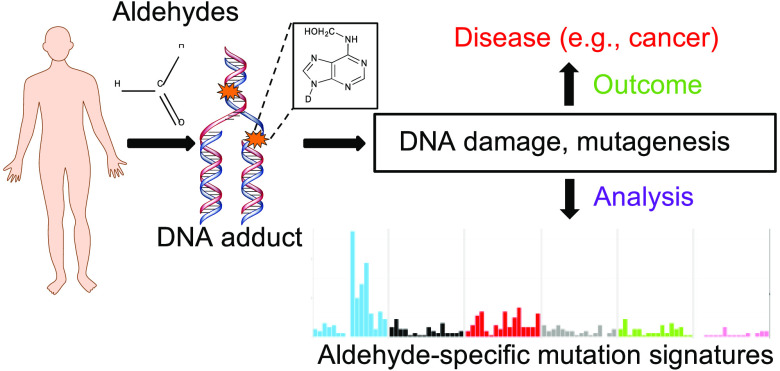
Abstract
Aldehydes are widespread in the environment, with multiple sources such as food and beverages, industrial effluents, cigarette smoke, and additives. The toxic effects of exposure to several aldehydes have been observed in numerous studies. At the molecular level, aldehydes damage DNA, cross-link DNA and proteins, lead to lipid peroxidation, and are associated with increased disease risk including cancer. People genetically predisposed to aldehyde sensitivity exhibit severe health outcomes. In various diseases such as Fanconi’s anemia and Cockayne syndrome, loss of aldehyde-metabolizing pathways in conjunction with defects in DNA repair leads to widespread DNA damage. Importantly, aldehyde-associated mutagenicity is being explored in a growing number of studies, which could offer key insights into how they potentially contribute to tumorigenesis. Here, we review the genotoxic effects of various aldehydes, focusing particularly on the DNA adducts underlying the mutagenicity of environmentally derived aldehydes. We summarize the chemical structures of the aldehydes and their predominant DNA adducts, discuss various methodologies, in vitro and in vivo, commonly used in measuring aldehyde-associated mutagenesis, and highlight some recent studies looking at aldehyde-associated mutation signatures and spectra. We conclude the Review with a discussion on the challenges and future perspectives of investigating aldehyde-associated mutagenesis.
Introduction
Aldehydes are a ubiquitous class of chemicals that are widely present in our diets, the immediate environment, as well as the intracellular milieu. Because of their high reactivity, these molecules can chemically modify all major biomolecules and impede their function. As a result, aldehyde exposure, especially from miscellaneous environmental sources, poses a high risk to human health. Unsurprisingly, aldehyde-mediated toxicity underlies several human diseases, as noted in the upcoming sections.
Aldehydes belong to a group of chemicals referred to as reactive carbonyls. These molecules typically have a polarized carbon–oxygen (C=O) double bond, which imparts a substantial dipole moment to aldehydes. This difference in electronegativity makes the carbonyl carbon a strong electrophile and therefore readily reactive toward nucleophilic molecules like amino groups of proteins and nucleobases of DNA. Depending on their chemical complexity, aldehydes can be classified into various chemical categories (Figure 1). These range from unbranched simple aldehydes, such as formaldehyde and acetaldehyde, to α,β-unsaturated aldehydes, such as acrolein and crotonaldehyde, and aromatic aldehydes, such as benzaldehyde, cinnamaldehyde, and vanillin.1 Due to their short chain lengths, shorter unbranched simple aldehydes like formaldehyde are strongly hydrophilic, which greatly enhances their toxicity.
Figure 1.
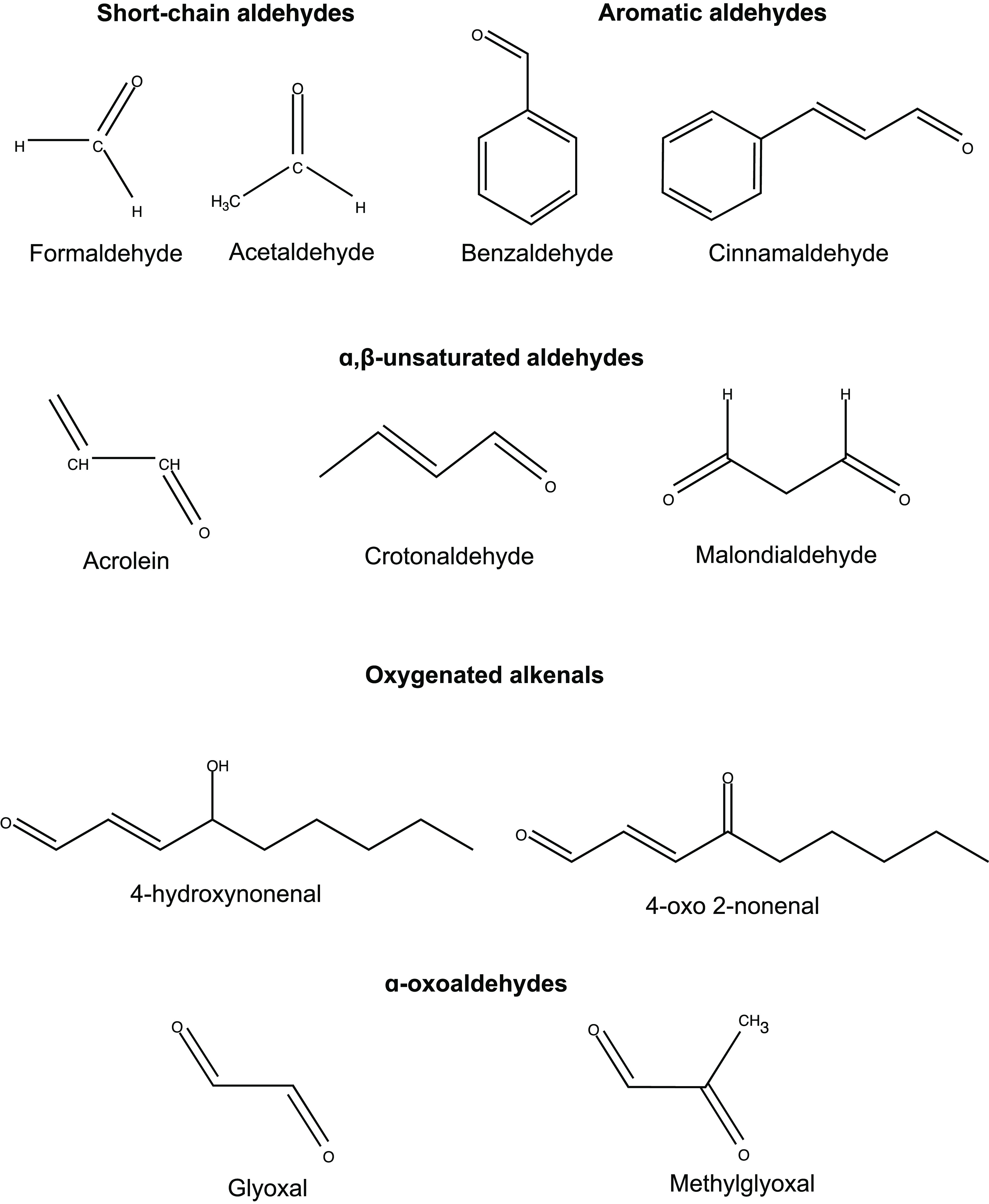
Chemical classification and structures of common environmental and endogenous aldehydes.
Given the variety in chemical structures and properties of diverse aldehydes, the molecular mechanism of how these molecules impact the genome, the underlying chemical modifications, adducts, and downstream repair pathways for exposure to most aldehydes remain incompletely understood. The purpose of this Review is to highlight recent advances in understanding aldehyde-associated mutagenesis. Readers will be provided a short primer on the extant knowledge on the various classes of aldehydes, their sources, mechanisms of toxicity, and the corresponding detoxification machinery. The primary focus of the Review is to highlight studies that explored aldehyde mutagenicity and to discuss future avenues to investigate aldehyde-associated mutagenesis.
A Brief Overview of the Major Aldehydes and Their Sources
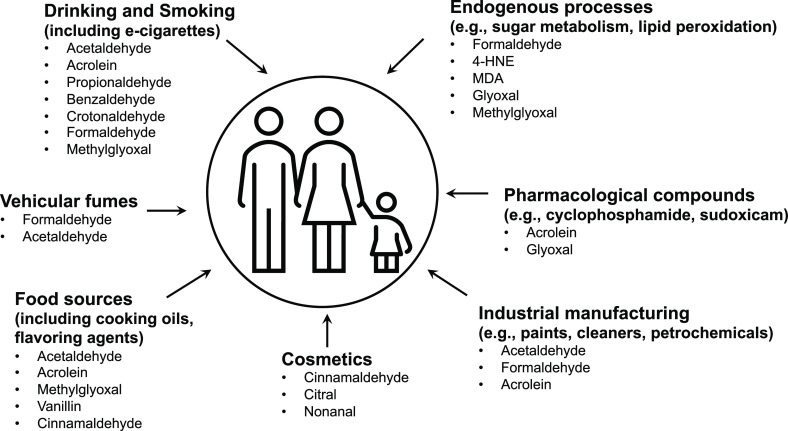
Aldehydes can occur in the environment from a multitude of natural sources (Figure 2). A variety of day-to-day human activities either use or release aldehydes into the ambient environment, including air and soil. Several excellent reviews provide an in-depth analysis of aldehyde sources, mechanism of cytotoxicity, and aldehyde clearance systems.1−3 A summary of the main environmental sources for well-known aldehydes is listed below.
Figure 2.
Common environmental and endogenous sources of aldehydes.
Smoking and alcohol consumption carry the highest risk of individual exposure to aldehydes. Cigarette smoke is particularly enriched for several different types of aldehydes, chiefly acetaldehyde, acrolein, and crotonaldehyde.4 Cumulatively, the concentration of aldehydes in tobacco smoke is reportedly ∼1000 times higher than polycyclic aromatic compounds and tobacco-specific nitrosamines.5 E-cigarettes also result in exposure of users to a variety of reactive carbonyl compounds including acetaldehyde, formaldehyde and acrolein, and methylglyoxal.6−9 These aldehydes can lead to the formation of DNA adducts; for example, tobacco smoke exposure induced increased production of the aldehyde adduct γ-hydroxypropanodeoxyguanosine (γ-OH-PrdG) in mice and humans.10
Alcoholic beverages are consumed worldwide and represent one of the largest and most ubiquitous sources of acetaldehyde exposure. Acetaldehyde toxicity is thought to be the primary contributor to alcoholic liver disease and likely the key underlying factor in the genotoxicity of ethanol and the associated cancers of the esophagus, upper respiratory tract, gastrointestinal tract, and liver.11−15
Fuel combustion is an additional major contributor to environmental aldehydes, chiefly via the photooxidative conversion of emitted hydrocarbons to aldehydes. A variety of aldehydes are released to the environment as volatile organic compounds (VOCs) in this manner, including formaldehyde, acetaldehyde, acrolein, as well as aromatic aldehydes such as benzaldehyde and tolualdehyde.2,16 Newer modifications to fuel sources such as the addition of ethanol have been shown to further contribute to overall environmental acetaldehyde levels.17,18 Aldehydes are additionally generated as intermediate chemicals during many chemical manufacturing processes.19
Diet and cooking processes release multiple complex aldehydes into the environment. Aldehyde emissions are associated with cafes and coffee-roasting facilities.20 The artificial sweetener aspartame is metabolized to formaldehyde in the gastrointestinal tract,21 and multiple dairy products, fruits, vegetables, and meats contain formaldehyde and acetaldehyde. Food processing, especially deep-frying, can greatly increase generation of aldehydes through prolonged cooking times and high-temperature exposure of polyunsaturated fats in edible oils.22−24
Many commonly available pharmacological drugs are metabolized to produce aldehydes. For example, the antineoplastic agent cyclophosphamide is metabolized by cytochrome P450 enzymes to generate acrolein, which is associated with an elevated risk of renal toxicity.25 Similarly, the enol-carboxamide drug sudoxicam—the first nonsteroidal anti-inflammatory drug shown to have anti-inflammatory effects in animals—is metabolized to glyoxal and is associated with acute hepatotoxicity.26
Hair treatment reagents often contain formaldehyde, resulting in high indoor concentrations in locations like hair salons.27 Cinnamaldehyde is found in components of deodorants and is associated with allergic contact dermatitis.28 Other cosmetic products such as skin care treatments, colognes, nail polish removers, and fragrant hand sanitizers can be a source of toxic aldehydes like benzaldehyde and acetaldehyde, further increasing the risk of topical allergies and other skin-related diseases with prolonged contact and usage.29
Lastly, several endogenous cellular pathways produce toxic aldehydes that can damage DNA. DNA metabolism can yield furfural.30 Oxidative demethylation reactions involving RNA and DNA can generate formaldehyde as a byproduct, as can neutrophilic myeloperoxidase enzymes.31,32 The oxo-aldehyde methylglyoxal is commonly formed from triose phosphate intermediates during respiration,33 while glyoxal is the common byproduct of sorbitol and ascorbate metabolism.2 α,β-unsaturated aldehydes including acrolein and 4-hydroxynonenal (4-HNE) are commonly generated during lipid peroxidation.34
Aldehyde Exposure and Human Health
Environmental aldehydes put millions of individuals at risk of aldehyde-associated health problems. Both formaldehyde and acetaldehyde are listed as Class I agents by the IARC35−38 and are associated with multiple cancer types including nasopharyngeal cancer, esophageal carcinoma, hepatocellular carcinoma, head-and-neck cancers, and blood cancers.39−45 Defects in the aldehyde dehydrogenase gene ALDH2 are responsible for “flushed face” syndrome.46 This phenomenon occurs in roughly 36% of all individuals of East Asian descent, is marked by facial redness, palpitations, and muscle weakness upon alcohol consumption,47 and additionally puts such individuals at increased risk for esophageal cancer.48
Formaldehyde is a driver of Cockayne syndrome, which is a rare autosomal recessive genetic disorder marked by cachexia, renal failure, and neurodevelopmental defects such as microencephaly.49,50 The transcription-coupled nucleotide excision repair factor ERCC6 and aldehyde dehydrogenase factor ADH5 synergize to prevent DNA damage induced by endogenous formaldehyde, and defects in both genes phenocopy symptoms of Cockayne syndrome.50 Digenic defects in ALDH2 and ADH5 underlie the recently described AmeD (“aplastic anemia, mental retardation, dwarfism) syndrome and are likely linked to inefficient clearance of endogenous formaldehyde.51 Fanconi anemia, an aplastic anemia that impairs bone marrow function, is prevalent among individuals of Ashkenazi Jewish ancestry.52−54 Defects in formaldehyde-induced DNA damage repair,55 as well as defects in the aldehyde dehydrogenase gene ALDH2,56 predispose individuals to Fanconi anemia. Failure to efficiently clear formaldehyde in ALDH2-deficient mothers, combined with FANC deficiencies in the embryo, likely exposes fetal genomes to toxic aldehydes and produce early embryonic defects in neurodevelopment and hematopoiesis, thus providing a molecular basis for the disease.57
Other aldehydes have been implicated in human diseases. Lipid peroxidation-associated aldehydes such as malondialdehyde (MDA) and 4-HNE can be detected as protein–aldehyde adducts in atherosclerotic lesions from human aortas,58 and serum levels of isopentanaldehyde were increased in samples from patients of cardiovascular disease (CVD).59 Furthermore, high plasma concentrations of 4-HNE were detected in patients of chronic kidney disease prior to hemodialysis.60 In addition, aldehydes have been proposed as key mediators in several neuropathologies; aldehydes such as 3-aminopropanal are derived from the metabolism of polyamines (e.g., spermine), and polyamine levels are greatly elevated in oligodendrocytes from multiple sclerosis patients.61 Additionally, aldehyde–protein adducts are elevated in amyotrophic lateral sclerosis (ALS) and Alzheimer’s disease.60 In neurons from Parkinson’s disease patients, the metabolism of neuroendocrine factors such as dopamine and epinephrine as well as increased lipid peroxidation produces elevated levels of biogenic aldehydes such as MDA, 4-HNE, and 3,4-dihydroxyphenylacetaldehyde (DOPAL).62 Oxo-aldehydes like glyoxal and methylglyoxal can react with amino acid residues on proteins and precipitate the formation of advanced glycation end products (AGEs) and additionally make nucleobase adducts with guanines. Complications associated with diabetes, including vascular damage, are often attributed to decreased protein functions resulting from AGEs, likely formed due to elevated blood glucose levels that lead to a net increase in methylglyoxal levels.2
Air concentrations of formaldehyde and acetaldehyde are significantly elevated in occupational settings such as indoor nail salons, manufacturing plants for carpets, paints, furniture, and fabrics, healthcare locations, chemical laboratories, and funeral parlors.29,63 As such, workers in these settings are at an increased risk for respiratory and dermal problems. In combination with the prevalence of aldehydes in day-to-day emissions such as cigarette smoke, motor vehicle exhaust, and cooking oil fumes, elevated environmental aldehyde levels pose a major and constant threat to human health.
Mechanisms of Detoxification
The toxic effects of aldehydes are mitigated in large part owing to robust detoxification machinery. These consist of alcohol dehydrogenases, aldehyde dehydrogenases, aldehyde oxidases, and the cytochrome P450 reductase family of enzymes, among other proteins. Their mechanisms are briefly discussed below.
Aldehyde dehydrogenases (ALDH) are a class of enzymes that rely on NAD(P)-dependent oxidation of aldehydes, with varying substrate specificities. In humans, aldehyde dehydrogenases are roughly divided into three classes, ALDH1–3, with members typically functioning as homodimers or tetramers. Conserved residues line the catalytic pocket and participate in NAD binding, substrate alignment and deprotonation, and subsequent oxidation steps.64ALDH2 deficiencies are associated with multiple disease states. In particular, the ALDH2*2 variant, which loses the ability to bind the NAD cofactor, results in a loss of catalytic activity and accumulation of toxic levels of intracellular acetaldehyde. This results in dire health consequences including alcohol toxicity, cardiovascular complications, and cancers.42,43,56ALDH7A1 is involved in the metabolism of the amino acid lysine, and dysfunction is correlated with developmental defects and seizures, such as pyridoxine-associated epilepsy (PDE).65,66
Cytochrome P450 (CYP450) enzymes are a class of membrane-bound enzymes present in the mitochondria and endoplasmic reticulum and are ubiquitous in the animal kingdom. These enzymes are monooxygenases and have a heme molecule as a prosthetic group that is sulfenylated via a cysteine–thiolate bond in the protein. In addition to performing multiple detoxifying reactions, including metabolism of drugs and xenobiotics and fat-soluble vitamins, CYPs play a key role in the metabolic activation of many compounds, steroid hormone synthesis, and breakdown of unsaturated fatty acids (reviewed in ref (67)). CYPs catalyze the oxidation of aldehydes to their corresponding carboxylic acids using oxygen and NADPH. In addition to simple aldehydes such as acetaldehyde, CYPs act on a range of aldehydes including acrolein, monoterpenoid aldehydes such as citronellal, benzaldehydes, and α,β-unsaturated aldehydes such as 4-HNE, which is derived from lipid peroxidation and is associated with chronic inflammation and neurodegeneration.68,69
α-Oxoaldehydes are detoxified by the glyoxylase system that consists of glutathione-S-transferase (GSH)-dependent enzymes. The most-well studied members of this system are the Glo1 isomerase and Glo2 thioesterase, which are highly conserved glyoxylases that are critical for metabolizing methylglyoxal (MG) and glyoxal within tissues,.270 Glo1 acts on glutathione conjugates of oxoaldehydes such as the MG-derived hemithioacetal and the glyoxal-derived glycolate, while Glo2 acts in the later steps of the pathway, for example, hydrolysis of the hemiacetal-derived thioester to D-lactate.71,72
Moreover, monomeric aldo-keto reductases (AKR) act via general acid–base catalysis reactions that reduce aldehydes and ketones to primary and secondary alcohols. Currently three AKR families comprising 14 members are described for humans, which utilize NADP(H) as a cofactor. Endogenous aldehydes resulting from lipid peroxidation and sugar metabolism, as well as xenobiotic-derived aldehydes (e.g., aflatoxin), are among the substrates for detoxification by AKRs.73
Aldehyde oxidases (AOX) are a small group of enzymes that require molybdenum, iron–sulfur clusters, and FAD as cofactors. Humans have a single AOX1 enzyme, which follows general base catalysis to oxidize aldehydes, first via the generation of activated molybdenum (mo-O–), nucleophilic attack on the substrate by activated molybdenum (mo-O–), and generation of a stable intermediate, followed by release of product, reoxidation of molybdenum, and transfer of reducing equivalents via Fe–S clusters.74 AOX enzymes are largely confined to the hepatic tissues, where they help in the metabolism of several antitumor, immunosuppressive drugs such as methotrexate, as well as antiviral compounds. Crotonaldehyde, benzaldehyde, the aromatic aldehyde vanillin, and retinal are among the common aldehyde substrates of AOX.75
In humans, there are five classes of alcohol dehydrogenases (ADHI–V). This group of enzymes typically acts as homo-or heterodimers, has two zinc atoms in the active site, and uses NADP(H) as a cofactor. Class I ADHs are primarily expressed in the stomach and are involved in the metabolism of alcohol-derived aldehydes like acetaldehyde, whereas class II, III, and IV metabolize benzaldehyde, formaldehyde, and lipid peroxidation-derived aldehydes and are expressed in the liver and other tissues including brain.76 ADHs can reversibly reduce aldehydes to primary alcohols, when physiological concentrations of aldehydes and reduced cofactors (NADP(H)) remain higher than alcohol and NADP+.77
In addition to the mechanisms listed above, there are various nonenzymatic routes for the detoxification of aldehydes. For example, 4-HNE can be conjugated to glutathione, which links with aldehyde molecules through its sulfhydryl moiety and aids in their detoxification by aldol reductases.78 Methylglyoxal can be efficiently scavenged by hydrazone-forming compounds such as hydrazine and its derivatives and by aminoguanidine.79,80 The histidine dipeptide carnosine is endogenously present across different organs including skeletal tissues, the brain, and the gastrointestinal system; it can nonenzymatically scavenge reactive oxygen species (ROS), prevent glycation, and react with several aldehyde and carbonyl compounds including methylglyoxal, lipid-peroxidation-derived malondialdehyde, and acetaldehyde.81−83
Aldehyde Adducts and the Genome
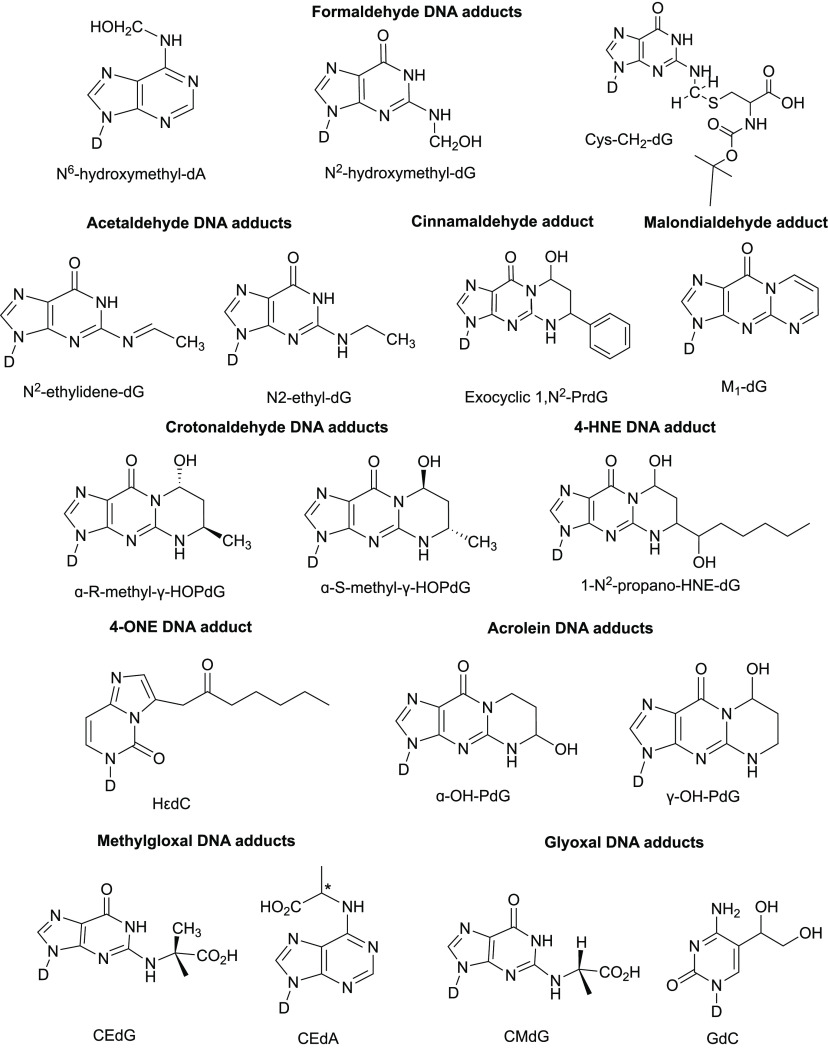
Covalent adduct formation is a hallmark of many different aldehydes due to the presence of a highly electrophilic carbonyl which tends to readily react with strong nucleophiles like the N2 nitrogen on deoxyguanosine in DNA molecules and the ϵ-amino groups on lysine residues in proteins. Conversely, unsaturated α,β-unsaturated aldehydes are less electrophilic and tend to target sulfhydryl thiolate sites that are often present on cysteine residues within proteins.84 This becomes even more critical given that the catalytic centers of many key enzymes harbor cysteine residues, implying that adduct formation on such residues is highly detrimental to many key cellular processes. The following sections summarize specific types of DNA adducts associated with aldehyde exposure. The best-described adducts for each of the aldehydes listed below are illustrated in Figure 3.
Figure 3.
Chemical structures of the major DNA adducts associated with common environmental and endogenous aldehydes. All adducts are shown on deoxyribonucleosides (nucleobase-linked sugar moiety labeled D in all the above figures). dA, dG, and dC in adduct names indicate whether the nucleoside is in deoxyriboseadenosine, deoxyriboseguanosine, or deoxyribosecytidine, respectively. Refer to the section Aldehyde Adducts and the Genome for details.
1. Acetaldehyde Adducts
Acetaldehyde reacts with the N2 group on deoxyguanosine bases to make an unstable adduct N2-ethylidene-deoxyguanosine.85 Detection of this adduct in vitro requires a subsequent reduction step to convert it to a more stable N2-ethyl-deoxyguanosine. Recent studies have shown that the reduced form is also detected in human tissues in oral mucosa, urine, and leukocytes from drinkers’ samples.86,87 Additionally, N2-ethyl-deoxyguanosinehas been biochemically shown to stall elongation by the replicative polymerase Pol α but not the translesion polymerase Pol η.88,89 Furthermore, aldh2–/– mice that were exposed to ethanol had elevated levels of N2-ethylidene-dG compared to wildtype mice.90
Interaction of acetaldehyde with an intermediate adduct in the presence of positively charged amino groups, such as those present on the basic amino acid lysine, can make DNA–DNA or DNA–protein cross-links, both of which are highly detrimental to genome stability.91−93 Hodskinson et al. elegantly showed using Xenopus egg extracts that when DNA is treated with acetaldehyde to produce hydroxy-PrdG adducts (see Figure 3 for example structures). The resulting interstrand cross-links are efficiently repaired by two separate repair pathways, including the Fanconi anemia pathway.94 High-throughput sequencing showed that such adducts lead to mutagenic repair across the damaged guanine base, leading to C → A and C → T substitutions in a Rev1-dependent manner.94 Besides acetaldehyde, other types of aldehydes such as crotonaldehyde and malondialdehyde can form PrdG adducts upon reacting with DNA.94−97 To a lesser extent acetaldehyde can also react with deoxyadenosine and deoxycytosine residues, as measured by reverse-phase HPLC analysis, to produce lower yield adducts Aa-dAdo and Aa-dCyd, respectively.85
2. Formaldehyde Adducts
Formaldehyde is a potent cross-linking agent through its ability to form methylene bridges between the exocyclic amino groups of nucleoside bases like deoxyguanosine and the N/S groups in the side chains of amino acids like lysine, histidine, and cysteine.98,99 DNA–protein cross-links (DPCs) formed in this manner are relatively stable and major impediments to genome stability.92,100 Specialized proteases have evolved to digest DPCs and protect the genome in lower and higher eukaryotes, and their absence has been shown to make cells more susceptible to DNA damage by formaldehyde.101−103 In addition to cross-links, formaldehyde can make hydroxymethyl DNA adducts with N2 of deoxyguanosine residues (N2-HOMeG) and with N6 of deoxyadenine residues (N6-HOMeA).104,105 Because hydroxymethyl adducts are unstable, their in vitro analysis relies upon a reduction step with sodium cyanoborohydride (NaBH3CN) to generate stable methyl-deoxynucleosides.106,107 Consistent with tobacco smoke being a primary source of exogenous formaldehyde, methyl deoxyadenosine adducts are greatly enriched in DNA samples from smokers compared to nonsmokers.108 Notably, mice lacking ALDH2 and ADH5 dehydrogenase aldehyde genes had 10-fold higher levels of blood formaldehyde and >20-fold increase in N2-MeG in tissue samples including brain, kidney, and liver compared to wildtype mice.32
3. Methylglyoxal and Glyoxal Adducts
Methylglyoxal (MG) and glyoxal belong to a class of α-oxoaldehydes that are produced as endogenous byproducts of sugar metabolism and from other dietary sources such as meats and high-sugar snacks.109,110 Both molecules react with and irreversibly modify various biomolecules. In particular, the sequential modification of arginine residues on proteins by these compounds (referred to as glycation) generates advanced glycation end products (AGEs), which reduce the cellular half-life of proteins and greatly destabilize the proteome (reviewed in refs (111 and 112)). As such, AGEs contribute to several pathologies including neurodegenerative diseases like Parkinson’s, diabetes, aging, cardiovascular disease, and cancer. Both methylglyoxal and glyoxal can react with DNA, and the resulting reactions have been found genotoxic and/or mutagenic in human cells, reporter systems, as well as Salmonella typhimurium.113,114 Yeast cells deleted for DNA damage repair genes RAD23 and RAD50 are sensitive to methylglyoxal.115 Carbonyl stress induced by methylglyoxal and glyoxal treatments was shown to cause DNA breaks and protein–DNA cross-links in human skin cells.116 The primary DNA adducts of MG are N2-(1-carboxyethyl)-2′-deoxyguanosine (CEdG) and N6-(1-carboxyethyl)-2′-deoxyadenosine and have been shown to induce genotoxicity in the form of single-strand breaks (Figure 3).117,118 Similarly, glyoxal reacts with dG and dC residues in DNA to make N2-carboxymethyl-deoxyguanosine (CMdG) and 5-glycolyldeoxycytidine (gdC) adducts.119 Further, glyoxal treatment increases G → C and G → T substitutions in reporter systems in mammalian cells.120 Interestingly, wildtype Escherichia coli strains treated with methylglyoxal exhibited a higher mutation frequency and a different spectrum from uvrC strains that were deficient in nucleotide-excision repair; this phenomenon may be explained by the pro-mutagenic nature of NER under conditions of oxidative stress due to erroneous gap filling by DNA Pol I.121 Finally, methylglyoxal treatment inflicted greater DNA damage in cells deficient in folic acid122 and is able to induce chromosomal instability even at low concentrations,123 which suggests that the genotoxicity of oxoaldehydes is further elevated from errors in metabolism.
4. Malondialdehyde Adducts
Malondialdehyde (MDA) has been shown to make adducts with deoxyguanosine, deoxyadenosine, and deoxycytidine molecules. Pyrimido[1,2α] purin-10(3H)-one (M1G) is the major adduct formed by the reaction between MDA and dG (Figure 3). Other adducts such as N6-(3-oxo-propenyl) deoxyadenosine (M1A) are also formed, although less frequently.124 Additionally, the enol moiety on MDA uniquely allows it to make oligomeric adducts, although these are not typically observed under physiological conditions.124 MDA adducts have been shown to be mutagenic in assays in E. coli using a lacZα containing M13 shuttle vector, whereby elevated mutagenicity correlated with increasing concentrations of MDA.125 The predominant mutation class observed was single-base substitution, with G → T transversions as the main mutation type, along with G → A transitions and frameshift mutations. Similar studies done with genome-incorporated adducts showed M1G-associated mutagenicity in E. coli.126 Human studies have shown that M1G is present in transplant liver samples127 and breast tissue.128 Finally, dietary fat intake has been correlated with increased MDA-associated adducts in humans.86
5. Crotonaldehyde Adducts
The principal DNA adducts of crotonaldehyde are enantiomers of α-methyl-γ-hydroxypropanodeoxyguanosine (α-R-methyl-γ-HOPdG and α-S-methyl-γ-HOPdG) (Figure 3).129 These adducts are mutagenic due to their ability to inhibit topoisomerase-I mediated DNA cleavage,130 and by generating interstrand cross-link formation in a CG context.131 Using a single-stranded shuttle vector system containing ligated oligonucleotides with crotonaldehyde-derived adducts, it was shown that in mammalian cells both stereoisomers of the adduct were equally mutagenic, with G→ T transversions being the main mutation type, as evaluated via autoradiography.132 However, these adducts are predominantly reported to adopt an open-ring confirmation in duplex DNA, which favors correct base pairing and could account for their low mutagenicity compared to other aldehyde adducts.131
6. 4-HNE Adducts
Peroxidation of linoleic acid generates 4-hydroxy-2-nonenal (4-HNE), which is an oxygenated alkenal and a potent biomarker of oxidative stress.133 Much like the short-chain aldehydes, reactions involving alkenals and DNA bases result in exocyclic adducts, the most common being cyclic 1,N2-dG (Figure 3).134 Two out of the four reported stereoisomers of 4-HNE-derived adducts are more mutagenic in human cells but not strongly miscoding, resulting in G → N base substitutions (N = T, C, or A) on tandem (GG) bases.135,136 Urine samples from hepatitis B virus (HBV)-infected patients of chronic liver diseases had >70 fold higher concentrations of secreted ethenobase adducts such as the 4-HNE associated adduct N6-etheno-2′-deoxyadenosine, suggesting that robust inflammation and lipid peroxidation may contribute to aldehyde-mediated DNA damage in the cells of such patients.137 Mouse embryonic fibroblasts deficient in the Y-family error-prone polymerase iota (Pol ι) are sensitized to 4-HNE treatment, suggesting that Pol ι is required for the efficient bypass of 4-HNE-derived genomic lesions.138
7. 4-ONE Adducts
Linoleic acid hydroxyperoxides are broken down to oxynonenals. These aldehydes are highly electrophilic; they can undergo Michael-type addition reactions with sulfhydryl groups, such as those present on GSH, to form Schiff bases or react with nucleobases, creating etheno adducts such as heptanone-etheno-2′-deoxycytidine (HεdC) (Figure 3).139−142 The occurrence of such adducts could be as frequent as 100 adducts/108 bases, as suggested by a liquid chromatography-based adductome analysis of >60 autopsy samples from cardiopulmonary, hepatic, and gastrointestinal tissues of deceased individuals.142
8. Benzaldehyde and Cinnamaldehyde Adducts
Despite evidence that aldehydes are respiratory irritants, flavoring agents such as benzaldehyde and cinnamaldehyde are widespread in e-cigarettes, present in >70% and >50% of all e-cigarettes, respectively.143 Toxicological assessments show that both agents are highly cytotoxic and genotoxic.144,145 Intriguingly, several studies show an apparent antimutagenic activity for cinnamaldehyde in the presence of other mutagens, although it is possible that such effects are dependent on various extraneous factors such as the metabolic state of the cell, genetic background, and varying repair mechanisms.146−148 Although benzaldehyde adducts are poorly understood in the literature, in silico models predict possible adduct formation by cinnamaldehyde on N2-dG residues, creating exocyclic substituted N2-PrdG adducts (Figure 3).149,150
9. Acrolein Adducts
The major DNA adducts of acrolein are α- and γ-OH-PdG (α- and γ-hydroxy-1,N2-propano-2′-deoxyguanosine (Figure 3). The adducts are mutagenic and capable of forming both DNA interstrand cross-links as well as DNA–protein cross-links.151−155 Moreover, regions of the genome enriched in guanine residues, such as CpG sites, preferentially act as mutational hotspots for acrolein-association especially where such sites are methylated.156
Mutagenicity of Aldehydes
The electrophilic properties of alkanals and alkenals make them particularly strong mutagens because of the ease with which they react with nucleobases. The resulting adducts are remarkably stable and can impede genome stability in a variety of ways, including replication fork stalling, sister chromatid exchanges, interstrand cross-links, and DNA single- and double-stranded breaks.3,157 While such phenotypes provide a good measure of bulk genome instability, genotoxic agents often exert more subtle effects in the form of point mutations or small insertions and deletions (InDels). Genomes exposed to toxic agents often gradually accumulate such mutations and can result in distinct mutational signatures, which serve as molecular imprints of the past or prevailing insults faced by genomes (reviewed in ref (158)). A summary of studies assessing aldehyde mutagenesis is presented below. In addition, these studies are listed in Table 1, along with the proposed mechanism(s) underlying mutagenesis by the listed aldehydes.
Table 1. Studies Exploring Aldehyde-Associated Mutagenesisa.
| agent | system | chemical agent | reporter | mutagenicity | sequencing | signature | proposed mechanism | ref |
|---|---|---|---|---|---|---|---|---|
| acetaldehyde (AA) | SV-40 transformed fibroblasts | direct AA treatment of reporter plasmid | supF tRNA gene | yes | Sanger | GG → TT (CC → AA) | ICL formation impaired NER | (159) |
| peripheral T lymphocytes | AA | HPRT | yes | Sanger | G → A | transcription-associated NER | (161) | |
| HEK 293 cells | N2-,O6-dG adduct-containing oligos | supF tRNA gene | yes | Sanger | G:C → A:T | error-prone TLS, postreplicative MMR | (163) | |
| human XPA cells | site-specific plasmid-borne adduct | bsd resistance (survival) | yes | Sanger | G → T | ICL formation, TLS | (164) | |
| human fibroblasts | AA | TP53 | yes | Sanger | G → A, G → T | ICL formation, TLS | (165) | |
| mice | AA | HSCs | yes | WGS | aldh2–/–fancd2–/– cells had more base substitutions, microhomology (MH)-mediated deletions, rearrangements, likely stochastic damage | (168) | ||
| human iPSC | AA | no | WGS | DNA damage response induction without mutagenesis | (169) | |||
| Saccharomyces cerevisiae | AA, EtOH | CAN1 | none for AA | Sanger | increased C → T with EtOH | EtOH-induced replication stress, followed by error-prone DNA repair but no AA-specific mutations | (197) | |
| Xenopus egg extracts | AA-ICL adduct plasmid replication | yes | high-throughput sequencing | G → T | Rev1, pol ζ-mediated TLS, ICL repair via FA pathway | (94) | ||
| S. cerevisiae | AA | CAN1 ADE2 | yes | WGS | gCn → gAn (nGc → nTc), ssDNA-specific | TLS mediated by pol ζ, ssDNA-specific signature also observed in alcohol-associated cancers | (170) | |
| S. cerevisiae | AA | CAN1 | yes | WGS | C:G → T:A, T:A → C:G, small deletions | likely TLS | (171) | |
| formaldehyde (FA) | S. cerevisiae | FA | CAN1, lys2ΔA746, NR | yes | Sanger | large deletions in direct repeats, complex insertions | lesion bypass by NER, Pol ζ-mediated TLS | (172) |
| mice | methanol | endogenous N2-hydroxymethyl-dG | DNA damage | increased yH2AX, p53 induction | FA cross-link repair pathway and ADH5 mediated protection | (99) | ||
| human iPSC | FA | no | WGS | no effect on DDR pathway or mutagenesis | (169) | |||
| mice | FA | yes | WGS (HPSCs) | C → T, T → A | SBS signatures 3, 5, 25, 40 observed. age-associated damage, FA pathway defect | (32) | ||
| FA SCCs | likely elevated aldehyde levels from smoking, alcohol | yes | PacBio, WGS | multiple COSMIC SBS and ID signatures | CIN is high in FA cells, likely drives mutagenesis, complex SV formation | (186) | ||
| S. cerevisiae | FA | CAN1 | yes | WGS | C:G → T:A, T:A → C:G, no indels | likely TLS | (171) | |
| acrolein (Acr) | S. typhimurium | Acr | S9 fraction enzyme activation | yes | (175) | |||
| COS-7 cells | pMS2 shuttle vector with adduct (γ-HO-PdG) | yes | Sanger | G → T, G → A, G → C | lesion bypass by error-prone polymerases, DNA:DNA, DNA: protein cross-links | (176) | ||
| NHLF cells | Acr | supF, p53 | yes | Sanger | G → T, G → A, G → C | Acr in cigarette smoke can mutate cancer drivers, NER-mediated bulky lesion repair | (153) | |
| mouse embryonic fibroblasts, human XPA fibroblasts | Acr | cII transgene, supF | no | dose-dependent, chromosome context-dependent mutagenicity of Acr, error-free lesion bypass | (198) | |||
| human CCL-202 lung fibroblasts | Acr | supF | yes | Sanger | G → T, G → A, G → C | Acr mutations scale proportionately with Acr-DNA adducts, NER-dependent repair. | (177) | |
| human iPSC | Acr | no | WGS | no effect on DDR pathway or mutagenesis | (169) | |||
| oxoaldehydes | E. coli | methylglyoxal | lacI | yes | Sanger | G:C → C:G, G:C → T:A | NER-dependent repair | (181) |
| COS-7 | methylglyoxal | supF | yes | Sanger | G:C → C:G, G:C → T:A | NER-dependent repair | (114) | |
| human fibroblasts (XP+ and XP−) | CEdG-adducted shuttle vector | supF | yes | Sanger | A:T → G:C | NER-dependent adduct removal and DNA repair | (182) | |
| COS-7 | glyoxal | supF | yes | Sanger | G:C → C:G, G:C → T:A | NER-dependent repair | (120) | |
| crotonaldehyde | COS-7 cells | N2-,O6-dG adduct-containing vector | GFP vector (transfection efficiency) | yes | probe hybridization | G → T | replication blockage, NER | (132) |
| human XPA-cells | shuttle vector with site-specific ICL | blasticidin S | yes | probe hybridization | G → T | NER-independent repair at replication forks, TLS | (199) | |
| malondialdehyde (MDA) | E. coli | ssDNA M13 vector replication | lacZ alpha forward mutation | yes | Sanger | G → T, A → G, C → T | (125) | |
| M1G adduct in ssDNA vector | yes | probe hybridization | G → A, G → T | NER, adduct blocks replication fork passage | (126) | |||
| human fibroblasts | MDA-treated pSP189 shuttle vector | supF reporter mutations | yes | Sanger | small indels, GC → AT, GC → TA, GC → CG | NER | (200) | |
Refer to main text for details of reporter systems and additional references.
Acetaldehyde Mutagenesis
Early studies of acetaldehyde mutagenicity came from analyzing the replication of acetaldehyde-treated plasmids carrying the supF tRNA reporter gene in human fibroblasts. Using both single- and double-stranded plasmids, tandem-base substitutions were found to be the predominant mutation type, especially GG → TT transversions.159 Based on these studies, in liver cancers, a dinucleotide base signature was identified and attributed to acetaldehyde exposure, comprised primarily of CC → AA changes and additionally lower levels of CC → AG and CC → AT changes.160 Interestingly, CC → AA mutations are the predominant double-base substitutions present in the COSMIC signature DBS2, which is widely observed in tobacco-smoking associated lung cancers.160 Whether this mutation subtype is linked to acetaldehyde exposure in this signature has not been formally explored but nevertheless remains a tantalizing possibility. In another study, T lymphocytes treated with acetaldehyde reported a higher mutation frequency in the HPRT gene.161 Like the prior study, G → A transitions as well as A → T transversions were identified, with a preference for a 5′-AAG-3′ or 5′-AGG-3′ motif. Both the above studies are consistent with N2-Eth-dG-based mutagenesis. Interestingly, mutations in the TP53 gene isolated from esophageal carcinoma patients display a preponderance of G → A transitions, with more mutations associated with smokers and drinkers.162 Such studies provided an early indication that acetaldehyde-induced mutagenesis might lie at the core of carcinogenesis associated with alcohol and smoking. Several other studies using similar reporters have corroborated the predominant in vivo mutation spectrum of acetaldehyde to be comprised of mutations in guanines (Table 1).163−165
An obvious caveat to the above studies is their reliance on a single gene or reporter to measure mutations. Several studies have shown that mutation frequencies and spectra can greatly vary with several factors including cell type, replication timing, local chromatin context, and the presence of nearby DNA-bound transcription factors (reviewed in ref (166)). Moreover, the dynamic interplay between DNA damage and DNA repair processes can further determine mutagenicity.160,167 Therefore, whole-genome analysis provides an unbiased view of the full landscape of mutations associated with a given mutagen. In an elegant study, somatic mutation loads of engrafted hematopoietic stem cells from the bone marrow of wildtype and aldh2–/–fancd2–/– mice were compared.168aldh2–/–fancd2–/– mice displayed an increased frequency of base substitutions, rearrangements, and indels and displayed evidence of stochastic DNA damage. However, low mutation loads in this study precluded accurate mutation signature analysis. Follow-up mechanistic studies revealed that cells rely on a two-step response to mitigate aldehyde-associated damage—first via an excision repair-dependent pathway that removes and repairs interstrand DNA cross-links (ICLs) and second a mutagenic translesion synthesis pathway that requires DNA replication and involves cutting within the cross-link itself to unhook the ICL.94 Using Xenopus egg extracts, researchers monitored the replication of plasmids carrying acetaldehyde-derived cross-links and showed that both the above repair pathways were mutagenic. Unlike prior reports showing G → A transitions, the Xenopus experiment G → T transversions were the most common base substitutions. In agreement with the role of excision repair in mutagenesis, ablation of the FA pathway via p97 inhibition removed this mutagenic signature, while depletion of the TLS factor REV1 inhibited bypass of the ICL lesion in a strand-specific fashion.94 On the other hand, acetaldehyde treatment elicited a DNA damage response without any associated mutagenesis in vivo with human-induced pluripotent stem cells (iPSCs) (Table 1).169
Recent work from our group demonstrated that acetaldehyde is highly mutagenic; however, its primary genomic substrate is single-stranded DNA (ssDNA).170 In line with earlier studies, mutagenesis was found to be TLS-dependent and gave rise to a preponderance of C → A (G → T) base substitutions. Further, we observed that the ssDNA-associated mutations were present in a gCn (nGc) motif, which revealed a ssDNA-specific gCn → A (nGc → T) mutation signature for acetaldehyde (Table 1).170 Even more remarkably, mutation loads in this genomic context were enriched in whole-genome- and whole-exome-sequenced cancers associated with smoking/alcohol, especially liver cancers. Interestingly, the enriched mutations observed in cancers strongly associated with the nontranscribed strand of genes, which suggests that ssDNA formed during increased transcription in cancers likely acts as an ideal substrate for acetaldehyde-induced mutagenesis.170 A related study demonstrated the mutagenesis of ssDNA in response to acetaldehyde, albeit with different base substitutions, i.e., more C → T and T → A changes; interestingly, acetaldehyde treatment led to an increase in the proportion of ssDNA-associated deletions of ≥5 bp but without any associated microhomology at break points.171
Formaldehyde Mutagenesis
In budding yeast, formaldehyde-mutated CAN1 mutants and lys2 frameshift revertants were sequenced and found to contain frameshifts consisting of NER-dependent large deletions as well as complex insertions in hotspots of the LYS2 gene. Comparison of mutational spectra from strains lacking REV3, RAD30, or RAD14 showed that these complex mutations are dependent on NER as well as mutagenic bypass via Pol ζ-mediated translesion synthesis.172 A recent study demonstrated that formaldehyde, like acetaldehyde, was mutagenic to ssDNA in yeast. Formaldehyde treatment generated C → T and T → A transversions and revealed a mutational signature akin to COSMIC SBS40, which is a remarkably common signature among most cancer types. However, unlike acetaldehyde, no appreciable increase in indels was observed with formaldehyde exposure (Table 1).171
Beside yeast, mice serve as excellent models for aldehyde-associated human diseases such as Fanconi anemia and Cockayne syndrome, owing to the conservation of hematopoietic cell lineages and physiological aldehyde response systems between rodents and humans.32,94,168,173,174 In mice lacking CSB5 (ERCC6), an excision repair gene, and ADH5, which is involved in endogenous formaldehyde clearance, formaldehyde-associated adducts (N2-methyl-dG) are elevated in kidneys and brain, and cells display transcriptional stress.50 In response to methanol treatment, adh5–/– mice accumulated DNA damage, including increased frequency of SCE events.32 Further, γH2AX levels are elevated in hematopoietic stem cells from adh5–/–fancd2–/– mice, indicating DNA damage, with animals displaying sensitivity to exogenous methanol. aldh2–/–adh5–/– mice display sensitivity to acetaldehyde and formaldehyde and confer a variety of deficiencies including lymphoid cancers. DNA from several organs of aldh2–/–adh5–/– mice also showed increased N2-meth-dG adduction, with bone marrow from these animals displaying a >two-fold increase in both single-base substitutions and indels. Whole-genome sequencing of hematopoietic stem and progenitor cells (HSPC) from bone marrow of aldh2–/–adh5–/– mice show increased formaldehyde-associated base substitutions, with a predominant T → A and T → C bias, and a preference for adenines on the transcribed strand.32fancd2–/–aldh2–/– mice have similarly been utilized to model the pathogenesis of fetal alcohol syndrome, which is related to maternal alcohol consumption.173
Acrolein Mutagenesis
Sublethal doses of acrolein were shown to be mutagenic in S. typhimurium assays.175 Subsequent work with human cell lines similarly noted acrolein-associated mutagenesis in reporter genes. Plasmids incorporated with acrolein-derived γHOPdG adducts permitted relatively efficient elongation by replicative polymerases in human COS-7 cells, albeit with a small percentage of predominantly G → T base substitutions.176 Other studies examined the impact of acrolein-derived adducts on TP53 mutations in lung fibroblasts and observed that the site of adduct formation within TP53 correlated with mutational hotspots that are commonly seen with smoking-associated lung cancer.153supF reporter analyses show that G → T transversions are greatly enhanced in acrolein-treated samples (Table 1).153,177 However, acrolein was not found to be mutagenic in a whole-genome analysis of treated human iPSC cells nor did it elicit a DNA damage response.169 Therefore, the molecular mechanism(s) underlying acrolein-induced mutagenicity and any associated mutation signatures are still relatively unclear and warrant further investigation.
Mutagenicity of Oxoaldehydes
Endogenous metabolic processes such as lipid peroxidation and glycolysis can produce oxoaldehydes such as methylglyoxal and glyoxal, which can rapidly cross-react with amino acid side chains and give rise to advanced glycation end products (AGEs). In addition to proteins, studies have shown oxoaldehydes to react with nucleobases, chiefly guanosine and produce adducts.178−180 Even so, the biological relevance and mutagenicity of such adducts is an open question. Initial studies in E. coli and human COS-7 cells showed that methylglyoxal can induce G → C and G → T mutations and 20–300 bp deletions.114,181 Reporter gene studies with glyoxal yielded similar results.120 In subsequent studies, the primary methylglyoxal adduct N2-(1-carboxyethyl)-2′-deoxyguanosine (CEdG) led to increased mutation frequencies, with G:C → T:A transverions being the major mutation type.182 Interestingly, although repair-deficient cells had a higher mutation frequency, they nevertheless had an unchanged mutation spectrum compared to repair-proficient cells. A role for the Y-family error-prone polymerase Pol iota (pol ι) in the gap-filling repair step of NER has been suggested as a possible mechanism for the observed mutation spectrum.182,183In vitro studies also suggest frequent purine incorporation opposite CEdG adducts.184 Treatment of keratinocytes and fibroblasts suggests that distinct DNA damage can accumulate in response to glyoxal and methylglyoxal. While the former was seen to induce a higher frequency of double-strand breaks, the latter caused a higher incidence of DNA–protein cross-links.116 Glyoxal and methylglyoxal have also been shown to make imidazopurinone derivatives with deoxyguanosine bases, which have been shown to increase the frequency of double-strand breaks in HL60 cells treated with methylglyoxal185 and might contribute to differential oxoaldehyde-associated mutagenesis.
Similar reporter-based mutagenesis studies have been performed for other aldehydes and are summarized in Table 1.
Other Mutational Events Associated with Aldehyde Exposure
Finally, recent work explored the mutational landscape of squamous cell carcinoma patients with defects in the Fanconi anemia pathway, which is characterized at the molecular level by increased DNA interstrand cross-links (ICLs), a common occurrence in response to toxic aldehydes. The major mutational event in these cells was chromosome instability in the form of large structural variants in the range of 1–100 kb and MMEJ-mediated rearrangements. Such events are thought to contribute to overall oncogenic activation and provide insights into how genomes exposed to toxic aldehydes through smoking, drinking, or other environmental pollutants undergo mutagenesis and accumulate genome instability.186
A General Model for Aldehyde-Associated Genome Instability and Mutagenesis
Historically, the lesions most associated with aldehyde exposure are DNA:DNA cross-links (ICLs) as well as DNA:protein cross-links.92,94,102,154,159,187 Unless resolved, such lesions can destabilize the genome in multiple ways, including stalling replication forks, blocking transcription, and initiating double-strand breaks. We showed that in yeast single-stranded DNA, while acetaldehyde exposure resulted in an uptick in G → T mutations, there was no substantial enrichment for GG → TT double-base substitutions, which is the preferred genomic context for acetaldehyde-induced intrastrand cross-links.159 This implies that additional mechanisms could drive aldehyde-associated mutagenesis.
Surveys of aldehyde-associated mutagenesis listed so far seem to share certain commonalities. The overall mutation spectra look remarkably similar, with guanines being the preferred base for adduction, and NER appears to be the preferred pathway for minimizing aldehyde-associated DNA damage (Table 1). Although such observations are not exclusive to aldehydes, they nevertheless hint at a potential mechanistic link through which different aldehydes target the genome and stimulate mutagenesis.
One such link could be mutagenesis of single-stranded DNA. The genome is transiently single-stranded during several processes such as DNA replication, transcription, and DNA repair (Figure 4). ssDNA is highly mutagenizable due to the lack of any associated repair pathways. Work from our lab and others has shown that ssDNA is mutagenized by both formaldehyde and acetaldehyde in yeast, which raises the possibility of other aldehydes following a similar mechanism. If ssDNA is in fact the preferred genomic substrate for aldehydes, perhaps that would explain why most prior research has yielded mixed results on classifying aldehydes as mutagenic in vivo. Most studies using cell lines or mouse models would not have sufficient ssDNA formed as the substrate for aldehyde mutagenesis, leading them to conclude somewhat erroneously that the given aldehyde is not mutagenic. When genomes are repeatedly exposed to toxic aldehydes, the presence of vulnerable substrates (e.g., ssDNA) would enhance aldehyde genotoxicity, generating several types of lesions (such as cross-links) and bulky adducts. Erroneous processing of such lesions (e.g., in repair-deficient genetic backgrounds) would result in marked genome instability in the form of replication/transcriptional stress, accumulation of mutations, and widespread DNA damage, which would ultimately contribute to diseases such as carcinogenesis. (Figure 4). In fact, most cancers are marked by replication stress, replication–transcription collisions and R-loop formation, and hypertranscription, all events that generate copious ssDNA.188,189 As such, mutations in ssDNA regions could represent a genomic record of aldehyde exposures, such as what we and others have demonstrated.170,171
Figure 4.
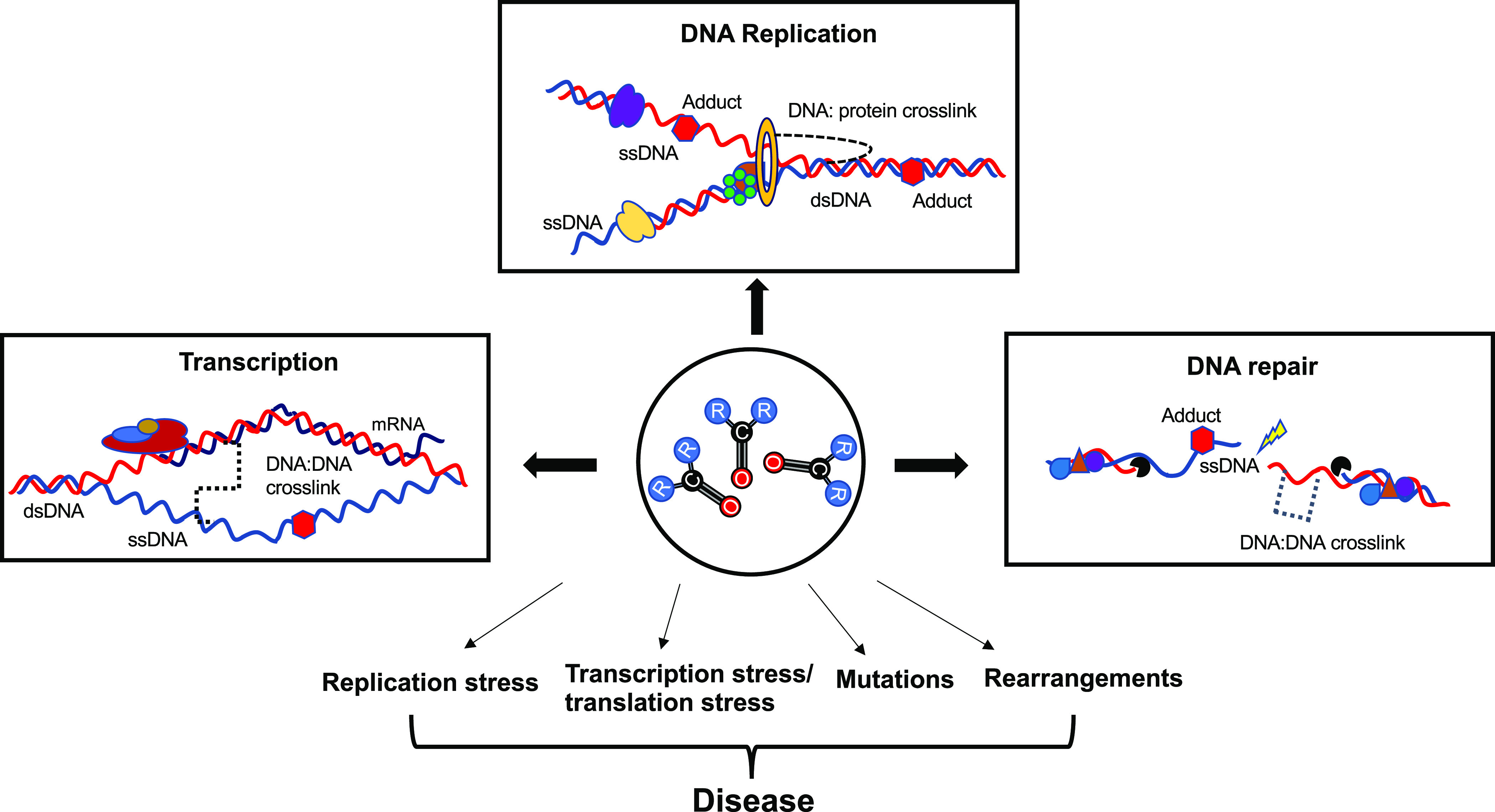
Mechanisms of aldehyde-associated genome instability. Major genome-associated pathways are illustrated in open boxes. All the listed processes involve unwinding of the double helix, leading to the generation of single-stranded DNA (ssDNA). In the presence of reactive carbonyls (ball and stick molecules), open circle, genomes can accumulate a variety of lesions on both ssDNA as well as double-stranded DNA (dsDNA), including DNA:protein and DNA:DNA cross-links (broken connectors) and adducts (red hexagons). Failure to repair such lesions or erroneous bypass can result in severe genome instability, which can contribute to aldehyde-related diseases.
Conclusions and Future Perspectives
Decades of elegant research have led to the discovery of the main adducts of several environmental and endogenous aldehydes and shaped the field of aldehyde mutagenesis. However, several challenges remain. First, a direct cause-and-effect relationship between aldehyde-associated mutagenesis and the offending adduct is not straightforward. Mutagenesis could result from the processing of either DNA adducts or a combination of DNA, RNA, and protein adducts. Second, depending on the cell type, local genomic context, DNA accessibility, and variable DNA repair efficiencies, aldehydes could be processed to one or several adducts in vivo. Third, different adducts might have varying degrees of stability in vivo, as well as strand preferences. Consequently, it is possible to have varying degrees of mutagenesis even with the same aldehyde. Discrepancies between in vivo and in vitro adduct chemistries could partly explain why reports of mutagenesis for several aldehydes drastically vary between different studies (Table 1).
Several unresolved questions remain that would help broaden our understanding of aldehyde-associated mutagenesis. For instance, is single-stranded DNA the preferred in vivo genomic substrate for other aldehydes? What are the determinants of the genomic distribution of aldehyde-associated DNA adducts? Are there other specialized repair pathways that are dedicated to specific aldehydes, much like acetaldehyde-associated FANCJ? Depending on the cell or tissue type, metabolic state, and genetic background, do aldehydes have multiple mutation signatures? Do aldehydes broadly damage the genome, or are there damage and mutational hotspots? On a related note, can localized aldehyde-associated damage generate clustered mutations (kategis) that are often a hallmark of many different types of cancers?
Exploring the above themes can help determine if certain genomic contexts are “at-risk” of aldehyde-associated damage and allow us to identify which genomic lesions are mutagenic.
A combination of cutting-edge adduct-mapping techniques and high-throughput sequencing now provides researchers means to extensively survey the genotoxic impacts of aldehyde exposure. Precision methods, such as LC-NSI-HRMS-based mass spectrometry, have been successfully used for in vivo mapping of acrolein-derived adducts.190 Similar applications would enable the generation of high-resolution adductome maps, predict which chemicals acted as precursors to the adducts, and can eventually help trace back to the source environmental chemicals that are responsible for genotoxicity.187,191 Similarly, newer sequencing technologies can help define a much broader mutational landscape for a given aldehyde. Newer versions of single-molecule real-time (SMRT) sequencing improve significantly upon their predecessors to provide single-base resolution of DNA damage with multiple types of modified bases such as alkylated purines (O6-methyl guanine, 6-methyladenine), hydroxylated bases (e.g., 5-hydroxycytosine), and UV-damaged cyclobutene dimers and can be potentially leveraged to study aldehyde-derived adducts.192,193 Similarly, PCR-free amplification methods like nanopore sequencing (NPS) are greatly suited to adduct mapping. NPS has been used for mapping a wide range of adducts associated with many common exogenous genotoxins.194−196 Finally, elegant computational pipelines, including NMF- and/or knowledge-based signature analyses, and machine learning/AI can refine the key genomic coordinates that drive mutagenesis and lead to development of prognostic genomic biomarkers for aldehyde exposure, thus greatly informing on therapeutics and leading to better health outcomes.
Aldehyde exposure is intricately tied to human health. From a clinical standpoint, it is imperative to explore research avenues that clearly outline the genomic risk factors for aldehyde-associated toxicity to formulate better strategies for health risk mitigation. Although significant advances have been made toward determining aldehyde genotoxicity, these only represent the tip of the iceberg. As such, several additional avenues should be explored to comprehend the full spectrum of aldehyde-associated genome instability. Such efforts are vital for providing well-defined, publicly available data sets that link distinct aldehydes to specific mutational patterns and eventually diseases.
Acknowledgments
We would like to thank T. Blouin, J. McCollum, and A. Ruggiero for their critical reading of the manuscript and helpful feedback.
Glossary
Abbreviations
- 4-HNE
4-hydroxynonenal
- 4-ONE
4-oxynonenal
- γ-HOPdG
γ-hydroxy-1,N-2-propano-2′-deoxyguanosine
- β-HOPdG
β-hydroxy-1,N-2-propano-2′-deoxyguanosine
- AA
acetaldehyde
- AI
artificial intelligence
- Acr
acrolein
- AGE
advanced glycation end product
- ALS
amyotrophic lateral sclerosis
- CEdG
N2-(1-carboxyethyl)-2′-deoxyguanosine
- CMdG
N2-carboxymethyl-deoxyguanosine
- COSMIC
Catalog of Somatic Mutations in Cancers
- CPD
cyclobutane pyrimidine dimers
- CRC
colorectal cancer
- CVD
cardiovascular disease
- CYP450
cytochrome P450
- DNA
deoxyribonucleic acid
- DOPAL
3,4-dihydroxyphenylacetaldehyde
- DPC
DNA–protein cross-link
- EMS
ethylmethanesulfonate
- FA
Fanconi anemia
- FAD
flavin adenine dinucleotide
- gdC
5-glycolyldeoxycytidine
- GSH
glutathione S-transferase
- H2AX
gamma H2AX
- HBV
hepatitis B virus
- HεdC
heptanone-etheno-2′-deoxycytidine
- HPLC
high-performance liquid chromatography
- HPRT
hypoxanthine-guanine phosphoribosyltransferase
- LC-NSI-HRMS/MS)
liquid chromatography-nanoelectrospray ionization-high-resolution tandem mass spectrometry
- HSPC
hematopoietic stem and progenitor cells
- IARC
International Agency for Research on Cancer
- ICL
inter/intrastrand cross-link
- InDels
insertions and deletions
- ISC
intestinal stem cells
- LC
liquid chromatography
- M1A
N6-(3-oxo-propenyl) deoxyadenosine
- M1G
pyrimido[1,2α]purin-10(3H)-one
- MG
methylglyoxal
- MDA
malondialdehyde
- MMEJ
microhomology-mediated end joining
- MMS
methylmethanesulfonate
- MS
mass spectrometry
- N2-Eth-dG
N2-ethylidenedeoxyguanosine
- N2-MeG
N2-hydroxymethyl-deoxyguanosine
- N6-MeA
N6-hydroxymethyl-deoxyadenosine
- NAD
nicotinamide adenine dinucleotide phosphate
- NER
nucleotide excision repair
- NMF
non-negative matrix factorization
- NMR
nuclear magnetic resonance
- NPS
nanopore sequencing
- PCR
polymerase chain reaction
- PDE
pyridoxine-associated epilepsy
- PrdG
1,N2-propanodeoxyguanosine
- iPSC
induced pluripotent stem cells
- RNA
ribonucleic acid
- ROS
reactive oxygen species
- SBS
single-base substitution
- SCE
sister chromatid exchange
- SMRT
single-molecule real time
- SNP
single-nucleotide polymorphism
- SNV
single-nucleotide variant
- ssDNA
single-stranded DNA
- TLC
thin layer chromatography
- TLS
translesion synthesis
- VOC
volatile organic compounds
Biographies
Sriram Vijayraghavan: Dr. Vijayraghavan received his Ph.D. from the University of Pittsburgh, where his research focused on the interplay of DNA replication and repair using yeast as a model system. He did his postdoctoral research at the Duke University Medical Center, North Carolina, studying mitochondrial DNA variation and RNA viruses in natural yeast populations, using a combination of genomics and molecular biology. He is currently a staff scientist in Dr. Natalie Saini’s lab at the Medical University of South Carolina, where he is interested in analyzing the genome-wide mutagenicity of various aldehydes, using yeast and tissue culture models.
Natalie Saini: Dr. Saini received her Ph.D. from the Georgia Institute of Technology, where she studied the impact of fragile DNA repeats on genome stability. She proceeded to do her postdoctoral work at the National Institute of Environmental Health Sciences, North Carolina, where her research focused on how the accumulation of somatic mutations impacts normal cells. She obtained a K99/R00 Pathway to Independence grant and transitioned to her current position as an Assistant Professor in the Department of Biochemistry at the Medical University of South Carolina, where she continues to study the impact of environmental and endogenous DNA damage on genome stability.
Author Contributions
CRediT: Sriram Vijayraghavan conceptualization, writing-original draft, writing-review & editing; Natalie Saini conceptualization, funding acquisition, resources, supervision, writing-original draft, writing-review & editing.
Research support to N.S. is through NIH grant 5R00ES028735–03 via the National Institute for Environmental and Health Sciences (NIEHS).
The authors declare no competing financial interest.
References
- LoPachin R. M.; Gavin T. Molecular mechanisms of aldehyde toxicity: a chemical perspective. Chem. Res. Toxicol. 2014, 27 (7), 1081–91. 10.1021/tx5001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien P. J.; Siraki A. G.; Shangari N. Aldehyde sources, metabolism, molecular toxicity mechanisms, and possible effects on human health. Crit Rev. Toxicol 2005, 35 (7), 609–62. 10.1080/10408440591002183. [DOI] [PubMed] [Google Scholar]
- Voulgaridou G. P.; Anestopoulos I.; Franco R.; Panayiotidis M. I.; Pappa A. DNA damage induced by endogenous aldehydes: current state of knowledge. Mutat. Res. 2011, 711 (1–2), 13–27. 10.1016/j.mrfmmm.2011.03.006. [DOI] [PubMed] [Google Scholar]
- Smith C. J.; Hansch C. The relative toxicity of compounds in mainstream cigarette smoke condensate. Food Chem. Toxicol. 2000, 38 (7), 637–46. 10.1016/S0278-6915(00)00051-X. [DOI] [PubMed] [Google Scholar]
- Hoffmann D.; Djordjevic M. V.; Hoffmann I. The changing cigarette. Prev Med. 1997, 26 (4), 427–34. 10.1006/pmed.1997.0183. [DOI] [PubMed] [Google Scholar]
- Wang P.; Chen W.; Liao J.; Matsuo T.; Ito K.; Fowles J.; Shusterman D.; Mendell M.; Kumagai K. A Device-Independent Evaluation of Carbonyl Emissions from Heated Electronic Cigarette Solvents. PLoS One 2017, 12 (1), e0169811 10.1371/journal.pone.0169811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogunwale M. A.; Li M.; Ramakrishnam Raju M. V.; Chen Y.; Nantz M. H.; Conklin D. J.; Fu X. A. Aldehyde Detection in Electronic Cigarette Aerosols. ACS Omega 2017, 2 (3), 1207–1214. 10.1021/acsomega.6b00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X.; Zeng X.; Xiao F.; Chen R.; Sinharoy P.; Gross E. R. E-cigarette aerosol exacerbates cardiovascular oxidative stress in mice with an inactive aldehyde dehydrogenase 2 enzyme. Redox Biol. 2022, 54, 102369. 10.1016/j.redox.2022.102369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facchinetti F.; Amadei F.; Geppetti P.; Tarantini F.; Di Serio C.; Dragotto A.; Gigli P. M.; Catinella S.; Civelli M.; Patacchini R. Alpha,beta-unsaturated aldehydes in cigarette smoke release inflammatory mediators from human macrophages. Am. J. Respir. Cell Mol. Biol. 2007, 37 (5), 617–23. 10.1165/rcmb.2007-0130OC. [DOI] [PubMed] [Google Scholar]
- Weng M. W.; Lee H. W.; Park S. H.; Hu Y.; Wang H. T.; Chen L. C.; Rom W. N.; Huang W. C.; Lepor H.; Wu X. R.; Yang C. S.; Tang M. S. Aldehydes are the predominant forces inducing DNA damage and inhibiting DNA repair in tobacco smoke carcinogenesis. Proc. Natl. Acad. Sci. U. S. A. 2018, 115 (27), E6152–E6161. 10.1073/pnas.1804869115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boffetta P.; Hashibe M. Alcohol and cancer. Lancet Oncol 2006, 7 (2), 149–56. 10.1016/S1470-2045(06)70577-0. [DOI] [PubMed] [Google Scholar]
- Chang J.; Tan W.; Ling Z.; Xi R.; Shao M.; Chen M.; Luo Y.; Zhao Y.; Liu Y.; Huang X.; Xia Y.; Hu J.; Parker J. S.; Marron D.; Cui Q.; Peng L.; Chu J.; Li H.; Du Z.; Han Y.; Tan W.; Liu Z.; Zhan Q.; Li Y.; Mao W.; Wu C.; Lin D. Genomic analysis of oesophageal squamous-cell carcinoma identifies alcohol drinking-related mutation signature and genomic alterations. Nat. Commun. 2017, 8, 15290. 10.1038/ncomms15290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letouze E.; Shinde J.; Renault V.; Couchy G.; Blanc J. F.; Tubacher E.; Bayard Q.; Bacq D.; Meyer V.; Semhoun J.; Bioulac-Sage P.; Prevot S.; Azoulay D.; Paradis V.; Imbeaud S.; Deleuze J. F.; Zucman-Rossi J. Mutational signatures reveal the dynamic interplay of risk factors and cellular processes during liver tumorigenesis. Nat. Commun. 2017, 8 (1), 1315. 10.1038/s41467-017-01358-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. C.; Wang M. Y.; Yang M.; Dai H. J.; Zhang B. F.; Wang W.; Chu X. L.; Wang X.; Zheng H.; Niu R. F.; Zhang W.; Chen K. X. A mutational signature associated with alcohol consumption and prognostically significantly mutated driver genes in esophageal squamous cell carcinoma. Ann. Oncol 2018, 29 (4), 938–944. 10.1093/annonc/mdy011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz H. K.; Stickel F. Molecular mechanisms of alcohol-mediated carcinogenesis. Nat. Rev. Cancer 2007, 7 (8), 599–612. 10.1038/nrc2191. [DOI] [PubMed] [Google Scholar]
- Zhang J.; Lioy P. J.; He Q. Characteristics of aldehydes: concentrations, sources, and exposures for indoor and outdoor residential microenvironments. Environ. Sci. Technol. 1994, 28 (1), 146–52. 10.1021/es00050a020. [DOI] [PubMed] [Google Scholar]
- Gaffney J. S.; Marley N. A. Atmospheric chemistry and air pollution. ScientificWorldJournal 2003, 3, 199–234. 10.1100/tsw.2003.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean D.; Grosjean E. Airborne carbonyls from motor vehicle emissions in two highway tunnels. Res. Rep. Health Eff. Inst. 2002, 107, 57–78. [PubMed] [Google Scholar]; Discussion 79–92.
- Jonsson A.; Persson K. A.; Grigoriadis V. Measurements of some low molecular-weight oxygenated, aromatic, and chlorinated hydrocarbons in ambient air and in vehicle emissions. Environ. Int. 1985, 11 (2), 383–392. 10.1016/0160-4120(85)90033-9. [DOI] [Google Scholar]
- LeBouf R. F.; Blackley B. H.; Fortner A. R.; Stanton M.; Martin S. B.; Groth C. P.; McClelland T. L.; Duling M. G.; Burns D. A.; Ranpara A.; Edwards N.; Fedan K. B.; Bailey R. L.; Cummings K. J.; Nett R. J.; Cox-Ganser J. M.; Virji M. A. Exposures and Emissions in Coffee Roasting Facilities and Cafes: Diacetyl, 2,3-Pentanedione, and Other Volatile Organic Compounds. Front Public Health 2020, 8, 561740. 10.3389/fpubh.2020.561740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob S. E.; Stechschulte S. Formaldehyde, aspartame, and migraines: A possible connection. Dermatitis 2008, 19 (3), E10-1. [PubMed] [Google Scholar]
- Katragadda H. R.; Fullana A.; Sidhu S.; Carbonell-Barrachina Á. A. Emissions of volatile aldehydes from heated cooking oils. Food Chem. 2010, 120 (1), 59–65. 10.1016/j.foodchem.2009.09.070. [DOI] [Google Scholar]
- Guillen M.; Uriarte P. Aldehydes contained in edible oils of a very different nature after prolonged heating at frying temperature: Presence of toxic oxygenated α,β unsaturated aldehydes. Food Chem. 2012, 131, 915–926. 10.1016/j.foodchem.2011.09.079. [DOI] [Google Scholar]
- Zhang Q.; Qin W.; Lin D.; Shen Q.; Saleh A. S. The changes in the volatile aldehydes formed during the deep-fat frying process. J. Food Sci. Technol. 2015, 52 (12), 7683–96. 10.1007/s13197-015-1923-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox P. J. Cyclophosphamide cystitis--identification of acrolein as the causative agent. Biochem. Pharmacol. 1979, 28 (13), 2045–9. 10.1016/0006-2952(79)90222-3. [DOI] [PubMed] [Google Scholar]
- Obach R. S.; Kalgutkar A. S.; Ryder T. F.; Walker G. S. In vitro metabolism and covalent binding of enol-carboxamide derivatives and anti-inflammatory agents sudoxicam and meloxicam: insights into the hepatotoxicity of sudoxicam. Chem. Res. Toxicol. 2008, 21 (9), 1890–9. 10.1021/tx800185b. [DOI] [PubMed] [Google Scholar]
- Golden R.; Valentini M. Formaldehyde and methylene glycol equivalence: Critical assessment of chemical and toxicological aspects. Regul. Toxicol. Pharmacol. 2014, 69 (2), 178–186. 10.1016/j.yrtph.2014.03.007. [DOI] [PubMed] [Google Scholar]
- Smith C. K.; Moore C. A.; Elahi E. N.; Smart A. T.; Hotchkiss S. A. Human skin absorption and metabolism of the contact allergens, cinnamic aldehyde, and cinnamic alcohol. Toxicol. Appl. Pharmacol. 2000, 168 (3), 189–99. 10.1006/taap.2000.9025. [DOI] [PubMed] [Google Scholar]
- Sinharoy P.; McAllister S. L.; Vasu M.; Gross E. R. Environmental Aldehyde Sources and the Health Implications of Exposure. Adv. Exp. Med. Biol. 2019, 1193, 35–52. 10.1007/978-981-13-6260-6_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barciszewski J.; Siboska G. E.; Pedersen B. O.; Clark B. F.; Rattan S. I. A mechanism for the in vivo formation of N6-furfuryladenine, kinetin, as a secondary oxidative damage product of DNA. FEBS Lett. 1997, 414 (2), 457. 10.1016/S0014-5793(97)01037-5. [DOI] [PubMed] [Google Scholar]
- Headlam H. A.; Davies M. J. Beta-scission of side-chain alkoxyl radicals on peptides and proteins results in the loss of side-chains as aldehydes and ketones. Free Radic Biol. Med. 2002, 32 (11), 1171–84. 10.1016/S0891-5849(02)00814-6. [DOI] [PubMed] [Google Scholar]
- Dingler F. A.; Wang M.; Mu A.; Millington C. L.; Oberbeck N.; Watcham S.; Pontel L. B.; Kamimae-Lanning A. N.; Langevin F.; Nadler C.; Cordell R. L.; Monks P. S.; Yu R.; Wilson N. K.; Hira A.; Yoshida K.; Mori M.; Okamoto Y.; Okuno Y.; Muramatsu H.; Shiraishi Y.; Kobayashi M.; Moriguchi T.; Osumi T.; Kato M.; Miyano S.; Ito E.; Kojima S.; Yabe H.; Yabe M.; Matsuo K.; Ogawa S.; Gottgens B.; Hodskinson M. R. G.; Takata M.; Patel K. J. Two Aldehyde Clearance Systems Are Essential to Prevent Lethal Formaldehyde Accumulation in Mice and Humans. Mol. Cell 2020, 80 (6), 996. 10.1016/j.molcel.2020.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y.; Edwards L. G.; Thornalley P. J. Effect of methylglyoxal on human leukaemia 60 cell growth: modification of DNA G1 growth arrest and induction of apoptosis. Leuk Res. 1996, 20 (5), 397–405. 10.1016/0145-2126(95)00162-X. [DOI] [PubMed] [Google Scholar]
- Marnett L. J. Oxy radicals, lipid peroxidation and DNA damage. Toxicology 2002, 181–182, 219–22. 10.1016/S0300-483X(02)00448-1. [DOI] [PubMed] [Google Scholar]
- Acetaldehyde. IARC Monogr. Eval. Carcinog. Risk Chem. Hum. 1985, 36, 101. [PubMed] [Google Scholar]
- Formaldehyde. IARC Monogr. Eval. Carcinog. Risk Chem. Hum. 1982, 29, 345–89. [PubMed] [Google Scholar]
- Smith C. J.; Perfetti T. A.; Rumple M. A.; Rodgman A.; Doolittle D. J. ″IARC group 2A Carcinogens″ reported in cigarette mainstream smoke. Food Chem. Toxicol. 2000, 38 (4), 371–83. 10.1016/S0278-6915(99)00156-8. [DOI] [PubMed] [Google Scholar]
- Wilbourn J.; Heseltine E.; Moller H. IARC evaluates wood dust and formaldehyde. International Agency for Research on Cancer. Scand J. Work Environ. Health 1995, 21 (3), 229–232. 10.5271/sjweh.1368. [DOI] [PubMed] [Google Scholar]
- Salaspuro M. Acetaldehyde and gastric cancer. J. Dig Dis 2011, 12 (2), 51–9. 10.1111/j.1751-2980.2011.00480.x. [DOI] [PubMed] [Google Scholar]
- Baan R.; Straif K.; Grosse Y.; Secretan B.; El Ghissassi F.; Bouvard V.; Altieri A.; Cogliano V. Carcinogenicity of alcoholic beverages. Lancet Oncol 2007, 8 (4), 292–293. 10.1016/S1470-2045(07)70099-2. [DOI] [PubMed] [Google Scholar]
- Seitz H. K.; Becker P. Alcohol metabolism and cancer risk. Alcohol Res. Health 2007, 30 (1), 38–41. 44–47. [PMC free article] [PubMed] [Google Scholar]
- Boccia S.; Hashibe M.; Galli P.; De Feo E.; Asakage T.; Hashimoto T.; Hiraki A.; Katoh T.; Nomura T.; Yokoyama A.; van Duijn C. M.; Ricciardi G.; Boffetta P. Aldehyde dehydrogenase 2 and head and neck cancer: a meta-analysis implementing a Mendelian randomization approach. Cancer Epidemiol Biomarkers Prev 2009, 18 (1), 248–54. 10.1158/1055-9965.EPI-08-0462. [DOI] [PubMed] [Google Scholar]
- Seitz H. K.; Stickel F. Acetaldehyde as an underestimated risk factor for cancer development: role of genetics in ethanol metabolism. Genes Nutr 2010, 5 (2), 121–8. 10.1007/s12263-009-0154-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenberg J. A.; Moeller B. C.; Lu K.; Rager J. E.; Fry R. C.; Starr T. B. Formaldehyde carcinogenicity research: 30 years and counting for mode of action, epidemiology, and cancer risk assessment. Toxicol Pathol 2013, 41 (2), 181–9. 10.1177/0192623312466459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D. S.; Kim H. S.; Jung J. H.; Lee C. M.; Ahn Y. S.; Seo Y. R. Formaldehyde exposure and leukemia risk: a comprehensive review and network-based toxicogenomic approach. Genes Environ. 2021, 43 (1), 13. 10.1186/s41021-021-00183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.; Borinskaya S.; Yoshimura K.; Kal’ina N.; Marusin A.; Stepanov V. A.; Qin Z.; Khaliq S.; Lee M. Y.; Yang Y.; Mohyuddin A.; Gurwitz D.; Mehdi S. Q.; Rogaev E.; Jin L.; Yankovsky N. K.; Kidd J. R.; Kidd K. K. Refined geographic distribution of the oriental ALDH2*504Lys (nee 487Lys) variant. Ann. Hum Genet 2009, 73, 335. 10.1111/j.1469-1809.2009.00517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada S.; Agarwal D. P.; Goedde H. W. Aldehyde dehydrogenase deficiency as cause of facial flushing reaction to alcohol in Japanese. Lancet 1981, 318, 982. 10.1016/S0140-6736(81)91172-7. [DOI] [PubMed] [Google Scholar]
- Brooks P. J.; Enoch M. A.; Goldman D.; Li T. K.; Yokoyama A. The alcohol flushing response: an unrecognized risk factor for esophageal cancer from alcohol consumption. PLoS Med. 2009, 6 (3), e1000050 10.1371/journal.pmed.1000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nance M. A.; Berry S. A. Cockayne syndrome: review of 140 cases. Am. J. Med. Genet 1992, 42 (1), 68–84. 10.1002/ajmg.1320420115. [DOI] [PubMed] [Google Scholar]
- Mulderrig L.; Garaycoechea J. I.; Tuong Z. K.; Millington C. L.; Dingler F. A.; Ferdinand J. R.; Gaul L.; Tadross J. A.; Arends M. J.; O’Rahilly S.; Crossan G. P.; Clatworthy M. R.; Patel K. J. Aldehyde-driven transcriptional stress triggers an anorexic DNA damage response. Nature 2021, 600 (7887), 158–163. 10.1038/s41586-021-04133-7. [DOI] [PubMed] [Google Scholar]
- Oka Y.; Hamada M.; Nakazawa Y.; Muramatsu H.; Okuno Y.; Higasa K.; Shimada M.; Takeshima H.; Hanada K.; Hirano T.; Kawakita T.; Sakaguchi H.; Ichimura T.; Ozono S.; Yuge K.; Watanabe Y.; Kotani Y.; Yamane M.; Kasugai Y.; Tanaka M.; Suganami T.; Nakada S.; Mitsutake N.; Hara Y.; Kato K.; Mizuno S.; Miyake N.; Kawai Y.; Tokunaga K.; Nagasaki M.; Kito S.; Isoyama K.; Onodera M.; Kaneko H.; Matsumoto N.; Matsuda F.; Matsuo K.; Takahashi Y.; Mashimo T.; Kojima S.; Ogi T.. Digenic mutations in ALDH2 and ADH5 impair formaldehyde clearance and cause a multisystem disorder, AMeD syndrome. Sci. Adv. 2020, 6 ( (51), ). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green A. M.; Kupfer G. M. Fanconi anemia. Hematol Oncol Clin North Am. 2009, 23 (2), 193–214. 10.1016/j.hoc.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Winter J. P.; Joenje H. The genetic and molecular basis of Fanconi anemia. Mutat. Res. 2009, 668 (1–2), 11–9. 10.1016/j.mrfmmm.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Wang M.; Dingler F. A.; Patel K. J. Genotoxic aldehydes in the hematopoietic system. Blood 2022, 139 (14), 2119–2129. 10.1182/blood.2019004316. [DOI] [PubMed] [Google Scholar]
- Rosado I. V.; Langevin F.; Crossan G. P.; Takata M.; Patel K. J. Formaldehyde catabolism is essential in cells deficient for the Fanconi anemia DNA-repair pathway. Nat. Struct Mol. Biol. 2011, 18 (12), 1432–4. 10.1038/nsmb.2173. [DOI] [PubMed] [Google Scholar]
- Hira A.; Yabe H.; Yoshida K.; Okuno Y.; Shiraishi Y.; Chiba K.; Tanaka H.; Miyano S.; Nakamura J.; Kojima S.; Ogawa S.; Matsuo K.; Takata M.; Yabe M. Variant ALDH2 is associated with accelerated progression of bone marrow failure in Japanese Fanconi anemia patients. Blood 2013, 122 (18), 3206–9. 10.1182/blood-2013-06-507962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberbeck N.; Langevin F.; King G.; de Wind N.; Crossan G. P.; Patel K. J. Maternal aldehyde elimination during pregnancy preserves the fetal genome. Mol. Cell 2014, 55 (6), 807–817. 10.1016/j.molcel.2014.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida K. Role of reactive aldehyde in cardiovascular diseases. Free Radic Biol. Med. 2000, 28 (12), 1685–96. 10.1016/S0891-5849(00)00226-4. [DOI] [PubMed] [Google Scholar]
- Liao S.; Zhang J.; Shi S.; Gong D.; Lu X.; Cheang I.; Zhang H.; Li X. Association of aldehyde exposure with cardiovascular disease. Ecotoxicol Environ. Saf 2020, 206, 111385. 10.1016/j.ecoenv.2020.111385. [DOI] [PubMed] [Google Scholar]
- Soulage C. O.; Pelletier C. C.; Florens N.; Lemoine S.; Dubourg L.; Juillard L.; Guebre-Egziabher F. Two Toxic Lipid Aldehydes, 4-hydroxy-2-hexenal (4-HHE) and 4-hydroxy-2-nonenal (4-HNE), Accumulate in Patients with Chronic Kidney Disease. Toxins (Basel) 2020, 12 (9), 567. 10.3390/toxins12090567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood P. L.; Khan M. A.; Moskal J. R. The concept of ″aldehyde load″ in neurodegenerative mechanisms: cytotoxicity of the polyamine degradation products hydrogen peroxide, acrolein, 3-aminopropanal, 3-acetamidopropanal and 4-aminobutanal in a retinal ganglion cell line. Brain Res. 2007, 1145, 150–6. 10.1016/j.brainres.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Cagle B. S.; Crawford R. A.; Doorn J. A. Biogenic Aldehyde-Mediated Mechanisms of Toxicity in Neurodegenerative Disease. Curr. Opin Toxicol 2019, 13, 16–21. 10.1016/j.cotox.2018.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen L. H.; Hamaji C.; Allen H. L.; Parker G. H.; Ennis J. S.; Kreider M. L. Assessment of formaldehyde exposures under contemporary embalming conditions in U.S. funeral homes. J. Occup Environ. Hyg 2022, 19 (7), 425–436. 10.1080/15459624.2022.2076861. [DOI] [PubMed] [Google Scholar]
- Shortall K.; Djeghader A.; Magner E.; Soulimane T. Insights into Aldehyde Dehydrogenase Enzymes: A Structural Perspective. Front Mol. Biosci 2021, 8, 659550. 10.3389/fmolb.2021.659550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughlin C. R. 2nd; Swanson M. A.; Spector E.; Meeks N. J. L.; Kronquist K. E.; Aslamy M.; Wempe M. F.; van Karnebeek C. D. M.; Gospe S. M. Jr.; Aziz V. G.; Tsai B. P.; Gao H.; Nagy P. L.; Hyland K.; van Dooren S. J. M.; Salomons G. S.; Van Hove J. L. K. The genotypic spectrum of ALDH7A1 mutations resulting in pyridoxine dependent epilepsy: A common epileptic encephalopathy. J. Inherit Metab Dis 2019, 42 (2), 353–361. 10.1002/jimd.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X.; Gong P.; Niu Y.; Zhang Y.; Yang Z. A Rare Presentation Characterized by Epileptic Spasms in ALDH7A1, Pyridox(am)ine-5′-Phosphate Oxidase, and PLPBP Deficiency. Front Genet 2022, 13, 804461. 10.3389/fgene.2022.804461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed Laskar A.; Younus H. Aldehyde toxicity and metabolism: the role of aldehyde dehydrogenases in detoxification, drug resistance and carcinogenesis. Drug Metab Rev. 2019, 51 (1), 42–64. 10.1080/03602532.2018.1555587. [DOI] [PubMed] [Google Scholar]
- Shoeb M.; Ansari N. H.; Srivastava S. K.; Ramana K. V. 4-Hydroxynonenal in the pathogenesis and progression of human diseases. Curr. Med. Chem. 2013, 21 (2), 230–7. 10.2174/09298673113209990181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Zhao T.; Li J.; Xia M.; Li Y.; Wang X.; Liu C.; Zheng T.; Chen R.; Kan D.; Xie Y.; Song J.; Feng Y.; Yu T.; Sun P. Oxidative Stress and 4-hydroxy-2-nonenal (4-HNE): Implications in the Pathogenesis and Treatment of Aging-related Diseases. J. Immunol Res. 2022, 2022, 2233906. 10.1155/2022/2233906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornalley P. J. Protein and nucleotide damage by glyoxal and methylglyoxal in physiological systems--role in ageing and disease. Drug Metabol Drug Interact 2008, 23 (1–2), 125. 10.1515/DMDI.2008.23.1-2.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange J. N.; Wood K. D.; Knight J.; Assimos D. G.; Holmes R. P. Glyoxal formation and its role in endogenous oxalate synthesis. Adv. Urol 2012, 2012, 819202. 10.1155/2012/819202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornalley P. J. Glutathione-dependent detoxification of alpha-oxoaldehydes by the glyoxalase system: involvement in disease mechanisms and antiproliferative activity of glyoxalase I inhibitors. Chem. Biol. Interact 1998, 111–112, 137–51. 10.1016/S0009-2797(97)00157-9. [DOI] [PubMed] [Google Scholar]
- Jin Y.; Penning T. M. Aldo-keto reductases and bioactivation/detoxication. Annu. Rev. Pharmacol Toxicol 2007, 47, 263–92. 10.1146/annurev.pharmtox.47.120505.105337. [DOI] [PubMed] [Google Scholar]
- Terao M.; Garattini E.; Romao M. J.; Leimkuhler S. Evolution, expression, and substrate specificities of aldehyde oxidase enzymes in eukaryotes. J. Biol. Chem. 2020, 295 (16), 5377–5389. 10.1074/jbc.REV119.007741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terao M.; Romao M. J.; Leimkuhler S.; Bolis M.; Fratelli M.; Coelho C.; Santos-Silva T.; Garattini E. Structure and function of mammalian aldehyde oxidases. Arch. Toxicol. 2016, 90 (4), 753–80. 10.1007/s00204-016-1683-1. [DOI] [PubMed] [Google Scholar]
- Holmes R. S. Alcohol dehydrogenases: a family of isozymes with differential functions. Alcohol Alcohol Suppl. 1994, 2, 127. [PubMed] [Google Scholar]
- Edenberg H. J.; McClintick J. N. Alcohol Dehydrogenases, Aldehyde Dehydrogenases, and Alcohol Use Disorders: A Critical Review. Alcohol.: Clin. Exp. Res. 2018, 42 (12), 2281–2297. 10.1111/acer.13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S.; Dixit B. L.; Cai J.; Sharma S.; Hurst H. E.; Bhatnagar A.; Srivastava S. K. Metabolism of lipid peroxidation product, 4-hydroxynonenal (HNE) in rat erythrocytes: role of aldose reductase. Free Radic Biol. Med. 2000, 29 (7), 642–51. 10.1016/S0891-5849(00)00351-8. [DOI] [PubMed] [Google Scholar]
- Desai K.; Wu L. Methylglyoxal and advanced glycation endproducts: new therapeutic horizons?. Recent Pat Cardiovasc Drug Discov 2007, 2 (2), 89–99. 10.2174/157489007780832498. [DOI] [PubMed] [Google Scholar]
- Brown B. E.; Mahroof F. M.; Cook N. L.; van Reyk D. M.; Davies M. J. Hydrazine compounds inhibit glycation of low-density lipoproteins and prevent the in vitro formation of model foam cells from glycolaldehyde-modified low-density lipoproteins. Diabetologia 2006, 49 (4), 775–83. 10.1007/s00125-006-0137-3. [DOI] [PubMed] [Google Scholar]
- Hipkiss A. R. Carnosine and its possible roles in nutrition and health. Adv. Food Nutr Res. 2009, 57, 87–154. 10.1016/S1043-4526(09)57003-9. [DOI] [PubMed] [Google Scholar]
- Jukic I.; Kolobaric N.; Stupin A.; Matic A.; Kozina N.; Mihaljevic Z.; Mihalj M.; Susnjara P.; Stupin M.; Curic Z. B.; Selthofer-Relatic K.; Kibel A.; Lukinac A.; Kolar L.; Kralik G.; Kralik Z.; Szechenyi A.; Jozanovic M.; Galovic O.; Medvidovic-Kosanovic M.; Drenjancevic I. Carnosine, Small but Mighty-Prospect of Use as Functional Ingredient for Functional Food Formulation. Antioxidants (Basel) 2021, 10 (7), 1037. 10.3390/antiox10071037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z.; Baba S. P.; Sweeney B. R.; Barski O. A. Detoxification of aldehydes by histidine-containing dipeptides: from chemistry to clinical implications. Chem. Biol. Interact 2013, 202 (1–3), 288–97. 10.1016/j.cbi.2012.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoPachin R. M.; Barber D. S.; Gavin T. Molecular mechanisms of the conjugated alpha,beta-unsaturated carbonyl derivatives: relevance to neurotoxicity and neurodegenerative diseases. Toxicol. Sci. 2008, 104 (2), 235–49. 10.1093/toxsci/kfm301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaca C. E.; Fang J. L.; Schweda E. K. Studies of the reaction of acetaldehyde with deoxynucleosides. Chem. Biol. Interact 1995, 98 (1), 51–67. 10.1016/0009-2797(95)03632-V. [DOI] [PubMed] [Google Scholar]
- Fang J. L.; Vaca C. E. Detection of DNA adducts of acetaldehyde in peripheral white blood cells of alcohol abusers. Carcinogenesis 1997, 18 (4), 627–32. 10.1093/carcin/18.4.627. [DOI] [PubMed] [Google Scholar]
- Matsuda T.; Terashima I.; Matsumoto Y.; Yabushita H.; Matsui S.; Shibutani S. Effective utilization of N2-ethyl-2’-deoxyguanosine triphosphate during DNA synthesis catalyzed by mammalian replicative DNA polymerases. Biochemistry 1999, 38 (3), 929–35. 10.1021/bi982134j. [DOI] [PubMed] [Google Scholar]
- Perrino F. W.; Blans P.; Harvey S.; Gelhaus S. L.; McGrath C.; Akman S. A.; Jenkins G. S.; LaCourse W. R.; Fishbein J. C. The N2-ethylguanine and the O6-ethyl- and O6-methylguanine lesions in DNA: contrasting responses from the ″bypass″ DNA polymerase eta and the replicative DNA polymerase alpha. Chem. Res. Toxicol. 2003, 16 (12), 1616–23. 10.1021/tx034164f. [DOI] [PubMed] [Google Scholar]
- Brooks P. J.; Theruvathu J. A. DNA adducts from acetaldehyde: implications for alcohol-related carcinogenesis. Alcohol 2005, 35 (3), 187–93. 10.1016/j.alcohol.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Nagayoshi H.; Matsumoto A.; Nishi R.; Kawamoto T.; Ichiba M.; Matsuda T. Increased formation of gastric N(2)-ethylidene-2’-deoxyguanosine DNA adducts in aldehyde dehydrogenase-2 knockout mice treated with ethanol. Mutat. Res. 2009, 673 (1), 74–7. 10.1016/j.mrgentox.2008.11.009. [DOI] [PubMed] [Google Scholar]
- Wang M.; McIntee E. J.; Cheng G.; Shi Y.; Villalta P. W.; Hecht S. S. Identification of DNA adducts of acetaldehyde. Chem. Res. Toxicol. 2000, 13 (11), 1149–57. 10.1021/tx000118t. [DOI] [PubMed] [Google Scholar]
- Ruggiano A.; Ramadan K. DNA-protein crosslink proteases in genome stability. Commun. Biol. 2021, 4 (1), 11. 10.1038/s42003-020-01539-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clauson C.; Scharer O. D.; Niedernhofer L. Advances in understanding the complex mechanisms of DNA interstrand cross-link repair. Cold Spring Harb Perspect Biol. 2013, 5 (10), a012732. 10.1101/cshperspect.a012732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodskinson M. R.; Bolner A.; Sato K.; Kamimae-Lanning A. N.; Rooijers K.; Witte M.; Mahesh M.; Silhan J.; Petek M.; Williams D. M.; Kind J.; Chin J. W.; Patel K. J.; Knipscheer P. Alcohol-derived DNA crosslinks are repaired by two distinct mechanisms. Nature 2020, 579 (7800), 603–608. 10.1038/s41586-020-2059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao H.; Schnetz-Boutaud N. C.; Weisenseel J. P.; Marnett L. J.; Stone M. P. Duplex DNA catalyzes the chemical rearrangement of a malondialdehyde deoxyguanosine adduct. Proc. Natl. Acad. Sci. U. S. A. 1999, 96 (12), 6615–20. 10.1073/pnas.96.12.6615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz A. J.; Lloyd R. S. 1,N2-deoxyguanosine adducts of acrolein, crotonaldehyde, and trans-4-hydroxynonenal cross-link to peptides via Schiff base linkage. J. Biol. Chem. 2003, 278 (8), 5970–6. 10.1074/jbc.M212012200. [DOI] [PubMed] [Google Scholar]
- Kozekov I. D.; Nechev L. V.; Moseley M. S.; Harris C. M.; Rizzo C. J.; Stone M. P.; Harris T. M. DNA interchain cross-links formed by acrolein and crotonaldehyde. J. Am. Chem. Soc. 2003, 125 (1), 50–61. 10.1021/ja020778f. [DOI] [PubMed] [Google Scholar]
- Lu K.; Ye W.; Zhou L.; Collins L.; Chen X.; Gold A.; Ball L.; Swenberg J. Structural Characterization of Formaldehyde-Induced Cross-Links Between Amino Acids and Deoxynucleosides and Their Oligomers. J. Am. Chem. Soc. 2010, 132, 3388–99. 10.1021/ja908282f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontel L. B.; Rosado I. V.; Burgos-Barragan G.; Garaycoechea J. I.; Yu R.; Arends M. J.; Chandrasekaran G.; Broecker V.; Wei W.; Liu L.; Swenberg J. A.; Crossan G. P.; Patel K. J. Endogenous Formaldehyde Is a Hematopoietic Stem Cell Genotoxin and Metabolic Carcinogen. Mol. Cell 2015, 60 (1), 177–88. 10.1016/j.molcel.2015.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide H.; Nakano T.; Salem A. M. H.; Shoulkamy M. I. DNA-protein cross-links: Formidable challenges to maintaining genome integrity. DNA Repair (Amst) 2018, 71, 190–197. 10.1016/j.dnarep.2018.08.024. [DOI] [PubMed] [Google Scholar]
- Stingele J.; Bellelli R.; Alte F.; Hewitt G.; Sarek G.; Maslen S. L.; Tsutakawa S. E.; Borg A.; Kjaer S.; Tainer J. A.; Skehel J. M.; Groll M.; Boulton S. J. Mechanism and Regulation of DNA-Protein Crosslink Repair by the DNA-Dependent Metalloprotease SPRTN. Mol. Cell 2016, 64 (4), 688–703. 10.1016/j.molcel.2016.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingele J.; Jentsch S. DNA-protein crosslink repair. Nat. Rev. Mol. Cell Biol. 2015, 16 (8), 455–60. 10.1038/nrm4015. [DOI] [PubMed] [Google Scholar]
- Lopez-Mosqueda J.; Maddi K.; Prgomet S.; Kalayil S.; Marinovic-Terzic I.; Terzic J.; Dikic I.. SPRTN is a mammalian DNA-binding metalloprotease that resolves DNA-protein crosslinks. Elife 2016, 5. 10.7554/eLife.21491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller B. C.; Lu K.; Doyle-Eisele M.; McDonald J.; Gigliotti A.; Swenberg J. A. Determination of N2-hydroxymethyl-dG adducts in the nasal epithelium and bone marrow of nonhuman primates following 13CD2-formaldehyde inhalation exposure. Chem. Res. Toxicol. 2011, 24 (2), 162–4. 10.1021/tx1004166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng G.; Wang M.; Upadhyaya P.; Villalta P. W.; Hecht S. S. Formation of formaldehyde adducts in the reactions of DNA and deoxyribonucleosides with alpha-acetates of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK), 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL), and N-nitrosodimethylamine (NDMA). Chem. Res. Toxicol. 2008, 21 (3), 746–51. 10.1021/tx7003823. [DOI] [PubMed] [Google Scholar]
- Cheng G.; Shi Y.; Sturla S. J.; Jalas J. R.; McIntee E. J.; Villalta P. W.; Wang M.; Hecht S. S. Reactions of formaldehyde plus acetaldehyde with deoxyguanosine and DNA: formation of cyclic deoxyguanosine adducts and formaldehyde cross-links. Chem. Res. Toxicol. 2003, 16 (2), 145–52. 10.1021/tx025614r. [DOI] [PubMed] [Google Scholar]
- Beland F. A.; Fullerton N. F.; Heflich R. H. Rapid isolation, hydrolysis and chromatography of formaldehyde-modified DNA. J. Chromatogr. 1984, 308, 121–31. 10.1016/0378-4347(84)80202-9. [DOI] [PubMed] [Google Scholar]
- Wang M.; Cheng G.; Balbo S.; Carmella S. G.; Villalta P. W.; Hecht S. S. Clear differences in levels of a formaldehyde-DNA adduct in leukocytes of smokers and nonsmokers. Cancer Res. 2009, 69 (18), 7170–4. 10.1158/0008-5472.CAN-09-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treibmann S.; Spengler F.; Degen J.; Lobner J.; Henle T. Studies on the Formation of 3-Deoxyglucosone- and Methylglyoxal-Derived Hydroimidazolones of Creatine during Heat Treatment of Meat. J. Agric. Food Chem. 2019, 67 (20), 5874–5881. 10.1021/acs.jafc.9b01243. [DOI] [PubMed] [Google Scholar]
- Degen J.; Hellwig M.; Henle T. 1,2-dicarbonyl compounds in commonly consumed foods. J. Agric. Food Chem. 2012, 60 (28), 7071–9. 10.1021/jf301306g. [DOI] [PubMed] [Google Scholar]
- Kold-Christensen R.; Johannsen M. Methylglyoxal Metabolism and Aging-Related Disease: Moving from Correlation toward Causation. Trends Endocrinol Metab 2020, 31 (2), 81–92. 10.1016/j.tem.2019.10.003. [DOI] [PubMed] [Google Scholar]
- Lai S. W. T.; Lopez Gonzalez E. J.; Zoukari T.; Ki P.; Shuck S. C. Methylglyoxal and Its Adducts: Induction, Repair, and Association with Disease. Chem. Res. Toxicol. 2022, 35 (10), 1720–1746. 10.1021/acs.chemrestox.2c00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliore L.; Barale R.; Bosco E.; Giorgelli F.; Minunni M.; Scarpato R.; Loprieno N. Genotoxicity of methylglyoxal: cytogenetic damage in human lymphocytes in vitro and in intestinal cells of mice. Carcinogenesis 1990, 11 (9), 1503–7. 10.1093/carcin/11.9.1503. [DOI] [PubMed] [Google Scholar]
- Murata-Kamiya N.; Kamiya H.; Kaji H.; Kasai H. Methylglyoxal induces G:C to C:G and G:C to T:A transversions in the supF gene on a shuttle vector plasmid replicated in mammalian cells. Mutat. Res. 2000, 468 (2), 173–82. 10.1016/S1383-5718(00)00044-9. [DOI] [PubMed] [Google Scholar]
- Pavin S. S.; Prestes A. S.; Dos Santos M. M.; de Macedo G. T.; Ferreira S. A.; Claro M. T.; Dalla Corte C.; Vargas Barbosa N. Methylglyoxal disturbs DNA repair and glyoxalase I system in Saccharomyces cerevisiae. Toxicol Mech Methods 2021, 31 (2), 107–115. 10.1080/15376516.2020.1838019. [DOI] [PubMed] [Google Scholar]
- Roberts M. J.; Wondrak G. T.; Laurean D. C.; Jacobson M. K.; Jacobson E. L. DNA damage by carbonyl stress in human skin cells. Mutat. Res. 2003, 522 (1–2), 45–56. 10.1016/S0027-5107(02)00232-4. [DOI] [PubMed] [Google Scholar]
- Frischmann M.; Bidmon C.; Angerer J.; Pischetsrieder M. Identification of DNA adducts of methylglyoxal. Chem. Res. Toxicol. 2005, 18 (10), 1586–92. 10.1021/tx0501278. [DOI] [PubMed] [Google Scholar]
- Pischetsrieder M.; Seidel W.; Munch G.; Schinzel R. N(2)-(1-Carboxyethyl)deoxyguanosine, a nonenzymatic glycation adduct of DNA, induces single-strand breaks and increases mutation frequencies. Biochem. Biophys. Res. Commun. 1999, 264 (2), 544–9. 10.1006/bbrc.1999.1528. [DOI] [PubMed] [Google Scholar]
- Kasai H.; Iwamoto-Tanaka N.; Fukada S. DNA modifications by the mutagen glyoxal: adduction to G and C, deamination of C and GC and GA cross-linking. Carcinogenesis 1998, 19 (8), 1459–65. 10.1093/carcin/19.8.1459. [DOI] [PubMed] [Google Scholar]
- Murata-Kamiya N.; Kamiya H.; Kaji H.; Kasai H. Glyoxal, a major product of DNA oxidation, induces mutations at G:C sites on a shuttle vector plasmid replicated in mammalian cells. Nucleic Acids Res. 1997, 25 (10), 1897–902. 10.1093/nar/25.10.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvajal-Garcia J.; Samadpour A. N.; Viera A. J. H.; Merrikh H. Nucleotide excision repair is universally mutagenic and transcription-associated. bioRxiv 2022, 10.1101/2022.06.28.497968. [DOI] [Google Scholar]
- Donnellan L.; Simpson B. S.; Dhillon V. S.; Costabile M.; Fenech M.; Deo P. Folic acid deficiency increases sensitivity to DNA damage by glucose and methylglyoxal. Mutagenesis 2022, 37 (1), 24–33. 10.1093/mutage/geac003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnellan L.; Simpson B.; Dhillon V. S.; Costabile M.; Fenech M.; Deo P. Methylglyoxal induces chromosomal instability and mitotic dysfunction in lymphocytes. Mutagenesis 2021, 36 (5), 339–348. 10.1093/mutage/geab028. [DOI] [PubMed] [Google Scholar]
- Marnett L. J. Lipid peroxidation-DNA damage by malondialdehyde. Mutat. Res. 1999, 424 (1–2), 83–95. 10.1016/S0027-5107(99)00010-X. [DOI] [PubMed] [Google Scholar]
- Benamira M.; Johnson K.; Chaudhary A.; Bruner K.; Tibbetts C.; Marnett L. J. Induction of mutations by replication of malondialdehyde-modified M13 DNA in Escherichia coli: determination of the extent of DNA modification, genetic requirements for mutagenesis, and types of mutations induced. Carcinogenesis 1995, 16 (1), 93–9. 10.1093/carcin/16.1.93. [DOI] [PubMed] [Google Scholar]
- Fink S. P.; Reddy G. R.; Marnett L. J. Mutagenicity in Escherichia coli of the major DNA adduct derived from the endogenous mutagen malondialdehyde. Proc. Natl. Acad. Sci. U. S. A. 1997, 94 (16), 8652–7. 10.1073/pnas.94.16.8652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary A. K.; Nokubo M.; Marnett L. J.; Blair I. A. Analysis of the malondialdehyde-2’-deoxyguanosine adduct in rat liver DNA by gas chromatography/electron capture negative chemical ionization mass spectrometry. Biol. Mass Spectrom 1994, 23 (8), 457–64. 10.1002/bms.1200230802. [DOI] [PubMed] [Google Scholar]
- Wang M.; Dhingra K.; Hittelman W. N.; Liehr J. G.; de Andrade M.; Li D. Lipid peroxidation-induced putative malondialdehyde-DNA adducts in human breast tissues. Cancer Epidemiol Biomarkers Prev. 1996, 5 (9), 705–710. [PubMed] [Google Scholar]
- Wang M.; McIntee E. J.; Cheng G.; Shi Y.; Villalta P. W.; Hecht S. S. A Schiff base is a major DNA adduct of crotonaldehyde. Chem. Res. Toxicol. 2001, 14 (4), 423–30. 10.1021/tx000234w. [DOI] [PubMed] [Google Scholar]
- Dexheimer T. S.; Kozekova A.; Rizzo C. J.; Stone M. P.; Pommier Y. The modulation of topoisomerase I-mediated DNA cleavage and the induction of DNA-topoisomerase I crosslinks by crotonaldehyde-derived DNA adducts. Nucleic Acids Res. 2008, 36 (12), 4128–36. 10.1093/nar/gkn334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y. J.; Wang H.; Kozekov I. D.; Kurtz A. J.; Jacob J.; Voehler M.; Smith J.; Harris T. M.; Lloyd R. S.; Rizzo C. J.; Stone M. P. Stereospecific formation of interstrand carbinolamine DNA cross-links by crotonaldehyde- and acetaldehyde-derived alpha-CH3-gamma-OH-1,N2-propano-2’-deoxyguanosine adducts in the 5′-CpG-3′ sequence. Chem. Res. Toxicol. 2006, 19 (2), 195–208. 10.1021/tx050239z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes P. H.; Kanuri M.; Nechev L. V.; Harris T. M.; Lloyd R. S. Mammalian cell mutagenesis of the DNA adducts of vinyl chloride and crotonaldehyde. Environ. Mol. Mutagen 2005, 45 (5), 455–9. 10.1002/em.20117. [DOI] [PubMed] [Google Scholar]
- Zarkovic N. 4-hydroxynonenal as a bioactive marker of pathophysiological processes. Mol. Aspects Med. 2003, 24 (4–5), 281–91. 10.1016/S0098-2997(03)00023-2. [DOI] [PubMed] [Google Scholar]
- Huang H.; Kozekov I. D.; Kozekova A.; Wang H.; Lloyd R. S.; Rizzo C. J.; Stone M. P. DNA cross-link induced by trans-4-hydroxynonenal. Environ. Mol. Mutagen 2010, 51 (6), 625–34. 10.1002/em.20599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minko I. G.; Kozekov I. D.; Harris T. M.; Rizzo C. J.; Lloyd R. S.; Stone M. P. Chemistry and biology of DNA containing 1,N(2)-deoxyguanosine adducts of the alpha,beta-unsaturated aldehydes acrolein, crotonaldehyde, and 4-hydroxynonenal. Chem. Res. Toxicol. 2009, 22 (5), 759–78. 10.1021/tx9000489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z.; Hu W.; Tang M. S. Trans-4-hydroxy-2-nonenal inhibits nucleotide excision repair in human cells: a possible mechanism for lipid peroxidation-induced carcinogenesis. Proc. Natl. Acad. Sci. U. S. A. 2004, 101 (23), 8598–602. 10.1073/pnas.0402794101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair J.; Srivatanakul P.; Haas C.; Jedpiyawongse A.; Khuhaprema T.; Seitz H. K.; Bartsch H. High urinary excretion of lipid peroxidation-derived DNA damage in patients with cancer-prone liver diseases. Mutat. Res. 2010, 683 (1–2), 23–8. 10.1016/j.mrfmmm.2009.10.002. [DOI] [PubMed] [Google Scholar]
- Temviriyanukul P.; Meijers M.; van Hees-Stuivenberg S.; Boei J. J.; Delbos F.; Ohmori H.; de Wind N.; Jansen J. G. Different sets of translesion synthesis DNA polymerases protect from genome instability induced by distinct food-derived genotoxins. Toxicol. Sci. 2012, 127 (1), 130–8. 10.1093/toxsci/kfs074. [DOI] [PubMed] [Google Scholar]
- Rindgen D.; Nakajima M.; Wehrli S.; Xu K.; Blair I. A. Covalent modifications to 2’-deoxyguanosine by 4-oxo-2-nonenal, a novel product of lipid peroxidation. Chem. Res. Toxicol. 1999, 12 (12), 1195–204. 10.1021/tx990034o. [DOI] [PubMed] [Google Scholar]
- Pollack M.; Oe T.; Lee S. H.; Silva Elipe M. V.; Arison B. H.; Blair I. A. Characterization of 2’-deoxycytidine adducts derived from 4-oxo-2-nonenal, a novel lipid peroxidation product. Chem. Res. Toxicol. 2003, 16 (7), 893–900. 10.1021/tx030009p. [DOI] [PubMed] [Google Scholar]
- Kawai Y.; Nuka E. Abundance of DNA adducts of 4-oxo-2-alkenals, lipid peroxidation-derived highly reactive genotoxins. J. Clin Biochem Nutr 2018, 62 (1), 3–10. 10.3164/jcbn.17-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou P. H.; Kageyama S.; Matsuda S.; Kanemoto K.; Sasada Y.; Oka M.; Shinmura K.; Mori H.; Kawai K.; Kasai H.; Sugimura H.; Matsuda T. Detection of lipid peroxidation-induced DNA adducts caused by 4-oxo-2(E)-nonenal and 4-oxo-2(E)-hexenal in human autopsy tissues. Chem. Res. Toxicol. 2010, 23 (9), 1442–8. 10.1021/tx100047d. [DOI] [PubMed] [Google Scholar]
- Behar R. Z.; Luo W.; Lin S. C.; Wang Y.; Valle J.; Pankow J. F.; Talbot P. Distribution, quantification and toxicity of cinnamaldehyde in electronic cigarette refill fluids and aerosols. Tob Control 2016, 25 (Suppl 2), ii94–ii102. 10.1136/tobaccocontrol-2016-053224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulker Z.; Alpsoy L.; Mihmanli A. Assessment of cytotoxic and apoptotic effects of benzaldehyde using different assays. Hum Exp Toxicol 2013, 32 (8), 858–64. 10.1177/0960327112470271. [DOI] [PubMed] [Google Scholar]
- Behar R. Z.; Davis B.; Wang Y.; Bahl V.; Lin S.; Talbot P. Identification of toxicants in cinnamon-flavored electronic cigarette refill fluids. Toxicol In Vitro 2014, 28 (2), 198–208. 10.1016/j.tiv.2013.10.006. [DOI] [PubMed] [Google Scholar]
- Neudecker T. The genetic toxicology of cinnamaldehyde. Mutat. Res. 1992, 277 (3), 173–85. 10.1016/0165-1110(92)90042-8. [DOI] [PubMed] [Google Scholar]
- King A. A.; Shaughnessy D. T.; Mure K.; Leszczynska J.; Ward W. O.; Umbach D. M.; Xu Z.; Ducharme D.; Taylor J. A.; Demarini D. M.; Klein C. B. Antimutagenicity of cinnamaldehyde and vanillin in human cells: Global gene expression and possible role of DNA damage and repair. Mutat. Res. 2007, 616 (1–2), 60–9. 10.1016/j.mrfmmm.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Silva H. V.; Shankel D. M. Effects of the antimutagen cinnamaldehyde on reversion and survival of selected Salmonella tester strains. Mutat. Res. 1987, 187 (1), 11–9. 10.1016/0165-1218(87)90071-1. [DOI] [PubMed] [Google Scholar]
- Kiwamoto R.; Ploeg D.; Rietjens I. M.; Punt A. Dose-dependent DNA adduct formation by cinnamaldehyde and other food-borne alpha,beta-unsaturated aldehydes predicted by physiologically based in silico modelling. Toxicol In Vitro 2016, 31, 114–25. 10.1016/j.tiv.2015.11.014. [DOI] [PubMed] [Google Scholar]
- Kang J.; Valerio L. G. Investigating DNA adduct formation by flavor chemicals and tobacco byproducts in electronic nicotine delivery system (ENDS) using in silico approaches. Toxicol. Appl. Pharmacol. 2020, 398, 115026. 10.1016/j.taap.2020.115026. [DOI] [PubMed] [Google Scholar]
- Minetti C. A.; Remeta D. P.; Johnson F.; Iden C. R.; Breslauer K. J. Impact of alpha-hydroxy-propanodeoxyguanine adducts on DNA duplex energetics: opposite base modulation and implications for mutagenicity and genotoxicity. Biopolymers 2009, 93 (4), 370–382. 10.1002/bip.21355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone M. P.; Cho Y. J.; Huang H.; Kim H. Y.; Kozekov I. D.; Kozekova A.; Wang H.; Minko I. G.; Lloyd R. S.; Harris T. M.; Rizzo C. J. Interstrand DNA cross-links induced by alpha,beta-unsaturated aldehydes derived from lipid peroxidation and environmental sources. Acc. Chem. Res. 2008, 41 (7), 793–804. 10.1021/ar700246x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z.; Hu W.; Hu Y.; Tang M. S. Acrolein is a major cigarette-related lung cancer agent: Preferential binding at p53 mutational hotspots and inhibition of DNA repair. Proc. Natl. Acad. Sci. U. S. A. 2006, 103 (42), 15404–9. 10.1073/pnas.0607031103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minko I. G.; Kozekov I. D.; Kozekova A.; Harris T. M.; Rizzo C. J.; Lloyd R. S. Mutagenic potential of DNA-peptide crosslinks mediated by acrolein-derived DNA adducts. Mutat. Res. 2008, 637 (1–2), 161–72. 10.1016/j.mrfmmm.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghe A.; Ghare S.; Lamoreau B.; Mohammad M.; Barve S.; McClain C.; Joshi-Barve S. Molecular mechanisms of acrolein toxicity: relevance to human disease. Toxicol. Sci. 2015, 143 (2), 242–55. 10.1093/toxsci/kfu233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. T.; Weng M. W.; Chen W. C.; Yobin M.; Pan J.; Chung F. L.; Wu X. R.; Rom W.; Tang M. S. Effect of CpG methylation at different sequence context on acrolein- and BPDE-DNA binding and mutagenesis. Carcinogenesis 2013, 34 (1), 220–7. 10.1093/carcin/bgs323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks P. J.; Zakhari S. Acetaldehyde and the genome: beyond nuclear DNA adducts and carcinogenesis. Environ. Mol. Mutagen 2014, 55 (2), 77–91. 10.1002/em.21824. [DOI] [PubMed] [Google Scholar]
- Kim Y. A.; Leiserson M. D. M.; Moorjani P.; Sharan R.; Wojtowicz D.; Przytycka T. M. Mutational Signatures: From Methods to Mechanisms. Annu. Rev. Biomed Data Sci. 2021, 4, 189–206. 10.1146/annurev-biodatasci-122320-120920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T.; Kawanishi M.; Yagi T.; Matsui S.; Takebe H. Specific tandem GG to TT base substitutions induced by acetaldehyde are due to intra-strand crosslinks between adjacent guanine bases. Nucleic Acids Res. 1998, 26 (7), 1769–74. 10.1093/nar/26.7.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov L. B.; Kim J.; Haradhvala N. J.; Huang M. N.; Tian Ng A. W.; Wu Y.; Boot A.; Covington K. R.; Gordenin D. A.; Bergstrom E. N.; Islam S. M. A.; Lopez-Bigas N.; Klimczak L. J.; McPherson J. R.; Morganella S.; Sabarinathan R.; Wheeler D. A.; Mustonen V.; Group P. M. S. W.; Getz G.; Rozen S. G.; Stratton M. R.; Consortium P. The repertoire of mutational signatures in human cancer. Nature 2020, 578 (7793), 94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noori P.; Hou S. M. Mutational spectrum induced by acetaldehyde in the HPRT gene of human T lymphocytes resembles that in the p53 gene of esophageal cancers. Carcinogenesis 2001, 22 (11), 1825–30. 10.1093/carcin/22.11.1825. [DOI] [PubMed] [Google Scholar]
- Montesano R.; Holestein M.; Hainaui P. Genetic alterations in esophageal cancer and their relevance to etiology and pathogenesis: a review. Int. J. Cancer 1996, 69 (3), 225–235. . [DOI] [PubMed] [Google Scholar]
- Upton D. C.; Wang X.; Blans P.; Perrino F. W.; Fishbein J. C.; Akman S. A. Replication of N2-ethyldeoxyguanosine DNA adducts in the human embryonic kidney cell line 293. Chem. Res. Toxicol. 2006, 19 (7), 960–7. 10.1021/tx060084a. [DOI] [PubMed] [Google Scholar]
- Stein S.; Lao Y.; Yang I. Y.; Hecht S. S.; Moriya M. Genotoxicity of acetaldehyde- and crotonaldehyde-induced 1,N2-propanodeoxyguanosine DNA adducts in human cells. Mutat. Res. 2006, 608 (1), 1–7. 10.1016/j.mrgentox.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Paget V.; Lechevrel M.; Sichel F. Acetaldehyde-induced mutational pattern in the tumour suppressor gene TP53 analysed by use of a functional assay, the FASAY (functional analysis of separated alleles in yeast). Mutat. Res. 2008, 652 (1), 12–9. 10.1016/j.mrgentox.2007.11.010. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Perez A.; Sabarinathan R.; Lopez-Bigas N. Local Determinants of the Mutational Landscape of the Human Genome. Cell 2019, 177 (1), 101–114. 10.1016/j.cell.2019.02.051. [DOI] [PubMed] [Google Scholar]
- Alexandrov L. B.; Nik-Zainal S.; Wedge D. C.; Campbell P. J.; Stratton M. R. Deciphering signatures of mutational processes operative in human cancer. Cell Rep 2013, 3 (1), 246–59. 10.1016/j.celrep.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garaycoechea J. I.; Crossan G. P.; Langevin F.; Mulderrig L.; Louzada S.; Yang F.; Guilbaud G.; Park N.; Roerink S.; Nik-Zainal S.; Stratton M. R.; Patel K. J. Alcohol and endogenous aldehydes damage chromosomes and mutate stem cells. Nature 2018, 553 (7687), 171–177. 10.1038/nature25154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucab J. E.; Zou X.; Morganella S.; Joel M.; Nanda A. S.; Nagy E.; Gomez C.; Degasperi A.; Harris R.; Jackson S. P.; Arlt V. M.; Phillips D. H.; Nik-Zainal S. A Compendium of Mutational Signatures of Environmental Agents. Cell 2019, 177 (4), 821–836 e16. 10.1016/j.cell.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayraghavan S.; Porcher L.; Mieczkowski P. A.; Saini N. Acetaldehyde makes a distinct mutation signature in single-stranded DNA. Nucleic Acids Res. 2022, 50 (13), 7451–7464. 10.1093/nar/gkac570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapa M. J.; Fabros R. M.; Alasmar S.; Chan K.. Analyses of mutational patterns induced by formaldehyde and acetaldehyde reveal similarity to a common mutational signature. G3 (Bethesda) 2022, 12 ( (11), ). 10.1093/g3journal/jkac238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grogan D.; Jinks-Robertson S. Formaldehyde-induced mutagenesis in Saccharomyces cerevisiae: molecular properties and the roles of repair and bypass systems. Mutat. Res. 2012, 731 (1–2), 92–8. 10.1016/j.mrfmmm.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langevin F.; Crossan G. P.; Rosado I. V.; Arends M. J.; Patel K. J. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature 2011, 475 (7354), 53–8. 10.1038/nature10192. [DOI] [PubMed] [Google Scholar]
- Kemp C. J. Animal Models of Chemical Carcinogenesis: Driving Breakthroughs in Cancer Research for 100 Years. Cold Spring Harb Protoc 2015, 2015 (10), 865–874. 10.1101/pdb.top069906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent R. A.; Caravello H. E.; San R. H. Mutagenic activity of acrolein in S. typhimurium and E. coli. J. Appl. Toxicol 1996, 16 (2), 103–8. . [DOI] [PubMed] [Google Scholar]
- Kanuri M.; Minko I. G.; Nechev L. V.; Harris T. M.; Harris C. M.; Lloyd R. S. Error prone translesion synthesis past gamma-hydroxypropano deoxyguanosine, the primary acrolein-derived adduct in mammalian cells. J. Biol. Chem. 2002, 277 (21), 18257–65. 10.1074/jbc.M112419200. [DOI] [PubMed] [Google Scholar]
- Wang H. T.; Zhang S.; Hu Y.; Tang M. S. Mutagenicity and sequence specificity of acrolein-DNA adducts. Chem. Res. Toxicol. 2009, 22 (3), 511–7. 10.1021/tx800369y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nukaya H.; Inaoka Y.; Ishida H.; Tsuji K.; Suwa Y.; Wakabayashi K.; Kosuge T. Modification of the amino group of guanosine by methylglyoxal and other alpha-ketoaldehydes in the presence of hydrogen peroxide. Chem. Pharm. Bull. (Tokyo) 1993, 41 (4), 649–53. 10.1248/cpb.41.649. [DOI] [PubMed] [Google Scholar]
- Vaca C. E.; Fang J. L.; Conradi M.; Hou S. M. Development of a 32P-postlabelling method for the analysis of 2’-deoxyguanosine-3′-monophosphate and DNA adducts of methylglyoxal. Carcinogenesis 1994, 15 (9), 1887–94. 10.1093/carcin/15.9.1887. [DOI] [PubMed] [Google Scholar]
- Papoulis A.; al-Abed Y.; Bucala R. Identification of N2-(1-carboxyethyl)guanine (CEG) as a guanine advanced glycosylation end product. Biochemistry 1995, 34 (2), 648–55. 10.1021/bi00002a032. [DOI] [PubMed] [Google Scholar]
- Murata-Kamiya N.; Kaji H.; Kasai H. Deficient nucleotide excision repair increases base-pair substitutions but decreases TGGC frameshifts induced by methylglyoxal in Escherichia coli. Mutat. Res. 1999, 442 (1), 19–28. 10.1016/S1383-5718(99)00054-6. [DOI] [PubMed] [Google Scholar]
- Tamae D.; Lim P.; Wuenschell G. E.; Termini J. Mutagenesis and repair induced by the DNA advanced glycation end product N2–1-(carboxyethyl)-2’-deoxyguanosine in human cells. Biochemistry 2011, 50 (12), 2321–9. 10.1021/bi101933p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissier A.; McDonald J. P.; Frank E. G.; Woodgate R. poliota, a remarkably error-prone human DNA polymerase. Genes Dev. 2000, 14 (13), 1642–50. 10.1101/gad.14.13.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuenschell G. E.; Tamae D.; Cercillieux A.; Yamanaka R.; Yu C.; Termini J. Mutagenic potential of DNA glycation: miscoding by (R)- and (S)-N2-(1-carboxyethyl)-2’-deoxyguanosine. Biochemistry 2010, 49 (9), 1814–21. 10.1021/bi901924b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornalley P. J.; Waris S.; Fleming T.; Santarius T.; Larkin S. J.; Winklhofer-Roob B. M.; Stratton M. R.; Rabbani N. Imidazopurinones are markers of physiological genomic damage linked to DNA instability and glyoxalase 1-associated tumour multidrug resistance. Nucleic Acids Res. 2010, 38 (16), 5432–42. 10.1093/nar/gkq306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster A. L. H.; Sanders M. A.; Patel K.; Dietrich R.; Noonan R. J.; Lach F. P.; White R. R.; Goldfarb A.; Hadi K.; Edwards M. M.; Donovan F. X.; Hoogenboezem R. M.; Jung M.; Sridhar S.; Wiley T. F.; Fedrigo O.; Tian H.; Rosiene J.; Heineman T.; Kennedy J. A.; Bean L.; Rosti R. O.; Tryon R.; Gonzalez A. M.; Rosenberg A.; Luo J. D.; Carroll T. S.; Shroff S.; Beaumont M.; Velleuer E.; Rastatter J. C.; Wells S. I.; Surralles J.; Bagby G.; MacMillan M. L.; Wagner J. E.; Cancio M.; Boulad F.; Scognamiglio T.; Vaughan R.; Beaumont K. G.; Koren A.; Imielinski M.; Chandrasekharappa S. C.; Auerbach A. D.; Singh B.; Kutler D. I.; Campbell P. J.; Smogorzewska A. Genomic signature of Fanconi anaemia DNA repair pathway deficiency in cancer. Nature 2022, 612 (7940), 495–502. 10.1038/s41586-022-05253-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke M. S.; Chang Y. J.; Chen Y. R.; Hu C. W.; Chao M. R. Nucleic acid adductomics - The next generation of adductomics towards assessing environmental health risks. Sci. Total Environ. 2023, 856 (Pt 2), 159192. 10.1016/j.scitotenv.2022.159192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowry A.; Kelly R. D. W.; Petermann E. Hypertranscription and replication stress in cancer. Trends Cancer 2021, 7 (9), 863–877. 10.1016/j.trecan.2021.04.006. [DOI] [PubMed] [Google Scholar]
- Roberts S. A.; Gordenin D. A. Hypermutation in human cancer genomes: footprints and mechanisms. Nat. Rev. Cancer 2014, 14 (12), 786–800. 10.1038/nrc3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiano V.; Maertens L.; Guidolin V.; Yang J.; Balbo S.; Hecht S. S. Quantitative Liquid Chromatography-Nanoelectrospray Ionization-High-Resolution Tandem Mass Spectrometry Analysis of Acrolein-DNA Adducts and Etheno-DNA Adducts in Oral Cells from Cigarette Smokers and Nonsmokers. Chem. Res. Toxicol. 2020, 33 (8), 2197–2207. 10.1021/acs.chemrestox.0c00223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalta P. W.; Balbo S. The Future of DNA Adductomic Analysis. Int. J. Mol. Sci. 2017, 18 (9), 1870. 10.3390/ijms18091870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eid J.; Fehr A.; Gray J.; Luong K.; Lyle J.; Otto G.; Peluso P.; Rank D.; Baybayan P.; Bettman B.; Bibillo A.; Bjornson K.; Chaudhuri B.; Christians F.; Cicero R.; Clark S.; Dalal R.; Dewinter A.; Dixon J.; Foquet M.; Gaertner A.; Hardenbol P.; Heiner C.; Hester K.; Holden D.; Kearns G.; Kong X.; Kuse R.; Lacroix Y.; Lin S.; Lundquist P.; Ma C.; Marks P.; Maxham M.; Murphy D.; Park I.; Pham T.; Phillips M.; Roy J.; Sebra R.; Shen G.; Sorenson J.; Tomaney A.; Travers K.; Trulson M.; Vieceli J.; Wegener J.; Wu D.; Yang A.; Zaccarin D.; Zhao P.; Zhong F.; Korlach J.; Turner S. Real-time DNA sequencing from single polymerase molecules. Science 2009, 323 (5910), 133–8. 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- Clark T. A.; Spittle K. E.; Turner S. W.; Korlach J. Direct detection and sequencing of damaged DNA bases. Genome Integr 2011, 2, 10. 10.1186/2041-9414-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An N.; Fleming A. M.; White H. S.; Burrows C. J. Nanopore detection of 8-oxoguanine in the human telomere repeat sequence. ACS Nano 2015, 9 (4), 4296–307. 10.1021/acsnano.5b00722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Price N. E.; Fang X.; Yang Z.; Gu L. Q.; Gates K. S. Characterization of Interstrand DNA-DNA Cross-Links Using the alpha-Hemolysin Protein Nanopore. ACS Nano 2015, 9 (12), 11812–9. 10.1021/acsnano.5b03923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera R. T.; Fleming A. M.; Johnson R. P.; Burrows C. J.; White H. S. Detection of benzo[a]pyrene-guanine adducts in single-stranded DNA using the alpha-hemolysin nanopore. Nanotechnology 2015, 26 (7), 074002. 10.1088/0957-4484/26/7/074002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voordeckers K.; Colding C.; Grasso L.; Pardo B.; Hoes L.; Kominek J.; Gielens K.; Dekoster K.; Gordon J.; Van der Zande E.; Bircham P.; Swings T.; Michiels J.; Van Loo P.; Nuyts S.; Pasero P.; Lisby M.; Verstrepen K. J. Ethanol exposure increases mutation rate through error-prone polymerases. Nat. Commun. 2020, 11 (1), 3664. 10.1038/s41467-020-17447-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. I.; Pfeifer G. P.; Besaratinia A. Lack of mutagenicity of acrolein-induced DNA adducts in mouse and human cells. Cancer Res. 2007, 67 (24), 11640–7. 10.1158/0008-5472.CAN-07-2528. [DOI] [PubMed] [Google Scholar]
- Liu X.; Lao Y.; Yang I. Y.; Hecht S. S.; Moriya M. Replication-coupled repair of crotonaldehyde/acetaldehyde-induced guanine-guanine interstrand cross-links and their mutagenicity. Biochemistry 2006, 45 (42), 12898–905. 10.1021/bi060792v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedernhofer L. J.; Daniels J. S.; Rouzer C. A.; Greene R. E.; Marnett L. J. Malondialdehyde, a product of lipid peroxidation, is mutagenic in human cells. J. Biol. Chem. 2003, 278 (33), 31426–33. 10.1074/jbc.M212549200. [DOI] [PubMed] [Google Scholar]