ABSTRACT
Chlamydia trachomatis is an obligate intracellular pathogen that replicates in a host-derived vacuole termed the inclusion. Central to pathogenesis is a type III secretion system that translocates effector proteins into the host cell, which are predicted to play major roles in host cell invasion, nutrient acquisition, and immune evasion. However, until recently, the genetic intractability of C. trachomatis hindered identification and characterization of these important virulence factors. Here, we sought to expand the repertoire of identified effector proteins and confirm they are secreted during C. trachomatis infection. Utilizing bioinformatics, we identified 18 candidate substrates that had not been previously assessed for secretion, of which we show four to be secreted, using Yersinia pseudotuberculosis as a surrogate host. Using adenylate cyclase (CyaA), BlaM, and glycogen synthase kinase (GSK) secretion assays, we identified nine novel substrates that were secreted in at least one assay. Interestingly, only three of the substrates, shown to be translocated by C. trachomatis, were similarly secreted by Y. pseudotuberculosis. Using large-scale screens to determine subcellular localization and identify effectors that perturb crucial host cell processes, we identified one novel substrate, CT392, that is toxic when heterologously expressed in Saccharomyces cerevisiae. Toxicity required both the N- and C-terminal regions of the protein. Additionally, we show that these newly described substrates traffic to distinct host cell compartments, including vesicles and the cytoplasm. Collectively, our study expands the known repertoire of C. trachomatis secreted factors and highlights the importance of testing for secretion in the native host using multiple secretion assays when possible.
KEYWORDS: Chlamydia trachomatis, effector functions, protein secretion
INTRODUCTION
Chlamydia trachomatis is an obligate intracellular bacterium that is the most common sexually transmitted bacterium and the leading cause of infectious blindness worldwide (1). Over 1.7 million chlamydia cases are reported in the United States each year, with the global burden approaching 131 million cases annually (2). While most infections are asymptomatic, ascension into the upper genital tract can lead to pelvic inflammatory disease, ectopic pregnancy, and infertility (3). Significantly, chlamydial infection does not result in long-term immunity, leading to high reinfection rates (4).
C. trachomatis has a biphasic developmental cycle where the infectious, elementary body (EB) is internalized by a host cell into a membrane-bound compartment termed the inclusion (5). The inclusion avoids lysosome fusion and utilizes microtubules to traffic to the peri-Golgi region (6, 7). Here, the EB differentiates into the noninfectious, metabolically active form termed the reticulate body (RB) and initiates replication (8, 9). After multiple rounds of replication, RBs convert back into EBs and are released by host cell lysis or extrusion (10).
From the confines of its inclusion, C. trachomatis must interact with various host cell organelles and manipulate signaling pathways to obtain nutrients for replication, inhibit host defense mechanisms, and promote host cell viability. To achieve these feats, C. trachomatis secretes effector proteins into the host cell throughout its infection cycle via a type III secretion system (T3SS) (11). C. trachomatis is predicted to secrete over 100 T3SS effectors into its host cell (12), including inclusion membrane (Inc) proteins and conventional T3SS (cT3SS) effector proteins.
The presence of one or more bilobed hydrophobic domains has been used to identify 58 potential Inc proteins; however, only 38 have been shown to localize to the inclusion membrane during infection (1, 13–15). In contrast, cT3SS proteins lack a bilobed hydrophobic domain and have been shown to localize to the plasma membrane, nucleus, and cytoplasm or to associate with the inclusion (8, 16, 17). Based on their localization, these proteins are predicted to play vital roles in host cell invasion, nutrient acquisition, and subversion of host defense mechanisms (9, 18–20). While cT3SS effectors likely play important roles in chlamydial infection, identification of cT3SS effector proteins has been challenging as most lack readily identifiable secretion signals, functional domains indicative of effector function, or homology with other proteins. Furthermore, the genetic intractability of C. trachomatis has significantly hampered efforts to identify these important factors. To date, over 45 candidate C. trachomatis effector proteins have been shown to be secreted using Yersinia pseudotuberculosis, Shigella flexneri, or Salmonella enterica serovar Typhimurium as surrogate hosts (10, 17, 21). While research is still challenging, advances in chlamydial genetics, including the ability to transform C. trachomatis serovar L2 with a shuttle vector and the adaption of multiple reporters, now allow for confirmation of effector protein secretion during infection (15, 22–24).
In this study, we identify three novel secretion substrates and an additional six substrates that are possibly secreted. The putative secretion substrates were selected using bioinformatic analysis through identification of eukaryotic-like domains, a predicted T3SS signal, or homology to known secreted effectors. By initially screening each candidate in Yersinia pseudotuberculosis, followed by validation of secretion in C. trachomatis, we demonstrate that several substrates are not capable of being secreted by a surrogate host, highlighting the importance of testing for secretion in the native organism. Of the effectors that were confirmed to be secreted or possibly secreted by C. trachomatis, we identified two with unique localization patterns when ectopically expressed in mammalian cells. One secretion substrate induced toxicity when expressed in Saccharomyces cerevisiae, implying it targets a crucial eukaryotic cellular pathway. Collectively, our study expands the list of known C. trachomatis secretion substrates and highlights the power of genetic approaches to identifying putative secreted factors.
RESULTS
Identification of previously undescribed Chlamydia trachomatis type III secreted effector proteins using Yersinia pseudotuberculosis as a surrogate host.
To expand the list of chlamydial T3SS substrates, we employed a bioinformatics-guided approach to identify any putative chlamydial proteins that possess eukaryotic-like domains as predicted by the Simple Modular Architecture Research Tool (SMART), a T3SS signal (pEffect and Effective T3S), or homology to a known secreted effector protein (BLAST). In total, we identified 18 candidate substrates (Table 1). We first tested for secretion using Y. pseudotuberculosis as a surrogate host due to its ease of manipulation and culture. Furthermore, it would allow us to directly compare the rigor of a surrogate system to that of the endogenous host. To assess secretion, the first 40 amino acids (aa), representing the putative secretion signal, were fused to neomycin phosphotransferase (NPT) and constructs were transformed into Y. pseudotuberculosis. Cultures were grown under secretion-inducing (lacking calcium) or secretion-inhibiting (possessing calcium) conditions, and cells were fractionated into supernatant (broth) and pellet (bacterial cell) fractions. Expression and secretion were determined by western blotting. Using this approach, we identified six putative substrates that are capable of being secreted when using Y. pseudotuberculosis as a surrogate host: CTL0037 (CT668), CTL0060 (CT691), CTL0386 (CT131), CTL0137 (CT768), CTL0065 (CT696), and CTL0105 (CT736) (Fig. 1). As is convention in the field, we used serovar L2 for all experiments but we will refer to the serovar D nomenclature. Four of these six (CT131, CT768, CT696, and CT736) were detected in the supernatant only under T3SS-inducing conditions, whereas two (CT668 and CT691) were present in the supernatant under both T3SS-inducing and -inhibiting conditions, suggesting that these two substrates could be secreted through a different mechanism. Collectively, we identified four new candidate substrates that can be secreted by Yersinia and may be secreted by C. trachomatis.
TABLE 1.
Candidate type III secreted effectors evaluated for secretion using Y. pseudotuberculosisa
| D/UW-3/CX | L2/434/Bu | Eukaryotic-like domain | Predicted T3S signal | Expressed | Secreted under conditions: |
|
|---|---|---|---|---|---|---|
| Inducing | Inhibitory | |||||
| CT051 | CTL0307 | Coiled-coil | Yes | Yes | No | No |
| CT131 | CTL0386 | TMD | Yes | Yes | Yes | No |
| CT149 | CTL0404 | Alpha/beta hydrolase fold | Yes | Yes | No | No |
| CT350 | CTL0604 | HEAT repeat | No | No | No | No |
| CT360 | CTL0614 | Coiled-coil | No | Yes | No | No |
| CT392 | CTL0648 | DUF1207 (specific to Chlamydia) | No | No | No | No |
| CT460 | CTL0720 | SWIB (ATP-dependent chromatin-remodeling) proteins | No | Yes | No | No |
| CT503 | CTL0765 | Coiled-coil | No | Yes | No | No |
| CT537 | CTL0799 | PFAM: TsaE | Yes | Yes | No | No |
| CT584 | CTL0847 | Coiled-coil | Yes | Yes | No | No |
| CT627 | CTL0891 | PFAM: Rhodanese_C, RHOD | Yes | No | No | No |
| CT668 | CTL0037 | Coiled-coil | No | Yes | Yes | Yes |
| CT683 | CTL0052 | TPR | Yes | Yes | No | No |
| CT691 | CTL0060 | PFAM: PhoU_div | Yes | Yes | Yes | Yes |
| CT696 | CTL0065 | None | No | Yes | Yes | Yes |
| CT736 | CTL0105 | PFAM: PBP | No | Yes | Yes | Yes |
| CT744 | CTL0113 | Coiled-coil | Yes | Yes | No | No |
| CT768 | CTL0137 | Coiled-coil | Yes | Yes | Yes | No |
Inducing conditions contain medium without Ca2+. Inhibitory conditions contain medium with Ca2+.
FIG 1.
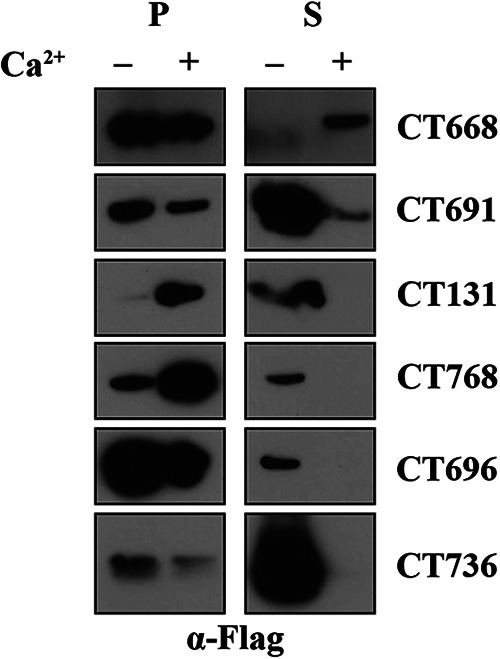
Novel C. trachomatis type III secreted effector proteins are secreted using Y. pseudotuberculosis as a surrogate host. Y. pseudotuberculosis was transformed with each C. trachomatis gene of interest, and transformants were grown in secretion-inducing (− Ca2+) or secretion-inhibiting (+ Ca2+) medium. Pellets (P) and supernatants (S) were separated by centrifugation and normalized by optical density at 600 nm. Samples were resolved by SDS-PAGE expression, and immunoblots were probed with anti-FLAG antibodies. Data are representative of three independent experiments.
Identification of previously undescribed type III secreted effector proteins in Chlamydia trachomatis.
To determine whether these candidate substrates are capable of being secreted by C. trachomatis, we utilized three secretion assays. Each candidate protein was cloned into a pBomb4-tet vector and expressed as a C-terminal fusion to the tag of interest. Each construct was transformed into C. trachomatis serovar L2, and expression was confirmed by western blotting. We sought to test all 18 candidate substrates for secretion in C. trachomatis; however, CT051 and CT350 were excluded from our list as we were unable to generate a clone for any of the three assays.
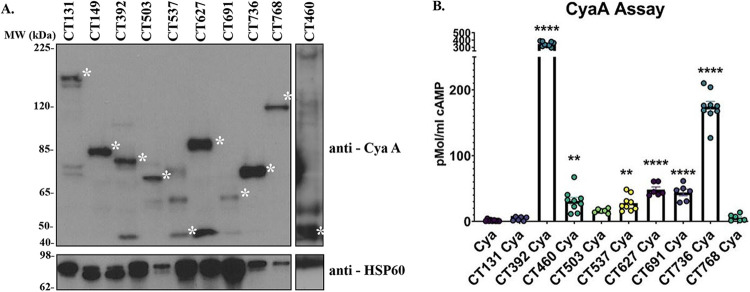
The CyaA assay utilizes the Bordetella pertussis calmodulin-dependent adenylate cyclase toxin CyaA to monitor effector secretion. It has been used to identify type IV and type III secreted effector proteins from Legionella pneumophila, Coxiella burnetii, Salmonella spp., Yersinia spp., and recently C. trachomatis (22, 25, 26–28). In this assay, if the effector protein of interest is released into the host cell cytosol, the secreted CyaA will interact with host calmodulin to convert ATP to cAMP, the levels of which can be monitored by enzyme-linked immunosorbent assay (ELISA). Each effector protein (Table 2) was expressed as a C-terminal fusion to CyaA, constructs were transformed into C. trachomatis, and expression of the fusion protein was confirmed by western blotting. As shown in Fig. 2A, CyaA fusions of CT131, CT149, CT392, CT503, CT537, CT627, CT691, CT736, CT768, and CT460 were readily expressed and subsequently evaluated for secretion. All CyaA fusions matched predicted molecular weights except for CT503-CyaA. It is possible that CT503-CyaA is posttranscriptionally modified, resulting in the higher observed molecular weight. CyaA tag alone was used as a negative control. Significantly elevated levels of cAMP were noted in cells infected with CT392, CT460, CT537, CT627, CT691, and CT736 relative to CyaA alone, indicating secretion (Fig. 2B; Table 2).
TABLE 2.
Candidate secretion substrates evaluated for secretion in C. trachomatisa
| D/UW-3/CX | L2/434/Bu | CyaA assay | BlaM assay | GSK assay | Final designation |
|---|---|---|---|---|---|
| CT051 | CTL0307 | NAb | NAb | NAb | Unable to conclude |
| CT131 | CTL0386 | Not secreted | NAd | Secreted | Possibly secreted |
| CT149 | CTL0404 | NAd | Secreted | Secreted | Secreted |
| CT350 | CTL0604 | NAb | NAb | NAb | Unable to conclude |
| CT360 | CTL0614 | NAd | NAd | NAd | Unable to conclude |
| CT392 | CTL0648 | Secreted | NAd | NAd | Possibly secreted |
| CT460 | CTL0720 | Secreted | NAd | NAd | Possibly secreted |
| CT503 | CTL0765 | Not secreted | NAc | Not secreted | Not secreted |
| CT537 | CTL0799 | Secreted | NAd | NAd | Possibly secreted |
| CT584 | CTL0847 | NAd | Secreted | Secreted | Secreted |
| CT627 | CTL0891 | Secreted | Not secreted | Secreted | Secreted |
| CT668 | CTL0037 | NAd | NAd | NAd | Unable to conclude |
| CT683 | CTL0052 | NAd | NAd | NAd | Unable to conclude |
| CT691 | CTL0060 | Secreted | NAd | NAd | Possibly secreted |
| CT696 | CTL0065 | NAd | NAd | NAd | Unable to conclude |
| CT736 | CTL0105 | Secreted | Not secreted | Not secreted | Possibly secreted |
| CT744 | CTL0113 | NAb | NAd | NAc | Unable to conclude |
| CT768 | CTL0137 | Not secreted | Not secreted | NAc | Not secreted |
Candidates secreted in at least two assays were considered secreted, and those positive in only one assay were considered possibly secreted. Those with negative results in at least two assays were designated “not secreted” whereas those that were unable to be evaluated in at least two assays were designated “unable to conclude.”
Not assayed (NA) due to lack of a clone.
Not assayed (NA) due to lack of transformants.
Not assayed (NA) because of inability to confirm expression.
FIG 2.
Identification of novel C. trachomatis secreted effector proteins via an adenylate cyclase (CyaA) secretion assay. Candidate secretion substrates were expressed as C-terminal fusions to the CyaA tag and transformed into C. trachomatis. (A) Transformants were used to infect HeLa cells at an MOI of 5 for 24 h, after which expression of the fusion protein was confirmed by immunoblotting with anti-CyaA antibodies. C. trachomatis heat shock protein (HSP) was used as a loading control. The asterisk on the right of each band indicates the molecular weight (MW) of each CyaA fusion protein. Predicted molecular weights are as follows: CT131-CyaA, 151 kDa; CT149-CyaA, 59 kDa; CT392-CyaA, 66 kDa; CT503-CyaA, 42 kDa; CT537-CyaA, 42 kDa; CT627-CyaA, 63 kDa; CT691-CyaA, 50 kDa; CT736-CyaA, 41 kDa; CT768-CyaA, 89 kDa; CT460-CyaA, 35 kDa. (B) HeLa cells were infected with each candidate at an MOI of 5, and after 24 h, cytosolic levels of cAMP were measured by competitive ELISA. The levels of cAMP present in C. trachomatis expressing CyaA alone were compared to those in C. trachomatis expressing the candidate secretion substrate. CT392, CT460, CT537, CT627, CT691, and CT736 exhibited elevated levels of cAMP relative to C. trachomatis expressing CyaA alone and were thus considered secreted. Error bars represent the standard deviations from the means. Statistical significance was determined using one-way ANOVA. ****, P < 0.0001; **, P < 0.01. Data are representative of three independent experiments with three replicates (A and B).
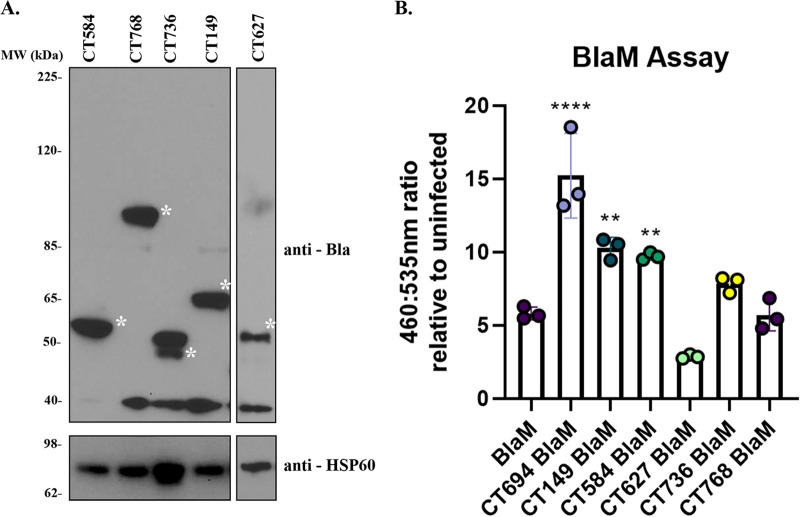
The TEM/BlaM assay has also been used previously to identify bacterial factors that are translocated into the host cell cytosol (23, 25, 29). The substrate, CCF4-AM, possesses two dye molecules that are in close proximity and undergo a fluorescence resonance energy transfer (FRET) event. However, the two dye molecules are separated by a beta-lactam ring. If the candidate substrate is secreted from C. trachomatis into the host cell cytosol, beta-lactamase will cleave the beta-lactam ring, disrupting the FRET event, causing the substrate to fluoresce blue (460 nm) instead of green (535 nm). Prior to assaying the candidates, western blotting was employed to confirm effector secretion. CT694/TmeA, which was previously shown to be secreted in this assay, was used as a positive control (23). Of those substrates tested, CT584, CT768, CT736, CT149, and CT627 were expressed (Fig. 3A), and only CT149 and CT584 had a higher 460:535 ratio than C. trachomatis expressing BlaM alone (negative control) (Fig. 3B; Table 2), suggesting they are released to the host cell cytosol.
FIG 3.
Identification of novel secreted proteins from C. trachomatis via a beta-lactamase (BlaM) secretion assay. Candidate secretion substrates were fused to the BlaM tag and transformed into C. trachomatis. (A) HeLa cells were infected at an MOI of 5 for 24 h with transformants. Expression of the BlaM fusion protein was confirmed by immunoblotting with anti-BlaM antibodies, and anti-HSP was used as a loading control. The asterisk indicates the molecular weight of each BlaM fusion protein. Predicted molecular weights are as follows: CT584-BlaM, 54 kDa; CT768-BlaM, 94 kDa; CT736-BlaM, 46 kDa; CT149-BlaM, 64 kDa; CT627, 68 kDa. (B) Secretion was determined by infecting HeLa cells at an MOI of 5 for 24 h. Infected cells were loaded with CCF4-AM for 1 h, and the change in 460-/535-nm fluorescence was monitored on a plate reader. A shift from 535-nm fluorescence to increased 460-nm fluorescence is indicative of secretion into the host cell cytosol. Ratios associated with the candidate substrate-BlaM fusions were compared to those of C. trachomatis expressing BlaM alone. CT694 was included as a positive control. Error bars represent standard deviations from the means. Statistical significance was determined using one-way ANOVA. ****, P < 0.0001; **, P < 0.01. (B). Data are representative of three experiments (A and B).
We also employed a third assay that makes use of a small, 13-residue epitope tag, glycogen synthase kinase (GSK), which is phosphorylated by eukaryotic kinases when relocated to the cytosol (30, 31). Due to the relatively low abundance of the secreted proteins and many background bands associated with the antibody (32), we added a FLAG tag which allowed us to immunoprecipitate the samples to concentrate our candidate effector proteins. To test for secretion, cells were infected with C. trachomatis expressing the candidate of interest fused to the GSK FLAG tag. The fusion protein was immunoprecipitated and analyzed by western blotting with GSK and phospho-GSK (P-GSK) antibodies. As shown in Fig. 4, CT149, CT131, CT627, and CT584 were secreted, whereas phosphorylation of CT503 and CT736 was not detected, suggesting they are not secreted in this assay.
FIG 4.
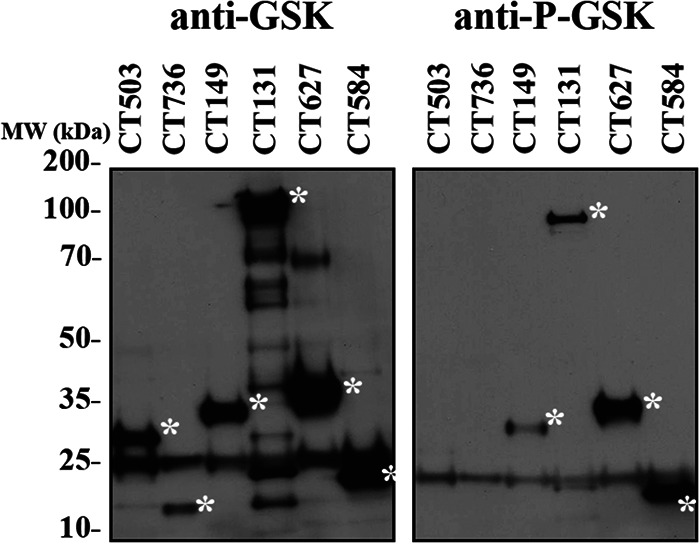
Evaluation of C. trachomatis candidate secretion using a GSK FLAG secretion assay. Candidate secretion substrates were fused to a GSK FLAG tag and transformed into C. trachomatis. HeLa cells were infected at an MOI of 5 with each strain, and after 24 h, the candidate peptide was immunoprecipitated using anti-FLAG beads. Samples were resolved by SDS-PAGE, and immunoblots were probed with anti-GSK or anti-P-GSK to evaluate expression and secretion, respectively. CT149, CT131, CT627, and CT584 were determined to be secreted using this assay. The asterisk indicates the molecular weight of each GSK FLAG fusion. Predicted molecular weights are as follows: CT503-GSK-FLAG, 30 kDa; CT736-GSK-FLAG, 19 kDa; CT149-GSK-FLAG, 37 kDa; CT131-GSK-FLAG, 129 kDa; CT627-GSK-FLAG, 41 kDa; CT584-GSK-FLAG, 24 kDa. Data are representative of 3 independent experiments.
CT392 and CT691 have unique localization patterns when ectopically expressed in HeLa cells.
Upon release into a host cell, bacterial effector proteins traffic to host cell organelles where they perturb host cell processes. To determine whether these newly identified secreted factors target specific host cell compartments, we ectopically expressed each secreted or possibly secreted effector protein in HeLa cells. CT131, CT149, and CT584 were previously evaluated for subcellular localization (33); thus, we excluded them from our analysis. As shown in Fig. 5, CT460, CT537, CT627, and CT736 resulted in a localization pattern similar to that of green fluorescent protein (GFP) alone and thus were designated as having no localization pattern. In contrast, CT392 localized in cytoplasmic puncta whereas CT691 was cytoplasmic and excluded from the nucleus. Collectively, these results demonstrate that several of the newly identified effector proteins traffic to distinct subcellular compartments when ectopically expressed in eukaryotic cells.
FIG 5.
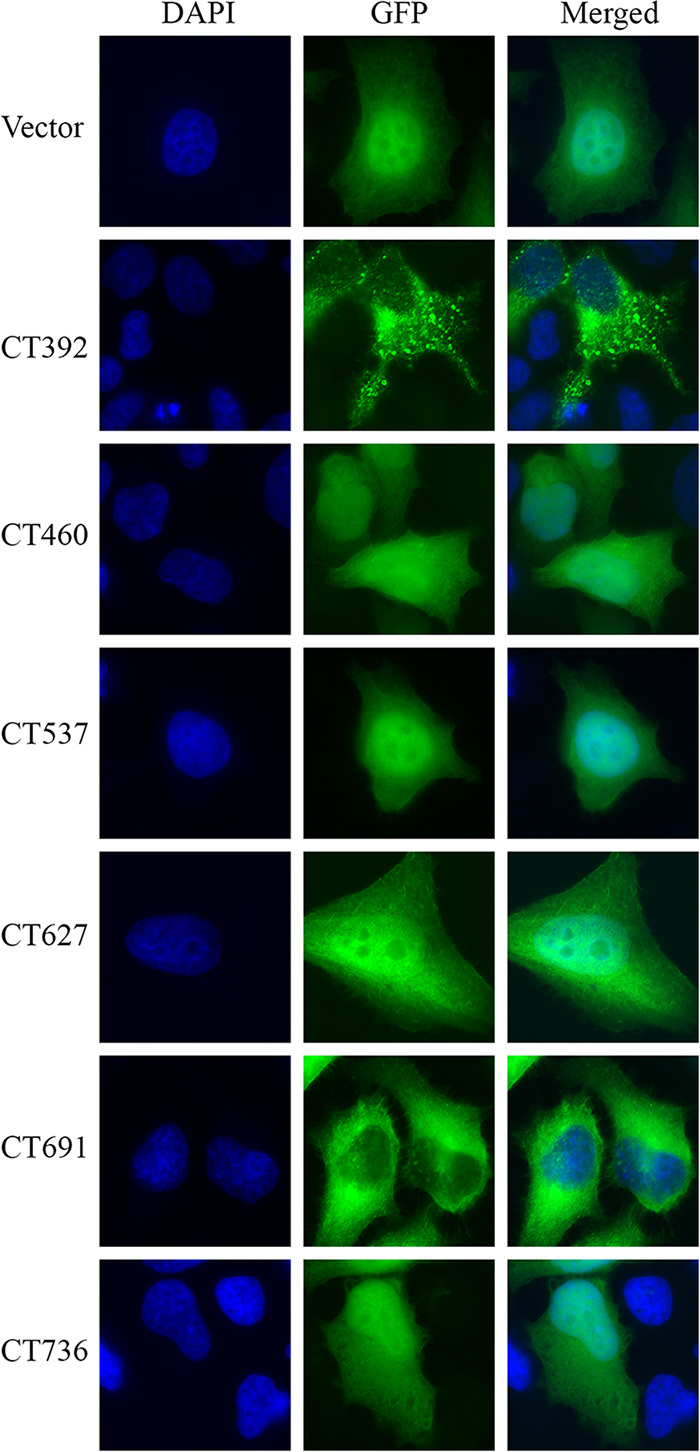
C. trachomatis secretion substrates traffic to distinct subcellular compartments. Proteins confirmed to be secreted were fused to a GFP tag and used to transiently transfect HeLa cells. Cells were fixed in 4% formaldehyde, and DNA was stained with DAPI (blue) to visualize the nucleus. Samples were imaged for unique localization patterns in comparison to vector alone using immunofluorescence microscopy. Data are representative of three independent experiments.
CT392 displays a toxic phenotype in Saccharomyces cerevisiae.
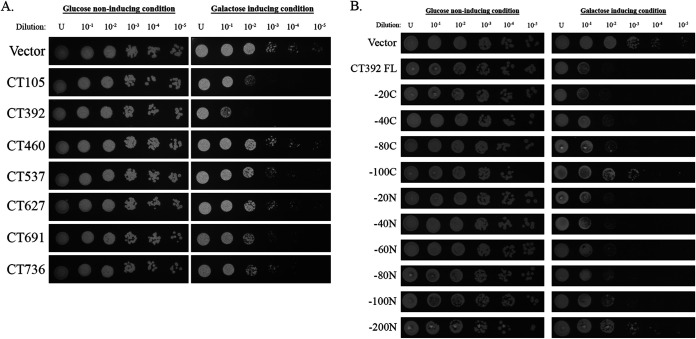
To determine if C. trachomatis secreted substrates perturb critical eukaryotic cell pathways, we expressed all the secreted and possibly secreted effector proteins (Table 2) under the control of a galactose-inducible promoter in S. cerevisiae. CT131, CT149, and CT584 were previously evaluated for toxicity in yeast (33); thus, we excluded them from our analysis. After the samples were diluted and spotted on galactose (inducing) medium, each sample was evaluated for a growth defect, denoted as a toxic phenotype, by comparing it to growth obtained on glucose (noninducing) plates as well as to vector alone. If overexpression of the C. trachomatis effector of interest causes a toxic phenotype, then that effector is most likely disrupting an important pathway in yeast (29, 34, 35). Since many of the same cellular processes are conserved between yeast and mammals, this is a simple assay that can determine whether the effector of interest is disrupting a critical pathway in human cells. CT105/CteG was used as a positive control as we previously demonstrated it has a toxic phenotype in this system (36). Of the six effectors tested here, we demonstrate that only CT392 displayed a toxic phenotype, suggesting that it could disrupt an essential eukaryotic host cell pathway (Fig. 6A). Toxicity was specific to our yeast assay as no growth difference or toxicity was noted in C. trachomatis overexpressing CT392 (see Fig. S1 and S2 in the supplemental material). To identify a region of CT392 required for toxicity, we generated C- and N-terminal truncations of CT392. While deletion of a region between amino acids 80 and 100 from the C terminus modestly reduced toxicity, deletion of a region between amino acids 100 and 200 from the N terminus abolished toxicity (Fig. 6B), suggesting this is the region required for CT392 toxicity in yeast.
FIG 6.
C. trachomatis secretion substrates perturb normal host cell processes. (A) Secretion substrates were expressed in S. cerevisiae under a galactose-inducible promoter. Each was serially diluted to 10−5 and spotted on uracil dropout medium containing glucose (noninducing) or galactose (inducing). Vector alone was used as a negative control, and CT105 was used as a positive control. (B) CT392 truncations were expressed in S. cerevisiae under a galactose-inducible promoter, serially diluted, and spotted. Loss of toxicity is noted for −100C and −200N truncations. FL, full length. Data are representative of 3 independent experiments (A and B).
DISCUSSION
As an obligate intracellular pathogen, C. trachomatis must have methods to manipulate host organelles and signaling pathways from the confines of the inclusion. C. trachomatis achieves this by releasing an array of cT3SS effector proteins into the host cell. It is hypothesized that many of these effector proteins play roles in promoting host cell viability, inhibiting host defense mechanisms, facilitating nutrient acquisition, altering the cell cycle, and initiating host cell invasion (8, 18, 19, 36, 37). To date, only a few cT3SS effectors have been characterized. One major limitation to understanding how these proteins contribute to chlamydial infection is the lack of a list of bona fide cT3SS substrates that have been confirmed to be secreted. Our study directly compares the efficiency of a surrogate host to that of the native organism for evaluating candidate secretion substrates and highlights the importance of validation in the native host. Importantly, we expand on the repertoire of known secretion substrates.
Several prior studies have sought to identify C. trachomatis secretion substrates by measuring their ability to be translocated using a surrogate host (8, 10, 16, 21, 37–39). However, we hypothesized that novel secretion substrates might still be uncovered. With the advent of tools for genetic manipulation of C. trachomatis, we now have the unprecedented opportunity to perform a side-by-side comparison of a surrogate system to that of the native organism. Using multiple bioinformatic predictions, we identified 18 candidate secretion substrates that, to the best of our knowledge (with the exception of CT696 [23]), had not been previously assessed for secretion. While whether a specific factor will be secreted can be hard to predict, some attributes increase the odds that a candidate might be a secreted protein. Many secreted substrates possess eukaryotic-like domains such as coiled-coil motifs, ankyrin repeats, tetratricopeptide repeats (TPRs), or F-box domains to mediate interactions with their eukaryotic binding partners (25, 29, 40–42). Of the 18 candidates we tested here, seven possessed a coiled-coil motif and one harbored a TPR domain (Table 1). Additionally, we identified one candidate, CT392/CTL0648, that possesses a domain of unknown function (DUF1207) that is unique to Chlamydia spp. Prior studies identified a class of proteins that possess another domain of unknown function that is unique to Chlamydia spp., DUF582, and each of these candidates was subsequently shown to be secreted using Shigella flexneri as a surrogate host (39). While the function of DUF1207 and DUF582 domains remains unknown, the fact that these unique domains are found in several secreted proteins may suggest that they have evolved to mediate chlamydia-specific interactions with the host cell.
In the past 2 decades, several machine learning algorithms have been developed to predict candidate secretion substrates. These approaches typically work by comparing a large set of known effectors and noneffectors to define attributes that distinguish these two groups (43–45). Here, we used the pEffect and Effective T3S databases. These databases identified 10 candidate substrates that are bioinformatically predicted to possess a T3SS signal, of which three were shown to be secreted by Y. pseudotuberculosis. Of our 18 total candidates identified by bioinformatics, we found that six were secreted by Y. pseudotuberculosis, confirming previous assertions that bioinformatic predictions need to be confirmed experimentally due to the possibility of false positives. Of these, only four were secreted under T3SS-inducing conditions. Surprisingly, CT691 and CT668 were secreted under both T3SS-inducing and -inhibiting conditions, indicating they are likely secreted through a T3SS-independent mechanism.
Until 2011, C. trachomatis was recalcitrant to genetic manipulation, necessitating the use of a surrogate host to determine whether a candidate factor was capable of being secreted. Using Yersinia spp., Shigella spp., or Salmonella spp., ~35 conventional T3SS effector proteins have been described (8, 10, 16, 21, 37–39). While robust secretion by a surrogate host may suggest a factor is secreted, it does not always correlate with secretion by the native organism. Indeed, a recent study using C. trachomatis to define Chlamydia pneumoniae secreted factors was unable to confirm the secretion of numerous substrates that had been previously reported as secreted (32). One possible explanation is that since the T3SS requires chaperones to stabilize effectors, C. trachomatis likely has specific chaperones that are not present in surrogate systems to aid in the secretion of some of its effector proteins (46–48). While Inc proteins can readily be visualized by immunofluorescence microscopy to affirm secretion (15), conventional secretion substrates tend to be present at lower abundance and diffusely localized throughout the cell, requiring signaling amplification to detect. Thus, to rigorously test each candidate substrate for secretion by C. trachomatis, we employed three distinct secretion assays that have previously been used to monitor effector secretion in other organisms (23, 25, 28–31) and have been recently adapted for use in C. trachomatis (22, 23, 32). Not surprisingly, we saw differences in which substrates were secreted based on the secretion assay that was used. Notably, six, two, and four were positive for secretion in the CyaA, BlaM, and GSK assays, respectively (Table 2). The CyaA and BlaM assays both utilized large tags (~30 kDa), which may hinder certain proteins from being secreted. The GSK tag is smaller (~3 kDa) and so should be less inhibitory toward secretion and could potentially produce more reliable results. Surprisingly though, we found the highest number of effectors to be secreted using the CyaA assay, suggesting that this assay provides high sensitivity and that the large tag may not be an impediment to secretion as predicted.
As previously noted, differences between secretion in a surrogate and that in the native host have been recently described (32). Y. pseudotuberculosis was capable of secreting CT668, CT691, CT131, CT768, CT696, and CT736, with CT668 and CT691 being released into the supernatant independent of the T3SS. We were successfully able to confirm secretion by C. trachomatis for CT691, CT736, and CT131 in at least one assay. Surprisingly, while CT149, CT392, CT460, CT537, CT584, and CT627 were not secreted by Y. pseudotuberculosis, they were translocated by C. trachomatis in at least one assay. This difference in secretion between Yersinia- and Chlamydia-based experiments may be explained by multiple factors. As with previous studies, we fused the first 30 amino acids of our candidate protein to NPT to assay secretion in Y. pseudotuberculosis. Our C. trachomatis assays used the full-length protein, giving a more accurate measure of secretion. While the N terminus is required for secretion, chaperones are also necessary for translocation, and it is possible that differences in available T3SS chaperones may account for differences in secretion noted between the organisms. Collectively, these results highlight the importance of evaluating secretion in the native host.
While CT668 and CT696 were secreted by Y. pseudotuberculosis, we were unable to confirm their secretion in C. trachomatis. CT668 secretion was noted under T3SS-inhibiting conditions and thus could be due to a different secretion method or could be due to bacterial lysis. CT696 is a hypothetical protein that is located immediately downstream from the T3SS effector proteins CT694/TmeA and CT695/TmeB. All three proteins are expressed at the mid-cycle of infection; however, TmeA and TmeB are cotranscribed while CT696 is transcribed separately (23). Prior studies using Y. pseudotuberculosis as a surrogate host sought to determine whether CT696 is secreted. CT696 was detected in the supernatant, but the molecular weight was significantly lower than anticipated (23). Intriguingly, the molecular weight of CT696 isolated from the pellet fraction ran at the anticipated molecular weight on the SDS-PAGE gel, suggesting it could be processed during secretion in the native host (21). Due to the low level detected in the supernatant and the inconsistent molecular weight, whether CT696 was secreted or not could not be confirmed. Here, we detected CT696 in the supernatant when the first 30 amino acids were expressed in Y. pseudotuberculosis and fused to NPT. Unfortunately, we were unable to confirm its secretion by C. trachomatis due to a lack of detectable expression in each of the assays. Similarly, Mueller and Fields (23) attempted to confirm CT696 secretion using the BlaM/TEM1 assay but were unable to confirm expression. While a precise cause is unknown, it is possible that the levels of CT696, even using the overexpression system, are too low to detect without immunoprecipitation.
Large-scale screens such as ectopic expression and yeast toxicity assays have been used extensively for preliminary characterization of secretion substrates (25, 29, 33, 40, 49). While a C. trachomatis open reading frame (ORF) library was constructed previously with the goal of expressing each ORF in S. cerevisiae to evaluate subcellular localization and toxicity, some were not tested due to a lack of a clone or of expression. Of the newly described secreted or possibly secreted substrates identified here (Table 2), only CT131, CT149, and CT584 were previously evaluated (33). Here, we report that CT392 localizes to cytoplasmic puncta and CT691 localizes to the cytoplasm. Of the effectors tested, only CT392 expression resulted in a growth defect in S. cerevisiae (Fig. 6A). Due to CT392’s unique phenotypes, we further explored if there was a specific region of the protein that was responsible for the toxic phenotype. Bioinformatic analysis using SMART predicted a coiled-coil region at 85 aa to 116 aa. The −100N truncation still retained toxicity, suggesting it is dispensable for toxicity. Interestingly, removal of 200 aa from the N terminus abolished toxicity, suggesting residues 100 to 200 of the N terminus are important for toxicity. Using the eukaryotic-linear motif (ELM) resource, we identified an F and H motif at 177 to 189 aa, a motif that has been shown to be required for interactions with inositol polyphosphate 5-phosphatases (OCRL1). OCRL1 is an important regulator of membrane trafficking and is recruited to the chlamydia inclusion (50). While we were unable to demonstrate binding between CT392 and OCRL1 (data not shown), it is possible that this motif may serve another, unknown function. Regardless, CT392 has been shown previously to be important for chlamydial infection, and further exploration to elucidate its function is warranted (51). In conclusion, we have identified three novel secreted effectors and an additional six proteins that may be secreted by C. trachomatis. Four of these secreted effectors were secreted in a surrogate system and further confirmed using C. trachomatis. Intriguingly, several proteins were translocated by C. trachomatis but not Y. pseudotuberculosis, highlighting differences between the native organism and the surrogate host. Collectively, our study highlights the importance of validating secretion in the native organism when possible and using multiple secretion assays.
MATERIALS AND METHODS
Bioinformatic analysis.
To identify novel secretion substrates, open reading frames from the Chlamydia trachomatis L2 (LGV 434/Bu) genome were subjected to multiple Graphical User Interface (GUI) bioinformatic analyses. Considering that cT3SS effectors likely interact with eukaryotic host cell factors, eukaryotic-like domains were predicted by the Simple Modular Architecture Research Tool (SMART). Though T3SS effectors do not always contain a known secretion signal, some secretion signals have been characterized. The presence of a T3SS signal was assessed by pEffect and Effective T3S databases. As several C. trachomatis proteins are known to be secreted into the host cell, homology to a known secreted effector protein was determined via the Basic Local Alignment Search Tool (BLAST) to determine proteins that would likely have roles within the eukaryotic host.
Bacterial and cell culture.
C. trachomatis serovar L2 (LGV 434/Bu) was cultivated using HeLa 229 cells (American Type Culture Collection). A gastrografin density gradient was performed to purify EBs as previously described (52). HeLa cells were grown in RPMI 1640 medium (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (Gibco) at 37°C with 5% CO2. Y. pseudotuberculosis was maintained in heart infusion broth (HIB) (Millipore Sigma) at 37°C. S. cerevisiae was grown in yeast-peptone-dextrose medium at 30°C.
Cloning.
To assess secretion of each candidate effector protein, a modified version of pBomb4-tet-mCherry (52) was employed by cloning the unique secretion assay tag into the KpnI/SalI site, creating pBomb4 CyaA, pBomb4 BlaM, and pBomb4 GSK FLAG. The coding sequence for cyaA and blaM was amplified from pJB-Kan-TetRPA-cyaA and pUC57, respectively. The GSK FLAG tag was synthesized by GenScript. Each C. trachomatis gene was amplified from L2/434/Bu genomic DNA via PCR using gene-specific primers (see Table S1 in the supplemental material) and was subsequently cloned into the NotI/KpnI site to generate a C-terminal fusion to CyaA, BlaM, or GSK FLAG. For yeast toxicity and ectopic expression, each effector was cloned into the KpnI/XhoI site of pYesNTA-Kan (34) or pcDNA3.1 GFP, respectively. Each construct was analyzed, and sequence was confirmed via DNA sequencing (McLab).
Yersinia T3SS assay.
To assess secretion of new candidate effector proteins, the first 120 nucleotides of each C. trachomatis gene of interest were PCR amplified and cloned into pFLAG-CTC. Plasmids were transformed into wild-type Y. pseudotuberculosis, and transformants were selected on brain heart infusion agar with carbenicillin as previously described (15, 22). To assess secretion, transformants were inoculated into secretion-inducing medium (HIB, 100 mM EGTA, and 1 M MgCl2) and secretion-inhibiting medium (HIB and 500 mM CaCl2). Each culture was incubated with shaking at 26°C for 2 h, after which expression of the effector-neomycin phosphotransferase (NPT) fusion and T3SS was induced with 1 mM isopropyl-d-thiogalactopyranoside (IPTG) and a temperature shift to 37°C for 4 h, respectively. After incubation, each culture was separated into supernatant and pellet fractions and trichloroacetic acid (TCA) was added to precipitate the proteins overnight on ice at 4°C. Pelleted fractions were resuspended in Tris and 1× phosphate-buffered saline (PBS) with 4× lithium dodecyl sulfate (LDS) sample buffer. Both fractions were analyzed by western blotting using an anti-FLAG antibody (Millipore Sigma).
Transformation of Chlamydia.
C. trachomatis serovar L2 (LGV 434/Bu) was transformed as previously described with minor modifications (53, 54). Fresh lysates of C. trachomatis EBs were mixed with 5 μg of plasmid DNA and 10 μL of 5× transformation mix (50 mM Tris, pH 7.5, 250 mM CaCl2) in a final volume of 50 μL. After 30 min, 4 mL of RPMI medium was added and cultures were applied to 2 wells of a 6-well plate containing confluent HeLa cell monolayers. Plates were centrifuged at 900 × g for 30 min at 15°C and incubated at 37°C with 5% CO2 for 18 h. After 18 h, selection medium containing 0.3 μg/mL penicillin G was added, and cells were incubated for an additional 24 h. Every 48 h, C. trachomatis lysates were passaged and used to infect new HeLa cell monolayers using RPMI medium containing 0.3 μg/mL penicillin G and 1 μg/mL cycloheximide. Infectious progenies were passaged until there were viable inclusions (~P2) as evident by expression of GFP.
CyaA secretion assay.
Expression of the fusion protein was first confirmed by western blotting as described below, and only those cells with positive expression were tested in the adenylate cyclase (CyaA) assay. HeLa cells were seeded in 96-well clear-bottom white plates (Greiner) and incubated at 37°C with 5% CO2 overnight. Cells were infected with C. trachomatis harboring pBomb4 CyaA using a multiplicity of infection (MOI) of 5. Expression of the fusion protein was induced using 10 ng/mL anhydrous tetracycline HCl (aTc). At 24 h postinfection (hpi), a competitive ELISA (Abcam) was used to measure the amount of cAMP in host cells. Levels of cAMP in cells infected with C. trachomatis expressing the candidate effectors were compared to those of a negative-control vector (pBomb4 CyaA plasmid with no fused effector protein) to calculate effector secretion. Data are representative of three independent experiments with three technical replicates per experiment.
Beta-lactamase assay.
Confirmation of expression of each effector-BlaM fusion protein was first confirmed via western blotting as described below, and only those cells that expressed the fusion protein were assayed. HeLa cells were seeded into black clear-bottom 96-well plates (Greiner) and incubated at 37°C with 5% CO2 until confluent. Cells were infected with each BlaM-transformed C. trachomatis strain at an MOI of 5, and expression was induced with 10 ng/mL aTc. At 24 hpi, cells were washed twice with 1× PBS and loaded with CCF4-AM following the manufacturer’s instructions (Thermo Fisher Scientific) using the alternative loading protocol as previously described (23, 25, 29). Plates were placed in the dark for 1 h at room temperature. Fluorescence was read on a Tecan plate reader. For quantification, background was subtracted, the ratio of 460 nm to 535 nm (blue to green) was quantified, and expression relative to uninfected cells was calculated as previously described (55). Data are representative of three experiments with three technical replicates.
GSK FLAG immunoprecipitation.
HeLa cells were seeded in T175 cell culture flasks and infected at an MOI of 5 with C. trachomatis. Expression of the GSK FLAG-effector fusion was induced with 10 ng/mL aTc. After 24 h, infected cells were washed with ice-cold 1× PBS and scraped in 800 μL ice-cold eukaryotic lysis solution (ELS) (50 mM Tris-HCl, 150 mM sodium chloride, 1 mM EDTA, and 1% Triton X-100) containing 1× Halt cocktail protease and phosphatase solution (Thermo Fisher Scientific) and 10 μM GSK inhibitor {1-(7-methoxyquinolin-4-yl)-3-[6-(trifluoromethyl)pyridin-2-yl]urea (Tocris)}. Lysates were transferred to a prechilled microcentrifuge tube on ice for 20 min and were then pelleted at 12,000 × g for 20 min at 4°C. Supernatants were applied to anti-FLAG magnetic beads (Thermo Fisher Scientific) and were placed on a rotator at 4°C for 1 h. Samples were then washed 5 times with ice-cold ELS without Triton X-100 to remove unbound proteins. The GSK FLAG fusion protein was eluted using 4× NuPAGE LDS sample buffer (Thermo Fisher Scientific), and samples were analyzed via western blotting. Data were collected from three independent experiments.
Western blotting.
HeLa cells in 6-well plates were infected with C. trachomatis CyaA- or BlaM-effector transformants at an MOI of 5 to confirm expression. Cells were lysed at 24hpi with ELS and Halt cocktail protease inhibitor. Samples were resolved using 4 to 12% Bis-Tris gels and transferred to a polyvinylidene difluoride (PVDF) membrane. Membranes were probed with anti-CyaA (Santa Cruz) or anti-BlaM (QED Bioscience) primary antibodies and anti-mouse or anti-rabbit horseradish peroxidase (HRP) (Bio-Rad) secondary antibodies, respectively.
Ectopic expression.
HeLa cells, seeded in 24-well plates with coverslips, were transfected using Lipofectamine LTX with Plus reagent (Thermo Fisher Scientific) and incubated at 37°C per the manufacturer’s instructions. After 4 h, the medium was removed and replaced with 1 mL RPMI medium. Cells were fixed 18 h posttransfection in 4% formaldehyde and permeabilized with 0.1% Triton X-100. Samples were blocked in 2% bovine serum albumin (BSA), and DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI).
Yeast toxicity.
S. cerevisiae was transformed with the yeast toxicity plasmid using the lithium acetate method as previously described (34). Transformants were serially diluted and spotted on synthetic dropout medium lacking uracil and supplemented with glucose (noninducing conditions) or galactose (inducing conditions) as the carbon source. Plates were incubated for 48 h at 30°C in triplicate to assess toxicity.
Growth curve.
HeLa cells, seeded on 24-well plates, were chilled on ice at 4°C for 10 min. Cells were infected with C. trachomatis overexpressing CT392-FLAG or CT865-FLAG at an MOI of 1. Cells were chilled on ice at 4°C for 30 min, after which the medium was changed and samples were incubated at 37°C. At 48 hpi, cells were scraped, serially diluted, and plated onto fresh HeLa cell monolayers. GFP-expressing infectious forming units (IFUs) were enumerated by immunofluorescence microscopy.
LDH assay.
HeLa cells, seeded on 24-well plates, were infected with C. trachomatis overexpressing CT392-FLAG or CT865-FLAG at an MOI of 2.5. Expression of the fusion protein was induced with 10 ng/mL aTc. Cells were incubated for either 24, 48, or 72 h. At each time point, cells were rocked for 5 to 10 min, 100 μL of supernatant was centrifuged at 600 × g for 10 min, and 100 μL of lactate dehydrogenase (LDH) mix (BioVision) was added to a black clear-bottom 96-well plate in triplicate. Ten microliters of supernatant was added to each well, and the plate was incubated at room temperature in the dark for 30 min. Absorbance was read on a Tecan plate reader at 450 nm with a reference wavelength of 650 nm.
Statistics.
Statistical analysis was performed using GraphPad Prism 9.3.0 software. One-way analyses of variance (ANOVAs) were used followed by Dunnett’s multiple comparisons with P values shown as follows: P < 0.05, *; P < 0.01, **; P < 0.001, ***; P < 0.0001, ****.
ACKNOWLEDGMENTS
We acknowledge grant support from the NIH (M.M.W., R01 AI150812 and R01 AI155434; B.S., T32 AI007511) and the University of Iowa Stead Family Scholars to M.M.W. S.E.A. was supported through the University of Iowa Stinski Undergraduate Research Fellowship.
We thank Weber lab members Lanci Bulman, Alix McCullough, and Grace Vebi for assistance in cloning, as well as Jabeena CA, Steve Huang, and Alix McCullough for critical review of the manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Mary M. Weber, Email: mary-weber@uiowa.edu.
Nancy E. Freitag, University of Illinois Chicago
REFERENCES
- 1.Elwell C, Mirrashidi K, Engel J. 2016. Chlamydia cell biology and pathogenesis. Nat Rev Microbiol 14:385–400. doi: 10.1038/nrmicro.2016.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.CDC. 2020. Sexually transmitted disease surveillance 2020. CDC, Atlanta, GA. [Google Scholar]
- 3.Malhotra M, Sood S, Mukherjee A, Muralidhar S, Bala M. 2013. Genital Chlamydia trachomatis: an update. Indian J Med Res 138:303–316. [PMC free article] [PubMed] [Google Scholar]
- 4.Starnbach MN. 2018. Action needed on Chlamydia vaccines. Trends Microbiol 26:639–640. doi: 10.1016/j.tim.2018.05.006. [DOI] [PubMed] [Google Scholar]
- 5.Scidmore MA, Rockey DD, Fischer ER, Heinzen RA, Hackstadt T. 1996. Vesicular int eractions of the Chlamydia trachomatis inclusion are determined by chlamydial early protein synthesis rather than route of entry. Infect Immun 64:5366–5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grieshaber SS, Grieshaber NA, Hackstadt T. 2003. Chlamydia trachomatis uses host cell dynein to traffic to the microtubule-organizing center in a p50 dynamitin-independent process. J Cell Sci 116:3793–3802. [DOI] [PubMed] [Google Scholar]
- 7.Grieshaber SS, Grieshaber NA, Miller N, Hackstadt T. 2006. Chlamydia trachomatis causes centrosomal defects resulting in chromosomal segregation abnormalities. Traffic 7:940–949. [DOI] [PubMed] [Google Scholar]
- 8.Abdelrahman Y, Ouellette SP, Belland RJ, Cox JV. 2016. Polarized cell division of Chlamydia trachomatis. PLoS Pathog 12:e1005822. doi: 10.1371/journal.ppat.1005822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee JK, Enciso GA, Boassa D, Chander CN, Lou TH, Pairawan SS, Guo MC, Wan FYM, Ellisman MH, Sütterlin C, Tan M. 2018. Replication-dependent size reduction precedes differentiation in Chlamydia trachomatis. Nat Commun 9:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hybiske K, Stephens RS. 2007. Mechanisms of host cell exit by the intracellular bacterium Chlamydia. Proc National Acad Sci 104:11430–11435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dai W, Li Z. 2014. Conserved type III secretion system exerts important roles in Chlamydia trachomatis. Int J Clin Exp Pathol 7:5404–5414. [PMC free article] [PubMed] [Google Scholar]
- 12.Bugalhão JN, Mota LJ. 2019. The multiple functions of the numerous Chlamydia trachomatis secreted proteins: the tip of the iceberg. Microb Cell 6:414–449. doi: 10.15698/mic2019.09.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scidmore-Carlson MA, Shaw EI, Dooley CA, Fischer ER, Hackstadt T. 1999. Identification and characterization of a Chlamydia trachomatis early operon encoding four novel inclusion membrane proteins. Mol Microbiol 33:753–765. doi: 10.1046/j.1365-2958.1999.01523.x. [DOI] [PubMed] [Google Scholar]
- 14.Shaw EI, Dooley CA, Fischer ER, Scidmore MA, Fields KA, Hackstadt T. 2000. Three temporal classes of gene expression during the Chlamydia trachomatis developmental cycle. Mol Microbiol 37:913–925. doi: 10.1046/j.1365-2958.2000.02057.x. [DOI] [PubMed] [Google Scholar]
- 15.Weber MM, Bauler LD, Lam J, Hackstadt T. 2015. Expression and localization of predicted inclusion membrane proteins in Chlamydia trachomatis. Infect Immun 83:4710–4718. doi: 10.1128/IAI.01075-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pennini ME, Perrinet S, Dautry-Varsat A, Subtil A. 2010. Histone methylation by NUE, a novel nuclear effector of the intracellular pathogen Chlamydia trachomatis. PLoS Pathog 6:e1000995. doi: 10.1371/journal.ppat.1000995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pais SV, Key CE, Borges V, Pereira IS, Gomes JP, Fisher DJ, Mota LJ. 2019. CteG is a Chlamydia trachomatis effector protein that associates with the Golgi complex of infected host cells. Sci Rep 9:6133. doi: 10.1038/s41598-019-42647-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keb G, Ferrell J, Scanlon KR, Jewett TJ, Fields KA. 2021. Chlamydia trachomatis TmeA directly activates N-WASP to promote actin polymerization and functions synergistically with TarP during invasion. mBio 12:e02861-20. doi: 10.1128/mBio.02861-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faris R, McCullough A, Andersen SE, Moninger TO, Weber MM. 2020. The Chlamydia trachomatis secreted effector TmeA hijacks the N-WASP-ARP2/3 actin remodeling axis to facilitate cellular invasion. PLoS Pathog 16:e1008878. doi: 10.1371/journal.ppat.1008878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clifton DR, Fields KA, Grieshaber SS, Dooley CA, Fischer ER, Mead DJ, Carabeo RA, Hackstadt T. 2004. A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc Natl Acad Sci USA 101:10166–10171. doi: 10.1073/pnas.0402829101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.da Cunha M, Milho C, Almeida F, Pais SV, Borges V, Maurício R, Borrego MJ, Gomes JP, Mota LJ. 2014. Identification of type III secretion substrates of Chlamydia trachomatis using Yersinia enterocolitica as a heterologous system. BMC Microbiol 14:40. doi: 10.1186/1471-2180-14-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bauler LD, Hackstadt T. 2014. Expression and targeting of secreted proteins from Chlamydia trachomatis. J Bacteriol 196:1325–1334. doi: 10.1128/JB.01290-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mueller KE, Fields KA. 2015. Application of β-lactamase reporter fusions as an indicator of effector protein secretion during infections with the obligate intracellular pathogen Chlamydia trachomatis. PLoS One 10:e0135295. doi: 10.1371/journal.pone.0135295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, Hybiske K, Stephens RS. 2018. Direct visualization of the expression and localization of chlamydial effector proteins within infected host cells. Pathog Dis 76:fty011. doi: 10.1093/femspd/fty011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen C, Banga S, Mertens K, Weber MM, Gorbaslieva I, Tan Y, Luo Z-Q, Samuel JE. 2010. Large-scale identification and translocation of type IV secretion substrates by Coxiella burnetii. Proc Natl Acad Sci USA 107:21755–21760. doi: 10.1073/pnas.1010485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagai H, Cambronne ED, Kagan JC, Amor JC, Kahn RA, Roy CR. 2005. A C-terminal translocation signal required for Dot/Icm-dependent delivery of the Legionella RalF protein to host cells. Proc Natl Acad Sci USA 102:826–831. doi: 10.1073/pnas.0406239101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geddes K, Worley M, Niemann G, Heffron F. 2005. Identification of new secreted effectors in Salmonella enterica serovar Typhimurium. Infect Immun 73:6260–6271. doi: 10.1128/IAI.73.10.6260-6271.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sory MP, Boland A, Lambermont I, Cornelis GR. 1995. Identification of the YopE and YopH domains required for secretion and internalization into the cytosol of macrophages, using the cyaA gene fusion approach. Proc Natl Acad Sci USA 92:11998–12002. doi: 10.1073/pnas.92.26.11998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weber MM, Chen C, Rowin K, Mertens K, Galvan G, Zhi H, Dealing CM, Roman VA, Banga S, Tan Y, Luo Z-Q, Samuel JE. 2013. Identification of Coxiella burnetii type IV secretion substrates required for intracellular replication and Coxiella-containing vacuole formation. J Bacteriol 195:3914–3924. doi: 10.1128/JB.00071-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartra SS, Plano G. 2017. Measurement of effector protein translocation using phosphorylatable epitope tags and phospho-specific antibodies. Methods Mol Biol 1531:111–119. doi: 10.1007/978-1-4939-6649-3_10. [DOI] [PubMed] [Google Scholar]
- 31.Garcia JT, Ferracci F, Jackson MW, Joseph SS, Pattis I, Plano LRW, Fischer W, Plano GV. 2006. Measurement of effector protein injection by type III and type IV secretion systems by using a 13-residue phosphorylatable glycogen synthase kinase tag. Infect Immun 74:5645–5657. doi: 10.1128/IAI.00690-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yanatori I, Miura K, Chen Y-S, Valdivia RH, Kishi F. 2021. Application of a Chlamydia trachomatis expression system to identify Chlamydia pneumoniae proteins translocated into host cells. J Bacteriol 203:e00511-20. doi: 10.1128/JB.00511-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sisko JL, Spaeth K, Kumar Y, Valdivia RH. 2006. Multifunctional analysis of Chlamydia-specific genes in a yeast expression system. Mol Microbiol 60:51–66. doi: 10.1111/j.1365-2958.2006.05074.x. [DOI] [PubMed] [Google Scholar]
- 34.Faris R, Weber MM. 2019. Identification of host pathways targeted by bacterial effector proteins using yeast toxicity and suppressor screens. J Vis Exp doi: 10.3791/60488. [DOI] [PubMed] [Google Scholar]
- 35.Weber MM, Faris R, van Schaik EJ, Samuel JE. 2018. Identification and characterization of arginine finger-like motifs, and endosome-lysosome basolateral sorting signals within the Coxiella burnetii type IV secreted effector protein CirA. Microbes Infect 20:302–307. doi: 10.1016/j.micinf.2017.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steiert B, Icardi CM, Faris R, Klingelhutz AJ, Yau PM, Weber MM. 2023. The Chlamydia trachomatis type III secreted effector protein CteG induces centrosome amplification through interactions with centrin-2. Proc Natl Acad Sci USA 120:e2303487120. doi: 10.1073/pnas.2303487120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chellas-Géry B, Linton CN, Fields KA. 2007. Human GCIP interacts with CT847, a novel Chlamydia trachomatis type III secretion substrate, and is degraded in a tissue-culture infection model. Cell Microbiol 9:2417–2430. doi: 10.1111/j.1462-5822.2007.00970.x. [DOI] [PubMed] [Google Scholar]
- 38.Subtil A, Delevoye C, Balañá M-E, Tastevin L, Perrinet S, Dautry-Varsat A. 2005. A directed screen for chlamydial proteins secreted by a type III mechanism identifies a translocated protein and numerous other new candidates. Mol Microbiol 56:1636–1647. doi: 10.1111/j.1365-2958.2005.04647.x. [DOI] [PubMed] [Google Scholar]
- 39.Muschiol S, Boncompain G, Vromman F, Dehoux P, Normark S, Henriques-Normark B, Subtil A. 2011. Identification of a family of effectors secreted by the type III secretion system that are conserved in pathogenic Chlamydiae. Infect Immun 79:571–580. doi: 10.1128/IAI.00825-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.VieBrock L, Evans SM, Beyer AR, Larson CL, Beare PA, Ge H, Singh S, Rodino KG, Heinzen RA, Richards AL, Carlyon JA. 2014. Orientia tsutsugamushi ankyrin repeat-containing protein family members are type 1 secretion system substrates that traffic to the host cell endoplasmic reticulum. Front Cell Infect Microbiol 4:186. doi: 10.3389/fcimb.2014.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pan X, Lührmann A, Satoh A, Laskowski-Arce MA, Roy CR. 2008. Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science 320:1651–1654. doi: 10.1126/science.1158160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo T, Kuriakose JA, Zhu B, Wakeel A, McBride JW. 2011. Ehrlichia chaffeensis TRP120 interacts with a diverse array of eukaryotic proteins involved in transcription, signaling, and cytoskeleton organization. Infect Immun 79:4382–4391. doi: 10.1128/IAI.05608-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wagner N, Teper D, Pupko T. 2022. Predicting type III effector proteins using the Effectidor web server. Methods Mol Biol 2427:25–36. doi: 10.1007/978-1-0716-1971-1_3. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, Zhang Y, Xiong Y, Wang H, Deng Z, Song J, Ou H-Y. 2022. T4SEfinder: a bioinformatics tool for genome-scale prediction of bacterial type IV secreted effectors using pre-trained protein language model. Brief Bioinform 23:bbab420. doi: 10.1093/bib/bbab420. [DOI] [PubMed] [Google Scholar]
- 45.Burstein D, Zusman T, Degtyar E, Viner R, Segal G, Pupko T. 2009. Genome-scale identification of Legionella pneumophila effectors using a machine learning approach. PLoS Pathog 5:e1000508. doi: 10.1371/journal.ppat.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ghosh P. 2004. Process of protein transport by the type III secretion system. Microbiol Mol Biol Rev 68:771–795. doi: 10.1128/MMBR.68.4.771-795.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee SH, Galán JE. 2004. Salmonella type III secretion-associated chaperones confer secretion-pathway specificity. Mol Microbiol 51:483–495. doi: 10.1046/j.1365-2958.2003.03840.x. [DOI] [PubMed] [Google Scholar]
- 48.Muschiol S, Bailey L, Gylfe A, Sundin C, Hultenby K, Bergström S, Elofsson M, Wolf-Watz H, Normark S, Henriques-Normark B. 2006. A small-molecule inhibitor of type III secretion inhibits different stages of the infectious cycle of Chlamydia trachomatis. Proc Natl Acad Sci USA 103:14566–14571. doi: 10.1073/pnas.0606412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carey KL, Newton HJ, Lührmann A, Roy CR. 2011. The Coxiella burnetii Dot/Icm system delivers a unique repertoire of type IV effectors into host cells and is required for intracellular replication. PLoS Pathog 7:e1002056. doi: 10.1371/journal.ppat.1002056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Faris R, Merling M, Andersen SE, Dooley CA, Hackstadt T, Weber MM. 2019. Chlamydia trachomatis CT229 subverts rab GTPase-dependent CCV trafficking pathways to promote Chlamydial infection. Cell Rep 26:3380–3390.e5. doi: 10.1016/j.celrep.2019.02.079. [DOI] [PubMed] [Google Scholar]
- 51.LaBrie SD, Dimond ZE, Harrison KS, Baid S, Wickstrum J, Suchland RJ, Hefty PS. 2019. Transposon mutagenesis in Chlamydia trachomatis identifies CT339 as a ComEC homolog important for DNA uptake and lateral gene transfer. mBio 10:e01343-19. doi: 10.1128/mBio.01343-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Faris R, Weber MM. 2019. Propagation and purification of Chlamydia trachomatis serovar L2 transformants and mutants. Bio Protoc 9:e3459. doi: 10.21769/BioProtoc.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weber MM, Lam JL, Dooley CA, Noriea NF, Hansen BT, Hoyt HF, Carmody AB, Sturdevant GL, Hackstadt T. 2017. Absence of specific Chlamydia trachomatis inclusion membrane proteins triggers premature inclusion membrane lysis and host cell death. Cell Rep 19:1046–1417. doi: 10.1016/j.celrep.2017.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weber MM, Faris R. 2019. Mutagenesis of Chlamydia trachomatis using TargeTron. Methods Mol Biol 2042:165–184. doi: 10.1007/978-1-4939-9694-0_12. [DOI] [PubMed] [Google Scholar]
- 55.Newton P, Latomanski EA, Newton HJ. 2016. Applying fluorescence resonance energy transfer (FRET) to examine effector translocation efficiency by Coxiella burnetii during siRNA silencing. J Vis Exp 113:54210. doi: 10.3791/54210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 and S2 and Table S1. Download iai.00491-22-s0001.pdf, PDF file, 0.2 MB (156.8KB, pdf)