Abstract
1,4,5,6-Tetrahydro-2-methyl-4-pyrimidinecarboxylic acid (ectoine) is an excellent osmoprotectant. The biosynthetic pathway of ectoine from aspartic β-semialdehyde (ASA), in Halomonas elongata, was elucidated by purification and characterization of each enzyme involved. 2,4-Diaminobutyrate (DABA) aminotransferase catalyzed reversively the first step of the pathway, conversion of ASA to DABA by transamination with l-glutamate. This enzyme required pyridoxal 5′-phosphate and potassium ions for its activity and stability. The gel filtration estimated an apparent molecular mass of 260 kDa, whereas molecular mass measured by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was 44 kDa. This enzyme exhibited an optimum pH of 8.6 and an optimum temperature of 25°C and had Kms of 9.1 mM for l-glutamate and 4.5 mM for dl-ASA. DABA acetyltransferase catalyzed acetylation of DABA to γ-N-acetyl-α,γ-diaminobutyric acid (ADABA) with acetyl coenzyme A and exhibited an optimum pH of 8.2 and an optimum temperature of 20°C in the presence of 0.4 M NaCl. The molecular mass was 45 kDa by gel filtration. Ectoine synthase catalyzed circularization of ADABA to ectoine and exhibited an optimum pH of 8.5 to 9.0 and an optimum temperature of 15°C in the presence of 0.5 M NaCl. This enzyme had an apparent molecular mass of 19 kDa by SDS-PAGE and a Km of 8.4 mM in the presence of 0.77 M NaCl. DABA acetyltransferase and ectoine synthase were stabilized in the presence of NaCl (>2 M) and DABA (100 mM) at temperatures below 30°C.
Halotolerance is of considerable interest scientifically and from the perspective of wide application in fermentation industries and in agriculture. When eubacteria are exposed to hyperosmotic stress, they accumulate various low-molecular-weight organic compounds, the so-called “compatible solutes” such as polyols, amino acids, sugars, and betaines (7–9, 13, 19, 48), because maintenance of turgor pressure is a prerequisite for growth under the conditions of elevated external osmotic pressure. Since Galinski et al. (14) discovered 1,4,5,6-tetrahydro-2-methyl-4-pyrimidinecarboxylic acid (ectoine) as a compatible solute in Ectothiorhodospira halochloris, an extremely halophilic phototrophic eubacterium, ectoine has been found to be distributed widely in nature, largely in moderately halophilic eubacteria (3, 11, 12, 26, 38, 50). In addition, ectoine has been investigated as a new excellent universal osmoprotectant in this decade, since incorporation of external ectoine under hyperosmotic stress has been observed to confer protection on various nonhalotolerant eubacteria (16, 21, 44).
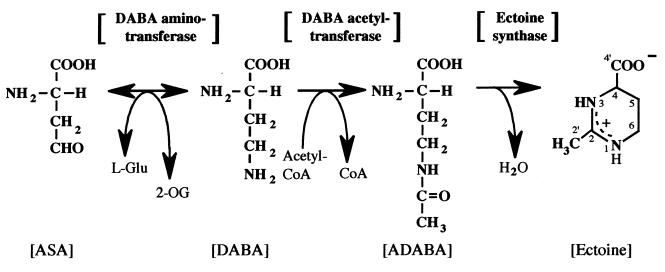
We previously isolated a moderately halophilic eubacterium, Halomonas elongata (31), from dry salty land in Thailand. We identified ectoine and γ-N-acetyl-α,γ-diaminobutyric acid (ADABA), which is one of the cleavage structures of ectoine, as osmotically responding compounds in the cells grown in a glucose-mineral medium containing NaCl in a concentration range of 3 to 15% (31). To understand the accumulation mechanism of the intracellular ectoine, characterization of enzymes involved in the biosynthesis of ectoine is indispensable. Therefore, we have focused on the biosynthetic enzyme of ectoine in this organism. We observed that radioactivity from [1-14C]aspartate was most efficiently incorporated into ectoine and that the signal intensity was enriched preferentially from [1-13C]acetate into the methyl carbon at position 2′ and from [2-13C]acetate into the methine carbon at position 2 of the ectoine skeleton, respectively, in 13C nuclear magnetic resonance (NMR) spectroscopy (22). From these findings, we also hypothesized the following pathway essentially similar to that described by Peters et al. (34): aspartic β-semialdehyde (ASA) is converted to 2,4-diaminobutyric acid (DABA) by transamination, and DABA is converted to ADABA by acetylation with acetyl coenzyme A (CoA), which in turn yields ectoine by circularization (Fig. 1). The three enzymes involved in this pathway are DABA aminotransferase, DABA acetyltransferase, and ectoine synthase in order of the reactions to ectoine. Peters et al. (34) detected the activity of the first and the second of the three steps by using crude extracts of E. halochloris and H. elongata. However, the characterization of these enzymes was limited; in particular, their responses to various salt concentrations remained unknown.
FIG. 1.
Proposed biosynthetic pathway of ectoine in H. elongata OUT30018.
In this study, we confirmed the biosynthetic pathway of ectoine by using purified enzymes in H. elongata OUT30018 and characterized the three enzymes involved in the conversion of ASA to ectoine for the first time.
MATERIALS AND METHODS
Bacterial strain and growth conditions.
A moderately halophilic and halotolerant eubacterium, H. elongata OUT30018 (Osaka University type culture), formerly designated strain KS3, isolated from a salty soil in Thailand (31), was used in this study. For stock culture, Luria-Bertani agar medium (39) consisting of 1.0% tryptone (Difco, Detroit, Mich.), 0.5% yeast extract (Difco), and 1.0% NaCl, was modified by the addition of NaCl at a final concentration of 15% (wt/vol). A mineral-glucose medium, M63, consisting of 100 mM KH2PO4, 75 mM KOH, 15 mM (NH4)2SO4, 1 mM MgSO4, 3.9 μM FeSO4, and 22 mM glucose, pH 7.2 (33), was modified by the addition of NaCl at a concentration of 3 or 15% and designated M63S-3 or M63S-15, respectively. Cells grown overnight in M63S-3 medium supplemented with 0.25% yeast extract were transferred into prewarmed fresh M63S-3 medium in Sakaguchi flasks with a working volume of 30% at 2% inoculum size and cultivated by reciprocal shaking at 37°C. The concentration of NaCl in the medium was upshifted from 3 to 15% by the addition of NaCl crystals at an optical density at 660 nm of 2.0, and the cultivation was continued. When necessary, an antifoaming agent, Adekanol LG-109 (Asahi Denka Kogyo Ltd., Tokyo, Japan), was added. The cells were harvested at an optical density at 660 nm of 4.0 and washed once with ice-cold 50 mM Tris-HCl buffer (pH 8.0), containing 2.86 M NaCl and 5 mM EDTA, and the cell pellets were stored at −80°C until use.
Preparation of crude enzyme extract.
Unless otherwise stated, enzymes were prepared at 0 to 4°C. For small-scale preparation, the cells obtained from a 100-ml culture were resuspended in 1 ml of 50 mM Tris-HCl buffer (pH 8.0) and treated with 20 μg of lysozyme at 37°C for 5 min and thereafter with 0.5 mg of DNase I and 2.0 mg of RNase A at 37°C for 1 min. After addition of 1 mM phenylmethylsulfonyl fluoride (PMSF), 0.5 mM EDTA, and 0.1 M NaCl (final concentrations), the cell lysate was centrifuged at 11,000 × g for 20 min. To remove endogenous low-molecular-weight metabolites, the supernatant was passed through a column (12 by 40 mm) of Sephadex G-25 (Pharmacia LKB, Biotechnology AB, Uppsala, Sweden) equilibrated with 50 mM Tris-HCl buffer, pH 9.0, and crude proteins recovered by elution with the same buffer were used for the radiotracer experiment with [14C]acetyl-CoA.
For large-scale preparation, cell pellets obtained from a 5-liter culture were resuspended in 50 ml of 50 mM Tris-HCl buffer (pH 8.0) containing 5 mM EDTA, treated with lysozyme at 50 μg ml−1 at 30°C for 1 min, and disrupted with a French pressure cell press (SLM Aminco Instruments, Inc., Urbana, Ill.) at 15,000 lb/in2, once. After the addition of PMSF to a final concentration of 1 mM, the lysate was centrifuged at 15,000 × g for 20 min. The supernatant was diluted with the same buffer to 500 ml, treated with 0.25% protamine sulfate to remove nucleic acids, and centrifuged at 22,000 × g for 20 min. The ratio of the absorbance at 260 nm to that at 280 nm of this supernatant was in the range of 1.2 to 1.4. This supernatant was used for the purification of each enzyme as described below.
Purification of enzymes.
For purification of DABA aminotransferase, the protein fraction precipitating between 60 and 80% (NH4)2SO4 saturation (10) was recovered from the crude enzyme solution by centrifugation. The precipitate was dissolved in 50 mM Tris-HCl (pH 9.0) and readjusted to 60% (NH4)2SO4 saturation. The solution was subjected to hydrophobic interaction column chromatography (HIC) on a column (2.5 by 39 cm) of Sepharose CL-6B (Pharmacia) equilibrated with 50 mM Tris-HCl (pH 9.0) buffer containing 1.73 M (NH4)2SO4. Unbound proteins were washed off with the same buffer, and the bound proteins were eluted by counterdirected linear gradients of (NH4)2SO4 from 1.73 to 0 M and of NaCl from 0 to 2 M at a flow rate of 1.0 ml min−1. The active fractions were collected, and (NH4)2SO4 was added to saturation. The HIC was repeated twice, and the active fractions were collected and concentrated by using an Advantec ultrafiltration apparatus fitted with a UP-20 membrane (Advantec, Tokyo, Japan). The concentrated fraction was loaded on a column (0.9 by 79 cm) of Toyopearl HW-65 (Tosoh, Tokyo, Japan) equilibrated with 50 mM Tris-HCl (pH 9.0) containing 50 mM KCl and 10 μM pyridoxal 5′-phosphate (PLP) and eluted with the same buffer at a flow rate of 0.2 ml min−1. The active fractions were collected, concentrated, and stored at 0°C until use.
For purification of DABA acetyltransferase, the protein fraction precipitating between 40 and 80% (NH4)2SO4 saturation was recovered from the crude enzyme solution by centrifugation. The precipitate was dissolved in 50 mM Tris-HCl (pH 8.5) containing 1.5 M (NH4)2SO4, 0.02% Brij 35 (Nacalai Tesque, Kyoto, Japan), 10 mM 2-mercaptoethanol, 1 mM DABA, 1 mM EDTA, and 1 mM PMSF, and subjected to HIC on a column of Sepharose CL-6B equilibrated with the same buffer. After the column was washed with the same buffer at a flow rate of 1.0 ml min−1, bound proteins were eluted by counterdirected linear gradients of (NH4)2SO4 from 1.5 to 0 M and of NaCl from 0 to 2 M. The active fractions were collected, desalted, and concentrated by ultrafiltration with a Diaflo YM30 membrane (Amicon Co., Danvers, Mass.) with 50 mM Tris-HCl (pH 8.5) containing 0.02% Brij 35, 1 mM 2-mercaptoethanol, 10 mM DABA, 1 mM EDTA, and 1 mM PMSF. The solution was subjected to affinity chromatography with a Blue Sepharose 6FF (Pharmacia LKB) column (2 ml) equilibrated with the same buffer and eluted with a linear gradient of NaCl from 0 to 2 M in the same buffer, except for the concentration of DABA at 100 mM, at a flow rate of 0.2 ml min−1. The active fractions were collected, applied to the second Blue Sepharose chromatography column, and eluted with a linear gradient of NaCl from 0 to 0.3 M. The active fractions were collected, concentrated, and stored at 0°C until use.
For ectoine synthase, (NH4)2SO4 was dissolved in the crude enzyme solution to give 60% saturation and stirred slowly for 2 h. The supernatant obtained by centrifugation at 22,000 × g for 20 min was subjected to HIC on a column (3 by 43.5 cm) of Sepharose CL-6B equilibrated with 1.86 M (NH4)2SO4 and 1 mM DABA in 50 mM Tris-HCl (pH 9.0). Unbound proteins were washed with the same buffer, and bound proteins were eluted by counterdirected linear gradients of (NH4)2SO4 from 1.86 M to 1.2 M and of NaCl from 0 to 2 M at a flow rate of 1.0 ml min−1. The active fractions were collected, and (NH4)2SO4 was removed by ultrafiltration with a UP-10 membrane (Advantec). The concentrated solution was loaded on a column (1.83 by 11.5 cm) of hydroxyapatite (HA) (Wako Pure Chemicals Ltd., Osaka, Japan) equilibrated with a buffer containing 50 mM Tris-HCl (pH 9.0), 2 M NaCl, and 1 mM DABA (buffer A). The enzyme was eluted by a linear gradient of K2HPO4 from 0 to 50 mM in buffer A at a flow rate of 0.33 ml min−1. This HA chromatography was repeated, if necessary. The active fractions were collected, concentrated, and stored at −80°C until use.
Protein concentration was determined according to the method of Bradford (6) or by the UV absorption method at 280 nm with bovine serum albumin as the standard.
Assays for enzymatic activities.
DABA aminotransferase activity was determined routinely by the measurement of the amount of glutamate produced in the reverse reaction. DABA aminotransferase was reacted with a 100-μl mixture composed of 5 mM 2-oxoglutarate (2-OG), 10 mM DABA, 10 μM PLP, 50 mM Tris-HCl (pH 8.5), and 25 mM KCl at 25°C for 30 min, and the reaction was stopped by boiling the mixture for 5 min. The concentration of the glutamate produced was determined by a colorimetric method with the F kit for l-glutamic acid (Boehringer Mannheim GmbH, Mannheim, Germany). Activity of the forward reaction leading to ectoine synthesis was determined by detection of DABA produced from l-glutamate and dl-ASA. The reaction was performed at 15°C for 30 min in a 100-μl mixture consisting of 10 mM dl-ASA, 20 mM sodium glutamate, 50 mM Tris base, 25 mM KCl, 0.1 mM EDTA, 50 μM PLP, and enzyme solution, at pH 8.5. The reaction was stopped by the addition of trifluoroacetic acid (TFA) to a final concentration of 0.3%. The reaction mixtures were filtered through a 0.2-μm-pore-size membrane filter (LCR4-LG; Millipore Co., Bedford, Mass.), and 50 μl of each was used for amino acid analysis with a CCP-8000 system (Tosoh) connected to a TSKgel AminoPak column (4.6 by 120 mm; Tosoh). Amino acids and DABA were detected at 570 nm as ninhydrin reaction-positive compounds. For quantitative analysis, 12.5 nmol of l-alanine and 20 nmol of l-arginine were used as internal standards for ASA and DABA, respectively.
Activity of DABA acetyltransferase was determined by detection of ADABA at 210 nm by high-performance liquid chromatography (HPLC) with the Shimadzu SPD-6A system connected to a reverse-phase column of TSKgel ODS-80Tm (4.6 by 300 mm; Tosoh). The reaction was performed in a 100-μl mixture consisting of 30 mM DABA, 10 mM acetyl-CoA, 0.4 M NaCl, and 60 mM Tris-HCl (pH 8.5) at 20°C for 20 min and stopped by the addition of TFA to a final concentration of 0.5%. DABA was used immediately after neutralization with NaOH. Elution was performed with 0.1% TFA at a flow rate of 0.9 ml min−1. Chemically synthesized and purified ADABA was used as a standard.
The activity of ectoine synthase was determined by detection of ectoine by HPLC with elution conditions similar to those for ADABA analysis. The reaction was performed in a 45-μl mixture consisting of 10 mM ADABA, 0.77 M NaCl, 1 mM DABA, 50 mM Tris-HCl (pH 9.5), and enzyme solution at 15°C for 10 min and stopped by the addition of TFA to a final concentration of 0.3%. To estimate the activity of enzymes at various temperatures, the reaction mixture was sampled every 30 s and the initial velocity of the reaction was calculated. Purified ectoine from H. elongata OUT30018 was used as a standard. One unit of the enzyme activities of 2,4-DABA aminotransferase for the forward reaction, 2,4-DABA aminotransferase for the reverse reaction, 2,4-DABA acetyltransferase, and ectoine synthase was defined as the amount of enzyme which yielded 1 μmol of the products DABA, l-glutamate, ADABA, and ectoine, respectively, for 1 min.
Substrate specificity of enzymes.
To determine the substrate specificity of DABA aminotransferase, DABA acetyltransferase, and ectoine synthase, incubation mixtures each consisted of test compounds for substrate at 10 mM each, and other conditions were similar to the standard methods described above. For the estimation of Km of DABA aminotransferase and ectoine synthase, the same incubation conditions as those for the standard methods described above were used, except that the amounts of substrate in the reaction mixtures changed. The ranges of amounts of each substrate were as follows: with DABA aminotransferase, for estimation of Km of dl-ASA, l-glutamate, 2-OG, and DABA, dl-ASA concentration of 0.5 to 5 mM with 10 mM sodium glutamate, sodium glutamate concentration of 0.5 to 20 mM with 10 mM ASA, DABA concentration of 1 to 20 mM with 5 mM 2-OG, and 2-OG concentration of 0.5 to 10 mM with 10 mM DABA, respectively. With ectoine synthase, for estimation of Km of ADABA, 1 to 20 mM ADABA was used. To confirm the linearity of the reaction, each reaction mixture was sampled every 30 s and the products were determined.
Determination of effect of pH on enzyme activity and stability.
For the determination of effect of pH on activity and stability of the enzymes, the following buffers were used at 50 mM: citrate-Na2HPO4, pH 4.0 to 7.6; Tris-HCl, pH 7.0 to 9.3; and glycine-NaOH, pH 8.0 to 10.0. Replacement of buffers was performed by ultrafiltration at 0°C.
Evaluation of the effects of metals and other various compounds on enzyme activity.
Metal ions were added in the incubation mixture as chloride salts at a final concentration of 1 mM, and other conditions were similar to those for the standard assay. Dithiothreitol, 2-mercaptoethanol, and N-ethylmaleimide, at a final concentration of 10 mM, were used in the standard assay.
Evaluation of the effects of salts and amino acids on the enzyme stability.
After addition of salts and/or amino acids to each enzyme solution and the incubation of enzyme solution under the conditions of indicated temperature and time, these test compounds were replaced with each standard buffer by ultrafiltration, and then residual activity was determined by the standard assay. The final concentrations used were as follows: NaCl and KCl, 10 mM to 3 M; amino acids, 1 mM to 0.5 M.
Radiotracer experiment.
[1-14C]acetyl-CoA (2.22 GBq mmol−1; Amersham International plc, Little Chalfont, Buckinghamshire, England) was added to the reaction mixture consisting of 67 mM Tris-HCl (pH 9.5), 3.3 mM DABA, 4.5 mM acetyl-CoA, 2.2 mM MgCl2, 90 mM NaCl, and crude enzyme solution. The reaction mixture was analyzed by HPLC (System Gold, version 4; Beckman Instruments, Inc., San Ramon, Calif.) by using two TSKgel ODS-80Tm columns connected in series (4.6 by 250 mm by 2). Elution was performed with 0.1% TFA at a flow rate of 0.5 ml min−1. The eluted compounds were monitored by spectrometry at 210 nm, and radioactivities of the eluted substances were detected by scintillation counting.
Preparation of ASA, ADABA, and ectoine.
ASA was synthesized chemically and purified by the modified method of Black (5). Ozone, produced with an Ozonator YO-6B (Yanagimoto Co., Ltd., Kyoto, Japan), was passed through a solution of 0.5 g of dl-allylglycine (Sigma Chemicals, St. Louis, Mo.) in 1 N HCl at 0°C for 6 h. Dimethyl sulfide was added to remove the excess ozone and to cleave ozonide to ASA and formaldehyde under conditions of stirring and cooling in an ice bath in a ventilation chamber overnight. The products were loaded on an ion-exchange column, AG50W-X8 (H+) (Bio-Rad, Richmond, Calif.; 1.0 by 28 cm), washed with a large volume of distilled water, and then eluted with 4 N HCl. Fractions of 5 ml each were collected. The concentration of l-ASA was determined enzymatically by the method of Black (4) by using a partially purified homoserine dehydrogenase from commercial pressed baker’s yeast (Oriental Yeast, Tokyo, Japan). The amount of residual formaldehyde was analyzed by the colorimetric method of Small and Tanes (40) with 4-amino-3-hydrazino-5-mercapto-1,2,4-triazole (AHMT; Nacalai Tesque, Inc.), and it was confirmed that there was no typical absorption peak of formaldehyde at 550 nm. The ASA fraction was lyophilized with an FDU-810 freeze-dryer (EYELA, Tokyo, Japan), and dried ASA was dissolved in a minimum amount of 0.1 N HCl immediately after drying. The ASA solution was kept at −20°C in a tightly closed container until use. It remained stable for at least 6 months. Aspartic acid as a major impurity was assayed by using the amino acid analyzer connected to a TSKgel AminoPak column as described above, and l-aspartic acid contamination was found to be limited to less than 5 mol%. Preparation of ADABA was performed according to the method of Benoiton and Leclerc (2). Purification of ADABA synthesized and of ectoine extracted from cells of H. elongata OUT30018 was performed as described previously (31).
Determination of molecular mass.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed according to Laemmli’s method (23) with protein molecular weight markers obtained from Bio-Rad. Proteins were stained with Coomassie brilliant blue. Gel filtration chromatography with a Toyopearl HW-65 column (1.54 by 97 cm) was used for the estimation of molecular mass. The buffers for equilibration and elution were buffer A for DABA acetyltransferase and ectoine synthase and a buffer containing 0.1 M KCl instead of the 2 M NaCl of buffer A for DABA aminotransferase. Size marker proteins were obtained from Boehringer Mannheim.
Determination of isoelectric point.
Isoelectric focusing (IEF) electrophoresis was performed by semidry electrophoresis with the Resolute HMP chamber with IsoGel agarose IEF plates (pH3-10; FMC BioProducts, Rockland, Maine). Acetic acid (0.5 M) for the anode solution and NaOH (1.0 M) for the cathode solution were used, and electrofocusing was carried out at 5°C for 10 min at 1 W and then for approximately 50 min at 1 kV. pI markers used were as follows; horse heart acetylated cytochrome c (pI, 4.1, 4.9, 6.4, 8.3, 9.7, and 10.6; Oriental Yeast Co., Ltd.), β-lactoglobulin A (pI, 5.1), and carbonic anhydrase I (pI, 6.6; Sigma). Proteins were stained with Coomassie brilliant blue.
Determination of the amino acid composition.
Ten micrograms of desalted and lyophilized enzyme was hydrolyzed in 6 N twice-distilled HCl plus 0.2% phenol at 110°C for 24 h. Amino acid analysis was performed with a Hitachi L-8500 system (Hitachi, Ltd., Tokyo, Japan).
Other analyses.
For mass spectroscopy, samples were applied to a fast atom bombardment mass spectrometer, JMS-DX303 (JEOL, Tokyo, Japan). For 1H NMR spectroscopy, the samples were dissolved in D2O and the spectra were obtained with a JNM-GSX-400 spectrometer (JEOL).
RESULTS
Incorporation of radioactivity from acetyl-CoA into ADABA and ectoine.
To determine the biosynthetic pathway of ectoine in H. elongata OUT30018, 14C-radioactive compounds resulting from [1-14C]acetyl-CoA in the reaction mixture containing the crude enzyme were analyzed by reverse-phase HPLC. 14C radioactivity from acetyl-CoA shifted significantly to the ADABA peak in the first 5 min of the reaction and was incorporated into the ectoine fraction in the next 5 min. The peak compounds produced in the reaction were isolated and identified as ADABA and ectoine by fast atom bombardment mass spectrometry and 1H NMR spectroscopy. This result indicates that the acetyl group of ADABA was supplied by acetyl-CoA and that ADABA was converted to ectoine. Thus, we attempted to purify the enzymes involved in ectoine synthesis in H. elongata OUT30018. The enzymes which catalyzed the three reactions were predicted to be DABA aminotransferase, DABA acetyltransferase, and ectoine synthase, in order of steps from ASA to ectoine.
Purification of enzymes and confirmation of their catalytic reactions.
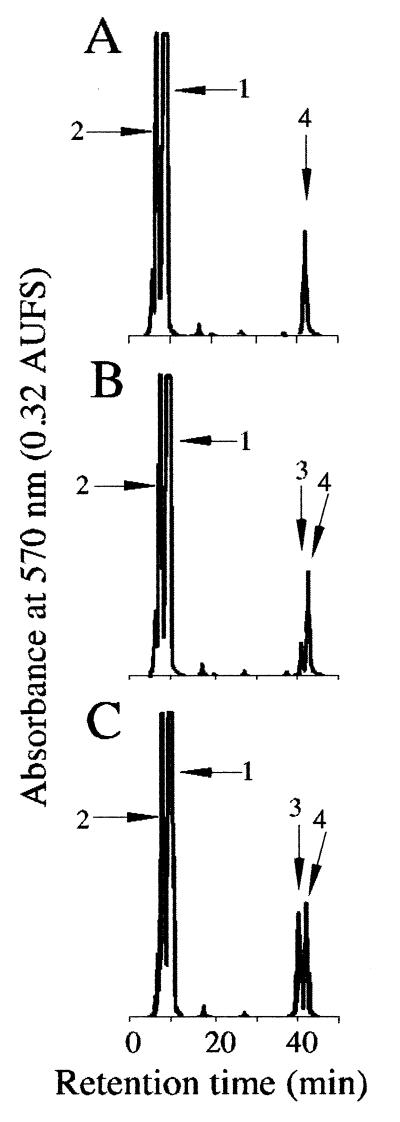
2,4-DABA aminotransferase was purified by monitoring the activities of the reverse reaction, because the predicted substrate, ASA, as an amino group acceptor could not be obtained commercially and because the detection of l-glutamate as a product of the reverse reaction was much easier. DABA aminotransferase was purified to give a major band at 44 kDa on SDS-PAGE (Fig. 2), whereas the apparent molecular mass of the native enzyme was estimated to be approximately 260 kDa by gel filtration. From these results, this enzyme could be a homohexamer in solution. The isoelectric point of this enzyme was estimated to be 6.2 by IEF electrophoresis. The typical purification steps are summarized in Table 1. The specific activity at the final step was 12 U mg of protein−1 in the reverse reaction. The enzyme activity decreased greatly during the last two purification steps in the absence of PLP and/or potassium, and the apparently inactivated enzyme was reactivated partially by the addition of 10 μM PLP and 10 mM KCl. This stabilizing effect of KCl was similar to those of other kinds of potassium salts such as sulfate or phosphate. These results suggested that DABA aminotransferase might be a PLP-dependent enzyme similar to other aminotransferases and requiring K+ for its activity and stability. To confirm the catalysis of the reaction from ASA to DABA by this enzyme, the reaction products from l-glutamate and ASA were analyzed with an amino acid analyzer (Fig. 3). A peak was detected at a retention time similar to that of standard DABA, and this peak area increased with the incubation time proportionally. Since only l-homoserine was accepted as an amino group donor to ASA with 5% of the activity with l-glutamate to ASA among l-glutamine, l-alanine, l-aspartate, l-lysine, and l-homoserine, DABA aminotransferase seemed to use l-glutamate specifically as the amino group donor to ASA in the presence of PLP (Table 2). On the other hand, in the reverse reaction, γ-aminobutyric acid (GABA) and l-ornithine were also used as amino group donors to 2-OG, although DABA showed the highest activity among the amino group compounds tested (Table 2).
FIG. 2.

SDS-PAGE of the purified DABA aminotransferase and ectoine synthase of H. elongata OUT30018. Lane 1, molecular mass standards (in kilodaltons): phosphorylase b, 97.4; serum albumin, 66.2; ovalbumin, 45.0; carbonic anhydrase, 31.0; trypsin inhibitor, 21.5; lysozyme, 14.4 (top to bottom, respectively); lane 2, purified DABA aminotransferase; lane 3, purified ectoine synthase.
TABLE 1.
Purification steps of DABA aminotransferase from H. elongata OUT30018
| Purification step | Total protein (mg) | Total activityb (U) | Sp act (U mg−1) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|
| Crude extracta | 1,060 | 48.1 | 0.045 | 1.0 | 100 |
| Ammonium sulfate | 170 | 45.8 | 0.27 | 6.0 | 95 |
| HIC I | 36.3 | 35.2 | 0.97 | 22 | 73 |
| HIC II | 3.50 | 21.5 | 6.1 | 136 | 45 |
| Gel filtration | 0.31 | 3.72 | 12 | 267 | 13 |
Cells from a 5-liter culture were used.
Activities were determined by the reverse reaction as described in Materials and Methods.
FIG. 3.
HPLC chromatogram of amino acids in the mixture reacted with ASA, l-glutamate, and purified DABA aminotransferase from H. elongata OUT30018. Ninhydrin-positive compounds were monitored at 570 nm with an amino acid analyzer. The numbered peaks are as follows: 1, l-glutamate; 2, dl-ASA; 3, DABA; 4, NH4+ ion. The minor peak before the dl-ASA peak is l-aspartate as a contaminant of dl-ASA. AUFS indicates an absorbance value of the full scale.
TABLE 2.
| Substrate | Relative activity (%) |
|---|---|
| Forward reaction | |
| l-Glutamic acid (control) | 100 |
| l-Glutamine | <1 |
| l-Alanine | <1 |
| l-Aspartic acid | <1 |
| l-Homoserine | 5 |
| l-Lysine | <1 |
| Reverse reaction | |
| DABA (control) | 100 |
| l-Aspartic acid | <0.3 |
| GABA | 60 |
| l-Ornithine | 6 |
| l-Lysine | <0.3 |
The reaction buffer was composed of 10 μM PLP, 20 mM KCl, and 50 mM Tris-HCl, pH 8.5. Various amino compounds at 10 mM were used as amino group donors. As amino group acceptors, 10 mM ASA and 5 mM 2-OG were used for the forward reaction and reverse reaction, respectively. Eight milliunits of purified DABA aminotransferase was used, and the reaction mixture was incubated at 25°C for 30 min. Activities were determined by the detection of ASA for the forward reaction and of l-glutamate for the reverse reaction as described in Materials and Methods. The detection limit was 0.80 nmol of DABA for the forward reaction and 0.68 nmol of l-glutamate for the reverse reaction.
DABA acetyltransferase was purified to a specific activity of 50 U mg of protein−1 (Table 3). Although the enzyme was purified by more than 400-fold, we failed to obtain the enzyme as a major band in SDS-PAGE because of its instability. The apparent molecular mass was 45 kDa by gel filtration (data not shown). To determine the substrate specificity, several amino acids, GABA, l-ornithine, l-lysine, and l-aspartate, were tested instead of DABA. However, no new peaks were observed upon reverse-phase HPLC at 210 nm.
TABLE 3.
Purification steps of DABA acetyltransferase from H. elongata OUT30018
| Purification step | Total protein (mg) | Total activity (U) | Sp act (U mg−1) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|
| Crude extracta | 1,100 | 130 | 0.12 | 1.0 | 100 |
| Ammonium sulfate | 850 | 120 | 0.14 | 1.2 | 96 |
| HIC | 60 | 75 | 1.3 | 11 | 58 |
| Blue Sepharose I | 0.90 | 10 | 11 | 94 | 8 |
| Blue Sepharose II | 0.05 | 2.5 | 50 | 420 | 2 |
Cells from a 5-liter culture were used.
Ectoine synthase was purified in the presence of 1 mM DABA and 2 M or higher NaCl to homogeneity with a specific activity of 16 U mg of protein−1 (Table 4). The apparent molecular mass was approximately 19 kDa by SDS-PAGE (Fig. 2), whereas the peak position of activity showed 35 kDa by gel filtration in the presence of 2 M NaCl. In the gel filtration with a buffer containing NaCl at a lower concentration, 0.5 M, the detection of activity failed because of inactivation. We could not clarify whether the 35-kDa size of ectoine synthase was the result of a homodimer as the native form or of nonspecific aggregation due to the high (2 M) concentration of NaCl. To determine the substrate specificity, several N-acetylated amino acids, N-α-acetyl-l-asparagine, N-α-acetyl-l-ornithine, N-α-acetyl-l-lysine, and N-ɛ-acetyl-l-lysine, were tested. No new peaks were observed upon reverse-phase HPLC at 210 nm, although circularized products from these test compounds were expected to be detected by this method. N-δ-Acetyl-l-ornithine remains a candidate for a substrate, if ectoine synthase recognizes a skeleton one carbon longer than that of ADABA. Since H. elongata OUT30018 could utilize ectoine as a sole carbon source in the presence of NaCl at a concentration of 3 to 15% (unpublished data) and Peters et al. (34) stated the reversibility of ectoine synthase reaction with crude extracts of E. halochloris, reversibility of the reaction was tested in a range of 10 mM to 1 M ectoine as the substrate in the presence of 50 mM or 0.5 M NaCl. However, formation of ADABA was not detected by HPLC. Considering the threshold for detection of ADABA, the ratio of the concentration of ADABA to that of ectoine was 3,000−1 or lower. To clarify whether ectoine synthase is involved in the reaction of assimilation of ectoine in this organism, further investigation is necessary. The isoelectric point of ectoine synthase was 4.2 to 4.4 by IEF electrophoresis. The amino acid composition of ectoine synthase was determined, and a high content of acidic amino acids, l-aspartate and l-glutamate, was noted (data not shown).
TABLE 4.
Purification steps of ectoine synthase from H. elongata OUT30018
| Purification step | Total protein (mg) | Total activity (U) | Sp act (U mg−1) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|
| Crude extracta | 912 | 45.6 | 0.050 | 1.0 | 100 |
| Ammonium sulfate | 684 | 40.6 | 0.059 | 1.2 | 89 |
| HIC | 6.4 | 20.5 | 3.2 | 64 | 45 |
| HAb | 0.81 | 11.4 | 14 | 280 | 25 |
| Gel filtration | 0.50 | 8.0 | 16 | 320 | 18 |
Cells from a 5-liter culture were used.
HA column chromatography.
Characteristics of the reactions involved in ectoine synthesis.
Characteristics of the three enzymes involved in ectoine synthesis are summarized in Table 5. All three enzymes exhibited similar properties with respect to the optimal reaction conditions of pH (8.2 to 9.0), temperature (15 to 25°C), and concentration of NaCl (0.4 to 0.5 M). The temperature was lower than the optimal growth temperature for this strain, 37°C. However, the apparent temperature optimum for the activity of ectoine synthase was shifted from 0 to 30°C with increasing NaCl concentrations. In regard to the other two enzymes, a similar shift of the optimum temperature occurred but only within a 5°C range. The optimal pH and temperature for the reverse reaction of DABA aminotransferase were also similar to those for the forward reaction.
TABLE 5.
Characterizations of ectoine synthetic enzymes from H. elongata OUT30018
| Property | Value for enzyme:
|
||
|---|---|---|---|
| DABA aminotransferasea | DABA acetyltransferase | Ectoine synthase | |
| Optimum conditions for activity | |||
| pH | 8.6–8.7 | 8.2 | 8.5–9.0 |
| Temp (°C) | 25 | 20 | 0–10 (NaCl, 0.05 M) |
| 15 (NaCl, 0.77 M) | |||
| 30 (NaCl, 3.0 M) | |||
| Concn of NaCl (M) | 0.5b | 0.4 | 0.5 (15°C) |
| Kinetic constants | |||
| Km (mM) | 9.1 (l-Glu) | NDc | 11 (NaCl, 0.05 M) |
| 4.5 (dl-ASA) | 8.4 (NaCl, 0.77 M) | ||
| Vmax (μmol min−1 mg−1) | 12 (dl-ASA) | ND | 85 (NaCl, 0.05 M) |
| 56 (NaCl, 0.77 M) | |||
Forward reaction.
Although activities of both forward and reverse directions decreased with increasing concentrations of NaCl, the decrease in the activity for the reverse reaction occurred more rapidly than that for the forward reaction up to 1 M NaCl. The optimal concentration of NaCl for the forward reaction was determined as the concentration at which the ratio of the activity for the forward reaction to that for the reverse reaction was maximum.
ND, not determined.
Chloride salts of various cations (Mg2+, Ca2+, Mn2+, Li+, K+, and NH4+) (1 mM) did not influence the activity of ectoine synthase. These cations except for Mn2+ did not influence the activity of DABA aminotransferase, and in the case of Mn2+, it was found that ASA used as a substrate vanished in the mixture during the reaction. N-Ethylmaleimide (10 mM) inhibited the activity of ectoine synthase by 15%. This result suggests that free thiol groups may be involved in the catalytic activity.
Effect of DABA on the stability of DABA acetyltransferase and ectoine synthase.
DABA stabilized DABA acetyltransferase and ectoine synthase, but the concentration necessary to achieve stabilization was different for the two enzymes. Ectoine synthase became stable to some extent in the presence of 1 mM DABA at 0°C and extremely stable in the presence of both DABA (1 mM) and NaCl (2 M) at −80°C, at which it was stored without decrease in the activity for at least 6 months. At an elevated temperature, however, higher concentrations of DABA were necessary for stabilization; the residual activities after a 3-h incubation at 37°C were 85% in the presence of 100 mM DABA and 2 M NaCl and 3% in the presence of 1 mM DABA and 2 M NaCl. In contrast, DABA acetyltransferase required 100 mM DABA for sufficient stabilization at 0°C. These stabilizing effects of DABA were not replaced by other basic amino acids such as GABA, l-lysine, or l-ornithine. Also, we could not find a significant stabilizing effect for the ectoine synthesis enzymes by glycerol, mannitol, ectoine, and bovine serum albumin.
Effect of salt on the activity and stability of the three enzymes.
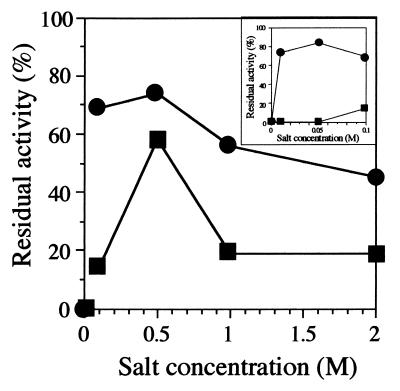
Many halophilic enzymes are stable in the presence of salt at high concentrations (3 to 4 M) at room temperature rather than at 4°C (24). Since ectoine synthesis was enhanced in vivo under external hyperosmotic conditions (31), we suspected that the ectoine synthetic enzymes might be activated or stabilized by an increase in the concentration of salts or other solutes. Thus, the effects of concentrations of salt on the activity and stability of these enzymes, especially those of a combination of salt concentrations and temperatures, were investigated in detail. The stability of DABA aminotransferase was very low in the presence of 0 to 50 mM NaCl at 37°C; however, it increased linearly with increasing concentrations of NaCl in the range of 50 mM to 0.5 M and decreased thereafter above 0.5 M (Fig. 4). The activities of this enzyme for both forward and reverse reactions decreased with increasing concentrations of NaCl, above 50 mM; however, the degree of decrease was different for the two reactions. The activity ratio for the forward to the reverse reactions was the highest at 0.5 M NaCl (approximately 3). This observation suggested that the production of DABA by DABA aminotransferase might be correlated with the concentration of NaCl in the range of 50 mM to 0.5 M, and the optimal concentration of NaCl for the forward reaction of DABA aminotransferase was 0.5 M in vitro. In contrast, KCl in the range of 10 mM to 0.5 M exhibited greater protective effects on this enzyme than did NaCl, and the protective effect of KCl seemed to be independent of its concentration in the range of physiological concentrations (Fig. 4).
FIG. 4.
Salt dependence of the stability of DABA aminotransferase from H. elongata OUT30018. An expanded figure for the concentration of salt from 0 to 0.1 M is insert. The purified enzyme was incubated in 50 mM Tris-HCl buffer, pH 8.5, containing various concentrations of NaCl (closed squares) or KCl (closed circles), at 37°C for 30 min. After being chilled on ice, the enzyme solutions were replaced with 50 mM Tris-HCl buffer (pH 8.5), containing 10 μM PLP and 25 mM KCl by ultrafiltration. The DABA aminotransferase activity was determined by the reverse reaction described in Materials and Methods. One hundred percent residual activity was defined as the activity of the purified DABA aminotransferase immediately before replacement with a buffer containing salt at an indicated concentration.
The activity of DABA acetyltransferase increased linearly with increasing concentrations of NaCl up to 0.4 M at 20°C and decreased thereafter above 0.4 M NaCl. DABA acetyltransferase was stabilized in the presence of NaCl, and residual activity after a 3-h incubation was 100% in the presence of 2 M NaCl at 0°C. However, the residual activities decreased gradually with increasing temperature and were 62% at 30°C and 35% at 37°C.
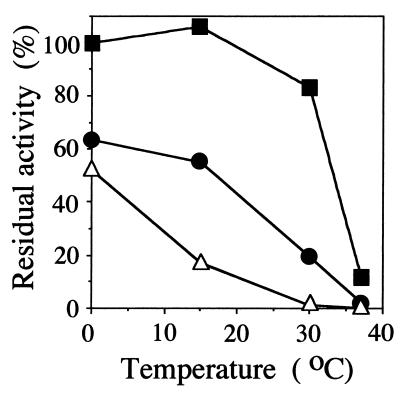
Ectoine synthase also exhibited salt dependence, and the stabilizing effect of NaCl was greater than that of KCl at the same concentration. Although the residual activity after a 3-h incubation in the presence of 3 M NaCl was greater than 80% in the temperature range of 0 to 30°C, it decreased to 13% at 37°C, the temperature optimum for the growth of the strain (Fig. 5). In addition, it was found that the effect of NaCl on the activity of ectoine synthase was affected by temperature and that maximal activities were obtained at different temperatures with different concentrations of NaCl (Table 5 and Fig. 6). The linearity of the reaction was obtained for 10 min at a temperature up to 15°C and for only 1 min at a higher temperature. The temperature at which ectoine synthase exhibited maximum activity increased with the increase in concentration of NaCl.
FIG. 5.
Effect of salts on the thermal stability of ectoine synthase from H. elongata OUT30018. Purified ectoine synthase was incubated in 50 mM Tris-HCl buffer (pH 9.0) containing 1 mM DABA (no salt; open triangles) plus 3 M KCl (closed circles) or 3 M NaCl (closed squares) at a designated temperature for 3 h. After replacement of the treatment buffer with the basal buffer A by ultrafiltration, the residual activities were determined as described in Materials and Methods. One hundred percent residual activity was defined as the activity of the purified ectoine synthase which was kept in buffer A before replacement with a buffer containing KCl or NaCl at 3 M.
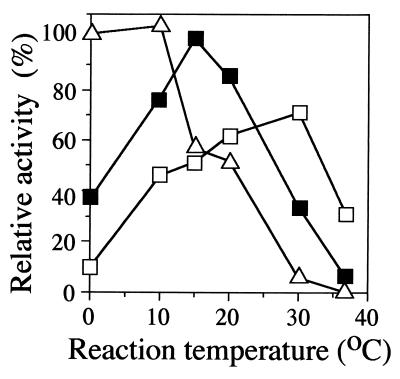
FIG. 6.
Effects of NaCl concentration on the activity of ectoine synthase from H. elongata OUT30018. The purified enzyme in 50 mM Tris-HCl buffer (pH 9.0) containing NaCl at 0.05 M (open triangles), 0.77 M (closed squares), or 3 M (open squares) was reacted at the indicated temperatures and sampled every 30 s for 10 min, and the level of ectoine produced was determined. The activity was calculated from the initial velocity. The amounts of ectoine produced were determined as described in Materials and Methods. One hundred percent activity was defined as that obtained with 0.77 M NaCl, at 15°C.
DISCUSSION
DABA exists in nature as a moiety of the cell wall peptidoglycans of gram-positive bacteria (32, 42, 43) or of peptide antibiotics such as colistin (20) or polymyxin (35). Despite such wide distribution, knowledge of the metabolic pathways and enzymes involved in the production of DABA is limited (18, 30, 51, 52). Two metabolic pathways for DABA synthesis have been proposed to date: the first is from l-aspartate to DABA via l-homoserine in Lathyrus sylvestris, reported by Nigam and Ressler (30), and another is the direct conversion of ASA to DABA in Xanthomonas sp. (36) and Acinetobacter baumannii (18), similar to that for H. elongata DABA aminotransferase as shown in this study. Although DABA aminotransferase from Xanthomonas sp. was probably different from the H. elongata enzyme since the Xanthomonas sp. enzyme preferred l-alanine as an amino group donor, the A. baumannii enzyme involved in the 1,3-diaminopropane production pathway (18) was similar to the H. elongata enzyme with respect to several properties such as substrate specificity, pH optimum for the activity, and the Km values for DABA and 2-OG. Both DABA aminotransferases, from H. elongata and A. baumannii, acted on ASA as an amino group acceptor and l-glutamate as an amino group donor to produce DABA. From the pattern of transamination, between the distal position of the ω-amino acid and the α position of the α-amino acid, we concluded that DABA aminotransferase belongs to subgroup II of aminotransferases, which consists of acetylornithine aminotransferase (EC 2.6.1.11), ornithine aminotransferase (EC 2.6.1.13), ω-amino acid aminotransferase (EC 2.6.1.18), 4-aminobutyrate (GABA) aminotransferase (EC 2.6.1.19), and diaminopelargonate aminotransferase (EC 2.6.1.62) (28). H. elongata DABA aminotransferase showed a large molecular mass corresponding to a homohexamer by gel filtration, whereas A. baumannii DABA aminotransferase, which was overproduced and purified from Escherichia coli, was in the form of a homotetramer with a molecular mass of 188 kDa (18). Although the sizes of each subunit are in a narrow range of 40 to 50 kDa in most aminotransferases, the native molecules exist largely as homodimers (17, 41, 47, 53) or homotetramers (18, 45, 49). As a rare example, ornithine aminotransferase from rats was reported to be a homohexamer composed of a dimer of trimers, with a molecular mass of 256 kDa in the solution containing NaCl (27). At present, we cannot rule out the possibility that DABA aminotransferase from H. elongata also exists in a form like that of the rat ornithine aminotransferase.
Among the three enzymes involved in the ectoine synthetic pathway, only DABA aminotransferase preferred K+ to Na+. Although K+-dependent properties have been reported for many halophilic enzymes from halophilic archaebacteria, in such extremely halophilic enzymes, the degrees of specificity for K+ vary, ranging from little or no preference to fairly high selectivities, and also there may be no specific cation-binding sites (24). However, existence of a specific binding site for K+ was exemplified in dialkylglycine decarboxylase from Pseudomonas cepacia (46), which requires K+ for its activity and stability (1). The K+ specificity of DABA aminotransferase might be due to the presence of a specific binding site(s) for alkali metal ion, similar to dialkylglycine decarboxylase.
From the results of substrate specificity for DABA acetyltransferase and ectoine synthase, we concluded that these two enzymes were novel as Peters et al. reported (34). The results of analysis of substrate specificity of ectoine synthase suggested that the enzyme was highly specific for ADABA and that the N-acetyl group at the α position seemed not to contribute to circularization.
In the overall characterization of the three enzymes, some common features were observed, such as the requirement for a pH of 8.2 to 9.0, a temperature of 15 to 25°C, and the presence of 0.4 to 0.5 M NaCl, for ectoine synthesis (Table 5). Although the temperature optimum for growth of H. elongata OUT30018 was 37°C, the cells grown at 30°C accumulated higher amounts of ectoine than did the cells grown at 37°C (unpublished data). Wohlfarth et al. (50) reported that the cells of H. elongata DSM2581 grown at 20°C accumulated more ectoine than did the cells grown at 40°C. These phenomena, in regard to the intracellular level of ectoine, could be explained by the temperature dependence of the ectoine synthetic enzymes as shown in vitro in this study. The cytoplasmic membranes of cells grown at high temperatures have a higher phase-transition temperature, in general, which confers the advantage of homeostatic permeability of materials under hyperosmotic conditions on the cells. Such differences in membrane functions might reflect the differences between the temperature optimum for growth and that for accumulation of ectoine. The salt concentration optimum for activity of the three enzymes, lower than that expected from the salt tolerance of this strain, might be due to the intracellular concentration of free salt ions. Low intracellular levels of free Na+ in a range of 0.04 to 0.2 M were determined by 23Na NMR spectroscopy in the cells of halophilic eubacteria, Vibrio costicola (15) and Brevibacterium sp. (29), grown in the presence of high concentrations of NaCl.
Since we could not purify DABA acetyltransferase to homogeneity, the kinetic constants of DABA acetyltransferase could not be determined. Therefore, we could not complete the entire scheme of regulation of the ectoine biosynthetic pathway; however, it was suggested that the rate-limiting step in the ectoine biosynthetic pathway was probably DABA production by DABA aminotransferase, considering the values of Vmax for DABA aminotransferase and ectoine synthase, and of specific activity of DABA acetyltransferase obtained at the last purification step. Considering the toxicity of free cytoplasmic DABA (37), it would be deleterious to cells if the rate-limiting step was that catalyzed by DABA acetyltransferase or ectoine synthase. DABA must be converted rapidly to nontoxic compounds. Our previous results (31) that DABA was not detected in the cell extracts support this possibility. To summarize the characteristics of the ectoine synthesis enzymes, it was suspected that DABA might contribute to stabilizing the system of ectoine biosynthesis under hyperosmotic conditions. However, we recognize the disparity between the effective concentration of DABA in vitro and the in vivo concentration in H. elongata. There needs to be further investigation to clarify the surroundings of these enzymes in vivo.
ACKNOWLEDGMENTS
We thank Yoshiko Yagi, Institute for Protein Research, Osaka University, for assistance with amino acid analysis.
This study was supported partially by a grant-in-aid for scientific research from the Ministry of Education, Science, Sports and Culture of Japan, no. 05650800.
REFERENCES
- 1.Aaslestad H G, Bouis P J, Jr, Philips A T, Larson A D. Characterization of a decarboxylation-dependent transaminase. In: Snell E E, Braunstein A E, Severin E S, Torchinsky Y M, editors. Pyridoxal catalysis: enzymes and model systems. New York, N.Y: Wiley-Interscience; 1968. pp. 479–490. [Google Scholar]
- 2.Benoiton L, Leclerc J. An improved synthesis of ɛ-N-acetyl-l-lysine and similar compounds. Can J Chem. 1965;43:991–993. [Google Scholar]
- 3.Bernard T, Jebbar M, Rassouli Y, Himdi-Kabbab S, Hamelin J, Blanco C. Ectoine accumulation and osmotic regulation in Brevibacterium linens. J Gen Microbiol. 1993;139:129–136. [Google Scholar]
- 4.Black S. Conversion of aspartic acid to homoserine. Methods Enzymol. 1962;5:820–827. [Google Scholar]
- 5.Black S. β-Aspartyl phosphate and aspartic-β-semialdehyde. Methods Enzymol. 1963;6:622–624. [Google Scholar]
- 6.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7.Brown A D. Microbial water stress. Bacteriol Rev. 1976;40:803–846. doi: 10.1128/br.40.4.803-846.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christian J H B, Waltho J A. Solute concentrations within cells of halophilic and non-halophilic bacteria. Biochim Biophys Acta. 1962;65:506–508. doi: 10.1016/0006-3002(62)90453-5. [DOI] [PubMed] [Google Scholar]
- 9.Csonka L N. Physiological and genetic responses of bacteria to osmotic stress. Microbiol Rev. 1989;33:121–147. doi: 10.1128/mr.53.1.121-147.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dawson R M C, Elliot D C, Elliot W H, Jones K M. Data for biological research. 3rd ed. Oxford, England: Clarendon Press; 1986. [Google Scholar]
- 11.Del Moral A, Severin J, Ramos-Cormenzana A, Truper H G, Galinski E A. Compatible solutes in new moderately halophilic isolates. FEMS Microbiol Lett. 1994;122:165–172. [Google Scholar]
- 12.Farwick M, Siewe R M, Kramer R. Glycine betaine uptake after hyperosmotic shift in Corynebacterium glutamicum. J Bacteriol. 1995;177:4690–4695. doi: 10.1128/jb.177.16.4690-4695.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galinski E A. Osmoadaptation in bacteria. Adv Microb Physiol. 1995;37:273–328. [PubMed] [Google Scholar]
- 14.Galinski E A, Pfeiffer H-P, Trupper H G. 1,4,5,6-Tetra-hydro-2-methyl-4-pyrimidinecarboxylic acid: a novel cyclic amino acid from halophilic phototrophic bacteria of the genus Ectothiorhodospira. Eur J Biochem. 1985;149:135–139. doi: 10.1111/j.1432-1033.1985.tb08903.x. [DOI] [PubMed] [Google Scholar]
- 15.Gilboa H, Kogut M, Chalamish S, Regev R, Avi-Dor Y, Russell N J. Use of 23Na nuclear magnetic resonance spectroscopy to determine the true intracellular concentration of free sodium in a halophilic eubacterium. J Bacteriol. 1991;173:7021–7023. doi: 10.1128/jb.173.21.7021-7023.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gouesbet G, Trautwetter A, Bonnassie S, Wu L F, Blanco C. Characterization of the Erwinia chrysanthemi osmoprotectant transporter gene ousA. J Bacteriol. 1996;178:447–455. doi: 10.1128/jb.178.2.447-455.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammer T, Bode R. Purification and characterization of an inducible l-lysine: 2-oxoglutarate 6-aminotransferase from Candida utilis. J Basic Microbiol. 1992;32:21–27. doi: 10.1002/jobm.3620320106. [DOI] [PubMed] [Google Scholar]
- 18.Ikai H, Yamamoto S. Identification and analysis of a gene encoding l-2,4-diaminobutyrate:2-ketoglutarate 4-aminotransferase involved in the 1,3-diaminopropane production pathway in Acinetobacter baumannii. J Bacteriol. 1997;179:5118–5125. doi: 10.1128/jb.179.16.5118-5125.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imhoff J F. Osmoregulation and compatible solutes in eubacteria. FEMS Microbiol Rev. 1986;39:57–66. [Google Scholar]
- 20.Ito M, Aida K, Uemura T. Studies on the bacterial formation of a peptide antibiotic, colistin. III. On the biosynthetic pathway of α,γ-diaminobutyric acid and relationship between colistin formation and amino acids metabolism in Bacillus colistinus KOYAMA. Agric Biol Chem. 1969;33:949–958. [Google Scholar]
- 21.Jebbar M, Talibart R, Gloux K, Bernard T, Blanco C. Osmoprotection of Escherichia coli by ectoine: uptake and accumulation characteristics. J Bacteriol. 1992;174:5027–5035. doi: 10.1128/jb.174.15.5027-5035.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khunajakr N, Shinmyo A, Takano M. Compatible solutes in a halotolerant bacterium. Annu Rep ICBiotech. 1989;12:157–169. [Google Scholar]
- 23.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Lanyi J K. Salt-dependent properties of proteins from extremely halophilic bacteria. Bacteriol Rev. 1974;38:272–290. doi: 10.1128/br.38.3.272-290.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leah J M, Palmer T, Billett E E, Williams C R. A comparison of ornithine aminotransferase from human and rat sources. Biochem Int. 1987;15:629–634. [PubMed] [Google Scholar]
- 26.Malin G, Lapidot A. Induction of synthesis of tetrahydropyrimidine derivatives in Streptomyces strains and their effect on Escherichia coli in response to osmotic and heat stress. J Bacteriol. 1996;178:385–395. doi: 10.1128/jb.178.2.385-395.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Markovic-Housley Z, Kania M, Lustig A, Vincent M G, Jansonius J N. Quaternary structure of ornithine aminotransferase in solution and preliminary crystallographic data. Eur J Biochem. 1987;162:345–350. doi: 10.1111/j.1432-1033.1987.tb10607.x. [DOI] [PubMed] [Google Scholar]
- 28.Mehta P K, Hale T I, Christen P. Aminotransferases: demonstration of homology and division into evolutionary subgroups. Eur J Biochem. 1993;214:549–561. doi: 10.1111/j.1432-1033.1993.tb17953.x. [DOI] [PubMed] [Google Scholar]
- 29.Nagata S, Adachi K, Shirai K, Sano H. 23Na NMR spectroscopy of free Na+ in the halotolerant bacterium Brevibacterium sp. and Escherichia coli. Microbiology. 1995;140:729–736. doi: 10.1099/13500872-141-3-729. [DOI] [PubMed] [Google Scholar]
- 30.Nigam S N, Ressler C. Biosynthesis of 2,4-diaminobutyric acid from l-[3H]homoserine and dl-[1-14C]aspartic acid in Lathyrus sylvestris W. Biochemistry. 1966;5:3426–3431. doi: 10.1021/bi00875a006. [DOI] [PubMed] [Google Scholar]
- 31.Ono H, Okuda M, Tongpim S, Imai K, Shinmyo A, Sakuda S, Kaneko Y, Murooka Y, Takano M. Accumulation of compatible solutes, ectoine and hydroxyectoine, in a moderate halophile, Halomonas elongata KS3, isolated from dry salty land in Thailand. J Ferment Bioeng. 1998;85:362–368. [Google Scholar]
- 32.Perkins H R, Cummins C S. Ornithine and 2,4-diaminobutyric acid as components of the cell walls of plant pathogenic Corynebacteria. Nature. 1964;202:1105–1107. doi: 10.1038/2011105a0. [DOI] [PubMed] [Google Scholar]
- 33.Perroud B, Le Rudulier D. Glycine betaine transport in Escherichia coli: osmotic modulation. J Bacteriol. 1985;161:393–401. doi: 10.1128/jb.161.1.393-401.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peters P, Galinski E A, Truper H G. The biosynthesis of ectoine. FEMS Microbiol Lett. 1990;71:157–162. [Google Scholar]
- 35.Pichard B, Larue J P, Thouvenot D. Gavaserin and saltavalin, new peptide antibiotics produced by Bacillus polymyxa. FEMS Microbiol Lett. 1995;133:215–218. doi: 10.1111/j.1574-6968.1995.tb07887.x. [DOI] [PubMed] [Google Scholar]
- 36.Rao D R, Hariharan K, Vijayalakshmi K R. A study of the metabolism of l-alpha gamma-diaminobutyric acid in a Xanthomonas species. Biochem J. 1969;114:107–115. doi: 10.1042/bj1140107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rasmussen M A, Allison M J, Foster J G. Flatpea intoxication in sheep and indications of ruminal adaptation. Vet Hum Toxicol. 1993;35:123–127. [PubMed] [Google Scholar]
- 38.Ronit R, Peri I, Gilboa H, Avi-Dor Y. 13C NMR study of the interrelation between synthesis and uptake of compatible solutes in two moderately halophilic eubacteria: bacterium Ba1 and Vibrio costicola. Arch Biochem Biophys. 1990;278:106–112. doi: 10.1016/0003-9861(90)90237-s. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Small W C, Tanes M E. A quantitative colorimetric assay for semialdehyde. Anal Biochem. 1990;185:156–159. doi: 10.1016/0003-2697(90)90272-b. [DOI] [PubMed] [Google Scholar]
- 41.Stoner G L, Eisenberg M A. Purification and properties of 7,8-diaminopelargonic acid aminotransferase. J Biol Chem. 1975;250:4020–4035. [PubMed] [Google Scholar]
- 42.Suzuki K, Sasaki J, Uramoto M, Nakase T, Komagata K. Agromyces mediolanus sp. nov., nom. rev., comb. nov., a species for “Corynebacterium mediolanum” Mamoli 1939 and for some aniline-assimilating bacteria which contain 2,4-diaminobutyric acid in the cell wall peptidoglycan. Int J Syst Bacteriol. 1996;46:88–93. doi: 10.1099/00207713-46-1-88. [DOI] [PubMed] [Google Scholar]
- 43.Takeuchi M, Weiss N, Schumann P, Yokota A. Leucobacter komagatae gen. nov., sp. nov., a new aerobic gram-positive, nonsporulating rod with 2,4-diaminobutyric acid in the cell wall. Int J Syst Bacteriol. 1996;46:967–971. doi: 10.1099/00207713-46-4-967. [DOI] [PubMed] [Google Scholar]
- 44.Talibart R, Jebbar M, Gouesbet G, Himdi-Kabbab S, Wroblewski H, Blanco C, Bernard T. Osmoadaptation in rhizobia: ectoine-induced salt tolerance. J Bacteriol. 1994;176:5210–5217. doi: 10.1128/jb.176.17.5210-5217.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toney M D, Hohenester E, Cowan S W, Jansonius J N. Dialkylglycine decarboxylase structure: bifunctional active site and alkali metal sites. Science. 1993;261:756–759. doi: 10.1126/science.8342040. [DOI] [PubMed] [Google Scholar]
- 46.Toney M D, Hohenester E, Keller J W, Jansonius N. Structural and mechanistic analysis of two refined crystal structures of the pyridoxal phosphate-dependent enzyme dialkylglycine decarboxylase. J Mol Biol. 1995;245:151–179. doi: 10.1006/jmbi.1994.0014. [DOI] [PubMed] [Google Scholar]
- 47.Umemura I, Yanagiya K, Komatsubara S, Sato T, Tosa T. Purification and some properties of alanine aminotransferase from Candida maltosa. Biosci Biotechnol Biochem. 1994;58:283–287. doi: 10.1271/bbb.58.283. [DOI] [PubMed] [Google Scholar]
- 48.Vreeland R H, Mierau B D, Litchfield C D, Martin E L. Relationship of the internal solute composition to the salt tolerance of Halomonas elongata. Can J Microbiol. 1983;29:407–414. [Google Scholar]
- 49.Watanabe N, Sakabe K, Sakabe N, Higashi T, Sasaki K, Aibara S, Morita Y, Yonaha K, Toyama S, Fukutai H. Crystal structure analysis of ω-amino acid: pyruvate aminotransferase with a newly developed Weissenberg camera and an imaging plate using synchrotron radiation. J Biochem. 1989;105:1–3. doi: 10.1093/oxfordjournals.jbchem.a122600. [DOI] [PubMed] [Google Scholar]
- 50.Wohlfarth A, Severin J, Galinski E A. The spectrum of compatible solutes in heterotrophic halophilic eubacteria of the family Halomonadaceae. J Gen Microbiol. 1990;136:705–712. [Google Scholar]
- 51.Yamamoto S, Suemoto Y, Seito Y, Nakao H, Shinoda S. The presence of l-2,4-diaminobutyric acid decarboxylase activity in Vibrio species: a new biosynthetic pathway for 1,3-diaminopropane. FEMS Microbiol Lett. 1986;35:289–293. [Google Scholar]
- 52.Yamamoto S, Tsuzaki Y, Tougou K, Shinoda S. Purification and characterization of l-2,4-diaminobutyric decarboxylic acid from Acinetobacter calcoaceticus. J Gen Microbiol. 1992;138:1461–1465. doi: 10.1099/00221287-138-7-1461. [DOI] [PubMed] [Google Scholar]
- 53.Yonaha K, Suzuki K, Toyama S. 4-Aminobutyric acid: 2-oxoglutarate aminotransferase of Streptomyces griseus. Eur J Biochem. 1985;146:101–106. doi: 10.1111/j.1432-1033.1985.tb08625.x. [DOI] [PubMed] [Google Scholar]