Abstract
Methods for profiling genes at the single-cell level have revolutionized our ability to study several biological processes and systems including development, differentiation, response programs and disease progression. In many of these studies, cells are profiled over time in order to infer dynamic changes in cell states and types, sets of expressed genes, active pathways, and key regulators. However, time-series single-cell RNA sequencing (scRNA-seq) also raises several new analysis and modelling issues. These issues range from determining when and how deep to profile cells, linking cells within and between time points, learning continuous trajectories and integrating bulk and single-cell data for reconstructing models of dynamic networks. In this Review, we discuss several approaches for the analysis and modelling of time-series scRNA-seq, highlighting their steps, key assumptions, and the types of data and biological questions they are most appropriate for.
Table of contents blurb
In this Review, Ding, Sharon and Bar-Joseph discuss how dynamic features can be incorporated into single-cell transcriptomics studies, using both experimental and computational strategies to provide biological insights.
Introduction
Biological processes and systems are dynamic. To fully understand the molecular and cellular components and networks that are activated as part of these processes researchers often collect data over time. The duration of the process or system being studied varies considerably among studies: from a few hours in immune response and drug treatment studies1 to days, months and even years in development, cell differentiation and ageing studies2. However, a unifying theme in all such studies is that the temporal data sets, which are often collected at discrete intervals, need to be analyzed, visualized, combined and integrated with other time-series and snapshot data to fully reconstruct dynamic models.
Over the past few years single-cell RNA sequencing (scRNA-seq) has become the method of choice for profiling the expression of genes in molecular studies3. There are several obvious advantages for scRNA-seq over bulk RNA-seq data including the ability to characterize the set of cells and the frequency of cell types in each sample4, the ability to identify the genes and networks activated within each cell or cell type5, and the ability to study relationships among cells or cell types6. However, this data type also raises new challenges, some of which apply to both single-timepoint (‘snapshot’) data and time-series data (for example, how many cells to profile, or how to group cells and assign cell types), whereas others are unique to time-series studies. For example, in bulk studies it is easy to relate the expression of genes at one time point to their expression in the previous time point, but for scRNA-seq data it is not trivial to link individual cells between two consecutive time points. An additional challenge in scRNA-seq studies is that cells collected at the same time point can represent a relatively wide range of different stages or cell states7 and so assigning all of them to the same point in the process is likely to be wrong. Several related computational issues arise when analyzing these data, including how to represent the large number of cells collected over time, how to infer the networks and pathways activated, and how to determine the exact timing of specific events.
Although several methods for the analysis and modelling of time-series bulk data have been developed8, many are not directly applicable to scRNA-seq data because they cannot address the challenges mentioned above. In addition, unlike for bulk data, even snapshot scRNA-seq data from a single sample can provide information on the dynamics of the process, either through trajectory [G] inference or RNA velocity9 (see below). This led to the development of several experimental and computational methods that are focused on studying the dynamics of biological processes using time-series scRNA-seq data. These methods provide information on the timing and ordering of events and enable researchers to take full advantage of scRNA-seq data. Such methods include experimental and computational methods to improve the way the data are collected, to obtain complementary information to aid in the data analysis, methods for visualizing the very large number of cells being profiled at each time point, their trajectories and ordering, and methods for integrating time-series scRNA-seq data with other time-series and snapshot data to reconstruct models of gene regulation and cell differentiation over time.
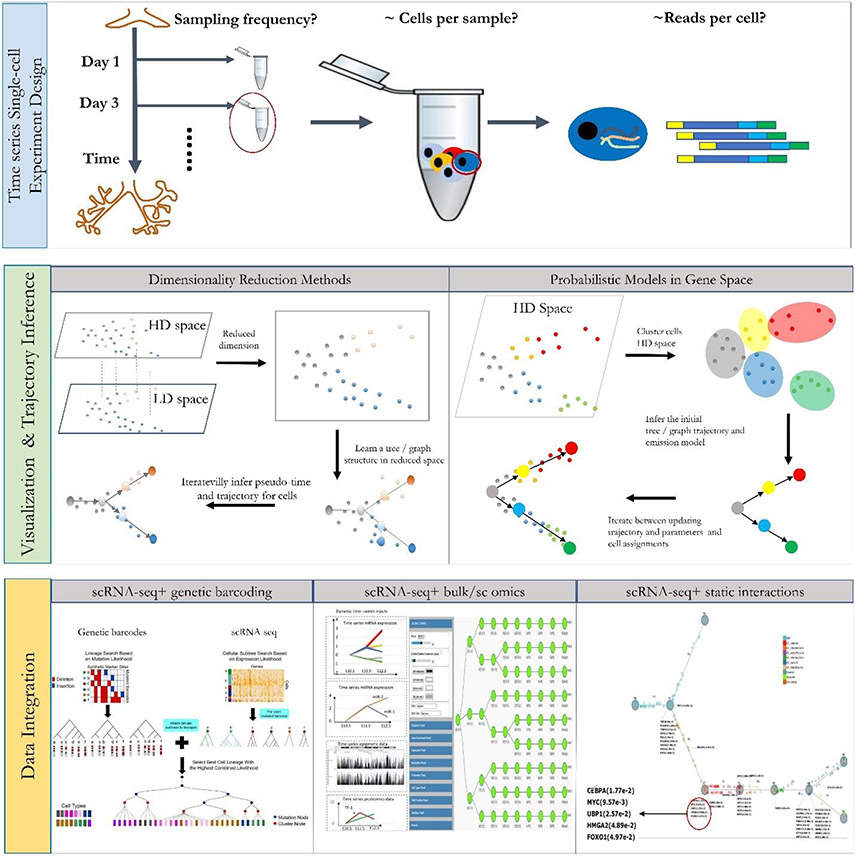
In this Review, we discuss both the experimental and computational approaches that have been developed for studying time-series scRNA-seq data. While several computational methods have been developed for pseudotime [G] ordering of scRNA-seq data, these are not always able to correctly reconstruct the temporal ordering and developmental trajectory of cells. We thus start by discussing experimental methods that can be used to explicitly infer such ordering or that can be integrated with scRNA-seq data to improve pseudotime inference. We next discuss computational methods that can be applied to any scRNA-seq time-series data and methods that integrate this data type with other snapshot and time-series data. For each of the methods we present, we mention the assumptions and requirements and discuss their input, output, and goals. We also discuss how different methods can be combined to perform end-to-end modelling of dynamic biological processes. Figure 1 presents an overview of the common time-series scRNA analysis pipeline, which we discuss in detail below.
Figure 1. Overview of time-series single-cell RNA-seq data analysis.
Top: Experimental design of time-series single-cell studies. Although many of the issues involved in designing time-series single-cell RNA sequencing (scRNA-seq) studies are similar to issues involved in designing single-timepoint (snapshot) scRNA-seq studies, additional consideration should be given to the sampling rates and the number of cells per sample (Box 1). The optimal sampling rate is impacted by the expected change in cell states and cell types, whereas the number of cells per sample is dependent on the distribution of cell types. Middle: Visualization and initial analysis of time-series data. Most methods for the analysis of time-series scRNA-seq data attempt to visualize the trajectory and pseudotime order of cells, both within each time point and between time points. Many different methods have been developed for this and these differ in the way they use the data, in the type of models they reconstruct and in how they assign the pseudotime. Bottom: Data integration. Several methods have been developed to complement time-series scRNA-seq by integrating it with other types of omics and interaction data. Examples include genetic barcoding methods (left), time-series bulk data (middle) and protein–DNA interaction data (right).
Experimental techniques for dynamic inference
Several experimental approaches have recently been developed to provide empirical information on the timing and order of molecular events at the single cell level. These methods provide “anchors” that increase the accuracy of the computational methods, and the two types of approaches can be combined to improve the analysis of time-series scRNA-seq data, as we discuss later. As even the most sophisticated computational methods for ordering and trajectory inference using scRNA-seq data require validation by experimental means, these experimental methods will remain an important part of the toolbox for single-cell analysis.
Metabolic labelling of RNAs.
Inferring the relative age of different mRNA transcripts can improve the accuracy of pseudotime analysis methods, as it reveals the actual order of transcriptional events within a cell. This layer of information can be deduced computationally from the relative abundance of intronic sequences, which are present only in nascent mRNA molecules9 (see below). However, several methods introduced recently manage to distinguish old transcripts from new in a straight-forward manner, through metabolic labelling of nascent RNA. 4-thiouridine (s4U) is a nucleotide analogue which can transport through the cell membrane and incorporate into nascent RNA as a substitute to uridine. Its alkylation following a reaction with iodoacetamide (IAA) results in misincorporation of a guanine in the corresponding site at the complementary strand during reverse transcription, leading to it being read as a T to C substitution in the original RNA transcript upon sequencing10. Herzog et al.10 developed the method SLAM-seq (thiol(SH)-linked alkylation for the metabolic sequencing of RNA), in which s4U is administered to cells in culture for a limited time; allowing for distinction of old RNA molecules from new ones based on higher T-to-C conversions rates in the latter. Several methods published recently combine this approach with various scRNA-seq techniques. For example scSLAM-seq11 and NASC-seq12, follow s4U incorporation and alkylation with smartseq-based library preparation, whereas sci-fate developed by the Shendure laboratory uses combinatorial double barcode labelling of fixed cells13. scNT-seq14 enables the use of droplet-based microfluidics for single-cell library preparation by using the alternative TimeLapse15 chemical reaction which, rather than alkylating s4U, transforms s4U into a cytosine analogue (trifluoroethylcytosine). Unlike IAA-mediated alkylation, TimeLapse chemistry increases the abundance of truncated mRNA molecules15, but it also allows the use of 6-thioguanine to introduce G to A substitutions16, thereby providing a hypothetical tool to label nascent RNA molecules over two time intervals. Overall, the main differences between the methods derive from the approaches they adopt for library preparation and not from the labelling chemistry. scSLAM-seq and NASC-seq are fit to handle low numbers of cells (several hundreds) and provide full-transcript sequencing, whereas sci-fate and scNT-seq which allow cost-effective sequencing of thousands of cells are based on 3′-end sequencing, with the added accuracy provided by unique molecular identifiers [G] (UMIs).
It should be noted that focusing on newly synthesized transcripts alone may not be enough to detect the slight underlying differences between cells over time. To improve trajectory reconstruction one should combine old and new transcripts, and determine their ratio13. A gene which showed high rates of C to T conversion during the labelling period may represent a gene which was recently turned on, but it could also be a gene with rapid turn-over. Measurement of the ratio between the abundance of new and old transcripts can identify those genes that underwent a change in their expression during the experimental time window. By highlighting the dynamic elements in the system, metabolic labelling methods efficiently increase the resolution of scRNA-seq based methods for trajectory reconstruction. Indeed, studies of both scNT-seq14 and scSLAM-seq11 show that s4U incorporation outperforms splicing-based RNA velocity (see below) in the ability to identify temporal directionality. This is most likely due to the metabolic labelling of newly synthesized transcripts being independent both from the number of introns in the gene and from the speed of the splicing process14.
A potential downside of metabolic labelling methods when compared to computational-only methods is that they were only demonstrated for cell cultures in vitro. It should be noted, however, that scRNA-seq on in vivo labelled RNA seems highly feasible. In SLAM-ITseq17 (a variation on SLAM-seq), RNA sequencing is performed on tissue extracted from mice engineered to express uracil phosphoribosyltransferase (UPRT) in a cell-type specific manner. This protozoan enzyme, which does not have an equivalent active form in mammalian cells, transforms 4-thiouracil into 4- thiouridine monophosphate, and enables the incorporation of the modified uridine into nascent RNA molecules specifically in UPRT+ cells. Although sequencing of these cells was performed in bulk, there is no reason to suspect this cannot work for scRNA-seq as well.
Cell-type specific reporters.
Another experimental approach that complements time-series trajectory inference from scRNA-seq data is the use of cell-type specific reporters or markers with a temporal expression pattern — which enables sequencing of only a subset of the cells18. Gehart et. al took this approach a step further, when they combined scRNA-seq with a fluorescent time-recording reporter to gain an additional layer of data that assists in the construction of time- rdered trajectories19. To study the dynamics of enteroendocrine cell development, they inserted a sequence that codes for two fluorescent proteins — red tdTomato and a destabilized form of mNeonGreen — immediately downstream of Neurog3, which is a transcription factor (TF) gene that is transiently expressed during early differentiation of enteroendocrine cells. Because the gene and both reporters are found on the same transcript, the Neurog3Chrono mice generated equimolar amounts of the endogenous gene and of the red and green fluorescent reporters. However, due to the faster decay of mNeoGreen relative to tdTomato, red:green fluorescence ratios measured at continuous intervals could serve as a standard-clock that measures the actual time elapsed since Neurog3 expression in each cell. Based on this information, the authors sorted single neurog3Chrono cells into multi-well plates, and combined the transcriptional profile of each cell with its fluorescence status (as established by fluorescence-activated cell sorting (FACS)) to obtain an actual-time (rather than pseudotime) developmental map. An interesting outcome was the realization that two cell populations that seemed at first to appear in parallel, had actually formed at consecutive intervals, indicating that one had arisen from the other, and not with it. In addition, whereas the length of branches on pseudotime plots cannot provide any information about the actual time needed for the described process to occur, scRNA-seq in combination with a time-recording reporter provided this type of information. Altogether, Chrono labelling generates real-time anchors to which the computational time-series analysis has to comply, thereby improving the accuracy of scRNA-seq-based temporal dynamic trajectories.
Genetic barcoding.
Genetic lineage tracking is a third experimental approach which aspires to set anchors that direct the calculated trajectory, but on a longer time scale than those established by the Chrono method discussed above. Conceptually, this approach extends from previous endeavours to deduce cell lineages by identifying genetic mutations shared among different cells20. With the advent of CRISPR–Cas9, numerous mutations can now be generated deliberately at specific loci targeted by guide RNAs (gRNAs). After their introduction, these mutations remain as ‘scars’ in the genome, and sequencing the ‘scar-recording’ locus in cells of different tissues can serve to construct lineage maps (such as in the genome editing of synthetic target arrays for lineage tracing (GESTALT) approach)21. Recently, several methods were developed for combining genomic scarring with scRNA-seq, thereby providing fate maps with higher resolution compared to that obtained through scar analysis alone; and with increased validity and accuracy relative to those obtained only through scRNA-seq based pseudotime trajectory reconstruction. ScarTrace22 uses a tandem repeat sequence of GFP introduced into the zebrafish genome, to record mutations caused by Cas9 and a gRNA injected into the early embryo. Cells are sorted into multi-well plates, and during the preparation of the expression library, the scar-recording region is amplified from the genomic DNA. Similarly, LINNAEUS23 records scars on RFP sequences in the fish, but reads them directly from the transcriptome library, thus allowing the use of droplet-based sample preparation. scGESTALT24 introduced an inducible form of Cas9 with a constitutively expressed gRNA, thus enabling ‘scarring’ the genome at later stages of fish development, and revealing complex lineage relationships within the brain.
Bowling et al. introduced Cas9-based scarring into mice by generating the CARLIN mouse25. In this mouse, 10 different constitutively expressed gRNAs are designed to target a constitutively expressed cassette upon the inducible expression of Cas9. This elaborate design provides a comprehensive system that allows combining ‘scarring’-based lineage tracing with scRNA-seq of any tissue in the mammalian body, at any stage. Using this approach, Bowling et al. were able to combine expression-based trajectories with clonal analysis to follow the dynamics of the haematopoietic system during embryonic development and regeneration. This line of analysis adds an additional layer to scRNA-seq-based developmental maps, as the clonal analysis is able to detect past events including cell population bottlenecks which are almost impossible to detect using methods that rely solely on transcriptional similarity between cells. Additionally, combining lineage tracing with scRNAseq can provide unique insights into areas other than embryonic development. For example, Quinn et al. used piggybac transposition to insert a lineage-tracing cassette into lung cancer cells, which allowed them to follow tumour development upon transplantation into mice26. In this framework, each cassette they used contained one of 10 stable barcodes introduced upstream of the cas9 edited region27. This strategy provided an additional layer to their lineage tracing: initial clones could be identified by the composition of their stable barcodes, whereas the scars generated in the editable region defined the sub lineages of each initial clone. By comparing the mutations registered on the main tumour to its metastases, they were able to outline the spatial distribution of the tumour mutations across cell generations.
Pseudotime and trajectory inference methods
After designing and performing a time-series scRNA-seq experiment (Box 1 and Figure 2), a key analysis challenge is linking the cells within and across time points. Several computational methods have been developed to address this challenge and these largely fall into three major categories. The first two (dimensionality [G] reduction and gene-space methods) use the expression of genes in cells to link them over time, whereas the third (RNA velocity) uses information about spliced and unspliced genes. Dimensionality reduction methods rely on a low-dimensional representation of the cells to infer a spanning tree or another graph representation on which cells are projected to reconstruct trajectories. Gene-space methods work in regular gene space without dimensionality reduction and utilize probabilistic models (often probabilistic graph representations) to infer discrete or continuous assignment for cells. RNA velocity does not rely on the levels of genes to connect cells but instead attempts to determine the next state for each cell based on the differences between unspliced and spliced transcripts for each gene.
Box 1. Experimental design considerations for time-series scRNA-seq data.
Several issues should be considered when designing time-series single-cell RNA sequencing (scRNA-seq) experiments. Some of these arise in all scRNA-seq studies (for example, which technology to use) whereas others either require special consideration for time-series studies (for example, the number of cells to profile at each time point) or are unique to time-series studies (sampling rates). The number of cells needed, and the reads per cell are major issues in the design of any single-cell experiments69. A common suggestion is to consider the expected frequency of the rarest cell type and determine the number of cells based on that74. A few methods have been proposed to recommend the number of cells for single-cell experiments, including single-cell empirical Bayes (sceb)69 and howmanycells (https://satijalab.org/howmanycells/). Note that the number can probably be reduced for time-series studies if one expects to see the same cell types at some or all of the time points, especially if the sampling rate is high enough to overcome noise at each point70. Sequencing depth (how many reads per cell) often varies among different single-cell sequencing platforms. A list of recommended sequencing depth for various single-cell sequencing platforms is available in REF75.
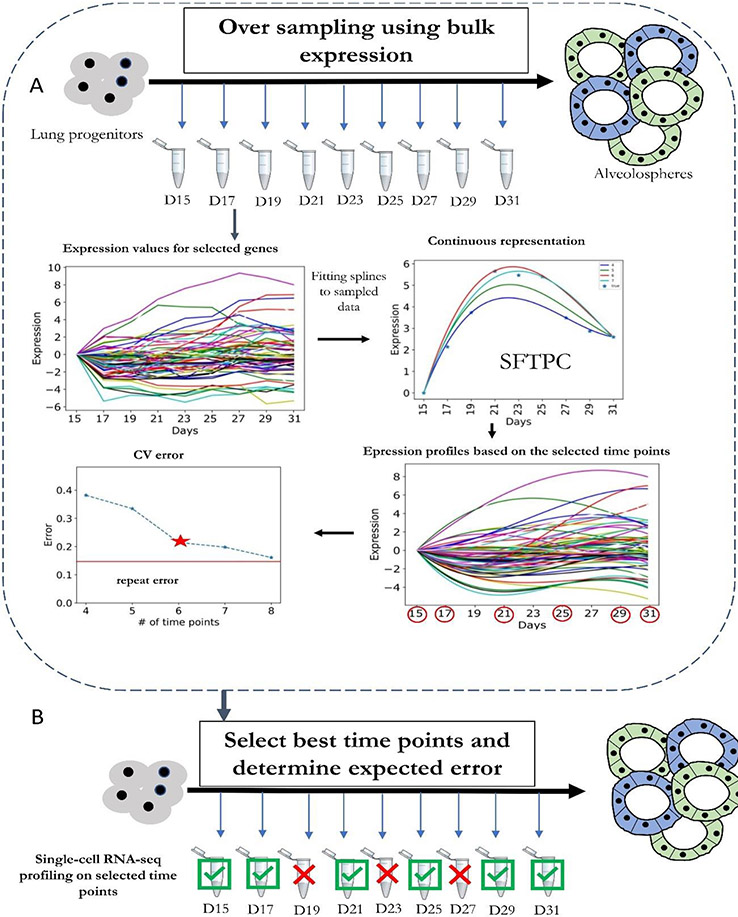
How to choose the best time points to profile is a unique challenge in time-series studies. Most current studies rely on knowledge of the biological process to determine both the duration of the study and the sampling rate. For example, if one expects a uniform process (such as for the cell cycle76) then the sampling should be uniform. In other cases — for example, various responses to stimuli — it may be better to sample more densely at the beginning and less frequently later8. However, there are cases where the dynamics of the process being studied are unknown or when assumptions are based on the phenotypic behaviour, which may not reflect the underlying molecular dynamics that are being profiled by scRNA-seq70. To address this, Kleyman et al. developed the Time Point Selection (TPS) method (Figure 2)70. TPS was originally developed and applied to bulk RNA-seq but can also be used for scRNA-seq. It works by initially oversampling bulk-level RNA-seq using cheap array methods. Next, spline curves are used to fit the profiled data, which enables the method to predict values for unobserved time points. A heuristic optimization function is then used to select the most informative time points, those points that if sampled provide enough information to reconstruct the entire expression trajectory for all genes. TPS can also provide a measure of the error expected from using only a subset of time points, allowing researchers to balance cost and accuracy. Application of TPS to a time-series scRNA-seq study of induced pluripotent stem cells (iPSCs) differentiating into lung cells resulted in a somewhat surprising selection of time points that focused more on the end of the process rather than the more traditional approach that mainly focuses on the beginning. Selected time points were validated using the complete dataset54.
Note that although TPS works well for some single-cell studies, it uses bulk data to select the optimal time points. This may lead to missing critical transition points for cell types that are not well represented in the sample (rare cell types). One potential way to address this issue is to place more emphasis on specific markers for rare cells (if they are known) when computing the reconstruction error for TPS.
Another challenge is batch effects. Most methods that have been developed for dealing with batch effects in scRNA-seq studies (for example, when analyzing data from multiple individuals or conditions) can also be applied to data from multiple time points. Single-cell multiplexing77 could help mitigate the impact of batch effects by pooling single-cell samples from different time points and sequencing them together. Housekeeping genes78 or spike-ins79 could also improve normalization between time points (or batches). At the bulk level, many tools have been developed to correct batch effects between different samples, batches or experiments, including ComBat80 and limma81. These methods have been extended to use single-cell data. A recent benchmarking study of single-cell batch effect correction methods82 provides useful comparison between several popular methods.
Figure 2. Selecting time points to sample in single-cell RNA-seq experiments.
a ∣ Oversampling of bulk RNA sequencing (RNA-seq) data. The bulk data are then used to determine the expected error for each potential subset of time points used. A heuristic search is then performed to select the optimal set of time points given cost or error constraints. b ∣ Selected time points are then used to profile single-cell RNA sequencing (scRNA-seq) data. Errors computed in (a) for this subset of time points can be used to bound the expected difference between reconstructed and underlying expression levels. CV, coefficient of variation; D, days; SFTPC, pulmonary surfactant-associated protein C (a marker of AT2 alveolar stem cells).
Dimensionality reduction based methods for trajectory inference.
These are likely to be the most popular and most widely used methods since they allow for both inference of the trajectories and representation of all cells in a visually appealing and interoperable manner. They are often composed of three key steps, although each can use a variety of different approaches. In the first key step, the high-dimensional single-cell dataset (e.g., scRNA-seq) is embedded in a lower dimension. Low-dimensional representation is a very popular technique in data analysis which is useful for both removing noise (by focusing on the most abundant and consistent signal) as well as for visualization purposes. A commonly used method is to first reduce the high-dimensional data into a mid-range number of dimensions (e.g., 50) using linear methods such as principal components analysis (PCA)28 and perform the trajectory inference using this representation. The data set is then further projected into two or three dimensions for visualization. Here, numerous dimensionality reduction methods including t-distributed stochastic neighbour embedding (t-SNE29), uniform manifold approximation and projection (UMAP)30 and neural-network-based auto-encoders [G] 31 have been used. In the second key step, a graph structure (usually a trajectory tree) is learned on the low-dimensional manifold to best connect all cells. Here the definition of ‘best’ differs between methods. For example, Monocle32 uses independent component analysis for dimensionality reduction and then infers the trajectory directly on the cells by constructing a minimal spanning tree (MST). Qiu et al. later developed Monocle 233, which improves Monocle by using cell centroids (milestones) to learn the tree. The method then iterates between moving the cells towards the nodes of the current tree graph and updating the tree structure until convergence. Monocle 2 maintains an invertible map between the high-dimensional and low-dimensional space, which simultaneously reduces the data dimensionality and learns the trajectory. A few other methods such as ‘tools for single-cell analysis’ (TSCAN)34 and Slingshot35 utilize a similar approach: constructing the MST on cell centroids, which represent cell types and states. Another commonly used method, partition-based graph abstraction (PAGA)36, reconstructs the trajectory using a different strategy: PAGA partitions, prunes, and connects the clusters based on a statistical connectivity among the clusters in a general graph. Monocole 337 improves on PAGA by learning a principal graph on each of the PAGA partitions, leading to a trajectory with a higher resolution. One of the most popular tools in this category is Seurat3, a comprehensive tool that contains various methods for each step in the trajectory inference (e.g., dimensionality reduction, data imputation, and clustering). Seurat also contains modules to integrate different single-cell datasets. In the third key step, a root cell (or a list of root cells) is determined to seed the trajectory (i.e., the starting point on the differentiation tree). A pseudotime is then assigned to cells based on their location. Different strategies have been developed to estimate the pseudotime. One commonly used strategy is using the distance (e.g., Euclidean or correlation) to the root cell(s) to represent the pseudotime38. Another group of popular pseudotime inference methods including single-cell lineage inference using cell expression similarity and entropy (SLICE)39 do not require the user to define root cells. Instead, these methods calculate the entropy for each cell and use it to represent the pseudotime. This works best in cases where the entropy is expected to drop as time passes, for example, during development40.
Several methods based on optimal transport have also been applied to infer trajectories form scRNA-seq data. These methods, which include Waddington-OT41, ImageAEOT42, and LineageOT43 do not make strong assumptions about specific placing of cells along the developmental trajectory but rather assume a distribution for possible placement of each cell along the trajectory. The method learns these distributions and can make inference about underlying molecular events that drive the observed trajectories.
Finally, learned trajectories from different methods described above can be evaluated using automatic scoring functions. For example, dynverse44 provides a set of guidelines to help users select the best result for their dataset. The single-cell data and reconstructed trajectories can be visualized by several developed tools such as STREAM45, Cellxgene46, and the commercial package BioTuring (https://bioturing.com/).
Pseudotime inference in gene space.
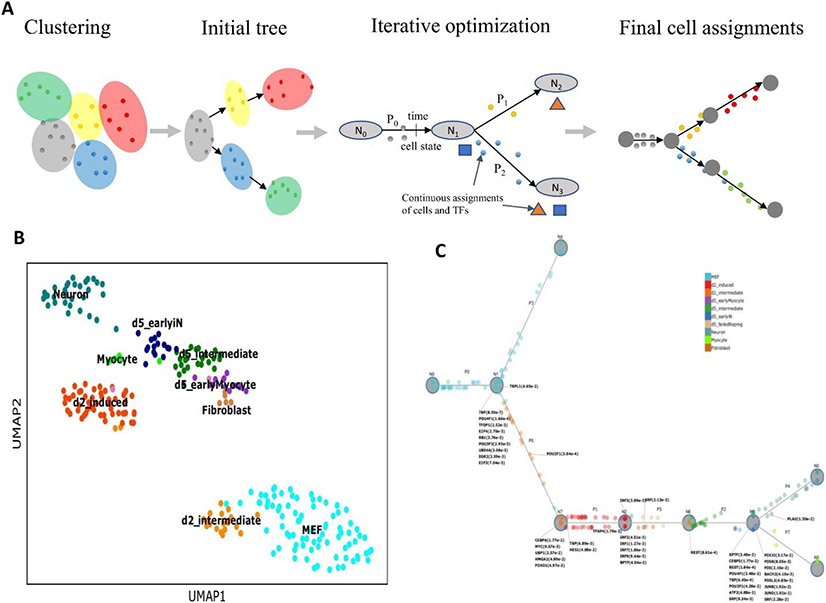
A potential downside of dimensionality reduction is that the trajectory is inferred based on a subset of the most abundant differentially expressed genes. This could make it hard to distinguish and accurately reconstruct clusters and trajectories for cell states that are represented by fewer cells, especially for time-series studies where the last time point may include several different cell types. In addition, dimensionality reduction limits the ability to rely on additional information as part of the trajectory inference as we discuss below. To overcome this problem, several methods attempt to infer pseudotime and trajectories based on the gene space itself. These methods usually adopt probabilistic models and attempt to construct a graph that represents both the expression levels of genes within each cell and relationships between cells. In addition to representing branching information, the graph provides an emission model which captures the expected expression of all genes in each state. Most probabilistic trajectory-inference methods based on graphical models [G] iterate between learning/updating the graph structure and the emission parameters and assigning cells to locations on the graph, until convergence. Early methods used a discrete trajectory graph, in which cells are assigned to a small number of discrete nodes. Examples methods include temporal assignment of single cells (TASIC)47, scdiff38 and single-cell clustering using bifurcation analysis (SCUBA)48. More recent models extended this approach to continuous trajectory. For example, continuous-state hidden Markov models (CSHMMs)49 can be used to assign cells to any positions of the trajectory graph (Figure 3). CSHMMs start by clustering all the cells in the full gene space. An initial tree-structured trajectory is learned by connecting all clusters based on their distances to the root cells. The parameters to define any states in the trajectory are also estimated from the initial trajectory. All cells are then re-assigned to the state that represents the largest assignment probability. The method iterates between updating the trajectory graph and re-assigning all the cells until a stopping criterion is met. Unlike standard hidden Markov models (HMMs), which are defined using a discrete set of states, CSHMMs can have infinitely many states, which allows for continuous assignment of cells along developmental trajectories.
Figure 3. Dynamic regulatory network inference using CSHMM.
a ∣ A scheme for continuous-state hidden Markov model (CSHMM) and cell assignment learning. The method is initialized using clustering in gene space. Relationships between clusters are analyzed to obtain an initial branching model. Next the method iterates between cell assignment along the branches of the branching model and learning model parameters including structure and emission probabilities. Cell assignment is also determined based on predicted transcription factors (TFs) for each branching point and their targets allowing the method to infer key TFs and their activation time. b ∣ A Standard uniform manifold approximation and projection (UMAP) plot of cells profiled to study neuron differentiation73. c ∣ CSHMM reconstructed trajectory for the same cells. Cells are assigned to different locations along the branches based on their inferred pseudotime. The model also includes parameters for the expected expression levels for all genes at each time. Key TFs and their p-values are associated with each of the branching points in the model.
RNA velocity for trajectory inference.
All strategies discussed so far rely only on the expression of the exons of the genes in the cells profiled. This is based on the assumption that expression levels change gradually and that enough cells were sampled from all intermediate states to enable the reconstruction of continuous trajectories. Although this works well for highly sampled data sets, it may not be enough for data sets that do not contain cells from all such states. An alternative is to use RNA velocity9, which captures transcriptional dynamics within a cell rather than directly between two cells. RNA velocity is based on the relationship between spliced and unspliced transcripts in the same cell. To quantity the RNA velocity, expression level derivatives are determined by the balance between the abundance of the spliced mRNAs and unspliced mRNAs, as well as mRNA degradation. The steady states are reached asymptotically when the abundance of spliced and unspliced molecules is constrained to a fixed-slope relationship. This equilibrium slope combines degradation and splicing rates to capture gene-specific regulatory properties. Specifically, during gene upregulation, transcripts are skewed towards unspliced transcripts, whereas during gene downregulation the skew is towards spliced transcripts. Hence this ratio can provide insight into how transcript levels are changing over time and thus where cells might be ‘headed’ in the future state50. The RNA velocity method assumes that the transcriptional phases of gene expression induction and repression last sufficiently long to reach either an actively transcribing or inactive silenced steady-state equilibrium. This assumption limits application to transient cell states, in which the steady states are often not reached as induction might terminate before mRNA-level saturation. To overcome this problem and enable the use of the RNA velocity framework for transient cell states, Bergen et al.51 developed scVelo, a likelihood-based dynamical model, which infers gene-specific reaction rates of transcription, splicing, degradation, and an underlying latent time in an expectation-maximization [G] (EM) framework. This inferred latent time describes the cell position in the underlying biological process.
Several methods have recently combined RNA velocity with expression similarity for trajectory inference. These methods benefit from the ability of RNA velocity to infer direction and the ability of traditional pseudotime inference methods to cluster cells based on expression similarity. For example, CellRank52 uses the RNA velocity information to infer the ordering of different groups of cells and uses expression similarities within groups to further refine their orderings. This feature enables CellRank to uncover putative lineage drivers and visualize lineage-specific gene expression trends. CellPath53 is another trajectory inference method that utilizes RNA velocity information to identify root cells that are assigned to the first path in the reconstructed model.
Data integration for modelling dynamics
Reconstructed temporal trajectories from scRNA-seq data can be used to address several different questions. However, they do not provide information on other molecular aspects of the process including changes to the epigenome and the set of regulators that are activated at specific time points. In addition, given their strong dependence on the assumption of gradual change in the expression of genes within or between time points they may not be appropriate for studies that need to sample at lower frequencies. To overcome these problems several computational methods have been developed to integrate time-series scRNA-seq with other bulk or single-cell data.
Integrating time-series single-cell expression with genetic information.
Computational methods have been developed to integrate time-series and snapshot scRNA-seq data with the genetic barcoding and CRISPR–Cas9 data. Such integration can be used to improve the trajectories reconstructed from each method separately. Hurley et al. projected the expression of barcoded single cells on a dynamic model for cell differentiation, enabling them to validate predictions about the timing of cell fate commitment for induced pluripotent stem cells (iPSCs) differentiating to alveolar epithelial type 2 cells (AEC2s)54. They then identified genes whose expression differs, at the time of commitment, between lung epithelium and non-lung endoderm cells. Many of these genes were associated with the WNT pathway, and by withdrawing the GSK3 inhibitor CHIR they were able to significantly increase the percentage of lung AEC2 cells. Barcoding also showed that the resulting cells exhibited a stable phenotype and nearly limitless self-renewal capacity.
Weinreb et al55 used lentiviral constructs to express random barcodes in haematopoietic progenitors, and sequenced both barcodes and mRNA at the single-cell level shortly after and at later time points. By consecutively sampling cells from the same culture plate, they could examine how each clone expanded and differentiated over time, providing a partial ‘ground truth’ for the pseudotime path. As expected, their findings confirmed the ability of scRNA-seq-based lineage maps to describe the transcriptional changes a cell undergoes during differentiation. However, they also concluded that scRNA-seq was insufficient to capture the point at which a cell’s fate had been determined. Specifically, when clones were split into separate wells, their separated daughters mostly acquired the same fate, even though scRNA-seq could not identify the original clone’s poised status. This finding presents a notable challenge to the field, as it suggests that in spite of the immense power provided by scRNA-seq analysis, the major question time-series analysis is expected to answer — how cells decide between two possible fates — lies in processes that may not be observed at the transcriptional level in some cases; such processes might include chromatin structure or protein abundance and regulation.
Other methods were designed to combine scRNA-seq with lineage tracing data from CRISPR–Cas9 mutations. scRNA-seq data can be used to help overcome the problem of scar saturation which can lead to inability to infer the exact set of sequential edits that led to the observed set of mutations56. Zafar et al57 developed LinTIMaT, a general method for combining scRNA-seq data with scar data. LinTIMaT reconstructs cell lineages using a maximum-likelihood framework which combines mutation and expression agreement along the branches. When applied to zebrafish scar data, the method was able to resolve the ambiguities arising when only using the scar data and it identified additional cell subtypes that could not be resolved without using the expression data. Furthermore, LinTIMaT also enables the integration of different individual lineages for the reconstruction of an invariant lineage tree leading to better cell type coherence and new insights on progenitors and differentiation pathways.
Integrating multiple types of time-series data with single-cell data.
In addition to using genetic information, several studies also integrated other types of time-series molecular data. For example, the interactive dynamic regulatory events miner (iDREM)58 was used to project single-cell lung developmental data on a detailed human lung development model constructed using bulk expression data to infer the cell types activated at each stage59. Similarly, PhenoPath, a statistical analysis method that incorporates the impact of environmental and genetic covariates, was used to analyze time-series bulk and single-cell transcriptomics data for inferring pseudotime trajectories60. Other methods integrate time-series assay for transposase-accessible chromatin sequencing (ATAC-seq) data with time-series scRNA-seq data. For example, TimeReg61 was recently applied to combine gene expression and chromatin accessibility at the single-cell level. The method first infers context-specific regulatory interactions from ATAC-seq and RNA-seq data at a single time point and then uses dimensionality reduction to extract core regulatory interactions across the time points. These interactions are then used to identify regulators that drive the changes in expression observed. TimeReg was applied to study retinoic acid (RA)-induced development and was able to identify several novel regulatory elements for cerebellar development, synapse assembly, and hindbrain morphogenesis.
Integrating time-series single-cell data with interaction data.
Although the integration of complementary time-series datasets can lead to much better models for regulatory networks, these approaches require additional experiments and are not always feasible. By contrast, general protein–DNA and protein–protein interaction data can be integrated with any single-cell data leading to the improved reconstruction of the networks and pathways activated in the study. Although interactions are obviously changing over time, almost all large interaction datasets are measured as static, single-timepoint data (or inferred from DNA motif information which is obviously not changing over the time the experiments are conducted). Several methods have been developed to incorporate such interaction information and these can be largely divided into two major categories. The first uses these interactions for post-processing of trajectories learned by other methods, whereas the second uses them as an integral part of the trajectory inference.
An example for the use of post-processing methods is the work by Sanchez-Castillo et al62 which presented a Bayesian method utilizing autoregressive moving-average model (GRNVBEM) to infer gene regulatory networks from pseudotime ordered cells. Similarly, Hamey et al.63 used diffusion maps to order blood stem and progenitor cells. They next applied Boolean network learning to the ordered cells to infer transcriptional regulatory network models that recapitulated differentiation of haematopoietic stem cells (HSCs) into progenitor cell types. This enabled them to identify and experimentally validate the regulation of Nfe2 and Cbfa2t3h by the GATA2 TF. Although Hamey et al63. used a specific ordering method (diffusion maps), their Boolean network analysis can also be applied to orderings obtained by other algorithms, which allows users to flexibly match ordering and network inference methods. For further information we refer readers to a review of bulk network inference methods64 that can be applied to ordered single cell data, and a comprehensive survey of gene regulatory network inference methods for single-cell data65. Overall, the advantage of post-processing methods is that the trajectory inference and interaction evaluation are decoupled. Thus, most post-processing network inference methods can use ordering from several different trajectory inference methods and similarly, most methods for trajectory inference can be matched to network analysis methods, thus providing much better flexibility for users. As mentioned above, several packages, including dynverse44 can transform the output of several popular trajectory inference and pseudotime ordering methods to a common output. Using this output allows several different methods to be used for inferring regulatory networks.
The second set of methods uses the interaction information when reconstructing the trajectories. This may improve the ability to focus on the most important genes and regulators when learning the model and, if the models are generative, can help in the prediction of the impact of various perturbations. For example, SimiC jointly infers the gene regulatory networks (GRNs) and the cell states enabling the grouping of cells to clusters representing unique stages66. The GRN inference problem is modelled as a LASSO optimization problem and the model is constrained by requiring that cells be assigned to contiguous states, which uses the interaction information to enforce smooth transitions between states. Other methods utilize a probabilistic model for the integration of interaction and expression data. Ding et al. relied on graphical models (extending HMMs) to integrate static protein–DNA interaction information with time-series scRNA-seq data38. The algorithm iteratively identifies key TFs and uses their known targets to assign cells and TFs to specific branches on the trajectory. This provides both a pseudotime model for profiled cells and, for each branching point, the set of TFs predicted to regulate the split in cell trajectories observed. This method was used to model lung development in mice and identified hypoxia-inducible factor (HIF1A), CREB1, and HMGA2 as key TFs controlling the differentiation of type 1 to type 2 epithelial lung cells38. Knockout and overexpression experiments at the time predicted by the model indicated that perturbing the expression of these TFs indeed impacts cell fate decisions and can lead to decreased sacculation and a lower fraction of type 2 cells. This method was also used to model iPSC differentiation into cardiomyocytes and identified the dysregulation of HOPX during differentiation as a mechanism underlying the failure of in vitro-derived cardiomyocyte maturation67. Figure 3c presents an extension of these methods, termed Continuous State HMM-TF (CSHMM-TF), which enables better identification of the TF activation time and the interactions between TFs68.
Conclusions and perspectives
As biological processes are dynamic, many studies include the profiling of samples over several time points. scRNA-seq has now become the method of choice when studying the expression levels of genes in such samples. While this technology offers several advantages, it is also raising new challenges. Many of these challenges can be addressed by combining experimental and computational methods to design (Box 1), process, analyze, visualize and integrate time-series scRNA-seq data.
Experimental methods have largely focused on complementary approaches to infer the timing of specific events beyond the actual sampling time. These include metabolic labelling and genetic barcoding methods that provide direct information on the time of specific transcription events and relationships between cells. This can provide valuable information for both inferring the exact ordering of different cellular states and accurately linking cells and their states over time. Although the experimental approaches can help in some studies, they may not be enough to correctly order cells for other studies (for example, cases when no cell division occurs for genetic barcoding methods). Several computational pseudotime inference methods have been developed to address these issues. Such methods differ in the assumptions they make about the importance of specific genes in subsets of cells or all of the cells, their ability to infer complicated trajectories and their ability to overcome issues related to sampling. When the expected trajectory is largely unknown, and sampling is believed to be adequate, it may be best to use the dimensionality reduction based methods. When there is reason to believe that small populations of cells may be important, especially in later stages, it may be best to use the graphical model approaches. RNA velocity is often the method of choice when trying to infer dynamics from snapshot (single time-point) samples since it provides information on future events not yet seen in the expression levels themselves.
Integrating time-series scRNA-seq data with other types of omics and interaction data is a very active area of research. While most approaches perform such analysis as a post-processing step, integration as part of the trajectory inference provides a number of advantages. First, it may lead to better trajectories and models. Second, it provides information on the exact timing of various regulatory events. Other approaches integrate data collected from the same cells using different experimental platforms, most notably single-cell ATAC-seq. The joint analysis is beneficial for both types of data in that case, helping identify cell types for the ATAC data and infer active regulators for the RNA data. Going forward, as we discuss in Box 2, we expect to see many studies integrating time-series and spatial data which would open the door to inferring not just the dynamics of regulatory networks but also signaling networks that are often upstream of these and temporal impacts of cell–cell interactions. Together, these will greatly improve our ability to reconstruct cell- and tissue-based models of biological processes.
Box 2. Spatio-temporal analysis of single-cell data.
Several methods have been recently developed to profile genes and proteins spatially. While most of these extend fluorescence in situ hybridization (FISH) methods some also utilize sequencing technologies recording the location of where the sequences are captured83. Some of these methods enable the quantification of expression levels for several genes at single-cell resolution (for example MERFISH84,85, seqFISH86, seqFISH+87, osmFISH88, and the 3D transcriptomics record STARmap89), whereas others provide a more transcriptome-wide survey but at lower spatial resolution that groups expression from several cells for each profile90,91. Spatial transcriptomics techniques have now been applied to study the organization of cells in several different organs and tissues including lung92 and kidney93.
An interesting direction that is still in its infancy is to perform such spatial studies over time. This would enable the determination of both intracellular networks (for example regulatory networks) and intercellular networks (for example intercellular signalling networks) over time. As the two network types are highly dependent, data that enable joint modelling provide valuable information on the drivers of specific regulatory events and on the impact of gene expression on cell–cell interactions94. The use of time-series data for such studies would provide much better information on the causal relationships between different molecular events, which are critical for development95, various responses to stimuli96,97, and several other biological processes98.
To date, most work on spatio-temporal analysis involved the use of single-cell RNA sequencing (scRNA-seq) or other expression information obtained over time from different locations99-101. While this provides important information about the cells and trajectories in different regions, such data are not sufficient to provide information on the cell–cell interactions. More recently, a few studies profiled spatial transcriptomics data over time102. SlideSeq, a high-resolution spatial transcriptomics method, was applied to study the brain’s response to traumatic injury over time, identifying genes with unique spatial expression patterns at different time points91. However, to date, the focus of such studies has mainly been on the dynamics of expression changes rather than on reconstructing underlying signalling and regulatory networks. Still, the combination of spatial and time-series studies at the single-cell level promises to be the next leading technology for studying biological systems at the molecular level. Such data are likely to require the development of novel computational methods that can infer relationships across space and time and connect levels of genes within cells to their impact on internal and external cell states.
Table 1.
A summary table for all discussed methods
| Method name | Category | Input | Output | Suitability / assumptions | Implementation / access |
Software link | Ref. |
|---|---|---|---|---|---|---|---|
| sceb | ED | 1) Budget 2) Pilot data |
1) Number of cells to profile 2) Sequencing depth |
Determining number of cells and read coverage for any single-cell experiment, including time-series and snapshot studies | Python / Open | https://github.com/martinjzhang/single_cell_eb | 69 |
| howmanycells | ED | 1) Expected number of cell types 2) Minimum fraction of rarest cell type 3) Minimum number of expected cells per type |
1) Number of cells to profile | Determining the number of cells to profile for studies in which rare cell types are either of interest or expected to be important. | HTML JavaScript / Open |
https://satijalab.org/howmanycells/ | NA |
| TPS | ED | 1) Pilot data 2) A level of reconstruction error |
1) The best time points to profile | Cases in which the sampling rate is expected to be able to recover all major molecular events occurring in the process being studied. | Python / Open | http://sb.cs.cmu.edu/TPS/ | 70 |
| Monocle | TI | Cells by genes matrix | 1) Clusters 2) Trajectory graph 3) Pseudotime for all cells |
Pseudotime inference for time-series or single-time point (unsynchronized) studies in which the profiled cells are expected to span the entire duration of the process. Clustering can be used for cases in which it is not clear if the sampling rate covers all major biological transitions. Time-series information is not utilized by the method, and so inference is based on the expression levels only. | R / Open | https://cole-trapnell-lab.github.io/monocle3/ | 37 |
| TSCAN | TI | Cells by genes matrix | 1) Clusters 2) Trajectory graph 3) Pseudotime for all cells |
Similar to Monocle in terms of assumptions and suitability. | R / Open | https://github.com/zji90/TSCAN | 34 |
| Slingshot | TI | Cells by genes matrix | 1) Clusters 2) Trajectory graph 3) Pseudotime for all cells |
Similar to Monocle in terms of assumptions and suitability. | R / Open | https://github.com/kstreet13/slingshot | 35 |
| SLICE | TI | Cells by genes matrix | 1) Clusters 2) Trajectory graph 3) Pseudotime for all cells |
Relies on entropy-based analysis and so is most suitable for developmental studies. It can infer the starting set of cells on its own without using the time information. | R / Open | https://github.com/xu-lab/SLICE | 39 |
| PAGA | TI | Cells by genes matrix | 1) Clusters 2) Trajectory graph 3) Pseudotime for all cells |
Suitable for learning complex trajectory structure with multiple branching. Efficient and often fairly fast. | Python / Open | https://github.com/theislab/paga | 36 |
| Seurat | TI, IM | Cells by genes matrix | 1) Clusters 2) Trajectory graph 3) Pseudotime for all cells |
An inclusive suite of tools for the analysis of scRNA-seq data. It provides implementation of several methods. It is very efficient and fast but may be less accurate for complex trajectories. | R / Open | https://satijalab.org/seurat/ | 53,71 |
| SCUBA | TI | Cells by genes matrix | 1) Clusters 2) Trajectory graph 3) Pseudotime for all cells |
Does not infer continuous trajectories, only states and their relationships. It is mainly successful for cases where the trajectory is linear or includes few branches. | MATLAB / Open | https://github.com/gcyuan/SCUBA | 48 |
| scdiff | TI, GRN IM |
Cells by genes matrix | 1) Clusters 2) Trajectory graph 3) Pseudotime for all cells 4) Regulatory networks |
Does not infer continuous trajectories, only states and their relationships. It is suitable for learning complex branching models. It works in gene space and so is suitable for cases in which several cell types are expected. | Python, JavaScript /Open | https://github.com/phoenixding/scdiff | 38 |
| CSHMM | TI, GRN, IM | Cells by genes matrix | 1) Clusters 2) Trajectory graph 3) Pseudotime for all cells 4) Regulatory networks |
Infers continuous trajectories in gene space and so can generate complex branching models for cases in which several cell types are expected. It is slower than most methods that work on reduced dimension space. | Python / Open | https://github.com/jessica1338/CSHMM-for-time-series-scRNA-Seq | 49 |
| RNA velocity and scVelo | TI | scRNA-seq data (all reads) | RNA velocity vectors for all cells | These methods do not rely on expression similarity and so may be better suited for data sampled at intervals that do not fully capture all possible molecular events. As the methods are based on identifying splicing status, they may be problematic for any scRNA-seq datasets where the reads do not sufficiently cover both intronic and exonic regions. They are often a valuable complement to pseudotime inference methods. | Python / Open | https://github.com/theislab/scvelo | 9,51,72 |
| LinTIMaT | TI, IM | 1) Cells by barcodes matrix 2) Cells by genes matrix |
Trajectories and barcodes that mark different cell fates | Assumes the existence of CRISPR-based mutation information for cells. Learns linage models by combining these with scRNA-seq expression data. | Python / Open | https://jessica1338.github.io/LinTIMaT/ | 57 |
| PhenoPath | TI, IM | 1) Cells by genes matrix 2) Vectors of covariates |
1) Clusters 2) Trajectory graph 3) pseudotime for all cells |
Suitable for cases in which data from multiple individuals, perhaps at different diseases or development stages, are integrated. Does not assume or require a clear ordering of the samples. | R / Open | https://bioconductor.org/packages/release/bioc/html/phenopath.html | 60 |
| TimeReg | GRN, IM | 1) Expression matrix for RNA-seq data 2) Bam files for epigenetics data |
1) Regulatory network (TFs and target genes) | Requires information about TF–gene interactions. | R / Open | https://github.com/SUwonglab/TimeReg | 61 |
| GRNVBEM | GRN, IM | 1) Cells by genes matrix | 1) Regulatory network | Can either use time series or pseudotime-inferred ordering to reconstruct the GRN. | MATLAB / Open | https://github.com/mscastillo/GRNVBEM | 62 |
CSHMM, continuous-state hidden Markov model; ED, experimental design; GRN, gene regulatory network; GRNVBEM, a gene regulatory network (GRN) inference method using a variational Bayesian expectation-maximization (VBEM) framework; IM, integrative model; LinTIMaT, lineage tracing by integrating mutation and transcriptomic data; PAGA, partition-based graph abstraction; RGN, regulatory network inference; RNA-seq, RNA sequencing; sceb, single-cell empirical Bayes; scRNA-seq, single-cell RNA sequencing; SCUBA, single-cell clustering using bifurcation analysis; SLICE, single-cell lineage inference using cell expression similarity and entropy; TF, transcription factor; TI, trajectory inference; TPS, time point selection; TSCAN, tools for single-cell analysis.
Acknowledgements
Work partially supported by NIH grants 1R01GM122096, OT2OD026682, 1U54AG075931 and 1U24CA268108 to ZB-J.
Glossary:
- Pseudotime
Partial ordering of cells in single-cell RNA sequencing (scRNA-seq) data that represents predecessor and descendent cell state information.
- Trajectory
A graph (often tree) structure which represents the states and their order during the biological process being studied. Cells are assigned to points on this graph.
- Unique molecular identifiers
UMIs. Sequence indices (often randomly generated) which are added to sequencing libraries before PCR amplification and enable the identification of PCR duplicates.
- Dimensionality
In single-cell analyses, dimensionality typically refers to the high versus low number of dimensions of the data. When working with large samples where each is composed of tens of thousands of features (for example cells and their gene expression levels) the high dimension corresponds to the original values whereas low dimension is a compact, though lossy, way to represent the data with many fewer values. Several low-dimension representation methods have been developed and they differ in the function they attempt to optimize (such as minimizing reconstruction loss, or minimizing differences in distance between the high- and low-dimensional spaces).
- Auto-encoders
Neural networks whose goal is to reconstruct the input values. These networks are used for dimensionality reduction since they compress all input values through a small intermediate layer and then reconstruct them from the information in that layer.
- Graphical models
Computational methods that are used to represent joint probability distributions in a compact manner. These include Bayesian networks, hidden Markov models and more.
- Expectation maximization
(EM). A widely used computational method that can be used to fill in missing data while simultaneously learning model parameters. The method iterates between the expectation (E) step which determines expected values for missing data and the maximization (M) step which infers parameters using the values obtained by the E step.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Gasch AP et al. Single-cell RNA sequencing reveals intrinsic and extrinsic regulatory heterogeneity in yeast responding to stress. PLoS biology 15, e2004050 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zou Z. et al. A single-cell transcriptomic atlas of human skin aging. Developmental cell 56, 383–397. e388 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Stuart T & Satija R Integrative single-cell analysis. Nature Reviews Genetics 20, 257–272 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Alavi A, Ruffalo M, Parvangada A, Huang Z & Bar-Joseph Z A web server for comparative analysis of single-cell RNA-seq data. Nature communications 9, 1–11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dumitrascu B, Villar S, Mixon DG & Engelhardt BE Optimal marker gene selection for cell type discrimination in single cell analyses. Nature communications 12, 1–8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Efremova M, Vento-Tormo M, Teichmann SA & Vento-Tormo R CellPhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes. Nature protocols 15, 1484–1506 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Bendall SC et al. Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell 157, 714–725 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bar-Joseph Z, Gitter A & Simon I Studying and modelling dynamic biological processes using time-series gene expression data. Nature Reviews Genetics 13, 552–564 (2012). [DOI] [PubMed] [Google Scholar]
- 9.La Manno G. et al. RNA velocity of single cells. Nature 560, 494–498 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herzog VA et al. Thiol-linked alkylation of RNA to assess expression dynamics. Nature methods 14, 1198–1204 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erhard F. et al. scSLAM-seq reveals core features of transcription dynamics in single cells. Nature 571, 419–423 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Hendriks G-J et al. NASC-seq monitors RNA synthesis in single cells. Nature communications 10, 1–9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao J, Zhou W, Steemers F, Trapnell C & Shendure J Sci-fate characterizes the dynamics of gene expression in single cells. Nature biotechnology 38, 980–988 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiu Q et al. Massively parallel and time-resolved RNA sequencing in single cells with scNT-seq. Nature methods 17, 991–1001 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schofield JA, Duffy EE, Kiefer L, Sullivan MC & Simon MD TimeLapse-seq: adding a temporal dimension to RNA sequencing through nucleoside recoding. Nature methods 15, 221 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiefer L, Schofield JA & Simon MD Expanding the nucleoside recoding toolkit: revealing RNA population dynamics with 6-thioguanosine. Journal of the American Chemical Society 140, 14567–14570 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsushima W. et al. SLAM-ITseq: sequencing cell type-specific transcriptomes without cell sorting. Development 145 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Byrnes LE et al. Lineage dynamics of murine pancreatic development at single-cell resolution. Nature communications 9, 1–17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gehart H. et al. Identification of enteroendocrine regulators by real-time single-cell differentiation mapping. Cell 176, 1158–1173. el 116 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Reizel Y. et al. Colon stem cell and crypt dynamics exposed by cell lineage reconstruction. PLoS Genet 7, e1002192, doi: 10.1371/journal.pgen.1002192 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKenna A. et al. Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science 353 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alemany A, Florescu M, Baron CS, Peterson-Maduro J & Van Oudenaarden A Whole-organism clone tracing using single-cell sequencing. Nature 556, 108–112 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Spanjaard B. et al. Simultaneous lineage tracing and cell-type identification using CRISPR-Cas9-induced genetic scars. Nature biotechnology 36, 469–473 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raj B. et al. Simultaneous single-cell profiling of lineages and cell types in the vertebrate brain. Nature biotechnology 36, 442–450 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bowling S. et al. An engineered CRISPR-Cas9 mouse line for simultaneous readout of lineage histories and gene expression profiles in single cells. Cell 181, 1410–1422. e1427 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quinn JJ et al. Single-cell lineages reveal the rates, routes, and drivers of metastasis in cancer xenografts. Science 371 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan MM et al. Molecular recording of mammalian embryogenesis. Nature 570, 77–82 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pearson K. LIII. On lines and planes of closest fit to systems of points in space. The London, Edinburgh, and Dublin Philosophical Magazine and Journal of Science 2, 559–572 (1901). [Google Scholar]
- 29.Maaten L. v. d. & Hinton G Visualizing data using t-SNE. Journal of machine learning research 9, 2579–2605 (2008). [Google Scholar]
- 30.Becht E. et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nature biotechnology 37, 38–44 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Lin C, Jain S, Kim H & Bar-Joseph Z Using neural networks for reducing the dimensions of single-cell RNA-Seq data. Nucleic acids research 45, e156–e156 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trapnell C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nature biotechnology 32, 381 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qiu X. et al. Reversed graph embedding resolves complex single-cell trajectories. Nature methods 14, 979 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ji Z & Ji H TSCAN: Pseudo-time reconstruction and evaluation in single-cell RNA-seq analysis. Nucleic acids research 44, e117–e117 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Street K. et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC genomics 19, 1–16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolf FA et al. PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome biology 20, 1–9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao J. et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ding J. et al. Reconstructing differentiation networks and their regulation from time series single-cell expression data. Genome research 28, 383–395 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo M, Bao EL, Wagner M, Whitsett JA & Xu Y SLICE: determining cell differentiation and lineage based on single cell entropy. Nucleic acids research 45, e54–e54 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Halbritter F. et al. Epigenomics and single-cell sequencing define a developmental hierarchy in Langerhans cell histiocytosis. Cancer discovery 9, 1406–1421 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schiebinger G. et al. Optimal-transport analysis of single-cell gene expression identifies developmental trajectories in reprogramming. Cell 176, 928–943. e922 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang KD et al. Predicting cell lineages using autoencoders and optimal transport. PLoS computational biology 16, e1007828 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Forrow A & Schiebinger G LineageOT is a unified framework for lineage tracing and trajectory inference. Nature Communications 12, 1–10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saelens W, Cannoodt R, Todorov H & Saeys Y A comparison of single-cell trajectory inference methods. Nature biotechnology 37, 547–554 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Chen H. et al. Single-cell trajectories reconstruction, exploration and mapping of omics data with STREAM. Nature communications 10, 1–14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Megill C. et al. cellxgene: a performant, scalable exploration platform for high dimensional sparse matrices. bioRxiv (2021). [Google Scholar]
- 47.Rashid S, Kotton DN & Bar-Joseph Z TASIC: determining branching models from time series single cell data. Bioinformatics 33, 2504–2512 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Marco E. et al. Bifurcation analysis of single-cell gene expression data reveals epigenetic landscape. Proceedings of the National Academy of Sciences 111, E5643–E5650 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin C & Bar-Joseph Z Continuous-state HMMs for modeling time-series single-cell RNA-Seq data. Bioinformatics 35, 4707–4715 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Consortium TM Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367–372 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bergen V, Lange M, Peidli S, Wolf FA & Theis FJ Generalizing RNA velocity to transient cell states through dynamical modeling. Nature biotechnology 38, 1408–1414 (2020). [DOI] [PubMed] [Google Scholar]
- 52.Lange M. et al. CellRank for directed single-cell fate mapping. BioRxiv (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stuart T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902. e1821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hurley K. et al. Reconstructed Single-Cell Fate Trajectories Define Lineage Plasticity Windows during Differentiation of Human PSC-Derived Distal Lung Progenitors. Cell Stem Cell (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weinreb C, Rodriguez-Fraticelli A, Camargo FD & Klein AM Lineage tracing on transcriptional landscapes links state to fate during differentiation. Science 367, doi: 10.1126/science.aaw3381 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hwang B. et al. Lineage tracing using a Cas9-deaminase barcoding system targeting endogenous L1 elements. Nature communications 10, 1–9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zafar H, Lin C & Bar-Joseph Z Single-cell lineage tracing by integrating CRISPR-Cas9 mutations with transcriptomic data. Nature communications 11, 1–14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ding J, Hagood JS, Ambalavanan N, Kaminski N & Bar-Joseph Z iDREM: Interactive visualization of dynamic regulatory networks. PLoS computational biology 14, e1006019 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ding J. et al. Integrating multiomics longitudinal data to reconstruct networks underlying lung development. American Journal of Physiology-Lung Cellular and Molecular Physiology 317, L556–L568 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Campbell KR & Yau C Uncovering pseudotemporal trajectories with covariates from single cell and bulk expression data. Nature communications 9, 1–12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Duren Z, Chen X, Xin J, Wang Y & Wong WH Time course regulatory analysis based on paired expression and chromatin accessibility data. Genome research 30, 622–634 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sanchez-Castillo M, Blanco D, Tienda-Luna IM, Carrion M & Huang Y A Bayesian framework for the inference of gene regulatory networks from time and pseudo-time series data. Bioinformatics 34, 964–970 (2018). [DOI] [PubMed] [Google Scholar]
- 63.Hamey FK et al. Reconstructing blood stem cell regulatory network models from single-cell molecular profiles. Proceedings of the National Academy of Sciences 114, 5822–5829 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Todorov H, Cannoodt R, Saelens W & Saeys Y in Gene regulatory networks 235–249 (Springer, 2019). [Google Scholar]
- 65.Nguyen H, Tran D, Tran B, Pehlivan B & Nguyen T A comprehensive survey of regulatory network inference methods using single cell RNA sequencing data. Briefings in bioinformatics 22, bbaa190 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peng J. et al. SimiC: A Single Cell Gene Regulatory Network Inference method with Similarity Constraints. BioRxiv (2020). [Google Scholar]
- 67.Friedman CE et al. Single-cell transcriptomic analysis of cardiac differentiation from human PSCs reveals HOPX-dependent cardiomyocyte maturation. Cell stem cell 23, 586–598. e588 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin C, Ding J & Bar-Joseph Z Inferring TF activation order in time series scRNA-Seq studies. PLoS computational biology 16, e1007644 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang MJ, Ntranos V & Tse D Determining sequencing depth in a single-cell RNA-seq experiment. Nature communications 11, 1–11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kleyman M. et al. Selecting the most appropriate time points to profile in high-throughput studies. Elife 6, doi: 10.7554/eLife.18541 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Butler A, Hoffman P, Smibert P, Papalexi E & Satija R Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nature biotechnology 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tritschler S. et al. Concepts and limitations for learning developmental trajectories from single cell genomics. Development 146 (2019). [DOI] [PubMed] [Google Scholar]
- 73.Treutlein B. et al. Dissecting direct reprogramming from fibroblast to neuron using single-cell RNA-seq. Nature 534, 391–395 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Davis A, Gao R & Navin NE SCOPIT: sample size calculations for single-cell sequencing experiments. BMC bioinformatics 20, 1–6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Haque A, Engel J, Teichmann SA & Lönnberg T A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome medicine 9, 1–12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schwabe D, Formichetti S, Junker JP, Falcke M & Rajewsky N The transcriptome dynamics of single cells during the cell cycle. Molecular systems biology 16, e9946 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kang HM et al. Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nature biotechnology 36, 89–94 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.de Kok JB et al. Normalization of gene expression measurements in tumor tissues: comparison of 13 endogenous control genes. Laboratory investigation 85, 154–159 (2005). [DOI] [PubMed] [Google Scholar]
- 79.Lun AT, Calero-Nieto FJ, Haim-Vilmovsky L, Göttgens B & Marioni JC Assessing the reliability of spike-in normalization for analyses of single-cell RNA sequencing data. Genome research 27, 1795–1806 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Johnson WE, Li C & Rabinovic A Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, 118–127 (2007). [DOI] [PubMed] [Google Scholar]
- 81.Ritchie ME et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids research 43, e47–e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tran HTN et al. A benchmark of batch-effect correction methods for single-cell RNA sequencing data. Genome biology 21, 1–32 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moter A & Göbel UB Fluorescence in situ hybridization (FISH) for direct visualization of microorganisms. Journal of microbiological methods 41, 85–112 (2000). [DOI] [PubMed] [Google Scholar]
- 84.Chen KH, Boettiger AN, Moffitt JR, Wang S & Zhuang X Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Moffitt JR et al. High-throughput single-cell gene-expression profiling with multiplexed error-robust fluorescence in situ hybridization. Proceedings of the National Academy of Sciences 113, 11046–11051 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shah S, Lubeck E, Zhou W & Cai L In situ transcription profiling of single cells reveals spatial organization of cells in the mouse hippocampus. Neuron 92, 342–357 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Eng C-HL et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH+. Nature 568, 235–239 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Codeluppi S. et al. Spatial organization of the somatosensory cortex revealed by osmFISH. Nature methods 15, 932–935 (2018). [DOI] [PubMed] [Google Scholar]
- 89.Wang X. et al. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 361 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bergenstråhle J, Larsson L & Lundeberg J Seamless integration of image and molecular analysis for spatial transcriptomics workflows. BMC genomics 21, 1–7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rodriques SG et al. Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution. Science 363, 1463–1467 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schiller HB et al. The Human Lung Cell Atlas: a high-resolution reference map of the human lung in health and disease. American journal of respiratory cell and molecular biology 61, 31–41 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Park J, Liu CL, Kim J & Susztak K Understanding the kidney one cell at a time. Kidney international 96, 862–870 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gitter A, Carmi M, Barkai N & Bar-Joseph Z Linking the signaling cascades and dynamic regulatory networks controlling stress responses. Genome Res 23, 365–376, doi: 10.1101/gr.138628.112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wagner DE et al. Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science 360, 981–987 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Su Y. et al. Single-cell analysis resolves the cell state transition and signaling dynamics associated with melanoma drug-induced resistance. Proceedings of the National Academy of Sciences 114, 13679–13684 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Russell AB, Elshina E, Kowalsky JR, Te Velthuis AJ & Bloom JD Single-cell virus sequencing of influenza infections that trigger innate immunity. Journal of virology 93 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rozenblatt-Rosen O. et al. The Human Tumor Atlas Network: charting tumor transitions across space and time at single-cell resolution. Cell 181, 236–249 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Delile J. et al. Single cell transcriptomics reveals spatial and temporal dynamics of gene expression in the developing mouse spinal cord. Development 146 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Huisman SM et al. BrainScope: interactive visual exploration of the spatial and temporal human brain transcriptome. Nucleic acids research 45, e83–e83 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhu Y. et al. Spatiotemporal transcriptomic divergence across human and macaque brain development. Science 362 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Maniatis S. et al. Spatiotemporal dynamics of molecular pathology in amyotrophic lateral sclerosis. Science 364, 89–93 (2019). [DOI] [PubMed] [Google Scholar]