Abstract
After shifting an oxygen-respiring culture of Pseudomonas stutzeri to nitrate or nitrite respiration, we directly monitored the expression of the nirS gene by mRNA analysis. nirS encodes the 62-kDa subunit of the homodimeric cytochrome cd1 nitrite reductase involved in denitrification. Information was sought about the requirements for gene activation, potential regulators of such activation, and signal transduction pathways triggered by the alternative respiratory substrates. We found that nirS, together with nirT and nirB (which encode tetra- and diheme cytochromes, respectively), is part of a 3.4-kb operon. In addition, we found a 2-kb monocistronic transcript. The half-life of each of these messages was approximately 13 min in denitrifying cells with a doubling time of around 2.5 h. When the culture was subjected to a low oxygen tension, we observed a transient expression of nirS lasting for about 30 min. The continued transcription of the nirS operon required the presence of nitrate or nitrite. This anaerobically manifested N-oxide response was maintained in nitrate sensor (NarX) and response regulator (NarL) knockout strains. Similar mRNA stability and transition kinetics were observed for the norCB operon, encoding the NO reductase complex, and the nosZ gene, encoding nitrous oxide reductase. Our results suggest that a nitrate- and nitrite-responsive regulatory circuit independent of NarXL is necessary for the activation of denitrification genes.
Denitrification is a facultative trait whose manifestation depends on environmental factors. Physiological studies have shown that the expression of denitrification genes usually occurs in the absence of oxygen, or at least at a low oxygen tension (pO2), and requires the simultaneous presence of an N-oxide (reviewed in reference 33). The expression of denitrification in the strict sense is sequential in Pseudomonas stutzeri with respect to nitrate respiration, since activation of the narGHJI operon occurs at a higher partial oxygen pressure than that at which the other reductase genes are activated (24, 25). By monitoring the production of narH and nirS transcripts of Paracoccus denitrificans, it has been shown that induction of nitrate reductase precedes that of nitrite reductase (8).
In Escherichia coli, the transcription factor FNR mediates the oxygen response while NarL, as part of a NarXL two-component sensor-response regulator system, mediates the nitrate signal. Both FNR and NarL are required for the expression of the narGHJI operon. The numerous studies directed at these transcription factors have provided a detailed mechanistic picture of anaerobic and nitrate-dependent gene activation in a nitrate-respiring bacterium (reviewed in references 19 and 20). Moreover, a crystal structure for the nitrate response regulator NarL has been reported recently (7).
It is unknown which N-oxide-sensitive regulatory system controls the genes encoding the reductases of denitrifying bacteria that act on the substrates nitrite, nitric oxide, and nitrous oxide and to what extent their activation depends on coordinately and/or sequentially acting regulators. Advances in mRNA methodology prompted us to study gene expression in P. stutzeri by monitoring the kinetics of individual transcripts rather than by using reporter gene fusions. In the present study, we investigated the signal and regulator requirements for the transcriptional activation of nirS, norCB, and nosZ (i.e., the structural genes for the three reductases involved in nitrite denitrification) following a shifting of the respiratory metabolism from oxygen to nitrate or nitrite. By studying narX and narL deletion mutants, we obtained evidence for the existence of a second nitrate- and nitrite-responsive regulatory system in P. stutzeri that is specific for denitrification. A preliminary account of this work has appeared previously (21).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Wild-type traits of P. stutzeri ATCC 14405 are represented in this study by the spontaneously streptomycin-resistant strain MK21 (36). P. stutzeri was grown on synthetic asparagine-citrate medium at 30°C (36). For aerated cultures, 500 ml of medium in a 1-liter flask was inoculated with an overnight culture to an optical density at 660 nm of about 0.2 and incubated in a gyratory shaker at 240 rpm. Denitrification was induced by adding sodium nitrate (0.1%) or sodium nitrite (0.05%) and simultaneously shifting to strongly O2-limited conditions by reducing the shaker speed to 120 rpm. When necessary, kanamycin, ampicillin, or streptomycin was added at a final concentration of 50, 100, or 200 μg ml−1, respectively. Nitrite in the culture medium was measured as an azo dye (32).
Isolation of RNA and Northern blot analysis.
Total RNA was extracted from batch cultures by the hot-phenol method, with 10- or 20-ml samples being taken at appropriate time intervals during the transition from aerobic to denitrifying growth conditions (1). Twenty micrograms of RNA from each sample was denatured by glyoxal-dimethyl sulfoxide treatment and separated on a 1.2% agarose gel (26). Equal loading of samples onto the gel was verified by acridine orange staining of the rRNA. Transfer of RNA to a positively charged nylon membrane (Boehringer Mannheim) was achieved by upward capillary action. DNA probes were labeled with dUTP-digoxigenin by random priming.
The nirS probe was derived by KpnI digestion of plasmid pNIR44, a 1.9-kb PstI-BglII fragment of plasmid pNOR161 served as the norCB probe, and the nosZ probe was obtained as a 500-bp PstI fragment from plasmid NS220. Details of these probes are described elsewhere (31). DNA labeling and detection kits were from Boehringer Mannheim; they were used in accordance with the specifications of the manufacturer. The dioxetane reagent CDP-Star [disodium 2-chloro-5-(4-methoxyspiro{1,2-dioxetane-3,2′-(5′-chloro)tricyclo[3.3.1.13,7]decan}-4-yl)-1-phenyl phosphate] was used as the chemiluminescent substrate for membrane-based detection of alkaline phosphatase conjugates. Biomax MR (Kodak) exposures were quantitated by scanning laser densitometry with an ImageMaster scanner and software (Pharmacia).
Primer extension analysis.
The 5′ end of the nirS transcript was mapped by primer extension (6). Reverse transcription was initiated from the γ-32P 5′-labeled primer 5′-ATCAGGCCAGCCAGGATAGG-3′, which was complementary to the coding strand at positions 267 to 286 of the published nirS sequence (23). The nucleotide sequence was obtained by the dideoxy chain termination method, using a Thermo-Sequenase kit (United States Biochemical Corp.), [35S]dATP (Amersham), and the same primer. The primer extension and sequencing reaction products were analyzed on a 6% denaturing polyacrylamide gel.
Construction of narL and narX deletion strains.
The narXL gene region was obtained from cosmid clones of Sau3A-digested genomic DNA of P. stutzeri by using the vectors pJA1 (12) and SuperCos1 (Stratagene). An 892-bp fragment of cosmid c164 with the complete narL gene (22) was amplified with the primers 5′-GCAAAGCTTCGGCCTGAAGAACAGCG-3′ and 5′-TCGAATTCTTGAGCGATTGCGCACAG-3′. The primers added HindIII and EcoRI restriction sites (nucleotides in boldface) to allow cloning into pUC18. The pUC derivative with the narL gene was designated pUCnarL. The kanamycin resistance (Kmr) cassette was excised from plasmid pBSL15 (2) and used to replace a 358-bp AvaI-HincII fragment in narL, resulting in vector pUCnarL::Kmr. The narX-carrying plasmid pBSXL was constructed by inserting a 2.4-kb AvaI fragment of the narX cosmid g279 into pBluescriptII SK(−). The narX locus was mutated by replacing a 520-bp internal Eco47III-HindIII fragment with the Kmr cassette from pBSL15 to give plasmid pBSXL::Kmr. Plasmids pUCnarL::Kmr and pBSXL::Kmr were transferred to MK21 by electroporation (Gene Pulser; Bio-Rad). The narL mutant MRL118 and the narX mutant MRX119, resulting from double-crossover events, were obtained by selection for kanamycin resistance and ampicillin sensitivity. Mutational inactivation of the genes was verified by sequencing and Southern hybridization. MRL118 was negative with the AvaI-HincII narL fragment as a probe and gave a single hybridizing 2.8-kb fragment with the Kmr probe in a genomic PstI digest. The mutated 3.1-kb narX fragment (wild-type size, 2.4 kb) was detected in a genomic AvaI digest of DNA from MRX119 by hybridization, using a 153-bp PCR fragment generated from the 3′ end of the gene as the probe (primer pair 5′-CAAGCATGCGGAAGCGAACC-3′ and 5′-GGCGCGTTCTTGCAGG-3′).
RESULTS
nirS is part of an operon.
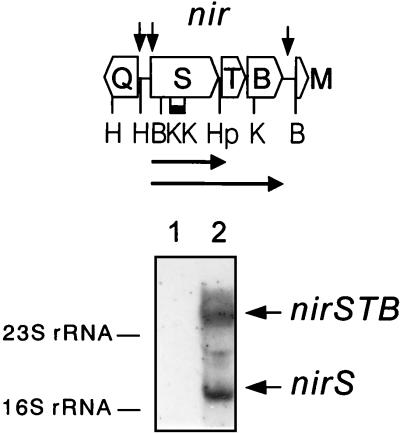
We identified the nirS transcript in the total-RNA fraction of cells grown under denitrifying conditions to monitor the expression pattern of nitrite reductase at the mRNA level. A 0.5-kb KpnI fragment from the central region of nirS was used as hybridization probe (Fig. 1). In Northern blot analyses, two transcripts, of 2 and 3.4 kb, were found which were absent from aerobic cells. We interpret them to be the monocistronic transcript of the nirS gene and the polycistronic message from the nirSTB operon. Transcription upstream of nirS, beginning with nirQ, proceeds in the opposite direction (Fig. 1); thus, the large transcript cannot originate from upstream genes. The monocistronic transcript may result from early termination of transcription or from posttranscriptional processing of the polycistronic message. Several inverted repeats, possible stem-loop-forming mRNA structures, at the 3′ ends of nirS and nirB (23) are candidates for affecting transcription and/or processing. The resulting increase of the nirS transcript relative to that of nirTB may be necessary to control the amounts of NirS, NirT, and NirB proteins so that they are in the appropriate balance for the denitrification process. The genes nirT and nirB encode tetraheme and diheme c-type cytochromes, respectively. Mutagenesis of nirT demonstrated the requirement of the encoded heme protein for an in vivo-functional nitrite-reducing system (23). NirT is similar to the NapC proteins encoding the putative electron-transferring c-type cytochromes of periplasmic nitrate reductases (33). Homologs of NirT have also been found in nondenitrifying bacteria and ascribe a broader significance to this protein (11, 13, 18, 27). Transcription of nirB together with nirS indirectly provides support for a role for the encoded diheme protein in nitrite reduction. A proteolytically shortened NirB was shown to have peroxidase activity (17).
FIG. 1.
nirS is transcribed as an operon with two downstream nir genes. Shown at the top are the physical map and organization of the nir operon of P. stutzeri. The horizontal arrows indicate the extents of the two nirS transcripts. The black bar shows the position of the 0.5-kb KpnI fragment used as the nirS probe in Northern blotting. Vertical arrows point to the location of FNR boxes. Restriction site abbreviation are as follows: B, BamHI; H, HindIII; Hp, HpaI; and K, KpnI. At the bottom of the figure is a Northern blot of 20-μg quantities of total RNA from cells aerobically cultivated without nitrate addition (lane 1) and from nitrate-denitrifying cells (lane 2). Hybridization was done with the digoxigenin-labeled KpnI fragment. The 2.0-kb monocistronic nirS transcript and the 3.4-kb transcript of the nirSTB operon are marked by arrows; the 16S and 23S rRNA species served as standards.
Mapping the nirS promoter.
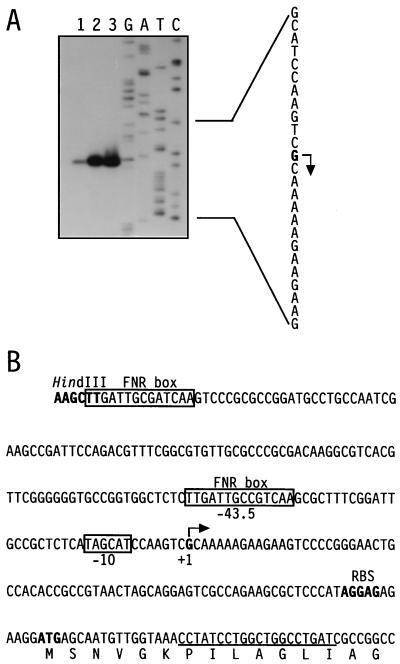
The promoter of nirS was mapped by primer extension analysis (Fig. 2). The sequence TAGCAT at position −10 shows similarity to sigma factor ς70-dependent promoters. A potential FNR-binding site, TTGAT-N4-GTCAA, that is almost identical to the FNR-binding consensus sequence of E. coli is positioned at −43.5. FNR of E. coli binds to the partially palindromic nucleotide sequence TTGAT-N4-ATCAA, known as the FNR box, of FNR-activated promoters that is centered preferentially −41.5 bp from the start site of transcription (20). Expression of nirS depends on DnrD (formerly termed FnrD), which is a member of the greater FNR-CRP family of regulators (30). We presume that the FNR box of the nirS promoter is the target of DnrD-dependent regulation. The second FNR box, located further upstream, is considered to be required for the expression of nirQ. In Pseudomonas aeruginosa, this gene was shown to be expressed anaerobically and to possibly be regulated from the single FNR box shared by the nirS and nirQ genes (4).
FIG. 2.
The nirS promoter. (A) Primer extension analysis. Total RNA was extracted from cells cultured under nitrate-denitrifying conditions. The primer extension products from 10, 30, and 50 μg of RNA (lanes 1 to 3, respectively) were separated on a sequencing gel together with a sequence ladder generated with the primer shown in panel B. The complementary sequence of the transcription initiation site is represented; the 5′ end of the nirS transcript is indicated by the arrow. (B) The nirS promoter. The 5′ end of the transcript is labeled +1. Putative polymerase (−10) and FNR (−43.5) binding sites are boxed. The oligonucleotide that was used for primer extension is underlined. The first few N-terminal amino acids of the NirS protein are shown in one-letter code. RBS, ribosome-binding site.
Stability of transcripts.
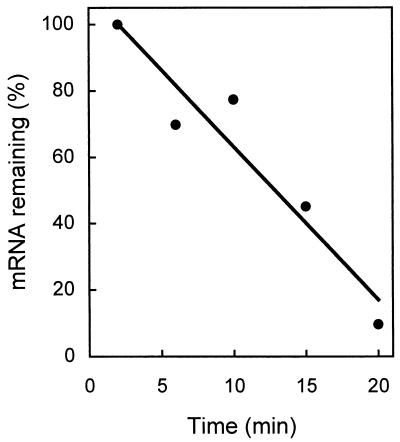
The balance between mRNA synthesis and decay has a profound effect on prokaryotic and eukaryotic gene expression (10). A fast rate of turnover of mRNA is crucial for a rapid response with a pattern of gene expression altered by changing external growth conditions. The half-life of the nirS transcript was determined with MK21 which had been induced for denitrification over a 30-min period. Rifampin (200 μg ml−1 final concentration) was then added to prevent de novo mRNA synthesis, and the amount of nirS transcript remaining was determined by Northern blot analysis. We determined a half-life of approximately 13 min for both the monocistronic and polycistronic nirS messages (Fig. 3). Also, the transcripts of norCB and nosZ were monitored under the same conditions. The genes norCB, encoding the NO reductase complex, form an operon with a single 2-kb transcript (35). nosZ, the N2O reductase structural gene, is transcribed from six promoters, but in each case as a monocistronic unit (14). The norCB and nosZ mRNAs each exhibited the same half-life as the nirS transcripts in the denitrifying cell (data not shown). Hence, a differential stability of transcripts is apparently not an element in the regulation of the structural genes encoding the oxidoreductases of denitrification.
FIG. 3.
Stability of nirS mRNA. A culture of P. stutzeri was induced for denitrification under O2-limited conditions (shaker speed, 120 rpm) with sodium nitrate (1 g liter−1). At 2, 10, 15, and 20 min after inhibition of transcription by addition of rifampin (200 μg μl−1 final concentration), total RNA was isolated and separated electrophoretically, and transcripts were detected by Northern blotting. Least-squares analysis gave a half-life of 12.6 min for the nirS mRNA.
The stability of mRNA can be affected by the growth rate (28). A half-life of 13 min was determined for mRNA in denitrifying cells with a doubling time of about 2.5 h. Overall, this qualifies the transcripts of denitrification genes as being of intermediate to high stability. Reported half-life values for bacterial mRNAs range from 0.5 to 50 min, typically being between 2 and 4 min (9). An extremely long half-life of 5 to 7 h has been reported for hoxS mRNA (encoding a soluble hydrogenase) of Ralstonia (Alcaligenes) eutropha cells exhibiting a doubling time of 20 h (29). ompA mRNA, encoding a porin, is another example of a rather stable mRNA, with a half-life of 13 to 25 min in cells doubling every 40 min. However, this mRNA species becomes short-lived in slow-growing cells (28).
Temporal changes of nirS mRNA in the transition to denitrification.
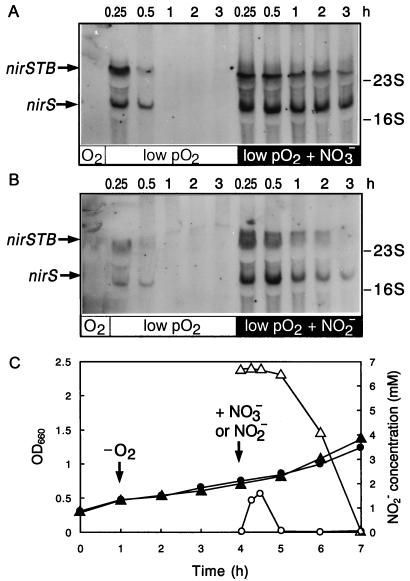
The time course of expression of nirS in response to nitrate and nitrite was monitored by mRNA analysis during the shift from aerobiosis to oxygen-limited and denitrifying conditions. We have previously shown that complete anaerobiosis is not required for expression of the denitrification system of P. stutzeri (25). The denitrification reductases appear when the level of saturation of the medium with air falls below 17% and an N-oxide is present. In a well-aerated culture (shaker speed, 240 rpm), no transcripts from the nirS operon were detected (Fig. 4). When the oxygen tension was subsequently lowered by reducing the shaker speed to 120 rpm and excluding the N-oxide, both nirS transcripts appeared within 15 min. At about 30 min after the onset of oxygen limitation, the nirS transcripts had nearly disappeared again, and they were not detected at subsequent sampling points extending to 3 h (Fig. 4A). This shows that although oxygen limitation alone causes a transient induction of nirS in P. stutzeri, it is not sufficient for long-term induction in the absence of an N-oxide. It is important to note that on addition of nitrate or nitrite to well-aerated cells, no activation of nirS transcription took place (data not shown).
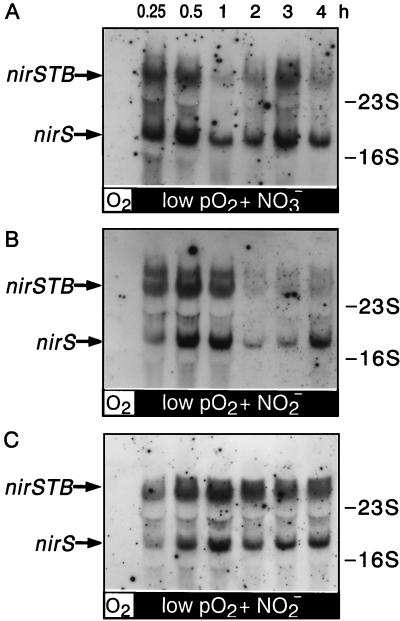
FIG. 4.
Kinetics of nirS induction by nitrate or nitrite under low-oxygen-tension conditions. MK21 was grown aerobically (shaker speed, 240 rpm) and checked for the absence of nirS mRNA (left lanes, panels A and B). (A) The culture was shifted first to low-oxygen-tension conditions (shaker speed, 120 rpm; lanes labeled low pO2 in panels A and B) and then induced for denitrification by addition of sodium nitrate (1 g liter−1). (B) As in panel A, but induction was with sodium nitrite (0.5 g liter−1). At the indicated time intervals after shifting to low-pO2 conditions and adding the respiratory substrate, the appearance of nirS transcripts was monitored by Northern blot analysis. (C) Increases in cell mass, as determined by measurement of optical densities at 600 nm (closed symbols), and changes in nitrite concentrations (open symbols) were monitored in both experiments; data were plotted on the absolute time scale. Circles correspond to panel A; triangles correspond to panel B. Vertical arrows point to the effected changes in growth conditions.
Addition of nitrate to oxygen-limited cells rapidly induced nirS transcription and cytochrome cd1 synthesis (Fig. 4A). A short period of accumulation of nitrite in the culture medium accompanied the transition phase (Fig. 4C); this can be rationalized from the sequential induction of nitrate reduction and nitrite reduction (8, 24, 25). The level of nirS mRNA reached a maximum within 15 min and remained constant during the 3-h period of observation (Fig. 4A). Since the half-life of nirS mRNA is about 13 min, persistence of those transcripts for hours can occur only if the nirS operon is continuously activated. We repeated the experiment using nitrite instead of nitrate and found the same temporal response of the nirS transcripts (Fig. 4B). Toward the end of this experiment, the amount of nirS mRNA was decreased in the 3-h measurement, which coincided with the disappearance of nitrite (Fig. 4C). In parallel experiments lasting up to 5 h, we correlated the disappearance of the nirS transcripts with exhaustion of nitrite in the medium, i.e., lack of the inducer. The amount of nir message decreased within the time frame of the mRNA half-life.
We have performed the oxygen respiration-to-denitrification shift experiment in an identical manner with the probes for norCB and nosZ. The induction patterns observed for norCB and nosZ were the same as those seen for the nirS operon (data not shown). In both cases there was a short period of gene activation in response to lowering of the pO2, whereas continuing transcription of norCB and nosZ again required the presence of nitrate or nitrite. Within the limits of the temporal resolution obtained with 15-min intervals of the initial sampling points, coordinate expression of nirS, norCB, and nosZ was observed. This is in agreement with the results from a previous immunochemical study (24).
Evidence for a regulatory system other than NarXL that mediates nitrate and nitrite induction of denitrification.
We have recently found that P. stutzeri possesses a nitrate response regulator, NarL. This regulator acts in concert with the sensor NarX as part of a two-component system that mediates nitrate and nitrite response in the expression of respiratory nitrate reductase (22). The narL and narX genes were mutated to generate the deletion strains MRL118 and MRX119, respectively, as described in Materials and Methods. Under aerobic growth conditions, no nirS transcripts were detected, but under conditions of oxygen limitation, transcription of nirS was induced by nitrate or nitrite in both mutants (Fig. 5A and B). The nitrate-challenged narL strain MRL118 induced nirS mRNA within 15 min, and this mRNA then remained present at approximately the same level during the observation period of 4 h. The same result was obtained when the mutant was challenged with nitrite. The activation kinetics of nirS transcription in the narL mutant in response to nitrate or nitrite were indistinguishable from that of the wild type. Transcription of the nirS operon in the narX mutant MRX119 in response to nitrate and nitrite exhibited again the same kinetic pattern as the narL mutant or the wild type. Figure 5C shows exemplarily the activation of nirS transcription by nitrite in an experiment involving a shift from aerobic respiration to denitrification. Also, the expression of norCB and nosZ was not altered in the mutants MRL118 and MRX119. Since neither narX nor narL inactivation affected expression of the reductases involved in denitrification in the strict sense, we postulate the existence of one or more signal transduction pathways triggered by nitrate or nitrite, independent of the two-component sensor regulator system NarXL.
FIG. 5.
Nitrate- or nitrite-induced nirS expression is independent of NarL or NarX. The nitrate response regulator mutant MRL118(ΔnarL) (A and B) and the nitrate sensor mutant MRX119(ΔnarX) (C) were grown aerobically (shaker speed, 240 rpm), checked for the absence of nirS transcripts by Northern blotting (data points labeled O2), and then induced for denitrification by a shift to low pO2 (shaker speed, 120 rpm) and the addition of nitrate (1 g of NaNO3 per liter) (A) or nitrite (0.5 g of NaNO2 per liter) (B and C). The production of transcripts were monitored in each case for 4 h after the shift.
DISCUSSION
As shown in Fig. 4, oxygen withdrawal alone caused only a transient effect on the nirS operon, whereas its continuing expression and likewise that of the norCB and nosZ genes, required activation by nitrate or nitrite. Transcriptional activation of denitrification genes induced by low-oxygen conditions cannot as yet be ascribed to a distinct regulator in P. stutzeri. Although FnrA, an FNR-type regulator, of this bacterium has been found earlier (15), it does not act as a global regulator, which would induce the structural genes for denitrification reductases in response to oxygen withdrawal, nor does it act on DnrD, which is also necessary for the expression of nirS and norCB (30). We presume that the FNR box of the nirS promoter (Fig. 2) is the target of DnrD. The dnrD gene itself is part of an operon that shows a complex pattern of transcriptional response to oxygen and nitrate (30). DnrD and its homologs from other bacteria belong into a separate phylogenetic branch within the greater FNR family. This group is unlikely to comprise oxygen-responsive elements, since all lack the cysteine residues needed for complexing the 4Fe-4S clusters of redox-active FNR proteins. In this respect, the situation in P. stutzeri is clearly different from that in P. aeruginosa, for which the FNR-type regulator ANR was suggested to respond to oxygen and function in a hierarchical relationship with the DNR regulator, targeting both nirS and norCB (5).
The observations made in the studies with the narX and narL mutants provide evidence for a pathway of nitrate and nitrite regulation which we ascribe to a new regulatory circuit. Since inactivation of narX did not produce a phenotype with respect to the induction of the denitrification genes proper, it is clear that NarX functions independently from this system; i.e., the latter has a nitrate- and nitrite-sensory element distinct from NarX. The genetic organization of narXL is suggestive of an operon structure (22), which means that the narX mutation is polar on narL, but this will not affect the conclusions noted above. Further work is needed to determine whether the postulated new system belongs to the two-component paradigm, which may comprise a system homologous to NarXL, such as NarQP of E. coli (16), but specific for denitrification, or whether it comprises a novel type of nitrate- and nitrite-responsive system.
In an immunochemical study, we have previously found that the mutant strain MK137, which lacks nitrate metabolism, requires nitrite for full induction of cytochrome cd1. This observation suggests the presence of a nitrite-responsive element (34), which we can now attribute to the new regulatory system. The activator component of this system may directly recognize a promoter element of nirS or act indirectly via a further transcriptional regulator. Within this context, it is of interest that the nirS promoter of P. aeruginosa was demonstrated to be nitrite responsive by the use of lacZ fusions (3). The regulator for this effect and the cognate promoter element, however, were not identified. Experiments to isolate and characterize the genes encoding the nitrate- and nitrite-responsive components necessary for denitrification in P. stutzeri are in progress.
ACKNOWLEDGMENTS
We thank M. F. Alexeyev for kindly supplying plasmid pBSL15 and U. Schiek for providing a gene bank and the narL mutant.
The work was supported by the Deutsche Forschungsgemeinschaft and Fonds der Chemischen Industrie.
REFERENCES
- 1.Aiba H, Adhya S, de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli. J Biol Chem. 1981;256:11905–11910. [PubMed] [Google Scholar]
- 2.Alexeyev M F. Three kanamycin resistance gene cassettes with different polylinkers. BioTechniques. 1995;18:52–55. [PubMed] [Google Scholar]
- 3.Arai H, Igarashi Y, Kodama T. Nitrite activates the transcription of the Pseudomonas aeruginosa nitrite reductase and cytochrome c-551 operon under anaerobic conditions. FEBS Lett. 1991;288:227–228. doi: 10.1016/0014-5793(91)81040-f. [DOI] [PubMed] [Google Scholar]
- 4.Arai H, Igarashi Y, Kodama T. Structure and ANR-dependent transcription of the nir genes for denitrification from Pseudomonas aeruginosa. Biosci Biotechnol Biochem. 1994;58:1286–1291. doi: 10.1271/bbb.58.1286. [DOI] [PubMed] [Google Scholar]
- 5.Arai H, Kodama T, Igarashi Y. Cascade regulation of the two CRP/FNR-related transcriptional regulators (ANR and DNR) and the denitrification enzymes in Pseudomonas aeruginosa. Mol Microbiol. 1997;25:1141–1148. doi: 10.1046/j.1365-2958.1997.5431906.x. [DOI] [PubMed] [Google Scholar]
- 6.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1995. [Google Scholar]
- 7.Baikalov I, Schröder I, Kaczor-Grzeskowiak M, Grzeskowiak K, Gunsalus R P, Dickerson R E. Structure of the Escherichia coli response regulator NarL. Biochemistry. 1996;35:11053–11061. doi: 10.1021/bi960919o. [DOI] [PubMed] [Google Scholar]
- 8.Baumann B, Snozzi M, Zehnder A J B, van der Meer J R. Dynamics of denitrification activity of Paracoccus denitrificans in continuous culture during aerobic-anaerobic changes. J Bacteriol. 1996;178:4367–4374. doi: 10.1128/jb.178.15.4367-4374.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belasco J G. mRNA degradation in prokaryotic cells: an overview. In: Belasco J G, Brawerman G, editors. Control of messenger RNA stability. San Diego, Calif: Academic Press, Inc.; 1993. pp. 3–12. [Google Scholar]
- 10.Belasco J G, Higgins C F. Mechanisms of mRNA decay in bacteria: a perspective. Gene. 1988;72:15–23. doi: 10.1016/0378-1119(88)90123-0. [DOI] [PubMed] [Google Scholar]
- 11.Bergmann D J, Arciero D M, Hooper A B. Organization of the hao gene cluster of Nitrosomonas europaea: genes for two tetraheme c cytochromes. J Bacteriol. 1994;176:3148–3153. doi: 10.1128/jb.176.11.3148-3153.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braun C, Zumft W G. The structural genes of the nitric oxide reductase complex from Pseudomonas stutzeri are part of a 30-kilobase gene cluster for denitrification. J Bacteriol. 1992;174:2394–2397. doi: 10.1128/jb.174.7.2394-2397.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Camilli A, Mekalanos J J. Use of recombinase fusions to identify Vibrio cholerae genes induced during infection. Mol Microbiol. 1995;18:671–683. doi: 10.1111/j.1365-2958.1995.mmi_18040671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cuypers H, Berghöfer J, Zumft W G. Multiple nosZ promoters and anaerobic expression of nos genes necessary for Pseudomonas stutzeri nitrous oxide reductase and assembly of its copper centers. Biochim Biophys Acta. 1995;1264:183–190. doi: 10.1016/0167-4781(95)00128-4. [DOI] [PubMed] [Google Scholar]
- 15.Cuypers H, Zumft W G. Anaerobic control of denitrification in Pseudomonas stutzeri escapes mutagenesis of an fnr-like gene. J Bacteriol. 1993;175:7236–7246. doi: 10.1128/jb.175.22.7236-7246.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darwin A J, Stewart V. The NAR modulon systems: nitrate and nitrite regulation of anaerobic gene expression. In: Lin E C C, Lynch A S, editors. Regulation of gene expression in Escherichia coli. New York, N.Y: Chapman & Hall; 1996. pp. 343–359. [Google Scholar]
- 17.Denariaz C M, Liu M-Y, Payne W J, LeGall J, Marquez L, Dunford H B, van Beeumen J. Cytochrome c peroxidase activity of a protease-modified form of cytochrome c-552 from the denitrifying bacterium Pseudomonas perfectomarina. Arch Biochem Biophys. 1989;270:114–125. doi: 10.1016/0003-9861(89)90013-1. [DOI] [PubMed] [Google Scholar]
- 18.Dolata M M, van Beeumen J J, Ambler R P, Meyer T E, Cusanovich M A. Nucleotide sequence of the heme subunit of flavocytochrome c from the purple phototrophic bacterium, Chromatium vinosum. A 2.6-kilobase pair DNA fragment contains two multiheme cytochromes, a flavoprotein, and a homolog of human ankyrin. J Biol Chem. 1993;268:14426–14431. [PubMed] [Google Scholar]
- 19.Gennis R B, Stewart V. Respiration. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: ASM Press; 1996. pp. 217–261. [Google Scholar]
- 20.Guest J R, Green J, Irvine A S, Spiro S. The FNR modulon and FNR-regulated gene expression. In: Lin E C C, Lynch A S, editors. Regulation of gene expression in Escherichia coli. New York, N.Y: Chapman & Hall; 1996. pp. 317–342. [Google Scholar]
- 21.Härtig E, Zumft W G. Nitrate and nitrite response of denitrification genes studied by mRNA analysis. BIOspektrum. 1996;2:88. [Google Scholar]
- 22.Härtig E H, Zumft W G. Respiratory nitrate reductase of Pseudomonas stutzeri is regulated by a two-component system, NarXL, that is ineffective in nitrate control of denitrification sensu stricto. BIOspektrum. 1998;4:51. [Google Scholar]
- 23.Jüngst A, Wakabayashi S, Matsubara H, Zumft W G. The nirSTBM region coding for cytochrome cd1-dependent nitrite respiration of Pseudomonas stutzeri consists of a cluster of mono-, di-, and tetraheme proteins. FEBS Lett. 1991;279:205–209. doi: 10.1016/0014-5793(91)80150-2. [DOI] [PubMed] [Google Scholar]
- 24.Körner H. Anaerobic expression of nitric oxide reductase from denitrifying Pseudomonas stutzeri. Arch Microbiol. 1993;159:410–416. [Google Scholar]
- 25.Körner H, Zumft W G. Expression of denitrification enzymes in response to the dissolved oxygen level and respiratory substrate in continuous culture of Pseudomonas stutzeri. Appl Environ Microbiol. 1989;55:1670–1676. doi: 10.1128/aem.55.7.1670-1676.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McMaster G K, Carmichael G G. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc Natl Acad Sci USA. 1977;74:4835–4838. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Méjean V, Iobbi-Nivol C, Lepelletier M, Giordano G, Chippaux M, Pascal M-C. TMAO anaerobic respiration in Escherichia coli: involvement of the tor operon. Mol Microbiol. 1994;11:1169–1179. doi: 10.1111/j.1365-2958.1994.tb00393.x. [DOI] [PubMed] [Google Scholar]
- 28.Nilson G, Belasco J G, Cohen N S, von Gabain A. Growth-rate dependent regulation of mRNA stability in Escherichia coli. Nature. 1984;312:75–77. doi: 10.1038/312075a0. [DOI] [PubMed] [Google Scholar]
- 29.Oelmüller U, Schlegel H G, Friedrich C G. Differential stability of mRNA species of Alcaligenes eutrophus soluble and particulate hydrogenases. J Bacteriol. 1990;172:7057–7064. doi: 10.1128/jb.172.12.7057-7064.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vollack, K.-U., E. Härtig, H. Körner, and W. G. Zumft. Multiplicity of transcription factors of the FNR family in denitrifying Pseudomonas stutzeri: characterization of four fnr-like genes, regulatory responses, and cognate metabolic processes. Submitted for publication. [DOI] [PubMed]
- 31.Vollack K-U, Xie J, Härtig E, Römling U, Zumft W G. Localization of denitrification genes on the chromosomal map of Pseudomonas aeruginosa. Microbiology. 1998;144:441–448. doi: 10.1099/00221287-144-2-441. [DOI] [PubMed] [Google Scholar]
- 32.Werner W. Nachweis von Nitrit und Nitrat über die Bildung von Azofarbstoffen. Fresenius Z Anal Chem. 1980;304:117–124. [Google Scholar]
- 33.Zumft W G. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev. 1997;61:533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zumft W G, Blümle S, Braun C, Körner H. Chlorate resistant mutants of Pseudomonas stutzeri affected in respiratory and assimilatory nitrate utilization and expression of cytochrome cd1. FEMS Microbiol Lett. 1992;91:153–158. [Google Scholar]
- 35.Zumft W G, Braun C, Cuypers H. Nitric oxide reductase from Pseudomonas stutzeri: primary structure and gene organization of a novel bacterial cytochrome bc complex. Eur J Biochem. 1994;219:481–490. doi: 10.1111/j.1432-1033.1994.tb19962.x. [DOI] [PubMed] [Google Scholar]
- 36.Zumft W G, Döhler K, Körner H, Löchelt S, Viebrock A, Frunzke K. Defects in cytochrome cd1-dependent nitrite respiration of transposon Tn5-induced mutants from Pseudomonas stutzeri. Arch Microbiol. 1988;149:492–498. doi: 10.1007/BF00446750. [DOI] [PubMed] [Google Scholar]