ABSTRACT
Intellectual disability is a neurodevelopmental disorder that affects 2-3% of the general population. Syndromic forms of intellectual disability frequently have a genetic basis and are often accompanied by additional developmental anomalies. Pathogenic variants in components of TATA-binding protein associated factors (TAFs) have recently been identified in a subset of patients with intellectual disability, craniofacial hypoplasia, and congenital heart disease. This syndrome has been termed as a TAFopathy and includes mutations in TATA binding protein (TBP), TAF1, TAF2, and TAF6. The underlying mechanism by which TAFopathies give rise to neurodevelopmental, craniofacial, and cardiac abnormalities remains to be defined. Through a forward genetic screen in zebrafish, we have recovered a recessive mutant phenotype characterized by craniofacial hypoplasia, ventricular hypoplasia, heart failure at 96 h post-fertilization and lethality, and show it is caused by a nonsense mutation in taf5. CRISPR/CAS9 mediated gene editing revealed that these defects where phenocopied by mutations in taf1 and taf5. Mechanistically, taf5-/- zebrafish displayed misregulation in metabolic gene expression and metabolism as evidenced by RNA sequencing, respiration assays, and metabolite studies. Collectively, these findings suggest that the TAF complex may contribute to neurologic, craniofacial, and cardiac development through regulation of metabolism.
Keywords: TAFopathy, TAF1, TAF5, Heart development, Craniofacial development, Metabolism
Summary: Pathogenic variants in components of TATA-binding protein associated factors (TAFs) have been identified in patients with intellectual disability, craniofacial hypoplasia, and congenital heart disease. Our findings demonstrate that the TAF complex contributes to neurologic, craniofacial, and cardiac development through regulation of metabolism.
INTRODUCTION
Birth defects are a major cause of morbidity and mortality worldwide. Between 3-5% of live born infants have birth defects including cardiac, craniofacial, and neurodevelopmental abnormalities that each contribute to adverse perinatal outcomes (Webber et al., 2015). While significant progress has been made identifying environmental contributors to birth defects, our understanding of their genetic basis remains incomplete (Kirby, 2017).
Transcriptional dysregulation represents an important mechanism leading to birth defects (Prescott and Wilkie, 2007). Gene regulatory networks that establish cell states, pattern tissues, and regulate cell differentiation and organ maturation are controlled by thousands of transcription factors, cofactors, and chromatin regulators. Failure to regulate these programs leads to a diverse array of developmental syndromes, including but not limited to, growth and intellectual disability, limb deformities, craniofacial anomalies, and congenital heart defects (Jenkins et al., 2007; de Soysa et al., 2019). It is increasingly recognized that pathogenic variants in key transcription factors are responsible for many developmental syndromes. These ‘general regulator’ transcription factors control gene expression and chromatin accessibility through selective recruitment of RNA Polymerase II and epigenetic machinery to specific genomic loci (Clark et al., 2006; Chen et al., 2022).
Recently, pathogenic variants in TATA Binding Protein Associated Factors (TAFs) key components of transcription factor TFIID have been identified in a subset of patients with intellectual disability, craniofacial hypoplasia, and congenital heart disease (O'Rawe et al., 2015). This syndrome has been termed as a ‘TAFopathy’ and includes mutations in TATA Binding Protein (TBP), TAF1, TAF2, TAF6, TAF8, and TAF13 (Rooms et al., 2006; Tawamie et al., 2017; Hellman-Aharony et al., 2013; Alazami et al., 2015). The underlying mechanism by which TAFopathies give rise to neurodevelopmental, craniofacial, and cardiac abnormalities is not understood.
TFIID is a large complex comprised of multiple proteins including TBP and multiple TAFs. The complete TFIID complex and holo-TFIID (lacking TBP) bind to distinct DNA sequences (Wu and Chiang, 2001). Previous studies have identified specific DNA sequences to which TAFs bind within initiator and downstream promoter elements (Juven-Gershon et al., 2006; Juven-Gershon and Kadonaga, 2010). TAFs also interact with enhancers through bromodomain motifs (Lehmann et al., 2012). TAF1 and TAF5 are core components of TFIID complexes. TAF1 binds to DNA and TBP, forming a complex that recruits RNA Polymerase II at sites of transcription. TAF5 acts as a scaffold protein important for the assembly of TFIID components (Bieniossek et al., 2013; Wang et al., 2014). Further adding to the complexity, distinct TFIID complexes consisting of unique TAF combinations may mediate the effect of different transcriptional activators (Brou et al., 1993; Asturias, 2009; Louder et al., 2016).
Here, we report a novel zebrafish model of TAFopathies caused by nonsense mutations in taf5. taf5−/− zebrafish display reduced survival, heart failure, and facial and cardiac hypoplasia and fail to thrive. These findings were recapitulated in taf1−/− zebrafish. Mechanistically, we reveal roles for TAF1 and TAF5 in coordinating metabolic programs essential for craniofacial and cardiac development.
RESULTS
corastl325/stl325 embryos show signs of early-onset heart failure
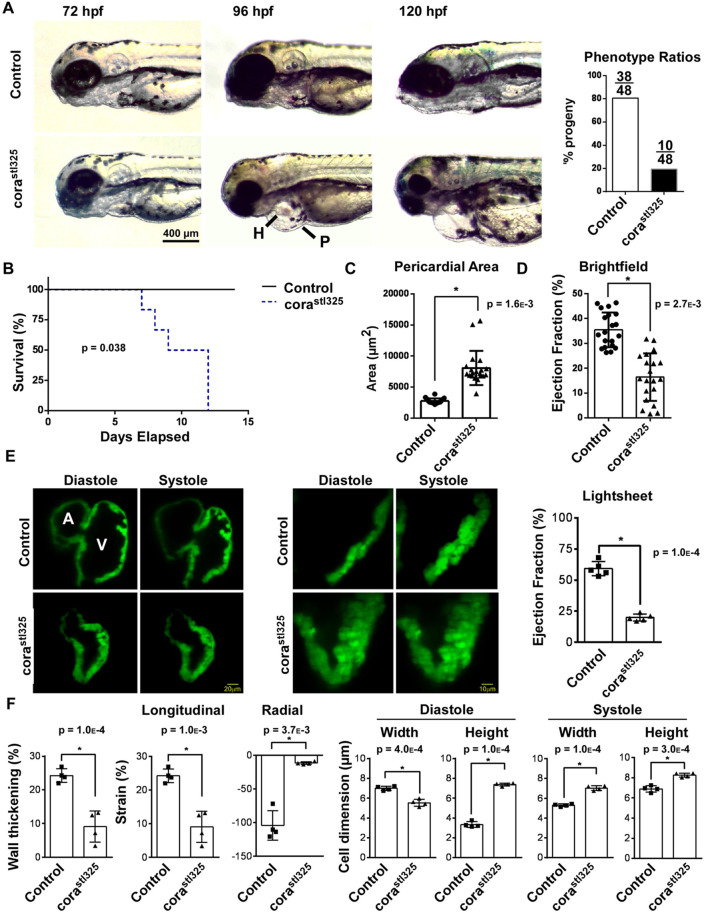
We recovered a mutation from an ENU mutagenesis screen that displayed a small heart and craniofacial hypoplasia and lethality, which we referred to as corazoncitostl325 (corastl325) (Gray et al., 2021). Affected embryos demonstrate evidence of heart failure in Mendelian ratios suggestive of a recessive allele (Fig. 1A). To characterize the cardiac phenotype in detail, we first observed the temporal progression of heart failure in progeny obtained from a corastl325/+ carrier self-cross. corastl325/stl325 embryos displayed a collapsed heart and pericardial edema beginning at 96 h post-fertilization (hpf). corastl325/stl325 hearts were visually indistinguishable from unaffected clutch mates at 72 hpf (Fig. 1A). We observed 100% mortality of embryos that displayed the corazoncito phenotype within 2 weeks post-fertilization (Fig. 1B). Quantitatively, corastl325/stl325 embryos had a nearly three-fold increase in pericardial surface area compared to unaffected clutch mates (Fig. 1C). Measurement of ejection fraction by bright field imaging indicated that corazoncito embryos displayed evidence of systolic ventricular dysfunction (Fig. 1D).
Fig. 1.
corastl325/stl325 embryos display reduced survival and heart failure. (A) Light microscopy showing the emergence of pericardial edema between 72 and 120 h post fertilization (hpf). P, pericardium; H, heart. Phenotype ratios are presented on the right. (number of trials=4; number of samples per trial=48) (B) Survival curve of corastl325 embryos relative to their unaffected clutchmates (number of trials=4; number of samples per trial=48). (C) Quantification of light microscopy images measuring area of the pericardium (number of trials=4; number of samples per trial=48). (D) Quantification of light microscopy images measuring ejection fraction (number of trials=4; number of samples per trial=48). (E) Lightsheet microscopy generated images of 96 hpf embryos harboring the cmlc::GFP transgenic reporter. Hearts are shown in systole and diastole. A, atrium; V, ventricle. Quantification of ejection fraction (n=4). (F) Quantification of changes in cell dimension using lightsheet-generated videos (n=4).
To further characterize defects in cardiac function, we crossed corastl325/+ to the cardiomyocyte-specific fluorescent reporter, Tg(cmlc2: GFP) (Huang et al., 2003). Lightsheet microscopy performed at 96 hpf confirmed reduced ejection fraction in corazoncito embryos (Fig. 1E,F; Movies 1,2). Lightsheet microscopy afforded us sufficient resolution to visualize changes in wall thickness and cellular dimensions throughout the cardiac cycle. This technique revealed that corastl325/stl325 embryos displayed significant decreases in systolic wall thickening, longitudinal, and radial strain. Measurement of cardiomyocyte dimensions throughout the cardiac cycle indicated that corastl325/stl325 cardiomyocytes failed to shorten and thicken in systole suggestive of systolic dysfunction. Furthermore, we observed that corastl325/stl325 cardiomyocytes were shorter and thicker in diastole compared to controls indicative of concomitant diastolic dysfunction (Fig. 1F).
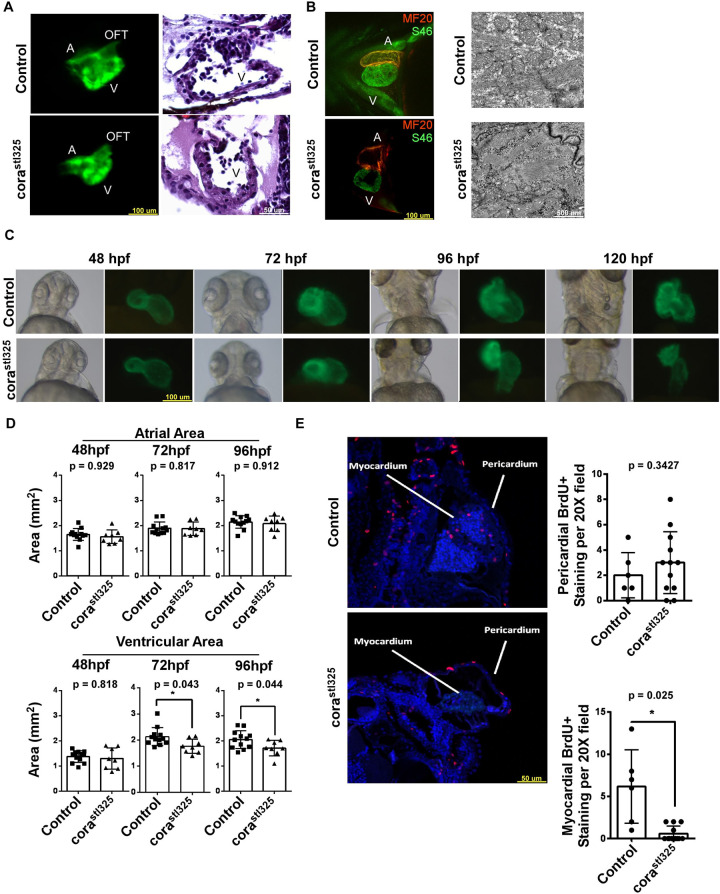
corastl325/stl325 hearts show normal atrial and ventricular architecture, chamber patterning, and sarcomere formation (Fig. 2A,B). Serial imaging of corastl325/stl325 Tg(cmlc2:GFP) hearts revealed diminished ventricular area compared to clutch mates beginning at 72 hpf. Atrial area did not differ between experimental groups (Fig. 2C,D). Measurement of bromodeoxyuridine (BrdU) incorporation – a synthetic nucleoside analogue used to study cell proliferation in living tissues – showed reduced abundance of replicating myocardial cells in corastl325/stl325 embryos compared to unaffected siblings. We did not observe differences in abundance of replicating pericardial cells as indicated by the number of BrdU positive cells present in the pericardium (Fig. 2E). Together, these data support the hypothesis that corastl325/stl325 embryos show signs of embryonic heart failure and ventricular hypoplasia.
Fig. 2.
corastl325 hearts display hypoplasia and reduced proliferation. (A) Fluorescent microscopy images of 96 hpf embryos (left, cmlc2::GFP; n=8) and H&E staining (right; n=4). (B) Fluorescent microscopy images of 96 hpf embryos stained with the SF46 (orange) and MF20 (green) showing normal atrioventricular patterning (left). Electron microscopy showing intact sarcomeres in control and corastl325 cardiomyocytes (right; n=4). (C) Brightfield and fluorescence microscopy time-course of cmlc2::GFP expression between 48 and 120 hpf (Control (n=12) and corastl325 (n=8). (D) Quantification of atrial and ventricular cross-sectional areas obtained from C [Control (n=12) and corastl325 (n=8)]. (E) BrdU immunostaining (red) of 96 hpf embryos embedded in paraffin [Control (n=6) and corastl325 (n=12)].
corastl325/stl325 embryos have craniofacial deformities and neuroanatomical defects
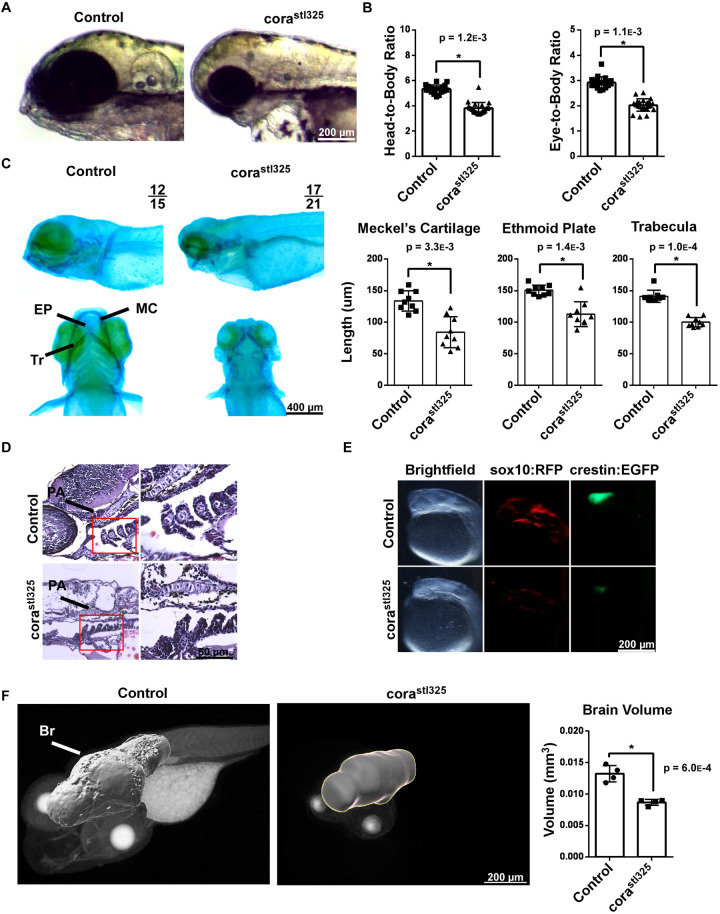
In addition to cardiac defects, we observed evidence of craniofacial hypoplasia in corastl325/stl325 embryos (Fig. 3A). At 96 hpf, corazoncito mutants displayed a 33% reduction in head-to-body and eye-to-body ratios despite slight reductions in body length seen in corastl325/stl325 embryos (Fig. 3B). We performed Alcian Blue staining to evaluate cartilaginous structures, which revealed hypoplastic jaw cartilage (Meckel's Cartilage, Ethmoid plate, Trabecula) in corastl325/stl325 embryos (Fig. 3C). Histology of 96 hpf sections suggest that chondrocytes that were specified appear morphologically similar between cora and wild-type clutchmates (Fig. 3D). Imaging of Tg(crestin::GFP) (migratory neural crest cell marker) and Tg(sox10::RFP) (cartilage progenitor marker) embryos demonstrated evidence of delayed neural crest cell migration and cartilage specification in corastl325/stl325 embryos (Fig. 3E) (Kaufman et al., 2016). X-ray microscopy further demonstrated reduced brain volume in corastl325/stl325 embryos compared to unaffected siblings (Fig. 3F). Together, these data indicate that corastl325/stl325 embryos display impaired craniofacial and neurodevelopment.
Fig. 3.
corastl325 embryos have craniofacial deformities and neuroanatomical defects. (A) Head morphology of control and corastl325 embryos at 96 hpf. (B) Head-to-body and head-to-eye ratios. Control (n=13), corastl325 (n=9). (C) Alcian Blue staining of control and corastl325 embryos at 96 hpf (left), with quantification of major jaw elements (right). EP, Ethmoid plate; MC, Meckel's Cartilage; T, Trabeculae. n=9 per experimental group. (D) H&E staining of the jaw of control and corastl325 embryos at 96 hpf. (E) Fluorescence microscopy images of Tg(crestin::GFP) and Tg(sox10::RFP) control and corastl325 embryos. (F) X-ray microscopy-generated images of 96 hpf embryos (left) and quantification of tissue volume (right). Br, brain Control (n=4), corastl325 (n=4).
corastl325/stl325 is a nonsense mutation in taf5
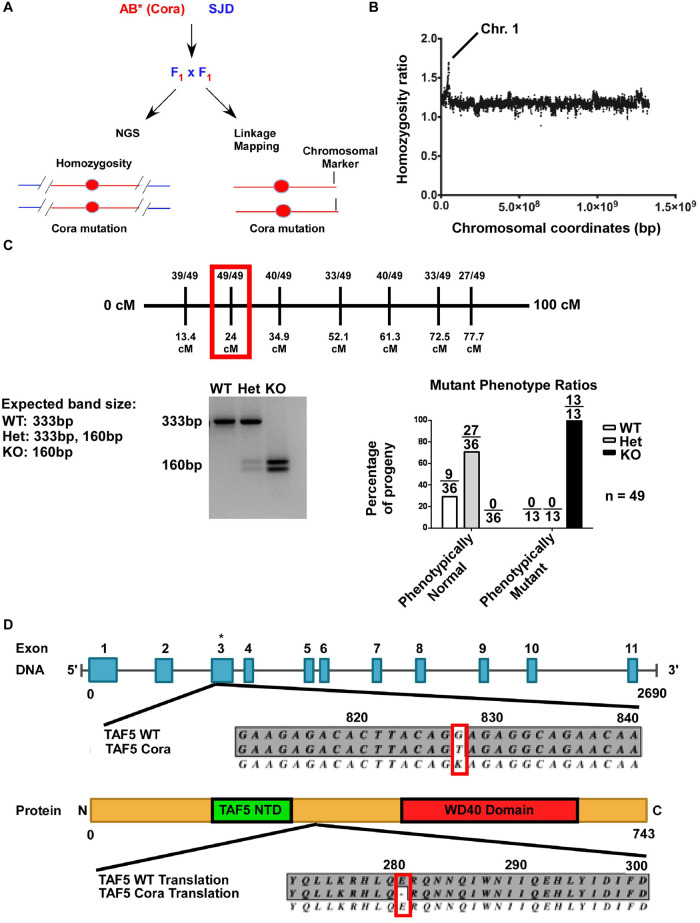
To map the corastl325/stl325 mutation, we crossed corastl325/stl325 carriers (SAT background) into the SJD mapping background as described in Gray et al. (2021) (Kaufman et al., 2016). corastl325/stl325/SJD F1 carriers were identified and self-crossed. F2 embryos were collected, pooled into corastl325/stl325 and unaffected clutch mate groups, genomic DNA isolated, and whole genome sequencing performed (Fig. 4A). We identified an area on Chromosome 1 that retained homozygosity for the AB* background that was found exclusively in embryos displaying the corastl325/stl325 phenotype (Fig. 4B). Fine chromosomal mapping identified a region on Chromosome 1 (24cM) that was closely linked with the corazoncito phenotype (Fig. 4C). By curating mutations found in the mapped region, we discovered a nonsense mutation in exon 3 of taf5 (Fig. 4D). We developed a genotyping assay that specifically targeted the mutant base-pair substitution by HpyCH4III restriction digest. Genotyping of 49 embryos demonstrated 100% association between the mutated base pair and corazoncito phenotype, although Sanger sequencing of individual mutant embryos has not been performed (Fig. 4C).
Fig. 4.
corastl325 encodes a nonsense mutation in taf5. (A) Workflow for mapping corastl325 using whole genome next generation sequencing. (B) Manhattan plot displaying homozygosity ratio for the AB* background (y-axis) as a function of chromosome location (x-axis). A value of 2.0 indicates 100% homozygosity for the AB* background. Chr. 1: chromosome 1. (C) Fine chromosomal mapping identified a 24 cM region (red box) that was linked with the corastl325 phenotype (top). Genotyping to identify the point mutation leading to a nonsense mutation in taf5. The nonsense mutation was linked to the corastl325 phenotype (bottom). (D) Graphical representation of the taf5 genomic locus and TAF5 protein including major functional domains. Red box indicates the mutated base and resulting nonsense mutation.
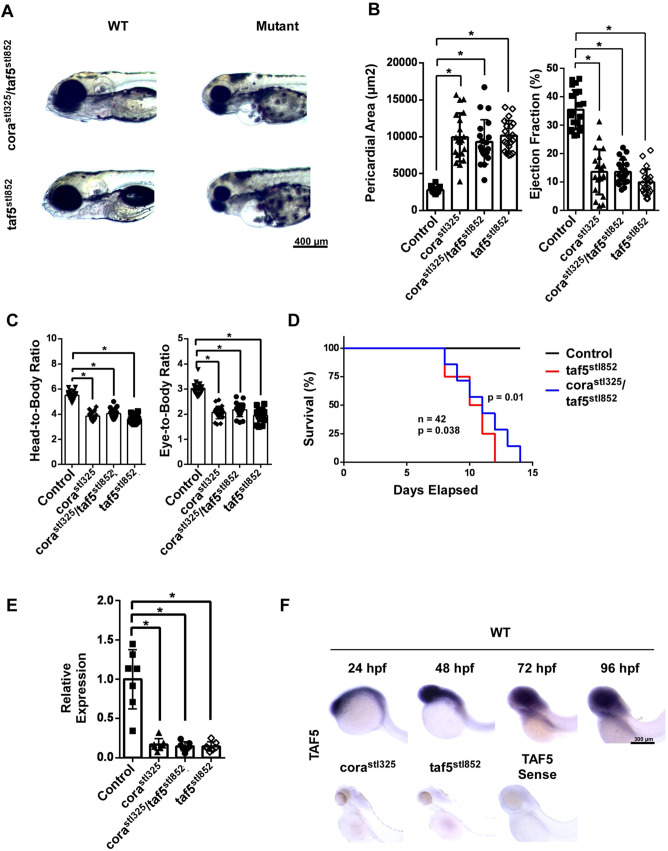
To rigorously implicate taf5 loss of function as the cause of the corazoncito phenotype, we generated a stable taf5 null allele using CRISPR/Cas9-mediated mutagenesis (referred to as taf5stl852). taf5stl852/stl852 embryos recapitulated the corazoncito phenotype of heart failure and craniofacial hypoplasia. Furthermore, corastl325 failed to complement the taf5stl852 allele (Fig. 5A). Quantitative measurements of pericardial edema, ejection fraction, head-to-body, and eye-to-body ration were consistent across corastl325/stl325, taf5stl852/stl852, and corastl325/taf5stl852 groups (Fig. 5B,C). Measurement of taf5 mRNA expression by quantitative RT-PCR revealed marked reductions in taf5 expression in corastl325/stl325, taf5stl852/stl852, and corastl325/taf5stl852 embryos compared to controls (Fig. 5E). WISH further revealed that taf5 was expressed in the whole head and whole heart region. No taf5 mRNA was detected in corastl325/stl325 or taf5stl852/stl852 embryos (Fig. 5F). Collectively, these findings support the conclusion that taf5 loss of function is responsible for the corazoncito phenotype.
Fig. 5.
corastl325 is a null allele of taf5. (A) Brightfield images of control, corastl325, taf5stl852, and corastl325/ taf5stl852 embryos at 96 hpf. (B) Measurement of pericardial area and ejection fraction in control, corastl325, taf5stl852, and corastl325/ taf5stl852 embryos at 96 hpf. n=22. (C) Measurement of head-to-body and head-to-eye ratios in control, corastl325, taf5stl852, and corastl325/ taf5stl852 embryos at 96 hpf. n=22. (D) Survival of control, taf5stl852, and corastl325/taf5 CRISPR embryos. n=42. (E) RTPCR for taf5 mRNA in control, corastl325, taf5stl852, and corastl325/ taf5stl852 embryos at 96 hpf. n=7. (F) Time course of taf5 mRNA expression by whole-mount in situ hybridization (top) and taf5 mRNA expression in corastl325 and taf5stl852 embryos at 96 hpf. Note: all brackets marked with ‘*’ represent P<1E-4.
taf1 deletion recapitulates the corazoncito phenotype
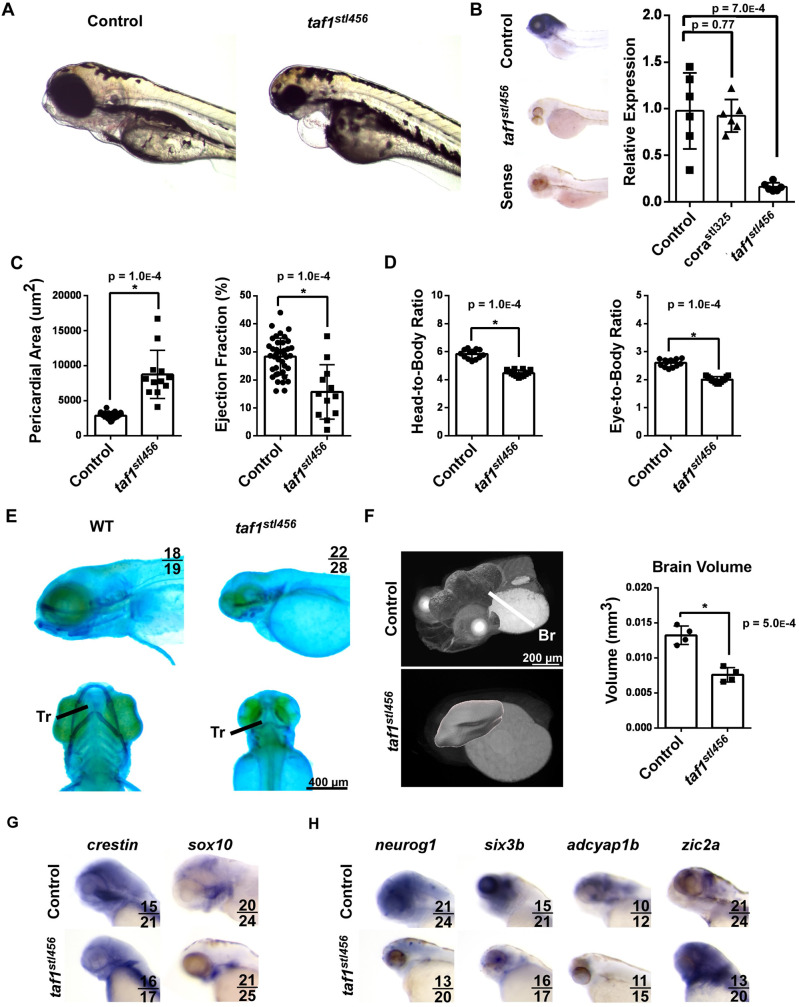
TAF5 acts as a scaffold protein important for the assembly of TFIID components (Bieniossek et al., 2013). Coincidentally, studies on TAF1, the catalytic subunit of TFIID, have shown that knockdown of TAF1 results in a similar phenotype of aberrant neurodevelopment (Jacobson et al., 2000). To directly compare phenotypes elicited by deletion of taf5 and taf1, we utilized Cas9/CRISPR gene editing to generate a new null allele of taf1 (referred to as taf1stl456/stl456). We observed evidence of pericardial edema, reduced ejection fraction, and craniofacial hypoplasia in taf1stl456/stl456 mutant embryos relative to unaffected siblings. Consistent with these phenotypes taf1 mRNA was expressed in the head and heart regions at 96 hpf (Fig. 6A-D). Alcian Blue staining demonstrated severe hypoplasia and loss of craniofacial cartilaginous structures (Fig. 6E), whereas X-ray microscopy showed reduced brain volume (Fig. 6F). We next performed whole mount in situ hybridization for markers of craniofacial development and brain patterning. WISH indicated that crestin expression was similar between control and taf1stl456/stl456 embryos, suggesting that neural crest cells specification is not impacted by taf1 inactivation. In contrast, sox10 expression was markedly reduced indicating a defect in the differentiation of cartilage progenitors from neural crest cells (Fig. 6G). We also observed decreased expression of pan-neural (neurog1), forebrain (six3b), and midbrain (adcyap1b) markers and an expansion of the hindbrain marker, zic2a (Fig. 6H). Collectively, these findings indicate an overlapping phenotype between taf1 and taf5 mutant embryos that recapitulates key features of TAFopathies. The phenotype of taf1 mutant embryos was comparable to that of taf5 mutant embryos.
Fig. 6.
taf1 deletion recapitulates the corazoncito phenotype. (A) Brightfield images of control and taf1stl456/stl456 embryos at 96 hpf. (B) taf1 mRNA expression measured by in situ hybridization (left) and RT-PCR (right). N=6 per experimental group. (C) Quantification of pericardial area (left) and ejection fraction (right) in control (n=30) and taf1stl456/stl456 (n=12) embryos at 96 hpf. (D) Quantification of head-to-body ratio (left) and head-to-eye ratio (right) in control (n=21) and taf1stl456/stl456 (n=20) embryos at 96 hpf. (E) Alcian Blue staining of control and taf1 knockout (KO) embryos. (F) X-ray microscopy-generated images of control and taf1 KO embryos at 96 hpf (left). Quantification of brain volume (right). N=4 per experimental group. Br, brain (red), heart (blue). (G) Whole-mount in situ hybridization of crestin and sox10 mRNA expression in control and taf1 KO embryos at 96 hpf. (H) Whole-mount in situ hybridization of neuronal (neurog1) and brain region markers (six3b, Adcyap1b, zic2a).
taf5 l is dispensable for embryonic development
taf5 is closely related (46% amino acid identity) to an evolutionarily conserved homolog, taf5l (Fig. S1A). To ascertain its function and examine potential redundancy between taf5 and taf5l, we generated a null allele in exon 2 of taf5l, taf5lstl851, WISH revealed that taf5l was expressed ubiquitously throughout WT embryos, whereas taf5l RNA was not detected in embryos homozygous for our nonsense allele (Fig. S1B). taf5lstl851/stl851 mutant embryos were viable and did not display any evidence of craniofacial hypoplasia, pericardial edema, or reduced cardiac function at 96 hpf and grew into fertile adults. Crosses of taf5lstl851/stl851 fish to test for possible maternal effects produced normal progeny, indicating that taf5l is dispensable for development. Furthermore, compound mutant analyses indicated that loss of taf5l function did not worsen craniofacial and cardiac phenotypes observed in taf5 mutant (corazoncito) embryos (Fig. S1C,D) suggesting that taf5 and taf5l are not redundant.
taf5 regulates oxidative metabolism
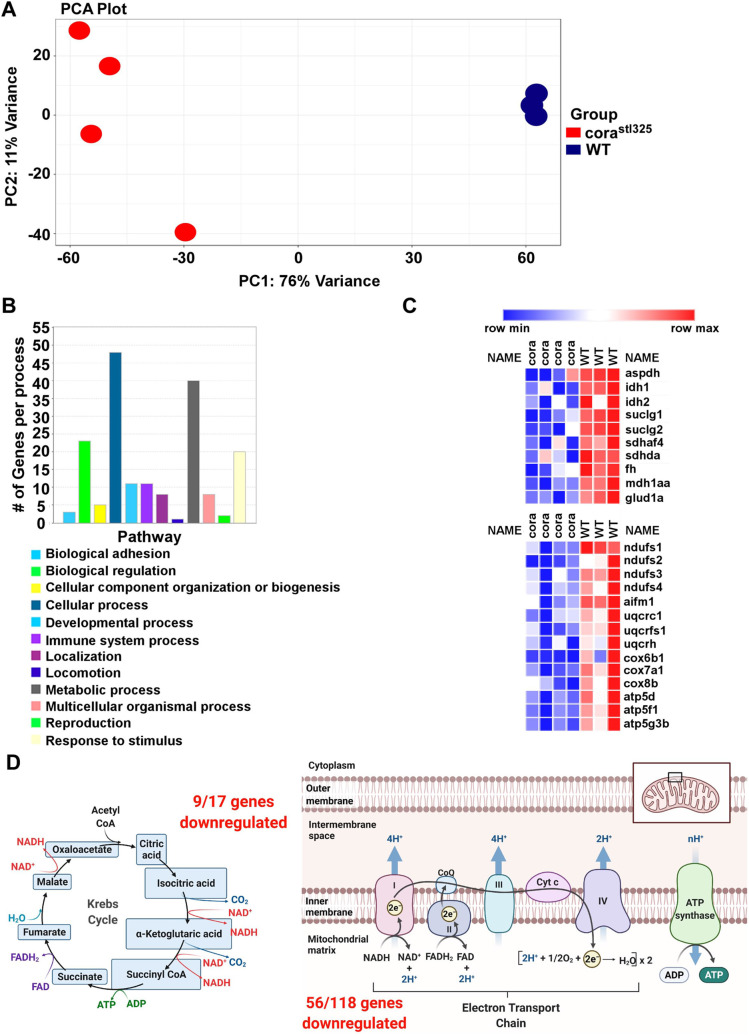
As taf5 is a member of the TFIID complex (Patel et al., 2018) and likely regulates transcription, we performed RNAseq to investigate potential mechanisms by which taf5 loss of function contributes to the observed cardiac phenotypes. We isolated RNA from embryonic hearts recovered from corastl325/stl325; Tg(cmlc2::gfp) embryos and their unaffected Tg(cmlc2::gfp) clutchmates at 96 hpf. Control and corastl325/stl325 sequencing libraries were generated from pooled samples each containing 100 hearts. Principal component analysis (PCA) revealed markedly distinct transcriptional profiles between control and corastl325/stl325 hearts, with 2274 genes showing an log2 fold value of 1 or greater in expression (Fig. 7A). Pathway analysis suggested alterations in several distinct biological processes including metabolism (Fig. 7B). corastl325/stl325 hearts displayed downregulation of numerous genes involved in the TCA cycle (9/17 genes) and the Electron Transport Chain (56/118 genes) (Fig. 7C,D). Interestingly, other major pathways, including glycolysis (7/20), amino acid (10/24), and lipid (20/26) metabolism were significantly impacted.
Fig. 7.
RNA sequencing of control and corastl325 identifies derangements metabolic gene expression. (A) Principal component analysis (PCA) plot of pooled sequencing libraries generated from control and corastl325 embryos at 96 hpf. Each dot represents a pool of 100 embryonic hearts. (B) Bar graph of the top Gene Ontology (GO) pathways associated with genes differentially expressed between control and corastl325 hearts. (C) Heat map of metabolic genes significant with significantly reduced expressed in corastl325 hearts. (D) Graphical representation of the number of differentially expressed metabolic genes associated with the TCA cycle (left) and the electron transport chain (right). Made with BioRender.
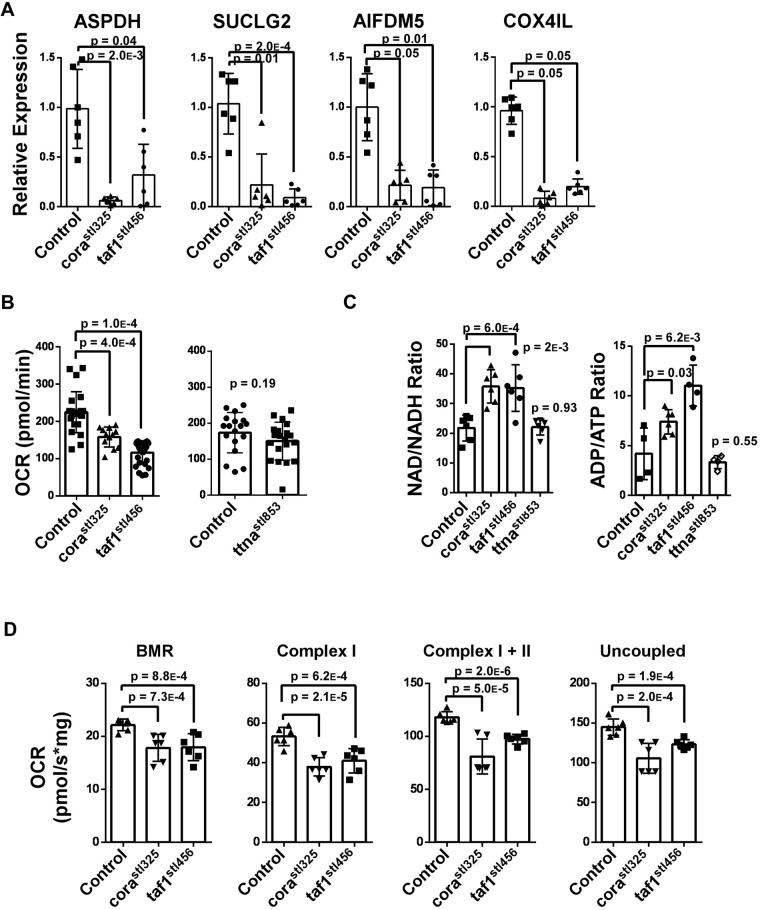
To determine whether metabolic genes were globally downregulated, we collected RNA from control, corastl325/stl325, and taf1stl456/stl456 whole embryos at 96 hpf. Quantitative RT-PCR confirmed reduced aspdh, suclg2, aidm5, and cox4il mRNA expression in corastl325/stl325 and taf1stl456/stl456 embryos relative to unaffected siblings (Fig. 8A). To discern whether altered metabolic gene expression affected metabolic function, we measured whole embryo basal metabolic rate using a Seahorse XF24 Bioanalyzer at 96 hpf. These studies showed diminished oxygen consumption rate (OCR) in corastl325/stl325 and taf1stl456/stl456 embryos relative to control clutch mates. To verify that decreased basal metabolic rate was not a secondary result of heart failure, we measured basal OCR in embryos lacking titin (ttnastl853), an established model of profound heart failure (Fig. S2) (Nechiporuk et al., 1999). Basal OCR did not differ between control and ttnastl853/stl853 embryos (Fig. 8B). Reduced metabolic activity was further supported by increased NAD/NADH and ADP/ATP ratios in corastl325/stl325 and taf1stl456/stl456 embryos relative to control siblings and N2:ttna embryos (Fig. 8C). Mitochondrial function was assessed using an Oroboros O2k-FluoRespirometer at 96 hpf. Consistent with the Seahorse data, corastl325/stl325 mitochondrial respiration showed significant perturbations at all states of respiration (Fig. 8D). Together, these results suggest that taf1 and taf5 are essential regulators of oxidative metabolism during embryonic development.
Fig. 8.
Defective oxidative metabolism in corastl325 and taf1 knockout embryos. (A) RTPCR showing the reduced expression of genes involved in the TCA cycle (Asdph, Suclg2) and the electron transport chain (Aifdm5, Cox4il) in corastl325 and taf1stl456 embryos at 96 hpf. Each data point represents a pool of 20 embryos. (B) Reduced basal oxygen consumption rates (OCR) in corastl325 (n=12) and taf1stl456 (n=12) embryos compared to controls at 96 hpf (left). Preserved basal OCR in ttnstl853/stl853 (n=20) compared to controls (n=18) embryos at 96 hpf (right). Data were obtained from a Seahorse XF24 Bioanalyzer. (C) Targeted metabolite assays measuring NAD/NADH (left) and ADP/ATP (right) in control, corastl325 taf1stl456, and ttnstl853/stl853 embryos at 96 hpf. n=4 pools of 100 embryos per genotype. (D) Oxygen consumption rates of isolated mitochondria for fatty acid metabolism. n=4 pools of 100 embryos per genotype
DISCUSSION
While the biochemical properties of TAFs and the TFIID complex are well understood, much remains to be learned regarding their in vivo functions, including roles during embryonic development and tissue homeostasis as well as how pathogenic variants in TAFs contribute to diseases. Here, we leveraged several genetic zebrafish models we generated to identify a causal link between mutations in taf1 and taf5, TAFopathy phenotypes, and derangements in metabolism. These studies suggest that the TAF complex does not only globally regulate transcription, but rather acts as a selective modulator of specific transcriptional programs. Furthermore, our findings provide new insights implicating defects in metabolism in the pathogenesis of TAFopathies.
TAFs are general transcription factors that are TFIID complex components which recruit RNA polymerase II to gene promoters to form the pre-initiation complex (PIC) (Patel et al., 2018). Many TAFs modulate interactions between gene-specific transcriptional activators and general transcription machinery by either stabilizing or inducing conformation chances to the PIC (Zou et al., 2015; Rhee and Pugh, 2012). Structurally, TAF5 forms a homodimer and acts as a scaffold for TFIID complex formation. Its C-terminus contains a WD40 domain, which mediates protein-protein interactions and may be important for TAF-TAF interactions (Roeder, 1996). TAF1 is the largest subunit of TFIID and contains multiple domains with enzymatic activity and chromatin interaction capabilities (Oelgeschläger et al., 1998; Wang et al., 2014). Recent studies have revealed that the TFIID complex is a three-lobed, asymmetric structure and that the TAF1 and TAF5 subunits contribute to forming this characteristic structure (Bhattacharya et al., 2007; (Oelgeschläger et al., 1998).
The alleles generated in this study for taf5, taf5l, and taf1 all contained nonsense mutations that represent loss of function phenotypes. Interestingly, while it is believed that taf5 and taf1 operate in the same complex, deletion of one did not impact the expression of the others. Phenotypically, we show that taf5 and taf1 are necessary for embryonic heart, craniofacial, and brain development in zebrafish. Brightfield and fluorescent microscopy revealed classical signs of heart failure in taf5 and taf1 mutant (loss of function, null alleles) as measured by reduced ejection fraction, dimensional cardiomyocyte strain, and pericardial edema. Despite these functional defects, neither taf5 nor taf1 were necessary for heart patterning, as atrioventricular staining revealed no obvious differences in chamber specification or structural architecture. Instead, we observed decreased myocardial proliferation in taf5 mutants. Proliferation of other structures such as the pericardium was not affected, suggesting a cell-specific role for taf5 in myocardial proliferation. Consistent, with this finding, TAF1 upregulates cyclin D and cyclin A expression through TAF1 histone acetyltransferase activity (Jacobson et al., 2000) and TAF1 has been shown to associate with the leukemia-promoting oncogene AML1-ETO, which promotes proliferation of AML1-ETO-expressing myeloid leukemia cells. Furthermore, TAF1 is required for leukemic cell self-renewal, and its reduction promotes the differentiation and apoptosis of AML1-ETO+ myeloid leukemia cells (Gangloff et al., 2000).
We also observed significant decreases in head size, brain volume, and disruption of defined brain regions in taf5 and taf1 mutants. Neurodevelopmental defects have been previously observed in taf1 mutants as evidenced by decreased head-to-body and eye-to-body ratios, and measurements of optic tectum size (Kloet et al., 2012). Loss of TAF1 in rats alters the morphology and function of the cerebellum and cerebral cortex, and leads to hypoplasia and loss of Purkinje cells, with behavioral abnormalities paralleling that seen in TAF1/MRSX33 intellectual disability syndrome (Xu et al., 2019). TAF1 has also been linked to X-linked dystonia parkinsonism, with an alternatively spliced transcript of TAF1 discovered in neurons (Gudmundsson et al., 2019; Grune et al., 2022; Capponi et al., 2021). These findings implicate TAF1 in early neurodevelopment and aging.
To illuminate mechanisms that contribute to TAFopathies in our zebrafish models, we performed RNA sequencing in taf5 mutant hearts. Surprisingly, we did not detect global downregulation of transcription. Instead, we identified specific reductions in the expression of genes involved in metabolism: fatty acid oxidation, TCA cycle, and electron transport chain. Derangements in metabolism were further supported by reduced expression of key metabolic genes throughout the embryo, increased NAD/NADH and ADP/ATP ratios, and measurements of oxygen consumption at the organismal and mitochondrial levels. Importantly, these metabolic derangements were absent in N2:TTNa mutants with akinetic hearts indicating that metabolic impairments found in taf1 and taf5 embryos were not secondary to heart failure. Indeed, metabolic remodeling plays an essential role in cardiac hypertrophy and heart failure (Al Ali et al., 2021; Aneichyk et al., 2018; Ashrafian et al., 2007). Consistent with previous findings in taf1 mutants, we observed downregulation of the cell cycle regulator cyclin D (ccnd1) in taf5 mutant hearts along with reduced expression of many other cyclins (ccna1, ccna2, ccnb2, ccnb3, ccne1). Whether heart, craniofacial, and/or brain hypoplasia is primarily caused by metabolic remodeling or a direct effect on the transcription of cell cycle regulators represents an avenue for future investigation that will require dedicated tools that dissect the gene regulatory networks controlled by TAFs and the TFIID complex.
Our study is not without limitations. We primarily focused on zebrafish models and validation of our findings pertaining to the phenotypic impact of taf1 and taf5 mutations and impact on metabolism in mammalian systems is an important next step. In support of our findings, previous studies have suggested that perturbations in zebrafish taf1 result in overlapping phenotypes including reduced embryo length, underdeveloped cartilage, short pectoral fins, edema, and body axis deformations (Kloet et al., 2012). Our work adds to the understanding of the requirement for taf1 during zebrafish development and provides new information implicating a similar requirement for taf5 in craniofacial, neurodevelopmental, and cardiac development. The precise mechanism by which taf1 and taf5 specifically regulate transcriptional programs such as metabolism remains to be defined. An interesting possibility is that the TAF complex may optimize interactions between promoter and enhancer regions through the BRD complex, as TAF1 contains both TATA binding protein and BRD binding sites (Kundu et al., 2015; Doukas et al., 2007).
In conclusion, our findings establish a causal link between null mutations in taf1 and taf5, phenotypes present in TAFopathy patients, and perturbations in metabolism and cell cycle-related pathways. These findings provide exciting and novel insights into how components of general transcription machinery selectively regulate specific transcriptional programs and contribute to tissue and organ maturation during embryonic development.
MATERIALS AND METHODS
Establishment and maintenance of zebrafish lines
corastl325 was generated through an ENU mutagenesis screen (de Bruijn et al., 2009). Transgenic reporter lines such as Tg(cmlc2:GFP) were obtained from other labs (Huang et al., 2003). taf5stl852/stl852, taf5lstl851/stl851 taf1stl456/stl456 lines were generated using CRISPR/Cas9-mediated mutagenesis. Using a guide RNA targeting exon 1 of TAF5 (gRNA sequence: GGCTGCGGTGAGTGGCGATGAGG), we deleted a ∼2.5 kb region spanning exons 1 and 2. taf5lstl851 was generated using CRISPR/Cas9 (gRNA sequence: GGTGTCCGCGGCCCCGTGTCAGG) to create a 17 bp deletion that resulted in a nonsense mutation. Lastly, taf1stl456 was generated by using a gRNA targeting exon 7 that created a 19 bp insertion resulting in a nonsense mutation (gRNA sequence: GGTGTCCGCGGCCCCGTGTCAGG). These larvae were grown and selected based on genotype, all of which was based on existing protocols (Li et al., 2016). Genotyping of corazoncito exon 3 was performed using the primers 5′-ATGCGTCATGACGTAATCACATCCAGC-3′ and 5′- TCACTGGGAGTTGTAGGAGCCTGCGGC-3′, which amplified a 333-bp region including the corazoncito mutation, using the restriction enzyme HpyCh4III (NEB; cat. No. R0618) that bifurcates the amplicon into ∼161-bp doublets. All lines used in this study were raised and maintained by Washington University's Fish Facility. A summary of their zebrafish husbandry guidelines can be found here (https://zebrafishfacility.wustl.edu/facility-documents/).
Microscopy experiments
Brightfield microscopy images were taken using a Leica M80 Microscope with a mounted Leica IC80 HD camera. Fish were anesthetized in 20-30 mg/L of Tricaine in E3 water and positioned for imaging. Measurements of cartilaginous structures, head and eye components, ejection fraction and pericardium were performed with ImageJ (Collins, 2007). Ejection fraction was measured by measuring the area of the ventricle at systole and diastole. We used these area measurements to find the difference in area, which was then divided by the ventricle area during diastole.
Fluorescence microscopy was performed using a Zeiss Lightsheet 7 in the Washington University Center for Cellular imaging (WUCCI). Sample preparation protocol can be found here (https://www.iob.uu.se/digitalAssets/576/c_576400-l_1-k_protocol-for-lightsheet-z.1-using-zebrafish.pdf). For Zeiss lightsheet microscopy, larvae were anesthetized in 20-30 mg/L of Tricaine in E3 water, and then placed in a 1% low-melting agarose solution. This larvae/agarose combination was then inserted into a black glass capillary, after which larvae hearts were imaged.
X-ray microscopy was performed using a Zeiss Xradia Versa 520 XRM microscope at WUCCI (Bayguinov et al., 2020). Zebrafish larvae were fixed in 4% paraformaldehyde, then transferred to Lugol's Iodine for 3 weeks, after which samples were imaged. All imaging analysis was performed using Imaris.
Whole-mount staining
Zebrafish larvae were fixed overnight at 4°C using 4% paraformaldehyde, then washed in PBS+0.1% Tween. These embryos were then placed in 100% methanol and stored at −20°C. Alcian Blue staining was performed as previously described (Walker and Kimmel, 2007). Zebrafish larvae were imaged and staged in 100% glycerol+KOH using a Zeiss Discovery V.12 stereomicroscope.
Whole-mount in situ hybridization was performed according to established protocols (Cunningham and Monk, 2018). Briefly, probes were generated through PCR amplification of the coding sequences of the genes of interest using AB* cDNA libraries, followed by standard TOPO cloning (Invitrogen; cat. No. K4500) (Table S1). Transformation was performed using TOP10 chemocompetent cells (Thermo Fisher Scientific; cat. No. C505003), with confirmation of probe generation and correct orientation performed through Sanger sequencing. Once probes were validated, fixed embryos were rehydrated in a methanol/PBS gradient, permeabilized at room temperature (RT) with 10 μg/ml proteinase K, and left to fix in 4% paraformaldehyde, after which they were transferred to hybridization buffer (65% formamide, 5× SSC, 0.1% Tween, 50 μg ml−1 of heparin, 500 μg ml−1 of RNase-free tRNA adjusted to pH 6.0 by adding citric acid) containing 3 ng/μl probe overnight at 70°C. These embryos were then washed with hybridization buffer/PBS, followed by PBS washes, then incubated at 4°C overnight with 1:5000 anti-digoxigenin antibody (Roche, Catalog # 11093274910). Embryos were then washed with PBS and stained using BM Purple until expression could be clearly observed (Roche, Catalog # 11442074001).
Next generation sequencing and RNA sequencing
Genomic DNA and cDNA obtained from Corazoncito and their unaffected clutchmates was submitted to the Genome Technology Access Center for whole genome sequencing and RNA sequencing. For whole genome sequencing and mutation mapping, we used an existing pipeline as described here (Sanchez et al., 2017). Briefly, 1 µg larvae gDNA was extracted and submitted to the Genome Technology Access Center (GTAC) at Washington University for whole genome sequencing where they were bar-coded and pooled, and paired-end sequencing was performed in a single lane of a HiSeq2500 or 3000 (Illumina). Reads were aligned using NovoAlign (Novocraft), and variants were called using SAMtools. Our pipeline involved using three scripts ‘ChromSplit’, ‘Allele Ratio Calculator’ (ARC), and ‘SNPFilter’ to split variants into chromosome-specific data files, calculate mutant allele frequency at each point of variation between groups, and omit SNPs that have been previously annotated. Manhattan plots were generated using GraphPad Prism). Chromosomal fine mapping was performed using established markers for chromosome 1 (Table S2). RNA sequencing pathway analysis was performed using the Shiny Transcriptome Analysis Resource Tool (Nelson et al., 2017). Pathway analysis was performed using Gene Ontology Resource (http://geneontology.org/). Heat maps were generated using Phantasus (https://artyomovlab.wustl.edu/phantasus/). FASTQ files are available (GEO GSE230183).
RT-PCR
Primers were generated through Integrated DNA Technologies' PrimerQuest tool (https://www.idtdna.com/pages/tools/primerquest) (Table S3). RT-PCR was performed with standard conditions. RNA was extracted from zebrafish larval tissue using the RNeasy RNA mini kit and Tissue Lyser II (Qiagen). RNA concentration was measured using a nanodrop spectrophotometer (Thermo Fisher Scientific). cDNA synthesis was performed using the High Capacity RNA to cDNA synthesis kit (Applied Biosystems). cDNA was synthesized using the iScript™ Reverse Transcription Supermix (Bio-Rad) and pre-amplified using the Sso Advanced PreAmp Supermix kit (Bio-Rad). Quantitative real time PCR reactions were prepared with sequence-specific primers (IDT) with PowerUP™ Syber Green Master mix (Thermo Fisher Scientific) in a 20 μl volume. Real time PCR was performed using QuantStudio 3 (Thermo Fisher Scientific). mRNA expression was normalized to HPRT. All RT-PCR assays were performed with appropriate quality controls including melt curves and negative controls.
Metabolism assays
Zebrafish larvae basal metabolic rate was measured in a Seahorse XF24e extracellular flux analyzer through modification of previously established protocols (Stackley et al., 2011). One 96 hpf embryo per well was used to measure the oxygen consumption rate at physiologically relevant temperature (28.5°C), with all other conditions held constant.
Targeted metabolite assays for NAD/NADH (Biovision; cat. No. K337) and ADP/ATP (Millipore-Sigma; cat. No. MAK081-1KT) were used. ATP extraction from tissues using PCA was performed according to the protocol supplied by the manufacturer. Larval tissue pools of 20 mg were flash frozen and homogenized with 200 μl of ice-cold homogenization buffer (0.25 mol L−1 sucrose and 10 mmol L−1 HEPES–NaOH, pH 7.4) by Tissue Lyser II (Qiagen) using one cycle of 30-s homogenization and 30-s cooling. After homogenization, the homogenate was centrifuged at 1000×g for 10 min at 4°C. One hundred microliters of the supernatant was quickly added to an equal volume of ice-cold 10% PCA and shaken for 20-s. The supernatant was then transferred into a 2.0-mL microtube for centrifugation (10,000×g for 10 min at 4°C), and 50 μl of supernatant was collected and added to 50 μl of 1 mol L−1 Tris–acetate buffer (pH 7.75) for neutralization. Ten microliters of aliquot from the supernatant was used in 96-well plates for luciferin–luciferase assay, which was performed using a TECAN m200 Infinite Pro plate reader.
NAD/NADH assay samples were prepared according to the manufacturer’s protocol. Twenty milligram samples of larval tissue were washed with ice cold PBS, after which 400 μl of NAD/NADH Extraction Buffer were added to the samples that were homogenized by Tissue Lyser II (Qiagen) using one cycle of 30-s homogenization and 30-s cooling. After homogenization, the homogenate was centrifuged at 24,000×g for 10 min at 4°C. Supernatant was transferred to a new tube, with 200 μl of this extract placed into a separate tube that was heated to 60°C for 30 minutes in a water bath. Both the non-decomposed and decomposed samples were added to a 96-well plate to be processed with kit reagents and OD450 read by a TECAN m200 Infinite Pro plate reader.
Isolation of mitochondria from zebrafish embryos
For mitochondrial isolation, anesthetized pools of 100 96 hpf embryos were collected and washed in mitochondrial isolation buffer (MIB 0.21 M mannitol, 70 mM sucrose, 0.1 mM potassium-EDTA, 1 mM EGTA, 10 mM Tris-HCL, pH 7.4) for each genotype and were homogenized at 70 rpm for 20 strokes on ice in a 4 ml glass homogenizer at a ratio of 10 mg tissue per mL of MIB. The homogenate was transferred to 1.7 ml microcentrifuge tubes and centrifuged for 10 min at 600×g. Subsequently, the supernatant was centrifuged for 10 min at 7200×g to pellet mitochondria. The pellet was then resuspended and washed twice in MIB, with the final pellet reconstituted in 500 μl of MIB.
Mitochondrial respiration assays for assessment of metabolic function
Protein concentration was determined using the BCA method per the manufacturer's instructions (Thermo Fisher Scientific). Mitochondria (300 ng protein) were placed in 2 ml mitochondria respiration buffer (MIR05: 0.5 mmol/L EGTA, 3 mmol/L MgCl2, 60 mmol/L K-lactobionate, 20 mmol/L taurine, 10 mmol/L KH2PO4, 20 mmol/L HEPES, 110 mmol/L sucrose, and 1 g/L BSA [pH 7.1]) and loaded into an Oxygraph-2k respirometer (Oxygraph-2k with O2k-Fluorescence module; Oroboros Instruments, Innsbruck, Austria) at 28.5°C with continuous stirring for the measurement of O2 flux. First, malate (1 mM) was added for depletion of endogenous substrates. Octanoylcarnitine (30 μM) was then added as a fatty acid substrate. Adenosine disphosphate (ADP; 5 mM) was added to induce state 3 respiration, with glutamate (10 mM) and succinate (10 mM) subsequently added for electron input in complex I and complex II, respectively. Cytochrome C (10 μM) was added to assess outer mitochondrial membrane integrity, and maximal oxygen flux rates were measured using chemical uncoupler carbonylcyanide-4-(trifluoromethoxy)-phenylhydrazone (FCCP), which was titrated (0.25 μM per addition) until no further stimulation of respiration could be detected.
Statistics
One-way ANOVA tests were used for ejection fraction, edema, qPCR, and metabolic experiments. Log-rank test was used for survival curves. RNA-Seq data was analyzed through START (https://nasqar.abudhabi.nyu.edu/STARTapp/) and Gene Ontology (http://geneontology.org/).
Supplementary Material
Acknowledgements
The authors are grateful to the Diabetes Research Center for providing access to their Seahorse XF24 Bioanalyzer (Supported by DRC, Grant No. P30 DK020579), Drs. Margot Williams and Diane Sepich for their kind provision of reagents, Drs. Gabe Haller and Martha Bagnell for their insights on brain anatomy, and Drs. Jun Zou and Rahul Deo for providing us with their titin mutant line. We thank the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine for help with genomic analysis. The Center is partially supported by NCI Cancer Center Support Grant #P30 CA91842 to the Siteman Cancer Center and by ICTS/CTSA Grant# UL1TR000448 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and NIH Roadmap for Medical Research. This publication is solely the responsibility of the authors and does not necessarily represent the official view of NCRR or NIH. Experiments, data, and analysis generated from electron microscopy, Lightsheet fluorescence microscopy, and X-Ray microscopy were performed in part through the use of Washington University Center for Cellular Imaging (WUCCI) supported by Washington University School of Medicine, The Children's Discovery Institute of Washington University and St. Louis Children's Hospital (CDI-CORE-2015-505 and CDI-CORE-2019-813) and the Foundation for Barnes-Jewish Hospital (3770 and 4642).
Footnotes
Author Contributions
Conceptualization: K.J.L.; Methodology: J.L., R.G., P.R., A.L.K., R.T., J.A.J.F., L.S.-K.; Formal analysis: J.L., R.G., P.R., A.L.K., R.T.; Investigation: J.L.; Resources: R.G., J.A.J.F., C.K., L.S.-K.; Data curation: J.L.; Writing - original draft: J.L., K.J.L.; Writing - review & editing: J.L., R.G., C.K., L.S.-K., K.J.L.; Visualization: J.L., J.A.J.F.; Supervision: L.S.-K., K.J.L.; Project administration: L.S.-K., K.J.L.; Funding acquisition: K.J.L.
Funding
K.J.L. is supported by funding provided from the Children's Discovery Institute of Washington University and St. Louis Children's Hospital, Foundation for Barnes-Jewish Hospital, Burroughs Foundation Welcome Fund, Leducq Foundation, National Institutes of Health (NIH) (HL138466, HL139714, HL151078, AI148877), and sponsored research agreements from Amgen and Novartis. J.L. is supported by an NIH F31 award (F31 HD106710-02). Open Access funding provided by Washington University School of Medicine in Saint Louis: Washington University in St Louis School of Medicine. Deposited in PMC for immediate release.
Data availability
All relevant data can be found within the article and its supplementary information.
References
- Al Ali, J., Vaine, C. A., Shah, S., Campion, L., Hakoum, A., Supnet, M. L., Acuña, P., Aldykiewicz, G., Multhaupt-Buell, T., Ganza, N. G. M.et al. (2021). TAF1 transcripts and neurofilament light chain as biomarkers for X-linked dystonia-parkinsonism. Mov. Disord. 36, 206-215. 10.1002/mds.28305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alazami, A. M., Patel, N., Shamseldin, H. E., Anazi, S., Al-Dosari, M. S., Alzahrani, F., Hijazi, H., Alshammari, M., Aldahmesh, M. A., Salih, M. A.et al. (2015). Accelerating novel candidate gene discovery in neurogenetic disorders via whole-exome sequencing of prescreened multiplex consanguineous families. Cell Rep. 10, 148-161. 10.1016/j.celrep.2014.12.015 [DOI] [PubMed] [Google Scholar]
- Aneichyk, T., Hendriks, W. T., Yadav, R., Shin, D., Gao, D., Vaine, C. A., Collins, R. L., Domingo, A., Currall, B., Stortchevoi, A.et al. (2018). Dissecting the causal mechanism of X-linked dystonia-parkinsonism by integrating genome and transcriptome assembly. Cell 172, 897-909.e21. 10.1016/j.cell.2018.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafian, H., Frenneaux, M. P. and Opie, L. H. (2007). Metabolic mechanisms in heart failure. Circulation 116, 434-448. 10.1161/CIRCULATIONAHA.107.702795 [DOI] [PubMed] [Google Scholar]
- Asturias, F. J. (2009). TFIID: a closer look highlights its complexity. Structure 17, 1423-1424. 10.1016/j.str.2009.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayguinov, P. O., Fisher, M. R. and Fitzpatrick, J. A. J. (2020). Assaying three-dimensional cellular architecture using X-ray tomographic and correlated imaging approaches. J. Biol. Chem. 295, 15782-15793. 10.1074/jbc.REV120.009633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya, S., Takada, S. and Jacobson, R. H. (2007). Structural analysis and dimerization potential of the human TAF5 subunit of TFIID. Proc. Natl. Acad. Sci. USA 104, 1189-1194. 10.1073/pnas.0610297104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniossek, C., Papai, G., Schaffitzel, C., Garzoni, F., Chaillet, M., Scheer, E., Papadopoulos, P., Tora, L., Schultz, P. and Berger, I. (2013). The architecture of human general transcription factor TFIID core complex. Nature 493, 699-702. 10.1038/nature11791 [DOI] [PubMed] [Google Scholar]
- Brou, C., Kuhn, A., Staub, A., Chaudhary, S., Grummt, I., Davidson, I. and Tora, L. (1993). Sequence-specific transactivators counteract topoisomerase II-mediated inhibition of in vitro transcription by RNA polymerases I and II. Nucleic Acids Res. 21, 4011-4018. 10.1093/nar/21.17.4011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capponi, S., Stöffler, N., Penney, E. B., Grütz, K., Nizamuddin, S., Vermunt, M. W., Castelijns, B., Fernandez-Cerado, C., Legarda, G. P., Velasco-Andrada, M. S.et al. (2021). Dissection of TAF1 neuronal splicing and implications for neurodegeneration in X-linked dystonia-parkinsonism. Brain Commun. 3, fcab253. 10.1093/braincomms/fcab253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z.-Z., Gao, Y.-Q., Xie, H., Huang, Y.-C., Chen, F. and Lei, Y.-P. (2022). Transcription factors dysregulated in three complex birth defects datasets. Reprod. Dev. Med. 6, 79-85. 10.1097/RD9.0000000000000018 [DOI] [Google Scholar]
- Clark, K. L., Yutzey, K. E. and Benson, D. W. (2006). Transcription factors and congenital heart defects. Annu. Rev. Physiol. 68, 97-121. 10.1146/annurev.physiol.68.040104.113828 [DOI] [PubMed] [Google Scholar]
- Collins, T. J. (2007). ImageJ for microscopy. BioTechniques 43, S25-S30. 10.2144/000112517 [DOI] [PubMed] [Google Scholar]
- Cunningham, R. L. and Monk, K. R. (2018). Whole mount in situ hybridization and immunohistochemistry for zebrafish larvae. Methods Mol. Biol 1739, 371-384. 10.1007/978-1-4939-7649-2_25 [DOI] [PubMed] [Google Scholar]
- De Bruijn, E., Cuppen, E. and Feitsma, H. (2009). Highly efficient ENU mutagenesis in zebrafish. Methods Mol. Biol. 546, 3-12. 10.1007/978-1-60327-977-2_1 [DOI] [PubMed] [Google Scholar]
- De Soysa, T. Y., Ranade, S. S., Okawa, S., Ravichandran, S., Huang, Y., Salunga, H. T., Schricker, A., Del Sol, A., Gifford, C. A. and Srivastava, D. (2019). Single-cell analysis of cardiogenesis reveals basis for organ-level developmental defects. Nature 572, 120-124. 10.1038/s41586-019-1414-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doukas, J., Wrasidlo, W., Noronha, G., Dneprovskaia, E., Hood, J. and Soll, R. (2007). Isoform-selective PI3K inhibitors as novel therapeutics for the treatment of acute myocardial infarction. Biochem. Soc. Trans. 35, 204-206. 10.1042/BST0350204 [DOI] [PubMed] [Google Scholar]
- Gangloff, Y.-G., Werten, S., Romier, C., Carré, L., Poch, O., Moras, D. and Davidson, I. (2000). The human TFIID components TAF(II)135 and TAF(II)20 and the yeast SAGA components ADA1 and TAF(II)68 heterodimerize to form histone-like pairs. Mol. Cell. Biol. 20, 340-351. 10.1128/MCB.20.1.340-351.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, R. S., Gonzalez, R., Ackerman, S. D., Minowa, R., Griest, J. F., Bayrak, M. N., Troutwine, B., Canter, S., Monk, K. R., Sepich, D. S.et al. (2021). Postembryonic screen for mutations affecting spine development in zebrafish. Dev. Biol. 471, 18-33. 10.1016/j.ydbio.2020.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grune, J., Lewis, A. JM., Yamazoe, M., Hulsmans, M., Rohde, D., Xiao, L., Zhang, S., Ott, C., Calcagno, D. M., Zhou, Y.et al. (2022). Neutrophils incite and macrophages avert electrical storm after myocardial infarction. Nat. Cardiovasc. Res 1, 649-664. 10.1038/s44161-022-00094-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsson, S., Wilbe, M., Filipek-Górniok, B., Molin, A.-M., Ekvall, S., Johansson, J., Allalou, A., Gylje, H., Kalscheuer, V. M., Ledin, J.et al. (2019). TAF1, associated with intellectual disability in humans, is essential for embryogenesis and regulates neurodevelopmental processes in zebrafish. Sci. Rep. 9, 10730. 10.1038/s41598-019-46632-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellman-Aharony, S., Smirin-Yosef, P., Halevy, A., Pasmanik-Chor, M., Yeheskel, A., Har-Zahav, A., Maya, I., Straussberg, R., Dahary, D., Haviv, A.et al. (2013). Microcephaly thin corpus callosum intellectual disability syndrome caused by mutated TAF2. Pediatr. Neurol. 49, 411-416.e1. 10.1016/j.pediatrneurol.2013.07.017 [DOI] [PubMed] [Google Scholar]
- Huang, C.-J., Tu, C.-T., Hsiao, C.-D., Hsieh, F.-J. and Tsai, H.-J. (2003). Germ-line transmission of a myocardium-specific GFP transgene reveals critical regulatory elements in the cardiac myosin light chain 2 promoter of zebrafish. Dev. Dyn. 228, 30-40. 10.1002/dvdy.10356 [DOI] [PubMed] [Google Scholar]
- Jacobson, R. H., Ladurner, A. G., King, D. S. and Tjian, R. (2000). Structure and function of a human TAFII250 double bromodomain module. Science 288, 1422-1425. 10.1126/science.288.5470.1422 [DOI] [PubMed] [Google Scholar]
- Jenkins, K. J., Correa, A., Feinstein, J. A., Botto, L., Britt, A. E., Daniels, S. R., Elixson, M., Warnes, C. A. and Webb, C. L. (2007). Noninherited risk factors and congenital cardiovascular defects: current knowledge: a scientific statement from the American Heart Association Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. Circulation 115, 2995-3014. 10.1161/CIRCULATIONAHA.106.183216 [DOI] [PubMed] [Google Scholar]
- Juven-Gershon, T. and Kadonaga, J. T. (2010). Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev. Biol. 339, 225-229. 10.1016/j.ydbio.2009.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven-Gershon, T., Cheng, S. and Kadonaga, J. T. (2006). Rational design of a super core promoter that enhances gene expression. Nat. Methods 3, 917-922. 10.1038/nmeth937 [DOI] [PubMed] [Google Scholar]
- Kaufman, C. K., Mosimann, C., Fan, Z. P., Yang, S., Thomas, A. J., Ablain, J., Tan, J. L., Fogley, R. D., Van Rooijen, E., Hagedorn, E. J.et al. (2016). A zebrafish melanoma model reveals emergence of neural crest identity during melanoma initiation. Science 351, aad2197. 10.1126/science.aad2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby, R. S. (2017). The prevalence of selected major birth defects in the United States. Semin. Perinatol. 41, 338-344. 10.1053/j.semperi.2017.07.004 [DOI] [PubMed] [Google Scholar]
- Kloet, S. L., Whiting, J. L., Gafken, P., Ranish, J. and Wang, E. H. (2012). Phosphorylation-dependent regulation of cyclin D1 and cyclin A gene transcription by TFIID subunits TAF1 and TAF7. Mol. Cell. Biol. 32, 3358-3369. 10.1128/MCB.00416-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu, B. K., Zhong, M., Sen, S., Davogustto, G., Keller, S. R. and Taegtmeyer, H. (2015). Remodeling of glucose metabolism precedes pressure overload-induced left ventricular hypertrophy: review of a hypothesis. Cardiology 130, 211-220. 10.1159/000369782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann, L., Ferrari, R., Vashisht, A. A., Wohlschlegel, J. A., Kurdistani, S. K. and Carey, M. (2012). Polycomb repressive complex 1 (PRC1) disassembles RNA polymerase II preinitiation complexes. J. Biol. Chem. 287, 35784-35794. 10.1074/jbc.M112.397430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M., Zhao, L., Page-Mccaw, P. S. and Chen, W. (2016). Zebrafish genome engineering using the CRISPR-Cas9 system. Trends Genet. 32, 815-827. 10.1016/j.tig.2016.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louder, R. K., He, Y., López-Blanco, J. R., Fang, J., Chacón, P. and Nogales, E. (2016). Structure of promoter-bound TFIID and model of human pre-initiation complex assembly. Nature 531, 604-609. 10.1038/nature17394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechiporuk, A., Finney, J. E., Keating, M. T. and Johnson, S. L. (1999). Assessment of polymorphism in zebrafish mapping strains. Genome Res. 9, 1231-1238. 10.1101/gr.9.12.1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, J. W., Sklenar, J., Barnes, A. P. and Minnier, J. (2017). The START App: a web-based RNAseq analysis and visualization resource. Bioinformatics 33, 447-449. 10.1093/bioinformatics/btw624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rawe, J. A., Wu, Y., Dörfel, M. J., Rope, A. F., Au, P. Y. B., Parboosingh, J. S., Moon, S., Kousi, M., Kosma, K., Smith, C. S.et al. (2015). TAF1 Variants Are Associated with Dysmorphic Features, Intellectual Disability, and Neurological Manifestations. Am. J. Hum. Genet. 97, 922-932. 10.1016/j.ajhg.2015.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelgeschläger, T., Tao, Y., Kang, Y. K. and Roeder, R. G. (1998). Transcription activation via enhanced preinitiation complex assembly in a human cell-free system lacking TAFIIs. Mol. Cell 1, 925-931. 10.1016/S1097-2765(00)80092-1 [DOI] [PubMed] [Google Scholar]
- Patel, A. B., Louder, R. K., Greber, B. J., Grünberg, S., Luo, J., Fang, J., Liu, Y., Ranish, J., Hahn, S. and Nogales, E. (2018). Structure of human TFIID and mechanism of TBP loading onto promoter DNA. Science 362, eaau8872. 10.1126/science.aau8872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott, K. R. and Wilkie, A. O. M. (2007). Genetic aspects of birth defects: new understandings of old problems. Arch. Dis. Child. Fetal Neonatal Ed. 92, F308-F314. 10.1136/adc.2004.062968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee, H. S. and Pugh, B. F. (2012). Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature 483, 295-301. 10.1038/nature10799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder, R. G. (1996). The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 21, 327-335. 10.1016/0968-0004(96)10050-5 [DOI] [PubMed] [Google Scholar]
- Rooms, L., Reyniers, E., Scheers, S., Van Luijk, R., Wauters, J., Van Aerschot, L., Callaerts-Vegh, Z., D'hooge, R., Mengus, G., Davidson, I.et al. (2006). TBP as a candidate gene for mental retardation in patients with subtelomeric 6q deletions. Eur. J. Hum. Genet. 14, 1090-1096. 10.1038/sj.ejhg.5201674 [DOI] [PubMed] [Google Scholar]
- Sanchez, N. E., Harty, B. L., O'Reilly-Pol, T., Ackerman, S. D., Herbert, A. L., Holmgren, M., Johnson, S. L., Gray, R. S. and Monk, K. R. (2017). Whole genome sequencing-based mapping and candidate identification of mutations from fixed zebrafish tissue. G3 7, 3415-3425. 10.1534/g3.117.300212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stackley, K. D., Beeson, C. C., Rahn, J. J. and Chan, S. S. L. (2011). Bioenergetic profiling of zebrafish embryonic development. PLoS One 6, e25652. 10.1371/journal.pone.0025652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawamie, H., Martianov, I., Wohlfahrt, N., Buchert, R., Mengus, G., Uebe, S., Janiri, L., Hirsch, F. W., Schumacher, J., Ferrazzi, F.et al. (2017). Hypomorphic pathogenic variants in TAF13 are associated with autosomal-recessive intellectual disability and microcephaly. Am. J. Hum. Genet. 100, 555-561. 10.1016/j.ajhg.2017.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, M. B. and Kimmel, C. B. (2007). A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech. Histochem. 82, 23-28. 10.1080/10520290701333558 [DOI] [PubMed] [Google Scholar]
- Wang, H., Curran, E. C., Hinds, T. R., Wang, E. H. and Zheng, N. (2014). Crystal structure of a TAF1-TAF7 complex in human transcription factor IID reveals a promoter binding module. Cell Res. 24, 1433-1444. 10.1038/cr.2014.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber, D. M., Macleod, S. L., Bamshad, M. J., Shaw, G. M., Finnell, R. H., Shete, S. S., Witte, J. S., Erickson, S. W., Murphy, L. D. and Hobbs, C. (2015). Developments in our understanding of the genetic basis of birth defects. Birth Defects Res. A Clin. Mol. Teratol. 103, 680-691. 10.1002/bdra.23385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, S. Y. and Chiang, C. M. (2001). TATA-binding protein-associated factors enhance the recruitment of RNA polymerase II by transcriptional activators. J. Biol. Chem. 276, 34235-34243. 10.1074/jbc.M102463200 [DOI] [PubMed] [Google Scholar]
- Xu, Y., Man, N., Karl, D., Martinez, C., Liu, F., Sun, J., Martinez, C. J., Martin, G. M., Beckedorff, F., Lai, F.et al. (2019). TAF1 plays a critical role in AML1-ETO driven leukemogenesis. Nat. Commun. 10, 4925. 10.1038/s41467-019-12735-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, J., Tran, D., Baalbaki, M., Tang, L. F., Poon, A., Pelonero, A., Titus, E. W., Yuan, C., Shi, C., Patchava, S.et al. (2015). An internal promoter underlies the difference in disease severity between N- and C-terminal truncation mutations of Titin in zebrafish. Elife 4, e09406. 10.7554/eLife.09406 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.