Abstract
Background
Emerging evidence from mouse models is beginning to elucidate the brain’s immune response to tau pathology, but little is known about the nature of this response in humans. In addition, it remains unclear to what extent tau pathology and the local inflammatory response within the brain influence the broader immune system.
Methods
To address these questions, we performed single-cell RNA sequencing (scRNA-seq) of peripheral blood mononuclear cells (PBMCs) from carriers of pathogenic variants in MAPT, the gene encoding tau (n = 8), and healthy non-carrier controls (n = 8). Primary findings from our scRNA-seq analyses were confirmed and extended via flow cytometry, droplet digital (dd)PCR, and secondary analyses of publicly available transcriptomics datasets.
Results
Analysis of ~ 181,000 individual PBMC transcriptomes demonstrated striking differential expression in monocytes and natural killer (NK) cells in MAPT pathogenic variant carriers. In particular, we observed a marked reduction in the expression of CX3CR1—the gene encoding the fractalkine receptor that is known to modulate tau pathology in mouse models—in monocytes and NK cells. We also observed a significant reduction in the abundance of nonclassical monocytes and dysregulated expression of nonclassical monocyte marker genes, including FCGR3A. Finally, we identified reductions in TMEM176A and TMEM176B, genes thought to be involved in the inflammatory response in human microglia but with unclear function in peripheral monocytes. We confirmed the reduction in nonclassical monocytes by flow cytometry and the differential expression of select biologically relevant genes dysregulated in our scRNA-seq data using ddPCR.
Conclusions
Our results suggest that human peripheral immune cell expression and abundance are modulated by tau-associated pathophysiologic changes. CX3CR1 and nonclassical monocytes in particular will be a focus of future work exploring the role of these peripheral signals in additional tau-associated neurodegenerative diseases.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13073-023-01205-3.
Keywords: Tau, Tauopathy, MAPT, CX3CR1, Dementia, Neurodegeneration, Nonclassical monocytes, Microglia, PBMCs, Single-cell RNA-seq
Background
Nearly 25 years after the discovery of pathogenic variants in MAPT (encoding the microtubule-associated protein tau) in familial frontotemporal dementia (FTD; [1, 2]; reviewed in [3]), there are still no effective therapeutics capable of halting or delaying tau-associated neurodegeneration [4, 5]. Diverse tau proteinopathies (tauopathies) also occur sporadically (i.e., in the absence of MAPT or other pathogenic variants) and are subdivided into primary tauopathies—which collectively fall under the umbrella term, frontotemporal lobar degeneration (FTLD)-tau—and secondary tauopathies, the most prominent example of which is Alzheimer’s disease (AD). Although much effort has gone into characterizing the natural history and longitudinal declines of MAPT pathogenic variant carriers (reviewed in [6]), we understand relatively little about the molecular mechanisms that impart risk for sporadic forms of tauopathy—whether primary or secondary. However, because it remains challenging to confidently diagnose sporadic tauopathy in vivo, familial tauopathy (identified via the presence of pathogenic MAPT variants in individuals with a family history of neurodegenerative disease) represents a powerful lens through which we can not only elucidate the pathophysiologic processes underlying hereditary tauopathy but also uncover shared processes that may contribute to sporadic tauopathy.
The last decade has witnessed a major revival in interest in immune mechanisms that may modulate risk for neurodegeneration, with a primary focus on microglia, the parenchymal macrophages of the brain (reviewed in [7]). Significantly less progress has been made in elucidating peripheral blood or cerebrospinal fluid (CSF) leukocyte perturbations in—and responses to—neurodegeneration, although this is beginning to change (reviewed in [8–10]). For example, we now know of changes to CD8+ T cells in AD and CD4+ T cells in Lewy body dementia [11, 12]. In addition, altered phospholipase C-ɣ2 signaling in peripheral lymphocytes has been reported in AD [13]. Beyond lymphocytes, changes in peripheral monocytes (particularly nonclassical [NC] monocytes) have been observed in Parkinson’s disease (PD) [14] and amyotrophic lateral sclerosis (ALS; [15, 16]). In addition, patients with the hereditary white-matter disorder, adult-onset leukoencephalopathy with axonal spheroids, and pigmented glia (ALSP) show striking reductions in peripheral NC monocytes [17]. Given that ALSP is thought to be driven by primary microglial defects (reviewed in [18]), the reduction in peripheral NC monocytes in this disorder suggests shared biology between these two cell types.
High-quality fluid biomarkers are being developed for AD (reviewed in [19, 20]) and FTD (reviewed in [21]), but those that can distinguish between the major neuropathologic division within FTD (i.e., FTLD-tau vs. FTLD due to TDP-43 pathology [FTLD-TDP]) have been lacking [21] until very recently [22]. We reasoned that an in-depth exploration of peripheral immune dysregulation in tauopathy may reveal novel, blood-based biomarkers associated with FTLD-tau. Therefore, in an effort to define the peripheral immune signatures of tauopathy, we carried out single-cell RNA sequencing (scRNA-seq) of peripheral blood mononuclear cells (PBMCs) in individuals with pathogenic MAPT variants—who have (or will develop) FTLD-tau pathology—and cognitively normal, non-carrier controls. We identified striking changes in NC monocytes—both in terms of cellular abundance and gene expression—as well as natural killer (NK) cells and other cell types. Moreover, we identified CX3CR1 as a potentially novel peripheral marker of tauopathy, suggesting parallel changes in CX3CR1 in microglia and peripheral leukocytes in the course of tau-mediated neurodegeneration. We also identified additional candidate genes whose expression may be altered in the periphery in tauopathy (e.g., FCGR3A and TMEM176A/B). Considering recent findings in ALS and PD, and given that NC monocytes can be detected in the human brain [23], our results add to the weight of evidence suggesting the importance of NC monocytes across a spectrum of neurodegenerative diseases. Taken together, our results indicate that PBMCs represent an accessible and informative tissue source not only for biomarker discovery but also for elucidation of peripheral immune responses in the context of tauopathy.
Methods
Study participants
All participants or their surrogates provided written informed consent prior to study participation; all aspects of the studies described here were approved by the University of California, San Francisco (UCSF) institutional review board. Sixteen individuals (n = 8 MAPT pathogenic variant carriers and n = 8 cognitively normal, non-carrier controls) participated in this study. Individuals were recruited as part of ongoing studies of FTD and related neurodegenerative diseases at the UCSF Memory and Aging Center (MAC). All participants underwent a multistep screening with an in-person visit at the MAC that included a neurologic exam, detailed cognitive assessment, medical history, and family history for neurodegenerative disease. Each participant’s study partner was interviewed regarding functional abilities. A consensus team of clinicians then reviewed all participants. In addition, all participants were screened for pathogenic variants in established FTLD genes, including MAPT. MAPT pathogenic variant carriers had clinical syndromes of bvFTD (n = 4), frontal AD (n = 1), subjective cognitive impairment (n = 1), or were clinically normal and presymptomatic (n = 2). Pathogenic MAPT variants represented within this study were p.P301L (n = 1), p.S305I (n = 1), p.S305S (n = 1), p.R406W (n = 3), and IVS10 + 16 (n = 2). Pathogenic variant carriers and cognitively normal controls were sex-matched, and equal numbers of female and male participants were included in the cohort. Age was not significantly different between carrier and control groups, as assessed using an unpaired t-test. Demographic information for study participants is included in Table 1.
Table 1.
Demographic characteristics of cohort
| Control | MAPT variant carrier | |
|---|---|---|
| n | 8 | 8 |
| Age, mean (SD) | 52.8 (10.0) | 54.4 (11.4) |
| Sex, n female | 4 | 4 |
| Clinical syndrome (n) | Clinically normal (8) | bvFTD (4), frontal AD (1), subjective cognitive impairment (1), presymp. (2) |
| MAPT variants (n) | N/A | p.P301L (1), p.S305I (1), p.S305S (1), p.R406W (3), IVS10 + 16 (2) |
| Splicing variants, n | N/A | 4 |
| CDR-SB, mean (SD) | 0.0 (0.0) |
5.8 (5.7) 7.8 (5.3)# |
Abbreviations: AD Alzheimer’s disease, bvFTD behavioral variant frontotemporal dementia, CDR-SB Clinical Dementia Rating scale Sum of Boxes, presymp. presymptomatic, SD standard deviation
#symptomatic carriers only
Clinical assessment
Study participants underwent a multistep screening process prior to an in-person clinical evaluation at the UCSF MAC, which included a neurological exam, cognitive assessment, and medical history [24]. Each participant’s study partner was interviewed to assess the participant’s functional abilities. A multidisciplinary team composed of a behavioral neurologist, a neuropsychologist, and a registered nurse established clinical diagnoses for cases according to consensus criteria for FTD and its subtypes [25] or frontal AD [26]. Controls and presymptomatic MAPT carriers had a normal neurological exam and a Clinical Dementia Rating scale Sum of Boxes (CDR-SB) [27] score of 0. All controls screened negative for disease-causing pathogenic variation in established AD and FTD genes.
Cell isolation
Human PBMCs were obtained from study participants at the UCSF MAC. Blood specimens were collected in yellow-top acid-citrate-dextrose vacutainer tubes (BD Biosciences) and processed within 5 h of collection. PBMCs were isolated via Ficoll density gradient centrifugation using Lymphosep separation medium (MP Biomedicals), washed with Ca2+- and Mg2+-free PBS (Thermo Fisher), and treated with one application of red blood cell lysis buffer (Biolegend). After a final wash step in PBS, PBMCs were diluted to a density of 1.5 × 106 cells/ml in freezing media composed of 10% DMSO in FBS and immediately frozen at − 80 °C. After 2 weeks, samples were transferred to liquid nitrogen for long-term storage. All PBMC samples used in our primary analyses were cryopreserved. Sensitivity analysis involving freshly isolated PBMCs was performed to compare gene expression in paired fresh vs. frozen and thawed samples.
Single-cell RNA-seq
PBMCs were thawed and prepared for scRNA-seq using the Chromium Single Cell 3’ v2 kit according to the manufacturer’s instructions (10 × Genomics). Samples were processed in two separate batches of eight samples each, with four MAPT variant carriers and four controls included in each batch. To further minimize the potential for batch effects, each batch contained equal numbers of samples from female and male participants. After sample thawing, counting, and dilution, PBMCs underwent standard 10 × processing, 3′ gene expression library construction steps, and next-generation sequencing at the UCSF Genomics CoLab and Institute for Human Genetics (IHG).
Sequencing data processing
For each of the two batches, single-cell 3′ libraries generated from eight samples were pooled and sequenced on one lane of a NovaSeq S4 flow cell. Raw sequencing reads were aligned to GRCh38, and feature-barcode matrices were generated using Cell Ranger version 3.0.2.
Quality control and clustering
We obtained a total of 7.3 × 109 reads and detected ~ 214,000 cells across the two independent 10 × and sequencing batches, yielding a moderate sequencing depth [28, 29] of ~ 34,000 mean reads/cell. We detected ~ 3700 median UMI counts/cell and ~ 1100 median genes/cell (Additional file 1: Table S1). There were no significant differences in the number of cells captured per sample, the number of reads per sample, or the mean read depth per sample when comparing the MAPT pathogenic variant carrier group to the non-carrier control group. Subsequent quality-control (QC) and downstream analysis steps were performed using Seurat v4.1 [30, 31]. QC filtering was applied to individual-sample feature-barcode matrices and consisted of the following steps: (i) genes detected in < 10 cells were removed; (ii) cells with ≤ 500 detected genes were removed; (iii) cells with ≤ 500 counts and those with ≥ 20,000 counts were removed; (iv) cells with mitochondrial mapping percentages ≥ 10 were removed; (v) doublets were identified and removed using DoubletFinder v2.0.3 [32, 33] using the recommended parameter settings. After stringent QC filtering, ~ 181,000 cells remained for downstream analysis (Additional file 2: Table S2).
After QC, we performed the following additional processing steps: (i) we applied sctransform [34]—a method for scRNA-seq count normalization and variance stabilization—at the individual-sample level, including mitochondrial mapping percentage as a covariate [34, 35], to minimize variability due to differences in sequencing depth between samples; (ii) the 16 individual samples were integrated with FindIntegrationAnchors and IntegrateData, specifying “sctransform” as the normalization method and reciprocal principal component analysis (PCA) as the reduction. Subsequently, PCA was performed followed by uniform manifold approximation and projection (UMAP) reduction using the first 30 PCs; clustering was performed using a resolution parameter of 0.5. This resulted in the generation of 21 distinct clusters that were annotated on the basis of marker gene expression, identified using FindMarkers and literature searches.
Differential expression analysis
Differential expression analysis was performed using limma [36–39] on individual clusters and sctransform-normalized data, with MAPT pathogenic variant carrier status as the contrast and age, sex, and scRNA-seq batch as covariates. To account for multiple testing, a false discovery rate-corrected p-value (pFDR) < 0.05 was considered statistically significant. For visualization of selected differentially expressed genes (DEGs), we used violin plots displaying normalized count data generated via Seurat’s NormalizeData function.
Cluster proportionality
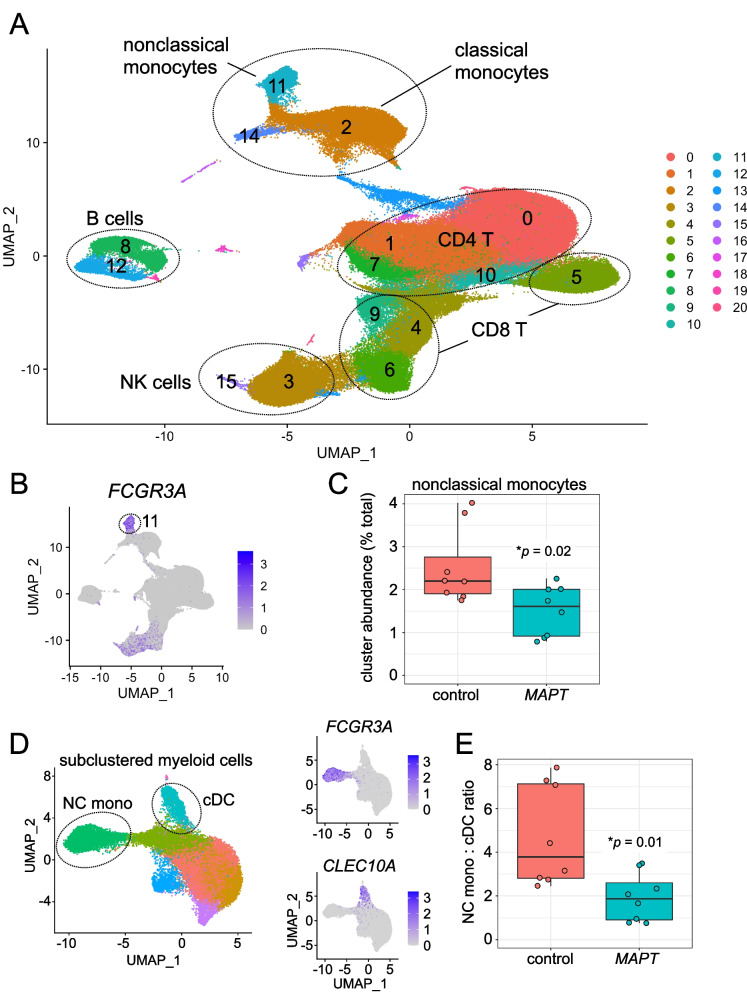
Cluster proportions were determined for individual samples by dividing the number of cells in a given cluster by the total number of cells in all clusters (after QC filtering) for each individual. Differences in cluster proportionality were assessed visually for all clusters according to MAPT variant carrier status. Only the NC monocyte cluster (cluster 11) showed a clear difference in proportionality between carriers and controls. Significance for cluster 11 proportionality, as well as NC monocyte subcluster ratios, was determined by linear modeling, covarying for age, sex, and scRNA-seq batch.
STRING network analysis
DEGs with pFDR < 0.05 and log2 fold-changes (LFC) > 0.1 for selected cell clusters were submitted for analysis via the STRING database (v11.5) [40] using the following parameters: the full STRING network was queried; network edge thickness indicated the confidence of the interaction; active interaction sources included databases, experiments, and text mining; a minimum interaction score of 0.4 was required; only query genes/proteins were displayed; disconnected nodes (i.e., DEGs with no interaction partners) were not displayed; gene modules were color-coded according to the results of Markov cluster algorithm (MCL) clustering [41], with modules color-coded within each panel according to their respective gene count. For clarity, within the “Results” section, we refer to specific MCL clusters (containing DEGs) as gene/protein “modules,” while reserving the term “cluster” to refer to cell clusters generated via scRNA-seq analysis.
Analysis of mouse RNA-seq data
Publicly available mouse RNA-seq data were downloaded from GEO (accession GSE93180) and originally published in [42]. Briefly, hippocampal microglia (Cd11b+ myeloid cells) were sorted from 6-month-old male hMAPT-P301S transgenic mice (n = 6) or non-transgenic littermates (n = 6) using BD FACSAria sorters, then RNA was extracted. RNA-seq libraries were prepared using the Ovation RNA-Seq System V2 (NuGEN). Reads were aligned to the GRCm38 genome using GSNAP and gene counts were acquired using the HTSeqGenie Bioconductor package. Average sequencing depth was 30 million total reads with 8% of reads aligning to exons. We downloaded the available counts data, reanalyzed it using DESeq2 [43], and plotted normalized counts for mouse Cx3cr1 using ggplot2 [44].
RNA extraction
To minimize biological variability and facilitate orthogonal validation, for droplet digital (dd)PCR experiments, we used PBMCs isolated from 15 of the 16 participants originally selected for scRNA-seq analysis. One additional cognitively normal control sample was used for ddPCR due to unavailability of one control sample used for scRNA-seq. RNA was extracted from the PBMCs using the RNeasy Micro Kit (Qiagen) and isolated RNA was quantified and its quality was assessed using the RNA 6000 Pico Bioanalyzer kit (Agilent). PBMC RNA samples had RNA integrity number (RIN) values ranging from 9.2 to 9.9, indicating high-quality RNA (Additional file 3: Table S3; [45]).
Droplet digital PCR
One nanogram of total RNA was used for single-tube reverse transcription (RT) and ddPCR using the One-Step RT-ddPCR Advanced kit (Bio-Rad). Droplets were generated and subsequently analyzed using the QX100 system (Bio-Rad) at the UCSF Center for Advanced Technology (CAT). Reactions were prepared and run essentially according to the manufacturer’s instructions. For steps in which a temperature range was specified, we used the following parameters: RT was performed at 50 °C, annealing/extension occurred at 55 °C, and samples were held at 12 °C in the C1000 thermocycler (Bio-Rad) prior to analysis on the droplet reader. To confirm specificity, we ran the following control reactions: wells lacking RNA but containing all other components and wells lacking reverse transcriptase but containing all other components. PrimePCR ddPCR Gene Expression primer–probe mixes coupled to FAM or HEX (Bio-Rad) were used to amplify specific genes.
Flow cytometry
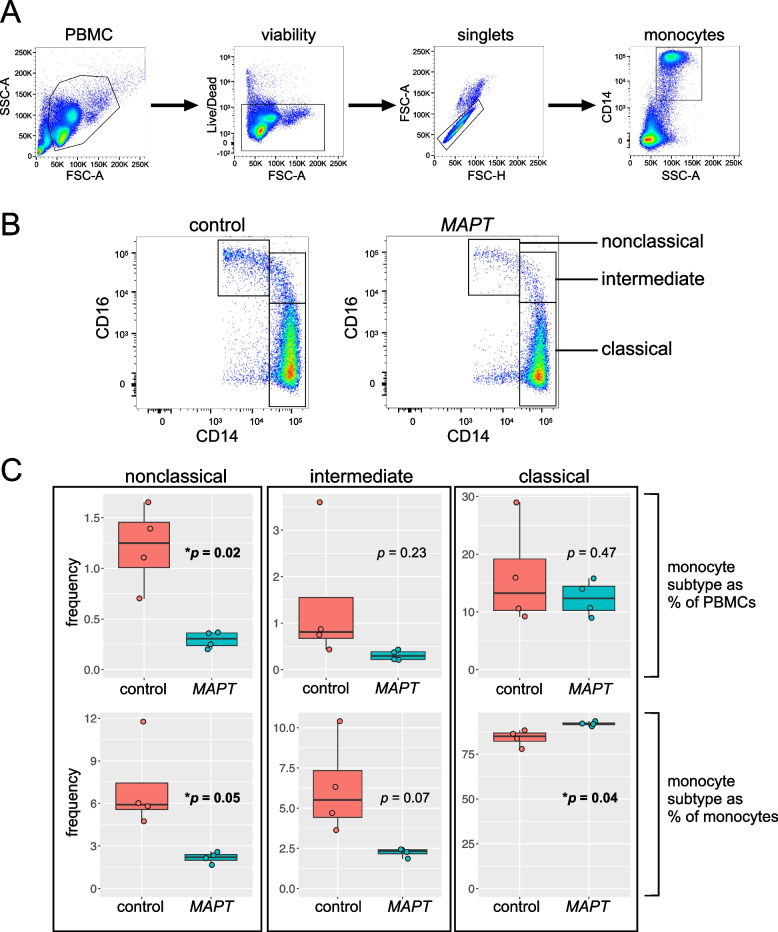
Multicolor flow cytometry was performed on thawed PBMC samples using an LSRFortessa (BD). Samples were stained using the following fluorochrome-conjugated antibodies: PE-CF594 mouse anti-human CD14 (BD), APC mouse anti-human CD16 (BD), BV 421 mouse anti-human CD56 (BioLegend), and PE rat anti-human CX3CR1 (BD). Viability was assessed using the LIVE/DEAD Aqua dye (Thermo Fisher). Cells were labeled at room temperature, covered from light for 30 min in a buffer consisting of 1% fetal bovine serum in MACS buffer containing PBS and EDTA (pH 7.2; Miltenyi). After staining, samples were washed three times in the above buffer, then kept on ice until analysis on the LSRFortessa. Compensation was performed using UltraComp eBeads (Thermo Fisher) for the antibodies and ArC amine-reactive compensation beads for LIVE/DEAD Aqua (Thermo Fisher). Control conditions included unstained PBMCs as well as “fluorescence minus one” (FMO) conditions in which a single antibody was omitted. For the primary samples to be analyzed, we acquired data on ~ 100,000 cells falling within the initially defined PBMC gate. All post-acquisition gating and analysis was performed in FlowJo v10 (BD). We used the following sequential gating scheme: (i) debris was excluded via an initial PBMC gate; (ii) live PBMCs were gated by low LIVE/DEAD Aqua fluorescence; (iii) singlets falling along the forward scatter (FSC)-height (-H) vs. FSC-area (-A) diagonal were gated next; (iv) finally, monocytes were gated based on their high side scatter (SSC-A) and CD14 positivity. Monocyte subtypes were gated from a starting population of all monocytes based on their characteristic CD14 vs. CD16 staining patterns. For quantifications, frequencies expressed as a percentage of all PBMCs used the number of cells in the live, singlet PBMC population as the denominator, while frequencies expressed as a percentage of all monocytes used the number of cells in the live, singlet monocyte population as the denominator.
Analysis of publicly available human brain scRNA-seq dataset
Publicly available human brain scRNA-seq data were downloaded from GEO (accession GSE137444) and originally published in [46]. Briefly, human microglia from patients (n = 7) who underwent amygdalohippocampectomy were isolated from the temporal cortex, FACS-sorted, processed for scRNA-seq using the Chromium Single Cell v2 kit (10 × Genomics), and sequenced on a HiSeq 4000 (Illumina). The downloaded counts data were processed and analyzed as described above for our PBMC scRNA-seq data. We used the marker genes identified in [23] to identify the cluster representing human brain CD16+ NC monocytes. After identification of the NC monocytes, we assessed cluster-specific expression of CX3CR1, FCGR3A, TMEM176A/B, and C3AR1.
Additional statistical analyses
For the analysis of ddPCR data, we performed linear modeling to assess whether MAPT carrier status predicted differences in gene expression while covarying for age and sex. Log2-transformed absolute concentration data for CX3CR1, FCGR3A, TMEM176A/B, and C3AR1 (or the ratios of these values with those of reference gene EEF2) were used for analyses assuming data normality, while non-transformed data are displayed in the plots. EEF2 levels can be leveraged as a normalization factor to reduce technical variation across samples. In our assays, we found that the results were unaffected by normalization. To test for differences in mitochondrial genome mapping percentages and differences in the proportions of monocyte subtypes calculated from flow cytometry data, we used Welch’s t-test. Differential gene expression for the mouse bulk RNA-seq data was determined using the Wald test implemented in DESeq2. Differences were considered significant at p ≤ 0.05 (ddPCR, mitochondrial mapping data, and flow cytometry) or pFDR < 0.05 (bulk RNA-seq data). Analyses were performed in R and plots were generated with ggplot2.
Results
Reduced nonclassical monocytes in MAPT pathogenic variant carriers
After QC filtering, clustering of ~ 181,000 PBMCs generated 21 primary clusters representing the major lymphoid and myeloid cell types, including CD4+ and CD8+ T cells, B cells, NK cells, monocytes, and dendritic cells (Fig. 1A; Additional file 4: Fig. S1). As expected, and in contrast to many other FTD-associated genes, MAPT expression was barely detectable in PBMCs (Additional file 4: Fig. S2). We reasoned, therefore, that any changes detected in PBMC cell-type proportionality or gene expression in MAPT pathogenic variant carriers would most likely represent a response to tau neuropathology or neurodegeneration rather than cell-intrinsic changes occurring directly downstream of variant MAPT. Of all PBMC clusters, only one showed a clear, consistent change in abundance in MAPT pathogenic variant carriers relative to controls. This cluster (11), which represents FCGR3A+ (CD16+) NC monocytes (Fig. 1A, B), localized in UMAP space near the more-abundant CD14+ classical monocyte cluster (2) and the CLEC10A+ conventional dendritic cell (cDC) cluster (14). In particular, NC monocytes, expressed as a percentage of total PBMCs for each participant, were significantly reduced in MAPT carriers (Fig. 1C). To gain more fine-grained insight into the nature of this change in abundance, we subsetted and re-clustered myeloid clusters 2, 11, and 14. Assessing the myeloid subclusters (Fig. 1D, left), we discovered that normalizing the NC monocyte subcluster to the CLEC10A+, CD1C+ cDC subcluster (corresponding to the most abundant blood cDC population, known as cDC2 [47]; Fig. 1D, lower right panel) yielded better separation between MAPT variant carriers and non-carrier controls (Fig. 1E). Further assessing the myeloid subclusters, we found that normalizing NC monocyte numbers to total cDCs (i.e., cDC1 + cDC2) gave a similar result (Additional file 4: Fig. S3A). On the other hand, expressing NC monocyte abundance as a fraction of all monocytes or all myeloid cells did not significantly differentiate carriers from controls (Additional file 4: Fig. S3B, C).
Fig. 1.
Single-cell RNA-seq reveals reductions in nonclassical monocytes in MAPT pathogenic variant carriers. A Two-dimensional UMAP plot of ~ 181,000 PBMCs from MAPT variant carriers and non-carrier controls, colored by cluster identity. Major cell types are labeled within the plot. B, C Cluster 11, marked by high FCGR3A expression and identified as NC monocytes, was significantly reduced in MAPT carriers (p = 0.02; data are expressed as percentage of total PBMCs for each sample). D Myeloid cells (clusters 2, 11, 14) were subset and re-clustered. NC monocyte and cDC2 subclusters were identified by FCGR3A and CLEC10A expression, respectively (D, right). E The ratio of NC monocytes to cDC2 was significantly reduced in MAPT variant carriers (p = 0.01)
Differential expression analysis
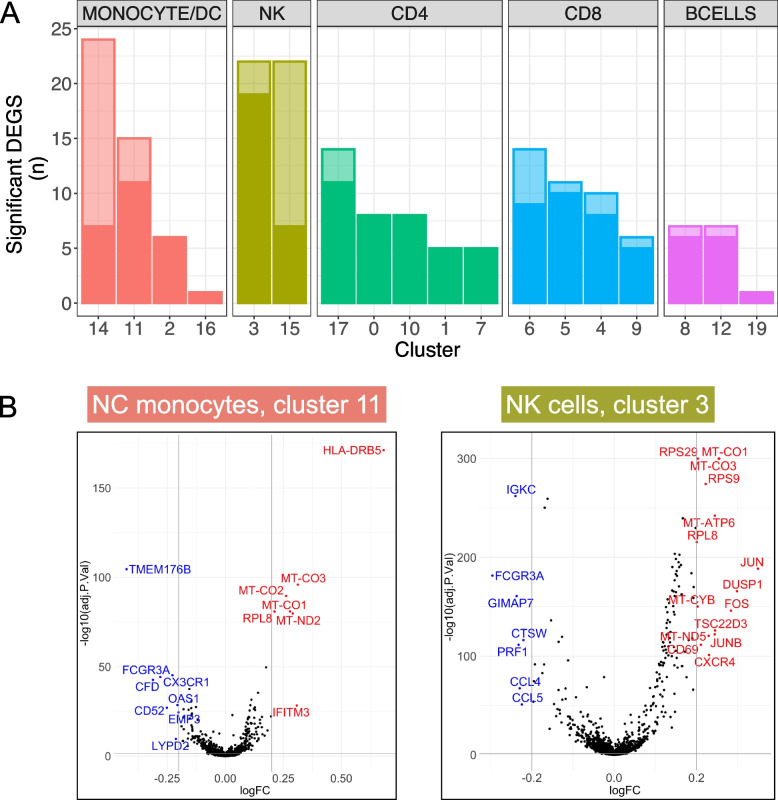
We next performed differential expression analysis on each of the primary cell clusters, comparing MAPT variant carriers to non-carrier controls and adjusting for age, sex, and scRNA-seq batch. Focusing initially on DEGs with absolute LFC > 0.2, we determined that cDC, NC monocyte, and NK cell clusters had the highest number of DEGs (Fig. 2A, B). We further characterized the DEGs of NC monocytes and NK cells by querying the STRING database [40] for functional and physical interactions, now using a more-permissive LFC cutoff of 0.1 to facilitate population of the gene interaction network (see Additional file 5: Table S4 for DEG lists for all clusters).
Fig. 2.
Differential expression in MAPT variant carriers by cell cluster. A Clusters are grouped by cell type and ranked by the number of DEGs with pFDR < 0.05 and absolute LFC > 0.2. Differential expression was determined in MAPT variant carriers relative to non-carrier controls while covarying for age, sex, and scRNA-seq batch. Solid-colored portions of the bars indicate DEGs shared by at least one other cluster, while the translucent portions indicate DEGs unique to a given cluster. cDCs (cluster 14), NK cells (clusters 3 and 15), and NC monocytes (cluster 11) had the highest numbers of DEGs with absolute LFC > 0.2. B Volcano plots of the NC monocyte cluster and major NK cell cluster; DEGs with absolute LFC > 0.2 are labeled in blue (downregulated) or red (upregulated). Several NK cell DEGs (right) with − log10(pFDR) values > 300 were set to 300 for visualization purposes
STRING network analysis
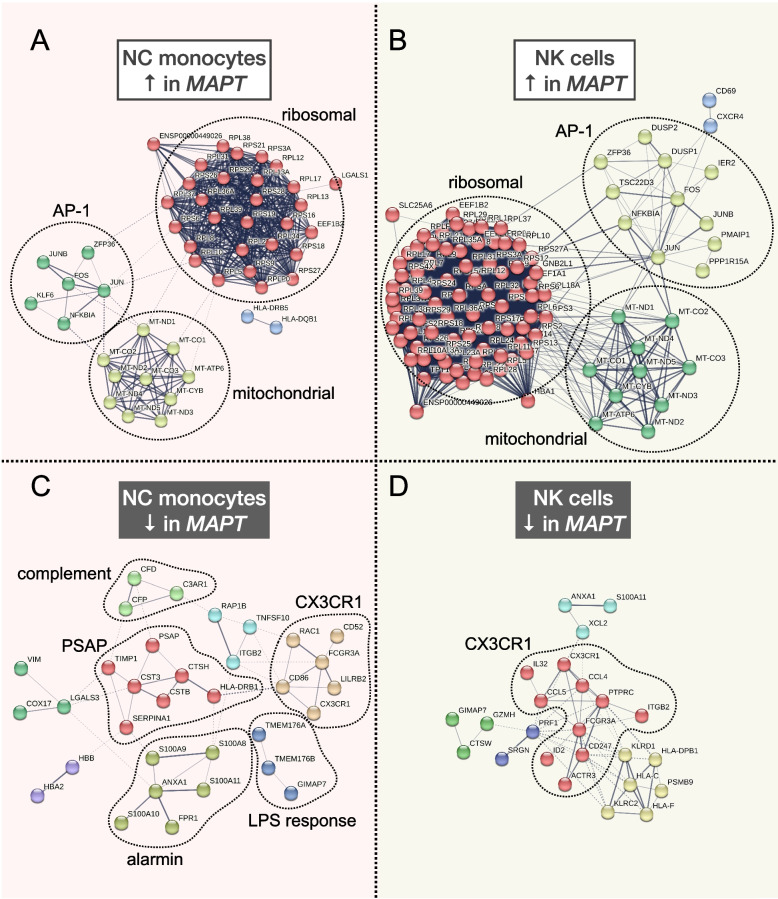
MAPT carriers showed striking upregulation of many ribosomal and mitochondrial genes that, respectively, formed large interaction modules (Fig. 3A, B). The biological significance of these coordinately upregulated DEGs is unclear, but tau is known to interact with and affect multiple ribosomal subunits [48–51] and mitochondrial proteins [51, 52]. Because the identified mitochondrial DEGs represent a subset of the mitochondrial genes used during QC (see “Methods”) to filter out putatively damaged cells [35], we considered the possibility that—despite the removal of cells with high mitochondrial mapping percentages (≥ 10%)—the apparent upregulation of mitochondrial DEGs may nevertheless be associated with higher mitochondrial mapping percentage in MAPT variant carriers. Consistent with this possibility, mitochondrial mapping percentage was subtly but significantly higher in MAPT carriers in the NC monocyte and NK cell clusters (11 and 3, respectively; Additional file 4: Fig. S4A, B). On the other hand, mitochondrial DEGs were absent from the cDC cluster (14; Additional file 4: Fig. S4D; Additional file 5: Table S4)—despite this cluster having the highest overall number of DEGs with LFC > 0.2 (Fig. 2A)—and mitochondrial mapping percentage was not significantly increased in variant carriers in this cluster (Additional file 4: Fig. S4C). This suggests (i) cell-type-specific and, presumably, biologically relevant dysregulation of mitochondrial genes in MAPT variant carriers, consistent with [51]; and (ii) that the upregulation of mitochondrial DEGs in the NC monocyte and NK cell clusters is associated with increased mitochondrial mapping percentage. To test the latter possibility, we next included mitochondrial mapping percentage as an additional covariate in the differential expression analyses, and, as expected, nearly all mitochondrial DEGs that previously had estimated LFCs > 0.1 no longer passed this threshold (not shown). Thus, the presence of mitochondrial DEGs is associated with increased mitochondrial mapping percentage. Importantly, coordinated upregulation of mitochondrial genes could lead to subtle shifts in mitochondrial mapping percentage, and therefore the causality underlying this relationship is unclear.
Fig. 3.
STRING interaction networks reveal relationships among nonclassical monocyte and natural killer cell differentially expressed genes. A, B Upregulated DEGs in NC monocytes and NK cells had similar overall network architecture, with large ribosomal and mitochondrial modules, and a third module containing members of the AP-1 transcription factor, among other genes. C The downregulated DEGs in NC monocytes contained a module featuring CX3CR1 and FCGR3A as members, in addition to modules harboring genes involved in LPS response, the alternative and classical complement cascades, the S100 alarmin molecules, as well as PSAP. D Downregulated DEGs in NK cells also featured a large module featuring both CX3CR1 and FCGR3A. All DEGs with pFDR < 0.05 and absolute LFC > 0.1 from clusters 3 and 11 were input into the STRING database as described in the “Methods” section. Modules are colored according to the results of MCL clustering
The other major upregulated STRING module found in both NC monocytes and NK cells contained members of the AP-1 transcription factor family [53], including FOS, JUN, and JUNB (Fig. 3A, B). Multiple members of this module (FOS, DUSP1) were previously found to be upregulated via bulk measurements of both PBMCs and hippocampus in AD [54], suggesting consistent dysregulation of AP-1 transcription factor genes in both primary and secondary tauopathies. NFKBIA, encoding the NF-κB inhibitor-ɑ, also appears in this module in both NC monocytes and NK cells, and this gene is highly upregulated by treatment of microglia with tau paired-helical filaments [55] and fibrils [56] and in the brain in late-onset AD [57].
In terms of significantly downregulated genes, both NC monocytes and NK cells contained a module populated by FCGR3A and CX3CR1 (Fig. 3C, D). Although FCGR3A is an established marker of NC monocytes, it is also expressed by a subset of NK cells (Fig. 1B). CX3CR1 is highly expressed by both NC monocytes and NK cells (Fig. 4A) and, strikingly, is a well-known modulator of tau pathophysiology in the brain [58–60]. Additional downregulated modules in NC monocytes included those containing components of the complement pathway (CFD, CFP, C3AR1 [61, 62]), members of the S100 alarmin family (S100A8-11 [63, 64]), highly lipopolysaccharide (LPS)-responsive microglial genes (TMEM176A, TMEM176B [65]), and the lysosomal gene PSAP, which promotes pro-inflammatory activity in monocytes [66] (Fig. 3C). Collectively, the downregulation of these gene modules in MAPT pathogenic variant carriers suggests possible dampening of latent pro-inflammatory pathways in NC monocytes. Of note, this apparent phenotypic change also occurred within a diminished population of circulating NC monocytes.
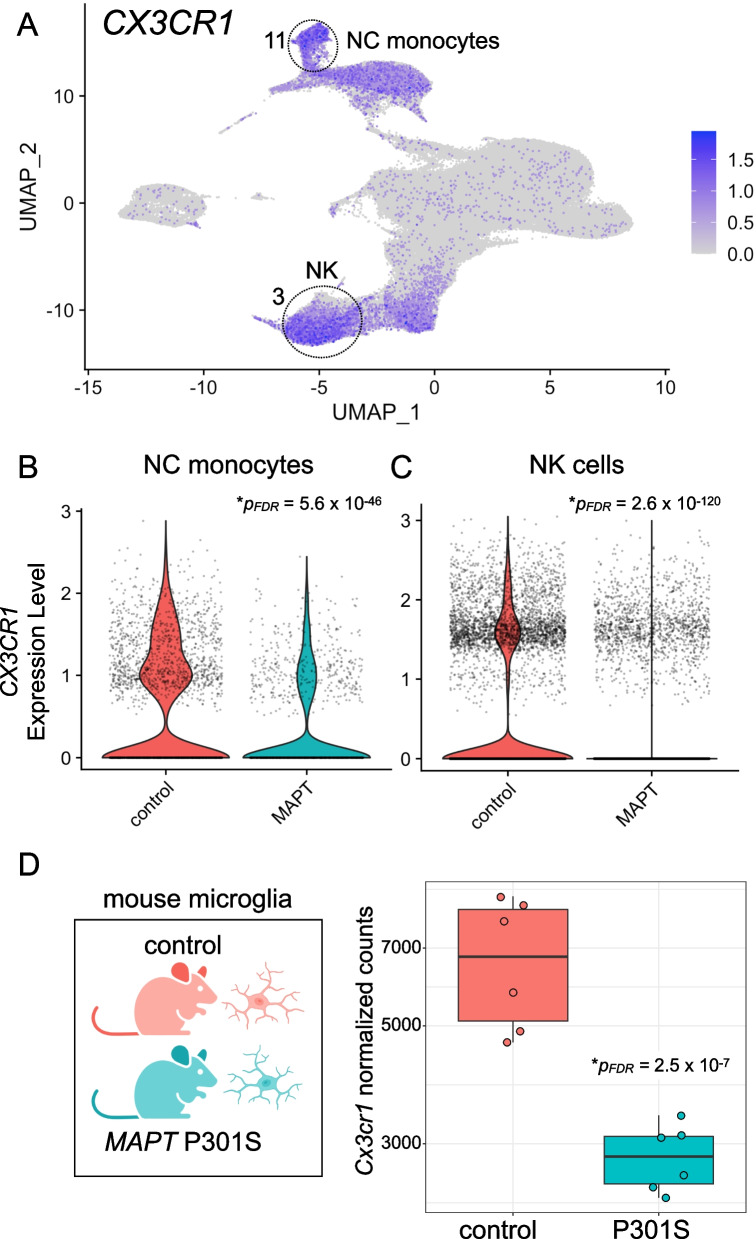
Fig. 4.
CX3CR1 expression is reduced in peripheral myeloid and lymphoid cells in familial tauopathy. A CX3CR1 is robustly expressed in both NC monocytes (cluster 11) and NK cells (cluster 3). NC monocytes (B; pFDR = 5.6 × 10−46) and NK cells (C; pFDR = 2.6 × 10−120) both show significantly reduced expression of CX3CR1 in MAPT pathogenic variant carriers. D Reanalysis of publicly available bulk RNA-seq data from mouse hippocampal CD11b+ microglia demonstrated a significant reduction (pFDR = 2.5 × 10−7) of Cx3cr1 in the MAPT P301S model
Validation of select differentially expressed genes
We next sought to validate a handful of DEGs which showed robust changes by differential expression analysis, often in multiple cell types, focusing on those deemed most likely to be biologically relevant to tau pathophysiology. In particular, we selected the following genes for validation via an orthogonal technique, ddPCR: CX3CR1, FCGR3A, TMEM176A, TMEM176B, and C3AR1. As noted above, CX3CR1 has many well-established connections to tau pathology via its role in microglia. However, to our knowledge, its role in peripheral myeloid cells has not been studied in the context of neurodegenerative disease. FCGR3A not only serves as a marker gene for the significantly reduced population of NC monocytes, but was also significantly downregulated in both NC monocytes and NK cells of MAPT carriers. TMEM176A/B are less well known but have been shown to be extremely responsive to LPS treatment in human microglia [65], are homologs of the MS4A gene family involved in risk for AD [67], and have also been shown to be dysregulated in PBMCs from AD patients [54]. Finally, C3AR1 has been implicated as a key player in the spread of tau neuropathology in mouse models [68], while the complement system more generally is thought to be a key regulator of neuronal loss in primary tauopathy as well as AD [69]. We first discuss validation of the CX3CR1 finding below.
Reduced CX3CR1 expression in familial tauopathy
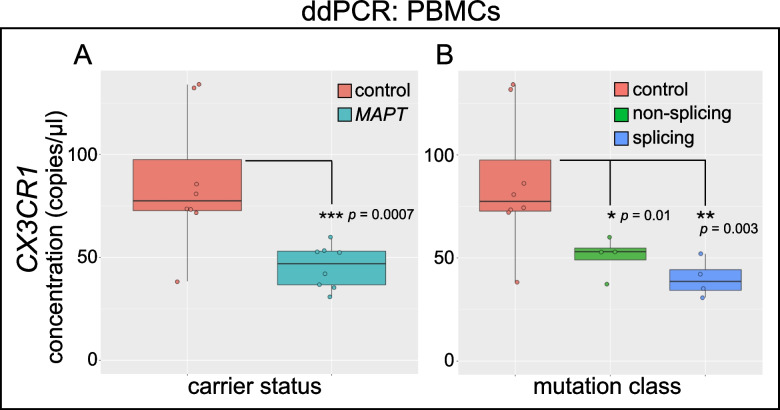
CX3CR1 showed robust expression in NC monocytes and NK cells (Fig. 4A), and its expression was significantly reduced in both cell types in MAPT pathogenic variant carriers (Fig. 4B, C). To determine whether downregulation in peripheral leukocytes was mirrored by changes in brain myeloid cells in the context of tauopathy, we analyzed a publicly available brain RNA-seq dataset [42] derived from MAPT P301S mouse hippocampal microglia. P301S microglia also displayed downregulation of Cx3cr1 (Fig. 4D), indicating consistent CX3CR1 dysregulation between mouse microglia and human peripheral immune cells in the context of tauopathy. Importantly, the P301S mutation used in this mouse model was not represented in our cohort, suggesting that the reduced peripheral CX3CR1 observed here may be a general feature of familial tauopathy and may therefore occur downstream of pathogenic MAPT variants beyond those directly assessed in this work. To validate the change in CX3CR1 expression in peripheral immune cells, we isolated PBMC RNA from MAPT pathogenic variant carriers and non-carrier controls and performed ddPCR. ddPCR confirmed the reduction in CX3CR1 (Fig. 5A). Normalizing the CX3CR1 signal to reference gene EEF2 gave similar results (Additional file 4: Fig. S5A, B). When MAPT variant carriers were stratified by variant class (splicing vs. non-splicing), both groups showed significant reductions in CX3CR1 (Fig. 5B), suggesting that multiple mechanistic forms of familial tauopathy converge on perturbation of CX3CR1 expression.
Fig. 5.
Confirmation of reduced CX3CR1 expression in MAPT variant carrier PBMCs via ddPCR. RNA was isolated from PBMCs from MAPT variant carriers and healthy, non-carrier controls; gene expression was determined by RT-ddPCR. A CX3CR1 was significantly reduced in MAPT carrier PBMCs (p = 0.0007) relative to controls. B Separation of samples according to MAPT variant class (non-splicing and splicing) reveals that CX3CR1 was significantly reduced in both groups, relative to controls (non-splicing, p = 0.01; splicing, p = 0.003)
Diminished FCGR3A expression in MAPT pathogenic variant carriers
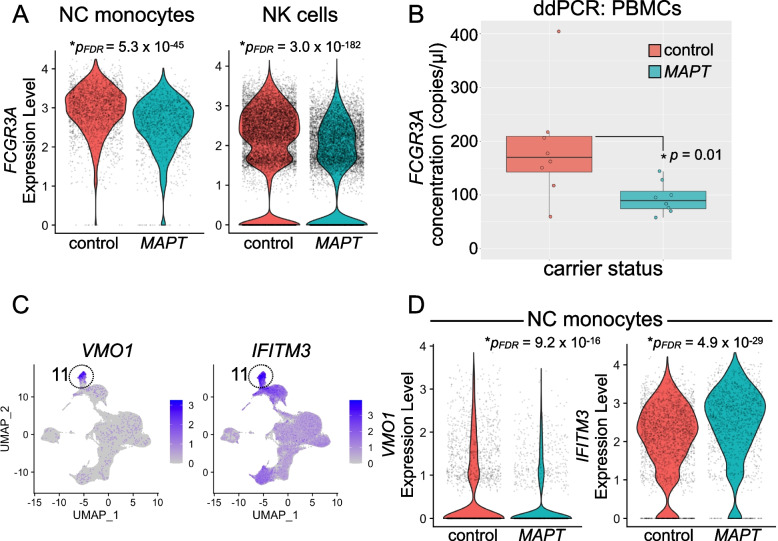
We next focused on validation of FCGR3A. We predicted that FCGR3A would show a robust decrease in expression in MAPT variant carriers in bulk PBMC RNA given that (i) NC monocytes are reduced in abundance in carriers (Fig. 1C), and (ii) FCGR3A expression was reduced in both NC monocytes and NK cells by scRNA-seq differential expression analyses (Figs. 6A and 2B). Indeed, ddPCR analysis showed a significant reduction in FCGR3A expression in MAPT pathogenic variant carrier PBMCs (Fig. 6B). The observed reduction likely reflects both reduced expression of FCGR3A and reduced levels of NC monocytes, which express FCGR3A at high levels. As with CX3CR1, normalization of the FCGR3A signal to reference gene EEF2 did not affect the results (Additional file 4: Fig. S5C). We also analyzed additional marker genes of the NC monocyte cluster, including VMO1 and IFITM3. VMO1, which is specifically expressed in cluster 11 (consistent with [17]), and IFITM3, which is enriched in cluster 11, both showed significant differential expression in MAPT variant carriers (Fig. 6C, D; Additional file 5: Table S4), indicating changes in multiple NC monocyte marker genes. Because the ddPCR and scRNA-seq studies described here depend on the use of cryopreserved PBMCs, we tested whether cryopreservation had a major effect on the measured expression levels of CX3CR1 and FCGR3A. Direct comparison by ddPCR of paired, fresh vs. frozen/thawed samples suggested that cryopreservation was associated with only a modest, ~ 15–20% reduction in the apparent expression of these transcripts (Additional file 4: Fig. S6A, B). Cryopreservation, therefore, was not associated with a major change in the levels of CX3CR1 and FCGR3A.
Fig. 6.
Analysis of nonclassical monocyte marker genes in MAPT variant carriers. A FCGR3A, the NC monocyte marker gene encoding CD16, is robustly expressed not only in NC monocytes but also in NK cells. FCGR3A is significantly reduced in both NC monocytes (left; pFDR = 5.3 × 10−45) and NK cells (right; pFDR = 3.0 × 10−182) in MAPT pathogenic variant carriers. B ddPCR confirmed a reduction in FCGR3A reduction in MAPT variant carrier PBMCs (p = 0.01). C Additional genes expressed specifically (VMO1, left) or enriched in (IFITM3, right) NC monocytes showed significant alterations (D) in MAPT variant carrier NC monocytes. D VMO1 (left) was significantly reduced (pFDR = 9.2 × 10−16), while IFITM3 (right) was significantly increased (pFDR = 4.9 × 10−29) in MAPT carriers
Analysis of TMEM176A/B in familial tauopathy
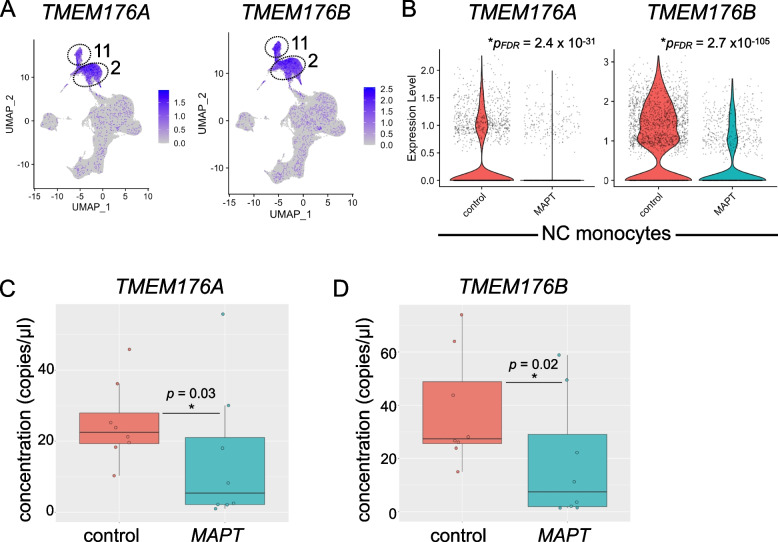
TMEM176A/B represent poorly characterized genes of the extended MS4A family [67] thought to be involved in microglial LPS response [65], antigen presentation [70], and inflammasome regulation [71]. TMEM176A/B were highly expressed in classical and NC monocytes (clusters 2 and 11; Fig. 7A) and were strongly downregulated in MAPT variant carrier NC monocytes (Fig. 7B). Reduced expression of TMEM176A and TMEM176B was confirmed via ddPCR of PBMC RNA (Fig. 7C, D). The levels of TMEM176A and TMEM176B, as detected by ddPCR, were tightly associated with one another, and a subset of MAPT pathogenic variant carriers (5 of 8) displayed lower expression of both genes than any non-carrier controls (Additional file 4: Fig. S5D).
Fig. 7.
Analysis of TMEM176A/B in MAPT pathogenic variant carriers. A TMEM176A/B are highly expressed in both classical (cluster 2) and NC (cluster 11) monocytes. B TMEM176A/B are significantly reduced in NC monocytes (TMEM176A, pFDR = 2.4 × 10−31; TMEM176B, pFDR = 2.7 × 10.−105) from MAPT carriers. These genes were similarly reduced in MAPT carrier classical monocytes (cluster 2; Additional file 5: Table S4). The reduction in TMEM176A (C) and TMEM176B (D) in MAPT variant carriers was confirmed using bulk PBMC RNA and ddPCR (TMEM176A, p = 0.03; TMEM176B, p = 0.02)
Analysis of C3AR1 in MAPT pathogenic variant carriers
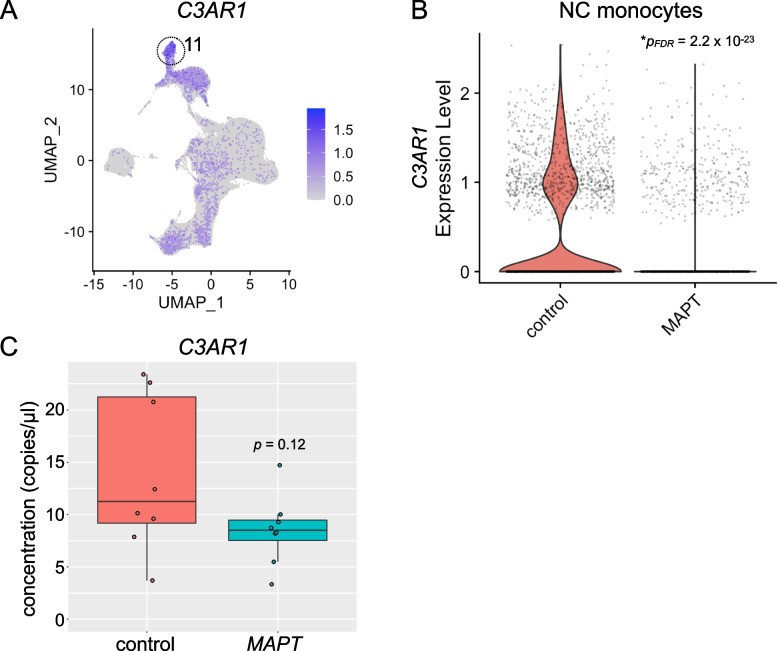
Finally, we examined the expression of C3AR1 given its importance in models of tauopathy [68]. C3AR1 expression was enriched in NC monocytes (Fig. 8A) and was strikingly reduced in this cell type in MAPT variant carriers (Fig. 8B). C3AR1 expression, as measured by ddPCR of PBMC RNA, trended toward reduction but did not achieve significance (Fig. 8C). However, given the importance of this gene and the complement pathway more generally in tauopathy and AD, analysis of peripheral C3AR1 expression in larger, better-powered cohorts is still warranted.
Fig. 8.
Potential dysregulation of C3AR1 in MAPT pathogenic variant carriers. A C3AR1 expression was enriched in the NC monocyte cluster (11). B C3AR1 expression in NC monocytes was significantly reduced in MAPT variant carriers (*pFDR = 2.2 × 10.−23). C ddPCR analysis of PBMC RNA revealed a trend toward reduced expression of C3AR1 in MAPT variant carriers which did not reach significance (p = 0.12)
Validation of the reduction in nonclassical monocytes via flow cytometry
To determine whether a central finding of the scRNA-seq analysis could be directly validated via an independent method, we performed multicolor flow cytometry analysis on PBMCs from MAPT pathogenic variant carriers and non-carrier controls. PBMCs, monocytes, and monocyte subtypes were gated as illustrated in Fig. 9A and B and as described in detail in the “Methods” section. Importantly, this analysis enabled us to confirm a significant reduction in NC monocytes (characterized by low CD14 expression and high CD16 expression) as a percentage of all PBMCs (p = 0.02; Fig. 9C, upper left). We also observed a marginally significant reduction in NC monocytes expressed as a percentage of all monocytes (p = 0.05; Fig. 9C; lower left). Intermediate monocytes (characterized by high CD14 levels and moderate-to-high CD16 levels) showed a trend toward reduced abundance, relative to either all PBMCs or all monocytes, but this difference did not reach significance (Fig. 9C, middle panels). Finally, classical monocytes (characterized by high CD14 levels and low expression of CD16) showed no change as a fraction of all PBMCs, but, as expected—given the relative reductions in NC and intermediate monocytes—showed a significant increase when expressed as a percentage of all monocytes (p = 0.04; Fig. 9C, lower right). No differences in viability were detected between MAPT pathogenic variant carriers and non-carrier controls (Additional file 4: Fig. S7). An alternative gating scheme that enabled the inclusion of additional NC monocytes with lower levels of CD14 expression resulted in very similar findings (Additional file 4: Fig. S8). Taken together, our observations are consistent with a bona fide decrease in the frequency of NC monocytes in MAPT pathogenic variant carriers, and this alteration does not appear to be secondary to an absolute increase in classical monocyte abundance. Finally, we also asked whether alterations in cell-surface expression of CX3CR1 and CD16 could be detected via flow cytometry on NC monocytes or NK cells. To do so, we measured the median fluorescence intensity (MFI) for CX3CR1 and CD16 in each cell population, but we did not observe a difference between MAPT carriers and non-carrier controls (data not shown). We infer from this that the ~ 50% reduction in transcript abundance for CX3CR1 and FCGR3A is insufficient to alter the cell-surface levels of their encoded proteins. Alternatively, there may be a more general divergence between the dysregulation of these transcripts and their encoded proteins. Future work will test these two possibilities.
Fig. 9.
Validation of the reduction in nonclassical monocytes via flow cytometry. A Gating scheme for identification and analysis of monocyte subtypes. PBMCs were gated as follows: debris was excluded, non-viable cells were excluded, then doublets were excluded. Next, monocytes were gated based on their high side scatter and CD14 expression. B Monocyte subtypes were gated based on their characteristic CD14 and CD16 expression, with classical monocytes having high CD14 expression and low CD16 expression, intermediate monocytes having high CD14 expression and moderate-to-high CD16 expression, and NC monocytes having low CD14 expression and high CD16 expression. C Quantification of the frequency of NC (left), intermediate (center), and classical monocytes (right), either as a percentage of PBMCs (top row) or all monocytes (bottom row). NC monocytes were reduced in MAPT pathogenic variant carriers as a fraction of PBMCs (upper left, p = 0.02) and as a fraction of monocytes (lower left, p = 0.05). Intermediate monocytes (center) showed a trend toward reduction relative to both PBMCs and monocytes. Classical monocytes (right) showed no change as a fraction of PBMCs but were significantly increased in MAPT pathogenic variant carriers as a fraction of all monocytes
Analysis of gene expression in human brain nonclassical monocytes
To begin to understand why peripheral NC monocytes are affected in familial tauopathy, we asked whether these cells have access to the human central nervous system (CNS). Indeed, very recent work has suggested an important role for human NC monocytes in the CSF during aging [72], and a recent analysis [23] of publicly available scRNA-seq data [46, 73] has also identified these cells in the human temporal cortex. We downloaded and reanalyzed this dataset (accession GSE137444, [46]) and similarly identified a clear NC monocyte population (cluster 13 in our reanalysis; Additional file 4: Fig. S9A), thus recapitulating the findings from [23]. This cluster was unique in its high expression of NC monocyte marker genes such as LYZ, S100A8, S100A9, and FCN1 (marker genes from [23]; Additional file 4: Fig. S9B). We next assessed expression levels of the DEGs that we focused on in this work—CX3CR1, FCGR3A, TMEM176A/B, and C3AR1. We detected robust expression of all genes except TMEM176A in the brain NC monocyte cluster (Additional file 4: Fig. S9C). Collectively, these findings suggest that NC monocytes have ready access to the human brain and potentially explain why robust changes in NC monocytes can be observed in the context of tauopathy—that is, they may have direct access to tissue harboring tau pathology. Future functional studies will be needed to determine NC monocytes’ potential to impact the course of tauopathy. In particular, depletion of NC monocytes in mouse models of tauopathy would enable direct testing of the role these cells play in the development of tau neuropathology.
Discussion
This study represents an effort to discover novel, blood-based biomarkers of tauopathy, and—more broadly—to begin to understand the nature of the peripheral leukocyte response to primary tauopathy. To this end, we conducted an unbiased scRNA-seq survey of ~ 181,000 PBMCs in carriers of pathogenic MAPT variants and healthy, non-carrier controls. In doing so, we uncovered novel, peripheral blood transcriptomic signatures of familial tauopathy at single-cell resolution. In particular, we observed a significant reduction in circulating NC monocytes and numerous DEGs that were particularly enriched in specific myeloid and NK cell clusters. Next, we validated changes in several candidate DEGs selected on the basis of plausible biological relevance. These included CX3CR1, FCGR3A, and TMEM176A/B. In addition, C3AR1, although not found to be significantly reduced in MAPT carriers by ddPCR, still trended toward reduced expression.
The differential expression of CX3CR1 observed by scRNA-seq was not only replicated via ddPCR analyses but also confirmed using a publicly available mouse microglia bulk RNA-seq dataset derived from the MAPT P301S model. This suggests CX3CR1 may have potential utility as a peripheral biomarker of tauopathy and warrants further study in larger cohorts. Intriguingly, mouse Cx3cr1 has been known for over a decade to control the levels of NC monocytes [74–76]. In addition, mouse Cx3cr1-mediated control of NC monocyte levels has more recently been suggested to modulate the innate immune response to traumatic brain injury [77]. Collectively, these findings suggest that the reduction in CX3CR1 expression we observe in MAPT pathogenic variant carrier NC monocytes (on a per-cell basis) may be directly related to the concomitant reduction in NC monocyte abundance also observed in MAPT carriers.
Quite aside from the role of Cx3cr1 in controlling circulating NC monocyte levels, Cx3cr1 exerts a well-established, microglia-mediated modulatory effect on tau neuropathology in mouse models [58–60, 78–84]. In particular, deletion of Cx3cr1 promotes hallmark neuropathological features of tauopathy including tau hyperphosphorylation and aggregation [58–60, 84]. More-recent studies on induced pluripotent stem cell-derived microglia-like cells have confirmed the importance of CX3CR1 in regulating human microglial homeostasis [85], consistent with Cx3cr1’s established role in promoting the homeostatic microglial phenotype in mice [86]. Given that microglial homeostasis is dysregulated in tauopathy [87–90], our novel findings—coupled with the well-defined relationship between CX3CR1 and tauopathy—provide a promising foundation for further investigation of this gene as a peripheral biomarker of tauopathy.
As noted above, we observed a significant reduction in NC monocytes in MAPT carriers relative to non-carrier controls. NC monocytes are recruited to sites of vascular damage, infection, or inflammation to patrol the local environment [76]. At these compromised sites, chemoattractant factors are released, and NC monocytes respond through the expression of cognate receptors, including CX3CR1 [76]. Strikingly, NC monocytes are thought to be reduced in peripheral blood in a variety of neurodegenerative diseases, including ALS [15, 16], PD [14], and the adult-onset, hereditary leukoencephalopathy, ALSP [17]. Conversely, NC monocyte levels may be increased in the CSF in PD, suggesting a possible shift of this monocyte population from blood to CSF in the context of neurodegeneration [91]. Considering these findings, and given that (i) ALS exists along a spectrum with FTD [92]; (ii) a portion of PD risk is mediated by variation near the MAPT locus [93, 94]; and (iii) ALSP can manifest clinically as FTD [18], our finding of reduced NC monocytes in familial tauopathy both strengthens and extends the purported relevance of this monocyte population in neurodegenerative disease.
We observed significantly reduced expression of the canonical NC monocyte marker gene FCGR3A as well as significant alterations in two additional NC monocyte marker genes (VMO1 and IFITM3) in MAPT pathogenic variant carriers by scRNA-seq. Given that FCGR3A expression was reduced not only in NC monocytes but also in NK cells and considering that NC monocyte abundance was simultaneously reduced, we reasoned that bulk assessment of PBMC RNA via ddPCR would be well suited to detect reduced expression of FCGR3A in MAPT carriers, and, indeed, this is what we found. This cellular phenotype suggests several possibilities. First, the reduced abundance of NC monocytes and diminished expression of FCGR3A on the remaining NC monocytes could reflect migration of mature NC monocytes (that express the highest levels of FCGR3A) out of the blood and into another compartment (e.g., CSF or brain). Second, our findings could reflect impaired survival of NC monocytes in tauopathy. Third, lower FCGR3A levels might reflect impaired differentiation of NC monocytes from classical or intermediate monocytes (the latter of which express intermediate levels of FCGR3A). We favor the first two possibilities, as the latter scenario would be expected to involve accumulation of other classes of monocytes, which we did not observe.
Our differential expression analyses identified cDCs (cluster 14) as the cell type with the highest number of DEGs with absolute LFC > 0.2. Although we did not focus on cDCs for our validation studies, we did find that normalizing NC monocyte abundance to cDC abundance enabled a clear separation of MAPT carriers from non-carrier controls. This finding begets the question: what is the biological link connecting NC monocytes to cDCs? Emerging literature has highlighted several intriguing connections between NC monocytes and particular subsets of DCs. For example, a putative DC subpopulation, identified transcriptomically and originally termed DC4 [95], is now considered to probably represent a subset of NC monocytes rather than DCs [47, 96, 97]. In addition, pathogenic variants in STAT3 have revealed this gene’s role in regulating the production of both NC monocytes and the less-abundant cDC population, cDC1 [98]. Although we could detect the rare cDC1 population in our dataset, it became apparent only upon myeloid cell re-clustering (myeloid subcluster 7, marked by CLEC9A expression), and these cells were too sparse to enable us to accurately gauge their abundance or use for normalization purposes. On the other hand, it remains unclear precisely how NC monocytes are biologically related to the more-abundant cDC2 population. Nevertheless, normalizing NC monocyte abundance to either cDC2 or total cDC abundance enabled a clear separation of MAPT variant carriers from healthy controls. Future studies in larger cohorts will be required to determine the precise quantification metric for NC monocytes that best differentiates MAPT carriers from controls. It will also be important to establish whether this finding extends to sporadic forms of tauopathy; this seems likely given that similar phenomena have been reported in disparate neurodegenerative diseases.
Recently published work indicates robust transcriptional changes in NC monocytes within the CSF during healthy aging and in the context of cognitive impairment [72]. An additional recent finding [23], which we confirm here (Additional file 4: Fig. S9), indicates that NC monocytes can also be found in the human temporal cortex. Given that the precise role of human NC monocytes in health and disease is still largely unknown [76], it remains unclear what a reduction in peripheral NC monocytes means vis-à-vis tauopathy. If the observed reduction in peripheral NC monocytes is accompanied by a corresponding increase in CNS NC monocytes—and if they contribute to heightened neuroinflammation—they are likely to play a net detrimental role in neurodegeneration. However, much work remains to be done to test these possibilities.
Beyond myeloid cells, our work also highlights a potentially novel role for NK cells in primary tauopathy. In particular, NK cells had a large number of DEGs with LFC > 0.2, and our findings implicating CX3CR1 expression not only in NC monocytes but also in NK cells as a candidate peripheral biomarker of familial tauopathy is complemented by recent research suggesting an important yet previously unappreciated role for NK cells in a mouse model of AD [99]. In addition, NK cell recruitment to the CNS has been observed in ALS as well as ALS models [100]. Collectively, the available data suggest a detrimental role for NK cells in ALS and AD. More broadly, in the experimental autoimmune encephalomyelitis model of multiple sclerosis, NK cell migration into the CNS is mediated in part by CX3CR1-dependent recruitment [101, 102], suggesting that the differential expression of CX3CR1 in NK cells that we observed in our study could plausibly affect NK cell recruitment to the CNS in primary tauopathy.
Reliable biomarkers can improve diagnostic acumen and enable elucidation of specific forms of neuropathology underlying clinical dementia syndromes. For example, examination of brain structure and function via neuroimaging is a powerful method for the determination of neurodegenerative disease etiology. The use of positron emission tomography (PET) imaging, in particular, with radiotracers that bind to aggregated forms of tau has facilitated the in vivo detection of tau neuropathology in individuals with AD (reviewed in [19, 103, 104]). However, tau-PET tracers do not bind strongly to most forms of FTLD-tau pathology and may exhibit off-target binding in individuals with FTLD-TDP pathology [103, 104]. Alternatively, the use of CSF- and blood-based protein biomarkers holds great promise for AD [19, 20, 105] and FTD [21, 106], although in the case of FTD, we still cannot discriminate between underlying FTLD-tau and -TDP pathology. Important limitations apply to several of these methods. In particular, PET imaging is costly and available only at specialized medical centers, and CSF collection requires invasive lumbar puncture. In contrast to these methods, peripheral blood biomarkers are easy to collect and, when coupled with analytic techniques such as ddPCR, may eventually enable low-cost alternatives to today’s better-developed biomarkers.
Despite numerous advances described above, our study has several important limitations. First, due to the significant expense of scRNA-seq and our desire to capture a relatively large number of PBMCs per individual, we necessarily used a small cohort for this study. We also opted to confirm gene expression findings via an orthogonal technique using bulk PBMC RNA measurements in essentially the same cohort that was used for the scRNA-seq analysis. While we employed this strategy to minimize the possibility that technical artifacts drove discovery of the candidate genes we characterized, it will be important to evaluate the generalizability of our findings in larger cohorts. Larger cohorts will also enable us to determine whether any of the dysregulated genes identified here are differentially sensitive to pathogenic MAPT variants that affect splicing, relative to those that do not, as potentially suggested for CX3CR1 (Fig. 5B). In addition, future work that measures absolute numbers of NC monocytes in familial tauopathy would be a valuable addition to the field, as the scRNA-seq and flow cytometry data reported here show relative reductions in NC monocyte frequency in MAPT pathogenic variant carriers. Another issue to be resolved in future studies is the biological significance of the upregulation of ribosomal and mitochondrial transcripts in myeloid and other cell types observed here in familial tauopathy. For example, do these changes in gene expression in NC monocytes primarily reflect increased fragility and/or decreased survival of these cells in tauopathy, or are they reflective of more specific changes in ribosomal and mitochondrial biology?
As alluded to above, it will also be important to determine which peripheral immune changes are conserved between familial tauopathy and diverse forms of sporadic primary and secondary tauopathy. In addition, given the complex temporal trajectories of brain myeloid responses in tauopathy [107], future research on large cohorts of presymptomatic MAPT pathogenic variant carriers will be needed to determine which peripheral changes observed here occur prior to disease onset. Finally, it will be important to ascertain whether the peripheral leukocyte changes discovered here are reflected by parallel changes in brain myeloid cells in individuals with tauopathy.
Conclusions
To our knowledge, this is the first scRNA-seq study of peripheral blood cells in primary tauopathy. Beyond our initial discoveries, we validated a handful of DEGs via an orthogonal technique, ddPCR. In particular, we have connected longstanding observations from mouse models regarding microglial Cx3cr1 and tau neuropathology to reduced CX3CR1 in peripheral leukocytes in individuals with familial tauopathy. Moreover, we discovered a significant reduction in the abundance of circulating NC monocytes, a cell type that is similarly reduced in several additional neurodegenerative diseases. We also discovered large numbers of DEGs in NK cells, including CX3CR1, which is thought to be involved in recruitment of NK cells to the CNS. Further studies are now required to investigate the generalizability of our findings through replication in larger cohorts and extension to other tauopathies and related neurodegenerative diseases. Analogous studies of PBMCs in GRN pathogenic variant carriers and C9orf72 hexanucleotide repeat expansion carriers should enable the discovery of peripheral biomarkers of FTLD-TDP. Ultimately, comparative studies should clarify the role of peripheral immune responses in distinct proteinopathies and enable discovery of novel peripheral biomarkers that can successfully discriminate between tau and TDP-43 neuropathology, providing critical new tools for diagnostics and clinical trials.
Supplementary Information
Additional file 1: Table S1. Cell Ranger metrics summarizing important characteristics of the detected cells for each sample.
Additional file 2: Table S2. Cell counts during quality-control steps for each sample.
Additional file 3: Table S3. RNA integrity number for PBMC samples.
Additional file 4: Figures S1-S9. All supplementary figures.
Additional file 5: Table S4. Cell-type specific differential expression analyses.
Acknowledgements
We thank the UCSF Genomics CoLab and IHG staff, including Catherine Chu, for expert technical assistance. In addition, we thank UCSF CAT staff, including Tyler Miyasaki, for expert advice on the use of ddPCR equipment. Finally, we are grateful to Collin Spencer in the UCSF Department of Neurology for his help and expert advice related to flow cytometry. The cartoon in Fig. 4D was created with BioRender.com.
Abbreviations
- AD
Alzheimer’s disease
- ALS
Amyotrophic lateral sclerosis
- ALSP
Adult-onset leukoencephalopathy with axonal spheroids and pigmented glia
- CAT
Center for Advanced Technology
- cDC
Conventional dendritic cell
- CDR-SB
Clinical Dementia Rating scale Sum of Boxes
- CNS
Central nervous system
- CSF
Cerebrospinal fluid
- dd
Droplet digital
- DEG
Differentially expressed gene
- FDR
False discovery rate
- FTD
Frontotemporal dementia
- FTLD
Frontotemporal lobar degeneration
- IHG
Institute for Human Genetics
- LFC
Log2 fold change
- LPS
Lipopolysaccharide
- MAC
Memory and Aging Center
- MCL
Markov cluster algorithm
- NC
Nonclassical
- NK
Natural killer
- PBMCs
Peripheral blood mononuclear cells
- PCA
Principal component analysis
- PD
Parkinson’s disease
- PET
Positron emission tomography
- QC
Quality control
- RIN
RNA integrity number
- RT
Reverse transcription
- scRNA-seq
Single-cell RNA sequencing
- UCSF
University of California, San Francisco
- UMAP
Uniform manifold approximation and projection
Authors’ contributions
DWS initiated the study; analyzed the scRNA-seq, ddPCR, and flow cytometry data; and wrote the manuscript. CWS analyzed the scRNA-seq data and contributed to writing the manuscript. TPJ isolated PBMCs and performed ddPCR and flow cytometry experiments with DWS. LWB analyzed scRNA-seq data and edited the manuscript. VES, SEL, KPR, HJR, ALB, WWS, and BLM were involved in research participant recruitment and clinical characterization. EGG was involved in early stages of the scRNA-seq analysis and provided guidance on study design. JSY initiated the study, oversaw the analysis, and edited the manuscript. All authors read and approved the final manuscript.
Funding
Funding to JSY and for this project comes from NIH-NIA K01AG049152, R01AG062588, R01AG057234, P30AG062422, P01AG019724; NIH-NINDS U54NS123985; the Rainwater Charitable Foundation; the Larry L. Hillblom Foundation; the Bluefield Project to Cure Frontotemporal Dementia; the Alzheimer’s Association; the Global Brain Health Institute; the French Foundation; and the Mary Oakley Foundation. VES is supported by R01AG052496. SEL is supported by R01AG058233. ALB receives research support from NIH (U19AG063911, R01AG038791, R01AG073482), the Tau Research Consortium, the Association for Frontotemporal Degeneration, Bluefield Project to Cure Frontotemporal Dementia, Corticobasal Degeneration Solutions, the Alzheimer’s Drug Discovery Foundation, and the Alzheimer’s Association, and has received research support from Biogen, Eisai and Regeneron. The content of this publication is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Availability of data and materials
The scRNA-seq dataset described here has been uploaded to the FAIR Data Sharing Portal within the Alzheimer’s Disease Workbench, which is supported by the Alzheimer’s Disease Data Initiative, and can be accessed at https://fair.addi.ad-datainitiative.org/#/data/datasets/single_cell_rna_seq_data_derived_from_mapt_carriers_and_controls.
Declarations
Ethics approval and consent to participate
All participants or their surrogates provided written informed consent prior to study participation; all aspects of the studies described here were approved by the UCSF institutional review board (committee number 10–03946). The research described herein conformed to the principles of the Helsinki Declaration.
Consent for publication
Not applicable.
Competing interests
JSY serves on the scientific advisory board for the Epstein Family Alzheimer’s Research Collaboration. ALB has served as a consultant for Aeovian, AGTC, Alector, Arkuda, Arvinas, AviadoBio, Boehringer Ingelheim, Denali, GSK, Life Edit, Humana, Oligomerix, Oscotec, Roche, Transposon, TrueBinding and Wave. The remaining authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Daniel W. Sirkis and Caroline Warly Solsberg have equal contribution.
References
- 1.Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H, et al. Association of missense and 5’-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- 2.Spillantini MG, Murrell JR, Goedert M, Farlow MR, Klug A, Ghetti B. Mutation in the tau gene in familial multiple system tauopathy with presenile dementia. Proc Natl Acad Sci U S A. 1998;95:7737–7741. doi: 10.1073/pnas.95.13.7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sirkis DW, Geier EG, Bonham LW, Karch CM, Yokoyama JS. Recent advances in the genetics of frontotemporal dementia. Curr Genet Med Rep. 2019;7:41–52. doi: 10.1007/s40142-019-0160-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.VandeVrede L, Boxer AL, Polydoro M. Targeting tau: Clinical trials and novel therapeutic approaches. Neurosci Lett. 2020;731:134919. doi: 10.1016/j.neulet.2020.134919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang CW, Shao E, Mucke L. Tau: Enabler of diverse brain disorders and target of rapidly evolving therapeutic strategies. Science. 2021;371:eabb8255. doi: 10.1126/science.abb8255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosen HJ, Boeve BF, Boxer AL. Tracking disease progression in familial and sporadic frontotemporal lobar degeneration: recent findings from ARTFL and LEFFTDS. Alzheimers Dement. 2020;16:71–78. doi: 10.1002/alz.12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song WM, Colonna M. The identity and function of microglia in neurodegeneration. Nat Immunol. 2018;19:1048–1058. doi: 10.1038/s41590-018-0212-1. [DOI] [PubMed] [Google Scholar]
- 8.Haage V, De Jager PL. Neuroimmune contributions to Alzheimer’s disease: a focus on human data. Mol Psychiatry. 2022;27:3164. doi: 10.1038/s41380-022-01637-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Y, Colonna M. Spontaneous and induced adaptive immune responses in Alzheimer’s disease: new insights into old observations. Curr Opin Immunol. 2022;77:102233. doi: 10.1016/j.coi.2022.102233. [DOI] [PubMed] [Google Scholar]
- 10.Sirkis DW, Bonham LW, Karch CM, Yokoyama JS. Immunological signatures in frontotemporal lobar degeneration. Curr Opin Neurol. 2019;32:272–278. doi: 10.1097/WCO.0000000000000665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gate D, Saligrama N, Leventhal O, Yang AC, Unger MS, Middeldorp J, et al. Clonally expanded CD8 T cells patrol the cerebrospinal fluid in Alzheimer’s disease. Nature. 2020;577:399–404. doi: 10.1038/s41586-019-1895-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gate D, Tapp E, Leventhal O, Shahid M, Nonninger TJ, Yang AC, et al. CD4 T cells contribute to neurodegeneration in Lewy body dementia. Science. 2021;374:868–874. doi: 10.1126/science.abf7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phongpreecha T, Fernandez R, Mrdjen D, Culos A, Gajera CR, Wawro AM, et al. Single-cell peripheral immunoprofiling of Alzheimer’s and Parkinson's diseases. Sci Adv. 2020;6:eabd5575. doi: 10.1126/sciadv.abd5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grozdanov V, Bliederhaeuser C, Ruf WP, Roth V, Fundel-Clemens K, Zondler L, et al. Inflammatory dysregulation of blood monocytes in Parkinson’s disease patients. Acta Neuropathol. 2014;128:651–63. doi: 10.1007/s00401-014-1345-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zondler L, Müller K, Khalaji S, Bliederhäuser C, Ruf WP, Grozdanov V, et al. Peripheral monocytes are functionally altered and invade the CNS in ALS patients. Acta Neuropathol. 2016;132:391–411. doi: 10.1007/s00401-016-1548-y. [DOI] [PubMed] [Google Scholar]
- 16.McGill RB, Steyn FJ, Ngo ST, Thorpe KA, Heggie S, Ruitenberg MJ, et al. Monocytes and neutrophils are associated with clinical features in amyotrophic lateral sclerosis. Brain Commun. 2020;2:fcaa013. doi: 10.1093/braincomms/fcaa013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hofer TP, Zawada AM, Frankenberger M, Skokann K, Satzl AA, Gesierich W, et al. slan-defined subsets of CD16-positive monocytes: impact of granulomatous inflammation and M-CSF receptor mutation. Blood. 2015;126:2601–2610. doi: 10.1182/blood-2015-06-651331. [DOI] [PubMed] [Google Scholar]
- 18.Sirkis DW, Bonham LW, Yokoyama JS. The role of microglia in inherited white-matter disorders and connections to frontotemporal dementia. Appl Clin Genet. 2021;14:195–207. doi: 10.2147/TACG.S245029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sirkis DW, Bonham LW, Johnson TP, La Joie R, Yokoyama JS. Dissecting the clinical heterogeneity of early-onset Alzheimer’s disease. Mol Psychiatry. 2022;27:2674–2688. doi: 10.1038/s41380-022-01531-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karikari TK, Ashton NJ, Brinkmalm G, Brum WS, Benedet AL, Montoliu-Gaya L, et al. Blood phospho-tau in Alzheimer disease: analysis, interpretation, and clinical utility. Nat Rev Neurol. 2022;18:400–418. doi: 10.1038/s41582-022-00665-2. [DOI] [PubMed] [Google Scholar]
- 21.Swift IJ, Sogorb-Esteve A, Heller C, Synofzik M, Otto M, Graff C, et al. Fluid biomarkers in frontotemporal dementia: past, present and future. J Neurol Neurosurg Psychiatry. 2021;92:204–215. doi: 10.1136/jnnp-2020-323520. [DOI] [PubMed] [Google Scholar]
- 22.Cousins KAQ, Shaw LM, Chen-Plotkin A, Wolk DA, Van Deerlin VM, Lee EB, et al. Distinguishing frontotemporal lobar degeneration tau from TDP-43 using plasma biomarkers. JAMA Neurol. 2022;79:1155. doi: 10.1001/jamaneurol.2022.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silvin A, Uderhardt S, Piot C, Da Mesquita S, Yang K, Geirsdottir L, et al. Dual ontogeny of disease-associated microglia and disease inflammatory macrophages in aging and neurodegeneration. Immunity. 2022;55:1448–65.e6. doi: 10.1016/j.immuni.2022.07.004. [DOI] [PubMed] [Google Scholar]
- 24.Miller ZA, Mandelli ML, Rankin KP, Henry ML, Babiak MC, Frazier DT, et al. Handedness and language learning disability differentially distribute in progressive aphasia variants. Brain. 2013;136:3461–3473. doi: 10.1093/brain/awt242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rascovsky K, Hodges JR, Knopman D, Mendez MF, Kramer JH, Neuhaus J, et al. Sensitivity of revised diagnostic criteria for the behavioural variant of frontotemporal dementia. Brain. 2011;134:2456–2477. doi: 10.1093/brain/awr179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ossenkoppele R, Pijnenburg YAL, Perry DC, Cohn-Sheehy BI, Scheltens NME, Vogel JW, et al. The behavioural/dysexecutive variant of Alzheimer’s disease: clinical, neuroimaging and pathological features. Brain. 2015;138:2732–2749. doi: 10.1093/brain/awv191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morris JC. The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology. 1993;43:2412–2414. doi: 10.1212/WNL.43.11.2412-a. [DOI] [PubMed] [Google Scholar]
- 28.Haque A, Engel J, Teichmann SA, Lönnberg T. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med. 2017;9:75. doi: 10.1186/s13073-017-0467-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen QH, Pervolarakis N, Nee K, Kessenbrock K. Experimental considerations for single-cell RNA sequencing approaches. Front Cell Dev Biol. 2018;6:108. doi: 10.3389/fcell.2018.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, 3rd, et al. Comprehensive integration of single-cell data. Cell. 2019;177:1888–902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hao Y, Hao S, Andersen-Nissen E, Mauck WM, 3rd, Zheng S, Butler A, et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–87.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGinnis CS, Murrow LM, Gartner ZJ. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 2019;8:329–37.e4. doi: 10.1016/j.cels.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xi NM, Li JJ. Benchmarking computational doublet-detection methods for single-Cell RNA sequencing data. Cell Syst. 2021;12:176–94.e6. doi: 10.1016/j.cels.2020.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hafemeister C, Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20:296. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ilicic T, Kim JK, Kolodziejczyk AA, Bagger FO, McCarthy DJ, Marioni JC, et al. Classification of low quality cells from single-cell RNA-seq data. Genome Biol. 2016;17:29. doi: 10.1186/s13059-016-0888-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soneson C, Robinson MD. Bias, robustness and scalability in single-cell differential expression analysis. Nat Methods. 2018;15:255–261. doi: 10.1038/nmeth.4612. [DOI] [PubMed] [Google Scholar]
- 39.Mou T, Deng W, Gu F, Pawitan Y, Vu TN. Reproducibility of methods to detect differentially expressed genes from single-cell RNA sequencing. Front Genet. 2019;10:1331. doi: 10.3389/fgene.2019.01331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:D605–D612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Dongen S, Abreu-Goodger C. Using MCL to extract clusters from networks. Methods Mol Biol. 2012;804:281–295. doi: 10.1007/978-1-61779-361-5_15. [DOI] [PubMed] [Google Scholar]
- 42.Friedman BA, Srinivasan K, Ayalon G, Meilandt WJ, Lin H, Huntley MA, et al. Diverse brain myeloid expression profiles reveal distinct microglial activation states and aspects of alzheimer’s disease not evident in mouse models. Cell Rep. 2018;22:832–847. doi: 10.1016/j.celrep.2017.12.066. [DOI] [PubMed] [Google Scholar]
- 43.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wickham H. ggplot2: elegant graphics for data analysis. Springer Science & Business Media; 2009. [Google Scholar]
- 45.Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol BioMed Central. 2006;7:1–14. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mancuso R, Van Den Daele J, Fattorelli N, Wolfs L, Balusu S, Burton O, et al. Stem-cell-derived human microglia transplanted in mouse brain to study human disease. Nat Neurosci. 2019;22:2111–2116. doi: 10.1038/s41593-019-0525-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cabeza-Cabrerizo M, Cardoso A, Minutti CM, da Pereira Costa M, e Reis Sousa C. Dendritic cells revisited. Annu Rev Immunol. 2021;39:131–166. doi: 10.1146/annurev-immunol-061020-053707. [DOI] [PubMed] [Google Scholar]
- 48.Evans HT, Benetatos J, van Roijen M, Bodea L-G, Götz J. Decreased synthesis of ribosomal proteins in tauopathy revealed by non-canonical amino acid labelling. EMBO J. 2019;38:e101174. doi: 10.15252/embj.2018101174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koren SA, Hamm MJ, Meier SE, Weiss BE, Nation GK, Chishti EA, et al. Tau drives translational selectivity by interacting with ribosomal proteins. Acta Neuropathol. 2019;137:571–583. doi: 10.1007/s00401-019-01970-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Evans HT, Taylor D, Kneynsberg A, Bodea L-G, Götz J. Altered ribosomal function and protein synthesis caused by tau. Acta Neuropathol Commun. 2021;9:110. doi: 10.1186/s40478-021-01208-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tracy TE, Madero-Pérez J, Swaney DL, Chang TS, Moritz M, Konrad C, et al. Tau interactome maps synaptic and mitochondrial processes associated with neurodegeneration. Cell. 2022;185:712–28.e14. doi: 10.1016/j.cell.2021.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu C, Song X, Nisbet R, Götz J. Co-immunoprecipitation with Tau isoform-specific antibodies reveals distinct protein interactions and highlights a putative role for 2N Tau in disease. J Biol Chem. 2016;291:8173–8188. doi: 10.1074/jbc.M115.641902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karin M, Liu Z g, Zandi E. AP-1 function and regulation. Curr Opin Cell Biol. 1997;9:240–6. doi: 10.1016/S0955-0674(97)80068-3. [DOI] [PubMed] [Google Scholar]
- 54.Leandro GS, Evangelista AF, Lobo RR, Xavier DJ, Moriguti JC, Sakamoto-Hojo ET. Changes in expression profiles revealed by transcriptomic analysis in peripheral blood mononuclear cells of Alzheimer’s disease patients. J Alzheimers Dis. 2018;66:1483–1495. doi: 10.3233/JAD-170205. [DOI] [PubMed] [Google Scholar]
- 55.Jiang S, Maphis NM, Binder J, Chisholm D, Weston L, Duran W, et al. Proteopathic tau primes and activates interleukin-1β via myeloid-cell-specific MyD88- and NLRP3-ASC-inflammasome pathway. Cell Rep. 2021;36:109720. doi: 10.1016/j.celrep.2021.109720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang C, Fan L, Khawaja RR, Liu B, Zhan L, Kodama L, et al. Microglial NF-κB drives tau spreading and toxicity in a mouse model of tauopathy. Nat Commun. 2022;13:1969. doi: 10.1038/s41467-022-29552-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li X, Long J, He T, Belshaw R, Scott J. Integrated genomic approaches identify major pathways and upstream regulators in late onset Alzheimer’s disease. Sci Rep. 2015;5:12393. doi: 10.1038/srep12393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bhaskar K, Konerth M, Kokiko-Cochran ON, Cardona A, Ransohoff RM, Lamb BT. Regulation of tau pathology by the microglial fractalkine receptor. Neuron. 2010;68:19–31. doi: 10.1016/j.neuron.2010.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cho S-H, Sun B, Zhou Y, Kauppinen TM, Halabisky B, Wes P, et al. CX3CR1 protein signaling modulates microglial activation and protects against plaque-independent cognitive deficits in a mouse model of Alzheimer disease. J Biol Chem. 2011;286:32713–32722. doi: 10.1074/jbc.M111.254268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maphis N, Xu G, Kokiko-Cochran ON, Jiang S, Cardona A, Ransohoff RM, et al. Reactive microglia drive tau pathology and contribute to the spreading of pathological tau in the brain. Brain. 2015;138:1738–1755. doi: 10.1093/brain/awv081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Whaley K. Biosynthesis of the complement components and the regulatory proteins of the alternative complement pathway by human peripheral blood monocytes. J Exp Med. 1980;151:501–516. doi: 10.1084/jem.151.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martin U, Bock D, Arseniev L, Tornetta MA, Ames RS, Bautsch W, et al. The human C3a receptor is expressed on neutrophils and monocytes, but not on B or T lymphocytes. J Exp Med. 1997;186:199–207. doi: 10.1084/jem.186.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang S, Song R, Wang Z, Jing Z, Wang S, Ma J. S100A8/A9 in Inflammation. Front Immunol. 2018;9:1298. doi: 10.3389/fimmu.2018.01298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Austermann J, Spiekermann C, Roth J. S100 proteins in rheumatic diseases. Nat Rev Rheumatol. 2018;14:528–541. doi: 10.1038/s41584-018-0058-9. [DOI] [PubMed] [Google Scholar]
- 65.Hasselmann J, Coburn MA, England W, Figueroa Velez DX, Kiani Shabestari S, Tu CH, et al. Development of a chimeric model to study and manipulate human microglia in vivo. Neuron. 2019;103:1016–33.e10. doi: 10.1016/j.neuron.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Leent MMT, Beldman TJ, Toner YC, Lameijer MA, Rother N, Bekkering S, et al. Prosaposin mediates inflammation in atherosclerosis. Sci Transl Med. 2021;13:1433. doi: 10.1126/scitranslmed.abe1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Naj AC, Jun G, Beecham GW, Wang L-S, Vardarajan BN, Buros J, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Litvinchuk A, Wan Y-W, Swartzlander DB, Chen F, Cole A, Propson NE, et al. Complement C3aR inactivation attenuates Tau pathology and reverses an immune network deregulated in tauopathy models and Alzheimer’s disease. Neuron. 2018;100:1337–53.e5. doi: 10.1016/j.neuron.2018.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu T, Dejanovic B, Gandham VD, Gogineni A, Edmonds R, Schauer S, et al. Complement C3 is activated in human AD brain and is required for Neurodegeneration in mouse models of Amyloidosis and Tauopathy. Cell Rep. 2019;28:2111–23.e6. doi: 10.1016/j.celrep.2019.07.060. [DOI] [PubMed] [Google Scholar]
- 70.Segovia M, Louvet C, Charnet P, Savina A, Tilly G, Gautreau L, et al. Autologous dendritic cells prolong allograft survival through Tmem176b-dependent antigen cross-presentation. Am J Transplant. 2014;14:1021–1031. doi: 10.1111/ajt.12708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Segovia M, Russo S, Jeldres M, Mahmoud YD, Perez V, Duhalde M, et al. Targeting TMEM176B enhances antitumor immunity and augments the efficacy of immune checkpoint blockers by unleashing inflammasome activation. Cancer Cell. 2019;35:767–81.e6. doi: 10.1016/j.ccell.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Piehl N, van Olst L, Ramakrishnan A, Teregulova V, Simonton B, Zhang Z, et al. Cerebrospinal fluid immune dysregulation during healthy brain aging and cognitive impairment. Cell. 2022;185:5028–39.e13. doi: 10.1016/j.cell.2022.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thrupp N, Sala Frigerio C, Wolfs L, Skene NG, Fattorelli N, Poovathingal S, et al. Single-nucleus RNA-Seq is not suitable for detection of microglial activation genes in humans. Cell Rep. 2020;32:108189. doi: 10.1016/j.celrep.2020.108189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Landsman L, Bar-On L, Zernecke A, Kim K-W, Krauthgamer R, Shagdarsuren E, et al. CX3CR1 is required for monocyte homeostasis and atherogenesis by promoting cell survival. Blood. 2009;113:963–972. doi: 10.1182/blood-2008-07-170787. [DOI] [PubMed] [Google Scholar]
- 75.Auffray C, Fogg DK, Narni-Mancinelli E, Senechal B, Trouillet C, Saederup N, et al. CX3CR1+ CD115+ CD135+ common macrophage/DC precursors and the role of CX3CR1 in their response to inflammation. J Exp Med. 2009;206:595–606. doi: 10.1084/jem.20081385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Narasimhan PB, Marcovecchio P, Hamers AAJ, Hedrick CC. Nonclassical monocytes in health and disease. Annu Rev Immunol. 2019;37:439–456. doi: 10.1146/annurev-immunol-042617-053119. [DOI] [PubMed] [Google Scholar]
- 77.Makinde HM, Cuda CM, Just TB, Perlman HR, Schwulst SJ. Non-classical monocytes mediate secondary injury, neurocognitive outcome, and neutrophil infiltration after Traumatic Brain Injury. J Immunol. 2017;199:3583. doi: 10.4049/jimmunol.1700896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee S, Xu G, Jay TR, Bhatta S, Kim K-W, Jung S, et al. Opposing effects of membrane-anchored CX3CL1 on amyloid and tau pathologies via the p38 MAPK pathway. J Neurosci. 2014;34:12538–12546. doi: 10.1523/JNEUROSCI.0853-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maphis N, Xu G, Kokiko-Cochran ON, Cardona AE, Ransohoff RM, Lamb BT, et al. Loss of tau rescues inflammation-mediated neurodegeneration. Front Neurosci. 2015;9:196. doi: 10.3389/fnins.2015.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Merino JJ, Muñetón-Gómez V, Alvárez M-I, Toledano-Díaz A. Effects of CX3CR1 and fractalkine chemokines in amyloid beta clearance and p-Tau accumulation in Alzheimer’s Disease (AD) rodent models: is Fractalkine a systemic biomarker for AD? Curr Alzheimer Res. 2016;13:403–412. doi: 10.2174/1567205013666151116125714. [DOI] [PubMed] [Google Scholar]
- 81.Bolós M, Llorens-Martín M, Perea JR, Jurado-Arjona J, Rábano A, Hernández F, et al. Absence of CX3CR1 impairs the internalization of Tau by microglia. Mol Neurodegener. 2017;12:59. doi: 10.1186/s13024-017-0200-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Guedes JR, Lao T, Cardoso AL, El Khoury J. Roles of microglial and monocyte chemokines and their receptors in regulating alzheimer’s disease-associated amyloid-β and Tau pathologies. Front Neurol. 2018;9:549. doi: 10.3389/fneur.2018.00549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fan Q, He W, Gayen M, Benoit MR, Luo X, Hu X, et al. Activated CX3CL1/Smad2 signals prevent neuronal loss and Alzheimer’s Tau pathology-mediated cognitive dysfunction. J Neurosci. 2020;40:1133–1144. doi: 10.1523/JNEUROSCI.1333-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Puntambekar SS, Moutinho M, Lin PBC, Jadhav V, Tumbleson-Brink D, Balaji A, et al. CX3CR1 deficiency aggravates amyloid driven neuronal pathology and cognitive decline in Alzheimer’s disease. Mol Neurodegener. 2022;17:47. doi: 10.1186/s13024-022-00545-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Murai N, Mitalipova M, Jaenisch R. Functional analysis of CX3CR1 in human induced pluripotent stem (iPS) cell-derived microglia-like cells. Eur J Neurosci. 2020;52:3667–3678. doi: 10.1111/ejn.14879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Deczkowska A, Keren-Shaul H, Weiner A, Colonna M, Schwartz M, Amit I. Disease-associated microglia: a universal immune sensor of neurodegeneration. Cell. 2018;173:1073–1081. doi: 10.1016/j.cell.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 87.Ishizawa K, Dickson DW. Microglial activation parallels system degeneration in progressive supranuclear palsy and corticobasal degeneration. J Neuropathol Exp Neurol. 2001;60:647–657. doi: 10.1093/jnen/60.6.647. [DOI] [PubMed] [Google Scholar]
- 88.Yoshiyama Y, Higuchi M, Zhang B, Huang S-M, Iwata N, Saido TC, et al. Synapse loss and microglial activation precede tangles in a P301S tauopathy mouse model. Neuron. 2007;53:337–351. doi: 10.1016/j.neuron.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 89.Bellucci A, Bugiani O, Ghetti B, Spillantini MG. Presence of reactive microglia and neuroinflammatory mediators in a case of frontotemporal dementia with P301S mutation. Neurodegener Dis S Karger AG. 2011;8:221–229. doi: 10.1159/000322228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Asai H, Ikezu S, Woodbury ME, Yonemoto GMS, Cui L, Ikezu T. Accelerated neurodegeneration and neuroinflammation in transgenic mice expressing P301L tau mutant and tau-tubulin kinase 1. Am J Pathol. 2014;184:808–818. doi: 10.1016/j.ajpath.2013.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schröder JB, Pawlowski M, Zu Meyer Hörste G, Gross CC, Wiendl H, Meuth SG, et al. Immune Cell Activation in the Cerebrospinal Fluid of Patients With Parkinson’s Disease. Front Neurol. 2018;9:1081. doi: 10.3389/fneur.2018.01081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Balendra R, Isaacs AM. C9orf72-mediated ALS and FTD: multiple pathways to disease. Nat Rev Neurol. 2018;14:544–558. doi: 10.1038/s41582-018-0047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chang D, Nalls MA, Hallgrímsdóttir IB, Hunkapiller J, van der Brug M, Cai F, et al. A meta-analysis of genome-wide association studies identifies 17 new Parkinson’s disease risk loci. Nat Genet. 2017;49:1511–1516. doi: 10.1038/ng.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Blauwendraat C, Nalls MA, Singleton AB. The genetic architecture of Parkinson’s disease. Lancet Neurol. 2020;19:170–178. doi: 10.1016/S1474-4422(19)30287-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Villani A-C, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017;356:eaah4573. doi: 10.1126/science.aah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Calzetti F, Tamassia N, Micheletti A, Finotti G, Bianchetto-Aguilera F, Cassatella MA. Human dendritic cell subset 4 (DC4) correlates to a subset of CD14CD16 monocytes. J Allergy Clin. Immunol. 2018;141:2276–9. doi: 10.1016/j.jaci.2017.12.988. [DOI] [PubMed] [Google Scholar]
- 97.Collin M, Bigley V. Human dendritic cell subsets: an update. Immunology. 2018;154:3–20. doi: 10.1111/imm.12888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Korenfeld D, Roussak K, Dinkel S, Vogel TP, Pollack H, Levy J, et al. STAT3 gain-of-function mutations underlie deficiency in human nonclassical CD16 monocytes and CD141 dendritic cells. J Immunol. 2021;207:2423–2432. doi: 10.4049/jimmunol.2000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang Y, Fung ITH, Sankar P, Chen X, Robison LS, Ye L, et al. Depletion of NK cells improves cognitive function in the Alzheimer disease mouse model. J Immunol. 2020;205:502–10. doi: 10.4049/jimmunol.2000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Garofalo S, Cocozza G, Porzia A, Inghilleri M, Raspa M, Scavizzi F, et al. Natural killer cells modulate motor neuron-immune cell cross talk in models of Amyotrophic lateral sclerosis. Nat Commun. 2020;11:1–16. doi: 10.1038/s41467-020-15644-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kaur G, Trowsdale J, Fugger L. Natural killer cells and their receptors in multiple sclerosis. Brain. 2013;136:2657–2676. doi: 10.1093/brain/aws159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hertwig L, Hamann I, Romero-Suarez S, Millward JM, Pietrek R, Chanvillard C, et al. CX3CR1-dependent recruitment of mature NK cells into the central nervous system contributes to control autoimmune neuroinflammation. Eur J Immunol. 2016;46:1984–1996. doi: 10.1002/eji.201546194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Leuzy A, Chiotis K, Lemoine L, Gillberg P-G, Almkvist O, Rodriguez-Vieitez E, et al. Tau PET imaging in neurodegenerative tauopathies—still a challenge. Mol Psychiatry. 2019;24:1112–1134. doi: 10.1038/s41380-018-0342-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Peet BT, Spina S, Mundada N, La Joie R. Neuroimaging in frontotemporal dementia: heterogeneity and relationships with underlying neuropathology. Neurotherapeutics. 2021;18:728–752. doi: 10.1007/s13311-021-01101-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zetterberg H, Bendlin BB. Biomarkers for Alzheimer’s disease-preparing for a new era of disease-modifying therapies. Mol Psychiatry. 2021;26:296–308. doi: 10.1038/s41380-020-0721-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gendron TF, Heckman MG, White LJ, Veire AM, Pedraza O, Burch AR, et al. Comprehensive cross-sectional and longitudinal analyses of plasma neurofilament light across FTD spectrum disorders. Cell Rep Med. 2022;3:100607. doi: 10.1016/j.xcrm.2022.100607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rexach JE, Polioudakis D, Yin A, Swarup V, Chang TS, Nguyen T, et al. Tau pathology drives dementia risk-associated gene networks toward chronic inflammatory States and Immunosuppression. Cell Rep. 2020;33:108398. doi: 10.1016/j.celrep.2020.108398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Cell Ranger metrics summarizing important characteristics of the detected cells for each sample.
Additional file 2: Table S2. Cell counts during quality-control steps for each sample.
Additional file 3: Table S3. RNA integrity number for PBMC samples.
Additional file 4: Figures S1-S9. All supplementary figures.
Additional file 5: Table S4. Cell-type specific differential expression analyses.
Data Availability Statement
The scRNA-seq dataset described here has been uploaded to the FAIR Data Sharing Portal within the Alzheimer’s Disease Workbench, which is supported by the Alzheimer’s Disease Data Initiative, and can be accessed at https://fair.addi.ad-datainitiative.org/#/data/datasets/single_cell_rna_seq_data_derived_from_mapt_carriers_and_controls.