Abstract
Objective
Social determinants of health (SDOH) are nonclinical, socioeconomic conditions that influence patient health and quality of life. Identifying SDOH may help clinicians target interventions. However, SDOH are more frequently available in narrative notes compared to structured electronic health records. The 2022 n2c2 Track 2 competition released clinical notes annotated for SDOH to promote development of NLP systems for extracting SDOH. We developed a system addressing 3 limitations in state-of-the-art SDOH extraction: the inability to identify multiple SDOH events of the same type per sentence, overlapping SDOH attributes within text spans, and SDOH spanning multiple sentences.
Materials and Methods
We developed and evaluated a 2-stage architecture. In stage 1, we trained a BioClinical-BERT-based named entity recognition system to extract SDOH event triggers, that is, text spans indicating substance use, employment, or living status. In stage 2, we trained a multitask, multilabel NER to extract arguments (eg, alcohol “type”) for events extracted in stage 1. Evaluation was performed across 3 subtasks differing by provenance of training and validation data using precision, recall, and F1 scores.
Results
When trained and validated on data from the same site, we achieved 0.87 precision, 0.89 recall, and 0.88 F1. Across all subtasks, we ranked between second and fourth place in the competition and always within 0.02 F1 from first.
Conclusions
Our 2-stage, deep-learning-based NLP system effectively extracted SDOH events from clinical notes. This was achieved with a novel classification framework that leveraged simpler architectures compared to state-of-the-art systems. Improved SDOH extraction may help clinicians improve health outcomes.
Keywords: SDOH, deep learning, machine learning, natural language processing, event extraction
INTRODUCTION
Social determinants of health (SDOH) are nonclinical, socioeconomic conditions that influence patient health and quality of life. SDOH may account for 80% of modifiable health factors, whereas medical care accounts for only the remaining 20%.1 For example, homelessness and living alone, substance abuse (drug, alcohol, or tobacco), and unemployment are linked to social isolation, depression, and increased morbidity and mortality.2–4 It is thus incumbent on healthcare stakeholders to identify SDOH to assist in healthcare interventions, public health reporting, and large-scale retrospective studies.
Electronic health records (EHR) capture SDOH in both structured fields and unstructured (narrative) notes. However, unstructured notes may capture SDOH more frequently and in a more nuanced fashion compared to structured data. For example, a study found that the prevalence of patients with identified social support increased from 0.4% to 16% when clinical notes were reviewed.5 Moreover, nonsystematically reported SDOH such as ethnicity and marital status are more often missing-not-at-random in structured data compared to unstructured data, which introduces selection bias.6
Manual extraction of SDOH from clinical notes is labor intensive and expensive due to the vast number of notes in any EHR system, the degree of variation and complexity of clinical natural language, and the need for human annotators. Therefore, the development of natural language processing (NLP) systems that automatically extract SDOH from clinical notes has been an active area of research over the past several years.7 While rule-based systems such as cTAKES and Moonstone are widely adopted,8,9 state-of-the-art SDOH extraction is now achieved by supervised machine learning (ML)-based systems. Traditional ML techniques (eg, SVM, k-NN, random forests) have been somewhat successful in SDOH extraction. However, deep-learning based NLP models, and especially those based on transformers like Bidirectional Encoder Representations from Transformers (BERT), are increasingly outperforming traditional ML approaches.10–14
The development of ML systems requires large, annotated datasets for training and evaluation. To this end, Lybarger et al15 curated and annotated the Social History Annotation Corpus (SHAC), which includes 4480 social history sections with detailed annotations for 12 SDOH characterizing the status, extent, and temporality of 18 000 distinct SDOH events in clinical notes from MIMIC-III16 and the University of Washington and Harborview Medical Centers (UW). Figure 1 shows an example of an annotated note from SHAC. The annotations include a trigger indicating the presence of an SDOH event (eg, “tobacco use” triggers a tobacco event), and arguments characterizing the event (eg, “quit” indicates a past temporal status of the tobacco event). Using SHAC, Lybarger et al15 trained a deep neural network to extract event triggers and their arguments for the most frequently annotated SDOH: substance use (tobacco, drugs, or alcohol), living status, and employment. This model featured a pretrained BERT, bidirectional long short-term memory (LSTM) layers, conditional random fields (CRF), and self-attention to achieve state-of-the-SDOH event extraction, which we describe in further detail in the “Materials and Methods” section. When trained on MIMIC and UW notes, their model achieved 0.95 F1 on trigger extraction, and 0.70–0.90 F1 for argument extraction on the test sets for MIMIC and UW notes.
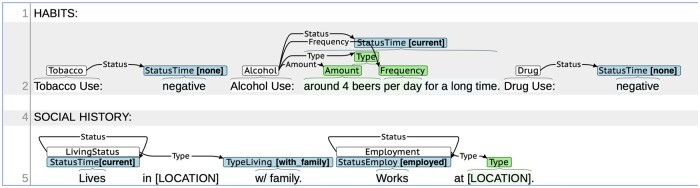
Figure 1.
Social determinants of health (SDOH) annotation example.
To further advance SDOH event extraction, Lybarger et al14 organized the 2022 n2c2 Track 2 shared task, Extracting Social Determinants of Health, in which participants developed new event extraction models from SHAC. We recently participated in this shared task, developing a novel deep neural network event extraction architecture that addressed 3 key limitations of the Lybarger et al15 model. First, Lybarger et al’s model (L1) cannot incorporate context from preceding or following sentences and cannot generate events that span multiple sentences; second, it (L2) cannot predict multiple event triggers of the same type (eg, alcohol) in a single sentence; third, it (L3) cannot identify overlapping spans for arguments that do not have predefined label categories (ie, span-only arguments). Supplementary Table S1 shows examples of all 3 limitations.
Our proposed NLP system, multilabel multitask BERT (MLTB), addresses the 3 above preceding limitations: it processes text at the note level (addressing L1), treats trigger detection as a token-level classification task (addressing L2), and treats argument detection as a multilabel token classification task, so each token can partake in multiple arguments (addressing L3). Indeed, we achieved between second and fourth place in the n2c2 competition across 3 different subtasks, performing within a 0.02 difference in F1 score compared to the highest scoring submissions. Our contribution is thus the development of a novel deep learning-based NLP system that can extract SDOH events in clinical notes with high accuracy, while also avoiding the limitations of the previous state-of-the-art model.
The rest of the paper is as follows. In the “Materials and Methods” section, we first describe SHAC and review Lybarger et al’s15 event-extraction model. Then, we describe the development of our own event-extraction architecture, which addresses the 3 limitations of the current-state-of-the-art model. In the “Results” section, we report our performance in the 3 subtasks of the 2022 n2c2 Track 2 shared task. Finally, we discuss these results, as well as their limitations and directions for future work.
MATERIALS AND METHODS
The shared task competition study was approved by the Internal Review Boards at the Massachusetts Institute of Technology and the University of Washington.
Data sets
SHAC consists of 4480 social history sections from MIMIC and UW notes. MIMIC notes included critical care discharge summaries dated from 2001 to 2012. Besides discharge summary notes from emergency department and inpatient visits, UW also included inpatient progress notes dated from 2008 to 2019. The original SHAC annotation guidelines included 12 SDOH categories (event types), ranging from substance abuse to environmental exposure and gender identity. However, the 5 most frequent SDOH categories—Drug, Alcohol, Tobacco, Employment, and Living Status—accounted for 97% of all SDOH events and were the focus of the n2c2 shared task.
Each annotated SDOH event included a trigger, that is, a span of text that anchors the event and identifies its event type (eg, employment), and one or more arguments characterizing the event’s duration, history, or other attributes (see Figure 1). Table 1 contains the event types and arguments that were used in the shared task. Arguments are divided into labeled arguments and span-only arguments. Labeled arguments like Status included an annotated span and a subtype category label, for example, “unemployed.” Span-only arguments like Duration or History included only an annotated span because normalizing these to a fixed, small set of labels is not practical. Figure 1 shows an example of 5 annotated events in a single note in SHAC using the BRAT rapid annotation tool.
Table 1.
Social determinants of health types and arguments
| Event type | Argument type | Argument subtype (label) | Span examples |
|---|---|---|---|
| Substance use (alcohol, drug, and tobacco) | Status* | {none, current, past} | “denies,” “smokes” |
| Duration | “for the past 8 years” | ||
| History | “7 years ago” | ||
| Type | “beer,” “cocaine” | ||
| Amount | “2 packs,” “3 drinks” | ||
| Frequency | “daily,” “monthly” | ||
| Employment | Status* | {employed, unemployed, retired, on disability, student, homemaker} | “works,” “unemployed” |
| Duration | “for five years” | ||
| History | “15 years ago” | ||
| Type | “nurse,” “office work” | ||
| Living status | Status* | {current, past, future} | “lives,” “lived” |
| Type* | {alone, with family, with others, homeless} | “with husband,” “alone” | |
| Duration | “for the past 6 months” | ||
| History | “until a month ago” |
Note: Labeled arguments, marked with a *, are mandatory. (Table adapted from Ref. 15.)
Notes were divided into training, development, and test sets for each of MIMIC and UW. The training samples were selected either randomly (29% of training notes) or actively selected15 (71%). All development and test data were randomly sampled.
State-of-the-art SDOH extraction approach and research gap
Lybarger et al proposed a deep-learning event extractor described in Reference 15. In their model, individual sentences are encoded with a pretrained, frozen Bio+Discharge summary BERT model.17,18 BERT encodings are then fed into a bi-LSTM, and the forward and backward output states of this bi-LSTM are concatenated, resulting in a matrix V, which feeds into separate trigger detection, labeled argument detection, and span-only argument detection layers. Using V as input, trigger detection is treated as a sentence-level, multilabel classification task with an attention mechanism to identify trigger spans. The output of this trigger detection layer is a matrix Pt of trigger probabilities. Using V and Pt as input, labeled argument detection is similarly treated as a sentence-level classification task, with attention used to identify argument spans. The output of this labeled argument detection layer is a matrix Ps of labeled argument probabilities. V and Ps are then used as inputs to a linear-chain CRF to predict span-only arguments. Span-only arguments are encoded with begin-inside-outside (BIO) encoding (eg, B-Duration, I-Duration, B-History, I-History, O), and a separate CRF is used for every event type. As Lybarger et al acknowledge, their model has 3 key limitations which we described in the “Introduction” section and which this study aims to address.
Proposed SDOH event extraction architecture
This section summarizes our proposed architecture which addresses the 3 key limitations of the current-state-of-the-art approach. Our MLTB architecture (1) incorporates context from the entire note when making predictions, (2) can predict multiple events of the same type per sentence, and (3) can predict span-only arguments with overlapping spans.
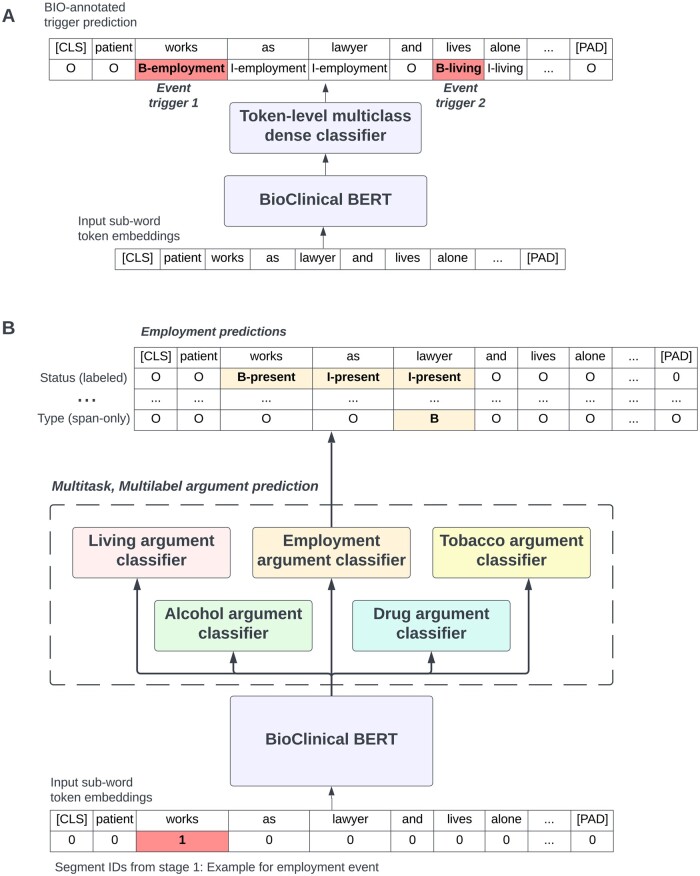
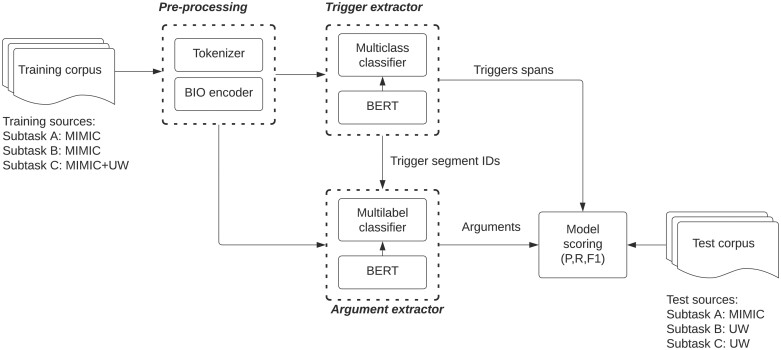
Figure 2 contains an overview of our pipelined 2-stage modeling approach. The first stage model extracts triggers (Figure 2A), and the second stage model extracts arguments for all triggers extracted in stage 1 (Figure 2B). Both stages involve fine-tuning a pretrained BioClinical BERT model trained on PubMed and MIMIC,17 and both are treated as token classification problems with BIO encoding of triggers and arguments. All models were implemented using Huggingface’s transformers library (v4.19.2) with a pytorch backend (v1.11.0).19,20Figure 3 describes our approach to model training and model inference, which we now discuss.
Figure 2.
Deep learning architecture for extraction of social determinants of health (SDOH). (A) Event trigger extraction via multiclass, token-level annotation of spans of text that anchor SDOH events. (B) Argument extraction via multitask, multilabel prediction of labeled and span-only arguments.
Figure 3.
Complete training and evaluation pipeline.
Trigger extraction
Text preprocessing
To train the trigger extraction model, we first converted the SHAC trigger annotations from BRAT format, which provides character-based offsets (positions) of all trigger spans, into BIO format using spaCy.21 The BIO format is used to tag tokens and indicates whether they are the beginning (B) of a chunk or span, or if they are inside (I) or outside (O) said span. Table 2A gives an example of the trigger spans of a note excerpt encoded in BIO format. Because there are 5 SDOH event types, we assigned “B” and “I” tags for every event type (eg, B-Alcohol, I-Alcohol, B-Tobacco, etc.) as well as a single “O” tag. As this encoding is token-based, corpus idiosyncrasies may affect the correct identification of spans. Therefore, we added custom regular expressions to spaCy’s tokenizer to improve the tokenization of training notes (code available upon request). For a few notes, we added the special token label “-” to our customized tokenizer to align the character offsets given by annotations. Retokenization with BioClinical BERT’s tokenizer was required to convert spaCy tokens into wordpieces (eg, the word “worked” was divided into 2 subwords: “work” and “ed”), which have pretrained embeddings in BioClinical BERT. Finally, we added a special character [NLSP] to replace nonword tokens ignored by the BioClinical BERT tokenizer. These tokens included new lines, white spaces, tabs, and bullet points, which may contain potentially useful information (eg, events may be somewhat unlikely to span 2 or more lines separated by new-line characters). The [NLSP] token was added to the tokenizer and initialized as a random embedding within the BERT model and was later tuned during training. The BERT tokenizer truncated notes to a maximum length of 512 wordpieces, padded shorter notes to this maximum, and pre-pended all sequences with the special token [CLS]. BIO labels were applied to the first wordpiece in each token. All other wordpieces, including [CLS] and [PAD] tokens, were given a special label of −100 indicating that they were ignored in the loss computation during backpropagation. The [PAD] tokens were added at the end of notes shorter than 512 wordpieces so that all documents had the same dimension. Similarly, [CLS] stands for classification, and is a special token introduced in the BERT architecture to allow the model to encode the meaning of an entire document in the hidden states of a single token. This is useful for document classification tasks, but since our task is token rather than document classification, this token is only necessary for compatibility with the pretrained BERT model.
Table 2.
Encoding of social determinants of health (SDOH) notes during training
| (A) BIO encoding of all trigger spans in a note | |
|---|---|
| Tokens | Triggers |
| He | O |
| is | O |
| a | B-Employment |
| bartender | I-Employment |
| and | O |
| lives | B-LivingStatus |
| alone | O |
| (B) BIO encoding of the trigger span and arguments for just the employment event in A | |||||
|---|---|---|---|---|---|
| Tokens | Trigger | StatusEmploy | Duration | History | Type |
| He | O | O | O | O | O |
| is | O | O | O | O | O |
| a | B-Employment | B-StatusEmployEmployed | O | O | B |
| bartender | I-Employment | I-StatusEmployEmployed | O | O | I |
| and | O | O | O | O | O |
| lives | O | O | O | O | O |
| alone | O | O | O | O | O |
| (C) Multi-label encoding of Status and Type arguments for the employment event in A and B — space prohibits display of other arguments | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Tokens | TokenTypeID | B-StatusEmploy=Employed | I-StatusEmploy=Employed | B-StatusEmploy=Retired | I-StatusEmploy=Retired | O-StatusEmploy | … | B-Type | I-Type | O-Type |
| He | 0 | 0 | 0 | 0 | 0 | 1 | … | 0 | 0 | 1 |
| is | 0 | 0 | 0 | 0 | 0 | 1 | … | 0 | 0 | 1 |
| a | 1 | 1 | 0 | 0 | 0 | 0 | … | 1 | 0 | 0 |
| bartender | 1 | 0 | 1 | 0 | 0 | 0 | … | 0 | 1 | 0 |
| and | 0 | 0 | 0 | 0 | 0 | 1 | … | 0 | 0 | 1 |
| lives | 0 | 0 | 0 | 0 | 0 | 1 | … | 0 | 0 | 1 |
| alone | 0 | 0 | 0 | 0 | 0 | 1 | … | 0 | 0 | 1 |
Note: The sequence “a bartender” participates in both the StatusEmploy argument, and in the Type argument.
Trigger extraction model
The trigger extraction model was implemented via the BertForTokenClassification class in Huggingface’s transformers library (see Figure 2A). This model’s architecture consisted of a BioClinical BERT transformer followed by a dense classification layer. The inputs to the classification layer were the final hidden state representation given by BERT for all tokens in the input documents. All layers, including the pretrained BERT transformer were fine-tuned during training. Final training hyperparameters included training batch size (16), evaluation batch size (16), number of epochs to train (12), learning rate (3.6e−5), and weight decay (0.2).
Argument extraction
Text preprocessing
For each SDOH event in the SHAC corpus, we converted its trigger span and arguments from BRAT format into a multilayered BIO format as illustrated in Table 2B. The first layer of this representation is the BIO encoding for the trigger span of the current event. All other layers correspond to individual span-only (eg, Amount) or labeled arguments (eg, StatusTime). Span-only arguments like Amount were encoded as B-Amount, I-Amount, or O. Labeled arguments were concatenated with their values. For example, StatusTime for Drug events contained BIO labels in the set {B-StatusTime=none, I-StatusTime=none, B-StatusTime=current, I-StatusTime=current, B-StatusTime=past, I-StatusTime=past, O}. Argument layers were then one-hot encoded and concatenated into a single matrix representing all arguments, as indicated in Table 2C. In this matrix, each token has multiple labels—one for each argument layer—reflecting the multilabel token classification formulation we have pursued.
We reused the modified spaCy tokenizer in the trigger extraction for this task. On a few notes, our tokenizer was unable to align its output tokens with the character offsets given by annotations. These cases were omitted in training for the argument extraction model. Our special token [NLSP] was again utilized for new line tokens and other tokens dropped by the BERT tokenizer. One-hot encoding values of 0 or 1 were applied to only the first wordpiece in every token. All other wordpieces, including [CLS] and [PAD] tokens, were given a value of −100 for every column, and were ignored in the loss computation during backpropagation.
The token type ID’s for all wordpieces in a trigger span were set to 1, and all other token type ID’s were set to 0 to provide the argument extractor with awareness of the location of trigger spans for each event. In brief, BERT input embeddings are the sum of 3 embeddings: token embeddings unique to a wordpiece, positional embeddings unique to an integer index in the sequence of tokens in a document, and token type embeddings unique to a token type ID. In our use case, we used token type ID’s to demarcate the span of text corresponding to the trigger span for a particular event (see Ref. 22 for a similar approach).
Argument extraction model
The argument extraction model was trained in a multitask framework (Figure 2B). A single BERT encoder was shared and fine-tuned for all event types, and 5 separate event-specific, linear classification layers were added on top of this shared BERT. The training set containing samples of different events was shuffled so that training batches would contain multiple event types. The logits of the classification layer were softmaxed within an argument layer (ie, softmax was applied to the logits for B-Duration, I-Duration, and O-Duration), and then converted back to logits. This normalization within an argument layer emphasized that a single label (B-Duration or I-duration or O-Duration) should be predicted for each argument layer. Because this token classification task is multilabel (eg, a token could be labeled B-Type and B-StatusEmployUnemployed), we used PyTorch’s BCEWithLogitsLoss as our loss function, which combines a sigmoid loss with binary cross entropy. Because we utilized multitask learning, the loss for the set of samples was reduced to a single quantity with sum rather than mean reduction (default behavior), so that event types with more argument types contributed more to the loss (see Table 1).
Model inference
At inference time, notes were tokenized with our customized spaCy tokenizer, and then propagated through our trigger extraction model. Predicted triggers (with corresponding token type ID’s) were then passed through a fine-tuned argument extractor. Finally, extracted triggers and arguments were converted to BRAT format to apply the scoring criteria described in Reference 23. Practical details of the processing of model predictions are available in Supplementary File S1.
Overcoming the limitations of the state-of-the-art event-extraction model
Having described our architecture, we now briefly explain how it overcomes the 3 limitations of Lybarger et al’s model.15 First, while their model processes text at the level of sentences, our model takes entire notes as input (in SHAC a very small number of notes are too long for BERT; see discussion on alternative transformers that could avoid this problem). This allowed our model’s event predictions to depend not just on the current sentence, but also on neighboring sentences. Our model’s event predictions can then span arbitrary distances in text. Second, Lybarger et al’s model performs trigger detection via sentence classification and can therefore only predict a single event of a given type per sentence. In our model, trigger detection is treated as a token classification task, and our model can therefore predict as many events of a given type as there are tokens in a sentence. Finally, Lybarger et al’s model has a single CRF to predict span-only arguments for each event type, allowing each token to have only one label and participate in one span-only argument. Our model, however, has a separate output layer for each argument, allowing tokens to be labeled in multiple spans (eg, Amount and Type spans).
Model evaluation and n2c2 subtasks
The 2022 n2c2 Track 2 shared task was divided into 3 subtasks, which varied primarily in the sites of the training and test sets. In Subtask A (Extraction), participants were given MIMIC training and development sets (, ), and performance was evaluated on a MIMIC test set (). In Subtask B (Generalizability), participants were provided the same MIMIC training and development sets as Subtask A, and performance was evaluated on the UW training and development sets in SHAC (, ). Finally, in Subtask C (Learning Transfer), participants were provided text and labels for all MIMIC and UW training and development sets, and performance was evaluated on a new UW test set (). Prediction performance was evaluated via precision, recall, and F1 scores for the extraction of events (triggers, arguments, and argument roles) relative to the gold standard annotations, using a scoring script made available to task participants.23
RESULTS
Table 3 describes our model performance across all subtasks. The bottom rows in Table 3 contain the microaverage performance for all model predictions (triggers and arguments) as well as the best values reported after the n2c2 competition. The test sets used to score our model’s predictions were released after the model had been trained. Thus, we have no leakage from any subtask’s test set. Across all 3 subtasks, we achieved F1 scores from 0.77 to 0.88. As expected, we obtained better performance when the notes’ sites overlapped across training and test (subtasks A and C, F1 = 0.88) and worse performance when they did not (subtask B, F1 = 0.77).
Table 3.
Social determinants of health extraction performance
| Subtask A | Subtask B | Subtask C | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Training: MIMIC | Training: MIMIC | Training: MIMIC and UW | ||||||||||||
| Test: MIMIC |
Test: UW |
Test: UW |
||||||||||||
| Field | Event type | Argument | No. | P | R | F1 | No. | P | R | F1 | No. | P | R | F1 |
| Trigger | Alcohol | – | 312 | 0.96 | 0.95 | 0.95 | 209 | 0.97 | 0.96 | 0.96 | 404 | 0.99 | 0.98 | 0.98 |
| Drug | – | 194 | 0.97 | 0.94 | 0.96 | 269 | 0.95 | 0.87 | 0.91 | 491 | 0.97 | 0.93 | 0.95 | |
| Tobacco | – | 324 | 0.97 | 0.96 | 0.96 | 223 | 0.97 | 0.92 | 0.94 | 434 | 0.97 | 0.97 | 0.97 | |
| Employment | – | 193 | 0.92 | 0.80 | 0.85 | 100 | 0.97 | 0.84 | 0.90 | 157 | 0.92 | 0.89 | 0.90 | |
| LivingStatus | – | 250 | 0.97 | 0.94 | 0.96 | 201 | 0.86 | 0.78 | 0.81 | 357 | 0.91 | 0.90 | 0.91 | |
| Labeled argument | Alcohol | Status | 306 | 0.88 | 0.89 | 0.89 | 204 | 0.91 | 0.92 | 0.92 | 400 | 0.95 | 0.96 | 0.95 |
| Drug | Status | 191 | 0.94 | 0.93 | 0.93 | 258 | 0.87 | 0.83 | 0.85 | 486 | 0.88 | 0.85 | 0.87 | |
| Tobacco | Status | 321 | 0.91 | 0.91 | 0.91 | 210 | 0.93 | 0.94 | 0.94 | 430 | 0.93 | 0.93 | 0.93 | |
| Employment | Status | 184 | 0.86 | 0.79 | 0.82 | 97 | 0.91 | 0.81 | 0.86 | 151 | 0.86 | 0.87 | 0.87 | |
| LivingStatus | Status | 245 | 0.96 | 0.95 | 0.95 | 194 | 0.81 | 0.76 | 0.78 | 352 | 0.90 | 0.90 | 0.90 | |
| LivingStatus | Type | 250 | 0.93 | 0.90 | 0.91 | 194 | 0.73 | 0.69 | 0.71 | 356 | 0.84 | 0.84 | 0.84 | |
| Span-only argument | Alcohol | Amount, duration, frequency, history, type | 161 | 0.69 | 0.69 | 0.69 | 93 | 0.79 | 0.75 | 0.77 | 188 | 0.74 | 0.70 | 0.72 |
| Drug | Amount, duration, frequency, history, type | 136 | 0.82 | 0.79 | 0.80 | 156 | 0.64 | 0.60 | 0.62 | 339 | 0.75 | 0.71 | 0.73 | |
| Tobacco | Amount, duration, frequency, history, type | 292 | 0.78 | 0.80 | 0.79 | 188 | 0.82 | 0.78 | 0.80 | 386 | 0.78 | 0.74 | 0.76 | |
| Employment | Duration, history, type | 163 | 0.69 | 0.60 | 0.64 | 60 | 0.64 | 0.53 | 0.58 | 99 | 0.76 | 0.74 | 0.75 | |
| LivingStatus | Duration, history | 7 | 0.17 | 0.14 | 0.15 | 6 | 0.00 | 0.00 | 0.00 | 10 | 0.36 | 0.40 | 0.38 | |
| Combined micro averagea | – | – | 3529 | 0.87 | 0.89 | 0.88 | 2662 | 0.76 | 0.77 | 0.77 | 5040 | 0.87 | 0.89 | 0.88 |
| Best scoring submissionb | – | – | 3529 | 0.91 | 0.91 | 0.90 | 2662 | 0.81 | 0.77 | 0.77 | 5040 | 0.92 | 0.89 | 0.89 |
Performance values reported by the n2c2 shared task organizers for our model.
Performance of the best scoring teams reported by the n2c2 shared task organizers.
P: precision; R: recall; UW: University of Washington (UW) and Harborview Medical Centers.
Table 3 also provides detailed performance metrics broken down by event and argument type. We observed considerable variation in performance across event types, with better performance in substance use events (drug, alcohol, tobacco) compared to living status or employment. For argument extraction, this might be attributed to our multitask framework, where the transfer learning is more beneficial between the events that are more alike (eg, drug vs alcohol) than those that are unrelated (eg, drug vs employment). As for variation in argument types, the greatest challenge is span-only arguments of LivingStatus, where we never achieved F1 scores above 0.38. This is likely due to the small number of gold standard events with annotations for these arguments (eg, n = 7 in subtask A). It is also possible that differences in lexical diversity among the different SDOH event types accounted for differences in model performance; Feller et al10 found that unstable housing was more challenging for their model, and was described in notes in a greater variety of ways (compared to drug use and sexual orientation, which were easier for their model to classify).
The performance metrics in Table 3 mirror those in Table 3 of Lybarger et al,15 which shows the performance of their event extraction model on the MIMIC and UW test sets when trained on the entire SHAC corpus. Thus, we can reasonably compare their performance on UW to our performance in subtask C, which trains and tests on the same data. As can be seen, we generally matched or outperformed their model in 12 out of 15 extraction categories.
Overcoming limitations of the state-of-the-art event-extraction model
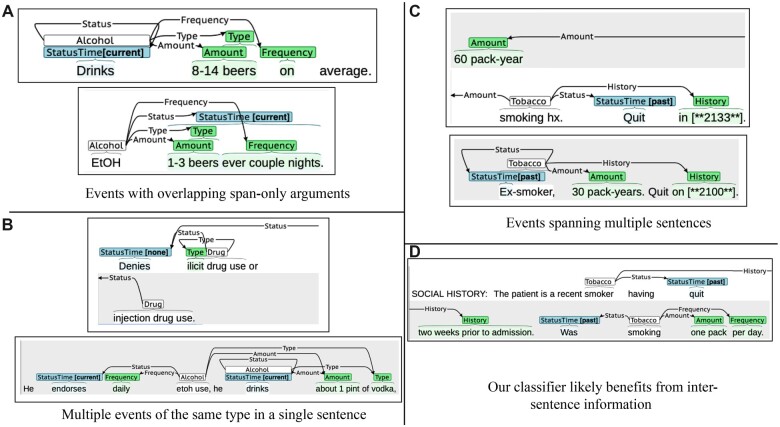
We identified notes and events susceptible to the limitations in the state-of-the-art model (described in the “Overcoming limitations of the state-of-the-art event-extraction model” section), and found that our model indeed overcomes these limitations. Figure 4 shows 2 examples of notes and events corresponding to each of these 3 cases. As can be seen, our approach can predict overlapping span-only arguments (Figure 4A), multiple events of the same type in the same sentence (Figure 4B), and events spanning multiple sentences (Figure 4C). Figure 4D shows that our approach succeeds on a note on which Lybarger et al’s model failed, wherein past tobacco use is described in 2 consecutive sentences. Lybarger et al’s model predicted past and current tobacco use in the first and second sentences, respectively. Lybarger et al suggest that this is because the first sentence includes a strong cue to past status (quit), which is less clear in the second sentence without this previous context. While their model makes predictions at the sentence level and thus cannot incorporate context from neighboring sentences, our model processes the entire note at once and incorporates relevant context. We note, however, that we cannot fully ascertain that this lack of context is the reason why Lybarger et al’s model failed in this particular instance.
Figure 4.
Overcoming limitations in state-of-the-art model. (A) Extraction of overlapping span-only arguments. (B) Extraction of multiple events of the same type in the same sentence. (C) Extraction of events spanning multiple sentences. (D) Leveraging context from preceding sentences.
DISCUSSION
SDOH have far-reaching effects on patient health, yet they are traditionally difficult to track and clinically intervene upon. This difficulty stems from the NLP challenges associated with (SDOH) information extraction from unstructured clinical notes. To address this, we developed a deep learning-based NLP system, MLTB, to extract SDOH events (ie, mentions and attributes of 5 SDOH categories). This system addressed 3 key limitations in the previous state-of-the-art model by Lybarger et al,15 and achieved excellent performance on MIMIC and UW notes in SHAC. Notably, our model achieved 0.88 F1 when trained and evaluated on data from the same site. Additionally, we found moderate generalizability of our model when tested on an external site with 0.77 F. Moreover, we found that transfer learning addressed this disparity and achieved 0.88 F1.
We believe the performance of our model is notable given its simplicity. We leveraged a pretrained, BERT-based transformer and added 2 stages of token classification systems feeding into one another. This is, in our view, much simpler than the event extractor model of Lybarger et al15—to which we performed comparably—as well as other state-of-the-art event extraction systems, which employ additional components besides BERT, including bi-LSTM’s, convolutional neural networks, CRFs, self-attention mechanisms, span representations, and more.24 We found that rather than pursuing higher complexity in the transformer and classification layers, we achieved improved performance by reframing the classification problem within a multitask, multilabel framework as shown in Figure 2. Moreover, in pipeline-based neural event extraction systems such as ours, classification errors from the trigger identification stage cascade into the argument identification stage.24 Given the simplicity of our model, and this known shortcoming of pipeline-based approaches, it may be surprising that our model performed so competitively. On the other hand, its simplicity may be a strength: there are fewer “moving parts” of our model that can fail, or overfit to our training data.25
Accurate extraction of SDOH may aid healthcare stakeholders address health inequities and outcomes via improved screening and healthcare interventions. Improved screening may have upstream implications in policymaking. A salient example is the State Innovation Models program, through which the Centers for Medicare and Medicaid Services awarded over US$622 million to fund healthcare transformation programs across 11 states with an emphasis in population health that recognizes the key role of SDOH.25 Furthermore, SDOH screening may be used during clinical encounters.26 Examples of interventions include the training of clinical staff to identify food and housing insecurity in primary care and wellness visits.27,28 These interventions showed that improvements in screenings during health encounters lead to increased retention in supplemental nutrition assistance program benefits which may help address food insecurity. We envision that an NLP system such as the one presented in this paper may be used to automate SDOH screening, thus reducing the need for training and clinician effort in identifying social and behavioral factors that affect patients’ health.
Future directions
We are considering several future directions. First, to improve our MLTB model, we plan to explore data augmentation techniques, where synthetic notes are generated and added to training and development data. For example, Yang et al22 took sentences annotated for event structure, and replaced the argument text (eg, “beer” for the Type argument of an alcohol event) with text that played the same role in other sentences (eg, “wine”). Additionally, it may be useful to introduce additional regional lexical variability, as previous research showed that some SDOH information in text is specific to a given clinical center or region.29 Second, BERT has been supplanted in many tasks by a more recent transformer named RoBERTa, which alters some details of pretraining and achieves higher performance on many tasks.30 We plan to replace BERT with a clinically pretrained RoBERTa, and retrain and test our model. Second, our own preliminary experiments suggested that adding a Bi-LSTM or CRF on top of BERT did not improve trigger extraction, but we plan to test this more rigorously. Third, and similarly, preliminary experiments showed that hyperparameter tuning did not improve performance beyond using defaults, but we need to show this systematically, as well. Fourth, preliminary experiments did show that our multitask framework for argument extraction outperformed fine-tuning of separate BERT-based argument extractors for every event type (single-task learning), but we plan to show this more systematically. Fifth, because we currently use BERT-based models, we are limited by the maximum sequence length of BERT (512 tokens), which forced us to truncate a few notes before processing them. To address this, notes could be split during preprocessing, but it is not clear how to do this without reintroducing (a version of) the first limitation of Lybarger et al’s15 model, which ignored context from preceding or following sentences when processing a given sentence. An alternative approach is to use a pretrained transformer like Clinical-Longformer,31 which has a longer maximum sequence length (4096 tokens).
CONCLUSIONS
In this study, we developed and evaluated a new, deep learning-based NLP system (MLTB) that extracts SDOH events from clinical notes. This system addresses 3 limitations of the previous state-of-the-art system,15 and performs quantitatively at least as well, if not better than, this system. Similarly, our system performed competitively in the 2022 n2c2 Track 2 shared task for extracting SDOH events. At the same time, the modeling approach we presented here is of lesser complexity than competing approaches, which allows several additional enhancements to be built on top or alongside it. Improvements in extraction of SDOH from clinical notes may aid healthcare stakeholders target clinical interventions and public health monitoring.
Supplementary Material
ACKNOWLEDGMENTS
Thanks to Kevin Lybarger and other organizers of the 2022 n2c2 Track 2 shared task. We also thank members of the Tsui lab for their feedback on this project.
Contributor Information
Russell Richie, Tsui Laboratory, Department of Biomedical and Health Informatics, Children’s Hospital of Philadelphia, Philadelphia, Pennsylvania, USA; MindCORE and Cognitive Science, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
Victor M Ruiz, Tsui Laboratory, Department of Biomedical and Health Informatics, Children’s Hospital of Philadelphia, Philadelphia, Pennsylvania, USA.
Sifei Han, Tsui Laboratory, Department of Biomedical and Health Informatics, Children’s Hospital of Philadelphia, Philadelphia, Pennsylvania, USA.
Lingyun Shi, Tsui Laboratory, Department of Biomedical and Health Informatics, Children’s Hospital of Philadelphia, Philadelphia, Pennsylvania, USA.
Fuchiang (Rich) Tsui, Tsui Laboratory, Department of Biomedical and Health Informatics, Children’s Hospital of Philadelphia, Philadelphia, Pennsylvania, USA; Department of Anesthesiology and Critical Care, University of Pennsylvania Perelman School of Medicine, Philadelphia, Pennsylvania, USA.
FUNDING
The National Institute of Mental Health grant number R21MH130853-01. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIMH.
AUTHOR CONTRIBUTIONS
Study concept and design: RR, VR, LS, SH, and FRT. Analysis and interpretation of data: RR, VR, and FRT. Collection or assembly of data: RR and VR. Drafting of the manuscript: RR, VR, and FRT. Critical revision of the manuscript for important intellectual content and approval of the final manuscript: RR, VR, LS, SH, and FRT. Funding: FRT. Study supervision and coordination: FRT.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
CONFLICT OF INTEREST STATEMENT
None declared.
DATA AVAILABILITY
The SHAC data underlying this article were provided by the University of Washington under license. Data will be shared on request to the corresponding author with permission of the University of Washington.
REFERENCES
- 1. Magnan S. Social determinants of health 101 for health care: five plus five. NAM Perspect 2017; 7 (10): 1–9. doi: 10.31478/201710c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Centers for Disease Control and Prevention (CDC). Annual smoking-attributable mortality, years of potential life lost, and productivity losses – United States, 1997–2001. MMWR Morb Mortal Wkly Rep 2005; 54 (25): 625–8. [PubMed] [Google Scholar]
- 3.Global status report on alcohol and health 2018.https://www.who.int/publications-detail-redirect/9789241565639. Accessed December 2, 2022.
- 4. Degenhardt L, Hall W.. Extent of illicit drug use and dependence, and their contribution to the global burden of disease. Lancet 2012; 379 (9810): 55–70. [DOI] [PubMed] [Google Scholar]
- 5. Navathe AS, Zhong F, Lei VJ, et al. Hospital readmission and social risk factors identified from physician notes. Health Serv Res 2018; 53 (2): 1110–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Goodday SM, Kormilitzin A, Vaci N, et al. Maximizing the use of social and behavioural information from secondary care mental health electronic health records. J Biomed Inform 2020; 107: 103429. [DOI] [PubMed] [Google Scholar]
- 7. Patra BG, Sharma MM, Vekaria V, et al. Extracting social determinants of health from electronic health records using natural language processing: a systematic review. J Am Med Inform Assoc 2021; 28 (12): 2716–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Conway M, Keyhani S, Christensen L, et al. Moonstone: a novel natural language processing system for inferring social risk from clinical narratives. J Biomed Semantics 2019; 10 (1): 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shoenbill K, Song Y, Gress L, et al. Natural language processing of lifestyle modification documentation. Health Informatics J 2020; 26 (1): 388–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Feller DJ, Bear Don’t Walk Iv OJ, Zucker J, et al. Detecting social and behavioral determinants of health with structured and free-text clinical data. Appl Clin Inform 2020; 11 (1): 172–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lituiev DS, Lacar B, Pak S, Abramowitsch P, De Marchis E, Peterson T. Automatic extraction of social determinants of health from medical notes of chronic lower back pain patients. medRxiv, 2022.03.04.22271541. 2022. 10.1101/2022.03.04.22271541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Han S, Zhang RF, Shi L, et al. Classifying social determinants of health from unstructured electronic health records using deep learning-based natural language processing. J Biomed Inform 2022; 127: 103984. [DOI] [PubMed] [Google Scholar]
- 13. Stemerman R, Arguello J, Brice J, et al. Identification of social determinants of health using multi-label classification of electronic health record clinical notes. JAMIA Open 2021; 4 (3): ooaa069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lybarger K, Yetisgen M, Uzuner Ö.. The 2022 n2c2/UW shared task on extracting social determinants of health. J Am Med Inform Assoc 2023; ocad012. doi: 10.1093/jamia/ocad012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lybarger K, Ostendorf M, Yetisgen M.. Annotating social determinants of health using active learning, and characterizing determinants using neural event extraction. J Biomed Inform 2021; 113: 103631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MIMIC-III, a freely accessible critical care database | Scientific Data.https://www.nature.com/articles/sdata201635. Accessed December 2, 2022. [DOI] [PMC free article] [PubMed]
- 17. Alsentzer E, Murphy JR, Boag W, et al. Publicly available clinical BERT embeddings. 2019. doi: 10.48550/arXiv.1904.03323, preprint: not peer reviewed. [DOI]
- 18. Devlin J, Chang M-W, Lee K, et al. BERT: pre-training of deep bidirectional transformers for language understanding. 2019. doi: 10.48550/arXiv.1810.04805, preprint: not peer reviewed. [DOI]
- 19. Wolf T, Debut L, Sanh V, et al. HuggingFace’s transformers: state-of-the-art natural language processing. 2020; doi: 10.48550/arXiv.1910.03771, preprint: not peer reviewed. [DOI]
- 20. Paszke A, Gross S, Massa F, et al. Desmaison A. Pytorch: An imperative style, high-performance deep learning library. Adv Neural Inf Process Syst 2019; 32: 1–12. [Google Scholar]
- 21. Honnibal M, Montani I, Van Landeghem S, et al. spaCy industrial-strength natural language processing in Python. 2020. doi: 10.5281/zenodo.1212303. [DOI]
- 22. Yang S, Feng D, Qiao L, et al. Exploring pre-trained language models for event extraction and generation. In: proceedings of the 57th Annual Meeting of the Association for Computational Linguistics. Florence, Italy: Association for Computational Linguistics; 2019: 5284–94. doi: 10.18653/v1/P19-1522. [DOI]
- 23. Lybarger K. BRAT scoring. 2022. https://github.com/Lybarger/brat_scoring/blob/9ea8004d9998d59dca3e9a08465ab6a66328763d/docs/sdoh_scoring.pdf. Accessed December 2, 2022.
- 24. Li Q, Li J, Sheng J, et al. A survey on deep learning event extraction: approaches and applications. 2022. doi: 10.48550/arXiv.2107.02126. [DOI] [PubMed]
- 25. Coughlin TA, Zuckerman S, Hill I, et al. State Innovation Models (SIM) Round 2: Model Test Annual Report Two. USA: Urban Institute; 2018. https://policycommons.net/artifacts/631075/state-innovation-models-sim-round-2/. Accessed January 29, 2023. [Google Scholar]
- 26. Eder M, Henninger M, Durbin S, et al. Screening and interventions for social risk factors: technical brief to support the US Preventive Services Task Force. JAMA 2021; 326 (14): 1416–28. [DOI] [PubMed] [Google Scholar]
- 27., Lane WG, Dubowitz H, Feigelman S, et al. The effectiveness of food insecurity screening in pediatric primary care. Int J Child Health Nutr 2014; 3 (3): 130–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stenmark SH, Steiner JF, Marpadga S, et al. Lessons learned from implementation of the food insecurity screening and referral program at Kaiser Permanente Colorado. Perm J 2018; 22: 18–093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Feller DJ, Zucker J, Yin MT, et al. Using clinical notes and natural language processing for automated HIV risk assessment. J Acquir Immune Defic Syndr 2018; 77 (2): 160–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu Y, Ott M, Goyal N, et al. RoBERTa: a robustly optimized BERT pretraining approach. 2019; doi: 10.48550/arXiv.1907.11692. [DOI]
- 31. Li Y, Wehbe RM, Ahmad FS, et al. Clinical-Longformer and Clinical-BigBird: transformers for long clinical sequences. 2022; doi: 10.48550/arXiv.2201.11838. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The SHAC data underlying this article were provided by the University of Washington under license. Data will be shared on request to the corresponding author with permission of the University of Washington.