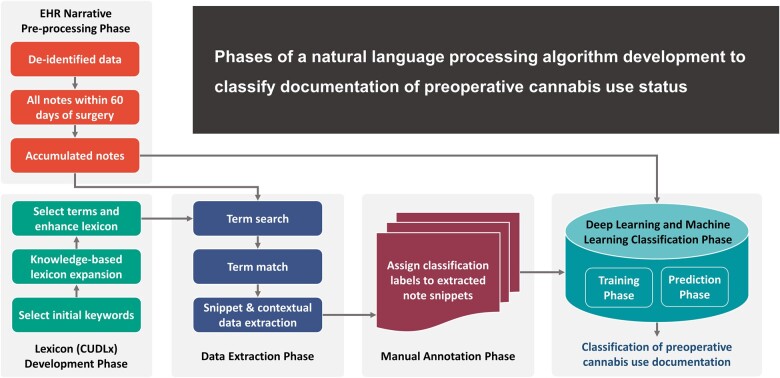
Abstract
Objective
This study aimed to develop a natural language processing algorithm (NLP) using machine learning (ML) techniques to identify and classify documentation of preoperative cannabis use status.
Materials and Methods
We developed and applied a keyword search strategy to identify documentation of preoperative cannabis use status in clinical documentation within 60 days of surgery. We manually reviewed matching notes to classify each documentation into 8 different categories based on context, time, and certainty of cannabis use documentation. We applied 2 conventional ML and 3 deep learning models against manual annotation. We externally validated our model using the MIMIC-III dataset.
Results
The tested classifiers achieved classification results close to human performance with up to 93% and 94% precision and 95% recall of preoperative cannabis use status documentation. External validation showed consistent results with up to 94% precision and recall.
Discussion
Our NLP model successfully replicated human annotation of preoperative cannabis use documentation, providing a baseline framework for identifying and classifying documentation of cannabis use. We add to NLP methods applied in healthcare for clinical concept extraction and classification, mainly concerning social determinants of health and substance use. Our systematically developed lexicon provides a comprehensive knowledge-based resource covering a wide range of cannabis-related concepts for future NLP applications.
Conclusion
We demonstrated that documentation of preoperative cannabis use status could be accurately identified using an NLP algorithm. This approach can be employed to identify comparison groups based on cannabis exposure for growing research efforts aiming to guide cannabis-related clinical practices and policies.
Keywords: cannabis, perioperative outcomes, natural language processing, NLP, substance use, social determinants of health
Graphical Abstract
INTRODUCTION
With nearly 50 million Americans aged 12 years or older using cannabis in 2020, a 60% increase from 2002,1 cannabis is the most widely used illicit substance in the United States. Unfortunately, knowledge gaps surrounding its health implications, including potential risks, of cannabis use remain significant.2,3 The National Institute on Drug Use declared research on cannabis use a top priority in 2021.4 While numerous research studies are investigating the potential therapeutic properties, social and behavioral impacts, patterns and trends in use, and mechanisms of action of substances in cannabis,5,6 very few studies are conducted in clinical settings to assess cannabis-related clinical outcomes. This results from the limitations on conducting clinical trials involving cannabis due to currently imposed federal regulations and restrictions7 and ethical considerations regarding its potential hazards.
The perioperative implications of cannabis use are a distinctively important area warranting additional research now that more preclinical and anecdotal evidence pointing to possible risks, including drug–drug interactions and interference with pain management,8–12 continues to emerge. The American Society of Regional Anesthesia and Pain Medicine has published its first Consensus Guidelines on managing perioperative patients on cannabis and cannabinoids. The guidelines could not recommend for or against tapering cannabis or cannabinoids in the perioperative period or performing toxicology screening, due to the lack of supporting evidence, highlighting the urgent need for additional research to guide safe and effective anesthesiology practices.13
Leveraging electronic health record (EHR) data in cannabis-related perioperative and other clinical research can help generate evidence highly needed to inform perioperative practices and policies and facilitate hypothesis generation for future controlled trials. Yet, leveraging EHR data to study cannabis-related health outcomes is prone to biases related to underestimating cannabis use, mainly when relying on structured data types.14 For example, ICD-CM codes for cannabis use disorder (CUD) fail to capture most patients who use cannabis, as they would not meet CUD diagnostic criteria. However, non-problematic cannabis use is still relevant for assessing potential risks and health outcomes.14 Meanwhile, performing toxicology screening tests for cannabinoids is not routine, and medication lists do not include cannabis products, except for prescription cannabinoids (eg, dronabinol) or products obtained through medical marijuana programs declared by patients to their health providers. Reliance on structured data to conduct cannabis-related research introduces a high probability of exposure misclassification and produces biased effect estimates. That said, most cannabis use-related information (>80%) is documented in unstructured narrative clinical notes.15 Hence, data extraction methods are needed to allow the utilization of unstructured narrative data in research.
Traditional approaches, such as manual chart review, can be tedious, time-consuming, irreproducible, and unfeasible when utilizing large-sized health databases.16 Artificial intelligence (AI) applications, such as machine learning (ML) techniques, including natural language processing (NLP) and deep learning (DL) techniques, are powerful tools used to extract medical concepts that can be employed to extract preoperative cannabis use status from unstructured narrative clinical notes. However, previous studies rarely utilized NLP techniques to extract information about cannabis use from clinical notes,15,17–20 and no NLP algorithms are currently available to identify and classify documentation of preoperative cannabis use status.
Nonetheless, even with AI applications, capturing narrative documentation of cannabis use can be challenging, given the complexity of the substance and the wide variability of semantics used to describe it, requiring a thorough understanding of cannabis-related clinical and linguistic concepts. Cannabis is a word that refers to all products derived from the complex plant Cannabis sativa,5 which contains over 120 active chemical compounds known as cannabinoids.21 Cannabis products are now abundant in different forms and can be consumed under different names, such as marijuana, pot, weed, CBD oil, to name a few. Furthermore, while the Food and Drug Administration (FDA) has not approved cannabis use for medical purposes, several approved drugs contain individual cannabinoids, such as dronabinol.5 A comprehensive approach covering a wide and exhaustive variety of cannabis concepts is needed to support NLP and ML pipelines involving keyword search strategies to identify documentation of cannabis use in clinical notes.
In this study, we developed an NLP algorithm to identify and classify documentation of preoperative cannabis use status using unstructured clinical notes within 60 days of surgery and created a comprehensive cannabis use documentation lexicon to support our NLP pipelines.
MATERIALS AND METHODS
Ethics approval
This study was approved by the University of Florida Institutional Review Board (UF IRB-01) as part of study protocol IRB201700747 with waived informed consent for secondary EHR data use.
Study cohort and EHR-narrative preprocessing phase
We used deidentified data from the PREsurgical Cognitive Evaluation via Digital clockfacEdrawing (PRECEDE) Bank from 22 478 patients between 2018 and 2020. PRECEDE is an ongoing databank that collects medical data of patients aged 65 years or older undergoing surgery at the University of Florida (UF)/Shands Hospital, including information from medical records (demographic, past medical/surgical history, surgery and anesthesia, pain intensity, and family medical history), questionnaires, and blood samples to assess biological markers of neurocognitive functioning.22 Data were retrieved from the UF Integrated Data Repository (IDR) via an honest data broker responsible for collecting and providing deidentified data to researchers. We included all clinical notes within 60 days of surgery. The mean (SD) note length was 26 (25.8) sentences; 142 notes had ≥500 sentences, and 5 notes had ≥800 sentences, the maximum being 1002 sentences. Identifying sentences/snippets of interest aimed to improve the efficiency of the methods by reducing computation time and manual annotation effort by allowing us to focus on snippets containing the search terms of interest. Data cleaning was performed using Python 3.
Lexicon development phase
We developed a Comprehensive Cannabis Use Documentation Lexicon (CUDLx) (Supplementary Appendix S1) that aims to provide a comprehensive and exhaustive list of concepts corresponding to cannabis use that may appear in clinical notes for NLP applications. We first identified an initial list of keywords from literature review15,17–19,23 and expert input (cannabis, cannabinoid, marijuana, marihuana, thc, cbd, tetrahydrocannabinol, cannabidiol, cannabigerol, cannabinol, epidiolex, dronabinol, nabilone, and nabiximols) to use as prompts for additional term generation using the National Library of Medicine Knowledge Sources and other search engines. Specifically, we used the Unified Medical Language Systems® (UMLS) Metathesaurus Browser (Version 2022AA),24 a large biomedical thesaurus linking synonymous names from over 200 different source vocabularies, including standard biomedical vocabularies, the UMLS Lexical Tools, the SPECIALIST Lexicon,25 and RxNav,26 to identify cannabis-related concepts and their synonyms in English under any semantic types. We also used RankWatch,27 a free online misspelling generation tool, to generate a list of possible misspellings for the most common keywords of cannabis or cannabinoids15,17,18 consisting of 5 letters or more (cannabis, cannabinoid, marijuana, marihuana, tetrahydrocannabinol, cannabidiol, cannabigerole, cannabinol), and all generic and brand names of approved synthetic prescription cannabinoids (Epidiolex®, dronabinol, Marinol®, Syndros®, Reduvo™, Cesamet™, nabilone, nabiximols, and Sativex®). Additionally, we included common misspellings of the words “THC” and “CBD” based on the literature review.17 Furthermore, we conducted an automated UMLS dictionary lookup using python to identify any additional search terms. The expanded search terms were reviewed by an expert who evaluated their appropriateness for inclusion and identified 85 additional search terms. Finally, we included a list of identified but excluded search terms, the source of their retrieval, and the reason for their exclusion. CUDLx included 3630 search terms that can be mapped into 13 higher keyword categories and 9 clinical concept categories based on the ULMS Semantic Groups and expert input for terms identified from other sources (Supplementary Appendix S1).
Data extraction phase
We searched for clinical notes containing CUDLx terms within 60 days of surgery using Python string matching and dictionary lookup, accounting for variations, such as spelling errors and morphological differences.
We included any clinical documentation within 60 days of surgery, including documentation of outpatient and inpatient visits. Among documented notes is the anesthesia preprocedure evaluation note, typically performed within 0–60 days before surgery and includes documentation of substance use, including cannabis. Outpatient and inpatient documentation typically include chief complaint, history of present illness, medical and family history, social history, objective data, and an assessment and plan.
We did not apply any section identification pipelines since substance use assessment is mostly performed without standardized templates, making section boundary definition difficult, in addition to expected lower generalizability.28 Moreover, we were interested in capturing any documentation of cannabis use to accurately ascertain cannabis use status in future steps involving patient-level classification.
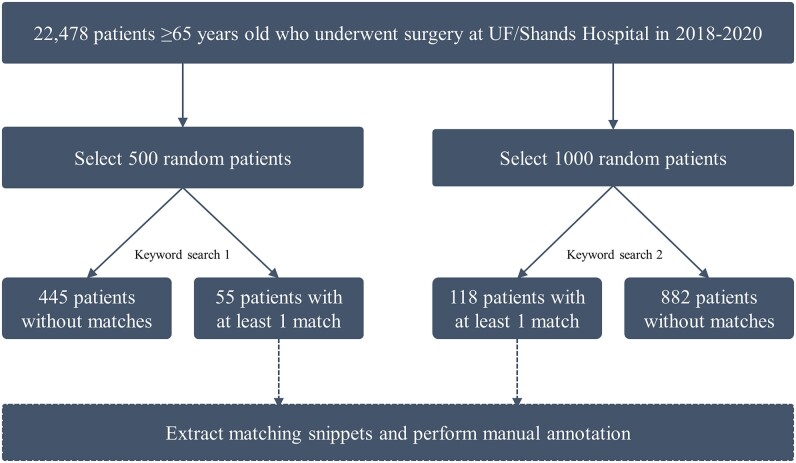
For rigorous keyword refinement and annotation, the keyword search was performed in 2 rounds allowing for improvement in keyword selection, data extraction, and annotation after the first round. Each search round would be annotated separately. In the first search, we selected 500 random patients, among whom 55 (11.00%) had at least one matching search term during the study period (448 total matches across 131 unique notes) (Figure 1). In the second search, we selected 1000 random patients, among whom 118 (11.80%) patients had at least one matching search term during the study period (1480 total matches across 332 unique notes).
Figure 1.
Flowchart summary of patient selection and keyword search to identify cannabis term matches.
For each match, we extracted a ± 10-word concordance window (“note snippet”), including punctuation, to provide textual context. Multiple snippets were generated from the same clinical note when multiple matching terms existed. The matching term was presented between 2 # marks in the extracted snippet. For additional clinical context, we also extracted the note type (eg, progress note) and the day the note was created relative to surgery day. The extracted data were exported into an Excel file for manual annotation.
Manual annotation phase
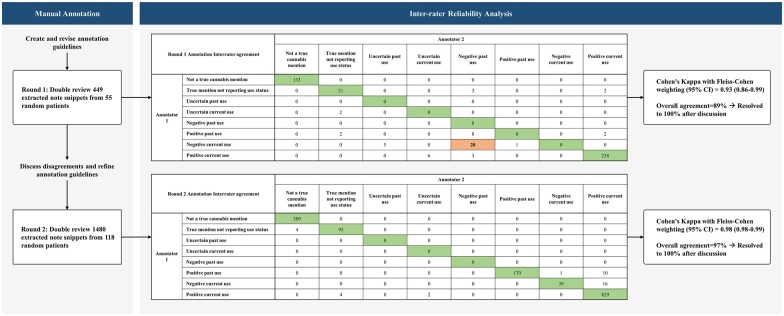
We developed a set of written guidelines supplemented with examples (Supplementary Appendix S2) and trained expert annotators to ensure the rigor and consistency of annotations.
We performed 2 manual annotation rounds using the notes identified in the extraction phase. Expert annotators were trained to assign accurate and consistent labels per guidelines by assessing 5 main properties of the matching snippet: Does the matching snippet represent a true cannabis mention? If it is a true cannabis mention, does the snippet report the patient’s cannabis use status? If the snippet reported the patient’s cannabis use status, does it refer to past or current use? Can cannabis use status be clearly ascertained from the snippet? And if cannabis use status is clear, is it positive or negative in the context of the time it was reported? The annotators assigned a multi-class label for each note snippets as (1) “Not a true cannabis mention,” (2) “True mention not reporting use status,” (3) “Uncertain past use,” (4) “Uncertain current use,” (5) “Negative past use,” (6) “Negative current use,” (7) “Positive past use,” and (8) “Positive current use.”
We double-reviewed 449 note snippets from 55 random patients in the first round and assigned note labels. The interrater reliability (IRR) analysis showed an 89% overall agreement and an overall Cohen’s Kappa with Fleiss–Cohen weighting (95% Confidence Interval [CI]) of 0.93 (0.86–0.99) (Figure 2). Discrepancies between annotators were resolved to 100% via consensus, and the annotation guidelines were revised to improve each property definition. We double-reviewed 1480 notes from 118 random patients in the second annotation round. The IRR analysis showed a 97% overall agreement and an overall Cohen’s Kappa with Fleiss–Cohen weighting (95% CI) of 0.98 (0.98–0.99). Discrepancies between annotators were also resolved to 100% via consensus in the second round.
Figure 2.
Summary of the manual annotation phase and inter-rater reliability analysis. Abbreviations: CI, confidence interval.
DL and ML classification phase
Training set
We combined the labeled note snippets from the 2 annotation rounds, a total of 1928 snippets from 463 unique notes of 173 patients, and used them to train ML classifiers to replicate human annotation for cannabis documentation in clinical notes.
Computational environment
DL and conventional ML experiments were conducted on the University of Florida Health (UF Health) DeepLearn platform, a high-performance, big-data platform. DeepLearn pulls data from the UF EHRs to support processing by ML and NLP technologies. Statistical analyses and manual annotation were performed via a UF Health virtual desktop that provides a secure platform for analyzing and sharing sensitive data (including protected health information) for UF investigators.
Classification
We used the annotated snippets to train and test a variety of multi-class ML classification models to replicate manual annotation for preoperative cannabis use status documentation.
Conventional ML models
The conventional ML models included logistic regression (LR)29 and linear support vector machines (L-SVM).30 The LR model assumes that one or more independent variables determine the outcome of the target variable. The SVM finds a hyperplane that separates training observations with the greatest margin possible. For SVM, we applied 3 kernels: linear, poly, and rbf and we found that the linear kernel performs best for our classification problem.
For both ML classifiers, we stemmed note snippets in the training set using the Porter stemmer and removed common stop words using the Natural Language Toolkit (NLTK) stop word list.31 To extract word ngram sequences (1–5 grams), we used the Python scikit-learn library32 and weighted the word ngram sequences by term frequency-inverse document frequency (TF-IDF).33 LR with L1 regularization was applied for feature vector dimensionality reduction.34
DL with non-contextualized word embedding models
Our next classification model is based on conventional non-contextualized word2vec embedding vectors,35 which perform well in CPU-only environments. These models have shown superior performance over conventional ML models and superior computational efficiency over transformer-based models.36 We integrated these word embeddings into a convolutional neural networks (CNNs) text classifier37 using Python spaCy 3.4 implementation.38 We tokenized each note snippet and created an embedding vector for each token using ScispaCy large models, which are trained on biomedical and clinical text and include nearly 785 000 vocabularies with 600 000-word word2vec vectors.39 For contextual token representation, the identified vectors were encoded into a sentence matrix by computing each token vector using a forward pass and a backward pass. A self-attention mechanism was applied to reduce sentence matrix representation dimensionality into a single context vector. Finally, we average-pooled the vectors used as features in a simple feed-forward network to predict labels for cannabis use status documentation. We used the spaCy 3.4 default network architecture and parameters for the CNN model.40 The model's predictions were compared against the reference annotations to estimate the gradient of the loss and change the weight values to improve the model's predictions over time.
Transformer-based models
The transformer-based pre-trained contextual embedding has achieved state-of-the-art results in various text classification tasks using limited labeled data.41 Since these language models have been trained with a large amount of data, they possess contextual knowledge, and fine-tuning them with a small amount of problem-specific data can achieve satisfactory results.
For our classification task, we used BERT-Base uncased42 and Bio_ClinicalBERT.43 We fine-tuned both transformer models based on cannabis use status documentation categorization using the HuggingFace library.44 Since the initial layers of the models only learn very general features, we kept them unchanged and only fine-tuned the last layers for our classification task. We tokenized and fed our input training data to fine-tune the models and then used the fine-tuned models for the test set classification. Using the 10-word window eliminated the need to truncate text segments in the transformer-based pipelines. A mini-batch size of 16 and a learning rate of 3×10−5 were used. The model was optimized using the Adam optimizer,45 and the loss parameter was set to sparse-categorical-cross-entropy. The model was trained for 5 epochs with early stopping.
Each model was evaluated via 10-fold cross-validation by randomly training using 90% of the data while keeping out 10% for testing to avoid overfitting. Each model’s F1 score was calculated based on its precision (P) and recall (R). Let c represent a particular class, and c′ refer to all other classes. The true positives (TP), false positives (FP), and false negatives (FN) for the class c are defined as follows:
TP = both true label and prediction refer a snippet to class c
FP = true label of a snippet is class c′, while prediction says it is class c
FN = true label marks a snippet as class c, while prediction refers to it as class c′
External validation
We replicated our data extraction, annotation, and ML methods using data from the Medical Information Mart for Intensive Care-III (MIMIC-III) database to test the robustness of our model and allow comparisons of our model with other existing and future models. MIMIC-III is a large, single-center database comprising information relating to patients admitted to critical care units at a large tertiary care hospital that has been contains EHR information from more than 50 000 hospital admissions. This database provides various types of data, including notes, medical history, diagnostic codes, demographics, and lab measurements for patients admitted to the Beth Israel Deaconess Medical Center in Boston, Massachusetts from 2001 to 2012.46 We extracted 1258 matching notes from 500 random patients with a cannabis search term match and utilized the same NLP methods applied on our original dataset. Our decision to utilize MIMIC-III was due to its accessibility under a data use agreement, and the possibility to easily reproduce and compare clinical studies. Our codes and Python scripts are available at https://github.com/masoud-r/cannabis-use-NLP.
RESULTS
Among 1500 randomly selected patients, 173 (11.53%) had at least one CUDLx term match. Among these patients, 70 (40.46%) had at least one documentation that was “Positive current use,” 60 (34.68%) had at least one documentation that was “True mention not reporting use status,” 38 (21.97%) had at least one match that was “Not a true cannabis mention,” 19 (10.98%) had at least one documentation that was “Positive past use,” and 12 (6.94%) had at least one documentation that was “Negative current use” (Supplementary Appendix S3, Table 1). Patients could have multiple matches with different labels (not mutually exclusive) (Supplementary Appendix S3, Table 2). Of the overall 1928 annotated snippets, 1085 (56.28%) were “Positive current use,” 442 (22.93%) were “Not a true cannabis mention,” 208 (10.79%) were “Positive past use,” 123 (6.38%) were “True mention not reporting use status,” and 70 (3.63%) were “Negative current use.” We did not identify any documentation that was “Uncertain past use,” “Uncertain current use,” or “Negated past use,” and we continue to display results related only to the identified labels. The matching keywords, their frequencies, and their categorization are summarized in Table 1.
Table 1.
Identified search terms included in search strategy for preoperative cannabis use concept extraction and frequency of keyword matches in 1928 extracted note snippets from 463 unique notes of 173 patients who had surgery at Shand's Hospital between 2018 and 2022 within 60 days of surgery
| Search terms | Total matches, n (%a) | Label assigned to matching snippet |
Keyword category | Concept category | ||||
|---|---|---|---|---|---|---|---|---|
| Not a true cannabis mention, n (%b) | True mention not reporting use status, n (%b) | Positive current use, n (%b) | Positive past use, n (%b) | Negated current use, n (%b) | ||||
| marijuana | 1086 (56.33) | 0 (0.00) | 112 (10.31) | 747 (68.78) | 191 (17.59) | 36 (3.31) | Cannabis | Cannabis |
| cbd | 538 (27.90) | 384 (71.38) | 6 (1.12) | 127 (23.61) | 6 (1.12) | 15 (2.79) | Cannabidiol | Cannabinoids |
| marinol | 52 (2.70) | 0 (0) | 4 (7.69) | 47 (90.38) | 1 (1.92) | 0 (0) | Dronabinol | Prescription Cannabinoids |
| dronabinol | 48 (2.49) | 0 (0) | 1 (2.08) | 47 (97.92) | 0 (0) | 0 (0) | Dronabinol | Prescription Cannabinoids |
| pot | 43 (2.23) | 43 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | Cannabis | Cannabis |
| thc | 29 (1.50) | 0 (0) | 0 (0) | 7 (24.14) | 7 (24.14) | 15 (51.72) | Tetrahydrocannabinol | Cannabinoids |
| epidiolex | 29 (1.50) | 0 (0) | 0 (0) | 29 (100) | 0 (0) | 0 (0) | Epidiolex | Prescription Cannabinoids |
| cannabidiol | 29 (1.50) | 0 (0) | 0 (0) | 29 (100) | 0 (0) | 0 (0) | Cannabidiol | Cannabinoids |
| mj | 23 (1.19) | 1 (4.35) | 0 (0) | 19 (82.61) | 0 (0) | 3 (13.04) | Cannabis | Cannabis |
| hemp | 16 (.83) | 0 (0) | 0 (0) | 16 (100.00) | 0 (0) | 0 (0) | Cannabis | Cannabis |
| weed | 11 (.57) | 11 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | Cannabis | Cannabis |
| marijaunac | 8 (.41) | 0 (0) | 0 (0) | 8 (100) | 0 (0) | 0 (0) | Cannabis | Cannabis |
| hash | 8 (.41) | 0 (0) | 0 (0) | 8 (100) | 0 (0) | 0 (0) | Cannabis | Cannabis |
| cannabis | 4 (.21) | 0 (0) | 0 (0) | 1 (25.00) | 3 (75.00) | 0 (0) | Cannabis | Cannabis |
| cbg | 2 (.10) | 2 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | Cannabigerol | Cannabinoids |
| tchc | 1 (.05) | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | Tetrahydrocannabinol | Cannabinoids |
| cannabinoid | 1 (.05) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | Cannabinoid | Cannabinoids |
| Total | 1928 (100) | 442 (22.93) | 123 (6.38) | 1085 (56.28) | 208 (10.79) | 70 (3.63) | ||
Note: Results are further stratified by the label assigned for the containing snippet during manual annotation and mapped into their corresponding keyword and concept categories. Search terms and snippet labels displayed in this table are only those for which a match was found in the term search strategy.
Column percent.
Row percent.
Misspelling.
Table 2.
Locations of matching search terms found in clinical notes during preoperative cannabis use search strategy of 1928 identified matches from 463 unique notes of 173 patients who had surgery at Shand's Hospital between 2018 and 2022 within 60 days of surgery, stratified by the assigned labels
| Note type | Total matches, n (%a) | Annotation labels assigned |
||||
|---|---|---|---|---|---|---|
| Not a true cannabis mention, n (%b) | True mention not reporting use status, n (%b) | Positive current use, n (%b) | Positive past use, n (%b) | Negative current use, n (%b) | ||
| Progress notes | 581 (30.13) | 142 (24.44) | 21 (3.61) | 302 (51.98) | 93 (16.01) | 23 (3.96) |
| Anesthesia preprocedure evaluation | 499 (25.88) | 47 (9.41) | 2 (.40) | 417 (83.75) | 0 (0) | 33 (6.61) |
| Letter | 236 (12.24) | 43 (18.22) | 9 (3.81) | 142 (60.17) | 34 (14.41) | 8 (3.39) |
| H&P | 96 (4.98) | 50 (52.08) | 0 (0) | 37 (38.54) | 9 (9.38) | 0 (0) |
| Telephone encounter | 89 (4.62) | 0 (0) | 87 (97.75) | 2 (2.25) | 0 (0) | 0 (0) |
| ED provider notes | 86 (4.46) | 20 (23.26) | 0 (0) | 38 (44.19) | 28 (32.56) | 0 (0) |
| H&P (view-only) | 86 (4.46) | 3 (3.49) | 0 (0) | 69 (80.23) | 14 (16.28) | 0 (0) |
| Discharge summary | 85 (4.41) | 55 (64.71) | 0 (0) | 28 (32.94) | 2 (2.35) | 0 (0) |
| Consults | 76 (3.94) | 32 (42.11) | 0 (0) | 12 (15.79) | 28 (36.84) | 4 (5.26) |
| Discharge instructions—AVS first page | 35 (1.82) | 33 (94.29) | 0 (0) | 2 (5.71) | 0 (0) | 0 (0) |
| IP AVS snapshot | 8 (0.41) | 0 (0) | 0 (0) | 8 (100) | 0 (0) | 0 (0) |
| Op note | 8 (0.41) | 4 (50.00) | 0 (0) | 4 (50.00) | 0 (0) | 0 (0) |
| Immediate post-operative note | 6 (0.31) | 6 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| MR AVS snapshot | 6 (0.31) | 0 (0) | 0 (0) | 4 (66.67) | 0 (0) | 2 (33.33) |
| Patient care transfer note | 4 (0.21) | 3 (75.00) | 0 (0) | 1 (25.00) | 0 (0) | 0 (0) |
| Patient instructions | 4 (0.21) | 0 (0) | 4 (100) | 0 (0) | 0 (0) | 0 (0) |
| Post admission assessment | 4 (0.21) | 0 (0) | 0 (0) | 4 (100) | 0 (0) | 0 (0) |
| Sedation documentation | 4 (0.21) | 0 (0) | 0 (0) | 4 (100) | 0 (0) | 0 (0) |
| Interval H&P note | 3 (0.16) | 0 (0) | 0 (0) | 3 (100) | 0 (0) | 0 (0) |
| Not included in original source | 3 (0.16) | 0 (0) | 0 (0) | 3 (100) | 0 (0) | 0 (0) |
| ED AVS snapshot | 2 (0.10) | 0 (0) | 0 (0) | 2 (100) | 0 (0) | 0 (0) |
| ED notes | 2 (0.10) | 2 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Nursing post-procedure note | 2 (0.10) | 0 (0) | 0 (0) | 2 (100) | 0 (0) | 0 (0) |
| Plan of care update | 2 (0.10) | 2 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Outpatient student | 1 (0.05) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 0 (0) |
| Total | 1928 (100) | 442 (22.93) | 123 (6.38) | 1085 (56.28) | 208 (10.79) | 70 (3.63) |
AVS: after-visit summary; ED: emergency department; H&P: history and physical examination; IP: in-patient; op: operation; MR: medical record.
Column percent.
Row percent.
Manual annotation revealed various linguistic patterns that did and did not represent documentation of the patient’s cannabis use status and can be assigned with different labels (Supplementary Appendix S3, Table 3). Most commonly, matches were found in the progress notes (30.15%) and the anesthesia preprocedure evaluation notes (25.90%). Note Types with search term matches are summarized in Table 2. A summary of note types with keyword matches relative to the total note types in the corpus is provided in Supplementary Appendix S3, Table 4.
Table 3.
Performance of conventional machine learning, deep learning, and transformer-based classifiers of preoperative cannabis use status documentation in unstructured narrative clinical notes
| Class | LR |
L-SVM |
W2V CNN |
BERT-Base |
Bio_ClinicalBERT |
Support (N)d | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pa | Rb | Fc | Pa | Rb | F c | Pa | Rb | Fc | Pa | Rb | Fc | Pa | Rb | Fc | ||
| Not a true cannabis mention | .99 | .98 | .98 | .80 | .95 | .87 | .99 | .98 | .98 | 1.0 | .96 | .98 | .98 | .99 | .99 | 44 |
| True mention not reporting use status | .83 | .72 | .77 | .77 | .77 | .77 | .89 | .71 | .79 | .89 | .71 | .79 | .84 | .76 | .80 | 12 |
| Positive past use | .37 | .35 | .36 | .56 | .41 | .47 | .71 | .54 | .61 | .71 | .56 | .63 | .70 | .56 | .62 | 10 |
| Negative current use | .63 | .77 | .69 | .80 | .83 | .81 | .74 | .70 | .72 | .64 | .67 | .65 | .84 | .83 | .84 | 20 |
| Positive current use | .92 | .94 | .93 | .95 | .95 | .95 | .91 | .96 | .93 | .91 | .99 | .95 | .94 | .99 | .96 | 108 |
| Weighted average | .88 | .90 | .89 | .90 | .91 | .91 | .91 | .91 | .91 | .90 | .92 | .91 | .93 | .95 | .94 | 192 |
BERT: Bidirectional Encoder Representations from Transformers; CNN: convolutional neural networks; LR: logistic regression; L-SVM: linear support vector machines.
Precision/positive predictive value.
Recall/sensitivity.
F score = 2×([Precision×Recall]/[Precision+Recall]).
Number of snippets included in model evaluation.
Table 4.
External validation results of the performance of conventional machine learning, deep learning, and transformer-based classifiers of preoperative cannabis use status documentation in unstructured narrative clinical notes using 1258 matching notes from 500 random patients in the MIMIC-III database
| Class | LR |
L-SVM |
W2V CNN |
BERT-Base |
Bio_ClinicalBERT |
Support (N)d | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pa | Rb | Fc | Pa | Rb | F c | Pa | Rb | Fc | Pa | Rb | Fc | Pa | Rb | Fc | ||
| Not a true cannabis mention | 1.00 | .99 | 1.00 | 1.00 | .99 | 1.00 | 0.97 | 0.99 | 0.98 | 0.99 | 1.00 | 0.99 | 1.00 | 0.99 | 1.00 | 135 |
| Positive current use | .85 | .82 | .84 | .83 | .82 | .83 | 0.84 | 0.81 | 0.82 | 0.87 | 0.90 | 0.88 | 0.87 | 0.91 | 0.89 | 67 |
| Positive past use | .73 | .80 | .77 | .73 | .78 | .75 | 0.77 | 0.83 | 0.80 | 0.89 | 0.83 | 0.86 | 0.89 | 0.78 | 0.83 | 41 |
| Negative current use | .78 | .70 | .74 | .78 | .70 | .74 | 0.75 | 0.60 | 0.67 | 0.89 | 0.80 | 0.84 | 0.62 | 0.80 | 0.70 | 10 |
| Weighted average | .91 | .91 | .91 | .90 | .90 | .90 | 0.90 | 0.90 | 0.90 | 0.94 | 0.94 | 0.94 | 0.93 | 0.93 | 0.93 | 253 |
BERT: Bidirectional Encoder Representations from Transformers; CNN: convolutional neural networks; LR: logistic regression; L-SVM: linear support vector machines.
Precision/positive predictive value.
Recall/sensitivity.
F score = 2×([Precision×Recall]/[Precision+Recall]).
Number of snippets included in model evaluation.
ML classifications of preoperative cannabis use documentation were close to human performance across all models, with a P range of 88–93% and an R range of 90–95%, as demonstrated in Table 4. Bio_ClinicalBERT achieved the highest overall performance replicating human annotations (P = 93%; R = 95%; F = 94%). Other models, including CNN with word2vec, BERT-Base, and linear SVM, had slightly lower performance (F for all = 91%). LR provides the lowest scores (P = 88%; R = 90%; F = 89%).
The external validation results were also consistent, acehiving a P range of 90–94% and an R range of 90–94% (Table 4.) However, BERT-Base achieved the highest performance replicating human annotations (P = 94%; R = 94%; F = 95%), followed by very close performance by Bio_ClinicalBERT (P = 93%; R = 93%; F = 93%). LR (P = 91%; R = 91%, F = 91%) had a slightly lower performance, followed by L-SVM and word2vec CNN (P = 90%; R = 90%; F = 90% for both).
DISCUSSION
We utilized NLP and ML techniques to identify and classify documentation of preoperative cannabis use status from unstructured narrative notes with up to 93% precision and 95% recall. All methods performed well, demonstrating that even with various documentation styles, preoperative cannabis use documentation can be identified using NLP pipelines. Conventional ML models, which do not consider the textual context, were outperformed by contextual models that consider the broader context. Bio_ClinicalBERT, which is trained on biomedical and clinical text, had a superior overall performance than all other models using notes from our original cohort, unlike previous works that reported superior performance of linear models.47,48 However, different models performed differently in specific classification tasks. For example, BERT-Base performed best for recognizing past use, while linear SVM was one of the best for predicting negation, comparable to Bio_ClinicalBERT. Although CNN with word2vec is not a fully contextualized model, it is trained on the clinical text and performed better than BERT-Base for negation cases. In the external validation using notes from MIMIC-III, we had consistent results showing superior performance of contextual models over conventional ML models. Nevertheless, BERT-Base had the best performance. Overall, all the models performed well (at least 89% and 90% F scores, using data from original cohort and MIMIC-III, respectively, with balanced P and R) in cannabis use documentation classification and can be used when system needs are met. Our study provides a baseline framework for identifying and classifying documentation of preoperative cannabis use status.
Of note, when searching for clinical documentation of cannabis use status within 60 days of surgery, most matches were found in the progress notes and the anesthesia preprocedure evaluation notes. However, meaningful documentation to ascertain cannabis use status was most frequently found in the anesthesia evaluation notes, while matches referring to concepts other than cannabis use status (ie, false matches, eg, “Not a true cannabis mention”) were most frequently found in the progress notes. While cannabis use assessment and documentation are not integrated into our healthcare system, anesthesiologists are trained to assess cannabis use, and other substances, in the preoperative anesthesia evaluation due to concerns regarding potential drug–drug interactions with anesthetic medications and perioperative adverse events.49–51 This fact should be considered when expanding our model. Furthermore, in our randomly selected sample, “marijuana” was the most frequent matching term and generated the highest number of documentations representing true cannabis mentions that report patients’ use status. On the contrary, “CBD,” the second most frequent matching term, generated the highest number of documentations that does not represent true cannabis mentions. “CBD” can also be used as an abbreviation for the “common bile duct,” a term that is likely to be seen in surgical patients.
Since our model is applied on the note level, some patients had multiple matching snippets with conflicting labels. This can be explained by different clinician assessment and documentation styles, the existence of false-positive matches, and potential changes in use status over time. We will apply expert-defined criteria for patient-level classification that account for conflicting labels at the note level as shown in Supplementary Appendix S3, Table 2, followed by integrating structured data types.
To our knowledge, this is the first NLP tool to identify and classify documentation of preoperative cannabis use status. Few studies applied NLP techniques specifically to extract information about cannabis use from EHRs.15,17–19 Still, most of these studies did not develop algorithms to classify cannabis use documentation based on time and context (eg, negation, past use), that can be then used to perform patient-level classification (cannabis users, nonusers, past users) for identifying comparison cohorts for clinical research purposes. Moreover, limitations of past approaches include relying on limited keywords and likely to miss a significant number of relevant search terms,15 focusing on limited aspects of cannabis use, such as only documentation of medical cannabis use17 or urine drug test results,19 becoming outdated,18 and being ungeneralizable/untransferable.15,17–19 The latter is due to development in settings where cannabis use screening and assessment is routine care,15,17–19 and documentation is performed using standardized templates,52 opposite to most other healthcare systems. Other previous models considering context and time in extracting information about social determinants of health and substance use in general exist.20,47,53–55 However, these works are not tailored to capturing cannabis use specifically,20,47,53–55 as opposed to our specialized model. We have demonstrated that concepts related to cannabis use can be complex, due to the complex nature of the substance. Tailoring our model to cannabis allowed it to deploy a more comprehensive approach capturing several key aspects related to documentation of cannabis use, including capturing different types of exposure to the substance in a clinically meaningful manner, and evaluating relevance to the patient, time, clarity, and negation. Our NLP model can successfully differentiate between different types of cannabis use-specific documentation, including negated and past use, utilizes a comprehensive lexicon, and is tested against a variety of documentation styles without relying on standardized templates, adding to applied NLP methods for clinical concept extraction and classification.56
Our NLP tool has multiple applications. First and most importantly, it can support clinical and epidemiological research investigating a wide array of health outcomes related to cannabis use by facilitating the use of EHR data in a reliable and reproducible fashion. Such research is a priority, given the continuous increase in cannabis use prevalence and the significant knowledge gaps surrounding its use.4,7,14 Second, our model can be deployed in operational settings, particularly perioperative clinical decision support, by simplifying the analytic process guiding provider practices and planning safe anesthetic care.49–51,57 For example, it can be used to identify documentation of perioperative cannabis use during anesthesia regimen planning instead of performing time-consuming chart reviews. This process would additionally strengthen the evaluation of quality improvement programs for perioperative cannabis use-related interventions and promote screening, assessment, and documentation of cannabis use status in medical records. Our methods should also be replicable to identify cannabis use status documentation in different clinical populations, such as adolescents, among whom targeted clinical interventions may be warranted.
Our study has several strengths. Our systematically developed lexicon (CUDLx) contains a comprehensive list of search terms covering a wide range of cannabis-related concepts, ensuring almost any documentation of cannabis use was captured. These terms can be mapped into keyword and clinical concept categories, to further identify subcategories of cannabis exposure. For example, patients using prescription cannabinoids (eg, dronabinol) can be distinguished since they have inherent differences from other cannabis users, potentially introducing biased and confounded effects if included in the same exposure group.58,59 Another example is patients using CBD-based products versus THC-based products who may experience different outcomes.2 While some of the included terms may seem redundant from a technical perspective (eg, “CBD” and “CBD cream”), they carry different meanings, can be classified into different categories with distinctive clinical significance, and provide different levels of specificity, with longer spans likely to hold more specific information. For example, “CBD” is generally used to describe the chemically active cannabinoid “cannabidiol,” which can exist in many forms, while “CBD cream” specifically means CBD products in topical cream preparations. Second, our rigorous manual annotation relied on written guidelines that provided clear definitions and examples for label assignment. Two experts performing manual annotation allowed standardizing the definition and classification of preoperative cannabis use documentation, leading to a high inter-rater agreement. Our NLP and ML pipelines can identify and classify documentation of the patient’s cannabis use status based on the time of exposure (past vs present), and the nature of documentation (confirmed vs negated), in addition to differentiating notes that do not truly represent documentation of cannabis use status. Finally, we tested our model using an external database, and achieved consistent performance results, demonstrating the robustness of our model.
Limitations of our study include using data from a single health system and only older adult patients. However, our healthcare system included patients of different socioeconomic levels, demographic characteristics, and surgical services, improving generalizability to older surgery populations. We also performed additional external validation using publicly accessible data from a second health system and achieved consistent performance results showing the robustness of our model. Still, providers at other institutions may use different styles to document cannabis use, possibly not considered in this study, limiting generalizability outside the home-trained institution. However, in addition to developing a comprehensive lexicon and conducting rigorous keyword refinement, our lexical search was applied in a healthcare system without standardized templates to document cannabis use, which diversified the linguistic styles and patterns included in the study. Second, while applying keyword search techniques generates a high P, it also leads to a lower R compared to completely manual annotation techniques. However, the comprehensive lexicon we developed aimed to minimize this limitation. Our future work will involve sampling n% of patients from the entire population and performing a thorough chart review to evaluate potentially missing positive cannabis use cases and report the resultant R score. Finally, even with applying NLP techniques to extract documentation of cannabis use status from EHRs, there remain several challenges and limitations contributing to under-documentation of cannabis use in medical records.14,60 Nonetheless, our model offers the opportunity to improve the quality of cannabis use data extracted from EHRs for clinical and epidemiological research.
The next steps include applying patient-level classification of preoperative cannabis use status, integrating other structured data forms, and creating a computable phenotype that can support reliable clinical research.
CONCLUSION
Our NLP pipeline successfully identified and classified documentation of preoperative cannabis use status in unstructured notes with precision approximating human annotation. The application of this NLP pipeline allows future identification of patient cohorts for clinical and epidemiological research investigating cannabis-related health outcomes aiming to inform clinical interventions and policymaking in the wake of the continuously increasing cannabis use prevalence.
Supplementary Material
ACKNOWLEDGMENTS
Research reported in this publication was supported by the University of Florida Clinical and Translational Science Institute, which is supported in part by the NIH National Center for Advancing Translational Sciences under award number UL1TR001427. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Contributor Information
Ruba Sajdeya, Department of Epidemiology, College of Public Health & Health Professions & College of Medicine, University of Florida, Gainesville, Florida, USA.
Mamoun T Mardini, Department of Health Outcomes & Biomedical Informatics, College of Medicine, University of Florida, Gainesville, Florida, USA.
Patrick J Tighe, Department of Anesthesiology, College of Medicine, University of Florida, Gainesville, Florida, USA.
Ronald L Ison, Department of Anesthesiology, College of Medicine, University of Florida, Gainesville, Florida, USA.
Chen Bai, Department of Health Outcomes & Biomedical Informatics, College of Medicine, University of Florida, Gainesville, Florida, USA.
Sebastian Jugl, Department of Pharmaceutical Outcomes & Policy, Center for Drug Evaluation and Safety (CoDES), University of Florida, Gainesville, Florida, USA.
Gao Hanzhi, Department of Biostatistics, University of Florida, Gainesville, Florida, USA.
Kimia Zandbiglari, Department of Pharmaceutical Outcomes & Policy, Center for Drug Evaluation and Safety (CoDES), University of Florida, Gainesville, Florida, USA.
Farzana I Adiba, Department of Pharmaceutical Outcomes & Policy, Center for Drug Evaluation and Safety (CoDES), University of Florida, Gainesville, Florida, USA.
Almut G Winterstein, Department of Pharmaceutical Outcomes & Policy, Center for Drug Evaluation and Safety (CoDES), University of Florida, Gainesville, Florida, USA.
Thomas A Pearson, Department of Epidemiology, College of Public Health & Health Professions & College of Medicine, University of Florida, Gainesville, Florida, USA.
Robert L Cook, Department of Epidemiology, College of Public Health & Health Professions & College of Medicine, University of Florida, Gainesville, Florida, USA.
Masoud Rouhizadeh, Department of Pharmaceutical Outcomes & Policy, Center for Drug Evaluation and Safety (CoDES), University of Florida, Gainesville, Florida, USA.
AUTHOR CONTRIBUTIONS
RS, MTM, PJT, RLI, CB, AGW, TAP, RLC, and MR conceptualized the manuscript. RS and MR drafted the initial manuscript. RS created figures and tables. RS, MTM, PJT, RLI, CB, SJ, HG, KZ, FIA, AGW, TAP, RLC, and MR edited, revised, and approved the final manuscript. RS developed the initial lexicon. RS, RLC, and MR revised, edited, and approved the final lexicon. RLI, CB, KZ, and FIA performed the data preprocessing. CB, KZ, FIA, and MR performed the data extraction. RS drafted the initial annotation guidelines. RS, SJ, and MR revised and approved the final annotation guidelines. RS and SJ performed the manual annotation. RS and HG performed the inter-rater reliability analysis. RS, KZ, FIA, and MR performed the data analysis. RS, MTM, PJT, RLI, CB, SJ, HG, KZ, FIA, AGW, TAP, RLC, and MR revised, and approved the results and conclusions.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
CONFLICT OF INTEREST STATEMENT
None declared.
DATA AVAILABILITY
This study utilizes both the publicly available MIMIC-III dataset and the UF Health dataset. The MIMIC-III dataset can be accessed upon obtaining permission from the PhysioNet team. Interested researchers must complete the required training and submit a data use agreement. Detailed information on the data access process and required documentation can be found on the PhysioNet website (https://physionet.org/content/mimiciii/1.4/). The UF Health dataset, on the other hand, cannot be shared publicly due to adherence to the Health Insurance Portability and Accountability Act (HIPAA) rules and regulations protecting the privacy of individuals who participated in the study. However, the data and models from the UF Health dataset may be shared upon reasonable request in the future, contingent on IRB approval. Specific requests for access to the UF Health dataset should be directed to the corresponding author, who will facilitate the process in accordance with the necessary ethical and institutional guidelines.
REFERENCES
- 1. 2020 National Survey of Drug Use and Health (NSDUH) Releases | CBHSQ Data. https://www.samhsa.gov/data/release/2020-national-survey-drug-use-and-health-nsduh-releases. Accessed July 12, 2022.
- 2. National Academies of Sciences, Engineering, and Medicine; Health and Medicine Division; Board on Population Health and Public Health Practice; Committee on the Health Effects of Marijuana: An Evidence Review and Research Agenda. Therapeutic Effects of Cannabis and Cannabinoids. Washington, DC: National Academies Press; 2017. https://www.ncbi.nlm.nih.gov/books/NBK425767/. Accessed February 29, 2020. [Google Scholar]
- 3. Jugl S, Okpeku A, Costales B, et al. A mapping literature review of medical cannabis clinical outcomes and quality of evidence in approved conditions in the USA from 2016 to 2019. Med Cannabis Cannabinoids 2021; 4 (1): 21–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. National Institute on Drug Abuse. Research Priorities. 2021. https://nida.nih.gov/international/research-funding/research-priorities. Accessed November 21, 2022.
- 5. NCCIH. Cannabis (marijuana) and cannabinoids: what you need to know. https://www.nccih.nih.gov/health/cannabis-marijuana-and-cannabinoids-what-you-need-to-know. Accessed November 18, 2022.
- 6. National Institute on Drug Abuse. NIDA research on cannabis and cannabinoids. 2020. https://nida.nih.gov/research-topics/marijuana/nida-research-cannabis-cannabinoids. Accessed November 18, 2022.
- 7. National Academies of Sciences, Engineering, and Medicine; Health and Medicine Division; Board on Population Health and Public Health Practice; Committee on the Health Effects of Marijuana: An Evidence Review and Research Agenda. Challenges and Barriers in Conducting Cannabis Research. Washington, DC: National Academies Press; 2017. https://www.ncbi.nlm.nih.gov/books/NBK425757/. Accessed February 29, 2020. [Google Scholar]
- 8. Goel A, McGuinness B, Jivraj NK, et al. Cannabis use disorder and perioperative outcomes in major elective surgeries: a retrospective cohort analysis. Anesthesiology 2020; 132 (4): 625–35. [DOI] [PubMed] [Google Scholar]
- 9. McGuinness B, Goel A, Elias F, Rapanos T, Mittleman MA, Ladha KS.. Cannabis use disorder and perioperative outcomes in vascular surgery. J Vasc Surg 2021; 73 (4): 1376–87.e3. [DOI] [PubMed] [Google Scholar]
- 10. Liu CW, Bhatia A, Buzon-Tan A, et al. Weeding out the problem: the impact of preoperative cannabinoid use on pain in the perioperative period. Anesth Analg 2019; 129 (3): 874–81. [DOI] [PubMed] [Google Scholar]
- 11. Zhang BH, Saud H, Sengupta N, et al. Effect of preoperative cannabis use on perioperative outcomes: a retrospective cohort study. Reg Anesth Pain Med 2021; 46 (8): 650–5. [DOI] [PubMed] [Google Scholar]
- 12. Sajdeya R, Ison RL, Tighe PJ. The anesthetic implications of cannabis use: a propensity-matched retrospective cohort study. In: Abstract Presented at: Anesthesiology 2021. 2021; San Diego, CA. http://asaabstracts.com/strands/asaabstracts/abstract.htm?year=2021&index=17&absnum=6156. Accessed November 18, 2022.
- 13. Shah S, Schwenk ES, Narouze S.. ASRA pain medicine consensus guidelines on the management of the perioperative patient on cannabis and cannabinoids. Reg Anesth Pain Med 2023; 48 (3): 119. [DOI] [PubMed] [Google Scholar]
- 14. Sajdeya R, Goodin AJ, Tighe PJ.. Cannabis use assessment and documentation in healthcare: priorities for closing the gap. Prev Med 2021; 153: 106798. [DOI] [PubMed] [Google Scholar]
- 15. Keyhani S, Vali M, Cohen B, et al. A search algorithm for identifying likely users and non-users of marijuana from the free text of the electronic medical record. PLoS One 2018; 13 (3): e0193706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Matt V, Matthew H.. The retrospective chart review: important methodological considerations. J Educ Eval Health Prof 2013; 10: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Carrell DS, Cronkite DJ, Shea M, et al. Clinical documentation of patient-reported medical cannabis use in primary care: toward scalable extraction using natural language processing methods. Subst Abus 2022; 43 (1): 917–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jackson RG, Ball M, Patel R, Hayes RD, Dobson RJB, Stewart R.. TextHunter—a user friendly tool for extracting generic concepts from free text in clinical research. AMIA Annu Symp Proc 2014; 2014: 729–38. [PMC free article] [PubMed] [Google Scholar]
- 19. Morasco BJ, Shull SE, Adams MH, Dobscha SK, Lovejoy TI.. Development of an algorithm to identify cannabis urine drug test results within a multi-site electronic health record system. J Med Syst 2018; 42 (9): 163. [DOI] [PubMed] [Google Scholar]
- 20. Lybarger K, Ostendorf M, Yetisgen M.. Annotating social determinants of health using active learning, and characterizing determinants using neural event extraction. J Biomed Inform 2021; 113: 103631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. National Academies of Sciences, Engineering, and Medicine; Health and Medicine Division; Board on Population Health and Public Health Practice; Committee on the Health Effects of Marijuana: An Evidence Review and Research Agenda. Cannabis. Washington, DC: National Academies Press; 2017. https://www.ncbi.nlm.nih.gov/books/NBK425762/. Accessed September 3, 2021. [Google Scholar]
- 22. (PRECEDE) PREsurgical Cognitive Evaluation via Digital clockfacEdrawing Bank. UF Health, University of Florida Health. 2019. https://ufhealth.org/research-study/precede-presurgical-cognitive-evaluation-digital-clockfacedrawing-bank. Accessed January 10, 2022.
- 23. CDC. Urine testing for detection of marijuana: an advisory. https://www.cdc.gov/mmwr/preview/mmwrhtml/00000138.htm. Accessed September 7, 2021.
- 24. Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res 2004; 32 (Database issue): D267–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lu CJ, Payne A, Mork JG.. The Unified Medical Language System SPECIALIST Lexicon and Lexical Tools: development and applications. J Am Med Inform Assoc 2020; 27 (10): 1600–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. RxNav. https://mor.nlm.nih.gov/RxNav/. Accessed November 16, 2022.
- 27. Keyword Misspelling Tool | Typo Generator | Misspelled Text Maker. https://www.rankwatch.com/free-tools/typo-generator. Accessed May 18, 2022.
- 28. Pomares-Quimbaya A, Kreuzthaler M, Schulz S.. Current approaches to identify sections within clinical narratives from electronic health records: a systematic review. BMC Med Res Methodol 2019; 19 (1): 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hosmer D, Lemeshow S, Sturdivant R.. Applied Logistic Regression. 3rd ed. Hoboken, NJ: John Wiley & Sons; 2013.
- 30. Cortes C, Vapnik V.. Support-vector networks. Mach Learn 1995; 20 (3): 273–97. [Google Scholar]
- 31. Loper E, Bird S. NLTK: the Natural Language Toolkit. In: proceedings of the ACL-02 Workshop on Effective Tools and Methodologies for Teaching Natural Language Processing and Computational Linguistics. Volume 1. ETMTNLP ’02. Association for Computational Linguistics; July 7, 2002; Philadelphia, PA; 2002: 63–70.
- 32. Varoquaux G, Buitinck L, Louppe G, Grisel O, Pedregosa F, Mueller A.. Scikit-learn: machine learning without learning the machinery. GetMobile: Mobile Comp and Comm 2015; 19 (1): 29–33. [Google Scholar]
- 33. Rouhizadeh M, Jaidka K, Smith L, Schwartz HA, Buffone A, Ungar L. Identifying locus of control in social media language. In: Proceedings of the 2018 Conference on Empirical Methods in Natural Language Processing. Association for Computational Linguistics; October 31–November 4, 2018; Brussels, Belgium; 2018: 1146–52.
- 34. Park MY, Hastie T.. L1-regularization path algorithm for generalized linear models. J R Stat Soc Series B Stat Methodol 2007; 69 (4): 659–77. [Google Scholar]
- 35. Mikolov T, Chen K, Corrado G, Dean J. Efficient estimation of word representations in vector space. [published online ahead of print September 6, 2013]. doi: 10.48550/arXiv.1301.3781. [DOI]
- 36. Alzubaidi L, Zhang J, Humaidi AJ, et al. Review of deep learning: concepts, CNN architectures, challenges, applications, future directions. J Big Data 2021; 8 (1): 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang Y, Wallace B. A sensitivity analysis of (and practitioners’ guide to) convolutional neural networks for sentence classification. [published online ahead of print April 6, 2016]. doi: 10.48550/arXiv.1510.03820. [DOI]
- 38. spaCy. Industrial-strength natural language processing in python. https://spacy.io/. Accessed November 16, 2022.
- 39. Neumann M, King D, Beltagy I, Ammar W. ScispaCy: fast and robust models for biomedical natural language processing. In: proceedings of the 18th BioNLP workshop and shared task; February 20, 2019; Cornell University; 2019: 319–27.
- 40. Explosion. Embed, encode, attend, predict: the new deep learning formula for state-of-the-art NLP models. https://explosion.ai/blog/deep-learning-formula-nlp. Accessed November 16, 2022.
- 41. Merritt R. What is a transformer model? NVIDIA Blog. 2022. https://blogs.nvidia.com/blog/2022/03/25/what-is-a-transformer-model/. Accessed November 29, 2022.
- 42. Devlin J, Chang MW, Lee K, Toutanova K. BERT: pre-training of deep bidirectional transformers for language understanding. [Published online ahead of print May 24, 2019]. doi: 10.48550/arXiv.1810.04805. [DOI]
- 43. Alsentzer E, Murphy JR, Boag W, et al. Publicly available Clinical BERT embeddings. [published online ahead of print June 20, 2019]. doi: 10.48550/arXiv.1904.03323. [DOI]
- 44. Wolf T, Debut L, Sanh V, et al. Transformers: state-of-the-art natural language processing. In: proceedings of the 2020 conference on empirical methods in natural language processing: system demonstrations. Association for Computational Linguistics; October 2020; Online; 2020: 38–45.
- 45. Kingma DP, Ba J. Adam: a method for stochastic optimization [published online ahead of print January 29, 2017]. doi: 10.48550/arXiv.1412.6980. [DOI]
- 46. Johnson AEW, Pollard TJ, Shen L, et al. MIMIC-III, a freely accessible critical care database. Sci Data 2016; 3 (1): 160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lybarger K, Yetisgen M, Uzuner Ö. The 2022 n2c2/UW shared task on extracting social determinants of health [published online ahead of print January 13, 2023]. doi: 10.48550/arXiv.2301.05571. [DOI] [PMC free article] [PubMed]
- 48. Schwartz JL, Tseng E, Maruthur NM, Rouhizadeh M.. Identification of prediabetes discussions in unstructured clinical documentation: validation of a natural language processing algorithm. JMIR Med Inform 2022; 10 (2): e29803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Echeverria-Villalobos M, Todeschini AB, Stoicea N, Fiorda-Diaz J, Weaver T, Bergese SD.. Perioperative care of cannabis users: a comprehensive review of pharmacological and anesthetic considerations. J Clin Anesth 2019; 57: 41–9. [DOI] [PubMed] [Google Scholar]
- 50. Narouze S, Strand N, Roychoudhury P.. Cannabinoids-based medicine pharmacology, drug interactions, and perioperative management of surgical patients. Adv Anesth 2020; 38: 167–88. [DOI] [PubMed] [Google Scholar]
- 51. Hepner D. Coming clean: your anesthesiologist needs to know about marijuana use before surgery. Harvard Health Blog. Published January 15, 2020. https://www.health.harvard.edu/blog/coming-clean-your-anesthesiologist-needs-to-know-about-marijuana-use-before-surgery-2020011518642. Accessed May 5, 2021.
- 52. Richards JE, Bobb JF, Lee AK, et al. Integration of screening, assessment, and treatment for cannabis and other drug use disorders in primary care: an evaluation in three pilot sites. Drug Alcohol Depend 2019; 201: 134–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Patra BG, Sharma MM, Vekaria V, et al. Extracting social determinants of health from electronic health records using natural language processing: a systematic review. J Am Med Inform Assoc 2021; 28 (12): 2716–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Uzuner Ö, South BR, Shen S, DuVall SL.. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc 2011; 18 (5): 552–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Harkema H, Dowling JN, Thornblade T, Chapman WW.. Context: an algorithm for determining negation, experiencer, and temporal status from clinical reports. J Biomed Inform 2009; 42 (5): 839–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sheikhalishahi S, Miotto R, Dudley JT, Lavelli A, Rinaldi F, Osmani V.. Natural language processing of clinical notes on chronic diseases: systematic review. JMIR Med Inform 2019; 7 (2): e12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Horvath CA, Dalley CB, Grass N, Tola DH.. Marijuana use in the anesthetized patient: history, pharmacology, and anesthetic considerations. AANA J 2019; 87 (6): 451–8. [PubMed] [Google Scholar]
- 58. Rowell-Cunsolo TL, Liu J, Shen Y, Britton A, Larson E.. The impact of HIV diagnosis on length of hospital stay in New York City, NY, USA. AIDS Care 2018; 30 (5): 591–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Silva FF, Bonfante GdS, Reis IA, Rocha HD, Lana AP, Cherchiglia ML.. Hospitalizations and length of stay of cancer patients: a cohort study in the Brazilian Public Health System. PLoS One 2020; 15 (5): e0233293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hersh WR, Weiner MG, Embi PJ, et al. Caveats for the use of operational electronic health record data in comparative effectiveness research. Med Care 2013; 51 (8 Suppl 3): S30–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study utilizes both the publicly available MIMIC-III dataset and the UF Health dataset. The MIMIC-III dataset can be accessed upon obtaining permission from the PhysioNet team. Interested researchers must complete the required training and submit a data use agreement. Detailed information on the data access process and required documentation can be found on the PhysioNet website (https://physionet.org/content/mimiciii/1.4/). The UF Health dataset, on the other hand, cannot be shared publicly due to adherence to the Health Insurance Portability and Accountability Act (HIPAA) rules and regulations protecting the privacy of individuals who participated in the study. However, the data and models from the UF Health dataset may be shared upon reasonable request in the future, contingent on IRB approval. Specific requests for access to the UF Health dataset should be directed to the corresponding author, who will facilitate the process in accordance with the necessary ethical and institutional guidelines.