Abstract
Strains of Pseudomonas aeruginosa isolated from the respiratory tracts of patients with cystic fibrosis often display a mucoid morphology due to high levels of expression of the exopolysaccharide alginate. The response regulator AlgB is required for full transcription of the alginate biosynthetic operon. Repeated attempts to demonstrate a direct interaction between AlgB and the promoter region of algD, the first gene in the alginate operon, have thus far been unsuccessful. The possibility that AlgB exerts its effect on algD indirectly exists. To identify putative genes under the control of AlgB which affect algD transcription, transposon mutagenesis of nonmucoid algB derivatives of the mucoid strain FRD1 was employed. Of approximately 3,000 transposon mutants screened, 6 were found to display phenotypes which were mucoid relative to the phenotype of the parental algB strain. The phenotypes of these mutants ranged from being only slightly mucoid to being indistinguishable from that of the original FRD1 strain. One of the particularly mucoid transposon mutants was chosen for further study. This strain was found to be disrupted in a previously uncharacterized open reading frame with 56% amino acid identity to PepA of Escherichia coli. PepA is classified as a leucine aminopeptidase, and homologs have been detected in a number of bacterial, plant, and animal species. This novel gene has been designated phpA (P. aeruginosa homolog of pepA). The insertional inactivation of phpA was found to correlate with the mucoid phenotype and an increase in algD transcription in the algB strain. Expression of phpA from an ectopic chromosomal locus compensated for the transposon insertion in the native phpA gene, restoring algD transcription to levels similar to those observed in the parental algB strain. While phpA expression did not appear to be under the control of AlgB at the transcriptional level, this study demonstrates that loss of phpA in an algB genetic background had a positive effect on alginate expression and, more specifically, on transcription of the alginate biosynthetic operon.
Pseudomonas aeruginosa has been and continues to be a formidable opportunistic pathogen. A ubiquitous gram-negative rod in soil and water environments, P. aeruginosa poses little threat to most healthy individuals. In certain clinical scenarios, however, P. aeruginosa becomes a pathogen of dire consequence. Among the patients for whom P. aeruginosa has life-threatening implications are individuals afflicted with the genetic disease cystic fibrosis (CF) (24).
A unique characteristic of many P. aeruginosa isolates from CF patients is a distinctive mucoid colony morphology. This mucoid morphology results from the production of the exopolysaccharide alginate, a linear polymer of l-guluronic and d-mannuronic acids (24). Alginate is an important virulence factor associated with chronic respiratory infections in CF patients (24). These patients are initially colonized with nonmucoid strains of P. aeruginosa, but over time mucoid strains emerge and predominate. This conversion occurs in vivo and seems to result from outgrowth of variants often harboring mutations in one of the muc genes (34). The mucoid clinical CF isolate FRD1 (mucA22) represents such a variant (34) and is the source of the strains used in this study. mucA encodes a product antagonistic to the activity of the alternative ς factor ς22, also referred to as AlgT or AlgU (40, 52). The dysregulation of ς22 activity leads to increased expression of the regulatory proteins AlgB, AlgR, and AlgZ (2, 51). Each of these factors has been shown to contribute positively to transcription of algD, the first gene in the alginate biosynthetic operon (2, 22, 24, 51).
AlgB is a member of the NtrC family of response regulators (50). As such, AlgB is expected to interact directly with DNA in order to regulate gene expression. However, repeated attempts to demonstrate binding of AlgB in the region extending from −570 to +1000 relative to the algD promoter have been unsuccessful. Despite this fact, transcriptional fusion studies indicate that levels of algD transcription are at least 20-fold lower in an algB::Tn501 insertion mutant than in the parental strain FRD1 (50, 51).
To reconcile this apparent discrepancy, we hypothesized that AlgB exerts its effect on algD transcription indirectly by altering the expression of an intermediate gene (Fig. 1). We reasoned that AlgB either activates or represses transcription of its direct target and that this regulation has a consequent positive effect on algD transcription. Since neither algR transcription nor AlgZ DNA-binding activity is affected in an algB mutant (2, 51), these factors are not likely to be the proposed intermediate of AlgB. To investigate our hypothesis and identify potential AlgB targets relevant to alginate expression, we performed transposon mutagenesis on nonmucoid algB mutants derived from the mucoid CF isolate FRD1. This approach has led to the discovery of a previously uncharacterized P. aeruginosa gene that affects transcription of algD and the consequent expression of alginate.
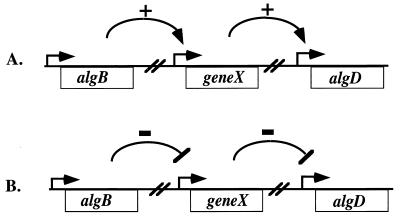
FIG. 1.
Two hypotheses for the mechanism of AlgB transcriptional activation of algD. (A) AlgB activates transcription of its direct target, gene X. The product of gene X then contributes to transcriptional activation of algD. (B) AlgB represses transcription of gene X. If expressed, the product of gene X inhibits transcription of algD.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. All P. aeruginosa strains except PA01 are ultimately derived from the mucoid clinical CF isolate FRD1. All enzymes used for construction of recombinant DNA were purchased from Promega unless otherwise noted. All plasmids were constructed by standard cloning techniques (32).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristic(s)a | Reference or source |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| HB101 | proAB leuB6 thi-1 lacY1 hsdR hsdM recA13 supE44 rpsL20 | 7 |
| JM109 | endA1 recA1 gyrA96 thi hsdR17 (rK− mK+) relA1 supE44 Δ(lac-proAB F′ traD36 proAB lacIqZΔM15 | 53 |
| Pseudomonas aeruginosa | ||
| PA01 | Prototrophic wild-type strain | 28 |
| FRD1 | CF isolate, Alg+ (due to mucA22 mutation) | 37 |
| FRD440 | mucA22 algT::Tn501 | 19 |
| FRD444 | mucA22 algB::Tn501 | 23 |
| FRD831 | mucA22 ΔalgR::ΩaacC1 | 31 |
| FRD840 | mucA22 ΔalgB::ΩaacC1 | 31 |
| FRD444.1 | mucA22 algB::Tn501 phpA::Tn5-B50 (Tcr) | This study |
| FRD444.2 | Mucoid Tn5-B50 mutant of FRD444 | This study |
| FRD444.3 | Mucoid Tn5-B50 mutant of FRD444 | This study |
| FRD875 | mucA22 algD::xylE aacC1 | This study |
| FRD879 | mucA22 algD::xylE aacC1 algB::Tn501 | This study |
| FRD906 | mucA22 algB::Tn501 phpA::Tn5-B50 (Tcs) | This study |
| FRD909 | mucA22 algB::Tn501 phpA::xylE aacC1, tandem transcription of phpA and xylE aacC1 | This study |
| FRD910 | mucA22 algB::Tn501 phpA::xylE ΩaacC1Ω, tandem transcription of phpA and xylE aacC1 | This study |
| FRD911 | mucA22 algB::Tn501 phpA::xylE aacC1, opposing transcription of phpA and xylE aacC1 | This study |
| FRD912 | mucA22 algB::Tn501 phpA::xylE ΩaacC1Ω, opposing transcription of phpA and xylE aacC1 | This study |
| FRD913 | mucA22 algB+ phpA::xylE aacC1, tandem transcription of phpA and xylE aacC1 | This study |
| FRD918 | Mucoid Tn5-B50 mutant of FRD840 | This study |
| FRD920 | mucA22 algD::xylE aacC1 algB::Tn501 phpA::Tn5-B50 | This study |
| FRD925 | Mucoid Tn5-B50 mutant of FRD840 | This study |
| FRD927 | mucA22 algD::xylE aacC1 algB::phpA phpA::Tn5-B50 | This study |
| FRD940 | mucA22 algB::Tn501 mucD::Tn5-B30 (Tcr) | This study |
| Plasmids | ||
| pDJW530 | pDJW482 + 2.2-kb SmaI fragment of pX1918G algD::xylE aacC1 (Materials and Methods) | This study |
| pEX100T | ColE1 AproriT sacB | 41 |
| pLAFR3 | IncP1 Tcr λcos oriT | 44 |
| pMMB67HE | IncQ AproriT | 20 |
| pRK2013 | ColE1-Tra(RK2)+ Kmr | 17 |
| pSUP::Tn5-B50 | P15A Cmr Tcr RP4 mob | 42 |
| pSUP::Tn5-B30 | P15A Cmr Tcr RP4 mob | 42 |
| pSW40 | pEX100T + 7.2-kb BamHI-SphI fragment of pSW50, phpA::Tn5-B50 (Tcr) | This study |
| pSW40.1 | pSW40 cut with HindIII and religated to delete a 3.5-kb fragment from the middle of Tn5-B50 (Tcs) | This study |
| pSW50 | 7.3-kb BamHI-XhoI fragment in pUC18 phpA::Tn5-B50 (Tcr) | This study |
| pSW90 | pEX100T + 1.95-kb BamHI-SphI fragment of pSW109, wild-type phpA | This study |
| pSW109 | pKS− + 6.8-kb BamHI-HindIII fragment of wild-type phpA | This study |
| pSW110 | pLAFR3 + 6.8-kb BamHI-HindIII fragment of pSW109, wild-type phpA | This study |
| pSW134 | pUS68 + 2.3-kb AseI fragment from pSW133, algB::phpA (Materials and Methods) | This study |
| pX1918G | ColE1 Apr Gmr | 41 |
| pX1918GT | ColE1 Apr ΩGmr | 41 |
Apr, Kmr, Cmr, and Gmr, ampicillin, kanamycin, chloramphenicol, and gentamicin resistance, respectively.
Media, antibiotics, and growth conditions.
Luria-Bertani (LB) broth (10.0 g of tryptone, 5.0 g of yeast extract, 5.0 g of NaCl per liter [pH 7.5]) and LB agar (Difco) were used for growth of all Escherichia coli strains. For growth of P. aeruginosa, modified LB broth and agar with no exogenous NaCl (LBNS and LANS, respectively) were used. Antibiotics were used at the following concentrations: tetracycline, 15 μg/ml (E. coli) or 100 μg/ml (P. aeruginosa); kanamycin, 30 μg/ml; ampicillin, 100 μg/ml; carbenicillin, 300 μg/ml; gentamicin, 15 μg/ml (E. coli) or 100 μg/ml (P. aeruginosa); and mercuric chloride, 18 μg/ml. Incubation was carried out at 37°C except for sucrose counterselection (see below), which was carried out at 30°C. All chemicals were purchased from Sigma unless otherwise specified.
Bacterial conjugation, allelic exchange, and transposon mutagenesis.
Triparental conjugation was carried out as described previously (51). Transconjugants were selected on LANS with appropriate antibiotics. Irgasan DP300 (Ciba Geigy) was included at 25 μg/ml to select against the E. coli donor strain. For allelic exchange, pEX100T (41) or pDJW525 (31) was used for construction of allele replacement vectors. These vectors contain an origin of transfer that allows for conjugation, a ColE1 origin of replication that allows episomal propagation in E. coli but not in P. aeruginosa, the selectable β-lactamase gene, and the counterselectable sacB gene, which is lethal in the presence of sucrose. Following conjugation of an allele replacement vector into P. aeruginosa, recombinants resulting from a single homologous recombination (merodiploids) were selected for on LANS containing carbenicillin and Irgasan at 37°C. To force a second recombination event resulting in excision of the appropriate allele and vector backbone, merodiploid strains were cultured overnight at 37°C in LBNS without selection and then plated at 30°C on LANS containing 5% (wt/vol) sucrose and, where appropriate, other antibiotics. Sucrose-resistant recombinants were screened for loss of carbenicillin resistance (Cbr) and either loss or acquisition of the appropriate selectable markers. Transposon mutagenesis was performed by conjugation of pSUP::Tn5-B50 or pSUP::Tn5-B30 into the nonmucoid algB strains FRD444 and FRD840 followed by selection on LANS containing tetracycline and Irgasan at 37°C. Tetracycline-resistant (Tcr) colonies were screened for a mucoid phenotype after 24 to 36 h of incubation.
Cloning of the transposon insertions.
Southern blot analysis (43) was used to determine suitable restriction fragments on which to recover the transposon-inactivated loci. Digested genomic DNAs from the transposon mutants were then size fractionated by agarose gel electrophoresis, purified from the gel with the Qiaquick gel purification system (Qiagen), and ligated into either pUC18 (48) or pBluescript KS(−) (Stratagene). Ligations were transformed into E. coli JM109, and the transformations were plated on LB containing tetracycline.
DNA sequence analysis, colony hybridization, and physical mapping.
DNA sequencing was performed on an ABI Prism 377 DNA sequencer (Perkin-Elmer). To determine the precise site of transposition in each of the cloned restriction fragments described in the paragraph above, each fragment was subcloned (by using unique restriction sites in the transposons) as two smaller fragments, each of which was comprised of one end of the transposon along with the adjacent P. aeruginosa chromosomal DNA. Each of these subclones was then sequenced with the oligonucleotide Tn5-OUT (5′CGGGAAAGGTTCCGTTCAGG3′), which hybridizes to either end of the Tn5 derivatives. Analysis and assembly of the final sequence shown in Fig. 2 were performed with Factura and AutoAssembler software (Perkin-Elmer). Homology searches were performed with the Gapped BLAST program (1) on the National Center for Biotechnology Information website (www.ncbi.nlm.nih.gov). The GAP program of the Wisconsin Package (version 9.1; Genetics Computer Group, Madison, Wis.) was used to determine the percentages of amino acid identity described in Table 2. The PILEUP and PRETTY programs of the Wisconsin Package were used to create the alignment depicted in Fig. 3. The sequences of the E. coli, Haemophilus influenzae, Rickettsia prowazekii, and Mycobacterium tuberculosis PepA homologs were obtained from the SwissProt database (accession no. P11648, P45334, P27288, and Q10401, respectively). Additional sequence information was obtained from the Pseudomonas Genome Project (www.pseudomonas.com, 15 June 1998 release). Colony hybridization was performed as described by Grunstein and Hogness (25). Probes for both Southern blot and colony hybridization were synthesized with the Prime-a-Gene system (Promega). Physical mapping experiments were conducted as described elsewhere (39) by pulsed-field gel electrophoresis of DpnI- and SpeI-cleaved PA01 chromosomal DNA followed by Southern hybridizations with the 6.8-kb BamHI-HindIII fragment from pSW109 as a probe.
FIG. 2.
Nucleotide and derived amino acid sequences for the P. aeruginosa homologs of pepA (phpA) and holC. The peptide sequence of PhpA is indicated with three-letter amino acid abbreviations, while the sequence of HolC is displayed with one-letter abbreviations. Potential start codons are underscored with solid lines. Stop codons are indicated by asterisks. Potential ribosome binding sites (RBS) are shown. The site of insertion of Tn5-B50 in phpA is shown by a dashed line, indicating the direct repeat generated by the transposition. ORF280 (see the text) is indicated by a dashed overline and extends 5′ of the sequence shown.
TABLE 2.
Mucoid transposon mutants isolated in this study
| Mutant | Parental strain | Transposon | Phenotypea | Tn-disrupted ORF similar to: | % Amino acid identityb |
|---|---|---|---|---|---|
| FRD444.1 | FRD444 | Tn5-B50 | Mucoid | E. coli PepA (P11648)c | 56 |
| FRD444.2 | FRD444 | Tn5-B50 | Mucoid | E. coli TrmD (P07020)c | 65 |
| FRD444.3 | FRD444 | Tn5-B50 | Slightly mucoid | E. coli Hnr (P37055)c | 25 |
| FRD918 | FRD840 | Tn5-B50 | Slightly mucoid | E. coli Orn (P39287)c | 67 |
| FRD925 | FRD840 | Tn5-B50 | Slightly mucoid | NDd | |
| FRD940 | FRD444 | Tn5-B30 | Mucoid | P. aeruginosa MucD (U32853)e | 100 |
Phenotype on LANS at 37°C compared to that of the parental strain.
Determined as described in Materials and Methods.
SwissProt database accession number.
ND, not determined.
GenBank database accession number.
FIG. 3.
Alignment of the peptide sequences of the PepA homologs from E. coli (E. col), H. influenzae (H. inf), P. aeruginosa (P. aer), R. prowazekii (R. pro), and M. tuberculosis (M. tub). Conserved residues (Con) are indicated where three or more sequences agree. The absolutely conserved (underscored) residues are believed to be involved in the active site and/or metal ion binding based on X-ray crystallographic studies of bovine lens leucine aminopeptidase (9).
Construction of the allele replacement vector pSW134.
The construction of pSW134, which was used for complementation studies, was accomplished as follows. First, pSW109 was digested with XhoI and religated to delete all sequence between the XhoI site shown in Fig. 2 and the XhoI site in the pKS− multiple-cloning site. The resulting construct, pSW127, was digested with BamHI, and the BamHI fragment from pGmΩ1 (H. Schweizer) bearing the gene encoding resistance to gentamicin (aacC1) was inserted to create pSW128. This plasmid was treated with XhoI, blunt ended, and ligated to a SmaI fragment from pHP45Ω-Tc (16), which bears the gene that encodes tetracycline resistance (tet). The resulting plasmid, pSW133, contained the 2,134-bp BamHI-XhoI fragment shown in Fig. 2 flanked at the 5′ end by an ΩaacC1 cassette and at the 3′ end by Ωtet. To create pSW134, pSW133 was digested with AseI (New England Biolabs) and blunt ended and the approximately 2.3-kb fragment containing phpA flanked by the transcription termination signals from the Ω elements was gel purified and ligated into BglII-digested Klenow fragment-treated pUS68. pUS68 is a pEX100T-based vector providing the sequence required for homologous recombination at algB (31). The original transcriptional terminators from the Ω elements are oriented so as to terminate transcription into phpA from either direction. Therefore, phpA should be expressed only from a promoter 3′ of the BamHI site shown (Fig. 2).
algD-xylE fusions.
The construction of pDJW530, which was used to generate chromosomal algD-xylE fusions, was performed as follows. An approximately 10-kb HindIII fragment containing algD plus flanking sequences was cloned into the gene replacement vector pDJW525 (31), resulting in pDJW482. A 2.2-kb SmaI fragment of pX1918G which contained a promoterless xylE gene and the aacC1 gene, which encodes resistance to gentamicin, was subcloned into XhoI-digested, Klenow fragment-treated pDJW482, thus placing xylE transcription under the control of the algD promoter. The resulting plasmid, pDJW530, was mobilized into P. aeruginosa strains, and gene replacements were performed as described above. For algD-xylE transcriptional fusion experiments, overnight shaken LBNS cultures were diluted 1:1,000 in fresh prewarmed LBNS and incubated in a 37°C shaker (250 rpm). Culture growth was monitored with 1-cm-path-length cuvettes in a Spectronic Genesys 5 spectrophotometer (Spectronic Instruments). Cultures were harvested at a final optical density at 540 nm (OD540) between 0.3 and 0.7. A 1.0-ml sample was pelleted at 20,800 × g for 5 min. Pellets were resuspended in 1 ml of assay buffer (50 mM potassium phosphate buffer [pH 7.5], 10% acetone), and samples were kept on ice until they were assayed. A 100-μl aliquot of each sample was mixed with 900 μl of assay buffer containing 1 mM catechol (Sigma), and the increase in OD375 was monitored at ambient temperature for 2 min against a 1-ml blank of the assay buffer-catechol solution. The rate of formation of the reaction product, 2-hydroxymuconic semialdehyde, was calculated from the rate of increase in OD375 and the molar extinction coefficient of the product, 4.4 × 104 M−1. Values were normalized by dividing them by the OD540 of the culture at the time of harvest.
Nucleotide sequence accession number.
The nucleotide sequence shown in Fig. 2 has been deposited in the GenBank database under accession no. AF054622.
RESULTS
Isolation of mucoid transposon mutants of nonmucoid algB strains.
We have hypothesized that AlgB exerts transcriptional control over algD indirectly (Fig. 1). This hypothesis can be reduced to two basic scenarios. The first possibility is that AlgB activates expression of an intermediate gene (gene X) (Fig. 1A), resulting in an overall increase in algD transcription. The second possibility is that AlgB increases algD transcription by repressing expression of the intermediate gene (Fig. 1B). In order to investigate either of these possibilities and identify potential AlgB targets, we performed transposon mutagenesis on the nonmucoid algB mutants FRD444 and FRD840, derivatives of the mucoid CF isolate FRD1. For this procedure, we used a set of Tn5 derivatives (42). One of these, Tn5-B50, contains an outwardly directed constitutive promoter of the nptII gene and has been found to be active in Pseudomonas (42). Therefore, this transposon should be able to activate transcription of genes normally requiring AlgB if it inserts upstream of this gene in the proper orientation (Fig. 1A). Alternatively, Tn5-B50 may insertionally inactivate AlgB-repressed genes (Fig. 1B). In either case, the requirement for AlgB may be bypassed and the original mucoid phenotype may be restored. A second Tn5 derivative used in this study was Tn5-B30, which contains a promoterless nptII gene. While Tn5-B30 is not likely to activate gene expression, it might still inactivate putative targets of AlgB. In addition, the promoterless nptII gene, conferring resistance to kanamycin when expressed, may be used to determine if the inactivated locus is regulated by AlgB, provided that the transposon has inserted in the correct orientation.
Transposon mutagenesis was performed on the nonmucoid algB strains FRD444 and FRD840 as described in Materials and Methods, and the resulting Tcr colonies were screened for the mucoid phenotype at 37°C. Of approximately 3,000 Tn5-B50 mutants screened, five isolates which displayed a mucoid phenotype relative to that of the parental strains were obtained. Interestingly, this phenotypic difference was apparent only in areas of heavy growth on LANS plates (i.e., in the first and second quadrants of streak plates, where growth was confluent). In addition, the phenotypes of these mutants ranged from only slightly mucoid to a degree of mucoid morphology indistinguishable from that of the original mucoid strain, FRD1. When grown on normal LB agar (containing 5 g of NaCl per liter), none of these mutants displayed a mucoid phenotype whereas FRD1 was still mucoid. A number of studies have suggested that alginate expression is enhanced by high osmolarity (3, 47), which seems to be in conflict with our observations. Whether the mucoid appearance of the transposon mutants on LANS versus LB agar results from differential gene expression is presently unknown. Of the approximately 250 Tn5-B30 mutants isolated from the only experiment using this transposon, one displayed a particularly mucoid phenotype on LANS and was even slightly mucoid on LB agar.
Data obtained from Southern blot analysis (data not shown) were used to clone the transposon-disrupted loci from the mutants. The cloned transposon insertions were subjected to sequence analysis, and similarity searches were performed as described in Materials and Methods. Table 2 summarizes the nature of the mutants isolated in this study in terms of that of the parental strain, the transposon used, the phenotype, and the similarity profiles of the inactivated loci to known sequences. While quantitative determination of the growth rates and levels of alginate have not been performed for these mutants, some qualitative observations bear mention. FRD444.1 and FRD940 appeared the most mucoid and produced colonies similar in size to the parental algB mutants. FRD444.2 was moderately mucoid but produced much smaller colonies. FRD444.3, FRD918, and FRD925 were the least mucoid and also produced smaller colonies. The mucoid phenotypes of these last three strains were particularly unstable, often reverting to the nonmucoid phenotype after a single passage. Although this article focuses on only one of these mutants, FRD444.1, a brief mention of the sequence information obtained for the other mutants is germane to our discussion.
FRD444.2 was found to be disrupted in an open reading frame (ORF) with 65% amino acid identity to TrmD of E. coli. trmD encodes the tRNA (m1G37) methyltransferase of E. coli and Salmonella typhimurium. This enzyme is required for methylation of guanosine at position 37 in all tRNAs that recognize codon CCN, CGN, or CUN (4). This base modification has been implicated in prevention of frameshifting during translation (4). trmD of E. coli is part of a four-gene operon containing rpsP, rimM, trmD, and rplS (10). This same arrangement is found in the P. aeruginosa PA01 chromosome as reported in the 15 June 1998 release of the P. aeruginosa Genome Sequencing Project.
The transposon in FRD444.3 was discovered to lie in an uncharacterized ORF with similarity to several members of the response regulator class of proteins. The highest degree of amino acid identity observed (25%) was to the predicted product of hnr of E. coli (5). Like most response regulators, the E. coli Hnr protein and the ORF inactivated in FRD444.3 contained an amino-terminal domain with conserved aspartates and lysine residues in the predicted phosphorylation acid pocket active site (46). The carboxy-terminal output domain of this ORF was not conserved with any other proteins in the database.
FRD918 contained a Tn5-B50 insertion in an ORF with 67% amino acid identity to orn of E. coli. orn, previously known as ORF o204a until it was renamed by Zhang et al. (54), encodes an oligoribonuclease which is highly conserved and highly specific for small oligoribonucleotides. Zhang et al. reported that they have been unsuccessful in preliminary attempts to inactivate orn, suggesting that this gene may be essential in E. coli (54). Our data suggest that, at least for P. aeruginosa, this gene is not absolutely essential. The insertion occurred approximately one-fifth of the way into the ORF. Although FRD918 and FRD925 were isolated independently in separate experiments, Southern blot analysis with three single-enzyme digests as well as three double-enzyme digests revealed the restriction pattern of FRD925 to be identical to that of FRD918. The alginate phenotypes of these strains were also similar, suggesting that orn expression is disrupted in both of these strains.
FRD940, the single mucoid Tn5-B30 mutant obtained in this study, was discovered to harbor the transposon in the intergenic region between mucC and mucD in the algT(algU)mucABCD cluster. The transposition occurred 12 bp upstream of the initiation codon for mucD and thus likely prevents the expression of mucD. Inactivation of mucD has been shown to cause conversion to the mucoid phenotype (6). MucD has significant amino acid identity (39%) to E. coli HtrA (DegP), a periplasmic serine protease. The orientation of Tn5-B30 was such that transcription of the promoterless nptII gene occurred in the same direction as the algT(algU)mucABCD cluster. FRD940 demonstrated growth on LB agar containing up to 700 μg of kanamycin per ml, while neither FRD1 nor FRD444 exhibited growth even on 300 μg/ml, the lowest concentration tested (data not shown). This indicated that a promoter upstream of the coding sequence of mucD drove expression of nptII. By allelic exchange, a wild-type copy of algB was provided to FRD940. Preliminary experiments revealed no discernible differences between the MICs of kanamycin for the algB+ and algB mutant mucD::nptII strains (data not shown). Since transcription of mucD did not appear to be AlgB dependent, we focused the remainder of this study on FRD444.1.
FRD444.1 harbors an insertion in a previously uncharacterized ORF with 56% amino acid identity to PepA of E. coli. PepA is classified as a leucine aminopeptidase (45, 49), and similarity searches revealed pepA to be a highly conserved gene in a variety of bacteria (Fig. 3) as well as plants and animals. PepA (also known as XerB) is also an accessory factor in the Xer-mediated site-specific recombination system, which acts to monomerize multimers of multicopy plasmids formed by homologous recombination (45). In addition, PepA is involved in transcriptional repression of the carAB operon, which encodes the subunits for carbamoylphosphate synthetase, as well as in repression of its own gene, pepA (13). Recently, Hauser et al. reported the identification of a secreted protein from P. aeruginosa which they designated PepA, for Pseudomonas exoprotein A (26). The sequence of the gene encoding this protein, pepA, and the predicted amino acid sequence showed no homology with known sequences and should not be confused with true homologs of the PepA aminopeptidase of E. coli. To prevent any confusion between pepA described by Hauser et al. and the gene described here, we have chosen the designation phpA (for P. aeruginosa homolog of pepA).
Nucleotide and predicted amino acid sequences of phpA.
In order to clone the wild-type phpA gene, genomic DNA from FRD1 was analyzed by a Southern blot analysis with XhoI-treated pSW50 as a probe template. The probe hybridized to an approximately 6.8-kb BamHI-HindIII fragment (data not shown). Size-fractionated BamHI- and HindIII-treated FRD1 genomic DNA was ligated into BamHI- and HindIII-treated pKS−, and the ligated plasmid was transformed into E. coli JM109. From this library, a clone designated pSW109 which contained an approximately-6.8-kb BamHI-HindIII fragment was identified by colony hybridization. The sequence of the first 2,278 bp of the insert in pSW109 was determined (Fig. 2).
There are two potential initiation codons for phpA, resulting in respective predicted products of 495 and 511 amino acids (Fig. 2). The second AUG codon is downstream of a potential ribosome binding site and best aligns with the initiation codons of the majority of the PepA homologs shown in Fig. 3. The predicted molecular mass of P. aeruginosa PhpA has been calculated to be either 52,298 or 53,892 Da, depending on which initiation codon is used. These values correlate well with the 54.8- and 53.5-kDa molecular masses predicted for the PepA homologs of E. coli and H. influenzae, respectively. There appears to be very strong amino acid conservation among the various PepA homologs depicted in Fig. 3, including the absolute conservation of seven amino acid residues shown to be in the active site or involved in metal binding of bovine lens leucine aminopeptidase (9) (Fig. 3).
Immediately 3′ and partially overlapping the PhpA ORF, a second ORF encoding a product with 31% amino acid identity to HolC of E. coli (11) and 33% identity to the HolC homolog from H. influenzae (18) was detected. HolC is the chi subunit of the DNA polymerase holoenzyme. Interestingly, the same genomic arrangement, pepA followed by holC, is also conserved in E. coli (11). Although there was no AUG initiation codon detected for P. aeruginosa holC, based on the alignment with the H. influenzae holC homolog, there was a potential GUG start codon (Fig. 2).
We used our sequence to scan the database of the Pseudomonas Genome Project (www.pseudomonas.com). We found a sequence containing phpA which revealed additional sequence 5′ of phpA. This 5′ region contained an uncharacterized ORF (encoding a potential product of 280 amino acids) which would be transcribed in the opposite direction from phpA (Fig. 2). Sequence comparison analysis indicated that the predicted product of this ORF (designated here ORF280) is 34% identical to a hypothetical protein from M. tuberculosis (data not shown). The genomic organization of the elements described above are represented schematically in Fig. 4.
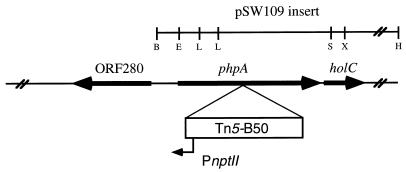
FIG. 4.
Genome organization of the P. aeruginosa phpA region. The region contained within the insert of pSW109 is indicated. Additional 5′ sequence information was obtained from the P. aeruginosa Genome Sequencing Project (15 June 1998 release). ORF280 displays 34% amino acid identity to an ORF of unknown function in M. tuberculosis. The orientation of Tn5-B50 enables transcription of ORF280 while at the same time inactivating phpA and possibly exerting a polar effect on holC. Restriction endonuclease sites correspond to those shown in Fig. 2. Abbreviations: B, BamHI; E, EcoRI; L, SalI; S, SphI; X, XhoI; H, HindIII.
Physical mapping of phpA on the P. aeruginosa PA01 chromosome.
Pulsed-field gel electrophoresis was used for chromosomal mapping of phpA. The approximately 6.8-kb BamHI-HindIII fragment from pSW109 hybridized to the 302-kb DpnI (H) fragment and the 517-kb SpeI (A) fragment of the P. aeruginosa PA01 chromosome. This places phpA on the approximately 100-kb linking fragment and localizes phpA to approximately 27 to 28.5 min on the PA01 chromosome. The gene exoS and the insertion sequence IS-PA-1 also map in this region of the chromosome (29).
The Tn5-B50 insertion in phpA causes conversion to the mucoid phenotype in an algB mutant but not in an algT or algR mutant.
Although FRD444.1 displayed a mucoid phenotype, it was not clear whether this was due to the transposon insertion or to secondary mutations which were Tn5-B50 independent. In addition, we wanted to determine if the Tn5-B50 insertion demonstrated gene specificity and restored the mucoid phenotype only in an algB mutant. To address these issues, it was necessary to create identical Tn5-B50 mutations in other P. aeruginosa strains and in a “clean” algB mutant background. We took advantage of the P. aeruginosa DNA flanking Tn5-B50 in pSW50 to construct an allele replacement vector, pSW40. This plasmid was conjugated into the mucoid CF isolate FRD1 and into the nonmucoid strains FRD444 (algB::Tn501), FRD440 (algT::Tn501), and FRD831 (ΔalgR::ΩaacC1) (Table 1). Recombinants were selected as described in Materials and Methods. The alginate phenotypes of the FRD1, FRD440, and FRD831 recombinants were not visibly altered, whereas the phenotype of the FRD444 recombinant had changed from nonmucoid to mucoid (data not shown). This confirmed that the mucoid phenotype of FRD444.1 was due to the Tn5-B50 insertion. In addition, this result indicated that the same insertion did not affect the visible alginate phenotype in FRD1 or the algR or algT genetic background.
Polar and nonpolar insertions in phpA result in a mucoid phenotype in an algB mutant background.
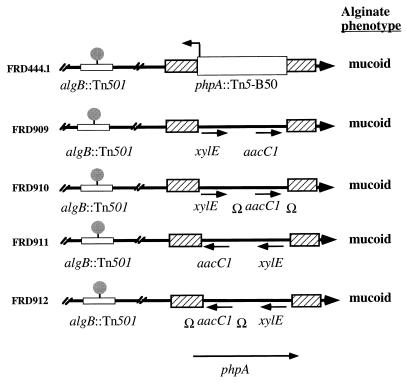
Due to the orientation of Tn5-B50 in phpA (Fig. 4) we postulated three possibilities for the cause of the mucoid phenotype of FRD444.1. The phenotype may result from Tn5-B50 driving transcription of ORF280 or some other gene 5′ of phpA (Fig. 4). Alternatively, the phenotype of FRD444.1 may result from the loss of phpA or a polar effect on the holC gene. In order to determine the cause of the mucoid phenotype of FRD444.1, we constructed a series of insertions in phpA in the chromosome of FRD444 (algB mutant). To accomplish this, pSW90 (Table 1) was digested with SalI, which removed an approximately 190-bp fragment from the coding region of phpA. The ends of the digested plasmid were rendered blunt and ligated to the xylE-aacC1-containing SmaI fragments of either pX1918GT or pX1918G (41). These fragments created polar and nonpolar insertions, respectively, in the direction of aacC1 transcription. Each insertion was obtained in both orientations in phpA, and the resulting plasmids were used to perform allelic exchange (Fig. 5). We reasoned that if expression of ORF280 5′ of phpA were the cause of the phenotype of FRD444.1, then only the nonpolar insertion in FRD911 would result in a mucoid phenotype (Fig. 5). However, if the phenotype of FRD444.1 resulted from a polar effect on holC, then all insertions shown in Fig. 5 except the nonpolar insertion in FRD909 would result in a mucoid phenotype. We observed that all insertions in phpA resulted in conversion to the mucoid phenotype (Fig. 5). These results are consistent with the inactivation of phpA as the cause of conversion.
FIG. 5.
Polar and nonpolar insertions in phpA result in a mucoid phenotype in algB::Tn501 P. aeruginosa. Allelic exchange was performed on the nonmucoid algB::Tn501 strain FRD444, replacing the wild-type phpA with the phpA::xylE aacC1 alleles shown. The alginate phenotypes of the resulting strains were then compared to the mucoid phenotype of the phpA::Tn5-B50 strain FRD444.1. Ω designates the original Ω element described by Prentki and Krisch (38), which contains the factor-independent transcriptional terminators of bacteriophage T4D gene 32 as well as translational stop codons in all six reading frames. The presence of Ω therefore generates an insertion polar on transcription and translation in either direction.
To confirm that it was indeed inactivation of phpA which was responsible for the mucoid phenotype, we attempted complementation of the lesion in phpA using both IncP (pLAFR3)- and IncQ (pMMB67HE)-type vectors. However, the results proved ambiguous. We repeatedly failed to obtain any transconjugants with pMMB67HE, despite the fact that the parental strain FRD444 maintained this plasmid quite well (data not shown). Using FRD906, a tetracycline-sensitive (Tcs) version of FRD444.1 generated by allelic exchange, we were able to obtain transconjugants with pLAFR3. However, the FRD906 strain formed colonies much smaller than those of the parental strain FRD444 harboring the same plasmid. The mucoid status of these colonies was impossible to determine due to their apparent growth defect. Interestingly, when pSW110 (pLAFR3 containing phpA) was conjugated into FRD906, colonies appeared similar in size and in nonmucoid morphology to FRD444 harboring pLAFR3.
Expression of phpA is not regulated by AlgB at the transcriptional level.
The xylE genes from pX1918GT and pX1918G are promoterless and thereby establish transcriptional fusions when they are inserted downstream of a promoter. In order to determine whether phpA was transcriptionally regulated by AlgB, we compared the XylE activity of FRD909 (Fig. 5) to that of the isogenic algB+ strain FRD913 (not shown). There was no discernible difference in the XylE activities between these two strains, indicating that phpA expression was not affected by AlgB at the transcriptional level (data not shown). Nevertheless the visible increase in alginate production in FRD444.1 (mutated algB and phpA) compared to the level of production in the parental algB mutant FRD444 warranted further investigation.
Inactivation of phpA in an algB::Tn501 strain results in increased algD transcription.
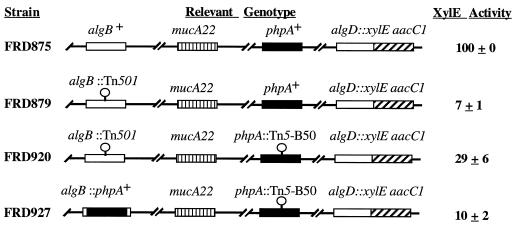
Data described above indicated that inactivation of phpA circumvented an algB mutation, restoring alginate production to the normally nonmucoid strain FRD444. Mutations in algB result in an approximately 20-fold reduction in algD transcription (51). To quantify the effect that inactivation of phpA had on algD expression, we used pDJW530 (Table 1) to construct xylE transcriptional fusions to the chromosomal algD promoters of FRD1 (mucA22), FRD444 (mucA22 algB::Tn501), and FRD444.1 (mucA22 algB::Tn501 phpA::Tn5-B50), which resulted in the strains FRD875, FRD879, and FRD920, respectively (Table 1; Fig. 6). The levels of XylE activity in these strains were measured, and the data from six independent experiments are summarized in Fig. 6. The algB::Tn501 strain FRD879 exhibited XylE activity equal to approximately 7% of that of the algB+ strain FRD875. However, the algB::Tn501 phpA::Tn5-B50 strain FRD920 expressed an approximately fourfold higher level of XylE activity than FRD879, reaching 29% of the value of FRD875 (Fig. 6). These data are in agreement with the visible alginate phenotypes of the corresponding algD+ strains (data not shown).
FIG. 6.
Inactivation of phpA in an algB::Tn501 strain results in increased algD expression. The four strains shown were constructed by replacing the chromosomal wild-type algD with algD::xylE by allelic exchange. XylE activity was assayed as detailed in Materials and Methods. Activity was normalized by dividing by the OD540 at the time of harvest. For each experiment, the activity of strain FRD875 was set at 100%. The activities of the remaining strains were then expressed as percentages of the activity of FRD875. The results of six independent experiments were used to determine the means and standard deviations shown.
Expression of phpA from an ectopic chromosomal locus complements the Tn5-B50 insertion in the native phpA gene.
Due to the effect of the phpA::Tn5-B50 insertion on the ability of the organism to acquire and/or properly maintain plasmids (see above), it was impossible to perform standard complementation analyses with plasmid-borne phpA alleles. To be certain that inactivation of phpA was the cause of the increase in algD transcription in the algB genetic background, we developed a strategy to complement the lesion in phpA by providing a wild-type copy of phpA elsewhere in the P. aeruginosa chromosome. This complementation would avoid any effects of phpA mutations on plasmid biology as well as the effects of any proteins encoded by plasmid-borne genes, antibiotic selection, or gene dosage. Since we were investigating the effects of phpA in an algB mutant background, the Tn501-marked algB allele provided a convenient locus for integration of a wild-type phpA allele. To carry out this experiment, we first constructed pSW134, an allele replacement vector containing the wild-type phpA gene. This copy of phpA was flanked at either end by factor-independent transcriptional terminators (38). These terminators were in turn bordered by sequences flanking the algB gene, thus providing the necessary homology for recombination of the vector with the chromosome at the algB locus (Materials and Methods). To isolate phpA+ gene replacements at the algB::Tn501 locus, sucrose-resistant colonies were screened for loss of mercuric chloride resistance (Hgr) (the marker on Tn501). The level of XylE activity of the resulting strain, FRD927 (phpA::Tn5-B50 algB::phpA), was found to be significantly lower than that observed in the parental strain FRD920 and was similar to that in FRD879 at 10% of wild-type XylE levels (Fig. 6). This result confirmed that loss of phpA was responsible for the phenotype of the original phpA::Tn5-B50 strain FRD444.1. In addition, the transcriptional terminators flanking phpA at the algB locus were oriented so as to block any incoming transcription from the adjacent chromosomal sequences. The observation of reduced algD transcription in FRD927 indicated that at least one promoter capable of expressing phpA was located between the start of phpA and the BamHI site depicted in Fig. 2.
DISCUSSION
In this study we have identified the P. aeruginosa homolog of pepA, which encodes leucine aminopeptidase in E. coli and a variety of other species. We have shown that insertional inactivation of this gene in the nonmucoid algB mutant FRD444 results in conversion to the mucoid phenotype. This is accompanied by a fourfold increase in algD transcription. We have designated this novel pepA homolog phpA to avoid confusion with P. aeruginosa pepA, recently described by Hauser et al. (26). Hermes et al. described the purification of an aminopeptidase from Pseudomonas putida which demonstrated activity toward a select group of l amino acid amides, particularly the l-leucine derivative, as well as all four dipeptide combinations of leucine and phenylalanine (27). Those authors determined the subunit molecular mass to be 53 kDa, which is in agreement with the predicted molecular mass of PhpA. However, the pI of the P. putida aminopeptidase was estimated to be 10.5 (27) whereas the predicted pI of PhpA is either 8.9 (first start codon) or 8.7 (second start codon). Whether phpA encodes the enzyme described by Hermes et al. remains to be determined, but there is little doubt that phpA is in fact the homolog of E. coli pepA. The extensive sequence similarity at the amino acid level, the conservation of predicted active-site residues, and the genomic clustering with holC in both organisms speak to the authenticity of the relationship between E. coli pepA and P. aeruginosa phpA.
A possible connection between an aminopeptidase activity of PhpA and transcriptional activation of algD involves ς22 (AlgT and AlgU). ς22 is a member of the ECF (extracytoplasmic function) family of alternative ς factors (14, 33, 35) and is homologous to ςE of E. coli (15, 35). This family of ς factors is believed to play an important role in stress response. ςE of E. coli is involved in the transcription of the extreme heat shock ς factor ςH (ς32) (15). ςE is also required for expression of HtrA (DegP), a periplasmic serine protease thought to function in the degradation of abnormal or denatured periplasmic proteins which may arise as a result of thermal or chemical insult (15). Consistent with this role, algT mutants of P. aeruginosa were found to be more susceptible to killing by heat and paraquat (35). Recently, Boucher et al. described two P. aeruginosa genes displaying similarity to htrA, namely, mucD and algW (6). MucD is encoded within the algTmucABCD cluster mapping near 67.5 min on the P. aeruginosa chromosome and is suggested to be subject to positive regulation by ς22 (6). MucD is believed to be a periplasmic protein and thus may play a role analogous to that of HtrA (6). AlgW, encoded at 69 min, lacks a conserved signal peptide and was postulated to localize to the cytoplasm, where it too may play a role in degrading denatured proteins (6). Accumulation of abnormal proteins has been shown to be a signal for ς32 activation in E. coli (8, 21, 30). If denatured proteins are indeed a signal for induction of stress response, loss of PhpA aminopeptidase function might be expected to further stimulate the ς22 regulon and partially overcome the requirement for AlgB by leading to the incomplete breakdown of senescent and misfolded proteins in the cytosol. The finding that a transposon insertion in mucD also suppressed the algB phenotype is entirely consistent with a hypothesis that increased expression or activity of AlgT overcomes the lack of AlgB and restores the mucoid phenotype. A similar argument could be made for the trmD mutant, i.e., that loss of TrmD leads to an increase in frameshifting during translation (4), in turn leading to abnormal or misfolded proteins which stimulate AlgT activity or expression. It should be noted, however, that only in the case of the phpA mutant have we ruled out the possibility that the phenotypes of the transposon mutants listed in Table 2 are due to polar effects. Nevertheless, according to this scenario many mutations may lead to partial suppression of algB as long as such mutations ultimately induce the stress response.
The effect of the phpA::Tn5-B50 insertion on algD transcription may or may not be due to the mechanism described above. In fact, PepA of E. coli is a remarkably multifunctional protein. In addition to the aminopeptidase activity of PepA, there at least two systems in which E. coli PepA participates as a site-specific DNA-binding protein involved in negative transcriptional regulation. PepA interacts with the promoter region of the pepA gene, repressing transcription from one of its three promoters (13). PepA has also been shown to bind within the promoter region of the carAB operon (13). The carAB genes encode the subunits of carbamoylphosphate synthetase. The upstream promoter of carAB, P1, is repressed in the presence of high concentrations of pyrimidines, a repression which requires site-specific DNA binding of PepA to the carAB control region (12). Therefore, by analogy, either PhpA may directly repress algD transcription or it may repress expression of a positive regulator of algD. E. coli PepA also plays a structural role in the Xer-mediated resolution of multimeric forms of multicopy plasmids such as ColE1 and pSC101 (45). Whether a site-specific recombination is involved in algD regulation is not known. It is also unknown whether PhpA plays a role in resolving plasmid multimers in P. aeruginosa, although this function would be consistent with our observations on the effects of phpA lesions on plasmid maintenance. Interestingly, the aminopeptidase activity of PepA is not required for its role in site-specific recombination or pyrimidine-specific regulation of carAB (13, 36). Experiments are under way to determine whether PhpA possesses aminopeptidase activity and whether such activity is required for the negative effect that PhpA has on algD transcription.
The original goal of these investigations was to identify any genes under AlgB control which participate in algD transcription. The fact that expression of phpA does not appear to be affected by AlgB at the transcriptional level suggests that phpA is not a true intermediate between AlgB and algD. However, since in E. coli, PepA represses transcription of its own gene (13), it remains a possibility that AlgB regulates phpA expression by a mechanism requiring the phpA gene product. Another indication that phpA is not the target of AlgB relevant to alginate expression is the observation that insertional inactivation of phpA in an algB mutant does not restore algD transcription to wild-type levels. In fact, the finding that transposon insertions in a variety of genes (Table 2) can at least partially suppress the algB phenotype raises new questions as to the mechanism whereby AlgB activates transcription of algD. Although transcription of phpA does not appear to be regulated by AlgB, it is clear from these studies that loss of phpA has a pronounced effect on algD transcription in the algB genetic background. Investigation of the mechanism of this effect as well as analysis of other algB-suppressing mutants should provide valuable insight into the basic physiological and genetic regulation of alginate expression.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant AI-35177 (D.J.W.) from the National Institutes of Allergy and Infectious Diseases.
We are grateful to S. Lory for advice and for providing the Tn5 derivatives. We thank H. Schweizer for providing expertise in allelic exchange and for providing pEX100T, pGmΩ1, and pHP45Ω-Tc. U. Selvaraj provided valuable technical assistance. We also thank K. Schmidt and B. Tummler for performing the physical mapping of phpA in PA01. Oligonucleotides were provided by E. Roesch at the DNA Synthesis Core Laboratory of the Cancer Center of Wake Forest University (CCWFU). DNA sequencing was performed by E. Jung of the DNA Sequencing Core Laboratory (CCWFU). Both facilities are supported in part by NIH grant CA-12197.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D L. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baynham P J, Wozniak D J. Identification and characterization of AlgZ, an AlgT-dependent DNA binding protein required for Pseudomonas aeruginosa algD transcription. Mol Microbiol. 1996;22:97–108. doi: 10.1111/j.1365-2958.1996.tb02659.x. [DOI] [PubMed] [Google Scholar]
- 3.Berry A, DeVault J D, Chakrabarty A M. High osmolarity is a signal for enhanced algD transcription in mucoid and nonmucoid Pseudomonas aeruginosa strains. J Bacteriol. 1989;171:2312–2317. doi: 10.1128/jb.171.5.2312-2317.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bjork G R, Wikstrom P M, Bystrom A S. Prevention of translational frameshifting by the modified nucleoside 1-methylguanosine. Science. 1989;244:986–989. doi: 10.1126/science.2471265. [DOI] [PubMed] [Google Scholar]
- 5.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 6.Boucher J C, Martinez-Salazar J, Schurr M J, Mudd M H, Yu H, Deretic V. Two distinct loci affecting conversion to mucoidy in Pseudomonas aeruginosa in cystic fibrosis encode homologs of the serine protease HtrA. J Bacteriol. 1996;178:511–523. doi: 10.1128/jb.178.2.511-523.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyer H W, Roullard-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 8.Bukau B. Regulation of the Escherichia coli heat-shock response. Mol Microbiol. 1993;9:671–680. doi: 10.1111/j.1365-2958.1993.tb01727.x. [DOI] [PubMed] [Google Scholar]
- 9.Burley S K, David P R, Sweet R M, Taylor A, Lipscomb W N. Structure determination and refinement of bovine lens leucine aminopeptidase and its complex with bestatin. J Mol Biol. 1992;224:113–140. doi: 10.1016/0022-2836(92)90580-d. [DOI] [PubMed] [Google Scholar]
- 10.Bystrom A S, Hjalmarsson K J, Wikstrom P M, Bjork G R. The nucleotide sequence of an Escherichia coli operon containing genes for the tRNA (m1G) methyltransferase, the ribosomal proteins S16 and L19 and a 21-K polypeptide. EMBO J. 1983;2:899–905. doi: 10.1002/j.1460-2075.1983.tb01519.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carter J, Franden M, Lippincott J, McHenry C. Identification, molecular cloning and characterization of the gene encoding the χ subunit of DNA polymerase III holoenzyme of Escherichia coli. Mol Gen Genet. 1993;241:399–408. doi: 10.1007/BF00284693. [DOI] [PubMed] [Google Scholar]
- 12.Charlier D, Gigot D, Huysveld N, Roovers M, Piérard A, Glansdorff N. Pyrimidine regulation of the Escherichia coli and Salmonella typhimurium carAB operons: CarP and integration host factor (IHF) modulate the methylation status of a GATC site present in the control region. J Mol Biol. 1995;250:383–391. doi: 10.1006/jmbi.1995.0384. [DOI] [PubMed] [Google Scholar]
- 13.Charlier D, Hassanzadeh G, Kholti A, Gigot D, Piérard A, Glansdorff N. carP, involved in pyrimidine regulation of the Escherichia coli carbamoylphosphate synthetase operon encodes a sequence-specific DNA-binding protein identical to XerB and PepA, also required for resolution of ColE1 multimers. J Mol Biol. 1995;250:392–406. doi: 10.1006/jmbi.1995.0385. [DOI] [PubMed] [Google Scholar]
- 14.Devries C A, Ohman D E. Mucoid-to-nonmucoid conversion in alginate-producing Pseudomonas aeruginosa often results from spontaneous mutations in algT, encoding a putative alternative sigma factor, and shows evidence for autoregulation. J Bacteriol. 1994;176:6677–6687. doi: 10.1128/jb.176.21.6677-6687.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erickson J W, Gross C A. Identification of the subunit of Escherichia coli RNA polymerase: a second alternate sigma factor involved in high-temperature gene expression. Genes Dev. 1989;3:1462–1471. doi: 10.1101/gad.3.9.1462. [DOI] [PubMed] [Google Scholar]
- 16.Fellay R, Frey J, Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of Gram-negative bacteria. Gene. 1987;52:147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- 17.Figurski D, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M E A. Whole-genome sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 19.Flynn J L, Ohman D E. Cloning of genes from mucoid Pseudomonas aeruginosa which control spontaneous conversion to the alginate production phenotype. J Bacteriol. 1988;170:1452–1460. doi: 10.1128/jb.170.4.1452-1460.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fürste J P, Pansegrau W, Frank R, Blöcker H, Scholz P, Bagdasarian M, Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- 21.Goff S A, Goldberg A L. Production of abnormal proteins in Escherichia coli stimulates transcription of lon and other heat shock genes. Cell. 1985;41:587–595. doi: 10.1016/s0092-8674(85)80031-3. [DOI] [PubMed] [Google Scholar]
- 22.Goldberg J B, Dahnke T. Pseudomonas aeruginosa AlgB, which modulates the expression of alginate, is a member of the NtrC subclass of prokaryotic regulators. Mol Microbiol. 1992;6:59–66. doi: 10.1111/j.1365-2958.1992.tb00837.x. [DOI] [PubMed] [Google Scholar]
- 23.Goldberg J B, Ohman D E. Construction and characterization of Pseudomonas aeruginosa algB mutants: role of algB in high-level production of alginate. J Bacteriol. 1987;169:1593–1602. doi: 10.1128/jb.169.4.1593-1602.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Govan J R W, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grunstein M, Hogness D. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc Natl Acad Sci USA. 1975;72:3961. doi: 10.1073/pnas.72.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hauser A R, Kang P J, Engel J N. PepA, a secreted protein of Pseudomonas aeruginosa, is necessary for cytotoxicity and virulence. Mol Microbiol. 1998;27:807–818. doi: 10.1046/j.1365-2958.1998.00727.x. [DOI] [PubMed] [Google Scholar]
- 27.Hermes H F M, Sonke T, Peters P J H, van Balken J A M, Kamphuis J, Dijkhuizen L, Meijer E M. Purification and characterization of an l-aminopeptidase from Pseudomonas putida ATCC 12633. Appl Environ Microbiol. 1993;59:4330–4334. doi: 10.1128/aem.59.12.4330-4334.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holloway B W. Genetics of Pseudomonas. Bacteriol Rev. 1969;33:419–443. doi: 10.1128/br.33.3.419-443.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holloway B W. Pseudomonas genetics and taxonomy. In: Nakazawa T, Furukawa K, Haas D, Silver S, editors. Molecular biology of pseudomonads. Washington, D.C: ASM Press; 1996. pp. 22–32. [Google Scholar]
- 30.Kanemori M, Mori H, Yura T. Induction of heat shock proteins by abnormal proteins results from stabilization and not increased synthesis of ς32 in Escherichia coli. J Bacteriol. 1994;176:5648–5653. doi: 10.1128/jb.176.18.5648-5653.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma S, Selvaraj U, Ohman D E, Quarless R, Hassett D J, Wozniak D J. Phosphorylation-independent activity of the response regulators AlgB and AlgR in promoting alginate biosynthesis in mucoid Pseudomonas aeruginosa. J Bacteriol. 1998;180:956–968. doi: 10.1128/jb.180.4.956-968.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 33.Martin D W, Holloway B W, Deretic V. Characterization of a locus determining the mucoid status of Pseudomonas aeruginosa: AlgU shows sequence similarities with a Bacillus sigma factor. J Bacteriol. 1993;175:1153–1164. doi: 10.1128/jb.175.4.1153-1164.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin D W, Schurr M J, Mudd M H, Govan J R W, Holloway B W, Deretic V. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc Natl Acad Sci USA. 1993;90:8377–8381. doi: 10.1073/pnas.90.18.8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin D W, Schurr M J, Yu H, Deretic V. Analysis of promoters controlled by the putative sigma factor AlgU regulating conversion to mucoidy in Pseudomonas aeruginosa: relationship to ςe and stress response. J Bacteriol. 1994;176:6688–6696. doi: 10.1128/jb.176.21.6688-6696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCulloch R, Burke M E, Sherratt D J. Peptidase activity of Escherichia coli aminopeptidase A is not required for its role in Xer site-specific recombination. Mol Microbiol. 1994;12:241–251. doi: 10.1111/j.1365-2958.1994.tb01013.x. [DOI] [PubMed] [Google Scholar]
- 37.Ohman D E, Chakrabarty A M. Genetic mapping of chromosomal determinants for the production of the exopolysaccharide alginate in a Pseudomonas aeruginosa cystic fibrosis isolate. Infect Immun. 1981;33:142–148. doi: 10.1128/iai.33.1.142-148.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prentki P, Kirsch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 39.Schmidt K D, Tummler B, Romling U. Comparative genome mapping of Pseudomonas aeruginosa PAO with P. aeruginosa C, which belongs to a major clone in cystic fibrosis patients and aquatic habitats. J Bacteriol. 1996;178:85–93. doi: 10.1128/jb.178.1.85-93.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schurr M J, Yu H, Martinez-Salazar J M, Boucher J C, Deretic V. Control of AlgU, a member of the ςE-like family of stress sigma factors, by the negative regulators MucA and MucB and Pseudomonas aeruginosa conversion to mucoidy in cystic fibrosis. J Bacteriol. 1996;178:4997–5004. doi: 10.1128/jb.178.16.4997-5004.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schweizer H P, Hoang T T. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene. 1995;158:15–22. doi: 10.1016/0378-1119(95)00055-b. [DOI] [PubMed] [Google Scholar]
- 42.Simon R, Quandt J, Klipp W. New derivatives of transposon Tn5 suitable for mobilization of replicons, generation of operon fusions and induction of genes in Gram-negative bacteria. Gene. 1989;80:161–169. doi: 10.1016/0378-1119(89)90262-x. [DOI] [PubMed] [Google Scholar]
- 43.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 44.Staskawicz B, Dahlbeck D, Keen N, Napoli C. Molecular characterization of cloned avirulence genes from race-0 and race-1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stirling C J, Colloms S D, Collins J F, Szatmari G, Sherratt D J. xerB, an Escherichia coli gene required for plasmid ColE1 site-specific recombination, is identical to pepA, encoding aminopeptidase A, a protein with substantial similarity to bovine lens leucine aminopeptidase. EMBO J. 1989;8:1623–1627. doi: 10.1002/j.1460-2075.1989.tb03547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stock J B, Surette M G, Levit M, Park P. Two-component signal transduction systems: structure-function relationships and mechanisms of catalysis. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: ASM Press; 1995. pp. 25–51. [Google Scholar]
- 47.Terry J M, Pina S E, Mattingly S J. Environmental conditions which influence mucoid conversion in Pseudomonas aeruginosa PAO1. Infect Immun. 1991;59:471–477. doi: 10.1128/iai.59.2.471-477.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 49.Vogt V M. Purification and properties of an aminopeptidase from Escherichia coli. J Biol Chem. 1970;245:4760–4769. [PubMed] [Google Scholar]
- 50.Wozniak D J, Ohman D E. Pseudomonas aeruginosa AlgB, a two-component response regulator of the NtrC family, is required for algD transcription. J Bacteriol. 1991;173:1406–1413. doi: 10.1128/jb.173.4.1406-1413.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wozniak D J, Ohman D E. Transcriptional analysis of the Pseudomonas aeruginosa genes algR, algB, and algD reveals a hierarchy of alginate gene expression which is modulated by algT. J Bacteriol. 1994;176:6007–6014. doi: 10.1128/jb.176.19.6007-6014.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xie Z, Hershberger D, Shankar S, Ye R W, Chakrabarty A M. Sigma factor–anti-sigma factor interaction in alginate synthesis: inhibition of AlgT by MucA. J Bacteriol. 1996;178:4990–4996. doi: 10.1128/jb.178.16.4990-4996.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yanisch-Perron C, Vieira J, Messing J. Improved M13 cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 54.Zhang X, Zhu L, Deutscher M P. Oligoribonuclease is encoded by a highly conserved gene in the 3′-5′ exonuclease superfamily. J Bacteriol. 1998;180:2779–2781. doi: 10.1128/jb.180.10.2779-2781.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]