosyl hydrolase, kinase, hemagglutinin-acting, and collagen-binding domains (35). The presence of a chitin-binding domain alongside domains with known pathogenic function indicates that interactions with chitin or chitin-like molecules within mammalian hosts may be important for infection. Various bacterial chitinases and chitin-binding proteins that have been implicated in or suggested to play a role in infection will be discussed in the following sections.
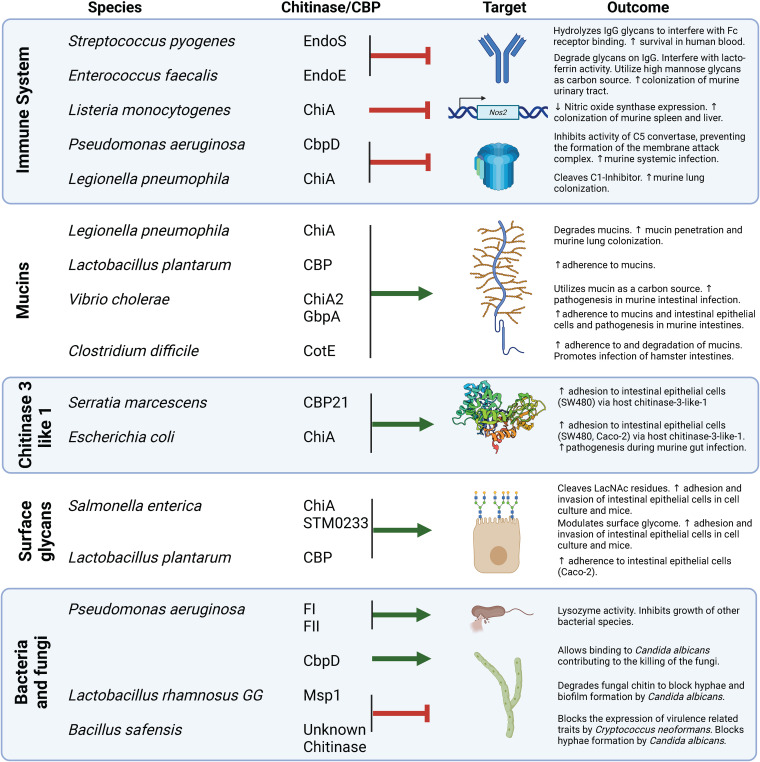
FIG 2.
Bacterial chitinases/CBPs with activity toward alternative molecular targets that are relevant to human infection.
SKIN COLONIZATION
The skin is an epithelial barrier that serves as vertebrate animals’ first line of protection against the outside world. The role of bacterial chitinases during skin colonization and infection has not been studied in great detail, but there is some evidence for their involvement. The opportunistic pathogen Staphylococcus epidermidis expresses the putative chitinase SE0760 during skin colonization but not during colonization of the nasal cavity (36). It has therefore been suggested that this chitinase is involved in the invasion of skin tissue (37). However, this hypothesis has not been confirmed yet by experimental studies. Future research needs to be performed to fully understand the role of this chitinase for skin colonization. While other bacterial skin pathogens, such as Staphylococcus aureus and Streptococcus pyogenes, carry chitinases, their role in skin infection has not been elucidated. These chitinases were studied for their role during systemic infection, which will be discussed in the following section.
SYSTEMIC INFECTION
Bacterial chitinases have been shown to directly contribute to bacterial pathogenesis during systemic infections. In numerous pathogens, chitinase expression leads to enhanced virulence. Specific targets and functions of chitinases in mammals have been described for multiple pathogens, as we will detail below. However, we will first describe chitinases with undefined targets that have been associated with bacterial pathogenesis. For example, a mouse model of Staphylococcus aureus sepsis revealed the chitinase-related protein SAUSA300_0964 as a potent virulence factor that is upregulated in a hypervirulent protease-deficient strain of S. aureus (38). SAUSA300_0964 expression is regulated by the Sae system alongside other virulence factors and is upregulated in the presence of the antibiotic colistin, but its function during infection is still unknown (39, 40). The pathogen Chromobacterium violaceum expresses the putative chitinase CV_4240, which is indirectly linked to virulence. Expression of virulence genes in C. violaceum is regulated by the CviR/CviI quorum sensing system. The same system was shown to regulate expression of CV_4240, indicating a potential role for the chitinase during infection (41). This putative chitinase was also found to be secreted and likely also contributes to chitin degradation by C. violaceum (42, 43). It is still unclear if chitinase activity contributes to C. violaceum pathogenesis or if it is only required for degradation of environmental chitin outside a human host. Additionally, the zoonotic pathogen Francisella tularensis type A upregulates a GH18 family chitinase in the spleen of infected mice (44). The chitinases ChiA and ChiB expressed by F. tularensis subsp. novicida have been reported to regulate biofilm production, affecting the ability of F. tularensis subsp. novicida to adhere to, invade, and replicate inside lung epithelial cells (32, 45).
Many bacterial chitinases seem to be involved in the modulation of the host immune response to infection, interfering with antibody function, the complement cascade, or phagocyte function. Their function therefore enables pathogenic microbes to efficiently infect their hosts, as we will describe in the following sections.
Targeting of glycans present on IgG and other immune system components.
A well-characterized GH18 glycosyl hydrolase that interferes with antibody function during systemic infection is endoglycosidase S (EndoS) of Streptococcus pyogenes. This enzyme hydrolyzes core GlcNAc residues on IgG glycans in human serum (46). EndoS contains a GH18 chitinase domain and a CBM. Mutation of the active residue of the GH18 chitinase domain blocks catalytic activity (47), while deletion of the CBM slows the rate of glycan degradation (48). The modification of IgG glycans by EndoS promotes survival of S. pyogenes in human blood by interfering with Fc receptor binding (49). A similar mechanism is exploited by the animal pathogen Corynebacterium pseudotuberculosis, which mainly infects sheep and goats. This bacterium expresses a GH18 chitinase domain-containing enzyme, CP40, that shares homology with EndoS of S. pyogenes (50). CP40 does not show activity toward chitin but, instead, degrades biantennary glycans on IgG (50). Enterococcus faecalis expresses one chitin-binding protein (EfCBM33A) and three chitinase domain-containing enzymes (EndoE, EfChi18A, and EfEndo18A). E. faecalis upregulates EfCBM33A and EfChi18A when exposed to horse blood and human urine (51, 52), but their functions during infection are still unclear. EndoE and EfEndo18A are better characterized and have been shown to interfere with different arms of the immune response. EndoE and EfEndo18A are characterized as β-1,4 endo-β-N-acetylglucosaminidases and demonstrate activity toward mammalian glycans. EndoE contains a GH18 chitinase domain and a GH20 domain. The GH18 domain of EndoE is responsible for cleaving high-mannose glycans, which was determined based on in vitro activity toward glycans present on RNase B, a model high-mannose protein (53, 54). EfEndo18A also demonstrates activity toward these high-mannose glycans. The liberated mannose can then be utilized as a carbon source by E. faecalis (55, 56). Complex glycans on IgG are also cleaved by EndoE, but there are conflicting reports on whether this activity is due to the GH18 or GH20 domain (53, 55). A recent study suggests that the GH18 and GH20 domains work in conjunction to degrade complex glycans, with GH20 targeting GlcNAc residues on branches and GH18 targeting core GlcNAcs (57). Finally, EndoE is capable of deglycosylating lactoferrin, thereby blocking its activity (58). This host protein normally interferes with bacterial biofilm production. E. faecalis can utilize the glycans released from lactoferrin as a carbon source (58). Deletion of all three E. faecalis GH18 enzymes results in reduced colonization of the pathogen in a murine urinary tract infection model (55).
Complement inactivation.
Pseudomonas aeruginosa secretes a chitin-binding protein, CbpD, which has now been demonstrated to have lytic polysaccharide monooxygenase activity (59, 60). The catalytic activity of CbpD is protective against the host complement cascade during systemic infection of mice. Activation of the host complement cascade on bacterial surfaces results in the deposition of a membrane attack complex and subsequent lysis of Gram-negative bacteria. CbpD inhibits a crucial step in this cascade by preventing the assembly of the C5 convertase and therefore the formation of the membrane attack complex. This study also suggests an additional role for CbpD in influencing the expression of multiple proteins, including other virulence factors (60). Studies have found that CbpD can carry various types of post-translational modifications, such as phosphorylation, succinylation, and acetylation (61, 62). Some of the lysine residues found in CbpD can exhibit more than one modification, and there are even changes in the overall levels of phosphorylation depending on if CbpD is intracellular or extracellular (62). This variation in post-translational modifications could lead to conformational changes and different protein interactions (62), which may explain the seemingly dual functions of CbpD in inhibiting C5 convertase and modulating protein expression (60).
Interference with macrophage function.
The gastrointestinal pathogen Listeria monocytogenes produces two chitinases (ChiA and ChiB) and a chitin-binding protein (lmo2467) that surprisingly do not seem to contribute to intestinal infection (63, 64). Instead, they are required for colonization of systemic sites such as the spleen and liver in an intravenous mouse model of infection (63). It appears that chitinase activity is intricately linked to virulence based on the fact that the Mour transcription factor regulates chitinase activity as well as the virulence-associated processes of biofilm formation and cell invasion (65). However, chitinase expression during infection could be regulated differently, since strains lacking Mour did not display a colonization defect during intravenous infection (65). This alternative regulatory system could involve the putative internalin/autolysin Lmo0327 and the putative transcriptional activator Lmo0325. The deletion of either of these genes blocks chitinase transcription (66). Other potential regulators of chitinase expression during L. monocytogenes infection are the virulence factor regulator PrfA, the sigma factor σB, the small RNA (sRNA) LhrA, and the agr quorum sensing system (64, 67, 68). ChiA is also upregulated during infection of murine macrophages (69). Further, it has been shown that ChiA promotes systemic infection through the downregulation of nitric oxide synthase expression (70). The role for ChiB during infection is still not well understood. An L. monocytogenes strain deficient in ChiB production was isolated from a clinical gastroenteritis infection, confirming data from intestinal epithelial cell (63, 64) infections that ChiB is likely not required for intestinal infection (71).
RESPIRATORY INFECTION
Chitinases are also emerging as virulence factors for respiratory pathogens. Pseudomonas aeruginosa expresses an active chitinase, ChiC, even though P. aeruginosa cannot use chitin as a carbon source in vitro (72, 73). ChiC production by P. aeruginosa is regulated by the GacS/GacA two-component system, which also regulates virulence in the lungs of mice with cystic fibrosis (74–76). ChiC is likely clinically relevant, as expression was upregulated in artificial-sputum medium, a model for cystic fibrosis sputum (77), and in hypervirulent strains isolated from cystic fibrosis patients (78, 79). ChiC is known to be secreted, but its actual functions during infection are still unknown (73). The chitin-binding protein CbpD, whose role for inhibiting the complement cascade during systemic infection we described in the previous section, was also found to be upregulated in artificial-sputum medium (80). CbpD is regulated by a quorum sensing system required for virulence in the lungs of rats, indicating that its role in complement cascade interference may extend to lung infection (81, 82). Further studies are required to explore if CbpD contributes to lung infection.
Legionella pneumophila expresses a chitinase (ChiA) that is secreted via the type II secretion system and is also contained within outer membrane vesicles (83, 84). While ChiA has activity toward chitin, it also has peptidase activity that degrades mammalian mucins to promote mucin layer penetration, and it cleaves the complement component human C1-inhibitor, potentially interfering with activation of the complement cascade (9, 83). This function could explain why a ChiA-deficient L. pneumophila strain does not persist as long as a wild-type strain in murine lungs (83). ChiA was also shown to be translocated to the cytosolic side of the Legionella-containing vacuole within infected macrophages (85). However, the function of this chitinase within a host cell is unclear since it was not required for intracellular replication (83).
GASTROINTESTINAL INFECTION
The roles of bacterial chitinases in mammals have usually been studied in the context of bacterial pathogens. However, some commensal bacteria also produce chitinases. For instance, the gut commensal Clostridium paraputrificum produces several active chitinases, one of which, Chit62J4, shows optimal activity at pH 5.5, which is relevant to the intestinal tract (86). This indicates that C. paraputrificum may target dietary chitin or intestinal glycans as a carbon source during gastrointestinal colonization. Another gut commensal, Lactobacillus plantarum, expresses a chitin-binding protein that promotes adhesion to mucins and intestinal epithelial cells (87).
Numerous gastrointestinal pathogens express chitinases that have been implicated in promoting intestinal infection. In Vibrio cholerae, two chitinases (ChiA1, ChiA2) and the chitin-binding protein GbpA have been fairly well characterized in their roles during infection. They are known to be secreted via the type II secretion system (88, 89). ChiA1 is conserved in V. cholerae strains isolated from pandemics but is not conserved in nonpandemic strains, strongly suggesting a potential role for this chitinase during infection (90). The chitinase ChiA2 was shown to be required for intestinal infection, as it releases nutrient sources from intestinal mucins and promotes intestinal survival (8). ChiS, the main regulator of chitinase expression, is activated not only in the presence of chitin (30, 31), but also by mucins, which leads to higher expression of ChiA2 (91). The expression of ChiS, ChiA1, and ChiA2 is ultimately regulated by the transcription factor CytR, which also regulates other virulence factors to promote intestinal pathogenicity (92). In addition to chitinases, the chitin-binding protein GbpA also plays a role during infection. Deletion of gpbA results in less adherence of V. cholerae to intestinal epithelial cells and less fluid buildup in the intestines of infected mice (89, 93). Further studies revealed that this phenotype is due to GbpA’s ability to bind GlcNAc residues present on intestinal mucins (93, 94). Interestingly, GbpA may only be required for early attachment and infection, since its expression is downregulated at high cell density (95). GbpA may also function as a toxin during infection, as treatment of colonic epithelial HT29 cells with purified GbpA induced necrosis (96). GbpA expression was found to be uniquely regulated by a complex interaction between the Vc1 riboswitch, c-di-GMP, and cAMP receptor protein (CRP) signaling systems (97, 98). Similar to some of the other bacterial chitin-binding proteins, GbpA demonstrates lytic polysaccharide monooxygenase activity (99).
We discussed the role of CBP21 in chitin degradation by Serratia marcescens; however, CBP21 has an additional role during infection. CBP21 can promote adhesion to colonic epithelial cells through interactions with the host chitin-binding protein chitinase-3-like-1 (100). The chitinase ChiA, produced by adherent-invasive Escherichia coli, also interacts with a glycan on chitinase-3-like-1 to promote adhesion to intestinal epithelial cells (101). Clostridium difficile incorporates the bifunctional enzyme CotE onto the surface of spores to promote colonization of the intestines (102). CotE demonstrates chitinase and peroxiredoxin activity, allowing C. difficile spores to bind to and degrade intestinal mucins, leading to enhanced pathogenicity in a hamster model of intestinal infection (102–104). CotE has been proposed as a therapeutic target, either for use in vaccines (105, 106) or as a potential target for inhibiting spore formation (107).
Chitinases produced by Salmonella enterica have recently been shown to contribute to intestinal infection (108, 109). Chitinase gene expression is upregulated during infection of HeLa cells, murine macrophages, and the gastrointestinal system of chickens (110–112). One of the chitinases, ChiA, demonstrates activity toward a common component of mammalian glycoproteins, N-acetyllactosamine (7, 113). Our group has shown that the chitinase ChiA and the putative chitinase STM0233 are upregulated in the gastrointestinal tract of mice and are required for the efficient invasion of small intestinal tissue by S. enterica serovar Typhimurium (108). This invasion phenotype is likely due to the targeting of surface glycans, since the presence of these chitinases induces specific changes in the abundance of GlcNAc-containing glycans (108). In a simultaneously published study, Chandra et al. confirmed the role for ChiA in promoting invasion and interaction with surface glycans and further showed that ChiA of S. enterica serovar Typhi has a similar function (109). That same group also revealed roles for ChiA in maintaining Salmonella-containing vacuoles during intracellular infection and modulating the host immune response during infection with S. Typhi and S. Typhimurium (109).
INTERACTIONS BETWEEN BACTERIAL CHITINASES AND OTHER MICROBES
We have discussed multiple examples of how bacterial chitinases interact with the host. However, there are also examples of bacterial chitinases that interact with other bacteria or fungi. Pseudomonas aeruginosa chitinases (FI and FII) demonstrate lysozyme activity, targeting the cell wall of other bacterial species and inhibiting their growth (114). Bacterial species have also demonstrated activity toward chitin present in the cell wall of fungal pathogens. P. aeruginosa has been shown to bind and kill Candida albicans hyphae (115). It was later shown that the chitin-binding protein CbpD allows binding of P. aeruginosa to C. albicans hyphae, likely contributing to this killing mechanism (116). The probiotic Lactobacillus rhamnosus GG also interacts with C. albicans by blocking hyphal morphogenesis and biofilm formation (117). It was further shown that the peptidoglycan hydrolase MspI blocks hyphal formation by degrading fungal chitin (117). The soil bacterium Bacillus safensis has also been shown to block biofilm production, capsulation, and melanization by Cryptococcus neoformans. Additionally, B. safensis can bind to and block hyphal formation by C. albicans. Both of these antifungal mechanisms were shown to be dependent on chitinase activity (118).
SUMMARY AND OUTLOOK
Despite the target limitation that is hinted at with the name “chitin”ase, bacterial chitinases interact with a broad range of molecular targets and play an important role in infection. We have highlighted evidence that chitinases interact with mammalian N-linked surface glycans, mucins, and immune system components to promote epithelial cell adhesion, liberate nutritional resources, or interfere with the host immune response. The activity of GH18 and GH19 glycosyl hydrolases toward GlcNAc-containing molecules explains their important but divergent roles during infection of different body sites with different pathogens.
While we continue to learn more about the roles of chitinases during pathogenesis, much remains unknown, such as the targets of many bacterial chitinases. Purified glycans are commonly used to assess the activity of bacterial chitinases, but recent advances in glycome analysis techniques can be applied to further understand the role of these chitinases in interacting with host glycans (119). Glycan arrays can be used to detect binding across a wide range of glycan structures (120–122). Fluorescently tagged lectins can be used to detect specific saccharide moieties on the surface of cells. Ma et al. have incorporated metal-conjugated lectin staining into a cytometry time of flight technique to detect surface glycans at a single-cell level (123). While mass spectrometry has been largely used to quantify glycan abundance in a total sample, mass spectrometry imaging can identify the spatial distribution of glycans in tissue (124). With the help of these novel methods, researchers might more easily and comprehensively understand the roles of chitinases during infection.
Targeting bacterial chitinases may be a useful method for designing antivirulence therapeutics. It has been proposed that the spore-inhibiting antibiotic fidaxomicin may target CotE on C. difficile (107). One potential strategy to limit bacterial infections is designing small molecules to inhibit chitinase activity in order to limit adhesion or block interactions with the immune system and treat active infections. Targeting chitinases may also be an effective way to limit the spread of foodborne pathogens in livestock. Humans express two chitinases, chitotriosidase 1 (CHIT1) and acid mammalian chitinase (AMCase), which are thought to contribute to the type 2 immune response to environmental chitin and chitin-containing pathogens, such as helminths (125, 126). Therefore, care must be taken to ensure that a potential inhibitor would not interfere with the ability of these host chitinases to contribute to an effective innate immune response.
ACKNOWLEDGMENTS
This work was supported by a grant from the National Institute of Health to J.B. (AI143641-01) and by startup funds from the Department of Microbiology and Immunology at the University of Illinois Chicago.
We thank Olivia A. Todd for critical reading of the manuscript. Figures were created at Biorender.com.
Biographies

Jason R. Devlin, Ph.D., was a graduate research assistant in the laboratory of Dr. Judith Behnsen at the University of Illinois Chicago and recently completed his Ph.D. in microbiology and immunology. His research interests involve understanding the interactions between pathogens, the microbiota, and the host and their contributions to infection. His Ph.D. research investigated the roles of Salmonella Typhimurium chitinases during gastrointestinal infection.

Judith Behnsen, Ph.D., is an assistant professor in the Department of Microbiology and Immunology at the University of Illinois Chicago. Research in her lab focuses on interspecies microbial interactions. Her lab is specifically interested in understudied cross-kingdom interactions in the gut environment, such as the role of commensal intestinal fungi during Salmonella Typhimurium pathogenesis. The lab’s interest also expands to novel bacterium-bacterium interactions, and they recently characterized how Proteus mirabilis reduces the viability of competitor bacteria.
Contributor Information
Judith Behnsen, Email: jbehnsen@uic.edu.
Karen M. Ottemann, University of California at Santa Cruz Department of Microbiology and Environmental Toxicology
REFERENCES
- 1.Shahidi F, Abuzaytoun R. 2005. Chitin, chitosan, and co-products: chemistry, production, applications, and health effects. Adv Food Nutr Res 49:93–135. doi: 10.1016/S1043-4526(05)49003-8. [DOI] [PubMed] [Google Scholar]
- 2.Adrangi S, Faramarzi MA. 2013. From bacteria to human: a journey into the world of chitinases. Biotechnol Adv 31:1786–1795. doi: 10.1016/j.biotechadv.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 3.Hamid R, Khan MA, Ahmad M, Ahmad MM, Abdin MZ, Musarrat J, Javed S. 2013. Chitinases: an update. J Pharm Bioallied Sci 5:21–29. doi: 10.4103/0975-7406.106559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Itoh T, Kimoto H. 2019. Bacterial chitinase system as a model of chitin biodegradation. Adv Exp Med Biol 1142:131–151. doi: 10.1007/978-981-13-7318-3_7. [DOI] [PubMed] [Google Scholar]
- 5.Veliz EA, Martínez-Hidalgo P, Hirsch AM. 2017. Chitinase-producing bacteria and their role in biocontrol. AIMS Microbiol 3:689–705. doi: 10.3934/microbiol.2017.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bokma E, van Koningsveld GA, Jeronimus-Stratingh M, Beintema JJ. 1997. Hevamine, a chitinase from the rubber tree Hevea brasiliensis, cleaves peptidoglycan between the C-1 of N-acetylglucosamine and C-4 of N-acetylmuramic acid and therefore is not a lysozyme. FEBS Lett 411:161–163. doi: 10.1016/s0014-5793(97)00682-0. [DOI] [PubMed] [Google Scholar]
- 7.Larsen T, Petersen BO, Storgaard BG, Duus JØ, Palcic MM, Leisner JJ. 2011. Characterization of a novel Salmonella Typhimurium chitinase which hydrolyzes chitin, chitooligosaccharides and an N-acetyllactosamine conjugate. Glycobiology 21:426–436. doi: 10.1093/glycob/cwq174. [DOI] [PubMed] [Google Scholar]
- 8.Mondal M, Nag D, Koley H, Saha DR, Chatterjee NS. 2014. The Vibrio cholerae extracellular chitinase ChiA2 is important for survival and pathogenesis in the host intestine. PLoS One 9:e103119. doi: 10.1371/journal.pone.0103119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rehman S, Grigoryeva LS, Richardson KH, Corsini P, White RC, Shaw R, Portlock TJ, Dorgan B, Zanjani ZS, Fornili A, Cianciotto NP, Garnett JA. 2020. Structure and functional analysis of the Legionella pneumophila chitinase ChiA reveals a novel mechanism of metal-dependent mucin degradation. PLoS Pathog 16:e1008342. doi: 10.1371/journal.ppat.1008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tamadoni Jahromi S, Barzkar N. 2018. Marine bacterial chitinase as sources of energy, eco-friendly agent, and industrial biocatalyst. Int J Biol Macromol 120:2147–2154. doi: 10.1016/j.ijbiomac.2018.09.083. [DOI] [PubMed] [Google Scholar]
- 11.Chen W, Jiang X, Yang Q. 2020. Glycoside hydrolase family 18 chitinases: the known and the unknown. Biotechnol Adv 43:107553. doi: 10.1016/j.biotechadv.2020.107553. [DOI] [PubMed] [Google Scholar]
- 12.Henrissat B, Davies G. 1997. Structural and sequence-based classification of glycoside hydrolases. Curr Opin Struct Biol 7:637–644. doi: 10.1016/s0959-440x(97)80072-3. [DOI] [PubMed] [Google Scholar]
- 13.Fujita K, Shimomura K, Yamamoto KI, Yamashita T, Suzuki K. 2006. A chitinase structurally related to the glycoside hydrolase family 48 is indispensable for the hormonally induced diapause termination in a beetle. Biochem Biophys Res Commun 345:502–507. doi: 10.1016/j.bbrc.2006.04.126. [DOI] [PubMed] [Google Scholar]
- 14.Terwisscha van Scheltinga AC, Armand S, Kalk KH, Isogai A, Henrissat B, Dijkstra BW. 1995. Stereochemistry of chitin hydrolysis by a plant chitinase/lysozyme and X-ray structure of a complex with allosamidin: evidence for substrate assisted catalysis. Biochemistry 34:15619–15623. doi: 10.1021/bi00048a003. [DOI] [PubMed] [Google Scholar]
- 15.Iseli B, Armand S, Boller T, Neuhaus JM, Henrissat B. 1996. Plant chitinases use two different hydrolytic mechanisms. FEBS Lett 382:186–188. doi: 10.1016/0014-5793(96)00174-3. [DOI] [PubMed] [Google Scholar]
- 16.Frederiksen RF, Paspaliari DK, Larsen T, Storgaard BG, Larsen MH, Ingmer H, Palcic MM, Leisner JJ. 2013. Bacterial chitinases and chitin-binding proteins as virulence factors. Microbiology (Reading) 159:833–847. doi: 10.1099/mic.0.051839-0. [DOI] [PubMed] [Google Scholar]
- 17.Churklam W, Aunpad R. 2020. Enzymatic characterization and structure-function relationship of two chitinases, LmChiA and LmChiB, from Listeria monocytogenes. Heliyon 6:e04252. doi: 10.1016/j.heliyon.2020.e04252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leisner JJ, Larsen MH, Jørgensen RL, Brøndsted L, Thomsen LE, Ingmer H. 2008. Chitin hydrolysis by Listeria spp., including L. monocytogenes. Appl Environ Microbiol 74:3823–3830. doi: 10.1128/AEM.02701-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paspaliari DK, Loose JSM, Larsen MH, Vaaje-Kolstad G. 2015. Listeria monocytogenes has a functional chitinolytic system and an active lytic polysaccharide monooxygenase. FEBS J 282:921–936. doi: 10.1111/febs.13191. [DOI] [PubMed] [Google Scholar]
- 20.Hunt DE, Gevers D, Vahora NM, Polz MF. 2008. Conservation of the chitin utilization pathway in the Vibrionaceae. Appl Environ Microbiol 74:44–51. doi: 10.1128/AEM.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keffeler EC, Parthasarathy S, Abdullahi ZH, Hancock LE. 2021. Metabolism of poly-β1,4-N-acetylglucosamine substrates and importation of N-acetylglucosamine and glucosamine by Enterococcus faecalis. J Bacteriol 203:e0037121. doi: 10.1128/JB.00371-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leisner JJ, Larsen MH, Ingmer H, Petersen BO, Duus JØ, Palcic MM. 2009. Cloning and comparison of phylogenetically related chitinases from Listeria monocytogenes EGD and Enterococcus faecalis V583. J Appl Microbiol 107:2080–2087. doi: 10.1111/j.1365-2672.2009.04420.x. [DOI] [PubMed] [Google Scholar]
- 23.Vaaje-Kolstad G, Bøhle LA, Gåseidnes S, Dalhus B, Bjørås M, Mathiesen G, Eijsink VGH. 2012. Characterization of the chitinolytic machinery of Enterococcus faecalis V583 and high-resolution structure of its oxidative CBM33 enzyme. J Mol Biol 416:239–254. doi: 10.1016/j.jmb.2011.12.033. [DOI] [PubMed] [Google Scholar]
- 24.Walter A, Friz S, Mayer C. 2021. Chitin, chitin oligosaccharide, and chitin disaccharide metabolism of Escherichia coli revisited: reassignment of the roles of ChiA, ChbR, ChbF, and ChbG. Microb Physiol 31:178–194. doi: 10.1159/000515178. [DOI] [PubMed] [Google Scholar]
- 25.Liu J, Xu Q, Wu Y, Sun D, Zhu J, Liu C, Liu W. 2023. Carbohydrate-binding modules of ChiB and ChiC promote the chitinolytic system of Serratia marcescens BWL1001. Enzyme Microb Technol 162:110118. doi: 10.1016/j.enzmictec.2022.110118. [DOI] [PubMed] [Google Scholar]
- 26.Vaaje-Kolstad G, Horn SJ, Sørlie M, Eijsink VGH. 2013. The chitinolytic machinery of Serratia marcescens: a model system for enzymatic degradation of recalcitrant polysaccharides. FEBS J 280:3028–3049. doi: 10.1111/febs.12181. [DOI] [PubMed] [Google Scholar]
- 27.Costa MDAA, Owen RA, Tammsalu T, Buchanan G, Palmer T, Sargent F. 2019. Controlling and co-ordinating chitinase secretion in a Serratia marcescens population. Microbiology (Reading) 165:1233–1244. doi: 10.1099/mic.0.000856. [DOI] [PubMed] [Google Scholar]
- 28.Hamilton JJ, Marlow VL, Owen RA, Costa MDAA, Guo M, Buchanan G, Chandra G, Trost M, Coulthurst SJ, Palmer T, Stanley-Wall NR, Sargent F. 2014. A holin and an endopeptidase are essential for chitinolytic protein secretion in Serratia marcescens. J Cell Biol 207:615–626. doi: 10.1083/jcb.201404127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Owen RA, Fyfe PK, Lodge A, Biboy J, Vollmer W, Hunter WN, Sargent F. 2018. Structure and activity of ChiX: a peptidoglycan hydrolase required for chitinase secretion by Serratia marcescens. Biochem J 475:415–428. doi: 10.1042/BCJ20170633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meibom KL, Li XB, Nielsen AT, Wu CY, Roseman S, Schoolnik GK. 2004. The Vibrio cholerae chitin utilization program. Proc Natl Acad Sci USA 101:2524–2529. doi: 10.1073/pnas.0308707101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X, Roseman S. 2004. The chitinolytic cascade in Vibrios is regulated by chitin oligosaccharides and a two-component chitin catabolic sensor/kinase. Proc Natl Acad Sci USA 101:627–631. doi: 10.1073/pnas.0307645100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Margolis JJ, El-Etr S, Joubert L-M, Moore E, Robison R, Rasley A, Spormann AM, Monack DM. 2010. Contributions of Francisella tularensis subsp. novicida chitinases and Sec secretion system to biofilm formation on chitin. Appl Environ Microbiol 76:596–608. doi: 10.1128/AEM.02037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tully BG, Huntley JF. 2020. A Francisella tularensis chitinase contributes to bacterial persistence and replication in two major U.S. tick vectors. Pathogens 9:1037. doi: 10.3390/pathogens9121037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tran HT, Barnich N, Mizoguchi E. 2011. Potential role of chitinases and chitin-binding proteins in host-microbial interactions during the development of intestinal inflammation. Histol Histopathol 26:1453–1464. doi: 10.14670/HH-26.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patel S, Rauf A, Meher BR. 2017. In silico analysis of ChtBD3 domain to find its role in bacterial pathogenesis and beyond. Microb Pathog 110:519–526. doi: 10.1016/j.micpath.2017.07.047. [DOI] [PubMed] [Google Scholar]
- 36.Teichmann P, Both A, Wolz C, Hornef MW, Rohde H, Yazdi AS, Burian M. 2022. The Staphylococcus epidermidis transcriptional profile during carriage. Front Microbiol 13:896311. doi: 10.3389/fmicb.2022.896311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y-Q, Ren S-X, Li H-L, Wang Y-X, Fu G, Yang J, Qin Z-Q, Miao Y-G, Wang W-Y, Chen R-S, Shen Y, Chen Z, Yuan Z-H, Zhao G-P, Qu D, Danchin A, Wen Y-M. 2003. Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228). Mol Microbiol 49:1577–1593. doi: 10.1046/j.1365-2958.2003.03671.x. [DOI] [PubMed] [Google Scholar]
- 38.Gimza BD, Jackson JK, Frey AM, Budny BG, Chaput D, Rizzo DN, Shaw LN. 2021. Unraveling the impact of secreted proteases on hypervirulence in Staphylococcus aureus. mBio 12:e03288-20. doi: 10.1128/mBio.03288-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cassat JE, Hammer ND, Campbell JP, Benson MA, Perrien DS, Mrak LN, Smeltzer MS, Torres VJ, Skaar EP. 2013. A secreted bacterial protease tailors the Staphylococcus aureus virulence repertoire to modulate bone remodeling during osteomyelitis. Cell Host Microbe 13:759–772. doi: 10.1016/j.chom.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haaber J, Friberg C, McCreary M, Lin R, Cohen SN, Ingmer H. 2015. Reversible antibiotic tolerance induced in Staphylococcus aureus by concurrent drug exposure. mBio 6:e02268-14. doi: 10.1128/mBio.02268-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stauff DL, Bassler BL. 2011. Quorum sensing in Chromobacterium violaceum: DNA recognition and gene regulation by the CviR receptor. J Bacteriol 193:3871–3878. doi: 10.1128/JB.05125-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ciprandi A, da Silva WM, Santos AV, de Castro Pimenta AM, Carepo MSP, Schneider MPC, Azevedo V, Silva A. 2013. Chromobacterium violaceum: important insights for virulence and biotechnological potential by exoproteomic studies. Curr Microbiol 67:100–106. doi: 10.1007/s00284-013-0334-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chernin LS, Winson MK, Thompson JM, Haran S, Bycroft BW, Chet I, Williams P, Stewart GS. 1998. Chitinolytic activity in Chromobacterium violaceum: substrate analysis and regulation by quorum sensing. J Bacteriol 180:4435–4441. doi: 10.1128/JB.180.17.4435-4441.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Twine SM, Mykytczuk NCS, Petit MD, Shen H, Sjöstedt A, Wayne Conlan J, Kelly JF. 2006. In vivo proteomic analysis of the intracellular bacterial pathogen, Francisella tularensis, isolated from mouse spleen. Biochem Biophys Res Commun 345:1621–1633. doi: 10.1016/j.bbrc.2006.05.070. [DOI] [PubMed] [Google Scholar]
- 45.Chung MC, Dean S, Marakasova ES, Nwabueze AO, van Hoek ML. 2014. Chitinases are negative regulators of Francisella novicida biofilms. PLoS One 9:e93119. doi: 10.1371/journal.pone.0093119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Collin M, Olsén A. 2001. EndoS, a novel secreted protein from Streptococcus pyogenes with endoglycosidase activity on human IgG. EMBO J 20:3046–3055. doi: 10.1093/emboj/20.12.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Allhorn M, Olsén A, Collin M. 2008. EndoS from Streptococcus pyogenes is hydrolyzed by the cysteine proteinase SpeB and requires glutamic acid 235 and tryptophans for IgG glycan-hydrolyzing activity. BMC Microbiol 8:3. doi: 10.1186/1471-2180-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dixon EV, Claridge JK, Harvey DJ, Baruah K, Yu X, Vesiljevic S, Mattick S, Pritchard LK, Krishna B, Scanlan CN, Schnell JR, Higgins MK, Zitzmann N, Crispin M. 2014. Fragments of bacterial endoglycosidase s and immunoglobulin g reveal subdomains of each that contribute to deglycosylation. J Biol Chem 289:13876–13889. doi: 10.1074/jbc.M113.532812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Collin M, Svensson MD, Sjöholm AG, Jensenius JC, Sjöbring U, Olsén A. 2002. EndoS and SpeB from Streptococcus pyogenes inhibit immunoglobulin-mediated opsonophagocytosis. Infect Immun 70:6646–6651. doi: 10.1128/IAI.70.12.6646-6651.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shadnezhad A, Naegeli A, Collin M. 2016. CP40 from Corynebacterium pseudotuberculosis is an endo-β-N-acetylglucosaminidase. BMC Microbiol 16:261. doi: 10.1186/s12866-016-0884-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vebø HC, Snipen L, Nes IF, Brede DA. 2009. The transcriptome of the nosocomial pathogen Enterococcus faecalis V583 reveals adaptive responses to growth in blood. PLoS One 4:e7660. doi: 10.1371/journal.pone.0007660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vebø HC, Solheim M, Snipen L, Nes IF, Brede DA. 2010. Comparative genomic analysis of pathogenic and probiotic Enterococcus faecalis isolates, and their transcriptional responses to growth in human urine. PLoS One 5:e12489. doi: 10.1371/journal.pone.0012489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Collin M, Fischetti VA. 2004. A novel secreted endoglycosidase from Enterococcus faecalis with activity on human immunoglobulin G and ribonuclease B. J Biol Chem 279:22558–22570. doi: 10.1074/jbc.M402156200. [DOI] [PubMed] [Google Scholar]
- 54.Roberts G, Tarelli E, Homer KA, Philpott-Howard J, Beighton D. 2000. Production of an endo-beta-N-acetylglucosaminidase activity mediates growth of Enterococcus faecalis on a high-mannose-type glycoprotein. J Bacteriol 182:882–890. doi: 10.1128/JB.182.4.882-890.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Keffeler EC, Iyer VS, Henderson AJ, Huck IL, Schwarting N, Cortez A, Hancock LE. 2021. Activity of CcpA-regulated GH18 family glycosyl hydrolases that contributes to nutrient acquisition and fitness in Enterococcus faecalis. Infect Immun 89:e0034321. doi: 10.1128/IAI.00343-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bøhle LA, Mathiesen G, Vaaje-Kolstad G, Eijsink VGH. 2011. An endo-β-N-acetylglucosaminidase from Enterococcus faecalis V583 responsible for the hydrolysis of high-mannose and hybrid-type N-linked glycans. FEMS Microbiol Lett 325:123–129. doi: 10.1111/j.1574-6968.2011.02419.x. [DOI] [PubMed] [Google Scholar]
- 57.García-Alija M, Du JJ, Ordóñez I, Diz-Vallenilla A, Moraleda-Montoya A, Sultana N, Huynh CG, Li C, Donahue TC, Wang L-X, Trastoy B, Sundberg EJ, Guerin ME. 2022. Mechanism of cooperative N-glycan processing by the multi-modular endoglycosidase EndoE. Nat Commun 13:1137. doi: 10.1038/s41467-022-28722-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garbe J, Sjögren J, Cosgrave EFJ, Struwe WB, Bober M, Olin AI, Rudd PM, Collin M. 2014. EndoE from Enterococcus faecalis hydrolyzes the glycans of the biofilm inhibiting protein lactoferrin and mediates growth. PLoS One 9:e91035. doi: 10.1371/journal.pone.0091035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Folders J, Tommassen J, van Loon LC, Bitter W. 2000. Identification of a chitin-binding protein secreted by Pseudomonas aeruginosa. J Bacteriol 182:1257–1263. doi: 10.1128/JB.182.5.1257-1263.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Askarian F, Uchiyama S, Masson H, Sørensen HV, Golten O, Bunæs AC, Mekasha S, Røhr ÅK, Kommedal E, Ludviksen JA, Arntzen MØ, Schmidt B, Zurich RH, van Sorge NM, Eijsink VGH, Krengel U, Mollnes TE, Lewis NE, Nizet V, Vaaje-Kolstad G. 2021. The lytic polysaccharide monooxygenase CbpD promotes Pseudomonas aeruginosa virulence in systemic infection. Nat Commun 12:1230. doi: 10.1038/s41467-021-21473-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ouidir T, Jarnier F, Cosette P, Jouenne T, Hardouin J. 2014. Extracellular Ser/Thr/Tyr phosphorylated proteins of Pseudomonas aeruginosa PA14 strain. Proteomics 14:2017–2030. doi: 10.1002/pmic.201400190. [DOI] [PubMed] [Google Scholar]
- 62.Gaviard C, Cosette P, Jouenne T, Hardouin J. 2019. LasB and CbpD virulence factors of Pseudomonas aeruginosa carry multiple post-translational modifications on their lysine residues. J Proteome Res 18:923–933. doi: 10.1021/acs.jproteome.8b00556. [DOI] [PubMed] [Google Scholar]
- 63.Chaudhuri S, Bruno JC, Alonzo F, Xayarath B, Cianciotto NP, Freitag NE. 2010. Contribution of chitinases contributes to Listeria monocytogenes pathogenesis. Appl Environ Microbiol 76:7302–7305. doi: 10.1128/AEM.01338-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Larsen MH, Leisner JJ, Ingmer H. 2010. The chitinolytic activity of Listeria monocytogenes EGD is regulated by carbohydrates but also by the virulence regulator PrfA. Appl Environ Microbiol 76:6470–6476. doi: 10.1128/AEM.00297-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pinheiro J, Lisboa J, Pombinho R, Carvalho F, Carreaux A, Brito C, Pöntinen A, Korkeala H, Dos Santos NMS, Morais-Cabral JH, Sousa S, Cabanes D. 2018. MouR controls the expression of the Listeria monocytogenes Agr system and mediates virulence. Nucleic Acids Res 46:9338–9352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Paspaliari DK, Kastbjerg VG, Ingmer H, Popowska M, Larsen MH. 2017. Chitinase expression in Listeria monocytogenes is influenced by lmo0327, which encodes an internalin-like protein. Appl Environ Microbiol 83:e01283-17. doi: 10.1128/AEM.01283-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nielsen JS, Larsen MH, Lillebæk EMS, Bergholz TM, Christiansen MHG, Boor KJ, Wiedmann M, Kallipolitis BH. 2011. A small RNA controls expression of the chitinase ChiA in Listeria monocytogenes. PLoS One 6:e19019. doi: 10.1371/journal.pone.0019019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Paspaliari DK, Mollerup MS, Kallipolitis BH, Ingmer H, Larsen MH. 2014. Chitinase expression in Listeria monocytogenes is positively regulated by the Agr system. PLoS One 9:e95385. doi: 10.1371/journal.pone.0095385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chatterjee SS, Hossain H, Otten S, Kuenne C, Kuchmina K, Machata S, Domann E, Chakraborty T, Hain T. 2006. Intracellular gene expression profile of Listeria monocytogenes. Infect Immun 74:1323–1338. doi: 10.1128/IAI.74.2.1323-1338.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chaudhuri S, Gantner BN, Ye RD, Cianciotto NP, Freitag NE. 2013. The Listeria monocytogenes ChiA chitinase enhances virulence through suppression of host innate immunity. mBio 4:e00617-12. doi: 10.1128/mBio.00617-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Halbedel S, Prager R, Banerji S, Kleta S, Trost E, Nishanth G, Alles G, Hölzel C, Schlesiger F, Pietzka A, Schlüter D, Flieger A. 2019. A Listeria monocytogenes ST2 clone lacking chitinase ChiB from an outbreak of non-invasive gastroenteritis. Emerg Microbes Infect 8:17–28. doi: 10.1080/22221751.2018.1558960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jagmann N, Brachvogel HP, Philipp B. 2010. Parasitic growth of Pseudomonas aeruginosa in co-culture with the chitinolytic bacterium Aeromonas hydrophila. Environ Microbiol 12:1787–1802. doi: 10.1111/j.1462-2920.2010.02271.x. [DOI] [PubMed] [Google Scholar]
- 73.Folders J, Algra J, Roelofs MS, van Loon LC, Tommassen J, Bitter W. 2001. Characterization of Pseudomonas aeruginosa chitinase, a gradually secreted protein. J Bacteriol 183:7044–7052. doi: 10.1128/JB.183.24.7044-7052.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kay E, Humair B, Dénervaud V, Riedel K, Spahr S, Eberl L, Valverde C, Haas D. 2006. Two GacA-dependent small RNAs modulate the quorum-sensing response in Pseudomonas aeruginosa. J Bacteriol 188:6026–6033. doi: 10.1128/JB.00409-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Faure E, Kwong K, Nguyen D. 2018. Pseudomonas aeruginosa in chronic lung infections: how to adapt within the host? Front Immunol 9:2416. doi: 10.3389/fimmu.2018.02416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Coleman FT, Mueschenborn S, Meluleni G, Ray C, Carey VJ, Vargas SO, Cannon CL, Ausubel FM, Pier GB. 2003. Hypersusceptibility of cystic fibrosis mice to chronic Pseudomonas aeruginosa oropharyngeal colonization and lung infection. Proc Natl Acad Sci USA 100:1949–1954. doi: 10.1073/pnas.0437901100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fung C, Naughton S, Turnbull L, Tingpej P, Rose B, Arthur J, Hu H, Harmer C, Harbour C, Hassett DJ, Whitchurch CB, Manos J. 2010. Gene expression of Pseudomonas aeruginosa in a mucin-containing synthetic growth medium mimicking cystic fibrosis lung sputum. J Med Microbiol 59:1089–1100. doi: 10.1099/jmm.0.019984-0. [DOI] [PubMed] [Google Scholar]
- 78.Salunkhe P, Smart CHM, Morgan JAW, Panagea S, Walshaw MJ, Hart CA, Geffers R, Tümmler B, Winstanley C. 2005. A cystic fibrosis epidemic strain of Pseudomonas aeruginosa displays enhanced virulence and antimicrobial resistance. J Bacteriol 187:4908–4920. doi: 10.1128/JB.187.14.4908-4920.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Manos J, Arthur J, Rose B, Bell S, Tingpej P, Hu H, Webb J, Kjelleberg S, Gorrell MD, Bye P, Harbour C. 2009. Gene expression characteristics of a cystic fibrosis epidemic strain of Pseudomonas aeruginosa during biofilm and planktonic growth. FEMS Microbiol Lett 292:107–114. doi: 10.1111/j.1574-6968.2008.01472.x. [DOI] [PubMed] [Google Scholar]
- 80.Scott NE, Hare NJ, White MY, Manos J, Cordwell SJ. 2013. Secretome of transmissible Pseudomonas aeruginosa AES-1R grown in a cystic fibrosis lung-like environment. J Proteome Res 12:5357–5369. doi: 10.1021/pr4007365. [DOI] [PubMed] [Google Scholar]
- 81.Nouwens AS, Beatson SA, Whitchurch CB, Walsh BJ, Schweizer HP, Mattick JS, Cordwell SJ. 2003. Proteome analysis of extracellular proteins regulated by the las and rhl quorum sensing systems in Pseudomonas aeruginosa PAO1. Microbiology (Reading) 149:1311–1322. doi: 10.1099/mic.0.25967-0. [DOI] [PubMed] [Google Scholar]
- 82.Wu H, Song Z, Givskov M, Doring G, Worlitzsch D, Mathee K, Rygaard J, Høiby N. 2001. Pseudomonas aeruginosa mutations in lasI and rhlI quorum sensing systems result in milder chronic lung infection. Microbiology (Reading) 147:1105–1113. doi: 10.1099/00221287-147-5-1105. [DOI] [PubMed] [Google Scholar]
- 83.DebRoy S, Dao J, Söderberg M, Rossier O, Cianciotto NP. 2006. Legionella pneumophila type II secretome reveals unique exoproteins and a chitinase that promotes bacterial persistence in the lung. Proc Natl Acad Sci USA 103:19146–19151. doi: 10.1073/pnas.0608279103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Galka F, Wai SN, Kusch H, Engelmann S, Hecker M, Schmeck B, Hippenstiel S, Uhlin BE, Steinert M. 2008. Proteomic characterization of the whole secretome of Legionella pneumophila and functional analysis of outer membrane vesicles. Infect Immun 76:1825–1836. doi: 10.1128/IAI.01396-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Truchan HK, Christman HD, White RC, Rutledge NS, Cianciotto NP. 2017. Type II Secretion substrates of Legionella pneumophila translocate out of the pathogen-occupied vacuole via a semipermeable membrane. mBio 8:e00870-17. doi: 10.1128/mBio.00870-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dohnálek J, Dušková J, Tishchenko G, Kolenko P, Skálová T, Novák P, Fejfarová K, Šimůnek J. 2021. Chitinase Chit62J4 essential for chitin processing by human microbiome bacterium Clostridium paraputrificum J4. Molecules 26:5978. doi: 10.3390/molecules26195978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sánchez B, González-Tejedo C, Ruas-Madiedo P, Urdaci MC, Margolles A. 2011. Lactobacillus plantarum extracellular chitin-binding protein and its role in the interaction between chitin, Caco-2 cells, and mucin. Appl Environ Microbiol 77:1123–1126. doi: 10.1128/AEM.02080-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sikora AE, Zielke RA, Lawrence DA, Andrews PC, Sandkvist M. 2011. Proteomic analysis of the Vibrio cholerae type II secretome reveals new proteins, including three related serine proteases. J Biol Chem 286:16555–16566. doi: 10.1074/jbc.M110.211078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kirn TJ, Jude BA, Taylor RK. 2005. A colonization factor links Vibrio cholerae environmental survival and human infection. Nature 438:863–866. doi: 10.1038/nature04249. [DOI] [PubMed] [Google Scholar]
- 90.Fennell TG, Blackwell GA, Thomson NR, Dorman MJ. 2021. gbpA and chiA genes are not uniformly distributed amongst diverse Vibrio cholerae. Microb Genom 7:000594. doi: 10.1099/mgen.0.000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chourashi R, Mondal M, Sinha R, Debnath A, Das S, Koley H, Chatterjee NS. 2016. Role of a sensor histidine kinase ChiS of Vibrio cholerae in pathogenesis. Int J Med Microbiol 306:657–665. doi: 10.1016/j.ijmm.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 92.Das S, Chourashi R, Mukherjee P, Gope A, Koley H, Dutta M, Mukhopadhyay AK, Okamoto K, Chatterjee NS. 2020. Multifunctional transcription factor CytR of Vibrio cholerae is important for pathogenesis. Microbiology (Reading) 166:1136–1148. doi: 10.1099/mic.0.000949. [DOI] [PubMed] [Google Scholar]
- 93.Bhowmick R, Ghosal A, Das B, Koley H, Saha DR, Ganguly S, Nandy RK, Bhadra RK, Chatterjee NS. 2008. Intestinal adherence of Vibrio cholerae involves a coordinated interaction between colonization factor GbpA and mucin. Infect Immun 76:4968–4977. doi: 10.1128/IAI.01615-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wong E, Vaaje-Kolstad G, Ghosh A, Hurtado-Guerrero R, Konarev PV, Ibrahim AFM, Svergun DI, Eijsink VGH, Chatterjee NS, van Aalten DMF. 2012. The Vibrio cholerae colonization factor GbpA possesses a modular structure that governs binding to different host surfaces. PLoS Pathog 8:e1002373. doi: 10.1371/journal.ppat.1002373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jude BA, Martinez RM, Skorupski K, Taylor RK. 2009. Levels of the secreted Vibrio cholerae attachment factor GbpA are modulated by quorum-sensing-induced proteolysis. J Bacteriol 191:6911–6917. doi: 10.1128/JB.00747-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mandal S, Chatterjee NS. 2016. Vibrio cholerae GbpA elicits necrotic cell death in intestinal cells. J Med Microbiol 65:837–847. doi: 10.1099/jmm.0.000298. [DOI] [PubMed] [Google Scholar]
- 97.Kariisa AT, Weeks K, Tamayo R. 2016. The RNA domain Vc1 regulates downstream gene expression in response to cyclic diguanylate in Vibrio cholerae. PLoS One 11:e0148478. doi: 10.1371/journal.pone.0148478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kariisa AT, Grube A, Tamayo R. 2015. Two nucleotide second messengers regulate the production of the Vibrio cholerae colonization factor GbpA. BMC Microbiol 15:166. doi: 10.1186/s12866-015-0506-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Loose JSM, Forsberg Z, Fraaije MW, Eijsink VGH, Vaaje-Kolstad G. 2014. A rapid quantitative activity assay shows that the Vibrio cholerae colonization factor GbpA is an active lytic polysaccharide monooxygenase. FEBS Lett 588:3435–3440. doi: 10.1016/j.febslet.2014.07.036. [DOI] [PubMed] [Google Scholar]
- 100.Kawada M, Chen CC, Arihiro A, Nagatani K, Watanabe T, Mizoguchi E. 2008. Chitinase 3-like-1 enhances bacterial adhesion to colonic epithelial cells through the interaction with bacterial chitin-binding protein. Lab Invest 88:883–895. doi: 10.1038/labinvest.2008.47. [DOI] [PubMed] [Google Scholar]
- 101.Low D, Tran HT, Lee I-A, Dreux N, Kamba A, Reinecker H-C, Darfeuille-Michaud A, Barnich N, Mizoguchi E. 2013. Chitin-binding domains of Escherichia coli ChiA mediate interactions with intestinal epithelial cells in mice with colitis. Gastroenterology 145:602–612.e9. doi: 10.1053/j.gastro.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Permpoonpattana P, Tolls EH, Nadem R, Tan S, Brisson A, Cutting SM. 2011. Surface layers of Clostridium difficile endospores. J Bacteriol 193:6461–6470. doi: 10.1128/JB.05182-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Permpoonpattana P, Phetcharaburanin J, Mikelsone A, Dembek M, Tan S, Brisson M-C, La Ragione R, Brisson AR, Fairweather N, Hong HA, Cutting SM. 2013. Functional characterization of Clostridium difficile spore coat proteins. J Bacteriol 195:1492–1503. doi: 10.1128/JB.02104-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hong HA, Ferreira WT, Hosseini S, Anwar S, Hitri K, Wilkinson AJ, Vahjen W, Zentek J, Soloviev M, Cutting SM. 2017. The Spore Coat Protein CotE Facilitates Host Colonization by Clostridium difficile. J Infect Dis 216:1452–1459. doi: 10.1093/infdis/jix488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pizarro-Guajardo M, Ravanal MC, Paez MD, Callegari E, Paredes-Sabja D. 2018. Identification of Clostridium difficile immunoreactive spore proteins of the epidemic strain R20291. Prot Clin Appl 12:1700182. doi: 10.1002/prca.201700182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Basak S, Deb D, Narsaria U, Kar T, Castiglione F, Sanyal I, Bade PD, Srivastava AP. 2021. In silico designing of vaccine candidate against Clostridium difficile. Sci Rep 11:14215. doi: 10.1038/s41598-021-93305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bassères E, Endres BT, Montes-Bravo N, Pérez-Soto N, Rashid T, Lancaster C, Begum K, Alam MJ, Paredes-Sabja D, Garey KW. 2021. Visualization of fidaxomicin association with the exosporium layer of Clostridioides difficile spores. Anaerobe 69:102352. doi: 10.1016/j.anaerobe.2021.102352. [DOI] [PubMed] [Google Scholar]
- 108.Devlin JR, Santus W, Mendez J, Peng W, Yu A, Wang J, Alejandro-Navarreto X, Kiernan K, Singh M, Jiang P, Mechref Y, Behnsen J. 2022. Salmonella enterica serovar Typhimurium chitinases modulate the intestinal glycome and promote small intestinal invasion. PLoS Pathog 18:e1010167. doi: 10.1371/journal.ppat.1010167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chandra K, Roy Chowdhury A, Chatterjee R, Chakravortty D. 2022. GH18 family glycoside hydrolase Chitinase A of Salmonella enhances virulence by facilitating invasion and modulating host immune responses. PLoS Pathog 18:e1010407. doi: 10.1371/journal.ppat.1010407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JCD. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol Microbiol 47:103–118. doi: 10.1046/j.1365-2958.2003.03313.x. [DOI] [PubMed] [Google Scholar]
- 111.Hautefort I, Thompson A, Eriksson-Ygberg S, Parker ML, Lucchini S, Danino V, Bongaerts RJM, Ahmad N, Rhen M, Hinton JCD. 2008. During infection of epithelial cells Salmonella enterica serovar Typhimurium undergoes a time-dependent transcriptional adaptation that results in simultaneous expression of three type 3 secretion systems. Cell Microbiol 10:958–984. doi: 10.1111/j.1462-5822.2007.01099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Harvey PC, Watson M, Hulme S, Jones MA, Lovell M, Berchieri A, Young J, Bumstead N, Barrow P. 2011. Salmonella enterica serovar typhimurium colonizing the lumen of the chicken intestine grows slowly and upregulates a unique set of virulence and metabolism genes. Infect Immun 79:4105–4121. doi: 10.1128/IAI.01390-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Frederiksen RF, Yoshimura Y, Storgaard BG, Paspaliari DK, Petersen BO, Chen K, Larsen T, Duus JØ, Ingmer H, Bovin NV, Westerlind U, Blixt O, Palcic MM, Leisner JJ. 2015. A diverse range of bacterial and eukaryotic chitinases hydrolyzes the LacNAc (Galβ1-4GlcNAc) and LacdiNAc (GalNAcβ1-4GlcNAc) motifs found on vertebrate and insect cells. J Biol Chem 290:5354–5366. doi: 10.1074/jbc.M114.607291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang SL, Chang WT. 1997. Purification and characterization of two bifunctional chitinases/lysozymes extracellularly produced by Pseudomonas aeruginosa K-187 in a shrimp and crab shell powder medium. Appl Environ Microbiol 63:380–386. doi: 10.1128/aem.63.2.380-386.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hogan DA, Kolter R. 2002. Pseudomonas-Candida interactions: an ecological role for virulence factors. Science 296:2229–2232. doi: 10.1126/science.1070784. [DOI] [PubMed] [Google Scholar]
- 116.Ovchinnikova ES, Krom BP, Harapanahalli AK, Busscher HJ, van der Mei HC. 2013. Surface thermodynamic and adhesion force evaluation of the role of chitin-binding protein in the physical interaction between Pseudomonas aeruginosa and Candida albicans. Langmuir 29:4823–4829. doi: 10.1021/la400554g. [DOI] [PubMed] [Google Scholar]
- 117.Allonsius CN, Vandenheuvel D, Oerlemans EFM, Petrova MI, Donders GGG, Cos P, Delputte P, Lebeer S. 2019. Inhibition of Candida albicans morphogenesis by chitinase from Lactobacillus rhamnosus GG. Sci Rep 9:2900. doi: 10.1038/s41598-019-39625-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mayer FL, Kronstad JW. 2017. Disarming fungal pathogens: Bacillus safensis inhibits virulence factor production and biofilm formation by Cryptococcus neoformans and Candida albicans. mBio 8:e01537-17. doi: 10.1128/mBio.01537-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cummings RD, Pierce JM. 2014. The challenge and promise of glycomics. Chem Biol 21:1–15. doi: 10.1016/j.chembiol.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sojitra M, Sarkar S, Maghera J, Rodrigues E, Carpenter EJ, Seth S, Ferrer Vinals D, Bennett NJ, Reddy R, Khalil A, Xue X, Bell MR, Zheng RB, Zhang P, Nycholat C, Bailey JJ, Ling C-C, Lowary TL, Paulson JC, Macauley MS, Derda R. 2021. Genetically encoded multivalent liquid glycan array displayed on M13 bacteriophage. Nat Chem Biol 17:806–816. doi: 10.1038/s41589-021-00788-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Geissner A, Reinhardt A, Rademacher C, Johannssen T, Monteiro J, Lepenies B, Thépaut M, Fieschi F, Mrázková J, Wimmerova M, Schuhmacher F, Götze S, Grünstein D, Guo X, Hahm HS, Kandasamy J, Leonori D, Martin CE, Parameswarappa SG, Pasari S, Schlegel MK, Tanaka H, Xiao G, Yang Y, Pereira CL, Anish C, Seeberger PH. 2019. Microbe-focused glycan array screening platform. Proc Natl Acad Sci USA 116:1958–1967. doi: 10.1073/pnas.1800853116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Oyelaran O, Gildersleeve JC. 2009. Glycan arrays: recent advances and future challenges. Curr Opin Chem Biol 13:406–413. doi: 10.1016/j.cbpa.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ma T, McGregor M, Giron L, Xie G, George AF, Abdel-Mohsen M, Roan NR. 2022. Single-cell glycomics analysis by CyTOF-Lec reveals glycan features defining cells differentially susceptible to HIV. Elife 11:e78870. doi: 10.7554/eLife.78870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Buchberger AR, DeLaney K, Johnson J, Li L. 2018. Mass spectrometry imaging: a review of emerging advancements and future insights. Anal Chem 90:240–265. doi: 10.1021/acs.analchem.7b04733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kumar A, Zhang KYJ. 2019. Human chitinases: structure, function, and inhibitor discovery. Adv Exp Med Biol 1142:221–251. doi: 10.1007/978-981-13-7318-3_11. [DOI] [PubMed] [Google Scholar]
- 126.Di Rosa M, Distefano G, Zorena K, Malaguarnera L. 2016. Chitinases and immunity: ancestral molecules with new functions. Immunobiology 221:399–411. doi: 10.1016/j.imbio.2015.11.014. [DOI] [PubMed] [Google Scholar]