Abstract
Alzheimer’s disease (AD) is the leading form of dementia worldwide, but its early detection and diagnosis remain a challenge. MicroRNAs (miRNAs) are a group of small endogenous RNA molecules that regulate mRNA expression. Recent evidence suggests miRNAs play an important role in the five major hallmarks of AD pathophysiology: amyloidogenesis, tauopathy, neuroinflammation, synaptic dysfunction, and neuronal death. Compared to traditional biomarkers of AD, miRNAs display a greater degree of stability in cerebrospinal fluid. Moreover, aberrant changes in miRNA expression can be measured over time to monitor and guide patient treatment. Specific miRNA profiles and combinations may also be used to distinguish AD subjects from normal controls and other causes of dementia. Because of these properties, miRNAs are now being considered as promising and potential biomarkers of AD. This review comprehensively summarizes the diagnostic potential and regulatory roles miRNAs play in AD.
Keywords: Alzheimer’s disease, neurodegenerative disorder, dementia, MicroRNAs, cerebrospinal fluid, diagnosis, biomarker, neuroinflammation
1. Introduction
Alzheimer’s disease (AD) is the leading form of dementia among the elderly and is currently estimated to affect 55 million people worldwide (Brito-Aguilar, 2019; Shin, 2022). AD is a multifactorial neurodegenerative disorder that is typically characterized by an insidious decline in cognitive function. The memory loss and behavioral impairment in AD patients can become so severe that they are unable to independently perform daily activities without external aid from others. Our current understanding of the pathophysiology underlying the development of AD is based on three main cardinal events: accumulation of amyloid-beta (Aβ), neurofibrillary tangle (NFT) formation, and sustained neuroinflammation (Ricciarelli and Fedele, 2017). These three events all eventually contribute to the synaptic dysfunction and neuronal loss primarily seen in the medial temporal lobe, frontal cortex, and hippocampus of AD patients (Shimanoe et al., 2019; Breijyeh and Karaman, 2020). Recent epidemiological studies highlight that the incidence of AD had increased by approximately 148% since 1990 with an estimated 7.7 million new cases occurring every year (Li et al., 2022; Shin, 2022). With the aging population expected to continue to rise worldwide, there is an increasing need to identify reliable diagnostic biomarkers so that patients receive timely intervention at early stages of AD.
The definitive diagnosis of AD is currently only possible by post-mortem microscopic examination of the patient’s brain tissue. However, several tools are nowadays available for clinicians to use in diagnosing living patients with a greater degree of certainty. These diagnostic modalities generally fall into one of the following three categories: clinical neuropsychological criteria, radiological brain assessment, and biochemical screening tests (Rajasekhar and Govindaraju, 2018; Weller and Budson, 2018). Although Aβ and Tau proteins are currently the only well-recognized biomarkers for AD diagnosis, recent evidence suggests that microRNAs (miRNAs) are a promising alternative (Angelucci et al., 2019; Liu et al., 2022). Alterations in miRNA expression can be readily detected in a patient’s serum, plasma, or cerebrospinal fluid (CSF). Compared to protein biomarkers, miRNAs form highly stable complexes that allow for their easier and more cost-effective detection (Angelucci et al., 2019). Additionally, since miRNAs have been demonstrated to directly regulate several pathways in AD, unique miRNA expression patterns can be used to diagnose AD with a high degree of specificity. It is currently reported that fluctuations in CSF proteins are not always AD-specific and may only reflect a general state of brain degeneration (Lee et al., 2019). Alterations in tau protein levels, for instance, have been observed to occur in acute brain insults such as stroke and trauma (Lee et al., 2019). While miRNA blood analysis is more convenient, the CSF as a medium better reflects changes in brain physiology and can more closely represent expression alterations of circulating miRNAs. In this review, we comprehensively summarize and present the current literature on the roles and molecular targets of differentially expressed CSF miRNAs in AD. We also explore the potential of CSF miRNAs as biomarkers for AD compared to more traditional diagnostic modalities.
2. Current issues with Alzheimer’s disease diagnosis
The early detection and prompt diagnosis of AD are important determinants of patient outcome. However, the neuropathological changes seen in AD can begin for up to a decade before the first signs of impairment are noticeable. Moreover, when symptoms are present, there is great variability in clinical presentation and patient symptoms may overlap with other forms of dementia. Research, therefore, continues to investigate new biomarkers that will allow for the identification of high-risk individuals and the detection of AD.
The current modalities used to diagnose AD fall into three categories: clinical neuropsychological criteria, radiological imaging, and biochemical tests (Dubois et al., 2021). First established in 1984, the criteria set by the National Institute on Aging and the Alzheimer’s Association (NIA-AA) have become the initial and most universally adopted tool by clinicians suspecting AD in a primary care setting. Although the criteria have been recently revised to account for the different stages of Alzheimer’s, they are still not reliable enough to establish a diagnosis alone. One of the main limitations of the criteria is that they cannot definitively distinguish AD from other forms of cognitive impairment (Gaugler et al., 2013). Studies suggest that up to 23% of all patients clinically diagnosed with AD are misdiagnosed and lack the characteristic pathological findings on autopsy (Gaugler et al., 2013). The misdiagnosis of AD can prevent timely intervention and management of dementia patients with conditions that are perhaps reversible. Another point of concern is that the criteria heavily rely on the subjective assessment of the healthcare provider. For instance, clinicians might not initially suspect AD in patients with coexisting medical conditions and thus unintentionally omit a thorough assessment of cognitive decline. Factors such as language, education, and socio-cultural background may also affect the attitudes of different patient populations toward aging and dementia which ultimately influence how signs of AD are reported. Various neuroimaging techniques may also be used to aid in the diagnosis of AD. Magnetic resonance imaging (MRI), for example, can detect cerebral atrophy and ventricular enlargement in dementia patients (Kim et al., 2022). Positron emission tomography (PET) can use radiolabeled biomarkers to detect reduced cerebral glucose metabolism, the presence of Aβ plaques, and hyperphosphorylated tau (Kim et al., 2022). Single-photon emission computerized tomography (SPECT) scans can reveal temporoparietal cerebral hypoperfusion, an important distinguishing factor of AD (Banerjee et al., 2020). While these imaging modalities are very sensitive and provide evidence of pathological changes in brain structure and function, they also have their limitations. Many of these techniques are time-consuming and may require the use of expensive equipment and tracers which limit their widespread availability. Moreover, the results may not be specific for AD as some changes, notably the presence of Aβ, can be seen in aging but cognitively normal individuals (Banerjee et al., 2020). Biochemical screening of AD is based on the detection of Aβ42, Aβ42/Aβ40 ratio, phosphorylated tau (p-tau), and total tau (t-tau) in body fluids. These biomarkers are major hallmarks of AD pathology and are thus believed to be a promising tool for early diagnosis of AD. However, according to recent research, only a combination of these biomarkers was sufficient to discriminate AD from healthy controls with a sensitivity and specificity of around 90% (Khoury and Ghossoub, 2019). When used individually, these biomarkers become less reliable as studies report inconsistent and varying results. Moreover, these biomarkers did not effectively distinguish AD from other causes of dementia (Khoury and Ghossoub, 2019). For example, while the sensitivity of using t-tau as a single biomarker remained at 81%, its specificity dramatically dropped to 57% (Hampel and Blennow, 2004). Although different samples from the same patient yield consistent results, studies report variations in the sensitivity and specificity of these biomarkers between different patient populations (Hampel and Blennow, 2004). The lack of threshold values and a standardized methodology present a major limitation to the use of these biomarkers. As of now, there is no single biomarker that can by itself diagnose AD. However, the effective and early diagnosis of AD may be achieved by a combination of biomarkers that cover different aspects of its pathophysiology.
3. Structure, function, and molecular characteristics of MicroRNAs
MicroRNAs are a group of small and endogenous, non-coding RNA molecules that are single-stranded and usually comprise 18–25 nucleotides (Alkowari et al., 2017). They were first discovered in the late 1990s in a nematode called Caenorhabditis elegans and have since been demonstrated to exist in most eukaryotic organisms, including humans. It is estimated that miRNAs account for nearly 1–5% of the human genome and can regulate up to a third of all protein-coding genes (Macfarlane and Murphy, 2010). To date, upward of 2200 miRNA genes have been identified around half of which are encoded in intergenic sequences, with their own promoters regulating transcription, and the remaining half in protein-coding genes, usually at untranslated regions (UTR) or introns (Hammond, 2015; Alkowari et al., 2017). Generally, miRNA biogenesis can be classified into two major pathways: canonical and non-canonical.
The canonical biogenesis pathway is the predominant mechanism by which our cells produce and process miRNAs. This pathway begins with the transcription of miRNA genes into primary miRNA transcripts (pri-miRNAs) by RNA polymerase II. Pri-miRNAs are then processed in the nucleus into precursor miRNAs (pre-miRNAs) by the microprocessor Drosha/DGCR8 complex (Alkowari et al., 2017). Once generated, pre-miRNAs are released into the nucleus via an XPO5/RanGTP complex and are catalyzed into mature miRNAs by the Dicer enzyme, a member of the RNase III family of endoribonucleases (Macfarlane and Murphy, 2010; Wahid et al., 2010; Alkowari et al., 2017). Although much of the cellular machinery remains the same, some unique miRNA families are synthesized by non-canonical routes. For example, pri-miRNAs produced from the splicing of mRNA introns, called mirtrons, bypass processing by Drosha and are instead directly exported to the cytoplasm. Similarly, pre-miRNAs generated from short introns, such as miR-451, are of insufficient length to act as substrates of Dicer and instead rely on Argonaute 2 (AGO2) for intracytoplasmic maturation (Alkowari et al., 2017). Regardless of the processing pathway, all mature miRNAs are eventually loaded onto AGO proteins to form a functional miRNA-induced silencing complex (miRISC). The specificity and mechanism of action of these complexes are primarily determined by their complementarity to miRNA response element (MRE) sequences on the target mRNAs (Alkowari et al., 2017).
The role of most miRNAs in gene regulation is inhibitory: they either directly bind and prevent the translation of the target mRNA or indirectly induce the mRNA’s early decay and degradation. However, some miRISC complexes are capable of binding the target mRNA’s promoter site to induce translation (Orang et al., 2014; Alkowari et al., 2017). An example of miRNA-mediated translational upregulation in AD is seen in the case of miR-125b. The induced overexpression of miR-125b in primary hippocampal neurons was reported to significantly activate translation of p44/42-MAPK (Erk1/2) (Banzhaf-Strathmann et al., 2014). The subsequent elevation in tau kinase activity was strongly associated with tau hyperphosphorylation and ultimately the impairment of working memory and learning in mice injected with miR-125b. Interestingly, some miRNAs can also interact with proteins other than AGO. A recent study demonstrated that miR-100-5p, for example, is involved in extracellular signaling and can bind Toll-like receptors (TLRs) causing neuronal apoptosis and microglial activation (Wallach et al., 2021). Regardless of their specific function, it is now well-recognized that miRNAs regulate several processes integral to cell proliferation, differentiation, and survival. Moreover, their documented dysregulation in many diseases presents an invaluable opportunity for research to investigate potential disease-specific biomarkers or novel therapeutic targets.
4. Molecular targets and roles of MicroRNAs in AD
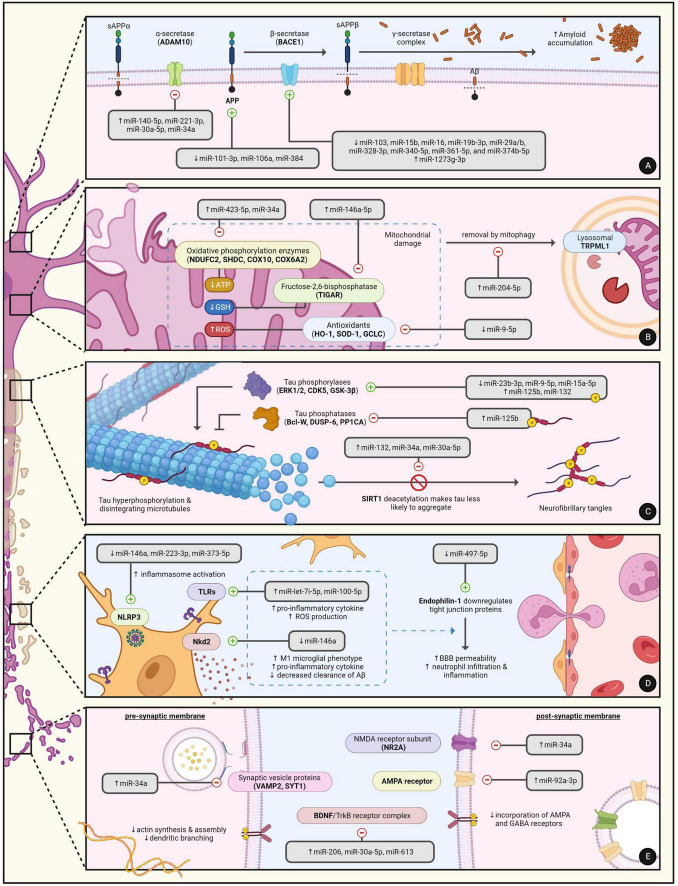
Over the past decade, the dysregulation of miRNA expression in AD has become well-documented in cell line experiments, animal models, and even human AD subjects. Much of our current understanding on the contribution of miRNAs to AD progression is rooted in experimental approaches where the over- or under-expression of the miRNA is induced. Although these study designs may indeed exaggerate the role of some miRNAs in certain aspects of AD, many of the findings are now being correlated with data obtained from human subjects. In general, miRNAs are believed to play a pivotal role in five major aspects of AD: amyloidogenesis, tauopathy, neuroinflammation, synaptic dysfunction, and neuronal death. Both Table 1 and Figure 1 summarize the targets and roles of the main differentially expressed miRNAs in the CSF of AD patients. Table 2 provides a more comprehensive review of the literature on all CSF miRNAs with their expression change and significance.
TABLE 1.
The major targets of differentially expressed miRNAs in the CSF of AD patients.
| Key targets | Related miRNA |
| Amyloidogenesis | |
| APP (amyloid precursor protein) | miR-101-3p, miR-106a, miR-384 |
| ADAM10 (a disintegrin and metalloprotease 10) | miR-140-5p, miR-221-3p, miR-30a-5p, miR-34a |
| BACE1 (β-site APP cleavage enzyme 1) | miR-103, miR-125b-5p, miR-1273g-3p, miR-15b, miR-16, miR-16-5p, miR-19b-3p, miR-29a/b, miR-29c-3p, miR-328-3p, miR-340-5p, miR-361-5p, miR-374b-5p |
| Tauopathy | |
| ERK1/2 (extracellular signal-regulated kinase 1 and 2) | miR-125b, miR-15a-5p |
| GSK-3β (glycogen synthase kinase-3β) | miR-23b-3p, miR-9-5p |
| CDK5 (cyclin dependent kinase 5) | miR-132 |
| SIRT1 (sirtuin-1) | miR-132, miR-30a-5p, miR-34a |
| CAV1 (caveolin-1) | miR-124-3p |
| DUSP6 (dual-specific phosphatase 6) | miR-125b |
| PPP1CA (protein phosphatase 1 catalytic subunit alpha isoform) | miR-125b |
| Bcl-W (Bcl-2-like protein 2) | miR-125b |
| PTPA (protein phosphatase 2 phosphatase activator) | miR-34a |
| Neuroinflammation | |
| Nkd2 (NKD inhibitor of WNT signaling pathway 2) | miR-146a |
| TLRs (toll-like receptors) | miR-100-5p, miR-let-7i-5p |
| STAT3 (signal transducer and activator of transcription 3) | miR-19b-3p, miR-29c-3p |
| Endophilin-1 | miR-497-5p |
| NLRP3 (NLR family pyrin domain containing 3) | miR-223-3p, miR-373-5p |
| Synaptic dysfunction | |
| BDNF (brain-derived neurotrophic factor) | miR-206, miR-30a-5p, miR-613 |
| NR2A (N-methyl-D-aspartate receptor 2A) | miR-34a |
| VAMP2 (vesicle associated membrane protein 2; synaptobrevin 2) | miR-34a |
| SYT1 (synaptotagmin 1) | miR-34a |
| AMPARs (α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor) | miR-92a-3p |
| Mitochondrial dysfunction and neuronal apoptosis | |
| NRF2 (nuclear factor-erythroid 2 p45-related factor 2) | miR-9-5p |
| KEAP1 (Kelch-like ECH-associated protein 1) | miR-9-5p |
| TIGAR (TP53-inducible glycolysis and apoptosis regulator) | miR-146a-5p |
| COX6A2 (cytochrome c oxidase subunit 6A2) | miR-423-5p |
| NDUFC2 (NADH: Ubiquinone oxidoreductase subunit C2) | miR-34a |
| SDHC (succinate dehydrogenase complex subunit C) | miR-34a |
| COX10 (cytochrome c oxidase assembly factor heme A:farnesyltransferase) | miR-34a |
| MFN2 (mitofusin-2) | miR-195 |
| BCL2L2 (Bcl-2-like protein 2) | miR-29b-3p |
| MCL-1 (induced myeloid leukemia cell differentiation protein 1) | miR-29b-3p |
| TRPML1 (transient receptor potential mucolipin-1) | miR-204-5p |
FIGURE 1.
The regulatory roles of miRNAs in the major hallmarks of AD pathophysiology including (A) MiRNAs in amyloidogenesis, (B) MiRNAs in mitochondrial dysfunction and neuronal apoptosis, (C) MiRNAs in tauopathy, (D) MiRNAs in neuroinflammation, and (E) MiRNAs in synaptic dysfunction. Created with BioRender.com.
TABLE 2.
Summary of the expression change, molecular target, and significance of miRNAs differentially expressed in the CSF of AD patients.
| Name | Change | Target | Effect on translation and significance | ||
| miR-100-5p | ↑ | Wallach et al., 2021 | TLR7/8 mTOR |
Increased binding and activation of microglial TLR7/8 promotes autonomous neurodegeneration and cortical accumulation of microglia; Increased inhibition of PI3K/Akt/mTOR pathway in late-stage AD leads to increased neuronal apoptosis | Ye et al., 2015; Wallach et al., 2021 |
| miR-101-3p | ↑ | Sørensen et al., 2016; Zhou et al., 2019 | APP | Although upregulated in CSF, decreased brain levels cause loss of APP inhibition increasing Aβ deposition | Vilardo et al., 2010; Zhou et al., 2019; Lee et al., 2021 |
| miR-103 | ↓ | Denk et al., 2015 | BACE1 | Decreased inhibition of BACE1 increases Aβ deposition and ROS production promoting neuronal loss | Denk et al., 2015; Lee et al., 2021 |
| miR-106a | ↑ | Denk et al., 2015 | APP | Although upregulated in CSF, decreased brain levels cause loss of APP inhibition increasing Aβ deposition | Lee et al., 2021 |
| miR-10b-5p | ↑ | Sørensen et al., 2016 | HODX10 | Increased inhibition of HODX10 and subsequent activation of the Rho/ROCK signaling pathway mediates hippocampal neuronal injury and inflammation | Ruan et al., 2021 |
| miR-124-3p | ↓ | Burgos et al., 2014 | CAV1 | Decreased inhibition of Caveolin-1 and subsequent modulation of the PI3K/Akt/GSK-3b pathway promotes tau hyperphosphorylation, neurofibrillary tangle formation, and cellular apoptosis | Kang et al., 2017; Samadian et al., 2021 |
| miR-125b | ↑ | Galimberti et al., 2014 | ERK1/2 Bcl-W DUSP6 PPP1CA |
Increased activation of ERK1/2 and downregulation of tau phosphatases (Bcl-W, DUSP6, PPP1CA) promotes tau hyperphosphorylation and reduces neuronal cell viability | Banzhaf-Strathmann et al., 2014 |
| miR-125b-5p | ↓ | Sørensen et al., 2016; Liu et al., 2022 | BACE1 | Decreased inhibition of BACE1 increases Aβ deposition and ROS production promoting neuronal loss | Li et al., 2020 |
| miR-1273g-3p | ↑ | Kim et al., 2021 | BACE1 Nicastrin TIMM13 GLRX5 MTCH1 |
Increased activation of the JNK pathway upregulates BACE1 and Nicastrin expression inducing Aβ and ROS production; Increased inhibition of mitochondrial genes (TIMM13, GLRX5, MTCH1) results in mitochondrial dysfunction and BACE1 upregulation | Kim et al., 2021 |
| miR-130a-3p | ↓ | Sørensen et al., 2016 | DAPK1 | Decreased inhibition of DAPK1 promotes hippocampal neurotoxicity and is associated with worse cognitive decline | Wang et al., 2021 |
| miR-132 | ↑ | Denk et al., 2015 | GTDC-1 CDK5 SIRT1 |
Increased inhibition of GTDC-1 promotes tau hyperphosphorylation and neuronal apoptosis by upregulation of Bax and downregulation of BCL2; Increased stimulation of CDK5 promotes tau hyperphosphorylation; Increased SIRT1 inhibition promotes tau hyperphosphorylation and amyloidogenesis | Hernandez-Rapp et al., 2016; Liu and Zhang, 2019 |
| miR-140-5p | ↓ | Lusardi et al., 2017 | ADAM10 SOX2 |
Although downregulated in CSF, increased brain levels inhibit expression of SOX2 and ADAM10 promoting the intraneural accumulation of Aβ and progression of AD | Akhter et al., 2018 |
| miR-142-3p | ↓ | Lusardi et al., 2017 | MAPT | Decreased regulation of MAPT compromises oligodendrocyte differentiation and cortical myelination exacerbating neurodegeneration | Hinman et al., 2021 |
| miR-143-3p | ↑ | Sørensen et al., 2016 | NRG1 | Increased inhibition of BACE1-dependent cleavage of NRG1 reduces neuronal cell viability and is associated with worse cognitive decline | Sun et al., 2020 |
| miR-146a | ↓ | Kiko et al., 2014; Liu et al., 2022 | Nkd2 | Overexpression of miRNA-146a in vivo has been demonstrated to induce microglial polarization (M1 pro-inflammatory to M2 phagocytic) and inhibit NLRP3 inflammasome formation enhancing Aβ clearance and reducing neuroinflammation, respectively, but the exact mechanisms remain unknown | Liang et al., 2021 |
| miR-146a-5p | ↑ | Sørensen et al., 2016 | TIGAR | Increased inhibition of TIGAR decreases NADPH production and GSH formation facilitating ROS production and neuronal pyroptosis (caspase-1-dependent cell death) | Lei et al., 2021 |
| miR-150-5p | ↑ | Sørensen et al., 2016 | PDCD4 | Associated with lower global cognitive scores, higher CSF t-tau, lower CSF Aβ42 levels, and lower blood PDCD4 | Chia et al., 2022 |
| miR-15a-5p | ↓ | Liu et al., 2022 | ERK1 | Decreased inhibition of ERK1 promotes tau phosphorylation and AD progression | Hébert et al., 2010 |
| miR-15b | ↑ | Denk et al., 2015 | BACE1 | Although upregulated in CSF, decreased brain levels cause loss of BACE1 inhibition increasing Aβ deposition | Gong et al., 2017; Liu et al., 2022 |
| miR-16 | ↓ | Denk et al., 2015 | BACE1 | Decreased inhibition of BACE1 increases Aβ deposition at the early stages of AD | Zhong et al., 2018; Manna et al., 2020 |
| miR-16-5p | ↓ | Lusardi et al., 2017 | BACE1 | Decreased inhibition of BACE1 increases Aβ deposition and ROS production promoting neuronal loss | Zhang et al., 2020 |
| miR-193a-3p | ↓ | Lusardi et al., 2017 | PTEN | Decreased inhibition of PTEN facilitates the dysregulation of the PI3K/AKT cell survival signaling pathway leading to neuronal apoptosis and AD progression | Takalkar et al., 2016 |
| miR-195 | ↓ | Cao et al., 2021; Liu et al., 2022 | SYNJ1 MFN2 |
Decreased inhibition of SYNJ1 facilitates the cleavage of neuronal membrane filaments and is positively correlated with the patient’s MMSE score while negatively correlated with CSF tau levels; Co-downregulation of MFN2 proteins facilitates mitochondrial dysfunction by fusion-fission imbalance | Zhang R. et al., 2016; Cao et al., 2021 |
| miR-19b-3p | ↓ | Sørensen et al., 2016 | BACE1 STAT3 |
Decreased inhibition of BACE1 leads to increased Aβ deposition; Loss of downregulation of STAT3 phosphorylation and activation leads to reactive gliosis and poor cognitive function | Wu et al., 2017; Zhang et al., 2020 |
| miR-204-5p | ↑ | Sørensen et al., 2016 | TRPML1 | Increased inhibition of TRPML1 disrupts lysosomal storage/transport and is associated with dysregulation of autophagy, increased accumulation of intraneural Aβ, decreased mitophagy, and decreased ROS removal | Zhang L. et al., 2021 |
| miR-206 | ↑ | Sørensen et al., 2016 | BDNF IGF1 |
Increased inhibition of BDNF results in synaptic dysfunction and impaired memory consolidation; Increased inhibition of IGF1 compromises microglial-mediated Aβ clearance and promotes LPS-induced neuroinflammation | Lee et al., 2012; Xing et al., 2016 |
| miR-210 | ↓ | Liu et al., 2022 | VEGF | Decreased stimulation of VEGF is associated with AD progression | Zhu et al., 2015 |
| miR-22-3p | ↓ | Sørensen et al., 2016 | SOX9 MAPK14 |
Decreased inhibition of SOX9 and MAPK14 indirectly promotes Aβ deposition and neuronal apoptosis | Ji Q. et al., 2019; Xia et al., 2022 |
| miR-221-3p | ↑ | Sørensen et al., 2016 | ADAM10 | Increased inhibition of ADAM10 promotes intraneural accumulation of Aβ | Manzine et al., 2018 |
| miR-223-3p | ↓ | Lusardi et al., 2017 | NLRP3 | Decreased inhibition of the NLRP3 protein leads to inflammasome formation and sustained neuroinflammation in AD and is positively correlated to the MMSE score | Bauernfeind et al., 2012; Jia and Liu, 2016 |
| miR-23b-3p | ↓ | Sørensen et al., 2016 | Gnt-III GSK-3β |
Decreased inhibition of GnT-III facilitates APP cleavage by BACE1 promoting Aβ production; Decreased inhibition of GSK-3b facilitates tau hyperphosphorylation and ROS production | Pan et al., 2021; Jiang et al., 2022 |
| miR-26b | ↓ | Galimberti et al., 2014 | Rb1/E2F NEP |
Although downregulated in CSF, increased brain levels upregulate the expression of Rb1/E2F promoting neuronal apoptosis; increased downregulation of NEP increases Aβ aggregation and impairs memory | Absalon et al., 2013; Chu et al., 2018 |
| miR-27a-3p | ↓ | Sala Frigerio et al., 2013 | NEAT1 | Decreased inhibition of NEAT1 increases BACE1 activity and Aβ deposition which promotes the progression of AD but no correlation with MMSE/MOCA score exists yet | He et al., 2022 |
| miR-27b-3p | ↓ | Lusardi et al., 2017 | CCL2/CCR2 | Impairment of the chemokine/chemokine receptor axis in blood-derived monocytes (BDMs) results in decreased clearance of Aβ aggregates | Wang et al., 2019 |
| miR-29a | ↑ | Kiko et al., 2014; Müller et al., 2016 | BACE1 | Although upregulated in CSF, decreased brain levels cause loss of BACE1 inhibition increasing Aβ deposition and ROS production promoting neuronal loss | Hébert et al., 2008 |
| miR-29a-3p | ↑ | Lusardi et al., 2017 | NAV3 ZNF346 LIF |
Increased activation of NAV3, ZNF346, and LIF affects neuron regeneration, survival, and differentiation, respectively | Peña-Bautista et al., 2022 |
| miR-29b | ↑ | Kiko et al., 2014 | BACE1 | Although upregulated in CSF, decreased brain levels cause loss of BACE1 inhibition increasing Aβ deposition and ROS production promoting neuronal loss | Hébert et al., 2008 |
| miR-29b-3p | ↑ | Sørensen et al., 2016 | BCL2L2 MCL-1 |
Increased inhibition of anti-apoptotic factors BCL2L2 and MCL-1 promotes neuronal cell death | Lungu et al., 2013; Satoh et al., 2015 |
| miR-29c | ↓ | Denk et al., 2015 | BACE1 | Decreased inhibition of BACE1 increases Aβ deposition and ROS production promoting neuronal loss | Lei et al., 2015; Manna et al., 2020 |
| miR-29c-3p | ↓ | Sørensen et al., 2016 | BACE1 STAT3 |
Decreased inhibition of BACE1 leads to increased Aβ deposition; Loss of downregulation of STAT3 phosphorylation and activation leads to reactive gliosis and poor cognitive function | Wu et al., 2017 |
| miR-30a-5p | ↑ | Sørensen et al., 2016 | ADAM10 SIRT1 BDNF |
Increased SIRT1 inhibition promotes tau hyperphosphorylation and accelerates neuronal damage; Increased inhibition of ADAM10 promotes intraneural accumulation of Aβ; Increased inhibition of BDNF results in synaptic dysfunction and impaired memory consolidation | Croce et al., 2013; Sun et al., 2022 |
| miR-328-3p | ↓ | Tan et al., 2021; Liu et al., 2022 | BACE1 | Decreased inhibition of BACE1 increases Aβ deposition and ROS production promoting neuronal loss | Thangavelu et al., 2020; Lee et al., 2021 |
| miR-331-3p | ↓ | Lusardi et al., 2017 | SQSTM1 OPTN |
Although downregulated in early stage AD, this miRNA increases later on promoting Aβ plaque formation and impairing its autophagic clearance | Chen et al., 2021 |
| miR-335-5p | ↓ | Sørensen et al., 2016 | JNK3 | Decreased inhibition of the JNK3 signaling pathway results in increased Aβ deposition | Denk et al., 2015; Wang et al., 2020 |
| miR-340-5p | ↓ | Lusardi et al., 2017 | BACE1 | Decreased inhibition of BACE1 increases Aβ deposition and ROS production promoting neuronal loss | Liu et al., 2022 |
| miR-34a | ↑ | Denk et al., 2015; Liu et al., 2022 | NDUFC2 SHDC COX10 SIRT1 PTPA NR2A VAMP2 SYT1 ADAM10 |
Increased inhibition of electron transport chain complexes (NDUFC2, SHDC, COX10) compromises mitochondrial function and ATP formation leading to neuronal cell death; Increased SIRT1 and PTPA inhibition promotes tau hyperphosphorylation; Increased NR2A inhibition on post-synaptic membranes compromises synaptic plasticity impairing working memory; Increased VAMP2 and SYT1 inhibition on pre-synaptic membranes compromises vesicle exo/endocytosis; Increased inhibition of ADAM10 promotes intraneural accumulation of Aβ and rapid cognitive decline | Sarkar et al., 2016, 2019; Liu et al., 2022 |
| miR-361-5p | ↓ | Sørensen et al., 2016 | BACE1 | Decreased inhibition of BACE1 increases Aβ deposition and ROS production promoting neuronal loss | Ji Y. et al., 2019 |
| miR-373-5p | ↓ | Sørensen et al., 2016 | NLRP3 | Decreased inhibition of the NLRP3 protein leads to inflammasome formation and sustained neuroinflammation in AD | Taşdelen et al., 2022 |
| miR-374b-5p | ↑ | Sørensen et al., 2016 | BACE1 | Although upregulated in CSF, decreased brain levels cause loss of BACE1 inhibition increasing Aβ deposition | Liu et al., 2022 |
| miR-384 | ↓ | Liu et al., 2022 | APP BACE1 |
Decreased regulation of APP increases Aβ production; Loss of downregulation of BACE1 leads to increased Aβ deposition | Liu et al., 2014 |
| miR-423-5p | ↑ | Sørensen et al., 2016 | COX6A2 | Increased inhibition of COX6A2 dramatically decreases ATP production levels compromising cell survival and energy metabolism | Siengdee et al., 2015 |
| miR-433 | ↓ | Burgos et al., 2014 | JAK2/STAT3 | Decreased inhibition of the JAK2/STAT3 pathway allows for increased neuronal proliferation and apoptosis, reactive gliosis, synaptic dysfunction, and Aβ aggregation | Wang and Zhang, 2020; Liu et al., 2022 |
| miR-497-5p | ↓ | Sørensen et al., 2016 | Endophilin-1 | Decreased inhibition of Endophilin-1 allows for the sustained downregulation of tight junction proteins (ZO-1, occludin, claudin-5) increasing BBB permeability and contributing to AD neuroinflammation | Zhu et al., 2019 |
| miR-613 | ↑ | Liu et al., 2022 | BDNF | Increased inhibition of BDNF results in synaptic dysfunction and is associated with decreased hippocampal neuron survival and proliferation | Li et al., 2016 |
| miR-9-3p | ↑ | Sørensen et al., 2016 | Dmd SAP97 REST |
Modulates dendritic growth and synaptic plasticity of hippocampal neurons by targeting long-term potentiation genes (Dmd, SAP97) and the REST protein; Although initially upregulated, CSF miRNA-9-3p levels are reported to decrease with AD progression | Giusti et al., 2014; Sim et al., 2016 |
| miR-9-5p | ↓ | Burgos et al., 2014 | GSK-3β NRF2/KEAP1 |
Decreased inhibition of GSK-3b facilitates ROS production, mitochondrial dysfunction, and neuronal apoptosis; Decreased activation of NRF2/KEAP1 signaling pathways results in downregulation of antioxidant enzymes (HO-1, SOD-1, GCLC) | Yang et al., 2017; Liu et al., 2020 |
| miR-92a-3p | ↑ | Sørensen et al., 2016 | AMPARs | Decreased translation and incorporation of GluA1-containing AMPA receptors into the synaptic membranes of hippocampal neurons is associated with defective synaptic transmission and plasticity | Letellier et al., 2014 |
| miR-let-7i-5p | ↑ | Sørensen et al., 2016 | TLR4 | Increased activation of extracellular TLR4 contributes to widespread neuronal damage | Sørensen et al., 2016 |
4.1. MicroRNAs in amyloidogenesis
The amyloid cascade hypothesis is one of the leading principles in our current understanding of the development and progression of neurodegeneration in AD (Ricciarelli and Fedele, 2017). The amyloid precursor protein (APP), a key mediator of neuronal growth and repair, is normally broken down by α-secretase and γ-secretase into soluble peptides that can be recycled. However, cleavage of APP at aberrant sites can yield insoluble Aβ peptides of varying lengths, with Aβ40 and Aβ42 being the most common (Ricciarelli and Fedele, 2017). The increased formation of Aβ is usually due to the imbalance, mutation, or genetic dysregulation of several key enzymes: (1) PSEN, the catalytic protein subunit of γ-secretase, (2) ADAM10, the major α-secretase in neurons, and (3) BACE, the major β-secretase in neurons. If not effectively cleared, Aβ accumulation ultimately leads to NFT formation, synaptic dysfunction, and neuronal apoptosis. Hence, it becomes clear that miRNAs that regulate the production and clearance of Aβ contribute to the disease and can serve as potential CSF biomarkers for AD.
Compared to healthy controls, CSF levels of miR-103, miR-16, miR-19b-3p, miR-328-3p, miR-340-5p, and miR-361-5p were all decreased in AD patients (Hébert et al., 2008; Denk et al., 2015; Sørensen et al., 2016; Gong et al., 2017; Lusardi et al., 2017; Zhong et al., 2018; Ji Y. et al., 2019; Zhang et al., 2020; Liu et al., 2022). These miRNAs modulate Aβ metabolism by directly targeting BACE1. Since alterations to the CSF often reflect brain changes, the downregulation of these miRNAs and the subsequent loss of inhibition on BACE1 expression promotes the deposition of Aβ. For example, the expression of miR-15b was reported to be decreased in the frontal cortices of sporadic AD patients by qRT-PCR analysis and was negatively correlated with BACE1 mRNA levels (Gong et al., 2017). Furthermore, the induced overexpression of miR-15b in transfected SH-SY5Y cells was observed to significantly downregulate BACE1 expression and protect against Aβ-induced neuronal apoptosis (Gong et al., 2017). Some miRNAs such as miR-1273g-3p can also regulate BACE1 indirectly. In contrast with the aforementioned miRNAs, CSF levels of miR-1273g-3p were reported to be constantly elevated in patients with early stage AD (Kim et al., 2021). MiR-1273g-3p is a key regulator of numerous molecular targets and primarily inhibits the mitochondrial genes TIMM13, MTCH1, and GLRX5. TIMM13 is of special interest because its knockdown in H4-APPswe cells not only compromised mitochondrial function but was also discovered to directly upregulate BACE1 expression and promote Aβ42 production (Kim et al., 2021). Although the altered expression of miR-1273g-3p was not tested for in post-mortem examination of AD brain tissue, TIMM13 was confirmed to be downregulated in the hippocampus.
ADAM10 is primarily responsible for the normal proteolytic cleavage of APP into soluble peptides. However, loss-of-function mutations or deficiencies of ADAM10 force APP to undergo amyloidogenic processing which ultimately promotes the formation of Aβ (Sarkar et al., 2019). In a recent study by Sarkar et al. (2019), the induced overexpression of miR-34a in transgenic mice was associated with a decrease in ADAM10 levels and rapid cognitive decline. Immunohistochemistry also revealed significant intraneural accumulation of Aβ and the development of AD-like neuropathology across several regions of the mice’s brains. When measured in human AD subjects, CSF levels of miR-34a were found to be elevated (Liu et al., 2022). Both miR-221-3p and miR-30a-5p were also significantly upregulated in the CSF and negatively correlated with the expression of ADAM10 (Sørensen et al., 2016; Manzine et al., 2018; Sun et al., 2022). In contrast, the current evidence on the precise expression levels of miR-140-5p in AD remains conflicting. MiR-140-5p is similar to miR-34a, miR-221-3p, and miR-30a-5p in that it is overexpressed in AD hippocampal tissue and inhibits the translation of ADAM10 (Manzine et al., 2018). However, it is reported that miR-140-5p CSF levels are remarkably decreased (Lusardi et al., 2017). Since the reasons for this discrepancy are not fully elucidated, further replication of these studies is needed to establish a clearer understanding of the role miR-140-5p plays in AD progression.
Amyloid precursor protein itself is the last major notable target of differentially expressed CSF miRNAs that have been discovered to play a role in amyloidogenesis. For example, miR-101-3p, miR-106a, and miR-384 have all been demonstrated to directly downregulate the expression of APP by binding the 3′UTR sequence of its mRNA transcript (Liu et al., 2014; Wang et al., 2019; Zhou et al., 2019). If this function is lost, the unregulated overexpression of APP will in turn increase activation of amyloidogenic pathways that are closely related with the occurrence of sporadic AD (Zhou et al., 2019). When measured in the CSF, miR-384 levels were reduced in patients with AD compared to normal controls. The expression of miR-384, in particular, was further shown to be decreased in the hippocampal tissue of APP/PSI transgenic mice as well as the serum and plasma of AD patients (Liu et al., 2014). Interestingly, these patients also displayed a significant negative correlation between CSF levels of miR-384 and Aβ42. These findings suggest that some miRNAs possess therapeutic and neuroprotective properties that can be used to slow AD development.
4.2. MicroRNAs in tauopathy
The aberrant hyperphosphorylation of tau is another major hallmark of AD. Tau, a microtubule-associated protein (MAP), is most abundantly expressed by mature neurons in the hippocampus and cerebral cortex of human brains. The phosphorylation of tau, under normal physiologic conditions, is essential for its activation and biological function: promotion of microtubule assembly and stability, preservation of cytoskeletal integrity, and maintenance of axonal transport (Iqbal et al., 2005). Although the exact underlying mechanisms are not yet fully understood, the extracellular deposition of Aβ and the imbalance between tau kinases/phosphatases have been shown to abnormally promote tau hyperphosphorylation. Hyperphosphorylated tau proteins generally exhibit lower binding affinity to axonal microtubules and are more prone to polymerize forming cytotoxic aggregates called neurofibrillary tangles. Several miRNAs have been implicated in the regulation of tau phosphorylation and are believed to significantly contribute to tau pathology in AD.
Of the major tau kinases, miRNAs are currently known to modulate the expression of ERK1/2, CDK5, and GSK-3β. MiR-15a-5p, a potent negative regulator of ERK1, is significantly decreased in the CSF and brains of AD patients (Hébert et al., 2010; Liu et al., 2022). Without the regulation of miR-15a-5p, increased ERK1 activity has been directly associated with tau hyperphosphorylation in primary cortical neurons and is believed to equally contribute to neurofibrillary pathology in humans (Hébert et al., 2010). In sharp contrast to miR-15a-5p, miR-125b overexpression increased ERK1/2 activation and subsequently promoted the phosphorylation of tau (Banzhaf-Strathmann et al., 2014). It is reported that, compared to healthy controls, the expression of miR-125b is elevated by approximately 1.6-fold in AD brains and is similarly high in the CSF (Banzhaf-Strathmann et al., 2014; Liu et al., 2022). Interestingly, the induced overexpression of miR-125b in primary hippocampal neurons was also seen to alter the levels of tau phosphatases such as Bcl-W, DUSP-6, and PP1CA. Translational inhibition of these phosphatases, which normally regulate ERK1/2 by negative feedback loops, significantly increased the phosphorylation of tau by fivefold compared with ERK1/2 upregulation alone (Banzhaf-Strathmann et al., 2014). Taken together, the concomitant inhibition of phosphatases and promotion of kinases by elevated miR-125b suggests that some miRNAs can modulate different targets that produce additive effects. Indeed, a recent study by Liu and Zhang (2019) reveals that the overexpression of miR-132 in AD upregulates CDK5 but downregulates GTDC-1, with both changes promoting the phosphorylation of tau. Similarly, miR-34a can inhibit the translation of PTPA and SIRT1 resulting in tau hyperphosphorylation (Sarkar et al., 2019). PTPA is normally an activator of protein phosphatase 2A (PP2A), the major serine/threonine phosphatase expressed in the brain. When activated, the PP2A complex enhances dephosphorylation of tau and consequently preserves synaptic function (Luo et al., 2013). However, PP2A has been shown to be downregulated in the brains of AD patients promoting tau phosphorylation, and this effect is perhaps further compounded by translational inhibition of PTPA via miR-34a (Wei et al., 2020). On the other hand, Sirtuin-1 is a NAD-dependent deacetylase that normally reverses the pathogenic acetylation of hyperphosphorylated tau proteins making them more prone to degradation by ubiquitin ligases and less likely to accumulate (Manjula et al., 2021). Even if aggregates do form, Sirtuin-1 was demonstrated to suppress the cell-to-cell propagation of abnormal tau limiting the spread of tauopathy into surrounding brain parenchyma (Min et al., 2018). In tauP3018 mice, the brain-specific deletion of Sirt1 was observed to directly result in tau-mediated synaptic loss causing behavioral and cognitive deficits (Min et al., 2018). In parallel, Sirtuin-1 is downregulated by miR-132, miR-30a-5p, and miR-34a, all of which are significantly elevated in the CSF of AD patients (Hernandez-Rapp et al., 2016; Sarkar et al., 2019; Sun et al., 2022). The last major kinase of interest is GSK-3β. MiR-23b-3p can directly bind and inhibit the translation of GSK-3β mRNA. Overexpression of miR-23b-3p in vitro reversed GSK-3β-mediated tau hyperphosphorylation and was described to be largely neuroprotective (Jiang et al., 2022). However, CSF levels of miR-23b-3p are downregulated in AD subjects and were found to be negatively correlated with tau phosphorylation as AD progressed into different stages (Jiang et al., 2022). Recent research also reveals that Caveolin-1 promotes the activity of GSK-3β by preventing its phosphorylation via the PI3K/Akt pathway (Kang et al., 2017). Thus, the decreased translational inhibition of Caveolin-1, as seen by low CSF miR-124-3p, allows for the continued activation of GSK-3β and phosphorylation of tau in AD (Kang et al., 2017).
4.3. MicroRNAs in neuroinflammation
Chronic low-grade inflammation of the brain parenchyma has long been demonstrated to play a central role in the development of AD. Neuroinflammation is characterized by the marked increase in the production and release of pro-inflammatory cytokines (such as IL-1β and TNF-α), chemokines (such as CCL1, CCL2, and CCL5), and reactive oxygen species (ROS). Although the primary mediators of neuroinflammation are innate immune cells, recent evidence indicates that peripheral blood cells can cross the blood-brain barrier (BBB) and contribute to the immune response seen in AD (Leng and Edison, 2021).
Microglia, resident macrophages of the brain, normally protect against neuronal loss by phagocytosing and preventing the accumulation of neurotoxic substances such as Aβ. In CNS injury or infection, microglia can migrate to the site of insult and become activated to one of two phenotypes, M1 (pro-inflammatory) or M2 (anti-inflammatory/phagocytic), whose different but balanced functions help facilitate tissue repair and healing (Onyango et al., 2021). However, with aging and in AD, microglia have been observed to assume a predominantly M1 phenotype (Onyango et al., 2021). The consequent decrease in amyloid clearance and increase in pro-inflammatory cytokine release are believed to accelerate AD pathogenesis and nerve damage. Moreover, the accumulation of Aβ has been linked to increased activation and recruitment of microglia further exacerbating neuroinflammation (Onyango et al., 2021). MiR-146a is believed to be a key regulator of the inflammatory response and microglial function. In a recent study by Liang et al. (2021), the overexpression of miR-146a in APP/PS1 transgenic mice was seen to reverse microglial polarization (i.e., phenotype transition) in favor of the M2 phenotype, which was associated with enhanced phagocytic clearance of Aβ. Moreover, miR-146a overexpression was observed to significantly reduce levels of pro-inflammatory cytokines (IL-1β, IL-6, TNF-α) and inflammasome markers (NLRP, ASC, caspase-1) indicating decreased NLRP3 activation (Liang et al., 2021). Collectively, these changes reduced Aβ accumulation, attenuated neuroinflammation, prevented neuronal apoptosis, and, most importantly, rescued cognition. Similar results were reported in a study where the intranasal administration of miR-146a in AD mice relieved hippocampal inflammation and ameliorated cognitive impairment (Mai et al., 2019). Although the exact molecular mechanisms have not yet been elucidated, the neuroprotective effects of miR-146a are currently thought to be mediated by negative regulation of Nkd2 (Liang et al., 2021). The knockdown of Nkd2 in vivo was reported to induce microglial phenotype switching and was negatively correlated with levels of miR-146a. Moreover, the 3′UTR sequence of its mRNA has been confirmed to be a target of miR-146a. In human AD subjects, CSF levels of miR-146a are significantly decreased (Liu et al., 2022). Without microglial phenotype switching and anti-inflammatory mediator release, the downregulation of miR-146a serves as a risk factor for sustained neuroinflammation in AD.
MiRNAs can also contribute to the AD inflammatory response by other mechanisms. MiR-100-5p, which is upregulated in the CSF of AD patients, can directly bind and activate endosomal TLR7/8 promoting cell-autonomous degeneration in neurons and pro-inflammatory cytokine release by microglia (Wallach et al., 2021). Similarly, increased activation of extracellular TLR4 by elevated CSF miR-let-7i-5p is associated with microglial ROS production and widespread neuronal damage (Sørensen et al., 2016). MiR-19b-3p and miR-29c-3p, both of which are downregulated in the CSF, lose their translational inhibition on STAT3 which in turn induces reactive gliosis and impaired cognitive function (Wu et al., 2017). MiR-497-5p, a direct negative regulator of Endophilin-1 expression, is decreased in AD allowing for Endophilin-1-mediated downregulation of tight junction proteins such as ZO-1, occludin, and claudin-5 (Zhu et al., 2019). The subsequent increase in BBB permeability augments the infiltration of peripheral blood cells further exacerbating neuroinflammation. Lastly, the downregulation of miR-223-3p and miR-373-5p in AD patients has been associated with increased NLRP3 inflammasome formation (Bauernfeind et al., 2012; Taşdelen et al., 2022).
4.4. MicroRNAs in synaptic dysfunction
Maintaining synapse structure is essential for the normal functioning of the brain. Synaptic plasticity refers to the ability of neuronal tissue to modify or strengthen synaptic connections in response to new information and stimuli. The neurochemical processes underlying synaptic plasticity are integral to learning and memory, and their derangement results in cognitive decline. In fact, the loss of synaptic transmission and activity are now recognized as the earliest events in AD pathology that precede clinical presentation. Thus, miRNAs that regulate synaptic plasticity and neuronal function are believed to greatly contribute to the cognitive impairment seen in AD patients.
Changes to synaptic plasticity, particularly in brain regions responsible for learning and memory, have been correlated with altered expression of neurotrophins, a group of regulatory proteins that promote the survival, growth, and function of neurons (Ng et al., 2019). BDNF is a potent neurotrophic factor that plays a key role in hippocampal long-term potentiation (LTP) and memory consolidation (Croce et al., 2013; Ng et al., 2019). When BDNF forms complexes with cell-surface tyrosine kinase B receptors (TrkB), major intraneural signaling pathways such as PI3K/Akt, MAPK, and PLC-γ are activated (Kowiański et al., 2017). Albeit by different mechanisms, these pathways will ultimately induce the synthesis of cytoskeletal proteins, polymerization of actin filaments, and branching of dendritic trees (Kowiański et al., 2017). The BDNF/TrkB complex can also modify the expression of AMPA and GABA receptors on post-synaptic membranes to promote LTP (Kowiański et al., 2017). MiR-206, miR-30a-5p, and miR-613, all of which are elevated in the CSF of AD patients, have been found to target and downregulate BDNF (Lee et al., 2012; Croce et al., 2013; Li et al., 2016). Without the effects of BDNF, the aging brain’s capacity for memory consolidation and synaptic plasticity is likely to become severely limited. Weinstein et al. (2014) found that in older adults, the risk for AD decreased by 33% for every increase in BDNF levels by one standard deviation increment. In parallel, the treatment of Tg2576 mice by a miR-206 antagomir restored brain levels of BDNF with direct improvements in hippocampal synaptic density and memory function (Lee et al., 2012).
Similar to BDNF, some miRNAs can directly alter the expression of surface receptors, channel proteins, or synaptic vesicle proteins involved in neurotransmission. For instance, elevated miR-34a has been shown to downregulate NR2A, a subunit of post-synaptic NMDA receptors, affecting LTP and memory formation (Sarkar et al., 2016). MiR-34a also affects pre-synaptic membranes by targeting VAMP2 and SYT1, key synaptic vesicle proteins involved in calcium-dependent neurotransmitter exocytosis (Sarkar et al., 2016). MiR-92a-3p, which is elevated in AD, also inhibits the translation and integration of AMPA receptors into post-synaptic membranes further contributing to synaptic dysfunction (Letellier et al., 2014).
4.5. MicroRNAs in mitochondrial dysfunction and neuronal apoptosis
The high metabolic demands of neural tissue make the brain particularly susceptible to injury following mitochondrial dysfunction. The energy generated by healthy mitochondria helps fuel vital intraneural processes such as ion transport, axonal development, and neurotransmitter biosynthesis. Mitochondria are also essential for intracellular calcium homeostasis and the generation of reactive oxygen species, both of which heavily impact cell integrity and viability. In AD, the derangement of mitochondrial function and the subsequent death of neural tissue have already been associated with the accumulation of amyloid-beta. However, increasing evidence suggests miRNAs that regulate the expression of mitochondrial proteins may also contribute to the development of AD.
miRNAs can cause mitochondrial damage by directly disrupting the oxidant-antioxidant balance. MiR-9-5p, an activator of the NRF2/KEAP1 pathway, plays a critical role in suppressing cancer and inflammation by promoting the expression of antioxidants such as HO-1, SOD-1, and GCLC (Liu et al., 2020). MiR-9-5p can also directly downregulate expression of GSK-3 β which has been linked to caspase activation and subsequent mitochondria-dependent neuronal apoptosis (Yang et al., 2017; Liu et al., 2020). In the CSF and frontal cortex of AD patients, however, miR-9-5p levels are reported to be decreased (Sørensen et al., 2016; Liu et al., 2020). Without the antioxidant and cytoprotective properties of miR-9-5p, oxidative stress can result in significant neuronal damage. This is further supported by the study of Liu et al. (2020) where the overexpression of miR-9-5p in an Aβ25-35-induced HT22 cell model reversed mitochondrial dysfunction and decreased cell apoptosis. MiR-146a-5p, on the other hand, is elevated in AD and has a pro-apoptotic effect as it targets TIGAR, a fructose-2,6-bisphosphatase (Lei et al., 2021). TIGAR’s activity normally shifts cellular glucose metabolism in favor of the pentose phosphate pathway which ultimately increases the availability of reduced glutathione (GSH), a potent antioxidant against mitochondrial hydrogen peroxide (Lei et al., 2021).
The dysregulation of miRNAs in AD may also compromise neuron survival by affecting mitochondrial function. For instance, miR-423-5p, which is upregulated in AD, has been observed to directly inhibit the translation of the mitochondrial cytochrome C mRNA, COX6A2, halting ATP production by the electron transport chain (Siengdee et al., 2015). Similarly, miR-34a downregulates the expression of NDUFC2, SHDC, and COX10 which have been correlated with severely decreased oxidative phosphorylation (Sarkar et al., 2016). Decreased miR-195 alters the expression of outer mitochondrial membrane mitofusin-2, a GTPase responsible for coordinating mitochondrial fusion-fission reactions (Zhang R. et al., 2016). Elevated miR-204-5p increases downregulation of TRPML1, or mucolipin-1, which is a lysosomal protein responsible for the removal of damaged or aged mitochondria by autophagy (Zhang X. et al., 2016; Zhang L. et al., 2021). Lastly, some miRNAs such as miR-29b-3p can promote neuronal apoptosis by inhibiting the anti-apoptotic factors BCL2L2 and MCL-1 (Lungu et al., 2013).
5. MicroRNAs as a potential diagnostic tool for AD
Biomarkers are objective indicators of patient health and disease progression. As our understanding of AD continues to expand, more reliable biomarkers are being identified. The differential expression of miRNAs and, by extension, their target mRNAs has been observed to correlate with the development of several diseases including AD. Although miRNAs are initially and most abundantly expressed by cells of the affected tissue (the brain parenchyma in the case of AD), they can also be found circulating in almost all body fluids. For these miRNAs to remain viable extracellularly, they either form complexes with argonaute proteins and high-density lipoproteins or are directly secreted into exosomes and microvesicles (Glinge et al., 2017). Compared to mRNA molecules, circulating miRNA complexes are remarkably stable and resistant to the effects of endogenous RNA-degrading enzymes as well as extreme changes in pH and temperature (Glinge et al., 2017).
According to the criteria set by the international consensus group on molecular and biochemical markers of AD, the ideal biomarker should have a minimum sensitivity of 85% and a specificity of 75% (Thies et al., 1999). In this regard, several studies present promising evidence. A recent meta-analysis of 18 studies that included a total of 826 AD patients and 658 normal controls indicated that the use of miRNA clusters had a sensitivity of 0.89 and a specificity of 0.84 (Zhang W. T. et al., 2021). A similar study showed that combinations of 2-4 CSF miRNAs distinguished AD patients from healthy controls with a specificity of 75–82% (Lusardi et al., 2017). Even the use of single miRNAs had good diagnostic accuracy compared to traditional AD biomarkers as their pooled sensitivity and specificity were 0.82 and 0.80, respectively (Zhang W. T. et al., 2021). Exosomal miRNA-384 was able to discriminate AD from Parkinson’s disease dementia (PDD) with a sensitivity of 97% and a specificity of 100% (Yang et al., 2018). Similarly, miR-384 differentiated AD from vascular dementia (VaD) patients with a sensitivity of 99.1% and a specificity of 100% (Yang et al., 2018). The combination of miR-125b/miR-29a, miR-125b/miR-874, and miR-107/miR-335-5p was found to have a sensitivity of 0.78 and a specificity of 0.76 when distinguishing AD from frontotemporal dementia (FTD) (Martinez and Peplow, 2022). If these results are confirmed by larger replication studies, it is possible that miRNA expression patterns unique to AD can be used in the future to help distinguish AD from other causes of dementia. MiRNAs may also help predict individuals with mild cognitive impairment (MCI) at risk of developing AD. In a large cohort study of 458 MCI patients, miR-206 levels successfully predicted the progression of MCI to AD with a sensitivity of 95.3% and a specificity of 77.8% (Ogonowski et al., 2022). Another study showed that miR-146a and miR-181 also predicted MCI conversion to AD although their specificity was not reported (Ogonowski et al., 2022).
6. Future directions
The abnormal expression of miRNAs has been proven to contribute to the pathophysiology of several diseases including neurodegenerative disorders. This paper reviews the different regulatory roles miRNAs play in the CNS as well as the major pathways by which they are involved in AD. Many of these targets are regulated by more than one miRNA and it is thus likely that future research can reveal a more complex network is at play. Although CSF collection is an invasive procedure, it represents the optimal source of miRNAs. The CSF is in direct contact with brain parenchyma and more accurately reflects real-time changes in miRNA expression (Liu et al., 2022). Moreover, the correlation between miRNA profiles of the brain and blood was found to be only moderately significant indicating that not all expression changes seen in blood are AD-specific (Kos et al., 2022). The use of miRNAs as biomarkers remains a novel area of research. Compared to traditional biomarkers, miRNAs have several properties that make them more favorable for the diagnosis and prognostic evaluation of AD patients. Changes in miRNA expression can also be measured over time to guide or monitor the effects of therapeutic intervention. However, some studies currently provide inconsistent or contradictory findings with regards to miRNA expression in AD. Although all miRNA targets are confirmed by luciferase reporter experiments, differences in sample collection, analysis, and contamination may influence study outcomes. Similarly, insufficiently sized patient populations and the lack of normalized cut-off values can impact how results are interpreted. Further large-scale replication studies are necessary to address these limitations.
Author contributions
ANE and AY: conceptualization. ANE, GA, MA-R, MA, and SAR: writing—original draft preparation. ANE, KA, and AY: writing—review and editing. KA and AY: supervision. All authors have read and approved the submitted version of the manuscript.
Acknowledgments
Figure were created using BioRender.com.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Absalon S., Kochanek D. M., Raghavan V., Krichevsky A. M. (2013). MiR-26b, upregulated in Alzheimer’s disease, activates cell cycle entry, tau-phosphorylation, and apoptosis in postmitotic neurons. J. Neurosci. 33 14645–14659. 10.1523/JNEUROSCI.1327-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhter R., Shao Y., Shaw M., Formica S., Khrestian M., Leverenz J., et al. (2018). Regulation of ADAM10 by miR-140-5p and potential relevance for Alzheimer’s disease. Neurobiol. Aging 63 110–119. 10.1016/j.neurobiolaging.2017.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkowari M., Vozzi D., Bhagat S., Krishnamoorthy N., Morgan A., Hayder Y., et al. (2017). Targeted sequencing identifies novel variants involved in autosomal recessive hereditary hearing loss in Qatari families. Mutat. Res. 80 29–36. [DOI] [PubMed] [Google Scholar]
- Angelucci F., Cechova K., Valis M., Kuca K., Zhang B., Hort J. (2019). MicroRNAs in Alzheimer’s disease: Diagnostic markers or therapeutic agents? Front. Pharmacol. 10:665. 10.3389/FPHAR.2019.00665/BIBTEX [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee D., Muralidharan A., Mohammed A. R. H., Malik B. H. (2020). Neuroimaging in Dementia: A Brief Review. Cureus 12:e8682. 10.7759/CUREUS.8682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banzhaf-Strathmann J., Benito E., May S., Arzberger T., Tahirovic S., Kretzschmar H., et al. (2014). MicroRNA-125b induces tau hyperphosphorylation and cognitive deficits in Alzheimer’s disease. EMBO J. 33 1667–1680. 10.15252/embj.201387576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauernfeind F., Rieger A., Schildberg F. A., Knolle P. A., Schmid-Burgk J. L., Hornung V. (2012). NLRP3 Inflammasome Activity Is Negatively Controlled by miR-223. J. Immunol. 189 4175–4181. 10.4049/JIMMUNOL.1201516 [DOI] [PubMed] [Google Scholar]
- Breijyeh Z., Karaman R. (2020). Comprehensive review on Alzheimer’s Disease: Causes and treatment. Molecules 25:24. 10.3390/MOLECULES25245789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito-Aguilar R. (2019). Dementia around the world and the Latin America and Mexican Scenarios. J. Alzheimers Dis. 71 1–5. 10.3233/JAD-190177 [DOI] [PubMed] [Google Scholar]
- Burgos K., Malenica I., Metpally R., Courtright A., Rakela B. (2014). Profiles of Extracellular miRNA in Cerebrospinal Fluid and Serum from Patients with Alzheimer’s and Parkinson’s Diseases Correlate with Disease Status and Features of Pathology Profiles of Extracellular miRNA in Cerebrospinal Fluid and Serum from Patients with Alzheimer. PLoS One 9:e94839. 10.1371/journal.pone.0094839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Huang M., Guo L., Zhu L., Hou J., Zhang L., et al. (2021). MicroRNA-195 rescues ApoE4-induced cognitive deficits and lysosomal defects in Alzheimer’s disease pathogenesis. Mol. Psychiatry 26 4687–4701. 10.1038/s41380-020-0824-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Hong C., Yue T., Li H., Duan R., Hu W., et al. (2021). Inhibition of miR-331-3p and miR-9-5p ameliorates Alzheimer’s disease by enhancing autophagy. Theranostics 11:2395. 10.7150/THNO.47408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia S., Vipin A., Ng K., Tu H., Bommakanti A., Wang B., et al. (2022). Upregulated blood miR-150-5p in Alzheimer’s Disease dementia is associated with cognition, cerebrospinal fluid amyloid-β, and cerebral atrophy. J. Alzheimers Dis. 88 1567–1584. 10.3233/JAD-220116 [DOI] [PubMed] [Google Scholar]
- Chu T., Shu Y., Qu Y., Gao S., Zhang L. (2018). miR-26b inhibits total neurite outgrowth, promotes cells apoptosis and downregulates neprilysin in Alzheimer’s disease. Int. J. Clin. Exp. Pathol. 11:3383. [PMC free article] [PubMed] [Google Scholar]
- Croce N., Gelfo F., Ciotti M., Federici G., Caltagirone C., Bernardini S., et al. (2013). NPY modulates miR-30a-5p and BDNF in opposite direction in an in vitro model of Alzheimer disease: a possible role in neuroprotection? Mol. Cell Biochem. 376 189–195. 10.1007/s11010-013-1567-0 [DOI] [PubMed] [Google Scholar]
- Denk J., Boelmans K., Siegismund C., Lassner D., Arlt S., Jahn H. (2015). MicroRNA Profiling of CSF Reveals Potential Biomarkers to Detect Alzheimer‘s Disease. PLoS One 10:e0126423. 10.1371/JOURNAL.PONE.0126423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois B., Villain N., Frisoni G., Rabinovici G., Sabbagh M., Cappa S., et al. (2021). Clinical diagnosis of Alzheimer’s disease: recommendations of the International Working Group. Lancet Neurol. 20 484–496. 10.1016/S1474-4422(21)00066-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galimberti D., Villa C., Fenoglio C., Serpente M., Ghezzi L., Cioffi S., et al. (2014). Circulating miRNAs as potential biomarkers in Alzheimer’s disease. J. Alzheimers Dis. 42 1261–1267. 10.3233/JAD-140756 [DOI] [PubMed] [Google Scholar]
- Gaugler J. E., Ascher-Svanum H., Roth D. L., Fafowora T., Siderowf A., Beach T. G. (2013). Characteristics of patients misdiagnosed with Alzheimer’s disease and their medication use: An analysis of the NACC-UDS database. BMC Geriatr. 13:137. 10.1186/1471-2318-13-137/TABLES/2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giusti S., Vogl A., Brockmann M., Vercelli C., Rein M., Trümbach D., et al. (2014). MicroRNA-9 controls dendritic development by targeting REST. eLife 3:e02755. 10.7554/ELIFE.02755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glinge C., Clauss S., Boddum K., Jabbari R., Jabbari J., Risgaard B., et al. (2017). Stability of Circulating Blood-Based MicroRNAs - Pre-Analytic Methodological Considerations. PLoS One 12:e0167969. 10.1371/journal.pone.0167969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong G., An F., Wang Y., Bian M., Yu L., Wei C. (2017). miR-15b represses BACE1 expression in sporadic Alzheimer’s disease. Oncotarget 8 91551–91557. 10.18632/oncotarget.21177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond S. (2015). An overview of microRNAs. Adv. Drug Deliv. Rev. 87 3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampel H., Blennow K. (2004). CSF tau and β-amyloid as biomarkers for mild cognitive impairment. Dialogues Clin. Neurosci. 6 379–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Chen Z., Wang J., Feng H. (2022). Expression relationship and significance of NEAT1 and miR-27a-3p in serum and cerebrospinal fluid of patients with Alzheimer’s disease. BMC Neurol. 22:203. 10.1186/S12883-022-02728-9/FIGURES/1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hébert S., Papadopoulou A., Smith P., Galas M., Planel E., Silahtaroglu A., et al. (2010). Genetic ablation of Dicer in adult forebrain neurons results in abnormal tau hyperphosphorylation and neurodegeneration. Hum. Mol. Genet. 19 3959–3969. 10.1093/HMG/DDQ311 [DOI] [PubMed] [Google Scholar]
- Hébert S. S., Horré K., Nicolaï L., Papadopoulou A., Mandemakers W., Silahtaroglu A., et al. (2008). Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/β-secretase expression. Proc. Natl. Acad. Sci. U. S. A. 105:6415. 10.1073/PNAS.0710263105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Rapp J., Rainone S., Goupil C., Dorval V., Smith P., Saint-Pierre M., et al. (2016). microRNA-132/212 deficiency enhances Aβ production and senile plaque deposition in Alzheimer’s disease triple transgenic mice. Sci. Rep. 6:30953. 10.1038/srep30953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinman J., Ngo K., Kim D., Chen C., Abraham C., Ghanbari M., et al. (2021). miR-142-3p regulates cortical oligodendrocyte gene co-expression networks associated with tauopathy. Hum. Mol. Genet. 30:103. 10.1093/HMG/DDAA252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal K., Alonso Adel C., Chen S., Chohan M., El-Akkad E., Gong C., et al. (2005). Tau pathology in Alzheimer disease and other tauopathies. Biochim. Biophys. Acta 1739 198–210. [DOI] [PubMed] [Google Scholar]
- Ji Y., Wang D., Zhang B., Lu H. (2019). MiR-361-3p inhibits β-amyloid accumulation and attenuates cognitive deficits through targeting BACE1 in Alzheimer’s disease. J. Integr. Neurosci. 18 285–291. [DOI] [PubMed] [Google Scholar]
- Ji Q., Wang X., Cai J., Du X., Sun H., Zhang N. (2019). MiR-22-3p Regulates Amyloid β Deposit in Mice Model of Alzheimer’s Disease by Targeting Mitogen-activated Protein Kinase 14. Curr. Neurovasc. Res. 16 473–480. 10.2174/1567202616666191111124516 [DOI] [PubMed] [Google Scholar]
- Jia L. H., Liu Y. N. (2016). Downregulated serum miR-223 servers as biomarker in Alzheimer’s disease. Cell Biochem. Funct. 34 233–237. 10.1002/cbf.3184 [DOI] [PubMed] [Google Scholar]
- Jiang H., Liu J., Guo S., Zeng L., Cai Z., Zhang J., et al. (2022). miR-23b-3p rescues cognition in Alzheimer’s disease by reducing tau phosphorylation and apoptosis via GSK-3β signaling pathways. Mol. Ther. Nucleic Acids 28 539–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Q., Xiang Y., Li D., Liang J., Zhang X., Zhou F., et al. (2017). MiR-124-3p attenuates hyperphosphorylation of Tau protein-induced apoptosis via caveolin-1-PI3K/Akt/GSK3β pathway in N2a/APP695swe cells. Oncotarget 8 24314–24326. 10.18632/oncotarget.15149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury R., Ghossoub E. (2019). Diagnostic biomarkers of Alzheimer’s disease: A state-of-the-art review. Biomark Neuropsychiatry 1:100005. [Google Scholar]
- Kiko T., Nakagawa K., Tsuduki T., Furukawa K., Arai H., Miyazawa T. (2014). MicroRNAs in plasma and cerebrospinal fluid as potential markers for Alzheimer’s disease. J. Alzheimers Dis. 39 253–259. 10.3233/JAD-130932 [DOI] [PubMed] [Google Scholar]
- Kim J., Jeong M., Stiles W. R., Choi H. S. (2022). Neuroimaging Modalities in Alzheimer’s Disease: Diagnosis and Clinical Features. Int. J. Mol. Sci. 23:6079. 10.3390/IJMS23116079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Choi K., Park Y., McLean C., Park J., Lee J., et al. (2021). Enhanced expression of microrna-1273g-3p contributes to alzheimer’s disease pathogenesis by regulating the expression of mitochondrial genes. Cells 10:2697. 10.3390/CELLS10102697/S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos M., Puppala S., Cruz D., Neary J., Kumar A., Dalan E., et al. (2022). Blood-Based miRNA Biomarkers as Correlates of Brain-Based miRNA Expression. Front. Mol. Neurosci. 15:89. 10.3389/FNMOL.2022.817290/BIBTEX [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowiański P., Lietzau G., Czuba E., Waśkow M., Steliga A., Moryś J. (2017). BDNF: A Key Factor with Multipotent Impact on Brain Signaling and Synaptic Plasticity. Cell. Mol. Neurobiol. 38 579–593. 10.1007/S10571-017-0510-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. Y., Ryu I. S., Ryu J. H., Cho H. J. (2021). miRNAs as Therapeutic Tools in Alzheimer’s Disease. Int. J. Mol. Sci. 22:22. 10.3390/IJMS222313012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. C., Kim S. J., Hong S., Kim Y. S. (2019). Diagnosis of Alzheimer’s disease utilizing amyloid and tau as fluid biomarkers. Exp. Mol. Med. 51 1–10. 10.1038/s12276-019-0250-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-T., Chu K., Jung K-H., Kim J. H., Huh J-Y., Yoon H, et al. (2012). miR-206 regulates brain-derived neurotrophic factor in Alzheimer disease model. Ann. Neurol. 72 269–277. 10.1002/ANA.23588 [DOI] [PubMed] [Google Scholar]
- Lei B., Liu J., Yao Z., Xiao Y., Zhang X., Zhang Y., et al. (2021). NF-κB-Induced Upregulation of miR-146a-5p promoted hippocampal neuronal oxidative stress and pyroptosis via TIGAR in a Model of Alzheimer’s Disease. Front. Cell Neurosci. 15:653881. 10.3389/fncel.2021.653881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei X., Lei L., Zhang Z., Zhang Z., Cheng Y. (2015). Downregulated miR-29c correlates with increased BACE1 expression in sporadic Alzheimer’s disease. Int. J. Clin. Exp. Pathol. 8:1565. [PMC free article] [PubMed] [Google Scholar]
- Leng F., Edison P. (2021). Neuroinflammation and microglial activation in Alzheimer disease: where do we go from here? Nat. Rev. Neurol. 17 157–172. 10.1038/s41582-020-00435-y [DOI] [PubMed] [Google Scholar]
- Letellier M., Elramah S., Mondin M., Soula A., Penn A., Choquet D., et al. (2014). miR-92a regulates expression of synaptic GluA1-containing AMPA receptors during homeostatic scaling. Nat. Neurosci. 17 1040–1042. 10.1038/nn.3762 [DOI] [PubMed] [Google Scholar]
- Li P., Xu Y., Wang B., Huang J., Li Q. (2020). miR-34a-5p and miR-125b-5p attenuate Aβ-induced neurotoxicity through targeting BACE1. J. Neurol. Sci. 413:116793. [DOI] [PubMed] [Google Scholar]
- Li W., Li X., Xin X., Kan P. C., Yan Y. (2016). MicroRNA-613 regulates the expression of brain-derived neurotrophic factor in Alzheimer’s disease. Biosci. Trends 10 372–377. [DOI] [PubMed] [Google Scholar]
- Li X., Feng X., Sun X., Hou N., Han F., Liu Y. (2022). Global, regional, and national burden of Alzheimer’s disease and other dementias, 1990–2019. Front. Aging Neurosci. 14:1120. 10.3389/FNAGI.2022.937486/BIBTEX [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C., Zou T., Zhang M., Fan W., Zhang T., Jiang Y., et al. (2021). MicroRNA-146a switches microglial phenotypes to resist the pathological processes and cognitive degradation of Alzheimer’s disease. Theranostics 11 4103–4121. 10.7150/thno.53418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. G., Wang J. L., Li L., Wang P. C. (2014). MicroRNA-384 regulates both amyloid precursor protein and β-secretase expression and is a potential biomarker for Alzheimer’s disease. Int. J. Mol. Med. 34 160–166. [DOI] [PubMed] [Google Scholar]
- Liu D. Y., Zhang L. (2019). MicroRNA-132 promotes neurons cell apoptosis and activates Tau phosphorylation by targeting GTDC-1 in Alzheimer’s disease. Eur. Rev. Med. Pharmacol. Sci. 23 8523–8532. [DOI] [PubMed] [Google Scholar]
- Liu J., Zuo X., Han J., Dai Q., Xu H., Liu Y., et al. (2020). MiR-9-5p inhibits mitochondrial damage and oxidative stress in AD cell models by targeting GSK-3β. Biosci. Biotechnol. Biochem. 84 2273–2280. 10.1080/09168451.2020.1797469 [DOI] [PubMed] [Google Scholar]
- Liu S., Fan M., Zheng Q., Hao S., Yang L., Xia Q., et al. (2022). MicroRNAs in Alzheimer’s disease: Potential diagnostic markers and therapeutic targets. Biomed. Pharmacother. 148:112681. [DOI] [PubMed] [Google Scholar]
- Lungu G., Stoica G., Ambrus A. (2013). MicroRNA profiling and the role of microRNA-132 in neurodegeneration using a rat model. Neurosci. Lett. 553 153–158. [DOI] [PubMed] [Google Scholar]
- Luo Y., Nie Y., Shi H., Ni Z., Wang Q., Wang J., et al. (2013). PTPA activates protein phosphatase-2A through reducing its phosphorylation at tyrosine-307 with upregulation of protein tyrosine phosphatase 1B. Biochim. Biophys. Acta 1833 1235–1243. [DOI] [PubMed] [Google Scholar]
- Lusardi T. A., Phillips J., Wiedrick J., Harrington C., Lind B., Lapidus J., et al. (2017). MicroRNAs in human cerebrospinal fluid as biomarkers for Alzheimer’s Disease. J. Alzheimers Dis. 55:1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane L.-A., Murphy P. R. (2010). MicroRNA: Biogenesis, Function and Role in Cancer. Manchester: University of Manchester. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai H., Fan W., Wang Y., Cai Y., Li X., Chen F., et al. (2019). Intranasal Administration of miR-146a Agomir rescued the pathological process and cognitive impairment in an AD Mouse Model. Mol. Ther. Nucleic Acids 18 681–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjula R., Anuja K., Alcain F. J. (2021). SIRT1 and SIRT2 activity control in neurodegenerative diseases. Front. Pharmacol 11:1899. 10.3389/fphar.2020.585821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna I., de Benedittis S., Quattrone A., Maisano D., Iaccino E., Quattrone A. (2020). Exosomal miRNAs as potential diagnostic biomarkers in Alzheimer’s disease. Pharmaceuticals 13 1–16. 10.3390/ph13090243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzine P., Pelucchi S., Horst M., Vale F., Pavarini S., Audano M., et al. (2018). microRNA 221 Targets ADAM10 mRNA and is Downregulated in Alzheimer’s Disease. J. Alzheimers Dis. 61 113–123. 10.3233/JAD-170592 [DOI] [PubMed] [Google Scholar]
- Martinez B., Peplow P. V. (2022). MicroRNA biomarkers in frontotemporal dementia and to distinguish from Alzheimer’s disease and amyotrophic lateral sclerosis. Neural Regen. Res. 17:1412. 10.4103/1673-5374.330591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min S., Sohn P., Li Y., Devidze N., Johnson J., Krogan N., et al. (2018). SIRT1 Deacetylates Tau and Reduces Pathogenic Tau Spread in a Mouse Model of Tauopathy. J. Neurosci. 38:3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M., Jäkel L., Bruinsma I. B., Claassen J. A., Kuiperij H. B., Verbeek M. M. (2016). MicroRNA-29a Is a Candidate Biomarker for Alzheimer’s Disease in Cell-Free Cerebrospinal Fluid. Mol. Neurobiol. 53:2894. 10.1007/S12035-015-9156-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng T., Ho C., Tam W., Kua E., Ho R. (2019). Decreased Serum Brain-Derived Neurotrophic Factor (BDNF) Levels in Patients with Alzheimer’s Disease (AD): A Systematic Review and Meta-Analysis. Int. J. Mol. Sci. 20:257. 10.3390/IJMS20020257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogonowski N., Salcidua S., Leon T., Chamorro-Veloso N., Valls C., Avalos C., et al. (2022). Systematic Review: microRNAs as Potential Biomarkers in Mild Cognitive Impairment Diagnosis. Front. Aging Neurosci. 13:959. 10.3389/FNAGI.2021.807764/BIBTEX [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onyango I. G., Jauregui G. V., . Èarná M., Bennett J. P., Stokin G. B. (2021). Neuroinflammation in Alzheimer’s Disease. Biomedicines 9:50524. 10.3390/BIOMEDICINES9050524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orang A. V., Safaralizadeh R., Kazemzadeh-Bavili M. (2014). Mechanisms of miRNA-mediated gene regulation from common downregulation to mRNA-specific upregulation. Int. J. Genom. 2014:970607. 10.1155/2014/970607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan K., Chen S., Wang Y., Yao W., Gao X. (2021). MicroRNA-23b attenuates tau pathology and inhibits oxidative stress by targeting GnT-III in Alzheimer’s disease. Neuropharmacology 196:108671. [DOI] [PubMed] [Google Scholar]
- Peña-Bautista C., Tarazona-Sánchez A., Braza-Boils A., Balaguer A., Ferré-González L., Cañada-Martínez A., et al. (2022). Plasma microRNAs as potential biomarkers in early Alzheimer disease expression. Sci. Rep. 12:15589. 10.1038/s41598-022-19862-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasekhar K., Govindaraju T. (2018). Current progress, challenges and future prospects of diagnostic and therapeutic interventions in Alzheimer’s disease. RSC Adv. 8 23780–23804. 10.1039/c8ra03620a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricciarelli R., Fedele E. (2017). The Amyloid Cascade Hypothesis in Alzheimer’s Disease: It’s Time to Change Our Mind. Curr. Neuropharmacol. 15 926–935. 10.2174/1570159X15666170116143743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Z., Li Y., He R., Li X. (2021). Inhibition of microRNA-10b-5p up-regulates HOXD10 to attenuate Alzheimer’s disease in rats via the Rho/ROCK signalling pathway. J. Drug Target 29 531–540. [DOI] [PubMed] [Google Scholar]
- Sala Frigerio C., Lau P., Salta E., Tournoy J., Bossers K., Vandenberghe R., et al. (2013). Reduced expression of hsa-miR-27a-3p in CSF of patients with Alzheimer disease. Neurology 81 2103–2106. 10.1212/01.WNL.0000437306.37850.22 [DOI] [PubMed] [Google Scholar]
- Samadian M., Gholipour M., Hajiesmaeili M., Taheri M., Ghafouri-Fard S. (2021). The Eminent Role of microRNAs in the Pathogenesis of Alzheimer’s Disease. Front. Aging Neurosci. 13:641080. 10.3389/fnagi.2021.641080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S., Engler-Chiurazzi E., Cavendish J., Povroznik J., Russell A., Quintana D., et al. (2019). Over-expression of miR-34a induces rapid cognitive impairment and Alzheimer’s disease-like pathology. Brain Res. 1721:146327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S., Jun S., Rellick S., Quintana D. D., Cavendish J. Z., Simpkins J. W. (2016). Expression of microRNA-34a in Alzheimer’s disease brain targets genes linked to synaptic plasticity, energy metabolism, and resting state network activity. Brain Res. 1646 139–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh J. I., Kino Y., Niida S. (2015). MicroRNA-Seq Data Analysis Pipeline to Identify Blood Biomarkers for Alzheimer’s Disease from Public Data. Biomark Insights 10:21. 10.4137/BMI.S25132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimanoe H., Ko S., Jeon Y., Nakabayashi K., Miyawaki J., Yoon A. (2019). Shortening Stabilization Time Using Pressurized Air Flow in Manufacturing Mesophase Pitch-Based Carbon Fiber. Polymers 11:1911. 10.3390/polym11121911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J. (2022). Dementia Epidemiology Fact Sheet 2022. Ann. Rehabil. Med. 46 53–59. 10.5535/arm.22027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siengdee P., Trakooljul N., Murani E., Schwerin M., Wimmers K., Ponsuksili S. (2015). MicroRNAs Regulate Cellular ATP Levels by Targeting Mitochondrial Energy Metabolism Genes during C2C12 Myoblast Differentiation. PLoS One 10:e0127850. 10.1371/JOURNAL.PONE.0127850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim S., Lim C., Kim J., Seo D., Chun H., Yu N., et al. (2016). The Brain-Enriched MicroRNA miR-9-3p Regulates Synaptic Plasticity and Memory. J. Neurosci. 36 8641–8652. 10.1523/JNEUROSCI.0630-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen S. S., Nygaard A.-B., Christensen T. (2016). miRNA expression profiles in cerebrospinal fluid and blood of patients with Alzheimer’s disease and other types of dementia-an exploratory study. Transl. Neurodegener. 5:6. 10.1186/s40035-016-0053-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C., Jia N., Li R., Zhang Z., Zhong Y., Han K. (2020). miR-143-3p inhibition promotes neuronal survival in an Alzheimer’s disease cell model by targeting neuregulin-1. Folia Neuropathol. 58 10–21. [DOI] [PubMed] [Google Scholar]
- Sun T., Zhao K., Liu M., Cai Z., Zeng L., Zhang J., et al. (2022). miR-30a-5p induces Aβ production via inhibiting the nonamyloidogenic pathway in Alzheimer’s disease. Pharmacol. Res. 178:106153. [DOI] [PubMed] [Google Scholar]
- Takalkar S., Xu H., Chen J., Baryeh K., Qiu W., Zhao J., et al. (2016). Gold Nanoparticle Coated Silica Nanorods for Sensitive Visual Detection of microRNA on a Lateral Flow Strip Biosensor. Anal. Sci. 32 617–622. 10.2116/analsci.32.617 [DOI] [PubMed] [Google Scholar]
- Tan Y., Wong B., Vaidyanathan R., Sreejith S., Chia S., Kandiah N., et al. (2021). Altered Cerebrospinal Fluid Exosomal microRNA Levels in Young-Onset Alzheimer’s Disease and Frontotemporal Dementia. J. Alzheimers Dis. Rep. 5:805. 10.3233/ADR-210311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taşdelen E., . Kizil E. T. Ö, Aydemir S. T., Yalap ÖE., Bingöl A. P., Kutlay N. (2022). Determination of miR-373 and miR-204 levels in neuronal exosomes in Alzheimer’s disease. Turk. J. Med. Sci. 52 1458–1467. 10.55730/1300-0144.5484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangavelu B., Wilfred B. S., Johnson D., Gilsdorf J. S., Shear D. A., Boutté A. M. (2020). Penetrating Ballistic-Like Brain Injury Leads to MicroRNA Dysregulation, BACE1 Upregulation, and Amyloid Precursor Protein Loss in Lesioned Rat Brain Tissues. Front. Neurosci. 14:915. 10.3389/FNINS.2020.00915/FULL [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thies B., Truschke E., Morrison-Bogorad M., Hodes R. J. (1999). Consensus report of the Working Group on: molecular and biochemical markers of Alzheimer’s disease. Neurobiol. Aging 20:247. 10.1385/1-59259-005-5:329 [DOI] [PubMed] [Google Scholar]
- Vilardo E., Barbato C., Ciotti M. T., Cogoni C., Ruberti F. (2010). MicroRNA-101 Regulates Amyloid Precursor Protein Expression in Hippocampal Neurons. J. Biol. Chem. 285:18344. 10.1074/JBC.M110.112664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahid F., Shehzad A., Khan T., Kim Y. Y. (2010). MicroRNAs: Synthesis, mechanism, function, and recent clinical trials. Biochim. Biophys. Acta 123:1803. [DOI] [PubMed] [Google Scholar]
- Wallach T., Mossmann Z., Szczepek M., Wetzel M., Machado R., Raden M., et al. (2021). MicroRNA-100-5p and microRNA-298-5p released from apoptotic cortical neurons are endogenous Toll-like receptor 7/8 ligands that contribute to neurodegeneration. Mol. Neurodegener. 16:80. 10.1186/s13024-021-00498-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Fei Z., Luo S., Wang H. (2020). MiR-335-5p Inhibits β-Amyloid (Aβ) Accumulation to Attenuate Cognitive Deficits Through Targeting c-jun-N-terminal Kinase 3 in Alzheimer’s Disease. Curr. Neurovasc. Res. 17 93–101. 10.2174/1567202617666200128141938 [DOI] [PubMed] [Google Scholar]
- Wang M., Qin L., Tang B. (2019). MicroRNAs in Alzheimer’s Disease. Front. Genet. 10:153. 10.3389/FGENE.2019.00153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Zhang J. (2020). Clinical significance of miR-433 in the diagnosis of Alzheimer’s disease and its effect on Aβ-induced neurotoxicity by regulating JAK2. Exp. Gerontol. 141:111080. 10.1016/j.exger.2020.111080 [DOI] [PubMed] [Google Scholar]
- Wang Y., Shi M., Hong Z., Kang J., Pan H., Yan C. (2021). MiR-130a-3p Has Protective Effects in Alzheimer’s Disease via Targeting DAPK1. Am. J. Alzheimers Dis. Other Demen. 36:15333175211020572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H., Zhang H., Xie J., Meng D., Wang X., Ke D., et al. (2020). Protein Phosphatase 2A as a Drug Target in the Treatment of Cancer and Alzheimer’s Disease. Curr. Med. Sci. 40 1–8. 10.1007/s11596-020-2140-1 [DOI] [PubMed] [Google Scholar]
- Weinstein G., Beiser A., Choi S., Preis S., Chen T., Vorgas D., et al. (2014). Serum brain-derived neurotrophic factor and the risk for dementia: the Framingham Heart Study. JAMA Neurol. 71 55–61. 10.1001/jamaneurol.2013.4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller J., Budson A. (2018). Open Peer Review Current understanding of Alzheimer’s disease diagnosis and treatment. F1000Res 7:F1000FacultyRev–1161. 10.12688/f1000research.14506.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Xu J., Xu J., Cheng J., Jiao D., Zhou C., et al. (2017). Lower Serum Levels of miR-29c-3p and miR-19b-3p as Biomarkers for Alzheimer’s Disease. Tohoku J. Exp. Med. 242 129–136. 10.1620/tjem.242.129 [DOI] [PubMed] [Google Scholar]
- Xia P., Chen J., Liu Y., Cui X., Wang C., Zong S., et al. (2022). MicroRNA-22-3p ameliorates Alzheimer’s disease by targeting SOX9 through the NF-κB signaling pathway in the hippocampus. J. Neuroinflammation 19:180. 10.1186/s12974-022-02548-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing H., Guo S., Zhang Y., Zheng Z., Wang H. (2016). Upregulation of microRNA-206 enhances lipopolysaccharide-induced inflammation and release of amyloid-β by targeting insulin-like growth factor 1 in microglia. Mol. Med. Rep. 14 1357–1364. 10.3892/MMR.2016.5369/HTML [DOI] [PubMed] [Google Scholar]
- Yang K., Chen Z., Gao J., Shi W., Li L., Jiang S., et al. (2017). The Key Roles of GSK-3β in Regulating Mitochondrial Activity. Cell Physiol. Biochem. 44 1445–1459. 10.1159/000485580 [DOI] [PubMed] [Google Scholar]
- Yang T. T., Liu C. G., Gao S. C., Zhang Y., Wang P. C. (2018). The Serum Exosome Derived MicroRNA-135a, -193b, and -384 Were Potential Alzheimer’s Disease Biomarkers. Biomed. Environ. Sci. 31 87–96. 10.3967/bes2018.011 [DOI] [PubMed] [Google Scholar]
- Ye X., Luo H., Chen Y., Wu Q., Xiong Y., Zhu J., et al. (2015). MicroRNAs 99b-5p/100-5p Regulated by Endoplasmic Reticulum Stress are Involved in Abeta-Induced Pathologies. Front. Aging Neurosci. 7:210. 10.3389/fnagi.2015.00210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Fang Y., Zhao X., Zheng Y., Ma Y., Li S., et al. (2021). miR-204 silencing reduces mitochondrial autophagy and ROS production in a murine AD model via the TRPML1-activated STAT3 pathway. Mol. Ther. Nucleic Acids 24 822–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W. T., Zhang G. X., Gao S. S. (2021). The potential diagnostic accuracy of circulating microRNAs for Alzheimer’s disease: A meta-analysis. Neurología 10.1016/J.NRL.2021.06.001 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Zhang N., Li W.-W., Lv C.-M., Gao Y.-W., Liu X.-L., Zhao L. (2020). miR-16-5p and miR-19b-3p prevent amyloid β-induced injury by targeting BACE1 in SH-SY5Y cells. Neuroreport 31 205–212. 10.1097/WNR.0000000000001379 [DOI] [PubMed] [Google Scholar]
- Zhang R., Zhou H., Jiang L., Mao Y., Cui X., Xie B., et al. (2016). MiR-195 dependent roles of mitofusin2 in the mitochondrial dysfunction of hippocampal neurons in SAMP8 mice. Brain Res. 1652 135–143. 10.1016/j.brainres.2016.09.047 [DOI] [PubMed] [Google Scholar]
- Zhang X., Cheng X., Yu L., Yang J., Calvo R., Patnaik S., et al. (2016). MCOLN1 is a ROS sensor in lysosomes that regulates autophagy. Nat. Commun. 7:12109. 10.1038/ncomms12109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z., Yuan K., Tong X., Hu J., Song Z., Zhang G., et al. (2018). MiR-16 attenuates β-amyloid-induced neurotoxicity through targeting β-site amyloid precursor protein-cleaving enzyme 1 in an Alzheimer’s disease cell model. Neuroreport 29 1365–1372. 10.1097/WNR.0000000000001118 [DOI] [PubMed] [Google Scholar]
- Zhou Q., Luo L., Wang X., Li X. (2019). Relationship between single nucleotide polymorphisms in the 3’UTR of amyloid precursor protein and risk of Alzheimer’s disease and its mechanism. Biosci. Rep. 39:20182485. 10.1042/BSR20182485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Lin M., Ma J., Liu W., Gao L., Wei S., et al. (2019). The role of LINC00094/miR-224-5p (miR-497-5p)/Endophilin-1 axis in Memantine mediated protective effects on blood-brain barrier in AD microenvironment. J. Cell Mol. Med. 23 3280–3292. 10.1111/jcmm.14214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Li C., Sun A., Wang Y., Zhou S. (2015). Quantification of microRNA-210 in the cerebrospinal fluid and serum: Implications for Alzheimer’s disease. Exp. Ther. Med. 9 1013–1017. 10.3892/ETM.2015.2179/HTML [DOI] [PMC free article] [PubMed] [Google Scholar]