ABSTRACT
The emergence of the Omicron variant of SARS-CoV-2 represented a challenge to the treatment of COVID-19 using monoclonal antibodies. Only Sotrovimab maintained partial activity, allowing it to be used in high-risk patients infected with the Omicron variant. However, reports of resistance mutations to Sotrovimab demand efforts to better understand the intra-patient emergence of Sotrovimab resistance. A retrospective genomic analysis was conducted on respiratory samples from immunocompromised patients infected with SARS-CoV-2 who received Sotrovimab at our hospital between December 2021 and August 2022. The study involved 95 sequential specimens from 22 patients (1 to 12 samples/patient; 3 to 107 days post-infusion; threshold cycle [CT] ≤ 32). Resistance mutations (in P337, E340, K356, and R346) were detected in 68% of cases; the shortest time to detection of a resistance mutation was 5 days after Sotrovimab infusion. The dynamics of resistance acquisition were highly complex, with up to 11 distinct amino acid changes in specimens from the same patient. In two patients, the mutation distribution was compartmentalized in respiratory samples from different sources. This is the first study to examine the acquisition of Sotrovimab resistance in the BA.5 lineage, enabling us to determine the lack of genomic or clinical differences between Sotrovimab resistance in BA.5 relative to that in BA.1/2. Across all Omicron lineages, the acquisition of resistance delayed SARS-CoV-2 clearance (40.67 versus 19.5 days). Close, real-time genomic surveillance of patients receiving Sotrovimab should be mandatory to facilitate early therapeutic interventions.
KEYWORDS: COVID-19
INTRODUCTION
The emergence of the Omicron variant of SARS-CoV-2 had a major impact on COVID-19 treatment, as this lineage was resistant to most monoclonal antibodies (MAb). Despite this, Sotrovimab, which targets an epitope in the receptor-binding domain (RBD) of the spike protein, a highly conserved region in sarbecoviruses (1, 2), maintains partial in vitro activity against different Omicron sublineages and has been used until recently in high-risk patients infected with the Omicron variant of SARS-CoV-2 (3, 4).
In Spain, in December 2021, the administration of Sotrovimab 19 was approved in the context of early treatment for high-risk patients, immunocompromised or for compassionate use in hospitalized, immunocompromised, seronegative patients with severe COVID-19 (5). This prompted an evaluation of the possible acquisition of resistance in patients who had been receiving it at our institution since its approval.
A retrospective study was conducted in 48 COVID-19 patients who received Sotrovimab (500 mg) between December 2021 and August 2022, supported by a longitudinal genomic analysis of their sequential positive samples. Twenty-eight patients (all immunocompromised) had at least one positive sample available (with sufficient viral load to be sequenced, threshold cycle [CT] ≤ 32; Supplemental File 1) ≥ 3 days after Sotrovimab infusion, and these were included in our analysis. Diagnostics and CT determination were performed on respiratory specimens by reverse transcription-PCR (RT-PCR) (TaqPath COVID-19 CE-IVD RT-PCR kit; Thermo Fisher Scientific, Waltham, MA, USA). Whole-genome sequencing was carried out as described elsewhere (6). Sequence analysis was performed using an in-house bioinformatics pipeline (https://github.com/MG-IiSGM/covid_multianalysis), giving sequencing data with quality scores above the threshold for 95 specimens from 22 patients (1 to 12 specimens per patient, 3 to 107 days post-infusion). The requirements for single-nucleotide polymorphism (SNP) calls were as follows: frequency ≥ 5%, total depth ≥ 10×; a SNP was considered fixed when its frequency was >80% and non-fixed when its frequency was between 5% and 80%. Minority SNP calls (<5%) always required visual inspection by Integrative Genomics Viewer (IGV) software. When necessary, short tandem repeat (STR) analysis was performed to ensure that the specimens belonged to the same individual (6).
The time interval between COVID diagnosis and Sotrovimab infusion was 1 to 4 days in 8 cases where it was administered early due to the risk of developing severe COVID-19; 1 to 51 days in 13 hospitalized patients with severe COVID; and 68 days in 1 patient with persistent infection.
EMERGENCE OF RESISTANCE MUTATIONS
Resistance mutations, among those previously described for Sotrovimab resistance in vitro assays (7–9) and clinical studies (10–12), were detected in 15 patients (68%), which places our findings above the highest frequencies (55% to 60%) reported to date (10, 12). The mutations identified (frequency > 5%) were distributed among the four codons previously reported to encode Sotrovimab resistance (P337, E340, R346, and K356) and included 14 different substitutions (P337S/R/T/L/A/H, E340Q/A/D/K/V/G, R346T, and K356T). Mutations in these spike protein residues have been shown to be associated with reduced neutralization by this MAb (10–13). This wide diversity of substitutions and their distribution among four codons is in agreement with other studies (10, 12), while two studies from the Netherlands (11, 14) found more homogeneous behavior (mutations only in P337 and E340 and a low diversity of substitutions).
Resistance mutations (frequency > 5%) were identified for the first time between 5 and 18 days post-treatment (Supplemental File 1). We explored the presence of minority mutations (<5%) in nine cases which had positive samples prior to the first mutation detected. We identified mutations at lower frequencies (1.6%, data not shown) in only one case (patient 15), preceding the initial detection of mutations by 8 days. On the other hand, the longest period until the first detection of a resistance mutation, among the cases without mutations during the 1-to-5-day period, was 16 days (patient 4). Other studies have reported later onset of resistance, in the ranges of 14 to 21 days (12, 13), 6 to 13 days (9), and up to day 31 (14) post-treatment. However, the robustness of some of these dates is limited by the availability of sequenced specimens prior to the first one with a mutation.
DYNAMICS OF RESISTANCE MUTATIONS
Given that Sotrovimab has a median terminal half-life of 56.5 days post-infusion (2), a prolonged analysis of the evolution of mutations is required as soon as the first one has been identified. Our study is the most exhaustive within-patient sampling effort to date, covering an observation window of up to 107 days post-infusion and including up to 12 sequential positive specimens per patient.
In 12 cases, at least 1 of the mutations observed was eventually fixed (Fig. 1). All 14 mutations that became fixed corresponded to either P337 or E340 (5 and 9, respectively), with the most frequent being E340K (4 times), followed by E340D and P337S (3 times each), E340Q (twice), and P337L and P337R (identified only once each). These substitutions are also found at high rates in other studies (9–13). More than one different fixed substitution never co-occurred in the same specimen. However, in two cases (patients 1 and 10), two different fixed substitutions at the same amino acid were observed at two different time points (E340D/K and P337S/R, respectively, Fig. 1), a phenomenon previously described by other authors (10, 13).
FIG 1.
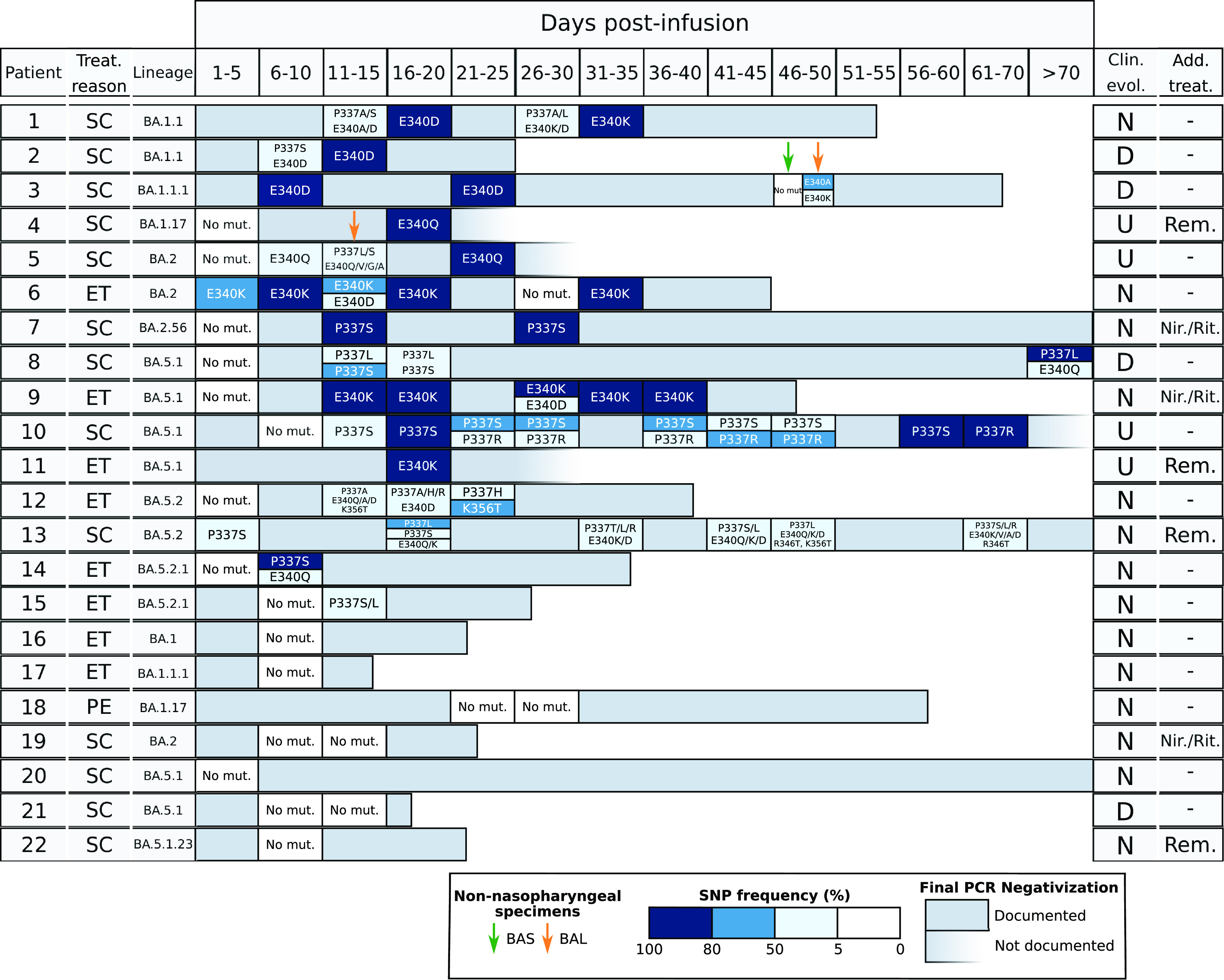
Sotrovimab resistance mutations identified in the patients in study. Each horizontal bar corresponds to the reverse transcription-PCR (RT-PCR) positivity period for each patient. For the specimens which were sequenced, boxed cells indicate whether or not mutations were detected; color gradient indicates the allele frequencies at which the single-nucleotide polymorphisms (SNPs) were identified. The reasons for Sotrovimab treatment (Treat. reason) are indicated as follows: SC, severe COVID; ET, early treatment; PE, SARS-CoV-2 persistence. Clinical evolution (Clin. evol.) is shown as follows: D, died; N, negativization documented by RT-PCR; and U, undefined negativization due to lack of a negative PCR. Additional antiviral treatments (Add. Treat) other than Sotrovimab are shown as follows: Rem, remdesivir; Nir./Rit., nirmatrelvir/ritonavir (Paxlovid). Bronchoalveolar lavage fluid (BAL fluid)/bronchial aspirate (BAS) specimens are indicated by yellow and green arrows, respectively.
Non-fixed substitutions (frequencies of 5% to 80%) were found in some specimens from 8 of the 12 cases in which some mutations eventually became fixed, and in another 3 cases in which only non-fixed mutations were detected throughout the analysis. The diversity of non-fixed substitutions was remarkably high and involved all four codons where mutations were identified (P337A/H/S/L/T/R, E340Q/A/V/D/K, K356T, and R346T). The homogenous composition observed in most of the specimens with fixed mutations contrasted with the greater within-sample diversity often identified in non-fixed substitutions (either in the same codon or even in more than one codon, Fig. 1). This diversity was highest in two of the patients (patients 12 and 13) with non-fixed mutations during the positivity period; 7 and 11 different amino acid changes, respectively, were identified (Fig. 1). For both fixed and non-fixed mutations, we observed either a transient presence (in only one specimen) or a more prolonged presence over several sequential specimens (Fig. 1). These data, taken together, illustrate the highly dynamic clonal behavior of SARS-CoV-2 under Sotrovimab pressure.
Our study is the first to include strains of the BA.5 lineage in genomic analysis of Sotrovimab resistance, with most cases under study (50%) belonging to this lineage (7 to BA.5.1* and 4 to BA.5.2*); the other variants involved were BA.1 and BA.2 (7 and 4 cases, respectively; Fig. 1). The lack of differences in the dynamics of Sotrovimab resistance acquisition found between BA.1 and BA.2 in previous studies can now be extended, based on our initial observations, to BA.5 variants. The Delta lineage has also been found to share similar patterns of acquisition of Sotrovimab resistance mutations (9). Our data support the inclusion of cases infected with the BA.5 lineage among those that could face therapeutical challenges when receiving Sotrovimab because the genomic findings for BA.1/BA.2 are equivalent to those for BA.5 based on general patterns, such as the percentage of cases that acquired resistance (73% in BA.5 cases and 64% non-BA.5 cases); and other qualitative findings, such as the fixation of mutations, coexistence of several different substitutions, complexity of the clonal dynamics, and rapid detection of the emergence of resistance (5 days for both BA.5 and BA.1/BA.2).
RESISTANCE IN NON-NASOPHARYNGEAL SPECIMENS
In addition to the predominantly nasopharyngeal specimens included in our study, we had the opportunity to analyze lower respiratory specimens (bronchoalveolar lavage fluid [BAL fluid] or bronchial aspirate [BAS]) from two of the patients with resistance detected in nasopharyngeal (NP) specimens (patients 5 and 3). In patient 5, E340Q was the only mutation identified in two sequential NP specimens, whereas a greater diversity of coexisting mutations was observed in a single BAL fluid sample (two different P337 substitutions and another four in E340; Fig. 1). In patient 3, with E340D fixed in two sequential NP specimens, another two different non-fixed substitutions in the same codon (E340K [29%] and E340A [67%]) were observed in the BAL fluid specimen (Fig. 1). Unexpectedly, no mutations were identified in the BAS specimen. The compartmentalized microevolution of microorganisms in different respiratory samples has also been described for SARS-CoV-2 (15), but this is its first description in Sotrovimab resistance.
MICROBIOLOGICAL AND CLINICAL EVOLUTIONARY ASPECTS
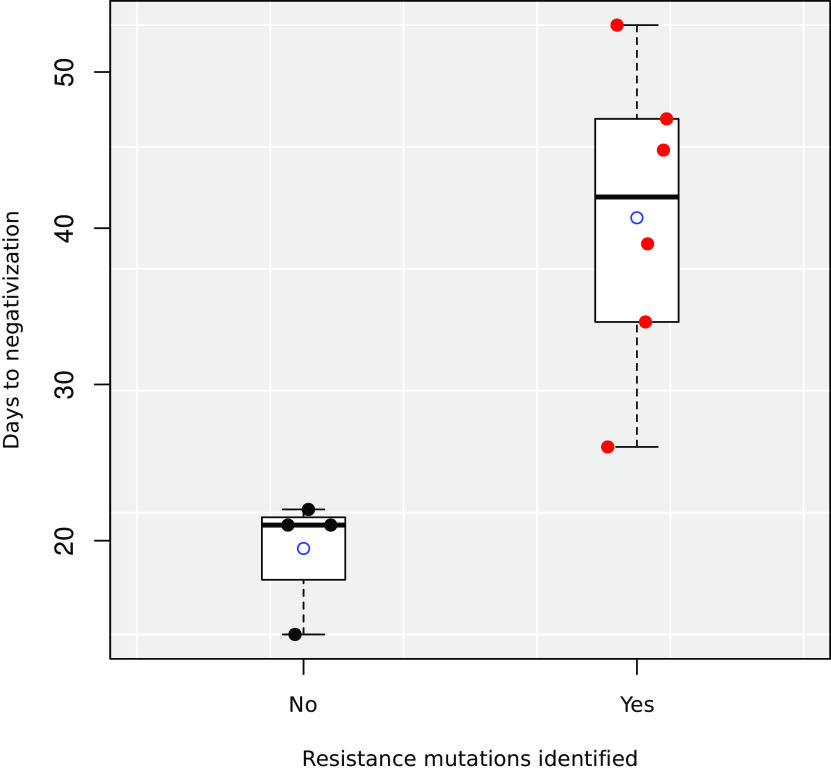
When we analyzed the effect of the acquisition of resistance on patient evolution and outcomes, we observed a significant increase in the mean number of days to viral clearance for patients in whom resistance mutations had been identified (40.67 days) versus those without resistance mutations (19.5 days; P = 0.003, analysis of variance on the linear model after removing outliers detected by the Grubbs test), as reported in other studies (11, 14) (Fig. 2).
FIG 2.
Boxplot of the time until viral clearance (days) for patients in whom resistance mutations were or were not identified. Average values of each group are shown by empty circles. Individual values are shown as scattered points.
Three of the patients who acquired resistance mutations (patients 2, 3, and 8; all three received Sotrovimab due to severe COVID-19) died from COVID. The time from treatment to death for each patient was 25, 65 and 114 days, respectively. In all three patients, specimens with a diversity of substitutions and others with a fixed mutation were identified (P337L in one case and E340D in the remaining two) (Fig. 1). Patients 2 and 8 died 12 and 7 days, respectively, after their last positive RT-PCR test and both had high viral loads (CT ~ 16; Supplemental File 1), suggesting that they probably did not clear the virus. Patient 3 died 43 days after the last NP positive RT-PCR test (CT = 26; Supplemental File 1) and no subsequent RT-PCRs were performed, so viral clearance before death was not evaluated.
The BA.5 lineage was not associated with a differential pattern in either patient outcome (18% of patients died in both groups) or mean time to viral clearance (for BA.1/2 lineages: 49 days for patients with resistance versus 19 days for patients without resistance; for BA.5 lineages: 36.5 and 21 days for patients with and without resistance, respectively).
Seven patients received additional treatments for COVID-19 other than Sotrovimab: three received nirmatrelvir/ritonavir and four received remdesivir (in both treatments, two patients received antivirals as early treatment, and viral persistence was maintained). None of these patients died from COVID and no decrease in time to viral clearance (20 to 124 days) was observed. In four of the five cases where mutations were identified, one evolved to fixation; in two of the remaining cases, no mutations were detected. The limited number of patients in the study who received treatment with two antiviral drugs limits the statistical power of our observations. In addition, most of these patients received treatment as sequential therapy, with Sotrovimab used as salvage therapy after microbiological failure of early treatment with nirmatrelvir/ritonavir or remdesivir. Thus, the possible benefits of combination treatments of small-molecule antiviral drugs associated with Sotrovimab previously suggested in other studies cannot be evaluated in this patient cohort (13).
New lineages such as BQ.1 and XBB.1.5 have become predominant in recent months, following our study period. Sotrovimab was not administered to patients infected with BQ.1 because its efficacy is compromised, with a 100-times reduction in its neutralization capacity (4, 16, 17). The prevalence of XBB.1.5 has only recently increased in our context and the lower COVID-19 general incidence, together with the milder episodes of the newly diagnosed patients, are responsible for our lack of data on this lineage.
CONCLUSIONS
Our study found the highest rates of acquisition of resistance mutations reported to date in immunocompromised patients receiving Sotrovimab. The increased diversity of lineages in our analysis includes, for the first time, Omicron BA.5, as well as BA.1 and BA.2, as being among the variants that make treatment with Sotrovimab challenging. Our genomic analysis of many sequential specimens offers a detailed snapshot of the highly dynamic and complex mutational pathways used by SARS-CoV-2 under selective pressure from Sotrovimab, which even lead to an asymmetrical, compartmentalized distribution of mutations at different sites. Acquisition of resistance leads to a delay in the time needed to clear the virus, with all the potential clinical implications that this implies. Close real-time genomic surveillance of patients receiving Sotrovimab should be mandatory to allow timely therapeutical intervention as soon as resistance is detected.
Ethical statement.
The study was approved by the research ethics committee of Gregorio Marañón Hospital (REF: MICRO.HGUGM.2020-042). Due to the retrospective nature of the study, informed content was not required by the Ethics Committee from our institution.
ACKNOWLEDGMENTS
This work was supported by the Instituto de Salud Carlos III (PI21/01823) together with the FEDER fund “A way of making Europe,” and by the ECDC (2021/PHF/23776) Darío García de Viedma and IiSGM (2021-II-PI-01) Darío García de Viedma. L.P.-L. was supported by a Miguel Servet contract (CPII20/00001 Laura Pérez-Lago). This research was supported by CIBER -Consorcio Centro de Investigación Biomédica en Red- (CB06/06/0058), Instituto de Salud Carlos III, Ministerio de Ciencia e Innovación and Unión Europea - European Regional Development Fund.
We thank the Genomics Unit of our institution for all the support in the sequencing runs and Janet Dawson for her help in editing and proofreading.
Gregorio Marañón Microbiology-ID COVID 19 Study Group members.
Luis Alcalá, Teresa Aldámiz, Roberto Alonso, Ana Álvarez-Uría, Elena Bermúdez, Emilio Bouza, Sergio Buenestado-Serrano, Almudena Burillo, Raquel Carrillo, Pilar Catalán, Emilia Cercenado, Alejandro Cobos, Cristina Díez, Pilar Escribano, Agustín Estévez, Chiara Fanciulli, Alicia Galar, Ma Dolores García, Darío García de Viedma, Paloma Gijón, Helmuth Guillén, Jesús Guinea, Marta Herranz, Álvaro Irigoyen, Martha Kestler, Juan Carlos López, Marina Machado, Mercedes Marín, Pablo Martín-Rabadán, Andrea Molero-Salinas, Pedro Montilla, Patricia Muñoz, Belén Padilla, Rosalía Palomino-Cabrera, María Palomo, María Jesús Pírez-Granda, Daniel Peñas-Utrilla, Laura Pérez-Lago, Leire Pérez, Elena Reigadas, Cristina Rincón, Belén Rodríguez, Sara Rodríguez, Cristina Rodríguez-Grande, Adriana Rojas, María Jesús Ruiz-Serrano, Carlos Sánchez, Mar Sánchez, Amadeo Sanz-Pérez, Julia Serrano, Francisco Tejerina, Maricela Valerio, Cristina Veintimilla, Lara Vesperinas, Teresa Vicente, and Sofía de la Villa.
Footnotes
Supplemental material is available online only.
Contributor Information
Darío García de Viedma, Email: dgviedma2@gmail.com.
Laura Pérez-Lago, Email: lperezg00@gmail.com.
Collaborators: Luis Alcalá, Teresa Aldámiz, Roberto Alonso, Ana Álvarez-Uría, Elena Bermúdez, Emilio Bouza, Sergio Buenestado-Serrano, Almudena Burillo, Raquel Carrillo, Pilar Catalán, Emilia Cercenado, Alejandro Cobos, Cristina Díez, Pilar Escribano, Agustín Estévez, Chiara Fanciulli, Alicia Galar, Ma Dolores García, Darío García de Viedma, Paloma Gijón, Helmuth Guillén, Jesús Guinea, Marta Herranz, Álvaro Irigoyen, Martha Kestler, Juan Carlos López, Marina Machado, Mercedes Marín, Pablo Martín-Rabadán, Andrea Molero-Salinas, Pedro Montilla, Patricia Muñoz, Belén Padilla, Rosalía Palomino-Cabrera, María Palomo, María Jesús Pérez-Granda, Daniel Peñas-Utrilla, Laura Pérez-Lago, Leire Pérez, Elena Reigadas, Cristina Rincón, Belén Rodríguez, Sara Rodríguez, Cristina Rodríguez-Grande, Adriana Rojas, María Jesús Ruiz-Serrano, Carlos Sánchez, Mar Sánchez, Amadeo Sanz-Pérez, Julia Serrano, Francisco Tejerina, Maricela Valerio, Ma Cristina Veintimilla, Lara Vesperinas, Teresa Vicente, and Sofía de la Villa
REFERENCES
- 1.Burnett DL, Jackson KJL, Langley DB, Aggrawal A, Stella AO, Johansen MD, Balachandran H, Lenthall H, Rouet R, Walker G, Saunders BM, Singh M, Li H, Henry JY, Jackson J, Stewart AG, Witthauer F, Spence MA, Hansbro NG, Jackson C, Schofield P, Milthorpe C, Martinello M, Schulz SR, Roth E, Kelleher A, Emery S, Britton WJ, Rawlinson WD, Karl R, Schäfer S, Winkler TH, Brink R, Bull RA, Hansbro PM, Jäck H-M, Turville S, Christ D, Goodnow CC. 2021. Immunizations with diverse sarbecovirus receptor-binding domains elicit SARS-CoV-2 neutralizing antibodies against a conserved site of vulnerability. Immunity 54:2908–2921.e6. doi: 10.1016/j.immuni.2021.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.European Medicines Agency (EMA). Xevudy product information. https://www.ema.europa.eu/en/documents/product-information/xevudy-epar-product-information_en.pdf. EMA, Amsterdam, The Netherlands. [Google Scholar]
- 3.Aleem A, Akbar Samad AB, Vaqar S. 2023. Emerging variants of SARS-CoV-2 and novel therapeutics against coronavirus (COVID-19). [PubMed]
- 4.Touret F, Giraud E, Bourret J, Donati F, Tran-Rajau J, Chiaravalli J, Lemoine F, Agou F, Simon-Lorière E, van der Werf S, de Lamballerie X. 2023. Enhanced neutralization escape to therapeutic monoclonal antibodies by SARS-CoV-2 Omicron sub-lineages. iScience 26:106413. doi: 10.1016/j.isci.2023.106413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agencia Española de Medicamentos y Productos Sanitarios (AEMPS). Criterios para valorar la administración de las nuevas alternativas terapéuticas antivirales frente a la infección por SARS-CoV-2. [In Spanish.] Available from https://www.aemps.gob.es/medicamentos-de-uso-humano/acceso-a-medicamentos-en-situaciones-especiales/criterios-para-valorar-la-administracion-de-las-nuevas-alternativas-terapeuticas-antivirales-frente-a-la-infeccion-por-sars-cov-2. AEMPS, Madrid, Spain. [Google Scholar]
- 6.Rodríguez-Grande C, Alcalá L, Estévez A, Sola-Campoy PJ, Buenestado-Serrano S, Martínez-Laperche C, De La Cueva VM, Alonso R, Andrés-Zayas C, Adán-Jiménez J, Losada C, Rico-Luna C, Comas I, González-Candelas F, Catalán P, Muñoz P, Pérez-Lago L, De Viedma DG, Gregorio Marañón Microbiology-ID COVID 19 Study Group 2 . 2022. Systematic genomic and clinical analysis of severe acute respiratory syndrome coronavirus 2 reinfections and recurrences involving the same strain. Emerg Infect Dis 28:85–94. doi: 10.3201/eid2801.211952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin WS, Chen IC, Chen HC, Lee YC, Wu SC. 2021. Glycan masking of epitopes in the NTD and RBD of the spike protein elicits broadly neutralizing antibodies against SARS-CoV-2 variants. Front Immunol 12:795741. doi: 10.3389/fimmu.2021.795741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cathcart AL, Havenar-Daughton C, Lempp FA, Ma D, Schmid MA, Agostini ML, Guarino B, Di Iulio J, Rosen LE, Tucker H, Dillen J, Subramanian S, Sloan B, Bianchi S, Pinto D, Saliba C, Culap K, Wojcechowskyj JA, Noack J, Zhou J, Kaiser H, Lee S, Farhat N, Chase A, Montiel-Ruiz M, Dellota E, Jr, Park A, Spreafico R, Sahakyan A, Lauron EJ, Czudnochowski N, Cameroni E, Ledoux S, Kawaoka Y, Werts A, Colas C, Soriaga L, Telenti A, Purcell LA, Hwang S, Snell G, Virgin HW, Corti D, Hebner CM. 2022. The dual function monoclonal antibodies VIR-7831 and VIR-7832 demonstrate potent in vitro and in vivo activity against SARS-CoV-2. bioRxiv. doi: 10.1101/2021.03.09.434607. [DOI]
- 9.Rockett R, Basile K, Maddocks S, Fong W, Agius JE, Johnson-Mackinnon J, Arnott A, Chandra S, Gall M, Draper J, Martinez E, Sim EM, Lee C, Ngo C, Ramsperger M, Ginn AN, Wang Q, Fennell M, Ko D, Lim HL, Gilroy N, O’Sullivan MVN, Chen SC-A, Kok J, Dwyer DE, Sintchenko V. 2022. Resistance mutations in SARS-CoV-2 Delta variant after Sotrovimab use. N Engl J Med 386:1477–1479. doi: 10.1056/NEJMc2120219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andrés C, González-Sánchez A, Jiménez M, Márquez-Algaba E, Piñana M, Fernández-Naval C, Esperalba J, Saubi N, Quer J, Rando-Segura A, Miarons M, Codina MG, Ruiz-Camps I, Pumarola T, Abrisqueta P, Antón A. 2023. Emergence of Delta and Omicron variants carrying resistance-associated mutations in immunocompromised patients undergoing Sotrovimab treatment with long-term viral excretion. Clin Microbiol Infect 29:240–246. doi: 10.1016/j.cmi.2022.08.021. [DOI] [PubMed] [Google Scholar]
- 11.Huygens S, Oude Munnink B, Gharbharan A, Koopmans M, Rijnders B. 2023. Sotrovimab resistance and viral persistence after treatment of immunocompromised patients infected with the severe acute respiratory syndrome coronavirus 2 Omicron variant. Clin Infect Dis 76:e507–e509. doi: 10.1093/cid/ciac601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vellas C, Trémeaux P, Del Bello A, Latour J, Jeanne N, Ranger N, Danet C, Martin-Blondel G, Delobel P, Kamar N, Izopet J. 2022. Resistance mutations in SARS-CoV-2 Omicron variant in patients treated with Sotrovimab. Clin Microbiol Infect 28:1297–1299. doi: 10.1016/j.cmi.2022.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gliga S, Lübke N, Killer A, Gruell H, Walker A, Dilthey AT, Thielen A, Lohr C, Flaßhove C, Krieg S, Pereira JV, Seraphin TP, Zaufel A, Däumer M, Orth H-M, Feldt T, Bode JG, Klein F, Timm J, Luedde T, Jensen B-EO. 2023. Rapid selection of Sotrovimab escape variants in severe acute respiratory syndrome coronavirus 2 Omicron-infected immunocompromised patients. Clin Infect Dis 76:408–415. doi: 10.1093/cid/ciac802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Birnie E, Biemond JJ, Appelman B, De Bree GJ, Jonges M, Welkers MRA, Wiersinga WJ. 2022. Development of resistance-associated mutations after Sotrovimab administration in high-risk individuals infected with the SARS-CoV-2 Omicron variant. JAMA 328:1104–1107. doi: 10.1001/jama.2022.13854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rueca M, Bartolini B, Gruber CEM, Piralla A, Baldanti F, Giombini E, Messina F, Marchioni L, Ippolito G, Di Caro A, Capobianchi MR. 2020. Compartmentalized replication of SARS-CoV-2 in upper vs. lower respiratory tract assessed by whole genome quasispecies analysis. Microorganisms 8:1302–1312. doi: 10.3390/microorganisms8091302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arora P, Kempf A, Nehlmeier I, Schulz SR, Jäck HM, Pöhlmann S, Hoffmann M. 2023. Omicron sublineage BQ.1.1 resistance to monoclonal antibodies. Lancet Infect Dis 23:22–23. doi: 10.1016/S1473-3099(22)00733-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tzou PL, Tao K, Pond SLK, Shafer RW. 2022. Coronavirus Resistance Database (CoV-RDB): SARS-CoV-2 susceptibility to monoclonal antibodies, convalescent plasma, and plasma from vaccinated persons. PLoS One 17:e0261045. doi: 10.1371/journal.pone.0261045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Download aac.00266-23-s0001.xlsx, XLSX file, 0.02 MB (20.9KB, xlsx)