Abstract
The bacterial strain RB1 has been isolated by enrichment cultivation with 2,4-dinitrophenol as the sole nitrogen, carbon, and energy source and characterized, on the basis of 16S rRNA gene sequence comparison, as a Rhodococcus species closely related to Rhodococcus opacus. Rhodococcus sp. strain RB1 degrades 2,4-dinitrophenol, releasing the two nitro groups from the compound as nitrite. The release of nitro groups from 2,4-dinitrophenol occurs in two steps. First, the 2-nitro group is removed as nitrite, with the production of an aliphatic nitro compound identified by 1H nuclear magnetic resonance and mass spectrometry as 3-nitroadipate. Then, this metabolic derivative is further metabolized, releasing its nitro group as nitrite. Full nitrite assimilation upon reduction to ammonia requires that an additional carbon source be supplied to the medium.
Nitroaromatic compounds are common pollutants of natural environments due to abusive industrial and agricultural practices (7). These xenobiotic compounds are produced by industries manufacturing explosives, herbicides, pesticides, and dyes, and degradative pathways of these compounds are widely distributed among bacteria (for a review, see reference 15). Among these microorganisms, some Rhodococcus strains exhibit a high potential for the degradation of polynitroaromatic compounds (4). The biological hydride transfer to the aromatic ring with the concomitant formation of a Meisenheimer complex was shown to occur during the metabolism of picric acid by Rhodococcus erythropolis HL 24-2 (9). Further evidence for a reductive metabolic route was given in the same paper, showing the accumulation of 2,4-dinitrophenol (2,4-DNP) from picric acid, and in an accompanying communication reporting 4,6-dinitrohexanoate as the metabolite that accumulated under anaerobic conditions (11). A similar mechanism might account for the degradation of trinitrotoluene (TNT) by a Pseudomonas sp. strain able to convert TNT into toluene; the mineralization of the chemical was achieved by transferring the TOL catabolic plasmid to the recipient strain (3).
In this work, we report the isolation and characterization of a gram-positive bacterium, characterized as a new Rhodococcus strain on the basis of its 16S rRNA gene sequence, with a high biodegradative potential. As far as we know, there is, beside the identification of 4,6-dinitrohexanoate (11), no evidence for the metabolic fate of the putative Meisenheimer complex formed from 2,4-DNP. On the basis of our results and those cited above, we propose a pathway for the mineralization of 2,4-DNP with 3-nitroadipate as a key metabolic intermediate.
MATERIALS AND METHODS
Bacterial isolation, growth conditions, and biodegradative potential.
Rhodococcus sp. strain RB1 was isolated by aerobic enrichment cultivation of an activated sludge from a wastewater plant in Alicante, Spain. The medium used was M9 mineral medium (13) with 0.5 mM 2,4-DNP as the sole nitrogen and carbon source. After enrichment (four reinoculation steps), samples of the culture were plated on the same medium, which was solidified with 1.8% (wt/vol) Bacto Agar (Difco). One colony was purified and proved capable of 2,4-DNP mineralization in liquid culture; it was used for further studies. Where indicated, acetate and/or ammonium or nitrite were added as the carbon and nitrogen sources.
Anaerobic conditions were obtained by completely filling screw-capped bottles with the culture medium (10-ml volume). For strict anaerobiosis, high-purity argon was bubbled through the cultures.
R. erythropolis HL 24-2 was kindly provided by H.-J. Knackmuss (10).
Taxonomic position and analysis of the 16S rRNA gene sequence of Rhodococcus sp. strain RB1.
The phylogenetic position of strain RB1 was estimated as described in detail previously (6) with the evolutionarily conserved primary sequence and secondary structure as the reference (5). Cluster analyses were carried out with programs contained in the Phylogeny Inference Package (PHYLIP), version 3.5c (J. Felsenstein, University of Washington, Seattle).
Analytical determinations.
Bacterial growth was monitored turbidimetrically at 600 nm. Nitrite, ammonium, and protein concentrations were estimated as described previously (1).
Nitrophenol compounds and possible transformation products were analyzed with a Beckman System Gold high-performance liquid chromatograph (HPLC) equipped with SC columns (125 by 4.6 mm) filled with 5-mm Lichrospher 100 RP8 particles (Bischoff, Leonberg, Federal Republic of Germany). 2,4-DNP was quantified in an isocratic elution program of an aqueous solvent containing 35% (vol/vol) methanol and 0.1% (vol/vol) H3PO4 in Milli-Q water (flow rate, 1 ml/min). Usually, 20-ml samples were analyzed. For the detection of the Meisenheimer complex of picric acid, a gradient elution program using 10 mM phosphate buffer, pH 6.8 (solvent A), and acetonitrile (solvent B) was performed as follows: a flow rate of 1 ml/min consisting of 3 ml of gradient (0 to 45% solvent B), followed by a 5-ml smooth gradient (45 to 100% solvent B) and 1 ml at 100% solvent B. The column effluent was simultaneously monitored at 210 and 270 nm with a diode array detector, and the scans between 200 and 600 nm of the significant peaks were stored by the system.
Extraction and analysis of metabolites.
The cells were cultured with 50 mM acetate and 1 mM 2,4-DNP as the carbon and nitrogen sources, respectively. Immediately after 2,4-DNP was exhausted completely, the cells were removed by centrifugation and the supernatant from a 1-liter culture was acidified with HCl to reach pH 2. The acidified supernatant was extracted twice with 100 ml of diethyl ether, and the organic phases were pooled, dried over anhydrous Na2SO4, and concentrated at 40°C under vacuum. The solid concentrate was dissolved in 1 ml of methanol and chromatographed though a semi-preparative HPLC column. The effluent was monitored at 210 nm. Fractions (1 ml each) were collected, and those corresponding to the main HPLC peak were pooled and dried at 40°C under vacuum. The solid was dissolved in 1 ml of deuterated methanol and stored at −20°C for further analysis.
The purified sample was injected directly into a Carlo Erba/MEGA SERIES gas chromatograph (GC) equipped with a 30-m DB1 capillary column with helium as the carrier gas. The temperature program was 80°C for 2 min and then a linear gradient of 10°C per min up to 300°C. The GC was directly coupled to a Kratos MS 50 mass spectrometer (MS). For electron ionization, a potential of 70 eV was used. Dynamic high-resolution MS of the molecular ion and the major fragment ions was performed with perfluorokerosene as an internal reference.
High-resolution one- and two-dimensional correlated spectroscopy 1H nuclear magnetic resonance (NMR) spectra were recorded on a Bruker WM 400 NMR spectrometer locked to the major deuterium resonance of the solvent, CD3OD. Chemical shifts are given in parts per million relative to tetramethylsilane and constants in Hertz.
Chemicals.
All reagents were of the maximal purity commercially available.
RESULTS AND DISCUSSION
(i) Isolation and characterization of the strain.
A bacterium, strain RB1, capable of degrading 2,4-DNP was isolated from activated sludge after incubation with this compound as the sole carbon, nitrogen, and energy source. Amplification by PCR of the 16S rRNA gene sequence and its comparison with reference 16S rRNA sequence data (12, 17) indicated that strain RB1 clustered phylogenetically with species of Rhodococcus rRNA group IV (14), particularly with the sequence of Rhodococcus opacus (DSM 43205T) (4 nucleotide differences). The genus Rhodococcus is the subject of intense investigation due to its implication in the degradation of a broad range of organic pollutants (8, 18). An independent research group using a similar approach to enrich cultures with 2,4-DNP as the target compound also isolated two Rhodococcus strains (11).
The biodegradative potential of Rhodococcus sp. strain RB1 was analyzed on gradient-agar plates and in liquid cultures. In addition to 2,4-DNP (Fig. 1), Rhodococcus sp. strain RB1 was also able to use acetate, benzoate, naphthalene, 3-hydroxybenzoate, 4-hydroxy-3-methoxycinnamate (ferulate), 3,4-dihydroxybenzoate (protocatechuate), and 4-methoxybenzoate (p-anisate) as the sole carbon source (2 mM ammonium as the nitrogen source). These chemicals were also used by the bacterium as the sole carbon source (5 mM concentration) in liquid culture experiments with 2 mM ammonium as the nitrogen source.
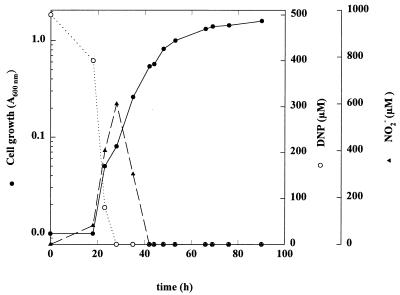
FIG. 1.
Time course of cell growth, 2,4-DNP uptake, and nitrite release by Rhodococcus strain RB1 cultured with 2,4-DNP as the sole nitrogen source. Cells were grown under aerobic conditions with 50 mM sodium acetate and 0.5 mM 2,4-DNP. A600 (•), 2,4-DNP concentration (○), and nitrite concentration (▴) were measured at the indicated times.
The bacterium used as nitrogen sources 2,4-DNP, ammonium, nitrate, and nitrite, with 50 mM acetate as the carbon source. Mononitrophenols were not used by the bacterium as either a carbon or nitrogen source. In contrast, both 2-nitrophenol and 4-nitrophenol strongly inhibited 2,4-DNP mineralization by Rhodococcus sp. strain RB1 (data not shown), even though Rhodococcus sp. strain RB1 partially transformed 4-nitrophenol into 4-nitrocatechol as a dead-end product (data not shown).
Rhodococcus sp. strain RB1 grew aerobically in liquid mineral medium with 2,4-DNP as the nitrogen source at concentrations of up to 5 mM (50 mM acetate as the carbon source). The maximal concentration of 2,4-DNP tolerated by the bacterium under the same conditions with 2,4-DNP as the sole carbon and nitrogen source was 2 mM. No growth was observed with 2,4-DNP under anaerobic conditions.
The degradation of 2,4-DNP as the sole carbon and nitrogen source in liquid cultures was complete and took place with an almost stoichiometric nitrite release (1 mol of 2,4-DNP to 1.65 mol of nitrite). The nitrite released into the medium was not taken up by the bacterium unless acetate was added to the carbon-starved cells (data not shown). The increment in optical density at 600 nm (ΔA600) observed in cultures with 1 mM 2,4-DNP as the sole carbon and nitrogen source was 0.18 ± 0.02. The ΔA600 in cultures growing with 3 mM acetate as the carbon source and 2 mM nitrite as the nitrogen source was 0.2 ± 0.03, thus indicating that 2,4-DNP was fully used as a carbon source without major energy losses.
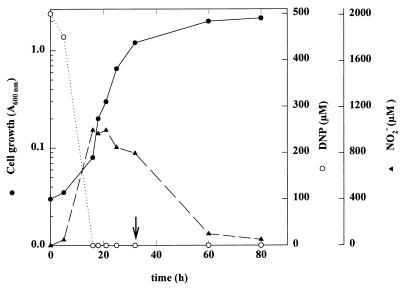
The utilization of 2,4-DNP as a nitrogen source by Rhodococcus sp. strain RB1 showed a diauxic growth curve (Fig. 1). After an initial adaptation period (cells precultured on acetate and nitrite), the first phase is characterized by a minute growth (doubling time, approximately 21 h) and a rapid transformation of 2,4-DNP with simultaneous release of nitrite (almost 1 mol of nitrite per mol of 2,4-DNP consumed). During the second phase, the strain grew more rapidly (doubling time, approximately 11 h) and the nitrite was taken up. Nevertheless, Rhodococcus sp. strain RB1 used both nitro groups as the nitrogen source. This conclusion was substantiated by comparing the ΔA600 obtained with an excess of carbon source (50 mM acetate) and that obtained with limiting amounts of either 2,4-DNP or equivalent amounts of nitrite as the nitrogen source (ΔA600 = 1.6 ± 0.05 units/mol of nitrite and ΔA600 = 3.15 ± 0.05 units/mol of 2,4-DNP). In order to further confirm that both nitro groups were used by Rhodococcus sp. strain RB1, experiments were performed in the presence of ammonium as an inhibitor of nitrite assimilation (2). As expected, ammonium inhibited nitrite uptake. As a matter in fact, in the presence of a nonlimiting concentration of ammonium (20 mM), up to 1 mM nitrite accumulated in the medium from 0.5 mM 2, 4-DNP (data not shown). When limiting ammonium concentrations (2 mM) were used in cell cultures containing 0.5 mM 2,4-DNP and 50 mM acetate, up to 1 mM nitrite appeared in the medium, but it was taken up again once ammonia was completely assimilated (Fig. 2). Ammonium did not inhibit 2,4-DNP uptake (Fig. 2) as it does in Rhodobacter capsulatus (1).
FIG. 2.
Time course of cell growth, 2,4-DNP uptake, and nitrite release by Rhodococcus strain RB1 cultured with 2,4-DNP and ammonium as nitrogen sources. Cells were grown under aerobic conditions with 50 mM sodium acetate, 0.5 mM 2,4-DNP, and 2 mM ammonium chloride. At the times indicated, A600 (•), 2,4-DNP concentration (○), and nitrite concentration (▴) were measured. The arrow indicates the time at which the concentration of ammonium in the medium was undetectable.
(ii) Degradation pathway of 2,4-DNP by Rhodococcus sp. strain RB1.
An initial reductive pathway has been demonstrated in some gram-positive bacteria that degrade nitroaromatic compounds (4). The transfer of a hydride to the aromatic ring of picric acid and 2-chloro-4,6-DNP provokes the release of the 2-nitro or 2-chloro group with the concomitant formation of 2,4-DNP (9, 10). We have also observed the formation of 2,4-DNP from picric acid in anaerobic resting-cell experiments with Rhodococcus sp. strain RB1 (data not shown), thus suggesting a similar reaction. In addition, Rhodococcus sp. strain RB1 cells induced with 2,4-DNP produced a brilliant red compound when transferred to medium with picric acid (0.5 mM), indicating the putative formation of a Meisenheimer complex. The formation of this compound from picric acid was unambiguously demonstrated in R. erythropolis HL 24-2 (9), and therefore, we have used it as the reference strain. The product formed by both R. erythropolis HL 24-2 and Rhodococcus sp. strain RB1 from picric acid coeluted during HPLC analysis and exhibited the same UV-visible light spectrum (data not shown). As mentioned above, the hydrogenation of the aromatic ring may result in the rearomatization and concomitant liberation of nitrite or chloride (9, 10). Nevertheless, it is unlikely that this mechanism (rearomatization) accounts for the degradation of 2,4-DNP by Rhodococcus sp. strain RB1, since it should yield a mononitrophenol derivative (probably 4-nitrophenol) which cannot be mineralized by the bacterium. In spite of that, this mechanism cannot be excluded in other bacteria, and in fact, 4-nitrophenol has been detected very recently in small amounts during the degradation of picric acid by Nocardioides simplex (16).
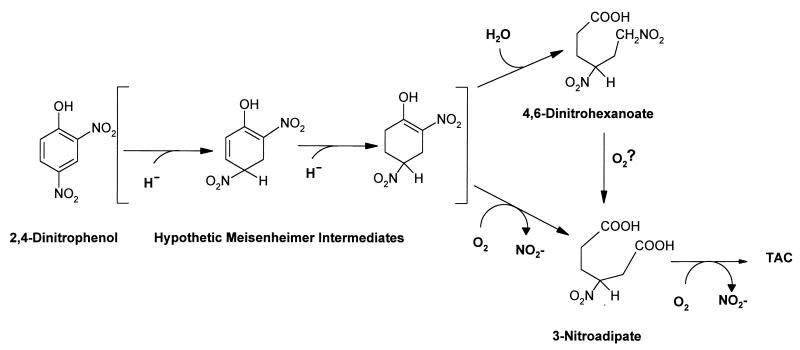
The ratio of nitrite released to 2,4-DNP consumed showed a stoichiometry of 1/1 instead of 2/1 during the first growth phase of Rhodococcus strain RB1 with 2,4-DNP as the nitrogen source (Fig. 1). As very little growth had occurred at the end of this growth phase, it can be assumed that the second nitrogen is bound to an intermediate. However, analysis of the supernatants of the culture medium from this phase by HPLC showed only minute peaks absorbing at 210 nm. To amplify the signals, these supernatants were extracted and concentrated as described in Materials and Methods. A complex mixture of compounds absorbing at 210 nm was detected by HPLC. The main compound (approximately 70% in relative units, at 5 min net retention time) was purified by preparative HPLC and showed a single absorption maximum at 204 nm. The product contained in this fraction was identified as 3-nitroadipate dimethyl ester according to GC–high-resolution MS analysis (Table 1). Nevertheless, we assume that a partial dimethylation of the chemical occurred due to the heating in the presence of methanol during the concentration step (see Materials and Methods). This assumption is based on the need for acidic conditions (pH 2) for the full extraction of the chemical from the supernatant. In addition, the 1H-NMR spectrum of the extracted compound corresponded to that of the free acid (Table 2) with a minute amount of the diester (estimated to be around 2% according to the two singlet signals at 3.71 and 3.73 ppm corresponding to the methyl groups), even though, as the free acid is not volatile, only the diester was detected by GC-MS. The 3-nitroadipate is obviously more oxidized than the proposed initial intermediate (Meisenheimer complex). The nature of the oxidant is unknown, but we suggest that it should be O2. The reason is that, under anaerobic conditions, R. erythropolis accumulates mainly 4,6-dinitrohexanoate from 2,4-DNP (11) or 1,3,5-trinitropentane from picric acid (9). The formation of 4,6-dinitrohexanoate may be achieved by the hydration of the double bond of a putative dihydro-2,4-DNP intermediate, whereas the dioxygenolytic cleavage of the double bond would produce 3-nitroadipate (Fig. 3).
TABLE 1.
Characteristic signals in the mass spectrum of 3-nitroadipate dimethyl ester
| Ion mass (m/z)
|
Intensity (%) | Molecular structure
|
||
|---|---|---|---|---|
| Experimental | Theoretical | Ion | Structural interpretation | |
| 219.0739 | C8H13O6N | [M]+ | ||
| 188.0541 | 188.0556 | 21 | C7H10O5N | [M-OCH3]+ |
| 170.0451 | 170.0451 | 12 | C7H8O4N | [M-OCH3-H2O]+ |
| 141.0514 | 141.0549 | 53 | C7H9O3 | [M-OCH3-HNO2]+ |
| 128.0364 | 128.0346 | 12 | C5H6O3N | [M-OCH3-HCO2CH3]+ |
| 113.0621 | 113.0600 | 52 | C6H9O2 | [M-CO2CH3-HNO2]+ |
| 109.0315 | 109.0288 | 28 | C6H5O2 | [M-OCH3-HOCH3-HNO2]+ |
| 81.0367 | 81.0339 | 25 | C5H5O | [M-OCH3-HCO2CH3-HNO2]+ |
| 71.0515 | 71.0495 | 100 | C4H7O | [C4H7O]+ |
| 59.0157 | 59.0132 | 60 | C2H3O2 | [CO2CH3]+ |
TABLE 2.
Structure and 1H-NMR data of 3-nitroadipate
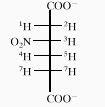 |
| Proton | Chemical shift (ppm) | Coupling constanta (Hz) |
|---|---|---|
| 1 | 3.13 | J1,2 = 17.7 Hz, J1,3 = 10.0 Hz |
| 2 | 2.87 | J2,3 = 3.5 Hz |
| 3 | 4.99 | J3,4 = 7.0 Hz, J3,5 = 7.0 Hz |
| 4 | 2.20 | J4,6 = 6.3 Hz, J4,7 = 6.3 Hz |
| 5 | 2.21 | J5,6 = 8.1 Hz, J5,7 = 8.1 Hz |
| 6 | 2.46 | |
| 7 | 2.44 |
The spectrum form indicated that H-4/H-5 and H-6/H-7 were nonequivalent. Spectral simulation and iteration using LAOCOON III allowed the shift differences shown to be estimated. As all the lower-intensity lines in the observed spectrum could not be determined accurately, only estimates (±0.3 Hz) of the couplings J3,4, J3,5, J4,6, J4,7, J5,6, and J5,7 were possible. The remaining couplings are ±0.1 Hz.
FIG. 3.
Proposed pathway for the degradation of 2,4-DNP by Rhodococcus. Compounds in brackets are hypothetical intermediates. The formation of 4,6-dinitrohexanoate has been described previously (12). TAC, tricarboxylic acid cycle.
These results may be summarized in the degradation pathway depicted in Fig. 3. We propose that 2,4-DNP mineralization by Rhodococcus takes place by a metabolic pathway including three different phases: (i) reduction of the aromatic ring, as previously demonstrated (10–12), by two successive hydride transfers; (ii) aerobic ortho ring fission, production of 3-nitroadipate, and concomitant release of the ortho nitro group as nitrite (nevertheless, the reductive formation of 4,6-dinitrohexanoate [11] and subsequent oxidation to 3-nitroadipate cannot be excluded); and (iii) further metabolism of 3-nitroadipate with release of a second mole of nitrite.
Experiments are in progress to determine if enzymes of the 3-oxoadipate pathway, actually involved in benzoate metabolism by Rhodococcus strain RB1 (data not shown), are also involved in the metabolism of 3-nitroadipate by this bacterium.
ACKNOWLEDGMENTS
We thank the Spanish Dirección General de Investigación Científica y Técnica (DGICYT grant PB95 0554 CO2 02) and Plan Andaluz de Investigación for financial support. Postdoctoral fellowships from the Ministerio de Educación y Ciencia and European Environmental Research Organization to R.B. and from Alexander von Humboldt to F.C. are also gratefully acknowledged.
We also thank Rolf Wittich for critically reading the manuscript, R. Blanco for his help in HPLC analysis, and Manfred Nimtz for performing GC-MS analysis. We also thank H.-J. Knackmuss for kindly supplying R. erythropolis HL 24-2.
REFERENCES
- 1.Blasco R, Castillo F. Light-dependent degradation of nitrophenols by the phototrophic bacterium Rhodobacter capsulatus E1F1. Appl Environ Microbiol. 1992;58:690–695. doi: 10.1128/aem.58.2.690-695.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caballero F J, Moreno-Vivián C, Castillo F, Cárdenas J. Nitrite uptake system in photosynthetic bacterium Rhodopseudomonas capsulata E1F1. Biochim Biophys Acta. 1986;848:16–23. [Google Scholar]
- 3.Duque E, Haidour A, Godoy F, Ramos J L. Construction of a Pseudomonas hybrid strain that mineralizes 2,4,6-trinitrotoluene. J Bacteriol. 1993;175:2278–2283. doi: 10.1128/jb.175.8.2278-2283.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gottschalk G, Knackmuss H-J. Bacteria and the biodegradation of chemicals achieved naturally, by combination, or by construction. Angew Chem Int Ed Engl. 1993;32:1398–1408. [Google Scholar]
- 5.Gutell R R, Weiser B, Woese C R, Noller H F. Comparative anatomy of 16S-like ribosomal RNA. Prog Nucleic Acid Res Mol Biol. 1985;32:155–216. doi: 10.1016/s0079-6603(08)60348-7. [DOI] [PubMed] [Google Scholar]
- 6.Karlson U, Dwyer D F, Hooper S W, Moore E R B, Timmis K N, Eltis L D. Two independently regulated cytochromes P-450 in a Rhodococcus rhodochrous strain that degrades 2-ethoxyphenol and 4-methoxybenzoate. J Bacteriol. 1993;175:1467–1474. doi: 10.1128/jb.175.5.1467-1474.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keith L H, Telliard W A. Priority pollutants. I. A perspective view. Environ Sci Technol. 1979;13:416–423. [Google Scholar]
- 8.Klatte S, Kroppenstedt R M, Rainey F A. Rhodococcus opacus sp. nov., an unusual nutritionally versatile Rhodococcus species. Syst Appl Microbiol. 1994;17:355–360. [Google Scholar]
- 9.Lenke H, Knackmuss H-J. Initial hydrogenation during catabolism of picric acid by Rhocococcus erythropolis HL 24-2. Appl Environ Microbiol. 1992;58:2933–2937. doi: 10.1128/aem.58.9.2933-2937.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lenke H, Knackmuss H-J. Initial hydrogenation and extensive reduction of substituted 2,4-dinitrophenols. Appl Environ Microbiol. 1996;62:784–790. doi: 10.1128/aem.62.3.784-790.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lenke H, Pieper D-H, Bruhn C, Knackmuss H-J. Degradation of 2,4-dinitrophenol by two Rhodococcus strains, HL 24-1 and HL 24-2. Appl Environ Microbiol. 1992;58:2928–2932. doi: 10.1128/aem.58.9.2928-2932.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–110. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 14.Rainey F A, Burghardt J, Kroppenstedt R M, Klatte S, Stackebrandt E. Phylogenetic analysis of the genera Rhodococcus and Nocardia and evidence for the evolutionary origin of the genus Nocardia from within the radiation of Rhodococcus species. Microbiology. 1995;141:523–528. [Google Scholar]
- 15.Spain J C. Biodegradation of nitroaromatic compounds. Annu Rev Microbiol. 1995;49:523–555. doi: 10.1146/annurev.mi.49.100195.002515. [DOI] [PubMed] [Google Scholar]
- 16.Valli K, Chivukula M, Armstrong E, Sariaslani S. Abstracts of the 97th General Meeting of the American Society for Microbiology 1997. Washington, D.C: American Society for Microbiology; 1997. Mechanism of degradation of picric acid by Nocardioides simplex, abstr. Q-346; p. 513. [Google Scholar]
- 17.Van de Peer Y, Jansen J, De Rijk P, De Wachter R. Database on the structure of small ribosomal subunit RNA. Nucleic Acids Res. 1997;25:111–116. doi: 10.1093/nar/25.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warhurst A M, Fewson C A. Biotransformations catalyzed by the genus Rhodococcus. Crit Rev Biotechnol. 1994;29:29–73. doi: 10.3109/07388559409079833. [DOI] [PubMed] [Google Scholar]