Abstract
Objective
The impact of social determinants of health (SDoH) on patients’ healthcare quality and the disparity is well known. Many SDoH items are not coded in structured forms in electronic health records. These items are often captured in free-text clinical notes, but there are limited methods for automatically extracting them. We explore a multi-stage pipeline involving named entity recognition (NER), relation classification (RC), and text classification methods to automatically extract SDoH information from clinical notes.
Materials and Methods
The study uses the N2C2 Shared Task data, which were collected from 2 sources of clinical notes: MIMIC-III and University of Washington Harborview Medical Centers. It contains 4480 social history sections with full annotation for 12 SDoHs. In order to handle the issue of overlapping entities, we developed a novel marker-based NER model. We used it in a multi-stage pipeline to extract SDoH information from clinical notes.
Results
Our marker-based system outperformed the state-of-the-art span-based models at handling overlapping entities based on the overall Micro-F1 score performance. It also achieved state-of-the-art performance compared with the shared task methods. Our approach achieved an F1 of 0.9101, 0.8053, and 0.9025 for Subtasks A, B, and C, respectively.
Conclusions
The major finding of this study is that the multi-stage pipeline effectively extracts SDoH information from clinical notes. This approach can improve the understanding and tracking of SDoHs in clinical settings. However, error propagation may be an issue and further research is needed to improve the extraction of entities with complex semantic meanings and low-frequency entities. We have made the source code available at https://github.com/Zephyr1022/SDOH-N2C2-UTSA.
Keywords: information extraction, social determinants of health, neural networks, natural language processing, NLP, machine learning
BACKGROUND AND SIGNIFICANCE
Social determinants of health (SDoH) are nonclinical factors influencing health, functioning, and quality of life outcomes and risks. For example, SDoH factors include where people are born, live, learn, work, play, worship, and their age.1–3 Decades of studies have shown that medical care accounts for only 10%–20% of an individual’s health status. However, social, behavioral, and genetic factors also significantly influence health risks, outcomes, access to health services, and adherence to prescribed care.4,5 Thus, addressing SDoH is critical for increasing healthcare quality, decreasing health disparities, and informing clinical decision-making.6
Unfortunately, electronic health records (EHRs) do not generally code SDoH information in structured data, for example, not in ICD-10 codes.7 Instead, healthcare organizations and professionals typically record SDoH in unstructured narrative clinical notes. Thus, this critical patient information is not easily accessible. Healthcare practitioners need to translate them into structured data to support downstream secondary use applications, like disease surveillance and clinical decision support.8 Traditionally, medical practitioners have to manually collect information from unstructured data, such as medical records, in order to make diagnoses and treatment plans. This process, known as medical record review, can be challenging and time-consuming. The extensive paperwork burden can increase fatigue, reduce job satisfaction, and contribute to medical errors and adverse events.9 Automating the extraction of SDoH from unstructured clinical notes using natural language processing (NLP) techniques can help to reduce the workload for medical practitioners; improve the accuracy and efficiency of the information collection process; and generate a comprehensive representation of the patient about their social, behavioral, and environmental information for downstream tasks.10,11 This approach has been shown to be effective in previous research, as demonstrated in prior work.12–14
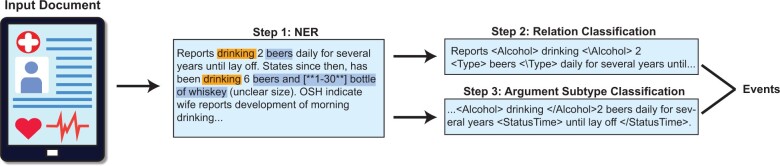
Previous studies on leveraging NLP to automate the extraction of SDoH information have included lexicons/rule-based methods13,15,16 and deep learning approaches.14,17–21 In this work, we introduce a novel system with 3 main components (see Figure 1) to extract event-based SDoH information from clinical notes: named entity recognition (NER),22 relation extraction (RE),23 and text classification (TC).24 We use the Social History Annotation Corpus (SHAC) developed for the 2022 N2C2 Shared Task—which is based on the work by Lybarger et al.14 One of the main challenges in extracting SDoH from text is a large number of overlapping entities. For example, Lybarger et al14 define smoking status as an SDoH. In their corpus, the span of the text “2–3 cig per day” includes 4 entities: the StatusTime argument (“2–3 cig per day”), the Amount argument (“2–3 cig”), the frequency (“per day”), and the type (“cig”). Even worse, entities with the exact same spans can refer to 2 separate entities. For example, “marijuana” represents the entity Drug and it represents the entity Type (ie, because it refers to a type of drug) in the Lybarger et al14 corpus.
Figure 1.
Overview of our marker-based pipeline.
Recently, several methods have been proposed for handling overlap in NER tasks.25–28 Some papers have designed different tagging schemes29,30 by combining token-level classes to deal with overlapping NER, which may cause data sparsity issues (eg, a word can be labeled as B-ORG-I-PER if it is the start of an organization span and the inner part of a person’s name). However, if 2 entity types overlap infrequently, this can cause a data sparsity issue. Span-based models are another approach for handling overlapping entities.31,32 These models follow a 2-stage framework, first extracting all possible text spans from the text and then using filters to reduce the total search space and computational complexity.31,32 Rojas et al33 show that simply training an individual model for every entity type (assuming overlapping entities only appear across entity types) produces better performance than more complex prior methods. However, training a single model for every entity type can be wasteful regarding memory usage. Moreover, if the number of entity types is large, the deployment of many models can be difficult.
To address limitations in prior work for extracting SDoH information from text using the NER approach, we propose a unified marker-based sequence labeling model for the simultaneous extraction of triggers and arguments in a single NER model. This model is then used as part of a larger event extraction system, which outperforms recent methods introduced in the 2022 N2C2 shared task. Our method is inspired by the success of prefix-based prompt-learning27,32,34 and the work by Rojas et al33 that shows individual models for each entity outperforming more complex overlapping NER systems. Intuitively, our approach simulates individual models trained for every entity type into a single system. Lybarger et al14 recognized 2 additional limitations of current SDoH extraction methods. First, prior methods lacked the ability to classify relationships between entities that span multiple sentences. Second, the methods were incapable of incorporating context from adjacent sentences when labeling various aspects of the SDoH event. Our system addresses the 2 limitations by working at the note level instead of the sentence level for the relation and subtype classification components of our pipeline.
In summary, this article makes the following contributions:
We propose a simple yet novel system for SDoH information extraction. Our system achieves state-of-the-art performance compared with other competitive systems submitted to the National NLP Clinical Challenges (n2c2) shared task.
We propose a novel marker-based sequence labeling method for extracting all possible triggers and argument entities while handling overlap. The method is shown to outperform more complex methods developed for overlapping NER. Moreover, our note-level components are able to identify relations across entities in separate sentences in the EHR note and incorporate cross-sentence context to improve subtype classification.
We conduct an ablation-like analysis to understand which components of our system have the greatest potential for improving SDoH extraction. Moreover, we perform an error analysis to provide future avenues of research.
METHODOLOGY
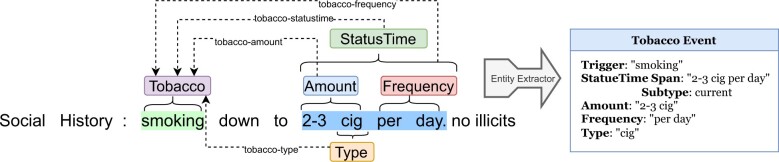
The SDoH extraction task aims to extract “triggers” and “arguments.” Triggers are mentions of SDoH factors (eg, Alcohol, Drug, Tobacco, Living Status, and Employment). Arguments link to the triggers to provide further context. An example is provided in Figure 2. The trigger extracted is “smoking” which was assigned the trigger entity Tobacco. Next, 4 argument entities are extracted: StatusTime, Amount, Frequency, and Type. Note that these entities can be nested (overlapping), as discussed in the “Background and significance” section. Intuitively, the argument entities provide information about the trigger entity, for example, what they were smoking and how often they smoked. Some arguments (eg, StatusTime) are also classified into specific subtypes to provide a standardized format for important information. In this case, we see that the person is a “current” smoker. Overall, while our main methodological advances come from the NER component (which we justify via a careful analysis), each piece works together to extract SDoH information to overcome several of the challenges described by Lybarger et al14 (eg, detecting cross-sentence relations). Finally, we describe the exact entity types for triggers, arguments, and all of the subtypes in their respective subsections below.
Figure 2.
An example for the SDoH extraction task.
Named entity recognition
The first stage of our SDoH system is to extract all trigger and argument entities within the text. There are 5 unique trigger entities: Drug, Alcohol, Tobacco, Employment, and LivingStatus. Likewise, there are 9 unique argument entity types: StatusTime, StatusEmploy, TypeLiving, Type, Method, History, Duration, Frequency, and Amount. Every argument type does not match every trigger type. For instance, TypeLiving refers to text spans that mention how a person lives (eg, whether they are homeless), which is not directly applicable to the other triggers such as Drug and Employment.
Formally, we frame this as a traditional NER sequential labeling task, where a sequence S consisting of n tokens , where n denotes the length of the sequence is classified into a sequence of labels L defined as . Specifically, we model where each label represents an entity type in Beginning–Inside–Outside (BIO) format (eg, B-Drug, I-Drug, and B-Type).35 Outside, or O, represents a token not classified into one of the SDoH trigger or argument entities. This traditional approach does not handle overlapping entities. In the SDoH corpus, overlapping entities appear across entity types. Generally, an entity does not overlap with an entity of the same type. This assumption is also used in prior overlapping entity work.33 However, to overcome this prior work, Rojas et al33 train an independent classifier for every entity type. For instance, a single model would predict all Drug entities, while another model would be dedicated to Employment. This approach could result in 14 unique models in our corpus (ie, a model for each trigger and argument entity type), for example, , , etc. Moreover, there may be information about one entity that can help improve the prediction of another. However, using independent models will overcome the issue of overlapping entities, but it will also cause the loss of access to cross-entity information.
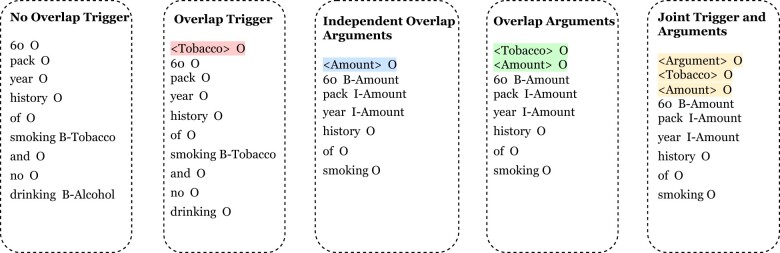
To overcome the limitations of training a separate model for each entity type, we explore methods of handling overlap without training multiple models by exploring different types of entity type markers, which have been shown to be effective at injecting information into the model.32,36,37 We explore 2 unique methods of training a joint NER model for the trigger entities, 2 models for the arguments, and 1 joint model for triggers and arguments. The summary of each marker-based system for each variation is shown in Figure 3. We describe each model below.
Figure 3.
Examples of the markers used for each of our NER systems.
Trigger Model 1 (no overlap trigger)
First, for triggers, we explore the use of the traditional flat NER, where we ignore overlap between trigger entities. We found that there is not substantial overlap between trigger entities, though it does appear within the dataset. Specifically, given the input sentence, we will simultaneously predict all trigger entity types by modeling where represents the NER model for all triggers. Each token will be assigned one, and only one, BIO formatted label .
Trigger Model 2 (overlap trigger)
Next, we explore a trigger model that can handle overlap. Intuitively, we simulate training a single model for every trigger entity type using a marker k. Intuitively, instead of predicting all trigger entity types in a single pass of the sentence and, thus, only assigning a single class to each token, we make predictions by first conditioning on the entity type we want to predict. Formally, we model which will only make predictions for each token for trigger k or not k. As we change k, the predictions will change. We implement this model by prepending a trigger type marker k to the start of each sequence that is formatted as TriggerName (eg, Tobacco), which transforms a sequence of tokens to . An example is provided in Figure 3.
Argument Model 1 (independent overlap arguments)
For the arguments, there is substantial overlap between entities. Hence, completely ignoring overlap is not feasible. The first argument model we explore involves training an Overlapping Argument model for each trigger. Specifically, we train a model similar to Overlap Trigger for arguments, but the model is trained for each trigger’s arguments. For instance, train a model for all of the Tobacco trigger’s arguments, StatusTime, Amount, Frequency, and Type. Likewise, we do the same for the other triggers, resulting in 5 models. Formally, we train a model where q represents an argument for trigger k. Similar to the Overlap Trigger model, we implement this by prepending the marker q to the sequence of tokens to form . Again, at inference time, we only predict one entity type where Q is the set of arguments for trigger k. To generate a different argument entity, we change q (eg, we prepend Type to predict the Type argument and Frequency to predict the frequency entity).
Argument Model 2: Overlap arguments
Instead of learning a joint argument model across all 5 triggers, we also experiment with a single argument model across all triggers. Formally, we model which conditions on trigger k and argument q. Again, we implement this by prepending both an argument and a trigger marker, transforming the tokens to .
Joint triggers and arguments
The final model we explore is a single joint model for Triggers and Arguments. Note that there is a substantial overlap between trigger and argument entities. Hence, this joint model tests the complete ability to handle the overlap of our marker-based system. This model is an extension of the Overlap Trigger and Overlap Arguments models. Specifically, we change what is prepended depending on what should be predicted. Formally, we model where k is a trigger marker (eg, Drug), q is an argument marker (eg, Type), and z is a marker that indicates whether we should predict a trigger or an argument (eg, Trigger or Argument). If we are predicting a trigger, then q is set to the empty string. Specifically, we transform the input sequence into . As an example, if we want to predict trigger Drug entities, we would modify the input sequence to start with “TriggerDrug.” To predict different entities, we modify the inputs to the system as appropriate.
Combinations
In our experiments, we explore 5 combinations of the models above: “No Overlap Trigger + Ind. Overlap Arguments,” “No Overlap Trigger + Overlap Arguments,” “Overlap Trigger + No Overlap Arguments,” “Overlap Trigger + Overlap Arguments,” and “Joint Trigger and Arguments.”
Relation classification
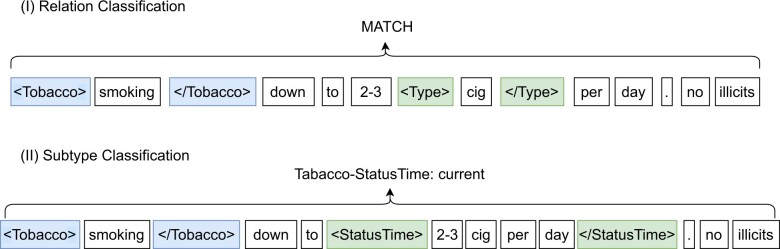
In our models for NER, we can map an extracted argument to a trigger of the correct type. However, there may be multiple triggers of the same type (eg, multiple Alcohol types in Figure 1). Therefore, matching arguments to an associated trigger instance is not possible with the NER models alone. Hence, we propose a relation classification (RC) framework to match arguments to their respective triggers. To follow a similar framework as our marker-based NER system, we applied the traditional RC (Matching the Blanks) approach.27,38–41 Specifically, we model the probability that an argument should map to a trigger as where represents the trigger entity and represents the argument entity. We model this classification task by wrapping the entities with markers. For example, given the sentence “smoking down to 2–3 cig per day,” if we want to check if the type argument “cig” maps to the trigger “smoking,” then the text is modified as “Tobacco smoking Tobacco down to 2–3 Type cig Type per day.” See another example in Figure 4. Our approach is able to detect relationships between entities that span different sentences by passing the entire clinical note to the RC model with the 2 entities and marked.
Figure 4.
Examples for the RC and subtype classification. (I) RC is a binary classification task that determines whether a relation exists between trigger and argument. The 2 possible classes are “match” and “not match.” (II) For subtype classification, a labeled argument is classified into one of several predefined subtypes, where each has a specific semantic meaning (eg, “current” drug user).
Argument subtype classification
The final piece of our SDoH extraction framework involves subtype classification. There are arguments (eg, Employment Status) that provide important information. However, it is generally stated in a wide array of formats. For instance, “John was just laid off work” and “John is not working” both mention that a person is unemployed. There are 6 arguments that are categorized into subtypes: Alcohol StatusTime, Drug StatusTime, Tobacco StatusTime, Employment Status, LivingStatus StatusTime, and LivingStatus TypeLiving. Each StatusTime subtype can take 1 of 3 categories: current, past, and future. Employment Status can be employed, unemployed, retired, on disability, student, or homemaker. LivingStatus TypeLiving can be alone, current, and past.
To detect subtypes, we use a similar framework as our RC component. Specifically, we model where represents the status argument we are subtyping and is its respective trigger entity matched via the RC model. s represents the subtype. Again, we model this via markers within the text, just like the RC task. For instance, given the sentence, “smoking down to 2–3 cig per day,” the StatusTime argument and Tobacco trigger are marked as “Tobacco smoking Tobacco down to StatusTime 2–3 cig per day StatusTime,” where the correct subtype would be “current.” We train a single model to capture all subtypes across the 6 arguments. See another example in Figure 4. Moreover, the entire clinical note is passed to the subtype model with markers such that contextual information from document can be used to improve performance. By using the entire note, we are able to overcome prior limitations of lacking contextual information mentioned by Lybarger et al.14
Implementation details
For the NER models, we train a Bi-directional Long Short-Term Memory (BiLSTM) network with conditional random fields.42 We explore 2 types of input embeddings for the model: Flair,43 BioBert,44 and T5-3B.45 For the Flair embedding model, we trained a marker-based NER model using a sample dropout of 0.4, a hidden layer size of 128, a learning rate of 0.1, and 25 epochs with a mini-batch size of 16. We save the model after each epoch and use the best version based on the validation dataset. The BioBert and T5-3B embedding models were trained in a similar fashion, with the exception of a sample dropout of 0.3, a hidden layer size of 1024, a maximum of 15 epochs, and a learning rate of 0.025. Both models fine-tuned the embedding layers. All NER models were implemented using the Flair software framework developed by Akbik et al43 (https://github.com/flairNLP/flair). For the RC and subtype classification models, we use a RoBERTa-base model46 with an Adam optimizer47 and the CosineAnnealingLR scheduler,48 a learning rate of 1e−5, and train for a maximum of 20 epochs. Again, the best epoch is chosen using the validation data. Finally, all experiments were performed on 4 NVidia GeForce GTX 1080 Ti GPUs and one NVidia A6000.
EXPERIMENTAL RESULTS
In this section, we describe the data, evaluation metrics, and report results, and an error analysis.
Datasets
We conducted our experiments on the 2022 N2C2 shared task version of the SHAC14 corpora. The dataset consists of 4480 annotated social history sections (70% train, 10% development, and 20% test) from MIMIC-III and the University of Washington Harborview Medical Centers data (UW). The systems are evaluated for 3 scenarios. First, Task A involves training and evaluating on the MIMIC-III data (ie, MIMIC-III MIMIC-III). Task B measures generalizability which involves training on the MIMIC-III and evaluating on UW data (ie, MIMIC-III UW). Finally, Task C involves training on MIMIC-III and UW data and evaluating on UW data (ie, MIMIC-III + UW UW). Table 1 presents basic information about the datasets.
Table 1.
Dataset statistics for the MIMIC-III and UW datasets
| Dataset | Subset | Number of documents | Max words | AVG words |
|---|---|---|---|---|
| MIMIC-III | Train | 1316 | 229 | 65.34 |
| Dev | 188 | 82 | 44.34 | |
| Test | 373 | 192 | 44.50 | |
| UW | Train | 1751 | 437 | 54.22 |
| Dev | 259 | 99 | 37.47 | |
| Test | 518 | 288 | 37.16 |
Note: Statistics include the number of examples/documents in each subset, max words in a document, and the average words per document.
Evaluation metrics
Performance is evaluated using the following metrics: overall precision (P), recall (R), and F1-score (F1), which is a microaverage of all trigger types, argument types, and argument subtypes (ie, true positives, false positives, and false negatives are summed across all categories). In all of our analysis, we use the evaluation tools provided by the N2C2 shared task organizers (https://github.com/Lybarger/brat_scoring).
Overall results
Table 2 shows the overall performance of our systems compared with the best models in the 2022 N2C2 shared task among the 15 participating teams. Although our model is simple, the marker-based system approach outperforms prior work. Specifically, our system with flair word embeddings and joint NER model achieves similar performance to the best-performing systems without using an ensemble or manually curated rules (0.8829 vs 0.9008 for Task A, 0.7523 vs 0.7738 for Task B, and 0.8788 vs 0.8886 for Task C). Additionally, we evaluated the BioBert model that was trained in the healthcare domain, PubMed. This improved our Flair model slightly and the results were close to the best-performing systems in the competition (0.8948 vs 0.9008 for Task A, 0.7586 vs 0.7738 for Task B, and 0.8889 vs 0.8886 for Task C). Our results are further improved using a larger pretrained model T5-3B. The T5-3B embeddings and Joint Trigger + Argument NER model achieve an absolute F1 score improvement compared with best competition results of 0.0104, 0.0354, and 0.0156 for Task A, Task B, and Task C, respectively. The improvements demonstrate the effectiveness of prefixing entity type markers in front of each sentence to handle overlapping NER. We also find that using joint models generally outperforms using more models. For example, Flair + RoBERTa Joint Trigger and Argument has an F1 of 0.7523 for Task B, while Flair + RoBERTa No Overlap Trigger + Ind. Overlap Argument has an F1 of 0.7189. One possible reason for the excellent performance is that when we train joint models, more cross-entity information is shared, similar to what happens with multi-task learning.
Table 2.
Overall performance across the 3 tasks: Task A (MIMIC MIMIC), Task B (MIMIC UW), and Task C (MIMIC+UW UW)
| Task A |
Task B |
Task C |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Representations | NER Method | P | R | F1 | P | R | F1 | P | R | F1 |
| Best Competition Models | 0.9093 | 0.8925 | 0.9008 | 0.8108 | 0.7400 | 0.7738 | 0.8906 | 0.8867 | 0.8886 | |
| Flair + RoBERTa | Joint Trigger and Argument | 0.9073 | 0.8597 | 0.8828 | 0.8016 | 0.7088 | 0.7523 | 0.8926 | 0.8642 | 0.8781 |
| Overlap Trigger + Overlap Argument | 0.9010 | 0.8655 | 0.8829 | 0.7967 | 0.7036 | 0.7473 | 0.8837 | 0.8741 | 0.8788 | |
| Overlap Trigger + Ind. Overlap Argument | 0.9001 | 0.8643 | 0.8818 | 0.7856 | 0.7040 | 0.7425 | 0.8870 | 0.8650 | 0.8759 | |
| No Overlap Trigger + Overlap Argument | 0.8915 | 0.8594 | 0.8752 | 0.7835 | 0.6714 | 0.7231 | 0.8707 | 0.8677 | 0.8692 | |
| No Overlap Trigger + Ind. Overlap Argument | 0.8890 | 0.8580 | 0.8732 | 0.7733 | 0.6717 | 0.7189 | 0.8739 | 0.8581 | 0.8659 | |
| BioBERT + RoBERTa | Joint Trigger and Argument | 0.8914 | 0.8983 | 0.8948 | 0.7827 | 0.7359 | 0.7586 | 0.8943 | 0.8835 | 0.8889 |
| Overlap Trigger + Overlap Argument | 0.8879 | 0.8897 | 0.8888 | 0.7775 | 0.7354 | 0.7559 | 0.8894 | 0.8904 | 0.8899 | |
| Overlap Trigger + Ind. Overlap Argument | 0.8855 | 0.8865 | 0.8860 | 0.7757 | 0.7174 | 0.7454 | 0.8881 | 0.8849 | 0.8865 | |
| No Overlap Trigger + Overlap Argument | 0.8645 | 0.8784 | 0.8714 | 0.7464 | 0.7434 | 0.7449 | 0.8819 | 0.8770 | 0.8795 | |
| No Overlap Trigger + Ind. Overlap Argument | 0.8617 | 0.8744 | 0.8680 | 0.7479 | 0.7241 | 0.7358 | 0.8794 | 0.8705 | 0.8749 | |
| T5-3B + RoBERTa | Joint Trigger and Argument | 0.9035 | 0.9167 | 0.9101 | 0.8144 | 0.7964 | 0.8053 | 0.9002 | 0.9049 | 0.9025 |
| Overlap Trigger + Overlap Argument | 0.9132 | 0.9092 | 0.9112 | 0.8194 | 0.7992 | 0.8092 | 0.9036 | 0.9049 | 0.9042 | |
| Overlap Trigger + Ind. Overlap Argument | 0.9036 | 0.9020 | 0.9028 | 0.8029 | 0.7800 | 0.7913 | 0.8982 | 0.9005 | 0.8994 | |
| No Overlap Trigger + Overlap Argument | 0.9009 | 0.8980 | 0.8994 | 0.8165 | 0.7780 | 0.7968 | 0.8969 | 0.9049 | 0.9009 | |
| No Overlap Trigger + Ind. Overlap Argument | 0.8924 | 0.8914 | 0.8919 | 0.8014 | 0.7575 | 0.7788 | 0.8916 | 0.9009 | 0.8962 | |
Note: Best scores are bolded for the best model(s) for each set of embedding types (Flair + Roberta and T5-3B + RoBERTa).
Analysis of system component importance
There are 3 major components to our SDoH extraction system: NER, RC, and subtype classification. For future work, which piece can provide the most benefit if improved? To understand each component better, we run an ablation-like experiment where we replace each component with the ground-truth predictions. Intuitively, we are trying to understand if we improved a single component, which has the most potential impact on the entire system. Table 3 shows the results of the study for Task A and Task C. By comparing, we find that using ground truth for argument-level NER yields the largest potential improvement (0.0433 for Task A and 0.0403 for Task C). The next largest potential improvement comes from the RC model. The component with the lowest potential impact on the overall performance is subtype classification, with an improvement of 0.0193 for Task A and 0.0162 for Task C.
Table 3.
Analysis of system component importance for Tasks A and C using their respective development sets
| Task A |
Task C |
|||||||
|---|---|---|---|---|---|---|---|---|
| Model | P | R | F1 | Diff F1 | P | R | F1 | Diff F1 |
| Joint Trigger and Argument | 0.8994 | 0.9074 | 0.9034 | — | 0.9064 | 0.9189 | 0.9126 | — |
| + Perfect NER-Trigger | 0.9410 | 0.9207 | 0.9308 | 0.0274 | 0.9391 | 0.9363 | 0.9377 | 0.0251 |
| + Perfect NER-Argument | 0.9482 | 0.9451 | 0.9467 | 0.0433 | 0.9579 | 0.9480 | 0.9529 | 0.0403 |
| + Perfect Subtype Classification | 0.9186 | 0.9268 | 0.9227 | 0.0193 | 0.9225 | 0.9352 | 0.9288 | 0.0162 |
| + Perfect RC | 0.9639 | 0.9046 | 0.9333 | 0.0300 | 0.9671 | 0.9152 | 0.9404 | 0.0278 |
Note: The biggest differences are bolded.
Comparison to a state-of-the-art span-based model
As mentioned in the “Background and significance” section, there has been significant progress in developing models that can handle overlapping spans. While some research has shown that training independent models outperform many of the recent methods,33 it is important to compare them as a baseline. Hence, we applied a recent span-based method Triaffine28 to using publicly available source code on the N2C2 shared task data (https://github.com/GanjinZero/Triaffine-nested-ner). This approach allows the model to capture complex dependencies and interactions between different elements in the input text, potentially improving its performance on tasks such as overlapped NER. Triaffine is currently a state-of-the-art method in this area.28 We compare 2 versions of the model, one that trains triggers and arguments jointly (Joint Trigger + Argument) and one that trains a model for the triggers separately from the arguments (Independent Trigger + Argument). We report the results in Table 4. Overall, we find that the independent model substantially outperforms the joint model across all 3 tasks (eg, 0.8594 vs 0.5942 for Task A). The Joint model potentially suffers because it cannot handle cases where the triggers overlap exactly with the span of an argument. Our method is capable of handling this by predicting each entity one at a time using markers. We also compare with assuming a perfect RC because the span-based model does not have information about matches between arguments and trigger types. Our models contain this information by including a marker for the trigger and the argument for argument prediction. Yet, even with a perfect RC model, it still underperforms our best approach without a perfect model.
Table 4.
Comparison to the Triaffine28 span-based model for overlapping entities
| Task A |
Task B |
Task C |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Model | P | R | F1 | P | R | F1 | P | R | F1 |
| Triaffine: Independent Trigger + Argument | 0.9050 | 0.8182 | 0.8594 | 0.7889 | 0.6641 | 0.7211 | 0.8876 | 0.8462 | 0.8664 |
| + Perfect RC | 0.9561 | 0.8222 | 0.8841 | 0.8108 | 0.7400 | 0.7738 | 0.8906 | 0.8867 | 0.8886 |
| Triaffine: Joint Trigger + Argument | 0.8585 | 0.4543 | 0.5942 | 0.8326 | 0.4377 | 0.5738 | 0.9101 | 0.5555 | 0.6899 |
| T5-3B + RoBERTa Joint Trigger + Argument (ours) | 0.9035 | 0.9167 | 0.9101 | 0.8144 | 0.7964 | 0.8053 | 0.9002 | 0.9049 | 0.9025 |
Notes: Results are on the test data. The largest numbers are bolded.
Detailed results for trigger and argument types
Table 5 provides a detailed analysis of performance based on event type and argument type using our best model, which is calculated using microaveraged F1 scores. The performance for Substance use (Alcohol, Drug, and Tobacco) and Employment triggers is consistent between Task A and Task C, with scores greater than 0.93, despite Task C having more training data. However, the Living Status trigger performance in Task C is lower compared with Task A due to the more complex living status descriptions in the UW dataset, such as “living in a specific Shelter” (0.9294 vs 0.9636). The labeled argument performance is similar in Task A and Task C for Tobacco and Employment. However, there are differences in Alcohol, Drug, and Living Status labeled arguments. Interestingly, the Drug Status argument’s performance decreases when more training data are available (0.9418 on Task A vs 0.8946 on Task C). This may be because more drug events are in the test dataset, providing a better performance estimate (189 on Task A vs 473 on Task C). For span-only arguments, the performance is comparable for Alcohol, Drug, Tobacco, and Status. However, there is a significant decrease in the performance for the Living Status, which is potentially due to the complex living history descriptions in the UW data or the small test dataset (6 on Task A vs 11 on Task C).
Table 5.
Overall performance for the 3 tasks based on event type and argument type—Task A (MIMIC MIMIC), Task B (MIMIC UW), and Task C (MIMIC+UW UW)
| Task A |
Task B |
Task C |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Event type | Argument | #Train | #Test | F1 | #Test | F1 | #Train | #Test | F1 | |
| Trigger | Alcohol | – | 1295 | 308 | 0.9776 | 1828 | 0.9540 | 2917 | 403 | 0.9865 |
| Drug | – | 987 | 189 | 0.9583 | 2263 | 0.9151 | 3004 | 473 | 0.9623 | |
| Tobacco | – | 1232 | 321 | 0.9721 | 1824 | 0.9394 | 1767 | 434 | 0.9655 | |
| Employment | – | 982 | 168 | 0.9388 | 872 | 0.8477 | 2390 | 153 | 0.9325 | |
| Living Status | – | 959 | 242 | 0.9636 | 1613 | 0.7925 | 2845 | 354 | 0.9294 | |
| Labeled argument | Alcohol | Status | 1295 | 308 | 0.9064 | 1828 | 0.8465 | 2917 | 403 | 0.9499 |
| Drug | Status | 987 | 189 | 0.9418 | 2263 | 0.8111 | 3004 | 473 | 0.8946 | |
| Tobacco | Status | 1232 | 321 | 0.9216 | 1824 | 0.8694 | 1767 | 434 | 0.9292 | |
| Employment | Status | 982 | 168 | 0.9059 | 872 | 0.7707 | 2390 | 153 | 0.8903 | |
| Living Status | Status | 959 | 242 | 0.9553 | 1611 | 0.7358 | 2845 | 354 | 0.9073 | |
| Type | 959 | 242 | 0.9309 | 1613 | 0.6497 | 2845 | 354 | 0.8759 | ||
| Span-only argument | Alcohol | Amount, duration, frequency, history, type, method | 1078 | 162 | 0.7262 | 1180 | 0.6928 | 2169 | 178 | 0.7865 |
| Drug | 1037 | 165 | 0.7915 | 2389 | 0.6699 | 3233 | 418 | 0.7910 | ||
| Tobacco | 1548 | 300 | 0.8508 | 1926 | 0.7918 | 3293 | 375 | 0.8194 | ||
| Employment | Duration, history, type | 806 | 140 | 0.7518 | 591 | 0.6209 | 1347 | 96 | 0.7389 | |
| Living Status | Duration, history | 56 | 6 | 0.5714 | 80 | 0.4364 | 133 | 11 | 0.4545 | |
Note: Task B uses the training data from Task A.
In Task B, which was trained on MIMIC and tested on the UW dataset, there is a slight decrease in the performance of Substance use triggers due to the difference between the training and test domains. Additionally, the performance of Employment and Living Status triggers and arguments decreases substantially, especially for the Living Status Type argument (0.6497 on Task B vs 0.9309 on Task A and 0.8759 on Task C). These may be due to the more intricate employment and living histories of patients in the UW dataset compared with those in MIMIC. Specifically, the UW dataset has a unique format of templated information, including details on substance use, which differs from the format present in the MIMIC data. Additionally, the writing style in the UW dataset is distinct from that in MIMIC.
Error analysis
We analyze common errors made by our Joint Trigger and Argument model. First, when there are direct mentions of different (unique) types of drugs that have different StatusTime (eg, current vs past), annotators will label each as separate triggers. For instance,
“Illicit drugs: current marijuana use, cocaine quit 5 years ago.”
has 2 Drug triggers: “marijuana use” and “cocaine.” Yet, our model only predicts the more general “Illicit drugs” as the trigger entity. We hypothesize that our model does not differentiate general concepts (eg, “Illicit drugs”) from more specific instances of the concept (eg, “marijuana” and “cocaine.” This is because it is not modeled explicitly in the architecture; moreover, the data generally contain more instances of generic mentions than more specific mentions. Another example of this is found in the example
“She drinks 2–3 alcoholic beverages per week.”
where our model predicts “drinks” as trigger, while the ground truth is “alcoholic.” Based on the criteria of Lybarger et al,14 the phrase describing a general substance (ie, alcohol, tobacco, or drug) or substance-related verb, such as drink can be a trigger. When both appear, a more specific concept should be used. Yet, again, our model fails to understand this underlying semantic meaning and does not differentiate instances from generic types. This error is very common for other trigger types. For example, our model incorrectly predicts “smokes” as a trigger instead of the ground-truth “cigarettes” often. Likewise, for the employment trigger, our model will predict “worked” as a trigger instead of “retired” in some examples. Another common error type happens for uncommon noun phrases. For instance, in the example,
“Currently at a rehab facility, but previously living with his wife at home.”
the ground truth for the LivingStatus trigger is “a rehab facility,” but our model fails to detect it. Another example of this error type includes
“Works in finance at Mass Eye & Ear.”
where our model predicts the Type argument for the Employment trigger as “finance,” while the ground-truth is “finance at Mass Eye & Ear.” Again, this indicates our models struggle with novel noun phrases, particularly when they include prepositional phrases. A future interesting research avenue would explore methods for incorporating external knowledge bases into transformer models. This could potentially help the model make more accurate predictions and avoid errors. One way to incorporate external knowledge into transformer models is through the use of external memory networks, which have been shown to be effective at incorporating common sense into language models.49
CONCLUSION
This article presents our approach for extracting SDoH events from clinical notes using the N2C2-2022 Task 2 shared task dataset. We introduce a novel NER system to extract overlapped entities and propose a multiple pipeline system to extract SDoH events, including NER, RC, and Subtype Classification models, which results in a new state-of-the-art performance for the N2C2 data. In future efforts, we aim to enhance our NER model by utilizing structured knowledge bases through demonstration-based learning,50 such as providing the sentence of task demonstrations or entity type descriptions instead of just using simple entity type markers for in-context learning. This can easily be integrated into our framework and we hypothesize that it would help low-resource entities.
Contributor Information
Xingmeng Zhao, Information Systems and Cyber Security, The University of Texas at San Antonio, San Antonio, Texas, USA.
Anthony Rios, Information Systems and Cyber Security, The University of Texas at San Antonio, San Antonio, Texas, USA.
FUNDING
This material is based upon work supported by the National Science Foundation (NSF) under Grant No. 2145357.
AUTHOR CONTRIBUTIONS
XZ performed the experiments and drafted the initial manuscript. AR conceived of the study, oversaw the design, and reviewed and approved the manuscript.
CONFLICT OF INTEREST STATEMENT
None.
DATA AVAILABILITY
The data underlying this article are available in as part of the 2022 N2C2 Shared Task at https://n2c2.dbmi.hms.harvard.edu/2022-track-2 and can be accessed by contacting the organizers at n2c2_uw_2022_sdoh@googlegroups.com.
REFERENCES
- 1. World Health Organization, et al. Social determinants of health Technical report, WHO Regional Office for South-East Asia, 2008.
- 2. Marmot M, Allen J, Bell R, Bloomer E, Goldblatt P; Consortium for the European Review of Social Determinants of Health and the Health Divide. Who European review of social determinants of health and the health divide. Lancet 2012; 380 (9846): 1011–29. [DOI] [PubMed] [Google Scholar]
- 3. Gucciardi E, Vahabi M, Norris N, Monte JPD, Farnum C.. The intersection between food insecurity and diabetes: A review. Curr Nutr Rep 2014; 3 (4): 324–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Singh GK, Siahpush M, Kogan MD.. Neighborhood socioeconomic conditions, built environments, and childhood obesity. Health Aff (Millwood) 2010; 29 (3): 503–12. [DOI] [PubMed] [Google Scholar]
- 5. Yang X, Yelton B, Chen S, et al. Examining social determinants of health during a pandemic: Clinical application of z codes before and during covid-19. Front Public Health 2022; 10: 888459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Koh HK, Piotrowski JJ, Kumanyika S, Fielding JE.. Healthy people: A 2020 vision for the social determinants approach. Health Educ Behav 2011; 38 (6): 551–7. [DOI] [PubMed] [Google Scholar]
- 7. Quan H, Sundararajan V, Halfon P, et al. Coding algorithms for defining comorbidities in ICD-9-CM and ICD-10 administrative data. Med Care 2005; 43 (11): 1130–9. [DOI] [PubMed] [Google Scholar]
- 8. Karran EL, Grant AR, Moseley GL.. Low back pain and the social determinants of health: A systematic review and narrative synthesis. Pain 2020; 161 (11): 2476–93. [DOI] [PubMed] [Google Scholar]
- 9. Conway M, Keyhani S, Christensen L, et al. Moonstone: A novel natural language processing system for inferring social risk from clinical narratives. J Biomed Semantics 2019; 10 (1): 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Patra BG, Sharma MM, Vekaria V, et al. Extracting social determinants of health from electronic health records using natural language processing: A systematic review. J Am Med Inform Assoc 2021; 28 (12): 2716–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bompelli A, Wang Y, Wan R, et al. Social and behavioral determinants of health in the era of artificial intelligence with electronic health records: A scoping review. Health Data Sci 2021; 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jensen PB, Jensen LJ, Brunak S.. Mining electronic health records: Towards better research applications and clinical care. Nat Rev Genet 2012; 13 (6): 395–405. [DOI] [PubMed] [Google Scholar]
- 13. Hatef E, Rouhizadeh M, Tia I, et al. Assessing the availability of data on social and behavioral determinants in structured and unstructured electronic health records: A retrospective analysis of a multilevel health care system. JMIR Med Inform 2019; 7 (3): e13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lybarger K, Ostendorf M, Yetisgen M.. Annotating social determinants of health using active learning, and characterizing determinants using neural event extraction. J Biomed Inform 2021; 113: 103631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bejan CA, Angiolillo J, Conway D, et al. Mining 100 million notes to find homelessness and adverse childhood experiences: 2 case studies of rare and severe social determinants of health in electronic health records. J Am Med Inform Assoc 2018; 25 (1): 61–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chapman AB, Jones A, Kelley AT, et al. Rehoused: A novel measurement of veteran housing stability using natural language processing. J Biomed Inform 2021; 122: 103903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Feller DJ, Bear Don’t Walk Iv OJ, Zucker J.. Detecting social and behavioral determinants of health with structured and free-text clinical data. Appl Clin Inform 2020; 11 (1): 172–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stemerman R, Arguello J, Brice J, Krishnamurthy A, Houston M, Kitzmiller R.. Identification of social determinants of health using multi-label classification of electronic health record clinical notes. JAMIA Open 2021; 4 (3): ooaa069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yu Z, Yang X, Dang C, et al. A study of social and behavioral determinants of health in lung cancer patients using transformers-based natural language processing models. In: AMIA Annual Symposium Proceedings, Vol. 2021. American Medical Informatics Association; 2021: 1225. [PMC free article] [PubMed]
- 20. Yu Z, Yang X, Guo Y, Bian J, Wu Y.. Assessing the documentation of social determinants of health for lung cancer patients in clinical narratives. Front Public Health 2022; 10: 778463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Han S, Zhang RF, Shi L, et al. Classifying social determinants of health from unstructured electronic health records using deep learning-based natural language processing. J Biomed Inform 2022; 127: 103984. [DOI] [PubMed] [Google Scholar]
- 22. Tjong E, Sang K, De Meulder F. Introduction to the conll-2003 shared task: Language-independent named entity recognition. In: Proceedings of the Seventh Conference on Natural Language Learning at HLT-NAACL 2003; 2003: 142–7.
- 23. Zelenko D, Aone C, Richardella A.. Kernel methods for relation extraction. J Mach Learn Res 2003; 3 (Feb): 1083–106. [Google Scholar]
- 24. Garla VN, Brandt C.. Ontology-guided feature engineering for clinical text classification. J Biomed Inform 2012; 45 (5): 992–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sohrab MG, Miwa M. Deep exhaustive model for nested named entity recognition. In: Proceedings of the 2018 Conference on Empirical Methods in Natural Language Processing, October–November 2018. Brussels, Belgium: Association for Computational Linguistics; 2018: 2843–2849. (doi: 10.18653/v1/D18-1309) https://aclanthology.org/D18-1309 [DOI]
- 26. Wang B, Lu W. Combining spans into entities: A neural two-stage approach for recognizing discontiguous entities. In: Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP); 2019: 6216–24.
- 27. Zhong Z, Chen D. A frustratingly easy approach for entity and relation extraction. In: Proceedings of the 2021 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies; 2021: 50–61.
- 28. Yuan Z, Tan C, Huang S, Huang F. Fusing heterogeneous factors with triaffine mechanism for nested named entity recognition. In: Findings of the Association for Computational Linguistics: ACL 2022; 2022: 3174–86.
- 29. Wang J, Shou L, Chen K, Chen G. Pyramid: A layered model for nested named entity recognition. In: Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics; 2020: 5918–28.
- 30. Straková J, Straka M, Hajic J. Neural architectures for nested NER through linearization. In: Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics; 2019: 5326–31.
- 31. Yan H, Gui T, Dai J, Guo Q, Zhang Z, Qiu X. A unified generative framework for various NER subtasks. In: Proceedings of the 59th Annual Meeting of the Association for Computational Linguistics and the 11th International Joint Conference on Natural Language Processing (Volume 1: Long Papers); 2021: 5808–22.
- 32. Huang P, Zhao X, Hu M, Fang Y, Li X, Xiao W. Extract-select: A span selection framework for nested named entity recognition with generative adversarial training. In: Findings of the Association for Computational Linguistics: ACL 2022; 2022: 85–96.
- 33. Rojas M, Bravo-Marquez F, Dunstan J. Simple yet powerful: An overlooked architecture for nested named entity recognition. In: Proceedings of the 29th International Conference on Computational Linguistics; 2022: 2108–17.
- 34. Baldini Soares L, Fitzgerald N, Ling J, Kwiatkowski T. Matching the blanks: Distributional similarity for relation learning. In: Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics; 2019: 2895–905.
- 35. Ramshaw LA, Marcus MP.. Text chunking using transformation-based learning. In: Armstrong S, Church K, Isabelle P, Manzi S, Tzoukermann E, Yarowsky D, eds. Natural Language Processing Using Very Large Corpora. Text, Speech and Language Technology, Vol 11. Dordrecht: Springer; 1999: 157–76. [Google Scholar]
- 36. Van Nguyen M, Ngo Nguyen T, Min B, Nguyen TH. Crosslingual transfer learning for relation and event extraction via word category and class alignments. In: Proceedings of the 2021 Conference on Empirical Methods in Natural Language Processing; 2021: 5414–26.
- 37. Hsu I-H, Huang K-H, Boschee E, et al. Degree: A data-efficient generation-based event extraction model. In: Proceedings of the 2022 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies; 2022: 1890–908.
- 38. Baldini Soares L, FitzGerald N, Ling J, Kwiatkowski T. Matching the blanks: Distributional similarity for relation learning. In: ACL; 2019: 2895–905.
- 39. Lee Y, Son J, Song M.. Bertsrc: Transformer-based semantic relation classification. BMC Med Inform Decis Mak 2022a; 22 (1): 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang Z, Han X, Liu Z, Jiang X, Sun M, Liu Q. Ernie: Enhanced language representation with informative entities. In: Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics; 2019: 1441–51.
- 41. Peters ME, Neumann M, Logan R. Knowledge enhanced contextual word representations. In: Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP); 2019: 43–54.
- 42. Luo L, Yang Z, Yang P, et al. An attention-based BiLSTM-CRF approach to document-level chemical named entity recognition. Bioinformatics 2018; 34 (8): 1381–8. [DOI] [PubMed] [Google Scholar]
- 43. Akbik A, Bergmann T, Blythe D, Rasul K, Schweter S, Vollgraf R. FLAIR: An easy-to-use framework for state-of-the-art NLP. In: NAACL; 2019: 54–9.
- 44. Lee J, Yoon W, Kim S, et al. Biobert: A pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 2020; 36 (4): 1234–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Raffel C, Shazeer N, Roberts A, et al. Exploring the limits of transfer learning with a unified text-to-text transformer. J Mach Learn Res 2020; 21 (140): 1–67. http://jmlr.org/papers/v21/20-074.html.34305477 [Google Scholar]
- 46. Liu Y, Ott M, Goyal N, et al. Roberta: A robustly optimized bert pretraining approach. arXiv preprint arXiv:1907.11692; 2019.
- 47. Diederik PK, Ba J. Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980; 2014.
- 48. Loshchilov I, Hutter F. SGDR: Stochastic gradient descent with warm restarts. arXiv preprint arXiv:1608.03983; 2016.
- 49. Xing Y, Shi Z, Meng Z, Lakemeyer G, Ma Y, Wattenhofer R. Km-bart: Knowledge enhanced multimodal bart for visual commonsense generation. In: Proceedings of the 59th Annual Meeting of the Association for Computational Linguistics and the 11th International Joint Conference on Natural Language Processing (Volume 1: Long Papers); 2021: 525–35.
- 50. Lee D-H, Kadakia A, Tan K, et al. Good examples make a faster learner: Simple demonstration-based learning for low-resource NER. In: Proceedings of the 60th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers); 2022b: 2687–700.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data underlying this article are available in as part of the 2022 N2C2 Shared Task at https://n2c2.dbmi.hms.harvard.edu/2022-track-2 and can be accessed by contacting the organizers at n2c2_uw_2022_sdoh@googlegroups.com.