Abstract
Cell division in prokaryotes is mediated by the septal ring. In Escherichia coli, this organelle consists of several essential division proteins, including FtsZ, FtsA, and ZipA. To gain more insight into how the structure is assembled, we studied the interdependence of FtsZ, FtsA, and ZipA localization using both immunofluorescence and Gfp tagging techniques. To this end, we constructed a set of strains allowing us to determine the cellular location of each of these three proteins in cells from which one of the other two had been specifically depleted. Our results show that ZipA fails to accumulate in a ring shape in the absence of FtsZ. Conversely, depletion of ZipA does not abolish formation of FtsZ rings but leads to a significant reduction in the number of rings per unit of cell mass. In addition, ZipA does not appear to require FtsA for assembly into the septal ring and vice versa. It is suggested that septal ring formation starts by assembly of the FtsZ ring, after which ZipA and FtsA join this structure in a mutually independent fashion through direct interactions with the FtsZ protein.
Cell division in bacteria occurs by the coordinated invagination of the cell envelope layers that make up the cell wall. Work in the past few years has revealed that this process is mediated by a membrane-associated, cytoskeleton-like organelle which assembles at the prospective division site before the onset of septal invagination and which remains associated with the ingrowing cell wall until septal closure (24, 32). In Escherichia coli, septum formation requires at least nine gene products, FtsA, -I, -K, -L, -N, -Q, -W, -Z, and ZipA, which are specifically dedicated to this process. All these essential division proteins have recently been shown to be part of the septal ring structure in E. coli (2–4, 17, 25, 35, 39, 40) with the exception of FtsL and -Q. A distant homolog of FtsQ (DivIB), however, was recently localized to the septal ring of Bacillus subtilis (19), and it may be expected that FtsQ, as well as FtsL, is associated with the organelle during some stage of the division cycle in E. coli.
The phylogenetically highly conserved FtsZ protein is a major component of the septal ring and plays a key role in the division process. The protein is a tubulin-like GTPase which can form large protofilament-like polymers in vitro (9, 15, 23, 26–28, 31, 38, 41). In vivo, FtsZ moves from the cytoplasm to accumulate in a ring-like structure at the prospective division site early in the division cycle (4). Based on the in vitro properties of FtsZ, this ring is thought to be formed by a self-assembly reaction and to consist of a homopolymeric form of the protein. How formation of the FtsZ ring is initiated and by what mechanism it is normally restricted to certain sites on the cell envelope remains unclear. It is conceivable, however, that it involves the interaction of FtsZ with a hypothetical membrane factor (factor X) which marks potential division sites and stimulates polymerization of the protein at these sites.
We recently used an in vitro assay to search for factors in E. coli which interact directly with the FtsZ protein. This approach led to the discovery of a previously unknown division protein, which we called ZipA (17). ZipA is essential for cell division, and our results indicate not only that ZipA and FtsZ associate with each other in vitro but that this interaction also occurs in vivo and is required for cell constriction. Evidence suggests that ZipA is a bitopic integral membrane protein of type Ib of which the N terminus traverses the inner membrane once and of which the rest of the protein resides in the cytoplasm. Furthermore, localization studies indicate that ZipA and FtsZ interact primarily within the septal ring structure. Thus, like FtsZ, a ZipA-Gfp fusion protein was found to accumulate very early in the division cycle in a ring structure at the prospective division site and to remain associated with the invaginating septum during the cell constriction process (17).
In addition to its interaction with ZipA, recent two-hybrid experiments (14, 37) as well as localization studies (3, 25) have indicated that FtsZ also directly interacts with the FtsA protein, supporting previous genetic experiments suggesting such an interaction (8, 13). FtsA is a phosphoprotein which is peripherally associated with the membrane. It appears to belong to a family of ATPases which also includes actin, sugar kinases, and Hsp70 proteins (5, 33), but its role in the division process is presently not understood.
The observations that ZipA is an early component of the septal ring and that it binds to both FtsZ and the cytoplasmic membrane raise the possibility that the protein is directly involved in the assembly of FtsZ and/or other components, such as FtsA, into the septal ring organelle. In assessing the possible roles of ZipA in the division process, we have used both indirect immunofluorescence and Gfp tagging techniques to study the interdependence of ZipA, FtsZ, and FtsA localization. To this end, we constructed a set of strains allowing us to determine the cellular location of each of these three proteins in cells from which one of the other two had been specifically depleted.
Our results show that ZipA fails to accumulate in a ring shape in the absence of FtsZ. Conversely, depletion of ZipA does not abolish formation of FtsZ rings but leads to a significant reduction in the number of rings per unit of cell mass. In addition, ZipA does not appear to require FtsA for assembly into the septal ring and vice versa. It is suggested that septal ring formation starts by assembly of the FtsZ ring, after which ZipA and FtsA join this structure in a mutually independent fashion through direct interactions with the FtsZ protein.
MATERIALS AND METHODS
Strains.
Unless stated otherwise, cells were grown at 37°C in Luria-Bertani (LB) medium supplemented, where appropriate, with antibiotics at concentrations of 50 μg/ml (ampicillin, kanamycin, and spectinomycin), 25 μg/ml (chloramphenicol), and 12.5 μg/ml (tetracycline).
Strains GC13109 (his rpsL sulA366 leu::Tn10) (16), UT481 [met thy Δ(lac-pro) supD r− m+ Tn10/F′ traD36 proAB+ lacIq lacZ36ΔM15(Am)], PB103 (dadR trpE trpA tna), DX1 (dadR trpE trpA tna ΔminCDE recA::Tn10), CH3 (dadR trpE trpA tna recA::Tn10), CH5/pCH32 [dadR trpE trpA tna zipA::aph recA::Tn10/repA(Ts) ftsZ+ zipA+] (17), and PB143/pCX41 [dadR trpE trpA tna ftsZ0/repA(Ts) ftsZ+] (30) have been described previously. Strain BL21(λDE3)/plysS [ompT rB mB (PlacUV5::T7gene1)/T7lysS+)] was purchased from Novagen.
Cells used for the protein localization studies were all from PB103 or derivative strains, most of which contained a plasmid and/or a lysogenic phage. For clarity, the relevant genotypes of the host, plasmid, and phage components of these strains are listed separately in Table 1.
TABLE 1.
Hosts, plasmids, and phages used for localization studies
| Host, plasmid, or phage | Relevant genotype | Source or reference |
|---|---|---|
| Hosts | ||
| PB103 | dadR trpE trpA tna | 11 |
| CH3 | PB103 recA::Tn10 | 17 |
| PB143 | PB103 ftsZ0 recA::Tn10 | 30 |
| CH2 | PB103 ftsA0 recA::Tn10 | This work |
| CH5 | PB103 zipA::aph recA::Tn10 | 17 |
| Plasmids | ||
| pCX41 | CamrrepA(Ts) ftsZ+ | 36 |
| pDB280 | CamrrepA(Ts) ftsA+ | This work |
| pCH32 | aadA+repA(Ts) ftsZ+ zipA+ | 17 |
| pDB346 | aadA+repA+cI857(ts) PλR::ftsZ+ | 30 |
| pDB355 | aadA+repA+cI857(ts) PλR::ftsA+ | This work |
| pDB361 | aadA+ repA+cI857(ts) PλR::zipA+ | This work |
| Phages | ||
| λDR120 | imm21 bla+ lacIq+ Plac::gfp-ftsZ | This work |
| λCH75 | imm21 bla+ lacIq+ Plac::gfp-ftsA | This work |
| λCH50 | imm21 bla+ lacIq+ Plac::zipA-gfp | 17 |
To obtain CH5/pDB361, strain CH5/pCH32 was first transformed with pDB322 (Plac::zipA) (17) at 42°C in the presence of 5 μM IPTG (isopropyl-β-d-thiogalactopyranoside) selecting for ampicillin resistance (Apr), spectinomycin sensitivity (Specs) (indicating loss of pCH32), and an IPTG-dependent division phenotype. CH5/pDB322 was next transformed with pDB361 at 42°C, selecting for Specr (aadA+), Aps (loss of pDB322), and a cold sensitive (cs) division phenotype. Strain PB143/pDB346 was obtained by transformation of PB143/pCX41 with pDB346 at 42°C, and selection for aadA+, chloramphenicol sensitivity (Cams) (loss of pCX41), and a cs division phenotype. Host CH2 was obtained after several steps. First, the ftsA0 allele from plasmid pDB283 was crossed into the chromosome of strain UT481 by the method of Hamilton et al. (18) with selection for a temperature-sensitive (ts) division phenotype, yielding strain PB131/pDB280. In addition, we replaced the leu+ allele of PB103 with leu::Tn10 from GC13109 by P1-mediated transduction, yielding strain PB135. PB135 was transformed with pDB280, and strain CH1/pDB280 was obtained by P1-mediated transduction of the ftsA0 allele of PB131/pDB280 into the chromosome of PB135/pDB280, selecting for leu+ and ts cell division. Strain CH2/pDB280 was finally constructed by transduction of recA::Tn10 from DX1 to CH1/pDB280. To obtain CH2/pDB355, CH2/pDB280 was first transformed with a plasmid which expresses ftsA under control of the lac promoter (pDB297 [not shown]) and cured of pDB280 by growth at 37°C, selecting for Apr, Cams (loss of pDB280), and an IPTG-sensitive division phenotype. CH2/pDB297 was next transformed with pDB355 at 37°C and cured of pDB297, selecting for aadA+, Aps (loss of pDB297), and cs cell division. Where relevant, strains were lysogenized with λCH50, λCH75, or λDR120 as described previously (10).
Plasmids and phages.
Relevant features of the plasmids and phages used in this study are presented in Fig. 1 and Table 1. Plasmids pCX41, pDB280, pDB346, pDB355, pDB361, and pCH32 are all derivatives of pSC101. Plasmids pCX41 (36) and pCH32 (17) were described previously. Plasmid pDB280 was obtained by inserting a fragment containing the complete ftsA and partial ftsQ and ftsZ genes into pMAK705 (18). To create the ftsA0 allele, pDB280 was cut with BglII, and the ends were filled in using Klenow fragment. Religation yielded pDB283 and resulted in the replacement of the unique BglII site in ftsA with a ClaI site and a frameshift mutation in the ftsA open reading frame.
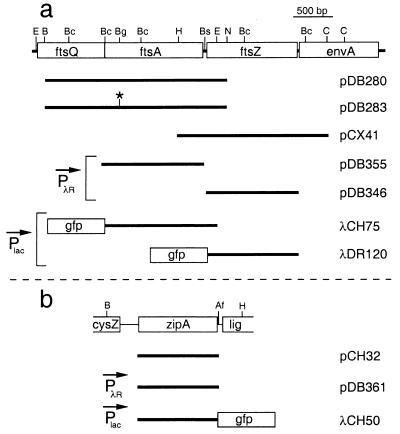
FIG. 1.
Chromosomal inserts of plasmids and phages. The physical maps of ftsA and ftsZ (a) and of zipA (b) are shown at the top of the panel. The positions of AflII (Af), BamHI (B), BclI (Bc), BglII (Bg), Bsu36I (Bs), ClaI (C), EcoRI (E), HindIII (H), and NsiI (N) restriction sites are indicated. Inserts of plasmids and phages are presented below each map. As indicated, the transcription of inserts in some of the constructs was placed under control of the lac promoter (Plac), or PλR. Note that in addition to the zipA fragment shown, pCH32 carries the ftsZ gene immediately downstream of this fragment (17).
To be able to express genes in a cs manner, we created vector pDB344, which was obtained by inserting a fragment of pXX747 (21), containing the rightward promoter of phage λ (PλR) as well as the ts allele of λ repressor (cI857) into pZC100 (42). Plasmid pDB344 expresses cI857 in the same orientation as the aadA gene, whereas PλR, which is located upstream of a number of unique restriction sites, directs transcription in the opposite orientation. Plasmids pDB346 (30), pDB355, and pDB361 were created by inserting a promoterless ftsZ, ftsA, and zipA fragment, respectively, downstream of PλR. Care was taken not to create fusions with the λ cro gene, a small portion of which was still present on pDB344.
Plasmids pDB273 and pCH15 are derivatives of pET21 (Novagen) and were used in the production of FtsA- and ZipA-specific antisera, respectively (see below).
Phage λCH50 was described previously (17). To obtain λDR120 and λCH75, we constructed plasmid derivatives of pMLB1113 (pDR120 and pCH75, respectively) and crossed these with phage λNT5 as described previously (10). Phage λDR120 encodes a 68.6-kDa Gfp-FtsZ fusion protein, in which the complete FtsZ peptide is separated from the complete Gfpmut2 peptide (7) by the linker peptide ASMTGGQQMGRGSH. Phage λCH75 encodes a 73.1-kDa Gfp-FtsA fusion protein, in which the N-terminal methionine residue of FtsA is replaced with the complete Gfpmut2 peptide and the linker peptide ASMTGGQQMGR. Full details of the preparation of genetic constructs can be obtained directly from the authors.
Antisera.
Polyclonal anti-FtsZ antiserum was raised against native FtsZ protein, which had been purified as described previously (9). Anti-FtsA antiserum was raised against a 35.6-kDa FtsA-His fusion consisting of the N-terminal 319 amino acids (aa) of FtsA fused to the peptide AAALE(H)6. Synthesis of this protein was induced by growth of strain BL21(λDE3)/plysS/pDB273 in the presence of IPTG. Virtually all of the FtsA-His protein was present in inclusion bodies. Cells were broken in a French pressure cell, and inclusion bodies were collected by low-speed centrifugation. The pellet fraction was treated with 4.0 M guanidine-HCl, and solubilized material was fractionated by nickel affinity chromatography. Fractions highly enriched for FtsA-His were pooled and further purified on a MonoQ column by fast protein liquid chromatography in the presence of 4.0 M urea. Anti-ZipA antiserum was raised against a soluble 35.9-kDa His-ZipA fusion protein in which the first 38 aa of ZipA, including its transmembrane domain, were replaced by the peptide MG(H)10SSGHIEGRHMASMTGGQQMGRI. Synthesis of this fusion protein was induced by growth of strain BL21(λDE3)/plysS/pCH15 in the presence of IPTG. The protein was purified in the absence of chaotropic agents by nickel affinity chromatography followed by anion exchange chromatography on a MonoQ column. Polyclonal antisera against FtsZ, FtsA, and ZipA were obtained by immunizing rabbits (12). For immunofluorescence staining, the FtsA and ZipA antisera were further affinity purified as previously described (3). Anti-FtsZ monoclonal antibodies have been described (34).
HID of FtsZ, FtsA, and ZipA.
Cultures of heat-induced depletion (HID) strains PB143/pCX41, CH2/pDB280, and CH5/pCH32 and the wild-type control strain CH3/pMAK700 were grown overnight at 30°C in LB medium with appropriate antibiotics. Cultures were diluted 1:200 in fresh LB medium without antibiotics, incubated at 30°C for 1.5 h, and then shifted to the nonpermissive temperature (42°C) for plasmid replication for 3 to 4 h. Optical densities at 600 nm (OD600) were between 0.4 and 0.8.
CID of FtsZ, FtsA, and ZipA.
For depletion of FtsZ, the cold-induced depletion (CID) strain PB143/pDB346 was grown overnight at 42°C. Cultures were diluted to an OD600 of 0.02 in LB medium and grown at 42°C for 1 h. Cultures were shifted to 30°C and further incubated for approximately 8 h until an OD600 of 0.7 to 0.9 was reached. For depletion of FtsA, strain CH2/pDB355 was grown overnight at 37°C and diluted to a starting OD600 of 0.04. Cultures were incubated immediately at 30°C for approximately 8 h to an OD600 of 0.7 to 0.9. For depletion of ZipA, strain CH5/pDB361 was grown overnight at 42°C and diluted to an OD600 of 0.01. Cultures were incubated immediately at 30°C for approximately 10 h to an OD600 of 0.8 to 1.0. For quantitative immunoblotting assays (see below), the wild-type control strains CH3/pDB346, CH3/pDB355, and CH3/pDB361 were treated identically to PB143/pDB346, CH2/pDB355, and CH5/pDB361, respectively.
Immunoblotting.
Strains were grown as described above. Cells were harvested by centrifugation, resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) buffer (12) to the equivalent of 20.0 OD600 units per ml, and incubated at 100°C for 5 min. Total protein concentration in each lysate was determined by the method of Bradford using bovine immunoglobulin G (IgG) as a standard. Each SDS-PAGE gel contained 70 μg of a depleted extract in one lane and different amounts (70, 35, 18, 9, 4, and 2 μg) of the appropriate control extract in adjacent lanes. Western blotting and antigen detection with rabbit polyclonal anti-FtsZ, anti-FtsA, and anti-ZipA antiserum were performed essentially as described (12) except that goat anti-rabbit IgG conjugated to alkaline phosphatase was used as secondary antibody. Blots were digitized by optical scanning, and the integrated density of each band of interest was measured using NIH-Image 1.60 software. Values corresponding to the control lanes were used to prepare a standard curve, on the basis of which the value corresponding to the depleted extract was calculated.
Immunofluorescence microscopy.
The wild-type strain PB103 was grown in LB medium at 37°C to an OD600 of 0.6. The HID strains PB143/pCX41, CH2/pDB280, and CH5/pCH32 were grown at nonpermissive temperature as described above. Cells were prepared for immunofluorescent staining as described previously (3). The samples were incubated overnight at 4°C with either a 1:2,000 dilution of an anti-FtsZ mouse monoclonal antibody cocktail consisting of equimolar amounts of monoclonal antibodies 4, 11, and 12 (34), or a 1:25 or 1:100 dilution of affinity-purified rabbit polyclonal antibody to FtsA or ZipA, respectively. Cy3-conjugated secondary antibody (Sigma) was used at a 1:4,000 (anti-mouse IgG) or 1:2,000 (anti-rabbit IgG) dilution. Cells were viewed and digitally imaged as previously described (17), with the exception that 535- to 585-nm excitation and 595- to 670-nm barrier filters were used.
Gfp localization.
Lysogenic derivatives of CID strains PB143/pDB346, CH2/pDB355, and CH5/pDB361 were grown at nonpermissive temperature as described above for their nonlysogenic parents, except that IPTG was present during incubation at the nonpermissive temperature. Lysogenic derivatives of wild-type strains PB103 and CH3 were also grown in the presence of IPTG at 30°C to an OD600 of 0.8 to 1.0. IPTG was used at 25 μM (λCH50 and λCH75) or 50 μM (λDR120). Cells were chemically fixed, viewed, and imaged as previously described (17).
Quantitation of cell lengths and fluorescent rings.
From each field of cells we obtained two digital images using fluorescence and differential interference contrast optics, respectively (17). The first image was used to count the number of ring structures per cell and the second was used to determine cell length by using Segmented Ruler (Tim Rand, Yale University) software. In case only a portion of a long filament was captured within the field of view, cell segments larger than 30 μm were counted as one cell. In each case, such cell segments made up less than 30% of the number of cells scored, but we note that the average length values of the populations containing long filaments in Tables 2 and 3 should be considered minima.
TABLE 2.
Quantitation of cell length and fluorescent rings after HID of FtsZ, FtsA, or ZipAa
| Strain | Depleted protein | Antibody | R− cell result
|
R+ cell result
|
R− + R+ cell result
|
||||
|---|---|---|---|---|---|---|---|---|---|
| % (n) | Avg length (μm) (range) | % (n) | Avg length (μm) (range) | L/R (μm) (range)b | Rings/cell (range) | L/R (μm) | |||
| PB103 | NA | Anti-FtsZ | 29 (21) | 1.5 (1.3–1.8) | 71 (52) | 2.1 (1.4–2.8) | 2.1 (1.4–2.8) | 0.71 (0–1) | 2.7 |
| CH2/pDB280 | FtsA | Anti-FtsZ | 0 (0) | NA | 100 (27) | 29.7 (10.0–55.6) | 6.2 (4.6–10.1) | 4.78 (1–10) | 6.2 |
| CH5/pCH32 | ZipA | Anti-FtsZ | 28 (41) | 45.4 (10.3–75.9) | 72 (103) | 37.7 (10.1–83.2) | 17.2 (3.3–89.5) | 1.56 (0–5) | 25.5 |
| PB103 | NA | Anti-FtsA | 18 (14) | 1.6 (1.3–1.8) | 82 (63) | 2.3 (1.3–3.7) | 2.3 (1.3–3.7) | 0.82 (0–1) | 2.6 |
| PB143/pCX41 | FtsZ | Anti-FtsA | 96 (90) | 14.5 (2.4–39.2) | 4 (4) | 2.9 (2.5–3.7) | 2.9 (2.5–3.7) | 0.04 (0–1) | 329.8 |
| CH5/pCH32 | ZipA | Anti-FtsA | 21 (9) | 40.8 (9.1–65.4) | 79 (34) | 27.6 (8.6–90.8) | 12.0 (5.8–40.8) | 1.81 (0–5) | 16.7 |
| PB103 | NA | Anti-ZipA | 25 (14) | 1.7 (1.3–1.90) | 75 (41) | 2.3 (1.3–3.6) | 2.3 (1.3–3.6) | 0.75 (0–1) | 2.9 |
| PB143/pCX41 | FtsZ | Anti-ZipA | 96 (78) | 17.3 (2.3–53.4) | 4 (3) | 8.1 (2.2–18.8) | 8.1 (2.2–18.8) | 0.04 (0–1) | 457.6 |
| CH2/pDB280 | FtsA | Anti-ZipA | 2 (1) | 16.1 (NA) | 98 (40) | 24.6 (7.1–50.0) | 5.5 (3.9–21.5) | 4.34 (0–10) | 5.6 |
Cells were treated and analyzed as described in the text. Some parameters were calculated separately for cells without fluorescent ring structures (R− cells), cells with one or more rings (R+ cells), and all cells combined (R− + R+ cells). NA, not applicable.
L/R, ratio of the combined length to the number of rings.
TABLE 3.
Quantitation of cell length and fluorescent rings after CID of FtsZ, FtsA, and ZipAa
| Strain | Depleted protein | Marker | R− cell result
|
R+ cell result
|
R− + R+ cell result
|
||||
|---|---|---|---|---|---|---|---|---|---|
| % (n) | Avg length (μm) (range) | % (n) | Avg length (μm) (range) | L/R (μm) (range) | Rings/cell (range) | L/R (μm) | |||
| PB103(λDR120) | NA | Gfp-FtsZ | 12 (14) | 2.1 (1.7–3.1) | 88 (104) | 2.7 (1.6–4.4) | 2.7 (1.6–4.4) | 0.88 (0–1) | 3.0 |
| CH2(λDR120)/pDB355 | FtsA | Gfp-FtsZ | 7 (4) | 56.1 (18.7–98.9) | 93 (50) | 34.4 (11.9–58.1) | 6.3 (4.7–35.7) | 5.04 (0–11) | 7.2 |
| CH5(λDR120)/pDB361 | ZipA | Gfp-FtsZ | 19 (12) | 36.1 (16.8–63.4) | 81 (52) | 35.5 (5.7–85.5) | 8.6 (4.9–40.2) | 3.36 (0–9) | 10.6 |
| PB103(λCH75) | NA | Gfp-FtsA | 15 (20) | 2.3 (1.6–4.0) | 85 (114) | 2.9 (1.8–6.3) | 2.9 (1.8–6.3) | 0.85 (0–1) | 3.3 |
| PB143(λCH75)/pDB346 | FtsZ | Gfp-FtsA | 100 (56) | 36.7 (5.3–66.3) | 0 (0) | NA | NA | 0 (NA) | >2,054.7 |
| CH5(λCH75)/pDB361 | ZipA | Gfp-FtsA | 8 (7) | 35.4 (15.0–56.3) | 92 (79) | 43.0 (16.8–84.0) | 9.4 (6.1–64.9) | 4.20 (0–9) | 10.1 |
| PB103(λCH50) | NA | ZipA-Gfp | 14 (16) | 2.0 (1.5–3.9) | 86 (100) | 2.7 (1.5–4.2) | 2.7 (1.5–4.2) | 0.86 (0–1) | 3.0 |
| PB143(λCH50)/pDB346 | FtsZ | ZipA-Gfp | 95 (127) | 19.7 (1.9–62.5) | 5 (6) | 10.5 (2.6–26.9) | 10.5 (2.6–26.9) | 0.05 (0–1) | 427.3 |
| CH2(λCH50)/pDB355 | FtsA | ZipA-Gfp | 7 (4) | 32.9 (14.0–55.4) | 93 (50) | 30.7 (10.2–57.9) | 6.4 (3.5–20.0) | 4.43 (0–9) | 7.0 |
Cells were treated and analyzed as described in the text. Columns list the same parameters as those described in the footnotes to Table 2 except that the third column (Marker) indicates the Gfp fusion that was used to mark septal rings. NA, not applicable.
RESULTS
Localization of ZipA, FtsZ, and FtsA.
Assembly of ZipA, FtsZ, and FtsA into the septal ring was monitored by the application of both indirect immunofluorescence and Gfp tagging techniques.
Immunofluorescence microscopy was performed essentially as described previously (1). To detect FtsZ, we used a mixture of monoclonal antibodies kindly provided by Jan Voskuil (34). For detection of ZipA and FtsA, we used affinity-purified polyclonal antibodies. Using this technique, all three division proteins were found to localize to the septal ring in normally dividing cells (Fig. 2a to c). The specificity of each antibody preparation was confirmed by strongly diminished signals after depletion of the corresponding antigen from cells, as judged by both Western analyses and immunofluorescence microscopy (results not shown) (see below).
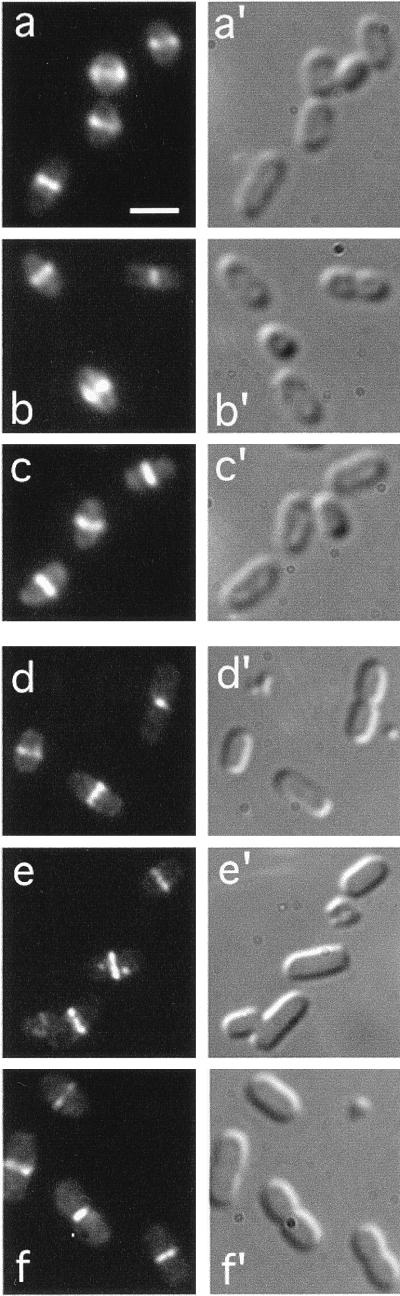
FIG. 2.
Localization of ZipA, FtsA, and FtsZ in wild-type cells. Fluorescence (a through f) and corresponding differential interference contrast (a′ through f′) micrographs showing the location in normally dividing cells of both native (a to c) and Gfp tagged (d to f) ZipA, FtsA, and FtsZ proteins, respectively. Cells shown are from strains PB103 (wild type [wt]) (a to c), CH3(λCH50)/pDB355 [wt(Plac::zipA-gfp)/cI857 PλR::ftsA+] (d), PB103(λCH75) [wt(Plac::gfp-ftsA)] (e), and CH3(λDR120)/pDB361 [wt(Plac::gfp-ftsZ)/cI857 PλR::zipA+] (f). Cells in panels a to c were grown at 37°C prior to immunofluorescence staining with the appropriate antibodies. Cells in panels d to f were grown at 30°C in the presence of 25 μM (d and e) or 50 μM (f) IPTG, and were observed immediately after chemical fixation. Results with a number of other normally dividing lysogens [e.g., PB103(λCH50), CH3(λCH75), and PB103(λDR120)] were similar to those shown in panels d to f. Bar in panel a represents 2.0 μm.
Gfp tagged derivatives of ZipA, FtsZ, or FtsA were expressed from lysogenic λ phages containing the appropriate gene fusion downstream of the lac promoter. Phage λCH50 [Plac::zipA-gfp] was described before (17) and encodes a ZipA-Gfp fusion in which GfpS65T (20) is fused to the C terminus of ZipA. In addition, we constructed λDR120 [Plac::gfp-ftsZ] and λCH75 [Plac::gfp-ftsA] encoding Gfpmut2 (7) fused to the N termini of FtsZ and FtsA, respectively. At sufficiently high levels of expression, all three Gfp tagged proteins prevented cell division, leading to the formation of filamentous cells (not shown). At the low levels of expression used in this study, in contrast, none of the fusion proteins interfered substantially with the division process, and all three localized to the septal ring in normally dividing cells (Fig. 2d to f).
Both when using each of the antibodies as well as when using each of the Gfp tagged proteins, we detected a single ring structure in the majority (71 to 88%) of exponentially grown cells with a normal division phenotype (Tables 2 and 3). In all cases, the subpopulation that lacked a clearly detectable ring structure consisted primarily of the smallest cells in the population, and the percentage of cells with more than one ring structure was less than 1%. We found that, particularly when using immunofluorescence detection, the exact percentage of cells with a clearly visible fluorescent ring could vary by as much as 20% between some repeated experiments (not shown). Differences in the 70 to 90 percentile range are, therefore, unlikely to be of biological significance, but rather due to inherent experimental variables.
We conclude that all three Gfp tagged division proteins used in this study are suitable markers for the septal ring structure.
Strains for HID or CID of division proteins.
Throughout this study, we used two types of strains which allowed for the specific, temperature-dependent depletion of ZipA, FtsZ, or FtsA (Table 1). For convenience, we use the acronyms HID (for heat-induced depletion) to indicate one type and CID (for cold-induced depletion) to indicate the other type.
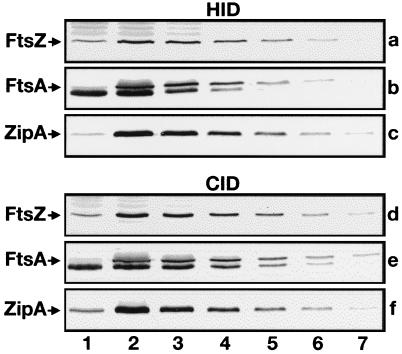
In HID strains, a chromosomal null allele of one of the division genes is complemented by a wild-type allele which is present on a repA(Ts) derivative of plasmid pSC101 (18). HID strains used for depletion of ZipA, FtsZ, and FtsA were CH5/pCH32 [zipA::aph/repA(Ts) zipA ftsZ], PB143/pCX41 [ftsZ0/repA(Ts) ftsZ+], and CH2/pDB280 [ftsA0/repA(Ts) ftsA+], respectively. Strains CH5/pCH32 (17) and PB143/pCX41 (30) were described previously. The chromosomal ftsA0 allele in strain CH2 contains a frameshift mutation and is predicted to encode an 86-aa peptide of which the first 76 aa correspond to the amino terminus of native FtsA. Cells of all three HID strains divided unimpeded at 30°C but formed long filaments at 42°C due to a failure of the complementing plasmids to replicate at this temperature. Furthermore, immunoblotting showed that the failure to divide correlated with the depletion of the corresponding division protein at 42°C (Fig. 3a to c). The HID strains were used to monitor ZipA, FtsZ, and/or FtsA localization by indirect immunofluorescence microscopy (see below).
FIG. 3.
Depletion of division proteins. Immunoblots showing cellular levels of FtsZ (a and d), FtsA (b and e), or ZipA (c and f) after growth of the corresponding HID, CID, and wild-type control strains at either 42°C (a to c) or 30°C (d to f). Lanes 1 contained 70 μg of total protein of depletion strains PB143/pCX41 (a), CH2/pDB280 (b), CH5/pCH32 (c), PB143/pDB346 (d), CH2/pDB355 (e), and CH5/pDB361 (f). Lanes 2 through 7 contained 70, 35, 18, 9, 4, and 2 μg of protein, respectively, of control strains CH3/pMAK700 (a to c), CH3/pDB346 (d), CH3/pDB355 (e), and CH3/pDB361 (f). Proteins were detected with polyclonal antisera. The anti-FtsA serum showed some reactivity to an abundant 40-kDa species (most likely EF-Tu) which migrated slightly faster than FtsA (b and e). Measurements of band intensities indicated that, in each case, the cellular concentration of the depleted division protein was less than 15% of its normal concentration.
In addition to using immunofluorescence techniques, we also wished to study the behavior of Gfp tagged derivatives of the division proteins in parallel experiments. Since the intensity of the Gfp signal diminished significantly at elevated temperatures (not shown), the HID strains were not suitable for these studies, prompting us to construct the CID strains. These strains are identical to the HID strains described above except that the chromosomal null allele of each is corrected by a plasmid in which expression of the corresponding wild-type allele is placed under control of PλR. The plasmid also produces the ts variant of λ repressor (CI857) such that gene expression is strongly repressed below 32°C but becomes gradually derepressed with increasing temperature. CID strains used for depletion of ZipA, FtsZ, and FtsA were CH5/pDB361 (zipA::aph/cI857 PλR::zipA), PB143/pDB346 (ftsZ0/cI857 PλR::ftsZ), and CH2/pDB355 (ftsA0/cI857 PλR::ftsA), respectively. Growth of these strains at 30°C caused the specific depletion of the corresponding division protein (Fig. 3d to f), resulting in the formation of long filamentous cells.
Localization of ZipA in FtsZ− and FtsA− filaments.
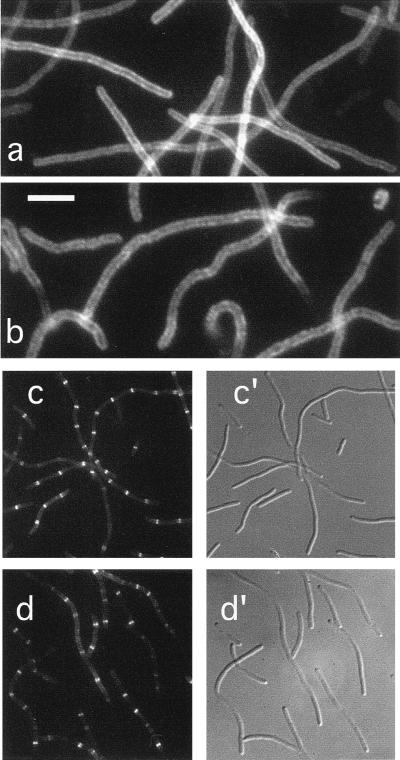
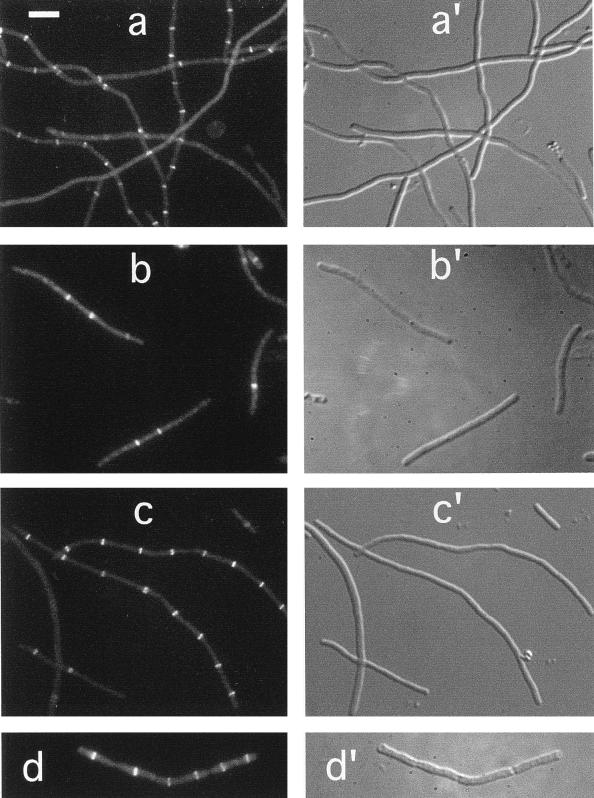
In a previous report, we showed that a ZipA-Gfp protein localized to the septal ring structure in wild-type dividing cells (17). As demonstrated above by immunofluorescence microscopy with affinity-purified anti-ZipA antibodies, the same is true for native ZipA. To study whether this localization pattern is dependent on the FtsZ protein, we first observed the distribution of ZipA-Gfp in cells from which FtsZ had been specifically depleted. For this experiment, the FtsZCID strain PB143/pDB346 (ftsZ0/cI857 PλR::ftsZ) was lysogenized with phage λCH50 (Plac::zipA-gfp), containing zipA-gfp downstream of the IPTG-inducible lac promoter (17). Cells were grown at 30°C in the presence of 25 μM IPTG and observed by fluorescence microscopy. As shown in Fig. 4a, cells formed long nonseptate filaments due to depletion of the FtsZ protein. Moreover, ZipA-Gfp failed to accumulate in ring structures but appeared to be evenly distributed along the periphery of the filaments. Of 133 filaments, representing a total cell length of 2,564 μm, only a single ring structure was seen in each of six relatively short filaments (Table 3). These results suggested that ZipA-Gfp was still associated with the cytoplasmic membrane, but required FtsZ in order to localize to the septal ring.
FIG. 4.
Localization of ZipA in FtsZ− and FtsA− filaments. Fluorescence (a to d) and differential interference contrast (c′ to d′) micrographs showing the location of ZipA-Gfp (a and c) or native ZipA (b and d) in filaments depleted for FtsZ (a and b) or FtsA (c and d). Cells shown are from the FtsZCID strain PB143(λCH50)/pDB346 [ftsZ0(Plac::zipA-gfp)/cI857 PλR::ftsZ+] (a), the FtsZHID strain PB143/pCX41 [ftsZ0/repA(Ts) ftsZ+] (b), the FtsACID strain CH2(λCH50)/pDB355 [ftsA0(Plac::zipA-gfp)/cI857 PλR::ftsA+] (c), and the FtsAHID strain CH2/pDB280 [ftsA0/repA(Ts) ftsA+] (d). Cells in panels b and d were grown at 42°C prior to staining with affinity-purified anti-ZipA antiserum. Cells in panels a and c were grown at 30°C in the presence of 25 μM IPTG and were observed immediately after chemical fixation. Bar represents 5.0 (a and b) or 11.7 (c and d) μm.
To confirm these observations, we used indirect immunofluorescence to determine the location of native ZipA protein in cells of strain PB143/pCX41 [ftsZ0/repA(Ts) ftsZ+] which had been depleted of FtsZ by growth at 42°C. Similar to the distribution of ZipA-Gfp in the experiment described above, native ZipA failed to accumulate into ring structures but was located along the periphery of the FtsZ− filaments (Fig. 4b). Also in this experiment, only a small minority (4%) of the filaments contained a single fluorescent ring (Table 2). We conclude that FtsZ is required for localization of ZipA to the septal ring structure.
To study whether FtsA is required for the localization of ZipA, we observed the distribution of ZipA-Gfp and native ZipA after depletion of FtsA in the FtsACID strain CH2(λCH50)/pDB355 [ftsA0(Plac::zipA-gfp)/cI857 PλR::ftsA] and the FtsAHID strain CH2/pDB280 [ftsA0/repA(Ts) ftsA+], respectively. As shown in Fig. 4c and d, the majority of both FtsACID and FtsAHID filaments showed multiple fluorescent rings, demonstrating that both ZipA-Gfp and native ZipA can still assemble into septal ring structures after depletion of the FtsA protein. Whereas over 90% of these filaments contained ring structures, the number of rings per unit of cell length was approximately half that found in wild-type cells. Thus, as indicated in Tables 2 and 3, one ZipA ring was present approximately every 3 μm on average in wild-type cells, whereas we detected on average only one ZipA ring approximately every 6 μm in FtsA-depleted filaments (CID or HID). This increase in the ratio of unit length per ZipA ring in these filaments parallels a similar increase in the length/FtsZ ring ratio after depletion of FtsA (see below and Tables 2 and 3) and presumably reflects the loss of a stabilizing effect of FtsA on the septal ring structure (1). These results indicate that ZipA joins the FtsZ ring in an FtsA-independent fashion.
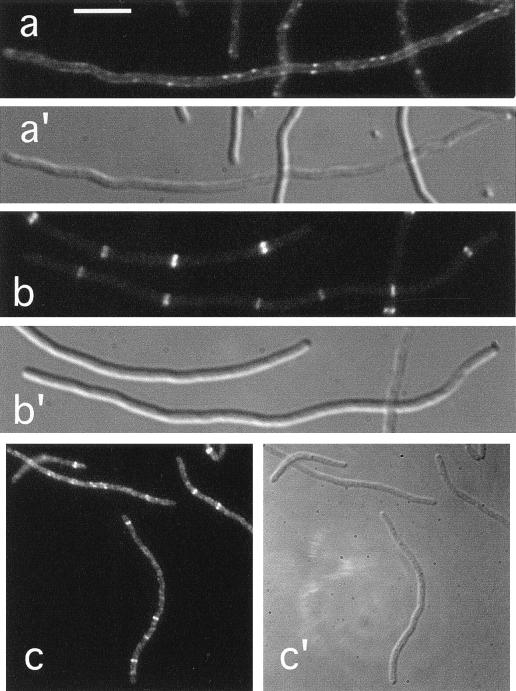
Localization of FtsZ in ZipA− and FtsA− filaments.
The finding that ZipA failed to localize to the septal ring after depletion of FtsZ suggested either that the incorporation of both proteins into the structure depended on the protein partner or that formation of the FtsZ ring can occur independently of ZipA. To assess which of these possibilities is most likely correct, we studied the localization of FtsZ in cells from which ZipA had been depleted. To this end, the ZipACID strain CH5/pDB361 (zipA::aph/cI857 PλR::zipA) was lysogenized with phage λDR120 (Plac::gfp-ftsZ), and cells of a resulting lysogen were shifted from 42 to 30°C in the presence of 50 μM IPTG. As illustrated in Fig. 5a, many of the resulting ZipA-depleted filaments showed multiple fluorescent ring structures, suggesting that FtsZ rings were still being formed. Not all filaments showed rings, however, and of 64 filaments examined, rings were absent in 12 (19%) cells. In addition, the majority of filaments contained fewer ring structures than one might expect. Thus, whereas in the ring-containing portion of a wild-type population the ratio of cell length to FtsZ ring was approximately 2.7 μm, this ratio was significantly higher (8.6 μm) in the portion of ZipACID filaments that contained ring structures. In addition, there was a large variation in the value of this ratio among individual filaments, ranging from 4.9 to 40.2 μm (Table 3).
FIG. 5.
Localization of FtsZ in ZipA− and FtsA− filaments. Fluorescence (a through d) and differential interference contrast (a′ to d′) micrographs showing the location of Gfp-FtsZ (a and c) or native FtsZ (b and d) in filaments depleted for ZipA (a and b) or FtsA (c and d). Cells shown are from the ZipACID strain CH5(λDR120)/pDB361 [zipA::aph(Plac::gfp-ftsZ)/cI857 PλR::zipA+] (a), the ZipAHID strain CH5/pCH32 (zipA::aph/repAts ftsZ+ zipA+) (b), the FtsACID strain CH2(λDR120)/pDB355 [ftsA0(Plac::gfp-ftsZ)/cI857 PλR::ftsA+] (c), and the FtsAHID strain CH2/pDB280 [ftsA0/repA(Ts) ftsA+] (d). Cells in panels b and d were grown at 42°C prior to staining with FtsZ-specific monoclonal antibodies. Cells in panels a and c were grown at 30°C in the presence of 50 μM IPTG and were observed immediately after chemical fixation. Bar in panel a represents 5.0 μm.
Quite similar results were obtained after staining native FtsZ in heat-induced filaments of the ZipAHID strain CH5/pCH32 [zipA::aph/repA(Ts) zipA ftsZ] (Fig. 5b; Table 2). In this case, 28% of the ZipA-depleted filaments showed no rings at all, and the length-to-ring ratio in the portion of filaments that did contain one or more rings was 17.2 μm.
Multiple FtsZ rings were previously also observed in filaments of a strain carrying a ts allele of ftsA, indicating that the formation of the FtsZ ring does not require fully functional FtsA (1). Consistent with those results, we found that FtsZ rings were also still present in filaments from which FtsA had been depleted. Our results are illustrated in Fig. 5c and d, showing filaments of, respectively, the FtsACID lysogen CH2(λDR120)/pDB355 [ftsA0(Plac::gfp-ftsZ)/cI857 PλR::ftsA], which was grown at 30°C in the presence of 50 μM IPTG, and of the FtsAHID strain CH2/pDB280 [ftsA0/repA(Ts) ftsA+], which was grown at 42°C and immunostained with FtsZ-specific antibodies. One or more FtsZ rings were present in over 90% of both the FtsACID and FtsAHID filaments, and the average length-to-ring ratio in both types was approximately 6.3 μm (Tables 2 and 3).
We conclude that FtsZ can still assemble into a ring structure under conditions in which the cellular concentration of either ZipA or FtsA is very low. It seems clear, however, that depletion of either protein leads to a significant reduction in the number of FtsZ ring structures per unit of cell mass.
Localization of FtsA in FtsZ− and ZipA− filaments.
In a previous study, it was shown that formation of the FtsZ ring is a prerequisite for the localization of FtsA to the septal ring structure (3). Consistent with the results of this study, we found that FtsA failed to localize in filaments from which FtsZ had been depleted. Thus, no fluorescent rings were observed in filaments of the FtsZCID strain PB143/pDB346 (ftsZ0/cI857 PλR::ftsZ), lysogenic for λCH75 (Plac::gfp-ftsA) and grown at 30°C in the presence of 25 μM IPTG (Fig. 6a; Table 3). Similarly, FtsA-specific antibodies failed to stain septal ring structures in the vast majority (96%) of FtsZHID filaments of strain PB143/pCX41 [ftsZ0/repA(Ts) ftsZ+] (Table 2).
FIG. 6.
Localization of FtsA in FtsZ− and ZipA− filaments. Fluorescence (a to c) and differential interference contrast (a′ to c′) micrographs showing the location of Gfp-FtsA (a and b) or native FtsA (c) in filaments depleted of FtsZ (a) or ZipA (b and c). Cells shown are from the FtsZCID strain PB143(λCH75)/pDB346 [ftsZ0(Plac::gfp-ftsA)/cI857 PλR::ftsZ+] (a), the ZipACID strain CH5(λCH75)/pDB361 [zipA::aph(Plac::gfp-ftsA)/cI857 PλR::zipA+] (b), and the ZipAHID strain CH5/pCH32 [zipA::aph/repA(Ts) ftsZ+ zipA+] (c). Cells in panel c were grown at 42°C prior to staining with affinity-purified anti-FtsA antiserum. Cells in panels a and b were grown at 30°C in the presence of 25 μM IPTG and were observed immediately after chemical fixation. Bar represents 5.0 (a and b) or 11.0 (c) μm.
Experiments described above indicated that the localization of ZipA to the septal ring requires FtsZ but occurs independently of the FtsA protein. To test whether FtsA localization is dependent on ZipA, we studied how depletion of ZipA affected the localization of FtsA. For this purpose, phage λCH75 (Plac::gfp-ftsA) was introduced into the ZipACID strain CH5/pDB361 (zipA::aph/cI857 PλR::zipA), and a resulting lysogen was incubated at 30°C in the presence of 25 μM IPTG and inspected by fluorescence microscopy. Many of the resulting filaments (92%) showed one or multiple fluorescent ring structures (Fig. 6b; Table 3), suggesting that depletion of ZipA does not prevent FtsA from associating with the septal ring. Again, similar results were obtained when heat-induced filaments of CH5/pCH32 (zipA::aph/repAts zipA ftsZ) were subjected to immunostaining with FtsA-specific antibodies (Fig. 6c; Table 2). As was observed for FtsZ rings above, the number and distribution of FtsA rings were quite heterogeneous among individual filaments of both the ZipACID and ZipAHID populations. These results indicate that FtsA does not require ZipA in order to associate with the septal ring.
DISCUSSION
The septal ring in E. coli is a complex structure consisting of at least seven different protein products. Important challenges concerning this organelle are to understand its precise molecular architecture, its mode of assembly, and the mechanism whereby it mediates the coordinated invagination of the cell envelope layers during septum formation. The order in which the different components assemble to form a mature septal ring has been studied in several laboratories by determining the localization of FtsA, -I, -N, -K, and -Z in filaments in which one of the essential division proteins has either lost function due to a conditional mutation or drug treatment or is largely lacking, due to a specific block in gene expression (2, 3, 6, 22, 29, 35, 39, 40). The picture emerging from these studies is that, first, FtsZ assembles at the prospective division site to form the FtsZ ring. FtsA then joins the FtsZ ring, followed by FtsI, FtsK and FtsN. The best evidence supporting early assembly of the FtsZ ring has come from the observations that FtsA (reference 3 and this study), FtsI (35), FtsK (40) and FtsN (2) completely failed to localize properly in FtsZ− filaments whereas, on the other hand, FtsZ rings were still present in FtsA− (reference 1 and this study), FtsI− (1, 29, 35, 39), FtsN− (2), FtsK− (40), FtsQ− (1), and FtsW− (6, 22) filaments.
We recently identified ZipA as a novel division factor and showed that it is an integral inner membrane protein which interacts directly with FtsZ. A ZipA-Gfp fusion protein, furthermore, localized to the septal ring at an early stage of the division cycle (17). Here, we confirmed this localization pattern for native ZipA by immunofluorescence microscopy.
The combined properties of ZipA raised the possibility that the protein plays a role in assembly of the FtsZ ring by attracting FtsZ to the prospective division site. The principal conclusion of this study is that this possibility is most likely incorrect. In FtsZ-depleted filaments, the bulk of both native ZipA or ZipA-Gfp clearly failed to accumulate at potential division sites but, rather, appeared to be evenly distributed over the cell membrane. Conversely, both native FtsZ and a Gfp-FtsZ fusion protein still assembled into ring structures in the majority of ZipA-depleted filaments. It is most likely, therefore, that the localization of ZipA to the septal ring is secondary to that of FtsZ.
In support of the conclusion that FtsA localization is also dependent on FtsZ (3), we found that FtsA and Gfp-FtsA failed to localize after depletion of FtsZ, but that FtsZ rings could still be formed in FtsA-depleted filaments. Interestingly, however, ZipA rings were still present in FtsA-depleted filaments, and FtsA rings were still present in ZipA-depleted filaments, indicating that the incorporation of ZipA into the septal ring does not require the prior localization of FtsA and vice versa. Combined with the knowledge that both ZipA and FtsA localize early in the division cycle and that both can bind FtsZ directly, we propose that both proteins become associated with the FtsZ ring either during or very soon after formation of the latter. In this regard, it is interesting that whereas the vast majority of FtsZ- or ZipA-depleted filaments appeared completely smooth, more than half of the FtsA-depleted filaments (HID or CID) showed one or more marked indentations of the cell wall (Fig. 4 to 6). This suggests that despite the early localization of FtsA, the early stages of septation are less sensitive to depletion of the protein than are later stages.
Although the scenario proposed above represents the most straightforward interpretation of our results, it is difficult to completely rule out an essential role for ZipA in the assembly of the FtsZ ring. Although the ZipA-depleted filaments used in this study clearly contained an insufficient level of ZipA to support cell division, it cannot be excluded that this level (∼10% of normal [Fig. 3]) may have been sufficient to actively stimulate formation of FtsZ rings. This same argument also applies to FtsA, -I, -N, -K, -Q, and -W in the studies in which FtsZ localization was observed after inactivation or depletion of one of these division proteins (1, 2, 6, 22, 29, 39, 40).
In these same studies, the ratio of cell length to the number of FtsZ rings in most types of Fts− filaments was much higher than expected, and a large variability in this ratio between individual filaments was observed. Similarly, we observed that the average length-to-FtsZ ring ratios in the ring-containing populations of both ZipA-depleted (17.2 μm in ZipAHID and 8.9 μm in ZipACID) and FtsA-depleted (6.2 μm in FtsAHID and 6.3 μm in FtsACID) filaments was significantly higher than that in wild-type cell (∼2 to 3 μm). In addition, this ratio varied widely between individual filaments. For instance, after heat-induced depletion of ZipA, 72% of the filaments (ranging in size from 10.1 to 83.1 μm) showed from one to five FtsZ rings per cell with a length-to-ring ratio ranging from 3.3 to 89.0 μm, and the remaining 28% (ranging in size from 10.3 to 76.9 μm) showed no ring at all (Table 2). After CID of ZipA, 81% of the population (ranging in size from 5.7 to 85.5 μm) contained from one to nine Gfp-FtsZ rings per cell with a ratio ranging from 4.9 to 40.2 μm, and the remaining 19% (ranging in size from 16.8 to 63.4 μm) were devoid of rings (Table 3). The length-to-ZipA ring ratio in FtsA− filaments and length-to-FtsA ring ratio in ZipA− filaments were also relatively high and variable which, given the high and variable length to FtsZ ring ratio in FtsA− and ZipA− filaments, is consistent with the interpretation that the incorporation of FtsA and ZipA into the septal ring is dependent on formation of the FtsZ ring component.
Why the numbers of FtsZ rings are so low and variable in ZipA-depleted cells (and other types of filaments) relative to wild-type cells is an intriguing question. The possibility that depletion of ZipA simply leads to a reduction in the cellular concentration of FtsZ was ruled out by quantitative immunoblot analyses showing that depletion of 90% of ZipA affected the FtsZ concentration by less than 5% (not shown). Inefficient fixation and/or staining of cells could lead to an underestimation of the number of ring structures by immunofluorescence, but we consider it unlikely that this was a determining factor in our experiments, especially since we obtained largely comparable results using Gfp tagged proteins as septal ring markers. One reasonable hypothesis is that binding of ZipA to the FtsZ ring stabilizes the structure, for instance, by providing an anchor to the cell membrane. If correct, and when combined with the results of previous studies (1, 2, 6, 22, 29, 39, 40), this would mean that most of the septal ring components contribute substantially to the stability of the structure. As pointed out by Pogliano et al. (29), an interesting alternative possibility is that the state of maturation or activity of a septal ring at one potential division site somehow affects the formation or stability of rings at additional sites in the cell. Clearly, more work is needed to test these possibilities.
The results of this study indicate that ZipA is not required for the initiation of FtsZ ring formation or for the recruitment of FtsA to the ring but suggest that it contributes significantly to the stability of the organelle. Many additional roles for the protein are possible. Because ZipA is very likely among the first factors to become associated with the FtsZ ring, it is conceivable that the protein helps recruit other components to the structure. It will, therefore, be interesting to determine the localization pattern of other Fts proteins in ZipA-depleted cells. Filaments lacking ZipA are smooth, which is consistent with a role for the protein throughout the cell wall invagination process. It is possible that ZipA simply ensures that the inner membrane remains anchored to the shrinking FtsZ ring. In addition, the protein may well play a more active role(s), such as stimulating constriction of the FtsZ ring or coordinating the activity of the FtsZ ring in the cytoplasm with the peptidoglycan synthesizing machinery in the periplasm. Additional experimentation is needed to define the role(s) of ZipA more precisely.
To date, assembly of the FtsZ ring at the prospective division site is the first recognizable step in the formation of the septal ring organelle. Although we cannot be absolutely certain, this and other studies have rendered it unlikely that any of the other known division proteins, including ZipA, perform an essential function prior to this step. To determine what defines a potential division site and how FtsZ assembly is initiated at this site remain important unanswered questions.
ACKNOWLEDGMENTS
Parts of this study were initiated in the laboratory of Larry Rothfield at the University of Connecticut Health Center, and we thank him for exceptional support, the gift of materials, and critical comments on the manuscript. We, furthermore, thank David Raskin, Robin Crossley, and Gregory Matera for technical assistance and advice and Jan Voskuil for the gift of monoclonal antibodies.
This work was supported by NIH grants GM-57059 (to P.D.B.) and GM-53276 (to L.R.), as well as by an NSF young investigator award (MCB94-58197) and by generous donations from Wyeth-Ayerst Research, The Elizabeth M. and William C. Treuhaft Fund, The Frank K. Griesinger Trust, Arline H. Garvin, James S. Blank, Charles E. Spahr, Alfred M. Taylor, and Theodore J. Castele (to P.D.B.).
REFERENCES
- 1.Addinall S G, Bi E, Lutkenhaus J. FtsZ ring formation in fts mutants. J Bacteriol. 1996;178:3877–3884. doi: 10.1128/jb.178.13.3877-3884.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Addinall S G, Cao C, Lutkenhaus J. FtsN, a late recruit to the septum in Escherichia coli. Mol Microbiol. 1997;25:303–309. doi: 10.1046/j.1365-2958.1997.4641833.x. [DOI] [PubMed] [Google Scholar]
- 3.Addinall S G, Lutkenhaus J. FtsA is localized to the septum in an FtsZ-dependent manner. J Bacteriol. 1996;178:7167–7172. doi: 10.1128/jb.178.24.7167-7172.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bi E, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 5.Bork P, Sander C, Valencia A. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc Natl Acad Sci USA. 1992;89:7290–7294. doi: 10.1073/pnas.89.16.7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyle D S, Khattar M M, Addinall S G, Lutkenhaus J, Donachie W D. ftsW is an essential cell-division gene in Escherichia coli. Mol Microbiol. 1997;24:1263–1273. doi: 10.1046/j.1365-2958.1997.4091773.x. [DOI] [PubMed] [Google Scholar]
- 7.Cormack B P, Valdivia R H, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 8.Dai K, Lutkenhaus J. The proper ratio of FtsZ to FtsA is required for cell division to occur in Escherichia coli. J Bacteriol. 1992;174:6145–6151. doi: 10.1128/jb.174.19.6145-6151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Boer P, Crossley R, Rothfield L. The essential bacterial cell-division protein FtsZ is a GTPase. Nature. 1992;359:254–256. doi: 10.1038/359254a0. [DOI] [PubMed] [Google Scholar]
- 10.de Boer P A J, Crossley R E, Rothfield L I. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell. 1989;56:641–649. doi: 10.1016/0092-8674(89)90586-2. [DOI] [PubMed] [Google Scholar]
- 11.de Boer P A J, Crossley R E, Rothfield L I. Isolation and properties of minB, a complex genetic locus involved in correct placement of the division site in Escherichia coli. J Bacteriol. 1988;170:2106–2112. doi: 10.1128/jb.170.5.2106-2112.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Boer P A J, Crossley R E, Rothfield L I. Roles of MinC and MinD in the site-specific septation block mediated by the MinCDE system of Escherichia coli. J Bacteriol. 1992;174:63–70. doi: 10.1128/jb.174.1.63-70.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dewar S J, Begg K J, Donachie W D. Inhibition of cell division initiation by an imbalance in the ratio of FtsA to FtsZ. J Bacteriol. 1992;174:6314–6316. doi: 10.1128/jb.174.19.6314-6316.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Din N, Quardokus E M, Sackett M J, Brun Y V. Dominant c-terminal deletions of FtsZ that affect its ability to localize in Caulobacter and its interaction with FtsA. Mol Microbiol. 1998;27:1051–1063. doi: 10.1046/j.1365-2958.1998.00752.x. [DOI] [PubMed] [Google Scholar]
- 15.Erickson H P, Taylor D W, Taylor K A, Bramhill D. Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc Natl Acad Sci USA. 1996;93:519–523. doi: 10.1073/pnas.93.1.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gottesman S, Halpern E, Trisler P. Role of sulA and sulB in filamentation by Lon mutants of Escherichia coli K-12. J Bacteriol. 1981;148:265–273. doi: 10.1128/jb.148.1.265-273.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hale C A, de Boer P A J. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell. 1997;88:175–185. doi: 10.1016/s0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- 18.Hamilton C M, Aldea M, Washburn B K, Babitzke P, Kushner S R. New method for generating deletions and gene replacements in Escherichia coli. J Bacteriol. 1989;171:4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harry E J, Wake R G. The membrane-bound cell division protein DivIB is localized to the division site in Bacillus subtilis. Mol Microbiol. 1997;25:275–283. doi: 10.1046/j.1365-2958.1997.4581822.x. [DOI] [PubMed] [Google Scholar]
- 20.Heim R, Cubitt A B, Tsien R Y. Improved green fluorescence. Nature. 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- 21.Hiraga S, Niki H, Ogura T, Ichinose C, Mori H, Ezaki B, Jaffe A. Chromosome partitioning in Escherichia coli: novel mutants producing anucleate cells. J Bacteriol. 1989;171:1496–1505. doi: 10.1128/jb.171.3.1496-1505.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khattar M M, Addinall S G, Stedul K H, Boyle D S, Lutkenhaus J, Donachie W D. Two polypeptide products of the Escherichia coli cell division gene ftsW and a possible role for FtsW in FtsZ function. J Bacteriol. 1997;179:784–793. doi: 10.1128/jb.179.3.784-793.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu C, Stricker J, Erickson H P. FtsZ from Escherichia coli, Azotobacter vinelandii, and Thermotoga maritima: quantitation, GTP hydrolysis, and assembly. Cell Motil Cytoskelet. 1998;40:71–86. doi: 10.1002/(SICI)1097-0169(1998)40:1<71::AID-CM7>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 24.Lutkenhaus J, Addinall S G. Bacterial cell division and the Z ring. Annu Rev Biochem. 1997;66:93–116. doi: 10.1146/annurev.biochem.66.1.93. [DOI] [PubMed] [Google Scholar]
- 25.Ma X, Ehrhardt D W, Margolin W. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc Natl Acad Sci USA. 1996;93:12998–13003. doi: 10.1073/pnas.93.23.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mukherjee A, Dai K, Lutkenhaus J. Escherichia coli cell division protein FtsZ is a guanine nucleotide binding protein. Proc Natl Acad Sci USA. 1993;90:1053–1057. doi: 10.1073/pnas.90.3.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mukherjee A, Lutkenhaus J. Dynamic assembly of FtsZ regulated by GTP hydrolysis. EMBO J. 1998;17:462–469. doi: 10.1093/emboj/17.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mukherjee A, Lutkenhaus L. Guanine nucleotide-dependent assembly of FtsZ into filaments. J Bacteriol. 1994;176:2754–2758. doi: 10.1128/jb.176.9.2754-2758.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pogliano J, Pogliano K, Weiss D S, Losick R, Beckwith J. Inactivation of FtsI inhibits constriction of the FtsZ cytokinetic ring and delays the assembly of FtsZ rings at potential division sites. Proc Natl Acad Sci USA. 1997;94:559–564. doi: 10.1073/pnas.94.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raskin D M, de Boer P A J. The MinE ring: an FtsZ-independent cell structure required for selection of the correct division site in E. coli. Cell. 1997;91:685–694. doi: 10.1016/s0092-8674(00)80455-9. [DOI] [PubMed] [Google Scholar]
- 31.RayChaudhuri D, Park J T. Escherichia coli cell-division gene ftsZ encodes a novel GTP-binding protein. Nature. 1992;359:251–254. doi: 10.1038/359251a0. [DOI] [PubMed] [Google Scholar]
- 32.Rothfield L I, Justice S S. Bacterial cell division: the cycle of the ring. Cell. 1997;88:581–584. doi: 10.1016/s0092-8674(00)81899-1. [DOI] [PubMed] [Google Scholar]
- 33.Sánchez M, Valencia A, Ferrándiz M-J, Sander C, Vicente M. Correlation between the structure and biochemical activities of FtsA, an essential cell division protein of the actin family. EMBO J. 1994;13:4919–4925. doi: 10.1002/j.1460-2075.1994.tb06819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Voskuil J L A, Westerbeek C A M, Wu C, Kolk A H J, Nanninga N. Epitope mapping of Escherichia coli division protein FtsZ with monoclonal antibodies. J Bacteriol. 1994;176:1886–1893. doi: 10.1128/jb.176.7.1886-1893.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang L, Khattar M K, Donachie W D, Lutkenhaus J. FtsI and FtsW are localized to the septum in Escherichia coli. J Bacteriol. 1998;180:2810–2816. doi: 10.1128/jb.180.11.2810-2816.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X, de Boer P A J, Rothfield L I. A factor that positively regulates cell division by activating transcription of the major cluster of essential cell division genes of Escherichia coli. EMBO J. 1991;10:3363–3372. doi: 10.1002/j.1460-2075.1991.tb04900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X, Huang J, Mukherjee A, Cao C, Lutkenhaus J. Analysis of the interaction of FtsZ with itself, GTP, and FtsA. J Bacteriol. 1997;179:5551–5559. doi: 10.1128/jb.179.17.5551-5559.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang X, Lutkenhaus J. The FtsZ protein of Bacillus subtilis is localized at the division site and has GTPase activity that is dependent upon FtsZ concentration. Mol Microbiol. 1993;9:435–442. doi: 10.1111/j.1365-2958.1993.tb01705.x. [DOI] [PubMed] [Google Scholar]
- 39.Weiss D S, Pogliano K, Carson M, Guzman L-M, Fraipont C, Nguyen-Distèche M, Losick R, Beckwith J. Localization of the Escherichia coli cell division protein FtsI (PBP3) to the division site and cell pole. Mol Microbiol. 1997;25:671–681. doi: 10.1046/j.1365-2958.1997.5041869.x. [DOI] [PubMed] [Google Scholar]
- 40.Yu X-C, Tran A H, Sun Q, Margolin W. Localization of cell division protein FtsK to the Escherichia coli septum and identification of a potential N-terminal targeting domain. J Bacteriol. 1998;180:1296–1304. doi: 10.1128/jb.180.5.1296-1304.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu X C, Margolin W. Ca2+-mediated GTP-dependent dynamic assembly of bacterial cell division protein FtsZ into asters and polymer networks in vitro. EMBO J. 1997;16:5455–5463. doi: 10.1093/emboj/16.17.5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao C-R, de Boer P A J, Rothfield L I. Proper placement of the Escherichia coli division site requires two functions that are associated with different domains of the MinE protein. Proc Natl Acad Sci USA. 1995;92:4314–4317. doi: 10.1073/pnas.92.10.4313. [DOI] [PMC free article] [PubMed] [Google Scholar]