Abstract
The products of the SOS-regulated umuDC operon are required for most UV and chemical mutagenesis in Escherichia coli, a process that results from a translesion synthesis mechanism. The UmuD protein is activated for its role in mutagenesis by a RecA-facilitated autodigestion that removes the N-terminal 24 amino acids. A previous genetic screen for nonmutable umuD mutants had resulted in the isolation of a set of missense mutants that produced UmuD proteins that were deficient in RecA-mediated cleavage (J. R. Battista, T. Ohta, T. Nohmi, W. Sun, and G. C. Walker, Proc. Natl. Acad. Sci. USA 87:7190–7194, 1990). To identify elements of the UmuD′ protein necessary for its role in translesion synthesis, we began with umuD′, a modified form of the umuD gene that directly encodes the UmuD′ protein, and obtained missense umuD′ mutants deficient in UV and methyl methanesulfonate mutagenesis. The D39G, L40R, and T51I mutations affect residues located at the UmuD′2 homodimer interface and interfere with homodimer formation in vivo. The D75A mutation affects a highly conserved residue located at one end of the central strand in a three-stranded β-sheet and appears to interfere with UmuD′2 homodimer formation indirectly by affecting the structure of the UmuD′ monomer. When expressed from a multicopy plasmid, the L40R umuD′ mutant gene exhibited a dominant negative effect on a chromosomal umuD+ gene with respect to UV mutagenesis, suggesting that the mutation has an effect on UmuD′ function that goes beyond its impairment of homodimer formation. The G129D mutation affects a highly conserved residue that lies at the end of the long C-terminal β-strand and results in a mutant UmuD′ protein that exhibits a strongly dominant negative effect on UV mutagenesis in a umuD+ strain. The A30V and E35K mutations alter residues in the N-terminal arms of the UmuD′2 homodimer, which are mobile in solution.
The gene products of the umuDC operon play important roles in the ability of Escherichia coli to tolerate damage to its DNA (10, 17, 30, 35, 36, 41). umuDC mutants were originally identified by screening directly for derivatives of E. coli that are nonmutable by treatment with UV and various other chemicals (13, 32). Mutagenesis by such agents occurs as the result of a translesion synthesis mechanism in which an incorrect nucleotide is inserted opposite the lesion and the chain is then subsequently extended (10, 24, 26, 33, 38).
Expression of the umuDC gene products is elaborately regulated, both transcriptionally and posttranslationally. The umuDC operon is expressed at a low basal level and is induced in response to DNA damage (1) as part of the cellular SOS response (10, 14, 30, 35, 37). A cell’s attempts to replicate damaged DNA generate regions of single-stranded DNA, which then permit the formation of RecA nucleoprotein filaments (RecA*). Interaction of LexA with RecA* then facilitates a latent capacity of LexA to autodigest. The resulting proteolytic inactivation of LexA leads to increased levels of expression of the umuDC operon and other LexA-repressed loci. The full-length UmuD protein that results from translation of the umuD gene is inactive in mutagenesis (19) and is converted to the form that is active in mutagenesis, UmuD′, by RecA*-mediated cleavage (5, 19, 27). This cleavage is mechanistically similar to the RecA*-mediated cleavage of LexA (16) and results in the removal of the N-terminal 24 amino acids of UmuD. Until recently, it appeared that intact UmuD was simply an inactive precursor of UmuD′ with respect to SOS mutagenesis. However, recent evidence suggests the possibility that UmuD, acting together with UmuC, may play a role that is functionally equivalent to a eukaryotic DNA damage checkpoint control system by regulating progression through the cell cycle after DNA damage (20, 21). Both UmuD and UmuD′ form homodimers (40), and the UmuD-UmuD′ heterodimer is more stable than either of the homodimers (2). The three dimeric forms of the umuD gene product differ from one another with respect to their susceptibility to specific cytoplasmic proteases (8). The UmuD′2 homodimer interacts with UmuC to form a UmuD′2C complex (4). After UmuD is cleaved to generate UmuD′, the UmuD′ and UmuC proteins participate in translesion synthesis (24, 26, 33) and may also play a role in modulating recombinational repair of damaged DNA (3, 31).
The crystal structure of the UmuD′ protein was recently determined, and the UmuD′ monomer was found to consist of a globular C-terminal domain with an extended N-terminal region (22). In the crystal structure that was determined, the UmuD′ monomers were arranged in a filament in which there were two different interfaces between UmuD monomers, one rich in aliphatic residues that involves interactions between the C termini (referred to by Peat et al. [22, 23] as the filament dimer interface) and one rich in aromatic residues (referred to by Peat et al. [22, 23] as the molecular dimer interface). Nuclear magnetic resonance (NMR) analyses of the UmuD′2 homodimer in solution (7) have indicated that the actual homodimer interface is the one involving contacts between the C termini as well as interactions involving N41 and L44 (using the same numbering system as for intact UmuD) rather than the interface rich in aromatic residues that was originally assigned on the basis of the crystallographic analyses. No evidence was seen in the NMR studies (7) for the UmuD filaments that have been hypothesized to be important for the role of UmuD′ in mutagenesis (22, 23). Structural information is not yet available for the UmuD2 homodimer, but cross-linking studies of monocysteine derivatives of UmuD (15) are consistent with the UmuD2 homodimer interface resembling the interface of the UmuD′2 homodimer. However, the UmuD′2 and UmuD2 homodimers differ in other respects. For example, monocysteine substitutions at positions 37 and 38 seem to be particularly well positioned to form disulfide bonds across the interface of the UmuD2 homodimer (11), while the N-terminal arms that contain the residues at positions 37 and 38 are free in solution in the UmuD′2 homodimer (7).
As part of an effort to understand the mechanism by which the umuD gene product exerts its biological roles, we had previously screened for umuD mutants that were defective in their ability to participate in SOS mutagenesis (2). However, the mutants obtained in that screen were found to produce derivatives of UmuD that were deficient in their ability to undergo the RecA*-facilitated autodigestion that gives rise to UmuD′, the form active in mutagenesis. Earlier examples of mutants of this class had been obtained by altering the Ser60 and Lys97 residues of UmuD (19), which play roles in UmuD cleavage that are analogous to those played by Ser119 and Lys156 in LexA cleavage (29). To gain insights into which residues of UmuD′ are required for its actual role in UV mutagenesis, we adopted an alternative strategy, namely mutagenizing a modified form of the umuD gene that directly encodes UmuD′ (19) and screening for umuD′ mutants with deficiencies in UV mutagenesis. While this paper was under review, a related study in which umuD′ mutants were identified on the basis of their deficiencies in promoting the umuD′C-dependent spontaneous mutagenesis observed in a lexA(Def) recA718 mutant was reported (18).
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and the plasmid used in this study have been described previously. E. coli GW3200 (6) is a umuD44 derivative of AB1157 (thr-1 leu-6 proA2 hisG4 thi-1 argE3 lacY1 galK2 ara-14 xyl-5 mtl-1 tsx-33 rpsL31 supE44). GW8025 (21) is a lexA300(Def)::spc Δ(umuDC)595::cat derivative of GW1000 [recA441 sulA11 ΔlacU169 thr-1 leu-6 hisG4 argE3 ilv(Ts) galK2 rpsL31]. The plasmid pGW2122 (19) carries the umuD′ gene under the control of the umuDC promoter. pGW2122 was constructed from the corresponding umuD+ plasmid pGW2020 (19) by using site-directed oligonucleotide mutagenesis to remove the nucleotides that encode amino acids 2 to 24 of UmuD.
Isolation of umuD′ mutants.
Plasmid pGW2122 DNA was purified by CsCl density gradient centrifugation and then treated with hydroxylamine as previously described (2). Mutagenized plasmids were transformed into the umuD44 strain GW3200 (19), and the resulting transformants were screened for failure to fully complement the deficiency of GW3200 in UV-induced argE3 reversion to an Arg+ phenotype (argE3→Arg+ reversion). Screening was carried out by patching colonies of GW3200 derivatives carrying the mutagenized plasmids in duplicate onto supplemented M9-glucose plates containing a trace of arginine (2), irradiating with a dose of 20 J of UV light/m2 and then incubating at 37°C for 2 days before scoring for Arg+ papillae. For plasmids encoding UmuD′ derivatives with deficiencies in UV mutagenesis, the umuD′ coding region and the upstream region were determined by dideoxynucleotide sequencing (19). In addition, several umuD′ mutants were isolated by site-directed oligonucleotide mutagenesis of pGW2122. All site-directed mutants were sequenced to confirm that the desired mutation had been successfully introduced.
UV and MMS mutagenesis.
The methods used to determine the susceptibility of various strains to mutagenesis by UV light (6) and methyl methanesulfonate (MMS) (34) have been described previously.
Measurements of UmuD′ in vivo levels and UmuD′2 homodimer formation.
Levels of UmuD′ in vivo were assessed by the following method. Overnight cultures of GW3200 bearing the mutant umuD′ plasmids were diluted 1:20 in M9 medium (39) supplemented with 0.1 mM CaCl2, 2 mM MgSO4, 0.2% glucose, 0.4% Casamino Acids, and 5 μg of thiamine per ml. At an optical density at 600 nm (OD600) of ca. 0.4, the cells were UV irradiated at 20 J/m2 after which they were grown at 37°C for 1 h. The cells were then pelleted and resuspended in sodium dodecyl sulfate (SDS) sample buffer (50 mM Tris-Cl [pH 6.8], 100 mM dithiothreitol, 2% SDS, 0.2% bromophenol blue, 10% glycerol), and 0.1 OD600 unit of cells was subjected to electrophoresis on an SDS–15% polyacrylamide gel. The proteins were then transferred to a polyvinylidene difluoride membrane (Immobilon-P). The membrane was incubated with affinity-purified antibodies raised against UmuD, and cross-reacting material was visualized by using chemiluminescence (Tropix). Homodimer formation was measured in vivo by formaldehyde cross-linking (28). GW8025 derivatives bearing the respective umuD′-expressing plasmids were grown in LB medium (39) at 42°C to an OD600 of ca. 0.8. The cells were harvested and resuspended in 100 mM sodium phosphate, pH 6.8, and the OD600 was adjusted to 0.5. Formaldehyde (Mallinckrodt) was added to a final concentration of 1%, and samples were incubated at room temperature for 30 min. After incubation, the cells were concentrated by centrifugation, lysed by addition of SDS sample buffer, and then subjected to electrophoresis and chemiluminescence detection as described above.
RESULTS
Isolation and generation of umuD′ mutants that are deficient in UV mutagenesis.
To identify mutations that affected the ability of the UmuD′ protein to participate in UV mutagenesis, the plasmid pGW2122 (19), which carries a modified form of the umuD gene that directly encodes UmuD′, was mutagenized with hydroxylamine and transformed into a umuD44 mutant. The resulting transformants were patched on minimal medium containing a trace of arginine, irradiated with UV light, and then screened for their proficiency in UV mutagenesis by a papillation assay based on argE3→Arg+ reversion. After screening several thousand derivatives, we identified 19 candidates that showed a significant reduction in the number of UV-induced Arg+ papillae relative to that of the umuD44 derivative carrying the parental umuD′ plasmid, pGW2122. Two of these grew slowly on supplemented minimal medium and were not characterized further, while restriction endonuclease analysis of the plasmid in a third derivative showed clearly that it had undergone a deletion of part of the umuD′ gene. The umuD′ coding and upstream regions of the remaining 16 plasmids were then sequenced, and the results are summarized in Table 1. Two independently isolated derivatives altered the −35 region of the umuDC promoter from TTGATA to TTAATA, and a mutant carrying this same promoter mutation was also isolated by McLenigan et al. (18) in a related study. The change of this critical promoter residue (25) severely reduces umuD′ expression (18). Another plasmid altered the initial ATG codon of umuD′, a change that would prevent translation of the umuD′ message. Two nonsense mutants were isolated, one with an ochre mutation affecting codon 100 and the other with an amber mutation affecting codon 52. Since the strain employed in our study carries the supE44 amber suppressor, which would lead to a substitution of glutamine for tyrosine 52 (12), it seems most likely that this substitution results in an inactive or unstable UmuD′ protein.
TABLE 1.
DNA sequence changes of umuD′ mutants
| Plasmid | Base substitution(s)a | Amino acid change(s)b | Source of mutationc |
|---|---|---|---|
| pGW5126 | 88GCA→GTA | A30V | HA |
| pGW5144 | 88GCA→GTA | A30V | HA |
| pGW5129 | 103GAA→AAA | E35K | HA |
| pGW5140 | 115GAT→GGT | D39G | HA |
| pGW5001 | 118CTG→CGG | L40R | SD |
| pGW5120 | 145AGC→AAC | S49N A50T | HA |
| 148GCG→ACG | |||
| pGW5130 | 151ACT→AAT | T51I | HA |
| pGW5003 | 223GAT→GCT | D75A | SD |
| pGW5141 | 385GGT→GAT | G129D | HA |
| pGW5124 | −60GAT→AAT | Promoter mutation | HA |
| pGW5142 | −60GAT→AAT | Promoter mutation | HA |
| pGW5127 | 298CAA→TAA | Ochre100d | HA |
| pGW5145 | 154TAC→TAG | Amber52e | HA |
| pGW5143 | 1ATG→ATA | M1I | HA |
| pGW5002 | 190GGT→GTT | G64V | SD |
| pGW5004 | 235ACG→GCC | T79A | SD |
| pGW5005 | 277GAG→GCG | E93A | SD |
| pGW5007 | 283ACG→GCG | T95A | SD |
| pGW5008 | 325CCC→CGC | P109R | SD |
The number indicates the position of the first base of the codon containing the base substitution. Nucleotide numbering is as for the umuD+ sequence, with number 1 corresponding to the first nucleotide of the umuD+ coding sequence.
Amino acids in UmuD′ have the same position number that they have in the intact UmuD protein.
The source of each mutation (either hydroxylamine treatment [HA] or site-directed mutagenesis [SD]) is indicated.
Ochre nonsense mutation at codon 100.
Amber nonsense mutation at codon 52.
The remaining mutated plasmids carried missense alleles that affect the ability of the UmuD′ protein to participate in UV mutagenesis. All caused single amino acid changes except for a double mutation that affected two adjacent amino acids (S49N A50T). The A30V mutant was isolated twice independently, but the other duplications in our collection of 16 mutants may not have been independent and are not listed in Table 1. As in our previous study (2), which also used hydroxylamine as a mutagen, the majority of the mutations resulted from G:C→A:T transitions. With the exception of the G129D mutation, all of the mutations affected amino acids in the N-terminal portion of the UmuD′ protein. By using site-directed mutagenesis, a few mutations that affected particular residues in the C-terminal portion of UmuD were also introduced (Table 1). A mutation resulting in the L40R change was also constructed because L40 is a highly conserved residue that lies within the UmuD′2 homodimer interface identified in our NMR studies (7).
Deficiencies of umuD′ mutants in mutagenesis induced by UV light and MMS.
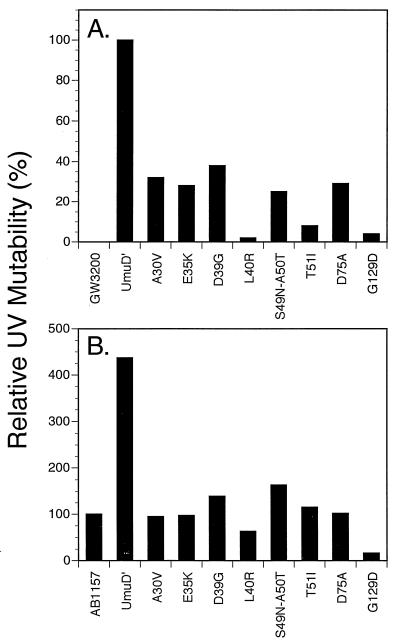
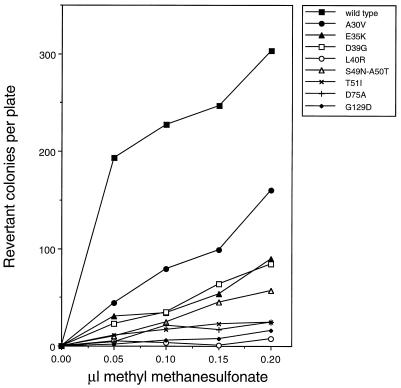
Derivatives of the umuD44 strain GW3200 carrying the umuD′ mutant plasmids described above were assayed for their abilities to be mutated in response to UV irradiation. Figure 1A compares the UV mutabilities of umuD44 derivatives carrying these missense mutant umuD′ plasmids to that of a derivative carrying the parental umuD′ plasmid. All of the mutants isolated in our screen had significant deficiencies in UV mutagenesis, with the G129D mutant being the most defective. Of the mutants constructed by site-directed mutagenesis, the L40R mutant had a severe deficiency in UV mutagenesis and the D75A mutant also was very significantly impaired; the other site-directed mutants showed less than a twofold reduction in UV mutagenesis (data not shown) and were not characterized further. These same derivatives were also tested for their ability to undergo the argE3→Arg+ reversion induced by the alkylating agent MMS (Fig. 2). The L40R and G129D mutants showed the most severe deficiencies in MMS mutagenesis, with the D75A and T51I mutants showing the next most severe. As mentioned above, one mutant umuD′ plasmid encodes two mutations, S49N and A50T, and was substantially deficient in MMS mutagenesis (Fig. 2). From our data alone, it is not possible to assess the relative contributions of the two alterations of UmuD′ towards the deficiency in MMS mutagenesis. However, in a related study, McLenigen et al. (18) isolated an A50T single mutant and found that it reduced MMS mutagenesis by less than a factor of two. Since we observed considerably more than a twofold reduction in MMS mutagenesis with the S49N A50T double mutant, the S49N alteration must also result in a reduction in mutagenesis proficiency. In fact, it seems likely that the reason that this derivative was detected in our screen was because the S49N and the A50T mutations combined to cause a phenotype that was readily detectable. Overall, the deficiencies of the mutant umuD′ strains in UV mutagenesis largely paralleled their deficiencies in MMS mutagenesis.
FIG. 1.
Effect of plasmids carrying umuD′ mutations on UV-induced (20 J/m2) reversion of argE3→Arg+ in an AB1157 umuD44 strain (GW3200) (A) and an isogenic umuD+ strain (AB1157) (B). UV mutability is expressed relative to the umuD44 strain carrying the umuD′ plasmid pGW2122 (17.8 induced Arg+ revertants/106 survivors) (A) and relative to the umuD+ strain AB1157 (8.3 induced Arg+ revertants/106 survivors) (B).
FIG. 2.
Effect of plasmids carrying umuD′ mutations on MMS-induced reversion of argE3→Arg+ in the AB1157 umuD44 strain, GW3200.
The plasmids producing umuD′ derivatives that were significantly impaired in UV and MMS mutagenesis were also introduced into an isogenic umuD+ strain to test whether the UmuD′ proteins they encoded were capable of interfering with the UV mutagenesis that is dependent on the chromosomal copy of the umuD+ gene. As shown in Fig. 1B, the introduction of the umuD′ derivative-producing plasmid into the umuD+C+ strain AB1157 substantially increases the level of UV mutagenesis that is observed, a finding indicating that UmuD′ is normally a limiting component for UV mutagenesis in UV-irradiated wild-type cells. In Fig. 1B, the level of UV mutagenesis observed with AB1157 derivatives carrying the various mutant umuD′ plasmids is expressed relative to that observed with the umuD+ strain, AB1157. Only two alleles, those encoding the L40R and G129D changes, exhibited a dominant negative effect over the chromosomal umuD+ allele even though the mutant UmuD′ derivatives were expressed at elevated levels from multicopy plasmids. Of the two, the G129D-encoding allele exhibited the strongest dominant negative effect. The somewhat higher level of UV mutagenesis observed in some other cases, for example with the plasmid carrying the D39G-encoding allele, is presumably due to the expression of elevated levels of a partially active UmuD′ protein.
In vivo levels of mutant UmuD′ derivatives.
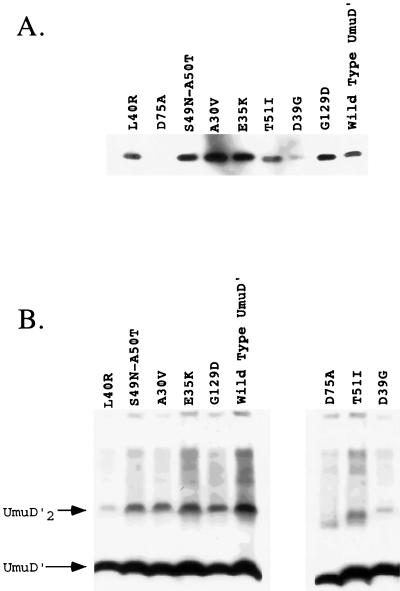
One possible explanation for the lack of activity of some of the UmuD′ mutants in UV and MMS mutagenesis could be that the mutant proteins were misfolded or structurally unstable and therefore subject to proteolytic degradation. To ascertain whether the UmuD′ mutants were present at levels sufficient to promote induced mutagenesis, their steady-state levels were compared to that of the plasmid-encoded wild-type UmuD′ in a lexA+ umuD44 strain (GW3200) following UV irradiation. No UmuD or UmuD′ was detected in the umuD44 strain in the absence of a umuD-expressing plasmid (data not shown). Repeated analyses indicated that with the exception of the mutants D39G and D75A and, possibly, T51I, all of the mutant UmuD′ proteins were present at levels comparable to or greater than that of wild-type UmuD′ (Fig. 3A).
FIG. 3.
The in vivo levels of the UmuD′ mutants (A) and their ability to form UmuD′ homodimers (B) were measured in crude lysates by using chemiluminescence immunodetection as described in Materials and Methods. (A) In vivo levels of the UmuD′ mutants relative to that of wild-type UmuD′ in a lexA+ strain (GW3200) following a 20-J/m2 UV dose are shown. (B) The abilities of the various UmuD′ mutants to be cross-linked as homodimers in vivo by formaldehyde are shown. The UmuD′ mutant proteins were expressed constitutively because of the lexA(Def) allele in strain GW8025. The positions of the UmuD′ monomer and UmuD′2 homodimer are indicated.
As is evident from the cross-linking experiments described in the next section, a strikingly different result was obtained when the levels of the mutant UmuD′ proteins were measured in a lexA(Def) strain. In this case, the SOS regulon is constitutively derepressed due to disruption of the lexA gene encoding the repressor of the SOS regulon. Whereas the D39G and D75A mutant proteins were barely detectable following UV irradiation of a lexA+ strain, they were nearly as abundant as wild-type UmuD′ when measured in a lexA(Def) strain (Fig. 3B). It is possible that the higher levels of RecA protein in the lexA(Def) strain are responsible for the stabilization of the mutant proteins (9). These results suggest that the poor activity of the mutants D39G, D75A, and, possibly, T51I in UV and MMS mutagenesis is due, at least in part, to their reduced abundance relative to that of wild-type UmuD′, whereas the poor mutability of the other UmuD′ mutants is due to a specific biochemical defect.
Ability of the mutant UmuD′ derivatives to form homodimers in vivo.
In addition to their in vivo levels, another explanation for the poor mutability of some of the UmuD′ mutants in UV and MMS mutagenesis could be that they have a defect in UmuD′2 homodimer formation. To determine if any of the UmuD′ mutants were poorly active in induced mutagenesis due to a defect in their ability to form UmuD′2 homodimers, we measured homodimerization in vivo by formaldehyde cross-linking. In these experiments, a lexA(Def) strain was used so that expression of the plasmid-encoded UmuD′ mutants was constitutive. Whereas the A30V, E35K, S49N, A50T, and G129D derivatives were comparable to wild-type UmuD′ with respect to their ability to form homodimers, the D39G, L40R, T51I, and D75A derivatives were severely impaired in this regard (Fig. 3B). Similar results were obtained when the plasmids carrying the mutant umuD′ alleles were examined in a lexA+ host in which umuD′ expression had been induced by a 20-J/m2 UV dose (data not shown), the only difference being that the D39G, T51I, and D75A derivatives were present at lower levels in the UV-irradiated cells. In light of these results, the impaired abilities of the D39G, L40R, T51I, and D75A derivatives to participate in UV and MMS mutagenesis are likely due, at least in part, to their reduced ability to form homodimers. With respect to the mutants D39G and D75A, their severely reduced steady-state levels relative to that of wild-type UmuD′ may be related to their reduced ability to form homodimers.
DISCUSSION
By a combination of direct screening and site-directed mutagenesis, we have identified several missense umuD′ mutants that are deficient in their ability to participate in UV-induced mutagenesis. By starting with an altered umuD gene that directly encodes UmuD′, we were able to obtain mutants that affect the role of UmuD′ in UV mutagenesis rather than mutants that interfere with UV mutagenesis indirectly by causing deficiencies in the RecA*-facilitated cleavage of UmuD, as in our earlier study (2). All of the mutants that we characterized had deficiencies in MMS mutagenesis that were largely similar to their deficiencies in UV mutagenesis, consistent with the UmuD′ protein playing similar mechanistic roles in the mutagenesis induced by both of these agents. Our observation that all of the mutants isolated by screening for deficiencies in UV mutagenesis were also deficient in MMS mutagenesis contrasts with the situation described by McLenigan et al. (18). Their primary screen was for plasmid-borne alleles of umuD′ that reduced the level of spontaneous mutagenesis observed in a lexA(Def) recA718 ΔumuDC strain that expressed UmuC from a medium-copy-number plasmid. In their case, only some of the umuD′ alleles that they identified exhibited substantial reductions in MMS mutagenesis; UV mutagenesis was not examined. Taken together, these two studies suggest that there are elements of UmuD′ structure that are important for the elevated umuD′C-dependent spontaneous mutation rate observed in a lexA(Def) recA718 genetic background that are unimportant, or of lesser importance, for the mutagenesis observed in wild-type cells whose DNA has been damaged by UV light or MMS.
Before discussing how particular amino acid changes might reduce the ability of the UmuD′ protein to participate in UV and MMS mutagenesis, it is worth pointing out that a mutationally induced change might (i) alter the core structure of the protein, rendering it nonfunctional or unable to bind to other proteins required for mutagenesis, or (ii) change the surface of the protein in a region that is involved in interactions with other proteins involved in the translesion synthesis mechanism that underlies SOS mutagenesis without substantially altering the overall structure of the protein. In some cases, a residue might be important for the structural integrity of UmuD′ and also lie on the surface, in which case either or both explanations for reduced functionality would apply.
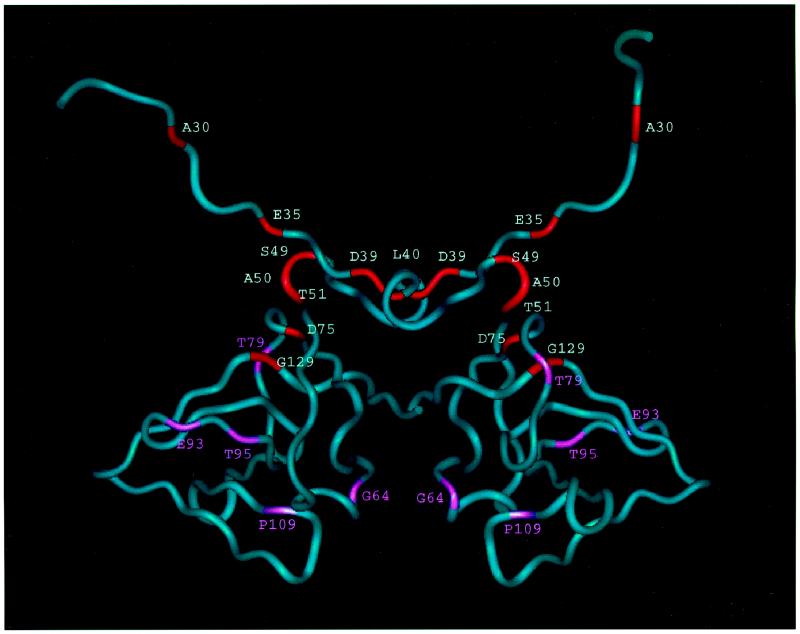
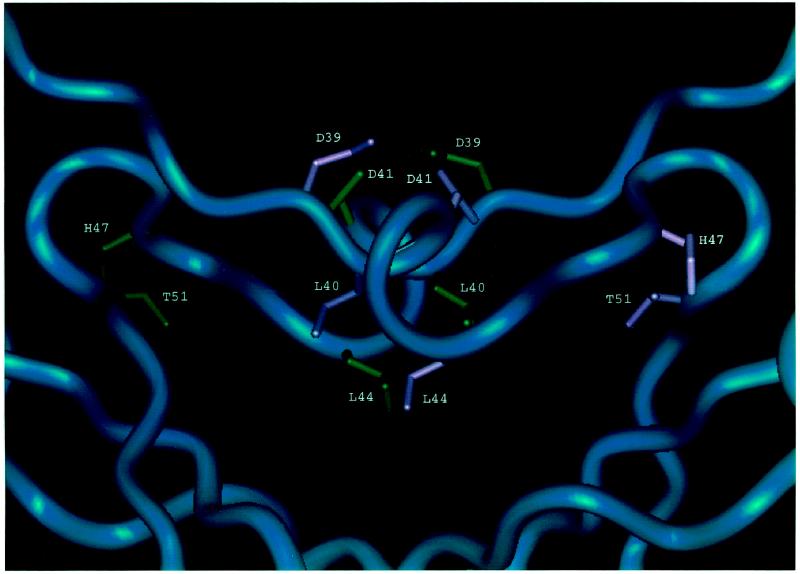
Four of the mutations that resulted in defective UV and MMS mutagenesis produced mutant UmuD′ proteins that had severe deficiencies in their abilities to form homodimers in vivo as determined by formaldehyde cross-linking: D39G, L40R, T51I, and D75A (Fig. 4 and 5). Three of these changes (D39G, L40R, and T51I) alter residues that are located at the homodimer interface that was identified by NMR studies of UmuD′2 in solution (7) and are not near the homodimer interface that had been assigned on the basis of crystallographic studies (22, 23). Thus, these mutant proteins provide additional support for our conclusion that the interface of the UmuD′2 homodimer in solution is the one involving contacts between the C termini and interactions involving N41 and L44 (7). As discussed below, the fourth alteration, D75A, appears to indirectly affect homodimer formation by causing a significant structural alteration of the UmuD′ monomer that interferes with its ability to dimerize. Furthermore, our observation that the mutant UmuD′ proteins displaying the most substantial reductions in homodimer formation in vivo also exhibited the lowest levels of UmuD′ protein in UV-irradiated lexA+ cells suggests that homodimer formation is an important factor in UmuD′ stability in vivo.
FIG. 4.
Worm representation of the UmuD′2 homodimer. Residues critical for UV mutagenesis are shown in red; those in pink are sites of missense mutations that caused less than a twofold reduction in UV mutagenesis.
FIG. 5.
Close-up view of the dimerization interface showing interactions of side chains that may be important for dimerization.
Although the UmuD′2 homodimer interface in solution may not exactly match the interface seen in the crystal that is rich in aliphatic contacts and involves interactions between the C termini (the one originally designated the filament dimer [22, 23]), it nevertheless appears to be a good approximation of the UmuD′2 homodimer interface that exists in solution (7). Part of this interface consists of interactions between the α-helices formed by residues 40 to 45 (Fig. 5). L40, the first amino acid of this helix, makes hydrophobic contacts with L44 of the other monomer so that the L40R mutation would be expected to disrupt these contacts and interfere with homodimer formation. Similarly, it appears that the D39G change also directly affects the homodimer interface since D39 normally hydrogen bonds to the amide of D41 in the opposite monomer. Residues 49 to 51 lie near the turn at residues 48 and 49 that poises the 40 to 45 α-helices so that they can interact with each other across the dimerization interface. The hydroxyl of T51 forms a hydrogen bond with the backbone carbonyl of H47 that is important for this turn. Mutating this residue to I51 would eliminate this bond and potentially be detrimental to the structure.
It is interesting that although in vivo formaldehyde cross-linking analyses indicated that the L40R UmuD′ derivative was impaired in UmuD′2 homodimer formation to approximately the same extent as the D39G, T51A, and D75A derivatives and is present in the cells at a higher level, it was markedly less proficient in UV and MMS mutagenesis. This observation suggests that the L40R mutation has an effect on UmuD′ function that goes beyond its influence on homodimer formation. This inference is additionally supported by our observation that the L40R derivative was only one of two umuD′ mutants that, when expressed from a multicopy-number plasmid, exerted a dominant negative effect on a chromosomal umuD+ gene with respect to UV mutagenesis. Thus, some feature of the structure of the L40R UmuD′ derivative enables it to interfere with the function of a wild-type protein.
The D75A alteration apparently interferes with homodimer formation indirectly by affecting the structure of the UmuD′ monomer and results in a UmuD′ derivative that is strikingly unstable in a UV-irradiated lexA+ host. D75 is located at one end of the central strand in a three-stranded β-sheet (Fig. 4) and is highly conserved across the family of mutagenesis and repressor proteins (10). Disruption of this three-stranded sheet by mutating D75 could prevent the protein from folding correctly and make it highly susceptible to proteolysis. In addition, the D75 side chain normally forms a hydrogen bond with the side chain of H47. A change of this residue to A75 would eliminate this interaction, which may be critical for holding together another part of the protein.
The G129D mutant protein, which was also identified by McLenigan et al. (18), is of particular interest because it is present in cells at levels comparable to that of the parental UmuD′ protein yet is severely deficient in its ability to participate in UV and MMS mutagenesis. Furthermore, the G129D mutant was the only derivative identified in our study that exerted a strongly dominant effect over a chromosomal umuD+ allele with respect to UV mutagenesis when it was expressed from a multicopy plasmid. Finally, the G129D mutant was identified in our earlier screen for umuD+ derivatives that were deficient in UV mutagenesis (2). In the context of the intact UmuD protein, the G129D mutation was found to cause a very severe defect in RecA*-mediated UmuD cleavage and to exert a strongly dominant negative effect on a chromosomal umuD+ allele with respect to UV mutagenesis (2). G129 is a critical structural residue, lying at the end of the long C-terminal β-strand and making hydrophobic contacts to residue V74 in the β-strand at positions 70 to 75 (Fig. 4). It is conserved across the family of mutagenesis proteins and repressors (10). Mutating the glycine to an aspartic acid would be expected to disrupt this interaction, causing some perturbation of the structure. It is intriguing that McLenigan et al. (18) found that the G129D UmuD′ derivative did not exert a dominant negative effect on the elevated levels of spontaneous mutagenesis observed in a recA730 mutant, a genetic background in which UmuD is constitutively cleaved to UmuD′ in the absence of exogenously induced DNA damage. One possible explanation is that there are requirements for UmuD′ function for the elevated spontaneous mutagenesis observed in a recA730 strain that are not identical to those required for UV and MMS mutagenesis in a wild-type strain, a possibility consistent with the properties of certain of the mutants identified by McLenigan et al. (18). An alternative explanation would be that the G129D UmuD′ derivative interferes with the RecA-mediated cleavage observed in a UV-irradiated cell but not with the RecA-mediated cleavage that occurs in a recA730 strain.
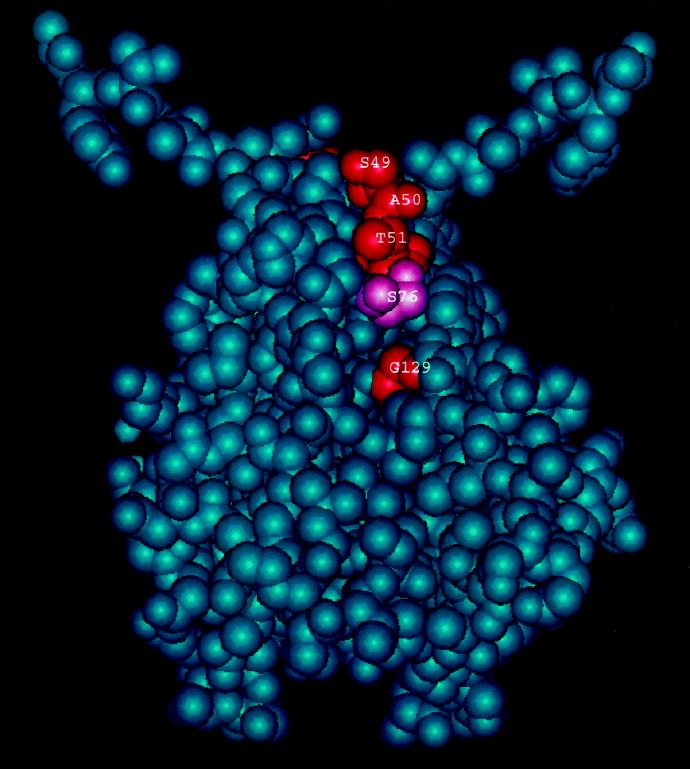
As discussed above, it appears that both the S49N and A50T mutations cause partial deficiencies in the ability of UmuD′ to participate in UV and MMS mutagenesis. S49 and A50 are located immediately next to the turn at residues 48 and 49 that positions the 40 to 45 α-helices at the homodimer interface, yet they do not cause an obvious impairment in homodimer formation. It is interesting that residues S49, A50, and T51 lie very near G129, with only residue D76 in between (Fig. 6). Thus, it seems that this region of UmuD′ is a likely candidate for a binding site for another protein.
FIG. 6.
Space-filling model of the UmuD′ dimer, rotated by 90° relative to the view shown in Fig. 4, and illustrating the spatial relationship of residues S49, A50, T51, S76, and G129.
Two of the most interesting mutants isolated in our screen for those deficient in UV mutagenesis were the ones that cause the A30V and E35K alterations in the N-terminal arms of the UmuD′2 homodimer. These arms have been shown to be mobile in solution (7). We have previously reported that derivatives of intact UmuD that carry monocysteine substitutions at positions 37 and 38 can be cross-linked by a disulfide bond with very high efficiency after oxidation with I2 (11), an observation suggesting that, in UmuD2, at least residues at these positions on the arms are constrained in a fashion that optimally positions them for rapid disulfide bond formation across the homodimer interface when I2 is added. Taken together with our observation that amino acids in the mobile N-terminal arms of UmuD′ are important for UV mutagenesis, these observations suggest that at least one purpose of RecA*-mediated cleavage of UmuD is to free up these arms so that they are able to interact with another protein in a fashion that was not possible while they were part of the UmuD2 homodimer. It will be interesting to learn how the changes in protein structure that occur after RecA*-mediated cleavage enable UmuD′ to participate in the process of translesion synthesis that is currently being analyzed in vitro (26, 33, 38).
ACKNOWLEDGMENTS
We thank the members of our laboratory, particularly Brad Smith, for their many helpful discussions and Leticia Vega and Lisa Lee for their assistance with the genetic screening. We also thank Roger Woodgate and his colleagues for sharing information with us in advance of publication.
This work was supported by Public Health Service grant CA21615 from the National Cancer Institute. A.E.F. was supported by an American Cancer Society postdoctoral fellowship and by the ONR Science Scholar program at the Bunting Institute of Radcliffe College. A.G. was supported by a postdoctoral fellowship from the Medical Research Council of Canada. S.C. carried out her research as part of the Undergraduate Research Opportunities Program (UROP) at the Massachusetts Institute of Technology.
REFERENCES
- 1.Bagg A, Kenyon C J, Walker G C. Inducibility of a gene product required for UV and chemical mutagenesis in Escherichia coli. Proc Natl Acad Sci USA. 1981;78:5749–5753. doi: 10.1073/pnas.78.9.5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Battista J R, Ohta T, Nohmi T, Sun W, Walker G C. Dominant negative umuD mutations decreasing RecA-mediated cleavage suggest roles for intact UmuD in modulation of SOS mutagenesis. Proc Natl Acad Sci USA. 1990;87:7190–7194. doi: 10.1073/pnas.87.18.7190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boudsocq F, Campbell M, Devoret R, Bailone A. Quantitation of the inhibition of Hfr x F− recombination by the mutagenesis complex UmuD′C. J Mol Biol. 1997;270:201–211. doi: 10.1006/jmbi.1997.1098. [DOI] [PubMed] [Google Scholar]
- 4.Bruck I, Woodgate R, McEntee K, Goodman M F. Purification of a soluble UmuD′C from Escherichia coli: cooperative binding of UmuD′C to single-stranded DNA. J Biol Chem. 1996;271:10767–10774. doi: 10.1074/jbc.271.18.10767. [DOI] [PubMed] [Google Scholar]
- 5.Burckhardt S E, Woodgate R, Scheuermann R H, Echols H. UmuD mutagenesis protein of Escherichia coli: overproduction, purification, and cleavage by RecA. Proc Natl Acad Sci USA. 1988;85:1811–1815. doi: 10.1073/pnas.85.6.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elledge S J, Walker G C. Proteins required for ultraviolet light and chemical mutagenesis: identification of the products of the umuC locus of Escherichia coli. J Mol Biol. 1983;164:175–192. doi: 10.1016/0022-2836(83)90074-8. [DOI] [PubMed] [Google Scholar]
- 7.Ferentz A E, Opperman T, Walker G C, Wagner G. Dimerization of the UmuD′ protein in solution and its implications for regulation of SOS mutagenesis. Nat Struct Biol. 1997;4:979–983. doi: 10.1038/nsb1297-979. [DOI] [PubMed] [Google Scholar]
- 8.Frank E G, Ennis D G, Gonzalez M, Levine A S, Woodgate R. Regulation of SOS mutagenesis by proteolysis. Proc Natl Acad Sci USA. 1996;93:10291–10296. doi: 10.1073/pnas.93.19.10291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frank E G, Hauser J, Levine A S, Woodgate R. Targeting of the UmuD, UmuD′ and MucA′ mutagenesis proteins to DNA by the RecA protein. Proc Natl Acad Sci USA. 1993;90:8169–8173. doi: 10.1073/pnas.90.17.8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedberg E C, Walker G C, Siede W. DNA repair and mutagenesis. Washington, D.C: The American Society for Microbiology; 1995. [Google Scholar]
- 11.Guzzo A, Lee M H, Oda K, Walker G C. Analysis of the region between amino acids 30 and 42 of intact UmuD by a monocysteine approach. J Bacteriol. 1996;178:7295–7303. doi: 10.1128/jb.178.24.7295-7303.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inokuchi H, Yamao F, Sakano H, Ozeki H. Identification of transfer tRNA suppressors in Escherichia coli. J Mol Biol. 1979;132:649–662. doi: 10.1016/0022-2836(79)90380-2. [DOI] [PubMed] [Google Scholar]
- 13.Kato T, Shinoura Y. Isolation and characterization of mutants of Escherichia coli deficient in induction of mutations by ultraviolet light. Mol Gen Genet. 1977;156:121–131. doi: 10.1007/BF00283484. [DOI] [PubMed] [Google Scholar]
- 14.Koch H K, Woodgate R. The SOS response. In: Nickoloff J A, Hoekstra M F, editors. DNA damage and repair. 1. DNA repair in prokaryotes and lower eukaryotes. Totowa, N.J: Humana Press; 1998. pp. 107–134. [Google Scholar]
- 15.Lee M H, Ohta T, Walker G C. A monocysteine approach for probing the structure and interactions of the UmuD protein. J Bacteriol. 1994;176:4825–4837. doi: 10.1128/jb.176.16.4825-4837.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Little J W. LexA cleavage and other self-processing reactions. J Bacteriol. 1993;175:4943–4950. doi: 10.1128/jb.175.16.4943-4950.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livneh Z, Cohen-Fix O, Skaliter R, Elizur T. Replication of damaged DNA and the molecular mechanism of ultraviolet light mutagenesis. Crit Rev Biochem Mol Biol. 1993;28:465–513. doi: 10.3109/10409239309085136. [DOI] [PubMed] [Google Scholar]
- 18.McLenigan M, Peat T S, Frank E G, McDonald J P, Gonzalez M, Levine A S, Hendrickson W A, Woodgate R. Novel Escherichia coli umuD′ mutants: structure-function insights into SOS mutagenesis. J Bacteriol. 1998;180:4658–4666. doi: 10.1128/jb.180.17.4658-4666.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nohmi T, Battista J R, Dodson L A, Walker G C. RecA-mediated cleavage activates UmuD for mutagenesis: mechanistic relationship between transcriptional derepression and posttranslational activation. Proc Natl Acad Sci USA. 1988;85:1816–1820. doi: 10.1073/pnas.85.6.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Opperman, T., S. Murli, and G. C. Walker. A DNA damage checkpoint control mechanism that regulates DNA synthesis and growth. Submitted for publication.
- 21.Opperman T, Murli S, Walker G C. The genetic requirements for UmuDC-mediated cold sensitivity are distinct from those for SOS mutagenesis. J Bacteriol. 1996;178:4400–4411. doi: 10.1128/jb.178.15.4400-4411.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peat T S, Frank E G, McDonald J P, Levine A S, Woodgate R, Hendrickson W A. Structure of the UmuD′ protein and its regulation in response to DNA damage. Nature. 1996;380:727–730. doi: 10.1038/380727a0. [DOI] [PubMed] [Google Scholar]
- 23.Peat T S, Frank E G, McDonald J P, Levine A S, Woodgate R, Hendrickson W A. The UmuD′ protein filament and its potential role in damage-induced mutagenesis. Structure. 1996;4:1401–1412. doi: 10.1016/s0969-2126(96)00148-7. [DOI] [PubMed] [Google Scholar]
- 24.Rajagopalan M, Lu C, Woodgate R, O’Donnell M, Goodman M F, Echols H. Activity of the purified mutagenesis proteins UmuC, UmuD′, and RecA in replicative bypass of an abasic DNA lesion by DNA polymerase III. Proc Natl Acad Sci USA. 1992;89:10777–10781. doi: 10.1073/pnas.89.22.10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Record M T, Jr, Reznikoff W S, Craig M L, McQuade K L, Schlax P J. Escherichia coli RNA polymerase (Eς70) promoters, and the kinetics of the steps of transcription initiation. In: Neidhardt F, Curtiss R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W, Riley M, Schaecter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: American Society for Microbiology; 1998. pp. 792–821. [Google Scholar]
- 26.Reuven N B, Tomer G, Livneh Z. The mutagenesis proteins UmuD′ and UmuC prevent lethal frameshifts while increasing base substitution mutations. Mol Cell. 1998;2:191–199. doi: 10.1016/s1097-2765(00)80129-x. [DOI] [PubMed] [Google Scholar]
- 27.Shinagawa H, Iwasaki H, Kato T, Nakata A. RecA protein-dependent cleavage of UmuD protein and SOS mutagenesis. Proc Natl Acad Sci USA. 1988;85:1806–1810. doi: 10.1073/pnas.85.6.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skare J T, Ahmer B M, Seachord C L, Darveau R P, Postle K. Energy transduction between membranes. TonB, a cytoplasmic membrane protein, can be chemically cross-linked in vivo to the outer membrane receptor FepA. J Biol Chem. 1993;268:16302–16308. [PubMed] [Google Scholar]
- 29.Slilaty S N, Little J W. Lysine-156 and serine-119 are required for LexA repressor cleavage: a possible mechanism. Proc Natl Acad Sci USA. 1987;84:3987–3991. doi: 10.1073/pnas.84.12.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith B T, Walker G C. Mutagenesis and more: umuDC and the Escherichia coli SOS response. Genetics. 1998;148:1599–1610. doi: 10.1093/genetics/148.4.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sommer S, Bailone A, Devoret R. The appearance of the UmuD′C protein complex in Escherichia coli switches repair from homologous recombination to SOS mutagenesis. Mol Microbiol. 1993;10:963–971. doi: 10.1111/j.1365-2958.1993.tb00968.x. [DOI] [PubMed] [Google Scholar]
- 32.Steinborn G. Uvm mutants of Escherichia coli K12 deficient in UV mutagenesis. I. Isolation of uvm mutants and their phenotypical characterization in DNA repair and mutagenesis. Mol Gen Genet. 1978;165:87–93. doi: 10.1007/BF00270380. [DOI] [PubMed] [Google Scholar]
- 33.Tang M, Bruck I, Eritja R, Turner J, Frank E G, Woodgate R, O’Donnell M, Goodman M F. Biochemical basis of SOS-mutagenesis in Escherichia coli: reconstitution of in vitro lesion bypass dependent on the UmuD′2C mutagenic complex and RecA protein. Proc Natl Acad Sci USA. 1998;95:9755–9760. doi: 10.1073/pnas.95.17.9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walker G C. Plasmid (pKM101)-mediated enhancement of repair and mutagenesis: dependence on chromosomal genes in Escherichia coli K-12. Mol Gen Genet. 1977;152:93–103. doi: 10.1007/BF00264945. [DOI] [PubMed] [Google Scholar]
- 35.Walker G C. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Rev. 1984;48:60–93. doi: 10.1128/mr.48.1.60-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walker G C. SOS-regulated proteins in translesion synthesis and mutagenesis. Trends Biochem Sci. 1995;10:416–420. doi: 10.1016/s0968-0004(00)89091-x. [DOI] [PubMed] [Google Scholar]
- 37.Walker G C. The SOS response of Escherichia coli. In: Neidhardt F, Curtiss R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W, Riley M, Schaecter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: American Society for Microbiology; 1996. pp. 1400–1416. [Google Scholar]
- 38.Walker G C. Skiing the black diamond slope: progress on the biochemistry of translesion synthesis. Proc Natl Acad Sci USA. 1998;95:10348–10350. doi: 10.1073/pnas.95.18.10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walker G C, Dobson P P. Mutagenesis and repair deficiencies of Escherichia coli umuC mutants are suppressed by the plasmid pKM101. Mol Gen Genet. 1979;172:17–24. doi: 10.1007/BF00276210. [DOI] [PubMed] [Google Scholar]
- 40.Woodgate R, Rajagopalan M, Lu C, Echols H. UmuC mutagenesis protein of Escherichia coli: purification and interaction with UmuD and UmuD′. Proc Natl Acad Sci USA. 1989;86:7301–7305. doi: 10.1073/pnas.86.19.7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woodgate R, Sedgwick S. Mutagenesis induced by bacterial UmuDC proteins and their plasmid homologues. Mol Microbiol. 1992;6:2213–2218. doi: 10.1111/j.1365-2958.1992.tb01397.x. [DOI] [PubMed] [Google Scholar]