Abstract
Objective
Social determinants of health (SDOH) are nonmedical factors that can influence health outcomes. This paper seeks to extract SDOH from clinical texts in the context of the National NLP Clinical Challenges (n2c2) 2022 Track 2 Task.
Materials and Methods
Annotated and unannotated data from the Medical Information Mart for Intensive Care III (MIMIC-III) corpus, the Social History Annotation Corpus, and an in-house corpus were used to develop 2 deep learning models that used classification and sequence-to-sequence (seq2seq) approaches.
Results
The seq2seq approach had the highest overall F1 scores in the challenge’s 3 subtasks: 0.901 on the extraction subtask, 0.774 on the generalizability subtask, and 0.889 on the learning transfer subtask.
Discussion
Both approaches rely on SDOH event representations that were designed to be compatible with transformer-based pretrained models, with the seq2seq representation supporting an arbitrary number of overlapping and sentence-spanning events. Models with adequate performance could be produced quickly, and the remaining mismatch between representation and task requirements was then addressed in postprocessing. The classification approach used rules to generate entity relationships from its sequence of token labels, while the seq2seq approach used constrained decoding and a constraint solver to recover entity text spans from its sequence of potentially ambiguous tokens.
Conclusion
We proposed 2 different approaches to extract SDOH from clinical texts with high accuracy. However, accuracy suffers on text from new healthcare institutions not present in the training data, and thus generalization remains an important topic for future study.
Keywords: social determinants of health, information extraction, natural language processing, clinical notes, deep learning
INTRODUCTION
The World Health Organization defines Social Determinants of Health (SDOH) as, “non-medical factors that can influence health outcomes”.1 Social factors like family support, economic factors like employment status, and health behaviors like substance use generally account for about 80% of the potentially controllable factors influencing health outcomes.2,3 For example, socioeconomic status is strongly associated with the prevalence of chronic disease such as diabetes4 and hypertension.5 Homeless people are at greater risk of COVID-19 infection due to crowded living conditions and lack of access to screening and testing.6
Large-scale use of SDOH is hindered because it is most often found in clinical note narrative text rather than in discrete fields of an electronic health record or in assigned billing codes.7,8 There is a critical need for automatic methods to extract and classify patient SDOH from clinical note text so that it can be used to improve interventions and to better understand outcomes.
Objective
We propose 2 deep learning approaches to the extraction of SDOH from clinical text that we evaluate in the context of the National NLP Clinical Challenges (n2c2) Track 2 challenge (https://n2c2.dbmi.hms.harvard.edu/2022-track-2). Systems competed to extract substance use, living status, and employment events. Performance was examined across 3 subtasks which focused on the extraction of SDOH, generalization to data from new healthcare facilities, and transfer learning adaptation to new facilities given additional training data.9
Background
Patra et al10 presented a systematic review of state-of-the-art natural language processing (NLP) approaches for the extraction of SDOH across 82 publications. They identify previous work that has addressed substance use,11 employment,12 and homelessness,13 among many other kinds of SDOH.
They found that homelessness and other less-studied SDOH (eg, education, financial problems, social isolation) are mostly identified using rule-based methods,14 while machine learning-based classification approaches are popular for identifying smoking status, substance use, and alcohol use. Some machine learning approaches relied on traditional models like support vector machines (SVMs)15 whereas others took advantage of deep learning models such as BiLSTM and BERT.16–18
For instance, Lybarger et al19 developed a novel deep-learning classification-based approach to help assess the benefits of the active learning technique used to create the SDOH corpus used in this work. Their model is based on a Bio+Discharge Summary BERT20 model, frozen to produce embeddings for subsequent trainable BiLSTM, self-attention, and conditional random field (CRF) layers. It conditions the extraction of event arguments on the extraction of event triggers and relies on self-attention and CRF output to determine token spans.
MATERIALS
Data
To train and evaluate the proposed SDOH models, we used 4 datasets: Medical Information Mart for Intensive Care (MIMIC), Social History Annotation Corpus (SHAC), In-House, and No-SDOH.
MIMIC-III (MIMIC)21 is a corpus of deidentified critical care patient data from the Beth Israel Deaconess Medical Center, collected from 2001 to 2012. The corpus contains a variety of clinical data, but only the clinical note text is used in this work.
The SHAC19 provides SDOH annotations for the social history sections from a subset of MIMIC (SHACM) and from a corpus of University of Washington Medical Center documents (SHACW).
An in-house corpus (In-House) of deidentified, mostly inpatient clinical notes from electronic health records was annotated for SDOH by following the same annotation guidelines (https://github.com/uw-bionlp/annotation_guidelines/blob/master/SDOH_annotation_guidelines.docx) used for the SHAC corpora.
A corpus with no annotatable SDOH (No-SDOH) was created in an ad-hoc attempt to reduce false-positives that were observed in non-SHAC MIMIC text. MIMIC documents were selected and then minimally edited by deleting or modifying text so that nothing rose to the level of annotatable SDOH.
Table 1 shows the number of documents in the corpora described above.
Table 1.
Corpora used for training, development, and evaluation
| # SDOH events |
||||
|---|---|---|---|---|
| Corpus | # Documents | Substance | Living status | Employment |
| MIMIC | 2 083 112 | – | – | – |
| No-SDOH | 2001 | 0 | 0 | 0 |
| In- House | 790 | 1332 | 354 | 421 |
| SHACM (train+dev) | 1504 | 3923 | 1075 | 1072 |
| SHACM (test) | 373 | 818 | 241 | 168 |
| SHACW (testB) | 2010 | 5913 | 1613 | 872 |
| SHACW (test) | 518 | 1309 | 354 | 153 |
Note: Documents in MIMIC and No-SDOH are entire clinical notes. Documents in SHAC and In-House are only the social history section of a clinical note. SHAC train, dev, and test splits were defined by the n2c2 challenge organizers. SHAC test splits were released the day before the corresponding subtask A, B, or C results were to be submitted. SHAC testB annotations were released before subtask C results were due, to support transfer learning
MIMIC: Medical Information Mart for Intensive Care; SDOH: social determinants of health; SHAC: Social History Annotation Corpus.
Annotations
A subset of the SDOH event type annotations provided by SHAC19 was used in the n2c2 challenge: Substance use (Alcohol, Drug, and Tobacco), LivingStatus, and Employment. An event is comprised of a trigger that indicates the event type and one or more arguments that provide further details. For example, an event may have the trigger Alcohol and an argument Status=none to denote that a patient does not use alcohol. Another event may have the trigger LivingStatus, an argument Status=current, and an argument Type=with_family to denote that a patient is currently living with family members.
Table 1 also shows the number of event type annotations in the corpora.
Pretrained models
Deep learning approaches benefit from large amounts of training data, in quantities far beyond what were provided by SHAC and even MIMIC. Since in-domain data are scarce, the approaches described in this paper were built on top of publicly available pretrained models: T5,22 RoBERTa,23 and XLM-RoBERTa24 that have been trained on enormous quantities of English language text.
METHODS
Sequence-to-sequence approach
The sequence-to-sequence (seq2seq) approach cast SDOH extraction as a translation task from text to a serialized, structured sequence of events (source available at https://github.com/romanows/SDOH-n2c2).
Representation
The source sequence was the full text of the social history section from a clinical note. The target sequence was the sequence of SDOH events, where each event was a sequence of arguments, and where each argument was a tuple of types, optional subtypes, and annotated tokens. Events, event arguments, and tokens were ordered left-to-right, according to the beginning token offset in the source text.
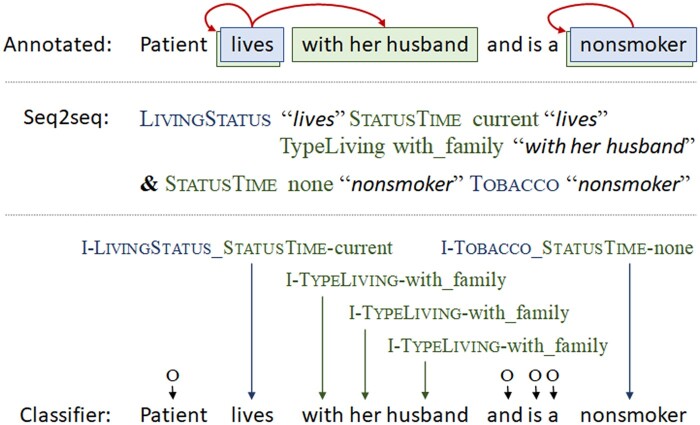
At the top of Figure 1, an example sentence with SDOH event annotations is shown. Here, the text “lives” is associated with both the trigger LivingStatus and a related argument of the type StatusTime with its subtype value current. The trigger is also related to the argument type TypeLiving with its subtype value with_family, and this argument is associated with the text “with her husband”. Our seq2seq serialized representation of this event is shown in the middle of Figure 1: ‘LivingStatus “lives” StatusTimecurrent “lives” TypeLivingwith_family “with her husband”’.
Figure 1.
Example sentence with SDOH event annotations, followed by the human-readable versions of the seq2seq and classification representations. SDOH: social determinants of health.
Training
The publicly available, pretrained T5 v1.1 large model (https://huggingface.co/google/t5-v1_1-large) was further pretrained using its standard unsupervised masked language modeling objective on a sliding window of all MIMIC and In-House text on one 80 Gb NVidia A100 GPU for about 60 h.
Next, that model was fine-tuned in a translation task with source text and target serialized event sequences from the SHACM,In-House, and No-SDOH corpora on 4 80Gb NVidia A100 GPUs for about 24 h. The model checkpoint, taken after each fine-tuning training epoch, that maximized the overall F1 performance on the competition-provided SHACM dev split was selected to compete in the extraction and generalization subtasks. That model was further fine-tuned on the SHACW train split, and the end-of-epoch model checkpoint that maximized the overall F1 performance on the SHACW dev split was selected to compete in the transfer learning subtask. We varied the learning rate, weight decay, and learning rate warmup parameters but did not find any that performed better than the defaults.
Augmented training data
Initially, fine-tuning did not produce a model that output meaningful target sequences. In a nod towards curriculum learning,25 a large number of shorter examples were automatically derived from the original training examples, and were used to augment the training data. However, unlike curriculum learning, no attempt was made to emphasize the shorter examples early in training and then to de-emphasize them later. Every fine-tuning training epoch used the entire augmented training dataset.
Shorter target sequences were constructed from contiguous subsets of events in the original examples. A new example was created by pairing a shorter target sequence with the shortest-possible contiguous span of source text that covered it (the tight-text-bound example). Another new example was created by pairing the same shorter target sequence with the longest-possible contiguous span of source text that covered it, without including text associated with other events (the loose-text-bound example). This is shown in Figure 2.
Figure 2.
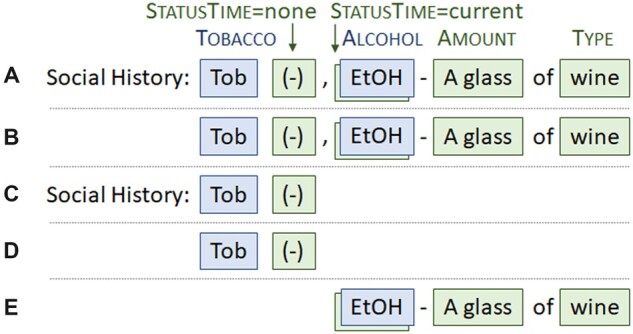
The top example (A) is the original training example. It has 2 SDOH events, where the event triggers are “Tob” and “EtOH”. Example (B) is a derived example that covers the same events as (A) but with source text created according to the tight-text-bound. Example (C) is a derived example covering only the first event in (A). Example (D) is the tight-text-bound version of example (C). The final derived example (E) covers only the second event in (A), and the loose-text-bound version is equivalent to the tight-text-bound version. SDOH: social determinants of health.
The augmentation procedure increased the number of training examples from 4,046 to 41,578. Fine-tuning on this dataset produced a model that successfully output usable target sequences.
Constrained decoding
During inference, greedy decoding occasionally produced ill-formed target sequences. Errors included invalid argument subtypes and target sequence text that did not match the source text. (In one amusing case, the T5 model helpfully corrected the misspelled source text “illicts” and output the target text “illicits” (as in “illicit drugs”). Unfortunately, the mismatch between source and target text caused problems when attempting to recover token offsets during postprocessing.)
Constrained greedy decoding prevented the production of most ill-formed target sequences. At each decoding step, candidates that would have caused the target sequence to become ill-formed were removed from consideration.
Some constraints enforced the argument type, subtype, and token event structure in the target sequence. Other constraints relied on the source text to enforce the strict left-to-right ordering of events, arguments, and tokens. A list of possible constraints, and the subset we considered to be worth implementing given our use of greedy decoding, are shown in Table 2.
Table 2.
Constraints on valid target sequences used in constrained decoding and constraint solving
| Use | Constraint |
|---|---|
| D | First output must be an argument type or the end-of-sequence (EOS) token |
| D | Argument types must be followed by a compatible subtype or by at least one token |
| D | Subtypes must be followed by at least one token |
| D | Tokens may be followed by other tokens, an argument type, the event separator, or EOS |
| D | Tokens must be subsets of the source text |
| D | Argument types in the same event must be compatible |
| DS | Within each argument , the tokens must be ordered left-to-right as relative to the original source text |
| DS | Within each event like it must be the case that |
| DS | Within each event sequence like it must be the case that |
| Events must contain exactly one trigger argument type | |
| Events must not contain duplicate argument tuples |
Note: In the “Use” column, a D indicates a constraint used in constrained decoding. An S indicates a constraint used in constraint solving.
Constraint solving
The n2c2 task required text offsets for each extracted argument, which necessitated further postprocessing.
First, the original text was searched for the target sequence tokens associated with triggers and arguments. However, target sequence tokens often matched multiple offsets in the original text. For example, “-” is often used to indicate an absence of tobacco and/or alcohol use, and so it may appear multiple times in a social history section.
To choose which of the candidate offsets in the original text should be associated with an argument, the OR-Tools constraint solver (https://developers.google.com/optimization, accessed September 2022) was used. This enforced the left-to-right ordering hard constraints listed in Table 2.
To further reduce ambiguity, soft constraints selected for compactness by minimizing the distance between the first and last characters of tokens in events and event arguments. These heuristics were motivated by an informal analysis of token span errors. For example, consider the original source text “Tobacco -, hypertension -” and the target sequence tokens “Tobacco” and “-”. The hyphen “-” in the target sequence tokens could be associated with either the first or second hyphen in the original text. The soft constraints bias the constraint solver towards a solution that chooses the first hyphen, because it minimizes the distance between “Tobacco” and “-” in the original text.
Related work
Our seq2seq approach was motivated by earlier success with deep learning encoder-decoder models in text-to-structured-sequence tasks such as Dong and Lapata’s conversion of text to its logical form.26
The use of constrained decoding has a long history in language processing, especially in automatic speech recognition. In NLP, an interesting application was Zhang and Lapata’s poetry generation system that enforced rhyming constraints during decoding.27
Our seq2seq approach happens to be quite similar to the Text2Event system developed by Lu et al.28 Apart from the use of a constraint solver to recover text offsets, the differences between the 2 systems are subtle: sentence-level processing versus our section-level processing, staged curriculum learning versus our all-at-once augmentation approach, trigger-first ordering versus our left-to-right ordering of triggers and arguments, and use of T5 versus our use of T5 v1.1.
Classification-based approach
The multi-label, multi-class classification approach cast the SDOH extraction problem as a token labeling task (source available at https://github.com/abachaa/SDOH-n2c2).
Representation
A token covered by SDOH annotations was assigned a class label that indicates the set of triggers and arguments associated with that token. These sets were encoded as strings by sorting the triggers and arguments and joining them together with underscores and dashes like: I-Trigger_ArgType-ArgSubtype. This approach is similar to the label powerset (LP) method, which transforms a multi-label problem to a multi-class problem by mapping every observed set of coinciding class labels to a single new class label.
Considering all possible combinations of triggers and arguments in the n2c2 challenge annotation guidelines, there are 904 potential class labels for tokens that are only associated with one SDOH event. This number is larger when tokens participate in multiple events. However, in practice, there were only 158 combinations observed in the SHACM training data.
Each token in the source text was tagged using the IO format (short for Inside and Outside) instead of the BIO tagging format (Beginning, Inside, and Outside)29 to reduce the number of classes. Tokens not associated with events were assigned the “O” label.
For example, an event about a nondrinker may have tokens labeled with the trigger class I-Alcohol and tokens labeled with the argument class I-Alcohol_Status-none. Or, the token “no” in text like “no alcohol, tobacco, or drugs” could have the class label I-Alcohol_Drug_Status-none_Tobacco. See Figure 1 for an additional example.
Modeling
A token-based classifier was built on top of a pretrained transformer-based model by adding a token classification head (a linear layer on top of the model hidden state outputs). We experimented with several pretrained models such as BERT,30 ClinicalBERT,31 and RoBERTa.23 After several experiments on the validation set, we selected and used RoBERTa-large without further pretraining. Models were trained on 4 24Gb NVidia K80 GPUs and hyperparameters were optimized with the Ray Tune library (https://docs.ray.io/en/latest/tune/index.html).
Ensemble modeling
Ensemble modeling has been shown to provide better performance than single models in tasks such as medical concept extraction32,33 and relation extraction.34,35 Ensemble models rely on multiple models to improve the overall performance either by combining their predictions/outputs or by combining their features. We designed several ensemble methods and training strategies (eg, fine-tuning the head layers with the models frozen for the first k epochs) and compared them on the validation set. The best ensemble relied on the concatenation of the final hidden states of a RoBERTa-base model and a XLM-R-base model. Both models were used without further pretraining.
Postprocessing
A postprocessing step generated the event relations between triggers and arguments from the sequence of class labels.
The SHACM training data was used to create a set of event templates. An event template specified which types of arguments could participate in an event with a particular type of trigger. For example, one event template was constructed to allow an association between Alcohol and Status types, since these kinds of events were seen in the training data. Similarly, an event template that allowed the association between Alcohol, Status, and Amount was included. However, an event template that allowed the association between Alcohol and Amount was not included, since such an event was never seen in the training data.
Each event template was considered for each target. An event template was satisfied when a mapping was found from the template trigger and argument slots to entities with types marked by class labels. When there were multiple entities that could satisfy one argument, the entity that was closest to the trigger entity’s text span was chosen.
From the set of satisfied event templates, only those with the greatest number of arguments were kept. For example, this would favor the event template with Alcohol, Status, and Amount over the event template with just Alcohol and Status. From this reduced set, the event template that occurred most often in the SHACM training data was chosen. For example, this would choose the Alcohol, Status, and Amount event template over the Alcohol, Status, and Type event template.
Related work
Token-level classification has been widely and efficiently used in several tasks such as named entity recognition (NER)36,37 and part-of-speech tagging.38 Our classification approach is similar to de Sousa et al,39 who developed BERT-based classification models30 to support multi-label clinical NER in Portuguese. They adopted the LP transformation-based method for multi-label classification and used the SemClinBr multi-label NER corpus of clinical notes manually labeled with Unified Medical Language System (UMLS) semantic types. Apart from the target domain involving SDOH, our system differs through its use of postprocessing to generate event relationships between the extracted entities.
RESULTS
Table 3 presents the official results evaluated with respect to our submissions on previously unseen SHAC data, released during the n2c2 competition week, and processed by our seq2seq and classifier systems. Participants could submit at most 3 runs per subtask. For the first 2 subtasks, we submitted the output of the seq2seq system, the output of the RoBERTa-based classifier system, and the output of the RoBERTa-ensemble classifier system. For the final subtask, we submitted the output of a seq2seq system where the model was first fine-tuned only on SHACM and then subsequently fine-tuned only on SHACW, a seq2seq system that was fine-tuned simultaneously on SHACM and SHACW together, and the output of the nonensemble variant of the classifier system. The seq2seq approach outperformed the classification approach and ranked above all other models in the competition.9
Table 3.
Runs submitted during the n2c2 competition week and their scores as reported by contest organizers
| Subtask | Run | System | Precision | Recall | F1 |
|---|---|---|---|---|---|
| A | 1 | Seq2seq | 0.9093 | 0.8925 | 0.9008 |
| 2 | Class-Ensemble | 0.8360 | 0.8692 | 0.8522 | |
| 3 | Class-RoBERTa | 0.8213 | 0.8580 | 0.8392 | |
| B | 1 | Seq2seq | 0.8108 | 0.7400 | 0.7738 |
| 2 | Class-RoBERTa | 0.6921 | 0.7256 | 0.7085 | |
| 3 | Class-Ensemble | 0.6916 | 0.7170 | 0.7041 | |
| C | 1 | Seq2seq (SHAC SHACW) | 0.8906 | 0.8867 | 0.8886 |
| 2 | Seq2seq (SHACM + SHACW) | 0.8800 | 0.8804 | 0.8802 | |
| 3 | Class-RoBERTa (SHACM + SHACW) | 0.7423 | 0.8468 | 0.7911 |
Note: Subtasks A, B, and C correspond to extraction, generalization, and transfer learning. “Class-Ensemble” is the ensemble of the classification-based approaches while “Class-RoBERTa” is the classification approach that used RoBERTa alone. In the transfer learning subtask C, “SHAC SHACW” means that a model was first fine-tuned on SHACM and then fine-tuned on SHACW. “SHACM + SHACW” means that a model was fine-tuned on both SHACM and SHACW together. The highest scores are bolded.
SHAC: Social History Annotation Corpus.
DISCUSSION
Representation benefits
While not required in the n2c2 challenge, it is worth noting that the seq2seq representation can capture spans covering any subset of the source text and that it can capture discontinuous annotation spans. It can also be trivially modified to output arguments that are not associated with any text span offsets, which may allow for cheaper annotation or more-reliable annotation when interannotator agreement on text spans is low.
Representation errors
Both the seq2seq and classification approaches output a sequence whose representation is lossy with respect to the ground truth.
The classification approach fails to capture relationships between entities in the same event. It cannot capture consecutive nor overlapping entities of the same type and cannot predict combinations of annotations that were not seen during training. Postprocessing heuristics that generate event relationships have a bias towards events with more arguments, whether this is warranted or not.
The seq2seq approach fails to unambiguously capture the association between entities and text spans. The constraint solver reduces the ambiguity but does not always completely resolve it. Soft constraints have a bias towards compact events, whether this is warranted or not. It is also possible for the constraint problem to be unsatisfiable if the model output even slightly violates one of the hard constraints.
To investigate the accuracy impact of the lossy seq2seq representation, we replace the model target sequence output with a target sequence derived directly from the ground truth annotations. This ideal target sequence is postprocessed and scored on all SHAC splits. The scores range from a low of 0.9844 on the SHACW dev split to a high of 1.0000 on the SHACM dev split. While it is possible that this representation is unnecessarily hard to learn for the seq2seq model, or that the postprocessing implementation is unduly affected by model output errors, we take the above results as evidence for the practical fidelity of the seq2seq representation.
Span recovery
Unlike the classification approach, the seq2seq approach required postprocessing to recover token spans. To investigate the impact of span recovery errors, we scored the competition model with and without considering token offsets on the SHACM test data. When token offsets are considered, the overall F1 is 0.9008, and when token offsets are ignored, the score is 0.9186. We take this as evidence that the postprocessing method is sufficiently accurate in recovering token spans for this task.
Scaling issues
The number of class labels in the classification approach may not scale well as new entity types and subtypes are introduced. In the worst case, the number of class labels can nearly double when new types are added. As the number of class labels grows, there will be fewer training examples available for each particular label.
The target sequence length in the seq2seq approach may not scale well as new entity types and subtypes are introduced. The classification approach always outputs the same number of class labels as there are input tokens, but the seq2seq approach can output sequences with more tokens that the source text when most of the source text is covered by many overlapping events. However, at the other end of the spectrum, source text with no SDOH only requires the output of a sequence with the end-of-sentence marker.
Model size
To examine the effect of model size in the seq2seq approach, we trained a new post-competition (after the competition, we found that smaller AdamW betas gave smoother learning curves for SHACM train and dev splits. We used these AdamW parameters to reduce the test split variance of postcompetition models selected based on dev split performance) T5 v1.1 large model (T5-large) and a new postcompetition T5 v1.1 base model (T5-base). On SHACM test, the T5-large overall F1 score was 0.9034 while the T5-base score was 0.8914 This difference is not significant according to a stratified approximate randomization hypothesis test (P = .1692).40 However, on the combined SHACW train, dev, and test splits, there is a significant difference between the T5-large score of 0.7738 and T5-base score of 0.7447 (P < .0001). The larger model helps with the generalization subtask.
Additional pretraining
To examine the effect of additional pretraining, we fine-tuned a T5 v1.1 large model that was not pretrained on MIMIC nor In-House data on SHACM train and compared it to the postcompetition model. The performance difference is statistically significant, with the former scoring 0.8754 overall F1 on SHACM test versus 0.9034 F1 (P = .005). The difference is larger when the models are scored on the combined SHACW train, dev, and test splits, with the former scoring 0.7308 overall F1 versus the baseline 0.7738 F1 (P < .0001). Additional pretraining for one or 2 epochs speeds convergence, leads to better performance on SHACM, and better generalizes to SHACW.
In-House advantage
To examine the benefits of the In-House data in pretraining and fine-tuning, we trained a postcompetition model that did not use the In-House corpus. On the SHACM test data, the performance difference was not significant: 0.9005 overall F1 versus the postcompetition baseline of 0.9034 F1 (P = .2915). However, the performance difference was significant on the combined SHACW train, dev, and test splits: 0.7540 overall F1 versus 0.7738 F1 (P < .0001). It seems that the non-MIMIC In-House data help with generalization to other non-MIMIC datasets for the seq2seq approach.
We also experimented with varying the data used to train the classification approach. Table 4 presents the results of fine-tuning the RoBERTa-based classifier on the SHACM, SHACM +SHACW, and SHACM +In-House data. The addition of the In-House dataset to training improves overall F1 performance for the classifier approach, although not as much as the addition of the SHACW dataset.
Table 4.
Results of the SDOH classifier on the SHACM test set when trained on the SHACM, SHACM+SHACW, and SHACM+In-House datasets
| Training set | Precision | Recall | F1 | |
|---|---|---|---|---|
| Overall (Triggers+Args) | SHACM | 0.8213 | 0.8580 | 0.8392 |
| SHACM+SHACW | 0.8417 | 0.8718 | 0.8565 | |
| SHACM+In-House | 0.8280 | 0.8698 | 0.8484 | |
| Alcohol (Trigger) | SHACM | 0.9830 | 0.9416 | 0.9619 |
| SHACM+SHACW | 0.9831 | 0.9448 | 0.9636 | |
| SHACM+In-House | 0.9796 | 0.9351 | 0.9568 | |
| Drug (Trigger) | SHACM | 0.9722 | 0.9259 | 0.9485 |
| SHACM+SHACW | 0.9832 | 0.9312 | 0.9565 | |
| SHACM+In-House | 0.9776 | 0.9259 | 0.9511 | |
| Tobacco (Trigger) | SHACM | 0.9804 | 0.9346 | 0.9569 |
| SHACM+SHACW | 0.9805 | 0.9408 | 0.9602 | |
| SHACM+In-House | 0.9772 | 0.9346 | 0.9554 | |
| Employment (Trigger) | SHACM | 0.9273 | 0.9107 | 0.9189 |
| SHACM+SHACW | 0.9167 | 0.9167 | 0.9167 | |
| SHACM+In-House | 0.9231 | 0.9286 | 0.9258 | |
| Living status (Trigger) | SHACM | 0.9620 | 0.9421 | 0.9520 |
| SHACM+SHACW | 0.9784 | 0.9380 | 0.9578 | |
| SHACM+In-House | 0.9588 | 0.9628 | 0.9608 |
SDOH: social determinants of health; SHAC: Social History Annotation Corpus. The highest scores are bolded.
False positives on non-social history section text
The No-SDOH corpus was created because we happened to observe spurious SDOH annotations on text not included in the SHAC corpus. To examine the effect of this additional fine-tuning data, a model was trained with and without the No-SDOH dataset. The difference in overall F1 on the SHACM test split between these 2 models was not statistically significant (P = .95).
To explore whether the No-SDOH corpus did indeed suppress false positives on some clinical text, a new dataset containing 6713 examples was sampled from MIMIC. No attempt was made to edit this dataset to remove SDOH; instead, we focused on the relative rates of SDOH annotations output by the 2 models. The model trained with No-SDOH produced annotations on 76 examples, while the model trained without No-SDOH produced annotations on all 6713 examples.
No-SDOH does reduce the amount of spurious SDOH annotation output; however, it is unknown whether the seq2seq model is broadly robust to false positives on more-general collections of text.
CONCLUSION
With sufficient training data, deep-learning approaches can leverage pretrained models to extract SDOH.
Future work could involve exploring data augmentation, soft constraints that take advantage of model attention activations during constraint solving, and explainable modeling approaches41 to recovering text spans for SDOH labels.
Generalization to new clinical text styles and conventions (and to nonclinical text42) is one of the more persistent problems in clinical NLP. Given the expense and expertise needed to annotate data, improvements should be pursued.
ACKNOWLEDGEMENTS
The authors thank the organizers of the n2c2 challenge for all their efforts in creating these high-quality datasets and organizing the challenge. The authors thank the contributors to the software and pretrained models used in this work. BRAT was used for the in-house annotations. The text-to-sequence models in particular depended upon jax 0.3.10, flax 0.4.2, huggingface transformers 4.19, pytorch 1.11, ortools 9.3.10497, and the pretrained T5 v1.1 large model. The classifier models depended upon stanza (https://stanfordnlp.github.io/stanza) 1.4.2, fairseq 0.9.0, pytorch-transformers 1.2.0, and the pretrained RoBERTa large model.
Contributor Information
Brian Romanowski, Nuance Communications, Burlington, Massachusetts, USA.
Asma Ben Abacha, Microsoft, Redmond, Washington, USA.
Yadan Fan, Nuance Communications, Burlington, Massachusetts, USA.
FUNDING
Research reported in this publication was supported by Microsoft and Nuance Communications Inc.
AUTHOR CONTRIBUTIONS
AB designed and coded the classification-based approach and was the n2c2 team lead. BR designed and coded the text-to-structured-sequence approach. YF experimented with the text-to-structured-sequence approach, annotated additional training data, and selected the final models for the competition.
CONFLICT OF INTEREST STATEMENT
None declared.
DATA AVAILABILITY
The MIMIC data are available (https://physionet.org/content/mimiciii/1.4) to researchers who pass a credentialing process. The SHAC data will be released by the n2c2 organizers. The authors will submit the No-SDOH data for publication on PhysioNet. Due to its proprietary nature, the In-House data cannot be made available.
REFERENCES
- 1. World Health Organization. Social Determinants of Health. https://www.who.int/health-topics/social-determinants-of-health. Accessed September 2022.
- 2. Remington P, Catlin B, Gennuso K.. The county health rankings: rationale and methods. Popul Health Metr 2015; 13: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hood C, Gennuso K, Swain G, et al. County health rankings: relationships between determinant factors and health outcomes. Am J Prev Med 2016; 250 (2): 129–35. [DOI] [PubMed] [Google Scholar]
- 4. Rabi D, Edwards A, Southern D, et al. Association of socio-economic status with diabetes prevalence and utilization of diabetes care services. BMC Health Serv Res 2006; 6: 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Colhoun H, Hemingway H, Poulter N.. Socio-economic status and blood pressure: an overview analysis. J Hum Hypertens 1998; 12 (2): 91–110. [DOI] [PubMed] [Google Scholar]
- 6. Tsai J, Wilson M.. COVID-19: a potential public health problem for homeless populations. Lancet Public Health 2020; 5 (4): e186–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen E, Manaktala S, Sarkar I, et al. A multi-site content analysis of social history information in clinical notes. AMIA Annu Symp Proc 2011; 2011: 227–36. [PMC free article] [PubMed] [Google Scholar]
- 8. Navathe A, Zhong F, Lei V, et al. Hospital readmission and social risk factors identified from physician notes. Health Serv Res 2018; 53 (2): 1110–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lybarger K, , YetisgenM, , Uzuner Ö.. The 2022 n2c2/UW shared task on extracting social determinants of health. J Am Med Inform Assoc2023. doi: 10.1093/jamia/ocad012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Patra B, Sharma M, Vekaria V, et al. Extracting social determinants of health from electronic health records using natural language processing: a systematic review. J Am Med Inform Assoc 2021; 28 (12): 2716–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang Y, Chen E, Pakhomov S, et al. Automated extraction of substance use information from clinical texts. AMIA Annu Symp Proc 2015; 2015: 2121–30. [PMC free article] [PubMed] [Google Scholar]
- 12. Dillahunt-Aspillaga C, Finch D, Massengale J, et al. Using information from the electronic health record to improve measurement of unemployment in service members and veterans with mTBI and post-deployment stress. PLoS One 2014; 9 (12): e115873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bejan C, Angiolillo J, Conway D, et al. Mining 100 million notes to find homelessness and adverse childhood experiences: 2 case studies of rare and severe social determinants of health in electronic health records. J Am Med Inform Assoc 2018; 25 (1): 61–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Conway M, Keyhani S, Christensen L, et al. Moonstone: a novel natural language processing system for inferring social risk from clinical narratives. J Biomed Semantics 2019; 10 (1): 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Feller DJ, Bear Don't Walk Iv OJ, Zucker J, et al. Detecting social and behavioral determinants of health with structured and free-text clinical data. Appl Clin Inform 2020; 11 (1): 172–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stemerman R, Arguello J, Brice J, et al. Identification of social determinants of health using multi-label classification of electronic health record clinical notes. JAMIA Open 2021; 4 (3): ooaa069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yu Z, Yang X, Dang C, et al. A study of social and behavioral determinants of health in lung cancer patients using transformers-based natural language processing models. AMIA Annu Symp Proc 2021; 2021: 1225–33. [PMC free article] [PubMed] [Google Scholar]
- 18. Han S, Zhang R, Shi L, et al. Classifying social determinants of health from unstructured electronic health records using deep learning-based natural language processing. J Biomed Inform 2022; 3127: 103984. [DOI] [PubMed] [Google Scholar]
- 19. Lybarger K, Ostendorf M, Yetisgen M.. Annotating social determinants of health using active learning, and characterizing determinants using neural event extraction. J Biomed Inform 2021; 1113: 103631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee J, Yoon W, Kim S, et al. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 2020; 936 (4): 1234–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Johnson A, Pollard T, Shen L, et al. MIMIC-III, a freely accessible critical care database. Sci Data 2016; 53 (1): 160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Raffel C, Shazeer N, Roberts A, et al. Exploring the limits of transfer learning with a unified text-to-text transformer. J Mach Learn Res 2020; 21 (140): 1–67.34305477 [Google Scholar]
- 23. Liu Y, Ott M, Goyal N, et al. Roberta: a robustly optimized bert pretraining approach. arXiv preprint arXiv:190711692, 2019. https://arxiv.org/abs/1907.11692. Accessed December 5, 2022.
- 24. Conneau A, Khandelwal K, Goyal N, et al. Unsupervised cross-lingual representation learning at scale. In: proceedings of the 58th annual meeting of the Association for Computational Linguistics. Association for Computational Linguistics; 2020, pp. 8440–51.
- 25. Bengio Y, Louradour J, Collobert R, et al. Curriculum learning. In: proceedings of the 26th Annual International Conference on Machine Learning. ICML ’09. New York, NY: Association for Computing Machinery; 2009, pp. 41–8.
- 26. Dong L, Lapata M. Language to logical form with neural attention. In: proceedings of the 54th annual meeting of the Association for Computational Linguistics (volume 1: long papers). Berlin, Germany: Association for Computational Linguistics; 2016, pp. 33–43.
- 27. Zhang X, Lapata M. Chinese poetry generation with recurrent neural networks. In: proceedings of the 2014 conference on empirical methods in natural language processing (EMNLP). Doha, Qatar: Association for Computational Linguistics; 2014, pp. 670–80.
- 28. Lu Y, Lin H, Xu J, et al. Text2Event: controllable sequence-to-structure generation for end-to-end event extraction. In: proceedings of the 59th annual meeting of the Association for Computational Linguistics and the 11th International Joint Conference on Natural Language Processing (volume 1: long papers). Association for Computational Linguistics; 2021, pp. 2795–806.
- 29. Ramshaw LA, Marcus MP. Text chunking using transformation-based learning. In: third workshop on very large corpora; 1995.
- 30. Devlin J, Chang MW, Lee K, et al. BERT: pre-training of deep bidirectional transformers for language understanding. In: proceedings of the 2019 conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, volume 1 (long and short papers). Minneapolis, MN: Association for Computational Linguistics; 2019, pp. 4171–86.
- 31. Huang K, Altosaar J, Ranganath R. ClinicalBERT: modeling clinical notes and predicting hospital readmission. arXiv preprint arXiv:190405342; 2019. https://arxiv.org/abs/1904.05342. Accessed December 2022.
- 32. Kang N, Afzal Z, Singh B, et al. Using an ensemble system to improve concept extraction from clinical records. J Biomed Inform 2012; 45 (3): 423–8. [DOI] [PubMed] [Google Scholar]
- 33. Kim Y, Meystre S.. Ensemble method-based extraction of medication and related information from clinical texts. J Am Med Inform Assoc 2020; 27 (1): 31–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yang D, Wang S, Li Z. Ensemble neural relation extraction with adaptive boosting. In: proceedings of the twenty-seventh international joint conference on artificial intelligence, IJCAI-18. Stockholm, Sweden: International Joint Conferences on Artificial Intelligence Organization; 2018, pp. 4532–38.
- 35. Wang L, Miller T, Bethard S, et al. Ensemble-based fine-tuning strategy for temporal relation extraction from the clinical narrative. In: proceedings of the 4th clinical natural language processing workshop. Seattle, WA: Association for Computational Linguistics; 2022, pp. 103–8.
- 36. Yadav V, Bethard S. A survey on recent advances in named entity recognition from deep learning models. In: proceedings of the 27th international conference on Computational Linguistics. Santa Fe, NM: Association for Computational Linguistics; 2018, pp. 2145–58.
- 37. Wang Y, Tong H, Zhu Z, et al. Nested named entity recognition: a survey. ACM Trans Knowl Discov Data 2022; 16 (6): 1–29. [Google Scholar]
- 38. Chiche A, Yitagesu B.. Part of speech tagging: a systematic review of deep learning and machine learning approaches. J Big Data 2022; 9 (1): 10. [Google Scholar]
- 39. de Souza J, Schneider E, Cezar J, et al. A multilabel approach to Portuguese clinical named entity recognition. In: proceedings of the XVII Congresso Brasileiro de Informática em Saúde (CBIS 2020). vol. 12; 2020, pp. 366–72. [Google Scholar]
- 40. Noreen E. Computer Intensive Methods for Testing Hypotheses. New York: John Wiley & Sons; 1989. [Google Scholar]
- 41. Saxena C, Garg M, Ansari G. Explainable causal analysis of mental health on social media sata. arXiv; 2022. https://arxiv.org/abs/2210.08430. Accessed January 2023.
- 42. Garg M. Quantifying the suicidal tendency on social media: a survey. arXiv; 2021. https://arxiv.org/abs/2110.03663. Accessed January 2023.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The MIMIC data are available (https://physionet.org/content/mimiciii/1.4) to researchers who pass a credentialing process. The SHAC data will be released by the n2c2 organizers. The authors will submit the No-SDOH data for publication on PhysioNet. Due to its proprietary nature, the In-House data cannot be made available.