Abstract
Objective
We applied natural language processing and inference methods to extract social determinants of health (SDoH) information from clinical notes of patients with chronic low back pain (cLBP) to enhance future analyses of the associations between SDoH disparities and cLBP outcomes.
Materials and Methods
Clinical notes for patients with cLBP were annotated for 7 SDoH domains, as well as depression, anxiety, and pain scores, resulting in 626 notes with at least one annotated entity for 364 patients. We used a 2-tier taxonomy with these 10 first-level classes (domains) and 52 second-level classes. We developed and validated named entity recognition (NER) systems based on both rule-based and machine learning approaches and validated an entailment model.
Results
Annotators achieved a high interrater agreement (Cohen’s kappa of 95.3% at document level). A rule-based system (cTAKES), RoBERTa NER, and a hybrid model (combining rules and logistic regression) achieved performance of F1 = 47.1%, 84.4%, and 80.3%, respectively, for first-level classes.
Discussion
While the hybrid model had a lower F1 performance, it matched or outperformed RoBERTa NER model in terms of recall and had lower computational requirements. Applying an untuned RoBERTa entailment model, we detected many challenging wordings missed by NER systems. Still, the entailment model may be sensitive to hypothesis wording.
Conclusion
This study developed a corpus of annotated clinical notes covering a broad spectrum of SDoH classes. This corpus provides a basis for training machine learning models and serves as a benchmark for predictive models for NER for SDoH and knowledge extraction from clinical texts.
Keywords: social determinants of health, natural language processing, natural language inference, machine learning, lower back pain, depression
INTRODUCTION
Adverse social determinants of health (SDoH), or social risk factors, such as food insecurity and housing instability, are recognized for their deleterious impacts on health outcomes and disparities.1 There is growing recognition of the role of social risks in chronic low back pain (cLBP), as highlighted in a recent systematic review that found strong associations of cLBP prevalence with educational attainment and socioeconomic status.2 Outcomes for cLBP, a leading cause of disability worldwide,3–5 are known to be worse in patients who are economically and socially disadvantaged.2,6–9 This can be attributed in part to treatment biases, including greater provision of non-evidence-based care,2,10,11 as well as patients’ prior experience of discrimination6,12 and beliefs about pain and pain treatment,2,13 which may influence engagement with the treatment offered. Much of the current research exploring disparities in cLBP care has been limited to stratifying analyses by socioeconomic status and race, as social risk data are not readily available in electronic health records (EHRs). While some social risk information exists in structured data fields, such as patient demographics and problem lists, these data are under-identified by clinical teams, under-reported by patients, and under-documented in structured fields.14–21 When clinical teams screen for and identify social risks, that information is more often documented within free-text fields, or unstructured data.17,20 Novel approaches to identifying SDoH in EHRs, such as natural language processing (NLP) and other machine learning (ML) techniques, leverage existing health information technology to scan unstructured data fields, resulting in automated extraction.22
Several studies have applied NLP methods to obtain SDoH information from clinical notes. Methods have included regular expressions,23 neural networks,24 and rule-based algorithmic approaches.25 Named entity recognition (NER), an NLP task of extracting phrases and their positions from texts, has been most frequently used. SDoH, such as housing situation, finances, and social support,26 have been less studied using NLP methods than behavioral determinants of health (BDoH), such as smoking status and substance and alcohol use.22 For BDoH, both rule-based and ML approaches to NER have been applied.22 SDoH, most notably housing situation, have most commonly been identified through rule-based approaches such as keyword matching.22 Most other SDoH domains, such as transportation access and finances, have been understudied by either technique. There has been no comprehensive comparison of the strengths and weaknesses of ML and rule-based methods when applied to SDoH on various dataset sizes. Also, we are not aware of any studies that apply NLP for SDoH for cLBP patients.
Both ML and rule-based systems are being used to extract clinical and SDoH concepts from healthcare narratives. Tried-and-true rule-based systems such as clinical Text Analysis and Knowledge Extraction System (cTAKES)27 could provide reasonable performance with no training data by leveraging standard ontologies (Unified Medical Language System, UMLS) and rule-based pattern matching.28,29 Modern ML techniques have revolutionized NLP and natural language understanding, with self-supervised methods such as BERT,30 allowing for efficient utilization of large amounts of unlabeled data. For optimal performance in supervised tasks in specialized domains, however, these techniques may require corpora of several thousands of documents of manually annotated text.31 While still providing important utility in domains where labeled data are scarce32 and being more interpretable than ML, rule-based systems often yield to ML methods in performance.33,34 In terms of development costs, both rule-based and ML-based models are resource intensive. Still, improvement of ML systems is normally less demanding in terms of technical development effort as compared to rule-based systems.
While several applications of natural language inference (NLI) methods, such as question answering (QA)35–37 and recognizing textual entailment (RTE),38–40 have been pursued in the wider medical domain, we were surprised to learn that these methods have not been used to study SDoH or BDoH. Given that these models have been trained on general knowledge texts (eg, Wikipedia and BooksCorpus), we would expect NLI models to generalize to SDoH and BDoH better than other clinical domains. This motivated application of NLI models in our study. Out of NLI approaches, we chose to use entailment (RTE) models instead of QA models,39 given our interest in off-line knowledge extraction (ie, without a need to present to a clinician for immediate decision making) with a pre-defined ontology. While extractive QA models (returning a piece of original text) may not be able to summarize text into a limited set of categories by design, abstractive QA models suffer from biases of characteristic to all generative models such as ability to learn toxic and biased attitudes41,42 as well as exhibit learned attitudes characteristic to mental illness and addiction.43
In this study, our aim was to create a dataset to evaluate and compare NLP tools to extract individual SDoH from free-text clinical notes for patients with cLBP. In this study, we make several contributions to this important topic: (1) we label and evaluate a dataset with social and BDoH in cLBP patients, (2) we tune and evaluate a rule-based NER system (cTAKES), (3) we train and evaluate 2 ML NER pipelines, and (4) we evaluate a previously trained common domain NLI entailment model. We believe this study is a step forward in both applying cutting-edge NLP technology to this important topic and in improving the structured ontology for identifying SDoH and BDoH.
MATERIALS AND METHODS
Study population
Our study population consisted of cLBP patients, defined by low back pain lasting at least 3 months,41 from an urban academic medical center at the University of California, San Francisco (UCSF) and was approved by institutional review (IRB #19-29016). All patients received care at the UCSF Integrated Spine Service.42 We extracted patients’ progress notes, history and physical (H&P) notes, emergency department (ED) provider notes, patient instructions, and telephone encounters (TE) from the UCSF clinical data warehouse between March 2017 and April 2020. The cohort demographics are shown in Supplementary Table S1.
Annotation ontology
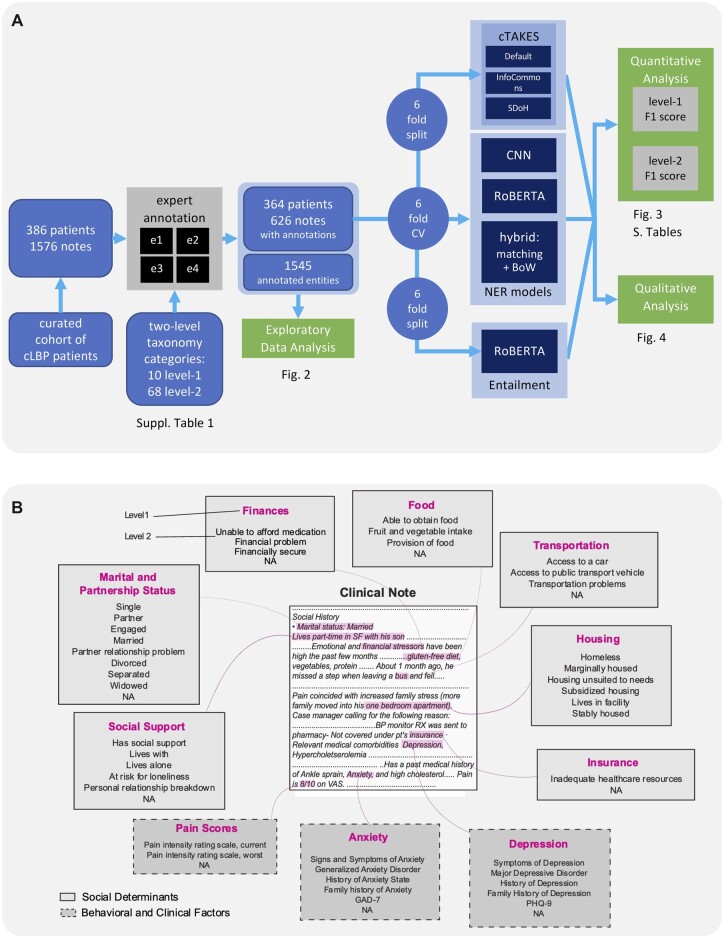
To create a model for extracting named entities related to SDoH from clinical free-text notes, we defined an ontology, annotated the dataset, and tuned and trained algorithms for NER (Figure 1). An ontology covering 8 SDoH domains (Figure 1) commonly screened in clinical practice43–47 was defined following an earlier study.48 During the study period, none of these SDoH were systematically screened for in our healthcare system. Additionally, we included mental health factors (anxiety and depression) as well as pain scores, as these are relevant for our ongoing work in characterizing cLBP population.49–54 Initial taxonomic ontology containing 68 second-level classes within 10 first-level classes was pruned based on annotation availability (next section), down to 52 second-level classes (Figure 1B and Supplementary Table S2). Entities within the first-level classes not captured by specific second-level labels due to ambiguity or low occurrence were assigned a second-level label of “NA” for “not applicable”.
Figure 1.
Study design. (A) Workflow of the study. (B). Annotation ontology. Clinical notes were annotated such that text relevant to the 7 studied social risk factors (solid border) or 3 clinical factors (dashed border) were marked. Two levels of labels were used, such that the second level was a subcategory of the first. Level 2 labels for each Level 1 annotation are shown in descending order of frequency. Level 2 annotations that comprised <1% of the group’s annotations are not shown. Text that can be classified to the first level but not the second due to ambiguity or low frequency is designated as “NA”. Examples of selected text are shown within the hypothetical clinical note.
Manual annotations
The notes were annotated according to our ontology by 4 trained annotators (physical therapy graduate students trained by domain experts supervised by a family medicine physician) using MAE labeling software55 for NER task by highlighting and assigning a class to the relevant phrases (Figure 1B). Each note was annotated by at least 2 annotators (Supplementary Figure S1A). The annotations were additionally spot-checked by the supervising physician and corrected. Initial annotations occasionally contained overlapping or duplicate labels that were resolved using the Python spacy package as described in Supplementary Methods. After all pre-processing steps, document-level inter-rater agreement was evaluated using span-level F1 and document-level Cohen’s kappa56 and Krippendorf’s alpha (with Jaccard index metric),57,58 and metrics were aggregated (see Supplementary Methods). We chose F1 as a span-level metric as it is both symmetric and less sensitive to true negatives, which dominate in NER tasks. In downstream applications (model training and evaluation), annotations from each expert were included. The data were split into 6 folds, while stratifying by the number of entities and number of annotators per note and keeping multiple annotations of the same note in the same fold.
The variance of annotation frequencies was analyzed using ANOVA in R software. Pairwise difference between model F1 scores was assessed using Wilcoxon test across folds.
cTAKES configuration
We used 3 configurations of cTAKES,27 a modular framework that leverages UMLS vocabulary.59 We used 3 configurations (see Supplementary Methods): (1) a default out-of-the box configuration, (2) an InfoCommons configuration60 developed for general purpose medical texts, and (3) a customized “SDoH” configuration that was tuned to identify our domains of interest. Tuning was done using an unlabeled set of notes and the labeling ontology definition.
NER model configuration
ML for the NER task was performed with spaCy software61 using a transition-based parser based on either convolutional neural network (CNN)62 or RoBERTa63 model. The “en_core_web_md” word embeddings were used for initialization. The model was trained with a variable batch size of 100–1000, 10% drop-out, learning rate of 0.0002, and ≤30 epochs with early stopping. Model was trained and evaluated in nested 6-fold cross-validation, with 4:1:1 for training:validation:test splits.
Hybrid model leveraging pattern matching and classification with Bag-of-Words
A classification model was applied to text extracted based on keyword matching patterns compiled for each first-level class based on subject matter identification and literature. The patterns are included in the code repository.64 Text was extracted around the matching pattern (±4 tokens), pre-processed by lemmatizing and removing stop words, then embedded using a term frequency-inverse document frequency vectorizer. Extracted text for each of the first-level set of patterns was then classified using a Bag-of-Words (BoW)65 multinomial logistic regression model into respective second-level subclasses according to manual labels, with unmatched text spans receiving a special exclusion label. Model was trained and evaluated in 6-fold cross-validation as described for the CNN model above.
Evaluation of NER models
The performance of the NER systems was evaluated using precision, recall, and F1 scores using scikit-learn. The metrics were calculated on first and second level of the labeling taxonomy. For cTAKES model, predicted UMLS terms overlapping ground truth labels were manually annotated as being matches (including synonyms and child terms), or mismatches of respective first-level and second-level ground truth categories, while accounting for negation, history, and family history modifiers. Labels with at least partial span overlap were scored as matches. Pairwise comparison between model performance metrics was performed using paired t-test across all cross-validation folds (for CNN and hybrid models) or respective data split folds (for cTAKES). Variation of model performance was analyzed using ANOVA and nested linear model ANOVA.
Evaluation of an entailment model
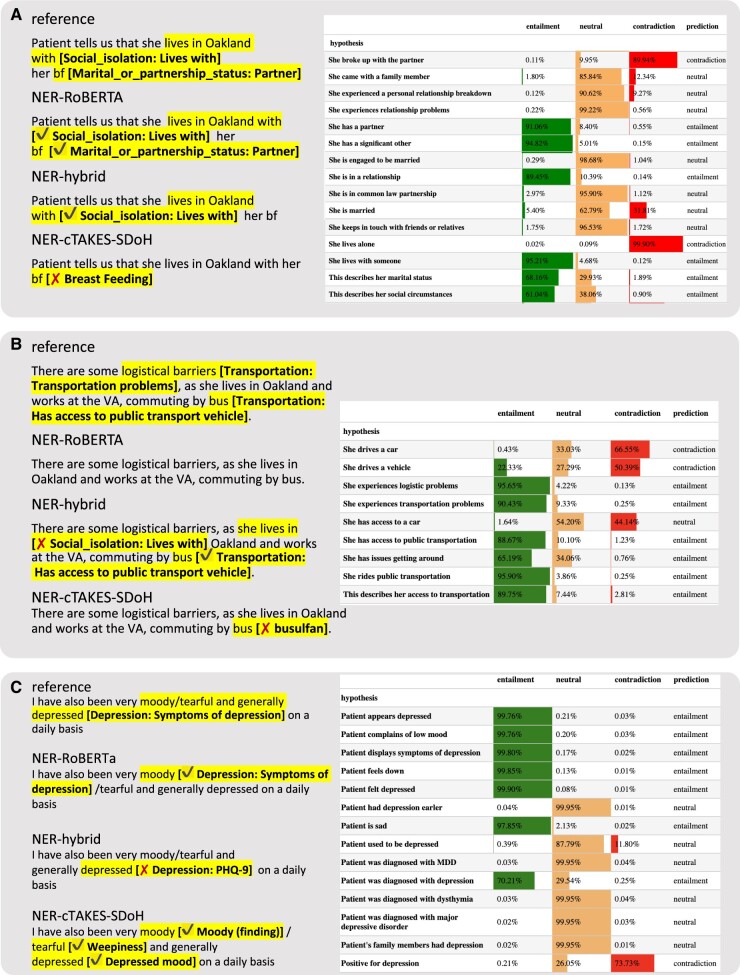
A RoBERTA-based63 entailment38 model, previously fine-tuned in an adversarial human-in-the loop setting,66 was evaluated without additional training on our data. NER labels, transformed into one or more hypotheses, and labeled sentences from the clinical notes taken as premises, served as model inputs to obtain entailment scores (see example in Figure 4). Pain scores, as well as PHQ-9 and GAD-7 scores were excluded from this analysis as they are not easily amenable for textual entailment task. Additional background and details are provided in Supplementary Methods. For computational expediency, only sentences with at least one NER label were examined. The results were aggregated per premise sentence and label combination either (1) by taking an average rate of entailment prediction over alternative hypotheses (“mean”) or (2) by scoring a match if at least one of the hypotheses was deemed to be entailed (“max”). Data were split into six folds matching the folds of NER analysis. Wherever positive prediction was missing and thus precision was undefined, F1 was assigned the value of recall.
Figure 4.
Examples of predictions from 4 best models per model class. Left: NER models. Right: RoBERTA entailment model. Probabilities of 3 possible relations are shown as shaded horizontal bars and numerically together with a final relation prediction.
RESULTS
Gold standard dataset annotation
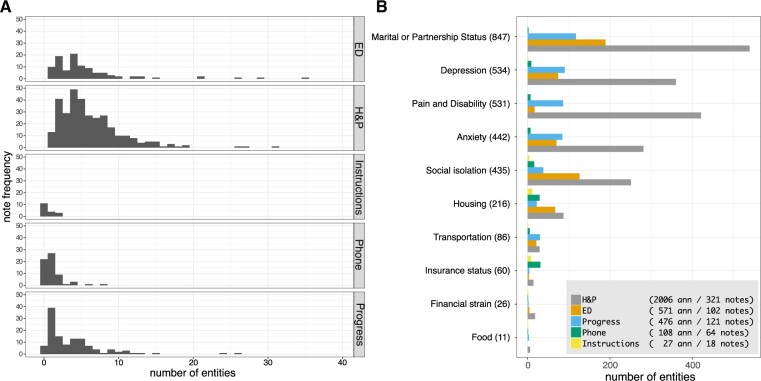
Four trained annotators annotated a set of 1576 clinical notes with an average inter-rater agreement of 95.3%, as measured by Cohen’s kappa and 83.4% as measured by Krippendorff’s alpha (pairwise agreement is shown in Supplementary Table S3) and weighted F1 of 91.2% and macro average F1 of 88.9% (Supplementary Table S4). The inter-rater agreement per each label is shown in Supplementary Table S5. Of the 1576 notes, 39.7% (626) contained at least one entity (Supplementary Figure S1A). Notes without annotations were discarded from further analysis. H&P notes contained on average the most annotations (6.2), followed by ED (5.6), progress notes (4.4), TE (1.8), and patient instructions (1.5). The number of annotations varied significantly with note type, but not with the annotator on aggregate (P = 5e−14 and P = 0.3 in ANOVA). The number of labels differed between note types (Figure 2). For example, Marital and partnership status and Pain Scores were frequently found in H&P and progress notes, but rare in ED provider notes; Insurance coverage was mainly present in TE and patient instructions notes. A detailed breakdown of variance P-value per note type and annotator is presented in Supplementary Figure S1B. All categories except Pain Scores, Depression, Finances, and Food were significantly associated with the note type (P < 0.05 in ANOVA). When annotations were present in a note, the association between the length of note and the number of annotations was significant (P < 0.001, linear regression R2 = 0.52, Supplementary Figure S1C).
Figure 2.
Exploratory data analysis. (A) Histogram of number of entities in different note types. (B) Number of entities per note type and first-level annotated domain. The pictorial legend contains the total number of notes and annotations per note type.
Tuning cTAKES tool for rule-based NER
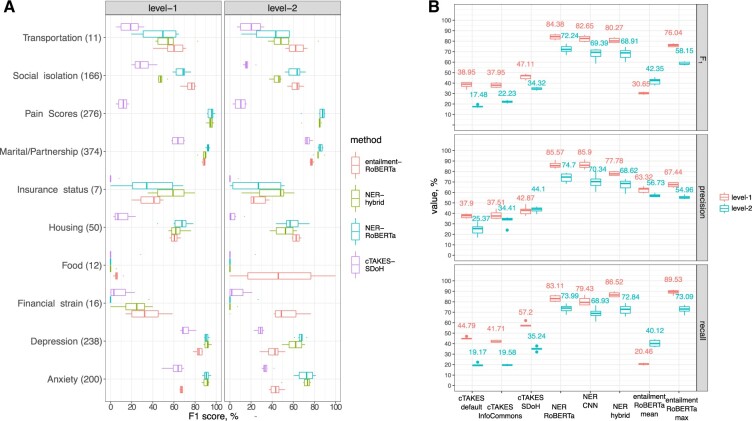
We tuned and assessed cTAKES for the task of NER of targeted SDoH domains, pain scores, and anxiety and depression. As the out-of-the-box version of cTAKES (“default”) has shown limitations,60 we used a version configured for high throughput NER at our institution (“InfoCommons”), and additionally customized cTAKES to identify SDoH for this study (“SDoH”). Tuning improved the performance (Figure 3B, Supplementary Tables S6 and S7), with weighted F1 achieving 38.9%, 37.9%, and 47.1% on first level for the default, InfoCommons, and SDoH versions, respectively. Performance at the second level of taxonomy was poor in all configurations (17.5%, 22.2%, and 34.3%, respectively). False negatives (resulting in suboptimal first-level cTAKES SDoH recall of 57.2%) were commonly due to free-text wording lacking common keywords that cTAKES is configured to match. Often, history or negation modifiers were wrongly attributed due to challenges with sentence segmentation. SDoH words and abbreviations are occasionally misinterpreted as abbreviations from other semantic domains (eg, “bus” and “bf” in Figure 4). Additional qualitative analysis is provided in Supplementary Results. In model comparisons below, we focus on the best performing SDoH configuration.
Figure 3.
Comparison of model performance. (A) Comparison of F1 performance in 4 best performing models per model class. Second-level metrics are aggregated using weighted average over first-level domains. (B) Comparison of F1, precision, and recall in all studied models. Metrics are aggregated using weighted average.
ML-based NER prediction with CNN and RoBERTa
Next, we trained and evaluated performance of a CNN NER model implemented using spaCy software. CNN NER and RoBERTa NER models achieved an average weighted F1 of 82.7% and 84.4%, respectively, on first-level and 69.4% and 72.2% on second-level (see Figure 3, Supplementary Tables S6 and S7). On a more granular level, RoBERTa NER significantly outperformed CNN only in Depression (second level, P = 0.043) and approached significance for Social Support (P = 0.08 and P = 0.06 for first and second level, respectively). Similar to cTAKES, some of the false positives in ML results were due to misinterpretation of context (eg, capture of “depressed” referencing an anatomical finding or “home” in “plan to discharge home” and “works as a home CNA” as housing-related concepts). In some cases, negation markers, such as “denies”, and misspellings, such as “derpession” for “depression”, were missed.
Hybrid text extraction and classification system
Next, we applied a hybrid approach, which extracts relevant text segments by pattern matching and classifies with a logistic regression BoW model. The hybrid model displayed a high recall (86.5% at first level), outperforming other methods (Figure 3, Supplementary Tables S6–S8), with an average first-level F1 = 80.3%. Several cases of failure may be attributed to fundamental limitations of the underlying BoW model, such as cases that refer to people other than the patient, eg, “her father lives with a partner” was interpreted as “Lives with” category pertaining to the patient.
Comparison of NER model performance
The ML and hybrid models significantly outperformed the cTAKES SDoH rule-based system in terms of F1 on the first level (P < 1e−5) and the second level (P < 0.03) of the taxonomy in nearly all classes (Figure 3, Supplementary Tables S6 and S8). While the NER-RoBERTa model achieved the highest first-level F1 = 84.38% among all models (P < 0.04), it scored on par (72.24%) with the hybrid model (68.91%) at the second level F1 (P = 0.13). The hybrid model had a consistently higher recall for all first-level categories but was outperformed by the RoBERTa-based NER in terms of first-level F1 in Marital status (P = 0.03) and in Social isolation (P < 0.001). All NER models performed poorly for the classes of Food and second-level domains with low F1 agreement (35%–60%, Supplementary Table S5), performed poorly in all methods (Supplementary Figure S2). A multifactor ANOVA revealed that both the method and the ontology class are significantly associated with the F1 performance (P < 2e−31 Supplementary Table S9). Additionally, per-class performance was correlated across methods (P < 2e−5, Supplementary Table S10). Inter-rater agreement F1 explains 6.76% of between-label variation in F1 model performance (P = 2e−20 in 1-way ANOVA).
Evaluation of an entailment model
We hypothesized that general domain NLI models (trained on books and Wikipedia) may be directly applied to SDoH and BDoH domains due to semantic overlap between SDoH and general domain texts. Thus, we evaluated performance of an entailment model66 without fine-tuning, whereby a subset of note-derived premise sentences containing named entities was passed together with a set of hypothesis sentences compiled to reflect semantics of interest. The aggregated metrics are shown in Figure 3 and examples of prediction are shown in Figure 4, while granular metrics are presented in Supplementary Figure S2. Max-aggregation (suggesting at least one of the hypotheses per class must hold) performed better than mean-aggregation (one assuming all hypotheses must hold), F1 = 76.0% versus F1 = 30.7% at the first level, respectively. This together with qualitative analysis suggests that predictions are sensitive to hypothesis wording. The entailment model outperformed the NER models evaluated above in the categories of Transportation, Finances, and Food (Figure 3). Entailment model performed poorly on long lists (eg, past diagnoses) partially due to sentence segmentation issues. Further qualitative details are provided in Supplementary Results.
DISCUSSION
In this study, we developed a corpus of annotated clinical notes covering a spectrum of SDoH domains, together with anxiety, depression, and pain scores for cLBP patients. This corpus provides a basis for training ML models and serves as a benchmark for predictive models for SDoH NER. By evaluating various NER pipelines, we identified strengths and weaknesses of both rule-based (cTAKES) and ML-based approaches for identifying SDoH. For most of our evaluated categories, the ML methods outperformed cTAKES. Some of cTAKES’ performance limitations appeared to be due to a failure to account for non-affirmative mentions (in questionnaires), missing rare keywords, or polysemous abbreviations.
Comparing all models, we noticed that some SDoH domains were consistently harder to detect across methods, which suggests constraints due to both fundamental variability of phrasing in free-text notes, as well as data availability. Additionally, levels of inter-rater variability across SDoH domains explain this pattern to a small degree. In our dataset, the least documented categories were Insurance status, Transportation access, Food security, and Finances. This may be due to low frequency of these risk factors in our population, lack of social risk screening, and/or under-documentation of these risks. Most of these rare categories were detected with low and highly variable performance. On average across categories, the CNN model performed best, yielding up to the hybrid and rule-based models for a few data-poor classes.
Evaluation of an out-of-the-box entailment RoBERTa67 model trained on general domain texts, yielded promising results. Without fine-tuning, this model performed similar to NER ML models trained on our corpus in most categories. Direct comparison of our study performance (F1 = 76%) to the original study67 (accuracy of 50%–93%) is not possible, as our data lack annotation for contradiction relation. However, our analysis suggests that the model generalizes reasonably well to the SDoH domain. Thus, we believe that entailment models may be readily deployed in user-facing tools for clinical text exploration and retrieval, such as EMERSE,67 thus allowing researchers to query data based on custom criteria. However, sensitivity of the model to the hypothesis wording observed in this study needs to be taken into consideration when creating fine-tuning data, directly applying to new data, or querying with different hypotheses. Additional fine-tuning on medical- and SDoH-related corpora may further increase performance and should be further explored.
Our study should be interpreted with consideration of its limitations. First, the generalizability of the models validated here may be affected by the fact that clinical notes came from an urban academic medical center, where fewer patients may experience—or fewer providers may document—social risk factors, versus the Veterans Affairs or safety-net hospitals. Our study may therefore be better generalizable to similar settings. Related, our study focused on notes for patients with chronic LBP specifically. We included, however, a diverse set of note types from different settings that may improve the generalizability to patients with other conditions. Further validation of the presented models across different medical centers and patient cohorts may be conducted. Second, as previously noted, some SDoH domains had particularly low counts of annotations, such as food security and finances. The strength and capacity of our models overall are limited by the quality of notes that were annotated. Third, dictionaries constructed during tuning of rule-based and hybrid models were composed based on the complete dataset (though before annotation); thus, data leakage may not be completely excluded. Finally, as noted above, our study’s resources limited our ability to fine tune cTAKES and further refine our models overall. Differences in performance between the models may therefore exaggerate the limitations of cTAKES for identifying SDoH as compared to other methods. This study, however, outlines how cTAKES, as well as the other models, can be improved upon.
CONCLUSION
This study is an important step toward understanding the differences between, and strengths and limitations of, a diverse set of NLP methods for detecting SDoH domains within clinical notes. To our knowledge, this is the first study to compare a broad panel of 10 domains of SDoH, BDoH, and pain scores with 52 granular sub-domains. This study lays the foundation for better detecting social risk factors in cLBP patients, which can advance our understanding of how social risks impact low back pain treatment access, utilization, and outcomes. Our findings and open-source methods can also be applied to other settings and patient populations, to fuel the growing momentum to apply ML techniques to the detection of SDoH in EHRs.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to acknowledge Madhumita Sushil and Hunter Mills for their advice and comments on this article, and Sara Jan, Matthew Lee, and Rafael Lee for their work as annotators.
Contributor Information
Dmytro S Lituiev, Bakar Computational Health Sciences Institute, University of California San Francisco, San Francisco, California, USA.
Benjamin Lacar, Bakar Computational Health Sciences Institute, University of California San Francisco, San Francisco, California, USA; Berkeley Institute for Data Science, University of California, Berkeley, California, USA.
Sang Pak, Department of Physical Therapy and Rehabilitation Science, University of California San Francisco, San Francisco, California, USA.
Peter L Abramowitsch, Bakar Computational Health Sciences Institute, University of California San Francisco, San Francisco, California, USA.
Emilia H De Marchis, Department of Family & Community Medicine, University of California San Francisco, San Francisco, California, USA.
Thomas A Peterson, Bakar Computational Health Sciences Institute, University of California San Francisco, San Francisco, California, USA; Department of Orthopaedic Surgery, University of California San Francisco, San Francisco, California, USA.
FUNDING
This work was supported by Back Pain Consortium (BACPAC) grant, UCSF Core Center for Patient-centric Mechanistic Phenotyping in Chronic Low Back Pain (UCSF REACH), UCSF Social Interventions Research and Evaluation Network (SIREN), the Innovate for Health program, including the UC Berkeley Institute for Data Science, the UCSF Bakar Computational Health Sciences Institute, and Johnson & Johnson (to BL).
AUTHOR CONTRIBUTIONS
DSL oversaw annotations, developed and validated models, analyzed data, developed data visualizations, and wrote and edited the article. BL developed and validated models, analyzed data, developed data visualizations, and wrote and edited the article. SP developed the annotation ontology and wrote and edited the article. PLA tuned and deployed the cTAKES models and edited the article. EHD designed the study, developed the annotation ontology, supervised manual annotation, and wrote and edited the article. TAP designed the study, developed data visualizations, and edited the article. All authors discussed the results, implications, and contributed to the article at all stages.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
CONFLICT OF INTEREST STATEMENT
DSL is a shareholder of Crosscope Inc and SynthezAI Corp and is currently employed by Johnson & Johnson. BL is supported by Innovate for Health Data Science Fellowship from Johnson & Johnson. PLA received funding from REAC RAP UCSF through UCSF. EDM received support from Hellman Fellows Fund Payment, and REAC RAP UCSF through UCSF. SP received support from Back Pain Consortium (BACPAC) grant through UCSF.
DATA AVAILABILITY
The data may not be shared due to patient privacy consideration. The model and analysis code is made available as a GitHub repository and weights hosted on Huggingface: https://github.com/BCHSI/social-determinants-of-health-clbp and https://huggingface.co/dlituiev/en_sdoh_roberta_cui.
REFERENCES
- 1. Hatef E, Predmore Z, Lasser EC, et al. Integrating social and behavioral determinants of health into patient care and population health at Veterans Health Administration: a conceptual framework and an assessment of available individual and population level data sources and evidence-based measurements. AIMS Public Health 2019; 6: 209–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anderson KO, Green CR, Payne R.. Racial and ethnic disparities in pain: causes and consequences of unequal care. J Pain 2009; 10: 1187–204. [DOI] [PubMed] [Google Scholar]
- 3. James SL, Abate D, Abate KH, et al. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 2018; 392: 1789–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. U.S. Burden of Disease Collaborators; Mokdad AH, Ballestros K, Echko M, et al. The State of US Health, 1990–2016: burden of diseases, injuries, and risk factors among US states. JAMA 2018; 319: 1444–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dutmer AL, Schiphorst Preuper HR, Soer R, et al. Personal and societal impact of low back pain: the Groningen Spine cohort. Spine (Phila Pa 1976) 2019; 44 (24): E1443–51. [DOI] [PubMed] [Google Scholar]
- 6. Trost Z, Sturgeon J, Guck A,. et al. Examining Injustice Appraisals in a Racially Diverse Sample of Individuals With Chronic Low Back Pain. J Pain 2019; 20 (1): 83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen Y, Campbell P, Strauss VY, et al. Trajectories and predictors of the long-term course of low back pain: cohort study with 5-year follow-up. Pain 2018; 159 (2): 252–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Batley S, Aartun E, Boyle E, et al. The association between psychological and social factors and spinal pain in adolescents. Eur J Pediatr 2019; 178: 275–86. [DOI] [PubMed] [Google Scholar]
- 9. Green CR, Anderson KO, Baker TA,. et al. The unequal burden of pain: confronting racial and ethnic disparities in pain. Pain Med 2003; 4 (3): 277–94. [DOI] [PubMed] [Google Scholar]
- 10. Tait RC, Chibnall JT, Andresen EM, Hadler NM.. Management of occupational back injuries: differences among African Americans and Caucasians. Pain 2004; 112 (3): 389–96. [DOI] [PubMed] [Google Scholar]
- 11. Gebauer S, Salas J, Scherrer JF.. Neighborhood socioeconomic status and receipt of opioid medication for new back pain diagnosis. J Am Board Fam Med 2017; 30 (6): 775–83. [DOI] [PubMed] [Google Scholar]
- 12. Ziadni MS, Sturgeon JA, Bissell D, et al. Injustice appraisal, but not pain catastrophizing, mediates the relationship between perceived ethnic discrimination and depression and disability in low back pain. J Pain 2020; 21: 582–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Suman A, Bostick GP, Schaafsma FG, Anema JR, Gross DP.. Associations between measures of socio-economic status, beliefs about back pain, and exposure to a mass media campaign to improve back beliefs. BMC Public Health 2017; 17 (1): 504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vest JR, Wu W, Mendonca EA.. Sensitivity and specificity of real-world social factor screening approaches. J Med Syst 2021; 45 (12): 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hong Y-R, Turner K, Nguyen OT, Alishahi Tabriz A, Revere L.. Social determinants of health and after-hours electronic health record documentation: a national survey of US physicians. Popul Health Manag 2022; 25: 362–6. [DOI] [PubMed] [Google Scholar]
- 16. Wang M, Pantell MS, Gottlieb LM, Adler-Milstein J.. Documentation and review of social determinants of health data in the EHR: measures and associated insights. J Am Med Inform Assoc 2021; 28: 2608–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hatef E, Rouhizadeh M, Tia I, et al. Assessing the availability of data on social and behavioral determinants in structured and unstructured electronic health records: a retrospective analysis of a multilevel health care system. JMIR Med Inform 2019; 7: e13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Arons A, DeSilvey S, Fichtenberg C, Gottlieb L.. Documenting social determinants of health-related clinical activities using standardized medical vocabularies. JAMIA Open 2019; 2 (1): 81–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cottrell EK, Dambrun K, Cowburn S, et al. Variation in electronic health record documentation of social determinants of health across a national network of community health centers. Am J Prev Med 2019; 57: S65–73. [DOI] [PubMed] [Google Scholar]
- 20. Beck AF, Klein MD, Kahn RS.. Identifying social risk via a clinical social history embedded in the electronic health record. Clin Pediatr (Phila) 2012; 51: 972–7. [DOI] [PubMed] [Google Scholar]
- 21. Torres JM, Lawlor J, Colvin JD, et al. ICD social codes: an underutilized resource for tracking social needs. Med Care 2017; 55: 810–6. [DOI] [PubMed] [Google Scholar]
- 22. Patra BG, Sharma MM, Vekaria V, et al. Extracting social determinants of health from electronic health records using natural language processing: a systematic review. J Am Med Inform Assoc 2021; 28: 2716–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen ES, Carter EW, Sarkar IN, Winden TJ, Melton GB.. Examining the use, contents, and quality of free-text tobacco use documentation in the electronic health record. AMIA Annu Symp Proc 2014; 2014: 366–74. [PMC free article] [PubMed] [Google Scholar]
- 24. Bejan CA, Angiolillo J, Conway D,. et al. Mining 100 million notes to find homelessness and adverse childhood experiences: 2 case studies of rare and severe social determinants of health in electronic health records. J Am Med Inform Assoc 2018; 25 (1): 61–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Conway M, Keyhani S, Christensen L, et al. Moonstone: a novel natural language processing system for inferring social risk from clinical narratives. J Biomed Semant 2019; 10: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stemerman R, Arguello J, Brice J, et al. Identification of social determinants of health using multi-label classification of electronic health record clinical notes. JAMIA Open 2021; 4. 10.1093/jamiaopen/ooaa069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010; 17: 507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Afshar M, Phillips A, Karnik N, et al. Natural language processing and machine learning to identify alcohol misuse from the electronic health record in trauma patients: development and internal validation. J Am Med Inform Assoc 2019; 26: 254–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shoenbill K, Song Y, Gress L, et al. Natural language processing of lifestyle modification documentation. Health Informatics J 2020; 26: 388–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Devlin J, Chang M-W, Lee K, Toutanova K. BERT: pre-training of deep bidirectional transformers for language understanding. Minneapolis, MN, USA: NAACL-HLT (1); 2019: 4171–86. 10.18653/v1/n19-1423. [DOI] [Google Scholar]
- 31. Rasmy L, Xiang Y, Xie Z, Tao C, Zhi D.. Med-BERT: pretrained contextualized embeddings on large-scale structured electronic health records for disease prediction. NPJ Digit Med 2021; 4: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chiticariu L, Li Y, Reiss FR. Rule-based information extraction is dead! Long live rule-based information extraction systems! In: Proceedings of the 2013 Conference on Empirical Methods in Natural Language Processing. Association for Computational Linguistics, Seattle, Washington, USA; 2013: 827–32.
- 33. Jorge A, Castro VM, Barnado A, et al. Identifying lupus patients in electronic health records: development and validation of machine learning algorithms and application of rule-based algorithms. Semin Arthritis Rheum 2019; 49: 84–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Topaz M, Murga L, Gaddis KM, et al. Mining fall-related information in clinical notes: comparison of rule-based and novel word embedding-based machine learning approaches. J Biomed Inform 2019; 90: 103103. [DOI] [PubMed] [Google Scholar]
- 35. Cairns BL, Nielsen RD, Masanz JJ, et al. The MiPACQ clinical question answering system. AMIA Annu Symp Proc 2011; 2011: 171–80. [PMC free article] [PubMed] [Google Scholar]
- 36. Pampari A, Raghavan P, Liang J, Peng J. emrQA: a large corpus for question answering on electronic medical records. In: Proceedings of the 2018 Conference on Empirical Methods in Natural Language Processing. Association for Computational Linguistics, Brussels, Belgium; 2018: 2357–68. doi: 10.18653/v1/D18-1258. [DOI]
- 37. Patrick J, Li M.. An ontology for clinical questions about the contents of patient notes. J Biomed Inform 2012; 45 (2): 292–306. [DOI] [PubMed] [Google Scholar]
- 38. Dagan I, Roth D, Sammons M, Zanzotto FM.. Recognizing textual entailment: models and applications. In: Hirst, G, ed. Synthesis Lectures on Human Language Technologies. Vol. 6. San Rafael, CA, USA: Morgan & Claypool; 2013: 1–220. [Google Scholar]
- 39. Ben Abacha A, Demner-Fushman D.. A question-entailment approach to question answering. BMC Bioinformatics 2019; 20 (1): 511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shivade C, Hebert C, Lopetegui M, et al. Textual inference for eligibility criteria resolution in clinical trials. J Biomed Inform 2015; 58Suppl: S211–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Deyo RA, Dworkin SF, Amtmann D, et al. Report of the NIH task force on research standards for chronic low back pain. Phys Ther 2015; 95: e1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. O’Neill C, Zheng P.. Integrated spine service: putting value into back pain care. Spineline 2019; 20: 12–4. [Google Scholar]
- 43. Institute of Medicine. Capturing Social and Behavioral Domains in Electronic Health Records: Phase 1. Washington (DC): The National Academies Press; 2014. doi: 10.17226/18709. [DOI] [PubMed] [Google Scholar]
- 44. Institute of Medicine. Capturing Social and Behavioral Domains and Measures in Electronic Health Records: Phase 2. Washington (DC): The National Academies Press; 2014. doi: 10.17226/18951. [DOI] [PubMed] [Google Scholar]
- 45. Hager ER, Quigg AM, Black MM, et al. Development and validity of a 2-item screen to identify families at risk for food insecurity. Pediatrics 2010; 126: e26–32. [DOI] [PubMed] [Google Scholar]
- 46.National Association of Community Health Centers, Inc (NACHC) & Association of Asian Pacific Community Health Organizations (AAPCHO). PRAPARE Screening Tool. https://prapare.org/the-prapare-screening-tool/. Accessed March 23, 2023.
- 47. Social Needs Screening Tool Comparison Table | SIREN 2019. https://sirenetwork.ucsf.edu/tools-resources/resources/screening-tools-comparison. Accessed March 23, 2023.
- 48. Arons A, DeSilvey S, Fichtenberg C, Gottlieb LM.. Compendium of Medical Terminology Codes for Social Risk Factors. University of California, San Francisco, CA, USA; Social Interventions Research and Evaluation Network; 2019.
- 49. Karran EL, Grant AR, Moseley GL.. Low back pain and the social determinants of health: a systematic review and narrative synthesis. Pain 2020; 161 (11): 2476–93. [DOI] [PubMed] [Google Scholar]
- 50. Pinheiro MB, Ferreira ML, Refshauge K, et al. Symptoms of depression as a prognostic factor for low back pain: a systematic review. Spine J 2016; 16: 105–16. [DOI] [PubMed] [Google Scholar]
- 51. Froud R, Patterson S, Eldridge S, et al. A systematic review and meta-synthesis of the impact of low back pain on people’s lives. BMC Musculoskelet Disord 2014; 15: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hong JH, Kim HD, Shin HH, Huh B.. Assessment of depression, anxiety, sleep disturbance, and quality of life in patients with chronic low back pain in Korea. Korean J Anesthesiol 2014; 66 (6): 444–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tsuji T, Matsudaira K, Sato H, Vietri J.. The impact of depression among chronic low back pain patients in Japan. BMC Musculoskelet Disord 2016; 17 (1): 447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pincus T, Burton AK, Vogel S, Field AP.. A systematic review of psychological factors as predictors of chronicity/disability in prospective cohorts of low back pain. Spine (Phila Pa 1976) 2002; 27 (5): E109–20. [DOI] [PubMed] [Google Scholar]
- 55. Rim K. MAE2: portable annotation tool for general natural language use. In: Proceedings 12th Joint ACL-ISO Workshop Interoperable Semantic Annotation; May 28, 2016; Portorož, Slovenia.
- 56. Cohen J. A coefficient of agreement for nominal scales. Educ Psychol Meas 1960; 20: 37–46. [Google Scholar]
- 57. Krippendorff K. Content Analysis: An Introduction to Its Methodology. Thousand Oaks, CA: SAGE Publications; 2013. [Google Scholar]
- 58. Braylan A, Alonso O, Lease M. Measuring annotator agreement generally across complex structured, multi-object, and free-text annotation tasks. In: Proceedings of the ACM Web Conference 2022, Virtual Event, Lyon France, ACM; 2022: 1720–30. doi: 10.1145/3485447.3512242. [DOI]
- 59. Unified Medical Language System (UMLS). https://www.nlm.nih.gov/research/umls/index.html. Accessed March 23, 2023.
- 60. Abramowitsch P. Apache cTAKES High Throughput Orchestration. 2020. Oral presentation at ApacheCon@Home 2020. https://www.youtube.com/watch?v=F5WCCPWz7Z0. Accessed March 23, 2023.
- 61. Hannibal M, Montani I, Van Landeghem S, Boyd A. spaCy: Industrial-strength Natural Language Processing in Python. 2020. 10.5281/zenodo.1212303. [DOI]
- 62. Honnibal M, Johnson M. An improved non-monotonic transition system for dependency parsing. In: Proceedings of the 2015 Conference on Empirical Methods in Natural Language Processing. Association for Computational Linguistics, Lisbon, Portugal; 2015: 1373–78. doi: 10.18653/v1/D15-1162. [DOI]
- 63. Liu Y, Ott M, Goyal N, et al. RoBERTa: a robustly optimized BERT pretraining approach. In: Proceedings of Chinese Computational Linguistics: 20th China National Conference, Hohhot, China, August 13–15, 2021: 471–84. 10.1007/978-3-030-84186-7_31. [DOI] [Google Scholar]
- 64.Accompanying code. https://github.com/BCHSI/social-determinants-of-health-clbp. Accessed March 23, 2023.
- 65. Harris ZS. Distributional structure. Word 1954; 10: 146–62. [Google Scholar]
- 66. Nie Y, Williams A, Dinan E, et al. Adversarial NLI: a new benchmark for natural language understanding. In: Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics. Association for Computational Linguistics; 2020. [Online]
- 67. Hanauer DA, Mei Q, Law J, Khanna R, Zheng K.. Supporting information retrieval from electronic health records: a report of University of Michigan’s nine-year experience in developing and using the Electronic Medical Record Search Engine (EMERSE). J. Biomed. Inform 2015; 55: 290–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data may not be shared due to patient privacy consideration. The model and analysis code is made available as a GitHub repository and weights hosted on Huggingface: https://github.com/BCHSI/social-determinants-of-health-clbp and https://huggingface.co/dlituiev/en_sdoh_roberta_cui.