Abstract
Organisms have evolved elaborate physiological pathways that regulate growth, proliferation, metabolism, and stress response. These pathways must be properly coordinated to elicit the appropriate response to an ever-changing environment. While individual pathways have been well studied in a variety of model systems, there remains much to uncover about how pathways are integrated to produce systemic changes in a cell, especially in dynamic conditions. We previously showed that deletion of Protein Kinase A (PKA) regulatory subunit BCY1 can decouple growth and metabolism in Saccharomyces cerevisiae engineered for anaerobic xylose fermentation, allowing for robust fermentation in the absence of division. This provides an opportunity to understand how PKA signaling normally coordinates these processes. Here, we integrated transcriptomic, lipidomic, and phospho-proteomic responses upon a glucose to xylose shift across a series of strains with different genetic mutations promoting either coupled or decoupled xylose-dependent growth and metabolism. Together, results suggested that defects in lipid homeostasis limit growth in the bcy1Δ strain despite robust metabolism. To further understand this mechanism, we performed adaptive laboratory evolutions to re-evolve coupled growth and metabolism in the bcy1Δ parental strain. The evolved strain harbored mutations in PKA subunit TPK1 and lipid regulator OPI1, among other genes, and evolved changes in lipid profiles and gene expression. Deletion of the evolved opi1 gene partially reverted the strain’s phenotype to the bcy1Δ parent, with reduced growth and robust xylose fermentation. We suggest several models for how cells coordinate growth, metabolism, and other responses in budding yeast and how restructuring these processes enables anaerobic xylose utilization.
Author summary
All organisms utilize an energy source to generate the cellular resources needed to grow and divide. Individual processes have been well studied, but the coordination and crosstalk between the process is not well understood. To study growth and metabolism coupling, we used a yeast strain that was genetically engineered to ferment the sugar xylose but lacked growth on the sugar. The decoupled growth and metabolism was caused by a deletion of a single gene in a highly conserved signaling pathway found in all eukaryotes. While our work is focused on xylose metabolism, we address the fundamental question of how cells coordinate growth with metabolism under non-ideal conditions. We identified changes in gene expression that implicated altered regulatory mechanisms involved in lipid metabolism correlating with decoupled growth and metabolism. Our work highlights the complexity of engineering new cellular functions and that global regulatory modifications, rather than altering individual pathways, may be required for broad cellular changes.
Introduction
Many physiological processes are essential for growth, but so too is the coordination of those processes to form an integrated cellular system. Actively dividing cells must coordinate metabolism and division with the synthesis and segregation of DNA, proteins, organelles, and other macromolecules, all within a precisely timed cell cycle. Failure to coordinate these processes can jeopardize fitness due to suboptimal cellular composition and energy expenditures. Mechanistically, much remains unknown about how cells coordinate cellular processes. One of the best studied examples is the intimate control of successive cell cycle phases, which depends on interconnected transcriptional and post-translational controls regulated by dispersed checkpoints along the way [1–8]. The cell cycle is also coordinated with metabolism; cell-cycle regulators coordinate metabolic flux with cell-cycle phases, which may be related to cell size checkpoints since cells must reach a critical size before a new cell cycle is initiated [2,3,6–8]. A critical feature of integrated cellular systems is thus balancing energy demands with division and replication.
Knowing how cells coordinate growth and division with other physiological processes is important for understanding how cells function on a fundamental level, but it also has practical applications. Microbes can be engineered to produce a variety of commodity chemicals and biofuels with high yields to maximize economic returns. Microbial design strategies have considered how cells allocate resources so as to redirect cellular energy toward making compounds of interest [9–15]. Redirecting resources away from other processes can improve cellular product yields and thus decrease costs [16,17]. However, an added complication is that many industrial processes are stressful for engineered microbes, which mount stress-defense systems that further deplete cellular resources from product formation. The interplay of growth, metabolism, division, and stress defense remain murky, limiting engineering efforts [18,19].
Here, we studied how growth, division, and metabolism are normally coupled in cells by investigating a strain in which these processes have been decoupled. We previously characterized a series of Saccharomyces cerevisiae strains engineered to produce biofuel products from xylose, a pentose sugar abundant in plant biomass but not recognized by S. cerevisiae as a fermentable sugar [17,20–22]. A major goal for sustainable biofuel production is to utilize xylose and other carbon sources to maximize biomass conversion to products. Past work in our center found that engineering S. cerevisiae to ferment xylose anaerobically requires core xylose metabolism genes (encoding xylose isomerase, xylulokinase, and transaldolase [23,24]); however, introducing these genes is not enough to enable fermentation. Many groups have combined strain engineering with adaptive laboratory evolution to evolve xylose fermentation [25–30]. Cells require additional null mutations in oxidoreductase GRE3, iron-sulfur (Fe-S) chaperone ISU1, and RAS signaling inhibitor IRA2 [27,31]. We previously showed that these mutations help to rewire cellular signaling to unnaturally upregulate the growth-promoting Protein Kinase A (PKA) pathway in conjunction with Snf1 that usually responds to poor carbon sources [32]. We proposed that activating PKA and Snf1 promotes growth in the context of an otherwise unrecognized carbon source [32]. Coordinated induction of PKA and Snf1 allows cells to recognize xylose as a fermentable carbon source while enhancing growth and metabolism signals.
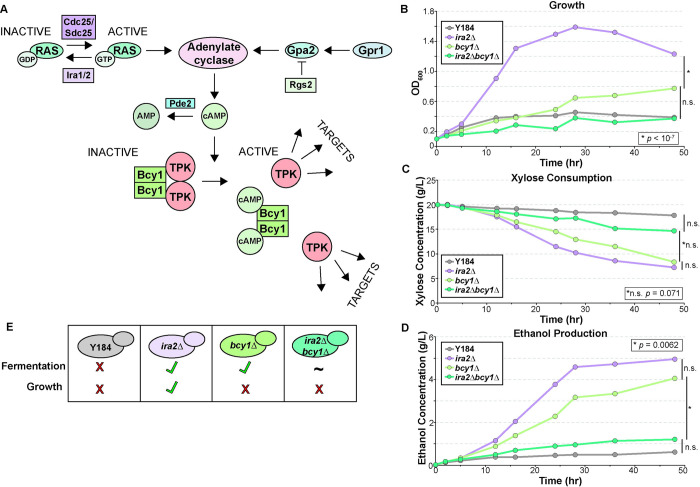
Although PKA activation is critical for anaerobic xylose growth and metabolism, during that study we made a surprising discovery: the mechanism of PKA up-regulation influences how growth and metabolism are coordinated. PKA can be activated by RAS activity, which stimulates adenylate cyclase to produce the allosteric regulator cAMP that binds and dissociates the PKA regulatory subunit Bcy1 (Fig 1A) [33]. In engineered yeast, activating PKA by IRA2 deletion, thus increasing RAS activity, enables rapid anaerobic xylose fermentation and growth on xylose as the sole carbon source. However, activating PKA by deleting the PKA regulatory subunit BCY1 allows rapid anaerobic xylose fermentation but with little to no growth (Fig 1B and 1C) [32]. In both strains, the effect is due to PKA upregulation since inhibition of PKA activity blocked both metabolism and growth [32]. Thus, deleting BCY1 in this strain background decouples growth and xylose metabolism for reasons that are not known. Importantly, other uncoupled biological processes related to PKA function have also been described [34], implying PKA’s central role in process coupling.
Fig 1. Activation of PKA is needed for xylose fermentation.
A. A brief overview of the PKA signaling pathway. B-C. Average (n = 6 biological replicates) growth (OD600, optical density) and (B) xylose concentration in the medium over time of parental Y184, ira2Δ, bcy1Δ, and ira2Δbcy1Δ strains grown anaerobically on rich medium containing xylose as a carbon source. Asterisks denote significant differences in profiles (p < 0.05, ANOVA); n.s. indicates ‘not significant’ (p > 0.05). D. Growth and fermentation capabilities of strains shown in B-C.
Here, we explored phenotypic consequences of IRA2 and BCY1 deletions to elucidate how cells normally coordinate growth and metabolism. We integrated transcriptomic, phospho-proteomic, and lipidomic analysis across a suite of strains with different mutations and growth/metabolism phenotypes. The results implicated the importance of lipid metabolism as a linchpin in the coordination of growth with metabolism: cells lacking BCY1 show unique transcriptomic and lipidomic responses that point to defects in lipid regulation. To uncover causal genes, we also performed adaptive evolution to re-evolve growth coordination in the bcy1Δ strain. Remarkably, the evolved strain acquired mutations in a PKA catalytic subunit TPK1 and phospholipid biosynthesis regulator OPI1, among other genes, and Opi1 was required for the growth-metabolism coupling in the evolved strain. These results suggest that PKA-dependent regulation of lipid metabolism is critical for growth, perhaps to coordinate membrane biogenesis and signaling with other cellular processes.
Results
We began by characterizing a suite of strains with different anaerobic xylose growth and fermentation capabilities. Parental strain Y184 harbors the xylose-metabolism gene cassette along with mutations in ISU1 and GRE3 but cannot grow on or metabolize xylose anaerobically (Fig 1B and 1C). Deleting IRA2 from this strain allows cells to grow on and metabolize xylose anaerobically. In contrast, deletion of BCY1 from Y184 permits rapid anaerobic xylose fermentation but with only minimal growth (Fig 1B, 1C, and 1E). Previous work studying growth over 90 hours validated limited growth of the bcy1Δ strain even after long periods [35]. We also investigated an ira2Δbcy1Δ double mutant. The double mutant was phenotypically similar to the bcy1Δ, although its xylose fermentation capabilities were highly variable across replicates, for reasons we do not understand but could pertain to extremely high PKA activity. As such, the double mutant was not statistically different from either the Y184 parent or the bcy1Δ strain. Nonetheless, we used it to investigate the genetics of PKA signaling through these different branches. The three strains grow indistinguishably on glucose (p> 0.05, S1A Fig) with similar glucose consumption (p > 0.05, S1B Fig) and ethanol production (p > 0.05, S1C Fig), indicating that these phenotypes are specific to anaerobic xylose conditions.
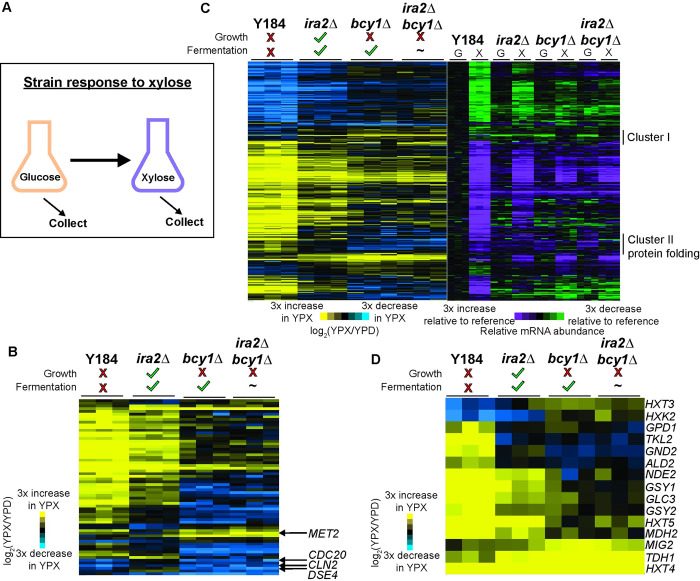
We started by comparing transcriptomic responses to identify transcripts whose abundance across the strain panel correlates with growth or anaerobic xylose metabolism. Cells were grown in an anaerobic chamber to mid-log phase on rich medium with glucose as a carbon source (YPD) then switched to rich medium containing only xylose (YPX) for three hours, long enough for the ira2Δ strain to resume growing (Fig 2A). We performed short-read sequencing to measure changes in transcript abundance after the glucose-to-xylose shift. To understand strain responses, we compared transcript abundances across strains grown under each condition; we also compared the fold change in transcript abundance within each strain responding to carbon shift. There were major differences in expression comparing the strains growing on xylose, whereas only mild expression differences were observed comparing strains grown on glucose (see Fig 2C, right panel). Correspondingly, strains do not differ substantially in their ability to grow anaerobically on glucose (S1 Fig) [35]. Thus, the differences in the fold-change expression response to the carbon shift are driven by differences in the xylose condition.
Fig 2. Few transcriptomic patterns correlate with anaerobic xylose growth.
A. Experimental overview. Strains were grown anaerobically in rich glucose medium to early/mid-log phase, then switched to anaerobic rich xylose medium for three hours. B. Expression of 65 genes whose log2(fold change) upon glucose to xylose shift is different (FDR < 0.05) in at least one of the three non-growing strains (Y184, bcy1Δ, ira2Δbcy1Δ) compared to ira2Δ. Genes (rows) were organized by hierarchical clustering across biological triplicates measured for each strain (columns). Genes discussed in the text are annotated on the figure. C. Hierarchical clustering of 292 genes whose log2(fold change) upon glucose to xylose shift is different (FDR < 0.05) between the non-fermenting Y184 strain and the two robustly xylose fermenting strains (ira2Δ, bcy1Δ). The blue-yellow heatmap on the left represents the log2(fold change) in expression upon glucose to xylose shift across biological triplicates (columns). The purple-green heatmap on the right represents the abundance of each transcript (rows) in each strain grown on glucose (G) or xylose (X), relative to the average (n = 3) abundance of that transcript measured in the Y184 YPD sample. Clusters I and II are described in the text. D. Expression of 15 genes from C) that have annotations linked to glycolysis, gluconeogenesis, TCA cycle, and carbohydrate storage.
Our expectation at the outset was two-fold. On the one hand, we expected to find expression changes common to the xylose fermenting ira2Δ and bcy1Δ mutants, but discordant in Y184 cells–these expression patterns may relate to xylose metabolism, since the Y184 strain is incapable of xylose metabolism [31,32]. On the other hand, expression patterns unique to the ira2Δ strain–the only strain capable of growing anaerobically on xylose–may reflect expression patterns related to growth.
Few gene expression patterns correlate strictly with growth phenotypes
Somewhat surprisingly, there were few genes whose expression correlated strictly with growth phenotypes. Only two genes showed xylose-responsive expression changes that were specific to ira2Δ cells compared to the other three strains analyzed as a group in the statistical model (FDR < 0.05; see Methods): daughter-cell-specific glucanase DSE4 and L-homoserine-O-acetyltransferase MET2. In fact, hierarchical clustering of all genes with a transcriptomic change in response to the carbon shift showed that the ira2Δ strain’s response to xylose shift was most similar to that of Y184 cells, even though one strain can grow on and anaerobically ferment xylose and the other cannot (S2A Fig and S1 Table). We next performed pairwise comparisons of the glucose-to-xylose fold-change responses between each strain and the ira2Δ strain, then combined the lists of genes identified in all three comparisons. This method identified 65 genes; however, investigating the expression patterns once again indicated that the ira2Δ response was most similar to Y184 cells but with weaker magnitudes of change (Fig 2B and S2 Table). This set of 65 genes was enriched for genes induced in the environmental stress response (iESR genes [36], p = 2x10-7, hypergeometric test). Many genes induced in the Y184 and ira2Δ strains, but largely not in bcy1Δ strains, included genes related to metabolism, including several in the mitochondrial TCA cycle and peroxisomal fatty-acid oxidation pathway, which may reflect that bcy1Δ strains are more likely to recognize xylose as a fermentable carbon source. We specifically investigated the set of 65 genes for those that encode cell-cycle regulators and kinases, since these may be involved in growth kinetics; however only three, six, or eight genes within these categories were differentially expressed in Y184, bcy1Δ, or ira2Δbcy1Δ cells, respectively, compared to ira2Δ cells in response to xylose shift (FDR < 0.05). The ira2Δ strain showed weakly lower expression of cyclin CLN2 and anaphase-promoting complex CDC20, whereas other strains showed strong reduction in expression (S3 Table). Transcript abundance for these genes is known to fluctuate during the cell cycle, thus while it is possible their expression influences growth arrest, it is likely that the expression of these genes reflects the expected difference between cycling (ira2Δ) and non-cycling (Y184, bcy1Δ, ira2Δbcy2Δ) cells. Expression of cell-cycle genes did not implicate arrest in a particular cell-cycle stage, consistent with early transcriptomic studies that showed that gene expression during cell-cycle arrest does not parallel expression of cells cycling through those phases [37].
Previous chemostat studies reported that repression of ribosomal protein (RP) and ribosome biogenesis (RiBi) genes is correlated with decreased growth, and these studies proposed that expression of these genes can predict cellular growth rate [38–41]. However, here we saw no correlation of RP and RiBi transcript abundance or response with growth phenotypes. The Y184 strain strongly repressed RP and RiBi genes upon xylose shift, which might be expected for a strain that arrests its growth, but so too did the ira2Δ strain, albeit with weaker magnitude of repression. Surprisingly, bcy1Δ and ira2Δbcy1Δ cells, whose growth is largely arrested after the xylose shift, showed little change in RP and RiBi transcripts compared to glucose-dependent growth (FDR <0.05, S2B Fig and S4 Table). These results reinforce past work from our lab that the expression of ribosome-associated genes does not necessarily parallel growth rate [42]. They further suggest that bcy1Δ strains cultured in xylose are unlikely limited by the abundance of RP and RiBi transcripts. Overall, while the non-growing strains have stronger repression of a few cell-cycle regulators when compared to the ira2Δ strain, there was not a clear gene expression pattern to describe why ira2Δ cells grow and bcy1Δ strains do not.
Few gene expression patterns correlate strictly with metabolism phenotypes
We next investigated shared gene expression changes related to robust anaerobic xylose fermentation. We compared expression in the Y184 strain responding to the xylose shift to the ira2Δ and bcy1Δ strains analyzed as a single group in the statistical model (we excluded the ira2Δbcy1Δ strain due to the variability of its fermentation phenotype, although it is capable of anaerobic xylose fermentation). This identified 292 differentially expressed genes (FDR < 0.05; S5 Table). Hierarchical clustering revealed that these genes typically had larger expression changes in Y184 and that those expression changes were progressively weaker across the strain series; once again, Y184 and the ira2Δ strain were more similar to one another than they were to the bcy1Δ strain (Fig 2C and S5 Table). Collectively, these genes were heavily enriched for genes in the ESR (p = 3.624x10-10, hypergeometric test).
Deeper interrogation revealed several small gene clusters of interest. Cluster I contained 19 genes induced in the ira2Δ and bcy1Δ strains but repressed in the Y184 strain. This group did not contain any functional enrichments; however, proteins encoded by several of these genes localize to the endoplasmic reticulum. Cluster II contained 31 genes induced in Y184 and either unchanged or repressed in both ira2Δ and bcy1Δ strains. This group was enriched for genes involved in protein folding (p = 9.49x10-6, hypergeometric test) and included the Hsp90 chaperone and cochaperone genes HSP82, STI1, and AHA1, as well as the mitochondrial matrix protein chaperone HSP10. Hsp90 can act as a signal transducer for alternative carbon source metabolism [43], again suggesting that Y184 does not recognize xylose as a fermentable carbon source.
We specifically interrogated the 292 genes for those involved in glycolysis, gluconeogenesis, TCA cycle, and carbohydrate storage, predicting that differences in expression would relate to altered xylose metabolism capabilities. This identified 15 genes with functional annotations linked to at least one of these processes (Fig 2D and S6 Table). The xylose fermenting strains shared expression at several hallmark genes. For example, Y184 cells strongly induced hexose transporter HXT5, normally induced by non-fermentable carbon sources, whereas the three mutant strains showed a weaker induction (FDR = 4.25x10-5). The glucose-repressed aldehyde dehydrogenease ALD2 was induced in Y184 and either did not change (ira2Δ cells) or was repressed (bcy1Δ cells) upon the switch to xylose (FDR = 4.32x107). Furthermore, glucose-induced transcriptional repressor MIG2 showed stronger induction in the xylose fermenting strains, and especially bcy1Δ strains, compared to Y184 (FDR = 0.047). These data are all consistent with the hypothesis that the xylose fermenters recognize xylose as a fermentable carbon, whereas Y184 activates a carbon-starvation response.
Regulatory analysis reveals strain-specific differences in carbon, iron, and lipid gene control
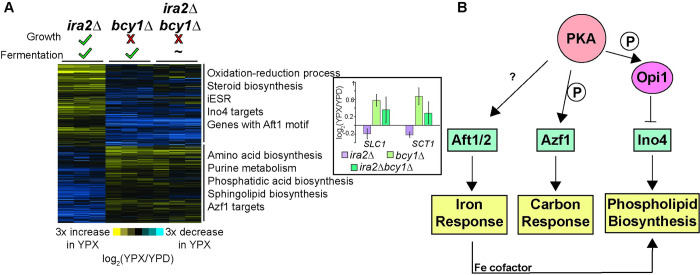
We next focused on understanding how growth and metabolism are decoupled in the bcy1Δ strain, and we thus directly compared its expression to that in ira2Δ cells. We focused on genes whose expression changes in response to the xylose shift were in opposing directions to implicate processes involved in decoupling growth and metabolism (Fig 3A and S7 Table, see Methods). Among the identified genes, we scored enrichment of functional terms as well as known targets of transcriptional regulators (S8 Table). We also used motif analysis to discover shared sequence motifs upstream of genes uniquely induced or repressed in the bcy1Δ strain, and then matched those to known transcription factor binding sites (see Methods). We identified 654 genes differentially expressed in bcy1Δ cells and with a fold-change in the opposite direction as ira2Δ cells upon the glucose-to-xylose shift (Fig 3A and S7 Table). Importantly, only 82 genes (12.5%) showed significant differences in basal gene expression when cells were grown on glucose (S3A Fig), indicating that the majority of genes are identified due to differences in response to xylose shift.
Fig 3. Genes uniquely expressed in the bcy1Δ strain implicate an integrated response to xylose metabolism and growth coupling.
A. Expression of 654 genes whose log2(fold change) upon glucose to xylose shift is different (FDR < 0.05) between the ira2Δ and bcy1Δ strains and whose expression change is in the opposite direction across the two strains (see Methods for details). Significant functional enrichments are annotated next to the two main clusters (p < 10−4, hypergeometric test). Bar graph inset represents the log2(fold change) of the two phosphatidic acid biosynthesis enzymes in this group, see text for details. B. Regulatory relationships between transcription factors whose targets or known binding sites were enriched in (A). Documented PKA-dependent phosphorylation is indicated by a P. See text for details.
The results implicated several regulators, some with prior connections to anaerobic xylose fermentation. 318 genes induced in the bcy1Δ strain shifted to xylose, but repressed in the ira2Δ cells, were enriched for amino acid and sphingolipid biosynthesis genes, as well as targets of the carbon-responsive Azf1 transcription factor (p < 10−4, hypergeometric test). Previous work from our lab implicated Azf1 in anaerobic xylose fermentation, and indeed, we showed that the over-expression of AZF1 in an ira2Δ strain enhances the rate of anaerobic xylose utilization [32]. Additionally, PKA has been implicated in Azf1 phosphorylation [44]; together with the fact that the AZF1 gene is uniquely induced in the bcy1Δ strain suggest its functional importance in xylose metabolism (see Discussion).
In contrast, several regulators were implicated by the 336 genes uniquely repressed in the bcy1Δ strain. These included genes harboring upstream binding sites of the iron-responsive Aft1/2 transcription factors (S3B Fig) and known targets of transcriptional activator Ino4 that responds to inositol for phospholipid biosynthesis (Fig 3A; see more below). Iron is an important cofactor of many enzymes, including those involved in mitochondrial respiration, lipid biogenesis, and amino acid biosynthesis, all of whose genes were among the differentially regulated genes studied here. Additionally, Aft1/2 regulation and the iron regulon have been linked with PKA activity; however, direct interactions remain to be identified [45]. Interestingly, Aft1/2 and Azf1 both are both connected to the regulator Mga2, which controls lipid and hypoxia genes and that we previously showed enhances anaerobic xylose fermentation when over-expressed in ira2Δ cells [32,46] (see Discussion). Targets of the Ino2/4 regulators that respond to inositol for phospholipid biosynthesis were also present in this gene set; while a majority of the targets identified here were repressed in the bcy1Δ strain, some of the known targets were repressed in the ira2Δ strain but induced in the bcy1Δ mutant (S3C Fig and S9 Table). This may reflect the complexities of the genes’ regulation by other factors. Nonetheless, Ino2/4 targets were enriched among the genes oppositely regulated in the bcy1Δ versus ira2Δ strain. Overall, these results provide an interesting link between PKA signaling, carbon and iron responses, and lipid metabolism (Fig 3B).
The presence of many lipid biosynthesis genes in this gene set and the highly regulated role of lipids in cell growth and proliferation prompted a deeper investigation of lipid metabolism genes. The bcy1Δ strain repressed genes involved in ergosterol biosynthesis and some targets of Ino2/4 that are involved in phospholipid metabolism (Figs 3A and S3C). This response is consistent with the model that Ino2/4 activity is reduced. However, the bcy1Δ strain also induced some genes involved in the synthesis of phosphatidic acid (PA) (Fig 3A inset), which normally promotes Ino4 activity by sequestering Ino4’s inhibitor Opi1 to the ER membrane [47]. This response suggests that some connection between PA, Opi1, and Ino4 is disrupted in the absence of BCY1. PKA is known to regulate the Ino2/4 pathway through direct phosphorylation of Opi1 to increase its inhibitory activity [48]. Together, these results raised the possibility that the bcy1Δ strain has important differences in lipid metabolism and perhaps composition, which could be modulated by differences in PKA activity in this strain.
Lipidomic and phosphoproteomic analyses show disrupted phospholipid metabolism in bcy1Δ cells
Since the transcriptomic responses implicated differences in lipid metabolism, we investigated the lipidomic composition of our strains. Strains were grown in a similar design as the transcriptomic analysis, where anaerobically glucose-grown Y184, ira2Δ, bcy1Δ, and ira2Δbcy1Δ cells were shifted to anaerobic xylose media for three hours before lipids were analyzed by mass spectrometry (see Methods). We detected over 4000 lipid species including 239 that were confidently assigned to a particular lipid class (S10 Table). All detected lipid species were included in the statistical analysis to obtain a wholistic understanding of lipidome differences between the strains. We again sought to find lipidomic profiles correlated with xylose metabolism and growth, xylose metabolism but no growth, and no xylose metabolism or growth.
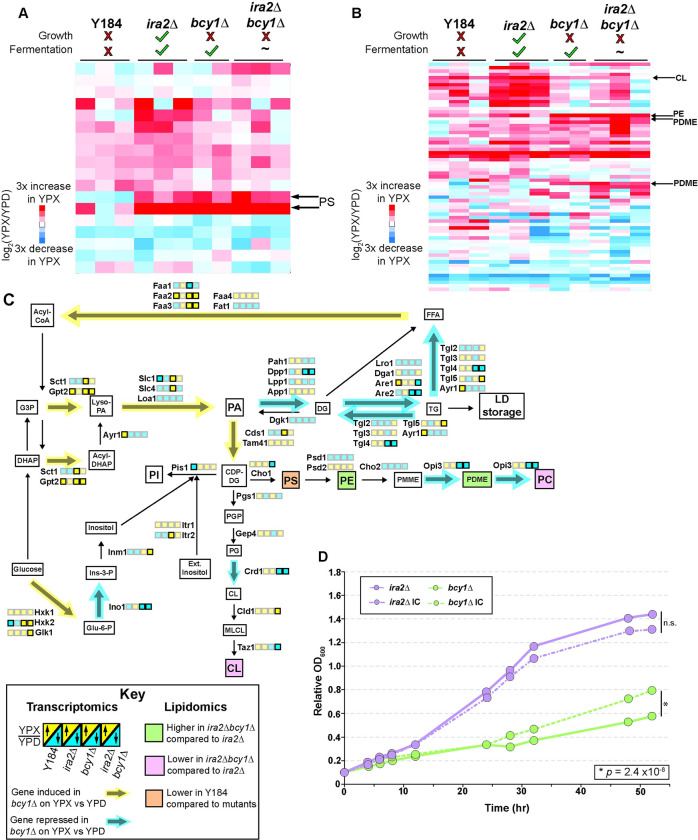
We compared the Y184 strain to the three strains with upregulated PKA activity and identified 18 lipids whose change in abundance upon a shift to xylose significantly differed in Y184 cells. This group included phosphatidylserine (PS) species (Fig 4A and S11 Table). Interestingly, all three mutants increased the abundance of these PS species when shifted to xylose, whereas Y184 cells decreased the abundance of one and failed to induce the other to the same degree as the mutants. The gene encoding the PS synthase CHO1 was strongly induced in Y184 cells, indicating that the decrease in PS in Y184 cells is unlikely due to decreased CHO1 expression. Instead, we analyzed previous phosphoproteomic data from our lab and discovered that Cho1 was phosphorylated to a much higher degree in the Y184 strain on serine 46 (|log2FC| > 1, Table 1), a known PKA site that inhibits Cho1 activity [49]. Together, these results indicate PKA-dependent inhibition of PS synthesis in Y184 cells.
Fig 4. bcy1Δ strains show altered phospholipids after anaerobic xylose shift.
A-B. Abundance of lipids (rows) with a significant difference in log2(fold change) upon anaerobic glucose-to-xylose shift in (A) Y184 compared to PKA pathway mutants (ira2Δ, bcy1Δ, ira2Δbcy1Δ) analyzed as a group in the statistical model or (B) ira2Δ cells compared to ira2Δbcy1Δ cells. Lipids of interest are annotated. C. Partial phospholipid biosynthesis pathway with transcriptomic and lipidomic data represented. Yellow-blue boxes next to each enzyme name represent the average log2(fold change) in transcript abundance upon glucose-to-xylose shift for each strain, as outlined in the key. Significant differences compared to the ira2Δ strain (FDR < 0.05) are represented in sharp, bolded boxes, whereas insignificant differences are translucent. Colorized pathway arrows (yellow: induced, blue: repressed) represent the predominant transcript patterns for that enzymatic step when comparing the bcy1Δ and ira2Δ strains. Lipids whose fold-change in abundance is different in specific strains are according to the key. Lipid abbreviations: FFA–free fatty acids; PA–phosphatidic acid; DG–diacylglycerol; TG–triacylglycerol; PI–phosphatidylinositol; PS–phosphatidylserine; PE–phosphatidylethanolamine; PMME–monomethyl-phosphatidylethanolamine; PDME–dimethyl-phosphatidylethanolamine; PC–phosphatidylcholine; CL–cardiolipin. D. Average (n = 4) change in OD600 of ira2Δ and bcy1Δ grown anaerobically in rich xylose medium either in the absence (solid lines) or presence (dashed lines, IC) of inositol (75 μM) and choline (10 mM) (* indicates p = 2.4 x 10−6, ANOVA).
Table 1. Phosphorylation changes of phospholipid biosynthetic enzymes.
| Protein | Residue | Av. logFC (bcy1Δ-ira2Δ) | Av. logFC (ira2Δbcy1Δ-ira2Δ) | Av. logFC (Y184-ira2Δ) | Av. logFC (Y184-bcy1Δ) |
|---|---|---|---|---|---|
| Are1 | S45 | 0.445 | 0.695 | 0.635 | 0.19 |
| Cho1 | S46 | -0.35 | -0.37 | 1.08 | 1.43 |
| Hxk1 | Y270 | 1.45 | 1.41 | 0.74 | -0.71 |
| Hxk1 | S262 | 1.57 | 1.28 | 1.1 | -0.47 |
| Hxk1 | S293 | 0.54 | 0.1 | 1.44 | 0.9 |
| Hxk1 | S158 | -1.3 | -0.74 | 0 | 1.3 |
| Hxk2 | S385 | -0.65 | -0.68 | 1.17 | 1.82 |
| Hxk2 | S158 | -0.58 | -0.38 | 0.01 | 0.59 |
| Pct1 | T59 | 0.19 | 0.94 | -0.24 | -0.43 |
| Pah1 | S166 | -0.52 | -0.5 | 0.3 | 0.82 |
| Pah1 | S823 | -1.53 | -1.49 | 0.84 | 2.36 |
| Pah1 | S168 | -0.33 | -0.44 | 0.3 | 0.62 |
| Are2 | S176 | 0.78 | 0.44 | 0.63 | -0.15 |
| Slc4 | S512 | 0.58 | 1.01 | 1.01 | 0.42 |
We next compared lipidomic profiles in the growing ira2Δ strain shifted to xylose to the bcy1Δ and ira2Δbcy1Δ strains that do not grow. Due to limited statistical power (caused by replicate variation in one of the three bcy1Δ strain replicates), we compared the ira2Δ response to ira2Δbcy1Δ cells, whose response was highly similar to two out of the three bcy1Δ strain replicates. One caveat of this analysis is that the ira2Δbcy1Δ strain displays a variable anaerobic-xylose fermentation profile; nonetheless, given the similarity to bcy1Δ phenotypes, the high reproducibility of the double mutant’s transcriptomic and lipidomic profiles suggests a good representation of hyper-active PKA signaling. It is possible that this analysis may miss some lipidomic changes related to the variation in ira2Δbcy1Δ metabolism profiles. Even so, we identified 67 lipids whose fold-change was significantly different in ira2Δbcy1Δ cells upon xylose shift versus ira2Δ cells (FDR < 0.05, Fig 4B and S12 Table). The analysis confidently classified six of the lipids, including phosphatidylethanolamines (PE), phosphatidyl dimethylethanolamines (PDME), and cardiolipins (CL).
PE and multiple PDME species were more abundant in the ira2Δbcy1Δ strain exposed to the shift compared to ira2Δ cells (FDR < 0.05, Fig 4B). These differences were particularly interesting because PE is further metabolized to PDME and then to phosphatidylcholine (PC), the most abundant phospholipid in the cell, through three consecutive methylation reactions by Cho2 and Opi3, respectively (Fig 4C) [50]. While the CHO2 transcript was not differentially expressed between ira2Δ and bcy1Δ strains, OPI3 was: ira2Δ cells shifted to xylose induced OPI3 expression, whereas bcy1Δ and ira2Δbcy1Δ cells repressed it (FDR = 2.45x10-12 and FDR = 6.22x10-13, respectively). Previous studies suggest that blocking PC synthesis through OPI3 deletion, but not CHO2 deletion, inhibits growth due to the accumulation of phosphatidyl monomethylethanolamine (PMME) and insufficient PC production [51]. To investigate effects on PC, we analyzed all PC lipid moieties in the dataset; PC lipids were reproducibly lower in abundance after the xylose shift in bcy1Δ cells when compared to ira2Δ cells (p = 0.000419, ANOVA; S4 Fig and S13 Table). We propose that the bcy1Δ strain experiences a bottleneck in that pathway leading to PC synthesis from PE, which may impact its ability to grow on xylose (see Discussion).
Among other lipids whose abundance was influenced by BCY1 deletion and xylose shift was cardiolipin, a major component of mitochondrial membranes critical for a variety of functions including acetyl coA synthesis, TCA cycle, iron metabolism, arginine metabolism, and protein import [52]. Interestingly, cardiolipin abundance was reduced in the ira2Δbcy1Δ strain upon xylose shift compared to ira2Δ cells. The difference is underscored by transcriptomic differences, since several cardiolipin biosynthetic genes were induced in ira2Δ cells but repressed or induced to a weaker extent in bcy1Δ and ira2Δbcy1Δ strains (FDR < 0.05). Additionally, production of PS, PE, and PC is dependent on properly functioning mitochondrial membranes as PS is shuttled into the mitochondria and converted to PE by the phosphatidylserine decarboxylase Psd1, before PE is shuttled back to the ER. Thus, the effects of cardiolipin reduction in bcy1Δ strains are further compounded by impacting other branches of phospholipid biosynthesis.
We expected to see differential abundance of PA in ira2Δbcy1Δ cells versus ira2Δ cells, since bcy1Δ and ira2Δbcy1Δ strains induced some PA biosynthesis genes whereas ira2Δ cells do not (Fig 3A). While there were no significant differences in PA moieties between the strains (FDR > 0.05), we did identify altered phosphorylation status of the PA phosphatase enzyme Pah1 (S823; Table 1). Pah1 converts PA to diacylglycerol, which is funneled into storage lipids [50]. Phosphorylation of serine 823 is significantly lower in the bcy1Δ and ira2Δbcy1Δ strains compared to the ira2Δ strain (log2(fold change) < -1). Interestingly, this serine has not been previously annotated as a phosphorylated residue (BioGRID version 4.4.213) [53], but it is within a potential PKA consensus site (RRxxS/T). PKA is known to phosphorylate Pah1 at another residue not captured in our dataset to inhibit its activity [54]. Our results raise the possibility that S823 regulates Pah1 activity in a manner that affects PA in these strains. Overall, the differences seen in PE, PDME, and PC abundances, as well as differences in transcript abundance and phosphorylation status of phospholipid biosynthesis enzymes, suggest a bottleneck in the pathway in the bcy1Δ strains that may inhibit their ability to proliferate on xylose (see Discussion).
Supplementation with phospholipid precursors only modestly improves growth
We questioned if supplementing xylose medium with phospholipid precursors, particularly inositol and choline that can be funneled into phospholipid biosynthesis via the Kennedy Pathway, may bypass a possible bottleneck and thus rescue the bcy1Δ strain’s growth. We therefore grew bcy1Δ and ira2Δ strains anaerobically in xylose medium with and without choline and inositol supplementation (we included inositol since the INO1 gene is repressed in bcy1Δ cells) (Fig 4C and S5 Table). After 52 hours of growth in supplementation, bcy1Δ cells experienced a very modest but statistically significant growth improvement (p = 2.4 x 10−6, ANOVA; Fig 4D), whereas the ira2Δ strain did not. While the bcy1Δ strain’s inability to grow anaerobically on xylose cannot be fully explained by a deficiency in phospholipid precursors, the modest improvement implicates it as a contributing factor to the phenotype.
Growth and metabolism can be genetically recoupled through directed evolution
We took a second approach to identify pathways and processes responsible for growth coordination in bcy1Δ strains by conducting adaptive laboratory evolutions to recouple xylose-dependent growth and metabolism. The bcy1Δ strain was first grown anaerobically in rich medium supplemented with 2% glucose to accumulate mutations [55], then the culture was seeded into fresh anaerobic medium containing 2% xylose and 0.1% glucose and passaged periodically for ~35 generations until the culture showed robust changes in cellular density over time (see Methods). Single colonies were isolated and characterized for their growth and fermentation capabilities, and genetic changes were identified through whole genome sequencing (see Methods). Three independent evolutions were performed, and several colonies were selected at different stages of the evolutions.
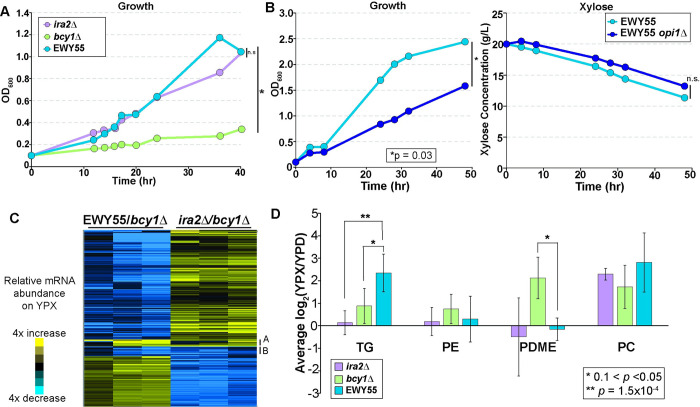
In all three experiments, we identified mutants with recoupled growth and metabolism despite the absence of BCY1, evident by their robust anaerobic growth on xylose medium compared to the ira2Δ strain (Figs 5A, S5 and S6). Interrogating the genome sequences identified multiple mutations in each strain, along with copy-number variations and aneuploidy in several of the evolved lines (Table 2). Only evolved mutations impacting the coding sequence of a gene were analyzed further. Interestingly, there was no genetic change common to all evolved strains, strongly suggesting multiple routes to recoupling growth and metabolism in the absence of BCY1. Four of the characterized strains from the three experiments regained growth rates comparable to and statistically indistinguishable from ira2Δ cells (p > 0.05, ANOVA), including EWY55 from the first culture, EWY87-1 and EWY87-3 from the second culture, and EWY89-3 from the third evolution culture (S6A and S6B, S6E Fig). Strains EWY89-1 and EWY89-2 showed modest growth on xylose but did not differ significantly from the bcy1Δ strain (p > 0.05, ANOVA; S6C and S6D Fig). Genetic changes for all evolved strains are listed in Table 2.
Fig 5. Directed evolution recoupled growth and metabolism on xylose.
A. Average (n = 3) change in OD600 of ira2Δ, bcy1Δ, and EWY55 strains grown anaerobically in rich xylose medium (*, p < 10−4, ANOVA; n.s., not significant). B. Change in OD600 (left panel) and xylose concentration (right panel) over 48 hours of EWY55 and EWY55 opi1Δ strains grown anaerobically on rich xylose medium. (*, p < 0.05, ANOVA). C. Expression of 233 genes whose transcript abundance during growth on xylose was significantly different in EWY55 and/or ira2Δ strains compared to the bcy1Δ strain (FDR < 0.05), visualized by hierarchical clustering. Data represent the log2 transcript abundance in each strain grown anaerobically in xylose compared to bcy1Δ strain. Cluster A (9 genes) and B (13 genes) are annotated, see text for details. D. Bar plot of the average and standard deviation log2(fold change) (n = 3) in lipid abundance of key lipids with reproducible differences 1.5-fold or greater in EWY55 compared to ira2Δ or bcy1Δ strains. Asterisks denote significant differences by ANOVA.
Table 2. Genetic changes in evolved bcy1Δ strains.
| Gene | Nucleotide change | Amino acid change | Chromosome Duplications | |
|---|---|---|---|---|
| EWY55 | OPI1 | T715G | S239A | Chr. I |
| TPK1 | G829C | A277P | ||
| TOA1 | T354A | N118K | ||
| RIM8 | C1591T | Q531* | ||
| EWY87-1 | - | - | - | Chr. VIII |
| EWY87-3 | RPA43 | A751G | S251N | - |
| EWY89-1 | HSC82 | G235C | D79H | Chr. X, XVI |
| EWY89-2 | - | Chr. I, X, XVI, XIV560217-625584 | ||
| EWY89-3 | HSC82 | G235C | D79H | Chr. I, IX, X, XVI, XIV560217-625585 |
Strain EWY55 was particularly interesting. This strain harbored nonsynonymous mutations in several genes, including PKA catalytic subunit TPK1, the negative regulator of phospholipid genes, OPI1, described above, RIM8 that is required for anaerobic growth [56], and TFIIA large subunit TOA1 (Table 2). The OPI1 mutation was especially interesting because Opi1 was implicated in the phospholipid transcriptomic analysis above (Fig 3) and because the mutation changes a known phosphorylation site, serine 239, to alanine (Table 2). CKII has been reported to phosphorylate this site and was previously shown to activate Opi1 [57]. This poses the question of whether Opi1 is aberrantly regulated in the bcy1Δ strain, and if this is responsible for its lack of growth on xylose.
To identify causal alleles responsible for recoupling growth and metabolism in the EWY55 strain, we performed single gene deletions and allele swaps in the bcy1Δ and EWY55 strains (see Methods). This strain background is derived from a wild isolate that is less genetically amenable than laboratory strains [58], and we were unable to recover TPK1 deletion in either strain despite many efforts. Deletion of RIM8 or TOA1 did not impact the growth of EWY55 cells, nor did substituting the parental alleles into the evolved strain (S5C Fig). However, deletion of the evolved opi1 gene partially but significantly reduced anaerobic xylose growth of the EWY55 strain in liquid medium (Fig 5B). Importantly, the strain retained robust xylose fermentation, indicating that Opi1 plays a role in the coupling of growth and metabolism (see Discussion). Complementation experiments to swap strain alleles were not successful, since introducing even the empty vector into this strain complemented anaerobic xylose growth on a plate for reasons that are not clear but may suggest that the cells grow differently during drug selection (S5C Fig). While we cannot be sure OPI1 is the causal gene, our results indicate that the genetics modulating this trait is complex and may result from different evolutionary paths, but at least in EWY55 is likely to include a role for evolved Opi1 function.
Transcriptomic and lipidomic analysis in the evolved strain reveals altered phospholipids
To further characterize the evolved EWY55 strain, we performed another transcriptomic and lipidomic experiment as described above (see Methods) with the main goal of identifying if the evolved EWY55 strain reverted its gene expression and lipid composition to that of the ira2Δ strain. Surprisingly, the EWY55 strain did not recapitulate the ira2Δ gene expression or lipid abundance profiles at most entities. We identified 297 transcripts less abundant in EWY55 growing anaerobically on xylose compared to the bcy1Δ strain (FDR < 0.05; S14 Table), and these were enriched for genes involved in mitochondrial functions, such as electron transport chain, oxidation-reduction, and targets of the HAP2/3/4/5 complex; genes involved in phospholipid metabolism; and genes involved in ergosterol synthesis (p < 0.05, hypergeometric test, see Methods). Many of these processes were significantly affected in our original comparison of the bcy1Δ and ira2Δ strains. Additionally, 93 genes with higher abundance in the EWY55 compared to the bcy1Δ cells (FDR < 0.05; S14 Table) were enriched for ribosomal protein genes and genes involved in translation and sulfate assimilation (p < 10−7, hypergeometric test), processes important for rapid growth. Since EWY55 cells recapitulated the xylose-dependent growth seen in ira2Δ cells, we next asked if its expression changes recapitulated ira2Δ patterns relative to bcy1Δ cells–surprisingly, most did not (Fig 5C). This indicates that the evolved EWY55 did not recouple growth and metabolism under anaerobic xylose conditions via reverting to the ira2Δ strain’s expression patterns. There were a few exceptions, including 22 transcripts of diverse functions (Fig 5C, Clusters A and B) in which expression differences in EWY55 recapitulated those seen in ira2Δ cells compared to the bcy1Δ strain (S15 Table; FDR < 0.05). While the role of these expression changes will require future study, it is intriguing that these clusters included several targets of the glucose-responsive transcription factor Rgt1 and the Sok2 regulator that responds to starvation and hypoxia; both genes have connections to PKA signaling [59–62]. We were particularly interested in phospholipid biosynthesis genes, given all the connections to this pathway throughout our studies. In general, EWY55 cells showed lower transcript abundances of phospholipid biosynthesis genes compared to the bcy1Δ strain grown anaerobically on xylose (S16 Table), making its expression even more divergent from the ira2Δ strain.
The phospholipid composition further supports the unique changes of the EWY55 strain that permit recoupled growth and metabolism on xylose. The EWY55 strain showed significantly greater abundance of the storage lipid triacylglycerol (TG; Fig 5D and S17 Table; p < 10−4, ANOVA). Importantly, EWY55 had significantly lower levels of PDME and trended towards higher levels of PC compared to bcy1Δ cells, recapitulating the pattern in ira2Δ cells (Fig 5D and S17 Table). These results are consistent with the hypothesis that the evolved EWY55 strain altered the pathway compared to bcy1Δ cells. Together, our results underscore the complexity of responses to xylose growth and metabolism across ira2Δ cells, the parental bcy1Δ strain, and EWY55 (see Discussion).
Discussion
We began this work with two primary goals: to identify signatures of xylose-dependent growth and metabolism across a suite of strains with varying capabilities and to elucidate the mechanism through which growth and metabolism are decoupled in cells lacking BCY1. One key result from our work is that there is no obvious gene expression signature associated with the ability to grow anaerobically on xylose (Fig 2). While we did identify a handful of cell-cycle genes whose expression was consistent with cycling in the ira2Δ strain, there were no clear signatures correlated with growth. This was especially interesting in the case of ribosome-related genes, since there has been much debate about whether the level of RP transcripts underlies growth rate [38–42]. In chemostat experiments where growth is limited by nutrient restriction, the abundance of RP and RiBi genes correlates with growth rate, consistent with one set of long-standing models of growth limitations in bacteria [63–68]. However, other seminal studies focusing on stress conditions suggest that growth during stress is not limited by ribosome production [42,69–73]. Our results show clearly that expression of RP and RiBi genes is higher in the non-growing bcy1Δ strains than dividing ira2Δ cells (S2B Fig). In contrast, the EWY55 strain that recovers anaerobic growth on xylose shows higher expression of ribosome-related genes, perhaps supporting rapid division. Together, our results add to a growing body of work that shows that, although production of ribosome components is often correlated with growth rate, division dynamics cannot be universally predicted by RP and RiBi transcript abundances.
However, transcriptomic patterns did implicate an interconnected network of expression differences specific to the bcy1Δ strain, and in turn the evolved EWY55 strain, connected to PKA signaling (Fig 3). The affected network implicates mitochondrial function, iron response, carbon metabolism, and phospholipids. We propose that these processes are normally coordinated by PKA signaling in a manner that requires the regulatory subunit Bcy1. Our results are consistent with prior implications that these processes are involved in anaerobic xylose fermentation. Targets of the carbon-responsive transcription factor Azf1 were altered in the bcy1Δ strain upon xylose shift compared to the ira2Δ strain (Fig 3A). We previously showed that altering expression of this transcription factor affects xylose fermentation rates and growth in an ira2Δ strain, and this was surprisingly connected to the ER-localized transcription factor Mga2 [32]. While Mga2 targets were not statistically enriched in comparisons here, 42% of the genes whose promoter is bound by Mga2 (11/26 genes) differed in expression between bcy1Δ and ira2Δ cells. Additionally, Aft1/2 activity and localization is dependent on Mga2 presence, thus adding another possible connection for Mga2 in our proposed regulatory network [74] (Fig 3B). Our results here strongly suggest that deletion of BCY1 naturally augments the transcription factors’ abundance and/or activity. These factors may indirectly alter mitochondrial and/or iron homeostasis. In fact, deletion of the iron-sulfur scaffold protein ISU1, an important sensor of iron availability, is required for anaerobic xylose metabolism [26,31]. Why ISU1 deletion is required for xylose fermentation remains unclear, but one possibility is that it aids in metabolic rewiring influenced by the iron regulon, the Aft1/2 transcription factors, and altered levels of PKA activity [45,75–77].
Remarkably, PKA is directly connected to all these processes. Past work implicated PKA in directly phosphorylating Azf1 [32,44]. While a direct link between PKA and Aft1/2 activity has yet to be identified, PKA catalytic subunit Tpk2 is required to repress the high-affinity iron uptake pathway under standard conditions [76]. Additionally, Ira2 can localize to mitochondria, suggesting that PKA can also localize to this organelle [78]. In fact, PKA is found at the mitochondria of higher eukaryotes [79], suggesting that yeast PKA may also localize to the mitochondria. Together, our results suggest that upregulated PKA activity is required for xylose fermentation and can occur via either IRA2 or BCY1 deletion, but deletion of BCY1 produces stronger effects that underscore its higher per-cell rate of xylose fermentation [32].
A fundamental aspect of BCY1 deletion is that cells can no longer grow robustly despite enhanced xylose metabolism. Our integrated analysis points to a defect in phospholipid flux or metabolism as a major contributor to this decoupling. First, the bcy1Δ strains showed altered
gene expression, including Ino2/4 targets such as INO1 (Figs 3A, 4C and S3C), that pointed to differences in phospholipid metabolism. Second, we found that bcy1Δ strains grown anaerobically on xylose display an altered lipid profile that implicates altered metabolism in the PE-PDME-PC pathway, along with phosphorylation differences on key phospholipid enzymes (Table 1). Finally, re-evolving a coupling between anaerobic-xylose growth and metabolism in the bcy1Δ parent implicated mutations in PKA subunit TPK1 and the Ino2/4 repressor OPI1 (Table 2), which is known to be directly regulated by PKA phosphorylation [48]. Opi1 has complex roles in regulating phospholipids, including during the switch to invasive growth depending on nutrients [80], a process also regulated by the RAS/PKA pathway [81–85]. While we were unable to elucidate the exact role of these alleles, our results suggest that the OPI1 mutation may alter Opi1 regulation, especially given that the identified mutation in Opi1 occurs at a known CKII kinase site that regulates Opi1 activity [57]. Our past network inference across this panel of engineered strains revealed altered phosphorylation of CKII targets [32]. Finding that complete deletion of the mutated OPI1 allele reduced growth of the evolved EWY55 strain on xylose (Fig 5B) suggests altered Opi1 activity in the bcy1Δ strain is somehow resolved by mutation of this CKII site. We propose that an interplay between PKA and possibly CKII affect Opi1 regulation in the bcy1Δ strain, and that this interplay is important for growth coupling.
Importantly, phospholipid metabolism is required for growth and division. Cells must generate enough phospholipids to support membrane biogenesis [4,86–91]. Furthermore, phospholipids function in inter-organelle communication, connecting the ER and mitochondria via the ER-mitochondria encounter structure (ERMES). Impairment of this structure and inter-organelle communication is known to cause diverse mitochondrial phenotypes and disrupt phospholipid biosynthesis [92,93], connecting phospholipid metabolism to mitochondrial functions, including xylose flux [31]. One possibility is that impaired regulation of Opi1 and Ino2/4 in bcy1Δ cells disrupt growth in the bcy1Δ strain due to insufficient levels of growth-supporting lipids (PC) via decreased production or increased recycling. But another possibility is that accumulation of methylated PE intermediates during the conversion to PC create a toxic buildup coupled with insufficient PC (Fig 4C). Ishiwata-Kimata et al. (2022) [51] found that accumulation of PMME leads to a growth defect by triggering the unfolded protein response and growth arrest. Accumulation of PDME in the bcy1Δ strain (Fig 4B and 4C) may also lead to ER stress, preventing growth paired with interfered ER-mitochondrial communication. Importantly, the evolved EWY55 strain does not share the bcy1Δ strain’s accumulation of PMDE, leading us to propose that EWY55 cells have overcome the possible bottleneck in PC synthesis (Fig 5D). Future studies analyzing pathway flux are needed to fully confirm the presence of and recovery from a bottleneck in phospholipid biosynthesis.
A major remaining question is how deletion of BCY1, but not IRA2, decouples growth from metabolism specifically under the conditions studied here. One possibility is that BCY1 deletion upregulates PKA activity to a higher level than deletion of IRA2, whose activation of PKA is indirect via cAMP regulation [33]. PKA activity over some threshold could cause decoupling, as deletion of BCY1 is well characterized to sensitize cells to environmental stressors [94]. An alternate model is that localized cAMP production could influence when and where PKA is active in ira2Δ cells. cAMP exists in concentration gradients in cells to control the subcellular location of active PKA [95–97]. It is possible areas with low cAMP concentration locally inactivate PKA in the ira2Δ strain, whereas BCY1 deletion leads to wholesale activation of PKA throughout the cell. Fitting with this model, BCY1 deletion inhibits growth and metabolism on non-fermentable carbon sources, causing cell death during the diauxic shift and stationary phase, likely from uninhibited PKA [98,99]. However, a third possibility is that loss of BCY1 leads to misdirection of PKA activity. PKA can be directed to subcellular targets in higher eukaryotes via A-kinase anchoring proteins (AKAP) that bind to and direct localization of PKA [100]. While yeast do not possess orthologs of AKAPs, functional analogs have been proposed including Bcy1 itself [79,101,102]. Anaerobic xylose growth and metabolism may be decoupled in bcy1Δ strains via disrupted subcellular localization and substrate interactions of PKA that are coordinated by Bcy1. Additionally, Bcy1 is reported to interact with fatty acid synthases subunits (Fas1/2) [102], implying a direct, physical connection between PKA and lipid biosynthesis. While future studies of PKA localization and substrate interactions are needed to confirm this model, our results show that Bcy1 plays a special role in coordinating PKA activity. It is also possible that other signaling pathways, such as TORC1, may be involved in modulating growth and metabolism phenotypes under anaerobic xylose conditions [103], though our work thus far has not investigated a role for TORC1 in these phenotypes. Finally, this study has been solely focused on the RAS branch of PKA activation, but it is also possible that the Gpa2 branch possesses an important role in growth and metabolism coupling [104]. It is clear that more studies are required to obtain a complete understanding of this mechanism.
It is evident from this and many other studies that cells have deeply intertwined the regulation of multiple processes, and disrupting one can have dramatic impacts on many others. Our results here and in previous work implicate the importance of regulatory rewiring in decoupling cellular processes. While engineering xylose metabolism pathways is essential to enable the process, anaerobic xylose fermentation is not enacted without rewiring the regulatory system to simultaneously activate Snf1 along with PKA [31,32]. Here, we propose roles for several regulators, including Opi1 and Bcy1, among downstream effectors like Azf1, Aft1/2, Mga2, and Ino2/4, in modulating growth and metabolism decoupling on anaerobic xylose. Our results strongly suggest that upstream regulatory tinkering rather than altering individual downstream effectors will be required to optimally engineer new cell functions.
Methods
Media and growth conditions
Cells were grown in YP media (10 g/L yeast extract, 20 g/L peptone) with 20g/L of either glucose or xylose. Aerobic cultures were grown at 30°C with vigorous shaking. Anaerobic cultures were grown in a Coy anaerobic chamber (10% CO2, 10% H2, 80% N2) at 30°C with a metal stir bar for mixing. All cultures were inoculated with cells grown aerobically to saturation in YP-glucose and washed one time with the desired growth medium. Anaerobic cultures were inoculated into media incubated in the anaerobic chamber for >16 hours before inoculation. Cell density was monitored by optical density at 600 nm (OD600) with an Eppendorf Spectrophotometer. Sugar and ethanol concentrations were measured with HPLC-RID (Refractive Index Detector) analysis [27]. Growth on solid media (Fig S5C) was performed by collecting 1 OD worth of cells from a saturated YP-glucose culture, washing cells with YP-xylose, and plating serial dilutions onto solid YP medium with 2% xylose, with or without 100 μg/mL of nourseothricin. Plates were grown in a Coy anaerobic chamber for seven days before imaging.
Strains and cloning
Saccharomyces cerevisiae strains used in this study are described in Table 3. Gene knockouts were created by homologous recombination with either KanMX or Hph cassettes [105,106] and confirmed with diagnostic PCRs. The KanMX cassette was rescued from the bcy1Δ and EWY55 strains with CRISPR-Cas9 using a gRNA specific for KanMX and a repair template containing the flanking sequence. The bcy1Δ or EWY55 strain’s allele of OPI1, RIM8, or TOA1 was cloned into the pKI plasmid, carrying a nourseothricin [NAT] resistance marker, using standard cloning techniques. Plasmids were verified with Sanger sequencing, then transformed into the appropriate bcy1Δ or EWY55 KAN marker rescued strain using NTC selection.
Table 3. Strains used in this study.
| Strain Name | Description | Ref |
|---|---|---|
| Y184 | CRB strain with xylose utilization genes (G418-R), gre3::MR isu1::loxP-Hyg (Hyg-R) | [32] |
| Y184 ira2Δ | Y184 ira2::MR | [31] |
| Y184 bcy1Δ | Y184 bcy1::KanMX (Hyg-R, G418-R) | [32] |
| Y184 ira2Δbcy1Δ | Y184 ira2::MR bcy1::KanMX (G418-R) | [35] |
| EWY55 | Y184 bcy1::KanMX (G418-R) opi1-T715G tpk1-G829C toa1-T354A rim8-C1591T chr. I dup. | This study |
| EWY87-1 | Y184 bcy1::KanMX (G418-R) chr. VIII dup. | This study |
| EWY87-3 | Y184 bcy1::KanMX (G418-R) rpa43A751G | This study |
| EWY89-1 | Y184 bcy1::KanMX (G418-R) hsc82G235C Chr. X, XVI dup. | This study |
| EWY89-2 | Chr. I, X, XVI, XIV560217-625584 dup. | This study |
| EWY89-3 | Y184 bcy1::KanMX (G418-R) hsc82G235C Chr. I, IX, X, XVI, XIV560217-625585 | This study |
| EWY55 opi1Δ | EWY55 opi1::KanMX (G418-R) | This study |
| EWY55 rim8Δ | EWY55 rim8::KanMX (G418-R) | This study |
| EWY55 toa1Δ | EWY55 toa1::KanMX (G418-R) | This study |
| EWY55 empty vector | EWY55 pJH1-NatMX | This study |
| EWY55 opi1Δ empty vector | EWY55 opi1::KanMX (G418-R) pJH1-NatMX | This study |
| EWY55 opi1Δ pOPI1-EWY55 | EWY55 opi1::KanMX (G418-R) pOPI1-EWY55-NatMX | This study |
| EWY55 opi1Δ pOPI1-bcy1Δ | EWY55 opi1::KanMX (G418-R) pOPI1-bcy1Δ-NatMX | This study |
| EWY55 rim8Δ empty vector | EWY55 rim8::KanMX (G418-R) pJH1-NatMX | This study |
| EWY55 rim8Δ pOPI1-EWY55 | EWY55 rim8::KanMX (G418-R) pRIM8-EWY55-NatMX | This study |
| EWY55 rim8Δ pOPI1-bcy1Δ | EWY55 rim8::KanMX (G418-R) pRIM8-bcy1Δ-NatMX | This study |
| EWY55 toa1Δ empty vector | EWY55 toa1::KanMX (G418-R) pJH1-NatMX | This study |
| EWY55 toa1Δ pOPI1-EWY55 | EWY55 toa1::KanMX (G418-R) pTOA1-EWY55-NatMX | This study |
| EWY55 toa1Δ pOPI1-bcy1Δ | EWY55 toa1::KanMX (G418-R) pTOA1-bcy1Δ-NatMX | This study |
RNA-seq sample collection, RNA extraction, library preparation, and sequencing
Cells from saturated cultures of Y184, ira2Δ, bcy1Δ, and ira2Δbcy1Δ were used to inoculate anaerobic YPD cultures at OD600 0.05. Cultures grew for five hours to early/mid-log phase. 50 mL of the culture was collected, washed with YPX, then used to inoculate anaerobic YPX cultures as described above. Cold 5% phenol/95% ethanol was added to the remaining 50mL YPD cultures, which were harvested by centrifuging at 3000 RPM for 3 minutes and flash frozen in liquid nitrogen. Cell pellets were stored at -80°C until further processing. The YPX cultures grew for 3.5 hours, when the ira2Δ strain resumed growth. Cold phenol/ethanol was added to the 50 mL cultures, which were harvested, flash frozen, and stored at -80°C. Samples were collected from three independent replicates performed on different days.
Total RNA was extracted using hot phenol lysis [107] and DNA was digested with Turbo-DNase (Life Technologies, Carlsbad, CA) for 30 minutes at 37°C. RNA was precipitated at 20°C in 2.5 M LiCl for 30 min. rRNA was depleted with EPiCenter Ribo-Zero Magnetic Gold Kit (Yeast) RevA kit (Illumina Inc, San Diego, CA), and the remaining RNA was purified using Agencourt RNACleanXP (Beckman Coulter, Indianapolis, IN) by following the manufacturers’ protocols. RNA-seq libraries were created with the Illumnia TruSeq stranded total RNA kit (Illumina) following the preparation guide (revision C), AMPure XP beads were used for PCR purification (Beckman Coulter, Indianapolis, IN), and cDNA generated with SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA) as described in the Illumina kit. Libraries were standardized to 2 μM and clusters were generated with standard Cluster kits (version 3) and the Illumina Cluster station. Paired-end 50-bp reads were generated using standard SBS chemistry (version 3) on an Illumina NovaSeq 6000 sequencer.
RNA-seq data processing and analysis
RNA-seq reads were processed with Trimmomatic version 0.3 [108] and mapped to the Y22-3 genome [58] using BWA-MEM version 0.7.17 with default settings. Read counts were calculated with HTSeq version 0.6.0 [109] using the Y22-3 gene annotations. All raw data were deposited in the NIH GEO database (GSE220465). Raw sequence counts were normalized using trimmed mean of M-values (TMM) method [110]. log2 fold changes (FC) between YP-xylose and YP-glucose samples for each strain and replicate were calculated, then hierarchical clustered using Gene Cluster 3.0 [111] and visualized with Java Treeview version 1.2.0 [112]. Differential expression was analyzed using linear modeling in edgeR version 4.0.3 [113] using pairwise and group comparisons, calling significance at < 0.05 Benjamini and Hochberg false discovery rate (FDR) [114]. Genes in Fig 2B were identified by pairwise comparisons between The Y184, bcy1Δ, and ira2Δbcy1Δ strains with the ira2Δ strain. Genes in Fig 2C were identified by comparing the ira2Δ, bcy1Δ, and ira2Δbcy1Δ strains as a group in the statistical model with the Y184 strain. Genes in Fig 3A were identified first by pairwise comparison between the ira2Δ and bcy1Δ strains, then subsequently further grouped by genes reproducibly expressed in opposing directions between the two strains (e.g. log2FC > 0 in bcy1Δ and log2FC < 0 in ira2Δ).
Genes differentially expressed between EWY55 and bcy1Δ strains grown anaerobically on xylose were identified using edgeR version 4.0.3 [113] at FDR [114] < 0.05. Genes were median centered, the log2 YPX abundance of EWY55 or ira2Δ transcripts, relative to log2 bcy1Δ YPX abundance were calculated, then hierarchically clustered Gene Cluster 3.0 [111] and visualized in Java Treeview version 1.2.0 [112].
Functional gene ontology (GO) term and transcriptional regulator enrichment was performed using SetRank [115]; an FDR cutoff of 0.05 was used for transcription target analysis and a Bonferroni corrected p-value cutoff of 10−4 was used to assess overlapping GO categories. Targets of transcription factors were downloaded from YeasTract [116] using only targets with DNA binding evidence. Upstream regulatory motifs were identified with MEME suite version 5.4.1 [117] and associated transcription factors were implicated using Tomtom [118].
Lipidomics sample collection and preparation
Cells were grown as described previously for the RNAseq collection, flash frozen in liquid nitrogen, then stored at -80°C. On the day of analysis, each sample was removed from -80°C and maintained on dry ice until time of extraction. 240 μL chilled methanol was added to cell pellet samples in their native tubes over dry ice. Native tubes were transferred to ice and then vortexed. Samples were then transferred to 2 mL microcentrifuge tubes over ice. Next 800 μL of chilled methyl tert-butyl ether (MTBE) was added to native tubes followed by vortexing; these samples were also transferred to the microcentrifuge tube. Microcentrifuge tubes were then vortexed for 10 seconds. A 1/32 teaspoon (0.15 mL) of 1,180 μm glass beads (16–25 US sieve) was added to each tube along with 200 μL LC-MS grade water. Tubes were vortexed for 10 seconds. All tubes were centrifuged at 4°C for 2 minutes at 5,000 x g to pellet cell debris. An extraction blank was prepared per sample preparation steps directly into a 2 mL microcentrifuge tube without yeast.
200 μL of the top (lipophilic) layer from each tube was aliquoted into a low volume amber borosilicate glass autosampler vial with tapered insert. For pooled YPD and pooled YPX samples, the 200 μL aliquot was performed in duplicate. Each vial was dried in a vacuum concentrator for approximately one hour. For pooled YPD and pooled YPX samples, resuspension was performed with 50 μL of a 9:1 MeOH:toluene solution on the first of two preparations (“1X”) while the second preparation was resuspended in 25 μL of 9:1 MeOH:toluene (“2X”). Remaining dried samples were resuspended in 50 μL of 9:1 MeOH:toluene. Each vial was vortexed vigorously for 10 seconds to ensure resuspension of the dried contents. Samples were placed in the instrument’s autosampler at 4°C to await injection.
Lipidomics LC-MS analysis
LC-MS/MS analysis was performed using an Acquity CSH C18 column (2.1 mm × 100 mm, 1.7 μm particle size, Waters) held at 50°C and a Vanquish Binary Pump (400 μL/mL flow rate; Thermo Scientific, Waltham, MA). Mobile phase A consisted of ACN:H2O (70:30, v/v) with 10 mM ammonium acetate and 0.025% acetic acid. Mobile phase B consisted of IPA:ACN (9:1, v/v) with 10 mM ammonium acetate and 0.025% acetic acid. Initially, mobile phase B was held at 2% for 2 min and increased to 30% over 3 min. In consecutive ramping steps, mobile phase B was increased to 50% over 1 minute, increased to 85% over 14 minutes, and increased to 99% over 1 minute. The gradient was held at 99% mobile phase B for 7 minutes, then decreased to 2% over 0.25 minutes. The column was equilibrated at 2% mobile phase B for 1.75 minutes before the next injection. 10 μL of each extract was injected by a Vanquish Split Sampler HT autosampler (Thermo Scientific, Waltham, MA) in a randomized order.
The LC system was coupled to a Q Exactive HF Orbitrap mass spectrometer (MS) through a heated electrospray ionization (HESI II) source (Thermo Scientific, Waltham, MA). Source conditions were as follows: HESI II and capillary temperature at 350°C, sheath gas flow rate at 25 units, aux gas flow rate at 15 units, sweep gas flow rate at 5 units, spray voltage at |3.5 kV|, and S-lens RF at 60.0 units. The MS was operated in a polarity switching mode acquiring positive and negative full MS and MS2 spectra (Top2) within the same injection. Acquisition parameters for full MS scans in both modes were 30,000 resolution, 1 × 106 automatic gain control (AGC) target, 100 ms ion accumulation time (max IT), and 200 to 2000 m/z scan range. MS2 scans in both modes were then performed at 30,000 resolution, 1 × 105 AGC target, 50 ms max IT, 1.0 m/z isolation window, stepped normalized collision energy (NCE) at 20, 30, 40, and a 10.0 s dynamic exclusion.
Lipidomics data analysis
The resulting LC–MS data were processed using Compound Discoverer 3.1 (Thermo Scientific, Waltham, MA) and LipiDex, an in-house-developed software suite [119]. All peaks between 0.4 min and 21.0 min retention time and between100 Da and 5000 Da MS1 precursor mass were aggregated into compound groups using a 10-ppm mass, 0.2 min retention time tolerance, a minimum peak intensity of 1x10^5, a maximum peak-width of 0.75 min, and a signal-to-noise (S/N) ratio of 3. Features were required to be 5-fold greater intensity in samples than blanks. MS/MS spectra were searched against an in-silico generated lipid spectral library. Spectral matches were required to have a dot product score greater than 500 and a reverse dot product score greater than 700. Lipid MS/MS spectra which contain acyl-chain specific fragments and contained no significant interference (<75%) from co-eluting isobaric lipids were identified at molecular species level. If individual fatty acid substituents were unresolved, then identifications were made with the sum of the fatty acid substituents. Lipid features were further filtered based on 1) presence in a minimum of two raw files, 2) a median absolute retention time deviation of 3.5, and 3) average pooled relative standard deviations of less than 30%.
Differential abundance of lipids was analyzed with linear modeling in edgeR version 4.0.3 using pairwise comparisons and a Benjamini and Hochberg [114] FDR < 0.05 to call significance. After the log2FC between YP-xylose and YP-glucose samples for each strain was calculated, lipids were hierarchically cluster in Gene Cluster 3.0 [111] and visualized in Java Treeview 1.2.0 [112]. For all phosphatidylcholine moieties, a paired ANOVA with a cutoff of p < 0.05 was performed between ira2Δ and bcy1Δ samples. All raw and processed lipidomics data files were deposited in MassIVE database under dataset number MSV000090868.
For EWY55 lipidomics data, differential abundance of lipids was analyzed by calculating the log2(fold change) ratio between YPX and YPD samples for each strain and replicate. The paired log2(fold change) differences between EWY55 and ira2Δ or bcy1Δ samples were calculated, and an absolute value difference greater than 1.5 on a log2 scale was called significant. Average log2(fold change) of classified, significant lipid classes of interest were calculated along with standard error, and significant differences in lipid classes were called by a paired ANOVA.
Phosphoproteomics data
Phosphoproteomics data from Myers et al. (2019) [32] was analyzed to compare the phosphorylation of phospholipid biosynthesis enzymes. Reproducible pairwise comparisons between YP-xylose samples of strains with a log2 fold-change >2 were called significant.
Inositol and choline supplementation
YP-xylose medium was prepared as described above. Myo-inositol (Sigma, Burlington, MA) was added to a final concentration of 75 μM and choline (Thermo Scientific, Waltham, MA) to a concentration of 10 mM. Anaerobic cultures were inoculated from saturated overnight cultures to an OD600 of 0.1. Growth, xylose concentration, and ethanol concentration was monitored over 44 hours. A paired ANOVA between YP-xylose and YP-xylose-inositol-choline cultures was performed to determine significant differences between growth using a p value cutoff of 0.05.
Adaptive laboratory evolutions
bcy1Δ cells were inoculated in anaerobic YP-glucose medium at an OD600 of 0.01 and grown for ~21 generations. This was used to seed a fresh anaerobic YP-glucose culture at an OD600 of 0.01, which grew for ~7 generations. From this, a YP-2% xylose 0.1% glucose culture was seeded at an OD600 of 0.01, then grown for ~7 generations. This process was repeated four more times before plating the culture on YP-xylose and collecting single colonies capable of growing anaerobically on xylose. Evolutions were performed in three independent cultures.
Evolved bcy1Δ strain genome sequencing and analysis
Evolved bcy1Δ strains were grown aerobically in YP-glucose and genomic DNA was extracted using the Qiagen (Hilden, Germany) Genomic-tip 20/G kit following manufacturer’s protocol. Genomic DNA was fragmented into ~200 bp fragments using a sonifier with four minutes on and one minute off while incubating on ice, repeated for a total of four cycles. DNA libraries were made using the NEBNext Ultra II DNA Library Prep Kit for Illumina protocol, using the NEBNext Multiplex Oligos for Illumina (Dual Index Primers Set 1) (New England Biolabs, Ipswich, MA). Paired-end 300 bp reads were generated on an Illumina MiSeq.
Variants in the parental bcy1Δ strain were identified with GATK version 4.2 (Broad Institute) and substituted into the Y22-3 reference genome as a mapping reference. Reads were mapped to the newly generated bcy1Δ strain genome, and variants were called using GATK version 4.2 and single nucleotide polymorphisms (SNPs) annotated with SnpEff version 5.0 and vcftools version v0.,1.12b. Only SNPs occurring in the coding region of a gene were considered for further analysis and verified with Sanger sequencing.
Supporting information
A-C. Average (n = 3 biological replicates) (A) growth (OD600, optical density), (B) glucose concentration, and (C) ethanol concentration of ira2Δ, bcy1Δ, and ira2Δbcy1Δ strains grown anaerobically on rich glucose medium (p > 0.05, ANOVA).
(TIF)
A. Expression of 5834 genes (rows) detected in all four strains (Y184, ira2Δ, bcy1Δ, ira2Δbcy1Δ), organized by hierarchical clustering of log2(fold change) upon glucose-to-xylose shift, as described in Fig 2. Each column represents one of three biological replicates of the denoted strain listed above. B. Expression of 135 ribosomal protein genes (rows) in all four strains (Y184, ira2Δ, bcy1Δ, ira2Δbcy1Δ), organized by hierarchical clustering of log2(fold change) upon glucose-to-xylose shift. The blue-yellow heatmap on the left represents the log2(fold change) in expression upon glucose to xylose shift across biological triplicates (columns). The purple-green heatmap on the right represents the abundance of each transcript (rows) in each strain grown on glucose (G) or xylose (X), relative to the average (n = 3) abundance of transcripts measured in the Y184 YPD sample.
(TIF)
A. Expression of 654 genes whose log2(fold change) upon glucose to xylose shift is different (FDR < 0.05) between the ira2Δ and bcy1Δ strains and whose change in expression is in the opposite direction (increased or decreased) across strains. (see Methods for details). The yellow-blue heatmap on the left represents the YPX/YPD log2(fold change). The green-purple heatmap on the right represents transcript (rows) abundance in anaerobic glucose (G) and anaerobic xylose (X) relative to the average (n = 3) abundance of transcript in the Y184 YPD sample. B. ~500 base pairs upstream of the ORF for genes repressed in the bcy1Δ strain upon shift to xylose were analyzed for enriched motifs (bottom motif; MEME Suite), analyzed for known transcription factor binding sites (TOMTOM), and identified the Aft1/2 consensus site (top motif; see Methods for details). C. Expression of 77 genes from A whose promoters are physically bound by Ino2 and/or Ino4, organized by hierarchical clustering.
(TIF)
A. The majority of phosphatidylcholine species (rows) identified show significantly lower log2(fold change) upon the shift from glucose to xylose in the bcy1Δ and ira2Δbcy1Δ compared to the ira2Δ strain (p = 0.0015082, ANOVA).
(TIF)
A-B. Average (n = 3 biological replicates) of (A) xylose concentration or (B) ethanol concentration for ira2Δ, bcy1Δ, and EWY55 strains anaerobically grown in rich xylose medium (p > 0.05, ANOVA). C. Representatives of multiple replicates of EWY55 or EWY55 cells lacking OPI1 (top panels), RIM8 (middle panels), or TOA1 (bottom panels) and complemented with an empty vector or parental or evolved allele grown anaerobically on solid xylose (left) or glucose (right) medium with NTC selection. D. Average (n = 3 biological replicates) of ethanol concentration for EWY55 and EWY55 opi1Δ strains anaerobically grown in rich xylose medium (p > 0.05).
(TIF)
A-E. Average (n = 3 biological replicates) growth (OD600, optical density) of ira2Δ, bcy1Δ, EWY55, and (A) EWY87-1, (B) EWY87-3, (C) EWY89-1, (D) EWY89-2, or (E) EWY89-3 strains grown anaerobically on rich xylose medium (p < 10−4, ANOVA).
(TIF)
Companion table for S2A Fig.
(XLSX)
Companion table for Fig 2B.
(XLSX)
(XLSX)
Companion table for S2B Fig.
(XLSX)
Data for ira2Δbcy1Δ cells is also shown. Companion table for Fig 2C.
(XLSX)
Data for ira2Δbcy1Δ cells is also shown. Companion table for Fig 2D.
(XLSX)
Companion table for Fig 3A.
(XLSX)
(XLSX)
Companion table for S3C Fig.
(XLSX)
(XLSX)
Companion table for Fig 4A.
(XLSX)
Companion table for Fig 4B.
(XLSX)
Companion table for S4 Fig.
(XLSX)
Transcript abundance differences (n = 3) of EWY55 and ira2Δ compared to bcy1Δ cells.
(XLSX)
Companion table for Fig 5C.
(XLSX)
(XLSX)
Companion table for Fig 5D.
(XLSX)
Acknowledgments
We thank Mike Place for computational help with transcriptomic data analysis, James Hose and Venera Bouriakov for help with generating RNAseq libraries, Kevin Myers for phosphoproteomic data, and members of the Gasch lab for constructive discussions.
Data Availability
Raw and processed RNAseq files have been submitted to the NIH GEO database under project number GSE220465. Raw and processed lipidomic files were deposited in MassIVE database under dataset number MSV000090868.
Funding Statement
This material is based upon work supported by the Great Lakes Bioenergy Research Center, U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research under Award Number DE-SC001840, funding APG. ERW is supported by the National Science Foundation Graduate Research Fellowship Program under Grant No. DGE-1747503. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation. ERW was also supported by the Graduate School and the Office of the Vice Chancellor for Research and Graduate Education at the University of Wisconsin-Madison with general funding from the Wisconsin Alumni Research Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, et al. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell [Internet]. 1998. Oct 13 [cited 2020 Jun 26];9(12):3273–97. Available from: https://www.molbiolcell.org/doi/abs/ doi: 10.1091/mbc.9.12.3273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ewald JC, Kuehne A, Zamboni N, Skotheim JM. The Yeast Cyclin-Dependent Kinase Routes Carbon Fluxes to Fuel Cell Cycle Progression. Mol Cell [Internet]. 2016. May 19 [cited 2022 Jun 16];62(4):532–45. Available from: http://www.cell.com/article/S1097276516001283/fulltext. doi: 10.1016/j.molcel.2016.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao G, Chen Y, Carey L, Futcher B. Cyclin-Dependent Kinase Co-Ordinates Carbohydrate Metabolism and Cell Cycle in S. cerevisiae. Mol Cell [Internet]. 2016. May 19 [cited 2019 Apr 26];62(4):546–57. Available from: https://www.sciencedirect.com/science/article/pii/S1097276516301022?via%3Dihub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blank HM, Papoulas O, Maitra N, Garge R, Kennedy BK, Schilling B, et al. Abundances of transcripts, proteins, and metabolites in the cell cycle of budding yeast reveal coordinate control of lipid metabolism. Mol Biol Cell [Internet]. 2020. May 1 [cited 2020 Sep 28];31(10):1069–84. Available from: https://www.molbiolcell.org/doi/abs/10.1091/mbc.E19-12-0708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blank HM, Perez R, He C, Maitra N, Metz R, Hill J, et al. Translational control of lipogenic enzymes in the cell cycle of synchronous, growing yeast cells. EMBO J [Internet]. 2017. Feb 15 [cited 2022 Jun 22];36(4):487–502. Available from: https://onlinelibrary.wiley.com/doi/full/10.15252/embj.201695050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fajas L. Re-thinking cell cycle regulators: The cross-talk with metabolism. Front Oncol. 2013;3 JAN:4. doi: 10.3389/fonc.2013.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Futcher B. Metabolic cycle, cell cycle, and the finishing kick to start. Genome Biol [Internet]. 2006. Apr 26 [cited 2022 Jun 17];7(4):1–5. Available from: https://genomebiology.biomedcentral.com/articles/10.1186/gb-2006-7-4-107. doi: 10.1186/gb-2006-7-4-107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Futcher B. Tgl4 Lipase: A Big Fat Target for Cell-Cycle Entry. Mol Cell. 2009. Jan 30;33(2):143–4. [DOI] [PubMed] [Google Scholar]
- 9.Liu J, Wang X, Dai G, Zhang Y, Bian X. Microbial chassis engineering drives heterologous production of complex secondary metabolites. Biotechnol Adv. 2022. Oct 1;59:107966. doi: 10.1016/j.biotechadv.2022.107966 [DOI] [PubMed] [Google Scholar]
- 10.Zhang C, Ottenheim C, Weingarten M, Ji LH. Microbial Utilization of Next-Generation Feedstocks for the Biomanufacturing of Value-Added Chemicals and Food Ingredients. Front Bioeng Biotechnol. 2022. Apr 11;10:486. doi: 10.3389/fbioe.2022.874612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shah AM, Yang W, Mohamed H, Zhang Y, Song Y. Microbes: A Hidden Treasure of Polyunsaturated Fatty Acids. Front Nutr. 2022. Mar 17;9:415. doi: 10.3389/fnut.2022.827837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joshi A, Verma KK, Rajput VD, Minkina T, Arora J. Recent advances in metabolic engineering of microorganisms for advancing lignocellulose-derived biofuels. https://doi.org/101080/2165597920222051856 [Internet]. 2022. Apr 1 [cited 2022 Apr 4];13(4):8135–63. Available from: https://www.tandfonline.com/doi/abs/10.1080/21655979.2022.2051856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Demeke MM, Dumortier F, Li Y, Broeckx T, Foulquié-Moreno MR, Thevelein JM. Combining inhibitor tolerance and D-xylose fermentation in industrial Saccharomyces cerevisiae for efficient lignocellulose-based bioethanol production. Biotechnol Biofuels [Internet]. 2013. Aug 26 [cited 2019 Jan 24];6(1):120. Available from: http://biotechnologyforbiofuels.biomedcentral.com/articles/10.1186/1754-6834-6-120. doi: 10.1186/1754-6834-6-120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Demeke MM, Dietz H, Li Y, Foulquié-Moreno MR, Mutturi S, Deprez S, et al. Development of a D-xylose fermenting and inhibitor tolerant industrial Saccharomyces cerevisiae strain with high performance in lignocellulose hydrolysates using metabolic and evolutionary engineering. Biotechnol Biofuels [Internet]. 2013. Jun 21 [cited 2019 Jan 24];6(1):89. Available from: http://biotechnologyforbiofuels.biomedcentral.com/articles/10.1186/1754-6834-6-89. doi: 10.1186/1754-6834-6-89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JW, Yook S, Koh H, Rao C V., Jin YS. Engineering xylose metabolism in yeasts to produce biofuels and chemicals. Curr Opin Biotechnol. 2021. Feb 1;67:15–25. [DOI] [PubMed] [Google Scholar]
- 16.Li X, Chen Y, Nielsen J. Harnessing xylose pathways for biofuels production. Curr Opin Biotechnol [Internet]. 2019. Jun 1 [cited 2019 Apr 1];57:56–65. Available from: https://www.sciencedirect.com/science/article/pii/S0958166918300892?via%3Dihub. doi: 10.1016/j.copbio.2019.01.006 [DOI] [PubMed] [Google Scholar]
- 17.van Maris AJA, Abbott DA, Bellissimi E, van den Brink J, Kuyper M, Luttik MAH, et al. Alcoholic fermentation of carbon sources in biomass hydrolysates by Saccharomyces cerevisiae: current status. Antonie Van Leeuwenhoek [Internet]. 2006. Nov 11 [cited 2019 Jan 24];90(4):391–418. Available from: http://link.springer.com/10.1007/s10482-006-9085-7. doi: 10.1007/s10482-006-9085-7 [DOI] [PubMed] [Google Scholar]
- 18.Joshi S, Mishra SD. Recent advances in biofuel production through metabolic engineering. Bioresour Technol. 2022. May 1;352:127037. doi: 10.1016/j.biortech.2022.127037 [DOI] [PubMed] [Google Scholar]
- 19.Piotrowski JS, Zhang Y, Bates DM, Keating DH, Sato TK, Ong IM, et al. Death by a thousand cuts: the challenges and diverse landscape of lignocellulosic hydrolysate inhibitors. Front Microbiol [Internet]. 2014. Mar 14 [cited 2019 Jan 24];5:90. Available from: http://journal.frontiersin.org/article/10.3389/fmicb.2014.00090/abstract. doi: 10.3389/fmicb.2014.00090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Almeida JRM, Runquist D, Sànchez Nogué V, Lidén G, Gorwa-Grauslund MF. Stress-related challenges in pentose fermentation to ethanol by the yeast Saccharomyces cerevisiae. Biotechnol J [Internet]. 2011. Mar 1 [cited 2019 Jan 24];6(3):286–99. Available from: http://doi.wiley.com/10.1002/biot.201000301. [DOI] [PubMed] [Google Scholar]
- 21.Kim SR, Lee KS, Choi JH, Ha SJ, Kweon DH, Seo JH, et al. Repeated-batch fermentations of xylose and glucose–xylose mixtures using a respiration-deficient Saccharomyces cerevisiae engineered for xylose metabolism. J Biotechnol. 2010. Nov 1;150(3):404–7. doi: 10.1016/j.jbiotec.2010.09.962 [DOI] [PubMed] [Google Scholar]
- 22.Kim SR, Lee KS, Kong II, Lesmana A, Lee WH, Seo JH, et al. Construction of an efficient xylose-fermenting diploid Saccharomyces cerevisiae strain through mating of two engineered haploid strains capable of xylose assimilation. J Biotechnol. 2013. Mar 10;164(1):105–11. doi: 10.1016/j.jbiotec.2012.12.012 [DOI] [PubMed] [Google Scholar]
- 23.Jin Y-S, Laplaza JM, Jeffries TW. Saccharomyces cerevisiae engineered for xylose metabolism exhibits a respiratory response. Appl Environ Microbiol [Internet]. 2004. Nov 1 [cited 2019 Mar 20];70(11):6816–25. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15528549. doi: 10.1128/AEM.70.11.6816-6825.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.KUYPER M, HARTOG M, TOIRKENS M, ALMERING M, WINKLER A, VANDIJKEN J, et al. Metabolic engineering of a xylose-isomerase-expressing strain for rapid anaerobic xylose fermentation. FEMS Yeast Res [Internet]. 2005. Feb 1 [cited 2019 Jan 24];5(4–5):399–409. Available from: https://academic.oup.com/femsyr/article-lookup/doi/10.1016/j.femsyr.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 25.Demeke MM, Foulquié-Moreno M íR, Dumortier F, Thevelein JM. Rapid Evolution of Recombinant Saccharomyces cerevisiae for Xylose Fermentation through Formation of Extra-chromosomal Circular DNA. PLOS Genet [Internet]. 2015. Mar 4 [cited 2022 Dec 22];11(3):e1005010. Available from: https://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1005010. doi: 10.1371/journal.pgen.1005010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.dos Santos LV, Carazzolle MF, Nagamatsu ST, Sampaio NMV, Almeida LD, Pirolla RAS, et al. Unraveling the genetic basis of xylose consumption in engineered Saccharomyces cerevisiae strains. Sci Rep [Internet]. 2016. Dec 21 [cited 2019 Jan 24];6(1):38676. Available from: http://www.nature.com/articles/srep38676. doi: 10.1038/srep38676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parreiras LS, Breuer RJ, Avanasi Narasimhan R, Higbee AJ, La Reau A, Tremaine M, et al. Engineering and Two-Stage Evolution of a Lignocellulosic Hydrolysate-Tolerant Saccharomyces cerevisiae Strain for Anaerobic Fermentation of Xylose from AFEX Pretreated Corn Stover. Zhang Y-HP, editor. PLoS One [Internet]. 2014. Sep 15 [cited 2019 Jan 24];9(9):e107499. Available from: https://dx.plos.org/10.1371/journal.pone.0107499. doi: 10.1371/journal.pone.0107499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim SR, Skerker JM, Kang W, Lesmana A, Wei N, Arkin AP, et al. Rational and Evolutionary Engineering Approaches Uncover a Small Set of Genetic Changes Efficient for Rapid Xylose Fermentation in Saccharomyces cerevisiae. Fong SS, editor. PLoS One [Internet]. 2013. Feb 26 [cited 2019 Jan 24];8(2):e57048. Available from: http://dx.plos.org/10.1371/journal.pone.0057048. doi: 10.1371/journal.pone.0057048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim SR, Park YC, Jin YS, Seo JH. Strain engineering of Saccharomyces cerevisiae for enhanced xylose metabolism. Biotechnol Adv. 2013. Nov 1;31(6):851–61. doi: 10.1016/j.biotechadv.2013.03.004 [DOI] [PubMed] [Google Scholar]
- 30.Turner TL, Kim H, Kong II, Liu JJ, Zhang GC, Jin YS. Engineering and evolution of saccharomyces cerevisiae to produce biofuels and chemicals. Adv Biochem Eng Biotechnol [Internet]. 2018. [cited 2022 Dec 22];162:175–215. Available from: https://link.springer.com/chapter/10.1007/10_2016_22. doi: 10.1007/10_2016_22 [DOI] [PubMed] [Google Scholar]
- 31.Sato TK, Tremaine M, Parreiras LS, Hebert AS, Myers KS, Higbee AJ, et al. Directed Evolution Reveals Unexpected Epistatic Interactions That Alter Metabolic Regulation and Enable Anaerobic Xylose Use by Saccharomyces cerevisiae. Caudy A, editor. PLOS Genet [Internet]. 2016. Oct 14 [cited 2019 Jan 24];12(10):e1006372. Available from: https://dx.plos.org/10.1371/journal.pgen.1006372. doi: 10.1371/journal.pgen.1006372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Myers KS, Riley NM, MacGilvray ME, Sato TK, McGee M, Heilberger J, et al. Rewired cellular signaling coordinates sugar and hypoxic responses for anaerobic xylose fermentation in yeast. Dudley AM, editor. PLOS Genet [Internet]. 2019. Mar 11 [cited 2019 Mar 29];15(3):e1008037. Available from: http://dx.plos.org/10.1371/journal.pgen.1008037. doi: 10.1371/journal.pgen.1008037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conrad M, Schothorst J, Kankipati HN, Van Zeebroeck G, Rubio-Texeira M, Thevelein JM. Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev [Internet]. 2014. Mar 1 [cited 2019 Jan 24];38(2):254–99. Available from: https://academic.oup.com/femsre/article-lookup/doi/10.1111/1574-6976.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Zeebroeck G, Rubio-Texeira M, Schothorst J, Thevelein JM. Specific analogues uncouple transport, signalling, oligo-ubiquitination and endocytosis in the yeast Gap1 amino acid transceptor. Mol Microbiol [Internet]. 2014. Jul 1 [cited 2022 Dec 22];93(2):213–33. Available from: https://onlinelibrary.wiley.com/doi/full/10.1111/mmi.12654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wagner ER, Myers KS, Riley NM, Coon JJ, Gasch AP. PKA and HOG signaling contribute separable roles to anaerobic xylose fermentation in yeast engineered for biofuel production. PLoS One [Internet]. 2019. May 1 [cited 2022 Dec 15];14(5):e0212389. Available from: https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0212389. doi: 10.1371/journal.pone.0212389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, et al. Genomic Expression Programs in the Response of Yeast Cells to Environmental Changes. Silver PA, editor. Mol Biol Cell [Internet]. 2000. Dec [cited 2019 Mar 19];11(12):4241–57. Available from: http://www.molbiolcell.org/doi/10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gasch AP, Huang M, Metzner S, Botstein D, Elledge SJ, Brown PO. Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast ATR Homolog Mec1p. Mol Biol Cell [Internet]. 2001. Oct 13 [cited 2022 Dec 13];12(10):2987–3003. Available from: https://www.molbiolcell.org/doi/10.1091/mbc.12.10.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brauer MJ, Huttenhower C, Airoldi EM, Rosenstein R, Matese JC, Gresham D, et al. Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol Biol Cell [Internet]. 2008. Jan 24 [cited 2022 Sep 8];19(1):352–67. Available from: https://www.molbiolcell.org/doi/10.1091/mbc.e07-08-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Castrillo JI, Zeef LA, Hoyle DC, Zhang N, Hayes A, Gardner DCJ, et al. Growth control of the eukaryote cell: A systems biology study in yeast. J Biol [Internet]. 2007. Apr 30 [cited 2022 Jun 16];6(2):1–25. Available from: https://jbiol.biomedcentral.com/articles/10.1186/jbiol54. doi: 10.1186/jbiol54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Regenberg B, Grotkjær T, Winther O, Fausbøll A, Åkesson M, Bro C, et al. Growth-rate regulated genes have profound impact on interpretation of transcriptome profiling in Saccharomyces cerevisiae. Genome Biol [Internet]. 2006. Nov 14 [cited 2022 Sep 8];7(11):1–13. Available from: https://genomebiology.biomedcentral.com/articles/10.1186/gb-2006-7-11-r107. doi: 10.1186/gb-2006-7-11-r107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Airoldi EM, Huttenhower C, Gresham D, Lu C, Caudy AA, Dunham MJ, et al. Predicting Cellular Growth from Gene Expression Signatures. PLOS Comput Biol [Internet]. 2009. [cited 2022 Sep 8];5(1):e1000257. Available from: https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1000257. doi: 10.1371/journal.pcbi.1000257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ho YH, Shishkova E, Hose J, Coon JJ, Gasch AP. Decoupling Yeast Cell Division and Stress Defense Implicates mRNA Repression in Translational Reallocation during Stress. Curr Biol. 2018. Aug 20;28(16):2673–2680.e4. doi: 10.1016/j.cub.2018.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bali M, Zhang B, Morano KA, Michels CA. The Hsp90 Molecular Chaperone Complex Regulates Maltose Induction and Stability of the Saccharomyces MAL Gene Transcription Activator Mal63p *. J Biol Chem [Internet]. 2003. Nov 28 [cited 2022 Jul 29];278(48):47441–8. Available from: http://www.jbc.org/article/S0021925820757662/fulltext. doi: 10.1074/jbc.M309536200 [DOI] [PubMed] [Google Scholar]
- 44.Ptacek J, Devgan G, Michaud G, Zhu H, Zhu X, Fasolo J, et al. Global analysis of protein phosphorylation in yeast. Nat 2005 4387068 [Internet]. 2005. Dec 1 [cited 2022 Jul 31];438(7068):679–84. Available from: https://www.nature.com/articles/nature04187. doi: 10.1038/nature04187 [DOI] [PubMed] [Google Scholar]
- 45.Martins TS, Costa V, Pereira C. Signaling pathways governing iron homeostasis in budding yeast. Mol Microbiol [Internet]. 2018. Aug 1 [cited 2020 Nov 5];109(4):422–32. Available from: http://doi.wiley.com/10.1111/mmi.14009. [DOI] [PubMed] [Google Scholar]
- 46.Jordá T, Puig S. Regulation of ergosterol biosynthesis in saccharomyces cerevisiae [Internet]. Vol. 11, Genes. MDPI AG; 2020. [cited 2020 Aug 27]. p. 1–18. Available from: /pmc/articles/PMC7397035/?report=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Loewen CJR, Gazpar ML, Jesch SA, Delon C, Ktistakis NT, Henry SA, et al. Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science (80-) [Internet]. 2004. Jun 11 [cited 2022 Sep 8];304(5677):1644–7. Available from: https://www.science.org/doi/10.1126/science.1096083. [DOI] [PubMed] [Google Scholar]
- 48.Sreenivas A, Carman GM. Phosphorylation of the Yeast Phospholipid Synthesis Regulatory Protein Opi1p by Protein Kinase A *. J Biol Chem [Internet]. 2003. Jun 6 [cited 2021 Oct 8];278(23):20673–80. Available from: http://www.jbc.org/article/S0021925820733648/fulltext. doi: 10.1074/jbc.M300132200 [DOI] [PubMed] [Google Scholar]
- 49.Choi HS, Han GS, Carman GM. Phosphorylation of Yeast Phosphatidylserine Synthase by Protein Kinase A: IDENTIFICATION OF SER46 AND SER47 AS MAJOR SITES OF PHOSPHORYLATION. J Biol Chem. 2010. Apr 9;285(15):11526–36. doi: 10.1074/jbc.M110.100727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klug L, Daum G. Yeast lipid metabolism at a glance. FEMS Yeast Res [Internet]. 2014. May 1 [cited 2021 Aug 11];14(3):369–88. Available from: https://academic.oup.com/femsyr/article/14/3/369/580500. doi: 10.1111/1567-1364.12141 [DOI] [PubMed] [Google Scholar]
- 51.Ishiwata-Kimata Y, Le QG, Kimata Y. Induction and Aggravation of the Endoplasmic-Reticulum Stress by Membrane-Lipid Metabolic Intermediate Phosphatidyl-N-Monomethylethanolamine. Front Cell Dev Biol. 2022. Jan 6;9:3783. doi: 10.3389/fcell.2021.743018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ji J, Greenberg ML. Cardiolipin function in the yeast S. cerevisiae and the lessons learned for Barth syndrome. J Inherit Metab Dis [Internet]. 2022. Jan 1 [cited 2022 Jul 13];45(1):60–71. Available from: https://onlinelibrary.wiley.com/doi/full/10.1002/jimd.12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oughtred R, Chatr-Aryamontri A, Breitkreutz BJ, Chang CS, Rust JM, Theesfeld CL, et al. Biogrid: A resource for studying biological interactions in yeast. Cold Spring Harb Protoc. 2016. Jan 1;2016(1):29–34. doi: 10.1101/pdb.top080754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Su WM, Han GS, Casciano J, Carman GM. Protein Kinase A-mediated Phosphorylation of Pah1p Phosphatidate Phosphatase Functions in Conjunction with the Pho85p-Pho80p and Cdc28p-Cyclin B Kinases to Regulate Lipid Synthesis in Yeast. J Biol Chem. 2012. Sep 28;287(40):33364–76. doi: 10.1074/jbc.M112.402339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng J, Payne JL, Wagner A. Cryptic genetic variation accelerates evolution by opening access to diverse adaptive peaks. Science (80-) [Internet]. 2019. Jul 26 [cited 2020 Dec 2];365(6451):347–53. Available from: http://science.sciencemag.org/. doi: 10.1126/science.aax1837 [DOI] [PubMed] [Google Scholar]
- 56.Ishtar Snoek I, Yde Steensma H. Why does Kluyveromyces lactis not grow under anaerobic conditions? Comparison of essential anaerobic genes of Saccharomyces cerevisiae with the Kluyveromyces lactis genome. FEMS Yeast Res [Internet]. 2006. May 1 [cited 2022 Dec 13];6(3):393–403. Available from: https://academic.oup.com/femsyr/article/6/3/393/563987. doi: 10.1111/j.1567-1364.2005.00007.x [DOI] [PubMed] [Google Scholar]
- 57.Chang YF, Carman GM. Casein Kinase II Phosphorylation of the Yeast Phospholipid Synthesis Transcription Factor Opi1p *. J Biol Chem [Internet]. 2006. Feb 24 [cited 2022 Jun 29];281(8):4754–61. Available from: http://www.jbc.org/article/S0021925819767702/fulltext. doi: 10.1074/jbc.M513064200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McIlwain SJ, Peris D, Sardi M, Moskvin O V., Zhan F, Myers KS, et al. Genome sequence and analysis of a stress-tolerant, wild-derived strain of Saccharomyces cerevisiae used in biofuels research. G3 Genes, Genomes, Genet [Internet]. 2016. Jun 1 [cited 2022 Dec 13];6(6):1757–66. Available from: https://academic.oup.com/g3journal/article/6/6/1757/6029942. doi: 10.1534/g3.116.029389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Palomino A, Herrero P, Moreno F. Rgt1, a glucose sensing transcription factor, is required for transcriptional repression of the HXK2 gene in Saccharomyces cerevisiae. Biochem J [Internet]. 2005. Jun 1 [cited 2023 Apr 24];388(2):697–703. Available from: /biochemj/article/388/2/697/93331/Rgt1-a-glucose-sensing-transcription-factor-is. doi: 10.1042/BJ20050160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Busti S, Coccetti P, Alberghina L, Vanoni M. Glucose Signaling-Mediated Coordination of Cell Growth and Cell Cycle in Saccharomyces Cerevisiae. Sensors 2010, Vol 10, Pages 6195–6240 [Internet]. 2010. Jun 21 [cited 2023 Apr 24];10(6):6195–240. Available from: https://www.mdpi.com/1424-8220/10/6/6195/htm. doi: 10.3390/s100606195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pan X, Heitman J. Sok2 Regulates Yeast Pseudohyphal Differentiation via a Transcription Factor Cascade That Regulates Cell-Cell Adhesion. Mol Cell Biol [Internet]. 2000. Nov 15 [cited 2020 Jul 14];20(22):8364–72. Available from: http://mcb.asm.org/. doi: 10.1128/MCB.20.22.8364-8372.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ward MP, Gimeno CJ, Fink GR, Garrett S. SOK2 may regulate cyclic AMP-dependent protein kinase-stimulated growth and pseudohyphal development by repressing transcription. Mol Cell Biol [Internet]. 1995. Dec [cited 2022 Jul 2];15(12):6854–63. Available from: https://journals.asm.org/doi/abs/10.1128/MCB.15.12.6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dennis PP, Bremer H. Regulation of ribonucleic acid synthesis in Escherichia coli Br: An analysis of a shift-up: 1. Ribosomal RNA chain growth rates. J Mol Biol. 1973. Mar 25;75(1):145–59. [DOI] [PubMed] [Google Scholar]
- 64.Dennis PP, Bremer H. Differential rate of ribosomal protein synthesis in Escherichia coli B/r. J Mol Biol. 1974. Apr 15;84(3):407–22. doi: 10.1016/0022-2836(74)90449-5 [DOI] [PubMed] [Google Scholar]
- 65.Young R, Bremer H. Polypeptide-chain-elongation rate in Escherichia coli B/r as a function of growth rate. Biochem J [Internet]. 1976. Nov 15 [cited 2022 Dec 13];160(2):185–94. Available from: /biochemj/article/160/2/185/11251/Polypeptide-chain-elongation-rate-in-Escherichia. doi: 10.1042/bj1600185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gourse RL, Gaal T, Bartlett MS, Ross W. rRNA Transcription and growth rate-dependent regulation of ribosome synthesis in Escherichia coli. Artic Annu Rev Microbiol [Internet]. 1996. [cited 2022 Dec 13]; Available from: https://www.researchgate.net/publication/14302128. doi: 10.1146/annurev.micro.50.1.645 [DOI] [PubMed] [Google Scholar]
- 67.Carrera J, Rodrigo G, Singh V, Kirov B, Jaramillo A. Empirical model and in vivo characterization of the bacterial response to synthetic gene expression show that ribosome allocation limits growth rate. Biotechnol J [Internet]. 2011. Jul 1 [cited 2022 Dec 13];6(7):773–83. Available from: https://onlinelibrary.wiley.com/doi/full/10.1002/biot.201100084. [DOI] [PubMed] [Google Scholar]
- 68.Scott M, Klumpp S, Mateescu EM, Hwa T. Emergence of robust growth laws from optimal regulation of ribosome synthesis. Mol Syst Biol [Internet]. 2014. Aug 1 [cited 2022 Dec 13];10(8):747. Available from: https://onlinelibrary.wiley.com/doi/full/10.15252/msb.20145379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dai X, Zhu M, Warren M, Balakrishnan R, Patsalo V, Okano H, et al. Reduction of translating ribosomes enables Escherichia coli to maintain elongation rates during slow growth. Nat Microbiol 2016 22 [Internet]. 2016. Dec 12 [cited 2022 Dec 13];2(2):1–9. Available from: https://www.nature.com/articles/nmicrobiol2016231. doi: 10.1038/nmicrobiol.2016.231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dai X, Zhu M, Warren M, Balakrishnan R, Okano H, Williamson JR, et al. Slowdown of translational elongation in Escherichia coli under hyperosmotic stress. MBio [Internet]. 2018. Jan 1 [cited 2022 Dec 13];9(1). Available from: https://journals.asm.org/doi/10.1128/mBio.02375-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Korem Kohanim Y, Levi D, Jona G, Towbin BD, Bren A, Alon U. A Bacterial Growth Law out of Steady State. Cell Rep. 2018. Jun 5;23(10):2891–900. doi: 10.1016/j.celrep.2018.05.007 [DOI] [PubMed] [Google Scholar]
- 72.Wu C, Balakrishnan R, Braniff N, Mori M, Manzanarez G, Zhang Z, et al. Cellular perception of growth rate and the mechanistic origin of bacterial growth law. Proc Natl Acad Sci U S A [Internet]. 2022. May 17 [cited 2022 Dec 13];119(20):e2201585119. Available from: https://www.pnas.org/doi/abs/10.1073/pnas.2201585119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Towbin BD, Korem Y, Bren A, Doron S, Sorek R, Alon U. Optimality and sub-optimality in a bacterial growth law. Nat Commun 2017 81 [Internet]. 2017. Jan 19 [cited 2022 Dec 13];8(1):1–8. Available from: https://www.nature.com/articles/ncomms14123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jordá T, Romero AM, Perea-García A, Rozès N, Puig S. The lipid composition of yeast cells modulates the response to iron deficiency. Biochim Biophys Acta—Mol Cell Biol Lipids. 2020. Aug 1;1865(8):158707. doi: 10.1016/j.bbalip.2020.158707 [DOI] [PubMed] [Google Scholar]
- 75.Chen OS, Crisp RJ, Valachovic M, Bard M, Winge DR, Kaplan J. Transcription of the yeast iron regulon does not respond directly to iron but rather to iron-sulfur cluster biosynthesis. J Biol Chem [Internet]. 2004. Jul 9 [cited 2022 Nov 14];279(28):29513–8. Available from: http://www.jbc.org/article/S0021925819712034/fulltext. doi: 10.1074/jbc.M403209200 [DOI] [PubMed] [Google Scholar]
- 76.Robertson LS, Causton HC, Young RA, Fink GR. The yeast A kinases differentially regulate iron uptake and respiratory function. Proc Natl Acad Sci U S A. 2000. May 23;97(11):5984–8. doi: 10.1073/pnas.100113397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schothorst J, Zeebroeck G Van, Thevelein JM. Identification of Ftr1 and Zrt1 as iron and zinc micronutrient transceptors for activation of the PKA pathway in saccharomyces cerevisiae. Microb Cell [Internet]. 2017. Mar 1 [cited 2021 Jan 26];4(3):74–89. Available from: /pmc/articles/PMC5349193/?report=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Belotti F, Tisi R, Paiardi C, Rigamonti M, Groppi S, Martegani E. Localization of Ras signaling complex in budding yeast. Biochim Biophys Acta—Mol Cell Res [Internet]. 2012. Jul 1 [cited 2019 Mar 13];1823(7):1208–16. Available from: https://www.sciencedirect.com/science/article/pii/S0167488912001073?via%3Dihub. doi: 10.1016/j.bbamcr.2012.04.016 [DOI] [PubMed] [Google Scholar]
- 79.Kocik RA, Gasch AP. Breadth and Specificity in Pleiotropic Protein Kinase A Activity and Environmental Responses. Front Cell Dev Biol. 2022. Feb 16;10:334. doi: 10.3389/fcell.2022.803392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vandermeulen MD, Cullen PJ. Gene by Environment Interactions reveal new regulatory aspects of signaling network plasticity. PLOS Genet [Internet]. 2022. Jan 4 [cited 2022 Dec 22];18(1):e1009988. Available from: https://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1009988. doi: 10.1371/journal.pgen.1009988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stanhill A, Schick N, Engelberg D. The yeast ras/cyclic AMP pathway induces invasive growth by suppressing the cellular stress response. Mol Cell Biol [Internet]. 1999. Nov 1 [cited 2019 Jan 24];19(11):7529–38. Available from: http://www.ncbi.nlm.nih.gov/pubmed/10523641. doi: 10.1128/MCB.19.11.7529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aun A, Tamm T, Sedman J. Dysfunctional mitochondria modulate cAMP-PKA signaling and filamentous and invasive growth of saccharomyces cerevisiae. Genetics. 2013. Feb 1;193(2):467–81. doi: 10.1534/genetics.112.147389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chow J, Starr I, Jamalzadeh S, Muniz O, Kumar A, Gokcumen O, et al. Filamentation Regulatory Pathways Control Adhesion-Dependent Surface Responses in Yeast. Genetics [Internet]. 2019. Jul 1 [cited 2022 Dec 22];212(3):667–90. Available from: https://academic.oup.com/genetics/article/212/3/667/5931226. doi: 10.1534/genetics.119.302004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cullen PJ, Sprague GF†. The Regulation of Filamentous Growth in Yeast. 2012; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chow J, Dionne HM, Prabhakar A, Mehrotra A, Somboonthum J, Gonzalez B, et al. Aggregate Filamentous Growth Responses in Yeast. mSphere [Internet]. 2019. Apr 24 [cited 2022 Dec 22];4(2). Available from: /pmc/articles/PMC6403458/. doi: 10.1128/mSphere.00702-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Young BP, Shin JJH, Orij R, Chao JT, Li SC, Guan XL, et al. Phosphatidic acid is a pH biosensor that links membrane biogenesis to metabolism. Science (80-). 2010. Aug 27;329(5995):1085–8. [DOI] [PubMed] [Google Scholar]
- 87.Anastasia SD, Nguyen DL, Thai V, Meloy M, MacDonough T, Kellogg DR. A link between mitotic entry and membrane growth suggests a novel model for cell size control. J Cell Biol [Internet]. 2012. Apr 2 [cited 2021 Aug 16];197(1):89–104. Available from: www.jcb.org/cgi/doi/10.1083/jcb.201108108JCB89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chauhan N, Han G, Somashekarappa N, Gable K, Dunn T, Kohlwein SD. Regulation of sphingolipid biosynthesis by the morphogenesis checkpoint kinase Swe1. J Biol Chem [Internet]. 2016. Jan 29 [cited 2022 Nov 21];291(5):2524–34. Available from: http://www.jbc.org/article/S0021925820360877/fulltext. doi: 10.1074/jbc.M115.693200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Clarke J, Dephoure N, Horecka I, Gygi S, Kellogg D. A conserved signaling network monitors delivery of sphingolipids to the plasma membrane in budding yeast. Mol Biol Cell [Internet]. 2017. Oct 1 [cited 2022 Nov 21];28(20):2589–99. Available from: https://www.molbiolcell.org/doi/10.1091/mbc.e17-01-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rao MJ, Srinivasan M, Rajasekharan R. Cell size is regulated by phospholipids and not by storage lipids in Saccharomyces cerevisiae. Curr Genet [Internet]. 2018. Oct 1 [cited 2022 Nov 21];64(5):1071–87. Available from: https://link.springer.com/article/10.1007/s00294-018-0821-0. doi: 10.1007/s00294-018-0821-0 [DOI] [PubMed] [Google Scholar]
- 91.Sommer RA, Dewitt JT, Tan R, Kellogg DR. Growth-dependent signals drive an increase in early g1 cyclin concentration to link cell cycle entry with cell growth. Elife. 2021. Oct 1;10. doi: 10.7554/eLife.64364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kornmann B, Currie E, Collins SR, Schuldiner M, Nunnari J, Weissman JS, et al. An ER-Mitochondria Tethering Complex Revealed by a Synthetic Biology Screen. Science [Internet]. 2009. Jul 7 [cited 2022 Nov 18];325(5939):477. Available from: /pmc/articles/PMC2933203/. doi: 10.1126/science.1175088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kornmann B, Walter P. ERMES-mediated ER-mitochondria contacts: molecular hubs for the regulation of mitochondrial biology. J Cell Sci [Internet]. 2010. May 1 [cited 2022 Nov 18];123(9):1389–93. Available from: https://journals.biologists.com/jcs/article/123/9/1389/31714/ERMES-mediated-ER-mitochondria-contacts-molecular. doi: 10.1242/jcs.058636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Toda T, Cameron S, Sass P, Zoller M, Scott JD, McMullen B, et al. Cloning and characterization of BCY1, a locus encoding a regulatory subunit of the cyclic AMP-dependent protein kinase in Saccharomyces cerevisiae. Mol Cell Biol [Internet]. 1987. Apr 1 [cited 2019 Mar 7];7(4):1371–7. Available from: http://www.ncbi.nlm.nih.gov/pubmed/3037314. doi: 10.1128/mcb.7.4.1371-1377.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bock A, Annibale P, Konrad C, Hannawacker A, Anton SE, Maiellaro I, et al. Optical Mapping of cAMP Signaling at the Nanometer Scale. Cell [Internet]. 2020. Sep 17 [cited 2020 Sep 23];182(0):1–12. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0092867420309430. doi: 10.1016/j.cell.2020.07.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Houslay MD. Underpinning compartmentalised cAMP signalling through targeted cAMP breakdown. Trends Biochem Sci [Internet]. 2010. Feb 1 [cited 2019 Mar 13];35(2):91–100. Available from: https://www.sciencedirect.com/science/article/pii/S0968000409001923?via%3Dihub. doi: 10.1016/j.tibs.2009.09.007 [DOI] [PubMed] [Google Scholar]
- 97.Vandamme J, Castermans D, Thevelein JM. Molecular mechanisms of feedback inhibition of protein kinase A on intracellular cAMP accumulation. Cell Signal [Internet]. 2012. Aug 1 [cited 2019 Jan 24];24(8):1610–8. Available from: https://www.sciencedirect.com/science/article/pii/S0898656812001167. doi: 10.1016/j.cellsig.2012.04.001 [DOI] [PubMed] [Google Scholar]
- 98.Cannon JF, Gitan R, Tatchell K. Yeast cAMP-dependent protein kinase regulatory subunit mutations display a variety of phenotypes. J Biol Chem [Internet]. 1990. Jul 15 [cited 2019 Mar 8];265(20):11897–904. Available from: http://www.ncbi.nlm.nih.gov/pubmed/2164021. [PubMed] [Google Scholar]
- 99.Peck VM, Fuge EK, Padilla PA, Gomez MA, Werner-Washburne M. Yeast bcy1 mutants with stationary phase-specific defects. Curr Genet 1997. 322 [Internet]. 1997 [cited 2022 Nov 18];32(2):83–92. Available from: https://link.springer.com/article/10.1007/s002940050251 [DOI] [PubMed] [Google Scholar]
- 100.Langeberg LK, Scott JD. A-kinase-anchoring proteins. J Cell Sci [Internet]. 2005. Aug 1 [cited 2022 Nov 11];118(15):3217–20. Available from: https://journals.biologists.com/jcs/article/118/15/3217/28454/A-kinase-anchoring-proteins. doi: 10.1242/jcs.02416 [DOI] [PubMed] [Google Scholar]
- 101.Griffioen G, Branduardi P, Ballarini A, Anghileri P, Norbeck J, Baroni MD, et al. Nucleocytoplasmic distribution of budding yeast protein kinase A regulatory subunit Bcy1 requires Zds1 and is regulated by Yak1-dependent phosphorylation of its targeting domain. Mol Cell Biol [Internet]. 2001. Jan 15 [cited 2019 Jan 25];21(2):511–23. Available from: http://www.ncbi.nlm.nih.gov/pubmed/11134339. doi: 10.1128/MCB.21.2.511-523.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Galello F, Moreno S, Rossi S. Interacting proteins of protein kinase A regulatory subunit in Saccharomyces cerevisiae. J Proteomics. 2014. Sep 23;109:261–75. doi: 10.1016/j.jprot.2014.07.008 [DOI] [PubMed] [Google Scholar]
- 103.Kunkel J, Luo X, Capaldi AP. Integrated TORC1 and PKA signaling control the temporal activation of glucose-induced gene expression in yeast. Nat Commun [Internet]. 2019. Dec 1 [cited 2020 Sep 28];10(1):1–11. Available from: 10.1038/s41467-019-11540-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li S, Li Y, Rushing BR, Harris SE, McRitchie SL, Jones JC, et al. Multi-omics analysis of glucose-mediated signaling by a moonlighting Gβ protein Asc1/RACK1. PLOS Genet [Internet]. 2021. Jul 2 [cited 2023 Apr 20];17(7):e1009640. Available from: https://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1009640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hadfield C, Jordan BE, Mount RC, Pretorius GHJ, Burak E. G418-resistance as a dominant marker and reporter for gene expression in Saccharomyces cerevisiae. Curr Genet 1990. 184 [Internet]. 1990 Nov [cited 2022 Dec 13];18(4):303–13. Available from: https://link.springer.com/article/10.1007/BF00318211. doi: 10.1007/BF00318211 [DOI] [PubMed] [Google Scholar]
- 106.Kaster KR, Burgett SG, Ingolia TD. Hygromycin B resistance as dominant selectable marker in yeast. Curr Genet 1984 85 [Internet]. 1984. Jul [cited 2022 Dec 13];8(5):353–8. Available from: https://link.springer.com/article/10.1007/BF00419824. doi: 10.1007/BF00419824 [DOI] [PubMed] [Google Scholar]
- 107.Gasch AP. Yeast genomic expression studies using DNA microarrays. Methods Enzymol. 2002. Jan 1;350:393–414. doi: 10.1016/s0076-6879(02)50976-9 [DOI] [PubMed] [Google Scholar]
- 108.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics [Internet]. 2014. Aug 1 [cited 2022 Dec 13];30(15):2114–20. Available from: https://academic.oup.com/bioinformatics/article/30/15/2114/2390096. doi: 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics [Internet]. 2015. Jan 15 [cited 2022 Dec 13];31(2):166–9. Available from: https://academic.oup.com/bioinformatics/article/31/2/166/2366196. doi: 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol [Internet]. 2010. Mar 2 [cited 2022 Sep 8];11(3):1–9. Available from: https://genomebiology.biomedcentral.com/articles/10.1186/gb-2010-11-3-r25. doi: 10.1186/gb-2010-11-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.de Hoon MJL, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics [Internet]. 2004. Jun 12 [cited 2021 May 21];20(9):1453–4. Available from: http://gcc.gnu.org. doi: 10.1093/bioinformatics/bth078 [DOI] [PubMed] [Google Scholar]
- 112.Saldanha AJ. Java Treeview—extensible visualization of microarray data. Bioinformatics [Internet]. 2004. Nov 22 [cited 2022 Dec 13];20(17):3246–8. Available from: https://academic.oup.com/bioinformatics/article/20/17/3246/186177. doi: 10.1093/bioinformatics/bth349 [DOI] [PubMed] [Google Scholar]
- 113.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics [Internet]. 2010. Jan 1 [cited 2019 Mar 25];26(1):139–40. Available from: https://academic.oup.com/bioinformatics/article-lookup/doi/10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Stat Soc Ser B [Internet]. 1995. Jan 1 [cited 2022 Dec 13];57(1):289–300. Available from: https://onlinelibrary.wiley.com/doi/full/10.1111/j.2517-6161.1995.tb02031.x. [Google Scholar]
- 115.Simillion C, Liechti R, Lischer HEL, Ioannidis V, Bruggmann R. Avoiding the pitfalls of gene set enrichment analysis with SetRank. BMC Bioinformatics [Internet]. 2017. Mar 4 [cited 2021 May 21];18(1):1–14. Available from: https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-017-1571-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Teixeira MC, Monteiro PT, Guerreiro JF, Gonçalves JP, Mira NP, Dos Santos SC, et al. The YEASTRACT database: an upgraded information system for the analysis of gene and genomic transcription regulation in Saccharomyces cerevisiae. Nucleic Acids Res [Internet]. 2014. Jan 1 [cited 2022 Dec 13];42(D1):D161–6. Available from: https://academic.oup.com/nar/article/42/D1/D161/1042148. doi: 10.1093/nar/gkt1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res [Internet]. 2009. Jul 1 [cited 2019 Mar 31];37(Web Server):W202–8. Available from: https://academic.oup.com/nar/article-lookup/doi/ doi: 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS. Quantifying similarity between motifs. Genome Biol [Internet]. 2007. Feb 26 [cited 2022 Dec 13];8(2):1–9. Available from: https://genomebiology.biomedcentral.com/articles/10.1186/gb-2007-8-2-r24. doi: 10.1186/gb-2007-8-2-r24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hutchins PD, Russell JD, Coon JJ. LipiDex: An Integrated Software Package for High-Confidence Lipid Identification. Cell Syst [Internet]. 2018. May 23 [cited 2020 Nov 30];6(5):621–625.e5. Available from: /pmc/articles/PMC5967991/?report=abstract. doi: 10.1016/j.cels.2018.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]