Abstract
To study the molecular evolution of flagellin, the protein subunit specifying flagellar (H) antigens, the fliC genes from 15 pathogenic strains of Escherichia coli were amplified by PCR and sequenced. Comparison of fliC sequences of H6 and H7 strains revealed that alleles have a mosaic structure indicating the occurrence of past horizontal transfer of DNA segments between strains. The close similarity of H7 sequences also indicates the exchange of an entire fliC H7 allele between distant clonal lineages. In addition, the ratio of silent substitutions to amino acid replacements suggests that a short segment in the central region of fliC has been under positive selection in the divergence of H6 and H7 alleles. Phylogenetic analysis demonstrates that the fliC sequences of O157:H7 and O55:H7 serotypes are nearly identical and highly divergent from those of E. coli strains expressing H6 and H2 flagellar antigens. A nonmotile clone of sorbitol-fermenting O157 has rapidly accumulated multiple mutations in fliC, presumably as a result of the silencing of flagellin expression.
For many years, the principal method of identifying pathogenic strains of Escherichia coli has been O:H serotyping based on the presence of the cell surface lipopolysaccharide O antigen and the flagellar H antigen (18). The H antigen of E. coli is specified by a single structural subunit (flagellin) encoded by the fliC gene (12). For the species as a whole, there are 53 recognized antigenic types (H types) (12), which, in combination with the O antigen, are often associated with bacterial clones that cause specific kinds of disease (1, 17, 20).
Clonal analysis of enteropathogenic E. coli (EPEC) strains associated with infantile diarrhea (14) has demonstrated that they represent two divergent clonal lineages, which can be recognized by their distinct electrophoretic types (ETs) (30) and the insertion site of a pathogenicity island (34). In addition, O:H serotypes of each group reflect an evolutionary conservation of H2 and H6 flagellar antigens (30). The two EPEC groups are only distantly related to other pathogenic E. coli strains and are also distinct from strains of an atypical EPEC clone, O55:H7, which is an immediate ancestor to the foodborne pathogen E. coli O157:H7 (31). These clonal relationships suggest specific hypotheses about the evolution of fliC sequences underlying the distribution of H antigens.
Comparison of the amino acid sequences of the flagellins of many bacterial species has revealed a distinctive domain structure of the protein (12, 35). The N- and C-terminal parts of the molecule, which are responsible for secretion and polymerization, are conserved among species, whereas the central region, which produces the surface-exposed antigenic part of the flagellar filament, is highly variable both within and among species (12, 35). From studies of Salmonella enterica, two hypotheses have been suggested to explain the extensive variability in the central domain. The first hypothesis proposes that this region undergoes “unconstrained evolution” through the rapid fixation of neutral mutations by genetic drift (28). The second hypothesis, for which there is considerable supporting evidence (22), states that lateral gene transfer and recombination of foreign DNA are major sources of variation that create new fliC alleles and generate antigenic diversity (23, 24). In either case, the role of natural selection is problematic: in some parts of the molecule, it operates as a conservative force against amino acid change, whereas elsewhere it can be a diversifying force promoting protein polymorphism and rapid evolution (29).
The objective of the study reported here was to examine the molecular basis of antigenic variation in pathogenic strains of E. coli representing several major clonal groups. To accomplish this, we sequenced fliC alleles from representative strains that express H2, H6, and H7, as well as the fliC gene from E. coli O157:H7 and its nonmotile relatives. The patterns of nucleotide polymorphism were used to test the hypothesis of neutral, unconstrained evolution in the central domain, to detect past recombination events involving segments of the flagellin gene, and to assess the action of natural selection in promoting diversity in different parts of the molecule.
MATERIALS AND METHODS
Bacterial strains.
The 15 strains of E. coli used as sources of DNA for fliC sequencing were originally isolated from patients with diarrheal disease (Table 1). Five strains represent classical serotypes of EPEC, including four strains expressing the H6 flagellar antigen (572-56, 3787-62, E2348/69, and E851/71) and one with the H2 antigen (E74/68). The sample also includes strains with the H7 antigen (or nonmotile relatives) of enterohemorrhagic E. coli (EHEC). The EHEC strains include the following bacteria of the O157:H7 clone complex (3): two Shiga toxin-producing O157:H7 strains (DEC 3a and CL 8), two Shiga toxin-producing nonmotile O157 variants (3204-92 and FDA 413), and two O55:H7 strains (DEC 5d and DEC 5f) that are close relatives of O157:H7. In addition, the sample includes a nonmotile O157 strain representing a clone implicated in disease outbreaks in Europe that has been shown to be an early diverging lineage in the O157:H7 clone complex (3). For comparison, two additional strains, DEC 6a (O111:H21) and DEC 13a (O128:H7), from the DEC collection of diarrheagenic E. coli were selected, along with five fliC sequences from GenBank: E. coli K-12 (M14358) (10); nondiarrheagenic strains expressing flagellar antigens H1 (L07387), H7 (L07388), and H12 (L07389) (19); and phase 1 flagellin (G217062) of S. enterica Typhimurium (7, 24).
TABLE 1.
E. coli strains and fliC PCR fragment sizes
| ET groupa and strain | Serotypeb | Originc | Fragment size (bp) |
|---|---|---|---|
| EPEC 1 | |||
| DEC 1a | O55:H6 | Pennsylvania | 1,569 |
| DEC 2a | O55:H6 | Congo | 1,563 |
| E2348/69 | O127:H6 | England | 1,551 |
| E851/71 | O142:H6 | Scotland | 1,524 |
| EHEC 1 | |||
| DEC 3a | O157:H7 | Washington | 1,678 |
| CL 8 | O157:H7 | Canada | 1,668 |
| 3204-92 | O157:[H7] | CDC | 1,665 |
| FDA 413 | O157:[H7] | FDA | 1,665 |
| DEC 3f | O157:[H7] | Germany | 1,668 |
| DEC 5d | O55:H7 | Sri Lanka | 1,665 |
| DEC 5f | O55:H7 | USDA | 1,665 |
| ECOR 37 | ON:[H7] | Washingtond | 1,665 |
| EPEC 2 E74/68 | O128:H2 | England | 1,680 |
| Other | |||
| DEC 6a | O111:H12 | New Jersey | 1,695 |
| DEC 13a | O128:H7 | Texas | 1,649 |
Based on ET defined by multilocus enzyme electrophoresis (30, 33). EPEC refers to strains that cause infant diarrhea, and EHEC refers to strains that cause hemorrhagic colitis (14).
Brackets denote nonmotile strains of an ET that are predicted to have the H type characteristic of the motile strains of the same ET. This notation for nonmotile variants was used in reference 25.
CDC, Centers for Disease Control and Prevention; FDA, Food and Drug Administration; USDA, U.S. Department of Agriculture.
Originally isolated from a marmoset in a zoo (16).
DNA preparation and PCR amplification.
E. coli strains were grown on Luria-Bertani agar overnight at 37°C. A single colony of each strain was transferred to 10 ml of nutrient broth (Difco, Detroit, Mich.) and grown overnight in a 37°C shaking water bath. Chromosomal DNA was isolated from each strain in accordance with the instructions in the Puregene DNA Isolation Kit from Gentra Systems, Inc. (Minneapolis, Minn.). Isolated DNA was stored at 4°C.
Oligonucleotide primers EcoH1 (5′-AATACCAACAGCCTCTCGCT-3′) and EcoH2 (5′-AGAGACAGAACCTGCTGC-3′) were designed based on the sequence of the fliC gene (formerly hag [6]) in E. coli K-12. EcoH1 is located on the plus strand, 15 bp downstream of the translational start site, and EcoH2 is located on the minus strand, 11 bp internal to the stop codon. Both primers were synthesized by a Beckman 1000 oligonucleotide synthesizer (Beckman, Fullerton, Calif.). Template DNA for cycle sequencing was obtained through amplification for 30 cycles as follows: 94°C for 1 min, 53°C for 2 min, and 72°C for 3 min with an initial denaturing step of 94°C for 5 min. The PCR products were purified with Qiaquick spin columns (QIAGEN Inc., Valencia, Calif.) and suspended in 10% Tris-EDTA buffer to suitable concentrations, as determined by agarose gel electrophoresis.
Nucleotide sequencing.
Cycle sequencing was performed with a Prism Ready Reaction DyeDeoxy Terminator Cycle Sequencing kit from Applied Biosystems. Sequencing gels were run on an Applied Biosystems 373A automated sequencer. Raw sequences of both DNA strands were analyzed and concatenated by DNASTAR with additional internal sequencing primers designed based on the generated sequence data. All conflicting and putative polymorphic nucleotide sites were sequenced at least three times to reduce sequencing error.
Statistical analysis.
Multiple-sequence alignment of the inferred amino acid sequences was performed with Clustal W (26), and gene trees were constructed with MEGA (8). The proportions of polymorphic synonymous (pS) and nonsynonymous (pN) sites were calculated by the method of Nei and Gojobori (15). To examine variation in the functional constraints of different parts of the molecule, these statistics were tabulated in a sliding-window analysis of 30 codons along the gene by the program PSWIN. The ratio of synonymous-to-nonsynonymous differences among alleles of the same H type was compared to the level of divergence between alleles of different H types by the method of Whittam and Nei (32). Estimates of the sampling variance of these statistics were made by Monte Carlo simulation.
To identify the ends of segments of a mosaic allele, we used MAXCHI, a computer program that implements the maximum chi-square method (13). The program compared each sequence to the consensus and found the point, kmax, at which the chi-square statistic achieved a maximum. The sequence was then divided into two segments, and a new maximum was found within each segment. This cycle was repeated four times so that 16 maxima were found. The significance of the kmax values for the nested segments was tested by a Monte Carlo procedure in which sites were placed randomly along the sequence 1,000 times and the null distribution of kmax was tabulated. Observed kmax values that exceeded values in the 5% tail of the null distribution were considered significant.
RESULTS
Size variation and sequence polymorphism in fliC.
The fliC gene from 15 strains of pathogenic E. coli was amplified by PCR with primers EcoH1 and EcoH2 (Table 1). The DNA fragments produced ranged in size from 1,524 bp for E851/71 (O142:H6) to 1,695 bp for strain DEC 6a (O111:H21). There was also variation in gene size among strains expressing the same H antigen (H6 ranges from 1,524 to 1,569 bp, and H7 ranges from 1,665 to 1,678 bp), although the extent of the variation is less than the average difference in gene size between H types (Table 1).
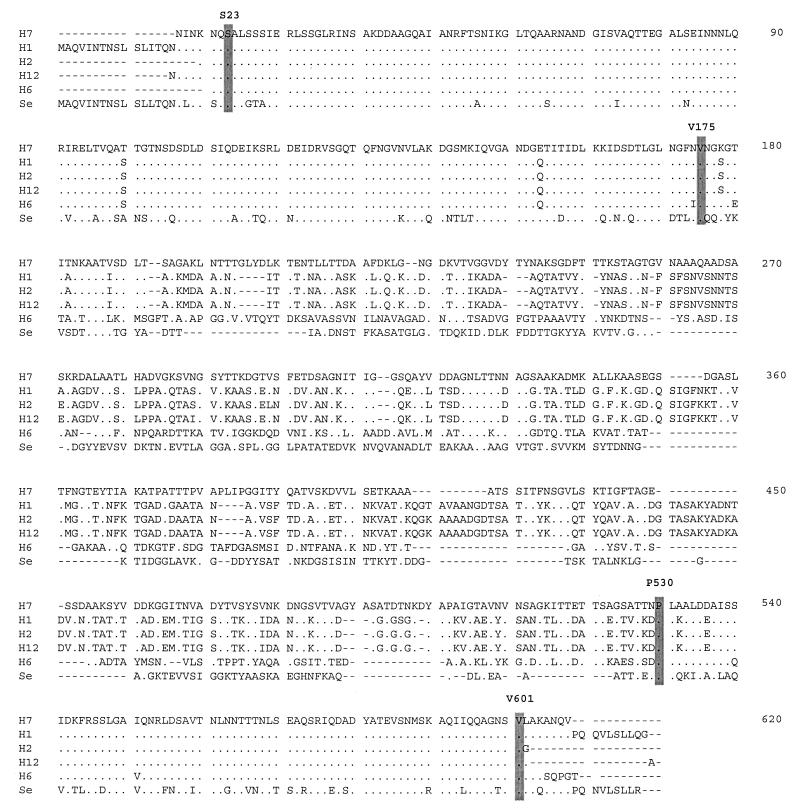
The multiple-sequence alignment of 20 inferred amino acid sequences of flagellin, including the 15 fliC sequences determined here, 4 published fliC sequences from nondiarrheagenic E. coli, and the phase I flagellin of S. enterica Typhimurium, by the Clustal W method (26) required the introduction of numerous alignment gaps, particularly in the central region of the gene. The alignment for representative alleles of five H types shows both the conservation of the primary sequence in the N- and C-terminal regions and the concentration of variability in length and amino acid content in the central region (Fig. 1). The alignment also revealed that the fliC sequence from one nonmotile O157 strain (DEC 3f) had two nucleotide insertions which required realignment to restore the proper reading frame for analysis (see below).
FIG. 1.
Amino acid alignment of five flagellin sequences representing five distinct flagellar antigens (H types) with the phase 1 flagellin sequence of S. enterica Typhimurium (Se). The O type and strains of representative serotypes are as follows: H7, O157 and DEC 3a; H1, O2 and Su 1242; H2, O128 and DEC 11f; H21, O111 and DEC 6a; H6, O55 and DEC 2a.
The aligned sequences were divided into three regions based on the presence of amino acids conserved among the E. coli and S. enterica Typhimurium flagellins. The first region is the N-terminal region of 153 codons, from the serine at position 23 to the valine at position 175 in the multiple-sequence alignment (Fig. 1). The C-terminal region is composed of 68 codons, from the conserved proline at position 530 to the valine at 601. The central variable region includes a variable number of codons located between positions 176 and 529 in the alignment (Fig. 1).
Phylogenetic analysis of fliC.
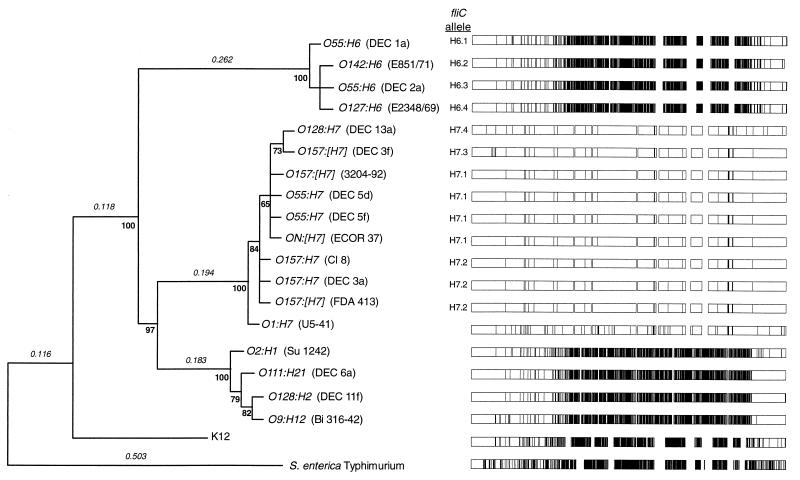
A phylogenetic tree, constructed from the amino acid sequences by the neighbor-joining algorithm (8) and rooted by the Typhimurium sequence, revealed three clusters of E. coli fliC alleles (Fig. 2). The first cluster contains alleles that specify the H6 antigen and occur in the EPEC 1 strains. There are four distinct H6 alleles, each of which differs from the others by two amino acids (Table 2). The most divergent sequence of the H6 alleles is H6.4, found in strain E2348/69, which also has three silent substitutions not seen in the other H6 alleles (Table 2).
FIG. 2.
Gene phylogeny for E. coli fliC rooted by the phase 1 flagellin of S. enterica Typhimurium. The phylogenetic tree was constructed by the neighbor-joining algorithm based on the gamma distance with α = 2. The gamma distance assumes that substitutions follow a gamma distribution with the α parameter specifying the amount of variation across amino acid positions. Branch lengths, in terms of amino acid substitutions per site, are given above the major branches. Bootstrap confidence levels are given under the nodes. At the right is a graph of the locations of polymorphic nucleotide sites, marked as vertical lines that plot nucleotides that differ from the consensus sequence at each position.
TABLE 2.
Polymorphic codons defining H6 alleles of flagellin (fliC) in the EPEC 1 clonal group
| Allele | Amino acid and codon in multiple-sequence alignment at codon positiona:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 76 | 159 | 174 | 220 | 229 | 287 | 347 | 505 | 555 | 570 | |
| Gln | Asp | Asn | Asn | Asp | Thr | Gly | Gly | Leu | Ser | |
| H6.1 | CAA | GAT | AAT | AAT | GAT | ACG | GGT | GGT | CTG | TCT |
| Gln | Glu | Asp | ||||||||
| H6.2 | CAG | GAA | — | GAT | — | — | — | — | — | — |
| Gln | Ile | Ala | ||||||||
| H6.3b | CAG | — | ATA | — | — | — | GCT | — | — | — |
| Gln | Asn | Thr | Gly | Leu | Phe | |||||
| H6.4 | CAG | — | — | — | AAT | ACA | — | GGG | TTG | TTT |
A dash means that the codon is identical to that of H6.1. Boldface type indicates amino acids that differ from those in the H6.1 sequence. Altered nucleotides are underlined.
Insert of six nucleotides at position of 348 (ACT GGT).
The second cluster of H alleles includes 10 isolates that are both motile and express H7 antigen or are nonmotile and are predicted to have an H7 allele, based on clonal relatedness. The H7 alleles have, on average, 13.8 (0.83%) nucleotide differences and 4.8 (0.87%) amino acid differences. Most of the sequence variation is due to the H7 allele of the O1:H7 strain, which differs by more than 3% of the nucleotides and more than 2% of the amino acids from the other H7 alleles. The remaining H7 genes have four distinct sequences that differ, on average, at 2.4 amino acid positions (Table 3). The most common H7 allele (H7.1) is found in O55 strains DEC 5d and DEC 5f, in a nonmotile O157 strain (3204-92), and in ECOR37 (Fig. 2). Three O157 strains (DEC 3a, CL 8, and FDA 413) have allele H7.2, which differs by a single amino acid (Asp-239) from the common H7.1 sequence. The H7 alleles of DEC 3f (H7.3) and DEC 13a (H7.4) are distinct from one another (seven amino acids) and differ by four to six amino acids from the other H7 alleles. These two alleles cluster together in the gene tree because they both have a serine replacement at position 45 (Table 3).
TABLE 3.
Polymorphic codons defining H7 alleles of flagellin (fliC) found in the O157:H7 clone complex
| Allele | Amino acid and codon in multiple-sequence alignment at codon positiona:
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 22 | 27 | 38 | 40 | 44 | 45 | 64 | 65 | 76 | 239 | 505 | 525 | 558 | 566 | 594 | 609 | |
| Gln | Ser | Ile | Ser | Asp | Ala | Ala | Ala | Gln | Asn | Gly | Ser | Ala | Thr | Ile | Pro | |
| H7.1 | CAG | AGT | ATT | AGC | GAC | GCC | GCG | GCG | CAG | AAT | GGT | TCT | GCA | ACT | ATT | CCG |
| Asp | ||||||||||||||||
| H7.2 | — | — | — | — | — | — | — | — | — | GAT | — | — | — | — | — | — |
| His | Leu | Arg | His | Ser | Arg | |||||||||||
| H7.3 | CAC | — | CTT | CGC | CAC | TCC | — | — | — | — | — | — | — | — | — | CGA |
| Ile | Ser | Ala | Ala | Gln | Gly | Phe | Ala | Asn | Ile | |||||||
| H7.4 | — | ATT | — | — | — | TCC | GCT | GCA | CAA | — | GGC | TTT | GCG | AAT | ATC | — |
A dash means that the codon is identical to that in the H7.1 sequence. Boldface type indicates that the amino acid differs from that in the H7.1 sequence. Altered nucleotides are underlined.
The third cluster is a mixed group of antigens, including H1, H12, H21, and H2. The fliC sequences specifying these distinct antigens differ, on average, by 30.2 (1.8%) nucleotides and 7.8 (1.4%) amino acids. The H2 and H21 flagellins are similar in sequence and differ by only two amino acids of the central region (Table 4). The most divergent member of this cluster is the H1 flagellin, which has six distinct amino acid replacements (Table 4).
TABLE 4.
Polymorphic codons predicting amino acid replacements among flagellin (fliC) sequences of four H types
| H type | Amino acid and codon in multiple-sequence alignment at codon positiona:
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 71 | 271 | 289 | 299 | 317 | 355 | 376 | 410 | 412 | 415 | 449 | 450 | 496 | 546 | 602 | |
| Gly | Glu | Ser | Leu | Lys | Lys | Asp | Lys | Ala | Asp | Lys | Ala | Thr | Ser | Gly | |
| H2 | GGT | GAA | AGT | TTG | AAA | AAA | GAT | AAA | GCT | GAC | AAA | GCT | ACG | TCT | GGC |
| Ile | Val | Leu | |||||||||||||
| H21 | — | — | ATT | GTG | — | — | — | — | — | — | — | — | — | — | CTG |
| Ala | Ala | Val | Lys | Val | Pro | Leu | |||||||||
| H12 | GCT | GCA | — | GTG | — | AAG | — | — | GTT | — | — | — | — | CCT | CTG |
| Ala | Val | Glu | Asn | Gly | Thr | Val | Asn | Asn | Thr | Ser | Ser | Leu | |||
| H1 | — | GCA | — | GTG | GAA | AAT | GGT | ACA | GTT | AAC | AAT | ACT | TCG | TCA | CTG |
A dash means that the codon is identical to that in the H2 sequence. Boldface type indicates that the amino acid differs from that in the H2 sequence. Nucleotides that differ are underlined.
Mosaic gene structure.
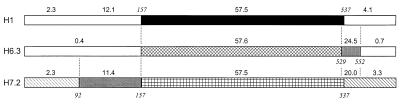
Genes with mosaic structures are composed of segments of DNA with different histories that have been brought together by horizontal transfer and recombination. To detect the mosaic structure of the fliC alleles, we used the maximum chi-square method (13) to compare pairs of nucleotide sequences and locate points of significant heterogeneity in levels of sequence divergence. We first removed extraneous alignment gaps and compared three alleles that represented the major clusters in the dendrogram. The pairwise comparison of the H1, H6.3, and H7.2 sequences revealed significant chi-square values and mosaic structure within each sequence. The locations of the breakpoints (kmax) and the percentage of sequence divergence between segments are diagrammed in Fig. 3. All three alleles show striking divergence in the central region, which extends from a breakpoint at position 157 to position 537, or to 529 in the case of H6.3 (Fig. 3). The 5′ end of H7.2 is further subdivided at position 92 into two segments that differ by about 2 and 12%, respectively, from the other two alleles. The 3′ end of H6.3 is also further subdivided by a breakpoint at position 552 (Fig. 3).
FIG. 3.
Locations of breakpoints (kmax) and percentages of sequence divergence between segments detected by the maximum chi-square method. The values in italics refer to the codon positions in the multiple-sequence alignment of Fig. 1. The values above the segments are percentages of nucleotide difference between pairs of sequences. The upper line of percentages is for the H1-H7.2 comparison, the second line is for the H1-H6.3 comparison, and the third line is for the H6.3-H7.2 comparison.
Although some of the heterogeneity along the sequence can be explained by relaxed functional constraints on the central region of flagellin, discrepancies in the pairwise comparisons suggest past recombination of different segments. The most convincing case of intragenic recombination is the close similarity of the sequences of the 5′ ends of H6.3 and H1, which differ by less than 0.4%, whereas their central regions differ by more than 50% of the nucleotides. The strains are only distantly related in chromosomal background, and the 5′-end sequence of H1 is more similar to H6 than to other alleles in its cluster. Together, these observations indicate that the segments of the mosaic alleles have independent histories.
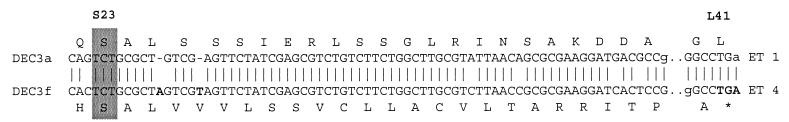
Frameshift mutations in nonmotile O157.
Nonmotile strain DEC 3f (O157:H−) is a divergent clone of the O157:H7 complex that fails to produce functional flagella and is distinct biotypically from O157:H7 (3). Nucleotide sequencing of its fliC gene revealed two insertions (positions 12 and 17) in the 5′ conserved region relative to the sequence of O157:H7 (Fig. 4). These insertions produce a shift in reading frame that introduces a premature stop codon at position 41 (Fig. 4). A comparison of the amino acid sequence predicted by the correct reading frame (realigned by removing the two base insertions) shows that the H7.3 allele of DEC 3f is similar to those of O55:H7 and O157:H7 (Table 3). Although there are five distinct replacements in the DEC 3f sequence, it has other amino acids, at positions 27, 239, 525, 566, and 609 (Table 3), that are identical to those of O157:H7 and O55:H7 and distinct from those of the H7.4 allele of the O128 strain (DEC 13a). In addition, the realigned fliC sequence of DEC 3f is most similar to those of the H7.1 and 7.2 alleles for silent substitutions at codons 64, 65, 76, 505, and 558 (Table 3).
FIG. 4.
Comparison of the 5′ ends of the fliC sequences of O157:H7 strain DEC 3a and its nonmotile relative DEC 3f. Two nucleotides that occur in the DEC 3f gene (in boldface) cause a frameshift and predict a premature stop codon (position 61 in the multiple-sequence alignment of Fig. 1).
Divergence of H6 and H7 alleles.
If evolution of the central region of flagellin results from the unconstrained accumulation of neutral mutations, the pattern of substitution, in terms of the ratio of synonymous to nonsynonymous changes, should be the same among alleles of the same H type as that between alleles specifying different H types (32). To test this prediction of the neutral-mutation hypothesis, we calculated the amounts of polymorphism and sequence divergence at synonymous and nonsynonymous sites separately for the three regions of the gene, both within and between alleles of H6 and H7 (Table 5). Three of the six comparisons, including polymorphisms in the N and C termini, have a ratio of pN to pS in the range of 0.11 to 0.17 (Table 5). Surprisingly, the degree of amino acid divergence between H6 and H7 alleles is highly constrained in the N-terminal region of the protein, as reflected in the pN/pS ratio of 0.02. As expected, the central region shows much less constraint on amino acid-altering mutations among alleles of the same H type, as well as between alleles of different H types. In fact, the ratio of nonsynonymous to synonymous mutations in the divergence of the central region between H6 and H7 alleles is nearly unity (0.93 ± 0.05), which is the ratio predicted for unconstrained, completely neutral variation.
TABLE 5.
Polymorphism within and divergence between fliC domains for H7 and H6 flagellar antigens
| Domain | No. of codons | Mean ± SE polymorphism within domain
|
pN/pS ratio | Mean ± SE divergence between domains
|
pN/pS ratio | ||
|---|---|---|---|---|---|---|---|
| pS (102) | pN (102) | pS (102) | pN (102) | ||||
| N terminus | 153 | 4.60 ± 0.87 | 0.52 ± 0.18 | 0.12 ± 0.05 | 21.70 ± 3.69 | 0.49 ± 0.32 | 0.02 ± 0.01 |
| Central region | 240 | 1.13 ± 0.33 | 0.61 ± 0.18 | 0.57 ± 0.27 | 69.57 ± 3.43 | 64.33 ± 2.14 | 0.93 ± 0.05 |
| C terminus | 68 | 2.69 ± 1.03 | 0.44 ± 0.25 | 0.17 ± 0.13 | 24.07 ± 5.23 | 2.55 ± 1.76 | 0.11 ± 0.08 |
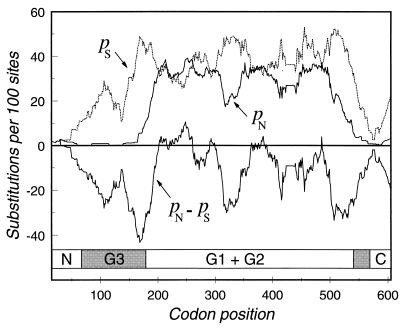
To determine how the level of selective constraint varies along the molecule, we calculated the pN and pS for subsets of 30 codons in a sliding window for the length of the gene (Fig. 5). The difference, pN − pS, is a measure of the degree of selective constraint: the more negative the value, the less the contribution of replacement substitutions and the greater the contribution of synonymous substitutions. The zero-difference line indicates selectively neutral variation, where the per-site rates of synonymous and nonsynonymous substitutions are equal. A positive difference, where amino acid replacements exceed the silent substitutions, suggests the action of diversifying (positive) selection. In general, the N-terminal region is highly constrained, with pN − pS reaching its maximum negative difference around codon 168, near V175, marking the end of the conserved region (Fig. 1). The C terminus is also highly conserved, as indicated by the negative value of pN − pS, which achieves a minimum between codons 507 and 531. There are three stretches within the central variable region where there is a positive difference between the nonsynonymous- and synonymous-substitution rates. The longest stretch with pN − pS > 0 is 88 codons in length and runs from position 187 to position 275. In this region, the rate of substitution per 100 sites (corrected for multiple hits) is dN = 125 ± 19 and dS = 109 ± 27. The second region extends from codon 276 to codon 312, the third region extends from 349 to 405, and the fourth region peaks with a positive difference at codon 486. None of these three regions has a significant excess of nonsynonymous substitutions. Thus, only the 88-codon segment between 187 and 275 is a likely target for diversifying selection.
FIG. 5.
Sliding window plot of the number of substitutions per 100 sites for synonymous (pS) and nonsynonymous (pN) sites between H6 (O55, DEC 2a) and H7 (O157, DEC 3a) fliC alleles. The difference (pN − pS) is a measure of the level of selective constraint on various parts of the molecule. A diagram of the five-domain model of flagellin (27) is shown at the bottom. The shaded regions denote the discontinuous coding regions predicted for the G3 domain.
DISCUSSION
Direct sequencing of the fliC gene in pathogenic E. coli strains with distinct H antigens shows that the N- and C-terminal regions of alleles are largely conserved, whereas the central region is polymorphic and highly diverse. The concentration of genetic variation in the central region of flagellin has been repeatedly found in many species of bacteria and has yielded two hypotheses about its cause. Wei and Joys (28) suggested that the central region of Salmonella phase I flagellin evolves by the rapid accumulation of neutral mutations by genetic drift, whereas amino acid replacements in the terminal domains are highly constrained. Selander and coworkers (11, 21, 23, 24) have demonstrated that horizontal gene transfer and recombination are significant sources of variation for the divergence of Salmonella flagellar antigens. Similarly, a central role for recombination in flagellin evolution has recently been reported for Campylobacter jejuni (5).
Evidence of recent lateral gene transfer.
Comparative sequence analysis of the fliC genes of E. coli revealed two types of recombination events underlying antigenic variation. The first type of event was the past exchange of an entire fliC gene between unrelated chromosomal backgrounds. The observation that the H7 alleles of O157:H7 and O128:H7 differ by only 11 nucleotides (0.67%) indicates that these sequences have diverged very recently from a common ancestral allele. In contrast, the genomic backgrounds of O157:H7 and O128:H7 strains are highly divergent, based on results obtained by multilocus enzyme electrophoresis (33), and belong to different clonal lineages. This finding strongly suggests that an ancestral H7 fliC allele was horizontally transferred after the divergence of the O157:H7 and O128:H7 lineages from a common ancestor. At this point, we cannot resolve the direction and timing of this transfer event. It is relevant, however, that the O128:H7 clone is closely related (data not shown) to an O76:H7 clone (2), which suggests that the transfer occurred sufficiently long ago for the descendants to have diverged in somatic antigens.
The second type of event is the recombination of a segment of fliC to create a new allele with a mosaic structure. The evidence for this type of intragenic recombination is based on the heterogeneity in divergence between sequences that cannot be explained simply by relaxed selective constraints. The maximum chi-square analysis found several significant breakpoints in the comparison of the H6 and H7 alleles. These breakpoints locate positions of significant heterogeneity in the divergence of the sequences and identify putative mosaic segments that comprise each allele.
Further evidence for the role of recombination in creating mosaic fliC alleles is the presence of a chi sequence (5′-GCTGGTGG-3′) beginning at codon 277 in the H7 alleles. The chi sequence is at the boundary of the second region of positive selection (pN > pS). The presence of a chi sequence identifies a potential target for exonuclease V that can generate a single-stranded stretch of DNA with a free 3′ end: this stimulates homologous recombination, particularly when chi sequences occur on both homologues (4). The fact that the chi sequences were not found in the other H alleles is not unexpected because these sites are often lost after a recombination event (4).
One consequence of the mosaic structure of the fliC alleles is that the past exchange of short segments of DNA between lineages obscures the evolutionary history of the alleles and invalidates a purely phylogenetic approach. This means that although the close relatedness of the alleles within a cluster in the dendrogram (Fig. 2) is valid, the divergence of the clusters does not fit a single branching gene phylogeny but, instead, reflects a reticulate pattern of molecular evolution.
Evidence of selection in the central domain.
The pattern of synonymous and nonsynonymous changes in the divergence of fliC alleles encoding H6 and H7 antigens shows that the N- and C-terminal regions are highly constrained, whereas the central region evolves, for the most part, by substitution with equal rates at synonymous and nonsynonymous sites. As pointed out by Smith and Selander (24), however, the central variable region of S. enterica Typhimurium flagellin is not completely free of selective constraints on amino acid replacement, for there are no cysteine or tryptophan residues and only a few histidines. In addition, they suggested that a moderate value of the codon adaptation index indicates weak selection against some synonymous change in this region (24).
Vonderviszt and colleagues (27) proposed that the central region of the flagellin of serovar Typhimurium is composed of three globular domains: G1 and G2 are β-sheeted structures with very little α-helical content; G1 and G2 are exposed parts of the molecule that encode antigenic determinants, and G3 is a compact, protease-resistant structure with both antiparallel β-sheets and helical segments. They concluded that G3 is important for the structural integrity of the G1 and G2 domains because if G3 is removed, the G1 and G2 domains are destabilized (27). In their structural model, G3 is specified by two distinct segments in the fliC gene that flank the central part encoding the G1 and G2 domains (segments 67 to 178 and 419 to 446).
Comparison of the G domains with the pattern of synonymous and nonsynonymous substitutions (Fig. 5) shows that the proposed G3-encoding segments of the Typhimurium flagellin designate areas in the E. coli H6 and H7 flagellins that are highly constrained, with deep valleys in the pN − pS coefficient. The comparison also suggests that the G3 domain of E. coli is encoded about 150 nucleotides upstream of the predicted location in the Typhimurium flagellin (Fig. 5).
Although the pattern of substitution in the G1 and G2 domains fits the model of unconstrained evolution (pN = pS; Fig. 5), there is a short segment where the per-site nonsynonymous-substitution rate exceeds the synonymous-substitution rate. This pattern suggests that this region of 88 codons is a target for diversifying selection, where natural selection has favored the substitution of amino acids. Because the short segment lies within the central domain that is the major antigenic component of flagellin (9), the evolutionary benefit to new variants may be the enhanced ability to escape herd immunity and colonize new hosts.
Flagellin alleles and divergence of EPEC.
Selander et al. (21) have noted that the high level of variability and rapid evolution of fliC provide a unique opportunity to test phylogenetic hypotheses. The fliC sequences of EPEC with the H6 antigen are 99% similar, with an average of eight nucleotide differences per 1,536 sites and an average of 4.3 amino acid differences between pairs of alleles. This observation supports the hypothesis that strains of the classical EPEC 1 serotypes with the H6 antigen (e.g., O55:H6, O127:H6, and O142:H6) are derived from a common ancestral strain and form a distinct clone complex (30). The degree of synonymous site divergence between the conserved regions of fliC H6 alleles and the fliC sequence of the H2 antigen (EPEC 2) is extensive, with an average of 9.8 differences per 100 synonymous sites. This is indicative of a long period of separation between these alleles and supports the hypothesis that the EPEC 1 and EPEC 2 lineages have evolved independently for millions of generations (30).
The results also support the model of the stepwise evolution of the O157:H7 complex from an ancestral O55:H7-like ancestor through several intermediate stages (3). This model, which is based, in part, on the results of multilocus enzyme electrophoresis, predicts that two branches of O157 strains emerged from an O55:H7 ancestor. One branch lost several metabolic functions (e.g., sorbitol fermentation) and led to the O157:H7 clone that has spread worldwide. The other branch retained the ability to ferment sorbitol, lost motility, and has emerged as a public health problem in central Europe. This scenario is supported by the accumulated mutations in the fliC alleles (Table 3). The common allele (H7.1) is found in both O55:H7 and sorbitol-negative O157:H7 strains. The five replacements in the H7.3 allele presumably were fixed after the clonal lineage lost motility and expression of the flagellin gene. The permanent loss of fliC expression would remove the selective constraints on amino acid-altering mutations.
ACKNOWLEDGEMENTS
This research was supported by Public Health Service grants AI 42391 (T.S.W) and AI 22144 (R.K.S) from the National Institutes of Health.
ADDENDUM IN PROOF
Our report of nearly identical flagellin sequences of E. coli O157:H7 and its close relatives (O55:H7 and nonmotile O157 strains) explains the observations of Fields et al. (P. I. Fields, K. Blom, H. J. Hughes, L. O. Helsel, P. Fang, and B. Swaminathan, J. Clin Microbiol. 35:1066–1070, 1997), who found that O55:H7 and O157:H7 strains gave indistinguishable patterns of fliC restriction digests.
REFERENCES
- 1.Achtman M, Pluschke G. Clonal analysis of descent of virulence among selected Escherichia coli. Annu Rev Microbiol. 1986;40:185–210. doi: 10.1146/annurev.mi.40.100186.001153. [DOI] [PubMed] [Google Scholar]
- 2.Bettelheim K A, Brown J E, Lolekha S, Echeverria P. Serotypes of Escherichia coli that hybridized with DNA probes for genes encoding Shiga-like toxin I, Shiga-like toxin II, and serogroup O157 enterohemorrhagic E. coli fimbriae isolated from adults with diarrhea in Thailand. J Clin Microbiol. 1990;28:293–295. doi: 10.1128/jcm.28.2.293-295.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feng P, Lampel K A, Karch H, Whittam T S. Genetic and phenotypic changes in the emergence of Escherichia coli O157:H7. J Infect Dis. 1998;177:1750–1753. doi: 10.1086/517438. [DOI] [PubMed] [Google Scholar]
- 4.Friedman-Ohana R, Karunker I, Cohen A. Chi-dependent intramolecular recombination in Escherichia coli. Genetics. 1998;148:545–557. doi: 10.1093/genetics/148.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harrington C S, Thomson-Carter F M, Carter P E. Evidence for recombination in the flagellin locus of Campylobacter jejuni: implications for the flagellin gene typing scheme. J Clin Microbiol. 1997;35:2386–2392. doi: 10.1128/jcm.35.9.2386-2392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iino T, Komeda Y, Kutsukake K, Macnab R M, Matsumura P, Parkinson J S, Simon M I, Yamaguchi S. New unified nomenclature for the flagellar genes of Escherichia coli and Salmonella typhimurium. Microbiol Rev. 1988;52:533–535. doi: 10.1128/mr.52.4.533-535.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joys T M. The covalent structure of the phase-1 flagellar filament protein of Salmonella typhimurium and its comparison with other flagellins. J Biol Chem. 1985;260:15758–15761. [PubMed] [Google Scholar]
- 8.Kumar S, Tamura K, Nei M. MEGA: molecular evolutionary genetics analysis, version 1.0. University Park, Pa: The Pennsylvania State University; 1993. [Google Scholar]
- 9.Kuwajima G. Flagellin domain that affects H antigenicity of Escherichia coli K-12. J Bacteriol. 1988;170:485–488. doi: 10.1128/jb.170.1.485-488.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuwajima G, Asaka J, Fujiwara T, Node K, Kondo E. Nucleotide sequence of the hag gene encoding flagellin of Escherichia coli. J Bacteriol. 1986;168:1479–1483. doi: 10.1128/jb.168.3.1479-1483.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J, Nelson K, McWhorter A C, Whittam T S, Selander R K. Recombinational basis of serovar diversity in Salmonella enterica. Proc Natl Acad Sci USA. 1994;91:2552–2556. doi: 10.1073/pnas.91.7.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Macnab R M. Flagella and motility. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: ASM Press; 1996. pp. 123–145. [Google Scholar]
- 13.Maynard Smith J. Analyzing the mosaic structure of genes. J Mol Evol. 1992;34:126–129. doi: 10.1007/BF00182389. [DOI] [PubMed] [Google Scholar]
- 14.Nataro J P, Kaper J B. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 16.Ochman H, Selander R K. Standard reference strains of Escherichia coli from natural populations. J Bacteriol. 1984;157:690–693. doi: 10.1128/jb.157.2.690-693.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ørskov F, Whittam T S, Cravioto A, Ørskov I. Clonal relationships among classic enteropathogenic Escherichia coli (EPEC) belonging to different O groups. J Infect Dis. 1990;162:76–81. doi: 10.1093/infdis/162.1.76. [DOI] [PubMed] [Google Scholar]
- 18.Ørskov I, Ørskov F, Jann B, Jann K. Serology, chemistry, and genetics of O and K antigens of Escherichia coli. Bacteriol Rev. 1977;41:667–710. doi: 10.1128/br.41.3.667-710.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schoenhals G, Whitfield C. Comparative analysis of flagellin sequences from Escherichia coli strains possessing serologically distinct flagellar filaments with a shared complex surface pattern. J Bacteriol. 1993;175:5395–5402. doi: 10.1128/jb.175.17.5395-5402.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Selander R K, Caugant D A, Whittam T S. Genetic structure and variation in natural populations of Escherichia coli. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1987. pp. 1625–1648. [Google Scholar]
- 21.Selander R K, Li J, Boyd E F, Wang F-S, Nelson K. DNA sequence analysis of the genetic structure of populations of Salmonella enterica and Escherichia coli. In: Priest F G, editor. Bacterial diversity and systematics. New York, N.Y: Plenum Press; 1994. pp. 17–49. [Google Scholar]
- 22.Selander R K, Li J, Nelson K. Evolutionary genetics of Salmonella enterica. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 2691–2707. [Google Scholar]
- 23.Smith N H, Beltran P, Selander R K. Recombination of Salmonella phase I flagellin genes generates new serovars. J Bacteriol. 1990;172:2209–2216. doi: 10.1128/jb.172.5.2209-2216.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith N H, Selander R K. Sequence invariance of the antigen-coding central region of the phase 1 flagellar filament gene (fliC) among strains of Salmonella typhimurium. J Bacteriol. 1990;172:603–609. doi: 10.1128/jb.172.2.603-609.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stenderup J, Ørskov F. The clonal nature of enteropathogenic Escherichia coli strains. J Infect Dis. 1983;148:1019–1024. doi: 10.1093/infdis/148.6.1019. [DOI] [PubMed] [Google Scholar]
- 26.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalities and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vonderviszt F, Uedaira H, Kidokoro S-I, Namba K. Structural organization of flagellin. J Mol Biol. 1990;214:97–104. doi: 10.1016/0022-2836(90)90149-g. [DOI] [PubMed] [Google Scholar]
- 28.Wei L N, Joys T M. Covalent structure of three phase-1 flagellar filament proteins of Salmonella. J Mol Biol. 1985;186:791–803. doi: 10.1016/0022-2836(85)90397-3. [DOI] [PubMed] [Google Scholar]
- 29.Whittam T S. Genetic population structure and pathogenicity in enteric bacteria. In: Baumberg S, Young J P W, Wellington E M H, Saunders J R, editors. Population genetics of bacteria. Cambridge, England: Cambridge University Press; 1995. pp. 217–245. [Google Scholar]
- 30.Whittam T S, McGraw E A. Clonal analysis of EPEC serogroups. Rev Microbiol Sao Paulo. 1996;27:7–16. [Google Scholar]
- 31.Whittam T S, McGraw E A, Reid S D. Pathogenic Escherichia coli O157:H7: a model for emerging infectious diseases. In: Krause R M, editor. Emerging infections. New York, N.Y: Academic Press, Inc.; 1998. pp. 163–183. [Google Scholar]
- 32.Whittam T S, Nei M. Neutral mutation hypothesis test. Nature. 1991;354:114–116. doi: 10.1038/354114e0. [DOI] [PubMed] [Google Scholar]
- 33.Whittam T S, Wolfe M L, Wachsmuth I K, Ørskov F, Ørskov I, Wilson R A. Clonal relationships among Escherichia coli strains that cause hemorrhagic colitis and infantile diarrhea. Infect Immun. 1993;61:1619–1629. doi: 10.1128/iai.61.5.1619-1629.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wieler L H, McDaniel T K, Whittam T S, Kaper J B. Insertion site of the locus of enterocyte effacement in enteropathogenic and enterohemorrhagic Escherichia coli differs in relation to the clonal phylogeny of strains. FEMS Microbiol Lett. 1997;156:49–53. doi: 10.1111/j.1574-6968.1997.tb12704.x. [DOI] [PubMed] [Google Scholar]
- 35.Winstanley C, Morgan J A W. The bacterial flagellin gene as a biomarker for detection, population genetics and epidemiological analysis. Microbiology. 1997;143:3071–3084. doi: 10.1099/00221287-143-10-3071. [DOI] [PubMed] [Google Scholar]