Abstract
Seven genes coding for small heat shock proteins (sHsps) in Bradyrhizobium japonicum have been identified. They are organized in five operons that are coordinately regulated by ROSE, a negatively cis-acting DNA element. The deduced sHsps can be divided into two separate classes: class A, consisting of proteins that show similarity to Escherichia coli IbpA and IbpB, and class B, whose members display significant similarity to other sHsps from prokaryotes and eukaryotes. Two-dimensional gel electrophoresis and Edman sequencing revealed the presence of at least 12 sHsps in B. japonicum, indicating a remarkable abundance of sHsps in this organism. Three additional members of class A and two potentially novel heat shock proteins were identified on the basis of their amino termini. The presence of multiple sHsps was also demonstrated for a variety of Rhizobium and Bradyrhizobium species by immunoblot analysis and two-dimensional gel electrophoresis. An extensive database survey revealed that, in contrast to the rhizobia, other bacteria contain maximally two sHsps whereas many plants have been reported to possess a sHsp superfamily.
All organisms so far examined respond to a sudden increase in growth temperature by inducing the synthesis of a number of heat shock proteins (Hsps). Some of these proteins are also important during normal growth. The regulation, structure, and function of several Hsps have been studied in great detail. The chaperone machineries GroES/GroEL and DnaK/DnaJ/GrpE, for example, are involved in diverse processes such as protein folding and protein degradation, assembly of protein complexes, and transport of proteins across membranes. Their function appears to be highly conserved between prokaryotes and eukaryotes (reviewed in references 7 and 40).
Comparatively little is known, however, about small Hsps (sHsps). In contrast to the highly conserved DnaK and GroEL proteins, sHsps show much less sequence similarity. This protein family is characterized by the following criteria: (i) a molecular mass typically between 12 and 30 kDa; (ii) a conserved central domain, referred to as the α-crystallin domain; (iii) formation of large oligomeric complexes, ranging from 150 to 800 kDa; (iv) ATP-independent chaperone activity (9, 27, 51). The latter concept, however, has been challenged by the observation that ATP enhances the molecular chaperone activity of αB-crystallin (36). According to the present model, sHsps bind to denatured proteins accumulated under stress conditions and maintain them in a folding-competent state (15, 32). Recently, the crystal structure of a sHsp from Methanococcus jannaschii has been solved (29). Twenty-four monomers form a hollow spherical complex with a total of 14 “windows” that might allow polypeptides to enter the complex.
Not surprisingly, most of the work on sHsps has been conducted with eukaryotic members of this superfamily because they are related to the α-crystallin proteins of the vertebrate eye lens (26). α-Crystallins play a structural role in maintaining lens stability and transparency but notably they are also expressed in nonlenticular tissues, e.g., in heart, muscle, and kidney (3). A remarkable abundance of sHsps was reported in heat-stressed plants. Up to 30 different sHsps comprising six different classes are induced after a temperature upshift, depending on the plant species. Each gene family encodes proteins localized in a distinct cellular compartment (51).
Most bacteria appear to have only a small number of heat shock proteins. The completed genome sequences indicate that Mycoplasma genitalium completely lacks any gene coding for sHsps (19). M. jannaschii encodes one and Escherichia coli encodes two sHsps (8, 17). The first hint that Rhizobiaceae may be an exception in that they possess a larger set of sHsps was provided by a two-dimensional gel analysis by Michiels et al. (34). The authors compared the induction of Hsps in a heat-tolerant and a heat-sensitive Rhizobium strain and observed eight heat-inducible protein spots in extracts from the temperature-sensitive strain. By contrast, the tropical, heat-tolerant strain induced only two sHsps.
In the process of elucidating the complex regulatory network that controls the heat shock response of Bradyrhizobium japonicum, the nitrogen-fixing root-nodule symbiont of soybean, we identified six genes encoding sHsps (37, 38). They are organized in four operons that are located in an extended heat-shock gene cluster. Each operon is preceded by a conserved DNA element of approximately 100 bp that is positioned between the transcription start and the start codon of the first gene. This element was designated ROSE (for Repression Of heat Shock gene Expression), and several lines of evidence suggest that it serves as a binding site for a putative repressor protein under non-heat shock conditions (37).
Here we report on a bacterial sHsp superfamily comprising at least 12 members. The seven B. japonicum sHsps identified so far can be grouped into two distinct classes. We monitored the induction of sHsps under various stress conditions and examined their heat shock induction by two-dimensional gel electrophoresis. Finally, we provide evidence that the presence of a sHsp superfamily is not restricted to B. japonicum but might be widespread in the Rhizobiaceae.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
B. japonicum 110spc4 was grown aerobically at 28°C in PSY medium (43) supplemented with 0.1% (wt/vol) arabinose and 100 μg of spectinomycin per ml. YEM medium (12) supplemented with 10 mM KNO3 was used for anaerobic B. japonicum cultures. Bradyrhizobium sp. (Parasponia) ANU289 and Bradyrhizobium sp. (Lupinus) ATCC 10319 were propagated in PSY medium with 0.1% (wt/vol) arabinose. TY medium (5) was used to grow Rhizobium leguminosarum bv. viciae 897, Rhizobium etli (formerly R. leguminosarum bv. phaseoli) 8002, R. leguminosarum bv. trifolii ATCC 14480, Rhizobium sp. strain NGR234, and Sinorhizobium meliloti 2011. E. coli cells were grown in Luria-Bertani medium (35) supplemented with ampicillin (200 μg/ml) if required.
DNA manipulations and sequence analysis.
Recombinant DNA techniques were performed according to standard protocols (45). Chromosomal DNA was isolated as described previously (23). Southern blot hybridizations using DIG (digoxigenin-11-dUTP)-labeled DNA probes were performed according to the manufacturer’s instructions (Boehringer GmbH, Mannheim, Germany). The 111-bp ROSE1 probe was produced by PCR using plasmid pRJ5035, a pUC18 derivative containing a 1.7-kb HindIII insert carrying the hspA gene region (38), and the oligonucleotides Sig36 (5′-CGCCGCGACAAGCGGTCC-3′) and Sig37 (5′-GTCCTCATAGCCAAATCCTCC-3′). Plasmid DNA was sequenced by the chain termination method (46) with a Model 373 DNA sequencer (Applied Biosystems, Foster City, Calif.). The DNA sequence was analyzed with the software package of the Genetics Computer Group of the University of Wisconsin—Madison (UWGCG) (version 8.0) or the National Center for Biotechnology Information network server. Multiple sequence alignments were generated with the PILEUP program provided by the UWGCG software.
Western blot (immunoblot) analysis.
Crude cell extracts were prepared, separated on sodium dodecyl sulfate–12% polyacrylamide gels, and transferred to nitrocellulose membranes as described previously (39). Bacteroid extracts were prepared as described elsewhere (16). Anti-E. coli IbpA serum (2) was kindly provided by A. Easton (St. Louis, Mo.) and was used in 1,500-fold dilution. Primary rabbit antibodies were detected with the Chemiluminescence Western Blotting Kit (Boehringer GmbH).
Two-dimensional gel electrophoresis and Edman sequencing.
Two-dimensional gel electrophoresis, protein elution, concentration, electrotransfer, and N-terminal sequencing were performed as described elsewhere (42).
Transcript mapping.
RNA isolation and primer extension analysis was performed as described elsewhere (4). The oligonucleotides AN1 (5′-CTGAACATAGTCTGCCAGGTTGAACTGC-3′) and AN18 (5′-CCGTTTCAACGAGGTCGAAAAGGC-3′) were used to determine the hspH transcription start site.
Nucleotide sequence accession numbers.
The nucleotide sequences described here have been deposited in the EMBL, GenBank, and DDBJ databases under the following accession numbers: U55047 (hspA, hspB, and hspC gene region), AJ003064 (hspD, hspE, and hspF gene region), and AJ010144 (hspH gene region).
RESULTS AND DISCUSSION
Two classes of sHsps in B. japonicum.
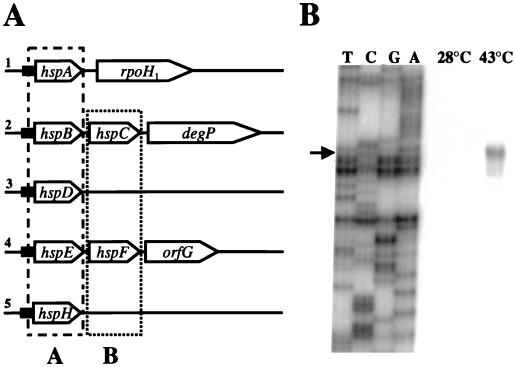
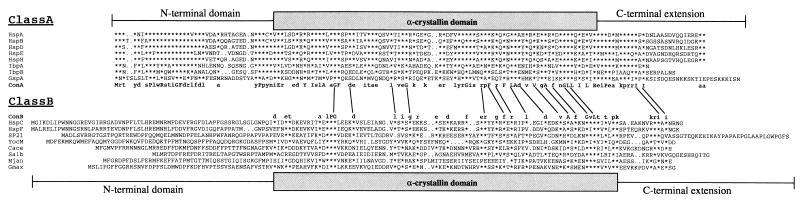
Six genes coding for small Hsps (hspA, -B, -C, -D, -E, and -F) have recently been identified in a heat shock gene cluster of B. japonicum (37, 38). They are organized in four operons together with some additional heat shock genes. Heat-inducible transcription of each operon is mediated by ROSE, a novel regulatory element that consists of approximately 100 bp and precedes the first gene of each operon (37). We identified a putative fifth ROSE-dependent operon by using a ROSE1 fragment as a probe in Southern hybridization experiments (data not shown). Two hybridizing fragments, a 5.8-kb BamHI fragment and a 5.6-kb SalI fragment, were subsequently cloned and found to contain the hspH gene region (Fig. 1A). No additional heat shock genes were present up- or downstream of hspH. An amino acid sequence comparison of the deduced small Hsps revealed that they fall into two distinct classes, as indicated in Fig. 1A and shown more precisely in Fig. 2. Class A contains only bacterial proteins, namely the B. japonicum proteins HspA, -B, -D, -E, and -H, E. coli IbpA and IbpB, and Legionella pneumophila GspA. It is evident from the alignment that proteins belonging to this class are highly similar to each other throughout their entire length (between 34 and 73% positional amino acid sequence identity). The similarity is not restricted to the α-crystallin domain but extends into the flanking amino- and carboxy-terminal regions. Class B proteins are much more divergent in length, sequence, and phylogenetic origin. They include prokaryotic as well as eukaryotic members from a wide variety of organisms. The similarity between class A proteins and class B proteins is rather low (around 20% amino acid sequence identity). Although the degree of homology within class B is significant (between 30 and 60% identical amino acids), only the B. japonicum proteins HspC and HspF reach the latter, highest score. The identity among the other members is generally between 30 and 35%. Identical amino acids are almost exclusively displayed in the α-crystallin domain, and the flanking regions are highly variable in length and sequence (with the exception of HspC and HspF).
FIG. 1.
Physical map of five ROSE-dependent heat shock operons of B. japonicum and determination of the transcription start site upstream of ROSE5. (A) Schematic representation of the ROSE-dependent operons. The ROSE elements (1 through 5) are represented by black boxes. Class A and class B small heat shock genes are indicated. No significant open reading frames were identified downstream of hspD and hspH. (B) Primer extension analysis to determine the transcription start site upstream of hspH. The extension product of primer AN18 is shown. The same primer was used for the corresponding sequencing reaction (TCGA).
FIG. 2.
Amino acid sequence alignment of sHsp representatives from class A and class B. Regions corresponding to the N-terminal domain, α-crystallin domain, and C-terminal extension are indicated (9, 33). The consensus sequences (ConA and ConB) are defined by capital letters when an amino acid is present in all eight aligned sequences and by lowercase letters when an amino acid is present in at least five of the eight sequences. Amino acids that conform with the consensus are indicated by asterisks. Amino acids appearing in both consensus sequences are connected by a line. The complete sequence of the deduced proteins is shown and designated as follows: HspA to HspH, B. japonicum HspA to HspH (references 37 and 38 and this work); IbpA and IbpB, E. coli IbpA and IbpB (2); GspA, L. pneumophila GspA (1); SP21, S. aurantiaca SP21 (24); YocM, B. subtilis YocM (31); Cace, C. acetobutylicum Hsp18 (47); Salb, S. albus Hsp18 (48); Mjan, M. jannaschii Hsp16.5 (8); Gmax, Glycine max Hsp17.5 (11).
The B. japonicum hspH gene is preceded by a typical ς70-type promoter and a ROSE element with high sequence similarity to all previously identified ROSE elements (37). With the exception of one nucleotide (a G instead of a C at the ROSE1-equivalent position +32), all previously described conserved ROSE nucleotides were conserved in the ROSE5 sequence. In particular, the nucleotides in the promoter-distal half of ROSE are highly conserved (data not shown). Transcription of hspH was heat inducible, and the transcription start site was located at the expected position just upstream of ROSE5, as determined by primer extension (Fig. 1B). Thus, all presently known sHsp genes of B. japonicum are coordinately regulated by ROSE, a negatively cis-acting DNA element that precedes each class A gene and presumably serves as a repressor binding site under normal growth conditions (37). The two-dimensional gel analysis revealed that the degree of induction varied from protein to protein (see below), suggesting that posttranscriptional or posttranslational mechanisms might contribute to their regulation. Heat-induced expression of class A genes in other organisms (the E. coli ibpAB operon and the L. pneumophila gspA gene) is dependent on a ς32-type promoter (1, 2) whereas several class B genes are under negative control. Transcription of the Streptomyces albus hsp18 gene is subject to repression by the OrfY protein at low temperatures (49). Transcriptional repression has also been proposed to control the expression of Clostridium acetobutylicum hsp18, Leuconostoc oenos hsp18, and Synechococcus vulcanus hspA (28, 44, 47). These genes are transcribed from a typical ς70-type housekeeping promoter, which implies that additional mechanisms must prevent their expression during normal growth. However, the exact control mechanisms have not been elucidated yet.
Induction of sHsps in B. japonicum.
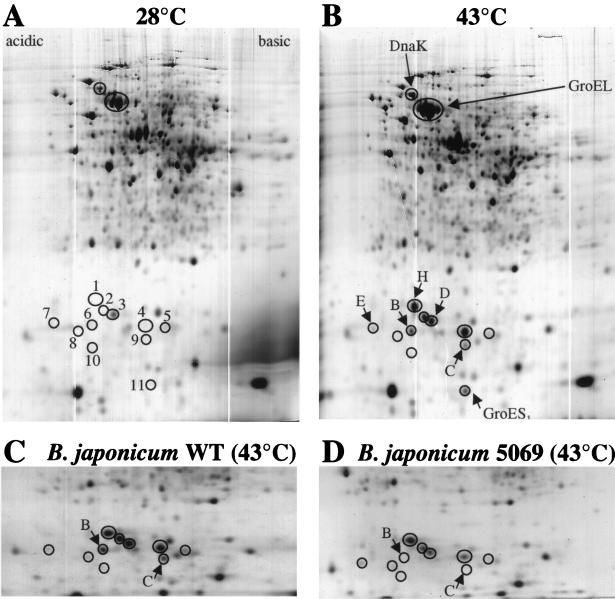
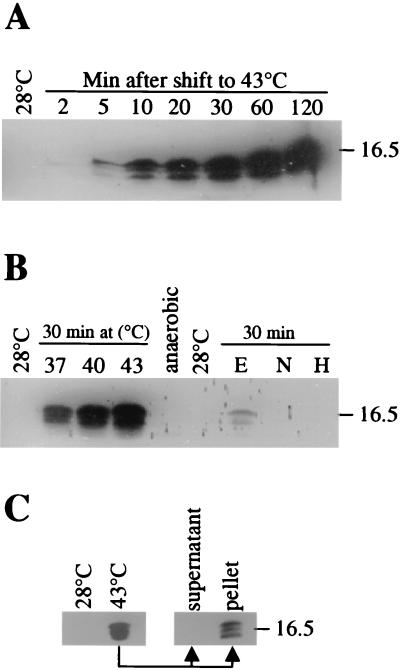
An as-yet-undefined set of B. japonicum sHsps was recognized by an antiserum raised against the 15 N-terminal amino acids of E. coli IbpA which is similar to the B. japonicum class A proteins (2, 39). Three cross-reacting bands were detected in extracts from heat-shocked B. japonicum cells, and the fastest migrating band was absent in a hspBC mutant (B. japonicum 5069) indicating that the antiserum specifically recognized the HspB protein (39). Immunoblots of two-dimensional gels revealed that the serum indeed recognizes several class A proteins (HspB, HspD, HspH, and spots 4, 8, and 10) (data not shown; compare with Fig. 4). We monitored the kinetics of sHsp induction in B. japonicum by using this antiserum. Extracts from cells harvested before and at different time points after a heat shock from 28 to 43°C were analyzed (Fig. 3A). The first faint signal was observed at 5 min after the heat shock. The accumulation of sHsps continued until the level reached a maximum approximately 60 min after the temperature upshift. This elevated level was maintained for at least another hour. In a separate experiment, we determined how a shift to various temperatures affected the induction of sHsps. The amount of sHsps increased in proportion to the severity of the shift (Fig. 3B). A shock from 28 to 37°C was sufficient to induce the complete set of immunodetectable sHsps, but a shift to 40°C and in particular to 43°C was much more efficient. Next, we analyzed whether other stress conditions could induce the synthesis of class A sHsps. Cultures grown at 28°C did not induce sHsps during the onset of, or in, stationary phase (data not shown). Extracts from bacteroids that had been isolated from soybean root nodules did not contain detectable amounts of sHsps (data not shown). Neither continuous growth under anaerobic conditions nor a shift of aerobically grown cultures to high-salt (0.3 M NaCl) conditions or to a highly oxidizing environment (0.001% H2O2) elicited a significant response (Fig. 3B). However, the addition of ethanol (5%) to a culture led to the production of sHsps, albeit to a much lesser extent than a heat shock. This result suggests that both a temperature shift and an ethanol shock trigger a signal that is finally transduced to induce the synthesis of sHsps in B. japonicum.
FIG. 4.
Two-dimensional gel electrophoresis of B. japonicum extracts. Crude extracts of cells grown at 28°C (A) or of cells shifted from 28 to 43°C for 30 min (B) were separated and stained with Coomassie blue. DnaK, a set of GroEL proteins, and sHsps are circled. The positions numbered 1 to 10 in panel A correspond to the proteins listed in Table 1. HspB, -C, -D, -E, and -H that were identified by amino-terminal sequencing are labeled in panel B as B, C, D, E, and H, respectively. Induction of sHsps in B. japonicum wild type (WT) (C) and the hspBCdegP mutant B. japonicum 5069 (D) (34). Relevant sections of two-dimensional Coomassie-blue stained gels are shown. The positions of HspB (labeled B) and HspC (labeled C) which are present in the wild type but missing in the mutant are indicated by an arrow. Other sHsps in panels C and D are circled.
FIG. 3.
Immunoblot analyses of B. japonicum extracts by using anti-E. coli IbpA serum. (A) B. japonicum was grown to mid-exponential phase at 28°C. After a reference sample (28°C) had been taken, the culture was shifted to 43°C and samples were collected at the time points indicated. (B) Experiment similar to that shown in panel A, but the cultures were shifted to the temperatures indicated (37, 40, or 43°C), or ethanol (E) (5%), NaCl (N) (0.3M), or H2O2 (H) (0.001%) were added. One extract (anaerobic) originated from a culture grown under anaerobic conditions at 28°C. (C) Fractionation of heat-shocked B. japonicum extracts. Normally grown cells (28°C) and heat-shocked cells (48°C) were passed four times through a French pressure cell at 110 MPa. The soluble (supernatant) and insoluble (pellet) fractions of the heat-shocked cells were separated by centrifugation at 12,000 × g for 30 min. The apparent molecular mass (in kilodaltons) of a reference protein (lysozyme) is indicated on the right.
Many studies indicate that certain sHsps in animals, plants, and bacteria are regulated by a variety of environmental and developmental cues. Developmental synthesis of sHsps in eukaryotes is often tissue-specific in contrast to the coordinate heat shock induction of sHsps in almost all tissues. Constitutive, but low expression of Hsp27, a mammalian sHsp, was observed in different cell types. This protein plays a role in regulating the dynamics of actin filaments and probably confers stability to actin fibers (reviewed in reference 3). Expression of plant sHsps during pollen development and seed and fruit maturation has been reported. Again, only a subset of the sHsps reacts to the developmental signals, and their expression is temporally and spatially controlled (51). A number of bacterial sHsps can also be induced by developmental signals, although heat shock often is the major elicitor. L. pneumophila GspA is expressed during intracellular infection of macrophages and mycobacterial Hsp16 might also be induced in response to stresses encountered during an infection process (1, 52). The Bacillus subtilis CotM protein is developmentally induced during sporulation and Stigmatella aurantiaca SP21 is synthesized during sporulation and fruiting body formation (24, 25). Induction of C. acetobutylicum Hsp18 was demonstrated during a metabolic shift from acid to solvent production (41, 47). By contrast, our investigation indicates that at least the immunodetectable B. japonicum sHsps are classical heat stress proteins.
Small Hsps aggregate after heat shock in vivo.
Extracts of heat-shocked B. japonicum cells were separated into a soluble and insoluble fraction. The immunodetectable sHps were almost exclusively found in the pellet fraction (Fig. 3C), indicating that they form insoluble aggregates after heat shock. Whether these aggregates consist only of sHsps (homo- or heterooligomers) or whether substrate proteins are bound to the sHsps cannot be determined at present.
Identification of B. japonicum sHsps by two-dimensional gel analysis.
The presence of at least seven genes coding for sHsps in B. japonicum prompted us to investigate the induction of such proteins by comparative two-dimensional gel electrophoresis (Fig. 4). The positions of DnaK and GroEL are indicated for comparison (Fig. 4A and B). Note that B. japonicum contains five groESL operons and that the GroEL spot represents a composite of several GroEL proteins (16, 36a). At least 11 small proteins were reproducibly upregulated after a heat shock and visible on Coomassie-stained two-dimensional gels. GroES1, HspB, -C, -D, -E, and -H were identified by N-terminal sequencing of the collected protein spots from several gels (Fig. 4B). A comparison of the sHsp pattern after heat shock in the wild type and the hspBCdegP mutant 5069 confirmed the identity of the HspB and HspC spots because they were in fact missing in the mutant (Fig. 4C and D). HspA and HspF could not be identified. HspA may not be detectable due to a cathodic drift in the first dimension (calculated isoelectric point of 8.42). The amount of HspF is probably too low to be detectable because HspE, the product of the first gene of the hspEForfG operon, is also barely visible. The amino termini of proteins 4, 8, and 10 (MRTYDLTP, MRTYDFLP, and MRSYDFSPLWRSTXTG, respectively; compare with Fig. 2) indicated that B. japonicum contains at least three additional class A sHsps whose structural genes and regulatory elements have yet to be identified. The amino-terminal sequence of two proteins (ALYEHVFL and AGTVEQKL for spots 2 and 5 in Fig. 4, respectively) did not show similarity to class A or class B proteins or any other proteins in the databases which suggests that there might be additional sHsp classes in B. japonicum. In summary, we predict that B. japonicum contains a total of at least 12 sHsps.
A set of sHsps is present in other rhizobia.
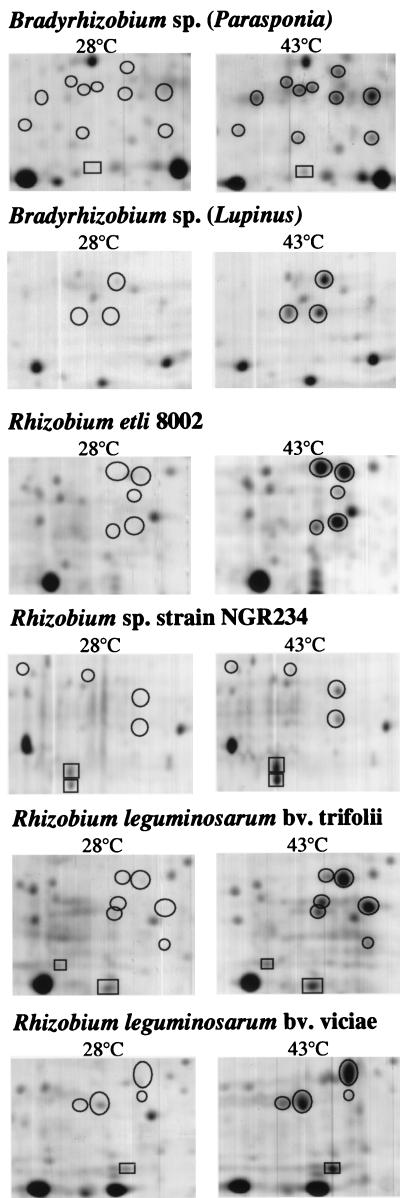
In order to test whether a superfamily of sHsps is present in other rhizobial species, we screened a variety of Bradyrhizobium and Rhizobium strains by immunoblot analysis using the anti-E. coli IbpA serum. Heat induction of one or several bands was observed in each case, indicating that all species tested possess class A-type sHsps (data not shown). To monitor the heat-induced proteins more accurately, we performed two-dimensional gel electrophoresis of extracts from six rhizobial species. Between 3 and 10 potential sHsps were observed in each strain (Fig. 5; Table 1). In summary, we conclude that the existence of a sHsp family is not restricted to B. japonicum but occurs in many rhizobial species.
FIG. 5.
Induction of sHsps in Bradyrhizobium and Rhizobium strains. Crude extracts of cells grown at 28°C or of cells shifted from 28 to 43°C for 30 min were separated by two-dimensional gel electrophoresis and stained with Coomassie blue. Only the section of the gel containing sHsps (in the range between approximately 10 and 20 kDa) is shown. Potential sHsps are circled. Spots marked by rectangles were not considered in Table 1. Based on their apparent molecular masses they might represent GroES proteins (cf. Fig. 4) or other proteins that do not belong to the sHsp family.
TABLE 1.
Number and classification of sHsps in various organisms
| Organism | No. of sHsps
|
Reference or database | ||
|---|---|---|---|---|
| Class A | Class B | Total | ||
| Bradyrhizobium japonicum | ≥8a | ≥2 | ≥12b | 37, 38, this work |
| Bradyrhizobium sp. (Parasponia) | n.d.c | n.d. | ≥10b | This work |
| Bradyrhizobium sp. (Lupinus) | n.d. | n.d. | ≥3b | This work |
| Rhizobium etli CNPAF512 | n.d. | n.d. | ≥8b | 34 |
| Rhizobium etli 8002 | n.d. | n.d. | ≥5b | This work |
| Rhizobium sp. strain NGR234 | n.d. | n.d. | ≥4b | This work |
| Rhizobium leguminosarum bv. trifolii | n.d. | n.d. | ≥6b | This work |
| Rhizobium leguminosarum bv. viciae | n.d. | n.d. | ≥4b | This work |
| Sinorhizobium meliloti | n.d. | n.d. | ≥3d | This work |
| Rhizobium tropici CIAT899 | n.d. | n.d. | ≥2b | 34 |
| Mycoplasma genitaliume | 0 | 0 | 0 | 19 |
| Haemophilus influenzaee | 0 | 0 | 0 | 17 |
| Helicobacter pylorie | 0 | 0 | 0 | 50 |
| Borrelia burgdorferie | 0 | 0 | 0 | 18 |
| Treponema pallidume | 0 | 0 | 0 | 20 |
| Escherichia colie | 2 | 0 | 2 | 6 |
| Methanococcus jannaschiie | 0 | 1 | 1 | 8 |
| Aquifex aeolicuse | 0 | 1 | 1 | 13 |
| Archaeoglobus fulgiduse | 0 | 2 | 2 | 30 |
| Bacillus subtilise | 0 | 2 | 2f | 31 |
| Mycobacterium tuberculosise | 0 | 2 | 2 | 10 |
| Saccharomyces cerevisiaee | 0 | 2 | 2 | 22 |
| Rhodobacter capsulatusg | 0 | 0 | 0 | Capsulapedia database |
| Legionella pneumophila | ≥1 | n.d. | ≥1 | 1 |
| Pseudomonas aeruginosa | ≥1 | n.d. | ≥1 | Pseudomonas Genome Project |
| Synechocystis sp. strain PCC6803 | n.d. | ≥1 | ≥1 | CyanoBase |
| Stigmatella aurantiaca | n.d. | ≥1 | ≥1 | 24 |
| Clostridium acetobutylicum | n.d. | ≥1 | ≥1 | 47 |
| Streptomyces albus | n.d. | ≥1 | ≥1 | 48 |
| Homo sapiens | 0 | 0 | ≥5h | 13 |
| Glycine max (soybean) | n.d. | ≥10 | ≥10 | 21 |
| Arabidopsis thaliana | n.d. | ≥12 | ≥12 | TIGR database |
| Oryza sativa (rice) | n.d. | ≥11 | ≥11 | TIGR database |
Based on DNA sequence analysis and Edman sequencing.
As determined by two-dimensional gel analysis.
Not determined or not known because the genome has not been sequenced.
As determined by one-dimensional gel electrophoresis and immunoblot with anti-IbpA serum (data not shown).
Complete genome sequence available.
YocM and CotM are defined as α-crystallin-type proteins in the corresponding SubtiList database. However, YdfT has also some similarity to class B proteins (around 20% overall sequence identity and approximately 25% identical amino acids in the α-crystallin domain) and might fall into this class.
67% of the genome sequence has been completed.
Human and animal sHsps appear to fall into a separate class distinct from classes A and B.
The presence of multiple sHsps in a bacterium is a rather uncommon feature. A literature and database survey that included the 35 microbial genomes that are completed or currently being sequenced, revealed that bacteria other than rhizobia encode either no or maximally two sHsps (Table 1). For example, no α-crystallin-like protein was found in the genomes of the pathogens M. genitalium, H. influenzae, Helicobacter pylori, and Borrelia burgdorferi. The available sequence of Rhodobacter capsulatus, an α-proteobacterium and close relative of rhizobia, also did not reveal any sHsp. One or two sHsps are encoded in the genome of a number of eubacteria and archaebacteria and in yeast. Interestingly, if one of these organisms contains two sHsps, they always belong to the same class.
The existence of a sHsp superfamily comprising defined classes is well established in plants (Table 1). For example, the sequences of 10 soybean (Glycine max) sHsps are deposited in the public databases. They clearly fall into class B but have been further subdivided in different subfamilies. Six groups were classified: two classes (class I and II) localized to the cytosol, and one class each localized to the chloroplast, endoplasmic reticulum, mitochondrium, and membrane compartment (51). The homology between individual members of these classes is restricted to only a few amino acids in the α-crystallin domain. An phylogenetic analysis suggested that the abundance of plant sHsps arose from an ancient gene duplication or amplification more than 150 million years ago that was followed by sequence divergence (51). A similar gene multiplication event with subsequent diversification might have occurred in B. japonicum, giving rise to the unusual broad spectrum of bacterial sHsps. The localization of six B. japonicum genes (hspA to hspE) encoding sHsps in a heat shock gene cluster probably supports this assumption. Five human sHsps have been described (14). The ongoing genome sequencing projects will reveal whether sHsp superfamilies are common in mammals.
It is unclear why the rhizobia analyzed in this work contain multiple sHsps whereas most other organisms do not. The relative abundance of rhizobial sHsps after a heat shock certainly implies an important cellular function offering an advantage in their natural environment. Short periods of intense sunlight, for example, might cause protein damage. When chaperones become temporarily overloaded with potential substrates, sHsps might play an important role as buffer for otherwise aggregation-prone enzymes. In agreement with a recent model (15, 32), one can imagine that this reservoir of folding-competent proteins will later be refolded by the cellular chaperone machineries under conditions when their capacity becomes available again. A tropical Rhizobium strain that is adapted to high temperatures apparently does not require multiple sHsps because it contains only two small heat-inducible proteins (34). The reason for the heat tolerance of this strain is unknown. Bacteria which thrive as mammalian pathogens live in an environment with more or less constant temperatures and may be able to cope without a sophisticated heat shock response. Their lifestyle is reflected by a comparatively small number of sHsp genes in their genome.
ACKNOWLEDGMENTS
We are grateful to Hauke Hennecke and Peter James for generous support, continuous interest in our work, and helpful comments on the manuscript. Hans-Martin Fischer and Evelyne Bauer are acknowledged for providing B. japonicum extracts. Rhizobium strains were obtained from Michael Göttfert. We thank Alan Easton for the generous gift of antisera and Wolfgang Weiglhofer for performing the experiment whose results are shown in Fig. 3A.
This study was supported by grants from the Swiss National Foundation for Scientific Research and the Swiss Federal Institute of Technology, Zürich.
REFERENCES
- 1.Abu Kwaik Y, Engleberg N C. Cloning and molecular characterization of a Legionella pneumophila gene induced by intracellular infection and by various in vitro stress conditions. Mol Microbiol. 1994;13:243–251. doi: 10.1111/j.1365-2958.1994.tb00419.x. [DOI] [PubMed] [Google Scholar]
- 2.Allen S P, Polazzi J O, Gierse J K, Easton A M. Two novel heat shock genes encoding proteins produced in response to heterologous protein expression in Escherichia coli. J Bacteriol. 1992;174:6938–6947. doi: 10.1128/jb.174.21.6938-6947.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arrigo A P, Landry J. Expression and function of the low-molecular-weight heat shock proteins. In: Morimoto R I, Tissiéres A, Georgopoulos C, editors. The biology of heat shock proteins and molecular chaperones. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. pp. 335–373. [Google Scholar]
- 4.Babst M, Hennecke H, Fischer H M. Two different mechanisms are involved in the heat-shock regulation of chaperonin gene expression in Bradyrhizobium japonicum. Mol Microbiol. 1996;19:827–839. doi: 10.1046/j.1365-2958.1996.438968.x. [DOI] [PubMed] [Google Scholar]
- 5.Beringer J E. R factor transfer in Rhizobium leguminosarum. J Gen Microbiol. 1974;84:188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- 6.Blattner F R, Plunkett G, Bloch C A, Perna N T, Burland V, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 7.Bukau B, Horwich A L. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 8.Bult C J, White O, Olsen G J, Zhou L X, Fleischmann R D, et al. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 9.Caspers G J, Leunissen J A M, de Jong W W. The expanding small heat-shock protein family, and structure predictions of the conserved ‘α-crystallin domain’. J Mol Evol. 1995;40:238–248. doi: 10.1007/BF00163229. [DOI] [PubMed] [Google Scholar]
- 10.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 11.Czarnecka E, Gurley W B, Nagao R T, Mosquera L A, Key J L. DNA sequence and transcript mapping of a soybean gene encoding a small heat shock protein. Proc Natl Acad Sci USA. 1985;82:3726–3730. doi: 10.1073/pnas.82.11.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daniel R M, Appleby C A. Anaerobic-nitrate, symbiotic and aerobic growth of Rhizobium japonicum: effects on cytochrome P450, other haemoproteins, nitrate and nitrite reductases. Biochim Biophys Acta. 1972;275:347–354. doi: 10.1016/0005-2728(72)90215-0. [DOI] [PubMed] [Google Scholar]
- 13.Deckert G, Warren P V, Gaasterland T, Young W G, Lenox A L, et al. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 14.De Jong W W, Caspers G J, Leunissen J A M. Genealogy of the α-crystallin–small heat-shock protein superfamily. Int J Biol Macromol. 1998;22:151–162. doi: 10.1016/s0141-8130(98)00013-0. [DOI] [PubMed] [Google Scholar]
- 15.Ehrnsperger M, Gräber S, Gaestel M, Buchner J. Binding of non-native protein to Hsp25 during heat shock creates a reservoir of folding intermediates for reactivation. EMBO J. 1997;16:221–229. doi: 10.1093/emboj/16.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischer H M, Babst M, Kaspar T, Acuña G, Arigoni F, Hennecke H. One member of a groESL-like chaperonin multigene family in Bradyrhizobium japonicum is co-regulated with symbiotic nitrogen fixation genes. EMBO J. 1993;12:2901–2912. doi: 10.1002/j.1460-2075.1993.tb05952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 18.Fraser C M, Casjens S, Huang W M, Sutton G G, Clayton R, et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 19.Fraser C M, Gocayne J D, White O, Adams M D, Clayton R A, et al. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 20.Fraser C M, Norris S J, Weinstock G M, White O, Sutton G G, et al. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science. 1998;281:375–388. doi: 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- 21.Gaestel M, Vierling E, Buchner J. The small heat shock protein (sHSP) family: an overview. In: Gething M J, editor. Guidebook to molecular chaperones and protein-folding catalysts. Oxford, England: Oxford University Press; 1997. pp. 269–272. [Google Scholar]
- 22.Goffeau, A., et al. 1997. The yeast genome directory. Nature 387(Suppl.):1–107. [PubMed]
- 23.Hahn M, Hennecke H. Localized mutagenesis in Rhizobium japonicum. Mol Gen Genet. 1984;193:46–52. [Google Scholar]
- 24.Heidelbach M, Skladny H, Schairer H U. Heat shock and development induce synthesis of a low-molecular-weight stress-responsive protein in the myxobacterium Stigmatella aurantiaca. J Bacteriol. 1993;175:7479–7482. doi: 10.1128/jb.175.22.7479-7482.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henriques A O, Beall B W, Moran C P. CotM of Bacillus subtilis, a member of the α-crystallin family of stress proteins, is induced during development and participates in spore outer coat formation. J Bacteriol. 1997;179:1887–1897. doi: 10.1128/jb.179.6.1887-1897.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ingolia T D, Craig E A. Four small Drosophila heat shock proteins are related to each other and to mammalian α-crystallin. Proc Natl Acad Sci USA. 1982;79:2360–2364. doi: 10.1073/pnas.79.7.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jakob U, Gaestel M, Engel K, Buchner J. Small heat shock proteins are molecular chaperones. J Biol Chem. 1993;268:1517–1520. [PubMed] [Google Scholar]
- 28.Jobin M-P, Delmas F, Garmyn D, Diviès C, Guzzo J. Molecular characterization of the gene encoding an 18-kilodalton small heat shock protein associated with the membrane of Leuconostoc oenos. Appl Environ Microbiol. 1997;63:609–614. doi: 10.1128/aem.63.2.609-614.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim K K, Kim R, Kim S H. Crystal structure of a small heat-shock protein. Nature. 1998;394:595–599. doi: 10.1038/29106. [DOI] [PubMed] [Google Scholar]
- 30.Klenk H P, Clayton R A, Tomb J F, White O, Nelson K E, et al. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 31.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 32.Lee G J, Roseman A M, Saibil H R, Vierling E. A small heat shock protein stably binds heat denatured model substrates and can maintain a substrate in a folding competent state. EMBO J. 1997;16:659–671. doi: 10.1093/emboj/16.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leroux M R, Melki R, Gordon B, Batelier G, Candido E P M. Structure-function studies on small heat shock protein oligomeric assembly and interaction with unfolded polypeptides. J Biol Chem. 1997;272:24646–24656. doi: 10.1074/jbc.272.39.24646. [DOI] [PubMed] [Google Scholar]
- 34.Michiels J, Verreth C, Vanderleyden J. Effects of temperature stress on bean-nodulating Rhizobium strains. Appl Env Microbiol. 1994;60:1206–1212. doi: 10.1128/aem.60.4.1206-1212.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 36.Muchowski P J, Clark J I. ATP-enhanced molecular chaperone functions of the small heat shock protein human αB crystallin. Proc Natl Acad Sci USA. 1998;95:1004–1009. doi: 10.1073/pnas.95.3.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36a.Münchbach, M., and H. M. Fischer. Unpublished results.
- 37.Narberhaus F, Käser R, Nocker A, Hennecke H. A novel DNA element that controls bacterial heat shock gene expression. Mol Microbiol. 1998;28:315–323. doi: 10.1046/j.1365-2958.1998.00794.x. [DOI] [PubMed] [Google Scholar]
- 38.Narberhaus F, Weiglhofer W, Fischer H-M, Hennecke H. The Bradyrhizobium japonicum rpoH1 gene encoding a ς32-like protein is part of a unique heat shock gene cluster together with groESL1 and three small heat shock genes. J Bacteriol. 1996;178:5337–5346. doi: 10.1128/jb.178.18.5337-5346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Narberhaus F, Weiglhofer W, Fischer H M, Hennecke H. Identification of the Bradyrhizobium japonicum degP gene as part of an operon containing small heat-shock protein genes. Arch Microbiol. 1998;169:89–97. doi: 10.1007/s002030050547. [DOI] [PubMed] [Google Scholar]
- 40.Netzer W J, Hartl F U. Protein folding in the cytosol: chaperonin-dependent and -independent mechanisms. Trends Biochem Sci. 1998;23:68–73. doi: 10.1016/s0968-0004(97)01171-7. [DOI] [PubMed] [Google Scholar]
- 41.Pich A, Narberhaus F, Bahl H. Induction of heat shock proteins during initiation of solvent formation in Clostridium acetobutylicum. Appl Microbiol Biotechnol. 1990;33:697–704. [Google Scholar]
- 42.Quadroni M, Staudenmann W, Kertesz M, James P. Analysis of global responses by protein and peptide fingerprinting of proteins isolated by two-dimensional gel electrophoresis: application to the sulfate-starvation response of Escherichia coli. Eur J Biochem. 1996;239:773–781. doi: 10.1111/j.1432-1033.1996.0773u.x. [DOI] [PubMed] [Google Scholar]
- 43.Regensburger B, Hennecke H. RNA polymerase from Rhizobium japonicum. Arch Microbiol. 1983;135:103–109. doi: 10.1007/BF00408017. [DOI] [PubMed] [Google Scholar]
- 44.Roy S K, Nakamoto H. Cloning, characterization, and transcriptional analysis of a gene encoding an α-crystallin-related, small heat shock protein from the thermophilic cyanobacterium Synechococcus vulcanus. J Bacteriol. 1998;180:3997–4001. doi: 10.1128/jb.180.15.3997-4001.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 46.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sauer U, Dürre P. Sequence and molecular characterization of a DNA region encoding a small heat shock protein of Clostridium acetobutylicum. J Bacteriol. 1993;175:3394–3400. doi: 10.1128/jb.175.11.3394-3400.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Servant P, Mazodier P. Characterization of Streptomyces albus 18-kilodalton heat shock-responsive protein. J Bacteriol. 1995;177:2998–3003. doi: 10.1128/jb.177.11.2998-3003.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Servant P, Mazodier P. Heat induction of hsp18 gene expression in Streptomyces albus G: transcriptional and posttranscriptional regulation. J Bacteriol. 1996;178:7031–7036. doi: 10.1128/jb.178.24.7031-7036.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 51.Waters E R, Lee G J, Vierling E. Evolution, structure and function of the small heat shock proteins in plants. J Exp Bot. 1996;47:325–338. [Google Scholar]
- 52.Yuan Y, Crane D D, Barry C E. Stationary phase-associated protein expression in Mycobacterium tuberculosis: function of the mycobacterial α-crystallin homolog. J Bacteriol. 1996;178:4484–4492. doi: 10.1128/jb.178.15.4484-4492.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]