Abstract
Azotobacter vinelandii produces the exopolysaccharide alginate, which is essential for the encystment process. In Pseudomonas aeruginosa, as well as in A. vinelandii, the ςE factor encoded by algU is required for transcription of algD, which encodes a key enzyme of the alginate biosynthetic pathway. The P. aeruginosa response regulator AlgR activates transcription of algD. fimS, located upstream algR, is proposed to encode the AlgR cognate sensor kinase. We have cloned and characterized the A. vinelandii algR gene; the deduced amino acid sequence of the protein encoded by this gene shows 79% identity with its P. aeruginosa homolog. Sequence analysis around the algR gene revealed the absence of a fimS homolog. Inactivation of A. vinelandii algR diminished alginate production by 50%, but did not affect algD transcription, and completely impaired the capacity to form mature cysts. Electron microscopy of the cyst structures formed by the algR mutant revealed that the encystment process is blocked at the step of exine formation. The transcriptional regulation of the A. vinelandii algR gene and the role of AlgR in alginate production differ significantly from those of its P. aeruginosa counterparts. These differences could be due to the fact that in A. vinelandii, alginate plays a role in encystment, a function not found in P. aeruginosa.
Azotobacter vinelandii is a soil bacterium that undergoes a process of cellular differentiation to form metabolically dormant cysts resistant to desiccation (for a review, see reference 34). Mucoid strains of A. vinelandii produce the extracellular polysaccharide alginate, a linear copolymer of d-mannuronic acid and its C-5 epimer l-guluronic acid. Alginate is a major component of the intine and exine layers of the cysts (31) and is essential to the encystment process, since nonmucoid strains fail to form cysts (2, 23, 29).
The alginate biosynthesis pathway in A. vinelandii has been elucidated (32). In this pathway, fructose-6-phosphate is converted by four enzymatic reactions to GDP-mannuronic acid, which is the substrate for polymerization. The resultant polymannuronic acid is secreted and modified by an O-acetylase and an extracellular C-5 epimerase to give the final product, alginate (32). A similar pathway operates in Pseudomonas aeruginosa (21).
The genetics of this process is well known in P. aeruginosa (7, 21), where alginate is an important virulence determinant and a major factor contributing to the intractability of P. aeruginosa lung infection in cystic fibrosis patients (11). With A. vinelandii, important advances have been made in the study of the genetics of alginate synthesis (for a review, see reference 10). These studies have been motivated by the role that this polysaccharide plays in the differentiation process and by the potential of this bacterium for use in the production of alginate for industrial purposes.
In A. vinelandii, as in P. aeruginosa, all of the alginate biosynthetic genes, except algC, are clustered (2, 16, 22, 23, 33). In P. aeruginosa, this cluster is organized in a polycistronic operon transcribed from a promoter located upstream of algD (3). In contrast, the A. vinelandii biosynthetic gene cluster is organized into three operons, one of which transcribes algD (2, 16, 22, 23).
In P. aeruginosa, transcription from the algD promoter is affected by the products of several regulatory genes. The algU-encoded alternative sigma factor, which is similar to the Escherichia coli and Salmonella typhimurium ςE factor (7, 13, 19), is required for algD transcription (36, 41). The products of the mucA and mucB genes counteract AlgU by suppressing its function (7, 36, 37, 43). The algR (5) and algB genes (40) encode response regulators belonging to the superfamily of two-component signal transduction elements. AlgR has been shown to bind to three sites (named RBS) within the algD promoter region and to activate transcription from the AlgU-dependent promoter (26); therefore, inactivation of algR abrogates alginate production (5). Another gene, fimS (also called algZ), which is located upstream of algR and encodes a protein involved in twitching motility (39), has been proposed to be the cognate AlgR sensor kinase, which is also involved in the control of alginate biosynthesis under certain conditions (44). Nonphosphorylated AlgB and AlgR response regulators were recently shown to be active as positive regulators of alginate synthesis (18).
In A. vinelandii, algD is transcribed from at least two promoters which are recognized by RNA polymerase with two different sigma factors: p1, recognized by ς70, and p2, recognized by the alternative ςE factor (AlgU). The algU-mucABCD operon has been characterized (20). These genes seem to be functionally equivalent to those of P. aeruginosa, since inactivation of algU abrogated alginate production, and introduction into strain ATCC 9046 of a plasmid containing the functional mucABCD genes abrogated transcription from the algDp2 promoter and diminished alginate production (29). AlgU activity has been shown to be essential for encystment, independently of its role in alginate synthesis (29). Transcription of the other two operons of the alginate biosynthetic cluster is AlgU independent (16, 22).
Evidence for the presence of algR in bacteria of the Azotobacteriaceae family was previously reported (9). In this study, we report the cloning and sequence of the A. vinelandii algR gene and show that its inactivation diminishes alginate production by 50% but does not affect the transcription of algD; in addition, inactivation of algR was shown to completely impair the encystment process.
MATERIALS AND METHODS
Microbiological procedures.
The bacterial strains and plasmids used in this study are listed in Table 1. The media and growth conditions used were as follows. A. vinelandii was grown at 30°C in Burk’s nitrogen-free salts supplemented with sucrose at 2% (15). Escherichia coli DH5α was grown on LB medium (24) at 37°C. The antibiotic concentrations (micrograms per milliliter) used for A. vinelandii and E. coli, respectively, were as follows: tetracycline, 20 and 20; kanamycin, 5 and 0; rifampin, 20 and 0; ampicillin, 0 and 100; nalidixic acid, 20 and 0; spectinomycin, 100 and 100.
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| A. vinelandii | ||
| ATCC 9046 | Highly mucoid | ATCCa |
| UW136 | Natural algU mutant strain, nonmucoid | 20 |
| AEIV | Mucoid | Svein Valla |
| ATR8 | ATCC 9045 with polar algR::Tcr mutation | This work |
| ATR801 | ATR8 derivative; algR+ Spr | This work |
| ATR9 | ATCC 9045 with Tcr mutation downstream of algR | This work |
| SMU88 | ATCC 9046 with algU::Kmr mutation | 29 |
| WI12 | ATCC 9046; algD-lacZ | 2 |
| WIR1 | WI12 with algR::Spr mutation | This work |
| P. aeruginosa | ||
| PAO1 | Wild type | 14 |
| FRD810 | algR::Smr; nonmucoid | 41 |
| 8852 | algR; nonmucoid | 4 |
| E. coli DH5α | supE44 ΔlacU169 hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 12 |
| Plasmids | ||
| pCP13 | RK2-derived cosmid vector; Tcr Kmr | 4 |
| pAD1039 | Cosmid containing P. aeruginosa DNA including algR; derived from pCP13; Tcr | A. Chakrabarty |
| pMSR1506 | Cosmid containing 25 kb of A. vinelandii DNA including algR, derived from pCP13; Tcr | This work |
| pMSR1507 | pMSR1506 derivative carrying algR::Spr mutation | This work |
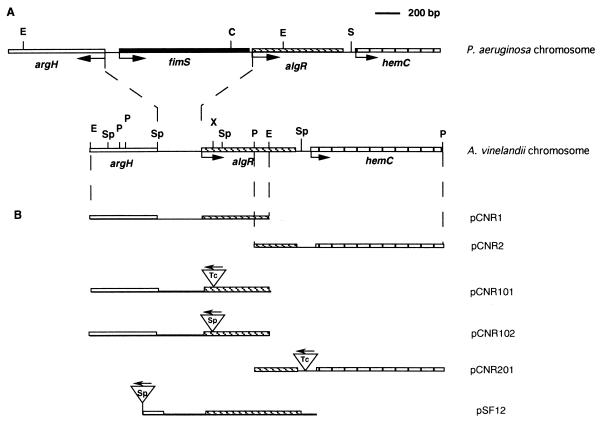
| pCNR1 | pUC18 derivative carrying 1.5-kb EcoRI fragment containing algR 5′ end | This work |
| pCNR101 | pCNR1 derivative containing algR::Tcr mutation | This work |
| pCNR102 | pCNR1 derivative containing algR::Spr mutation | This work |
| pCNR2 | pBluescriptSK+ derivative carrying 2-kb PstI fragment containing algR 3′ end and hemC | This work |
| pCNR201 | pCNR2 derivative containing tetracycline cassette ligated into unique SphI site located 80 nt downstream of algR TGA stop codon | This work |
| pSF12 | pBluescript SK+ derivative carrying entire algR gene amplified by PCR; contains spectinomycin cassette ligated into the polylinker | This work |
| pBluescriptSK+ | Used for subcloning of DNA to be sequenced; Apr | Stratagene |
| pUC18 | Used for subcloning of DNA to be sequenced; Apr | Stratagene |
ATCC, American Type Culture Collection.
Triparental and biparental matings were carried out as previously reported (15). A. vinelandii transformation was carried out as described by Bali et al. (1).
β-Galactosidase activity was measured as reported by Miller (24). One unit corresponds to 1 nmol of o-nitrophenyl-β-d-galactopyranoside hydrolyzed per min per μg of protein. Protein was determined by the method of Lowry et al. (17). All measurements were done in triplicate.
Alginate production was determined as previously described (23); all determinations were done in triplicate.
Encystment induction, desiccation resistance assays, and electron microscopy studies were carried out as previously described (2, 23).
Nucleic acid procedures.
RNA and DNA isolation and cloning, Southern blotting, and nick translation procedures were carried out as previously described (35). Plasmids pCNR1 and pCNR2 (Fig. 1) were used to determine the nucleotide sequence reported here. DNA sequencing was done with the Taq FS DNA polymerase and fluorescent dideoxy terminators by using a cycle sequencing method. Primer extension analysis of algR was carried out with 50 μg of RNA isolated from bacterial cultures grown for 48 h in Burk’s nitrogen-free salts supplemented with 2% sucrose. Primer extension of algD was carried out as previously described (2). Reactions were performed with a primer extension system (Amersham) as instructed by the manufacturer. The P. aeruginosa fimS probe was amplified by PCR using PAO1 chromosomal DNA as a template, as well as oligonucleotides fimS-5′ ACTCTGTCGATGCCTATCCG and fimS-3′ TAGCGTAGACAGGTGTAGTGC.
FIG. 1.
(A) Comparison of the P. aeruginosa and A. vinelandii chromosomal algR regions. (B) Physical map of the plasmids constructed in this study. Arrows indicate the direction of transcription. Antibiotic resistance cassettes are represented by inverted triangles. Abbreviation: C, ClaI; E, EcoRI; P, PstI; S, SmaI; Sp, SphI; X, XhoI.
Construction of plasmid pSF12.
The A. vinelandii algR gene was cloned by PCR using plasmid pMSR1506 as a template, as well as oligonucleotides algR-5′ AAGCTTGTGCAGCTTCTTGCCGGTGATGCC and algR-3′ AAGCTTCGACGGATTGGCGCGGATGATAGC. The resulting 1,458-nucleotide (nt) PCR product was cloned into pBluescript SK+. The resultant plasmid was used to introduce a spectinomycin resistance cassette into the vector polylinker to produce plasmid pSF12 (Fig. 1 and 2).
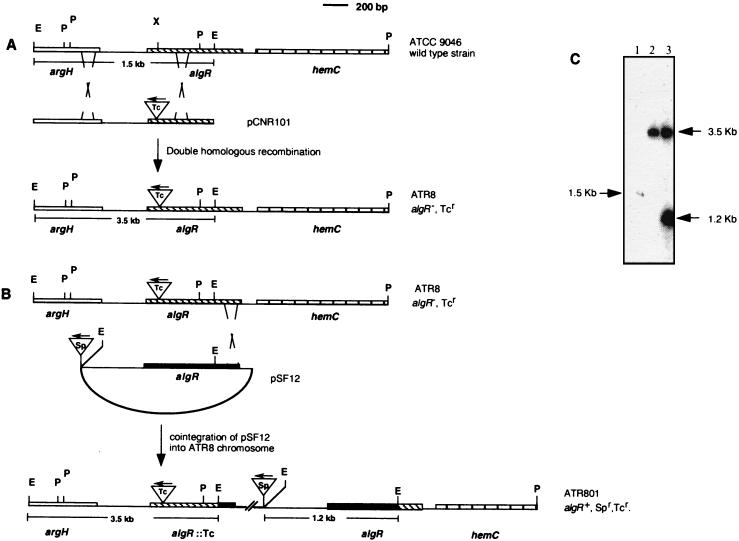
FIG. 2.
(A) Insertional inactivation of the algR gene in A. vinelandii ATCC 9046. (B) Integration of plasmid pSF12 into the ATR8 chromosome. (C) Southern blot hybridization of total genomic DNA digested with EcoRI endonuclease with plasmid pCNR1 as the probe. Lanes: 1, ATCC 9046; 2, ATR8; 3, ATR801.
Construction of strains ATR8, ATR9, and WIR1.
The 1.5-kb EcoRI fragment containing the algR 5′ end was cloned into plasmid pUC18, and the resultant plasmid, pCNR1, was used to introduce into the unique XhoI site either a 2.0-kb SmaI fragment containing an Ω-tetracycline cassette or an Ω-spectinomycin cassette (8). The resultant plasmids, pCNR101 and pCNR102 (Fig. 1B), were unable to replicate in A. vinelandii and were used to introduce the algR::Tcr mutation into strain ATCC 9046 and the algR::Spr mutation into strain WI12. ATR8, a tetracycline-resistant transformant, and WIR1, a spectinomycin-resistant transformant, were selected and confirmed by Southern blot analysis to carry the algR mutations (Fig. 2; data not shown for WIR1).
Plasmid pCNR2 (Fig. 1) was used to introduce the Ω-tetracycline cassette into the unique SphI site located 80 nt downstream of the TGA stop codon of the algR gene. The resultant plasmid, pCNR201 (Fig. 1), was transformed into ATCC 9046, rendering strain ATR9.
Isolation of plasmid pMSR1507.
To isolate a pMSR1506 derivative carrying an algR::Spr mutation, we transferred plasmid pMSR1506 by conjugation from strain WIR1 to E. coli. Selection of a plasmid derivative in which transfer of the algR::Spr mutation from the WIR1 chromosome to plasmid pMSR1506 had occurred by recombination was carried out by isolating E. coli transconjugants resistant to tetracycline and spectinomycin. One Spr Tcr transconjugant was shown by restriction analysis and Southern blotting to contain pMSR1506 with the algR::Spr mutation (data not shown). This plasmid was named pMSR1507.
Nucleotide sequence accession number.
The A. vinelandii algR sequence reported here has been assigned GenBank accession no. AF077237.
RESULTS
Cloning and sequencing of the A. vinelandii algR gene.
It has been previously reported that A. vinelandii has DNA sequences homologous to P. aeruginosa algR (9). The algR gene was cloned from strain ATCC 9046 on the basis of its homology to the corresponding P. aeruginosa gene. Southern blot analysis with an internal fragment of the P. aeruginosa algR gene used as a probe led to the identification of cosmid pMSR1506 carrying a 1.5-kb EcoRI fragment with algR-homologous sequences. The 1.5-kb EcoRI fragment and a 2.0-kb PstI fragment were subcloned into plasmids pUC18 and pBluescript, yielding plasmids pCNR1 and pCNR2, respectively (Fig. 1), which were used to determine the algR nucleotide sequence.
The A. vinelandii algR sequence codes for a 251-amino-acid polypeptide sharing 79% identity with its P. aeruginosa counterpart. As with other response regulators that are phosphorylated, A. vinelandii AlgR contains two highly conserved aspartate residues, Asp8 and Asp54 (Fig. 3).
FIG. 3.
Alignment of the predicted AlgR amino acid sequence from A. vinelandii (A. v.) and that of its homolog from P. aeruginosa (P. a.). Identical residues are shaded, and the conserved Asp8 and Asp54 residues are marked by asterisks.
The fimS gene is not present upstream of algR in A. vinelandii.
In P. aeruginosa, a 1.5-kb region located between argH and algR was recently characterized and shown to contain fimS, whose product is involved in twitching motility (39) (Fig. 1). FimS has also been postulated to be the AlgR cognate sensor kinase involved in the regulation of alginate production under certain conditions (44). Analysis of 1,000 nt of the DNA sequence upstream of the first ATG of algR in A. vinelandii revealed the presence of argH but the absence of a gene homologous to fimS (Fig. 1). Southern blot hybridization of EcoRI-digested DNA from A. vinelandii AEIV and UW136 with the 1.5-kb EcoRI fragment containing the 3′ end of argH and the 5′ portion of algR as a probe revealed a 1.5-kb EcoRI fragment (data not shown), indicating conservation of this region among different A. vinelandii strains. A 1,054-nt fragment corresponding to P. aeruginosa fimS, encompassing codons for amino acids 1 to 348, did not hybridize with the A. vinelandii chromosome, even under low-stringency conditions (data not shown), suggesting the absence of an fimS homolog in A. vinelandii.
Characterization of an algR mutant.
To determine whether the algR gene is involved in alginate regulation in A. vinelandii, strain ATR8 carrying an algR::Tcr mutation was constructed as described in Materials and Methods. In contrast to P. aeruginosa, where algR mutations totally abrogate alginate production, we found that in A. vinelandii, the algR mutation only diminished alginate production by 50% (Table 2).
TABLE 2.
Alginate production and encystment in different A. vinelandii strains
| Strain | Genotype | Mean alginate concn (μg/mg of protein) ± SEM
|
Mean encystment (%) ± SEM | |
|---|---|---|---|---|
| BSa | BBb | |||
| ATCC 9046 | Wild type | 3,612 ± 624 | 2,648 ± 322 | 5.16 ± 1.7 |
| ATR8 | algR::Tcr | 1,357 ± 92 | 920 ± 20 | 0.004 ± 0.002 |
| ATR801 | ATR8/algR+ | 4,171 ± 292 | NDc | 1.19 ± 0.13 |
| ATR9 | algR+ | ND | ND | 1.05 ± 0.19 |
| WI12 | algD::lacZ | 259 ± 45 | 1,121 ± 148 | 5.21 ± 1.0 |
BS, Burk’s nitrogen-free salts supplemented with 2% sucrose as a carbon source.
BB, Burk’s nitrogen-free salts supplemented with 0.2% n-butanol as a carbon source.
ND, not determined.
AlgR is not required for algD transcription.
In P. aeruginosa, AlgR activates transcription of algD from its AlgU-dependent promoter and has been shown to bind to three sites (RBS sites) upstream of algD (25, 26). We determined, by primer extension, algD transcription in ATCC 9046, as well as in the algR mutant. Transcription of algD initiates from the two previously reported promoters, AlgU (p2) and ς70 (p1), and also from a third site located 62 nt upstream the ATG start codon (Fig. 4). No consensus sequences similar to known promoters were found around the −10 and −35 regions of this transcription initiation site.
FIG. 4.
Primer extension analysis of algD transcription in strains ATCC 9046 and ATR8. (A) DNA sequence of the 5′ end of algD. Arrows indicate the start sites of algD transcription. The ATG initiation codon is overlined. (B) Primer extension of the algD gene in strains ATCC 9046 (lane 1) and ATR8 (lane 2).
We confirmed that the algR mutation has no effect on algD transcription by comparing the β-galactosidase activity of strain WI12, an ATCC 9046 derivative carrying an algD::lacZ gene fusion, with that of WIR1, a WI12 derivative carrying the algR::Spr mutation. As expected, the algR mutant and wild-type strains presented similar β-galactosidase activities (9.9 ± 0.1 and 11.4 ± 0.9 U/μg of protein, respectively) after 24 h of growth in Burk’s nitrogen-free salts supplemented with 2% sucrose, thus confirming that algD transcription is not affected by the algR mutation.
The A. vinelandii algR gene is functional in P. aeruginosa.
Cosmid pMSR1506 was transferred by conjugation into two P. aeruginosa algR mutants, FRD810 and 8852. As a positive control, we used plasmid pAD1039 carrying P. aeruginosa algR in the same vector. Plasmid pMSR1506 partially restored alginate production to both strains (Table 3). These data imply that the A. vinelandii AlgR protein is functional as an activator of the algD promoter in P. aeruginosa.
TABLE 3.
Complementation of P. aeruginosa algR mutants by A. vinelandii algR
| Strain | Genotype | Mucoidy | Mean alginate concn (μg/mg of protein)a ± SEM |
|---|---|---|---|
| FRD810 | algR | − | 51.8 ± 3 |
| FRD810/pAD1039 | P. aeruginosa algR+ | ++ | 7,607 ± 1,082 |
| FRD810/pMSR1506 | A. vinelandii algR+ | + | 648 ± 74 |
| FRD810/pCP13 | Vector | − | 113 ± 24 |
| 8852 | algR | − | 28 ± 8 |
| 8852/pAD1039 | P. aeruginosa algR+ | ++ | 9,218 ± 1,400 |
| 8852/pMSR1506 | A. vinelandii algR+ | + | 1,434 ± 162 |
| 8852/pCP13 | Vector | − | 10.3 ± 1.3 |
Alginate was determined in cells grown for 48 h on solid Pseudomonas isolation agar (PIA; Difco) medium.
Effect of the algR mutation on encystment.
We studied encystment in algR mutant strain ATR8. We measured the desiccation resistance of cultures induced for encystment with n-butanol. An encystment frequency reduction of more than 1,000-fold was observed in algR mutant strain ATR8 (Table 2).
We tested whether the failure of strain ATR8 to encyst was caused by the 50% decrease in alginate production. Alginate production under encystment conditions was determined for ATR8 and WI12. Table 2 shows that strain ATR8 is unable to form desiccation-resistant cysts despite the fact that under encysting conditions it produced alginate levels similar to those of encysting strain WI12.
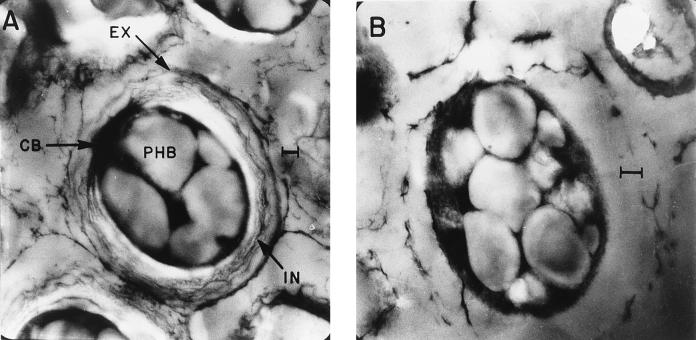
Electron microscopy of the cultures of the algR mutant induced for encystment is shown in Fig. 5. The morphology of a mature cyst, as has been described elsewhere (42), is observed in wild-type strain ATCC 9046; i.e., the central body is surrounded by two capsule-like layers, the intine and the exine. No mature cysts were seen in the algR mutant, where the central body was surrounded by an incipient exine, and no intine layer was observed. Since the exine is the first layer to be formed, this phenotype indicates that the encystment process stopped at an early stage in the differentiation process.
FIG. 5.
Electron micrographs of the cysts formed by strains ATCC 9046 (A) and ATR8 (B). Abbreviations: EX, exine; IN, intine; CB, central body; PHB, poly-β-hydroxybutyrate. Bars, 0.4 μm.
To rule out the possibility that the encystment-defective phenotype was caused by polarity of the algR mutation, strain ATR9, which carries an Spr cassette 80 nt downstream the algR TGA stop codon, was constructed as described in Materials and Methods. This strain was found to encyst (Table 2).
Complementation of the algR mutation.
To confirm that the algR mutation caused the cyst-defective phenotype, we constructed plasmid pSF12, a pSK derivative carrying the A. vinelandii algR gene (Fig. 2). This plasmid, which is unable to replicate in A. vinelandii, was transformed into the ATR8 mutant. Transformant ATR801, which is resistant to spectinomycin and carries plasmid pSF12 integrated into the chromosome, was selected. Integration of the plasmid was confirmed by Southern blot analysis (Fig. 2C). Strain ATR801 produced alginate in a manner similar to that of wild-type ATCC 9046 and was able to encyst. Encystment of ATR801 was reduced four times relative to that of the wild type; however, in our experience, differences in encystment below 10 times are not significant (Table 2).
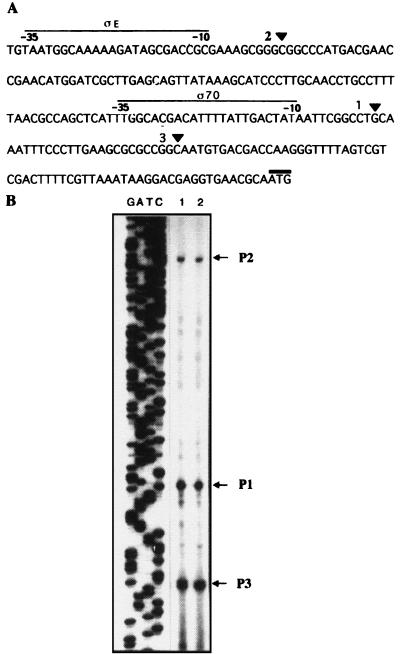
Transcription analysis of algR.
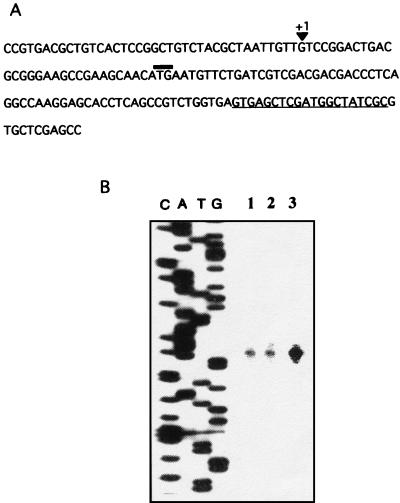
The mRNA start site for the algR gene was determined by primer extension (Fig. 6). A unique start site, 30 nt upstream the ATG codon, was found in strain ATCC 9046 (Fig. 6B, line 3). No consensus for known bacterial promoters was found at the −10 and −35 promoter algR region (Fig. 6A). In contrast to that in P. aeruginosa (41), algR transcription is not abrogated in algU mutants SMU88 and UW136 (Fig. 6B, lines 1 and 2).
FIG. 6.
Primer extension analysis of algR transcription in strains ATCC 9046, SMU88, and UW136. (A) DNA sequence of the 5′ end of algR. The arrow indicates the start site of algR transcription. The ATG initiation codon is overlined. The complementary sequence where the oligonucleotide used for primer extension analysis was generated is underlined. (B) Primer extension of the algR gene in strains SMU88 (lane 1), UW136 (lane 2), and ATCC 9046 (lane 3).
Plasmid pMSR1506 suppresses encystment in strain ATCC 9046.
Plasmid pMSR1506, harboring a copy of the wild-type algR gene, was found to reduce mucoidy and suppress encystment in wild-type strain ATCC 9046 (Table 4). The reduction in alginate production caused by plasmid pMSR1506 could be the cause for the cyst-defective phenotype. When overexpressed, P. aeruginosa algR has a negative effect on alginate synthesis (6). To investigate whether the negative effect of pMSR1506 on alginate production and encystment was due to algR, plasmid pMSR1507, a pMSR1506 derivative carrying an algR::Spr mutation, was isolated as described in Materials and Methods. This plasmid has the same effects on encystment and alginate production in ATCC 9046 (Table 4). Thus, the negative effect is not caused by algR.
TABLE 4.
Effect of plasmid pMSR1506 on alginate production and encystment in A. vinelandii
| Strain | Mean alginate concn (μg/mg of protein) ± SEM
|
Mean encystment (%) ± SEM | |
|---|---|---|---|
| BSa | BBb | ||
| ATCC 9046 | 3,612 ± 624 | 2,648 ± 322 | 5.6 ± 1.7 |
| ATCC 9046/pMSR1506 | 440 ± 50 | 265 ± 25 | 0.003 ± 0.001 |
| ATCC 9046/pMSR1507 | 215 ± 10 | 327 ± 35 | 0.002 ± 0.0008 |
| ATCC 9046/pCP13 | 3,070 ± 57 | 2,141 ± 270 | 6.4 ± 1.3 |
BS, Burk’s nitrogen-free salts supplemented with 2% sucrose as a carbon source.
BB, Burk’s nitrogen-free salts supplemented with 0.2% n-butanol as a carbon source.
DISCUSSION
In this study, we cloned and characterized the algR gene from A. vinelandii. We found a high degree of homology with P. aeruginosa algR, encoding the response regulator AlgR, which, together with AlgU, is absolutely required for activation of the alginate biosynthetic operon controlled by the algD promoter. AlgU is the alternative sigma factor required for transcription of algD in P. aeruginosa. In A. vinelandii, algD is transcribed from three promoters, one of which, p2, is an AlgU-dependent promoter (2, 29). Our results show that in contrast to P. aeruginosa, A. vinelandii does not require AlgR for activation of algD transcription from any of its promoters, including the p2 AlgU-dependent promoter. Partial complementation of P. aeruginosa algR mutants with the A. vinelandii algR gene, however, implies that A. vinelandii AlgR can bind the RBS sequences present upstream of the P. aeruginosa algD promoter and can interact with AlgU RNA polymerase. This finding is consistent with the lack in A. vinelandii of sequences homologous to P. aeruginosa RBS (2). We propose that a transcriptional regulator other than AlgR activates transcription of algD from the AlgU-dependent promoter.
In P. aeruginosa, transcription of algR is dependent on AlgU but not on AlgR (41); similarly, transcription of A. vinelandii algD from the p2 promoter depends on AlgU but not on AlgR; thus, there are promoters that are recognized by AlgU but not activated by AlgR.
Although AlgR does not appear to be required for transcription of algD in A. vinelandii, alginate production in the algR mutant is reduced by 50%, implying that AlgR may exert some control over other alginate biosynthetic or regulatory genes. Our previous studies have shown that an unidentified biosynthetic or regulatory alginate gene other than algD is under the control of AlgU (29). Thus, the above-mentioned alg gene could be activated by AlgR.
The ATR8 mutant reported here has a cyst-defective phenotype; encystment in ATR8 seems to stop at the step of exine organization. We previously showed that encystment is also impaired when plasmid pSMU865, carrying the mucABCD genes coding for negative regulators of AlgU activity, is introduced into strain ATCC 9046 (20, 29). However, in this case, encystment proceeds a step further, since cyst structures formed by ATCC 9046/pSMU865 lack the intine layer but show a well-structured exine. Both the AlgU and AlgR proteins are essential for encystment, so it is a possibility that AlgR can activate genes involved in the encystment process whose promoters are recognized by AlgU. The putative AlgR requirement for transcription of genes involved in encystment may facilitate the identification of such genes. Transcription of algR is not abrogated in the algU mutant strains; accordingly, the algR promoter does not have AlgU consensus sequences. This indicates the potential existence of another sigma factor involved in alginate and encystment control.
In P. aeruginosa, the algR gene is flanked by the hemC gene located immediately downstream (28) and the fimS gene coding for the putative cognate sensor kinase of AlgR, which is located immediately upstream of algR and is involved in twitching motility (39). The argH gene is located upstream of fimS (Fig. 1). Our characterization of the algR flanking regions in A. vinelandii identified the hemC gene immediately downstream of algR and the argH gene 230 nt upstream of algR (Fig. 1). No open reading frames or other recognizable features are found within these 230 nt. Thus, fimS was not present upstream of algR; we also found that sequences homologous to fimS were not present elsewhere in the A. vinelandii chromosome. Type 4 fimbriae are associated with twitching motility and are found in some pathogenic bacteria (38). There is no evidence that A. vinelandii possesses type 4 fimbriae; therefore, this may be the reason for the absence of fimS in this bacterium. The cognate sensor kinase of AlgR remains unidentified in A. vinelandii.
It was recently shown that in P. aeruginosa, phosphorylation of neither AlgR nor AlgB is needed for alginate production (18). Phosphorylation is essential for the activity of most of the response regulators so far studied; an exception is the nonphosphorylated response regulator DegU that activates the late competence genes comC and comG, whereas phosphorylated DegU is required for the expression of genes encoding degradative enzymes (30). Similarly, it was postulated that in P. aeruginosa, nonphosphorylated AlgR is involved in alginate production while phosphorylated AlgR may play a role in twitching motility (39).
Another hypothesis proposed to explain the lack of AlgR phosphorylation is that mucoid strains containing nonphosphorylated response regulators are not typical wild-type strains but carry mutations in the mucA gene and synthesize high levels of AlgU. Thus, mucoid P. aeruginosa mucA strains may have elevated levels of the response regulators AlgR and AlgB that bypass the need for phosphorylation.
Whether phosphorylation of A. vinelandii AlgR is necessary for activation of its target cyst promoters remains to be investigated. Further studies will help to clarify the role of a response regulator such as AlgR in signal transduction and its interaction with AlgU-RNA polymerase.
Finally, while trying to complement the A. vinelandii algR mutants with the cosmid pMSR1506, we detected an inhibition of alginate production and encystment caused by this plasmid. Inhibition of encystment could be a consequence of the reduction in alginate. However, we have previously shown that strains that produce alginate similar to that of ATCC 9046/pMSR1506 are able to encyst (29). We provided evidence indicating that this inhibition is not caused by algR itself. Thus, other loci that participate in alginate and encystment control lie in the algR chromosomal region.
ACKNOWLEDGMENTS
This work was supported by grant IN212096 from DGAPA-PAPIIT UNAM. C.N. thanks CONACYT and PADEP-UNAM for financial support during her Ph.D. studies.
We thank Rebeca Nájera and Josefina Guzmán for technical support and A. Chakrabarty, D. J. Wozniak, and J. Goldberg for providing the P. aeruginosa algR plasmid and strains.
REFERENCES
- 1.Bali A, Blanco G, Hill S, Kennedy C. Excretion of ammonium by a nifL mutant of Azotobacter vinelandii fixing nitrogen. Appl Environ Microbiol. 1992;58:1711–1718. doi: 10.1128/aem.58.5.1711-1718.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campos M-E, Martínez-Salazar J M, Lloret L, Moreno S, Núñez C, Espín G, Soberón-Chávez G. Characterization of the gene coding for GDP-mannose dehydrogenase (algD) from Azotobacter vinelandii. J Bacteriol. 1996;178:1793–1799. doi: 10.1128/jb.178.7.1793-1799.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chitnis C E, Ohman D E. Genetic analysis of the alginate biosynthetic gene cluster of Pseudomonas aeruginosa shows evidence of an operonic structure. Mol Microbiol. 1993;8:583–590. doi: 10.1111/j.1365-2958.1993.tb01602.x. [DOI] [PubMed] [Google Scholar]
- 4.Darzins A, Chakrabarty A M. Cloning of genes controlling alginate biosynthesis from a mucoid cystic fibrosis isolate of Pseudomonas aeruginosa. J Bacteriol. 1984;159:9–19. doi: 10.1128/jb.159.1.9-18.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deretic V, Dikshit R, Konyecsni W M, Chakrabarty A M, Misra T K. The algR gene, which regulates mucoidy in Pseudomonas aeruginosa, belongs to a class of environmentally responsive genes. J Bacteriol. 1989;171:1278–1283. doi: 10.1128/jb.171.3.1278-1283.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deretic V, Konyecsni W M. Control of mucoidy in Pseudomonas aeruginosa: transcriptional regulation of algR and identification of the second regulatory gene, algQ. J Bacteriol. 1989;171:3680–3688. doi: 10.1128/jb.171.7.3680-3688.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deretic V, Schurr M J, Boucher J C, Martin D W. Conversion of Pseudomonas aeruginosa to mucoidy in cystic fibrosis: environmental stress and regulation of bacterial virulence by alternative sigma factors. J Bacteriol. 1994;176:2773–2780. doi: 10.1128/jb.176.10.2773-2780.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fellay R, Frey J, Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of Gram-negative bacteria. Gene. 1987;52:147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- 9.Fialho A M, Zielinski N A, Fett W F, Chakrabarty A M, Berry A. Distribution of alginate gene sequences in the Pseudomonas rRNA homology group I-Azomonas-Azotobacter lineage of superfamily B procaryotes. Appl Environ Microbiol. 1990;56:436–443. doi: 10.1128/aem.56.2.436-443.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gacesa P. Bacterial alginate biosynthesis, recent progress and future prospects. Microbiology. 1998;144:1133–1143. doi: 10.1099/00221287-144-5-1133. [DOI] [PubMed] [Google Scholar]
- 11.Govan J R W. Alginate biosynthesis and other unusual characteristics associated with the pathogenesis of Pseudomonas aeruginosa in cystic fibrosis. In: Griffiths E, Donachie W, Stephen J, editors. Bacterial infections of respiratory and gastrointestinal mucosae. Oxford, England: IRL Press; 1988. pp. 67–96. [Google Scholar]
- 12.Hanahan D. Studies on transformation of E. coli. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 13.Hershberger C D, Ye R W, Parsek M R, Xie Z D, Chakrabarty A M. The algT (algU) gene of Pseudomonas aeruginosa, a key regulator involved in alginate biosynthesis, encodes an alternative ς factor (ςE) Proc Natl Acad Sci USA. 1995;92:7941–7945. doi: 10.1073/pnas.92.17.7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holloway B W. Genetic recombination in Pseudomonas aeruginosa. J Gen Microbiol. 1955;13:572–581. doi: 10.1099/00221287-13-3-572. [DOI] [PubMed] [Google Scholar]
- 15.Kennedy C, Gamal R, Hummprey R, Ramos J, Brigle K, Dean D. The nifH, nifM, and nifN genes of Azotobacter vinelandii: characterization by Tn5 mutagenesis and isolation from pLARF1 gene banks. Mol Gen Genet. 1986;205:318–325. [Google Scholar]
- 16.Lloret L, Barreto R, León R, Moreno S, Martínez-Salazar J, Espín G, Soberón-Chávez G. Genetic analysis of the transcriptional arrangement of Azotobacter vinelandii alginate biosynthetic genes: identification of two independent promoters. Mol Microbiol. 1996;21:449–457. doi: 10.1111/j.1365-2958.1996.tb02554.x. [DOI] [PubMed] [Google Scholar]
- 17.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 18.Ma S, Selvaraj U, Ohman D E, Quarless R, Hassett D J, Wozniak D J. Phosphorylation-independent activity of the response regulators AlgB and AlgR in promoting alginate biosynthesis in mucoid Pseudomonas aeruginosa. J Bacteriol. 1998;180:956–968. doi: 10.1128/jb.180.4.956-968.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin D W, Schurr M J, Yu H, Deretic V. Analysis of promoters controlled by the putative sigma factor AlgU regulating conversion to mucoidy in Pseudomonas aeruginosa: relationship to ςE and stress response. J Bacteriol. 1994;176:6688–6696. doi: 10.1128/jb.176.21.6688-6696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martínez-Salazar J M, Moreno S, Nájera R, Boucher J C, Espín G, Soberón-Chávez G, Deretic V. Characterization of the genes coding for the putative sigma factor AlgU and its negative regulators MucA, MucB, MucC, and MucD in Azotobacter vinelandii and evaluation of their roles in alginate biosynthesis. J Bacteriol. 1996;178:1800–1808. doi: 10.1128/jb.178.7.1800-1808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.May T, Chakrabarty A M. Pseudomonas aeruginosa: genes and enzymes of alginate synthesis. Trends Microbiol. 1994;2:151–157. doi: 10.1016/0966-842x(94)90664-5. [DOI] [PubMed] [Google Scholar]
- 22.Mejía-Ruíz H, Guzmán J, Moreno S, Soberón-Chávez G, Espín G. The Azotobacter vinelandii alg8 and alg44 genes are essential for alginate synthesis and can be transcribed from an algD-independent promoter. Gene. 1997;199:271–277. doi: 10.1016/s0378-1119(97)00380-6. [DOI] [PubMed] [Google Scholar]
- 23.Mejía-Ruíz H, Moreno S, Guzmán J, Nájera R, León R, Soberón-Chávez G, Espín G. Isolation and characterization of an Azotobacter vinelandii algK mutant. FEMS Microbiol Lett. 1997;156:101–106. doi: 10.1111/j.1574-6968.1997.tb12712.x. [DOI] [PubMed] [Google Scholar]
- 24.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 431–435. [Google Scholar]
- 25.Mohr C D, Hibler N S, Deretic V. AlgR, a response regulator controlling mucoidy in Pseudomonas aeruginosa, binds to the FUS sites of the algD promoter located unusually far upstream from the mRNA start site. J Bacteriol. 1991;173:5136–5143. doi: 10.1128/jb.173.16.5136-5143.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mohr C D, Leveau J H J, Krieg D P, Hibler N S, Deretic V. AlgR binding sites within the algD promoter make up a set of inverted repeats separated by a large intervening segment of DNA. J Bacteriol. 1992;174:6624–6633. doi: 10.1128/jb.174.20.6624-6633.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mohr C D, Martin D W, Konyecsni W N, Govan J R W, Lory S, Deretic V. Role of the far-upstream sites of the algD promoter and the algR and rpoN genes in environmental modulation of mucoidy in Pseudomonas aeruginosa. J Mol Biol. 1990;172:6576–6580. doi: 10.1128/jb.172.11.6576-6580.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mohr C D, Sonsteby S K, Deretic V. The Pseudomonas aeruginosa homologs of hemC and hemD are linked to the gene encoding the regulator of mucoidy algR. Mol Gen Genet. 1994;242:177–184. doi: 10.1007/BF00391011. [DOI] [PubMed] [Google Scholar]
- 29.Moreno S, Guzmán J, Nájera R, Soberón-Chávez G, Espín G. Role of the alternative ςE factor AlgU in encystment of Azotobacter vinelandii. J Bacteriol. 1998;180:2766–2769. doi: 10.1128/jb.180.10.2766-2769.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Msadek T, Kunst F, Rapoport G. A signal transduction network in Bacillus subtilis includes the DegS/DegU and ComP/ComA two-component systems. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: ASM Press; 1995. pp. 447–471. [Google Scholar]
- 31.Page W J, Sadoff H L. Relationship between calcium and uronic acids in the encystment of Azotobacter vinelandii. J Bacteriol. 1975;122:145–151. doi: 10.1128/jb.122.1.145-151.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pindar D F, Bucke C. The biosynthesis of alginic acid by Azotobacter vinelandii. Biochem J. 1975;152:617–622. doi: 10.1042/bj1520617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rehm H A, Ertesvag H, Valla S. A new Azotobacter vinelandii mannuronan C-5 epimerase gene (algG) is part of an alg gene cluster physically organized in a manner similar to that in Pseudomonas aeruginosa. J Bacteriol. 1996;178:5884–5889. doi: 10.1128/jb.178.20.5884-5889.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sadoff H L. Encystment and germination in Azotobacter vinelandii. Bacteriol Rev. 1975;39:516–539. doi: 10.1128/br.39.4.516-539.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Schurr M J, Martin D W, Mudd M H, Deretic V. Gene cluster controlling conversion to alginate-overproducing phenotype in Pseudomonas aeruginosa: functional analysis in a heterologous host and role in the instability of mucoidy. J Bacteriol. 1994;176:3375–3382. doi: 10.1128/jb.176.11.3375-3382.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schurr M J, Yu H, Martínez-Salazar J M, Boucher J C, Deretic V. Control of AlgU, a member of the ςE-like family of stress sigma factors, by the negative regulators MucA and MucB and Pseudomonas aeruginosa conversion to mucoidy in cystic fibrosis. J Bacteriol. 1996;178:4997–5004. doi: 10.1128/jb.178.16.4997-5004.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strom M S, Lory S. Structure-function and biogenesis of the type IV pili. Annu Rev Microbiol. 1993;47:565–596. doi: 10.1146/annurev.mi.47.100193.003025. [DOI] [PubMed] [Google Scholar]
- 39.Whitchurch C B, Alm R A, Mattick J S. The alginate regulator AlgR and an associated sensor FimS are required for twitching motility in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1996;93:9839–9843. doi: 10.1073/pnas.93.18.9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wozniak D J, Ohman D E. Pseudomonas aeruginosa AlgB, a two-component response regulator of the NtrC family, is required for algD transcription. J Bacteriol. 1991;173:1406–1413. doi: 10.1128/jb.173.4.1406-1413.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wozniak D J, Ohman D E. Transcriptional analysis of the Pseudomonas aeruginosa genes algR, algB, and algD reveals a hierarchy of alginate gene expression which is modulated by algT. J Bacteriol. 1994;176:6007–6014. doi: 10.1128/jb.176.19.6007-6014.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wyss O, Newmann M G, Socolofsky M D. Development and germination of the Azotobacter cyst. J Biophys Biochem Cytol. 1961;10:555–565. doi: 10.1083/jcb.10.4.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xie Z-D, Hershberger C D, Shankar S, Ye R W, Chakrabarty A M. Sigma factor–anti-sigma factor interaction in alginate synthesis: inhibition of AlgT by MucA. J Bacteriol. 1996;178:4990–4996. doi: 10.1128/jb.178.16.4990-4996.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu H, Mudd M, Boucher J C, Schurr M J, Deretic V. Identification of the algZ gene upstream of the response regulator algR and its participation in control of alginate production in Pseudomonas aeruginosa. J Bacteriol. 1997;179:187–193. doi: 10.1128/jb.179.1.187-193.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]